Abstract
Informed management and conservation efforts are vital to sustainable recreational fishing and biodiversity conservation. Because the taxonomic rank of species is important in conservation and management strategies, success of these efforts depends on accurate species delimitation. The Black Basses (Micropterus) are an iconic lineage of freshwater fishes that include some of the world’s most popular species for recreational fishing and world's most invasive species. Despite their popularity, previous studies to delimit species and lineages in Micropterus suffer from insufficient geographic coverage and uninformative molecular markers. Our phylogenomic analyses of ddRAD data result in the delimitation of 19 species of Micropterus, which includes 14 described species, the undescribed but well-known Altamaha, Bartram’s, and Choctaw basses, and two additional undescribed species currently classified as Smallmouth Bass (M. dolomieu). We provide a revised delimitation of species in the Largemouth Bass complex that necessitates a change in scientific nomenclature: Micropterus salmoides is retained for the Florida Bass and Micropterus nigricans is elevated from synonymy for the Largemouth Bass. The new understanding of diversity, distribution, and systematics of Black Basses will serve as important basis for the management and conservation of this charismatic and economically important clade of fishes.
Subject terms: Phylogenetics, Population genetics, Speciation, Taxonomy, Ichthyology
Introduction
Recreational fishing is globally among the most popular outdoor activities1 and has high socio-economic importance2. Approximately 47 billion fishes were landed recreationally in each year between the late 1990s to early 2000s worldwide3 and in the United States (USA) anglers spent $46.1 billion USD in 20164. While recreational fishing is expected to increase globally5, threats to the biodiversity that comprise recreational fisheries include habitat degradation, overfishing, invasive species, and climate change6–9. Informed management and conservation efforts are vital to sustainable recreational fishing and biodiversity conservation. Given that species is the taxonomic rank overwhelmingly targeted in conservation and management strategies10, success of these efforts depends on accurate species delimitation11.
Contemporary genomics and analytical methods complement traditional morphology-based species delimitation of fishes (e.g., Ref.12). The application of genomics to species delimitation facilitates the resolution of genetic population structure and the discovery of distinct evolutionary lineages13. Many groups of vertebrates are the subjects of genomic-based species discovery, including charismatic megafauna such as finless porpoises14, giraffes15, and orangutans16. Phylogenomics and population genomics provide important scientific insight on economically important species that includes revealing substantial intra- and interspecific differentiation among whitefishes (Coregonus) in Lake Superior17, the discovery of multiple distinct populations of Atlantic Cod (Gadus morhua) in northwestern Europe that challenges the current designations of management units18, and a complex history of secondary contact and linked selection in Atlantic Salmon (Salmo salar) across Europe and North America19. However, there is a surprising lack of genomic-based investigations of species delimitation and range-wide intraspecific genomic diversity for freshwater fishes of high international recreational importance. Failure to incorporate accurate species delimitation and information on population structure in fisheries management increases the risk of extinctions by overlooking rare and undiscovered species20 and the potential to disturb native genetic diversity through the introduction of non-native species or individuals from genetically divergent populations21.
Black Basses (Micropterus) are a lineage of freshwater fishes endemic to North American river basins east of the Rocky Mountains that includes species which are some of the world’s most popular targets for recreational fishing. The three species complexes of Micropterus most popular with anglers, Largemouth Bass (currently M. salmoides), Smallmouth Bass (M. dolomieu), and Spotted Bass (M. punctulatus) emerged as quintessential sportfishes in the nineteenth century22–24. Because of their popularity, species of Micropterus have been stocked throughout the world since the early 1800s23,25. Early stockings were poorly documented, but occurred outside the native ranges of species as well as across river basins within the range of a species, often without regard for species diversity within Micropterus23. Economic benefits stemming from recreational fishing and aquaculture have spurred introductions of the Largemouth Bass into at least 57 countries across every continent except Antarctica26, resulting in this species being recognized as both one of the world’s most popular freshwater sport fishes and among the world's most invasive species27; Global Invasive Species Database www.iucngisd.org/gisd/.
Although all of the three species complexes of Micropterus most popular with anglers have expansive native geographic distributions, they are not immune to the effects of stocking and mixing of distinct lineages or populations, which risks the erosion of local adaptation, interruption of co-adapted gene complexes, and species extinction. Present day management practices continue to promote stocking of the Florida Bass (currently recognized as Micropterus floridanus) into the range of the closely related Largemouth Bass to promote increased growth rate or maximum sizes28–31. Similarly, distinct lineages and populations of the Smallmouth Bass have been recognized since 1940 with the discovery and description of the Neosho Bass (M. dolomieu velox) as a subspecies of Smallmouth Bass32. Recent population genetic assessments illustrate that stocking of the more widespread Smallmouth Bass (M. dolomieu dolomieu) into impoundments has led to genetically admixed populations within the narrow native range of the Neosho Bass33,34. Despite the fact that the Alabama Bass (M. henshalli) was recognized as a distinct species nearly 15 years ago35, it is still introduced outside of its native distribution where it hybridizes with other species of Micropterus with the threat of regional extirpation of native species of Micropterus36,37.
In this study, we investigate the phylogeny and delimitation of species of Micropterus using a double digest restriction-site associated DNA (ddRAD) genomic dataset sampled with comprehensive geographic coverage of all recognized species in the clade. We rely on an operational species concept that views separately evolving metapopulation lineages as the sole criterion for the recognition of species38,39. Our analyses not only provide a perspective on species diversity in Micropterus that differs from those accepted by both ichthyologists and fisheries scientists24,40, but also dramatically reveal that the scientific names Micropterus salmoides41 and Micropterus floridanus42 have been incorrectly applied to the Largemouth Bass and Florida Bass over the past 75 years43. In addition to a new delimitation of Black Bass species, the genomic analyses provide a reconstruction of their evolutionary history and an important basis for the management and conservation of this economically and culturally important recreational fishery resource.
Materials and methods
Specimen sampling and ddRAD data collection and assembly
We analyzed 394 specimens that allowed representation of all recognized species and hypothesized undescribed species of Micropterus throughout their geographic ranges (Fig. 1 and Supplementary Table S1). Our sampling includes specimens collected by the authors and colleagues between 2002 and 2020, and tissue samples provided by museums and state agencies. Field-collected specimens were euthanized using MS-222 and the right-side pectoral fin was removed to provide tissue for DNA extraction. Fin-clips were stored in 100% non-denatured ethanol and the whole-body specimens were fixed in 10% formalin before being transferred to 70% ethanol for long-term preservation. Fin-clips and voucher specimens were cataloged and deposited at the Yale Fish Tissue Collection (YFTC) and Yale Peabody Museum of Natural History (YPM), respectively. All handling of animals was carried out in accordance with, and experimental protocols approved by, Yale University IACUC (No. 2018-10681) and our collecting permits. Our methods conform to ARRIVE guidelines. We obtained additional specimens for DNA extraction from several museum collections and government wildlife agencies (Supplementary Table S1).
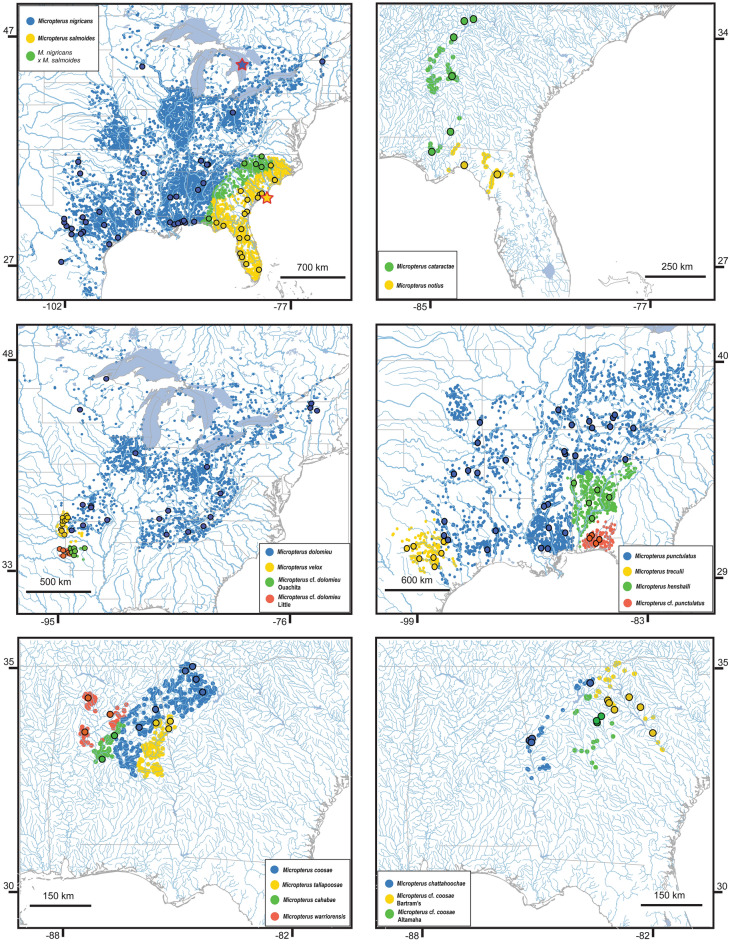
Figure 1.
