Abstract
Objective
Mitochondrial “retrograde” signaling may stimulate organelle biogenesis as a compensatory adaptation to aberrant activity of the oxidative phosphorylation (OXPHOS) system. To maintain energy-consuming processes in OXPHOS deficient cells, alternative metabolic pathways are functionally coupled to the degradation, recycling and redistribution of biomolecules across distinct intracellular compartments. While transcriptional regulation of mitochondrial network expansion has been the focus of many studies, the molecular mechanisms promoting mitochondrial maintenance in energy-deprived cells remain poorly investigated.
Methods
We performed transcriptomics, quantitative proteomics and lifespan assays to identify pathways that are mechanistically linked to mitochondrial network expansion and homeostasis in Caenorhabditis elegans lacking the mitochondrial calcium uptake protein 1 (MICU-1/MICU1). To support our findings, we carried out biochemical and image analyses in mammalian cells and mouse-derived tissues.
Results
We report that micu-1(null) mutations impair the OXPHOS system and promote C. elegans longevity through a transcriptional program that is independent of the mitochondrial calcium uniporter MCU-1/MCU and the essential MCU regulator EMRE-1/EMRE. We identify sphingosine phosphate lyase SPL-1/SGPL1 and the ATFS-1-target HOPS complex subunit VPS-39/VPS39 as critical lifespan modulators of micu-1(null) mutant animals. Cross-species investigation indicates that SGPL1 upregulation stimulates VPS39 recruitment to the mitochondria, thereby enhancing mitochondria-lysosome contacts. Consistently, VPS39 downregulation compromises mitochondrial network maintenance and basal autophagic flux in MICU1 deficient cells. In mouse-derived muscles, we show that VPS39 recruitment to the mitochondria may represent a common signature associated with altered OXPHOS system.
Conclusions
Our findings reveal a previously unrecognized SGPL1/VPS39 axis that stimulates intracellular organelle interactions and sustains autophagy and mitochondrial homeostasis in OXPHOS deficient cells.
Keywords: Autophagy, Caenorhabditis elegans, Longevity, MICU1, Mitochondria, Sphingosine signaling, VPS39
Graphical abstract
Highlights
-
•
micu-1(null) nematodes are long-lived mitochondrial mutants.
-
•
MICU-1/MICU1 deficiency stimulates VPS-39/VPS39 and SPL-1/SGPL1 upregulation.
-
•
VPS-39 sustains mitochondrial network expansion in micu-1(null) nematodes.
-
•
VPS39 and SGPL1 expression influences mitochondria-lysosome contact sites in MICU1 deficient cells.
-
•
VPS39/SGPL1 signaling may be a common signature of mitochondrial deficient cells.
1. Introduction
Mitochondrial Ca2+ uptake finely tunes intracellular signaling. The transient Ca2+ accumulation in the matrix depends on mitochondrial membrane potential and occurs through a high conductance, ruthenium red-sensitive channel spanning the inner mitochondrial membrane (IMM) [1,2]. In all metazoans with a well-characterized Ca2+ signaling [3,4], electrogenic Ca2+ entry in the mitochondrial matrix depends on a multiprotein complex comprising three evolutionarily conserved components [5]. The ion-conducting pore consists of mitochondrial Ca2+ uniporter (MCU) oligomers [6,7] that associate with the essential MCU regulator (EMRE), a single-pass transmembrane protein that influences MCU activity and its inner membrane localization [[8], [9], [10], [11]]. Within the mitochondrial intermembrane space, a membrane-embedded MCU-EMRE subcomplex interacts with the EF-hand containing mitochondrial calcium uptake protein 1 (MICU1) or its evolutionarily related paralogs MICU2 and MICU3 [[12], [13], [14], [15], [16], [17], [18]]. As MCU gatekeepers, MICU1 and its paralogs physically block MCU pore access and inhibit the electrogenic Ca2+ entry under resting conditions, thereby preventing mitochondrial Ca2+ overload [[17], [18], [19], [20]]. Beside its regulatory role of the MCU complex, MICU1 localizes in the inner boundary membrane (IBM) of the IMM, thereby stabilizing cristae junctions along with optic atrophy 1 (OPA1) and mitochondrial contact site and cristae organizing system (MICOS) [14].
The pathophysiological relevance of MCU and its components has become even more evident since the identification in humans of disease-causing loss-of-function mutations in MICU1 and MICU2 genes [[21], [22], [23], [24]]. As for other mitochondrial diseases [[25], [26], [27], [28]], MICU1 and MICU2 mutations lead to a variety of symptoms, including skeletal muscle weakness, fatigue, cognitive impairment, tremors and ataxia [[21], [22], [23], [24],29]. Patient-derived cells display aberrant mitochondrial OXPHOS and Ca2+ homeostasis along with other abnormalities, such as fragmented mitochondrial network, increased NAD(P)H levels [23] and enhanced sensitivity to oxidative stress [21]. A large number of in vivo studies have attempted to investigate the molecular mechanisms linked to dysfunctional MCU complex. Very surprisingly, MCU knockout mice are viable in a mixed background and display diminished pyruvate dehydrogenase (PDH) activity associated with reduced muscular strength [30]. To a similar extent, EMRE KO mice do not show obvious metabolic changes or altered skeletal muscle performance even under challenging conditions [9,15]. In contrast to MCU and EMRE KO mice, MICU1 KO mice show high perinatal lethality with a few escapers that weigh 50% less than wild type littermates and display skeletal muscle degeneration and neurological defects [15,16]. Importantly, MICU1 deficient muscles exhibit clear signatures of mitochondrial dysfunction, with decreased succinate dehydrogenase (SDH)/cytochrome c oxidase (COX) staining, increased production of reactive oxygen species (ROS), low ATP levels and higher amounts of lactate [15]. Consistent with the role of MICU-family members in mitochondrial bioenergetics, it was shown that a loss-of-function (lof) of MICU1, but not the other MCU complex components, compromises the survival of Drosophila melanogaster larvae [31]. Notably, neither MCU(lof) nor EMRE(lof) suppresses the fly lethal phenotype due to MICU1 deficiency [31]. Taken together, it seems that MICU1 may contribute to cellular homeostasis and metabolism through additional pathways that are MCU independent and partially uncoupled from mitochondrial Ca2+ uptake. However, further investigations in model organisms are necessary to identify evolutionarily conserved molecular signatures that are relevant to our understanding of syndromes associated with disease-causing MICU1 and MICU2 mutations.
To gain insights into MICU1 biology, we employed the nematode Caenorhabditis elegans. We herein report that MICU-1/MICU1 deficiency stimulates longevity independently of MCU. Using quantitative multiplexed tandem mass tags (TMT)-based mass spectrometry coupled to RNAi screenings in C. elegans, we identify sphingosine phosphate lyase SPL-1/SGPL1 and the HOPS complex subunit VPS-39/VPS39 as critical regulators of homeostatic processes mechanistically linked to the increased lifespan of micu-1 mutant nematodes. In mammalian cells, MICU1 deficiency stimulates VPS39 recruitment to mitochondria, thereby enhancing the number of mitochondria-lysosome contact sites without altering mitochondria-ER tethering. Together, our findings provide new evidence of an MCU-independent role of MICU1 in metabolic remodeling and intracellular organelle homeostasis, the latter being of potential biological relevance in other diseases associated with aberrant mitochondrial bioenergetics.
2. Materials and methods
2.1. Antibodies
The following antibodies were used in our work: rabbit anti-NDUFB8 (Proteintech, 1479-1-AP); mouse anti-MTCO1 (Abcam, ab14705); mouse anti-total OXPHOS antibody cocktail (Abcam, ab110413); mouse anti-GFP (Roche, 11814460001); rabbit anti-VPS39 (Proteintech, 16219-1-AP and Novus Biologicals, NBP1-76535); mouse anti-KDEL (Millipore, 10C3); mouse anti-TOM40 (Santa Cruz, sc-365467); rabbit anti-TOM20 (Proteintech, 11802-1-AP); rabbit anti-LC3B (Sigma, L7543); rabbit anti-MICU1 (Sigma, PA5-83371); guinea pig anti-p62 (Progen, GP62-C); rabbit anti-pS473-AKT (Cell Signaling, 4060S), rabbit anti-AKT (Cell Signaling, 4685); rabbit anti-pT246-PRAS40 (Cell Signaling, 13175); anti-PRAS40 (Cell Signaling, 2691); rabbit anti-GAPDH (Santa Cruz, sc-25778); mouse anti-actin (Sigma, A5316 and abcam, ab14128); mouse anti-tubulin (Sigma, T6074); rabbit anti-SGPL1 (Atlas Antibodies, HPA021125); rabbit anti-AIF (Cell Signaling, 5318).
2.2. C. elegans strains and maintenance
Nematodes were maintained at 20 °C following standard culture methods. The following strains were used in this study: wild type N2 (Bristol), atfs-1(tm4525)V, BAN126 zuIs178[his-72p::his-72::GFP]; BAN299 nuo-6(qm200)I;zcIs14[myo-3p::GFP(mit)], BAN338 mcu-1(ju1154)IV;zcIs13[hsp-6p::GFP], BAN362 micu-1(bon20)IV/nT1[qIs51](IV;V), BAN369 micu-1(bon20)IV/nT1[qIs51](IV;V);zcIs13[hsp-6p::GFP], BAN370 micu-1(bon20)IV/nT1[qIs51](IV;V);zuIs178[his-72p::his-72::GFP], BAN372 micu-1(bon20)IV/nT1[qIs51](IV;V);bcIs39[lim-7p::ced-1::GFP + lin-15(+)], BAN373 micu-1(bon20)IV/nT1[qIs51](IV;V);zcIs14[myo-3p::GFP(mit)], BAN378 micu-1(bon20)IV/nT1[qIs51](IV;V);mcu-1(ju1154)IV, BAN380 nuo-6(qm200)I;micu-1(bon20)IV/nT1[qIs51](IV;V), BAN382 micu-1(bon77)IV/nT1[qIs51](IV;V), BAN397 micu-1(bon20)IV/nT1[qIs51](IV;V);atfs-1(tm4525)V, BAN407 emre-1(bon78)X, BAN436 age-1(hx546)III;micu-1(bon20)IV/nT1[qIs51](IV;V), BAN448 micu-1(bon20)IV/nT1[qIs51](IV;V);sqIs11[lgg-1p::mCherry::GFP::lgg-1 + rol-6)], BAN449 micu-1(bon20)IV;vps-39(ok2442)V/nT1[qIs51](IV;V), BAN491 glp-1(bn18)III;micu-1(bon20)/nT1[[qIs51](IV;V), BAN500 micu-1(bon20)IV/nT1[qIs51](IV;V);emre-1(bon78)X, BAN501 micu-1(bon20)IV/nT1[qIs51](IV;V);mcu-1(ju1154)IV;emre-1(bon78)X, DG2389 glp-1(bn18)III, CZ19982 mcu-1(ju1154)IV, MAH215 sqIs11[lgg-1p::mCherry::GFP::lgg-1 +rol-6)], MD701 bcIs39[lim-7p::ced-1::GFP + lin-15(+)], MQ1333 nuo-6(qm200)I, SJ4100 zcIs13[hsp-6p::GFP], SJ4103 zcIs14[myo-3p::GFP(mit)], TJ1052 age-1(hx546)III. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
2.3. Cell culture
Human embryonic kidney HEK293 and HeLa cells were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (100 U/ml penicillin; 100 mg/ml streptomycin). For biochemical analysis, cells were seeded in 6-well plates at a density of 4 × 105 cells per well and collected on the following day. Cell pellets were then either processed directly for biochemical analysis or stored at −80 °C until further use. For immunostaining and PLA experiments, cells were seeded onto poly-l-lysine-coated coverslips in 12-well plates at a density of 6 × 104 cells per well and fixed with 4% PFA 48 h afterwards. Transient transfections were performed using transfection complexes. For plasmid and siRNA transfection, we used Turbofectin and 1 μg of plasmid DNA or RNAiMAX and siRNAs diluted in Opti-MEM, respectively. After 48 h, cells were fixed with 4% PFA and used for PLA experiments or immunostaining. The following plasmids were used: mEGFP-ER-5a (Addgene plasmid # 56455), mitochondrially targeted-GFP and mitochondrially targeted-DsRed. The following siRNAs were used at a final concentration of 50 nM: scramble (ID: AM4611), siMICU1 (ID: 135594), siSGPL1 (ID: 118700), siVPS39 (ID:136872).
2.4. CRISPR/Cas9 genome editing
Young adult hermaphrodites were injected with a customized injection mix consisting of specific sgRNAs against target genes, Cas9 protein and a plasmid encoding myo-2p::GFP (NemaMetrix Inc., Eugene OR, USA). F1 offspring were screened for GFP expression in the pharynx, while F2 were genotyped using specific oligonucleotides upstream and downstream the expected deletions. Mutants were backcrossed at least three times with wt N2 animals and, when necessary, stabilized using nT1[qIs51] balancer.
2.5. Imaging and image analysis
Imaging of nematodes was performed on a Zeiss LSM900 equipped with an Airyscan detector. All high-resolution images were acquired and processed using ZEN (Carl Zeiss). For gonad staining, gonads were extruded and fixed with 4% PFA for 5 min, washed with M9 buffer, stained with DAPI for 5 min, followed by further washing. Gonads were imaged with a 20x air objective. For imaging of mitochondria in zcIs14[myo-3p::gfp(mt)] transgenic strains, age-matched animals were fixed with 4% PFA for 5 min, washed with M9 buffer, mounted on agarose pads and imaged posterior to the pharynx, using a 63x oil immersion objective. For imaging of sqIs11[lgg-1p::mCherry::GFP::lgg-1 + rol-6)] transgenic strains, adult nematodes were immobilized in levamisole on agarose pads and imaged with a 40x oil immersion objective. Z-stacks were taken at a thickness of 1 μm between focal planes, for a total thickness of 10 μm. Image analysis was performed using ImageJ on maximum intensity projections of Z-stacks. Red and green puncta were counted in the metacorpus of the pharynx using the Find Maxima function. Statistical analysis was performed using GraphPad Prism.
Image analyses of immunofluorescence-labelled and PLA-stained cells and tissue sections were carried out using a Zeiss LSM800 equipped with an Airyscan detector or a Zeiss LSM880 equipped with an Airyscan detector, using a 63x oil immersion objective. Z-stacks were taken at a thickness of 0.5 μm between focal planes. High-resolution images were acquired with ZEN (Carl Zeiss) and processed with the automated Airyscan processing algorithm in ZEN. Image analysis was done in ImageJ. Mitochondrial morphology was calculated after auto-threshold processing. Circularity and mitochondrial area were evaluated via the analyze particles plugin. For co-localization indices, Pearson's and Mander's coefficients were determined using the co-localization analysis plugin JACoP. PLA-dots were semi-automatically quantified as previously described [32].
