Abstract
We cloned and sequenced the gene encoding an NADPH-dependent aldehyde reductase (ARII) in Sporobolomyces salmonicolor AKU4429, which reduces ethyl 4-chloro-3-oxobutanoate (4-COBE) to ethyl (S)-4-chloro-3-hydroxybutanoate. The ARII gene is 1,032 bp long, is interrupted by four introns, and encodes a 37,315-Da polypeptide. The deduced amino acid sequence exhibited significant levels of similarity to the amino acid sequences of members of the mammalian 3β-hydroxysteroid dehydrogenase–plant dihydroflavonol 4-reductase superfamily but not to the amino acid sequences of members of the aldo-keto reductase superfamily or to the amino acid sequence of an aldehyde reductase previously isolated from the same organism (K. Kita, K. Matsuzaki, T. Hashimoto, H. Yanase, N. Kato, M. C.-M. Chung, M. Kataoka, and S. Shimizu, Appl. Environ. Microbiol. 62:2303–2310, 1996). The ARII protein was overproduced in Escherichia coli about 2,000-fold compared to the production in the original yeast cells. The enzyme expressed in E. coli was purified to homogeneity and had the same catalytic properties as ARII purified from S. salmonicolor. To examine the contribution of the dinucleotide-binding motif G19-X-X-G22-X-X-A25, which is located in the N-terminal region, during ARII catalysis, we replaced three amino acid residues in the motif and purified the resulting mutant enzymes. Substrate inhibition of the G19→A and G22→A mutant enzymes by 4-COBE did not occur. The A25→G mutant enzyme could reduce 4-COBE when NADPH was replaced by an equimolar concentration of NADH.
Aldehyde reductase (EC 1.1.1.2), aldose reductase (EC 1.1.1.21), and carbonyl reductase (EC 1.1.1.184) catalyze NADPH-dependent reduction of a variety of carbonyl compounds and are widely distributed in mammalian and plant tissues. These enzymes are members of the aldo-keto reductase superfamily (4, 8); however, their physiological functions are not well understood. The amino acid sequences of aldose reductases and aldehyde reductases exhibit significant levels of similarity, but the amino acid sequences of carbonyl reductases do not (32).
In previous papers, we described purification and characterization of three NADPH-dependent aldehyde reductases (ARI, ARII, and ARIII) of the red yeast Sporobolomyces salmonicolor AKU4429 (9, 14, 34). ARI is the most abundant aldehyde reductase in this yeast and catalyzes asymmetric reduction of ethyl 4-chloro-3-oxobutanoate (4-COBE) to ethyl (R)-4-chloro-3-hydroxybutanoate (4-CHBE) {enantiomeric excess for (R) = [(R − S)/(R + S] × 100 and vice versa}, a promising chiral building block for organic synthesis. In contrast, ARII is produced in considerably smaller amounts but reduces 4-COBE to the (S) enantiomer (92.7% enantiomeric excess), which is also a useful chiral building block for chemical synthesis of pharmaceuticals. In addition to the stereoselectivity of activity against 4-COBE, the N-terminal amino acid sequences of these two aldehyde reductases are quite different. Based on the amino acid sequence deduced from the cDNA sequence, ARI belongs to the aldo-keto reductase superfamily (13). Recently, an NADPH-dependent aldehyde reductase (S1), which reduces 4-COBE to the (S) enantiomer (100% enantiomeric excess), was purified from Candida magnoliae AKU4643 (31). The substrate specificities, subunit structures, and N-terminal amino acid sequences of ARII and S1 are not similar. This indicates that the two enzymes belong to the different groups.
In this study, we cloned and analyzed a cDNA clone of the aldehyde reductase gene (ARII) in order to compare the catalytic mechanisms of ARI and ARII and to understand the molecular basis of the stereospecific reduction of 4-COBE.
MATERIALS AND METHODS
Microorganisms and culture conditions.
S. salmonicolor AKU4429 was used as the DNA donor. This organism was cultivated at 30°C in YPG medium containing 5% glucose, 1% peptone, and 1% yeast extract. Escherichia coli JM109 [recA supE endA hsdR gyrA relA thi Δ(lac-proAB)/F′ (traD proAB+ lacIq lacZΔM15)], E. coli LE392 (supE supF hsdR galK galT metB trpR lacY), and E. coli MV1184 [ara Δ(lac-proAB) rpsL thi (φ80lacZΔM15) Δ(srl-recA)306::Tn10 (tetr)/F′ (traD proAB+ lacIq lacZΔM15)] were used as host strains. E. coli cells were grown at 37°C in Luria-Bertani medium containing 1% Bacto-Tryptone (Difco Laboratories, Detroit, Mich.), 0.5% Bacto-Yeast Extract (Difco Laboratories), and 1% NaCl (pH 7.0). When necessary, ampicillin (100 μg/ml), streptomycin (30 μg/ml), and tetracycline (10 μg/ml) were added to the medium.
