Abstract
Background:
Early diagnosis and treatment of prostate cancer (PCa) can be curative, however prostate specific antigen is a suboptimal screening test for clinically significant PCa. While prostate MRI has demonstrated value for the diagnosis of PCa, the acquisition time is too long for a first-line screening modality.
Purpose:
To accelerate prostate MRI exams, utilizing a variational network (VN) for image reconstruction.
Study Type:
Retrospective
Subjects:
113 subjects (train/val/test: 70/13/30) undergoing prostate MRI.
Field Strength/Sequence:
3.0 T; A T2 Turbo spin echo (TSE) T2 weighted image (T2WI) sequence in axial and coronal planes, and axial echo-planar diffusion weighted imaging (DWI).
Assessment:
Four abdominal radiologists evaluated the image quality of VN reconstructions of retrospectively under-sampled biparametric MRIs (bpMRI), and standard bpMRI reconstructions for 20 test subjects (studies). The studies included axial and coronal T2WI, DWI B50 s/mm2 and B1000 s/mm (4-fold T2WI, 3-fold DWI), all of which were evaluated separately for image quality on a Likert scale (1: non-diagnostic to 5: excellent quality). In another 10 test subjects, three readers graded lesions on bi-parametric MRIs (bpMRI) – which additionally included calculated B1500 s/mm2, and apparent diffusion coefficient (ADC) map – according to the Prostate Imaging Reporting and Data System (PI-RADS v2.1), for both VN and standard reconstructions. Accuracy of PI-RADS ≥ 3 for clinically significant cancer was computed. Projected scan time of the retrospectively under-sampled bi-parametric exam was also computed.
Statistical tests:
One-sided Wilcoxon signed-rank test was used for comparison of image quality. Sensitivity, specificity, positive predictive value, and negative predictive value were calculated for lesion detection and grading. Generalized estimating equation with cluster effect was used to compare differences between standard and VN bpMRI. A p value of <0.05 was considered statistically significant.
Results:
Three of four readers rated no significant difference for overall quality between the standard and VN axial T2WI (Reader 1: 4.00 ±0.56 (Standard), 3.90 ±0.64 (VN) p=0.33; Reader 2: 4.35 ±0.74 (Standard), 3.80 ±0.89 (VN) p=0.003; Reader 3: 4.60 ±0.50 (Standard), 4.55 ±0.60 (VN) p=0.39; Reader 4: 3.65 ±0.99 (Standard), 3.60 ±1.00 (VN) p=0.38). All four readers rated no significant difference for overall quality between standard and VN DWI B1000 s/mm2 (Reader 1: 2.25 ±0.62 (Standard), 2.45 ±0.75 (VN) p=0.96; Reader 2: 3.60 ±0.92 (Standard), 3.55 ±0.82 (VN) p=0.40; Reader 3: 3.85 ±0.72 (Standard), 3.55 ±0.89 (VN) p=0.07; Reader 4: 4.70 ±0.76 (Standard); 4.60 ±0.73 (VN) p=0.17) and three of four readers rated no significant difference for overall quality between standard and VN DWI B50 s/mm2 (Reader 1: 3.20 ±0.70 (Standard), 3.40 ±0.75 (VN) p=0.98; Reader 2: 2.85 ±0.81 (Standard), 3.00 ±0.79 (VN) p=0.93; Reader 3: 4.45 ±0.72 (Standard), 4.05 ±0.69 (VN) p=0.02; Reader 4: 4.50 ±0.69 (Standard), 4.45 ±0.76 (VN) p=0.50). In the lesion evaluation study, there was no significant difference in the number of PI-RADS ≥ 3 lesions identified on standard versus VN bpMRI (p=0.92, 0.59, 0.87) with similar sensitivity and specificity for clinically significant cancer. The average scan time of the standard clinical bi-parametric exam was 11.8 mins, and this was projected to be 3.2 mins for the accelerated exam.
Conclusion:
Diagnostic accelerated bi-parametric prostate MRI exams can be performed using deep learning methods in < 4 mins, potentially enabling rapid screening prostate MRI.
Keywords: Prostate MRI, accelerated imaging, image reconstruction, deep learning
INTRODUCTION
Prostate cancer (PCa) is the second most frequent cancer diagnosed in men and the fifth leading cause of death worldwide (1). Early diagnosis and treatment can be curative, but currently there is no adequate screening test. The blood test for prostate specific antigen (PSA) is inexpensive but lacks specificity for clinically significant PCa; it is not recommended for men over 70 years old and is only recommended for selective screening in men aged 55–69 years (2,3).
The role of prostate MRI in the detection and localization of clinically significant PCa (csPCa), defined as Gleason 7 or higher prostate cancer (4–6), has become increasingly important (7–10). Many recent studies support the role of multi-parametric MRI (mp-MRI) in the diagnosis and management of PCa, not only for detection but also for guiding biopsy of csPCa (11–13). While prostate MRI has demonstrated value, the prohibitively long acquisition time poses a major challenge in the clinical implementation as a first-line screening tool. To address this challenge, an abbreviated protocol has been proposed, which omits dynamic contrast enhanced (DCE) imaging, leaving only T2-weighted (T2WI) and diffusion-weighted imaging (DWI) (14,15). A recent study demonstrated increased sensitivity in detecting csPCa with this bi-parametric MRI (bp-MRI) protocol (16). Although this approach reduces acquisition time and cost, it still requires approximately 15 minutes and thus does not sufficiently reduce scan time for practical routine screening.
Accelerating MRI acquisitions has been an active area of research for many years. The major developments that have contributed to faster imaging are parallel imaging (17,18) and compressed sensing (19), and more recently deep learning. Deep learning- based reconstruction approaches are a generalization of traditional regularized reconstruction and have been shown to outperform parallel imaging and compressed sensing in many applications (20–23). These techniques show great promise in achieving high image quality in short scan times. A recent study demonstrated that 4-fold accelerated knee exams, reconstructed with a variational network (VN) were interchangeable with clinical protocol (24). Another study, showed that VN reconstruction of pediatric abdominal images resulted in superior image quality compared to the traditional parallel imaging, compressed sensing (PICS) reconstruction (25).
Our work builds upon the VN described by Hammernik et al (23). With expanded model capacity, incorporation of multiple sets of coil sensitivities (26) and utilization of state-of-the-art optimizer (27), we extended the VN to reconstruction of accelerated clinical prostate images. With this novel VN implementation, we aim to accelerate biparametric prostate exams while maintaining the image quality of the current clinical protocol.
METHODS
Study Population
This study was approved by an institutional review board (IRB) and was HIPAA compliant. Given the retrospective nature of this study it received waiver of consent. Clinical prostate MRI exam performed previously in 113 male patients on a 3.0T MR scanner (MAGNETOM Vida, Siemens Healthineers, Germany) with a 30-channel body coil was utilized in this study. The mean age of the subjects was 68 ± 7 years. These clinically acquired data sets were used retrospectively to develop, test, and validate accelerated acquisitions.