Geographic ranges of all species in Micropterus as delimited in this study. Circles mark museum collections records with GPS coordinates retrieved from Fishnet2 (http://www.fishnet2.net/). Black outlined circles mark the collection localities of specimens used in this study. The type locality of Micropterus nigricans and Micropterus salmoides are marked with red-bordered stars. Maps created using personal R scripts. See Supplementary Table S1 for a complete list of specimens used in this study.
The preparation of ddRAD libraries followed protocols outlined in Peterson et al.44, using PstI/MspI restriction enzymes. The details of genomic DNA extraction, ddRAD library preparation, and sequencing are described in Kim et al.12. The demultiplexed reads were analyzed with ipyrad v.0.9.6845, using default setting with the following exceptions: ‘denovo’ for assembly method, ‘ddrad’ for datatype, ‘TGCAG, CCG’ for restriction overhang, and 0.96 for clustering threshold46. The minimum number of specimens sharing a locus, hereafter referred to as ‘min’, was set to various numbers depending on the specific analysis.
Phylogenomics and population structure analysis
Phylogenies were inferred with maximum likelihood using IQ-TREE 247 from the dataset of concatenated ddRAD loci (min300; 51,717 loci; ~ 16.5% missing portion in data matrix) for 394 individuals of Micropterus. The best-fit sequence substitution model (TVM + F + R7) was determined using ModelFinder48 implemented in IQ-TREE based on the Bayesian information criterion (BIC). During the IQ-TREE analysis, trees were rooted with the Suwannee Bass (M. notius), following its placement in the centrarchid species tree inferred from the dataset of 16 Sanger-sequenced nuclear loci and in preliminary concatenated ddRAD loci phylogenies of Micropterus and its sister lineage Lepomis12,49. We assessed topological support with 1000 ultrafast bootstrap replications50.
We performed a population structure analysis with a sparse non-negative matrix factorization algorithm using the ‘snmf’ function implemented in the R package LEA v.3.0.051. We reduced the dataset to only include those 356 specimens for which the missing data comprised less than 20% of the total retained loci. During the snmf analysis, we searched for the optimal number of ancestral populations (K) based on the cross-entropy criterion52, through a wide range (K = 1–40) with 10 replications for each scenario on a genotype data matrix for only biallelic, unlinked SNPs (9486 SNPs). With the R package Hierfstat53, we estimated Nei’s standard genetic distance (Ds) and fixation index (Fst) for all pairs among the 19 major lineages identified in the IQ-TREE analysis.
Species tree and divergence time analyses
We inferred a species-tree using the multispecies coalescent framework using SVDQuartets54 as implemented in PAUP* v.4.0a168. Because this implementation of the multispecies coalescent model assumes no gene flow among species55, we excluded individuals tentatively identified as having admixed ancestry in the snmf population structure analysis. We treated each of the 19 major IQ-TREE lineages of Micropterus as species and subsampled four specimens from geographically spaced sampling locations for each of these species. The dataset was reduced to include 234,131 unlinked SNPs for 76 individuals. We used an exhaustive quartet sampling, standard 1000 bootstrap replicates, and a multispecies coalescent tree model.
We estimated divergence times among the 19 lineages of Micropterus using a relaxed molecular clock method in BEAST v.2.5.256. The tree topology was fixed to the SVDQuartets species-tree for the relaxed clock analysis and the taxon sampling was reduced to one specimen per lineage (min19; 9920 loci; ~ 1.8% missing portion in data matrix). We applied an age prior with a normal distribution (mean, 3.11 Mya; SD, 1.3; offset, 5.0) to MRCA of the Suwannee Bass and all other lineages of Micropterus derived from Near and Kim (2021) [8.11 Mya; 95% highest posterior density (HPD): 5.84–10.26 Mya]. Two independent runs of 10 million generations were performed, sampling parameters every 2000 generations. Tracer v.1.7.157 was used to check stationarity, to determine the number of generations discarded as burnin, and to confirm that effective sample sizes (ESS) were over 200. The converged runs were combined with LogCombiner (https://www.beast2.org/programs/). Post-burnin tree samples were annotated for mean height using TreeAnnotator (https://www.beast2.org/programs) and the final trees were visualized in FigTree v.1.3.1 (http://tree.bio.ed.ac.uk/software/figtree).
Genomic species delimitation
We tested hypotheses of species delimitation using the genealogical divergence index (gdi)58 for each of the four major species complexes, including the Largemouth Bass complex (Micropterus salmoides and M. floridanus), the Smallmouth Bass complex (M. dolomieu dolomieu, M. dolomieu velox, M. cf. dolomieu Little, and M. cf. dolomieu Ouachita), the Spotted Bass complex (M. punctulatus, M. cf. punctulatus Choctaw Bass, M. henshalli, and M. treculii), and the Redeye Bass complex (M. coosae, M. cahabae, M. chattahoochae, M. cf. coosae Altamaha Bass, M. cf. coosae Bartram’s Bass, M. tallapoosae, and M. warriorensis). The gdi values were calculated using theta and tau parameter estimates resulting from the algorithm A00 of BPP v.4.159. We conducted a BPP analysis using four to ten specimens from geographically spaced sampling locations for each species complex in Micropterus, excluding individuals identified as those exhibiting signatures of gene flow in the snmf population structure analysis. The dataset for each species complex contained loci that are shared by all specimens (409–9819 loci). With the R package PopGenome60, we estimated average nucleotide diversity within and between species61 as a basis for the theta and tau priors. The species tree inferred from the SVDQuartets analysis was used as the guide species tree. For each species complex, we performed three to nine independent runs each for 150,000 generations, sampling parameters every 50 generations, with a burnin of 50,000 generations. Tracer was used to check stationarity and to confirm that ESS were over 200.
Population genomics and demographic analyses
The snmf population structure analysis identified populations of genetic admixture among the major lineages in the Largemouth Bass and Spotted Bass complexes. To investigate patterns of gene flow between species, we performed population genomic and demographic analyses aimed at the Largemouth Bass and Spotted Bass complexes. For the Largemouth Bass complex, a dataset consisting of 143 specimens of Largemouth Bass and Florida Bass was used to visualize genetic differences among the populations using a PCA implemented in ipyrad. For the PCA, we used a sample imputation method, 50% for ‘minmap’, and 90% for ‘mincov’ (35,270 unlinked SNPs after filtering). With the R package Hierfstat, we calculated pairwise Fst values between the sampled populations of Largemouth Bass and Florida Bass and among populations within each of the two species. To estimate directionality and relative magnitude of gene flow between Largemouth Bass and Florida Bass, we used TreeMix v.1.1362 implemented in ipyrad. From the dataset used in the PCA, we subsampled unlinked SNPs that are shared by at least 50% of the specimens in each of the populations. In the TreeMix analysis, we inferred the maximum likelihood tree of the lineages with a migration edge of zero and then compared the likelihoods of the migration edges up to 10. We identified the point where log-likelihood values begin to plateau, indicating the optimal number of migration edges.
Using TreeMix with the same settings as the analysis of the Largemouth Bass complex, we identified the patterns of gene flow among major lineages of the Spotted Bass complex that are resolved as well-supported clades in the phylogenetic analysis of the concatenated dataset. Because there is inconsistency in the relationships inferred from the concatenated, species-tree, and TreeMix analyses of the Spotted Bass complex, we additionally performed demographic model testing analyses with Fastsimcoal2 v.2.663,64 at two hierarchical levels. We first tested phylogenetic relationships of the populations in the absence of gene flow. We then added gene flow scenarios to a model that was identified as the best-fit model lacking gene flow. We converted each dataset in the variant call format to a multisite frequency spectrum with the python script derived from Marques et al.65; available at: https://github.com/marqueda/SFS-scripts, maximizing the number of segregating sites. Timing of initial population splits were constrained with mean divergence-time estimates inferred from a molecular clock BEAST2 analysis. We assumed a generation time of three years for species of Micropterus66,67. A log-uniform prior was used for effective population size parameters (1,000–500,000) and migration rate parameters (0–1.0E-9). We estimated parameters 50 times for each model per dataset, determined the best-fit model under Akaike information criterion (AIC), and then obtained 95% confidence intervals for parameters from 100 parametric bootstrap replications of the best-fit model.
