Abstract
To identify regulators of penicillin biosynthesis, a previously isolated mutant of Aspergillus nidulans (Prg-1) which carried the trans-acting prgA1 mutation was used. This mutant also contained fusions of the penicillin biosynthesis genes acvA and ipnA with reporter genes (acvA-uidA and ipnA-lacZ) integrated in a double-copy arrangement at the chromosomal argB gene. The prgA1 mutant strain exhibited only 20 to 50% of the ipnA-lacZ and acvA-uidA expression exhibited by the wild-type strain and had only 20 to 30% of the penicillin produced by the wild-type strain. Here, using complementation with a genomic cosmid library, we isolated a gene (suAprgA1) which complemented the prgA1 phenotype to the wild-type phenotype; i.e., the levels of expression of both gene fusions and penicillin production were nearly wild-type levels. Analysis of the suAprgA1 gene in the prgA1 mutant did not reveal any mutation in the suAprgA1 gene or unusual transcription of the gene. This suggested that the suAprgA1 gene is a suppressor of the prgA1 mutation. The suAprgA1 gene is 1,245 bp long. Its five exons encode a deduced protein that is 303 amino acids long. The putative SUAPRGA1 protein was similar to both the human p32 protein and Mam33p of Saccharomyces cerevisiae. Analysis of the ordered gene library of A. nidulans indicated that suAprgA1 is located on chromosome VI. Deletion of the suAprgA1 gene resulted in an approximately 50% reduction in ipnA-lacZ expression and in a slight reduction in acvA-uidA expression. The ΔsuAprgA1 strain produced about 60% of the amount of penicillin produced by the wild-type strain.
Penicillin is a β-lactam antibiotic that is produced only by filamentous fungi, most notably Aspergillus nidulans and Penicillium chrysogenum (4). Biosynthesis of penicillin starts from three amino acid precursors, l-α-aminoadipic acid, l-cysteine, and l-valine. This process is catalyzed by three enzymes, which are encoded by the following three genes: acvA (pcbAB), ipnA (pcbC), and aatA (penDE), which encode δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase, isopenicillin N synthase, and acyl coenzyme A:isopenicillin N acyltransferase, respectively. The genes have been cloned and sequenced, and they are organized in a cluster (for reviews see references 4 and 19). acvA and ipnA are divergently transcribed and in A. nidulans and are separated by an 872-bp intergenic region (15).
In A. nidulans, several regulators which are involved in the regulation of penicillin biosynthesis have been identified. These regulators include the pH-dependent regulator PACC (8, 17, 29) and the CCAAT-binding complex PENR1 (AnCF), which resembles a Hap-like complex (12, 18, 27). Both of these regulators have the ability to bind to the DNAs of at least some of the penicillin biosynthesis gene promoters. To identify additional proteins involved in the regulation of penicillin biosynthesis, a mutant approach was used (6). Briefly, we used strain AXTII9, an A. nidulans strain carrying acvA-uidA and ipnA-lacZ gene fusions integrated in a double-copy arrangement at the chromosomal argB gene locus. On Aspergillus minimal medium (AMM) agar plates supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), colonies of this strain stained blue, which indicated that ipnA-lacZ expression occurred. After mutagenesis with UV light, colonies were isolated on agar plates containing lactose as the carbon source, and these colonies produced only a faint blue color or no color at all. The mutants (designated Prg mutants, for penicillin regulation) most likely were defective in trans-acting genes. It was not likely that mutants carrying cis-acting mutations (i.e., mutations in the ipnA promoter or the lacZ gene) would appear, because such mutations would probably not be detected due to the second gene fusion located on the chromosome. This hypothesis was confirmed by a complementation analysis which showed that the mutants carried trans-acting mutations. Two mutants (Prg-1 and Prg-6) with a reproducible phenotype (white colonies on AMM agar plates containing X-Gal and lactose as the carbon source) were characterized in detail. In a fermentation experiment, mutants Prg-1 and Prg-6 exhibited only 20 to 50% of the ipnA-lacZ expression exhibited by the wild type and produced only 20 to 30% of the penicillin produced by the wild type. A Western blot analysis showed that these mutants contained reduced amounts of the ipnA gene product (i.e., isopenicillin N synthase). Both mutant Prg-1 and mutant Prg-6 also differed from the wild type in the level of acvA-uidA expression. A segregation analysis revealed that in both mutants the Prg phenotype resulted from mutation of a single gene. Two different complementation groups were identified; these groups were designated prgA1 and prgB1 (6).
Pérez-Esteban et al. (20) independently used a similar approach which resulted in identification and characterization of a recessive mutation designated npeE1 (impaired in penicillin biosynthesis). A genetic analysis showed that the mutation is located on linkage group IV. To date, whether npeE1 differs from prgA1 and prgB1 has not been determined.
In this paper, we describe cloning and characterization of a gene which complemented the prgA1 phenotype to the wild-type phenotype.
MATERIALS AND METHODS
Strains and plasmids.
The bacterial and fungal strains used in this study are listed in Table 1. Vectors and plasmids were propagated in Escherichia coli DH5α. Strain K103 was generated by crossing strain Prg-1 with G191 (Table 1). Ascospores were plated onto AMM agar plates containing X-Gal (50 μg/ml) and glucose as the carbon source. Uracil-auxotrophic colonies which stained the agar blue, which indicated that the ipnA-lacZ gene fusion was present, were analyzed to determine whether the acvA-uidA and ipnA-lacZ gene fusions were present in a double-copy arrangement at the chromosomal argB gene locus, as determined by Southern blotting as previously described (6). The presence of the prgA1 mutation was checked by plating conidia onto AMM agar plates containing X-Gal (35 μg/ml) and lactose as the carbon source. In contrast to wild-type strain AXTII9, the prgA1 mutants did not stain the lactose-containing agar blue, which was due to the reduced ipnA-lacZ expression under these conditions (6). One of the resulting progeny with the desired genotype was designated K103 (Table 1).
