Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the contagious coronavirus disease 2019 (COVID-19) which was first identified in Wuhan, China, in December 2019. Around the world, many researchers focused their research on identifying inhibitors against the druggable SARS-CoV-2 targets. The reported genomic mutations have a direct effect on the receptor-binding domain (RBD), which interacts with host angiotensin-converting enzyme 2 (ACE-2) for viral cell entry. These mutations, some of which are variants of concern (VOC), lead to increased morbidity and mortality rates. The newest variants including B.1.617.2 (Delta), AY.1 (Delta plus), and C.37 (Lambda) were considered in this study. Thus, an exhaustive structure-based virtual screening of a ligand library (in which FDA approved drugs are also present) using the drug-likeness screening, molecular docking, ADMET profiling was performed followed by molecular dynamics (MD) simulation, and Molecular Mechanics-Poisson Boltzmann Surface Area (MM-PBSA) calculation to identify compounds or drugs can be repurposed for inhibiting the wild type, Delta, Delta plus and Lambda variants of RBD of the spike protein. Based on the virtual screening steps, two FDA approved drugs, Atovaquone (atv) and Praziquantel (prz), were selected and repurposed as the best candidates of SARS-CoV-2 RBD inhibitors. Molecular docking results display that both atv and prz contribute in different interaction with binding site residues (Gln493, Asn501 and Gly502 in the hydrogen bond formation, Phe490 and Tyr505 in the π- π stacking and Tyr449, Ser494, and Phe497 in the vdW interactions) in the wild type, Delta, Delta plus and Lambda variants of RBD of the spike protein. MD simulations revealed that among the eight studied complexes, the wild type-atv and Delta-prz complexes have the most structural stability over the simulation time. Furthermore, MM-PBSA calculation showed that in the atv containing complexes, highest binding affinity is related to the wild type-atv complex and in the prz containing complexes, it is related to the Delta-prz complex. The validation of docking results was done by comparing with experimental data (heparin in complex with wild type and Delta variants). Also, comparison of the obtained results with the result of simulation of the k22 with the studied proteins showed that atv and prz are suitable inhibitors for these proteins, especially wild type t and Delta variant, respectively. Thus, we found that atv and prz are the best candidate for inhibition of wild type and Delta variant of the spike protein. Also, atv can be an appropriate inhibitor for the Lambda variant. Obtained in silico results may help the development of new anti-COVID-19 drugs.
Keywords: SARS-CoV-2, Variants, Spike protein, Drug repurposing, Virtual screening, Molecular dynamics simulation
Graphical abstract
1. Introduction
The human population of the 21st century was attacked by a large-scale epidemic of highly pathogenic viruses such as a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In December 2019, the first case has been manifested in Wuhan, Hubei province, China, and since then, it has spread across the world with fever, dry cough, dyspnoea, diarrhea, and typical sore throat symptoms [1]. As of the last update on 25 February 2022, a total of 432,176,247 confirmed cases have been reported worldwide, with 5,949,044 cases having lost their lives [2]. Since it had highly interpersonal transmission, the World Health Organization (WHO) proclaimed the COVID-19 as a public health emergency of international concern (PHEIC) on January 30, 2020, and a pandemic on March 11, 2020 [3].
In the past two decades, Coronaviruses were common pathogens highly infectious for humans like the severe acute respiratory syndrome (SARS-CoV) in 2002 and the Middle East respiratory syndrome (MERS-CoV) in 2012. SARS-CoV-2 has been identified with 79.5% sequence similarity and etiological factors to SARS-CoV [4,5]. However, SARS-CoV-2 is more infectious than the other two coronaviruses. They are the members of the subfamily Coronavirinae in the family of Coronaviridae of the order Nidovirales viruses and contain a positive-sense single-stranded RNA(+ssRNA) (∼30 kb in size), which includes 23 putative open reading frames (ORFs) [[4], [5], [6]]. ORFs encode at least four main structural proteins; Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N) proteins, along with 16 non-structural proteins, and some ORFs also encode an additional hemagglutinin-esterase (HE) protein [5,7]. A large number of spike glycoprotein that forms trimers on the surface of the virion, belongs to class-I viral fusion proteins and cleaved into the S1 and S2 subunits at the boundary between them via a furin-like cleavage site. Hence the fusion cell membrane and the viral genome's entry into the host cell facilitated [8]. The S1 subunit consists of an N-terminal domain (NTD) and a C-terminal domain (CTD), whereas the S2 subunit includes a fusion peptide (FP), heptad repeats (HR1 and HR2), and transmembrane helix (TM), and this subunit is responsible for the cellular membrane fusion [9]. The receptor-binding domain (RBD) is located on the S1 subunit. It contains a core and a main functional receptor-binding motif (RBM) which specifically interacts with the host angiotensin-converting enzyme 2 (ACE2) receptor, mediating viral cell entry [10]. Recent studies have indicated that RBD contains essential amino acid residues (Phe486, Asn487, Ala475, Glu484, Leu455, Gln493, Tyr453, Ser494, Tyr449, Gln498, Gly496, Gly446, Asn501, Tyr505, Lys417, Phe456, Tyr489, Tyr473, Pro462, Tyr475, Tyr442, Gly488, Thr486, Thr500, Gln474, Gly476, Gly502, and Gln486) which interact with the interfacial residues of ACE2 (Gln24, Ile21, Met82, Tyr83, Ala80, Leu79, Phe28, Phe72, Thr27, Asp30, Asp38, Glu35, Lys353, Phe32, Lys31, Leu45, Asp353, Tyr4, His34, Tyr41, and phe79) [[11], [12], [13]]. ACE2 receptors are present on the epithelial cell tissues of the lungs, heart, kidneys, intestine, and bladder of the host organism. ACE2 was highly expressed on lung epithelial cells exactly, in the alveolar space [14]. In addition to the ACE2 receptor, the S protein is activated via protease activators like transmembrane serine 2 (TMPRSS2), and lysosomal proteases near the junction cell entry [15,16].
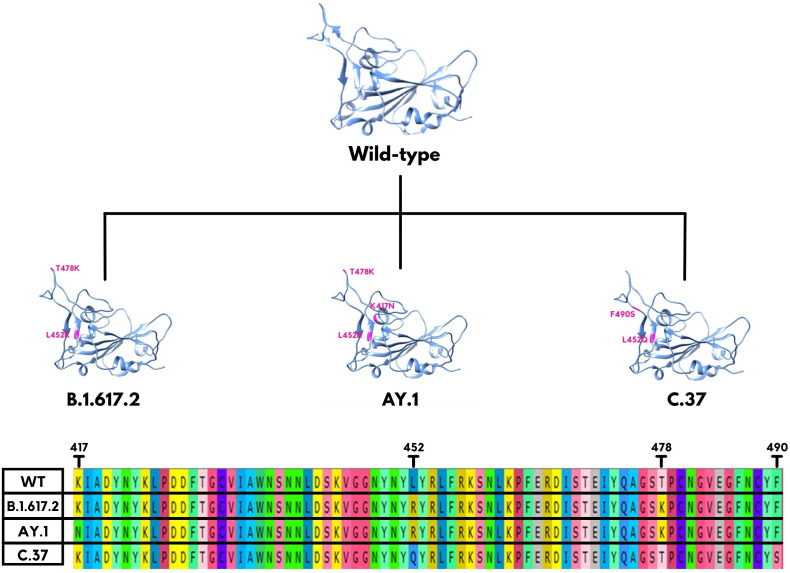
Like other viruses, the SARS-CoV-2 also has mutational changes in the genomic sequence and thus structural and functional changes in the proteins. These proteins are the essential drug targets and play the leading role in the development of pathogenic resistance. Mutations make viruses more adaptive to cell receptors, which, in turn, increases mortality rates in humans [17]. Deadly mutations have been documented as the Variants of Concern (VOC) including; The Alpha variant B.1.1.7 originating in the UK and associated with the N501Y RBD mutation, Beta variant B.1.351 was first coined in South Africa with the K417 N, E484K, and N501Y mutations in RBD, Gamma variant P.1 is one of Brazil's detected variants with the E484K, K417T, and N501Y RBD mutations, Delta variant B.1.617.2 was first seen on October 2020 in India, and at last Omicron variant B.1.1.529 with vast number of mutations. Meanwhile, WHO designated several Variants of Interest (VOI) like Lambda variant C.37 (originating in Peru), and Mu variant B.1.621 (originating in Colombia) currently [18,19]. The Lambda variant, first seen in August 2020, however, was identified by the WHO in June 2021. It has multiple mutations and a new deletion (G75V, T76I, L452Q, F490S, D614G, T859 N, and Δ246-252(in the S protein. Between these substitutes, the L452Q and F490S are present in the spike RBD [19,20]. The Delta variant has three sub-lineages (B.1.617.1, B.1.617.2, and B.1.617.3) with T19R, G142D, Δ156–157, R158G, L452R, T478K, D614G, P681R, and D950 N spike mutations. The critical RBD substitutions are Leu at position 452 and Thr at position 478, being replaced by Arg and Lys, respectively. E484Q substitution is present in B.1.617.1 and B.1.617.3 but not in B.1.617.2. Another variant AY.1 (Delta plus), has emerged with an additional RBD mutation Lys at position 417, being replaced by Asn, which is also present in the Beta variant [20,21]. In Fig. 1 , the RBD mutations in B.1.617.2, AY.1, and C.37 could be observed.
Fig. 1.
The receptor-binding domain (RBD) mutations of SARS-CoV-2 in B.1.617.2 (Delta), AY.1 (Delta plus), and C.37 (Lambda) variants.
From the first days of the pandemic, several options for COVID-19 therapy were considered. These include vaccines, monoclonal antibodies, oligonucleotide-based therapies, peptides.
Interferon therapies, and small molecules [22]. Many studies have reported the computational screening through molecular docking and structural dynamics techniques of natural molecules for various SARS-CoV-2 targets, such as nonstructural protein 1, 15, 16, RNA‐dependent RNA polymerase, and Spike protein [[23], [24], [25], [26], [27]]. However, most of the studies are based on in-silico repurposing techniques, from already approved substances such as anti-malarial, anticancer, antiviral, anti-inflammatory, etc. Since repurposing identification of drugs is an economical approach, and saves a lot of time [22,[28], [29], [30]]. For instance, a recent docking study exposed that the drugs Cytarabin, Raltitrexed, Tenofovir, Cidofovir, Fludarabine, and Lamivudine are possible inhibitors for the spike protein [30]. In other computational research studies, it was found that Atazanavir, Indinavir, Saquinavir, Lopinavir, Ritonavir, Ciluprevir, Glecaprevir, Viomycin, Bacampicillin, Remdesivir, and Hydroxychloroquine can serve as inhibitors of the main protease [31,32]. Since up to now no definite anti COVID-19 treatment has been developed, pharmacological researches are still ongoing.