Results and discussion
Phylogenomics: relationships among species of Micropterus
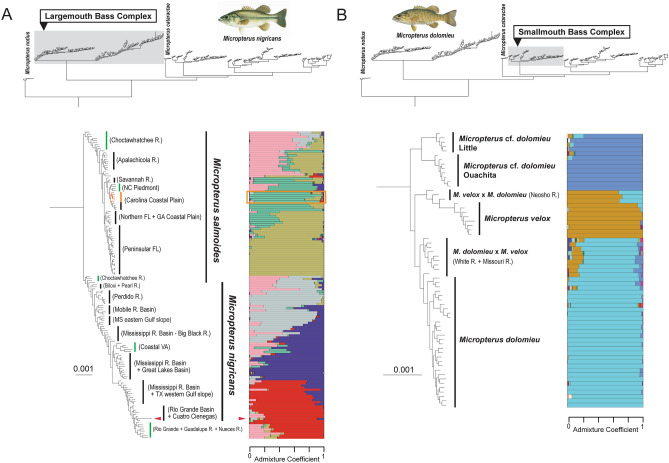
Maximum likelihood analysis of the concatenated ddRAD loci dataset using IQ-TREE strongly resolves each of morphologically defined species-complexes as monophyletic groups, places the Suwannee Bass (Micropterus notius) as the sister lineage of all other species of Micropterus, and resolves the Spotted Bass complex and Redeye Bass complex as sister lineages (Figs. 2, 3; Supplementary Fig. S1). There are two monophyletic groups within the Largemouth Bass complex that correspond to the current delimitation of the Largemouth Bass and the Florida Bass; however, specimens sampled from the type locality of Micropterus salmoides are phylogenetically nested well within the lineage currently delimited as the Florida Bass (Figs. 1, 2a), which we now refer to as Micropterus salmoides. Greater detail is provided below, but we now apply Micropterus nigricans Cuvier68 as the scientific name for the Largemouth Bass.
Figure 2.
Maximum likelihood phylogeny inferred from IQ-TREE analysis of concatenated ddRAD dataset for (a) the Largemouth Bass complex and (b) the Smallmouth Bass complex. Population structure (K = 20) inferred from snmf analysis for each species complex are shown to the right of the phylogeny. In the clade delimiting the Largemouth Bass complex (a), population structure assignments and branches leading to specimens from the vicinity of Charleston, South Carolina (type locality of Micropterus salmoides) are highlighted in orange. Green vertical bars indicate populations of genetic admixture between M. nigricans and M. salmoides that are identified in TreeMix analysis. Red arrows indicate the phylogenetic placement and population structure assignment of a specimen sampled from Cuatro Ciénegas. See Supplementary Fig. S1 for a completely annotated phylogeny tip labels and bootstrap supports values. Illustrations © Joseph R. Tomelleri, used with permission.
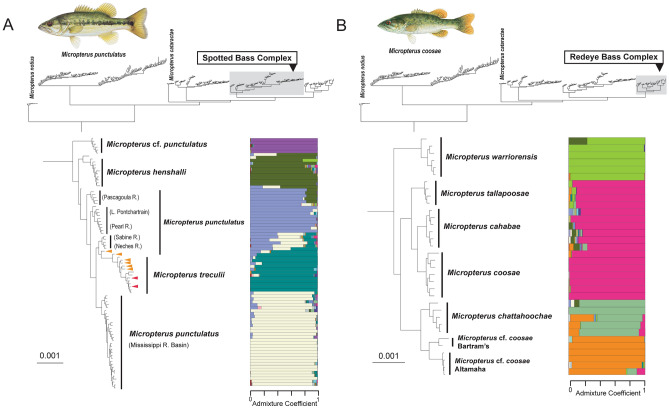
Figure 3.
Maximum likelihood phylogeny inferred from IQ-TREE analysis of concatenated ddRAD dataset for (a) the Spotted Bass complex and (b) the Redeye Bass complex. Population structure (K = 20) inferred from snmf analysis for each species complex are re shown to the right of the phylogeny. In the clade delimiting the Spotted Bass complex (a), orange and red arrows indicate specimens from the Brazos and Colorado rivers, respectively, that are identified morphologically as Micropterus punctulatus. See Supplementary Fig. S1 for a completely annotated phylogeny tip labels and bootstrap supports values. Illustrations © Joseph R. Tomelleri, used with permission.
The pairwise Fst and Nei’s Ds values among the 19 lineages of Micropterus were highest in all of the contrasts involving the Suwannee Bass and lowest among the more recently diverged sister lineages. The lowest pairwise Fst (0.24) is between the Cahaba Bass and Redeye Bass, while the highest (0.96) is between the Altamaha Bass and Suwannee Bass (Supplementary Table S2). Similarly, the Cahaba Bass and Redeye Bass exhibit the lowest pairwise sequence divergence (Ds = 0.01) and the highest pairwise genetic divergence is between the Florida Bass and Suwannee Bass (Ds = 0.12; Supplementary Table S2).
Species tree inference, relaxed molecular clock divergence time estimates, and genomic species delimitation
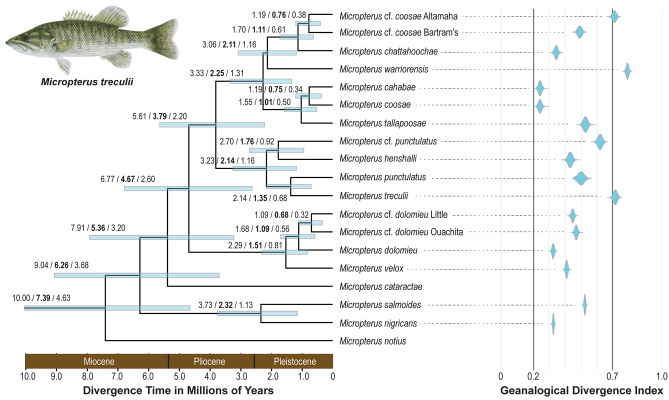
The phylogenetic relationships of Micropterus are congruent between the concatenated data IQ-TREE analysis and a species tree inferred with SVDQuartets, except for the phylogenetic resolution of the Neosho Bass (Micropterus velox), Choctaw Bass (M. cf. punctulatus), and Warrior Bass (M. warriorensis) (Fig. 4). In the concatenated analysis the Neosho Bass is resolved as the sister species of the Smallmouth Bass (M. dolomieu) (Fig. 2b), while Neosho Bass is the sister lineage to all remaining species of the Smallmouth Bass complex in the species tree analysis (Fig. 4). The Choctaw Bass is resolved as the sister species to the Alabama Bass (M. henshalli) in the species tree analysis (Fig. 4), although it is placed as the sister lineage of all other species in the Spotted Bass complex in the concatenated analysis (Fig. 3a). The Warrior Bass is resolved as the sister lineage to all remaining species of the Redeye Bass complex in the concatenated analysis (Fig. 3b); however, in the species tree analysis it is resolved as the sister lineage to a clade containing the non-Mobile species of the Redeye Bass complex (Fig. 4). Under certain conditions of phylogenomic analysis, including the presence of gene flow, elevated substitution rates, or a limited taxon sampling, coalescent methods (e.g., SVDQuartets) are more robust than concatenation methods69–71.
Figure 4.
Time-calibrated species tree for Micropterus, inferred from SVDQuartets and BEAST2. All nodes have bootstrap supports of 100%, except the node leading to M. cataractae and the Smallmouth, Spotted, and Redeye Bass complexes, to M. warriorensis, M. chattahoochae, and the undescribed Altamaha and Bartram’s basses, and to M. cahabae and M. coosae. Numbers at nodes indicate a mean and 95% highest posterior density (also depicted with light blue bars) of relaxed molecular clock divergence times. Violin plots for genealogical divergence index (gdi) of species delimitations are aligned with and shown on the right of species tree. Illustration © Joseph R. Tomelleri, used with permission.
The posterior age estimate from the relaxed molecular clock analysis for the most recent common ancestor (MRCA) of Micropterus is 7.39 Mya (95% HPD 4.63–10.00 Mya; Fig. 4). The posterior ages of the MRCA of the Largemouth Bass, Spotted Bass, and Redeye Bass complexes are similar, ranging between 2.32 and 2.14 Mya (Fig. 4). The estimated age of the MRCA of the Smallmouth Bass complex is 1.51 Mya (95% HPD: 0.81–2.29 Mya; Fig. 4). The mean ages of the most recent inferred speciation events range between 0.68 and 0.76 Mya (Fig. 4).
The BPP species delimitation analysis results in moderate to high genealogical divergence indices (gdi) that between 0.242 and 0.789, supporting the delimitation of 19 species of Micropterus (Table 1; Fig. 4). As suggested by Jackson et al.58, the gdi values estimated for the Warrior Bass, Altamaha Bass (M. cf. coosae), and Guadalupe Bass (M. treculii) were consistent with the expectations for distinct species (Fig. 4). The lowest gdi values were observed in Cahaba Bass and Redeye Bass (Fig. 4). All other species of Micropterus, including the undescribed Smallmouth Bass lineages in Little and Ouachita Rivers, Bartram’s Bass, Altamaha Bass, and Choctaw Bass, exhibit gdi values between 0.30 and 0.66 that fall within the range expected of distinct species that are supported with other lines of evidence such as morphological traits, differences in coloration and pigmentation, or behavior (Fig. 4).
Table 1.