TABLE 1.
Bacteria, fungi, plasmids, and cosmids used in this study
| Organism, plasmid, or cosmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| E. coli DH5α | F− φ80d/lacZ M15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK−, mK+) supE44 λ−thi-l gyrA96 relA1 | BRLa |
| Bacillus calidolactis | 5 | 5 |
| A. nidulans strains | ||
| R21 | yA2 pabaA1 | 9 |
| G191 | pabaA1 pyrG89 uaY fwA1 | 1 |
| AXB4A | biA1 bga0 argB2::pAXB4A ArgB+, WG355 derivative carrying pAXB4A | 5 |
| AXB4A2 | biA1 pyrG89 bga0 argB2::pAXB4A ArgB+, WG355 derivative carrying pAXB4A | 31 |
| AXTII9 | biA1 bga0 argB2::(pAXB4A)2 ArgB+, WG355 derivative carrying pAXB4A | 5 |
| Prg-1 | prgA1 biA1 bga0 argB2::(pAXB4A)2 ArgB+, AXTII9 derivative | 6 |
| Prg-6 | prgB1 biA1 bga0 argB2::(pAXB4A)2 ArgB+, AXTII9 derivative | 6 |
| K103 | prgA1 biA1 pyrG89 fwA1 argB2::(pAXB4A)2 ArgB+ | This study |
| PrgA1CA | K103 transformed with a cosmid derived from the pCAP2 library, PyrG+ PrgA1+ | This study |
| PrgA1CB | K103 transformed with a cosmid derived from the pCAP2 library, PyrG+ PrgA1+ | This study |
| PrgA1CC | K103 transformed with a cosmid derived from the pCAP2 library, PyrG+ PrgA1+ | This study |
| DE58 | ΔsuAprgA1 biA1 pyrG89 bga0 argB2::pAXB4A ArgB+ Pyr-4+ AXB4A2 carrying a deletion of the suAprgA1 gene | This study |
| PrgA1T | K103 transformed with a cosmid derived from the pCAP2 library, PyrG+ | This study |
| Plasmids and cosmids | ||
| pAXB4A | acvA-uidA ipnA-lacZ argBb | 5 |
| cosCA.1 | Cosmid rescued from PrgA1CA | This study |
| cosCA.2 | Cosmid rescued from PrgA1CA | This study |
| cosCA.3 | Cosmid rescued from PrgA1CA | This study |
| pKTB1 | pyr-4+ | K. Then Bergh |
| pKO1 | This study | |
| pKO2 | This study | |
| pKO3 | This study |
BRL, Bethesda Research Laboratories, Gaithersburg, Md.
The mutant argB allele can complement chromosomal argB mutations.
Gene libraries of A. nidulans.
A genomic pCAP2 cosmid library (2), an ordered genomic gene library (7) (available from the Fungal Genetics Stock Center, Kansas City, Kans.), and a cDNA library (constructed by R. Aramayo; available from the Fungal Genetics Stock Center) of A. nidulans were used in this study.
Media and cultivation of strains.
E. coli was grown in Luria-Bertani medium supplemented with ampicillin (50 μg/ml). AMM was prepared as previously described (6). A. nidulans fermentation was carried out in fermentation medium (FM) or AMM essentially as described previously (28). The seed cultures contained 4% (wt/vol) glucose and were cultivated for 24 h. Experimental cultures (20-ml portions of AMM or 20-ml portions of FM in 250-ml flasks) containing 4% (wt/vol) lactose as the carbon source were inoculated with 1 ml of a seed culture suspension, and the resulting preparations were incubated for various times, as indicated below. If required, biotin (0.3 μg/ml), p-aminobenzoic acid (15 μg/ml), l-arginine (0.5 mg/ml), uridine (1.2 mg/ml), or uracil (2.2 mg/ml) was added to the medium.
Penicillin bioassay and determination of dry weight.
We performed penicillin bioassays in which Bacillus calidolactis C953 was the indicator organism and determined the dry weights of cultures as previously described (5).
Genetic techniques.
Sexual crosses were performed and the resulting progeny were characterized as described by Pontecorvo et al. (21).
Standard DNA techniques.
For small-scale preparation of A. nidulans chromosomal DNA, the technique of Raeder and Broda (23) was used. Standard techniques for manipulation of DNA were performed as described by Sambrook et al. (24). A Southern blot analysis and colony hybridization were performed essentially as described previously (24). DNA probes were labelled with fluorescein-11-dUTP and hybridized fragments were detected by Southern blot analysis by using an enhanced chemiluminescence (ECL) random prime kit (Amersham Pharmacia Biotech, Freiburg, Germany) and an ECL chemiluminescence kit (Amersham Pharmacia Biotech) respectively, according to the manufacturer’s instructions. In vitro packaging in which chromosomal DNAs of the A. nidulans transformant strains were used was carried out by using a Gigapack III XL kit (Stratagene, La Jolla, Calif.).
Screening of the ordered gene library of A. nidulans.
To screen the ordered gene library, a suAprgA1-specific probe was generated by PCR by using chromosomal DNA of A. nidulans as the template and oligonucleotides RevC and Tet4 as the primers (Table 2). The DNA fragment which was obtained was 1.2 kbp long.
TABLE 2.
Oligonucleotides used
| Oligonucleotide | Sequence |
|---|---|
| RevC | 5′-GTTGAGGGGAAGAAACTGG-3′ |
| Tet4 | 5′-GGAGAATGGCGGATCTGG-3′ |
| PC81 | 5′-GAATAGTTTCGTCGGCGTC-3′ |
| PC82 | 5′-CCAGTTGCTAACCGCCCTG-3′ |
| suAKOSal | 5′-GTCGGCCTATCCAGAACTAGGTCGACGG-3′ |
| suAKOKpn | 5′-GCTCGGTACCTATAATTGAATGCATTGCTG-3′ |
| KOClaI | 5′-GACTACATCGATCTCGGTCAGGTCCGGCATCC-3′ |
| KONotI | 5′-GTGCTGGCGGCCGCGCCCAGCATCCCGAGGAC-3′ |
Generation of recombinant plasmids and probes for Southern blot analyses.
