Abstract
Thymic epithelial cells (TECs) are essential for the production of self-tolerant T cells. The newly identified thymic tuft cells are regulated by Pou2f3 and represent important elements for host type 2 immunity. However, epigenetic involvement in thymic tuft cell development remains unclear. We performed single-cell ATAC-seq of medullary TEC (mTEC) and established single-cell chromatin accessibility profiling of mTECs. The results showed that mTEC III cells can be further divided into three groups (Late Aire 1, 2, and 3) and that thymic tuft cells may be derived from Late Aire 2 cells. Pou2f3 is expressed in both Late Aire 2 cells and thymic tuft cells, while Pou2f3-regulated genes are specifically expressed in thymic tuft cells with simultaneous opening of chromatin accessibility, indicating the involvement of epigenetic modification in this process. Using the epigenetic regulator Sirt6-defect mouse model, we found that Sirt6 deletion increased Late Aire 2 cells and decreased thymic tuft cells and Late Aire 3 cells without affecting Pou2f3 expression. However, Sirt6 deletion reduced the chromatin accessibility of Pou2f3-regulated genes in thymic tuft cells, which may be caused by Sirt6–mediated regulation of Hdac9 expression. These data indicate that epigenetic regulation is indispensable for Pou2f3-mediated thymic tuft cell development.
Subject terms: Thymus, Lymphocyte differentiation
Thymic epithelial cells (TECs) are essential for the production of self-tolerant T cells. Single-cell ATAC-seq of medullary TECs suggests that Sirt6-mediated epigenetic regulation may regulate the development of thymic tuft cells from these TECs.
Introduction
The thymus is essential for the education and maturation of functionally competent and self-tolerant T cells, and diversely specialized thymic epithelial cells (TECs), including cortical TECs (cTECs) and medullary TECs (mTECs), play critical roles in these processes1. cTECs mainly coordinate the early stages and positive selection of thymocytes, while mTECs primarily carry out the later steps and negative selection of thymocytes (reviewed in ref. 2,3). The heterogeneity of mTECs in mice and humans has recently been recognized with lineage-tracing mouse models, bulk RNA sequencing (RNA-seq), and single-cell RNA-seq (scRNA-seq) assays4–8. Abramson and Amit’s teams identified four mTEC subgroups, including mTEC I-IV. In the adult thymus, immature Ccl21+Aire− mTEC I cells are considered the earliest mTEC cluster, which recruits CCR7+ thymocytes to migrate from the thymic cortex region into the medulla region. Mature Aire+ mTEC II cells, as a functional mature mTEC cluster, randomly express tissue-restricted antigens to eliminate autoreactive thymocytes and contribute to the establishment of central immune tolerance. The corneocyte-like post-Aire expression KRT10+ mTEC III population is a terminally differentiated mTEC cluster that is also known as the late Aire stage, and it downregulates the expression of Aire, CD80, and MHC class II. The double cortin-like kinase 1 (Dclk1)+ and/or transient receptor potential cation channel subfamily M member 5 (Trpm5)+ mTEC IV population is a recently identified terminally differentiated mTEC cluster4,5. The mTEC IV population is also called thymic tuft cells because it expresses a group of canonical taste transduction pathway-related genes, which are similar to intestinal tuft cells4,5,9,10. These thymic tuft cells account for 3–10% of the mTECs in the mouse and human thymus4,5. These observations are consistent with previous studies showing that a subset of eGFP+ mTECs express α-gustducin (GNAT3), Trpm5, and phospholipase Cβ2 (PLCβ2) and are present as terminally differentiated epithelia in Hassall corpuscle–like clusters in Chat-eGFP BAC transgenic mice11. These thymic tuft cells are also detectable in BAC transgenic mice expressing CreERT2 from the Tas2r143 promoter12. The expression of several bitter taste–associated receptors (Tas2r105, Tas2r108, and Tas2r131) in mTECs has been reported in taste receptor Tas2r131 reporter mice13. Recently, in vivo fate mapping and lineage-tracing results have shown that mTEC IV cells (thymic tuft cells) are derived from mTEC II and/or III cells or a common ancestor through an Aire-expressing stage and/or an Aire-independent pathway4,5. For the convenience of discussion, we use the term thymic tuft cells to describe the tuft cell-like mTEC IV subgroup.
Importantly, these thymic tuft cells express high levels of the type 2 cytokine IL-25, MHC class II molecules, and associated antigen presentation genes4,5. Pou2f3-deficient mice showed a marked decrease in the number of TCRβintCD1d+IL-4+ type 2 invariant natural killer T (NKT2) cells and IL-4-dependent EOMES+CD8+ thymocytes and an increased frequency of thymic Lin−TCR−CD127+GATA3+ ILC2s without a significant impact on CD4−CD8−, CD4+CD8+, CD4−CD8+, and CD4+CD8− cells in the thymus, companying with an absence of thymic tuft cells4,5. Thus, thymic tuft cells are immunologically important, highly differentiated epithelial cells in the thymic medulla4,5. POU class 2 homeobox 3 (Pou2f3, Skn-1a, and Oct11)- and Trpm5-deficient mice show a specific loss of thymic tuft cells4,5, indicating that taste transduction pathway genes, such as Pou2f3 and Trpm5, are required for the development and function of thymic tuft cells. However, the epigenetic regulation of thymic tuft cell development has not been clarified. In the present study, we performed scRNA-seq and scATAC-seq analyses of mouse TECs and applied a TEC-specific deletion of the Sirt6 mouse model to define the routine development of thymic tuft cells, the Pou2f3-regulated gene expressing landscape in different mTEC subsets and the roles of Sirt6 in the development of thymic tuft cells. Our results showed that thymic tuft cells may be derived from one subgroup of mTEC III cells (Late Aire 2) and that epigenetic regulation is indispensable for the development of Pou2f3-mediated thymic tuft cells. This work advances our understanding of epigenetic regulation of the development of mTECs, especially thymic tuft cells, and provides an unbiased scATAC-seq resource to study mTEC heterogeneity.
Results
Single-cell chromatin accessibility profiling of mouse mTECs
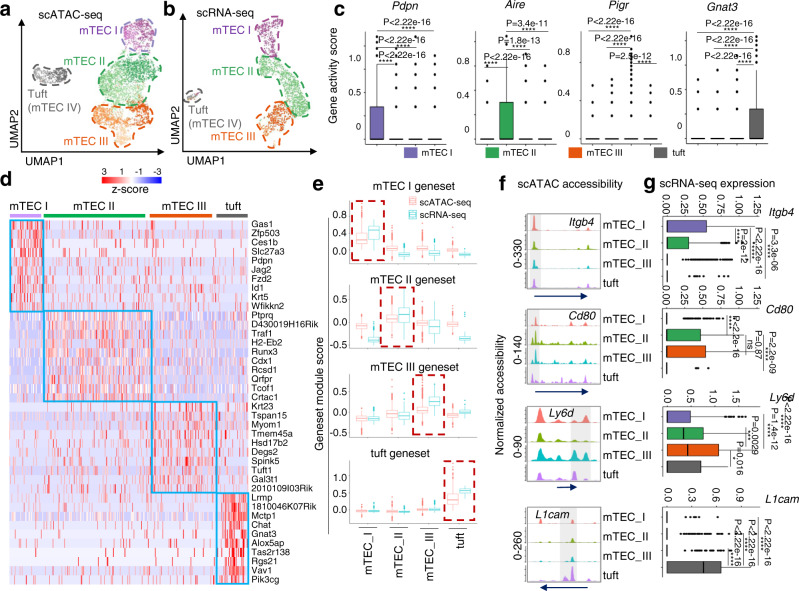
The single-cell assay for transposase-accessible chromatin with sequencing (scATAC-seq) method, which identifies active DNA regulatory elements by transposition of sequencing adapters into accessible chromatin in cells, has enabled mapping of cell-to-cell variability and epigenomic profiling of cells even in low-cell-number samples14–16. To investigate the chromatin accessibility characteristics of thymic tuft cells, 200000 CD45-EpCAM+ TECs were sorted from 4-week-old wild-type mice for scATAC-seq assays (10X Genomics Chromium) (Suppl. Fig. 1). After filtering, 4739 mTECs with high-quality data were analyzed (Method, Suppl. Fig. 2). To determine the cell type annotation of mTEC scATAC-seq, we first performed unbiased clustering on the downloaded scRNA-seq of WT17. According to the different molecular characteristics of mTEC clusters based on scRNA-seq assays5(Suppl. Figs. 3, 4), mTECs can be divided into 4 subgroups: (1) mTEC I highly expressed the indicator genes Pdpn, Ccl21, Ly6a, Itga6, and Itga4; (2) mTEC II highly expressed Aire, Fezf2, Cd80, Cd40, and Cd74; (3) mTEC III highly expressed Pigr, Ly6d, Ivl, and Krt10; and (4) thymic tuft cells highly expressed the indicator genes Lrmp, Gnat3, Dclk1, Trpm5, L1cam, and Avil.
To identify the genes accessible in each cell type, a gene activity matrix was established to serve as pseudoexpression data to identify the known mTEC subsets. Projection of gene expression (scRNA-seq) and gene activity scores (scATAC-seq) in the coembedded space shows that distinct clustering of unique cell types is preserved and that scATAC-seq and scRNA-seq cells of the same population cluster together16,18–20. scATAC-seq profiles were obtained by unbiased clustering on all peaks of WT mTECs using Signac, and the corresponding cell population annotation was predicted based on the gene activity matrix and coembedment analysis (Suppl. Fig. 5). Furthermore, we checked the subgroup-specific gene sets from scRNA-seq at different resolutions on scATAC-seq to determine the final scATAC-seq profile, which also proved the correlation between the identified cell type and the predicted id was consistent (Fig. 1a, b and Suppl. Figs. 6, 7).
Fig. 1. Chromatin accessibility profiling of mTECs was established according to the known single-cell transcriptional profile.
a UMAP visualization of scATAC-seq profiles of mTECs. Cell types are circled with dotted lines of different colors. b UMAP visualization of scRNA-seq profiles of mTECs. Cell types are circled with dotted lines of different colors. c Boxplots show marker genes for each mTEC cluster by gene accessibility matrix. Color represents specific clusters of mTECs. mTEC I: n = 638, mTEC II: n = 1982, mTEC III: n = 1262, thymic tuft cells: n = 495. d Heatmap of the gene expression profile of defined mTEC populations in scATAC-seq was generated by the top ten marker genes from the gene activity matrix. The color scale indicates the level of gene activity. e Marker genes of each mTEC population calculated by scRNA-seq were added to both scRNA-seq and scATAC-seq. Gene set module scores in scRNA-seq (blue) and scATAC-seq (red) were compared with boxplots. f Genome tracks of aggregate scATAC-seq data show marker chromatin accessibility peaks. The arrow indicates the length and direction of the genes detected by scATAC-seq. g Boxplots show normalized expression of the marker genes for each mTEC cluster in scRNA-seq data. mTEC I: n = 364, mTEC II: n = 991, mTEC III: n = 458, thymic tuft cells: n = 62. The statistics was determined using Wilcoxon signed-rank test. ****p < 0.001.