List of delimited species of Micropterus. Status indicates if delimitation is different from Taylor et al.24.
| Common name | Scientific name | Status |
|---|---|---|
| Alabama Bass | Micropterus henshalli Hubbs & Bailey, 1940 | As in Taylor et al.24 |
| Altamaha Bass | Micropterus cf. coosae Altamaha River | Discovered in Freeman et al.88 |
| Bartram's Bass | Micropterus cf. coosae Bartram's | Delimited in Freeman et al.88 |
| Cahaba Bass | Micropterus cahabae Baker, Johnston & Blanton, 2013 | As in Taylor et al.24 |
| Chattahoochee Bass | Micropterus chattahoochae Baker, Johnston & Blanton, 2013 | As in Taylor et al.24 |
| Choctaw Bass | Micropterus cf. punctulatus | Discovered in Tringali et al.85 |
| Florida Bass | Micropterus salmoides (Lacépède, 1802) | Delimitation changed. Most populations were previously delimited as M. floridanus |
| Guadalupe Bass | Micropterus treculii (Vaillant & Bocourt, 1874) | As in Taylor et al.24 |
| Largemouth Bass | Micropterus nigricans (Cuvier, 1828) | Delimitation changed. Previously delimited at M. salmoides, Largemouth Bass |
| Little River Bass | Micropterus cf. dolomieu Little River | Discovered in this study |
| Neosho Bass | Micropterus velox Hubbs & Bailey, 1940 | Elevated from synonymy with M. dolomieu |
| Ouachita Bass | Micropterus cf. dolomieu Ouachita River | Discovered in Stark and Echelle83 |
| Redeye Bass | Micropterus coosae Hubbs & Bailey, 1940 | As in Taylor et al.24 |
| Shoal Bass | Micropterus cataractae Williams & Burgess, 1999 | As in Taylor et al.24 |
| Smallmouth Bass | Micropterus dolomieu Lacépède, 1802 | As in Taylor et al.24 |
| Spotted Bass | Micropterus punctulatus (Rafinesque, 1819) | As in Taylor et al.24 |
| Suwannee Bass | Micropterus notius Bailey & Hubbs, 1949 | As in Taylor et al.24 |
| Tallapoosa Bass | Micropterus tallapoosae Baker, Johnston & Blanton, 2013 | As in Taylor et al.24 |
| Warrior Bass | Micropterus warriorensis Baker, Johnston & Blanton, 2013 | As in Taylor et al.24 |
Species delimitation of Largemouth Bass and Florida Bass, changes to scientific nomenclature, and geographic patterns of introgression
Micropterus salmoides was described by Lacépède41:716,717 based on an illustration of a specimen collected by Louis Bosc in the vicinity of Charleston, South Carolina32,72,73. The lack of type specimens and little detail in Lacépède’s41 description led to an 80-year period of nomenclatural confusion with Micropterus dolomieu, the Smallmouth Bass74,75. The application of the name Micropterus salmoides to the Largemouth Bass was suggested by Henshall22:110–132 and has remained unchanged to the present day. A study of geographic variation in morphological traits resulted in Cichla floridana Lesueur42 being recognized as the subspecies Micropterus salmoides floridanus, which was endemic to peninsular Florida43. Based on intermediate meristic traits it was hypothesized that M. s. salmoides and M. s. floridanus were hybridizing in a broad geographic area of intergradation that included nearly all of the Florida Panhandle and all of the rivers in Georgia that drain into the Gulf of Mexico and the Atlantic Ocean43: Map 1. Kassler et al.29 concluded that Micropterus salmoides and M. floridanus are each distinct species based on morphology, fixed differences at a number of allozyme loci, and deep reciprocal monophyly in mitochondrial gene trees. Subsequent studies recognize the Florida Bass as a distinct species relative to the Largemouth Bass (e.g., Refs.24,76,77); however, some authors and the Names of Fishes Committee (jointly hosted by the American Fisheries Society and the American Society of Ichthyologists and Herpetologists) continue to treat the Florida Bass and Largemouth Bass as subspecies (e.g., Refs.40,78,79).
Results of our ddRAD phylogenomic analyses do not support the hypothesis that Florida Bass and Largemouth Bass hybridize over a broad area of intergradation, but rather the Florida Bass has a much larger geographic distribution that ranges from peninsular Florida in the south and the Apalachicola River in the west to the Cape Fear River of North Carolina in the north along the Atlantic Coastal Plain (Figs. 1, 2A; Supplementary Figs. S1 and S2). Phylogenetic analysis of the ddRAD data strongly resolves populations from peninsular Florida and rivers in Georgia, South Carolina, and North Carolina that include the Savannah, Edisto, Ashley, Santee, Pee Dee, and Cape Fear Rivers as a monophyletic group that we delimit as the Florida Bass (Fig. 2a). The snmf population structure analysis identifies genetic variation associated with geography among populations of Florida Bass and the delimitation of populations in Georgia, South Carolina, and North Carolina as Florida Bass. The presence of Florida Bass in these areas is not likely result from stocking into areas previously occupied by Largemouth Bass (Figs. 1, 2a; Supplementary Fig. S2).
The identification of populations along the Atlantic Coastal Plain of Georgia, South Carolina, and North Carolina as Florida Bass highlights a need to dramatically revise the scientific nomenclature of both Largemouth Bass and Florida Bass. Bailey and Hubbs43 delimited Florida Bass as those populations with high counts and Largemouth Bass as populations with low counts in five meristic traits (e.g., number of scales along the lateral line). Populations just south of Charleston, South Carolina, which is the type locality for Micropterus salmoides, exhibit high or intermediate scale counts and were delimited by Bailey and Hubbs43: Map 1 as Florida Bass or intergrades between Florida Bass and Largemouth Bass. Our genomic analyses identify all specimens from the Ashley and Cooper Rivers that approximate the type locality of M. salmoides as conspecific with populations sampled from peninsular Florida and distinct from all other populations identified as Largemouth Bass (Fig. 2a; Supplementary Figs. S1 and S2).
Our results highlight that the scientific name Micropterus salmoides has been misapplied for the nearly 75 years following the delimitation of Largemouth Bass and Florida Bass outlined in Bailey and Hubbs43. In order to present a classification for species of Micropterus that is consistent with their phylogenetic relationships, we conclude that Lacépède’s41 description of Micropterus salmoides is the accurate and valid scientific name for the Florida Bass (Figs. 1, 2a). Micropterus floridanus42 which is currently applied to the Florida Bass (e.g., Refs.24,49) is a junior synonym of Micropterus salmoides41. The oldest available scientific name for the Largemouth Bass is Micropterus nigricans Cuvier68 with the type locality given as Lake Huron where the species is native31,32,80.
Both the Largemouth Bass and Florida Bass exhibit geographic population genetic structure. The ddRAD data identifies three genetic groups in Largemouth Bass: a southwestern group, ranging from the Rio Grande system in southern Texas and the Cuatro Ciénegas Basin in Coahuila, Mexico to the south to southwestern tributaries of the Mississippi River (e.g., Red and Canadian Rivers in Oklahoma), a northcentral group throughout the Mississippi River Basin and the Laurentian Great Lakes, and a southeastern group that is consistent with the proposed “Delta Bass81” that ranges from the southeastern tributaries of the Mississippi River to the Perdido River system in Alabama and Florida (Fig. 2a; Supplementary Fig. S2). There are three genetic groups in the Florida Bass, including one occurring throughout rivers in Florida and in the Atlantic Coastal Plain in the north to the Savannah River system of Georgia, a western group observed in the Choctawhatchee and Apalachicola River systems and rivers in the Atlantic Piedmont, and a northeastern group, ranging from the Satilla River of Georgia in the south to the Cape Fear River of North Carolina in the north along the Atlantic Coastal Plain (Fig. 2a; Supplementary Fig. S2).
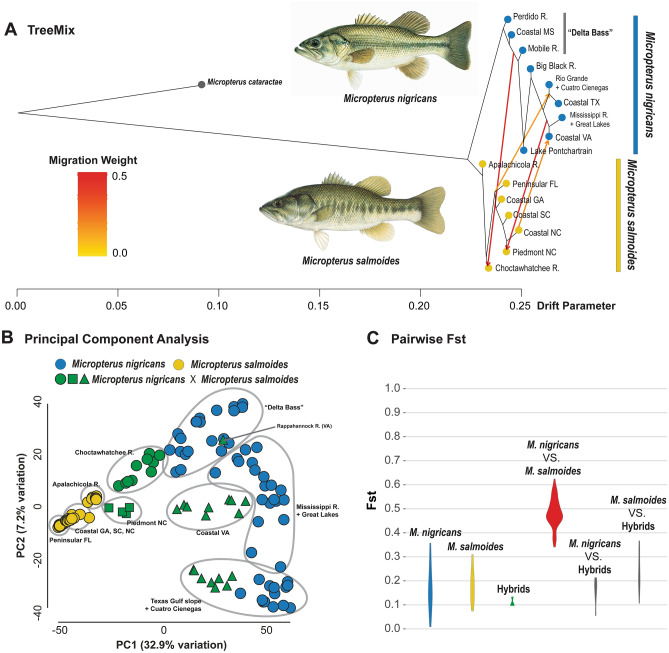
The snmf-inferred population structure and the TreeMix analyses identify signatures of genomic admixture between the Largemouth Bass and Florida Bass from two geographic regions, the Choctawhatchee River in northwestern Florida and rivers draining the Piedmont of North Carolina (Figs. 2a, 5a; Supplementary Fig. S2). Based on analyses of diagnostic nucleotide polymorphisms, we include the Piedmont of Georgia as an area of genomic admixture between Largemouth and Florida Bass81,82. Genetic admixture between the two species is also apparent in regions where Florida Bass were introduced to areas where Largemouth Bass are native, including the Rio Grande, Nueces, and Guadalupe River systems in Texas, and several river systems draining into the eastern Gulf of Mexico. Rivers draining into Chesapeake Bay in eastern Virginia also contain admixed individuals where both species were introduced (Figs. 2a, 5a; Supplementary Fig. S2). PCA of genomic diversity in the Largemouth Bass complex reveals the majority of the genetic variation is the result of genetic differentiation between Largemouth Bass and Florida Bass, placing specimens with genomic admixture intermediate along the PC1 axis (Fig. 5b). Pairwise Fst values are significantly higher between populations of Largemouth Bass and Florida Bass than those between populations within species or populations of genetic admixture between the two species (Fig. 5c; Supplementary Table S2).
