Abstract
Background: The gastrointestinal tract has been speculated to serve as a reservoir for Acinetobacter, however little is known about the ecological fitness of Acinetobacter strains in the gut. Likewise, not much is known about the ability of Acinetobacter to consume dietary, or host derived nutrients or their capacity to modulate host gene expression. Given the increasing prevalence of Acinetobacter in the clinical setting, we sought to characterize how A. calcoaceticus responds to gut-related stressors and identify potential microbe-host interactions.
Materials and Methods: To accomplish these aims, we grew clinical isolates and commercially available strains of A. calcoaceticus in minimal media with different levels of pH, osmolarity, ethanol and hydrogen peroxide. Utilization of nutrients was examined using Biolog phenotypic microarrays. To examine the interactions of A. calcoaceticus with the host, inverted murine organoids where the apical membrane is exposed to bacteria, were incubated with live A. calcoaceticus, and gene expression was examined by qPCR.
Results: All strains grew modestly at pH 6, 5 and 4; indicating that these strains could tolerate passage through the gastrointestinal tract. All strains had robust growth in 0.1 and 0.5 M NaCl concentrations which mirror the small intestine, but differences were observed between strains in response to 1 M NaCl. Additionally, all strains tolerated up to 5% ethanol and 0.1% hydrogen peroxide. Biolog phenotypic microarrays revealed that A. calcoaceticus strains could use a range of nutrient sources, including monosaccharides, disaccharides, polymers, glycosides, acids, and amino acids. Interestingly, the commercially available A. calcoaceticus strains and one clinical isolate stimulated the pro-inflammatory cytokines Tnf, Kc, and Mcp-1 while all strains suppressed Muc13 and Muc2.
Conclusion: Collectively, these data demonstrate that A. calcoaceticus is well adapted to dealing with environmental stressors of the gastrointestinal system. This data also points to the potential for Acinetobacter to influence the gut epithelium.
Keywords: Acinetobacter, intestine, organoid, Acinetobacter calcoaceticus, metabolism
Introduction
Acinetobacter is a Gram-negative nonfermenting coccobacillus that is widely distributed in nature. Certain Acinetobacter species are classified as opportunistic pathogens and are commonly associated with healthcare associated infections (Peleg et al., 2008; Nemec et al., 2015; Mancilla-Rojano et al., 2020). The Acinetobacter species that are considered to be pathogens include A. baumannii, A. pittii and A. nosocomialis (Mancilla-Rojano et al., 2020). These species cause infections like bacteremia, ventilator-associated pneumonia, urinary tract infection, meningitis, and wound infection. A. baumannii is the most frequently isolated and best studied of the Acinetobacter species. In contrast to A. baumannii, A. calcoaceticus has been considered to have a lower virulence since colonization is more frequently noted than infection (Glew et al., 1977). However, A. calcoaceticus can still cause infection and understanding the mechanism by which A. calcoaceticus interacts with the host remains an important topic.
Acinetobacter species are capable of occupying several ecological niches, including the mammalian intestine. Acinetobacter species have been identified in the human fecal microbiota and it has been speculated that the gut could serve as a potential reservoir for Acinetobacter infection (Timsit et al., 1993; Xavier et al., 1996; Ayats et al., 1997; Dijkshoorn et al., 2005; Roy et al., 2010; Pandey et al., 2012; Aljindan et al., 2015; Cheng et al., 2015; Li et al., 2015; Braun et al., 2017; Li et al., 2019). Consistent with this hypothesis, A. baumannii can bind to rabbit small intestinal glycosphingolipids (Madar Johansson et al., 2020) and colonize the mouse gastrointestinal tract in a secretory IgA dependent manner (Coron et al., 2017; Ketter et al., 2018). Moreover, ampicillin treatment of mice has been shown to elevate A. baumannii and A. calcoaceticus fecal levels, suggesting that antibiotic selection could allow the outgrowth of endogenous strains (Raplee et al., 2021). Apart from these studies, little is known regarding the factors that influence Acinetobacter colonization and little data exists on the effects of A. calcoaceticus. The importance of Acinetobacter in the gut is highlighted by the fact that Acinetobacter species are elevated in certain disease states including ulcerative colitis, a subset of inflammatory bowel disease (IBD) (Gophna et al., 2006; Lucke et al., 2006; Leung et al., 2014; Kevans et al., 2015; Tang et al., 2015; Sjoberg et al., 2017; Sekido et al., 2020; He et al., 2021; Qi et al., 2022). In this study, we sought to examine the ability of A. calcoaceticus strains to survive environmental stressors found in the gut, interact with other gut microbes and utilize dietary components. We also examined the reciprocal interactions of A. calcoaceticus strains on the host using intestinal organoids.