Image acquisition & reconstruction
T2-weighted images
For each patient a 2D T2 turbo spin echo (TSE) sequence was performed in both the axial and coronal planes. The scan parameters for the coronal acquisition were: TR = 4 s, TE = 100 ms Echo train length (ETL) = 25, in-plane resolution = 0.56 mm × 0.56 mm, matrix size = 320 × 320, slice thickness = 3 mm, under-sampling factor (R) = 2, Number of averages (NEX) = 2, and scan time 2.55 mins. For the axial acquisition, scan parameters were: TR = 4 s, TE =100 ms, ETL= 25, in-plane resolution = 0.56 mm × 0.56 mm, slice thickness = 3 mm, R = 2, NEX = 3, and scan time = 3.87 mins.
For each image, the coil sensitivity maps wer estimated using ESPIRIT (26). Similar to the soft-sense reconstruction described earlier (26), two sets of coil sensitivity maps were used for the reconstruction. This was necessitated by the extension of anatomy beyond the field of view.
The raw data were acquired with interleaved averages such that the target data was effectively fully sampled. Specifically, for the coronal acquisition, the first average sampled odd lines of k-space, while the second average sampled even lines. This is the clinical (standard) protocol at our institution. The ground truth/target image was then reconstructed by adding the two averages and performing an inverse fast Fourier transform (IFFT). For the axial acquisitions there were 3 averages. The odd averages sampled odd lines and the second average sampled even lines. The averages were then combined as: 0.5 × (average1+average3) + average2 before performing an IFFT.
The fully sampled raw k-space data were retrospectively under-sampled; the sampling pattern was consistent with an under-sampling rate (R) = 4 equidistant under-sampling, commonly used in parallel imaging, where every 4th line was sampled. In addition, we sampled 32 lines at the center of k-space for calibration of the receive coil sensitivity maps. The projected scan times for the accelerated protocol were calculated by dividing the standard scan time by the relative acceleration factor (4 or 6) and adding an additional TR for the extra center lines. The retrospectively under-sampled raw data were then reconstructed using the trained VN.
A soft-sense reconstruction was also performed for both the coronal (SScor) and axial (SSax) images to serve as a conventional parallel imaging comparison. This was essentially a relaxed sense reconstruction that utilized the same two sets of coil sensitivities as were used for the VN reconstruction. Structural similarity index (SSIM) (28) was calculated for all test set images in order to evaluate the relative performance of soft-sense and the VN.
Diffusion weighted images
In addition to T2-weighted images, echo planar imaging (EPI)-DWI were also acquired for each subject. The EPI-DWI scans were performed using tri-directional diffusion-sensitizing gradients with b values of 50 s/mm2 (B50), and 1000 s/mm2 (B1000), performed with 4, and 12 averages, respectively. Images were acquired with FOV 20 × 20 cm, matrix 90 × 90, R =2 under-sampling, ETL = 75, TR = 5.8 – 6.5 s, TE = 77 ms and scan time ~ 5.4 mins.
The standard B50 and B1000 images were reconstructed with an EPI-gridding method, followed by GRAPPA, with all the acquired averages. The averages were combined as the final step by combining the magnitude images in image space to avoid phase cancellation. The VN was used to reconstruct a single average for the B50 images and 4 averages for the B1000 images, resulting in a 4-fold and 3-fold acceleration respectively. The projected scan time for the accelerated protocol was calculated by multiplying the standard scan by the fraction of averages retained (5/16) and adding an additional TR for the extra calibration lines. A geometric mean of the tri-directional reconstructed images was then performed to yield diffusion trace images for B50 and B1000. Apparent diffusion coefficient (ADC) maps and an estimated B1500 image were also calculated from the tri-directional reconstructed images.
Variational network architecture and training
Our method was inspired by the VN – a model-based, deep learning reconstruction framework (18,20). The input to the network included the measured k-space samples, two sets of coil sensitivity maps calculated with ESPIRIT and the reconstructed zero-filled image. This data processing pipeline is illustrated in Figure 1.
Figure 1.
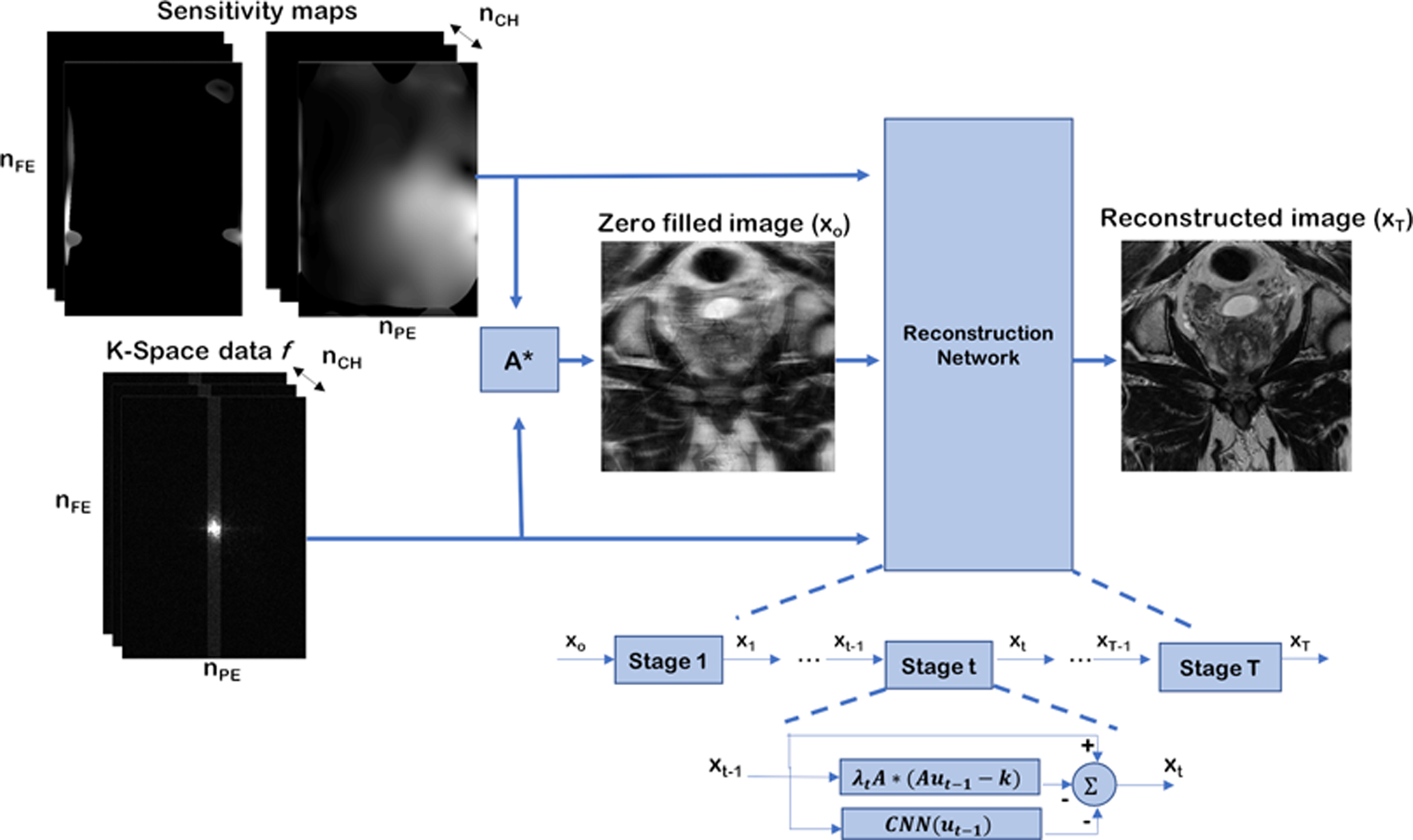
Data processing pipeline, and structure of the reconstruction network. First, a zero filled reconstruction is generated from the under-sampled k-space and coil sensitivity maps. Then, this reconstruction, along with the raw k-space data and sensitivity maps are passed as the input to the reconstruction network. The reconstruction network has 10 stages (T = 10), each stage applies data consistency and regularization, as described in equation (1). The T2 reconstruction networks and the B50 diffusion network reconstruct a single image from a single zero-filled input image, while the B1000 diffusion network reconstructs four images from multiple zero-filled images, and combines the averages as the last step.