Based on the expression levels of marker genes and gene sets, we identified four mTEC clusters (Fig. 1a), which showed clustering similar to that of the scRNA-seq data of mTECs5. Gene activity matrix data (pseudogene expression) enabled us to identify the highly expressed marker genes in different mTEC subgroups, as were recently reported5: Pdpn in mTEC I, Aire in mTEC II, Pigr in mTEC III, and Gnat3 in thymic tuft cells (Fig. 1c and Supplementary Data 1). We then enriched the highly accessible genes in different mTEC subgroups based on scATAC-seq data (Fig. 1d). After analyzing the specific gene set of each mTEC cluster, we found that the chromatin accessibility detected by scATAC-seq was highly consistent with the RNA expression detected by scRNA-seq in each cluster (Fig. 1e and Supplementary Data 1). Additionally, we found that the gene set of thymic tuft cells was more specific than the mTEC I-III gene set in terms of transcriptional signature and chromatin accessibility (Fig. 1e and Supplementary Data 1). To determine whether cell-type-specific chromatin accessibility correlated with gene expression, we compared chromatin accessibility and mRNA expression in the known characteristic genes of each mTEC subgroup, including Itgb4 in mTEC I, CD80 in mTEC II, Ly6d in mTEC III, and L1cam in thymic tuft cells. The openness of the target gene chromatin accessibility (Fig. 1f) was nicely associated with its mRNA expression in all mTEC subgroups (Fig. 1g, Suppl. Fig. 8, and Supplementary Data 1), indicating that there was a positive correlation between chromatin accessibility and transcriptional level in the same cell types. Analysis of ATAC-seq profiles revealed that mTEC I and thymic tuft cells possessed more open chromatin regions than mTEC II and III cells, as determined by the ATAC peak numbers (Suppl. Fig. 9). The distribution of the specific opened peaks in mTEC I and thymic tuft cells was located mainly in promoter and distal regions (Suppl. Fig. 9), which is consistent with the association of distal chromatin elements with the developing cell type specificity as reported21. These results collectively proved the consistency between chromatin accessibility obtained by scATAC-seq assays and its mRNA expression obtained by scRNA-seq assays in mTEC clusters, and they also indicated the rich diversity of mTECs is accompanied by global remodeling of the chromatin accessibility landscape.
Three subpopulations in the mTEC III subgroup
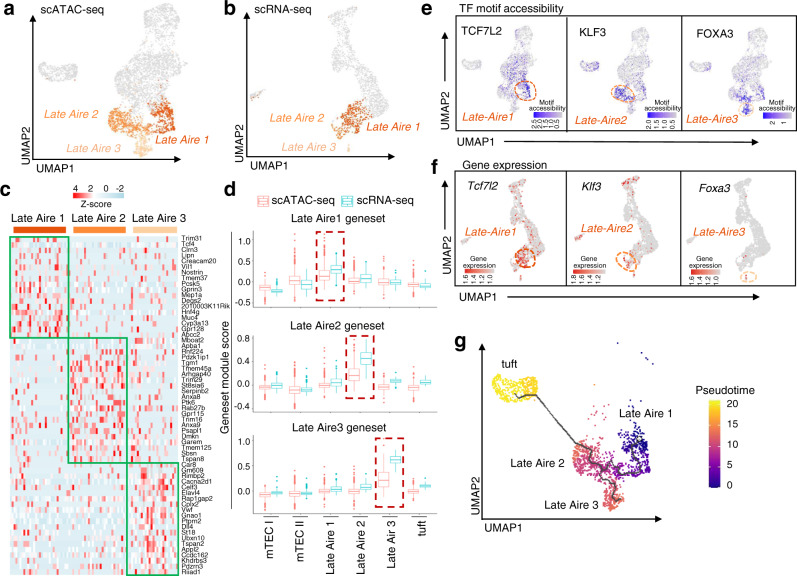
mTECs express autoantigens instantaneously and have many heterogeneous populations22, and although the properties have been studied, they are not completely understood. It is well known that fine cell types can be distinguished by different resolutions in single-cell sequencing22. mTEC III, which shows significant heterogeneity in scATAC-seq, can be divided into three subgroups even at different resolutions (Fig. 1a and Suppl. Figs. 10, 11). Therefore, we further divided mTEC II into Late Aire 1, Late Aire 2, and Late Aire 3 at resolution 1 (Fig. 2a, b). By analyzing the chromatin accessibility and transcriptome of the three identified mTEC III clusters, we found that Late Aire 1 (Hnf4g, Trim31, Nos2, and Gpa33), Late Aire 2 (Tspan8, Ly6d, Tmem45a, and Psapl1), and Late Aire 3 (Cd177, Fam174b, Car8, and Gm609) were relatively independent mTEC subgroups with distinct chromatin accessibility profiles (Fig. 2c and Suppl. Fig. 12). This observation was consistent with a recent report showing that mTECs might have as many as 14 subgroups as analyzed by scRNA-seq22, and Late Aire 1 corresponded to C7 (Gpa33 and Nos2), Late Aire 2 corresponded to C8 (Tspan8 and Ly6d), and Late Aire 3 corresponded to C9 (Cd177 and Car8). According to the marker genes determined by scATAC-seq, we found the corresponding three groups of cells by scRNA-seq (Suppl. Fig. 13). The highly expressed genes in Late Aire 1 to 3 of scRNA-seq were used to perform GO enrichment analyses (Suppl. Fig. 14), and calcium ion-regulated endocytosis was specifically enriched in Late Aire 3 cells. A new type of EPCAM+ cells has been identified in humans as neuro TECs8,23, which resemble neuroendocrine cells. In mice, Late Aire 3 cells did not express the markers Neurog1 and Neurod1 of neural TECs (Suppl. Fig. 15), indicating that Late Aire 3 cells might be a special subset of terminally differentiated cells. At the same time, the difference in the gene set module score in one of the Late Aire types (i.e., 1–3) further indicated the heterogeneity of mTEC III, as demonstrated by the scATAC-seq and scRNA-seq data (Fig. 2d and Supplementary Data 2). Compared with Late Aire 1 and 2, the major alteration of increased chromatin accessibility in Late Aire 3 was found in other intron regions (Suppl. Fig. 16).
Fig. 2. Chromatin accessibility, gene expression, and differentiation trajectory of three newly identified mTEC III subsets.
UMAP visualization of scATAC-seq profiles (a) and scRNA-seq profiles (b). Late Aire 1, Late Aire 2, and Late Aire 3 cells labeled with different colors are defined by the different chromatin accessibility in scATAC-seq and scRNA-seq profiles. c Heatmap of the gene expression profile of late Aire 1–3 populations in scATAC-seq was generated by the top 20 marker genes from the gene activity matrix. The color scale indicates the level of gene activity. d Marker genes of each Late Aire population calculated by scATAC-seq were added to both scRNA-seq and scATAC-seq. The gene set module scores of Late Aire 1-3 in scRNA-seq (blue) and scATAC-seq (red) were compared in boxplots. e UMAP visualization of TF motif accessibility for the indicated TFs in Late Aire 1, Late Aire 2, and Late Aire 3 cells. The blue scale in the UMAP plot indicates the level of TF motif accessibility in scATAC-seq analyzed by chromVAR. f UMAP visualization of scRNA-seq colored based on the expression of the indicated TFs in Late Aire 1, Late Aire 2, and Late Aire 3 cells. The red scale in UMAP plot indicate the level of gene expression in scRNA-seq. g UMAP visualization of scATAC-seq subsets of Late Aire 1, Late Aire 2, Late Aire 3, and thymic tuft cells. Colored based on the pseudotime, with blue corresponding to early cells and yellow corresponding to thymic tuft cells.
Chromatin accessibility analysis can be used to identify candidate transcriptional regulators of each cell state or cell type during cell differentiation24. We measured chromatin accessibility at cis-elements sharing a transcription factor (TF)-binding motif using the chromVAR analysis approach25 and cross-matched the data among enriched highly activated motifs, scATAC-seq data, and scRNA-seq data to identify the key transcription factors in each cell of the mTEC III subgroup. We found that Late Aire 1–3 were specifically regulated by different transcription factors (Suppl. Fig. 17). Late Aire 1 was predominantly and specifically regulated by TCF7L2, Late Aire 2 was mainly regulated by KLF3, and Late Aire 3 was selectively regulated by FOXA3 (Fig. 2e). In parallel, through the analysis of scRNA-seq data, we found that the genes Tcf7l2 and Foxa3 were specifically expressed in Late Aire 1 and Late Aire 3, respectively, while Klf3 was also partially expressed in mTEC I and thymic tuft cells in addition to Late Aire 2 (Fig. 2f).
Because lineage-tracing studies by two independent research teams showed that more than half of thymic tuft cells arise from mature mTEChigh cells4,5, Klf3 was partially expressed in thymic tuft cells and KLF3 regulatory motifs were partially located in thymic tuft cells, we speculated that Late Aire 2 cells have a strong developmental correlation with thymic tuft cells. The development path of thymic tuft cells could not be obtained in all cells based on scATAC-seq, which may be caused by the large difference in gene expression between tufts and other mTECs (Suppl. Fig. 18). Hence, we used scATAC-seq to reconstruct cellular developmental trajectories of the Late Aire 1, Late Aire 2, Late Aire 3, and thymic tuft cells in an unbiased manner14 to further prove their developmental relevance. Differentiation pseudotime analysis and RNA velocity analysis showed that the Late Aire 2 subgroup had a certain developmental correlation with thymic tuft cells, which was also verified by scRNA-seq (Fig. 2g and Suppl. Figs. 19, 20). Thus, we speculate that the Late Aire 2 subgroup may represent a crossroad for differentiation into either Late Aire 3 or thymic tuft cells.