Methods
General Bacterial Culturing Techniques
Acinetobacter calcoaceticus ATCC 23055 (American Type Culture Collection) and A. calcoaceticus CB1 (Carolina Biological Supply) were purchased from commercial sources. Four clinical isolates of A. calcoaceticus strains that were isolated from ventricular fluid of pediatric patients were provided by Dr. James Versalovic. All A. calcoaceticus strains were cultured aerobically in brain-heart-infusion (BHI) medium (ThermoFisher) at 37°C. Overnight cultures were sub-cultured into M9 minimal media containing glucose at an optical density (OD600nm) = 0.1. To model environmental stressors, M9 media was supplemented with NaCl (0.1, 0.5, or 1 M), H2O2 (0.05, 0.1, 0.2%), or ethanol (1, 2.5 or 5%). M9 was also adjusted to varying pH values (4, 5, 6, 7). Growth was monitored after 18 h incubation by OD600nm.
To examine nutrient uptake, A. calcoaceticus strains were added to M9 media lacking glucose at OD600nm = 0.1. Then 100 μL of this culture was added to each well of a 96-well Biolog Phenotype Microarrays for Microbial Cells (PM1 and PM2 plates). Growth was monitored at 18 h by OD600nm on a Synergy HT BioTek plate reader. Stool-based bioreactors were generated as previously described (Engevik et al., 2021). Briefly, stool samples were cultured anaerobically for 24 h to allow microbes to establish stable communities, then inoculated with A. calcoaceticus strains at an OD600nm = 0.05. After 48 h of incubation with A. calcoaceticus strains, samples were collected for gDNA isolation. For imaging purposes, A. calcoaceticus strains were fluorescently tagged with CFDA-SE (ThermoFisher) as previously described (Engevik et al., 2021). Briefly, A. calcoaceticus strains were washed 2x with sterile PBS and incubated for 1 h aerobically at 37°C with 10 μM CFDA-SE. After the incubation, bacteria were washed 3–5x with sterile PBS to remove any unused CFDA-SE. Bacteria were then ready for incubation with organoids.
Organoid Generation
Intestinal organoids were generated from four adult (8–12 weeks) C57B6/J mice as previously described (Engevik et al., 2013). The jejunum was rapidly dissected, washed with ice-cold PBS (PBS; Gibco) and opened longitudinally. The jejunum was then cut into ∼1-cm length pieces and placed in a 15-ml conical tube containing 5 ml of ice-cold PBS, 43.4 mM sucrose, 0.5 mM DTT and 3 mM EDTA (Gibco). The tissue was incubated at 4°C rocking for 30 min. Crypts were mechanically disrupted by shaking in 5 ml of ice-cold PBS with D-sorbitol and sucrose and collected following filtration with a 70-µm cell strainer (Corning cat# 431,751). Crypts were centrifuged, resuspended in Matrigel (Corning), and incubated with complex media containing WNT, R-spondin, Noggin and EGF. Organoids were passaged >2 times to ensure no tissue fragments remained. Organoids were differentiated and grown inside-out by adding split organoids directly to complex media without Wnt and incubated at 37°C with 5% CO2 for 5 days before use.
For imaging purposes, live fluorescently tagged A. calcoaceticus was incubated with inside-out organoids for 3 h. Organoids were then washed with PBS, fixed with 4% paraformaldehyde for 30 min, incubated in 30% sucrose/PBS overnight and frozen embedded. Sections were stained with phospho-ezrin, radixin, moesin (1:200 dilution, Rabbit Ab, Cell Signaling #3726S) overnight at 4°C. After washing, sections were incubated with donkey-anti-rabbit Alexa Fluor 555 (1:1,000 dilution, ThermoFisher # A31572) for 1 h at room temperature and counter stained with Hoechst 33,342 (Invitrogen) for 10 min at room temperature. Slides were cover-slipped with mounting media (Life Technologies) and imaged using a Zeiss AxioScan at ×20 (Zeiss). The relative fluorescence intensity of adhered bacteria was quantified using FIJI (Formerly ImageJ) software (NIH) and reported as relative fluorescence. Three regions of interests per image and four images per slide were used for semi-quantitation of stain intensity.
For gene expression analysis, inside-out organoids were incubated with live A. calcoaceticus strains adjusted to an OD600nm = 1.0 for 3 h at 37°C (n = 4 different mouse organoids; performed in replicates per mouse). After incubation, organoids were centrifuged at 300 × g for 5 min and the organoid pellet was resuspended in 400 μL TRIZOL. Organoids were stored in TRIZOL at −80°C until the RNA was isolated.