2.6. Immunocytochemistry, proximity ligation assay (PLA) and LysoTracker staining
For immunocytochemistry, PFA-fixed cells were first incubated in blocking solution (0.5% Triton X-100, 10% normal goat serum) for 30 min at room temperature, followed by overnight incubation with primary antibodies. On the next day, cells were washed in PBS and incubated with appropriate Alexa Fluor-conjugated secondary antibodies (Invitrogen) for 2 h at room temperature. As a last step, cells were counterstained with DAPI and mounted onto microscope slides.
PLA experiments were performed using Duolink® PLA fluorescence reagents (Sigma) according to the manufacturer's instructions and as previously described [32]. Briefly, cells and tissue samples were first permeabilized with 0.25% Triton X-100 in PBS, and then treated with Blocking solution for 1 h at 37 °C. Thereafter, samples were incubated with primary antibodies over night at 4 °C. Following three washes in Wash Buffer A, samples were then incubated with PLA probes for 1 h at 37 °C or, in case of tissue samples, for 24 h at 4 °C. Signal amplification was performed by incubation with amplification-polymerase solution for 100 min at 37 °C. After three washes in Wash Buffer B, cells were counterstained with DAPI and mounted onto microscope slides while tissue samples were incubated with anti-TOM20 antibody overnight. On the next day, sections were washed in PBS, incubated with appropriate secondary antibodies for 2 h at room temperature, and finally counterstained with DAPI. Imaging was then performed within 24 h.
For lysosomal labeling, mitochondria-targeted GFP-positive HEK293 and HeLa cells were seeded onto poly-l-lysine-coated coverslips. On the next day, medium was removed and replaced by pre-warmed growth medium containing LysoTracker Red DND-99 (Molecular probes, Life Technologies) at a concentration of 60 nM. Following 45 min of incubation at 37 °C, cells were fixed with 4% PFA, washed with PBS, mounted onto microscope slides and imaged immediately afterwards.
2.7. Lifespan assays and RNAi
All lifespan assays were carried out at 20 °C. RNAi experiments were performed by feeding with HT115 E. coli expressing dsRNA against target genes (Ahringer library, Source Bioscience LifeSciences). To obtain synchronized populations, gravid adult nematodes were bleached in hypochlorite solution and the resulting eggs transferred directly onto bacteria-seeded NGM plates. For lifespan-based RNAi screen, gravid micu-1(bon20)/nT1[qIs51] nematodes were bleached and eggs transferred onto HT115 E. coli expressing dsRNA against candidates. At L4/young adult stage, 50 micu-1(bon20) animals were transferred to a fresh plate for each RNAi condition. For all the other lifespan assays, nematodes were grown until L4/adult stage and then transferred to fresh plates in groups of 30 per plate. Animals were transferred every 2–4 days and scored at least every other day for touch-provoked movement until death. Animals that died abnormally (e.g., internal hatching, vulva protrusions) were scored as censored. Survival curves were generated using GraphPad Prism software (GraphPad Software Inc., San Diego, USA).
2.8. Mouse work
Aifm1(R200 del) knockin mice were generated as described previously [33]. Mice were housed in groups of two to four under a 12/12 h light/dark cycle with free access to food and tap water. All experiments were approved and performed in conformity to the guidelines of the State Agency for Nature, environment and Consumer Protection North Rhine Westphalia. At 6 months of age, animals were anaesthetized with an overdose of Ketamine: Xylazine and transcardially perfused with PBS followed by 4% PFA. Quadricep muscles were rapidly removed, post-fixed for 24 h in PFA, and stored in 30% sucrose solution until further processing. Muscle sections were cut on a cryostat at a thickness of 20 μM and stored at −20 °C until further use.
2.9. Next-generation RNA sequencing (NGS)
Adult nematodes were dissolved in 1 ml Trizol (Invitrogen) and total RNA was isolated via the miRNeasy Micro kit (Qiagen) according to the manufacturer's protocol. RNA concentration and integrity were determined by High Sensitivity RNA assay on a TapeStation 4200 system (Agilent). cDNA libraries were prepared from 5 ng total RNA using the SMART-seq2 protocol and tagmented with the Nextera XT kit (Illumina). Library purification and size selection was carried out with AMPure XP beads (Beckman–Coulter) and final library size distribution was measured via High Sensitivity D5000 assay on a TapeStation 4200 System (Agilent). Library concentration was determined using the HS dsDNA assay on a Qubit 3. Libraries were sequenced SR 75 cycles on a NextSeq500 system (Illumina) using High Output v2 chemistry. Base call files were converted to fastq format and demultiplexed using bcl2fastq v2.20. The 75 bp single-end reads were aligned to the C. elegans reference transcriptome WBcel235 by kallisto v0.44.0 using default parameters. Raw RNA seq data were deposited to GEO database under the common accession number GSE197286.
2.10. Oxygen consumption rate measurements (OCR)
OCR was measured using a Seahorse XFe24 Analyzer (Agilent). Synchronized animals were grown to first day of adulthood at 20 °C and transferred to heat-killed OP50 plates for 3 h to empty their gut of live bacteria. Each well of a Seahorse XFe24 Cell Culture Microplate was filled with 500 μl M9 buffer, and 50 animals transferred into each well, with a minimum of 3 wells per condition. OCR measurements were taken at basal conditions, and in response to addition of 20 μM FCCP, followed by 20 mM sodium azide (NaN3).
Cells were seeded on cell culture microplates (Agilent Seahorse XF24) in growth medium 24 h, or in case of siRNA experiments 48 h, before the assay. On the day of the experiment, growth medium was replaced with Seahorse XF base medium supplemented with 1 mM pyruvate, 2 mM glutamine and 10 mM glucose or galactose. Before starting the measurements, cells were equilibrated for 60 min in a CO2-free incubator at 37 °C. Following three baseline measurements, subsequent additions of 1 μM oligomycin, 1.5 μM FCCP and 0.5 μM rotenone/antimycin A were carried out as previously described. At the end of OCR and ECAR assessments, cells were collected and lysed in RIPA buffer (SIGMA), supplemented with protease and phosphatase inhibitors (Roche). Protein concentrations were determined via Bradford assay and OCR values were then normalized to the respective protein contents.
2.11. RNA extraction and quantitative real-time PCR
RT-PCR was used to quantify gene expression levels on synchronized populations. RNA extraction, purification and reverse transcription was performing using RNeasy RNA extraction kit (Qiagen) and qScript cDNA supermix (Quanta Biosciences). Quantitative RT-PCR was performed using Fast SYBR Green Master Mix (Applied Biosystems) in a Step One Plus Real Time PCR System (Applied Biosystems) and analyzed using comparative ΔΔCt method, normalized to mRNA levels of actin. Three technical replicates were performed for each biological replicate. The following oligonucleotides were used in this study: sod-3 5′- ccaaccagcgctgaaattcaatg-3′ and 5′-ggaaccgaagtcgcgcttaatag-3’; nhr-57 5′-tcggaatgaatccggaagt-3′ and 5′-atgcaggggaagatgaacag-3’; gst-4 5′-gatacttggcaagaaaatttggac-3′ and 5′-ttgatctacaattgaatcagcgtaa-3’; vps-39 5′-cgctttcccagacgcatttt-3′ and 5′-gtcgttttcggtagaggccc-3’; spl-1 5′-cagcgcctaacttcccatca-3′ and 5′-caggtatccggcgtcattca-3’; beta-actin 5′-tgtgatgccagatcttctccat-3′ and 5′-gagcacggtatcgtcaccaa-3’.
2.12. Sample preparation, LC-MS/MS measurements and database searching
Briefly, approximately 800 wt and micu-1(bon20)/nT1[qIs51] adult nematodes were collected at day 4 and 5 after hatching, respectively. Animals were washed twice with water and the pellets were kept at −80 °C. Samples were lysed in 200 μl Lysis buffer (50 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1.5% SDS, 1 mM DTT; supplemented with: 1 × protease and phosphatase inhibitor cocktail (ThermoScientific)). Lysis was aided by repeated cycles of sonication in a water bath (6 cycles of 1 min sonication (35 kHz) intermitted by 2 min incubation on ice). At least 40 μg of C. elegans protein lysates were reduced and alkylated prior to processing by a modified filter-aided sample preparation (FASP) protocol as previously described [34]. Samples were subjected to sequential overnight digestion with Lys-C (1:40 in 1.3 M Urea buffer) and Trypsin (1:20; in 50 mM ammonium bicarbonate) directly on the filters. Peptides were additionally precipitated using an equal volume of 2 M KCl for depletion of residual detergents, then cleaned and desalted on C18 stage tips, prior to re-suspension in 20 μl of 50 mM HEPES (pH 8.5). Then, wt (OP50), micu-1(bon20) (OP50), micu-1(bon20) (on control RNAi), micu-1(bon20) (on spl-1 RNAi), micu-1(bon20) (on vps-39 RNAi) C. elegans peptides were labelled with 25 μl of diluted (14.75 mM) 126; 127N; 128N, 128C, 129N; and 129C, 130N, 130C (batches 1 and 2) TMT 10plex, respectively, for 1 h at RT. TMT signal was quenched by addition of 2 μl of 5% hydroxylamine to the reaction, vortexing for 20 s and incubating for 15 min at 25 °C with shaking (1000 rpm). TMT-labelled peptides were acidified with 45% (vol/vol) of 10% FA in 10% ACN, prior to combining samples at equal amounts for drying in a concentrator. Dried peptides were re-suspended in 300 μl of 0.1% TFA for subsequent high pH reverse phase fractionation (Perce kit). 8 peptide fractions (10, 12.5, 15, 20, 22.5, 25, 50, and 80%) ACN were collected, concentrated and re-suspended in 20 μl 5% FA for LC-MS analysis. MS runs were performed in triplicates.
Tryptic peptides were analyzed on a Dionex Ultimate 3000 RSLC nanosystem coupled to an Orbitrap Exploris 480 MS. They were injected at starting conditions of 95% eluent A (0.1% FA in water) and 5% eluent B (0.1% FA in 80% ACN). Peptides were loaded onto a trap column cartridge (Acclaim PepMap C18 100 Å, 5 mm × 300 μm i.d., #160454, Thermo Scientific) and separated by reversed-phase chromatography on a 50 cm μPAC C18 column (PharmaFluidics) using a 120 min linear increasing gradient from 8% to 25% of eluent B for 85 min, followed by a 28 min linear increase to 50% eluent B. The mass spectrometer was operated in data dependent and positive ion mode with MS1 spectra recorded at a resolution of 120 k with an automatic gain control (AGC) target value of 300% (3 × 106) ions, maxIT set to Auto and an intensity threshold of 1 × 104, using a mass scan range of 350–1550. Precursor ions for MS/MS were selected using a top speed method with a cycle time of 2 ms and normalized collision energy (NCE) of 36% (High-energy Collision Dissociation (HCD)), to activate both the reporter and parent ions for fragmentation. MS2 spectra were acquired at 45 k resolution using an AGC target value of 200% (2 × 105), and maxIT set to 86 ms. Dynamic exclusion was enabled and set at 45 s. Isolation width was set at 0.7 m/z and the fixed first mass to 110 m/z to ensure reporter ions were detected. Peptide match was set to off, and isotope exclusion was on. Charge-state exclusion rejected ions that had unassigned charge states, were singly charged or had a charge state above 5. Full MS data were acquired in the profile mode with fragment spectra recorded in the centroid mode.
Raw data files were processed with Proteome Discoverer™ software (v2.4.0.305, Thermo Scientific) using SEQUEST® HT search engine against the Swiss-Prot® C. elegans database (v2020-05-08). Peptides were identified by specifying Lys-C and trypsin as the proteases, with up to 2 missed cleavage sites allowed. Precursor mass tolerance was set to 10 ppm, and fragment mass tolerance to 0.02 Da MS2. Static modifications were set as carbamidomethylated cysteine and TMT6plex (229.163 Da; N-terminal, K), while dynamic modifications included methionine, oxidation and N-terminal protein acetylation, for all searches. Resulting peptide hits were filtered for maximum 1% FDR using the Percolator algorithm. The TMT10plex quantification method within Proteome Discoverer software was used to calculate the reporter ratios with mass tolerance ± 10 ppm and applying isotopic correction factors. Only peptide spectra containing all reporter ions were designated as “quantifiable spectra”.
2.13. SDS-PAGE and western blotting
Adult nematodes were collected, washed in sterile H2O and frozen in liquid nitrogen. C. elegans and cell pellets were thawed on ice and sonicated in ice-cold RIPA buffer (Sigma) with protease and phosphatase inhibitors. Total lysates were spun down, quantified using Bradford reagent (Sigma), and boiled in 2X Laemmli buffer at 95 °C for 5 min. Samples were resolved on 10–15% poly-acrylamide gels and transferred onto nitrocellulose membranes using semi-dry transfer Trans-Blot Turbo™ (Bio-Rad). Immunoblots were developed in ECL and imaged using Chemidoc imaging system (Bio-Rad).
2.14. Statistics
GraphPad Prism Software was used for statistical analysis. Data were tested for normality distribution and statistical comparison of normal distributed data was performed with two-tailed Student's t test or one-way ANOVA, unless indicated differently. Non-normal distributed data were statistically compared using the Mann–Whitney U-test or Kruskal–Wallis test. Statistical significance was defined as ∗p < 0.05.
2.15. Thrashing assay
Synchronized nematodes at the first day of adulthood were used for thrashing assays. Individual animals were placed into a drop of M9 buffer, and each full back-and-forth lateral movement was counted for a total of 90 s. Thrashing was performed for 10 animals per strain, for each biological replicate.