Increased chromatin accessibility is essential for Pou2f3-induced thymic tuft cell development
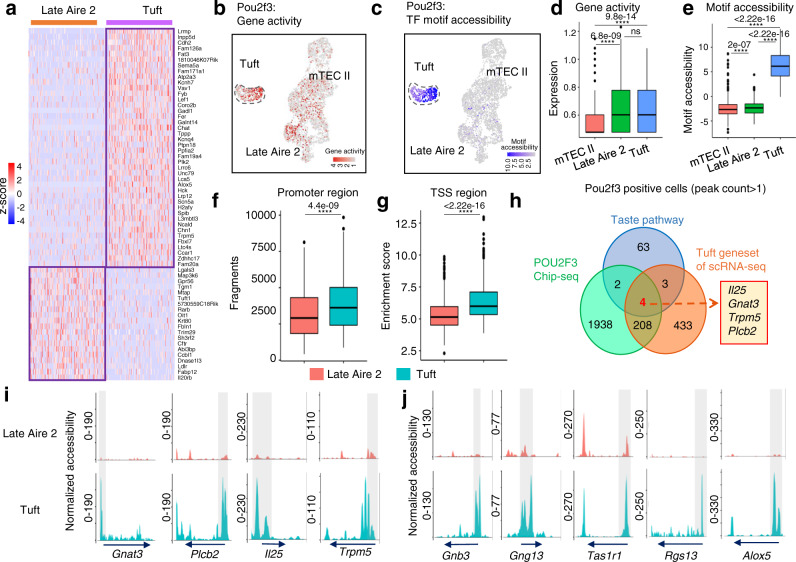
To further explore the developmental relationship between Late Aire 2 cells and thymic tuft cells, we analyzed their chromatin accessibility profiles by using scATAC-seq. The results showed that Late Aire 2 cells and thymic tuft cells had relatively specific chromatin accessibility profiles (Fig. 3a). Pou2f3, a well-known key transcription factor for the development of thymic tuft cells, may be accessible not only in thymic tuft cells but also in mTEC III5. Accordingly, we calculated the accessibility of Pou2f3 in scATAC-seq using pseudoexpression data (Fig. 3b). In mTEC I, Late Aire 1, and Late Aire 3, the peak count of Pou2f3 was less than 1 in 75% of the cells (upper edge of boxplot), while in both mTEC II and Late Aire 2, the peak count of Pou2f3 was more than 1 in 50% of the cells (boxplot median); moreover, Pou2f3 was accessible in 75% of the cells (boxplot lower margin) at a peak count of more than 1 in thymic tuft cells. This finding indicates that Pou2f3 is mainly accessible in mTEC II, Late Aire 2, and thymic tuft cells (Suppl. Fig. 19a). Among these three populations, Pou2f3 had the highest accessibility in thymic tuft cells (Suppl. Figs. 21, 22a). Except for thymic tuft cells, Pou2f3 was accessible on each exon in Late Aire 2 cells, while other populations mainly opened in exon 1 and exon 2 of Pou2f3 (Suppl. Fig. 22b). However, scRNA-seq data showed that the expression of Pou2f3 was detected only in Late Aire 2 cells and thymic tuft cells (Suppl. Fig. 23). The expression of Pou2f3 was not limited to thymic tuft cells, further suggesting that Late Aire 2 may have a stronger developmental correlation with thymic tuft cells than mTEC II. However, the peaks bound by Pou2f3 were detectable specifically in thymic tuft cells, while almost no detectable motif accessibility was detected in other mTEC subgroups (Fig. 3c and Suppl. Fig. 24), indicating the motif accessibility of Pou2f3 is critical for the differentiation of thymic tuft cells.
Fig. 3. Pou2f3 but not its target genes are expressed before Late Aire 2, and chromatin opening is required for target gene expression in thymic tuft cells.
a Heatmap of differentially accessible genes between Late Aire 2 cells and thymic tuft cells in scATAC-seq. The color scale indicates the level of gene activity. b UMAP visualization of the gene expression of Pou2f3 in scRNA-seq. c UMAP visualization of the motif accessibility score of Pou2f3 in scATAC-seq. d Gene expression of Pou2f3 in Pou2f3+ mTEC subsets in scATAC-seq was compared in a boxplot. Pou2f3+ mTEC II: n = 499, Pou2f3+ Late Aire 2 cells: n = 210, Pou2f3+ thymic tuft cells: n = 297. e Motif accessibility score of Pou2f3 in Pou2f3+ mTEC subsets in scATAC-seq was compared in a boxplot. Boxplots show the fragments at the promoter region (f) and TSS enrichment score (g) for Late Aire 2 cells (pink) and thymic tuft cells (blue). Late Aire 2 cells: n = 551, thymic tuft cells: n = 495. h Taste pathway-related gene (blue), Pou2f3 directly regulated gene sets (green), and tuft gene sets of scRNA-seq (orange) are calculated in a Venn diagram. The overlapping genes among them are shown on the right. The chromatin accessibility peaks of overlapping genes (i) and tuft-associated genes (j) are shown in scATAC-seq tracks. The arrow indicates the length and direction of the genes detected by scATAC-seq. The statistics was determined using Wilcoxon signed-rank test. ****p < 0.001.
Because the majority of mTECs did not express Pou2f3, we selected Pou2f3+ mTEC subsets for the motif accessibility analysis. The accessibility region of Pou2f3+ cells in each population did not change compared to that of all cells (Suppl. Figs. 19c, 25). As mentioned earlier, there was no significant difference (P > 0.05) in Pou2f3 accessibility between Pou2f3+ Late Aire 2 cells and Pou2f3+ thymic tuft cells in scATAC-seq (Fig. 3d and Supplementary Data 3). Furthermore, Pou2f3 regulatory motif accessibility analysis of Pou2f3+ cells showed that thymic tuft cells were significantly different from mTEC II and Late Aire 2 cells (P < 0.01, Fig. 3e and Supplementary Data 3). In general, the analysis of Pou2f3+ mTEC subsets showed that both Late Aire 2 cells and thymic tuft cells had the ability to express Pou2f3, whether in chromatin accessibility or RNA expression, although its regulatory motif was only opened in thymic tuft cells. In both scRNA-seq and scATAC-seq, more than 30% of the tuft markers were regulated by Pou2f3, while less than 20% were regulated in other populations (Suppl. Fig. 26). Together, these data indicated that the expression of Pou2f3 in mTECs alone is not sufficient to promote the expression of Pou2f3-regulated thymic tuft cell-related genes.
Furthermore, we found that the fragments of gene promoters and TSS enrichment in thymic tuft cells were higher than those in Late Aire 2 cells (Fig. 3f, g and Supplementary Data 3). To determine the specific role of increased chromatin accessibility in the development of Late Aire 2 cells to thymic tuft cells, we further analyzed the chromatin status of well-recognized important genes in thymic tuft cells. By analyzing the intersection of taste pathway-related genes (GO: 0050909, sensory perception of taste; KEGG: map04742, Taste transduction pathway), Pou2f3-regulated genes26 and genes highly expressed in thymic tuft cells, we identified that the overlapping genes included Gnat3, Plcb2, Il25, and Trpm5 (Fig. 3h and Supplementary Data 4). In addition, we also selected genes that have been reported to be important in the thymus and small intestine tuft cells, including Gnb3, Gng13, Tas1r1, Rgs13, and Alox5. Consistent with the gene expression patterns, the chromatin of these genes was basically closed in Late Aire 2 mTECs and specifically opened in thymic tuft cells (Fig. 3i, j and Suppl. Fig. 27). Combined with the significant difference in gene accessibility shown in Fig. 3a, we speculate that in addition to the expression of Pou2f3, epigenetic regulation is essential for the development of thymic tuft cells.
Deficiency of thymic tuft cell development in Sirt6 cKO mice
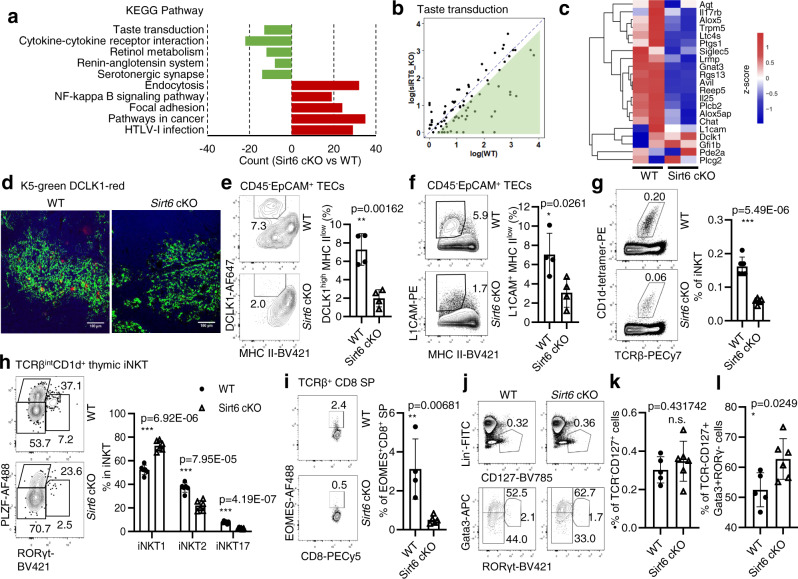
Chromatin accessibility is regulated by numerous epigenetic regulators27,28. Previous studies have proven that SIRT6, as a key chromatin regulator, remodels chromatin by promoting chromatin relaxation, although it has diverse biological functions, such as genome stability, glucose metabolism, and tumor suppression, through multiple pathways29–34. We, therefore, bred Foxn1-Cre mice with Sirt6flox/flox mice to obtain TEC-specific deletion of Sirt6 mice as reported recently35. TEC-specific Sirt6 deletion reduced the cell number of mTECs with an accelerated differentiation of CD80-Aire- immature mTECs to CD80+Aire- intermediate mature mTECs by promoting Spib expression35. However, we observed that the taste transduction signal pathway and taste-related genes were significantly downregulated in Sirt6-deficient mTECs, while endocytosis, NF-κB, and other pathways were enhanced, as detected by RNA-seq analysis (Fig. 4a, b and Supplementary Data 5). The known marker genes for thymic tuft cells compared with WT mTECs were also decreased after Sirt6 deletion (Fig. 4c). By detecting the proportion of DCLK1+ thymic tuft cells, we found that Sirt6 deletion blocked the development of thymic tuft cells, as determined by immunofluorescence staining and flow cytometry assays (P < 0.01, Fig. 4d, e and Supplementary Data 5). This conclusion was also confirmed by the decreased expression of L1CAM on thymic tuft cells in Sirt6 cKO mice (Fig. 4f and Supplementary Data 5). Consistent with the decreased cell number of thymic tuft cells in Pou2f3 knockout mice4,5, the functional deficiency of thymic tuft cells was evidenced by the abnormal development of NKT2 cells, EOMES+CD8+ single-positive thymocytes, and ILC2s in the thymus of Sirt6 cKO mice (Fig. 4g–j and Supplementary Data 5). TCRβintCD1d+ iNKT cells, especially RORγt−PLZF+ iNKT2 cells and RORγt+PLZF− iNKT17 cells, decreased significantly in Sirt6 cKO mice (P < 0.01, Fig. 4g, h and Supplementary Data 5). Meanwhile, EOMES+CD8+ single-positive cells were also markedly decreased after Sirt6 deletion (P < 0.01, Fig. 4i and Supplementary Data 5). The proportion of ILC2s increased slightly after Sirt6 ablation (Fig. 4j–l and Supplementary Data 5). These results clearly showed that Sirt6 is essential for the development of thymic tuft cells.