Enzymes and chemicals.
Restriction enzymes were purchased from Takara Shuzo Co., Ltd. (Kyoto, Japan) and Toyobo (Osaka, Japan). ExTaq DNA polymerase and LA Taq DNA polymerase were purchased from Takara Shuzo Co., Ltd.
Amino acid sequencing.
ARII was purified from cells of S. salmonicolor as described elsewhere (14) and was incubated with lysyl endopeptidase (Wako Pure Chemicals, Osaka, Japan) in 50 mM Tris-HCl (pH 9.0) containing 3 M urea for 12 h at 37°C at a substrate/enzyme molar ratio of 200:1. The resulting peptides were separated by high-performance liquid chromatography by using a Cosmosil 5C18-P column (4.6 by 150 mm; Nacalai Tesque, Kyoto, Japan) that previously had been equilibrated in 0.05% trifluoroacetic acid and was eluted with a linear acetonitrile gradient (0 to 100% acetonitrile in trifluoroacetic acid) at a flow rate of 0.6 ml/min. The amino acid sequences of peptides were determined with a model PPSQ-10 protein sequencer (Shimadzu, Kyoto, Japan). The phenylthiohydantoin amino acid derivatives were separated and identified with an on-line phenylthiohydantoin analyzer (model C-R7A; Shimadzu) as recommended in the instruction manual.
Screening of a genomic DNA library.
To amplify an ARII DNA fragment from S. salmonicolor chromosomal DNA by PCR, upstream and downstream primers were designed on the basis of the N terminus (14) and the internal amino acid sequence LYS, respectively. The sequences of the primers used were as follows: primer N1, 5′-GCIAA(A/G)AT(A/C/T)GA(C/T)AA(C/T)GCIGTI(C/T)T-3′; and primer LYS, 5′-TT(A/C)TC(A/G/T)ATICA(C/T)TCIA(A/G)(A/G)TTCCA-3′. Chromosomal DNA extracted from S. salmonicolor as described previously (13) was used as a template for amplification. The PCR mixture (100 μl) contained 50 pmol of each primer, each deoxynucleoside triphosphate (dNTP) at a concentration of 200 μM, 10 mM Tris-HCl (pH 8.3) (at 25°C), 1.5 mM MgCl2, 0.01% (wt/vol) gelatin, 500 ng of template DNA, and 10 U of ExTaq DNA polymerase. The reaction mixture was overlaid with mineral oil, and the reaction was carried out by using a Perkin-Elmer Cetus thermal cycler. The initial template denaturation step consisted of 2 min at 94°C. The amplification profile (1 min at 47°C, 1 min at 72°C, and 1 min at 94°C) was repeated for 35 cycles. The PCR product was purified and cloned into pGEM-T (Promega, Madison, Wis.).
The PCR product derived from the genomic DNA was then labeled by using a DIG DNA labeling kit (Boehringer Mannheim), and the resulting preparation was used to screen an S. salmonicolor genomic DNA library (24,000 plaques) (13).
Cloning of ARII cDNA.
First-strand cDNA was synthesized from mRNA isolated from S. salmonicolor (28) by using random hexamers and a GeneAmp RNA PCR kit (Perkin-Elmer). To amplify ARII cDNA, two primers, primer N2 (5′-ATGGCCAAAATCGACAACGCTGTG-3′) and primer C1 (5′-GGTTTCGGAGCCGACGAGGTC-3′), were synthesized based on the genomic DNA sequence. The PCR mixture (100 μl) contained 20 pmol of each primer, each dNTP at a concentration of 200 μM, 10 mM Tris-HCl (pH 8.3) (at 25°C), 1.5 mM MgCl2, 0.01% (wt/vol) gelatin, first-strand cDNA, and 2.5 U of ExTaq DNA polymerase. The reaction mixture was overlaid with mineral oil, and the reaction was carried out by using a Perkin-Elmer Cetus thermal cycler. The initial template denaturation step consisted of 2 min at 94°C. The amplification profile (1 min at 65°C, 1 min at 72°C, and 1 min at 94°C) was repeated for 35 cycles. The PCR product was purified and cloned into pGEM-T to obtain pGEM-AR2.
Subcloning and DNA sequencing.
Phage particles were purified with LambdaSorb phage adsorbent (Promega), and DNA was extracted with phenol-chloroform. The subfragments generated were cloned into pUC118 and pUC119 to provide templates. DNA sequencing was performed by the dideoxy chain termination method (20, 24) by using an automated DNA sequencer (model 373A; Applied Biosystems). The sequencing reaction was carried out as recommended in the manuals for Taq dye terminator cycle sequencing kits (Applied Biosystems). The sequences were deduced from the data obtained for both strands.