3. Results
3.1. MICU-1 deficiency extends C. elegans lifespan in an MCU-1 independent manner
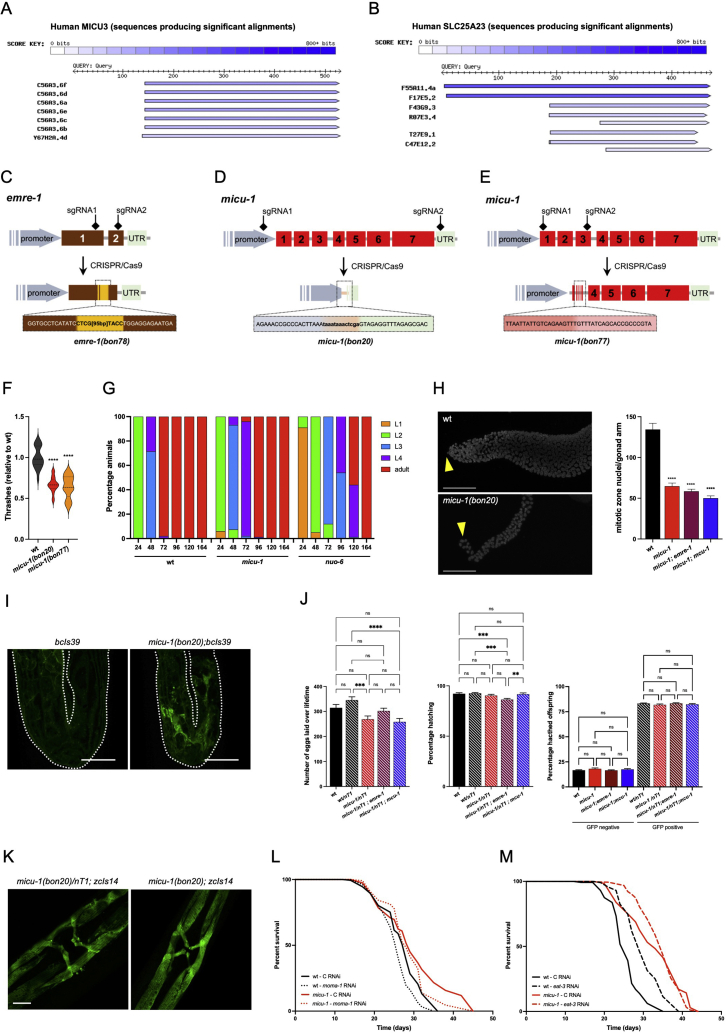
As in other multicellular organisms, C. elegans expresses the three core components of the MCU complex: MCU-1/MCU, EMRE-1/EMRE and MICU-1/MICU1 (Figure 1A). Sequence alignments indicate that the nematode genome may also encode a partially conserved MICU-3/MICU3 as well as two poorly characterized SLC25A23 orthologs with predicted Ca2+ binding activity (Supplementary Figs. S1A–B). To assess the contribution of the MCU complex to C. elegans survival, we initially studied the lifespan of hermaphrodites homozygous for an existing mcu-1(lof) allele [35]. To our surprise, we found that mcu-1 mutants lived as long as wild type (wt) nematodes (Figure 1B and Table S1), suggesting that MCU-1 is dispensable for C. elegans viability under standard laboratory conditions. Using CRISPR/Cas9-gene editing, we generated an emre-1(lof) allele consisting of an insertion in exon 2 that led to a truncated and potentially unstable protein lacking 38 amino acids (Supplementary Fig. S1C). We backcrossed this line, performed lifespan assays and observed that emre-1 mutant nematodes also lived as long as wt animals (Figure 1C and Table S1). To complete our genetic survey, we then manipulated the expression of the micu-1 gene by deleting a small portion of the promoter, the open reading frame and part of the 3′UTR (Supplementary Fig. S1D). An additional micu-1 mutant allele was obtained by targeting most of the region between exon 1 and 3 (Supplementary Fig. S1E), since this strategy also abrogates the expression of all predicted MICU-1 isoforms. Similar to previously described mitochondrial mutants [36,37], homozygous micu-1(null) nematodes exhibited diminished motility and larval developmental delays (Supplementary Figs. S1F–G). While mcu-1 and emre-1 mutant animals did not show any obvious phenotypes, micu-1(null) nematodes had reduced syncytial gonads and did not develop oocytes when compared to wt animals (Supplementary Figs. S1H–I). As for an earlier micu-1 mutant strain [38], we maintained our micu-1(null) mutations with the balancer nT1[qIs51](IV;V). Confocal image analysis showed that micu-1(null) mutants had far fewer nuclei in their germlines compared to wt gonads (Supplementary Fig. S1H). Moreover, micu-1 mutant gonads contained a large number of germ cells undergoing programmed cell death, as revealed by the intense CED-1:GFP fluorescence originating from sheath cells engulfing germ cell corpses in the pachytene and diplotene regions (Supplementary Fig. S1I). Of note, genetic manipulation of mcu-1 or emre-1 did not rescue the germline defects of micu-1 mutants (Supplementary Fig. S1H). We found that micu-1(bon20)(IV)/nT1[qIs51](IV;V) laid a comparable number of eggs to wt animals, with the population of hatched animals having genotypes that did not follow the Mendelian distribution (Supplementary Fig. S1J). Adult micu-1(null) mutants exhibited a fully differentiated vulva with no obvious abnormalities (Supplementary Fig. S1K).
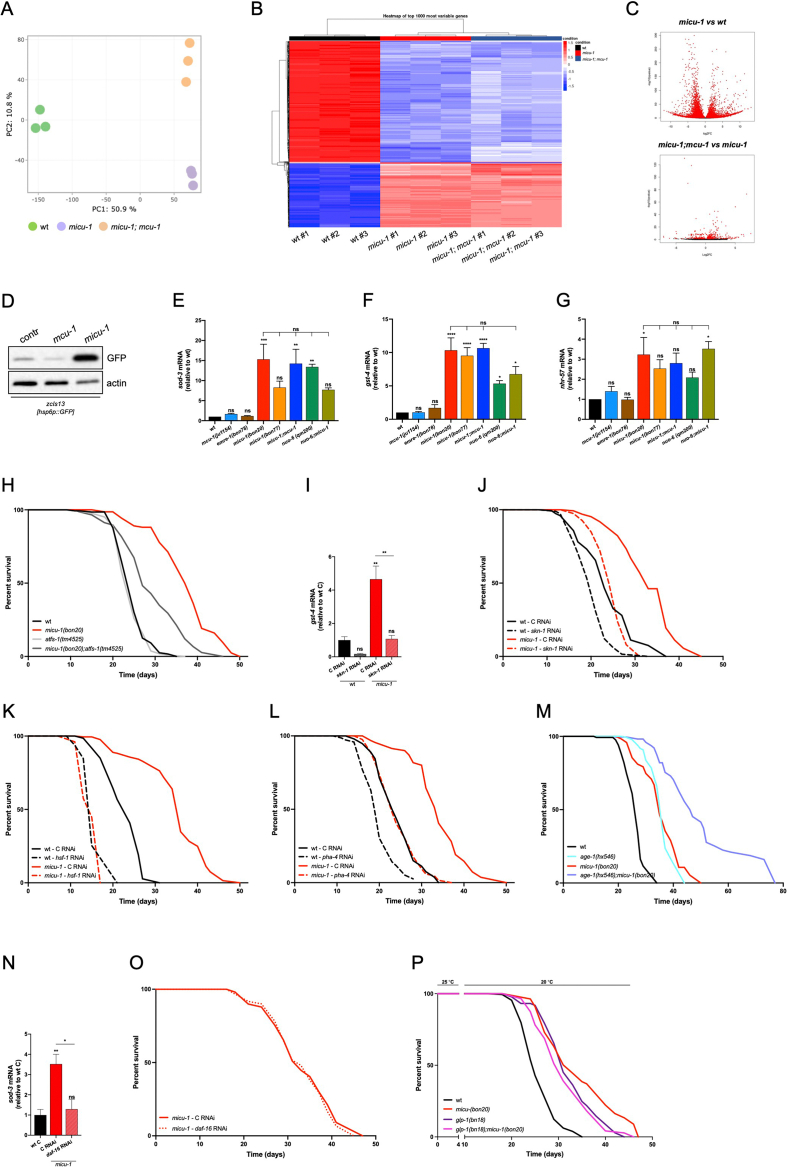
Figure 1.
micu-1(null) alleles extend C. elegans lifespan independently of MCU complex. (A) Schematic representation of the C. elegans mitochondrial calcium uniporter (MCU) complex. MICU-1 homo- or heterodimers (red) bind MCU-1 (blue) and EMRE-1 (brown) subunits. Upon Ca2+ binding, MICU-1 regulates MCU-1 permeability and Ca2+ influx into the mitochondrial matrix. (B–C) Representative survival curves of wt versus (B) mcu-1(ju1154) and (C) emre-1(bon78) mutant nematodes. (D–E) Representative survival curves of wt animals compared to (D) mcu-1(ju1154) and micu-1(bon20), (E) micu-1(bon20) and micu-1(bon77) nematodes. (F–G) Lifespan assay of micu-1(bon20) vs (F) micu-1(bon20);mcu-1(ju1154) and micu-1(bon77), (G) micu-1(bon20);emre-1(bon78) and micu-1(bon20);mcu-1(ju1154);emre-1(bon78). (H) Representative survival curves of micu-1(bon20) grown on control and RNAi against emre-1. (I) Confocal images of 9-day-old control and micu-1(bon20) animals expressing zcIs14[myo-3p::GFP(mit)] transgene. Mitochondria are visualized as GFP-positive structures (scale bar = 10 μm). (J) Quantification of mitochondrial elongation in zcIs14[myo-3p::GFP(mit)] expressing control (wt) and micu-1(bon20) animals at 2, 4 and 6 days after adulthood (Kruskal–Wallis ANOVA, Dunn's multiple comparison test: ns = not significant, ∗p < 0.05, n = 17–53 animals from at least 3 independent experiments). To be consistent with other experiments (see figure 3I), the assessment of mitochondrial elongation was performed on animals grown on control RNAi. (K) Seahorse OCR profiles of wt, mcu-1(ju1154), micu-1(bon20) and emre-1(bon78) animals. (L) Basal respiration and spare respiratory capacity of animals at day 1 of adulthood (mean ± S.E.M., Kruskal–Wallis test: ns = not significant, ∗p < 0.05, n = 3). (M) Immunoblot analyses of samples from wt and micu-1(bon20) animals using antibodies against CV subunit ATP5A, CIV subunit CTC-1/MTCO1, CI subunit NUO-2/NDUFS3 and actin (as loading control). Densitometry is reported on the right (mean ± S.E.M., unpaired Student's t-test: ns = not significant, ∗p < 0.05, ∗∗p < 0.01, n = 3). (N) Representative survival curve of nuo-6(qm200), nuo-6(qm200);micu-1(bon20) and micu-1(bon20);mcu-1(ju1154) mutant nematodes. (O) Representative lifespan assay of wt and micu-1(bon20) mutants grown on control (solid lines), eat-3 (dashed lines) and moma-1 (dotted lines) RNAi.
Consistent with the “mitochondrial threshold effect” theory [39], mitochondrial lesions can either extend or decrease C. elegans lifespan, according to the ability of the organism to compensate for aberrant mitochondrial OXPHOS through transcriptional programs regulating stress response and metabolism [[40], [41], [42], [43], [44], [45]]. Since MICU-1/MICU1 knockout compromises mitochondrial Ca2+ signaling and bioenergetics in mammalian and invertebrate cells [12,18,31,38], we reasoned that MICU-1/MICU1 deficiency could influence C. elegans survival similarly to other mitochondrial lesions [36,37,46]. In line with our hypothesis, we found that homozygous micu-1(null) hermaphrodites lived much longer than wt animals (Figure 1D–F and Table S1). Importantly, both mcu-1(lof) and/or emre-1 deficiency did not have a major impact on the lifespan extension of micu-1 mutants (Figure 1F–H and Table S1), strongly suggesting that MICU-1 longevity does not require a functional MCU complex and MCU-dependent Ca2+ uptake. Next, we performed high-resolution confocal microscopy of C. elegans body-wall muscle cells expressing a mitochondrially localized GFP. We observed that wt animals began to accumulate abnormally enlarged, as well as fragmented mitochondria a few days after reaching adulthood (Figure 1I,J), whereas age-matched micu-1(null) mutants appeared to have a well-organized mitochondrial network with highly interconnected tubular structures similar to those described in other long-lived mitochondrial mutants [44,47,48]. Using an adapted Seahorse protocol [44,49], we measured oxygen consumption rate (OCR) and found that micu-1(null) mutants had a lower spare respiratory capacity compared to wt nematodes (Figure 1K,L). To assess the mitochondrial OXPHOS system in micu-1 deficient nematodes, we ran immunoblot analyses and observed altered expression of mitochondrial complex I (CI) and complex IV (CIV) subunits in micu-1(null) mutants compared to wt (Figure 1M). Given this line of experimental evidence, we reasoned that the lifespan extension of micu-1 mutants was probably dependent on lesions altering the expression of electron transport chain (ETC) subunits. Thus, we generated nuo-6(qm200);micu-1(bon20) double mutants and observed a tendency toward further lifespan extension compared to both micu-1(bon20) and nuo-6(qm200) animals (Figure 1N and Table S1), suggesting that aberrant ETC composition may not be the only contributing factor underlying micu-1 longevity. Since MICU1 can be localized at cristae junctions and influences remodeling of the mitochondrial inner membrane [14], we assessed the survival of micu-1(null) mutants on bacteria expressing double strand RNAi against the mRNAs encoding the mitochondrial fusogen EAT-3/OPA1 [50] and MICOS complex subunit MOMA-1/APOOL/MIC27 [51]. We found that eat-3 downregulation had a minor, although statistically significant, effect on micu-1(null) lifespan, while moma-1 RNAi did not influence the survival of micu-1 mutant animals (Figure 1O, Supplementary Figure S1L-M and Table S1). These data suggest that additional IMM remodeling following EAT-3/OPA1 downregulation may further influence the longevity pathways linked to MICU-1 deficiency. In summary, our findings indicate that micu-1(null) mutations alter the maintenance and function of the OXPHOS system in C. elegans, recapitulating some of the mitochondrial defects observed in higher organisms carrying MICU1 or MICU2 lesions.
3.2. MICU-1 loss stimulates stress response pathways
Mitochondrial dysfunction during development can stimulate transcriptional regulation of stress response pathways that confer C. elegans longevity [40,41,[52], [53], [54]]. To explore the mechanisms underlying lifespan-extending properties of MICU-1 deficiency, we performed next-generation sequencing (NGS) of RNAs extracted from adult nematodes. Similar to other long-lived mitochondrial mutants [44,45], the micu-1(null) mutation led to profound transcriptional changes comprising of 3220 upregulated and 3132 downregulated genes compared to wt animals (Figure 2A–C). Many of the dysregulated genes encode for mitochondrial proteins, including ETC components (Supplementary Fig. S2A). Consistent with our lifespan assays, mcu-1(lof) had a negligible contribution to the gene expression profiles linked to MICU-1 deficiency (micu-1(bon20);mcu-1(ju1154) vs micu-1(bon20): 321 upregulated and 174 downregulated genes; Figure 2B–C), further emphasizing that MCU-1 participates only marginally in lifespan-extending programs established in micu-1(null) nematodes. Ingenuity pathway analysis (IPA) of up- and down-regulated genes further revealed protein synthesis and xenobiotic metabolism as significantly overrepresented pathways in the transcriptome of micu-1(bon20) compared to wt nematodes (Supplementary Figs. S2B–D), as reported in other C. elegans models of mitochondrial deficiency [44,55,56].