Fig. 4. Sirt6 deletion affects the cell number and function of thymic tuft cells.
a Downregulated genes or upregulated genes in Sirt6-deficient mTECs are enriched in KEGG pathways, and the top five pathways are ordered by p value. All pathways were selected under the standard of p < 0.05. b Scatter plot shows the difference in TPM values of taste transduction genes between WT and Sirt6-deficient mTECs. c Differential expression of thymic tuft cell-related genes between mTECs sorted from WT and Sirt6 cKO mice. The color scale indicates the level of gene expression. d Representative images of frozen thymic sections from WT and Sirt6 cKO mice (n = 4 per group). Green, KRT5. Red, DCLK1. Scale bars: 100 μm. e Representative flow cytometric profiles and frequency of thymic tuft cells (CD45–EpCAM+MHCIIlowDCLK1high) obtained from 4-week-old Sirt6 cKO mice and littermates (n = 4 per group). f Representative flow cytometric profiles and frequency of L1CAM+ thymic tuft cells from 4-week-old Sirt6 cKO mice and littermates (n = 4 per group). g Representative flow cytometric plots and frequency of TCRβintCD1d+ thymic iNKT cells in 4-week-old Sirt6 cKO mice and littermates (n = 6 per group). h Representative flow cytometric plots and frequencies of NKT1 (PLZF–RORγt–), NKT2 (PLZF+RORγt–), and NKT17 (PLZF–RORγt+) in 4-week-old Sirt6 cKO mice and littermates (n = 6 per group). i Representative flow cytometric plots and frequency of EOMES+TCRβ+CD8 single-positive thymocytes in 4-week-old WT (n = 4) and Sirt6 cKO (n = 5) mice. j Representative flow cytometric plots of Lin–TCR–CD127+ thymic ILCs (upper) and ILC subsets (lower) in Sirt6 cKO mice and littermates. k, l Frequencies of Lin–TCR–CD127+ thymic ILCs (k) and Lin–TCR–CD127+Gata3+Rorγt– ILC2s (l) of WT (n = 5) and Sirt6 cKO (n = 6) mice. *p < 0.05, **p < 0.01 and ***p < 0.001 compared with the identical WT control mice (Student’s t-test). The error bar represents one standard deviation.
Sirt6 deletion decreased the chromatin accessibility of Pou2f3-regulated genes in thymic tuft cells
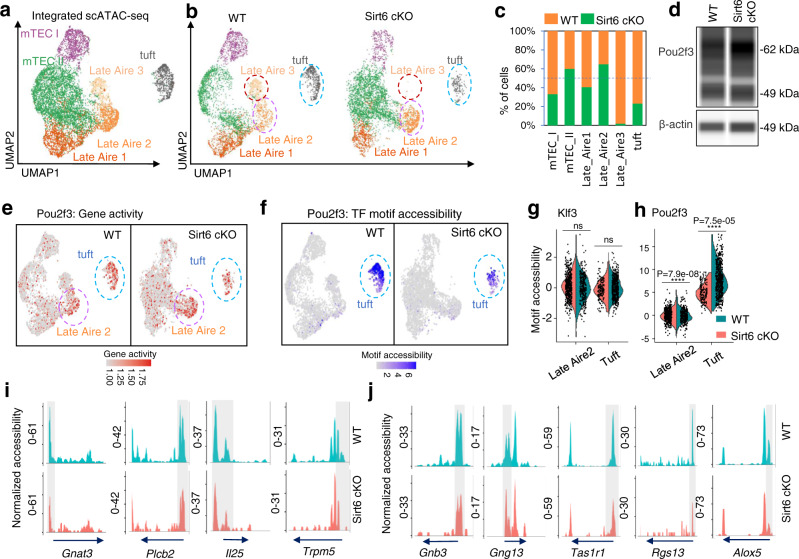
Sirt6 has been reported to regulate chromatin openness in many cells through its deacetylation34,36. By constructing Sirt6 cKO mice, it was found that the cell number and function of thymic tuft cells in the thymus were significantly affected (Fig. 4). Therefore, scATAC-seq was also performed on Sirt6 cKO mice to explore its possible effects on the developmental trajectory of thymic tuft cells. The sorting strategy and quality control rules are the same as those mentioned above (Suppl. Fig. 1, Method). Integrated scATAC-seq profiles were performed by unbiased clustering on all peaks of WT and Sirt6 cKO mTECs using Signac, and the corresponding cell population annotation was predicted based on a gene activity matrix and coembedded analysis (Suppl. Fig. 28). Furthermore, we checked the modular score of subgroup-specific genes from scRNA-seq to determine the integrated scATAC-seq profile, which also proved that the correlation between the identified cell type and the predicted id was consistent (Suppl. Figs. 29, 30). Therefore, we obtained the distribution of well-established mTEC populations (mTEC I, mTEC II, Late Aire 1, Late Aire 2, Late Aire 3, and thymic tuft cells) based on integrated scATAC-seq data from WT and Sirt6 cKO mice (Fig. 5a, b).
Fig. 5. Chromatin accessibility of the genes regulated by Pou2f3 decreased after Sirt6 deletion.
a, b scATAC-seq data of WT and Sirt6-deficient mTECs are coembedded into a single UMAP visualization (a) and are displayed separately (b). Cells are stained with the mTEC subset defined before. c Relative proportion of mTEC clusters between WT and Sirt6 cKO mice. d Simple Western analysis of POU2F3 expression in sorted mTECs (CD45−EpCAM+UEA-1+Ly51−) from 4-week-old WT and Sirt6 cKO mice. e UMAP visualization of the gene expression of Pou2f3 in WT (left) and Sirt6 cKO (right) scATAC-seq. The red scale in the UMAP plot indicates the level of Pou2f3 gene activity in scATAC-seq f UMAP visualization of the motif accessibility score of Pou2f3 in WT (left) and Sirt6 cKO (right) scATAC-seq. The blue scale in the UMAP plot indicates the level of Pou2f3 motif accessibility in scATAC-seq analyzed by chromVAR. g, h Motif accessibility scores of Klf3 (g) and Pou2f3 (h) in Late Aire 2 cells and thymic tuft cells of WT (Orange) and Sirt6 cKO mice (green) were compared in a violin plot. WT Late Aire 2 cells: n = 272, Sirt6 cKO Late Aire 2 cells: n = 500, WT thymic tuft cells: n = 501, Sirt6 cKO thymic tuft cells: n = 150. i, j Chromatin accessibility peaks of overlapping genes (i) and tuft-associated genes (j) mentioned in Fig. 3 in WT and Sirt6-deficient thymic tuft cells are shown in aggregated scATAC-seq tracks. The arrow indicates the length and direction of the genes detected by scATAC-seq. The statistics was determined using Wilcoxon signed-rank test. *p < 0.05, **p < 0.01, ***p < 0.005 and ****p < 0.001.
However, compared with WT control mice, Sirt6 cKO mice showed increased mTEC II and Late Aire 2 subgroups and decreased mTEC I, Late Aire 3, and thymic tuft cells (Fig. 5b, c and Supplementary Data 6), which is nicely consistent with the cell phenotype alteration of mTECs analyzed by flow cytometry (Fig. 4 and ref. 35). The integrated scATAC-seq data did not reveal the developmental trajectory of thymic tuft cells, which was consistent with the WT scATAC-seq results mentioned above. In particular, Late Aire 2 of WT mice has the potential to differentiate into Late Aire 3, while Late Aire 2 of Sirt6 cKO mice has the potential to develop into thymic tuft cells, but no Late Aire 3 (Suppl. Fig. 31). The increased Late Aire 2 proportion and decreased Late Aire 3 and thymic tuft cells in Sirt6 cKO mice suggested that Sirt6 deficiency may lead to the retardation of Late Aire 2 cells development to the subsequent stages (including Late Aire 3 cells and thymic tuft cells).
Since the transcription factor Pou2f3 is a key factor controlling the development of thymic tuft cell development4,5, we detected the expression of Pou2f3 in sorted WT and Sirt6 cKO mTECs by the Simple Western assay. Unexpectedly, the expression of Pou2f3 was not significantly affected after the Sirt6 deletion (Fig. 5d and Suppl. Fig. 32). This observation was further confirmed by scATAC-seq analysis, which showed that the accessibility of Pou2f3 in Late Aire 2 cells of WT and Sirt6 cKO mice was identical (Fig. 5e). Thus, Sirt6 deletion did not impact the accessibility and expression of Pou2f3 in the thymus.
By comparing the peaks of Klf3- and Pou2f3-regulated motifs in Late Aire 2 cells and thymic tuft cells between WT and Sirt6 cKO mice, we found that the peaks of Pou2f3-regulated motifs were reduced specifically in thymic tuft cells after Sirt6 deletion, whereas Klf3-regulated motifs in Late Aire 2 cells and thymic tuft cells were unimpaired after Sirt6 deletion (Fig. 5f–h, Suppl. Fig. 33, and Supplementary Data 6). Consistent with this result, the differentially accessible peaks of thymic tuft cells between WT and Sirt6 cKO mice were analyzed. A subsequent motif enrichment analysis conclusively showed that Pou2f3 was the most significant TF regulating the reduced accessible peak region after Sirt6 deficiency (Suppl. Fig. 34). The top three TFs of motif enrichment, namely, KLF15, EGR1, and E2F6, in the open peak region after Sirt6 deficiency were not necessary, and there was no significant increase in the motif activity score of those TFs (Suppl. Fig. 35). Therefore, Sirt6 knockout did affect thymic tuft cells by affecting the accessibility of Pou2f3 downstream genes. Furthermore, the chromatin accessibility of the identified key Pou2f3-regulated genes decreased in Sirt6 cKO thymic tuft cells (Fig. 5i, j). Thus, Sirt6 contributes to the development of thymic tuft cells by increasing the chromatin accessibility of Pou2f3-regulated motifs in the thymus.