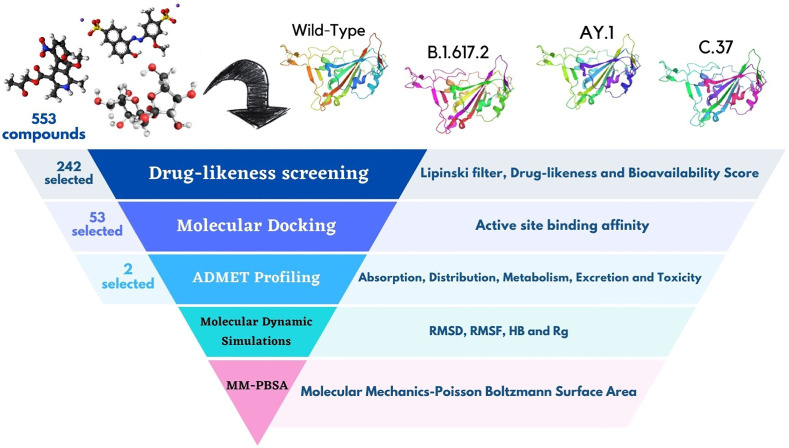
The present study focuses on the exhaustive structure-based virtual screening method. We mean the word “exhaustive” in this paper, the comprehensiveness of the steps applied in our study. Because in this study, molecular docking, ADMET profiling and molecular dynamics simulation sub methods were used for doing virtual screening [33]. Through the physical and chemical principles of 553 various compounds was tried to obtain the best inhibitors against the wild-type and mutated RBD of the spike glycoprotein of SARS-CoV-2. The RBD of the B.1.617.2 (Delta), AY.1 (Delta Plus), and C.37 (Lambda) variants were modeled as a receptor, and their interaction with ACE2 was noticed. Since different SARS-CoV-2 strains have different mutation patterns, it is important to understand their mutation impacts and infection changes accordingly. This work uses bioinformatics methods like drug-likeness prediction, molecular docking, ADMET profiling (Absorption, Distribution, Metabolism, Excretion, and Toxicity), molecular dynamic (MD) simulations, and MM-PBSA (Molecular Mechanics-Poisson Boltzmann Surface Area). Five virtual screening steps were accomplished by selecting the appropriate compounds in Fig. 2 .
Fig. 2.
A schematic flow chart of virtual screening's steps.
2. Materials and methods
2.1. Library preparation
A library containing 553 antiviral compounds derived from a collection of 700 molecules was selected for exploring novel antiviral agents against wild-type and mutated receptors. This database was expanded after our selection. In our structurally diverse collection, some compounds have been approved by the FDA, others target HCV protease, HIV protease, Integrase, Reverse Transcriptase, etc. (Available at https://www.selleckchem.com/screening/antiviral-compound-library.html). The SMILES and SDF format of the library were used in the following steps.
2.2. Drug-likeness screening
The first step of virtual screening used the SMILES format of the library as the inputs of SwissADME (http://www.swissadme.ch) [34] and Molsoft L.L.C online analysis servers (https://molsoft.com/mprop/). All of the 553 compounds were optimized based on Lipinski's rule of five [35] and bioavailability score [36] in SwissADME analysis server and drug-likeness model score in Molsoft L.L.C. online tool. As per drug-likeness screening, molecular weight (MW), number of hydrogen bond acceptors (NHBA), number of hydrogen bond donors (NHBD), octanol-water partition coefficient (MolLogP), water solubility Log (MolLogS), molecular polar surface area (MolPSA) which is defined as the sum of surfaces of oxygens, nitrogens and attached hydrogens, molecular volume (MolVol), and drug-likeness model score were investigated.
2.3. Receptor preparation and mutation
The 3D crystal structure of the SARS-CoV-2 spike receptor-binding domain bound with ACE2 (PDB ID 6M0J) was attained from the RCSB Protein Data Bank website (https://www.rcsb.org/). Since the resolution of this structure is 2.45 Å with x-ray diffraction method detection, it is an appropriate choice. 6M0J contains two macromolecule chains. Angiotensin-converting enzyme 2 shows chain A with 603 sequence length, and chain B is the S1 subunit of spike protein and has 229 sequence length. For pre-processing stage, first of all, chain A removed from a PDB file, and then based on AutoDock Vina documentation, release 1.2.0, native small molecules, redundant water molecules, and non-standard residues were deleted from the S1 subunit (chain B), however polar hydrogen atoms and Gasteiger charges [37] were added using UCSF Chimera 1.15 software [38]. Those residues of the RBD being in the interacting site of RBD-ACE2 were considered as active site and molecular docking grid box. NAG groups in the pdb file are located far from this interacting site, especially RBD residues. Thus, NAG groups were not considered. Another reason was that our goal was to repurpose drugs or compounds and not co-crystal molecules in pdb file.
The dual point mutations of the Delta and Lambda variants and multiple point mutations of the Delta plus variant were produced on prepared proteins by changing the amino acids to the mutated one using UCSF Chimera 1.15 software. During each mutation by UCSF Chimera, the best rotamer with the most probability was selected. Also, after the mutation, two consecutive steps of energy minimization were performed using the steepest descent and conjugate gradient algorithms to remove bad contacts and clashes. Finally, four receptors (wild-type, Delta, Delta plus, and Lambda) were obtained. These four RBDs (wild-type and mutants) were entered into the next steps for computational molecular docking and molecular dynamic simulations.
2.4. Molecular docking and validation
In the second step of virtual screening, 242 selected compounds from the previous level, were be docked to receptors (wild-type, Delta, Delta plus, and Lambda). PyRx is open-source software [39] for multiple docking, which was used in this study. PyRx uses the AutoDock Vina algorithm [40] to evaluate the binding affinities with molecular docking. Four receptors with PDB format and 242 ligands with SDF format were used as an input for PyRx, and changing the formats in PDBQT were done through Open Babel. Before running, all of the ligands stay on the lowest energy level with 200 total minimization steps and UFF force field [41]. The interacting site of RBD-ACE2 was analyzed with UCSF Chimera 1.15 software and the grid box enclosed in this section. This grid box of 24-40-20 points with a grid spacing value of 1 Å was constructed and centered on the X = −38.619, Y = 32.524, Z = 6.258 coordination. Then the protocol was run at exhaustiveness value of 8 and set on the favorable binding free energy pose from the top eight poses were retained for all compounds.
The docking validation was inspired by the Azza Hanif Harisna et al. study [42], which used decoy ligands similar to nelfinavir as a validator of the docking parameters. The decoy substances were gained from the DUD-E online server (http://dude.docking.org/) [43] and the docking parameters and process were just like the previously mentioned protocol. Also, to validate the in silico study (molecular docking and MD simulation), we found experimental data for the heparin in complexed with the wild type and Delta variants [44]. To this aim, the molecular docking as well as MD simulation of the wild type-heparin and delta-heparin complexes were performed. Only binding free energy results of wild type-heparin and delta-heparin complexes were reported in this study.
2.5. ADMET profiling
For 53 top compounds which showed the lowest dock score, ΔG < −7 kcal/mol or equal, using graph-based signature pkCSM online web server (http://biosig.unimelb.edu.au/pkcsm/) [45]. Since between these 53 molecules, 9 FDA approved drugs including: Imatinib, Bictegravir, Dolutegravir, Raltegravir, Atovaquone, Cabotegravir, Ciclesonide, Aprepitant, and Praziquantel are available, ADME and toxicity analysis is not required. However, entire compounds were analyzed. SMILES format of the ligands uploaded to predict lipophilicity (LogP), surface area and five main different pharmacokinetic properties such as absorption, distribution, metabolism, excretion, and toxicity (ADMET). Absorption property includes caco-2 permeability, water-solubility, intestinal absorption (human), skin permeability, P-glycoprotein substrate, P-glycoprotein I and II inhibitors; distribution: steady-state volume of distribution (VDss), fraction unbound (human), blood-brain barrier (BBB) permeability, central nervous system (CNS) permeability; metabolism: CYP2D6 and CYP3A4 substrates, CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4 inhibitors; excretion: total clearance, and renal OCT2 substrate; toxicity: AMES toxicity, maximum human tolerated dose, hERG I and II inhibitors, oral rat acute toxicity (LD50), oral rat chronic toxicity (LOAEL), hepatotoxicity, skin sensitization, T. pyriformis, and minnow toxicity.
2.6. Molecular dynamics simulation
The biomolecular software package GROMACS 2016 [46] was applied for the molecular dynamics simulations of the protein–ligand complexes. To this aim, the leapfrog integrator in a 2 fs time step was considered. MD simulation is a useful method for confirming molecular docking results and investigating the system dynamics. The gromacs topology files for protein and ligands were obtained using the AMBER99SB-ILDN [47] force field and GAFF [48] (in the Antechamber module of Amber Tools 14 [49]). AM1-BCC charges [50] were assigned to the ligand using the antechamber module from the Amber Tools 14. Each protein-ligand complex obtained from the best pose of molecular docking was placed in the center of a cubic box, with 1 nm distance to the edges, filled with a TIP3P model [51] of water molecules. The neutralization of the systems was done by the addition of a suitable number of sodium and chloride ions. Periodic boundary conditions (PBC) [52] in all three directions and the LINCS algorithm [53] for all bonds involving hydrogen atoms were considered. For long-range electrostatic interactions, Particle Mesh Ewald (PME) [54] was applied while for short-range electrostatics and van der Waals interactions, a cut off distance of 1.2 nm was used. The prepared systems were energy minimized using the steepest descent and conjugate gradient methods with position restraining on heavy atoms of protein and ligand and with force constant 1000 kJ mol−1 nm−. Afterward, two short equilibration steps (500 ps) in NVT and NPT ensembles were performed to relax the minimized systems. Temperature and pressure during the simulation were set to 310 K and 1 bar regulated by V-rescale thermostat [55] and Parrinello-Rahman barostat [56], respectively. Finally, a 120 ns long production run was performed in the NPT ensemble and the trajectory snapshots were saved every 15 ps for the analysis. The structural analysis such as root mean square deviation (RMSD), radius of gyration (Rg), root mean square fluctuation (RMSF) and hydrogen bonds were done using different GROMCAS modules. The molecular graphics of the simulated trajectory were created using Pymol software [57]. In this study, eight MD simulations were performed. For simplicity, the complex of the wild-type and mutated (Delta, Delta plus, and Lambda) RBD of the spike protein with Atovaquone and Praziquantel compounds were named as wt-atv, dlt-atv, pls-atv, lmb-atv, wt-prz, dlt-prz, pls-prz, lmb-prz, respectively. We consider a viral spike protein inhibitor, K22 [58], as a reference molecule and performed molecular docking and MD simulation with the wild type, Delta, Delta plus and Lambda variants of RBD of the spike protein to compare the results.