RNA Isolation, cDNA Generation, gDNA Isolation and qPCR
Total RNA was extractr’ed from organoids in TRIZOL according to manufactures instructions, with the addition of glycogen. cDNA was generated from 500 ng RNA via the Verso cDNA synthesis kit (ThermoFisher #AB-1453). gDNA was isolated from 1 ml aliquots of stool-based bioreactors using the Zymo Quick-DNA Fecal/Soil Microbe Kits according to the manufacturer’s instructions. Quantitative real time PCR (qPCR) was performed using a Bio-Rad CFX96 Real Time qPCR machine (Bio-Rad). Forward and reverse primers were added to SYBR Green mastermix (Genesee Scientific #17-501DP) and cDNA. Epithelial genes were normalized to the housekeeping gene 18S and relative expression was calculated using the ddCT method. Bacterial colony forming units (CFUs) were calculated from CT values based on standard curves as previously described (Engevik et al., 2013).
Statistics
Data are presented as mean ± standard deviation, with points representing individual bacteria strain growth rate with a n = 4 (repeated 3 independent times). Comparisons within groups were made with One-way Analysis of Variance (ANOVA) and comparisons between groups were made with a Two-way ANOVA (Table 1). All analyses were corrected for multiple comparisons by controlling the False Discovery Rate. GraphPad version 9.3 was used to generate graphs and statistics (GraphPad Software, Inc. La Jolla, CA). A *p < 0.05 value was considered significant while n is the number of experiments performed.
TABLE 1.
Two Way ANOVA statistics from bacterial growth in the presence of stressors. All comparisons were made against A. calcoaceticus ATCC 23055. Significant p values (p < 0.05) are colored in blue.
| NaCl | pH | ETOH | H202 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | 0 M | 0.1 M | 0.5 M | 1 M | pH 7 | pH 6 | pH 5 | pH 4 | 0% | 1% | 2.50% | 5% | 0% | 0.05% | 0.10% | 0.20% |
| CB1 | 0.1379 | 0.1097 | <0.0001 | <0.0001 | 0.0148 | 0.0003 | 0.4224 | 0.2573 | 0.079 | <0.0001 | <0.0001 | 0.0204 | 0.2819 | 0.163 | 0.2286 | 0.5514 |
| M3 | 0.9269 | 0.4824 | 0.0126 | 0.0136 | 0.1814 | 0.3755 | 0.0133 | 0.0827 | 0.31 | <0.0001 | 0.0023 | 0.0594 | 0.2781 | 0.0335 | <0.0001 | <0.0001 |
| T82 | 0.2879 | 0.0218 | 0.9543 | 0.0013 | 0.034 | 0.7245 | 0.6418 | 0.4756 | 0.8335 | 0.0001 | <0.0001 | 0.0003 | 0.2543 | 0.2975 | 0.0393 | 0.002 |
| M5 | 0.577 | 0.0131 | <0.0001 | <0.0001 | 0.8767 | 0.0002 | 0.2652 | 0.2672 | 0.1845 | <0.0001 | 0.0018 | <0.0001 | 0.0775 | 0.0083 | 0.0734 | 0.0007 |
| X75 | 0.1106 | 0.3661 | <0.0001 | 0.0002 | 0.2573 | 0.0002 | 0.0003 | 0.0017 | 0.4996 | <0.0001 | <0.0001 | <0.0001 | 0.0833 | 0.0199 | <0.0001 | <0.0001 |
Results
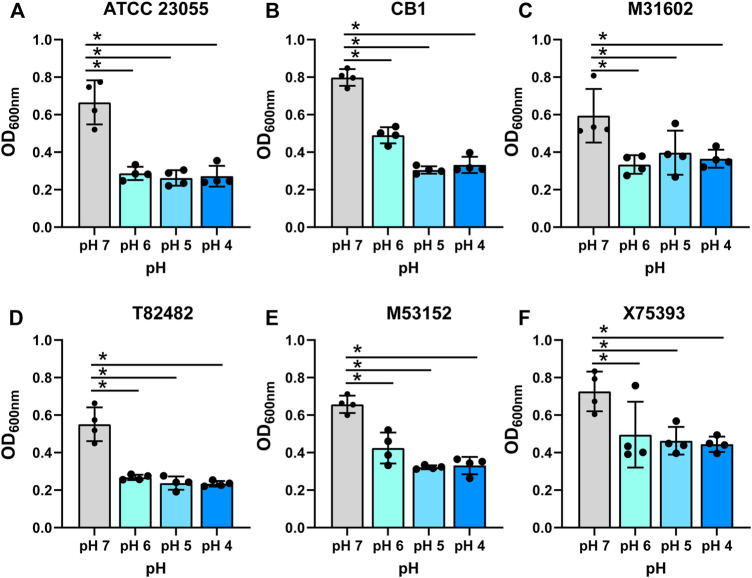
It has been speculated that the gut may be a site for Acinetobacter colonization and thus could be a source of endemic infections (Timsit et al., 1993; Xavier et al., 1996; Ayats et al., 1997; Dijkshoorn et al., 2005; Roy et al., 2010; Pandey et al., 2012; Aljindan et al., 2015; Cheng et al., 2015; Li et al., 2015; Braun et al., 2017; Li et al., 2019). To examine the efficiency of A. calcoaceticus strains to withstand the conditions of the gastrointestinal tract, we grew commercially available and clinical isolates of A. calcoaceticus in minimal media at pH 7, 6, 5, and 4. Cultures were seeded with OD600n = 0.1 and growth was considered to be above an OD600nm = 0.2. As expected, we observed robust growth of the commercially available A. calcoaceticus strains (ATCC 23055, CB1) and clinical isolates (M31602, T82482, M53152, and X75393) in pH 7 media (Figures 1A–F; Table 1). We observed growth (OD600nm > 0.2) of all strains at pH 6, pH 5, and pH 4; although growth was significantly reduced compared to pH 7. These findings suggest that A. calcoaceticus is well adapted to withstand varying intestinal pHs.