Sirt6 deletion increased Hdac9 expression to decrease the chromatin accessibility of Pou2f3-regulated genes
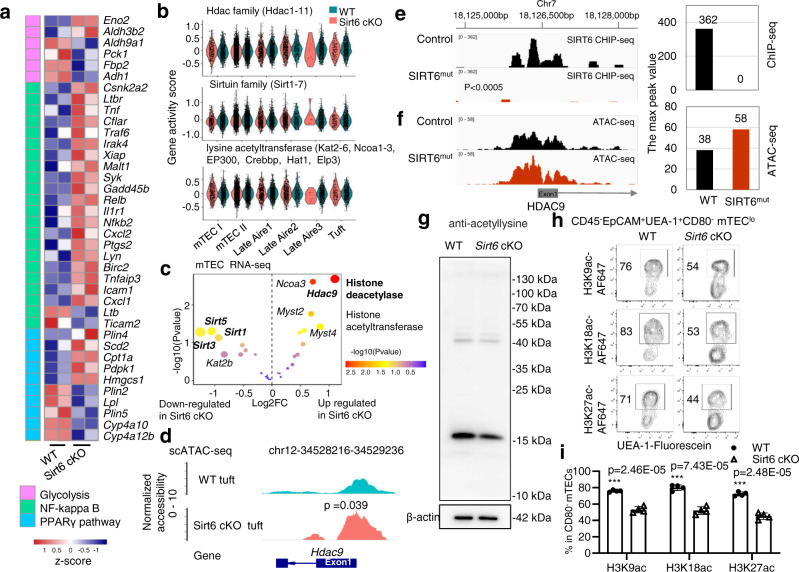
SIRT6 plays a pivotal role in regulating some fundamental cellular processes, such as cellular metabolism, DNA repair, gene expression, mitochondrial biology, and telemetric maintenance37,38. Deacetylation of specific sites on histone H3 (H3K9, H3K56, and H3K18) by SIRT6 can promote chromatin compaction and repress transcription of target genes39–44. SIRT6 predominantly represses the expression of various genes, such as glycolytic genes, Myc-target ribosomal protein genes, lipid metabolism genes, and NF-κB-dependent inflammatory genes29–33,45,46. In parallel with these studies, Sirt6-deficient mTECs expressed higher glycolysis-, NF-κB pathway-, and PPARγ pathway-related genes, as detected by bulk RNA-seq assays (Fig. 6a). However, the decreased Pou2f3 target gene accessibility caused by Sirt6 deletion is somehow contradictory to the general conception of the biofunction of Sirt6. Therefore, we observed increased gene expression profiles related to histone acetylation, including the families of histone deacetylase (HDAC), SIRTUIN, and histone lysine acetyltransferase (KAT), in Sirt6-deleted thymic tuft cells. The scATAC-seq assays showed that although the overall gene accessibility of the SIRTUIN, HDAC, and KAT families was unchanged in Sirt6-deleted thymic tuft cells (Fig. 6b and Supplementary Data 7), some genes, including the histone deacetylase gene (Hdac9) and histone acetyltransferase genes (Ncoa3, Myst2, and Myst4), were significantly upregulated in mTECs after Sirt6 deletion (Fig. 6c), while some histone deacetylase genes (Sirt5, Sirt3, and Sirt1) were significantly downregulated (Fig. 6c). In particular, Hdac9 was the only deacetylase gene with an accessible promoter region after deletion of Sirt6 in thymic tuft cells (Suppl. Fig. 36a). In addition, the peak of the promoter region of Hdac9 was significantly accessible in thymic tuft cells (Suppl. Fig. 36b). The chromatin accessibility of the Hdac9 promoter was also significantly increased in Sirt6-deleted thymic tuft cells (Fig. 6d), indicating that Sirt6 may promote chromatin accessibility of thymic tuft cells by inhibiting Hdac9.
Fig. 6. Decreased chromatin accessibility of Pou2f3-regulated genes was consistent with the decreased acetylation regulated by the SIRT6-HDAC9 axis.
a Glycolysis-, NF-kappaB pathway-, and PPARγ pathway-associated genes were compared between WT and Sirt6 cKO mTEC bulk RNA-seq by heatmap. The color scale indicates the level of gene expression. b Gene set module scores of the histone deacetylase gene family, sirtuin gene family, and histone lysine acetyltransferase gene family were compared between WT (green) and Sirt6 cKO mouse (red) mTEC populations. c Differential expression of the histone deacetylase gene family, sirtuin gene family, and histone lysine acetyltransferase gene family was calculated by DEseq2 using WT and Sirt6 cKO mTEC bulk RNA-seq. The x-axis represents Log2FC, and the y-axis represents −log10 (P value). The dot size is consistent with the |Log2FC| value. Histone deacetylase-related genes are labeled in bold, and histone acetyltransferase-related genes are not labeled in bold. d Chromatin accessibility peaks around the promoter of the Hdac9 gene between WT and Sirt6 cKO thymic tuft cells are shown in aggregated scATAC-seq tracks. The arrow indicates the length and direction of the peak around the promoter region of Hdac9 detected by scATAC-seq. e Capture of the UCSC (GRCh37/hg19) genome browser showing the human HDAC9 locus in ChIP-seq. f Capture of the UCSC (GRCh37/hg19) genome browser showing the human HDAC9 locus in ATAC-seq. g Western blot analysis of protein acetylation in mTECs (CD45−CD326+UEA-1+Ly51−) isolated from wild-type or Sirt6 cKO mice. h Representative flow cytometric profiles of H3K9ac, H3K18ac, and H3K27ac expressed in CD80− mTECs obtained from 4-week-old Sirt6 cKO mice and littermates. i Frequencies of H3K9ac, H3K18ac, and H3K27ac expression in CD80− mTECs of 4-week-old Sirt6 cKO mice and littermates (n = 4 per group). ***p < 0.001 compared with the identical WT control mice (Student’s t-test). The error bar represents one standard deviation.
Additionally, SIRT6 was found specifically bind the HDAC9 promoter region in SKMel-239 cell lines (Fig. 6e and Suppl. Fig. 37). This observation was further confirmed by the HDAC9 promoter had a tendency to increase chromatin accessibility in the SKMel-239 cell lines with a homozygous mutant SIRT636, although the significance cannot be calculated due to lack of replicates (Fig. 6f and Suppl. Fig. 37). Consistent with the enhanced histone deacetylase Hdac9 expression, acetylation in Sirt6-deficient mTECs, including H3K9ac, H3K18ac, and H3K27ac, was significantly decreased, as determined by Western blot and flow cytometry (P < 0.001, Fig. 6g–i, Suppl. Fig. 38, and Supplementary Data 7). Thus, deletion of histone deacetylase SIRT6 indirectly decreased the chromatin accessibility of Pou2f3-regulated motifs, likely by increasing histone deacetylase gene Hdac9 expression, and finally impaired Pou2f3-mediated thymic tuft cell development.
Discussion
We presented the scATAC-seq data of mouse mTECs showing the overall profiles from chromatin accessibility profiling of mTECs in all development stages. We found that the Late Aire-expressing mTEC III subgroup might include the Late Aire 1–3 subpopulations with distinctive gene accessibility profiles (Supplementary Data 8). Late Aire 1 mTECs likely differentiate into Late Aire 2 mTECs, which then differentiate into either Late Aire 3 cells or thymic tuft cells. Unexpectedly, the key transcription factor Pou2f3 was also widely expressed in pre- and Late Aire-expressing mTECs, but its regulated target genes were specifically expressed in thymic tuft cells, indicating critical roles of epigenetic modulation in Pou2f3-mediated thymic tuft cell development. Furthermore, Sirt6 deletion did not impact Pou2f3 expression in mTECs but significantly diminished the presence of thymic tuft cells by regulating chromatin accessibility of Pou2f3-regulated motifs. More importantly, Late Aire 3 cells almost completely disappeared after Sirt6 knockout, indicating that epigenetic regulation is indispensable in the development of Late Aire 2 cells to thymic tuft cells and Late Aire 3 cells.
Based on the scATAC-seq data of mouse mTECs, we confirmed the rich diversity of mTECs and could roughly divide mTECs into mTEC I-III and thymic tuft cells, as recently reported with scRNA-seq data5, indicating that promoter and distal region accessibility is linked to the expression of genes contributing to mTEC subgroup identity. Previous reports have shown that mTECs also contain a TAC-TEC cluster47, although our scATAC-seq data showed that the chromatin accessibility of the TAC-TEC characteristic gene set (cell cycle-related genes) was similar to that in other mTEC clusters. This finding is consistent with another report that chromatin accessibility of cell cycle-related genes has a limited contribution to gene expression in shared single-cell profiling48. Therefore, we did not conduct further chromatin accessibility analysis for the TAC-TEC cluster.
Among Late Aire-expressing mTEC III cells, we found heterogeneity and identified Late Aire 1-3 subgroups with distinct transcription factors in each subgroup, such as TCF7L2 in Late Aire 1 cells, KLF3 in Late Aire 2 cells, and FOXA3 in Late Aire 3 cells. mTEC III, as a terminally differentiated mTEC cluster, downregulates the expression of MHC II and Aire49 and highly expresses KRT1, KRT10, involucrin, desmogleins, SPINK5, Asprv1, cystatin, LY6D, and Pigr1,5–7,49–52. Late Aire 1 cells still expressed H2-DMb2, H2-Oa, H2-M2, and Aire (Suppl. Fig. 8), suggesting that Late Aire 1 has a strong developmental correlation with mTEC II, which is speculated to be the early stage of mTEC terminal differentiation. Late Aire 2 cells specifically expressed Spink5 and Ly6d, suggesting that Late Aire 2 is more consistent with the known terminally differentiated mTECs5,53. Late Aire 3 cells with high expression of Ascl1 may be similar to the reported Ascl1+ mTEClo 8; however, the specific function of Late Aire 3 needs to be further studied. Using scATAC-seq to reconstruct the cellular developmental trajectories of these mTECs in an unbiased manner, we found that Late Aire 1 cells likely differentiated to Late Aire 2 cells and finally to Late Aire 3 cells. On the other hand, some Late Aire 2 cells also differentiated into thymic tuft cells. Thus, Late Aire 2 mTECs may represent a crossroad for the differentiation into either Late Aire 3 cells or thymic tuft cells. Lineage-tracing studies by two independent research teams showed that more than half of thymic tuft cells arise from mature mTEChigh cells4,5, with the remaining half of the thymic tuft cells not appearing to pass through the AIRE-expressing mTEC stage. Specifically, diphtheria toxin treatment of mice, in which diphtheria toxin receptor expression is driven by the Aire promoter, resulted in only a partial deletion or even no significant impact of thymic tuft cells with no marked alterations in the gene expression profile of thymic tuft cells4,5. Thus, this thymic tuft cell differentiation pathway identified in the present study requires to be verified in further experiments.
Of note, Late Aire 2 cells exhibited similar accessibility levels of the key transcription factor of thymic tuft cells, namely, Pou2f3, as determined by transcriptional expression and open chromatin features. However, the Pou2f3-regulated genes were not expressed in Late Aire 2 cells but were specifically expressed in thymic tuft cells, and the loci of Pou2f3-target genes were inaccessible in Late Aire 2 cells, likely reflecting a requisite gain of accessible chromatin at that gene’s locus. These data strongly highlight the importance of chromatin accessibility and epigenetic modulation in Pou2f3-mediated thymic tuft cell differentiation; that is, Pou2f3 is essential but not sufficient for thymic tuft cell differentiation.
SIRT6, as a key chromatin regulator, predominantly represses the expression of various genes, such as glycolytic genes, Myc-target ribosomal protein genes, lipid metabolism genes, and NF-κB-dependent genes, to regulate some fundamental cellular processes, such as cellular metabolism, DNA repair, gene expression, mitochondrial biology, and telemetric maintenance29–33,45,46. The expression of these genes in mTECs and thymic tuft cells was upregulated after Sirt6 deletion. However, the expression and locus openness of Pou2f3-regulated genes were obviously decreased, which blocked the differentiation of Pou2f3-mediated thymic tuft cells. Unexpectedly, histone acetylation, including H3K9ac, H3K18ac, and H3K27ac, was decreased in Sirt6-deficient CD80− mTECs, which is inconsistent with the histone deacetylase function of SIRT6. We thus screened the expression of histone deacetylase and histone acetyltransferase and identified that histone deacetylase Hdac9 expression might be directly regulated by SIRT6 in thymic tuft cell differentiation. Enhanced histone deacetylase Hdac9 expression by Sirt6 deletion subsequently decreased the chromatin accessibility of Pou2f3-regulated motifs to block Pou2f3-mediated thymic tuft cell development. Therefore, epigenetic regulators are essential for the development of thymic tuft cells.