Figure 2.
micu-1(lof) induces a transcription-dependent stress response. (A) Principal component analysis of normalized RNA-seq data. Each dot represents an RNA-seq sample of the indicated genotype. Samples with similar gene expression profiles cluster together. (B) Heatmap showing the top 1000 differentially expressed genes in micu-1 and micu-1;mcu-1 mutants relative to wt animals. (C) Volcano plot of significantly dysregulated genes in micu-1(bon20) vs. wt (top panel) and micu-1(bon20);mcu-1(ju1154) vs. micu-1(bon20) (bottom panel). Compared to wt, 3220 genes were significantly up-regulated and 3132 genes significantly down-regulated in micu-1 mutants. In micu-1;mcu-1 double mutants compared to micu-1(null) animals, only a total of 495 genes were differentially expressed (321 upregulated and 174 downregulated genes). (D) Representative WB of samples from animals expressing the zcIs13[hsp-6p::gfp] transgene. Immunoblots were performed using antibodies against GFP and actin (as loading control). (E–G) RT-PCR of (E) sod-3, (F) gst-4 and (G) nhr-57 genes in animals at day 1 of adulthood (mean ± S.E.M., one-way ANOVA, Tukey's multiple comparisons test: ns = not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 ∗∗∗∗p < 0.0001, n = 3–5). (H) Representative survival curves of wt, micu-1, atsf-1 and micu-1;atfs-1 mutant nematodes. (I) RT-PCR of gst-4 expression in wt and micu-1(bon20) animals exposed to control and skn-1 RNAi constructs from hatching until adulthood (mean ± S.E.M., one-way ANOVA, Tukey's multiple comparisons test: ns = not significant, ∗∗p < 0.01, n = 3). (J–L) Representative survival curves of wt and micu-1(bon20) animals exposed to control (solid lines) and (J) skn-1, (K) hsf-1 and (L) pha-4 RNAi (dashed lines) from hatching. (M) Representative survival curves of wt, age-1, micu-1 and age-1;micu-1 animals. (N) RT-PCR of sod-3 gene expression in wt and micu-1(bon20) adult animals exposed to control and daf-16 RNAi (mean ± S.E.M., one-way ANOVA, Tukey's multiple comparisons test: ns = not significant, ∗p < 0.05, ∗∗p < 0.01, n = 3). (O) Lifespan assay of micu-1(bon20) animals grown on control (solid line) and daf-16 RNAi (dashed line) from hatching. (P) Representative survival curves of wt, micu-1(bon20), glp-1(bn18) and glp-1(bn18);micu-1(bon20) animals grown at 25 °C until adulthood, then shifted to 20 °C until all animals were dead. Exposure to 25 °C from hatching results in germline-less glp-1 animals and lifespan extension.
A vast body of literature indicates that decreased mitochondrial function evokes a nuclear encoded stress response associated with longevity [41,42]. Since micu-1 mutants show clear signatures of mitochondrial dysfunction (Figure 1K–M), we measured the expression of the unfolded protein response (UPRmt) reporter zcIs13[hsp-6p::gfp] and other stress-response markers (i.e., sod-3, gst-4 and nhr-57) using immunoblots and quantitative RT-PCR, respectively. Compared to control and mcu-1 mutant animals, we found that hsp-6p::gfp expression was strongly induced in micu-1(null) nematodes (Figure 2D). Similar to CI deficient nuo-6(qm200) mutants, MICU-1 loss led to enhanced expression of the DAF-16/ATFS-1 target sod-3 and the SKN-1 target gst-4, while the HIF-1 target nhr-57 was only marginally affected (Figure 2E–G). Consistent with a minor role of MCU-1 in the transcriptional profile of micu-1(null) nematodes, we observed that sod-3 and gst-4 genes were upregulated independently of MCU-1 activity (Figure 2E–F). Given these lines of evidence, we next tested the contribution of mitochondria-to-nucleus “retrograde” stress response to micu-1 longevity by genetically altering the expression of ATFS-1, an upstream transcriptional regulator of UPRmt [57,58], and the ROS responsive transcription factor SKN-1/NRF2 [59,60]. We showed that atfs-1(lof) as well as skn-1 RNAi significantly inhibited the lifespan extension of micu-1 mutants (Figure 2H–J and Table S1). To complete our epistatic analyses, we also assessed the contribution of other transcription factors that were previously reported to influence lifespan-extending programs in nematodes [54,[61], [62], [63]]. Specifically, we silenced the expression of heat shock factor HSF-1, which regulates proteostasis in mitochondrial deficient animals [64], and the diet-restriction regulator PHA-4/FOXA [65]. In line with their roles in longevity pathways linked to mitochondria, these two transcription factors contributed to the lifespan extension of micu-1 mutant nematodes (Figure 2K,L and Table S1). As was the case for other mitochondrial mutants [36,44], inhibition of the insulin/IGF-1 signaling pathway through hypomorphic age-1 mutation further extended micu-1 lifespan, while DAF-16/FOXO downregulation did not modify their survival (Figure 2M–O and Table S1). Together, these epistatic analyses suggest that micu-1(null) mutants have several transcriptional signatures commonly observed in other long-lived mitochondrial animals.
Since micu-1(null) nematodes have defective germline and are sterile (Supplementary Figs. S1H–J), we tested the contribution of germline deficiency on micu-1(null) lifespan. To do so, we employed a temperature-sensitive glp-1 mutation that impairs gonadogenesis in C. elegans hermaphrodites, stimulates DAF-16 activity (Supplementary Fig. S2E) and leads to longevity [66,67]. Upon exposure to a restrictive temperature that compromised the germline, we found that the median lifespan of glp-1;micu-1 double mutants was comparable to the survival of glp-1 single mutants and micu-1(null) nematodes (Figure 2P and Table S1). This non-addictive effect of glp-1 mutation suggests that signals from the germline may contribute to micu-1 longevity pathways.
3.3. SPL-1 and VPS-39 upregulation underlies cellular homeostasis and longevity of micu-1 mutant nematodes
To uncover novel regulatory pathways linked to MICU-1, we performed TMT-based mass spectrometry (MS) analyses of peptides extracted from wt and micu-1(bon20) mutant C. elegans samples from two distinct biological replicates (see materials and methods section for details). A total of 1563 and 1849 proteins were identified from the combined cytosolic and membrane fractions of the two biological replicates, with medium–high confidence (combined q value ≤ 0.05) (Supplementary Figs. S3A–D and Supplementary Tables S2–3). By integrating the two independent datasets, we found 56 down- and 49 up-regulated proteins in young micu-1(null) mutant animals compared to age-matched wt nematodes (Figure 3A; threshold = fold change 1.5, p value < 0.05). Gene ontology analysis of the overlapping significantly downregulated proteins in micu-1, by PANTHER (GO Consortium, http://geneontology.org/) revealed: protein refolding (GO:0080058, adjusted p-value: 2.19E-02); translation (GO:0006412, adjusted p-value: 1.24E-04); and determination of lifespan (GO:0008340, adjusted p-value: 5.78E-03), as the most enriched biological processes. Upregulated proteins were associated with enrichment of transsulfuration (GO:0019346, adjusted p-value: 4.86E-02). Among the significantly dysregulated proteins, we found 15 ribosomal components and two translation elongation factors (Supplementary Fig. S3E), implying a decreased protein synthesis as predicted by our IPA analysis of RNA sequencing data (Supplementary Figs. S2B–D). We detected significant expression changes of mitochondrial respiratory complexes (Supplementary Figs. S3E–F) and other mitochondrial proteins, including factors that contribute to cristae remodeling (i.e., F54A3.5/MICOS10, MOMA-1/APOOL), as well as mitochondrial or cytosolic enzymes involved in carbon metabolism, such as nucleotide biosynthesis (ADSS-1, AHCY-1), lipid and carbohydrate catabolism (i.e., MDH-2, ALDO-2, GPDH-2, GPD-1) (Supplementary Fig. S3E). Furthermore, micu-1 mutants had a diminished expression of voltage-dependent anion channel VDAC-1 and protein arginine N-methyltransferase PRMT-1 (Supplementary Fig. S3E). The former participates in mitochondrial Ca2+ uptake and ER coupling [68], while the latter is an important regulator of MICU1 activity and its affinity to Ca2+ and to the MCU/EMRE1 subcomplex [14]. Consistent with previous evidence in patient-derived samples [24], we observed aberrant expression of structural proteins, such as cytoskeleton components, extracellular matrix molecules (e.g., collagen) and proteins associated with the nuclear envelope (Supplementary Fig. S3E).
Figure 3.
SPL-1/SGPL1 and VPS-39/VPS39 expression contributes to the lifespan extension of micu-1(null) mutants. (A) Heatmap of statistically upregulated (left) and downregulated (right) proteins in micu-1(bon20) mutants compared to wt. Fold changes are color coded and are indicated on the right of each heatmap. (B–C) Lifespan assay of wt and micu-1(bon20) mutant animals exposed to control (solid lines), (B) vps-39 (dashed lines) and (C) spl-1 (dotted lines) RNAi from adulthood. For wt and micu-1(bon20) on control RNAi, representative curves come from the same experiment and figures were split for the sake of clarity (for each individual experiment, see Supplementary Table S1). (D) WB analysis of zcIs13[hsp-6p::gfp] transgene expression in control and micu-1(bon20) mutant nematodes exposed to the indicated RNAi. Immunoblots were developed using antibodies against GFP and actin (as loading control). Densitometry is reported on the right of each WB panel (Kruskal–Wallis ANOVA, Dunn's multiple comparison test, ∗p < 0.05, n = 3–4). (E) Confocal image of the head of a nematode expressing sqIs11[[lgg-1p::mCherry::GFP::lgg-1+ rol-6]. Statistical analysis of mCherry-positive (left violin graph) and GFP-positive (right violin graph) puncta in wt and micu-1(bon20) animals exposed to control, spl-1 and vsp-39 RNAi. Extracted eggs were grown on RNAi plates until the first day of adulthood. Animals were then exposed for 4 h to DMSO or 50 μg/ml clomipramine (Clo) and immediately imaged for mCherry-positive puncta (Kruskal–Wallis, Dunn's multiple comparison test: ns = not significant, ∗p < 0.05, ∗∗p < 0.01, n = 33–36 animals from 3 independent experiments). (F–G) Heatmaps of differentially regulated proteins in micu-1(bon20) animals exposed to (F) spl-1 and (G) vps-39 RNAi from hatching, compared to control RNAi. (H) Heatmap of common differentially expressed proteins in micu-1(bon20) animals grown on vps-39 or spl-1 RNAi. (I) High-resolution confocal images of 9-day-old control and 10-day-old micu-1(bon20) animals expressing zcIs14[myo-3p::GFP(mit)] transgene, with quantification of mitochondrial elongation. (Kruskal–Wallis ANOVA, Dunn's multiple comparison test: ns = not significant, ∗p < 0.05, ∗∗p < 0.01, n = 46–53 animals from 3 independent experiments, scale bar = 10 μm). RNAi treatment is indicated. (J–K) RT-PCR of (J) vps-39 and (K) spl-1 mRNA expression in micu-1(bon20) carrying atfs-1(tm4525) mutation (gray bars) or exposed to the indicated RNAi from hatching, relative to control RNAi (one-way ANOVA, Tukey's multiple comparisons test: ns = not significant, ∗p < 0.05, ∗∗p < 0.01, n = 3). (L) High-resolution confocal images of 7-day-old nuo-6 (qm200) animals expressing zcIs14[myo-3p::GFP(mt)] transgene, following exposure to control, vps-39, or spl-1 RNAi from hatching. Scale bar = 10 μm. (M) Lifespan assay of nuo-6 (qm200) mutant nematodes grown on control, vps-39 and spl-1 RNAi from hatching.
Taking advantage of C. elegans genetic tractability, we sought to determine the relevance of the upregulated proteins in micu-1 lifespan. For the available 37 RNAi clones, we carried out a small-scale screen and identified dyn-1, vps-39, spl-1 and lem-2 RNAi as potential modifiers of micu-1 survival (Supplementary Fig. S3G). To refine our investigation, we performed epistatic analysis on larger populations and with appropriate biological replicates. Since some of the RNAi (e.g., dyn-1 RNAi) altered C. elegans development, we adopted a RNAi protocol that interferes with gene expression starting at adulthood, even though this approach does not impact early programs in larvae that are often considered to be critical in establishing longevity of mitochondrial mutant nematodes [36,58]. We grew animals until adulthood on control E. coli and then moved adults to plates seeded with dsRNA-expressing bacteria (see materials and methods section for details). Despite the diminished RNAi effect of our experimental paradigm, we observed a significant lifespan reduction in micu-1(null) animals exposed to dyn-1, spl-1 and vps-39 RNAi, while lem-2 and RNAi starting from adulthood did not significantly alter animal survival (Figure 3B,C, Supplementary Figs. S3H–I and Table S1). We also tested these RNAi on wt animals and found that only dyn-1 downregulation considerably affected their survival (Figure 3B,C, Supplementary Figs. S3H–I and Table S1). Because DYN-1 deficiency had this confounding effect on wt, we focused our attention on sphingosine phosphate lyase (SPL-1) and vacuolar protein sorting-associated protein 39 homolog (VPS-39).
SPL-1 is a component of the sphingolipid degradative pathway and cleaves sphingosine 1-phosphase (S1P) into hexadecenal and phosphoethanolamine (Supplementary Fig. S3J), thereby regulating sphingolipid metabolism, intracellular calcium mobilization through S1P and stress response in a ceramide-dependent manner [55,69,70]. Yeast Vps39 is part of the homotypic fusion and vacuole protein sorting (HOPS) complex, which controls membrane tethering of yeast lysosome (i.e., vacuole)-late endosome and yeast lysosome-mitochondria [71]. Moreover, the VPS39-containing HOPS complex modulates intracellular autophagic flux [71] as well as mitochondria degradation (i.e., mitophagy) through the interaction between Vps39 and Tom40 at the outer membrane [72,73]. Based on these considerations, we initially set out to measure zcIs13[hsp-6p::gfp] expression in animals exposed to spl-1 and vps-39 RNAi, since sphingosine and ethanolamine pathways are highly upregulated in micu-1(null) mutants compared to wt (Supplementary Figs. S3J–K). Consistent with previous lines of evidence [55,69,70], spl-1 RNAi enhanced zcIs13[hsp-6p::gfp] expression in control animals (Figure 3D), possibly through increased ceramide availability that would sustain UPRmt [55]. Conversely, micu-1(null) mutants grown on vps-39 and spl-1 RNAi exhibited negligible changes in zcIs13[hsp-6p::gfp] expression compared to control RNAi (Figure 3D), suggesting that UPRmt is fully engaged as a result of SPTL-1 and SPL-1 upregulation (Figure 3A and Supplementary Figs. S3E and S3K), thereby overriding the effect of vps-39 and spl-1 RNAi.