Figure 5.
(a) Migration edges between Micropterus nigricans and M. salmoides, inferred from TreeMix analysis. Arrows show relative magnitude and direction of gene flow among populations of the Largemouth Bass complex. (b) Principal component analysis (PCA) of genetic variance for species of the Largemouth Bass complex. Grey ellipses indicate major geographic populations or lineages of the Largemouth Bass complex. We separate populations that are genetically admixed between Micropterus nigricans and Micropterus salmoides by origin of genetic admixture: presumably natural secondary contract (green circles); uncertain (green rectangles); and human-mediated origin (green triangles). (c) Violin plots for ranges of pairwise Fst values among populations within Micropterus nigricans, M. salmoides, and genetic admixtures (Choctawhatchee and North Carolina Piedmont) and between each pair of the three groups. Pairwise Fst values between populations of Micropterus nigricans and M. salmoides are considerably higher than other pairs in comparison. Illustrations © Joseph R. Tomelleri, used with permission.
Species discovery in the Smallmouth Bass complex
Since the description of the Smallmouth Bass by Lacépède41, twelve species and a subspecies were described between 1817 and 1940, but all of these scientific names were eventually synonymized with Micropterus dolomieu72,73. Among the taxa synonymized with the Smallmouth Bass, the Neosho Bass (M. dolomieu velox) is the subject of a debate in the systematics of Micropterus. Hubbs and Bailey32:29 described the Neosho Bass as a subspecies of the Smallmouth Bass based on external morphological traits and subsequent analysis observed a significant difference between the Neosho Bass and Smallmouth in head length33. The Neosho Bass is limited to a small area of the Arkansas River system, ranging from the Fourche La Fave River upstream to the Neosho River. Hubbs and Bailey32:Map 2 hypothesized that the White River system of Arkansas and Missouri, the Little River system in Arkansas and Oklahoma, and the upper Ouachita River in Arkansas are areas of intergradation between Neosho Bass and Smallmouth Bass. A range-wide allozyme study of the Smallmouth Bass complex identified several fixed allelic differences consistent with Hubbs and Bailey’s32 delimitation of the Neosho Bass and determined the populations of the Smallmouth Bass complex in the Little and Ouachita Rivers are genetically distinct and are not intergrades of Smallmouth Bass and Neosho Bass83. Studies using microsatellite loci convincingly identified the Neosho Bass as a distinct lineage and reveal signatures of genetic admixture as the result of human-mediated introductions of non-native Smallmouth Bass33,34.
Our phylogenomic analysis of the Smallmouth Bass complex resolves four major lineages: the Smallmouth Bass, the Neosho Bass, an undescribed species we call the Little River Bass (Micropterus cf. dolomieu Little), and another undescribed species native to the upper Ouachita River system we call the Ouachita Bass (M. cf. dolomieu Ouachita) (Figs. 1, 2b; Table 1; Supplementary Fig. S1). The BPP analysis resulted in moderate gdi values for each four lineages (Fig. 4), supporting the delimitation of each as a distinct species if additional layers of evidence such as phenotype, behavior, or ecology are available84. Based on the results of our phylogenomic analyses and documented morphological distinctiveness32,33, we elevate the Neosho Bass, Micropterus velox, to the species taxonomic rank. Although Stark and Echelle83 treated the Little River Bass and Ouachita Bass as single distinct genetic lineage, results from our phylogenomic analyses suggest that the two sister lineages are not only distinct from the Smallmouth Bass and Neosho Bass, they each have a gdi value which exceeds that observed for the other two species (Fig. 4). We consider both the Little River Bass and Ouachita Bass as distinct but undescribed species (Table 1).
The Spotted Bass complex: species delimitation and patterns of introgression
The Spotted Bass complex currently includes Spotted Bass, Guadalupe Bass, Alabama Bass, and the undescribed Choctaw Bass (Table 1). Maximum likelihood analysis of the concatenated ddRAD dataset resolves the Choctaw Bass as the sister lineage of all other species in the Spotted Bass complex (Fig. 3a; Supplementary Fig. S1), the species tree and demographic analyses support a clade containing the Choctaw and Alabama basses (Fig. 4; Supplementary Fig. S3a). The delimitation of both Choctaw Bass and Alabama Bass as distinct species is supported by moderate gdi values (Fig. 4) and morphological differences35,85.
The Spotted Bass is resolved as paraphyletic in the phylogeny inferred from the concatenated ddRAD dataset as the Guadalupe Bass is the sister lineage of populations sampled from the Sabine and Neches Rivers in Texas and Louisiana (Fig. 3a; Supplementary Fig. S1). Results of the population structure (Fig. 3a) and demographic analyses (Supplementary Fig. S3a,b) indicate that historical introgression from Alabama Bass and Guadalupe Bass may result in the paraphyly of Spotted Bass by resolving introgressed populations outside of a clade containing non-introgressed Spotted Bass plus Guadalupe Bass or as early diverging lineages of the Guadalupe Bass clade. This pattern is consistent with observations from the Longear Sunfish complex (e.g., Lepomis megalotis), where introgressed populations are resolved as early diverging or pectinate branches along stem lineages in phylogenetic analyses of concatenated datasets12.
Hubbs and Bailey32:Map 1 hypothesized that populations of Spotted Bass from Lake Pontchartrain, the Pearl River, and the Pascagoula River in Louisiana and Mississippi are intergrades between Spotted Bass and Alabama Bass. In the phylogeny inferred from the concatenated ddRAD dataset, these populations of the Spotted Bass resolve as early diverging lineages in a clade containing all other populations of the Spotted Bass and Guadalupe Bass (Fig. 3a; Supplementary Fig. S1). The Fastsimcoal2 demographic analyses suggests that the MRCA of the Spotted Bass populations from Lake Pontchartrain, the Pearl River, and the Pascagoula River diverged from the MRCA of all other populations of Spotted Bass in the late-Pleistocene. The Fastsimcoal2 and TreeMix analyses identify multiple instances of gene flow from Alabama Bass to the MRCA of the Lake Pontchartrain, the Pearl River, and the Pascagoula River populations (Supplementary Fig. S3a,b). These results provide support for the intergradation hypothesis of Hubbs and Bailey32, inferred from morphology. A list of tested models and parameter estimates of each model are presented in Supplementary Fig. S4 and Supplementary Table S3.
The snmf population structure analysis indicates that populations of the Spotted Bass in the Sabine and Neches Rivers in eastern Texas are the result of genetic admixture between the Spotted Bass and Guadalupe Bass (Fig. 3a). These populations of Spotted Bass are resolved as the sister lineage of the Guadalupe Bass in the phylogeny inferred from the concatenated dataset (Fig. 3a; Supplementary Fig. S1); however, the demographic analysis supports a model where the Sabine and Neches populations share common ancestry and are the sister lineage of Mississippi River Basin Spotted Bass populations (Supplementary Fig. S3a). Specimens of the Spotted Bass from the Brazos and Colorado River systems in central Texas are phylogenetically nested within a clade containing Guadalupe Bass in the phylogeny inferred from the concatenated dataset (Fig. 3a; Supplementary Fig. S1). The population structure and demographic analyses suggest specimens of Spotted Bass and Guadalupe Bass from the Brazos River system are genetic admixtures between the two species (Fig. 3a; Supplementary Fig. S3a). Two specimens in our analysis that were identified morphologically as Spotted Bass from the Colorado River system exhibit a genomic ancestry coefficient indicative of Guadalupe Bass (Fig. 3a; Supplementary Fig. S1). This observation is consistent with a study using Sanger-sequenced loci that detected Guadalupe Bass genes in specimens that were phenotypically identified as Spotted Bass86, highlighting that mismatch between phenotype and genotype for Spotted and Guadalupe basses in rivers of central Texas will be a challenge for the accurate species identification and conservation management of these populations.