By describing the chromatin landscape of mTECs, we identified previously unknown mTEC subpopulations and explored developmental trajectories and epigenetic regulation from mTEC III to thymic tuft cells. The difference between chromatin accessibility and gene expression contributes to an in-depth understanding of the development of mTECs.
Methods
Mice
Consistent with our previous reports35, the Foxn1-Cre mouse strain (a gift from Dr. Yu Zhang)54,55 was crossed with Sirt6loxp/loxp mouse strain (a gift from Dr. Zhenyu Ju)56,57 to generate the Foxn1-Cre Sirt6loxp/loxp mice and Foxn1-Cre negative littermate controls. Mice were maintained in the Institute of Zoology, Chinese Academy of Sciences (IOZ, CAS) specific pathogen-free conditions and approved by the Animal Ethics Committee of Institute of Zoology, Beijing, China.
Thymic stromal cell isolation
Thymi from 4-week-old wild-type and Sirt6 cKO mice were placed into 6 cm dishes, chopped into small pieces with surgical scissors, and suspended in DMEM (HyClone Laboratories, SH30022.01B) containing 2% fetal bovine serum (FBS; Gibco, 16000-044). After removing most thymocytes from the supernatant, the remaining thymus fragments were digested with a solution containing 20 mg/ml collagenase/dispase (Sigma-Aldrich, 11097113001) and 20 U/ml DNase I (Sigma-Aldrich, d5025) for 40 min in a water bath at 37 oC58. When the small pieces disappeared, the cell suspension was filtered through a 200-mesh filter to obtain a single-cell suspension.
scATAC-seq library generation and sequence alignment
The cell suspension mentioned above was incubated with CD45 microbeads (Miltenyi, mouse, 130-052-301) in the dark for 15 min on ice, and the CD45-negative cells were enriched by using a LS column (Miltenyi, 130-042-401). After incubation with antibodies, wild-type samples (150000 CD45-CD326+ TECs) and Sirt6 cKO samples (100000 CD45−CD326+ TECs plus 8400 CD45-CD326+L1CAM+MHC IIlow thymic tuft cells) were sorted by BD FACSAria™ Fusion Flow Cytometers (BD Biosciences, USA) for the 10× Genomics scATAC-seq analysis.
The nuclei were isolated and washed according to the method in the 10X Genomics Chromium Single-Cell ATAC Reagent Kit user guide (CG000169). Then, the nuclei were resuspended in chilled diluted nuclei buffer (10x Genomics; 2000153). The nuclei in bulk samples were partitioned into nanoliter-scale gel beads-in-emulsions (GEMs), a pool of ~750,000 10x barcodes was sampled to separately and uniquely index the transposed DNA of each individual nucleus, and libraries were generated (performed by CapitalBio Technology, Beijing). The libraries were sequenced using an Illumina NovaSeq sequencer with a sequencing depth of at least 25 k read pairs per nucleus using a paired-end 50 bp (PE50) reading strategy.
Raw sequencing data were converted to FASTQ format using Cell Ranger ATAC mkfastq. Alignment of scATAC-seq analyses was completed utilizing the 10x Genomics Cell Ranger ATAC pipeline (version 1.2.0). Each library was aligned to an indexed mm10 genome using Cell Ranger ATAC Count.
scRNA-seq data process and cell type clustering
The gene expression matrix data of TEC scRNA-seq were obtained from GSE13769917. We merged 2-week and 10-week wild-type control samples for a Seurat (4.0.1) analysis. The scRNA-seq project was constructed by the Seurat (4.0.1) R package with at least 200 genes per cell and at least three cells. Cells were filtered based on the number of gene features per cell, the number of reads per gene, the percent of ribosome-associated genes, and the percent of mitochondrial-associated genes, the value of which was restricted to 5 to 95%. After filtering, 19,014 features across 1875 cells remained for further analysis. Normalization and scale strategies were performed according to a previous method17 as follows. Quality control of scRNA-seq was based on a 5–95% threshold of the count of reads, a number of the feature genes, and the percent of mitochondria-related genes. The cutoff strategy of WT TEC scRNA-seq project was using the options “2274 < nCount_RNA < 3699, 1361 < nFeature_RNA < 6587, 0.007 < percent.mito < 0.014”. The scale and clustering strategy was using the options “ScaleData: scale.factor = 1000; FindVariableFeatures: mean.function = ExpMean, dispersion.function = LogVMR, x.low.cutoff = 0.0125, x.high.cutoff = 3, y.cutoff = 0.5; FindClusters: resolution = 0.6, random.seed = 0; RunUMAP: dims = 1:13”. For our analysis, we filtered Ly75+Psmb11+ cTECs for consistency with scATAC-seq data. The clustree (0.4.4) package was used to visualize the allocation of cells between clusters at different resolutions. The marker genes of each cluster were generated by Seurat’s default FindAllMarkers() function on the threshold of p_val_adj < 0.05 and avg_log2FC > 0.5. The initial cell population was defined by mTEC I (Itgb4, and Itga6), mTEC II (Aire and Cd80), mTEC III (Ly6d and Pigr), and tuft (L1cam and Gnat3) cells. The gene set module scores of the mTEC I, mTEC II, mTEC III, and thymic tuft cells were determined by the AddModuleScore() function.
Data processing and clustering analysis
The aggregated peak-by-cell data matrix was read into R (R version 4.0.3) and processed using Seurat (4.0.1)59 and Signac (1.1.1). The chromatin assay was constructed by CreateChromatinAssay and transferred to the Seurat object by CreateSeuratObject.
Blacklist regions indicate abnormal or high-signal areas in either of the next-generation sequencing experiments, and the exclusion of these regions can provide quality assurance for further analysis of functional genomic data. The nucleosome banding pattern, defined as a histogram of DNA fragment sizes (determined from the paired-end sequencing reads), should exhibit a strong nucleosome banding pattern corresponding to the length of DNA wrapped around a single nucleosome. Quality control of scATAC-seq was based on a 5–95% threshold of the fraction of fragments in peaks (pct_reads_in_peaks), the total number of fragments in the peak region (peak_region_fragments), the ratio of reads in the blacklist region (blacklist_ratio), the TSS enrichment score (TSS.enrichment) and the nucleosome binding pattern (nucleosome_signal). The cutoff strategy of WT TEC scATAC-seq project was using the options “60.01 < pct_reads_in_peaks < 88.95, 883 < peak_region_fragments < 17873, 0 < blacklist_ratio < 0.02, 2.30 < TSS.enrichment < 12.96, 0.03 < nucleosome_signal < 2.87” and the Sirt6 cKO TEC scATAC-seq project was using the options “60 < pct_reads_in_peaks < 79.9, 715 < peak_region_fragments < 10427, 0 < blacklist_ratio < 0.014, 2.81 < TSS.enrichment < 8.41, 0.02 < nucleosome_signal < 0.46”. Latent semantic indexing (LSI) was used to calculate the variable features, and the first ten components were used to generate a uniform manifold approximation and projection (UMAP) dimensionality reduction. To define the subsets of WT TECs, a shared-nearest-neighbor graph was generated from the 2:10 LSI components and used to cluster the cells via Seurat’s Louvain algorithm.
Notably, 50% of the Sirt6 cKO thymic tuft cells were randomly excluded based on the number of externally added cells to ensure that the cell proportion of the data was consistent with the true value in vivo. After that, WT TEC and Sirt6 cKO filtered TEC scATAC-seq data were merged into a combined object for further analysis.
Cell population and marker identification
The gene activity matrix was calculated for the filtered peak matrix by the Cicero analysis package (1.7.1)60 and monocle 3 alpha package (2.99.3)61, and it was added to the Seurat object based on RNA assays. Marker genes for peak-based clustering were generated using the FindAllMarkers() function on the gene activity matrix based on the threshold of p_val_adj <0.05 and avg_log2FC > 0.5. The shared characteristic genes between scATAC-seq and scRNA-seq of mTEC I, mTEC II, mTEC III and thymic tuft cells calculated by scRNA-seq mentioned before were added to the peak-by-cell data matrix through the AddModuleScore() function. The populations of mTEC I (Itgb4 and Itga6), mTEC II (Aire and Cd80), mTEC III (Ly6d and Pigr), and thymic tuft cells (L1cam and Gnat3) in WT scATAC-seq and combined scATAC-seq were all established by marker genes and the scRNA-seq clustering gene set module score.
Repopulation of the Late Aire subset
Late Aire repopulation was performed by WT scATAC-seq according to the population heterogeneity and unique gene expression profiles. The Late Aire 1, Late Aire 2, and Late Aire 3 marker genes calculated by FindMarkers() according to the threshold of p_val_adj <0.05 and avg_log2FC > 0.5 were derived from the gene activity matrix of scATAC-seq. The shared characteristic genes between WT scATAC-seq and scRNA-seq of Late Aire 1, Late Aire 2, and Late Aire 3 calculated by scATAC-seq were added to the scRNA-seq gene expression matrix through the AddModuleScore() function. The three subsets of mTEC III cells of the combined data were generated by the same method as mentioned above.
TF motif analysis
A motif enrichment analysis was performed using Signac (1.1.1) and ChromVAR (1.10.0)25. We created the motif class to store all the required information, including a list of position weight matrices (PWMs) or position frequency matrices (PFMs) and a motif occurrence matrix derived from the JASPAR2020 (species: 9606) database using the AddMotifs() function. Open chromatin peaks of each subset were defined by the FindAllMarkers() function on the peak assay of scATAC-seq based on the threshold of avg_log2FC > 0.5. The open chromatin peaks of each subset were used to perform DNA sequence motif analysis by FindMotifs(). Differentially accessible peaks between WT and Sirt6 cKO cells were calculated by FindMarkers(), and then FindMotifs() analysis was performed. All of the motif activity scores were run by chromVAR and visualized by FeaturePlot() and VlnPlot().
Pseudotemporal analyses
For the WT scATAC-seq analysis, subsets of cells pertaining to mTEC III (including Late Aire 1, Late Aire 2, and Late Aire 3) and thymic tuft cells were extracted for pseudotime analysis by Monocle3 (0.2.1). Using the UMAP cell positions of the extracted subset from the scATAC-seq analysis, we next employed monocle to learn a graph trajectory through this space. The start of pseudotime was chosen as the branch node that started within the Late Aire 1 subset.
Pou2f3 target gene selection
The Pou2f3 ChIP-seq data used in this article were downloaded from small intestine tissue (GSE116510). Reads were trimmed for Illumina adapter sequences using trim_galore (version 0.5.0) and aligned to mm10 using hisat2 (version 2.1.0). Pou2f3 significant peaks were identified using MACS2 (version 2.1.1) using the parameters--nomodel--shift-50--extsize 100, and matching input control was used to call peaks. ChIPpeakAnno (3.28.0) was used to annotate the peaks found by MACS2, matching the upstream and downstream 2 kb regions of genes in the mm10 genome to obtain 2152 predicted genes, which are provided in Supplementary Data 1.