2.7. Binding free energy calculation
The stability of protein-ligand complexes was investigated by calculating the binding free energy using Molecular Mechanics-Poisson-Boltzmann Surface Area (MM-PBSA) approach [59]. In this work, to compute the binding free energy, g_mmpbsa tool [60] was applied which is a widely used method for calculating binding free energy of a ligand to the complex [61,62]. ΔGbind of a protein-ligand complex can be calculated as follows:
| ΔGbind = ΔEMM + ΔGsolvation | (1) |
| ΔEMM = ΔEele + ΔEvdW | (2) |
| ΔGsolvation = ΔGpol + ΔGnon-pol | (3) |
| ΔGnon-pol =γA + b | (4) |
ΔEMM represents the molecular mechanical average energy. ΔG solvation is the change of free energy in the solvation process. ΔG pol describes the free energy changes of polar solvation calculated by Poisson Boltzmann equation, and ΔG non-pol represents the free energy changes of non-polar solvation due to ligand binding to the protein calculated via SASA (Solvent Accessible Surface Area). γ represents the coefficient of surface tension of the solvent, A represents SASA and b represents the fitting parameter.
3. Result and discussion
3.1. Drug-likeness screening
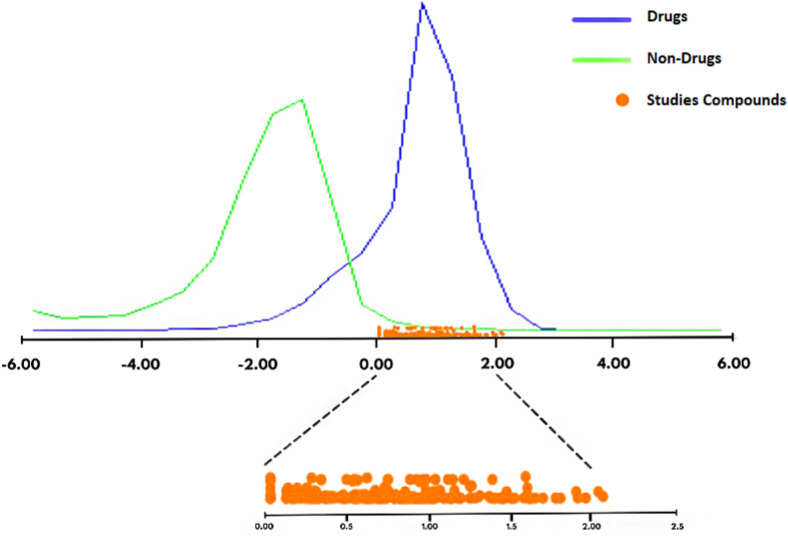
For the first sorting of 553 compounds, these were screened via Lipinski's rule of five, bioavailability score, and drug-likeness model score using online analysis server SwissADME and Molsoft L.L.C. According to the rule of five (RO5), the molecular weight (MW) should be less than 500 g/mol, the lipophilicity denoted by MLogP value should be lower than 4.15 or equal, the number of Hydrogen bond donors (NH or OH) and acceptors (N or O) should be less than 5 and 10 or equal, respectively. Compounds with two or more violations of the Lipinski's criteria were removed, and only ligands with yes; 0 violation or yes; 1 violation, were reported in Supplementary data, Table S1. Another separator parameter is Abbott bioavailability score (BS) which was category into the three percent of absorption ranges: 0.11, 0.17, 0.55, 0.56, and 0.85. These categories identify the range of drug absorption in human body. 0.11% or 0.17% is the lowest one, 0.55% or 0.56% is the average and 0.85% is the highest mode. Probability, medium to the high percentage of absorption is more favorable. In Supplementary data, Table S1, 0.55 or 0.56 and 0.85% of absorption were considered. To predict an overall drug-likeness model score (DLS) should evaluate; molecular weight (MW (g/mol)), number of hydrogen bond acceptors (NHBA), number of hydrogen bond donors (NHBD), MolLogP, MolLogS (Mol/L), MolPSA (A2), MolVol (A3), pKa of the most basic or acidic group, the blood-brain barrier (BBB) score, and the number of stereo centers. As seen in Fig. 3 , based on these parameters, a graph is drawn which shows the position of the studied compounds and in Supplementary data, Table S1, the just the most important parameters have been reported. The thresholds used to remove weak compounds are as follows: no Lipinski's rule of five with 2 or 3 violations, less than 0.55 bioavailability score (0.11 or 0.17), and mines or zero drug-likeness model score. Based on the drug-likeness analysis, we defined 187 structures with yes, 0 RO5 violation and 55 structures with yes; 1 violation. Two hundred and eleven, 22, and 9 compounds showed 0.55%, 0.56%, and 0.85% bioavailability score, respectively. 62 structures had 1.00 till 1.99 drug-likeness score and 2 of these with 2.1 and 2.07 score were stated. Two hundred and forty-two potential structures were finalized, tabulated in Supplementary data, Table S1, for further protein-ligand interaction study.
Fig. 3.
The position of the studied compounds based on drug-likeness model score.
3.2. Molecular docking calculations and validation
In this step, 242 selected compounds from the previous level were prepared to dock against wild-type and mutated (Delta, Delta plus, and Lambda) SARS-CoV-2 RBD. The purpose was based on identifying the least binding energy between the protein and the ligand that is shown with docking score and refers to the most stable binding mode. The interfacial residues between SARS-CoV-2 RBD and ACE2 receptor in the initial PDB file (6M0J) were selected as docking grid box in the molecular docking of wild-type RBD, Delta RBD, Delta plus RBD, and the Lambda RBD with atv and prz molecules. For the screening of 242 potential ligands using the virtual screening tool, PyRx. The molecular dockings were performed based on the protocol, mentioned in 2.4 section. After docking, 53 top hits were having the lowest binding affinity (ΔG ≤ −7 kcal/mol) were selected for more analysis. Docking score of screened compounds are displayed in Supplementary data, Table S2. Our docking method was validated using 102 decoy ligands matched with Atovaquone and Praziquantel compounds which were obtained from the DUD-E server. Decoys are similar compounds to the active ligands in physical properties but different chemically [63]. Therefore, assumed that they do not bind to the target [64]. This technique is based on overcoming false positives and enhance ligand enrichment. The result's binding affinity remained between −10.9 kcal/mol to −5 kcal/mol 18 decoy compounds showed higher binding energy compared to the wild-type RBD-atv and RBD-prz complexes. In addition, 51 decoy molecules showed no binding with wild-type RBD. This result confirms the docking and protocol efficiency. Moreover, as can be seen in Table 1 , the trends observed in the molecular docking results (binding energy), the molecular dynamics simulation results (ΔGbinding calculated by the MM-PBSA method) and the experimental data (binding affinity) for heparin in complex with wild type and Delta variants of RBD of the spike protein are the same. These studies confirm and validate the in silico method used.
Table 1.
The molecular docking, MD simulation result and in vitro data.
| Protein |
Ligand |
Docking results: |
Experimental data: |
MD simulation results (MM-PBSA): |
|---|---|---|---|---|
| Binding energy (kcal/mol) | Binding affinity (M) [44] | ΔGbinding (kcal/mol) | ||
| wt | heparin | -6.7 | 1.1 × 10 -7 | -21.55 |
| dlt | heparin | -6.5 | 4.1 × 10 -7 | -17.39 |
3.3. ADMET profiling
ADMET studies are essential to assess the safety and efficacy of promising compounds and their action in the body. Here we proposed a graph-based signature pkCSM online web server for 53 top hit compounds obtained from the previous section. Lipophilicity (LogP), and surface area were predicted along with five main different pharmacokinetic properties classes. As shown in Supplementary data, Table S3, each property is mentioned with a specific color. Absorption is evaluated with water solubility (log mol/L), caco-2 permeability (log Papp in 10-6 cm/s), and human intestinal absorption (% absorbed). The water solubility (LogS) indicates the solubility of a molecule in water at 25 °C. The caco-2 is an in vitro model of the human intestinal mucosa to predict the range of drug absorption. Distribution is evaluated with human fraction unbound (Fu), blood-brain barrier (BBB (log BB)) permeability, and central nervous system (CNS (log PS)) permeability. Since a drug more links to serum proteins, that is having less efficiently for traverse cellular membranes or diffuse. The ability of a drug to cross into the brain is an important parameter and measured as LogBB and LogPS to help improve the efficacy of drugs. Metabolism is evaluated with CYP2D6 substrate and inhibitor. The cytochrome P450′ isoforms are the important detoxification enzymes mainly found in the liver. They can oxidize xenobiotics to facilitate excretion. Therefore, it is important to assess the compound's ability to substrate or inhibit the cytochrome P450. Extinction is evaluated with total clearance (log ml/min/kg). Total clearance is measured as a combination of hepatic (liver metabolism with biliary clearance) and renal excretion. It is directly related to the bioavailability range. Toxicity is evaluated with AMES toxicity and hepatotoxicity. The AMES test indicates that mutagenic compounds and their action as a carcinogen. A compound was classed as hepatotoxic if there is at least one pathological or physiological liver complication which impairs normal liver function. ADMET results are expounded based on the marginal value compared with resultant value as high caco-2 permeability predicted value > 0.90, human intestinal absorption less than 30% is considered poorly absorbed, BBB permeability logBB >0.3 considered readily crosses BBB whereas logBB <1 are poorly distributed. CNS permeability interpreted through logPS > −2 penetrate CNS while logPS < −3 unable to penetrate [33]. For the given compounds, the predicted properties are mentioned in Supplementary data, Table S3. In ten hit compounds (Oleanonic acid, Betulonic acid, OSS_128167, Vidofludimus, Atovaquone, Eriodictyol, Megestrol, Ciclesonide, Praziquantel and, Naringenin), no metabolism and/or toxicity limitations were observed. Between those, Oleanonic acid, Betulonic acid, Atovaquone, Megestrol, Ciclesonide, Praziquantel, and Naringenin have the high caco-2 permeability value; besides they have an applicable intestinal absorption percentage. Based on LogBB and LogPS thresholds, we achieved two potential candidates (Atovaquone, and Praziquantel) for future analysis.