The insert of the plasmid derived from transformant PrgA1CC was amplified by PCR by using oligonucleotides PC81 and PC82 (Table 2). In the case of the plasmid rescued from transformant PrgA1CB, a 1.3-kbp DNA fragment derived from the chromosomal insert was isolated by restriction digestion with HindIII and NcoI. The DNA fragments which were isolated were labelled, and the probes generated were used for Southern blot analysis.
To construct the suAprgA1 knock-out plasmid, we determined the DNA sequences flanking the suAprgA1 gene upstream and downstream (data not shown). Based on the information obtained, primers suAKOSal (encoding a SalI site) and suAKOKpn (encoding a KpnI site) (Table 2) were used to generate a 3.3-kbp DNA fragment by PCR; this fragment contained the suAprgA1 gene and about 1 kbp of the flanking region upstream and downstream. The PCR fragment was cut with SalI and KpnI and cloned into KpnI-SalI-digested vector pUC18. The resulting plasmid was designated pKO1. To delete the suAprgA1 gene from this plasmid, we used inverse PCR performed with oligonucleotides KOClaI (encoding a ClaI site) and KONotI (encoding a NotI site) (Table 2). A 4.6-kbp DNA fragment lacking the suAprgA1 gene from nucleotide −515 to nucleotide 793 was obtained. The DNA fragment was phosphorylated by using polynucleotide kinase and ligated. E. coli cells were transformed with the ligated DNA fragment, and the resulting plasmid was designated pKO2. To introduce a selectable marker, plasmid pKO2 was cut with ClaI and NotI, and a 2-kbp ClaI-NotI DNA fragment obtained from plasmid pKTB1 (Table 1) carrying the pyr-4 gene of Neurospora crassa was ligated into the vector. The resulting plasmid was designated pKO3 (see Fig. 5A).
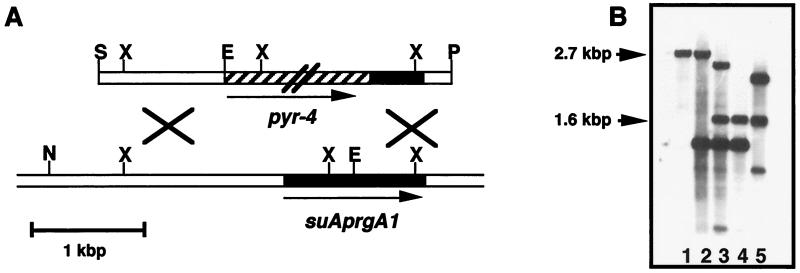
FIG. 5.
Construction of a suAprgA1 deletion strain. (A) Partial restriction map of the genomic region containing suAprgA1 and schematic representation of the gene replacement construct used. The arrows indicate the orientation of the genes. Restriction sites are indicated. The pyr-4 gene is not drawn to scale. Abbreviations: E, EcoRI; N, NarI; P, PvuII; S, SphI; X, XhoI. (B) Southern blot analysis. Chromosomal DNAs of wild-type strain AXB4A2 and transformant strains were cut with EcoRI and NarI. The DNAs were hybridized with a 1,150-bp DNA probe generated by PCR amplification performed with oligonucleotides suAKOSal and KOClal and chromosomal DNA of wild-type strain AXTII9 as the template. The 2.7-kbp band was obtained with the wild-type strain (lane 1, arrow). The band that characterized replacement of the suAprgA1 gene by the pyr-4 gene was the 1.6-kbp band (arrow). The thicker band obtained by tandem integration of the DNA fragment used for transformation at the suAprgA1 gene locus was the band at 1.5 kbp (lanes 2 through 4). Lane 1, wild-type AXB4A2; lane 2, transformant DE3; lane 3, transformant DE4; lane 4, transformant DE58; lane 5, transformant DE62.
PCR amplification, DNA sequence analysis, and computer programs.
For PCR amplification, sequence-specific oligonucleotides were synthesized (MWG Biotech, Ebersberg, Germany). The DNA fragments produced were purified by using standard methods. The sequences of DNA fragments were determined on both strands by the dideoxy chain termination method (25) by using fluorescent dyes that were analysed with a model ABI PRISM 377 automatic sequencer (PE Biosystems, Weiterstadt, Germany). Sequence data were edited with the program Sequence Navigator (Applied Biosystems, Foster City, Calif.). Similarities between amino acid sequences were analyzed by using the program Gene Works (IntelliGenetics, Inc., Mountain View, Calif.).
Isolation of cDNAs.
To identify cDNA clones encoding suAprgA1, DNAs from the A. nidulans λ cDNA library obtained by PCR were amplified by using internal primers specific for the suAprgA1 gene derived from the genomic DNA sequence and primer T3 (Stratagene) or T7 (Stratagene) essentially as previously described (32).
Northern blot analysis.
Mycelia of A. nidulans were harvested and broken by using liquid nitrogen as described previously (13). Total RNA was prepared by using an RNeasy total RNA purification kit (Qiagen, Hilden, Germany). The amount of RNA was determined both spectrophotometrically and by agarose gel electrophoresis. Northern blotting was performed as described by Sambrook et al. (24) by using the Gene Images system (Amersham Pharmacia Biotech).
Transformation of A. nidulans.
A. nidulans K103 was transformed to uracil prototrophy by using a method described previously (1). For transformation, plasmid DNA was purified by chromatography performed with NUCLEOBOND columns as recommended by the manufacturer (Macherey and Nagel).
β-GAL and β-GLU activity assays.