Epigenetic sequencing data analysis of the human SIRT6 knockout cell line
The interaction between SIRT6 and HDAC9 was analyzed by ChIP-seq (SIRT6) and ATAC-seq of WT and SIRT6 KO SKMel-239 cells derived from GSE10281336. The Bigwig file (.bw) containing the axis information and related scores of the peak region can be downloaded from the Gene Expression Omnibus (GEO) database. The visualization of peak regions was performed by Integrative Genomics Viewer62 software.
Bulk RNA-seq analysis
Bulk RNA-seq of WT and Sirt6 cKO mTECs was derived from our previously published studies, and the data are available in GSE16684035. The differentially expressed genes were calculated by the DEseq2 (1.28.1) package according to the threshold of adjusted p value <0.05 and |Log2FC | > 2. The KEGG analysis of the differentially expressed genes was performed on the KOBAS 3.0 website.
ChIP-seq analysis
Reads were trimmed for Illumina adapter sequences using trim_galore (v 0.5.0) and aligned to the mm10 using hisat2 (v 2.1.0). Pou2f3 significant peaks were identified using MACS2 (v 2.1.1) with parameters--nomodel--shift-50--extsize 100 and matching Input control was used to call peaks. ChIPpeakAnno (v 3.28.0) was used to annotate the peaks found by MACS2, matching the upstream and downstream 2 Kb regions of genes in EnsDb.Mmusculus.v79 genome to obtain 2152 predicted genes.
Flow cytometric analysis and antibodies
After the Fc receptor was blocked by 2.4G2, the cells were incubated with the designated antibody for 30 min at 4 °C63. For different intracellular staining, according to the instructions, fixation buffer (eBioscience, 00-5123-43 and 00-5223-56) and permeabilization buffer (eBioscience, 00-8333-56) were used for nuclear antibody staining, and BD Cytofix/Cytoperm™ Fixation and Permeabilization Solution (BD Biosciences, 554722) was used for cytoplasmic antibody staining. A BD LSRFortessa X-20 flow cytometer (BD Biosciences, USA) was used for the flow cytometry analysis.
The following antibodies were used: Fixable Viability Dye eFluor™ 506 (eBioscience, 65-0866-18, 1:400), CD45-BUV395 (BD Biosciences, clone 30-F11, 564279, 1:400), CD326-PE/Cy7 (Biolegend, clone G8.8, 188216, 1:800), UEA-I (Vector Laboratories, FL-1061, 1:300), Ly51-AF647 (Biolegend, clone 6C3, 108312, 1:300 dilution), Ly51-BV786 (BD Biosciences, clone BP-1, 740882, 1:300 dilution), I-A/I-E-BV421 (Biolegend, clone M5/114.15.2, 107632, 1:400 dilution), CD80-PE (Biolegend, clone 16-10A1, 104708, 1:2000 dilution), CD80-BV650 (Biolegend, clone 16-10A1, 104732, 1:300 dilution), L1CAM-PE (R&D, clone 555, FAB5674P, 1:20 dilution), rabbit anti-DCLK1 (Abcam, ab31704, 1:15000 dilution), TCR-β-PE-Cy7 (Biolegend, clone H57-597, 109222, 1:500 dilution), mouse CD1d loaded R-PE label (Proimmune, E001-2A-G, 1:600 dilution), RORγt-BV421 (BD Biosciences, clone Q31-378, 562894, 1:200 dilution), PLZF-AF488 (eBioscience, clone Mags.21F7, 53-9320-80, 1:200 dilution), CD4-PE (Biolegend, clone GK1.5, 100408, 1:1600 dilution), CD8a-PE/Cy5 (Biolegend, clone 53-6.7, 100710, 1:1600 dilution), EOMES-AF488 (eBioscience, clone Dan11mag, 53-4875-80, 1:200 dilution), CD127-BV785 (Biolegend, clone A7R34, 135037, 1:200 dilution), GATA3-APC (Biolegend, clone 16E10A23, 653805, 1:100 dilution), T-bet-PE (eBioscience, clone 4B10, 12-5825-82, 1:200 dilution); Lin--FITC: CD4-FITC (Biolegend, clone GK1.5, 100405, 1:1000 dilution); CD8-FITC (BD Biosciences, clone 53-6.7, 553031, 1:400 dilution); CD19-FITC (Biolegend, clone 1D3/CD19, 153404, 1:200 dilution); CD11c-AF488 (eBioscience, clone N418, 53-0114-82, 1:400 dilution); CD11b-FITC (eBioscience, clone M1/70, 11-0112-82, 1:500 dilution); F4/80-FITC (eBioscience, clone BM8, 11-4801-82, 1:500 dilution); TCRγδ-FITC (BD Biosciences, clone GL3, 553177, 1:200 dilution); NK1.1-FITC (eBioscience, clone PK136, 11-5941-81, 1:400 dilution); TER-119-FITC (Biolegend, clone TER-119, 116206, 1:400 dilution); Alexa Fluor 647 AffiniPure donkey anti-Rabbit IgG (H + L) (Jackson ImmunoResearch Laboratories, 703-545-155, 1:600 dilution). Acetyl-Histone H3 (Lys9) (C5B11) Rabbit mAb 9649 (1:1000 dilution), Acetyl-Histone H3 (Lys18) (D8Z5H) Rabbit mAb 13998 (1:1000 dilution) and Acetyl-Histone H3 (Lys27) (D5E4) XP® Rabbit mAb 8173 (1:200 dilution), all from Acetyl-Histone H3 Antibody Sampler Kit (Cell Signaling Technology, 9927).
Immunofluorescence staining
Frozen section samples of thymus tissue from 4-week-old WT and Sirt6 cKO mice were prepared according to previous methods4,5,35. Frozen slice samples (25 μm in thickness) were fixed with 4% polyoxymethylene (Solarbio, P1110) for 20 min and blocked in PBS containing 1% BSA and 0.3% Triton X-100 (Sigma–Aldrich) for 1 h64. Then, the sliced samples were stained with primary and secondary antibodies for 1 h. DAPI staining was performed for 10 min after the above steps. The following antibodies were used for staining: chicken anti-KRT5 (Biolegend, 905901; clone Poly9059) diluted by 1:300 and rabbit anti-DCLK1 (Abcam, ab31704) diluted by 1:250. Alexa Fluor 488-conjugated donkey anti-Chicken IgG (H + L) (Jackson ImmunoResearch Laboratories, 703-545-155) diluted by 1:300, Alexa Fluor 594-conjugated donkey anti-rabbit IgG (H + L) (Jackson ImmunoResearch Laboratories, 711-586-152) diluted by 1:300. All antibodies were diluted in PBS (0.5% BSA). Laser scanning confocal microscope (Zeiss LSM710, Oberkochen, Germany) were used to obtain images.
Simple Western
Referring to a previous method65, mTECs (CD45−CD326+UEA-1+Ly51−) isolated from 4-week-old WT and Sirt6 cKO mice were lysed with RIPA lysis buffer (Beyotime, P0013B) supplemented with 1 mM PMSF (Beyotime, ST506). According to the manufacturer’s instructions, a 12–230 kDa separation module (ProteinSimple, SM-W002-1) was used to detect the expression of POU2F3. The lysate was mixed with 5× Fluorescent master mix at a ratio of 4:1 and heated with a biotinylated ladder at 95 oC for 5 min. The primary antibody, anti-POU2F3 (Prosci, 7795), was diluted with Antibody Diluent II at 1:40, while the anti-rabbit secondary HRP antibody (042-206; ProteinSimple) was used directly. Lumino-S and peroxide were mixed and used as substrates. The above reagents were added to the plate according to the instructions and centrifuged at 2500 rpm for 5 min at room temperature. Finally, WES (ProteinSimple, USA) was used to detect the protein.
Western blot assay
Consistent with a previous description35, mTECs (CD45−CD326+UEA-1+Ly51−) sorted from 4-week-old WT and Sirt6 cKO mice were lysed with RIPA lysis buffer to detect protein acetylation. Protein was separated by 10% SDS–PAGE and transferred to PVDF membranes (Merck Millipore, IPFL00010). After blocking with 5% skim milk powder (Oxoid, LP0031) at room temperature for 60–90 min, the primary antibody was incubated overnight at 4 °C. The primary antibodies were anti-acetyllysine mouse mAb (PTM BIOLABS, clone Kac-01, PTM0101) diluted at 1:1000 and anti-actin (Sigma–Aldrich, A5441) diluted at 1:40000. Then, the corresponding secondary antibody was added at room temperature for 45 min. Chemiluminescence (Merck Millipore, WBKLS0500) was used to detect protein expression.
Statistics and reproducibility
The statistical significance of the gene expression and activity between mTEC populations visualized by boxplot and violin plot was determined using Wilcoxon signed-rank test. Boxplot and violin plots were visualized by ggpubr (v 0.4.0) and R (v4.0.3). The statistical significance of the difference in cell ratio of WT and Sirt6 cKO detected by flow cytometric was determined by Student’s t-test. The bar graphs were visualized using GraphPad Prism (v8) and the replication were in 4–6 independent experiments, as indicated in the figure legends. FDR-adjusted P values and fold changes of differential expressed genes between WT and Sirt6 cKO were derived from the DESeq2 package (v1.28.1). P values for KEGG pathway enrichment were derived from clusterProfiler package (v 3.16.1).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We gratefully acknowledge Mrs. Qing Meng and Xia Yang for the assistance in cell sorting, Mrs. Ling Li for her excellent lab assistance, and Mr. Yiming Jin for his assistance in some animal and cellular experiments. This study was funded by the National Key Research and Development Program of China (2017YFA0105002 and 2017YFA0104402, Y.Z.), the National Natural Science Foundation for General and Key Programs (31930041, Y.Z), and the Knowledge Innovation Program of the Chinese Academy of Sciences (XDA16030301, Y.Z.).
Author contributions
Q.Z. designed and completed the experiment, analyzed data, and wrote the manuscript; J.Z. designed and completed all bioinformatics analysis and wrote the manuscripts; T.L. performed the experiment and analyzed data; Z.L designed the experiment; X.D. performed a simple western experiment; L.S. supervised the research, discussed the results, and revised the manuscript; Y.Z. directed the study, supervised the research, and wrote the manuscript.
Peer review
Peer review information
Communications Biology thanks Rongxin Fang, Hélia Neves and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Eirini Trompouki and George Inglis.
Data availability
The gene expression matrix data of TEC scRNA-seq was obtained from GSE137699. The analysis of the interaction between SIRT6 and HDAC9 were performed by the ChIP-seq (SIRT6, H3K9ac, H3K27ac, and H3K56ac) and ATAC-seq of WT and SIRT6 mutant SKMel-239 cell line derived from GSE102813. The bulk RNA-seq of WT and Sirt6 cKO mTEC were derived from our recently published studies35, and the data was available on GSE166840. The genes regulated by POU2F3 were calculated by the ChIP-seq (POU2F3) of intestinal tuft derived from GSE116510. The raw data of scATAC-seq sequencing used in this article have been deposited at the National Genomics Data Center BioProject under number: PRJCA006664. Uncropped Western blots gel for Figs. 5c, 6g are presented in Suppl. Figs. 32, 38, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qian Zhang, Jiayu Zhang, Tong Lei.