Redeye Basses: genomic validation of recently described species
Hubbs and Bailey32 described the Redeye Bass (Micropterus coosae) with a geographic distribution that includes the Mobile Basin eastward to the upper portions of the Apalachicola, Altamaha, Savannah, and Saluda River systems. In a study of external morphology and a mitochondrial gene tree, Baker et al.87 described four species from populations of Redeye Bass: the Cahaba Bass (M. cahabae) endemic to the Cahaba River system, the Tallapoosa Bass (M. tallapoosae) endemic to the Tallapoosa River system, the Warrior Bass (M. warriorensis) endemic to the Black Warrior River system, and the Chattahoochee Bass (M. chattahoochae) endemic to the Chattahoochee River system. Baker et al.87 treated the Bartram’s Bass (M. cf. coosae) as a distinct and undescribed species distributed in the Savannah River system of Georgia and South Carolina. A subsequent study using morphological traits, DNA sequences from a single nuclear gene, and a mitochondrial gene tree delimited populations of the Redeye Bass complex in the Altamaha and Ogeechee River systems as a distinct and second undescribed species, the Altamaha Bass88.
Previous molecular phylogenetic studies of Micropterus prior to Near and Kim49 did not resolve the Redeye Bass complex as a clade88–90, as mitochondrial introgression resulted in an inference of paraphyly of the species comprising the Redeye Bass complex. The phylogenomic analyses of the ddRAD loci resolves the Redeye Bass complex as a monophyletic group with strong node supports and the sampled individuals from every species in the complex resolves as a reciprocally monophyletic group (Fig. 3b; Supplementary Fig. S1). The BPP species delimitation analyses resulted in moderate to high gdi values for the species in the Redeye Bass complex, ranging from the lowest among all species in Micropterus (0.20–0.27 for Cahaba Bass and Redeye Bass, Fig. 4) to values well within those expected among distinct species for the Altamaha Bass and Warrior Bass (Fig. 4). Given the reciprocal monophyly of all species of the Redeye Bass complex, the documented morphological differences, and the magnitude of genealogical divergence (Fig. 4), we consider the five described and the two undescribed species of the Redeye Bass complex as distinct evolutionary lineages.
Conclusions
The understanding of Micropterus phylogeny and species diversity have dramatically increased over the past 20 years with the application of molecular data and a reexamination of morphological differences29,33,35,83,85,87,88,91. The results of our phylogenomic analyses using ddRAD data result in the delimitation of 19 species of Micropterus (Fig. 4; Table 1), which includes 14 described species, two undescribed but well-known species in the Redeye Bass complex (Bartram’s Bass and Altamaha Bass), one undescribed but well-known species in the Spotted Bass complex (Choctaw Bass), and the recognition of two undescribed species currently classified as Smallmouth Bass (Little River Bass and Ouachita Bass).
Phylogeographic analysis of ddRAD data results in a revised distribution of the Largemouth Bass, Florida Bass, and hypothesized admixed populations of the two species. The native range of the Florida Bass spans from peninsular Florida northwards to North Carolina along the Atlantic Coastal Plain, including the type locality of Micropterus salmoides, which is currently recognized as the scientific name for the Largemouth Bass. Consequently, we synonymize Micropterus floridanus with Micropterus salmoides, which is retained for the Florida Bass, and elevate Micropterus nigricans out of synonymy with Micropterus salmoides as the valid scientific name for the Largemouth Bass. We delimit four genetically and geographically distinct evolutionary lineages in the Smallmouth Bass complex, elevating Micropterus velox (Neosho Bass) as a species and highlighting the distinctiveness of two undescribed species M. cf. dolomieu Little (Little River Bass) and M. cf. dolomieu Ouachita (Ouachita Bass). The phylogenomic analyses reveal a complicated history of interspecific introgression among species of the Spotted Bass complex. The species delimitation analyses support the recognition of seven species in the recently expanded Redeye Bass complex (Table 1).
The genomic based species delimitation outlined in our study provide not only novel understanding of diversity, distribution, and systematics of Micropterus, but also an important basis for the management and conservation of economically valuable recreational fishery resources. Human-mediated movement of species and distinct populations across natural boundaries poses one of the greatest threats to the conservation of Black Bass species diversity24. Although it may not be possible to ameliorate the consequences of past stockings of species of Micropterus that date back to the 1800’s, our results provide strong impetus to discontinue activities that ignore species and population boundaries, including stock enhancement programs and illegal interbasin transfers. Resource agencies and other stakeholders should prioritize conservation efforts towards the many narrow-ranged species to best protect of the unique diversity among the species of Micropterus.
Supplementary Information
Acknowledgements
We thank Tanya Darden and Larry Bowman (South Carolina Department of Natural Resources), Dean Hendrickson and Adam Cohen (Texas Natural History Collections, University of Texas), Matthew Wagner and Robert Ellwanger (Mississippi Museum of Natural Science), Lawrence Page, Robert Robins, and Pamela Soltis (Florida Museum Fish Collection, University of Florida), Kevin Conway and Heather Prestridge (Biodiversity Research and Teaching Collections, Texas A&M University), Byron Freeman (University of Georgia), Stephen Brown, Samantha Stelmack, Jeremy McCain, and Samantha Bergeron (Mississippi Department of Wildlife, Fisheries, and Parks), Matt Duplessis (Louisiana Department of Wildlife and Fisheries), Jeff Buckingham and Aaron Kern (Arkansas Game and Fish Commission), and Ben Neely (Kansas Department of Wildlife, Parks, and Tourism) for providing tissue samples used in this study. We thank Oliver Orr for assistance with laboratory procedures; Daniel MacGuigan, Kevin Conway, Ava Ghezelayagh, Richard Harrington, Cragen King, Amanda Kubicek, Kole Kubicek, Youl Kwon, Elyse Parker, Robert Robins, Randy Singer, Clay Tamburri, and Courtney Weyand for assistance in the field; Gregory Watkins-Colwell for assistance with research collections at the Yale Peabody Museum of Natural History; David Backeberg at the Yale High Performance Computing Systems for computational assistance; and Michael Donoghue and Erika Edwards for helpful discussions. The Bingham Oceanographic Fund maintained by the Peabody Museum of Natural History, Yale University, funded this work.
Author contributions
D.K. conceived the research, collected specimens in the field, collected the data, performed the research, and wrote the paper; A.T.T. collected specimens in the field and wrote the paper; and T.J.N. conceived the research, collected specimens in the field, and wrote the paper.
Data availability
Raw sequence data is available for download from the SRA (https://www.ncbi.nlm.nih.gov/sra/PRJNA796640). Data and supplementary materials are available from the Dryad Digital Repository: https://datadryad.org/stash/share/I4cSjJcrGRUPSI6uIZhivfBReyoCXLSIIDL89a3Qm20.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-11743-2.
References
- 1.Arlinghaus R, et al. Governing the recreational dimension of global fisheries. Proc. Nat. Acad. Sci. USA. 2019;116:5209–5213. doi: 10.1073/pnas.1902796116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowx IG, Gerdeaux D. The effects of fisheries management practises on freshwater ecosystems. Fish. Manag. Ecol. 2004;11:145–151. doi: 10.1111/j.1365-2400.2004.00411.x. [DOI] [Google Scholar]
- 3.Cooke SJ, Cowx IG. The role of recreational fishing in global fish crises. Bioscience. 2004;54:857–859. doi: 10.1641/0006-3568(2004)054[0857:TRORFI]2.0.CO;2. [DOI] [Google Scholar]
- 4.Gillespie N, et al. Socioeconomic benefits of recreational, commercial, and subsistence fishing associated with national forests. Fisheries. 2018;43:432–439. doi: 10.1002/fsh.10127. [DOI] [Google Scholar]
- 5.Cowx, I. G. Recreational fishing. In Handbook of Fish Biology and Fisheries, Vol. 2 (eds Hart, P. J. & Reynolds, J. D.). (Blackwell Publishing, 2002).
- 6.Arlinghaus, R. & Cooke, S. J. Recreational fisheries: Socioeconomic importance, conservation issues and management challenges. In Recreational Hunting, Conservation and Rural Livelihoods: Science and Practice (eds Dickson, B., Hutton, J. & Adams, W.). (Blackwell Publishing, 2009).