β-Galactosidase (β-GAL) and β-glucuronidase (β-GLU) activities were determined in crude extracts by using mycelia grown in FM as previously described (28). The β-GAL and β-GLU activities at each time point were determined with cell extracts obtained from three cultures that were incubated simultaneously. Protein concentrations were determined as described by Bradford (3). Specific activities of β-GLU and β-GAL were calculated as described previously (5).
Nucleotide sequence accession number.
The nucleotide sequence of the A. nidulans suAprgA1 gene and the deduced amino acid sequence have been deposited in the EMBL database under accession no. Y17330.
RESULTS
Cloning of a gene that complements the PrgA1 phenotype.
We attempted to clone a gene that complements the prgA1 phenotype by transformation by using an A. nidulans genomic library carrying the pyr-4 gene of N. crassa as the selectable marker. prgA1 mutant strain K103, which contained the pyrG89 mutation resulting in uracil auxotrophy, was used as the recipient. In addition, K103 carried the acvA-uidA and ipnA-lacZ gene fusions in a double-copy arrangement (see above). As previously shown, the growth of a strain carrying the prgA1 mutation in FM did not differ from the growth of a wild-type strain (6).
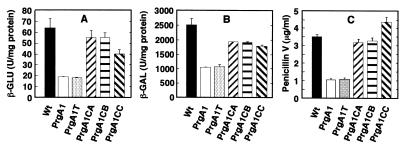
Strain K103 was transformed by using cosmid DNA from the genomic pCAP2 library. A total of 2,500 uracil-prototrophic transformants were isolated. Three of these (PrgA1CA, PrgA1CB, and PrgA1CC) complemented the prgA1 phenotype to the wild-type phenotype on agar plates containing X-Gal and lactose as the carbon source; i.e., the colonies stained the agar blue. In a penicillin fermentation analysis prgA1 mutant strain K103 exhibited reduced levels of acvA-uidA and ipnA-lacZ expression and produced less penicillin compared with wild-type strain AXTII9 (Wt) (Fig. 1A and B). The same results were obtained with a randomly chosen transformant, PrgA1T, which contained a noncomplementing cosmid. In contrast, in complemented transformants PrgA1CA, PrgA1CB, and PrgA1CC, the levels of acvA-uidA and ipnA-lacZ expression were greater than the levels observed in mutant strain K103 (Fig. 1A and B). In contrast to the prgA1 mutant, transformants PrgA1CA and PrgA1CB produced the same amount of penicillin as the wild-type strain produced (Fig. 1C). Transformant PrgA1CC even had a higher penicillin titer than the wild type (Fig. 1C).
FIG. 1.
Fermentation with 4% (wt/vol) lactose as the carbon source of A. nidulans wild-type strain AXTII9 (Wt), mutant strain K103 (PrgA1), and K103 transformed with a noncomplementing cosmid (PrgA1T) and complemented transformants PrgA1CA, PrgA1CB, and PrgA1CC. All strains carried plasmid pAXB4A (acvA-uidA ipnA-lacZ fusions) integrated in a double-copy arrangement at the chromosomal argB gene locus. Cultures were incubated in FM for 48 h at 26°C and 250 rpm. Data are the means and standard deviations of values obtained from three simultaneously harvested flasks. (A) Expression of acvA-uidA gene fusions, determined as β-GLU specific activity. (B) Expression of ipnA-lacZ gene fusions, determined as β-GAL specific activity. (C) Penicillin V titer.
To rescue the complementing cosmids, in vitro packaging of the chromosomal DNAs of the transformants was carried out. After E. coli DH5α was infected with DNA of transformant PrgA1CA, 11 ampicillin-resistant colonies were obtained. The cosmids of these colonies were isolated and analyzed by restriction analysis. Three different types of cosmids (cosCA.1, cosCA.2, and cosCA.3) were identified. With two of the cosmids (cosCA.1 and cosCA.3), 50% of the transformants of prgA1 mutant strain K103 exhibited the wild-type phenotype on agar plates, indicating that the cosmids encoded a gene which complemented the prgA1 phenotype (data not shown).
In order to reisolate the cosmid DNAs of transformants PrgA1CB and PrgA1CC, total chromosomal DNAs of the transformants were digested by HindIII and then ligated, which created circular plasmids. These plasmids contained the ampicillin resistance gene, the origin of replication in E. coli, and part of the A. nidulans chromosomal DNA derived from the cosmid up to the first HindIII site in the genomic insert. In the case of transformant PrgA1CB, an approximately 7-kbp plasmid was isolated, and this plasmid corresponded to an estimated 3-kbp A. nidulans chromosomal DNA insert. The border regions between the vector backbone and the insert were sequenced. The plasmid derived from transformant PrgA1CC in the same way contained a 220-bp insert (data not shown). A comparison of the DNA sequences obtained with sequences from databases did not reveal a major level of similarity to any previously described gene.
To analyze whether complementation of the different transformants was due to the same DNA fragment, we used the sequence information for the insert DNAs of both plasmids to design probes for Southern blot analyses. Since these fragments were at the borders of the recombinant parts of the cosmids, we expected that when the DNA fragments were used as probes, the sizes of the hybridizing bands corresponding to the chromosomal copy and additional bands would differ due to the integration of cosmids.
BamHI-digested chromosomal DNAs of two wild-type strains, DNAs of the three complemented transformants (PrgA1CA, PrgA1CB, and PrgA1CC), and BamHI-digested DNAs of the three cosmids rescued from transformant PrgA1CA were blotted onto a membrane and hybridized with the probes derived from PrgA1CC (Fig. 2A) and PrgA1CB (Fig. 2B). As expected, the PrgA1CC-derived probe (see above) hybridized with all of the chromosomal DNAs (Fig. 2A). The additional signal at 6.2 kbp observed with the chromosomal DNA of transformant PrgA1CC was due to integration of the cosmid into the genome (Fig. 2A, lane 3). No additional band was obtained with the chromosomal DNAs derived from transformants PrgA1CA and PrgA1CB (Fig. 2A, lanes 4 and 5), and consequently, there was no cross-reaction with the cosmid DNA isolated from transformant PrgA1CA (Fig. 2A, lanes 6 through 8).