Expression of ARII in E. coli.
A PCR was used to subclone the ARII gene. The two synthetic primers used for this procedure were primer N3 (5′-CCGGAATTCATAGGAGGGGATTATGGCCAAAATCGAC-3′), which contained a Shine-Dalgarno sequence (underlined nucleotides) (29) flanked by an EcoRI site, and primer C2 (5′-CGGAAGCTTTATCAAGCGGTTTCGGAGCCGACGAGG-3′), which was flanked by a HindIII site. Plasmid pGEM-AR2 was used as the template. The PCR mixture (100 μl) contained 20 pmol of each primer, each dNTP at a concentration of 200 μM, 10 mM Tris-HCl (pH 8.3) (at 25°C), 1.5 mM MgCl2, 0.01% (wt/vol) gelatin, 1 ng of pGEM-AR2, and 5 U of ExTaq DNA polymerase. The initial template denaturation step consisted of 5 min at 94°C. The amplification profile (1 min at 60°C, 1 min at 72°C, and 30 s at 94°C) was repeated for 25 cycles. The PCR-generated DNA fragment was ligated into pUC118 cleaved with EcoRI and HindIII and then was transformed into E. coli JM109. After ampicillin selection, several clones were picked, and the nucleotide sequence of the plasmid DNA was examined.
Purification of ARII expressed in E. coli.
E. coli JM109 cells carrying pUCAR2 were grown in 5 liters of Luria-Bertani medium containing ampicillin and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 9 h at 37°C. The cells (15 g, wet weight) were then harvested by centrifugation, resuspended in 100 ml of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA containing 500 μg of lysozyme per ml, and left on ice for 1 h. Then the cells were disrupted by sonication, and the debris was removed by centrifugation at 12,000 × g (30 min, 4°C). The supernatant was applied to a 200-ml DEAE-Sephacel (Pharmacia Biotech Inc., Tokyo, Japan) column (2.4 by 11 cm) that previously had been equilibrated with 10 mM Tris-HCl (pH 7.4)–0.1 mM dithiothreitol (buffer A). Proteins were eluted with a 500-ml linear 0 to 0.4 M NaCl gradient in buffer A. The fractions containing more than 10% of the maximum activity were combined. After the NaCl concentration was adjusted to 4 M with solid NaCl, the enzyme solution was applied to a Phenyl-Sepharose CL-4B (Pharmacia Biotech Inc.) column (2.4 by 11 cm) equilibrated with buffer A containing 4 M NaCl. After the column was washed with the same buffer, the enzyme was eluted by using a linear 4 to 0 M NaCl gradient and a simultaneous linear increase in the ethylene glycol concentration from 0 to 70% in buffer A. ARII was purified to electropheretic homogeneity by this procedure.
Site-directed mutagenesis and purification of ARII variants.
Site-directed mutagenesis of the ARII gene was carried out by using the method described by Hashimoto-Gotoh et al. (5) and a Mutan-Express Km kit (Takara Shuzo Co., Ltd.) as recommended by the manufacturer. The ARII gene was excised from pUCAR2 with EcoRI and HindIII and then ligated to pKF19k cleaved by the same restriction enzymes. The following oligonucleotides were used for mutagenic primers: 5′-AGCCGTTGGCAGCGGTGACGAC-3′ (G19→A), 5′-AAGCGACGAAAGCGTTGGCGCC-3′ (G22→A), and 5′-GACGTGCGAACCGACGAAGCC-3′ (A25→G) (the mismatched nucleotides are underlined). The presence of the mutations and the absence of unwanted mutations elsewhere in the gene were confirmed by sequencing both strands of the entire gene of each mutant. Each mutant gene was recovered by digestion with EcoRI and HindIII and then ligated to pUC118 cleaved with the same restriction enzymes. The plasmid was transformed into E. coli JM109, and then the mutant enzyme was purified from the recombinant E. coli cells as described above for the wild-type enzyme.
Analytical methods.
The activity of ARII and the optical purity of 4-COBE were determined as described elsewhere (34). The reductase activity was determined at 37°C by measuring the rate of decrease in the absorbance at 340 nm. The standard reaction mixture (1.0 ml) contained 0.2 mM 4-COBE, 125 μM NADPH, and 200 mM potassium phosphate buffer (pH 7.0). One unit of enzyme activity was defined as the amount of enzyme that catalyzed oxidation of 1 μmol of NADPH per min. Protein contents were measured by a Bio-Rad protein assay (Japan Bio-Rad Laboratories, Tokyo, Japan); bovine serum albumin was used as the standard. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed by the method of Laemmli (16). Western blotting (immunoblotting) was performed as described previously (13). The standard methods described by Sambrook et al. (23) were used for DNA manipulation.