FIGURE 1.
Growth of (A) calcoaceticus in various pHs. Growth of A. calcoaceticus strains as measured by OD600nm after 18 h of incubation. Growth was examined in the following strains: (A) ATCC 23055, (B) CB1; clinical isolates: (C) M31602, (D) T82482, (E) M53152, and (F) X75,393). *p < 0.05, One Way ANOVA.
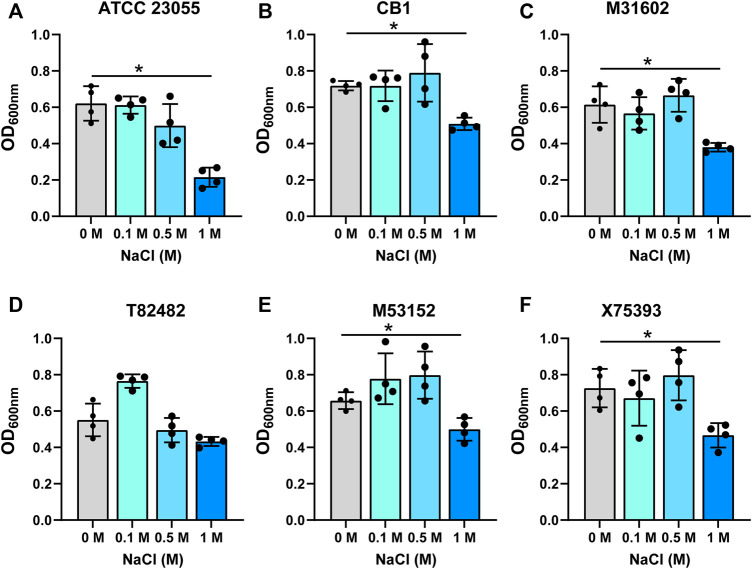
The intestine harbors varying ranges of osmolarity, with the small intestinal villi encountering 400–700 mM (Overduin et al., 2014). To model the gut, we added increasing concentrations of NaCl (0.1, 0.5 and 1 M) to minimal media and examined the growth of A. calcoaceticus (Figures 2A–F; Table 1). All strains were found to have similar growth at 0.1 and 0.5 M as media controls. Decreased growth was observed at 1 M NaCl with all but the T82482 strain. We observed that the ATCC 23055 strain exhibited the most significant decline in growth (Figure 2A). These data highlight that A. calcoaceticus can grow well in various osmolarities which mirror the gut environment.
FIGURE 2.
Growth of (A) . calcoaceticus at varying osmolarity. Growth of A. calcoaceticus strains as measured by OD600nm after 18 h of incubation. Growth was examined in the following strains: (A) ATCC 23055, (B) CB1; clinical isolates: (C) M31602, (D) T82482, (E) M53152, and (F) X75,393). *p < 0.05, One Way ANOVA.