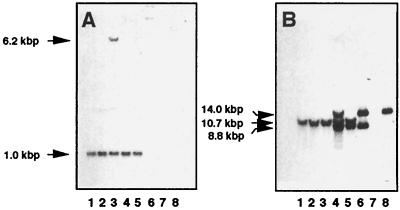
FIG. 2.
Southern blot analysis of BamHI-digested chromosomal DNAs from A. nidulans wild-type strains, mutant strain K103 complemented with cosmid DNA, and BamHI-digested cosmid DNA isolated from transformant PrgA1CA. Lane 1, wild-type strain R21; lane 2, wild-type strain AXTII9; lane 3, complemented K103 transformant PrgA1CC; lane 4, complemented K103 transformant PrgA1CA; lane 5, complemented K103 transformant PrgA1CB; lane 6, cosmid cosCA.1 isolated from PrgA1CA; lane 7, cosmid cosCA.2 isolated from PrgA1CA; lane 8, cosmid cosCA.3 isolated from PrgA1CA. (A) PrgA1CC-derived probe. The upper arrow indicates the 6.2-kbp band due to recombinant cosmid DNA integrated into the genome. (B) PrgA1CB-derived probe.
The 1.3-kbp probe derived from PrgA1CB also hybridized with all of the chromosomal DNAs, yielding a single band at 10.7 kb (Fig. 2B). However, additional signals were detected not only with the chromosomal DNA of transformant PrgA1CB but also with the DNA of PrgA1CA (Fig. 2B, lanes 4 and 5). The same bands were also detected with cosmids cosCA.1 and cosCA.3 derived from transformant PrgA1CA (Fig. 2B, lanes 6 and 8). This indicated that integration of these cosmids was responsible for the additional cross-reacting bands observed with PrgA1CA with the probe derived from PrgA1CB (Fig. 2B).
On the basis of this Southern blot analysis, we reached the following conclusions. (i) The independently isolated transformants PrgA1CA and PrgA1CB contained cosmids which carried at least in part identical DNA fragments, a finding which strongly supported the validity of the approach. (ii) The complementing cosmids cosCA.1 and cosCA.3 isolated from transformant PrgA1CA contained an overlapping fragment of chromosomal DNA; the probe derived from PrgA1CB was part of the overlapping region. These findings made it very likely that this DNA fragment was responsible for the ability to complement the mutant phenotype. (iii) The cosmid present in transformant PrgA1CC apparently differed from the cosmid found in PrgA1CA and PrgA1CB.
The DNA fragment present in the cosmids which were independently isolated from PrgA1CA and PrgA1CB was characterized further. A Southern blot analysis indicated that cosCA.1 and cosCA.3 had a 14-kbp DNA fragment in common (Fig. 2B); 8.5 kbp of this fragment was derived from the vector backbone of pCAP2. Therefore, the remaining 5.5 kbp should have contained the complementing gene. Different restriction fragments derived from this 5.5-kbp DNA fragment were used in complementation tests. A fragment which still had the ability to complement the prgA1 phenotype to the wild-type phenotype was 3.7 kbp long (data not shown); this fragment was cloned in plasmid pUC18 to give plasmid pST3.7.
suAprgA1 gene.
To identify open reading frames (ORFs) in the 3.7-kbp insert, Northern blotting (data not shown) and computer analyses were performed. These analyses showed that only a single ORF that resulted in a 1.2-kb mRNA transcript was fully encoded by this DNA fragment. The corresponding cDNA of the gene that was designated suAprgA1 (see below) was analyzed. The 5′ end of the suAprgA1 cDNA was located at position −35 with respect to the putative translational initiation codon. The gene contained four introns that were 159, 55, 79, and 49 bp long (EMBL accession no. Y17330).
The suAprgA1 gene is 1,245 bp long. Its five exons encode a 909-bp ORF. The predicted SUAPRGA1 polypeptide consists of 303 amino acids and has a molecular mass of 33,947 Da, and a deduced isoelectric point of pH 4.47.
Comparison of the deduced SUAPRGA1 amino acid sequence with sequences in databases.
A computer analysis in which the BLAST program was used revealed that the putative suAprgA1-encoded ORF was similar to a group of conserved proteins found in eukaryotes. This group includes the human p32 protein (11), Mam33p of Saccharomyces cerevisiae (16, 26), and a putative ORF of Schizosaccharomyces pombe (EMBL accession no. CAA22880). The overall levels of similarity at the amino acid level are 27% for SUAPRGA1 and Mam33p, 19% for SUAPRGA1 and p32, and 17% for Mam33p and p32. The region in these proteins that exhibited the highest level of similarity was the region at the C terminus (Fig. 3). The C-terminal parts of all of the proteins are rich in acidic and aliphatic amino acids (Fig. 3). All of the proteins have similar acidic isoelectric points. It is worth mentioning that a deletion of the S. cerevisiae Mam33 gene can be complemented by the human p32 protein gene (16). This suggests that despite the low overall level of similarity Mam33 and p32 are functional homologs.
FIG. 3.
Comparison of the deduced C-terminal amino acid sequence of A. nidulans SUAPRGA1 (A. n. SUAPRGA1) with the C-terminal sequences of the human p32 protein (SWISS-PROT accession no. Q07021) (H. s. p32), S. cerevisiae Mam33p (SWISS-PROT accession no. P40513) (S. c. Mam33p), and a putative homolog of S. pombe (EMBL accession no. CAA22880) (S. p. CAA22880). Amino acids that were the same in at least three of the four amino acid sequences are enclosed in boxes. The numbers on the right indicate the positions of the last amino acids shown for the proteins.
suAprgA1 appears to be a suppressor of the prgA1 mutation.