3.4. Protein-ligand interaction details
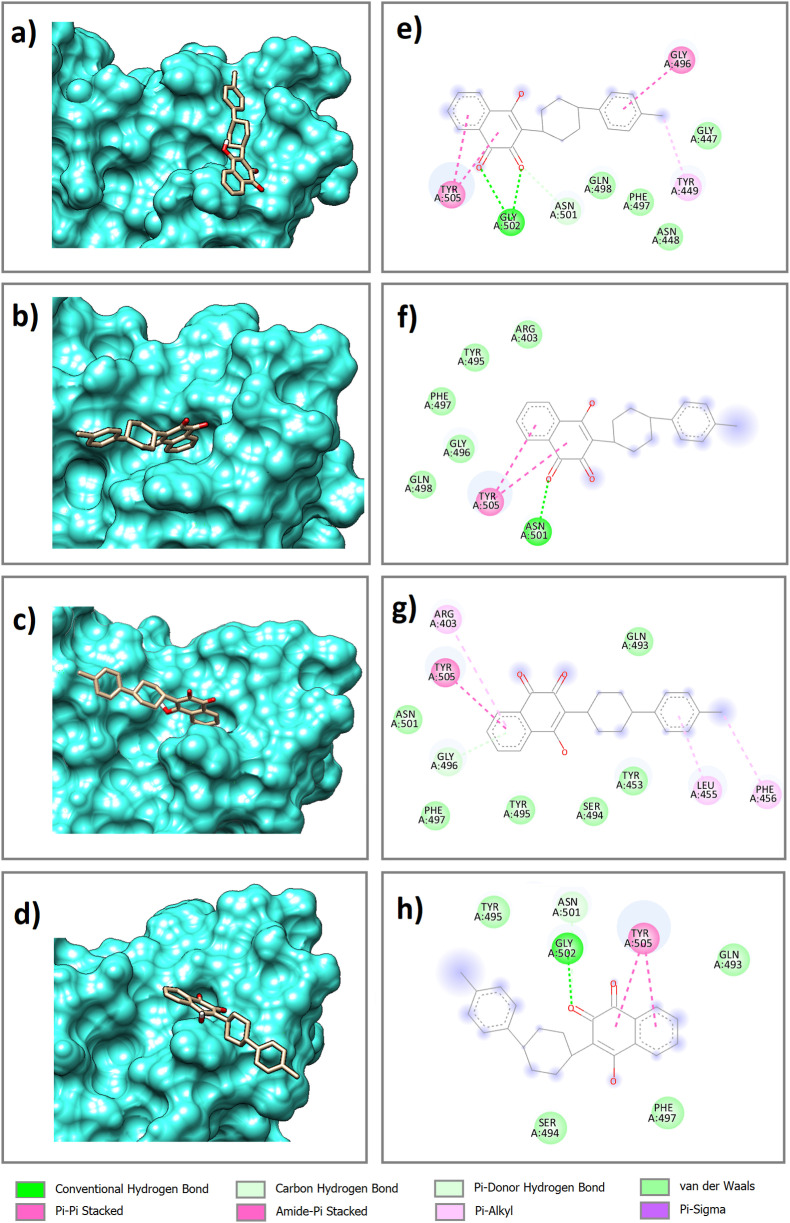
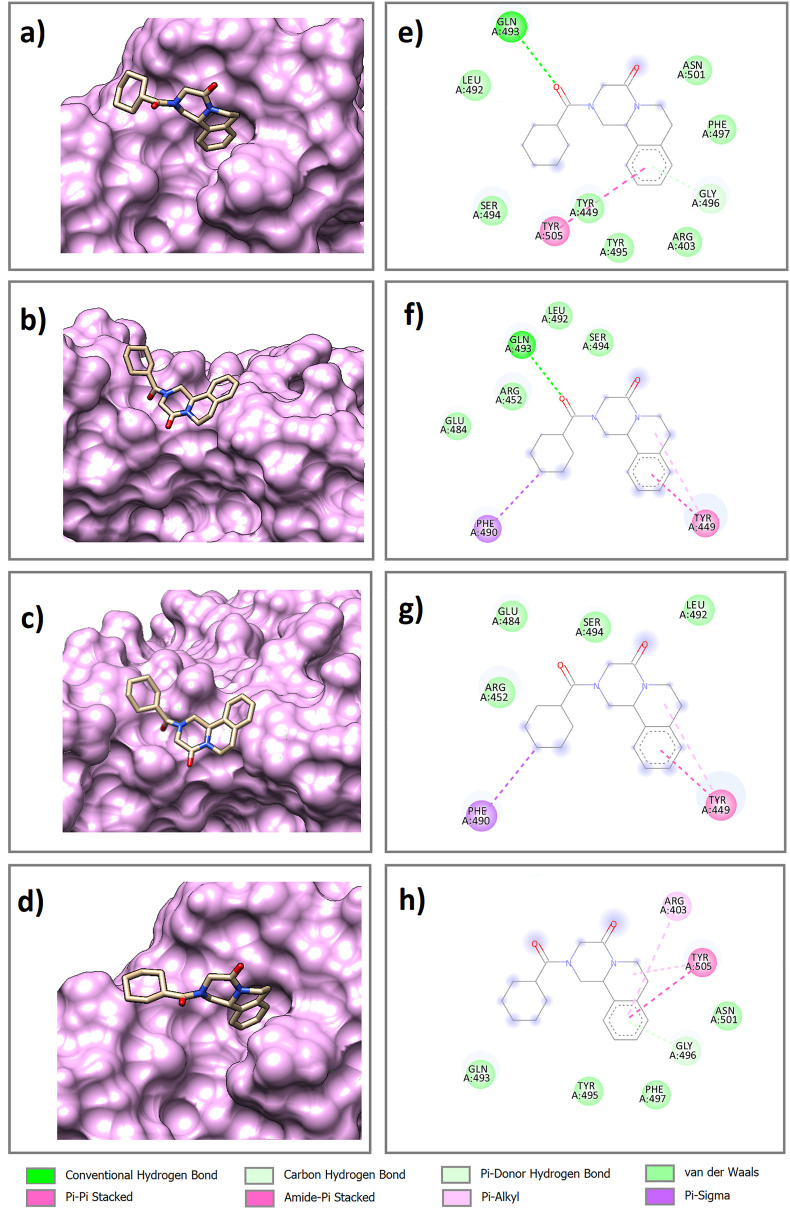
The best docking pose and orientation of atv and prz, the two final compounds obtained from the virtual screening, in the binding site of the wild-type and mutated (Delta, Delta plus, and Lambda) RBD of the spike protein were evaluated (Fig. 4, Fig. 5 ). As observed in Fig. 4a, b, 4c, 4d and Fig. 5a, b, 5c, 5d, in all complexes, atv and prz molecules are positioned in such a way that they occupy well the cavities in the binding site. Also, the 2D diagrams of docked complexes were plotted to investigate the key residues in protein-ligand interactions using Discovery Studio Visualizer [65].
Fig. 4.
The best docking pose and orientation of atv in the binding site of a) wild-type RBD and mutated RBD of spike: b) Delta, c) Delta plus, and d) Lambda variants. 2D diagram intermolecular interactions in e) wt-atv, f) dlt-atv, g) pls-atv, and h) lmb-atv complexes.
Fig. 5.
The best docking pose and orientation of prz in the binding site of a) wild-type RBD and mutated RBD of spike: b) Delta, c) Delta plus, and d) Lambda variants. 2D diagram intermolecular interactions in e) wt-prz, f) dlt-prz, g) pls-prz, and h) lmb-prz complexes.
In the wt-atv complex, two conventional hydrogen bonds (h-bonds), one carbon h-bond, four van der Waals interactions and three π- π stacking interactions were observed. In the dlt-atv complex, there are one conventional h-bond, five van der Waals interactions and two π- π stacking interactions. In the pls-atv complex, one carbon h-bond, six van der Waals interactions and one π- π stacking interaction were seen. In the wt-atv complex, one conventional h-bond, four van der Waals interactions and two π- π stacking interactions were observed. Residues involved in the intermolecular interaction include Gly502, Asn501 and Gly496 (h-bonds), Gly496 and Tyr505 (π- π stacking interactions) and Arg403, Gly447, Asn448, Tyr453, Gln493, Ser494, Tyr495, Gly496, Phe497, Gln498, Asn501. It was further observed that prz showed one conventional h-bond, one carbon h-bond, seven van der Waals interactions and one π- π stacking interaction in wt-prz complex, one conventional h-bond, four van der Waals interactions and one π- π stacking interaction in dlt-prz complex, four van der Waals interactions and one π- π stacking interaction in pls-prz complex and one carbon h-bond, four van der Waals interactions and one π- π stacking interaction in lmp-prz complex. Residues involved in these intermolecular interactions are as follow: Gln493 and Gly496 (h-bonds), Tyr449 and Tyr505 (π- π stacking interactions) and Arg403, Arg452, Glu484, Leu492, Gln493, Ser494, Tyr495, Gly496, Phe497, Asn501.