Nucleotide sequence accession number.
The nucleotide sequence of the ARII gene has been deposited in the GenBank database under accession no. AF160799.
RESULTS AND DISCUSSION
Isolation of a cDNA clone.
Purified S. salmonicolor ARII was proteolytically digested with lysyl endopeptidase, and six peptides were obtained by reverse-phase high-performance liquid chromatography. The amino acid sequences of these peptides were VRGTARSASK, LANLQK, DLVGSETA, QGAYDEVIK, TEA, and SWNLESIDK. To clone the ARII gene on the basis of these sequences, two oligonucleotide probes, primer N1 derived from the N-terminal amino acid sequence and primer LYS derived from the internal amino acid sequence (SWNLESIDK), were synthesized. PCR performed with primers N1 and LYS yielded a single product that was approximately 0.6 kb long. This PCR product was subcloned into pGEM-T, and its nucleotide sequence was determined. We concluded that the 0.6-kb fragment was a portion of the ARII gene, because the amino acid sequences of five of the peptide fragments were found in the amino acid sequence deduced from the nucleotide sequence of this PCR product. The N-terminal amino acid sequence was interrupted by an inserted unrelated DNA sequence. These results strongly suggested that introns were present in the ARII gene. The PCR fragment was then used to screen a genomic DNA library constructed in λEMBL3, and five positive clones were obtained. One of these clones, SAL-27, was analyzed further. The nucleotide sequence of a 2.3-kb SalI-PstI fragment of this clone was determined. Open reading frames in this sequence encoded all of the peptides derived from direct amino acid sequencing of the protein.
To amplify ARII cDNA by reverse transcriptase PCR, two primers, one derived from the N-terminal amino acid sequence and the other derived from the C-terminal amino acid sequence, were synthesized by using the genomic DNA sequence. A 1.0-kb fragment was synthesized and cloned into pGEM-T, and its nucleotide sequence was determined and compared to the genomic DNA sequence. The genomic DNA sequence covering the region encoding ARII was 1,375 bp long and was interrupted by four introns. This analysis revealed a single 1,032-bp open reading frame encoding a 344-amino-acid protein. The predicted molecular mass (37,184 Da, excluding the initial methionine) was nearly identical to the molecular mass estimated by SDS-PAGE (37,000 Da) (14).
Comparison of the ARII amino acid sequence.
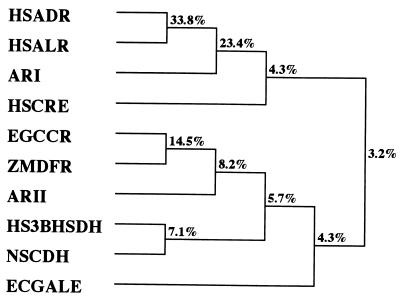
The deduced amino acid sequence of ARII was compared with other protein sequences in the GenBank database by using BLAST programs. The levels of identity with dihydroflavonol 4-reductases from higher plants (Zea mays, Arabidopsis thaliana, et al.), Eucalyptus gunnii cinnamyl alcohol dehydrogenase, and cinnamoyl coenzyme A (cinnamoyl-CoA) reductases were 30 to 34, 35, and 32 to 36%, respectively. Dihydroflavonol 4-reductase (EC 1.1.1.219) is required to convert dihydroflavonols to anthocyanin precursors (leucoanthocyanidins). Cinnamyl alcohol dehydrogenase (EC 1.1.1.195) catalyzes the conversion of p-hydroxycinnamaldehydes to the corresponding alcohols and is considered a key enzyme in lignin biosynthesis. Cinnamoyl-CoA reductase (EC 1.2.1.44) catalyzes the conversion of cinnamoyl-CoA esters to the corresponding cinnamaldehydes (i.e., the first specific step in the synthesis of lignin monomers). These enzymes are considered members of the mammalian 3β-hydroxysteroid dehydrogenase–plant dihydroflavonol 4-reductase superfamily (15). We compared the amino acid sequence of ARII with the amino acid sequences of other members of the superfamily by using CLUSTAL W programs. ARII exhibited significant levels of identity with bacterial UDP-galactose 4-epimerases (30%), Nocardia sp. cholesterol dehydrogenase (7) (30%), and mammalian 3β-hydroxysteroid dehydrogenases (30%). UDP-galactose 4-epimerase (EC 5.1.3.2) catalyzes the conversion of UDP-galactose to UDP-glucose through a mechanism involving transient reduction of NAD+. 3β-Hydroxysteroid dehydrogenase (EC 1.1.1.145) converts pregnenolone to progesterone, a key step in the synthesis of steroid hormones. NAD- or NADP-dependent cholesterol dehydrogenase from Nocardia sp. strain Ch 2-1 specifically oxidizes the 3β-OH group of cholesterol. In contrast, no similarity was found with carbonyl reductases or proteins belonging to the aldo-keto reductase superfamily, including ARI, which was isolated from S. salmonicolor. As shown in Fig. 1, ARII can be statistically considered a new member of the mammalian 3β-hydroxysteroid dehydrogenase–plant dihydroflavonol 4-reductase superfamily defined by Baker et al. (2) and Baker and Blasco (1). From the standpoint of molecular biology, we found the first instance of multiple 4-COBE-reducing enzymes with various stereoselectivities occurring in the same strain but belonging to different superfamilies.