Our DNA sequence analysis of the suAprgA1 gene of Prg-1 (prgA1) and Prg-6 (prgB1) did not reveal any mutation in either the coding region or flanking regions. Also, the transcript of suAprgA1 was detected in the Prg-1 (prgA1) mutant strain (data not shown). Taken together, these findings suggested that the gene isolated is a suppressor of the prgA1 phenotype.
suAprgA1 is located on chromosome VI.
To localize the suAprgA1 gene on one of the eight chromosomes of A. nidulans, the ordered A. nidulans gene library was screened by performing a Southern blot analysis (7, 22, 30). Using a suAprgA1-specific probe (see above), we obtained signals with cosmids L11A9, L11B9, L12B4, and W5E1 (data not shown) (9a). The cosmids were isolated, and PCR experiments confirmed that the suAprgA1 gene was present on these cosmids (data not shown). We found that all four of the cosmids carried chromosome VI DNA, indicating that the suAprgA1 gene is located on chromosome VI. In addition, cosmid L11A9 was fine mapped (9a). Interestingly, the penicillin biosynthesis gene cluster is located on the same chromosome (14) but in a different chromosomal region, suggesting that there is no physical linkage between suAprgA1 and the penicillin biosynthesis gene cluster.
Transcript of suAprgA1.
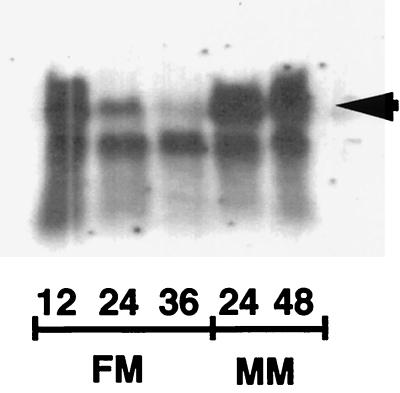
To further analyze the suAprgA1 gene, the steady-state level of its mRNA was determined by Northern blot analysis (Fig. 4). The transcript was 1.2 kb long, which corresponds to the length of the cDNA sequence obtained. The gene was transcribed during growth in both FM and AMM. In FM, the relative abundance of mRNA decreased from 12 to 36 h, whereas considerable levels of mRNA were still detected in AMM after 48 h.
FIG. 4.
Northern blot analysis. The band corresponding to the suAprgA1 transcript at about 1.2 kb is indicated by an arrow. mRNA was isolated from mycelia of wild-type strain R21 grown in FM supplemented with 2% (wt/vol) lactose for 12 h (12), 24 h (24), and 36 h (36) and in AMM (MM) supplemented with 2% (wt/vol) lactose for 24 h (24) and 48 h (48). The mRNA load was adjusted by determining the ratio of absorbance at 260 nm to absorbance at 280 nm and by staining an agarose gel. The band that migrated more quickly corresponded to a 0.9-kb transcript; it was encoded by a gene which was located downstream of suAprgA1, and it was also detected by using the 1.2-kb probe, which covered 150 bases downstream of the putative 3′ end of the suAprgA1 mRNA. However, when we used a probe that covered only an internal fragment of suAprgA1, it hybridized exclusively to the 1.2-kb mRNA (arrow), which corresponded to the suAprgA1 transcript (data not shown). Since the mRNA level of the unknown gene (lower band) did not change under the different conditions, it also served as a loading control.
Deletion of suAprgA1.
Since suAprgA1 appears to be a suppressor of the prgA1 phenotype, we decided to analyze the effect of a deletion of the suAprgA1 gene. Therefore, the knock-out plasmid pKO3 (see above) was cut with SphI and PvuII, which resulted in a linear fragment carrying the pyr-4 gene and on the two sides the 5′ and 3′ DNA sequences flanking the suAprgA1 gene (Fig. 5A). The linear fragment was used for transformation, which reduced the occurrence of transformants carrying ectopically integrated DNA.
For transformation of A. nidulans, we used uracil-auxotrophic strain AXB4A2 carrying the pyrG89 mutation, which could be complemented with the pyr-4 gene, and, in addition, acvA-uidA and ipnA-lacZ gene fusions integrated in a single-copy arrangement at the argB gene locus (Table 1). After transformation, 380 Pyr-4+ transformants were isolated. The transformants were analyzed by Southern blotting to determine whether the suAprgA1 gene was replaced by pyr-4 (Fig. 5B). Some of the transformants produced the typical hybridization band characteristic of replacement of the suAprgA1 gene by the pyr-4 gene via homologous recombination (Fig. 5B, lanes 3 through 5). However, additional signals were detected in all of these strains. The strong signal corresponding to a DNA fragment that was about 1.5 kbp long was due to tandem integration of the construct at the suAprgA1 gene locus (Fig. 5B, lanes 2 through 4). Only strain DE58 produced bands characteristic of deletion of the suAprgA1 gene by tandem integration without additional ectopically integrated DNA fragments (Fig. 5B, lane 4).
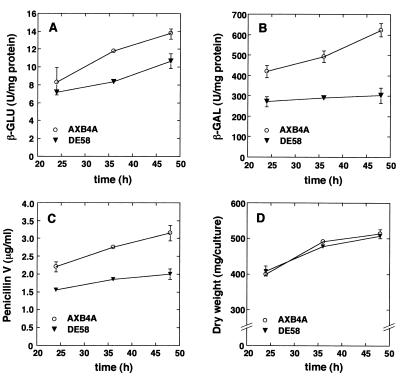
Expression of the acvA-uidA and ipnA-lacZ gene fusions was determined with ΔsuAprgA1 deletion strain DE58. The results are shown in Fig. 6. After 24 h there was only a slight difference between deletion mutant DE58 and wild-type strain AXB4A with respect to expression of the ipnA-lacZ fusion (Fig. 6B). However, in the later stages of fermentation ipnA-lacZ expression did not increase any more in the mutant strain, whereas it did increase in the wild-type strain (Fig. 6B). Hence, after 48 h the level of expression of the ipnA-lacZ gene fusion in the mutant was only about 50% of the level of expression observed in the wild-type strain. Deletion of the suAprgA1 gene resulted in only a slight reduction in the level of acvA-uidA expression (Fig. 6A). Determination of the penicillin titer revealed that ΔsuAprgA1 strain DE58 produced about 60% of the amount of penicillin produced by the wild-type strain (Fig. 6C). The reduction was not due to production of a smaller amount of mycelium because in FM the mutant strain grew as well as the wild-type strain and produced the same mycelial mass (Fig. 6D). However, it is interesting that in suAprgA1 deletion strain DE58 germination of conidia was delayed (data not shown). Taken together, the data indicated that the reduced levels of expression of both the acvA and ipnA gene fusions were paralleled by a reduction in the penicillin titer.