Contributor Information
Liguang Sun, Email: Sunliguang1717@vip.163.com.
Yong Zhao, Email: zhaoy@ioz.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03484-9.
References
- 1.Kadouri N, Nevo S, Goldfarb Y, Abramson J. Thymic epithelial cell heterogeneity: TEC by TEC. Nat. Rev. Immunol. 2020;20:239–253. doi: 10.1038/s41577-019-0238-0. [DOI] [PubMed] [Google Scholar]
- 2.Abramson J, Anderson G. Thymic epithelial cells. Annu Rev. Immunol. 2017;35:85–118. doi: 10.1146/annurev-immunol-051116-052320. [DOI] [PubMed] [Google Scholar]
- 3.Takahama Y, Ohigashi I, Baik S, Anderson G. Generation of diversity in thymic epithelial cells. Nat. Rev. Immunol. 2017;17:295–305. doi: 10.1038/nri.2017.12. [DOI] [PubMed] [Google Scholar]
- 4.Miller CN, et al. Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature. 2018;559:627–631. doi: 10.1038/s41586-018-0345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornstein C, et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature. 2018;559:622–626. doi: 10.1038/s41586-018-0346-1. [DOI] [PubMed] [Google Scholar]
- 6.Metzger TC, et al. Lineage tracing and cell ablation identify a post-Aire-expressing thymic epithelial cell population. Cell Rep. 2013;5:166–179. doi: 10.1016/j.celrep.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishikawa Y, et al. Biphasic Aire expression in early embryos and in medullary thymic epithelial cells before end-stage terminal differentiation. J. Exp. Med. 2010;207:963–971. doi: 10.1084/jem.20092144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bautista JL, et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat. Commun. 2021;12:1096. doi: 10.1038/s41467-021-21346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerbe F, Brulin B, Makrini L, Legraverend C, Jay P. DCAMKL-1 expression identifies Tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology. 2009;137:2179–2180. doi: 10.1053/j.gastro.2009.06.072. [DOI] [PubMed] [Google Scholar]
- 10.Gerbe F, et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J. Cell Biol. 2011;192:767–780. doi: 10.1083/jcb.201010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panneck AR, et al. Cholinergic epithelial cell with chemosensory traits in murine thymic medulla. Cell Tissue Res. 2014;358:737–748. doi: 10.1007/s00441-014-2002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, et al. Members of bitter taste receptor cluster Tas2r143/Tas2r135/Tas2r126 are expressed in the epithelium of murine airways and other non-gustatory tissues. Front. Physiol. 2017;8:849. doi: 10.3389/fphys.2017.00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soultanova A, et al. Cholinergic chemosensory cells of the thymic medulla express the bitter receptor Tas2r131. Int. Immunopharmacol. 2015;29:143–147. doi: 10.1016/j.intimp.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Satpathy AT, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 2019;37:925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;109:21 29 21–21 29 29. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cusanovich DA, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells, K. L. et al. Combined transient ablation and single-cell RNA-sequencing reveals the development of medullary thymic epithelial cells. Elife9, e60188 (2020). [DOI] [PMC free article] [PubMed]
- 18.Ranzoni AM, et al. Integrative single-cell RNA-seq and ATAC-seq analysis of human developmental hematopoiesis. Cell Stem Cell. 2021;28:472–487. doi: 10.1016/j.stem.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granja JM, et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 2019;37:1458–1465. doi: 10.1038/s41587-019-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pervolarakis N, et al. Integrated single-cell transcriptomics and chromatin accessibility analysis reveals regulators of mammary epithelial cell identity. Cell Rep. 2020;33:108273. doi: 10.1016/j.celrep.2020.108273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 22.Dhalla F, et al. Biologically indeterminate yet ordered promiscuous gene expression in single medullary thymic epithelial cells. EMBO J. 2020;39:e101828. doi: 10.15252/embj.2019101828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, J. E. et al. A cell atlas of human thymic development defines T cell repertoire formation. Science367, eaay3224 (2020). [DOI] [PMC free article] [PubMed]
- 24.Dravis C, et al. Epigenetic and transcriptomic profiling of mammary gland development and tumor models disclose regulators of cell state plasticity. Cancer Cell. 2018;34:466–482. doi: 10.1016/j.ccell.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods. 2017;14:975–978. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin, Y. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science356, eaaj2239 (2017). [DOI] [PMC free article] [PubMed]
- 27.Fan K, Moore JE, Zhang XO, Weng Z. Genetic and epigenetic features of promoters with ubiquitous chromatin accessibility support ubiquitous transcription of cell-essential genes. Nucleic Acids Res. 2021;49:5705–5725. doi: 10.1093/nar/gkab345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammelman J, Krismer K, Banerjee B, Gifford DK, Sherwood RI. Identification of determinants of differential chromatin accessibility through a massively parallel genome-integrated reporter assay. Genome Res. 2020;30:1468–1480. doi: 10.1101/gr.263228.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toiber D, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Etchegaray JP, et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat. Cell Biol. 2015;17:545–557. doi: 10.1038/ncb3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwer B, et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc. Natl Acad. Sci. USA. 2010;107:21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rezazadeh S, et al. SIRT6 promotes transcription of a subset of NRF2 targets by mono-ADP-ribosylating BAF170. Nucleic Acids Res. 2019;47:7914–7928. doi: 10.1093/nar/gkz528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kugel S, et al. Identification of and molecular basis for SIRT6 loss-of-function point mutations in cancer. Cell Rep. 2015;13:479–488. doi: 10.1016/j.celrep.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou T, et al. SIRT6 coordinates with CHD4 to promote chromatin relaxation and DNA repair. Nucleic Acids Res. 2020;48:2982–3000. doi: 10.1093/nar/gkaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, et al. Sirt6 regulates the development of medullary thymic epithelial cells and contributes to the establishment of central immune tolerance. Front. Cell Dev. Biol. 2021;9:655552. doi: 10.3389/fcell.2021.655552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strub T, et al. SIRT6 haploinsufficiency induces BRAF(V600E) melanoma cell resistance to MAPK inhibitors via IGF signalling. Nat. Commun. 2018;9:3440. doi: 10.1038/s41467-018-05966-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 2014;39:72–81. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang AR, Ferrer CM, Mostoslavsky R. SIRT6, a mammalian deacylase with multitasking abilities. Physiol. Rev. 2020;100:145–169. doi: 10.1152/physrev.00030.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu G, Chen H, Liu H, Zhang W, Zhou J. Emerging roles of SIRT6 in human diseases and its modulators. Med. Res. Rev. 2021;41:1089–1137. doi: 10.1002/med.21753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tasselli L, Zheng W, Chua KF. SIRT6: novel mechanisms and links to aging and disease. Trends Endocrinol. Metab. 2017;28:168–185. doi: 10.1016/j.tem.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michishita E, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tasselli L, et al. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016;23:434–440. doi: 10.1038/nsmb.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong L, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scialdone A, et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 48.Ma S, et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell. 2020;183:1103–1116. doi: 10.1016/j.cell.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, et al. Post-Aire maturation of thymic medullary epithelial cells involves selective expression of keratinocyte-specific autoantigens. Front. Immunol. 2012;3:19. doi: 10.3389/fimmu.2012.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishikawa Y, et al. Temporal lineage tracing of Aire-expressing cells reveals a requirement for Aire in their maturation program. J. Immunol. 2014;192:2585–2592. doi: 10.4049/jimmunol.1302786. [DOI] [PubMed] [Google Scholar]
- 51.Nuber UA, Schafer S, Stehr S, Rackwitz HR, Franke WW. Patterns of desmocollin synthesis in human epithelia: immunolocalization of desmocollins 1 and 3 in special epithelia and in cultured cells. Eur. J. Cell Biol. 1996;71:1–13. [PubMed] [Google Scholar]
- 52.Hale LP, Markert ML. Corticosteroids regulate epithelial cell differentiation and Hassall body formation in the human thymus. J. Immunol. 2004;172:617–624. doi: 10.4049/jimmunol.172.1.617. [DOI] [PubMed] [Google Scholar]
- 53.Galliano MF, et al. Characterization and expression analysis of the Spink5 gene, the mouse ortholog of the defective gene in Netherton syndrome. Genomics. 2005;85:483–492. doi: 10.1016/j.ygeno.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 54.Kim HS, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, et al. SIRT6 controls hematopoietic stem cell homeostasis through epigenetic regulation of Wnt signaling. Cell Stem Cell. 2016;18:495–507. doi: 10.1016/j.stem.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Soza-Ried C, Bleul CC, Schorpp M, Boehm T. Maintenance of thymic epithelial phenotype requires extrinsic signals in mouse and zebrafish. J. Immunol. 2008;181:5272–5277. doi: 10.4049/jimmunol.181.8.5272. [DOI] [PubMed] [Google Scholar]
- 57.Liu B, et al. Cbx4 regulates the proliferation of thymic epithelial cells and thymus function. Development. 2013;140:780–788. doi: 10.1242/dev.085035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang Z, et al. The development and survival of thymic epithelial cells require TSC1-dependent negative regulation of mTORC1 activity. J. Immunol. 2021;207:2039–2050. doi: 10.4049/jimmunol.2100463. [DOI] [PubMed] [Google Scholar]
- 59.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pliner HA, et al. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell. 2018;71:858–871. doi: 10.1016/j.molcel.2018.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu Z, Feng C, Sun C, Xu Y, Zhao Y. Primed macrophages gain long-term specific memory to reject allogeneic tissues in mice. Cell Mol. Immunol. 2021;18:1079–1081. doi: 10.1038/s41423-020-00521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian Q, et al. Skin and heart allograft rejection solely by long-lived alloreactive TRM cells in skin of severe combined immunodeficient mice. Sci. Adv. 2022;8:eabk0270. doi: 10.1126/sciadv.abk0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang Z, et al. mTORC2 negatively controls the maturation process of medullary thymic epithelial cells by inhibiting the LTbetaR/RANK-NF-kappaB axis. J. Cell Physiol. 2021;236:4725–4737. doi: 10.1002/jcp.30192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The gene expression matrix data of TEC scRNA-seq was obtained from GSE137699. The analysis of the interaction between SIRT6 and HDAC9 were performed by the ChIP-seq (SIRT6, H3K9ac, H3K27ac, and H3K56ac) and ATAC-seq of WT and SIRT6 mutant SKMel-239 cell line derived from GSE102813. The bulk RNA-seq of WT and Sirt6 cKO mTEC were derived from our recently published studies35, and the data was available on GSE166840. The genes regulated by POU2F3 were calculated by the ChIP-seq (POU2F3) of intestinal tuft derived from GSE116510. The raw data of scATAC-seq sequencing used in this article have been deposited at the National Genomics Data Center BioProject under number: PRJCA006664. Uncropped Western blots gel for Figs. 5c, 6g are presented in Suppl. Figs. 32, 38, respectively.