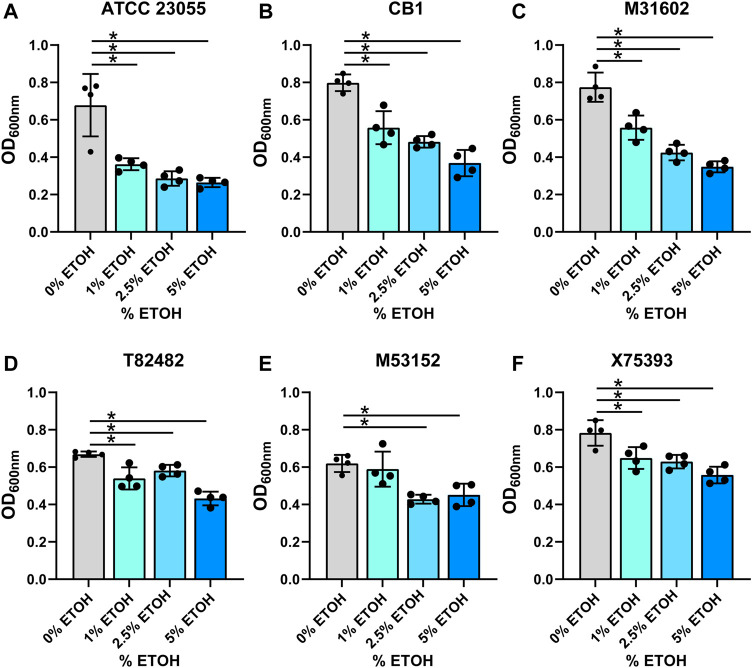
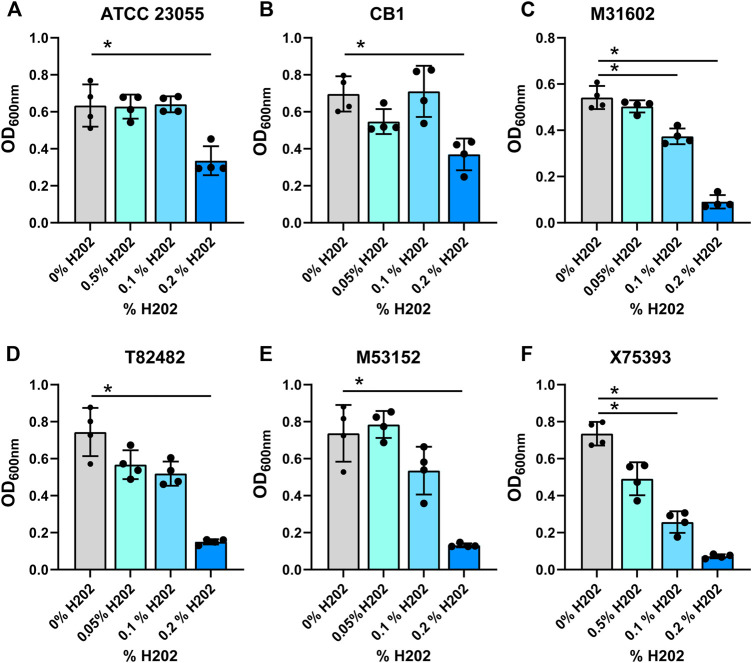
In the gut, commensal microbes such as Lactobacilli can generate ethanol and hydrogen peroxide (Engevik and Versalovic, 2017) and microbes occupying the same niche must adapt to these stressors. To model the production of localized ethanol and hydrogen peroxide, we supplemented minimal media with ethanol (1, 2.5 or 5%) (Figures 3A–F; Table 1) or hydrogen peroxide (0.05, 0.1 or 0.2%) (Figures 4A–F; Table 1). Impressively, all A. calcoaceticus strains could grow in up to 5% ethanol. Our commercially available ATCC 23055 strain was the most sensitive strain to ethanol, exhibiting a ∼2-fold decrease in growth in 1% ethanol compared to media controls (Figure 3A). In contrast, clinical isolates T82482 (Figure 3D), M53152 (Figure 3E), and X75,393 (Figure 3F) exhibited only a slight decline in growth at the higher ethanol concentrations; suggesting that these clinical isolates are highly resistant to ethanol. When A. calcoaceticus was treated with hydrogen peroxide (Figures 4A–F), all strains could grow with 0.05 and 0.1% hydrogen peroxide. The commercially available ATCC 23055 and CB1 were the most tolerant to 0.2% hydrogen peroxide (Figures 4A,B); exhibiting a less than 2-fold decrease in growth compared to the no hydrogen peroxide controls. In contrast, the clinical isolates M31602 (Figure 4C), T82482 (Figure 4D), M53152 (Figure 4E), and X75,393 (Figure 4F) were sensitive to 0.2% hydrogen peroxide and did not grow above the seeded density of OD600nm = 0.1. Collectively, these data with stressors indicate that A. calcoaceticus is well adapted for the environmental stressors associated with gut colonization.
FIGURE 3.
(A) calcoaceticus growth in percentages of ethanol. Growth of A. calcoaceticus strains as measured by OD600nm after 18 h of incubation. Growth was examined in the following strains: (A) ATCC 23055, (B) CB1; clinical isolates: (C) M31602, (D) T82482, (E) M53152, and (F) X75,393). *p < 0.05, One Way ANOVA.
FIGURE 4.