A previous genetic screen demonstrated that dyn-1 and vps-39 are in the same genetic pathway involved in the maturation of phagosomes during the engulfment and removal of dead corpses in adult hermaphrodite gonads [74]. Because of this role of VPS-39/Vps39 in phagosome biogenesis [74] and in yeast autophagy [73], we investigated the autophagic flux in control and micu-1 mutant animals expressing the transgene sqIs11[lgg-1p::mCherry::GFP::lgg-1+ rol-6] [75]. As originally described [75], tandem fluorescent tagged LGG1 is recruited to nascent phagophores and upon fusion with lysosomes, can be degraded into GFP-tagged LGG-1. We grew sqIs11[lgg-1p::mCherry::GFP::lgg-1+ rol-6] expressing animals on control, spl-1 and vps-39 RNAi. Before high-resolution confocal image analysis, adults were briefly exposed to DMSO or to the antidepressant clomipramine as a potent autophagic inhibitor [76,77]. Since GFP is quenched upon autophagosome fusion with lysosomes, we visualized mature autolysosomes stained as red structures [75]. We quantified the number of mCherry-positive puncta in the head of nematodes exposed from hatching to control or RNAi against spl-1 or vps-39 (Figure 3E). We found that downregulation of both spl-1 or vps-39 inhibited autophagosome formation and autophagic flux to the same extent of clomipramine (Figure 3E). To further corroborate our line of evidence, we generated micu-1(bon20)IV;vps-39(ok2442)V/nT1[qIs51](IV;V). However, we could retrieve only a few micu-1;vps-39 double mutant escapers out of thousands of nematodes, suggesting that a vps-39(null) mutation is incompatible with the survival of micu-1(null) C. elegans. Taken together, our findings suggest that SPL-1 and VPS-39 sustain autophagy, possibly by providing phosphatidylethanolamine to phagophore membranes as recently shown in other experimental settings [[78], [79], [80], [81]].
To further investigate the putative synergism between SPL-1 and VPS-39, we exposed micu-1(null) mutants to control, spl-1 and vps-39 RNAi starting from hatching. We then extracted proteins from 5-day-old animals and performed TMT-based MS analysis, using homogenates from control RNAi-treated nematodes as a reference. Based on MS analysis of three biological replicates run in duplicates, we identified 1394 proteins with medium–high confidence (p value ≤ 0.05). We found that spl-1 and vps-39 silenced micu-1 animals had 26 and 50 differentially expressed proteins (Figure 3F,G and Supplementary Table S4), respectively. Among the differentially expressed proteins uniquely found in spl-1 RNAi micu-1 mutants, we measured upregulation of EPPL-1/ETNPPL ethanolamine-phosphate phospho-lyase (i.e., an enzyme that catalyzes the breakdown of phosphoethanolamine) and downregulation of serine palmitoyl transferase SPTL-3/SPTLC2/SPTLC3 (i.e., an ER-localized enzyme that regulates sphingolipid biosynthesis) (Figure 3F–H and Supplementary Figs. S3J–K). This evidence suggests that SPL-1 downregulation decreases sphingolipid signaling and phosphatidylethanolamine synthesis (Supplementary Fig. S3K). Strikingly, ∼60% of the differentially expressed proteins in SPL-1 deficient nematodes had a similar trend in micu-1 mutants exposed to vps-39 RNAi (Figure 3H and Supplementary Figure S3L), further indicating that SPL-1 and VPS-39 act on common pathways. Among the differentially expressed proteins common in the two datasets (Figure 3H and Supplementary Figure S3L), we noticed changes of intracellular signaling cascade components (i.e., GCK-3/STK39, KGB-1/MAPK10/JNK3, PPH-4/PPP4C, PPH-6/PPP6C) along with the elongation factor 2 kinase EFK-1/EEF2K, the latter being an AMPK downstream target that, through inhibition of protein synthesis, can stimulate survival of cells and nematodes in response to nutrient deprivation and sustained energy crisis [82]. Additionally, we observed an increased expression of methionine synthase METR-1/MTR (Figure 3H and Supplementary Figure S3L), an enzyme that by connecting methionine folate cycles and transsulfuration pathway, can stimulate mitochondrial biogenesis as a compensatory mechanism to alleviate lysosomal dysfunction [83]. Other evident changes of evolutionarily conserved proteins included pitrilysin metallopeptidase PITRM1, a matrix-localized peptidase that contributes to mitochondrial proteome homeostasis by degrading aggregate-prone and pre-sequence peptides [84,85], the H+/sialic acid cotransporter SLC17A5, a H+/nitrate cotransporter causally linked to rare forms of lysosomal storage disorders [86], and the V-type proton ATPase UNC-32/ATP6V0A1, a subunit of the lysosomal ATPase that stimulates vacuolar acidification [87] (Figure 3H and Supplementary Figure S3L).
To complete our investigation and frame our findings in the context of mitochondria-to-nucleus “retrograde” signaling, we assessed mitochondrial morphology upon spl-1 and vps-39 silencing and found that spl-1 downregulation had a slight, though statistically not significant effect on mitochondrial network, whereas VPS-39 was required for the proper maintenance of the mitochondrial morphology in micu-1 mutant nematodes (Figure 3I). Next, we investigated the transcriptional regulation of vps-39 and spl-1 in micu-1(null) mutants. Following our previous epistatic analysis of the transcription factors involved in micu-1 longevity (Figure 2H–L), we found that ATFS-1 loss significantly compromised vps-39 expression in micu-1(null) animals, while downregulation of PHA-4, a transcription factor previously described as regulator of autophagy during dietary restriction [65,88], slightly enhanced vps-39 expression (Figure 3J). These data strongly suggest that vps-39 gene expression is under the transcriptional regulation of ATFS-1 and is therefore a component of the ATFS-1-dependent mitochondria-to-nucleus retrograde signaling. Conversely, neither atfs-1(lof) nor downregulation of other transcription factors influenced spl-1 expression (Figure 3K). Of note, a very mild spl-1 upregulation was detected in vps-39 deficient micu-1 mutant nematodes (Figure 3K). Neither vps-39 nor spl-1 RNAi could rescue the germline defects observed in micu-1(null) mutants (Supplementary Figs. S4A–B). Together, our findings describe a previously unknown SPL-1/VPS-39 signaling axis that, through sphingolipid metabolism and autophagy, may contribute to mitochondrial network expansion underlying lifespan-extending programs in micu-1 deficient C. elegans.
Finally, to determine whether spl-1 and vps-39 RNAi could influence mitochondrial maintenance also in other long-lived mitochondrial mutants, we employed nuo-6(qm200) expressing mitochondrial GFP in body wall muscle cells. In line with our data in micu-1(null) animals, we observed that SPL-1 and VPS-39 promoted the proper organization of the mitochondrial network in nuo-6 mutant nematodes (Figure 3L). Notably, spl-1 and vps-39 RNAi reduced nuo-6 longevity (Figure 3M), indicating that SPL-1/VPS-39 signaling may play a role in animals carrying genetic lesions of complex I.
3.4. VPS39 localization to the mitochondria is associated with enhanced autophagic flux in MICU1 deficient HEK293 cells
To gain deeper insights into evolutionarily conserved cellular processes mechanistically linked to MICU1 deficiency, we employed WT and MICU1 KO HEK293 cells as an in vitro mammalian model [89]. Conventional Seahorse measurements showed a clear decreased OCR in MICU1 KO cells grown in either glucose- or galactose-containing media (Supplementary Figs. S5A–B). The diminished mitochondrial respiration and the loss of mitochondrial respiratory complex subunits correlated with an increased extracellular acidification rate as an accepted proxy of enhanced glycolysis (Supplementary Figs. S5C–D). Using a newly validated proximity ligation assay (PLA)-based protocol [32], we performed in situ analysis of mitochondrial CI–CIV proximity as an indicator of mitochondrial respiratory supercomplexes (RSCs). We observed that MICU1 KO cells exhibited less CI, CIV-containing RSCs (Supplementary Fig. S5E), further confirming lesions of the mitochondrial OXPHOS system. Having characterized the bioenergetic profiles of these cells, we moved forward to substantiate our molecular evidence obtained in C. elegans. In WT and MICU1 KO HEK293 cells, we overexpressed mitochondria-targeted GFP and assessed mitochondrial morphology, thereby noticing that MICU1 KO mitochondria had slightly more elongated structures (Figure 4A). High-resolution confocal imaging showed no major changes of mitochondria-endoplasmic reticulum (ER) contacts in MICU1 KO cells compared to controls (Supplementary Fig. S5F). To support this evidence, we performed PLA between the mitochondrial translocase of the outer membrane TOM20 and ER localized KDEL-containing proteins. We quantified PLA dots and observed no differences between WT and MICU1 KO cells (Supplementary Fig. S5G), suggesting that MICU1 deficiency does not alter mitochondria-ER contact sites.
Figure 4.
MICU1 KO stimulates mitochondrial recruitment of VPS39. (A) Confocal images and quantification of mitochondrial elongation in WT and MICU1 KO HEK293 cells overexpressing mitochondrially targeted GFP (Student's t-test, ∗p < 0.05, n = 3, n > 20 cells/condition. Scale bar = 10 μm). (B) Western blot analyses of samples from WT and MICU1 KO cells. Immunoblots were performed using antibodies against MICU1, LC3B, p62, SGPL1 and actin (as loading control). Head arrow indicates lipidated LC3B at ∼16 kDa. Densitometry is reported on the right (fold differences relative to WT cells: Mann Whitney and Student's t-test, ∗p < 0.05, n = 3–5). Data are represented as mean ± S.E.M. (C) Confocal images and quantification of LC3B puncta/cell in WT and MICU1 KO HEK293 cells (Mann–Whitney test, ∗∗p < 0.01, n > 40 cells/condition). Scale bar = 10 μm. DAPI was used to stain nuclei (blue). (D) Confocal imaging of WT and MICU1 KO cells overexpressing mitochondrially targeted GFP (mtGFP) and stained with VPS39 antibody (scale bar = 10 μm). Quantification of VPS39 staining is shown on the right (Student's t-test, ∗∗p < 0.01, n = 28 cells/condition). (E) LysoTracker-stained WT and MICU1 KO HEK293 cells overexpressing mtGFP (scale bar = 10 μm). Lysosome-mitochondria overlap was quantified using Mander's and Pearson's correlation coefficient (Student's t-test, ∗∗p < 0.01, n = 3, WT:54 cells, KO:55 cells). (F) Proximity ligation assay (PLA) between VPS39 and TOM40 in WT and MICU1 KO HEK293 cells overexpressing mtGFP (scale bar = 10 μm). The number of PLA dots per cell was normalized to the mitochondrial area (Mann–Whitney U-test, ∗∗p < 0.01, n = 3, WT: 20 cells, KO: 21 cells).
We ran immunoblot analysis and found that, consistent with our data in nematodes, SGPL1 was considerably upregulated in MICU1 KO cells compared to WT ones (Figure 4B). Strikingly, these cells also had an increased basal level of autophagosome-associated lipidated LC3B [90] and a decreased amount of ubiquitin binding protein p62 (Figure 4B), a well-known receptor that promotes cargo delivery to the autophagosome which is selectively degraded during autophagy [91,92]. Such enhanced autophagic flux was neither linked to altered mTOR/Akt activity nor to obvious changes in AMPK signaling (Supplementary Figs. S6A–B). Consistent with our immunoblots (Figure 4B), high-resolution confocal image analysis revealed an increased number of LC3B-positive structures (Figure 4C) that were accompanied by more VPS39-positive dots in MICU1 KO cells compared to controls (Figure 4D). Next, we stained mitochondria and lysosomes in WT and MICU1 KO cells. We observed that MICU1 KO cells had an increased number of LysoTracker-positive vesicles, which may be interpreted as enhanced lysosomal content, with a consequent expansion of contacts between GFP-positive mitochondria and LysoTracker-stained vesicles (Figure 4E). Since Vps39 interacts with Tom40 and relocalizes to mitochondria in yeast during starvation [73,93], we set up a PLA between TOM40 and VPS39, then quantified PLA dots over the mitochondrial area. Using two independent sets of VPS39 antibodies, we found that VPS39 redistributed to mitochondria in MICU1 KO cells (Figure 4F and data not shown). Taken together, our findings reveal evolutionarily conserved molecular signatures that are mechanistically linked to MICU1 dysfunction. While MICU1 KO does not profoundly affect mitochondria-ER tethering, it increases the number of mitochondria-lysosome contact sites in HEK293 cells. Consistent with our data in C. elegans, MICU1 KO stimulates the autophagic flux in HEK293 cells grown in glucose, promoting VPS39 upregulation and its interaction with mitochondria as previously described in yeast [73].
3.5. SGPL1 controls mitochondria-tethered VPS39 in MICU1 deficient HeLa cells
To address the mechanistic contribution of SGPL1 and VPS39 to MICU1-dependent processes, we attempted to genetically modulate their expression using synthetic small interfering RNAs (siRNAs). However, we could not obtain sufficient downregulation of the two target genes due to the fast replication rate of HEK293 cells. To overcome this limitation, we employed tumorigenic HeLa cells, in which we manipulated gene expression in a highly efficient manner (Figure 5A,B and Supplementary Fig. S6C). To be consistent with our study in HEK293 cells, we carried out experiments in HeLa cells grown in glucose-containing medium. We found that MICU1 deficient HeLa cells exhibited a decreased OCR, although milder compared to cells grown in galactose-containing medium (Figure 5C). In standard growing conditions, MICU1 downregulation stimulated LC3B lipidation in HeLa cells, although it did not result in a significant decrease of p62 levels (Figure 5B). Notably, siRNA against VPS39 abrogated LC3B lipidation in MICU1 deficient cells, while it upregulated lipidated LC3B in control HeLa cells (Figure 5B). No significant differences of p62 levels were observed compared to controls, possibly because autophagy is not highly upregulated in cells grown in glucose- and serum-containing medium [94,95]. Our interpretation of this dataset is that VPS39 influences MICU1-dependent recruitment of LC3B to nascent phagophores. Having profiled the bioenergetics and autophagy levels of MICU1 deficient HeLa cells, we set out to confirm the SGPL1/VPS39 signaling axis and its role in cellular homeostasis (Figure 5D), as previously demonstrated in our experiments using C. elegans and HEK293 cells. To do so, we performed PLA (TOM40+VPS39) staining followed by high-resolution confocal image analysis. We quantified the number of PLA (TOM40+VPS39) dots and found that siRNA against SGPL1 reduced VPS39 proximity to TOM40 (Figure 5E,F), further supporting our hypothesis that SGPL1 activity is required for VPS39 association to mitochondria (Figure 5D). We transfected HeLa cells with mitochondrial GFP and siRNAs. We found that MICU1 downregulation enhanced the colocalization of LysoTracker-positive structures with GFP-labelled mitochondria (Figure 5G). However, VPS39 downregulation impaired lysosome-mitochondria contacts in MICU1 deficient HeLa cells (Figure 5G), resulting in aberrant mitochondrial morphology as observed in nematodes. Together, this experimental evidence mechanistically links VPS39 localization in proximity of the mitochondria with the remodeling of mitochondria-lysosome contact sites in MICU1 deficient cells.
Figure 5.