- 7.Brander K. Impacts of climate change on fisheries. J. Mar. Syst. 2010;79:389–402. doi: 10.1016/j.jmarsys.2008.12.015. [DOI] [Google Scholar]
- 8.Holmlund CM, Hammer M. Ecosystem services generated by fish populations. Ecol. Econ. 1999;29:253–268. doi: 10.1016/S0921-8009(99)00015-4. [DOI] [Google Scholar]
- 9.Lewin W-C, Arlinghaus R, Mehner T. Documented and potential biological impacts of recreational fishing: Insights for management and conservation. Rev. Fish. Sci. 2006;14:305–367. doi: 10.1080/10641260600886455. [DOI] [Google Scholar]
- 10.Agapow PM, et al. The impact of species concept on biodiversity studies. Q. Rev. Biol. 2004;79:161–179. doi: 10.1086/383542. [DOI] [PubMed] [Google Scholar]
- 11.Devitt TJ, Wright AM, Cannatella DC, Hillis DM. Species delimitation in endangered groundwater salamanders: Implications for aquifer management and biodiversity conservation. Proc. Nat. Acad. Sci. USA. 2019;116:2624–2633. doi: 10.1073/pnas.1815014116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D, Bauer BH, Near TJ. Introgression and species delimitation in the Longear Sunfish Lepomis megalotis (Teleostei: Percomorpha: Centrarchidae) Syst. Biol. 2022;72:273–285. doi: 10.1093/sysbio/syab029. [DOI] [PubMed] [Google Scholar]
- 13.Poelstra JW, et al. Cryptic patterns of speciation in cryptic primates: Microendemic mouse lemurs and the multispecies coalescent. Syst. Biol. 2021;70:203–218. doi: 10.1093/sysbio/syaa053. [DOI] [PubMed] [Google Scholar]
- 14.Zhou XM, et al. Population genomics of finless porpoises reveal an incipient cetacean species adapted to freshwater. Nat. Commun. 2018;9:1276. doi: 10.1038/s41467-018-03722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coimbra RTF, et al. Whole-genome analysis of giraffe supports four distinct species. Curr. Biol. 2021;31:2929–2938. doi: 10.1016/j.cub.2021.04.033. [DOI] [PubMed] [Google Scholar]
- 16.Nater A, et al. Morphometric, behavioral, and genomic evidence for a new Orangutan species. Curr. Biol. 2017;27:3487–3498. doi: 10.1016/j.cub.2017.09.047. [DOI] [PubMed] [Google Scholar]
- 17.Ackiss AS, Larson WA, Stott W. Genotyping-by-sequencing illuminates high levels of divergence among sympatric forms of coregonines in the Laurentian Great Lakes. Evol. Appl. 2020;13:1037–1054. doi: 10.1111/eva.12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansen T, et al. Genomic analysis reveals neutral and adaptive patterns that challenge the current management regime for East Atlantic Cod Gadus Morhua L. Evol. Appl. 2020;13:2673–2688. doi: 10.1111/eva.13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rougemont Q, Bernatchez L. The demographic history of Atlantic Salmon (Salmo salar) across its distribution range reconstructed from Approximate Bayesian Computations. Evolution. 2018;72:1261–1277. doi: 10.1111/evo.13486. [DOI] [PubMed] [Google Scholar]
- 20.Morrison WR, et al. The impact of taxonomic change on conservation: Does it kill, can it save, or is it just irrelevant? Biol. Conserv. 2009;142:3201–3206. doi: 10.1016/j.biocon.2009.07.019. [DOI] [Google Scholar]
- 21.Olver CH, Shuter BJ, Minns CK. Toward a definition of conservation principles for fisheries management. Can. J. Fish. Aquat. Sci. 1995;52:1584–1594. doi: 10.1139/f95-751. [DOI] [Google Scholar]
- 22.Henshall, J. A. Book of the Black Bass. (Robert Clarke & Co, 1881).
- 23.Long JM, Allen MS, Porak WF, Suski CD. A historical perspective of Black Bass management in the United States. Am. Fish. Soc. Symp. 2015;82:99–122. [Google Scholar]
- 24.Taylor AT, Long JM, Tringali MD, Barthel BL. Conservation of Black Bass diversity: An emerging management paradigm. Fisheries. 2019;44:20–36. doi: 10.1002/fsh.10187. [DOI] [Google Scholar]
- 25.Jenkins, R. E. & Burkhead, N. M. Freshwater Fishes of Virginia. (American Fisheries Society, 1994).
- 26.Froese, R. & Pauly, D. Fishbase www.Fishbase.Org (2021).
- 27.Near, T. J. & Koppelman, J. B. Species diversity, phylogeny, and phylogeography of Centrarchidae. In Centrarchid Fishes: Diversity, Biology, and Conservation (eds Cooke, S. J. & Philipp, D. P.). (Blackwell Science, 2009).
- 28.Barthel BL, Allen MS, Porak WF, Kerns J. Florida Bass Micropterus floridanus (Lesueur, 1822) Am. Fish. Soc. Symp. 2015;82:43–53. [Google Scholar]
- 29.Kassler TW, et al. Molecular and morphological analyses of the Black Basses (Micropterus): Implications for taxonomy and conservation. Am. Fish. Soc. Symp. 2002;31:291–322. [Google Scholar]
- 30.Philipp DP. Genetic implications of introducing Florida Largemouth Bass, Micropterus salmoides floridanus. Can. J. Fish. Aquat. Sci. 1991;48:58–65. doi: 10.1139/f91-304. [DOI] [Google Scholar]
- 31.Philipp DP, Childers WF, Whitt GS. A biochemical evaluation of the Northern and Florida subspecies of Largemouth Bass. Trans. Am. Fish Soc. 1983;112:1–20. doi: 10.1577/1548-8659(1983)112<1:ABGEOT>2.0.CO;2. [DOI] [Google Scholar]
- 32.Hubbs CL, Bailey RM. A revision of the Black Basses (Micropterus and Huro) with descriptions of four new forms. Misc. Publ. Mus. Zool. Univ. Mich. 1940;48:1–51. [Google Scholar]
- 33.Gunn JC, et al. Complex patterns of genetic and morphological differentiation in the Smallmouth Bass subspecies (Micropterus dolomieu dolomieu and M. d. velox) of the central interior highlands. Conserv. Genet. 2020;21:891–904. doi: 10.1007/s10592-020-01295-1. [DOI] [Google Scholar]
- 34.Taylor AT, Long JM, Schwemm MR, Brewer SK. Hybridization and genetic structure of Neosho Smallmouth Bass in the Ozark Highlands. N. Am. J. Fish. Manag. 2018;38:1226–1240. doi: 10.1002/nafm.10225. [DOI] [Google Scholar]
- 35.Baker WH, Johnston CE, Folkerts GW. The Alabama Bass, Micropterus henshalli (Teleostei: Centrarchidae), from the mobile river basin. Zootaxa. 2008;1861:57–67. doi: 10.11646/zootaxa.1861.1.6. [DOI] [Google Scholar]
- 36.Lewis MR, Ekema P, Holley M, Peatman EJ. Genetic evidence of introduced Redeye Bass and Alabama Bass and hybridization with native Micropterus spp. in Town Creek, Alabama, USA. N. Am. J. Fish. Manag. 2021;41:78–85. doi: 10.1002/nafm.10531. [DOI] [Google Scholar]
- 37.Moyer GR, Williams AS, Ml J, Ragland BJ, Churchill TN. Genetic confirmation and assessment of an unauthorized fish introduction in Parksville Reservoir, Tennessee. J. Southeast. Ass. Fish. Wildl. Agencies. 2014;1:64–69. [Google Scholar]
- 38.De Queiroz, K. The general lineage concept of species, species criteria, and the process of speciation: A conceptual unification and terminological recommendations. In Endless Forms: Species and Speciation (eds Howard, D. J. & Berlocher, S. H.). (Sinauer, 1998).
- 39.De Queiroz K. Species concepts and species delimitation. Syst. Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 40.Page, L. M. & Burr, B. M. Peterson Field Guide to Freshwater Fishes of North America North of Mexico, 2nd edn. (Houghton Mifflin Harcourt, 2011).
- 41.Lacépède, B. G. E. Histoire Naturelle Des Poissons V. 4 (1802).
- 42.Lesueur CA. Descriptions of the five new species of the genus Cichla of Cuvier. J. Acad. Nat. Sci. Phila. 1822;2:214–221. [Google Scholar]
- 43.Bailey RM, Hubbs CL. The Black Basses (Micropterus) of Florida with description of a new species. Occas. Pap. Mus. Zool. Univ. Mich. 1949;516:1–40. [Google Scholar]
- 44.Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 2012;7(5):E37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eaton DAR, Overcast I. ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics. 2020;36:2592–2594. doi: 10.1093/bioinformatics/btz966. [DOI] [PubMed] [Google Scholar]
- 46.Mccartney-Melstad E, Gidiş M, Shaffer HB. An empirical pipeline for choosing the optimal clustering threshold in RADseq studies. Mol. Ecol. Resour. 2019;19:1195–1204. doi: 10.1111/1755-0998.13029. [DOI] [PubMed] [Google Scholar]
- 47.Minh BQ, et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. Modelfinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Near TJ, Kim D. Phylogeny and time scale of diversification in the fossil-rich sunfishes and Black Basses (Teleostei: Percomorpha: Centrarchidae) Mol. Phylogenet. Evol. 2021;161:107156. doi: 10.1016/j.ympev.2021.107156. [DOI] [PubMed] [Google Scholar]
- 50.Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. Ufboot 2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frichot E, François O. Lea: An R package for landscape and ecological association studies. Methods Ecol. Evol. 2015;6:925–929. doi: 10.1111/2041-210X.12382. [DOI] [Google Scholar]
- 52.Alexander DH, Lange K. Enhancements to the admixture algorithm for individual ancestry estimation. BMC Bioinform. 2011;12:246. doi: 10.1186/1471-2105-12-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goudet J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes. 2005;5:184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- 54.Chifman J, Kubatko L. Quartet inference from SNP data under the coalescent model. Bioinformatics. 2014;30:3317–3324. doi: 10.1093/bioinformatics/btu530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rannala B, Yang ZH. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics. 2003;164:1645–1656. doi: 10.1093/genetics/164.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouckaert R, et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:E1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson ND, Carstens BC, Morales AE, O’Meara BC. Species delimitation with gene flow. Syst. Biol. 2017;66:799–812. doi: 10.1093/sysbio/syx001. [DOI] [PubMed] [Google Scholar]
- 59.Rannala B, Yang Z. Efficient Bayesian species tree inference under the multispecies coalescent. Syst. Biol. 2017;66:823–842. doi: 10.1093/sysbio/syw119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ. PopGenome: An efficient swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014;31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nei, M. Molecular Evolutionary Genetics. (Columbia University Press, 1987).