(A) calcoaceticus growth in hydrogen peroxide. Growth of A. calcoaceticus strains as measured by OD600nm after 18 h of incubation. Growth was examined in the following strains: (A) ATCC 23055, (B) CB1; clinical isolates: (C) M31602, (D) T82482, (E) M53152, and (F) X75,393). *p < 0.05, One Way ANOVA.
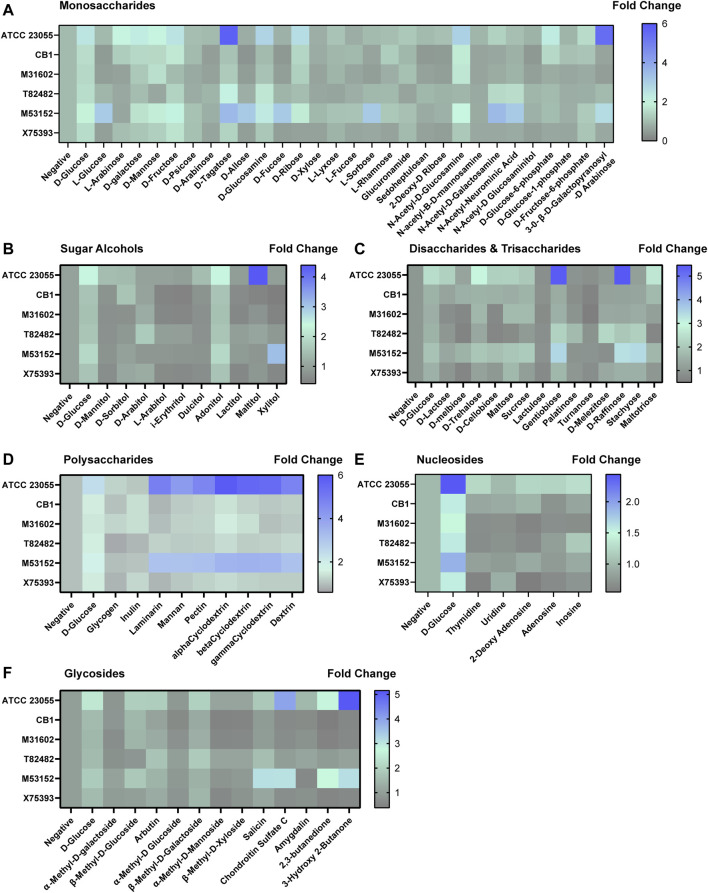
Next, we sought to examine the potential nutrient sources for A. calcoaceticus within the intestine. To address this, we grew A. calcoaceticus strains in minimal media lacking glucose in Biolog phenotypic microarrays (Figures 5, 6). Growth was considered to be > 1.5 fold change. Compared to growth in a media without a carbon source, A. calcoaceticus had improved growth in the presence of glucose (Figure 5A). Since A. calcoaceticus had improved growth with glucose, we first examined monosaccharides (Figure 5A). ATCC 23055 and CB1 grew well with L-arabinose, D-galactose, D-mannose, D-fructose, D-tagatose, D-glucosamine, D-ribose, and N-acetyl-D-glucosamine. Interestingly, clinical isolates M31602, T82482 and M53152 did not grow with arabinose and exhibited strain-dependent growth with D-mannose, D-fructose, S-tagatose, D-ribose and N-acetyl-D-glucosamine. M53152 alone grew with L-glucose, B-D-allose, D-fucose, and L-sorbose, highlighting strain-specific nutrient preferences. When we examined alcohol sugars (Figure 5B), we found that all strains could use adonitol and no strains could use D-mannitol, L-arabitol, i-erythritol, or dulcitol.
FIGURE 5.
Acinetobacter growth in varying carbon sources. Heat maps representing fold change in growth with the negative control (no added nutrients) set at 1. Growth was examined with (A) monosaccharides, (B) sugar alcohols, (C) disaccharides and trisaccharides, (D) polysaccharides, (E) nucleosides and (F) glycosides after 18 h incubation.
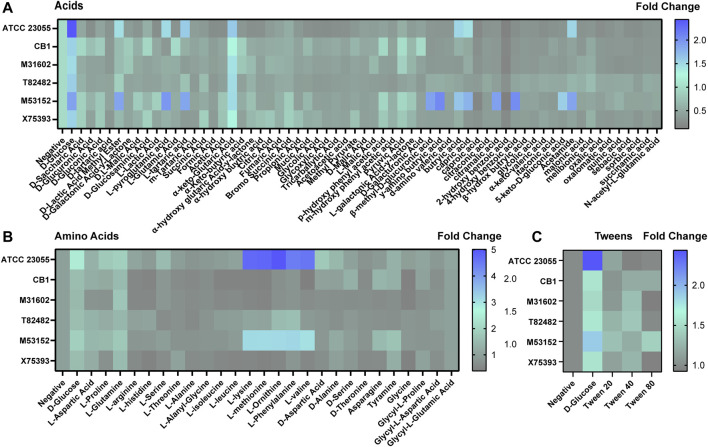
FIGURE 6.