SGPL1 influences mitochondrial recruitment of VPS39 in mitochondrial deficient cells. (A) Western blot analyses of samples from HeLa cells transfected with siRNAs against VPS39 (siVPS39) and MICU1 (siMICU1). Scramble siRNA was used as control. Immunoblots were developed using antibodies against MICU1 and actin (as loading control). (B) Western blot analyses of HeLa cells transfected with scramble or siRNA against MICU1 and/or VPS39. Immunoblots were developed using antibodies against LC3B (head arrow indicates lipidated LC3B), p62 and actin (as loading control). Densitometry is reported on the right (Student's t-test, ∗p < 0.05, n = 3). Data are represented as mean ± S.E.M. (C) OCR of HeLa cells transfected with siRNAs against MICU1 and grown in glucose- or galactose-containing medium (n = 1). (D) Schematic representation of SGPL1-dependent recruitment of VPS39 to the mitochondria and its expected effects on lysosome-mitochondria contacts, autophagy and mitochondrial homeostasis. (E) Representative immunoblots of HeLa cells transfected with siRNA against MICU1. Immunoblots were developed using antibodies against MICU1, SGPL1 and actin (as loading control). Densitometry is reported on the right (Student's t-test, ∗p < 0.05, n = 3). (F) Proximity ligation assay (PLA) of mtGFP-overexpressing HeLa cells transfected with scramble, siRNAs against MICU1 (siMICU1) and/or SGPL1 (siSGPL1). PLA dots per cell were normalized to the mitochondrial area (Kruskal–Wallis test, ∗∗∗∗p < 0.0001, n = 2, scramble: 38 cells, siMICU1: 36 cells, siMICU1 + siSGPL1: 40 cells). Scale bar = 10 μm. Representative WB analysis of siRNA transfected HeLa cells is reported on top right. (G) LysoTracker staining in mtGFP-overexpressing HeLa cells transfected with scramble, siMICU1 and/or siVPS39. Mitochondrial morphology was analyzed based on the mtGFP-signal and lysosome-mitochondria overlap was quantified via Mander's coefficient (Kruskal–Wallis test, ∗∗p < 0.01, n = 3, scramble: 42 cells, siMICU1: 49 cells, siMICU1+siVPS39: 54 cells). Scale bar = 10 μm. (H) Schematic representation of the transgenic mice and derived muscle tissues used in this work. Aifm1KI/Y are males carrying the disease-causing Aifm1(R200 del) mutation, whereas Aifm1wt/Y are wt littermates. (I) PLA (TOM40+VPS39) staining of muscle sections derived from 6-month-old mice. Mitochondria were counterstained using an antibody against TOM20 (scale bar = 10 μm). Quantification of the PLA dots/μm2 mitochondria is reported on the right (Mann–Whitney U-test, ∗p < 0.05, n = 2 mice/genotype with 10 muscle fibers/mouse).
Finally, to demonstrate that these newly identified molecular signatures may be commonly observed in cells carrying mitochondrial lesions, we employed muscle tissues from wt and transgenic mice expressing a disease-causing Aifm1(R200 del) mutation (Figure 5H) [28,33]. These knockin mice develop severe myopathy due to apoptosis-inducing factor (AIF) loss and consequent aberrant expression of ETC components [32,33,49,96]. We ran immunoblot analysis and showed that AIF loss stimulated VPS39 expression in 6-month-old muscles (Supplementary Fig. S6D). We carried out PLA staining and quantified TOM40-VSP39 proximity in control and AIF deficient muscle fibers. In symptomatic Aifm1(R200 del) knockin animals, we found that OXPHOS deficiency promoted VPS39 redistribution to the mitochondria (Figure 5I), which was reflected by a higher number of PLA dots compared to controls. Thus, our cross-species analysis indicates that SGPL1-dependent redistribution of VPS39 to mitochondria may be a detectable common signature in cells carrying mitochondrial lesions.
4. Discussion
Mitochondrial energy production is finely tuned to accurately interpret cellular needs. During embryogenesis or in tumors, dividing cells employ mitochondria to produce biomass, while they generate a large amount of ATP by redirecting glucose and other substrates to the glycolytic pathway. Conversely, ATP is mostly generated through mitochondrial OXPHOS in cells undergoing postmitotic differentiation or grown in presence of non-fermenting carbon substrates (e.g., galactose) [[97], [98], [99]]. Such plasticity in adjusting mitochondrial bioenergetics is a key adaptive mechanism that eukaryotic cells exploit in response to environmental changes, altered nutritional conditions, toxic stimuli and stress signals [41]. To coordinate mitochondrial bioenergetics, the nuclear genome must encode thousands of proteins that, upon correct import and folding, potentiate mitochondrial function and eventually scale up mitochondrial mass. This nucleus-to-mitochondria communication, generally known as anterograde signaling [100], depends on a sophisticated array of transcription factors regulating the functional properties of the organelles. To accurately and dynamically coordinate such broad metabolic rewiring, the expression of nuclear-encoded mitochondrial proteins triggers evolutionarily conserved mitochondria-to-nucleus “retrograde” signals that act as necessary inputs to adjust the intensity and temporal resolution of the anterograde signaling [41,42,52,101]. According to the physiological context, this highly interconnected regulatory network of communication between the nucleus and mitochondria ultimately determines cell division, growth, lineage commitment and differentiation. Notably, this bidirectional regulation of the mitochondrial proteome represents also a protective mechanism to adjust mitochondrial activity and homeostasis upon injury, ETC inhibition and stress due to the accumulation of unfolded proteins or other toxic species. In this regard, mitochondria-to-nucleus “retrograde” signals, including the unfolded protein response (UPRmt) [102,103], may confer healthspan and longevity of an organism when they occur within a certain time window during development [36,37,46]. Although UPRmt is not a predictor of C. elegans lifespan [104], its transcriptional inhibition can abrogate lifespan-extending programs in animals carrying genetic lesions or exposed to mitochondrial toxins [45,57,58,[105], [106], [107], [108]]. By failing in responding to the necessary adjustment of mitochondrial activity, cells lose their fitness and experience severe damage because protective compensatory mechanisms are not sufficient to prevent the accumulation of toxic species. While UPRmt is essential for the expansion of the mitochondrial network during development and in development-dependent processes underlying longevity [36,58,109], its contribution becomes less obvious in human pathology. Albeit these mechanisms help to overcome mitochondrial dysfunction in nematodes, the scenario is different in higher organisms, since these processes may become a maladaptive response that compromises cellular homeostasis in organs suffering from a chronic bioenergetic crisis [110]. As a consequence of increased mitochondrial mass due to uncontrolled biogenesis or impaired removal of defective organelles, cells are forced to balance their transcriptional output with compensatory processes that should prevent the progressive accumulation of damaged mitochondria, ultimately depleting the already scarce resources. Because of their clinical implications in metabolic disorders, these poorly elucidated processes are extremely relevant in biomedicine and deserve further attention.
We herein investigate novel molecular mechanisms linked to mitochondrial dysfunction by employing a C. elegans model of MICU1 deficiency, given the strong relevance of MICU1/MICU2 in human pathophysiology [[21], [22], [23], [24],29]. We demonstrate that micu-1(null) mutations lead to developmental defects, including abnormal cell death in the mitotic and pachytene zones of the gonads, impaired oocyte production and slow growth. As in higher organisms, we report that altered expression of OXPHOS subunits is linked to diminished mitochondrial respiration in C. elegans. Similar to other mitochondrial mutants, the significant lifespan extension of micu-1(null) nematodes requires the establishment of a mitochondria-to-nucleus stress response that relies on various transcription factors (i.e., ATFS-1, SKN-1, HSF-1 and PHA-4), whereas it is not dependent on components of the insulin/IGF-1 cascade (i.e., AGE-1 and DAF-16). While MICU-1 deficiency induces mitochondrial dysfunction and pro-longevity stress response, genetic ablation of micu-1 negatively affects the proper maintenance of the germline to a level which is not comparable to other mitochondrial mutants that have been previously described. Another additional intriguing aspect is that neither the lifespan extension of micu-1(null) nematodes nor the degenerative processes in the meiotic zone or the transcriptional control of pro-longevity genes depend on a functional MCU complex. Thus, the dispensable MCU contribution to the observed phenotypes prompted us to further investigate MICU-1/MICU1 function beyond its widely reported participation in Ca2+ signaling. In this regard, our quantitative proteomics and epistatic analyses unveil previously underestimated processes in which MICU-1/MICU1 is engaged. We report that micu-1(null) mutations stimulate the expression of sphingosine phosphate lyase SPL-1/SGPL1 (i.e., an enzyme involved in sphingolipid degradation), which possibly acts upstream the mitochondrial network expansion underlying longevity of mitochondrial mutant nematodes. Given that the basal mitochondrial matrix Ca2+ is comparable between control and micu-1(null) mutant animals under resting conditions [38], one possible explanation is that SPL-1 upregulation may also act as a compensatory mechanism to prevent excessive Ca2+ mobilization, since SPL-1-mediated degradation of S1P would attenuate Ca2+ release from intracellular stores (e.g., ER) [70]. As an alternative scenario, SPL-1 upregulation may stimulate phosphoethanolamine biosynthesis (as suggested by our proteomics) and trafficking between the ER and mitochondria, with the contact points between the two intracellular compartments acting as physical bridges necessary to sustain the transfer of phospholipids to lysosomes and/or nascent phagophores containing membrane anchored lipidated LGG-1/LC3 [79]. In support of this hypothesis and in line with previous evidence in other experimental models [78,80,81], genetic inhibition of spl-1 diminishes the autophagic flux in C. elegans, possibly because it exhausts mitochondrial membranes required to sustain autophagosome biogenesis in a system chronically under energy crisis and forced to recycle nutrients as shown in cultured cells [81]. It is certainly exciting to find that MICU-1/MICU1 deficiency induces SPL-1/SGPL1 and VPS-39/VPS39 upregulation in both C. elegans and mammalian cells. In this regard, our study conclusively describes how the mitochondria-to-nucleus “retrograde” program can expand mitochondrial mass and efficiently support their homeostasis [58,109]. We show that the bZIP transcription factor ATFS-1 regulates vps-39 expression, meaning that this gene is an integral component of those longevity programs associated with the “mitochondrial threshold effect” [39]. As unveiled by our cross-species investigation, we mechanistically establish that SGPL1 stimulates mitochondria-tethered VPS39 to contribute to functional remodeling of intracellular organelle interactions. Specifically, we conclusively elucidate that decreased SGPL1 expression inhibits the binding of VPS39 to TOM40, while VPS-39/VPS39 downregulation compromises the maintenance of the expanded mitochondrial network in MICU-1/MICU1 deficient cells, thereby altering the contacts between mitochondria and lysosomes. Of note, the increased VPS39 expression and localization to mitochondria does not have a profound effect on ER-mitochondria tethering, probably also because of mitochondrial elongation in response to sustained autophagy as previously reported [111]. We speculate that, as shown in Saccharomyces cerevisiae [73,93], mitochondria-tethered VPS39 may influence lipid exchange and eventually mitophagy. Consistent with the role of SPL-1/SGPL1 in autophagy (our data and [78]), we report that VPS-39/VPS39 downregulation diminishes the autophagosome numbers in MICU-1 deficient nematodes and the autophagic flux in cells grown in glucose- and serum-containing medium (i.e., not under starving conditions).
In the context of human pathophysiology, we speculate that MCU inhibition would not be sufficient as a therapeutic intervention for inherited mitochondrial diseases with altered MICU1/MICU2 function. Although difficult, it would be very informative to test whether manipulation of the sphingosine pathway by ethanolamine supplementation may have a beneficial effect in patients. Furthermore, our proof-of-principle evidence in nematodes and transgenic mice suggests that the importance of our findings may go beyond diseases linked to MICU1/MICU2 deficiency and be relevant to other metabolic disorders. Future studies will explore and clarify this possibility in detail.
5. Conclusion
In summary, we propose that the SGPL1/VPS39 axis contributes to mitochondria-nucleus “retrograde” signals and deciphers the energetic states of the cell, thereby attenuating maladaptive processes that are adopted to counteract aberrant mitochondrial OXPHOS.
Author contribution
Conceptualization, JJ, LW, DB; Methodology, JJ, LW, DB; Validation and Investigation, JJ, LW, ES, AnPe, YW, AnPi, DG, SS, MS, DB; Formal Analysis, JJ, LW, ES; Resources, KH, CEH, JW, AR, JLS, DE; Writing, DB; Visualization, LW, ES, DB; Supervision, LW, DB; Project Administration, DB; Funding Acquisition, PN and DB.
Acknowledgements
We wish to thank our DZNE colleagues at LMF; Prof. Ina Vorberg (DZNE), Prof. Donato Di Monte (DZNE) and Prof. Gerhild van Echten-Deckert (University of Bonn) for their useful suggestions; Ms. Christiane Bartling-Kirsch (DZNE), Ms. Anaïs Marsal-Cots (DZNE), Ms. Elena Schlimgen, Mr. Nick Atanelov, Ms. Elide Lo Cacciato and Dr. Tannaz Norizadeh Abbariki for their technical assistance. We thank Dr. Diego De Stefani and Dr. Agnese De Mario (University of Padova, Padova, Italy) for providing MICU1 KO HEK293 cells. This research was supported by the DZNE institutional budget, the CoEN (Carbon-Model, 3018) initiative and the Helmholtz cross-program topic “Aging and Metabolic Programming (AMPro)”. PN and DB are members of the DFG Cluster of Excellence ImmunoSensation funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC2151 – 390873048. JJ and DB are members of the Mitochondrial Dysfunction in Parkinson's Consortium (PD-MitoQUANT). PD-MitoQUANT has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No. 821522. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101503.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
Supplementary figure S1 (relative to figure 1). (A-B) Protein blast analysis of human (A) MICU3 and (B) SLC25A23 to C. elegans proteome. (C-E) CRISPR/Cas9 strategies for the generation of (C) emre-1(bon78), (D) micu-1(bon20) and (E) micu-1(bon77) alleles. The resulting mutated sequences were confirmed by DNA sequencing. (F) Thrashing assay of micu-1(bon20) and micu-1(bon77) compared to wt animals (one-way ANOVA, Tukey’s multiple comparisons test: ∗∗∗∗p<0.0001, ∗∗∗p<0.001, n=2). (G) Developmental timing of wt, micu-1 (bon20), and nuo-6(qm200) animals. Bars represent the percentage of animals in each stage of development at the indicated time point (n=150 animals per group). On the x axis, numbers are hours since eggs were transferred onto bacteria-seeded NGM plates. (H) Representative confocal images of wt and micu-1(bon20) gonads stained with DAPI. The distal tip cell is indicated with the yellow arrow (scale bar=50 m). Quantification of mitotic zone nuclei/gonad arm is reported on the right (one-way ANOVA, Tukey’s multiple comparisons test: ∗∗∗∗p<0.0001; n=8-13 animals). (I) Representative confocal images of micu-1(bon20);bcIs39 animals expressing CED-1::GFP fusion protein in the gonadal sheath cells, highlighting engulfed apoptotic cells within the gonad (scale bar=10 m). Dotted lines indicate the boundary of the gonad arm. (J) Quantification of number of eggs, percentage of hatching and hatched offspring for the indicated genotypes (Kruskal-Wallis ANOVA, Dunn’s multiple comparison test: ns=not significant, ∗∗p<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001; n=30 animals). For the hatched offspring, the genotype was scored based on the GFP expression by the nT1[qIs51] balancer. (K) Representative confocal image of animals expressing zcIs14[myo-3p::GFP(mit)] transgene (scale bar=10 m). The vulva muscles are visualized as green-positive strictures and show no major differences between micu-1(bon20) and micu-1(bon20)/nT1[qIs51] animals. (L-M) Representative survival curves of wt and micu-1(bon20) mutant animals grown on control, (L) moma-1 RNAi and (M) eat-3 RNAi.
figs2.