The reconstruction network consisted of multiple stages, each modeled after a single gradient descent step. In the t-th stage the image was updated from xt to xt+1 using:
| (1) |
where λt is a learned parameter that controls the relative weighting between data consistency and regularization, A is the linear forward operator that applies sensitivity maps, the 2D Fourier transform, and under-sampling, is the measured k-space data and CNN is a convolutional neural network that regularizes the reconstruction. The CNN used for this reconstruction was modelled after a UNet (29) and is illustrated in Figure 2. The encoder portion of the network was made up of convolution and max pooling layers, while the decoder was made up of convolution and up-sampling layers. Skip connections, concatenations of encoder and decoder layers, were also included. All convolution kernels were 3×3. Instance normalization and rectified linear unit (ReLU) were applied to the output of each convolution layer. The model consisted of approximately 8 million learnable parameters.
Figure 2.

Illustration of the CNN regularizer, based off the UNet model [24]
Network training for T2-weighted images
83 subjects were randomly divided into training (70) and validation (13) sets, the test set was an additional 20 datasets acquired from 20 consecutive subjects. Additional 10 subjects were included for the lesion detection experiment. The coronal image inputs were a single average with retrospective 4-fold under-sampling relative to Nyquist sampling, for an effective acceleration (R) = 4, relative to the clinical protocol. The axial image inputs were also a 4-fold retrospectively under-sampled single average, which resulted in an effective acceleration of R = 6, relative to the clinical protocol. The target images for both networks were the corresponding fully-sampled images. The two networks were trained with the Adam optimizer and a learning rate (LR) = 1 × 10−3. The weights of the model were updated after every forward pass through the network (batch size =1) to minimize the mean squared error (MSE) of the prediction relative to the fully-sampled target images. This process used the training data-set, while the validation set was used as stopping criteria to ensure the model was not over-fitting the training data.
Network training for diffusion-weighted images
The subjects described in the previous section were also used to train and evaluate the diffusion network. A separate model was trained for B50 and B1000 images. The image inputs were 1 average and 4 averages for the B50 and B1000, respectively, which results in an approximately 3-fold acceleration overall, relative to clinical protocol. The input and target images were 2-fold under-sampled relative to Nyquist sampling. The B1000 network had an extra channel dimension to accommodate the 4 averages. This dimension was retained throughout the network, and the averages were only combined at the final stage prior to calculation of the loss function. The target images were reconstructed with GRAPPA. The networks were again trained with the Adam optimizer, a learning rate (LR) = 1 × 10−3, and an MSE loss function.
Reader study
Image Quality:
The trained models were evaluated for image quality on an independent test data set from 20 consecutive patients. Four abdominal fellowship trained radiologists (with 6 (KM), 3 (PS), 1 (AD), and 1 (RP) years of clinical experience interpreting prostate MRIs) evaluated the images and were blinded to the acquisition/reconstruction schemes. The VN T2WI axial and coronal (VNT2ax and VNT2cor), standard T2WI axial and coronal (StandardT2Ax and StandardT2Cor), VN diffusion and standard clinical diffusion (VNDiffB50, VNDiffB1000, StandardDiffB50, StandardDiffB1000) images were evaluated for the following image quality metrics utilizing a five point Likert grading scale: overall quality, prostate capsule clarity, clarity of peripheral zone and transition zone boundary, clarity of periurethral region (T2WI only), and clarity in relation to artifacts (DWI only). The five-point Likert scale was applied to all the metrics by each of the readers and was defined as follows: 1 – non-diagnostic, 2 – poor quality, 3 – moderate quality, 4 – good quality, 5 – excellent quality (30). All four readers were trained with independent unlabeled example images of the same type of reconstructions prior to the study.
Lesion identification:
An additional 10 consecutive patients (new and not used in the above image quality experiment) who had undergone MRI of the prostate for suspicion for prostate cancer and who had prospectively identified lesions risk stratified as PI-RADS ≥ 3 were included. Each patient had a subsequent MRI/transrectal ultrasound (TRUS) fusion biopsy performed within six months of the MRI. The clinical histopathology results from the biopsies were obtained and reported as Gleason Grade Group scores with “clinically significant” prostate cancer (csPCa) defined as Gleason Grade Group 2 or greater (31). From these 10 patients, a total of 14 lesions were prospectively assigned PI-RADS ≥ 3 scores on multiparametric MRI and subsequently underwent MRI/TRUS fusion biopsy.
Three abdominal fellowship trained radiologists (with 6 (KM), 3 (PS), and 1 (RP) years of experience)blinded to the reconstruction scheme were presented independently and randomly, VN accelerated and standard bp-MRI data that consisted of T2WI and DWI (b50 s/mm2, b1000 s/mm2, calculated b1500 s/mm2, and ADC map). The VN accelerated exam consisted of axial and coronal VN T2WI and VN DWI. The conventional exam consisted of standard axial and coronal T2WI and standard axial DWI. The readers were asked to identify and assign scores to lesions according to Prostate Imaging Reporting and Data System (PI-RADS) v2.1. We used PI-RADS ≥ 3 as a cutoff for positive scoring for prostate cancer as it is used as a minimum score for targeted MRI targeted biopsy (5).
Statistical Analyses
One-sided paired Wilcoxon signed-rank test was used to compare image quality evaluations of standard versus reconstructed images with the null hypothesis that VN was at least as good as the standard. Significance was designated as having p < 0.05. Frequency of studies with difference of ±1 between standard and reconstructed images were also calculated.
Sensitivity, specificity, positive predictive value, and negative predictive value were calculated for each reader separated by standard and VN exams. The number of PI-RADS ≥ 3 lesions that were assigned by each reader on standard and VN exams were compared utilizing generalized estimating equation model with cluster effect.
RESULTS
T2WI results
The average acquisition time for the 20 coronal test set volumes was 2.55 mins, this scan time is projected to be reduced to 0.7 mins with the R = 4 acceleration compared to clinical protocol. The average scan time for the axial test set volumes was 3.87 mins, which would be reduced to 0.7 mins with the R = 6 acceleration compared to clinical protocol.