3.5. MD simulation analysis
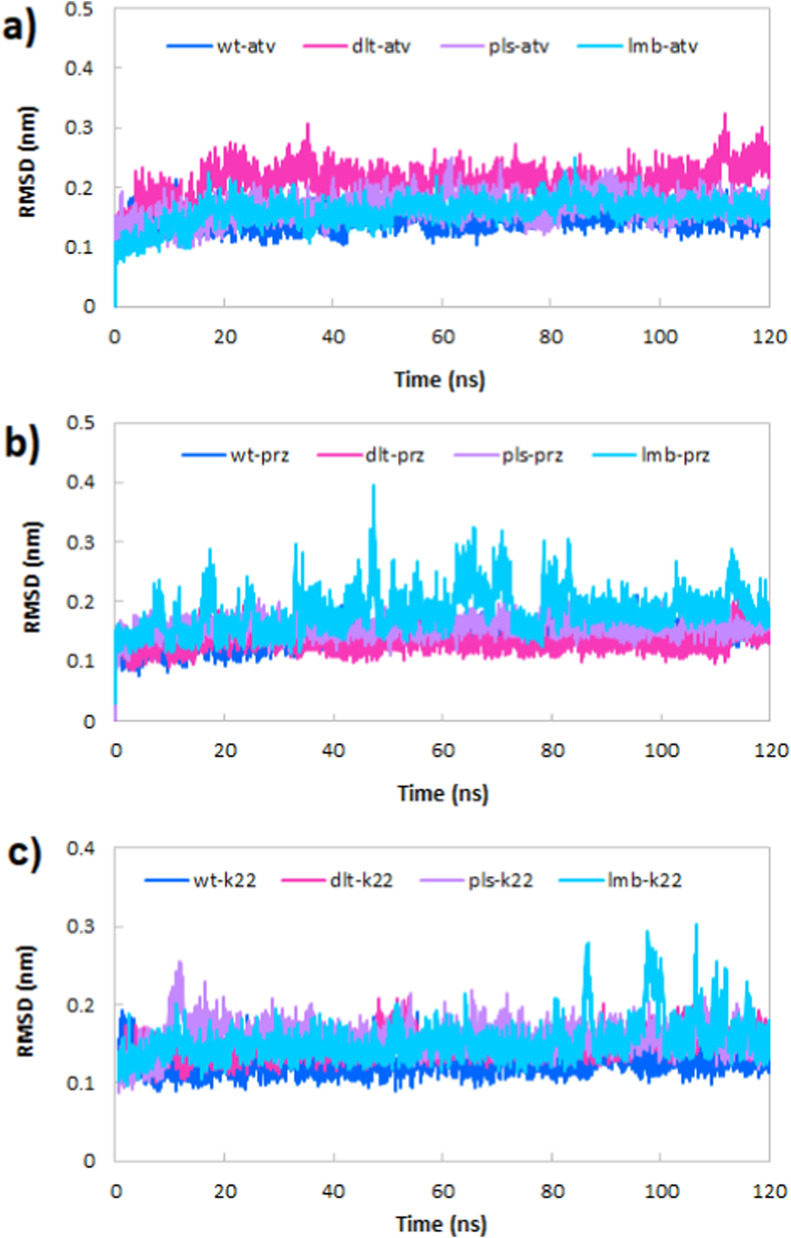
3.5.1. Root mean square deviations (RMSD)
Root mean square deviations (RMSD) of Cα atoms of all simulated systems were calculated with respect to the reference structure to investigate the conformational changes and structure stability. In Fig. 6 a, it is clear that the RMSD of all the complexes with atv equilibrated before 20 ns and remained stable until the end of the simulation time. All simulated trajectories show very low fluctuations and therefore stable trajectories. Also, RMSD curves and average values display that pls-atv (0.165 nm) and dlt-atv (0.208 nm) complexes have higher RMSD values than the wt-atv (0.146 nm) and the lmb-atv (0.155 nm) complexes, indicating more stability of wt-atv and lmb-atv complexes. Fig. 6b shows the time evolution of RMSD for the wild type, Delta, Delta plus and Lambda variants of RBD of the spike protein in complex with the prz. RMSD of all complexes show low fluctuation other than the lmb-prz complex. Among the simulated complexes, dlt-prz with the average RMSD 0.134 nm has the most structural stability. The wt-prz, pls-prz and lmb-prz complexes with average values 0.142, 0.157 and 0.183 nm are in the next order, respectively.
Fig. 6.
Root mean square deviations (RMSD) of Cα atoms of the wild type, delta, delta plus and lambda variants of RBD of spike in complex with the a) atv, b) prz and c) k22.
Also, Fig. 6c shows the time evolution of RMSD for the wild type, Delta, Delta plus and Lambda variants of RBD of the spike protein in complex with the k22, a viral spike protein inhibitor, as a reference molecule. This figure illustrates that RMSD of all complexes show low fluctuation other than the lmb-k22 complex. Moreover, all the complexes with k22 equilibrated before 10 ns and remained stable until the end of the simulation time. Furthermore, the RMSD of binding site residues and ligands heavy atoms were calculated (Table 2 ) to validate the stability of simulation. From the average RMSD values for the Cα atoms of proteins, binding site residues and ligands heavy atoms, it can be understood that the binding site residues have more stability than other residues which is related to the participation of these residues in interaction with ligands (atv, prz and k22). The same state (less RMSD values) can be seen for the RMSD of the ligand heavy atoms participation in the interaction with binding site residues.
Table 2.
The average value of structural parameters for all the complexes during 120-ns MD simulation.
| Simulated system | RMSD (nm) |
|||||
|---|---|---|---|---|---|---|
| Protein Cα |
Ligand heavy |
Binding site |
RMSF (nm) |
Rg (nm) |
No. of H-bonds |
|
| atoms | Atoms | Cα atoms | ||||
| wt-atv | 0.146 | 0.052 | 0.111 | 0.088 | 1.830 | 0.886 |
| dlt-atv | 0.208 | 0.066 | 0.149 | 0.088 | 1.860 | 0.043 |
| pls-atv | 0.165 | 0.098 | 0.152 | 0.092 | 1.850 | 0.052 |
| lmb-atv | 0.159 | 0.101 | 0.146 | 0.088 | 1.840 | 0.857 |
| wt-prz | 0.142 | 0.054 | 0.135 | 0.091 | 1.840 | 0.348 |
| dlt-prz | 0.134 | 0.077 | 0.130 | 0.083 | 1.830 | 0.395 |
| pls-prz | 0.157 | 0.110 | 0.148 | 0.089 | 1.840 | 0.250 |
| lmb-prz | 0.183 | 0.101 | 0.177 | 0.102 | 1.860 | 0.173 |
| wt-k22 | 0.127 | 0.067 | 0.114 | 0.085 | 1.840 | 0.605 |
| dlt-k22 | 0.146 | 0.088 | 0.140 | 0.090 | 1.850 | 0.695 |
| pls-k22 | 0.158 | 0.086 | 0.145 | 0.089 | 1.840 | 0.514 |
| lmb-k22 | 0.152 | 0.075 | 0.122 | 0.097 | 1.840 | 1.183 |
* RMSD values were calculated in protein-ligand complexes.
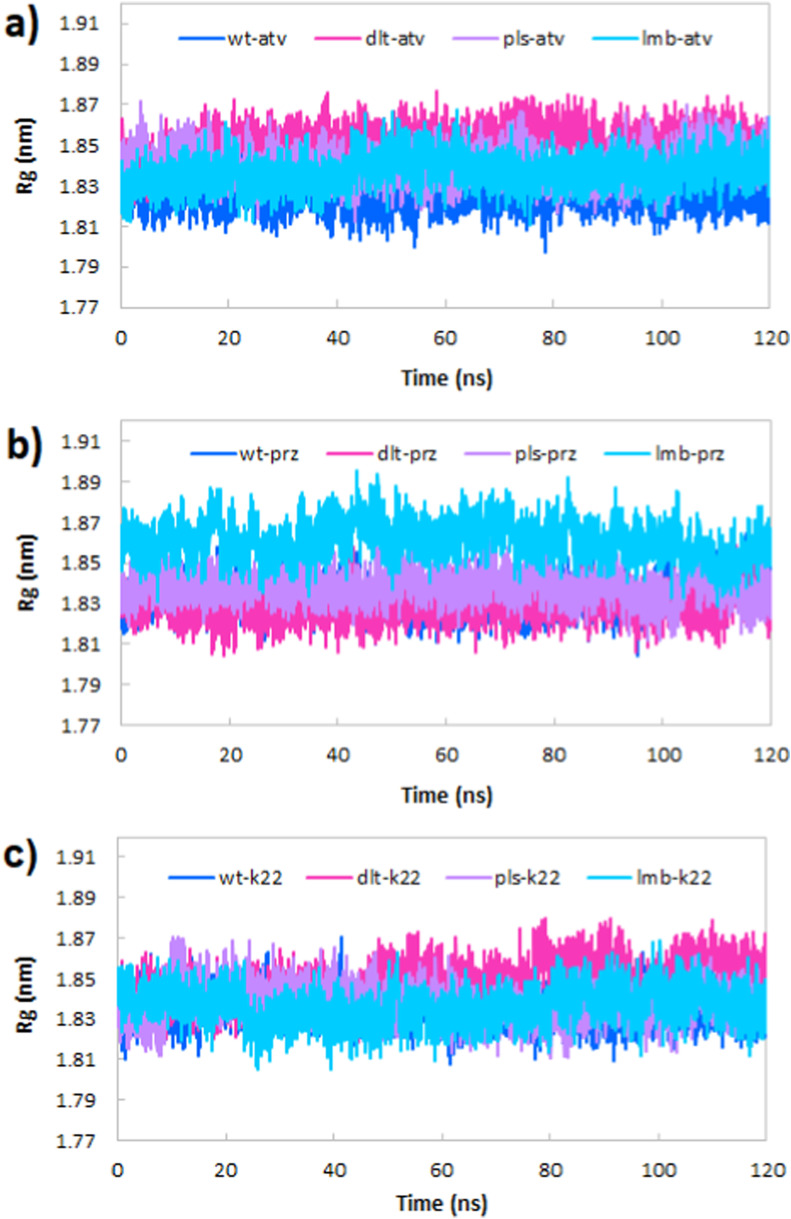
3.5.2. Radius of gyration (Rg)
The calculation of the radius of gyration (Rg) was done to assess the RBD of the spike overall compactness (Fig. 7 ). Time evolution of Rg curves in Fig. 7a and b displays that the fluctuation in Rg values is almost negligible in the complex of RBD of the spike protein with both atv and prz molecules. Moreover, the Rg values for all complexes were in the range of 1.80–1.90 nm. This depicts that protein was stable throughout the 120 ns simulation. Based on the average value of Rg in Table 2, it can be concluded that atv resulted in the more compactness in wild type of RBD of the spike protein relative to others, whereas prz has the more effect on the compactness of Delta variant of RBD of the spike. Also, Fig. 7c shows the slightly higher stability of proteins in complex with k22 relative to atv and prz.
Fig. 7.
Time dependence of the radius of gyration (Rg) of the wild type, Delta, Delta plus and Lambda variant of RBD of the spike.