FIG. 1.
Phylogenetic tree of ARII and related proteins. The tree was constructed by using the CLUSTAL V program (6). Abbreviations: HSADR, human aldose reductase (4); HSALR, human aldehyde reductase (4); ARI, S. salmonicolor ARI (13); HSCRE, human carbonyl reductase (32); ZMDFR, Z. mays dihydroflavonol 4-reductase (25); EGCCR, E. gunnii cinnamoyl-CoA reductase (15); ARII, S. salmonicolor ARII (this study); HS3BHSDH, human 3β-hydroxysteroid dehydrogenase (19); NSCDH, Nocardia sp. cholesterol dehydrogenase (7); ECGALE, E. coli UDP-galactose 4-epimerase (17).
As shown in Fig. 2, the N-terminal region of ARII between position 14 and position 49 is quite similar to the analogous regions of enzymes belonging to this superfamily. Most of these enzymes require NADPH; the only exception is UDP-galactose 4-epimerase, which requires NAD(H). The conserved N-terminal region is probably involved in the cofactor binding sites of the enzymes, since the secondary structure of E. coli UDP-galactose 4-epimerase (3, 30) and the predicted secondary structure of E. gunnii cinnamoyl-CoA reductase (15) correspond to the βαβ-dinucleotide binding fold of NADP(H)- and NAD(H)-dependent reductases and dehydrogenases (26, 33). The roles of the conserved amino acid residues in the N-terminal region were analyzed further, as described below. The four-amino-acid motif IPKS, which is thought to be involved in the interaction with NADPH in NADPH-dependent aldo-keto reductases, was not present in ARII.
FIG. 2.
Alignment of the N-terminal regions of ARII and related proteins. For an explanation of abbreviations see the legend to Fig. 1. The secondary structure of E. coli UDP-galactose 4-epimerase (30) and the calculated secondary structure of E. gunnii cinnamoyl-CoA reductase (15) are indicated below and above the amino acid sequence, respectively. The solid diamonds indicate amino acid residues which interact with NAD in E. coli UDP-galactose 4-epimerase. The numbers to the right and left of each sequence indicate the absolute locations within the whole sequence. The glycine residues in a putative nucleotide-binding motif are outlined in black. The positions conserved in cinnamoyl-CoA reductase and ARII are indicated by colons. α, α-helix; β, β-sheet.
Analysis of ARII purified from E. coli.
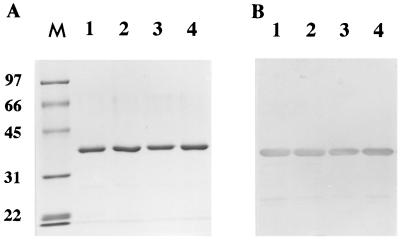
The ARII cDNA, to which a Shine-Dalgarno sequence was attached in the 5′-flanking region, was cloned into expression vector pUC118, and the resulting recombinant plasmid, pUCAR2, was transformed into E. coli JM109. Expression of the recombinant ARII was induced by adding IPTG; the enzyme was subsequently purified ninefold, and the yield was 61%. The purified enzyme produced a single band on SDS-PAGE gels (Fig. 3A) and on native PAGE gels (data not shown). The specific activity (15 U/mg), Km (1.82 mM), and Vmax (350 μmol min−1 mg−1) of the purified enzyme with 4-COBE were similar to the values obtained for the enzyme purified from S. salmonicolor (14). The sequence of the first 10 amino acids of the purified enzyme was determined by protein sequencing and was found to be identical to the sequence in the native enzyme. Also, the stereoselectivity [99.2% enantiomeric excess for (S)] for 4-COBE and the substrate specificity for typical aldehydes of the enzyme from E. coli were the same as those of the enzyme from S. salmonicolor.
FIG. 3.
SDS-PAGE of wild-type and mutant ARIIs purified from E. coli. Portions (1 μg) of wild-type (lane 1), G19→A (lane 2), G22→A (lane 3), and A25→G (lane 4) ARIIs were separated by 0.1% SDS–PAGE on a gel containing 10% polyacrylamide (A) and then analyzed by Western blotting (B) by using antibodies raised against wild-type ARII purified from E. coli. Lane M contained molecular mass standards, whose positions (in kilodaltons) are indicated on the left.