The mean SSIM to the fully sampled standard images, calculated over all slices of the test set was 0.86 ±0.05, and 0.90 ± 0.03 for the VNT2cor and VNT2ax images, respectively. For the soft-sense reconstructions (parallel imaging comparison) the mean SSIM was 0.61 ± 0.07 and 0.75 ± 0.07 for the SScor and SSax images, respectively. Two representative examples for the coronal and axial reconstructions are shown in Figures 3 and 4.
Figure 3.
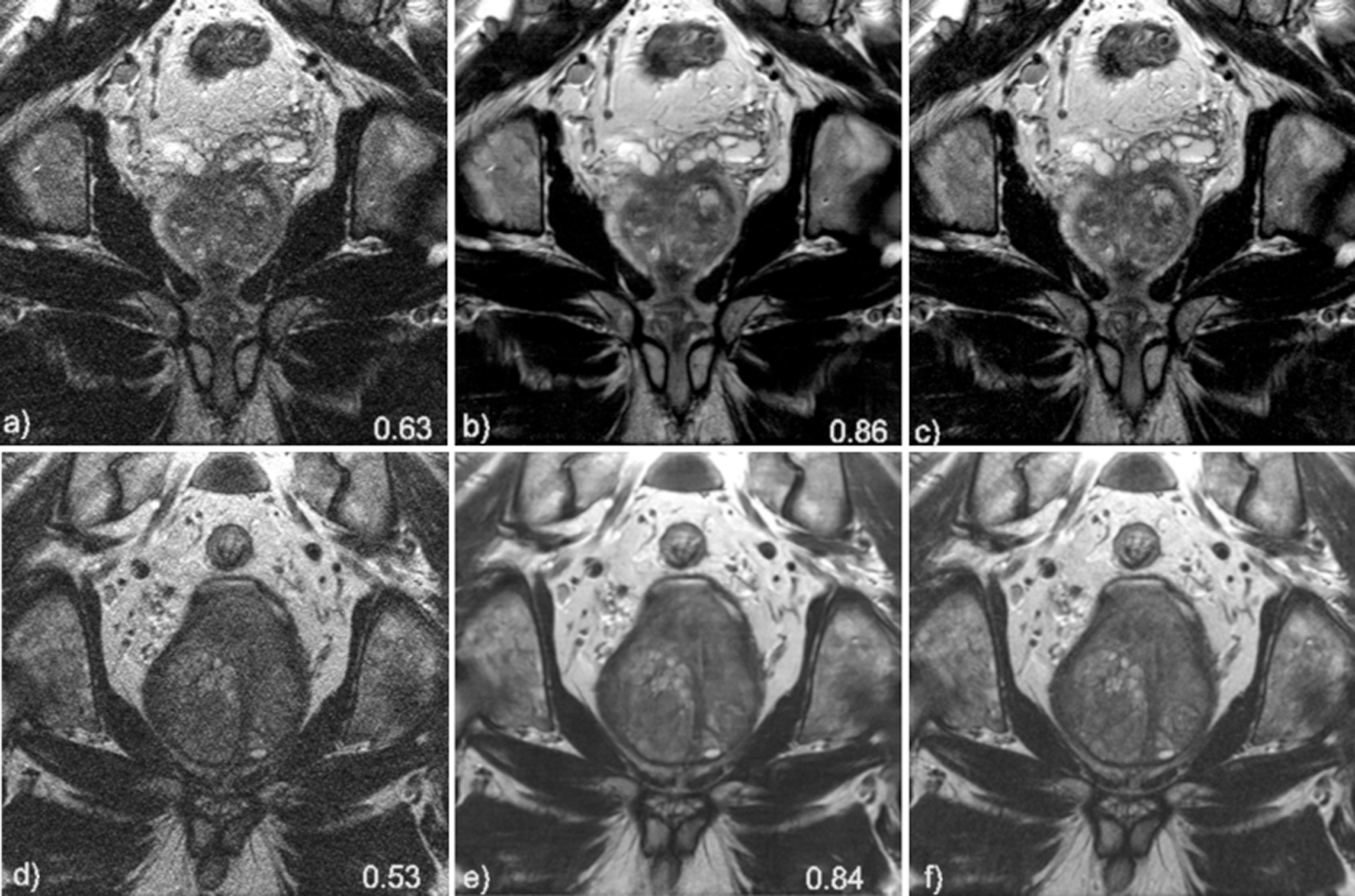
Coronal image results for subject 1 (top row) and subject 2 (bottom row). Soft-sense reconstructions are shown in a) and d). VN reconstructions are shown in b) and e) while the fully while the fully sampled, ground-truth images are shown in c) and f). The value indicated on the images is the calculated SSIM of the slice shown.
Figure 4.

Axial image results for subject 3 (top row) and subject 4 (bottom row). Soft-sense reconstructions are shown in a) and d). VN reconstructions are shown in b) and e) while the fully sampled, ground-truth images are shown in c) and f). The value indicated on the images is the calculated SSIM of the slice shown.
DWI results
The average acquisition time for the diffusion test set volumes was 5.4 mins, the projected scan time would be reduced to 1.8 mins with the described acceleration scheme. The mean SSIM to the fully sampled standard images, calculated over all slices of the test set was 0.93 ± 0.03 and 0.85 ± 0.07, for the B50 and B1000 images, respectively. For the GRAPPA reconstructions, the mean SSIM was 0.88 ± 0.05 and 0.77± 0.09 for the B50 and B1000 images, respectively. Representative results are shown in Figure 5.
Figure 5.