- 62.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:E1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Excoffier L, Foll M. Fastsimcoal: A continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics. 2011;27:1332–1334. doi: 10.1093/bioinformatics/btr124. [DOI] [PubMed] [Google Scholar]
- 64.Excoffier L, Dupanloup I, Huerta-Sanchez E, Sousa VC, Foll M. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013;9:e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marques DA, Lucek K, Sousa VC, Excoffier L, Seehausen O. Admixture between old lineages facilitated contemporary ecological speciation in Lake Constance stickleback. Nat. Commun. 2019;10:4240. doi: 10.1038/s41467-019-12182-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kelley JW. Sexual maturity and fecundity of the Largemouth Bass, Micropterus salmoides (Lacépède), in Maine. Trans. Am. Fish. Soc. 1962;91:23–28. doi: 10.1577/1548-8659(1962)91[23:SMAFOT]2.0.CO;2. [DOI] [Google Scholar]
- 67.Lorenzoni M, Martin Dörr AJ, Erra R, Giovinazzo G, Mearelli M, Selvi S. Growth and reproduction of Largemouth Bass (Micropterus salmoides Lacépède, 1802) in Lake Trasimeno (Umbria, Italy) Fish. Res. 2002;56:89–95. doi: 10.1016/S0165-7836(01)00309-5. [DOI] [Google Scholar]
- 68.Cuvier, G. & Valenciennes, A. Histoire naturelle des poissons. Tome Second. Livre Troisième. Des poissons de la famille des perches, ou des percoïdes. V. 2. F. G. Levrault (1828).
- 69.Xi Z, Liu L, Rest JS, Davis CC. Coalescent versus concatenation methods and the placement of Amborella as sister to water lilies. Syst. Biol. 2014;63:919–932. doi: 10.1093/sysbio/syu055. [DOI] [PubMed] [Google Scholar]
- 70.Liu L, Xi Z, Wu S, Davis CC, Edwards SV. Estimating phylogenetic trees from genome-scale data. Ann. N. Y. Acad. Sci. 2015;1360:36–53. doi: 10.1111/nyas.12747. [DOI] [PubMed] [Google Scholar]
- 71.Jiang X, Edwards SV, Liu L. The multispecies coalescent model outperforms concatenation across diverse phylogenomic data sets. Syst. Biol. 2020;69:795–812. doi: 10.1093/sysbio/syaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eschmeyer, W. N. & Fricke, R. Catalog of fishes. http://Research.Calacademy.Org/Research/Ichthyology/Catalog/Fishcatmain.Asp (2022).
- 73.Gibert CR. Type catalogue of recent and fossil North American freshwater fishes: Families Cyprinidae, Catostomidae, Ictaluridae, Centrarchidae, and Elassomatidae. Fla. Mus. Nat. Hist. Spec. Publ. 1998;1:1–284. [Google Scholar]
- 74.Jordan DS. Notes on certain typical specimens of American Fishes in the British Museum and in the Museum D'historie Naturelle at Paris. Proc. U.S. Natl. Mus. 1880;2:218–226. doi: 10.5479/si.00963801.81.218. [DOI] [Google Scholar]
- 75.Gill T. On the species of the genus Micropterus (Lac.) or Grystes (Auct.) Proc. Am. Assoc. Adv. Sci. 1873;23:55–72. [Google Scholar]
- 76.Near TJ, Kassler TW, Koppelman JB, Dillman CB, Philipp DP. Speciation in North American Black Basses, Micropterus (Actinopterygii: Centrarchidae) Evolution. 2003;57:1610–1621. doi: 10.1111/j.0014-3820.2003.tb00368.x. [DOI] [PubMed] [Google Scholar]
- 77.Barthel BL, et al. Genetic relationships among populations of Florida Bass. Trans. Am. Fish. Soc. 2010;139:1615–1641. doi: 10.1577/T09-185.1. [DOI] [Google Scholar]
- 78.Page LM, et al. Common and scientific names of fishes from the United States, Canada, and Mexico. Am. Fish. Soc. Spec. Publ. 2013;34:1–243. [Google Scholar]
- 79.Robins, R. H., Page, L. M., Williams, J. D., Randall, Z. S. & Sheehy, G. E. Fishes in the fresh waters of Florida: An identification guide and atlas. (University of Florida Press, 2018).
- 80.Bailey RM, Latta WC, Smith GR. An atlas of Michigan fishes with keys and illustrations for their identification. Misc. Publ. Mus. Zool. Univ. Mich. 2004;192:1–215. [Google Scholar]
- 81.Silliman K, Zhao H, Justice M, Thongda W, Bowen B, Peatman E. Complex introgression among three diverged largemouth bass lineages. Evol. Appl. 2021;14:2815–2830. doi: 10.1111/eva.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson, S. E. Utilization of diagnostic SNP markers for understanding genetic composition of Largemouth Bass populations in Georgia. [Masters Thesis] Auburn University (2019).
- 83.Stark WJ, Echelle AA. Genetic structure and systematics of Smallmouth Bass, with emphasis on interior highlands populations. Trans. Am. Fish. Soc. 1998;127:393–416. doi: 10.1577/1548-8659(1998)127<0393:GSASOS>2.0.CO;2. [DOI] [Google Scholar]
- 84.Leaché AD, Zhu T, Rannala B, Yang Z. The spectre of too many species. Syst. Biol. 2019;68:168–181. doi: 10.1093/sysbio/syy051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tringali MD, Barthel BL, Seyoum S, Knight JR. The Choctaw Bass: An undescribed species of Micropterus in the Gulf Coast Plain rivers of Florida. Am. Fish. Soc. Symp. 2015;82:421–448. [Google Scholar]
- 86.Lutz-Carrillo DJ, Husemann M, Bean PT, Williamson JC, De Jesús MJ, Ray JW. Hybridization and genetic structure in phenotypic Spotted Bass in Texas. Trans. Am. Fish. Soc. 2018;147:891–905. doi: 10.1002/tafs.10074. [DOI] [Google Scholar]
- 87.Baker WH, Blanton RE, Johnston CE. Diversity within the Redeye Bass, Micropterus coosae (Perciformes: Centrarchidae) species group, with descriptions of four new species. Zootaxa. 2013;3635:379–401. doi: 10.11646/zootaxa.3635.4.3. [DOI] [PubMed] [Google Scholar]
- 88.Freeman BJ, et al. Shoal Basses: A clade of cryptic diversity. Am. Fish. Soc. Symp. 2015;82:449–466. [Google Scholar]
- 89.Bagley JC, Mayden RL, Roe KJ, Holznagel W, Harris PM. Congeneric phylogeographical sampling reveals polyphyly and novel biodiversity within Black Basses (Centrarchidae: Micropterus) Biol. J. Linn. Soc. 2011;104:346–363. doi: 10.1111/j.1095-8312.2011.01720.x. [DOI] [Google Scholar]
- 90.Bangs MR, Oswald KJ, Greig TW, Leitner JK, Rankin DM, Quattro JM. Introgressive hybridization and species turnover in reservoirs: A case study involving endemic and invasive basses (Centrarchidae: Micropterus) in Southeastern North America. Conserv. Genet. 2018;19:57–69. doi: 10.1007/s10592-017-1018-7. [DOI] [Google Scholar]
- 91.Williams JD, Burgess GH. A new species of bass, Micropterus cataractae (Teleostei: Centrarchidae), from the Apalachicola River Basin in Alabama, Florida, and Georgia. Bull. Fla. Mus. Nat. Hist. 1999;42:80–114. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data is available for download from the SRA (https://www.ncbi.nlm.nih.gov/sra/PRJNA796640). Data and supplementary materials are available from the Dryad Digital Repository: https://datadryad.org/stash/share/I4cSjJcrGRUPSI6uIZhivfBReyoCXLSIIDL89a3Qm20.