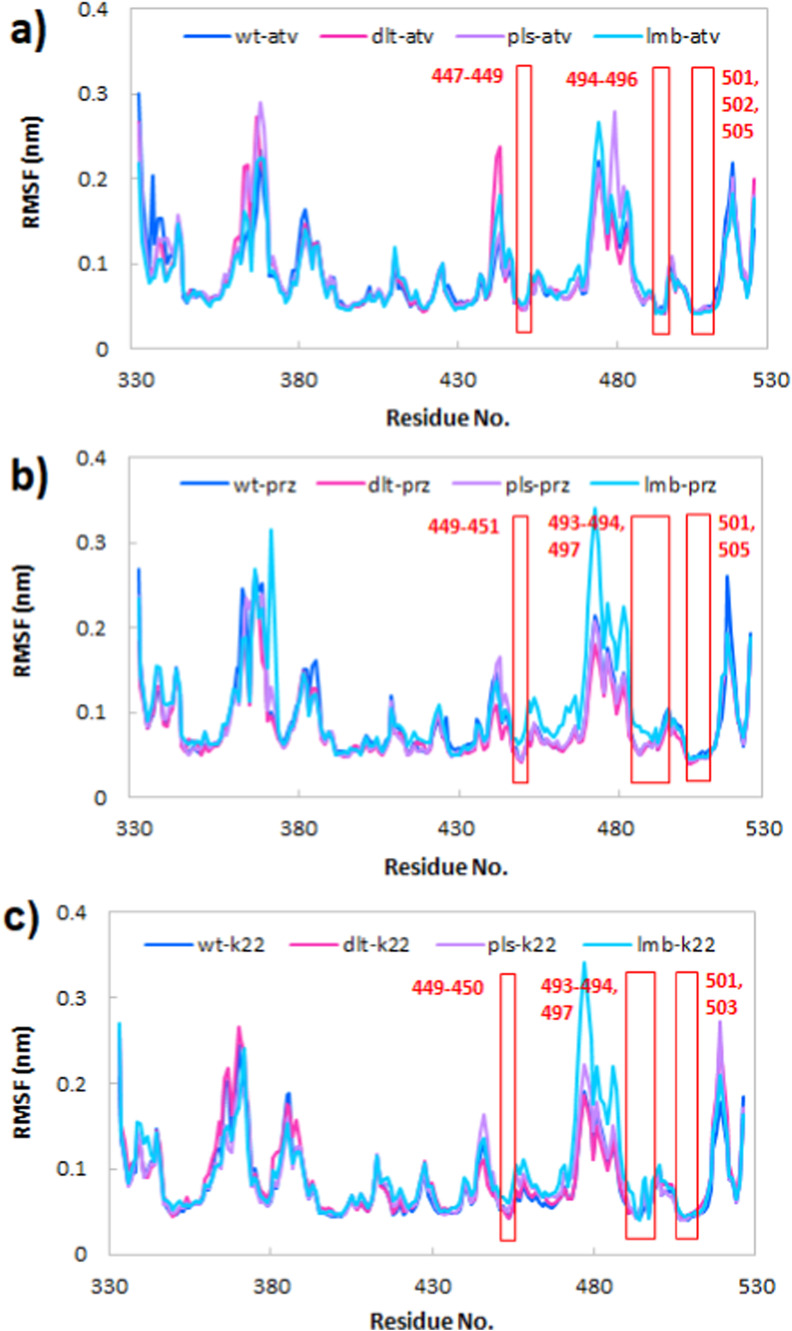
3.5.3. Root mean square fluctuation (RMSF)
The residual flexibility over the simulation time was explored by calculating the root mean square fluctuation (RMSF) of Cα atoms of all simulated systems. Per residue RMSFs were depicted in Fig. 8 . RMSF curves for all complexes with atv, prz and k22 indicate similar binding patterns. Based on this analysis, the least flexibility is related to those residues taking part in the interaction with atv and prz molecules, while the rest of the residues have higher mobility. These data are in good agreement with analysis discussed in Fig. 4, Fig. 5, Fig. 10. Residues with lower RMSF values are mostly in three regions; for complexes with atv molecule: 1) residues 447–449, 2) residues 494–496 and residues 501–505; for complexes with prz molecule: 1) residues 449–451, 2) residues 493–497 and residues 501–505 and for complexes with k22 molecule: 1) residues 449–450, 2) residues 493–494, 497 and residues 501, 503. As shown, there is a little difference between regions in Fig. 8a and b.
Fig. 8.
The root mean square fluctuation (RMSF) of Cα atoms of the wild type, Delta, Delta plus and Lambda variants of RBD of the spike protein in complex with the a) atv, b) prz and c) k22. The residues with the least RMSF values were marked with red rectangular boxes.
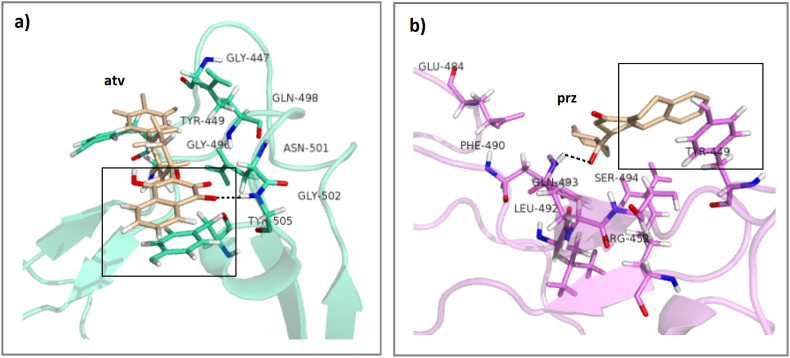
Fig. 10.
A representative snapshot of a) wt-atv and b) dlt-prz complexes. Compounds atv, prz and key residues of the binding site of RBD of the spike protein involved in the interaction are shown in sticks representation. The hydrogen bonds and the π-π stacking interaction are marked with black dash lines and black square, respectively.
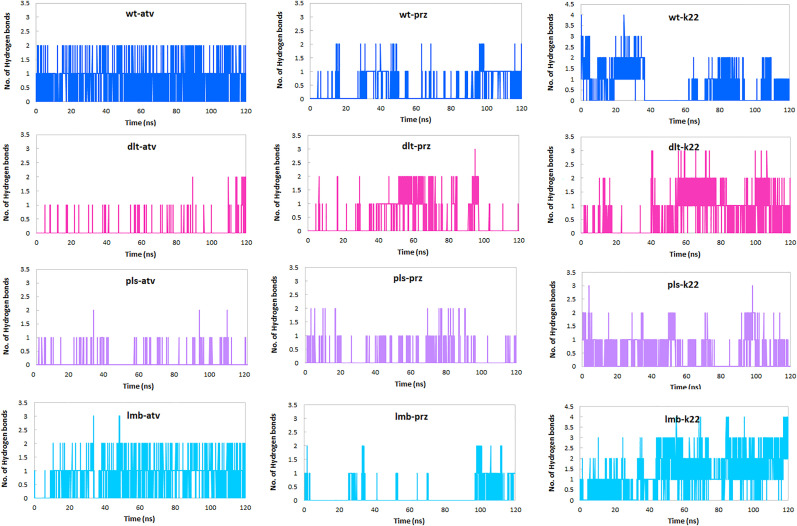
3.5.4. Hydrogen bond analysis
Due to the critical role of the hydrogen bonds in the stability of protein-ligands complexes and to have detailed information about intermolecular interactions between the wild type, Delta, Delta plus and Lambda variants of RBD of the spike protein and atv, prz and k22 molecules, the time evolution h-bonds was evaluated and depicted in Fig. 9 . The average number of h-bonds during the MD simulation for each complex summarized in Table 2. H-bond analysis shows that among the studied complexes, wt-atv, lmb-atv, dlt-prz and wt-prz complexes (with average number: 0.886, 0.857, 0.395 and 0.348) have the greatest number of h-bonds. As can be seen in Fig. 9, in wt-atv and lmb-atv complexes, two h-bonds are present during the most of the simulation time, whereas in pls-atv and dlt-atv complexes, one h-bond exists in some simulation frames. In case of complexes with prz, the time frames in which there are h-bonds are scattered. This scattering in the lmb-prz and pls-prz complexes is more obvious than the dlt-prz and wt-prz complexes. In case of complexes with k22, the average number of h-bonds is 0.605, 0.695, 0.514 and 1.183 for wt-k22, lmb-k22, dlt-k22 and wt-k22 complexes, respectively, indicating the importance of these bonds in the stability of these complexes. In some cases, the average number of h-bonds in complexes with atv is greater than that of complexes with k22.
Fig. 9.
Hydrogen bonds analysis. The average number of hydrogen bonds in wt-atv, dlt-atv, pls-atv, lmb-atv, wt-prz, dlt-prz, pls-prz, lmb-prz, wt-k22, dlt-k22, pls-k22, lmb-k22, complexes as a function of time.
Moreover, the h-bonds stability was assessed by the percentage of their presence during the simulation time. Also, the details of important h-bonds were briefed in Table 3 . As shown in Table, in the wt-atv complex, one h-bond with high occupancy (72.9%) is formed between the Gly502 N atom and the O2 atom of atv. Also, in the lmb-atv complex, Gly502 N atom contribute in one h-bond with O3 atom of atv with occupancy 71.5%. In case of dlt-prz and wt-prz complexes, NE2 atom Gln493 participates in h-bonds with O1 atom of prz with occupancies 23.8 and 22.1% respectively. We found that these h-bonds play a main role in binding of the atv and prz molecules to the RBD of the spike protein. In case of complexes with k22, two h-bonds with high occupancy (75.1% and 65.3%) are formed between the Gly502 N atoms (in dlt-k22 complex) and Ser 494 (lmb-k22 complex) OG atoms and the O3 atom of k22. Also, in the pls-k22 complex, Gly502 N atom contributes in one h-bond with O1 atom of k22 with occupancy 33.4%. The lowest occupancy of hydrogen bonds was observed between Ser 494 OG atoms and the O2 atom of k22 in the wt-k22 complex.
Table 3.
The hydrogen bonds formed between the wild-type and mutated (Delta, Delta plus, and Lambda) RBD of the spike protein and atv and prz molecules.
| Protein | Ligand | Occupancy (%)* |
|---|---|---|
| wt: GLY502 (N) | atv (O2) | 72.9 |
| lmb: GLY502 (N) | atv (O3) | 71.5 |
| dlt: GLN493(NE2) | prz (O1) | 23.8 |
| wt: GLN493(NE2) | prz (O1) | 22.1 |
| dlt: GLY502 (N) | k22 (O3) | 75.1 |
| lmb: SER494 (OG) | k22 (O3) | 65.3 |
| pls: GLY502 (N) | k22 (O1) | 33.4 |
| wt: SER494 (OG) | k22 (O2) | 25.6 |
*Only the hydrogen bonds with occupancy above 20% during the MD were included.