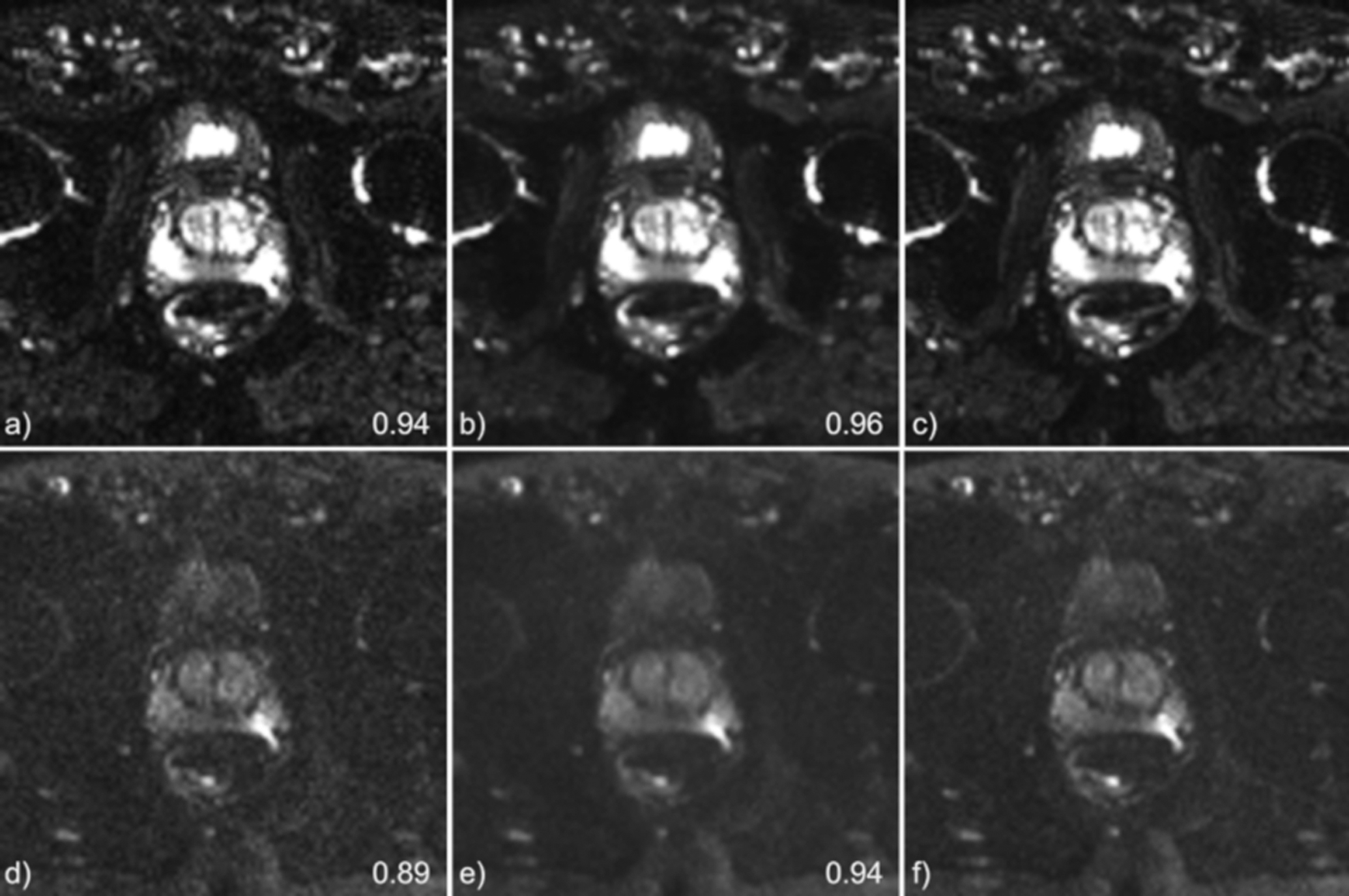
Diffusion B50 s/mm2 (top) and B1000 s/mm2 (bottom) results for a single subject. Grappa reconstructions are shown in a) and d). VN reconstructions are shown in b) and e) while the grappa reconstructed, ground-truth images are shown in c) and f). The value indicated on the images is the calculated SSIM of the slice shown.
Reader study results
Image Quality:
Table 1 includes the image quality comparison results of the T2WI from each reader. Three of four readers rated no significant difference of overall quality of VN axial T2WI compared to standard, although all readers rated decreased overall quality of VNT2Ax compared to StandardT2Ax (Reader 1: 4.00 ±0.56 (Standard), 3.90 ±0.64 (VN) p=0.33; Reader 2: 4.35 ±0.74 (Standard), 3.80 ±0.89 (VN) p=0.003; Reader 3: 4.60 ±0.50 (Standard), 4.55 ±0.60 (VN) p=0.39; Reader 4: 3.65 ±0.99 (Standard), 3.60 ±1.00 (VN) p=0.38).
Table 1.
Image Quality Comparison Between Standard and VN Reconstructed T2 weighted imaging
| Reader 1 | Reader 2 | Reader 3 | Reader 4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Imaging plane | Characteristic (1–5) | Standard (mean, std dev) | VN (mean, std dev) | p | Standard (mean, std dev) | VN (mean, std dev) | p | Standard (mean, std dev) | VN (mean, std dev) | p | Standard (mean, std dev) | VN (mean, std dev) | p |
| Axial | Overall Quality | 4.00, 0.56 | 3.90, 0.64 | 0.334 | 4.35, 0.74 | 3.80, 0.89 | 0.003 * | 4.60, 0.50 | 4.55, 0.60 | 0.386 | 3.65, 0.99 | 3.60, 1.00 | 0.383 |
| Prostate Capsule Clarity | 4.10, 0.64 | 3.90, 0.72 | 0.195 | 4.30, 0.80 | 3.80, 0.89 | 0.007 * | 4.60, 0.50 | 4.55, 0.60 | 0.386 | 4.95, 0.22 | 4.95, 0.22 | 0.681 | |
| Clarity of boundary of TZ and PZ | 4.05, 0.51 | 3.90, 0.64 | 0.258 | 4.40, 0.75 | 3.90, 0.97 | 0.012 * | 4.50, 0.51 | 4.45, 0.60 | 0.383 | 4.80, 0.52 | 4.55, 0.69 | 0.060 | |
| Clarity of peri-urethral area | 4.00, 0.46 | 3.85, 0.81 | 0.227 | 4.45, 0.76 | 3.70, 1.13 | 0.002 * | 4.50, 0.61 | 4.45, 0.69 | 0.386 | 4.80, 0.41 | 4.55, 0.60 | 0.018 * | |
| Coronal | Overall Quality | 3.75, 0.55 | 3.45, 0.60 | 0.072 | 4.20, 0.95 | 4.20, 0.89 | 0.531 | 4.30, 0.66 | 4.10, 0.55 | 0.065 | 3.20, 1.00 | 2.90, 0.97 | 0.021 * |
| Prostate Capsule Clarity | 3.80, 0.52 | 3.45, 0.60 | 0.049 * | 4.20, 0.81 | 4.20, 0.81 | 0.531 | 4.35, 0.66 | 4.10, 0.56 | 0.018 * | 3.90, 0.75 | 3.75, 0.91 | 0.117 | |
| Clarity of boundary of TZ and PZ | 3.70, 0.57 | 3.20, 0.52 | 0.010 * | 3.95, 0.95 | 4.25, 0.89 | 0.950 | 4.10, 0.59 | 3.85, 0.55 | 0.060 | 4.15, 1.07 | 3.70, 1.12 | 0.004 * | |
| Clarity of peri-urethral area | 3.75, 0.55 | 3.45, 0.60 | 0.072 | 4.20, 1.10 | 4.20, 1.02 | 0.531 | 4.30, 0.64 | 4.10, 0.49 | 0.065 | 3.20, 0.67 | 2.90, 0.92 | 0.021 * | |
Standard = Clinical Sequence; VN = Variational Network Sequence; PZ = Peripheral Zone; TZ = Transition Zone; std dev = standard deviation
significant p value
Three of four readers rated no significant difference in overall quality between VNT2cor and StandardT2Cor, including one reader who rated the exact same overall quality of VNT2cor compared to StandardT2Cor(Reader 1: 3.75 ±0.55 (Standard), 3.45 ±0.60 (VN) p=0.07; Reader 2: 4.20 ±0.95 (Standard), 4.20 ±0.89 (VN) p=0.53; Reader 3: 4.30 ±0.66 (Standard), 4.10 ±0.55 (VN) p=0.06; Reader 4: 3.20 ±1.00 (Standard), 2.90 ±0.97 (VN) p=0.02).