We noted that the amount of ARII produced was 5.8 mg/g of E. coli JM109 cells; this value was 2,000 times the amount produced in yeast cells (0.003 mg/g of cells). Since E. coli does not exhibit a detectable level of reducing activity with 4-COBE, this system could be useful for producing large amounts of ARII. We recently developed a method for asymmetric reduction of 4-COBE to (R)-CHBE involving E. coli cells expressing the ARI gene as a catalyst (10–12). A recombinant E. coli strain expressing the ARII gene could also be an economical way to produce (S)-CHBE.
Catalytic activity of ARII mutants.
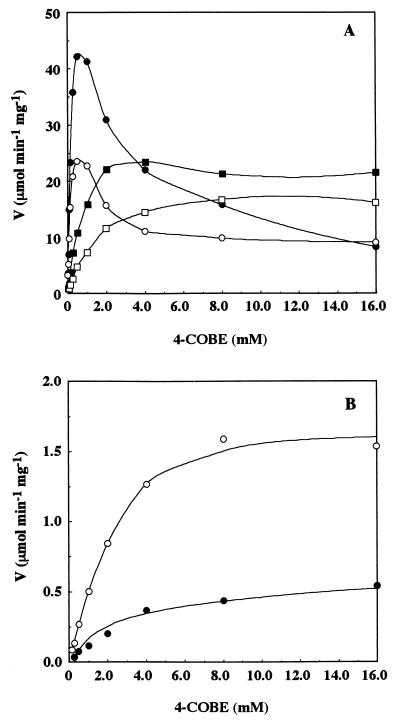
The amino acid sequence of ARII contains the dinucleotide-binding motif G19-X-X-G22-X-X-A25 in its N-terminal region. Since G7 and G10 in the same motif in the UDP-galactose 4-epimerase have been found to interact with NAD (30), we assumed that G19 and G22 in the motif in ARII might be involved in binding of NADP. Position 25 is of particular interest since there is a glycine residue at this position in most sequences, whereas the cinnamoyl-CoA reductases of E. gunnii (15), Saccharum officinarum (27), Z. mays (22), and Populus balsamifera subsp. trichocarpa (18) and ARII have an alanine residue at position 25. To determine the contributions of G19, G22, and A25 to the catalytic activity of ARII, we mutated these three amino acid residues and purified the mutant enzymes (G19→A, G22→A, and A25→G). The three mutant ARII enzymes were purified to homogeneity from E. coli lysates, and the yields were the same as the yield obtained for the wild-type enzyme (Fig. 3); they were then characterized by determining the NAD(P)H-dependent reductase activities. As expected, the G19→A and G22→A mutants both exhibited less than 20% of the activity of wild-type enzyme with NADPH in the presence of 0.2 mM 4-COBE, but the A25→G mutant exhibited the same level of activity as the wild-type enzyme. Initial velocity studies of 4-COBE reduction by the wild-type and mutant enzymes were performed by varying the concentration of 4-COBE and using a fixed concentration of NADPH (125 μM). The 4-COBE concentration was varied from 0.015 to 16 mM. The wild-type and A25→G mutant enzymes were strongly inhibited by excess concentrations of 4-COBE, but the G19→A and G22→A mutant enzymes were not (Fig. 4A). Wild-type ARII does not exhibit NADH-dependent reducing activity in the presence of 0.2 mM 4-COBE (14). We measured NADH-dependent 4-COBE reducing activity in the presence of a fixed concentration of NADH (125 μM). The 4-COBE concentration was varied from 0.015 to 16 mM. As shown in Fig. 4B, the wild-type and A25→G mutant enzymes exhibited significant activity with NADH in the presence of higher 4-COBE concentrations, but the G19→A and G22→A mutant enzymes did not (data not shown). The Km for 4-COBE (2.7 mM) was equivalent to the Km of the wild type (3.8 mM). The Vmax (1.7 μmol min−1 mg−1) of the A25→G mutant enzyme for 4-COBE when NADH was a cofactor was three times greater than the Vmax of the wild-type enzyme (0.66 μmol min−1 mg−1), which resulted in a 3.6-fold increase in the overall catalytic efficiency (kcat/Km, 21 min−1 mM−1) compared to the catalytic efficiency of the wild-type enzyme (5.9 min−1 mM−1). These results suggest that the amino acid residue at position 25 might be involved in recognition of the presence of a phosphate group at the 2′ position of the adenine ribose ring in nicotinamide nucleotide coenzymes and in the interaction of 4-COBE as well; i.e., some interaction between the substrate and NADPH might occur at residue 25. Changes in coenzyme specificity from NADP dependent to NAD dependent and vice versa have been observed with many enzymes (21, 26, 35). From a practical viewpoint, it is important to alter the coenzyme specificity of ARII, which is a useful catalyst for production of (S)-CHBE. Changing cofactor specificity was not possible with the replacement of one amino acid residue that interacted with the coenzyme. Replacement of a few residues or a module is required because a specific conformational change is induced by the binding of cofactor and substrate. We purified ARII in amounts sufficient for crystallization. Analysis of the three-dimensional structure of this enzyme should reveal a target region for replacement of amino acid residues for cofactor conversion.