3.5.5. MM-PBSA binding free energy calculation
To explain the energetics of the binding of atv and prz molecules to the wild type, Delta, Delta plus and Lambda variants of the RBD of the spike protein, the binding free energy for each complex calculated using the g_mmpbsa tool and its results tabulated in Table 4 . To increase the accuracy of the binding free energy calculations, based on the RMSD results (Fig. 6), snapshots related to the last 90 ns of the MD trajectories were considered. ΔG bind for studied complexes is as follows: wt-atv: −20.1 ± 2.5 kcal/mol, dlt-atv: −15.9 ± 2.1 kcal/mol, pls-atv: −16.9 ± 2.3 kcal/mol, lmb-atv: −17.6 ± 2.8 kcal/mol, wt-prz: −17.8 ± 2.6 kcal/mol, dlt-prz: −19.5 ± 2.3 kcal/mol, pls-prz: −12.1 ± 1.6 kcal/mol, lmb-prz: −7.5 ± 1.1 kcal/mol, wt-k22: −14.9 ± 1.4 kcal/mol, dlt-k22: −21.79 ± 2.2 kcal/mol, pls-k22: −16.8 ± 1.9 kcal/mol and lmb-k22: −19.7 ± 2.6 kcal/mol. Based on these results, it was found that atv has the most binding affinity to the wt and lmb, respectively, whereas the most binding affinity of prz is related to the dlt and wt, respectively. Also, k22 show the most.
Table 4.
Binding free energy (kcal/mol) of the simulated complexes calculated by the MM-GBSA method.
| System | ΔEvdw | ΔEele | ΔGpol | ΔGnonpol | ΔGbinding | ΔΔG* |
|---|---|---|---|---|---|---|
| wt-atv | -28.35 | -5.35 | 16.40 | -2.75 | -20.05 | 1.74 |
| dlt-atv | -26.79 | -3.75 | 15.02 | -2.75 | -15.88 | 5.91 |
| pls-atv | -23.27 | -3.29 | 11.12 | -2.67 | -16.92 | 4.87 |
| lmb-atv | -26.88 | -5.93 | 17.95 | -2.73 | -17.58 | 4.21 |
| wt-prz | -27.75 | -5.17 | 17.87 | -2.76 | -17.81 | 3.98 |
| dlt-prz | -30.24 | -4.14 | 17.52 | -3.38 | -19.52 | 2.27 |
| pls-prz | -21.10 | -3.96 | 13.16 | -2.41 | -12.16 | 9.63 |
| lmb-prz | -6.76 | -2.82 | 2.84 | -0.73 | -7.47 | 14.32 |
| wt-k22 | -26.81 | -0.40 | 14.82 | -2.57 | -14.95 | 6.84 |
| dlt-k22* | -40.28 | -1.69 | 23.57 | -3.39 | -21.79 | 0.00 |
| pls-k22 | -22.28 | -3.13 | 10.15 | -1.48 | -16.75 | 5.04 |
| lmb-k22 | -36.88 | -10.06 | 31.18 | -3.94 | -19.69 | 2.10 |
*ΔΔG is defined as the change of binding free energy using dlt-k22 as a reference.
Binding affinity to the dlt. However, for the other three complexes, significant binding free energy is seen. Given that k22 is a viral spike protein inhibitor, these findings are not surprising. But the important point is the proximity of some ΔG bind values of atv and prz complexes to k22 complexes. In all complexes, the van der Waals energy term (ΔEvdw) has the major contribution to the binding free energy. Next are the electrostatic (ΔEele) and non-polar solvation (ΔGnon-polar), whereas the polar solvation (ΔGpolar) term opposes binding.
Since the g_mmpbsa tool applied to calculate ΔGbinding in this study exclude the entropy contribution in the binding free energy, the binding free energy difference (ΔΔG) was generally used to assess the relative strength of activities in the similar systems with series ligands or mutated receptors. In several similar studies, this trend was applied [[66], [67], [68], [69], [70]].
Also, to gain better insight, per-residue binding free energy decomposition analysis was performed to get the energy contributions of the binding site residues to the total binding free energy (Table 5 ). As can be seen in this table, the residues with the most favorable contributions (lower than −0.4 kcal/mol) to the ΔGbinding, in most of the complexes, are bolded. These residues are located on the active sites (Gly 447, Asn 448, Tyr 449, Arg 452, Glu 484, Leu 492, Gln 493, Ser 494, Tyr 495, Gly 496, Phe 497, Gln 498, Asn 501, Gly 502, Tyr 505). According to Table 5, the contributions of Tyr 449, Phe 497, Gly 502 and Tyr 505 to ΔGbinding values for most of the systems are more than for other residues. These findings are in good agreement with RMSF results. Those residues having more contribution in ΔG bind, show less fluctuation because they participate in the interaction with the atv and prz molecules.
Table 5.
Energy contribution of the key residues to the binding free energy.
| atv |
prz |
|||||||
|---|---|---|---|---|---|---|---|---|
| Residues | wt | dlt | pls | lmb | wt | dlt | pls | lmb |
| Gly 447 | -0.572 | -0.089 | 0.004 | 0.245 | -0.118 | 0.002 | -0.007 | -0.003 |
| Asn 448 | -0.287 | -0.058 | 0.202 | -0.320 | -0.140 | 0.003 | -0.005 | -0.014 |
| Tyr 449 | -0.759 | -0.103 | -0.017 | -0.355 | -0.742 | -0.678 | -0.349 | -0.047 |
| Arg 452 | -0.001 | -0.149 | -0.010 | 0.019 | -0.002 | -0.451 | -0.692 | 0.002 |
| Glu 484 | -0.111 | 0.140 | -0.008 | -0.120 | -0.132 | -0.888 | -0.118 | 0.019 |
| Leu 492 | -0.011 | -0.019 | 0.002 | -0.003 | -0.411 | -0.403 | -0.447 | -0.015 |
| Gln 493 | -0.005 | -0.480 | -0.010 | -0.834 | 0.361 | -1.751 | -0.310 | -0.213 |
| Ser 494 | -0.118 | 0.326 | -0.711 | -0.512 | -0.397 | -0.512 | -0.226 | 0.000 |
| Tyr 495 | -0.140 | -0.542 | -0.403 | -0.951 | -0.551 | 0.003 | -0.009 | -0.403 |
| Gly 496 | -0.147 | -0.502 | -0.302 | -0.237 | -0.383 | 0.003 | -0.008 | -0.613 |
| Phe 497 | -0.709 | -1.059 | -0.902 | -0.789 | -0.996 | 0.002 | 0.003 | -0.701 |
| Gln 498 | -0.637 | -0.282 | 0.001 | -0.438 | -0.194 | -0.006 | -0.007 | -0.013 |
| Asn 501 | 0.129 | 0.710 | 0.005 | 0.175 | 0.262 | -0.003 | 0.009 | -0.004 |
| Gly 502 | -1.132 | 0.071 | -0.002 | -0.686 | -0.391 | -0.003 | -0.132 | -0.017 |
| Tyr 505 | -3.069 | -1.800 | -1.651 | -3.127 | -2.876 | -0.147 | 0.010 | -0.711 |
Residues with the most favorable contributions (lower than −0.4 kcal/mol) to the ΔGbinding, are bolded.
To investigate the intermolecular interactions in detail in the wt-atv and dlt-prz complexes, the most stable complexes based on ΔGbind, the representative frames of the wt-atv and dlt-prz complexes were displayed in Fig. 10. In the wt-atv complex, the atv is located in the region around which there are Gly447, Tyr 449, Gly496, Gln498, Asn501, Gly502 and Tyr505 residues (Fig. 10a). Among these residues, Gly502 contributes in h-bond with atv molecules and Tyr505 takes part in π-π stacking interaction. In case of the dlt-prz complex, residues Glu484, Phe490, Leu492, Gln493, Ser494 and Tyr449 are around the prz molecule. Gln493 and Tyr449 participate in h-bond and π-π stacking interactions (Fig. 10b). These results show good consistency with molecular docking results (Fig. 4, Fig. 5).
4. Conclusions
Considering that we are encountering with unpredictably different mutations in the SARS-CoV-2 genome, in this study, exhaustive structure-based virtual screening (including drug-likeness screening, molecular docking and ADMET parameters), MD simulation and MM-PBSA were performed to detect potent inhibitors for wild type, Delta, Delta plus and Lambda variants of RBD of the spike protein for COVID-19 treatment. Two compounds (Atovaquone and Praziquantel) were shortlisted based on drug-likeness screening, molecular docking and pharmacokinetic properties evaluation. Molecular docking analysis showed that there are several interactions such as hydrogen bond, hydrophobic and π- π stacking interactions between atv and prz and binding site residues (especially Tyr449, Phe490, Gln493, Asn501, Gly502, and Tyr505) in the wild type, Delta, Delta plus and Lambda variants of RBD of the spike protein. These interactions are more observed in the wt-atv, lmb-atv, dlt-prz and wt-prz complexes. The validation of docking results was done by comparing with experimental data (heparin in complex with wild type and Delta variants). Furthermore, MD simulations analysis showed that among the studied complexes, the wt-atv and dlt-prz complexes have the most structural stability during the simulation time. Furthermore, using MM-PBSA analysis, we found that in the atv containing complexes, highest binding affinity is related to the wt-atv complex and in the prz containing complexes, it is related to the dlt-prz complex. The lmb-atv and wt-prz are in the next orders, respectively. Since the final two compounds are FDA approved, there is no need to do long-term clinical trials. This is the result of using the drug repurposing approach applied in this study. Therefore, we identified the atv and prz molecules as the potent inhibitors targeting RBD of the spike protein to treat the COVID-19. Comparison of the obtained results with the result of simulation of the k22 with the studied proteins showed that atv and prz are suitable inhibitors for these proteins. Moreover, the molecular mechanism of inhibition studied in this work will be helpful for designing the new SARS-CoV-2 inhibitors.