Supplementary figure S2 (relative to figure 2). (A) Heatmap showing differentially expressed mitochondrial genes in micu-1 mutant nematodes. Mitochondrial genes were identified based on overlay of human homologues with MitoCarta. ETC components are highlighted in red. (B-D) IPA of the transcriptome of micu-1(bon20) vs wt nematodes reveals that (B) EIF2 signaling, (C) eIF4 and p70S6K signaling and (D) xenobiotic signaling are overrepresented in micu-1 mutants. In pink: genes identified in the transcriptome. In red: genes found to be significantly upregulated. In green: genes found to be significantly downregulated. In grey: genes identified in the transcriptome but which were not significantly altered. In white: genes which participate in the pathways but which were not identified in the transcriptome. Shades of red/green proportional to the log2(fold change), as calculated by IPA. (E) RT-PCR of sod-3 expression in glp-1 and glp-1;micu-1 mutants grown on control and daf-16 RNAi at 25 °C (Student’s t-test, ∗p<0.05, ∗∗∗p<0.001; n=3).
figs3.
Supplementary figure S3 (relative to figure 3). Volcano plot of differentially regulated proteins in micu-1(null) mutants compared to wt. (B) Chart depicting sample abundances after normalization of TMT Quantitation channels following LC-MS/MS. Wild type and micu-1 (bon20) samples were labelled with 126 and 127N tandem mass tags (TMT), respectively, and run in triplicate (F1-F3). (C) Evaluation of TMT labelling efficiency. Bar chart indicates that MS spectra from unlabeled peptides (No Quan Labels) constituted less than 2.5% of the signal and most of the peptide spectral matches (PSMs) were used for quantitation (<Not Set>). (D) Precision of masses for PSMs. Histogram shows the distribution of PSMs, most of which were identified with high precision (change of mass of≤5 ppm). (E) Summarizing table of dysregulated proteins in micu-1(bon20) animals compared to wt ones. Human orthologs are indicated where possible. Upregulated proteins are reported in red, whereas downregulated proteins are reported in green. The right column describes the annotated biological functions of the human proteins. (F) Heatmap of OXPHOS system components differentially regulated in micu-1(null) mutants compared to wt. In bold, ETC subunits for which the fold change reaches statistical significance. (G) Median lifespan (relative to control: black bar) of micu-1(bon20) animals grown on the indicated RNAi. Out of the 37 clones, 4 RNAi (red bars) decreased micu-1 survival by at least 25%. (H-I) Representative survival curves of wt and micu-1(bon20) animals exposed to control (solid lines), (H) dyn-1 RNAi (dotted line) and (I) lem-2 RNAi (dotted line) from adulthood. (J) Schematic representation of the sphingolipid signaling pathway. Enzymes under study are reported in red. (K) IPA of significantly dysregulated genes in micu-1(bon20) vs wt nematodes reveals enhanced ethanolamine/phosphoethanolamine biosynthesis. In red: upregulated genes. In green: downregulated genes. In blue/orange: molecules predicted to be decreased/increased, as calculated by IPA. (L) Summarizing table of common dysregulated proteins in micu-1(bon20) animals exposed to vps-39 or spl-1 RNAi from hatching, with human orthologs. Color code refers to the fold changes (as reported in Figure 3H).
figs4.
Supplementary figure S4 (relative to figure 3). (A) Quantification of mitotic nuclei/gonad arm in micu-1(bon20) animals grown on control, vps-39 and spl-1 RNAi bacteria (one way-ANOVA, Dunn’s multiple comparison test: ns=not significant; n=18-22 animals). (B) Quantification of total number of eggs, percentage of egg hatching and percentage of nT1[qIs51] negative offspring from micu-1(bon20)/nT1[qIs51] animals grown on control, vps-39 and spl-1 RNAi bacteria (Kruskal-Wallis test, Dunn’s multiple comparison test: ns=not significant, ∗p<0.05; n=30 animals).
figs5.
Supplementary figure S5 (relative to figure 4). (A-B) Representative Seahorse experiment reporting OCR of WT and MICU1 KO HEK293 cells in (A) glucose- or (B) galactose-containing medium. Statistical analyses of OCR at baseline and upon FCCP treatment are reported on the right (mean ± S.E.M; two-way RM ANOVA, Bonferroni’s multiple comparisons test, ns=not significant, ∗p<0.05, ∗∗p<0.01, n=3). (C) Extra cellular acidification rate for WT and MICU1 KO HEK293 cells (mean ± S.E.M; Mann-Whitney U-test, ∗∗p<0.01, n=3). (D) Immunoblots of samples from WT and MICU1 KO HEK293 cells. WBs were developed using the indicated antibodies. Densitometry is reported on the right (Student’s t-test, ∗∗∗p<0.001, n=5). (E) PLA between NDUFB8 and MTCO1 in WT and MICU1 KO HEK293 cells overexpressing mtGFP. PLA dots per cells were normalized to mitochondrial area (Student’s t-test, ∗∗p<0.01, n=3, WT: 24 cells, KO: 20 cells). Scale bar=5 m. (F) Assessment of ER-mitochondria co-localization in WT and MICU1 KO HEK293 cells overexpressing mitochondrially-targeted dsRed (mtDsRed) and ER-targeted EGFP (ER-GFP) using Mander’s and Pearson’s correlation coefficient (n=2, WT: 21 cells, KO: 23 cells. Scale bar=5 m). (G) PLA between KDEL and TOM20. ER-mitochondria contacts were quantified in WT and MICU1 KO HEK293 cells overexpressing mtGFP. PLA dots per cell were normalized to the mitochondrial area (n=4, WT: 36 cells, KO: 36 cells. Scale bar=5 m).
figs6.
Supplementary figure S6 (relative to figure 4-5). (A) Immunoblot analysis of phospho-Akt (Ser473), total AKT, phospho-PRAS40 (T246), total PRAS40, phospho-P70-S6K(Thr389), total P70-S6K, phospho-4E-BP1 (Thr37/Thr46) and total 4E-BP1 using samples from WT and MICU1 KO HEK293 cells. Densitometry is reported on the right (relative to WT, n=3, student’s t-test, ns=not significant). Data are represented as mean ± S.E.M. (B) Immunoblot analysis of phospho-acetyl-CoA carboxylase (Ser79) and phospho-AMPK (Thr172). Actin and tubulin were used as loading controls. Densitometry is reported on the right (relative to WT, n=3, student’s t-test, ns= not significant). Data are represented as mean ± S.E.M. (C) VPS39 immunofluorescence staining of TOM20 and VPS39 in HeLa cells treated with siRNAs against VPS39 and MICU1. DAPI was used to stain nuclei. Mean fluorescence is relative to VPS39 intensity (one-way ANOVA, Bonferroni’s multiple comparisons test, ∗∗∗p<0.001, n=1, scramble: 19 cells, siVPS39: 21 cells, siMICU1: 12 cells. Scale bar=10 m). (D) Western blot analysis of homogenates from muscle tissues from 6-month-old Aifm1KI/Y and Aifm1wt/Y mice. Immunoblots were developed using antibodies against VPS39, AIF and GAPDH (as loading control).
References
- 1.Kirichok Y., Krapivinsky G., Clapham D.E. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427(6972):360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 2.Vasington F.D., Murphy J.V. Ca ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. Journal of Biological Chemistry. 1962;237:2670–2677. [PubMed] [Google Scholar]
- 3.Bick A.G., Calvo S.E., Mootha V.K. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012;336(6083):886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pittis A.A., Goh V., Cebrian-Serrano A., Wettmarshausen J., Perocchi F., Gabaldon T. Discovery of EMRE in fungi resolves the true evolutionary history of the mitochondrial calcium uniporter. Nature Communications. 2020;11(1):4031. doi: 10.1038/s41467-020-17705-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Stefani D., Rizzuto R., Pozzan T. Enjoy the trip: calcium in mitochondria back and forth. Annual Review of Biochemistry. 2016;85:161–192. doi: 10.1146/annurev-biochem-060614-034216. [DOI] [PubMed] [Google Scholar]
- 6.Baughman J.M., Perocchi F., Girgis H.S., Plovanich M., Belcher-Timme C.A., Sancak Y., et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476(7360):341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Stefani D., Raffaello A., Teardo E., Szabo I., Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476(7360):336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancak Y., Markhard A.L., Kitami T., Kovacs-Bogdan E., Kamer K.J., Udeshi N.D., et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342(6164):1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J.C., Syder N.C., Ghorashi N.S., Willingham T.B., Parks R.J., Sun J., et al. EMRE is essential for mitochondrial calcium uniporter activity in a mouse model. JCI Insight. 2020 doi: 10.1172/jci.insight.134063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Nguyen N.X., She J., Zeng W., Yang Y., Bai X.C., et al. Structural mechanism of EMRE-dependent gating of the human mitochondrial calcium uniporter. Cell. 2019;177(5):1252–1261. doi: 10.1016/j.cell.2019.03.050. e1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vais H., Mallilankaraman K., Mak D.D., Hoff H., Payne R., Tanis J.E., et al. EMRE is a matrix Ca(2+) sensor that governs gatekeeping of the mitochondrial Ca(2+) uniporter. Cell Reports. 2016;14(3):403–410. doi: 10.1016/j.celrep.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perocchi F., Gohil V.M., Girgis H.S., Bao X.R., McCombs J.E., Palmer A.E., et al. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;467(7313):291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vais H., Payne R., Paudel U., Li C., Foskett J.K. Coupled transmembrane mechanisms control MCU-mediated mitochondrial Ca(2+) uptake. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(35):21731–21739. doi: 10.1073/pnas.2005976117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottschalk B., Klec C., Leitinger G., Bernhart E., Rost R., Bischof H., et al. MICU1 controls cristae junction and spatially anchors mitochondrial Ca(2+) uniporter complex. Nature Communications. 2019;10(1):3732. doi: 10.1038/s41467-019-11692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J.C., Liu J., Holmstrom K.M., Menazza S., Parks R.J., Fergusson M.M., et al. MICU1 serves as a molecular gatekeeper to prevent in vivo mitochondrial calcium overload. Cell Reports. 2016;16(6):1561–1573. doi: 10.1016/j.celrep.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antony A.N., Paillard M., Moffat C., Juskeviciute E., Correnti J., Bolon B., et al. MICU1 regulation of mitochondrial Ca(2+) uptake dictates survival and tissue regeneration. Nature Communications. 2016;7:10955. doi: 10.1038/ncomms10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Csordas G., Golenar T., Seifert E.L., Kamer K.J., Sancak Y., Perocchi F., et al. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca(2)(+) uniporter. Cell Metabolism. 2013;17(6):976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallilankaraman K., Doonan P., Cardenas C., Chandramoorthy H.C., Muller M., Miller R., et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell. 2012;151(3):630–644. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orrenius S., Zhivotovsky B., Nicotera P. Regulation of cell death: the calcium-apoptosis link. Nature Reviews Molecular Cell Biology. 2003;4(7):552–565. doi: 10.1038/nrm1150. [DOI] [PubMed] [Google Scholar]
- 20.Nicotera P., Orrenius S. Ca2+ and cell death. Annals of the New York Academy of Sciences. 1992;648:17–27. doi: 10.1111/j.1749-6632.1992.tb24520.x. [DOI] [PubMed] [Google Scholar]
- 21.Shamseldin H.E., Alasmari A., Salih M.A., Samman M.M., Mian S.A., Alshidi T., et al. A null mutation in MICU2 causes abnormal mitochondrial calcium homeostasis and a severe neurodevelopmental disorder. Brain. 2017;140(11):2806–2813. doi: 10.1093/brain/awx237. [DOI] [PubMed] [Google Scholar]
- 22.Lewis-Smith D., Kamer K.J., Griffin H., Childs A.M., Pysden K., Titov D., et al. Homozygous deletion in MICU1 presenting with fatigue and lethargy in childhood. Neurology Genetics. 2016;2(2):e59. doi: 10.1212/NXG.0000000000000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Logan C.V., Szabadkai G., Sharpe J.A., Parry D.A., Torelli S., Childs A.M., et al. Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nature Genetics. 2014;46(2):188–193. doi: 10.1038/ng.2851. [DOI] [PubMed] [Google Scholar]
- 24.Kohlschmidt N., Elbracht M., Czech A., Hausler M., Phan V., Topf A., et al. Molecular pathophysiology of human MICU1-deficiency. Neuropathology and Applied Neurobiology. 2021 doi: 10.1111/nan.12694. [DOI] [PubMed] [Google Scholar]
- 25.Frazier A.E., Thorburn D.R., Compton A.G. Mitochondrial energy generation disorders: genes, mechanisms, and clues to pathology. Journal of Biological Chemistry. 2019;294(14):5386–5395. doi: 10.1074/jbc.R117.809194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorman G.S., Chinnery P.F., DiMauro S., Hirano M., Koga Y., McFarland R., et al. Mitochondrial diseases. Nature Reviews Disease Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- 27.Koopman W.J., Beyrath J., Fung C.W., Koene S., Rodenburg R.J., Willems P.H., et al. Mitochondrial disorders in children: toward development of small-molecule treatment strategies. EMBO Molecular Medicine. 2016;8(4):311–327. doi: 10.15252/emmm.201506131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bano D., Prehn J.H.M. Apoptosis-inducing factor (AIF) in physiology and disease: the tale of a repented natural born killer. EBioMedicine. 2018;30:29–37. doi: 10.1016/j.ebiom.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilton K.M., Morales-Rosado J.A., Selcen D., Muthusamy K., Ewing S., Agre K., et al. Developmental brain abnormalities and acute encephalopathy in a patient with myopathy with extrapyramidal signs secondary to pathogenic variants in MICU1. JIMD Report. 2020;53(1):22–28. doi: 10.1002/jmd2.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan X., Liu J., Nguyen T., Liu C., Sun J., Teng Y., et al. The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nature Cell Biology. 2013;15(12):1464–1472. doi: 10.1038/ncb2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tufi R., Gleeson T.P., von Stockum S., Hewitt V.L., Lee J.J., Terriente-Felix A., et al. Comprehensive genetic characterization of mitochondrial Ca(2+) uniporter components reveals their different physiological requirements in vivo. Cell Reports. 2019;27(5):1541–1550. doi: 10.1016/j.celrep.2019.04.033. e1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertan F., Wischhof L., Scifo E., Guranda M., Jackson J., Marsal-Cots A., et al. Comparative profiling of N-respirasomes predicts aberrant mitochondrial bioenergetics at single-cell resolution. bioRxiv. 2020 [Google Scholar]
- 33.Wischhof L., Gioran A., Sonntag-Bensch D., Piazzesi A., Stork M., Nicotera P., et al. A disease-associated Aifm1 variant induces severe myopathy in knockin mice. Molecular Metabolism. 2018;13:10–23. doi: 10.1016/j.molmet.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scifo E., Szwajda A., Soliymani R., Pezzini F., Bianchi M., Dapkunas A., et al. Proteomic analysis of the palmitoyl protein thioesterase 1 interactome in SH-SY5Y human neuroblastoma cells. Journal of Proteomics. 2015;123:42–53. doi: 10.1016/j.jprot.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 35.Xu S., Chisholm A.D. C. elegans epidermal wounding induces a mitochondrial ROS burst that promotes wound repair. Developmental Cell. 2014;31(1):48–60. doi: 10.1016/j.devcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dillin A., Hsu A.L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A.G., et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298(5602):2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 37.Rea S.L., Ventura N., Johnson T.E. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biology. 2007;5(10):e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang N.H., Kim K.W., Xu S., Blazie S.M., Yee B.A., Yeo G.W., et al. The mRNA decay factor CAR-1/LSM14 regulates axon regeneration via mitochondrial calcium dynamics. Current Biology. 2020;30(5):865–876. doi: 10.1016/j.cub.2019.12.061. e867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossignol R., Faustin B., Rocher C., Malgat M., Mazat J.P., Letellier T. Mitochondrial threshold effects. Biochemical Journal. 2003;370(Pt 3):751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Hekimi S. Mitochondrial dysfunction and longevity in animals: untangling the knot. Science. 2015;350(6265):1204–1207. doi: 10.1126/science.aac4357. [DOI] [PubMed] [Google Scholar]