FIG. 6.
Fermentation with 4% (wt/vol) lactose as the carbon source of A. nidulans wild-type strain AXB4A and mutant DE58 (ΔsuAprgA1). Both strains carried plasmid pAXB4A (acvA-uidA ipnA-lacZ fusions) integrated in a single-copy arrangement at the chromosomal argB gene locus. Data are the means and standard deviations of values obtained from three simultaneously harvested flasks. (A) Expression of acvA-uidA gene fusions, determined as β-GLU specific activity. (B) Expression of ipnA-lacZ gene fusions, determined as β-GAL specific activity. (C) Penicillin V titer. (D) Dry weight.
DISCUSSION
Previously, we described the isolation of two types of trans-acting mutations, designated prgA1 and prgB1, which affect penicillin biosynthesis in A. nidulans. The mutants exhibited reduced levels of expression of the penicillin biosynthesis genes acvA and ipnA and reduced penicillin titers (6). Here, we cloned and characterized a gene designated suAprgA1, which complemented the prgA1 phenotype nearly to the wild-type phenotype. This was accomplished by complementing the K103 (prgA1) mutant with a cosmid library. We isolated 3 complemented transformants from the 2,500 transformants examined. Two of these contained cosmids which carried the suAprgA1 gene. Isolation of the same gene from two independently isolated transformants confirmed the validity of this approach.
The isolated gene was designated a suppressor of the prgA1 mutation (suAprgA1). This conclusion was based on the results of an analysis of the DNA sequence of the chromosomal version of the suAprgA1 gene in the Prg-1 (prgA1) mutant. Furthermore, we detected no difference in the steady-state mRNA levels of suAprgA1 in the Prg-1 mutant and the wild type. However, mapping of the prgA1 mutation is required to prove that prgA1 and suAprgA1 are located at different genetic loci.
Complementation of the prgA1 phenotype was accomplished with a single copy of the cosmid present in the genome. This finding is unusual because in general, expression of suppressor genes from a high-copy-number plasmid is required for suppression of a mutant phenotype. At this time, we assume that a certain threshold concentration of a factor is required for full expression of the penicillin biosynthesis genes and thus full production of penicillin. In strains carrying the prgA1 mutation, the concentration of the factor falls short of the threshold concentration. The threshold concentration was already exceeded when an additional copy of the suAprgA1 gene was present in the cell.
Computer analysis revealed that the putative suAprgA1-encoded protein was similar to a group of conserved proteins found in eukaryotes. This group includes the human protein designated p32 (11), Mam33p of S. cerevisiae (16, 26), and a putative ORF of S. pombe (EMBL accession no. CAA22880). The precise function of these proteins has not been clarified yet. The crystal structure of the human p32 protein was recently determined at 2.25 Å (10). Interestingly, the secondary structure of SUAPRGA1 predicted by using the computer program PredictProtein (EMBL database) suggested that this protein forms the same C-terminal α-helices and most of the β-strands found in the p32 protein (10). Thus, it is conceivable that SUAPRGA1 belongs to the p32 group of proteins. If this is true, it seems likely that SUAPRGA1 represents a general factor which is not specific for penicillin biosynthesis because it is also present in non-penicillin-producing organisms. Rather, SUAPRGA1 appears to be involved in generation of a physiological signal which is required for full expression of the penicillin biosynthesis genes and thus penicillin production. This hypothesis is supported by the observation that overexpression of the suAprgA1 gene in A. nidulans when the alcA promoter of A. nidulans was used did not result in an increase in penicillin production or expression of penicillin biosynthesis genes beyond the levels observed in wild-type strains (unpublished data).
Further analysis of the function of the suAprgA1 gene in the aerobically growing fungus A. nidulans might also help clarify the function of the p32 protein homologs in various organisms.
ACKNOWLEDGMENTS
We thank Reinhard Fischer for providing the ordered A. nidulans gene library and Sybille Traupe for excellent help during some of the experiments.
This work was supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 369).
REFERENCES
- 1.Ballance D J, Turner G. Development of a high-frequency transforming vector for Aspergillus nidulans. Gene. 1985;36:321–331. doi: 10.1016/0378-1119(85)90187-8. [DOI] [PubMed] [Google Scholar]
- 2.Borges-Walmsley M I, Turner G, Bailey A M, Brown J, Lehmbeck J, Clausen I G. Isolation and characterisation of genes for sulphate activation and reduction in Aspergillus nidulans: implications for evolution of an allosteric control region by gene duplication. Mol Gen Genet. 1995;247:423–429. doi: 10.1007/BF00293143. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–252. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Brakhage A A. Molecular regulation of beta-lactam biosynthesis in filamentous fungi. Microbiol Mol Biol Rev. 1998;62:547–585. doi: 10.1128/mmbr.62.3.547-585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brakhage A A, Browne P, Turner G. Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J Bacteriol. 1992;174:3789–3799. doi: 10.1128/jb.174.11.3789-3799.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brakhage A A, Van den Brulle J. Use of reporter genes to identify recessive trans-acting mutations specifically involved in the regulation of Aspergillus nidulans penicillin biosynthesis genes. J Bacteriol. 1995;177:2781–2788. doi: 10.1128/jb.177.10.2781-2788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brody H, Griffith J, Cuticchia A J, Arnold J, Timberlake W E. Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res. 1991;19:3105–3109. doi: 10.1093/nar/19.11.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espeso E A, Tilburn J, Sánchez-Pulido L, Brown C V, Valencia A, Arst H N, Jr, Peñalva M A. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J Mol Biol. 1997;274:466–480. doi: 10.1006/jmbi.1997.1428. [DOI] [PubMed] [Google Scholar]
- 9.Fantes P A, Roberts C F. β-Galactosidase activity and lactose utilization in Aspergillus nidulans. J Gen Microbiol. 1973;77:471–486. doi: 10.1099/00221287-77-2-417. [DOI] [PubMed] [Google Scholar]
- 9a.Fungal Genome Resource Website. David Hall. [Online.] http://fungus.genetics.uga.edu:5080/CH6_MIN.txt. [12 October 1999, last date accessed.]