Acinetobacter growth in acids, amino acids and tweens. Heat maps representing fold change in growth with the negative control (no added nutrients) set at 1. Growth was examined with (A) acids, (B) amino acids, and (C) tweens after 18 h incubation.
In terms of growth with disaccharides (Figure 5C), we found that all strains used gentiobiose and no strains used lactulose or turnanose. For the other disaccharides and trisaccharides, we observed strain-dependent growth. For example, M53152 grew with D-melbiose, D-trehalose, maltose, D-melibiose, sucrose, D-cellbiose, D-raffinose, stachyose and maltotriose, while T82482 only grew with D-palatinose, D-melezitose, D-raffinose and stachyose. When we examined polysaccharide utilization, we found very similar profiles between ATCC 23055 and M53152. Both strains were highly efficient at using laminarin, mannan, pectin, α-Cyclodextrin, β-Cyclodextrin, γ-cyclodextrin and dextrin (Figure 5D). In general, most strains grew well with dietary polysaccharides. In the nucleosides (Figure 5E) and glycosides (Figure 5F) classification, we found strain-dependent use of specific compounds. ATCC 23055 and M53152 exhibited the highest fold change in growth with chondroitin sulfate C, 2,3-butanedione and 3-hydroxy 2-butanone.
Of the acids (Figure 6A), we found that all strains could use a-keto-glutaric acid. ATCC 23055 and M53152 responded to the largest number of acids, including B-methyl-D-glucuronic acid, γ-amino butryic acid, butryic acid, capric acid, 4-hydroxy benzoic acid and acetamide. Unique profiles were observed with the other acids examined depending on the strain. Even without a carbon source, we found that all A. calcoaceticus strains had improved growth with L-glutamine and ATCC 23055 and M53152 used L-lysine, L-methionine, L-Ornithine, L-Phenylalanine and L-valine (Figure 6B), indicating that select amino acids could be used as an alternative to carbon. Tween can be employed as a stool emulsifier and tween enemas have been used to treat fecal mass obstructions (Wood and Katzberg, 1978). None of the A. calcoaceticus strains responded to tween with improved growth (Figure 6C). These data indicate that A. calcoaceticus can use a wide range of nutrients sources, ranging from sugars to amino acids.
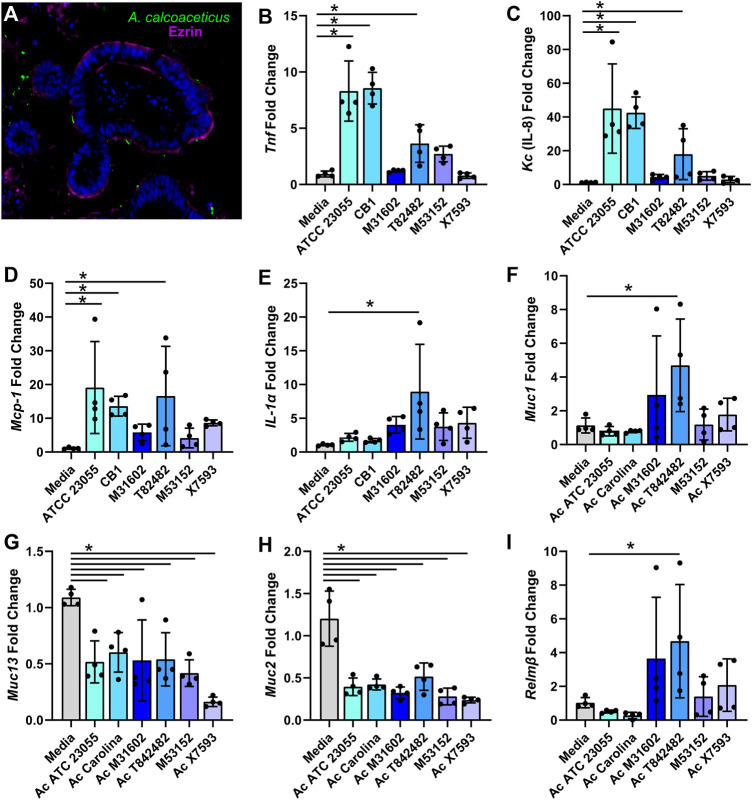
In addition to stressors and nutrients, Acinetobacter species encounter a complex community of micro-organisms when colonizing the gastrointestinal tract. To confirm that A. calcoaceticus could proliferate in this competitive environment, we cultured stool-based bioreactors and introduced A. calcoaceticus strains. After 48 h of culturing, we examined the presence of A. calcoaceticus in the bioreactors by qPCR (Supplementary Figure S1A). We found that all strains effectively colonized the bioreactors, indicating that A. calcoaceticus could be present within the intestinal milieu. Next, we sought to examine how A. calcoaceticus interacted with the intestinal epithelium. To test whether A. calcoaceticus was able to stimulate epithelial responses, we incubated live A. calcoaceticus strains with apical side-out intestinal organoids for 3 h. By immunostaining, we found that some A. calcoaceticus microbes adhered to the organoids (Figure 7A). A similar level of bacteria was observed on all organoids regardless of the strain (Supplementary Figure S1B). Analysis of pro-inflammatory cytokines by qPCR revealed that the commercially available strains ATCC 23055 and CB1 and one of our clinical isolates T82482 increased the expression of Tnf (Figure 7B), Kc (Figure 7C) and Mcp-1 (Figure 7D) compared to media controls. The T82482 strain also increased the expression of IL-1α (Figure 7E). Inflammation is known to regulate intestinal mucus, so we also examined adherent mucins Muc1 and Muc13 and secreted