FIG. 4.
Initial velocity (V) of 4-COBE reduction by the wild-type and mutant ARIIs. Symbols: ●, wild type; ■, G19→A mutant; □, G22→A mutant; ○, A25→G mutant. (A) Experiments performed with NADPH as a cofactor. (B) Experiments performed with NADH as a cofactor.
ACKNOWLEDGMENTS
This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan and by grant JSPS-RFTF 97I00302 from the Research for the Future program of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Baker M E, Blasco R. Expansion of the mammalian 3β-hydroxysteroid dehydrogenase/plant dihydroflavonol reductase superfamily to include a bacterial cholesterol dehydrogenase, a bacterial UDP-galactose 4-epimerase, and open reading frames in vaccinia virus and fish lymphocystis disease virus. FEBS Lett. 1992;301:89–93. doi: 10.1016/0014-5793(92)80216-4. [DOI] [PubMed] [Google Scholar]
- 2.Baker M E, Luu-The V, Simard J, Labri F. A common ancester for mammalian 3β-hydroxysteroid dehydrogenase and plant dihydroflavonol reductase. Biochem J. 1990;269:558–559. doi: 10.1042/bj2690558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer A J, Rayment I, Frey P A, Holden H M. The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5Å resolution. Proteins. 1992;12:372–381. doi: 10.1002/prot.340120409. [DOI] [PubMed] [Google Scholar]
- 4.Bohren K M, Bullock B, Wermuth B, Gabbay K H. The aldo-keto reductase superfamily. J Biol Chem. 1989;264:9547–9551. [PubMed] [Google Scholar]
- 5.Hashimoto-Gotoh T, Mizuno T, Ogasahara Y, Nakagawa M. An oligodeoxyribonucleotide-directed dual amber method for site-directed mutagenesis. Gene. 1995;152:271–275. doi: 10.1016/0378-1119(94)00750-m. [DOI] [PubMed] [Google Scholar]
- 6.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 7.Horinouchi S, Ishizuka H, Beppu T. Cloning, nucleotide sequence, and transcriptional analysis of the NAD(P)-dependent cholesterol dehydrogenase gene from a Nocardia sp. and its hyperexpression in Streptomyces spp. Appl Environ Microbiol. 1991;57:1386–1393. doi: 10.1128/aem.57.5.1386-1393.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jez J M, Bennett M J, Schlegel B P, Lewis M, Penning T M. Comparative anatomy for the aldo-keto reductase superfamily. Biochem J. 1997;326:625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kataoka M, Sakai H, Morikawa T, Katoh M, Miyoshi T, Shimizu S, Yamada H. Characterization of aldehyde reductase of Sporobolomyces salmonicolor. Biochim Biophys Acta. 1992;1122:57–62. doi: 10.1016/0167-4838(92)90127-y. [DOI] [PubMed] [Google Scholar]
- 10.Kataoka M, Rohani L P S, Yakamoto K, Wada M, Kawabata H, Kita K, Yanase H, Shimizu S. Enzymatic production of ethyl (R)-4-chloro-3-hydroxybutanoate: asymmetric reduction of ethyl 4-chloro-3-oxobutanoate by an Escherichia coli transformant expressing the aldehyde reductase gene from yeast. Appl Microbiol Biotechnol. 1997;48:699–703. doi: 10.1007/s002530051118. [DOI] [PubMed] [Google Scholar]
- 11.Kataoka M, Rohani L P S, Wada M, Kita K, Yanase H, Urabe I, Shimizu S. Escherichia coli transformant expressing the glucose dehydrogenase gene from Bacillus megaterium as a cofactor regenerator in a chiral alcohol production system. Biosci Biotechnol Biochem. 1998;62:167–169. doi: 10.1271/bbb.62.167. [DOI] [PubMed] [Google Scholar]
- 12.Kataoka M, Yamamoto K, Kawabata H, Wada M, Kita K, Yanase H, Shimizu S. Stereoselective reduction of ethyl 4-chloro-3-oxobutanoate by Escherichia coli transformant cells coexpressing the aldehyde reductase and glucose dehydrogenase genes. Appl Microbiol Biotechnol. 1999;51:486–490. doi: 10.1007/s002530051421. [DOI] [PubMed] [Google Scholar]
- 13.Kita K, Matsuzaki K, Hashimoto T, Yanase H, Kato N, Chung M C-M, Kataoka M, Shimizu S. Cloning of the aldehyde reductase gene from a red yeast, Sporobolomyces salmonicolor, and characterization of the gene and its product. Appl Environ Microbiol. 1996;62:2303–2310. doi: 10.1128/aem.62.7.2303-2310.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kita K, Nakase K, Yanase H, Kataoka M, Shimizu S. Purification and characterization of new aldehyde reductases from Sporobolomyces salmonicolor AKU4429. J Mol Catal B Enzym. 1999;6:305–313. [Google Scholar]