Table 2 includes the image quality comparison results of the DWI from each reader. One reader rated increased overall quality while three readers rated decreased overall quality of VN DWI B1000 s/mm2 compared to standard, all without significant difference (Reader 1: 2.25 ±0.62 (Standard), 2.45 ±0.75 (VN) p=0.96; Reader 2: 3.60 ±0.92 (Standard), 3.55 ±0.82 (VN) p=0.40; Reader 3: 3.85 ±0.72 (Standard), 3.55 ±0.89 (VN) p=0.07; Reader 4: 4.70 ±0.76 (Standard); 4.60 ±0.73 (VN) p=0.17). Two of four readers rated increased overall quality of VN DWI B50 s/mm2 compared to standard without significant difference while two readers rated decreased overall quality of VN DWI B50 s/mm2 compared to standard, one with significant difference (Reader 1: 3.20 ±0.70 (Standard), 3.40 ±0.75 (VN) p=0.98; Reader 2: 2.85 ±0.81 (Standard), 3.00 ±0.79 (VN) p=0.93; Reader 3: 4.45 ±0.72 (Standard), 4.05 ±0.69 (VN) p=0.02; Reader 4: 4.50 ±0.69 (Standard), 4.45 ±0.76 (VN) p=0.50).
Table 2.
Image Quality Comparison Between Standard and VN Reconstructed Diffusion Weighted Images
| Reader 1 | Reader 2 | Reader 3 | Reader 4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| b value (s//mm2) | Characteristic (1–5) | Standard (mean, std dev) | VN (mean, std dev) | p | Standard (mean, std dev) | VN (mean, std dev) | p | Standard (mean, std dev) | VN (mean, std dev) | p | Standard (mean, std dev) | VN (mean, std dev) | p |
| 50 | Overall Quality | 3.20, 0.70 | 3.40, 0.75 | 0.986 | 2.85, 0.81 | 3.00, 0.79 | 0.932 | 4.45, 0.72 | 4.05, 0.69 | 0.018 * | 4.50, 0.69 | 4.45, 0.76 | 0.500 |
| Prostate Capsule Clarity | 2.40, 0.60 | 2.55, 0.60 | 0.932 | 3.00, 0.65 | 3.05, 0.89 | 0.682 | 4.45, 0.67 | 4.10, 0.72 | 0.020 * | 4.80, 0.41 | 4.70, 0.57 | 0.500 | |
| Clarity of boundary of TZ and PZ | 3.70, 0.66 | 3.75, 0.44 | 0.725 | 3.10, 0.85 | 3.10, 0.91 | 0.546 | 4.40, 0.81 | 4.05, 0.69 | 0.020 * | 4.85, 0.37 | 4.80, 0.52 | 0.500 | |
| Clarity from artifacts | 3.25, 0.72 | 3.40, 0.75 | 0.932 | 2.75, 0.85 | 2.95, 0.69 | 0.868 | 4.85, 0.37 | 4.85, 0.37 | 0.681 | 4.60, 0.50 | 4.50, 0.69 | 0.173 | |
| 1000 | Overall Quality | 2.25, 0.62 | 2.45, 0.75 | 0.960 | 3.60, 0.92 | 3.55, 0.82 | 0.401 | 3.85, 0.72 | 3.55, 0.89 | 0.072 | 4.70, 0.76 | 4.60, 0.73 | 0.173 |
| Prostate Capsule Clarity | 3.50, 0.55 | 3.40, 0.60 | 0.212 | 3.75, 0.88 | 3.35, 1.00 | 0.033 * | 3.65, 0.67 | 3.40, 0.76 | 0.193 | 4.35, 0.47 | 4.15, 0.50 | 0.036 * | |
| Clarity of boundary of TZ and PZ | 3.20, 0.69 | 3.35, 0.75 | 0.890 | 3.65, 0.91 | 3.50, 1.09 | 0.232 | 4.85, 0.81 | 4.85, 0.82 | 0.681 | 4.60, 0.88 | 4.45, 0.81 | 0.074 | |
| Clarity from artifacts | 3.25, 0.62 | 3.40, 0.75 | 0.932 | 2.75, 0.81 | 2.95, 0.83 | 0.868 | 4.85, 0.37 | 4.85, 0.37 | 0.681 | 4.60, 0.50 | 4.50, 0.69 | 0.173 | |
Standard = Clinical Sequence; VN = Variational Network Sequence; PZ = Peripheral Zone; TZ = Transition Zone; std dev = standard deviation
significant p value
Lesion Identification.
A total of 5 of the 14 lesions demonstrated Gleason Grade Group 2 prostate cancer or higher. Table 3 shows the results of the reader study. PI-RADS ≥ 3 was assigned to 7 lesions on standard bp-MRI exams and to 8 lesions on VN bp-MRI exams by Reader 1 (p=0.92), assigned to 7 lesions on standard exams and to 9 lesions on VN exams by Reader 2 (p=0.59), and assigned to 8 lesions on standard exams and 8 lesions on VN exams by Reader 3 (p=0.87). The VN bp-MRI exam had sensitivity and specificity which were not significantly different, compared to the standard bp-MRI exam for csPCa as shown in the Table 3. Figure 6 shows an example of a lesion that is visible in both the standard and VN bp-MRI exams.
Table 3.
Comparison of sensitivity, specificity, positive predictive value, and negative predictive value of lesion identification and grading for readers individually and as a group.
| Reader 1 | Reader 2 | Reader 3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Standard | VN | P | Standard | VN | P | Standard | VN | P | ||
| PI-RADS >=3 criteria | Sensitivity | 0.40 | 0.60 | 0.53 | 0.60 | 1.00 | NA* | 0.60 | 0.60 | 1.00 |
| Specificity | 0.55 | 0.64 | 0.62 | 0.60 | 0.64 | 0.86 | 0.58 | 0.55 | 0.85 | |
| PPV | 0.29 | 0.38 | 0.72 | 0.43 | 0.56 | 0.62 | 0.38 | 0.38 | 1.00 | |
| NPV | 0.67 | 0.82 | 0.44 | 0.75 | 1.00 | NA* | 0.78 | 0.75 | 0.89 | |
NA = not applicable.
p value is not feasible when there is limited variation in data (e.g. when positive predictive value is 1 for one of the comparators). PI-RADS = prostate imaging reporting and data system; VN = Variational Network; PPV = positive predictive value; NPV = negative predictive value.
Figure 6.