- 41.Bar-Ziv R., Bolas T., Dillin A. Systemic effects of mitochondrial stress. EMBO Reports. 2020;21(6):e50094. doi: 10.15252/embr.202050094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Y.F., Haynes C.M. Metabolism and the UPR(mt) Molecular Cell. 2016;61(5):677–682. doi: 10.1016/j.molcel.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piazzesi A., Wang Y., Jackson J., Wischhof L., Zeisler-Diehl V., Scifo E., et al. CEST-2.2 overexpression alters lipid metabolism and extends longevity of mitochondrial mutants. EMBO Reports. 2022 doi: 10.15252/embr.202152606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gioran A., Piazzesi A., Bertan F., Schroer J., Wischhof L., Nicotera P., et al. Multi-omics identify xanthine as a pro-survival metabolite for nematodes with mitochondrial dysfunction. The EMBO Journal. 2019;38(6) doi: 10.15252/embj.201899558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piazzesi A., Papic D., Bertan F., Salomoni P., Nicotera P., Bano D. Replication-independent histone variant H3.3 controls animal lifespan through the regulation of pro-longevity transcriptional programs. Cell Reports. 2016;17(4):987–996. doi: 10.1016/j.celrep.2016.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng J., Bussiere F., Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Developmental Cell. 2001;1(5):633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 47.Lee S.S., Lee R.Y., Fraser A.G., Kamath R.S., Ahringer J., Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature Genetics. 2003;33(1):40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 48.Chaudhari S.N., Kipreos E.T. Increased mitochondrial fusion allows the survival of older animals in diverse C. elegans longevity pathways. Nature Communications. 2017;8(1):182. doi: 10.1038/s41467-017-00274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Troulinaki K., Buttner S., Marsal Cots A., Maida S., Meyer K., Bertan F., et al. WAH-1/AIF regulates mitochondrial oxidative phosphorylation in the nematode Caenorhabditis elegans. Cell Death & Disease. 2018;4:2. doi: 10.1038/s41420-017-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanazawa T., Zappaterra M.D., Hasegawa A., Wright A.P., Newman-Smith E.D., Buttle K.F., et al. The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genetics. 2008;4(2):e1000022. doi: 10.1371/journal.pgen.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Head B.P., Zulaika M., Ryazantsev S., van der Bliek A.M. A novel mitochondrial outer membrane protein, MOMA-1, that affects cristae morphology in Caenorhabditis elegans. Molecular Biology of the Cell. 2011;22(6):831–841. doi: 10.1091/mbc.E10-07-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shpilka T., Haynes C.M. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nature Reviews Molecular Cell Biology. 2018;19(2):109–120. doi: 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 53.Riera C.E., Dillin A. Tipping the metabolic scales towards increased longevity in mammals. Nature Cell Biology. 2015;17(3):196–203. doi: 10.1038/ncb3107. [DOI] [PubMed] [Google Scholar]
- 54.Troulinaki K., Bano D. Mitochondrial deficiency: a double-edged sword for aging and neurodegeneration. Frontiers in Genetics. 2012;3:244. doi: 10.3389/fgene.2012.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y., Samuel B.S., Breen P.C., Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508(7496):406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao K., Ji F., Breen P., Sewell A., Han M., Sadreyev R., et al. Mitochondrial dysfunction in C. elegans activates mitochondrial relocalization and nuclear hormone receptor-dependent detoxification genes. Cell Metabolism. 2019;29(5):1182–1191. doi: 10.1016/j.cmet.2019.01.022. e1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nargund A.M., Pellegrino M.W., Fiorese C.J., Baker B.M., Haynes C.M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337(6094):587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shpilka T., Du Y., Yang Q., Melber A., Uma Naresh N., Lavelle J., et al. UPR(mt) scales mitochondrial network expansion with protein synthesis via mitochondrial import in Caenorhabditis elegans. Nature Communications. 2021;12(1):479. doi: 10.1038/s41467-020-20784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tullet J.M., Hertweck M., An J.H., Baker J., Hwang J.Y., Liu S., et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132(6):1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker A.K., See R., Batchelder C., Kophengnavong T., Gronniger J.T., Shi Y., et al. A conserved transcription motif suggesting functional parallels between Caenorhabditis elegans SKN-1 and Cap'n'Collar-related basic leucine zipper proteins. Journal of Biological Chemistry. 2000;275(29):22166–22171. doi: 10.1074/jbc.M001746200. [DOI] [PubMed] [Google Scholar]
- 61.Kenyon C.J. The genetics of ageing. Nature. 2010;464(7288):504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 62.Fontana L., Partridge L., Longo V.D. Extending healthy life span--from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sen P., Shah P.P., Nativio R., Berger S.L. Epigenetic mechanisms of longevity and aging. Cell. 2016;166(4):822–839. doi: 10.1016/j.cell.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams R., Laskovs M., Williams R.I., Mahadevan A., Labbadia J. A mitochondrial stress-specific form of HSF1 protects against age-related proteostasis collapse. Developmental Cell. 2020;54(6):758–772. doi: 10.1016/j.devcel.2020.06.038. e755. [DOI] [PubMed] [Google Scholar]
- 65.Panowski S.H., Wolff S., Aguilaniu H., Durieux J., Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447(7144):550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 66.Austin J., Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51(4):589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 67.Arantes-Oliveira N., Apfeld J., Dillin A., Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295(5554):502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 68.Szabadkai G., Bianchi K., Varnai P., De Stefani D., Wieckowski M.R., Cavagna D., et al. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. The Journal of Cell Biology. 2006;175(6):901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim H.E., Grant A.R., Simic M.S., Kohnz R.A., Nomura D.K., Durieux J., et al. Lipid biosynthesis coordinates a mitochondrial-to-cytosolic stress response. Cell. 2016;166(6):1539–1552. doi: 10.1016/j.cell.2016.08.027. e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spiegel S., Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nature Reviews Molecular Cell Biology. 2003;4(5):397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 71.van der Beek J., Jonker C., van der Welle R., Liv N., Klumperman J. CORVET, CHEVI and HOPS - multisubunit tethers of the endo-lysosomal system in health and disease. Journal of Cell Science. 2019;132(10) doi: 10.1242/jcs.189134. [DOI] [PubMed] [Google Scholar]
- 72.Heo J.M., Harper N.J., Paulo J.A., Li M., Xu Q., Coughlin M., et al. Integrated proteogenetic analysis reveals the landscape of a mitochondrial-autophagosome synapse during PARK2-dependent mitophagy. Science Advances. 2019;5(11):eaay4624. doi: 10.1126/sciadv.aay4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonzalez Montoro A., Auffarth K., Honscher C., Bohnert M., Becker T., Warscheid B., et al. Vps39 interacts with Tom40 to establish one of two functionally distinct vacuole-mitochondria contact sites. Developmental Cell. 2018;45(5):621–636. doi: 10.1016/j.devcel.2018.05.011. e627. [DOI] [PubMed] [Google Scholar]
- 74.Kinchen J.M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C.D., et al. A pathway for phagosome maturation during engulfment of apoptotic cells. Nature Cell Biology. 2008;10(5):556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang J.T., Kumsta C., Hellman A.B., Adams L.M., Hansen M. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife. 2017;6 doi: 10.7554/eLife.18459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cavaliere F., Fornarelli A., Bertan F., Russo R., Marsal-Cots A., Morrone L.A., et al. The tricyclic antidepressant clomipramine inhibits neuronal autophagic flux. Scientific Reports. 2019;9(1):4881. doi: 10.1038/s41598-019-40887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rossi M., Munarriz E.R., Bartesaghi S., Milanese M., Dinsdale D., Guerra-Martin M.A., et al. Desmethylclomipramine induces the accumulation of autophagy markers by blocking autophagic flux. Journal of Cell Science. 2009;122(Pt 18):3330–3339. doi: 10.1242/jcs.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitroi D.N., Karunakaran I., Graler M., Saba J.D., Ehninger D., Ledesma M.D., et al. SGPL1 (sphingosine phosphate lyase 1) modulates neuronal autophagy via phosphatidylethanolamine production. Autophagy. 2017;13(5):885–899. doi: 10.1080/15548627.2017.1291471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Acoba M.G., Senoo N., Claypool S.M. Phospholipid ebb and flow makes mitochondria go. The Journal of Cell Biology. 2020;219(8) doi: 10.1083/jcb.202003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rockenfeller P., Koska M., Pietrocola F., Minois N., Knittelfelder O., Sica V., et al. Phosphatidylethanolamine positively regulates autophagy and longevity. Cell Death & Differentiation. 2015;22(3):499–508. doi: 10.1038/cdd.2014.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leprivier G., Remke M., Rotblat B., Dubuc A., Mateo A.R., Kool M., et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153(5):1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei W., Ruvkun G. Lysosomal activity regulates Caenorhabditis elegans mitochondrial dynamics through vitamin B12 metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(33):19970–19981. doi: 10.1073/pnas.2008021117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mossmann D., Vogtle F.N., Taskin A.A., Teixeira P.F., Ring J., Burkhart J.M., et al. Amyloid-beta peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metabolism. 2014;20(4):662–669. doi: 10.1016/j.cmet.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 85.Perez M.J., Ivanyuk D., Panagiotakopoulou V., Di Napoli G., Kalb S., Brunetti D., et al. Loss of function of the mitochondrial peptidase PITRM1 induces proteotoxic stress and Alzheimer's disease-like pathology in human cerebral organoids. Molecular Psychiatry. 2020 doi: 10.1038/s41380-020-0807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verheijen F.W., Verbeek E., Aula N., Beerens C.E., Havelaar A.C., Joosse M., et al. A new gene, encoding an anion transporter, is mutated in sialic acid storage diseases. Nature Genetics. 1999;23(4):462–465. doi: 10.1038/70585. [DOI] [PubMed] [Google Scholar]
- 87.Oka T., Toyomura T., Honjo K., Wada Y., Futai M. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. Journal of Biological Chemistry. 2001;276(35):33079–33085. doi: 10.1074/jbc.M101652200. [DOI] [PubMed] [Google Scholar]
- 88.Hansen M., Chandra A., Mitic L.L., Onken B., Driscoll M., Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genetics. 2008;4(2):e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamer K.J., Mootha V.K. MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO Reports. 2014;15(3):299–307. doi: 10.1002/embr.201337946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO Journal. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kirkin V., McEwan D.G., Novak I., Dikic I. A role for ubiquitin in selective autophagy. Molecular Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 92.Seibenhener M.L., Babu J.R., Geetha T., Wong H.C., Krishna N.R., Wooten M.W. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Molecular and Cellular Biology. 2004;24(18):8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iadarola D.M., Basu Ball W., Trivedi P.P., Fu G., Nan B., Gohil V.M. Vps39 is required for ethanolamine-stimulated elevation in mitochondrial phosphatidylethanolamine. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2020;1865(6):158655. doi: 10.1016/j.bbalip.2020.158655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nature Reviews Molecular Cell Biology. 2018;19(6):349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 96.Meyer K., Buettner S., Ghezzi D., Zeviani M., Bano D., Nicotera P. Loss of apoptosis-inducing factor critically affects MIA40 function. Cell Death & Disease. 2015;6:e1814. doi: 10.1038/cddis.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabolism. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Quiros P.M., Mottis A., Auwerx J. Mitonuclear communication in homeostasis and stress. Nature Reviews Molecular Cell Biology. 2016;17(4):213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 101.D'Amico D., Sorrentino V., Auwerx J. Cytosolic proteostasis networks of the mitochondrial stress response. Trends in Biochemical Sciences. 2017;42(9):712–725. doi: 10.1016/j.tibs.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 102.Zhao Q., Wang J., Levichkin I.V., Stasinopoulos S., Ryan M.T., Hoogenraad N.J. A mitochondrial specific stress response in mammalian cells. The EMBO Journal. 2002;21(17):4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haynes C.M., Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. Journal of Cell Science. 2010;123(Pt 22):3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 104.Bennett C.F., Vander Wende H., Simko M., Klum S., Barfield S., Choi H., et al. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nature Communications. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Labbadia J., Brielmann R.M., Neto M.F., Lin Y.F., Haynes C.M., Morimoto R.I. Mitochondrial stress restores the heat shock response and prevents proteostasis collapse during aging. Cell Reports. 2017;21(6):1481–1494. doi: 10.1016/j.celrep.2017.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benedetti C., Haynes C.M., Yang Y., Harding H.P., Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174(1):229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nargund A.M., Fiorese C.J., Pellegrino M.W., Deng P., Haynes C.M. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt) Molecular Cell. 2015;58(1):123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tian Y., Garcia G., Bian Q., Steffen K.K., Joe L., Wolff S., et al. Mitochondrial stress induces chromatin reorganization to promote longevity and UPR(mt) Cell. 2016;165(5):1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Palikaras K., Lionaki E., Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521(7553):525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- 110.Uittenbogaard M., Chiaramello A. Mitochondrial biogenesis: a therapeutic target for neurodevelopmental disorders and neurodegenerative diseases. Current Pharmaceutical Design. 2014;20(35):5574–5593. doi: 10.2174/1381612820666140305224906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gomes L.C., Di Benedetto G., Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nature Cell Biology. 2011;13(5):589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.