- 15.Lacombe E, Hawkins S, Van Doorsselaere J, Piquemal J, Goffner D, Poeydomenge O, Boudet A E, Grima-Pettenati J. Cinnamoyl-CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J. 1997;11:429–441. doi: 10.1046/j.1365-313x.1997.11030429.x. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lemaire H G, Muller-Hill B. Nucleotide sequences of the galE gene and the galT gene of E. coli. Nucleic Acids Res. 1986;14:7705–7711. doi: 10.1093/nar/14.19.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leple J C, Grima-Pettenati J, Montagu M, Boerjan W. A cDNA encoding cinnamoyl-CoA reductase from Populus trichocarpa. Plant Physiol. 1998;117:1126. [Google Scholar]
- 19.Lorence M C, Murry B A, Trant J M, Mason J I. Human 3β-hydroxysteroid dehydrogenase/δ 5-4 isomerase from placenta: expression in nonsteoidogenic cells of a protein that catalyzes the dehydrogenation/isomerization of C21 and C19 steroids. Endocrinology. 1990;126:2493–2498. doi: 10.1210/endo-126-5-2493. [DOI] [PubMed] [Google Scholar]
- 20.Messing J, Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982;19:269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- 21.Nishiyama M, Birktoft J J, Beppu T. Alteration of coenzyme specificity of malate dehydrogenase from Thermus flavus by site-directed mutagenesis. J Biol Chem. 1993;268:4656–4660. [PubMed] [Google Scholar]
- 22.Pichon M, Courbou I, Beckert M, Boudet A M, Grima-Pettenati J. Cloning and characterization of two maize cDNAs encoding cinnamoyl-CoA reductase (CCR) and differential expression of the corresponding genes. Plant Mol Biol. 1998;38:671–676. doi: 10.1023/a:1006060101866. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz-Sommer Z, Shepherd N, Tacke E, Gierl A, Rohde W, Leclercq L, Mattes M, Berndtgen R, Peterson P A, Saedler H. Influence of transposable elements on the structure and function of the A1 gene of Zea mays. EMBO J. 1987;6:287–294. doi: 10.1002/j.1460-2075.1987.tb04752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scrutton N S, Berry A, Perham R N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature (London) 1990;343:38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- 27.Selman-Housein, G., M. Lopez, D. Hernandez, L. Civardi, F. Miranda, J. Rigau, and P. Puigdomenech. 1998. Unpublished data.
- 28.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 29.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thoden J M, Frey P A, Holden H M. Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli. Biochemistry. 1996;35:2557–2566. doi: 10.1021/bi952715y. [DOI] [PubMed] [Google Scholar]
- 31.Wada M, Kataoka M, Kawabata H, Yasohara Y, Kizaki N, Hasegawa J, Shimizu S. Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction of ethyl 4-chloro-3-oxobutanoate, from Candida magnoliae. Biosci Biotechnol Biochem. 1998;62:280–285. doi: 10.1271/bbb.62.280. [DOI] [PubMed] [Google Scholar]
- 32.Wermuth B, Bohren K M, Heinemann G, Von Wartburg J-P, Gabbay K H. Human carbonyl reductase. Nucleotide sequence analysis of a cDNA and amino acid sequence of the encoded protein. J Biol Chem. 1988;263:16185–16188. [PubMed] [Google Scholar]
- 33.Wierenga R K, De Maeyer M C H, Hol W G J. Interaction of pyrophosphate moieties with α-helixes in dinucleotide binding proteins. Biochemistry. 1985;24:1346–1357. [Google Scholar]
- 34.Yamada H, Shimizu S, Kataoka M, Sakai H, Miyoshi T. A novel NADPH-dependent aldehyde reductase, catalyzing asymmetric reduction of β-keto acid esters, from Sporobolomyces salmonicolor: purification and characterization. FEMS Microbiol Lett. 1990;70:45–48. [Google Scholar]
- 35.Yaoi T, Miyazaki K, Oshima T, Komukai Y, Go M. Conversion of the coenzyme specificity of isocitrate dehydrogenase by module replacement. J Biochem. 1996;119:1014–1018. doi: 10.1093/oxfordjournals.jbchem.a021316. [DOI] [PubMed] [Google Scholar]