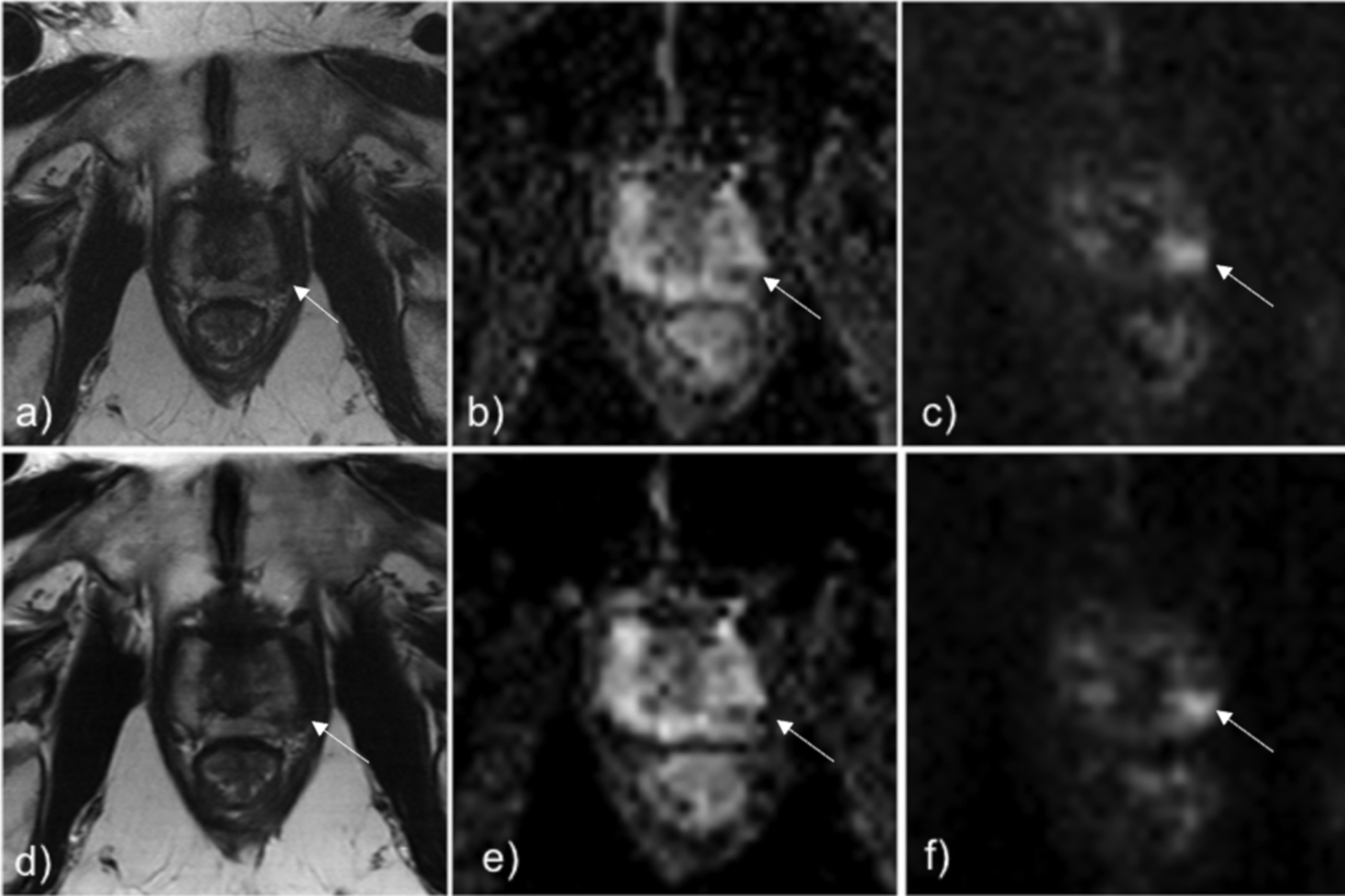
Standard T2 TSE (Fig a), ADC (Fig b), and DWI b=1500 s/mm2 (Fig c) shows lesion in the left posterolateral midgland prostate (white arrow), which was biopsied to be Gleason Grade Group 3. The same lesion can be seen identically on the DL-reconstructed T2 TSE (Fig d), ADC (Fig e), and DWI b=1500 s/mm2 (Fig f).
DISCUSSION
In this work, we developed and clinically evaluated a VN approach for accelerated T2WI and DWI of the prostate. The VN approach enabled reduced acquisition times from a standard protocol time of 11.8 mins to a projected scan time of 3.2 mins. Furthermore, compared with the parallel imaging reconstructions with equivalent acceleration, the VN- images had higher SSIM.
In our study, three of four readers rated no significant difference in image quality between Standard and VN in both axial and coronal planes. Although one reader scored VN reconstructions significantly lower than Standard sequences for the T2WI, the differences were small. A study by Gassenmaier et al (32), evaluated a deep learning reconstructed axial T2 of the prostate which found that readers rated the deep learning reconstructed axial T2 to be higher overall quality. However, in our reconstruction, we were able to decrease scan time by 6 fold compared to their 3 fold and 4 fold acceleration, which may account for the quality differences.
All four readers rated no significant difference in image quality between Standard and VN in B1000 DWI while three of four readers rated no significant difference in image quality between Standard and VN B50 DWI. As clinical interpretation of prostate MRI typically utilizes the high B values more heavily, these small differences are likely not to affect clinical interpretation.
This point is reiterated in our lesion identification and grading portion of the study, which showed no significant difference in the number of PI-RADS ≥ 3 lesions identified by any of the readers. Furthermore, the sensitivity and specificity of the VN bp-MRI exam for csPCa were also not significantly different compared to that of the standard bp-MRI exam. This preliminary study demonstrated promising results that VN reconstructed MRI may possibly be interchangeable with the clinical exams.
An abbreviated T2-weighted prostate MRI protocol that also utilizes deep learning reconstruction was recently proposed by O’Shea et al (33). This method used deep learning for noise removal and eliminated the need for multiple averages. Our proposed method builds upon this technique by utilizing deep learning for sparse sampling reconstruction in addition to denoising, enabling greater overall acceleration of the T2-weighted scans. Acceleration of the DWI scan was an additional novel element of our work and was critical in order to achieve scan times short enough for routine screening.
MRI is increasingly utilized as a tool for annual active surveillance in low and even intermediate risk PCa (34,35). In addition, there is potential for the use of prostate MRI as a screening tool for PCa (16). The same lengthy multiparametric MRI protocol is currently commonly used for all PCa indications, however with the acceleration of T2WI and DWI, a more patient tolerable, affordable, and accessible prostate MRI can be made available to patients who may not need a full diagnostic protocol.
While it is a critical step in achieving practical routine screening, reduction of scan time alone is unlikely to sufficiently reduces costs. An additional cost consideration is the need for highly trained radiologists with specific experience in the interpretation of prostate MRI. Several groups have been working on approaches for automated diagnosis of PCa (36,37). If the learned reconstruction presented in this work is paired with deep-learning methods for automated lesion detection and classification, this could further lower the cost and increase accessibility.
Limitations
While the presented study design allowed for the comparison of image quality between the conventional and accelerated image protocols, it was a small study evaluated on only 20 subjects on image quality evaluation and 10 subjects on lesion evaluation. Hence, we could not conclude that the protocols were interchangeable, though results were promising. An important future step will be correlating reader assessments of pathological findings with biopsy results in a larger cohort. Another limitation was that the under-sampling was done retrospectively rather than prospectively. Additionally, the exams were performed on a single 3T scanner from a single vendor. It will be important moving forward to establish a prospectively accelerated protocol that is robust across a variety of scanners and vendors and to correlate reader assessments of pathological findings with biopsy results.
CONCLUSIONS
Diagnostic quality highly accelerated bp-MRI prostate exams can be acquired and reconstructed using deep learning methods with < 4 mins of acquisition time, which can enable rapid screening prostate MRI.