- 10.Jiang J, Zhang Y, Krainer A R, Xu R-M. Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc Natl Acad Sci USA. 1999;96:3572–3577. doi: 10.1073/pnas.96.7.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 12.Litzka O, Papagiannopoulos P, Davis M A, Hynes M J, Brakhage A A. The penicillin regulator PENR1 of Aspergillus nidulans is a HAP-like transcriptional complex. Eur J Biochem. 1998;251:758–767. doi: 10.1046/j.1432-1327.1998.2510758.x. [DOI] [PubMed] [Google Scholar]
- 13.Litzka O, Then Bergh K, Brakhage A A. Analysis of the regulation of Aspergillus nidulans penicillin biosynthesis gene aat (penDE) encoding acyl coenzyme A:6-aminopenicillanic acid acyltransferase. Mol Gen Genet. 1995;249:557–569. doi: 10.1007/BF00290581. [DOI] [PubMed] [Google Scholar]
- 14.MacCabe A P, Riach M B R, Unkles S E, Kinghorn J R. The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J. 1990;9:279–287. doi: 10.1002/j.1460-2075.1990.tb08106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacCabe A P, van Liempt H, Palissa H, Unkles S E, Riach M B R, Pfeifer E, von Döhren H, Kinghorn J R. δ-(l-α-Aminoadipyl)-l-cysteinyl-d-valine synthetase from Aspergillus nidulans—molecular characterization of the acvA gene encoding the first enzyme of the penicillin biosynthetic pathway. J Biol Chem. 1991;266:12646–12654. [PubMed] [Google Scholar]
- 16.Muta T, Kang D, Kitajima S, Fujiwara T, Hamasaki N. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J Biol Chem. 1997;272:24363–24370. doi: 10.1074/jbc.272.39.24363. [DOI] [PubMed] [Google Scholar]
- 17.Orejas M, Espeso E A, Tilburn J, Sarkar S, Arst H N, Jr, Peñalva M A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 18.Papagiannopoulos P, Andrianopoulos A, Sharp J A, Davis M A, Hynes M J. The hapC gene of Aspergillus nidulans is involved in the expression of CCAAT-containing promoters. Mol Gen Genet. 1996;251:412–421. doi: 10.1007/BF02172369. [DOI] [PubMed] [Google Scholar]
- 19.Peñalva M A, Rowlands R T, Turner G. The optimization of penicillin biosynthesis in fungi. Trends Biotechnol. 1998;16:483–489. doi: 10.1016/s0167-7799(98)01229-3. [DOI] [PubMed] [Google Scholar]
- 20.Pérez-Esteban B, Gómez-Pardo E, Peñalva M A. A lacZ reporter fusion method for the genetic analysis of regulatory mutations in pathways of fungal secondary metabolism and its application to the Aspergillus nidulans penicillin pathway. J Bacteriol. 1995;177:6069–6076. doi: 10.1128/jb.177.21.6069-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pontecorvo G, Roper J A, Hemmons L M, MacDonald K D, Bufton A W J. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 22.Prade R A, Griffith J, Kochut K, Arnold J, Timberlake W E. In vitro reconstruction of the Aspergillus (= Emericella) nidulans genome. Proc Natl Acad Sci USA. 1997;94:14564–14569. doi: 10.1073/pnas.94.26.14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1:17–20. [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seytter T, Lottspeich F, Neupert W, Schwarz E. Mam33p, an oligomeric, acidic protein in the mitochondrial matrix of Saccharomyces cerevisiae, is related to the human complement receptor gC1q-R. Yeast. 1998;14:303–310. doi: 10.1002/(SICI)1097-0061(19980315)14:4<303::AID-YEA217>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 27.Steidl S, Papagiannopoulos P, Litzka O, Andrianopoulos A, Davis M A, Brakhage A A, Hynes M J. AnCF, the CCAAT binding complex of Aspergillus nidulans, contains products of the hapB, hapC and hapE genes and is required for activation by the pathway-specific regulatory gene amdR. Mol Cell Biol. 1999;19:99–106. doi: 10.1128/mcb.19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Then Bergh K, Litzka O, Brakhage A A. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J Bacteriol. 1996;178:3908–3916. doi: 10.1128/jb.178.13.3908-3916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Peñalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acidic- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Prade R A, Griffith J, Timberlake W E, Arnold J. A fast random cost algorithm for physical mapping. Proc Natl Acad Sci USA. 1994;91:11094–11098. doi: 10.1073/pnas.91.23.11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weidner G, d’Enfert C, Koch A, Mol P, Brakhage A A. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine monophosphate decarboxylase. Curr Genet. 1998;33:378–385. doi: 10.1007/s002940050350. [DOI] [PubMed] [Google Scholar]
- 32.Weidner G, Steffan B, Brakhage A A. The Aspergillus nidulans lysF gene encodes homoaconitase, an enzyme involved in the fungus-specific lysine biosynthesis pathway. Mol Gen Genet. 1997;255:237–247. doi: 10.1007/s004380050494. [DOI] [PubMed] [Google Scholar]