Declaration of competing interest
We have no conflict of interest to declare. This statement is to certify that all Authors have seen and approved the manuscript being submitted.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105709.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wang C., Horby P., Hayden F., Gao G. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/s0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.COVID Live Update: 432,176,247 Cases and 5,949,044 Deaths from the Coronavirus - Worldometer, Worldometers.Info. 2022. https://www.worldometers.info/coronavirus/ accessed. [Google Scholar]
- 3.Vardhan S., Sahoo S. Silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput. Biol. Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X., Ding Y., Du J., Fan Y. Update on human coronaviruses: one health, one world. Med. Novel Technol. Dev. 2020;8 doi: 10.1016/j.medntd.2020.100043. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L., Zhong L. Genomics functional analysis and drug screening of SARS-CoV-2. Genes & Diseases. 2020;7:542–550. doi: 10.1016/j.gendis.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., et al. The coding capacity of SARS-CoV-2. Nature. 2020;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 7.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirchdoerfer R., Cottrell C., Wang N., Pallesen J., Yassine H., Turner H., et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali A., Vijayan R. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-71188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muhseen Z., Hameed A., Al-Hasani H., Tahir ul Qamar M., Li G. Promising terpenes as SARS-CoV-2 spike receptor-binding domain (RBD) attachment inhibitors to the human ACE2 receptor: integrated computational approach. J. Mol. Liq. 2020;320 doi: 10.1016/j.molliq.2020.114493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulotta M., Lombino J., Perricone U., De Simone G., Mekni N., De Rosa M., et al. Targeting SARS‐CoV‐2 RBD interface: a supervised computational data‐driven approach to identify potential modulators. ChemMedChem. 2020;15:1921–1931. doi: 10.1002/cmdc.202000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108427. 108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mujwar S. Computational repurposing of tamibarotene against triple mutant variant of SARS-CoV-2. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104748. 104748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tracking SARS-CoV-2 variants. WHO.Int. 2022 https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ accessed. [Google Scholar]
- 19.Cosar B., Karagulleoglu Z., Unal S., Ince A., Uncuoglu D., Tuncer G., et al. Cytokine & Growth Factor Reviews; 2021. SARS-CoV-2 Mutations and Their Viral Variants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammadi M., Shayestehpour M., Mirzaei H. The impact of spike mutated variants of SARS-CoV2 [Alpha, Beta, Gamma, Delta, and Lambda] on the efficacy of subunit recombinant vaccines. Braz. J. Infect. Dis. 2021;25 doi: 10.1016/j.bjid.2021.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baral P., Bhattarai N., Hossen M., Stebliankin V., Gerstman B., Narasimhan G., et al. Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 Delta variant B.1.617.2 and implications for immune evasion. Biochem. Biophys. Res. Commun. 2021;574:14–19. doi: 10.1016/j.bbrc.2021.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yousefi H., Mashouri L., Okpechi S., Alahari N., Alahari S. Repurposing existing drugs for the treatment of COVID-19/SARS-CoV-2 infection: a review describing drug mechanisms of action. Biochem. Pharmacol. 2021;183 doi: 10.1016/j.bcp.2020.114296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh R., Bhardwaj V., Das P., Purohit R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma J., Kumar Bhardwaj V., Singh R., Rajendran V., Purohit R., Kumar S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against nonstructural protein-15 of SARS-CoV-2. Food Chem. 2021;346 doi: 10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh R., Bhardwaj V., Sharma J., Purohit R., Kumar S. In-silico evaluation of bioactive compounds from tea as potential SARS-CoV-2 nonstructural protein 16 inhibitors. Journal of Traditional and Complementary Medicine. 2022;12:35–43. doi: 10.1016/j.jtcme.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh R., Bhardwaj V., Purohit R. Potential of turmeric-derived compounds against RNA‐dependent RNA polymerase of SARS‐CoV‐2: an in-silico approach. Comput. Biol. Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104965. 104965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R., Bhardwaj V., Sharma J., Kumar D., Purohit R. Identification of potential plant bioactive as SARS-CoV-2 Spike protein and human ACE2 fusion inhibitors. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prieto Santamaría L., Díaz Uzquiano M., Ugarte Carro E., Ortiz-Roldán N., Pérez Gallardo Y., Rodríguez-González A. Drug Discovery Today; 2021. Integrating Heterogeneous Data to Facilitate COVID-19 Drug Repurposing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain R., Mujwar S. Repurposing metocurine as main protease inhibitor to develop novel antiviral therapy for COVID-19. Struct. Chem. 2020;31:2487–2499. doi: 10.1007/s11224-020-01605-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazniewski M., Dermawan D., Hidayat S., Muchtaridi M., Dawson W., Plewczynski D. Drug repurposing for identification of potential spike inhibitors for SARS-CoV-2 using molecular docking and molecular dynamics simulations. Methods. 2022 doi: 10.1016/j.ymeth.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhardwaj V., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021;128 doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahanta S., Chowdhury P., Gogoi N., Goswami N., Borah D., Kumar R., et al. Potential anti-viral activity of approved repurposed drug against main protease of SARS-CoV-2: an in silico based approach. J. Biomol. Struct. Dyn. 2020;39:3802–3811. doi: 10.1080/07391102.2020.1768902. [DOI] [PubMed] [Google Scholar]
- 33.Li Q., Shah S. 2017. Structure-Based Virtual Screening, Protein Bioinformatics; pp. 111–124. [DOI] [PubMed] [Google Scholar]
- 34.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7 doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipinski C., Lombardo F., Dominy B., Feeney P. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 36.Martin Y. A bioavailability score. J. Med. Chem. 2005;48:3164–3170. doi: 10.1021/jm0492002. [DOI] [PubMed] [Google Scholar]
- 37.Gasteiger J., Marsili M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. doi: 10.1016/0040-4020(80)80168-2. [DOI] [Google Scholar]
- 38.Pettersen E., Goddard T., Huang C., Couch G., Greenblatt D., Meng E., et al. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Dallakyan S., Olson A. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2014:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 40.Trott O., Olson A., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009 doi: 10.1002/jcc.21334. NA-NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rappe A., Casewit C., Colwell K., Goddard W., Skiff W. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992;114:10024–10035. doi: 10.1021/ja00051a040. [DOI] [Google Scholar]
- 42.Harisna A., Nurdiansyah R., Syaifie P., Nugroho D., Saputro K., Firdayani, et al. In silico investigation of potential inhibitors to main protease and spike protein of SARS-CoV-2 in propolis. Biochemistry And Biophysics Reports. 2021;26 doi: 10.1016/j.bbrep.2021.100969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mysinger M., Carchia M., Irwin J., Shoichet B. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J. Med. Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song Y., He P., Rodrigues A., Datta P., Tandon R., Bates J., et al. Anti-SARS-CoV-2 activity of rhamnan sulfate from monostroma nitidum. Mar. Drugs. 2021;19:685. doi: 10.3390/md19120685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pires D., Blundell T., Ascher D. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58:4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abraham M.J., van der Spoel D., Lindahl E. 2016. B. Hess, and the GROMACS Development Team.http://www.gromacs.org GROMACS User Manual Version. [Google Scholar]
- 47.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J., Dror R., et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Structure, Function, And Bioinformatics. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J., Wolf R., Caldwell J., Kollman P., Case D. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 49.Pearlman D., Case D., Caldwell J., Ross W., Cheatham T., DeBolt S., et al. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. doi: 10.1016/0010-4655(95)00041-d. [DOI] [Google Scholar]
- 50.Jakalian A., Bush B., Jack D., Bayly C. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method. J. Comput. Chem. 2000;21:132–146. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 51.Jorgensen W., Chandrasekhar J., Madura J., Impey R., Klein M. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 52.Wu W., Owino J., Al-Ostaz A., Cai L. SIMULIA Community Conference; 2014. Applying Periodic Boundary Conditions in Finite Element Analysis; pp. 707–719. [Google Scholar]
- 53.Hess B., Bekker H., Berendsen H., Fraaije J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(sici)1096-987x(199709)18:12<1463::aid-jcc4>3.0.co;2-h. [DOI] [Google Scholar]
- 54.Essmann U., Perera L., Berkowitz M., Darden T., Lee H., Pedersen L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 55.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 56.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 57.The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
- 58.Shagufta I. Ahmad. The race to treat COVID-19: potential therapeutic agents for the prevention and treatment of SARS-CoV-2. Eur. J. Med. Chem. 2021;213 doi: 10.1016/j.ejmech.2021.113157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kollman P., Massova I., Reyes C., Kuhn B., Huo S., Chong L., et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Accounts Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 60.Kumari R., Kumar R., Lynn A. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 61.Karami L., Saboury A., Rezaee E., Tabatabai S. Investigation of the binding mode of 1, 3, 4-oxadiazole derivatives as amide-based inhibitors for soluble epoxide hydrolase (sEH) by molecular docking and MM-GBSA. Eur. Biophys. J. 2016;46:445–459. doi: 10.1007/s00249-016-1188-0. [DOI] [PubMed] [Google Scholar]
- 62.Karami L., Tazikeh-Lemeski E., Saboury A. Molecular dynamics simulation and free energy studies on the interaction of salicylic acid with human serum albumin (HSA) Physical Chemistry Research. 2017;5:483–496. doi: 10.22036/PCR.2017.63757.1315. [DOI] [Google Scholar]
- 63.C S., S D., Ragunathan V., Tiwari P., A S., P B. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J. Biomol. Struct. Dyn. 2020;40:585–611. doi: 10.1080/07391102.2020.1815584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cereto-Massagué A., Guasch L., Valls C., Mulero M., Pujadas G., Garcia-Vallvé S. DecoyFinder: an easy-to-use python GUI application for building target-specific decoy sets. Bioinformatics. 2012;28:1661–1662. doi: 10.1093/bioinformatics/bts249. [DOI] [PubMed] [Google Scholar]
- 65.Biovia, Dassault Systèmes . Dassault Systèmes; San Diego: 2012. Discovery Studio Visualizer Software, Version 4.0. [Google Scholar]
- 66.Wang P., Zhang X., Fu T., Li S., Li B., Xue W., et al. Differentiating physicochemical properties between addictive and nonaddictive ADHD drugs revealed by molecular dynamics simulation studies. ACS Chem. Neurosci. 2017;8:1416–1428. doi: 10.1021/acschemneuro.7b00173. [DOI] [PubMed] [Google Scholar]
- 67.Wang P., Fu T., Zhang X., Yang F., Zheng G., Xue W., et al. Differentiating physicochemical properties between NDRIs and sNRIs clinically important for the treatment of ADHD. Biochim. Biophys. Acta Gen. Subj. 2017;1861:2766–2777. doi: 10.1016/j.bbagen.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 68.Fu T., Zheng G., Tu G., Yang F., Chen Y., Yao X., et al. Exploring the binding mechanism of metabotropic glutamate receptor 5 negative allosteric modulators in clinical trials by molecular dynamics simulations. ACS Chem. Neurosci. 2018;9:1492–1502. doi: 10.1021/acschemneuro.8b00059. [DOI] [PubMed] [Google Scholar]
- 69.Xue W., Fu T., Deng S., Yang F., Yang J., Zhu F. Molecular mechanism for the allosteric inhibition of the human serotonin transporter by antidepressant escitalopram. ACS Chem. Neurosci. 2022;13:340–351. doi: 10.1021/acschemneuro.1c00694. [DOI] [PubMed] [Google Scholar]
- 70.Xue W., Wang P., Tu G., Yang F., Zheng G., Li X., et al. Computational identification of the binding mechanism of a triple reuptake inhibitor amitifadine for the treatment of major depressive disorder. Phys. Chem. Chem. Phys. 2018;20:6606–6616. doi: 10.1039/C7CP07869B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.