Grant support:
NIH R01 EB024532, NIH/NCI R21 CA256324-01 and P41EB017183
REFERENCES
- 1.Rawla P Epidemiology of Prostate Cancer. World journal of oncology 2019;10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fenton J, Weyrich M, Durbin S, Liu Y, Bang H, Melnikow J. Prostate-Specific Antigen-Based Screening for Prostate Cancer: Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2018;319(18). [DOI] [PubMed] [Google Scholar]
- 3.Kilpeläinen T, Tammela T, Roobol M, et al. False-positive screening results in the European randomized study of screening for prostate cancer. European journal of cancer 2011;47(18). [DOI] [PubMed] [Google Scholar]
- 4.Epstein JI, Walsh PC, Carmichael M, Brendler CB. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. JAMA 1994;271(5):368–374. [PubMed] [Google Scholar]
- 5.Kasivisvanathan V, Rannikko AS, Borghi M, et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N Engl J Med 2018;378(19):1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed HU, El-Shater Bosaily A, Brown LC, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet 2017;389(10071):815–822. [DOI] [PubMed] [Google Scholar]
- 7.Rosenkrantz A, Deng F, Kim S, et al. Prostate cancer: multiparametric MRI for index lesion localization--a multiple-reader study. AJR American journal of roentgenology 2012;199(4). [DOI] [PubMed] [Google Scholar]
- 8.Johnson D, Raman S, Mirak S, et al. Detection of Individual Prostate Cancer Foci via Multiparametric Magnetic Resonance Imaging. European urology 2019;75(5). [DOI] [PubMed] [Google Scholar]
- 9.Thompson J, van Leeuwen P, Moses D, et al. The Diagnostic Performance of Multiparametric Magnetic Resonance Imaging to Detect Significant Prostate Cancer. The Journal of urology 2016;195(5). [DOI] [PubMed] [Google Scholar]
- 10.Panebianco V, Valerio M, Giuliani A, Pecoraro M, Ceravolo I, Barchetti G, Catalano C, Padhani A. Clinical Utility of Multiparametric Magnetic Resonance Imaging as the First-line Tool for Men with High Clinical Suspicion of Prostate Cancer. European urology oncology 2018;1(3). [DOI] [PubMed] [Google Scholar]
- 11.Brown L, Ahmed H, Faria R, et al. Multiparametric MRI to improve detection of prostate cancer compared with transrectal ultrasound-guided prostate biopsy alone: the PROMIS study. Health technology assessment 2018;22(39). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siddiqui M, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 2015;313(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasivisvanathan V, Rannikko A, Borghi M, et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. The New England journal of medicine 2018;378(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhl C, Bruhn R, Krämer N, Nebelung S, Heidenreich A, Schrading S. Abbreviated Biparametric Prostate MR Imaging in Men with Elevated Prostate-specific Antigen. Radiology 2017;285(2). [DOI] [PubMed] [Google Scholar]
- 15.van der Leest M, Israël B, Cornel E, et al. High Diagnostic Performance of Short Magnetic Resonance Imaging Protocols for Prostate Cancer Detection in Biopsy-naïve Men: The Next Step in Magnetic Resonance Imaging Accessibility. European urology 2019;76(5). [DOI] [PubMed] [Google Scholar]
- 16.Eldred-Evans D, Burak P, Connor M, et al. Population-Based Prostate Cancer Screening With Magnetic Resonance Imaging or Ultrasonography: The IP1-PROSTAGRAM Study. JAMA oncology 2021;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002;47(6):1202–1210. [DOI] [PubMed] [Google Scholar]
- 18.Pruessmann K, Weiger M, Scheidegger M, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magnetic resonance in medicine 1999;42(5). [PubMed] [Google Scholar]
- 19.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007;58(6):1182–1195. [DOI] [PubMed] [Google Scholar]
- 20.Aggarwal H, Mani M, Jacob M. MoDL: Model-Based Deep Learning Architecture for Inverse Problems. IEEE transactions on medical imaging 2019;38(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sriram A, Zbontar J, Tullie M, Defazio A, Zitnick CL, Yakubova N, Knoll F, Johnson PM. End-to-End Variational Networks for Accelerated MRI Reconstruction In: Martel AL et al. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 MICCAI 2020 Lecture Notes in Computer Science; 2020. [Google Scholar]
- 22.Lee D, Yoo J, Tak S, Ye J. Deep Residual Learning for Accelerated MRI Using Magnitude and Phase Networks. IEEE transactions on bio-medical engineering 2018;65(9). [DOI] [PubMed] [Google Scholar]
- 23.Hammernik K, Klatzer T, Kobler E, Recht MP, Sodickson DK, Pock T, Knoll F. Learning a Variational Network for Reconstruction of Accelerated MRI Data. Magn Reson Med 2018;79(6):3055–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Recht M, Zbontar J, Sodickson D, et al. Using Deep Learning to Accelerate Knee MRI at 3 T: Results of an Interchangeability Study. AJR American journal of roentgenology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen F, Taviani V, Malkiel I, et al. Variable-Density Single-Shot Fast Spin-Echo MRI with Deep Learning Reconstruction by Using Variational Networks. Radiology 2018;289(2):366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M. ESPIRiT — An Eigenvalue Approach to Autocalibrating Parallel MRI: Where SENSE meets GRAPPA. Magn Reson Med 2014;71(3):990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. arxiv 2014;1412.6980. [Google Scholar]
- 28.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 2004;13(4):600–612. [DOI] [PubMed] [Google Scholar]
- 29.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation In: Navab N, Hornegger J, Wells W, Frangi A (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 MICCAI 2015 Lecture Notes in Computer Science, vol 9351 2015. [Google Scholar]
- 30.Dickinson L, Ahmed HU, Allen C, et al. Scoring systems used for the interpretation and reporting of multiparametric MRI for prostate cancer detection, localization, and characterization: could standardization lead to improved utilization of imaging within the diagnostic pathway? Journal of Magnetic Resonance Imaging 2013;37(1):48–58. [DOI] [PubMed] [Google Scholar]
- 31.Pierorazio PM, Walsh PC, Partin AW, Epstein JI. Prognostic Gleason grade grouping: data based on the modified Gleason scoring system. BJU Int 2013;111(5):753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gassenmaier S, Afat S, Nickel D, Mostapha M, Herrmann J, Othman AE. Deep learning-accelerated T2-weighted imaging of the prostate: Reduction of acquisition time and improvement of image quality. Eur J Radiol 2021;137:109600. [DOI] [PubMed] [Google Scholar]
- 33.O’shea A, Guidon A, Lebel RM, Bayram E, Pierce T, Mojtahed A, Harisinghani MG. Initial experience in abbreviated T2-weighted Prostate MRI using a Deep Learning reconstruction. Proc Intl Soc Mag Reson Med 28 2020(2450). [Google Scholar]
- 34.Fascelli M, George A, Frye T, Turkbey B, Choyke P, Pinto P. The role of MRI in active surveillance for prostate cancer. Current urology reports 2015;16(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C S, B N, A R, et al. Long-Term Outcomes of Active Surveillance for Prostate Cancer: The Memorial Sloan Kettering Cancer Center Experience. The Journal of urology 2020;203(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sumathipala Y, Lay N, Turkbey B, Smith C, Choyke P, Summers R. Prostate cancer detection from multi-institution multiparametric MRIs using deep convolutional neural networks. Journal of medical imaging 2018;5(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoo S, Gujrathi I, Haider M, Khalvati F. Prostate Cancer Detection using Deep Convolutional Neural Networks. Scientific reports 2019;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]