mucins Muc2 in our organoid model. T82482 was the only strain that upregulated Muc1 (Figure 7F), but all strains suppressed Muc13 levels (Figure 7G) compared to media controls. Likewise, all strains suppressed Muc2 expression (Figure 7H). We also examined an antimicrobial protein secreted by goblet cells, Relmβ, and we observed that Relmβ expression was increased in response to T82482 (Figure 7I), suggesting that goblet cells were not decreased in this model despite decreased Muc2. Together these findings highlight that A. calcoaceticus strains are adept at dealing with environmental stressors, are able to consume multiple nutrient sources, colonize in the presence of other microbes, and can elicit pro-inflammatory signaling pathways.
FIGURE 7.
(A) calcoaceticus induces inflammatory responses in intestinal organoids. (A) Inside-out organoids incubated with live fluorescently tagged A. calcoaceticus and immunostained with the apical marker phospho-Ezrin. qPCR analysis of organoids after 3 h of incubation examining expression of (B) Tnf, (C) IL-18, (D) Mcp-1, (E) IL-1alpha, and (F) Muc1, (G) Muc13, (H) Muc2, and (I) Relmβ.
Discussion
The digestive tract is proposed to be a reservoir for Acinetobacter spp. (Timsit et al., 1993; Corbella et al., 1996; Agusti et al., 2002; Dijkshoorn et al., 2005; Thom et al., 2010; Lim et al., 2014). In this study, we confirmed the ability of A. calcoaceticus strains to withstand conditions that recapitulate the gastrointestinal tract luminal environment and use various nutritional sources found in the intestine. We identified that A. calcoaceticus strains are fairly resistant to changing pH, osmolarity, ethanol and hydrogen peroxide levels. We also found that the majority of A. calcoaceticus strains used glucose, L-arabinose, D-galactose, D-mannose, D-fructose and N-acetyl-D-glucosamine, D-trehalose, adonitol, mannan, pectin, α-Cyclodextrin, β-Cyclodextrin, γ-cyclodextrin, dextrin, D-ribose and α-keto-glutaric acid and L-glutamine. These data indicate that A. calcoaceticus can use a wide range of nutrients sources. Bioreactor experiments confirmed that A. calcoaceticus could colonize with other gut microbes. This work adds to existing research and suggests that Acinetobacter spp. are well adapted for survival in the gastrointestinal tract.
Entry of Acinetobacter spp. into the gut has recently been examined by Coron et al. (2017). The authors found that intranasal administration of A. baumannii in mice, which mimicked the major method by which ventilated patients in ICUs commonly become infected with Acinetobacter, resulted in digestive-tract colonization. This data suggests that patients could become colonized with Acinetobacter spp. in hospital settings and this gut colonization could be the precursor of severe infections. Consistent with this notion, Corbella et al. identified that patients colonized with A. baumannii in their digestive system had a positive association for blood infections with multidrug-resistant A. baumannii strains compared to patients without colonization (Corbella et al., 1996). Similarly, Medina et al. found that A. baumannii gut colonization was an independent risk factor for the development of A. baumannii respiratory infections (Medina-Presentado et al., 2013). Wisplinghof et al. reported that the portal of entry was not identified in 48.6% of the A. baumannii bloodstream infections, suggesting that a significant proportion of these infections could be due to intestinal carriage (Wisplinghoff et al., 2000; Coron et al., 2017). Our work complements these findings by demonstrating that Acinetobacter species are equipped to colonize the gastrointestinal tract, where they could serve as a reservoir for infection.
In animal models, A. baumannii colonized both the small and large intestine (Coron et al., 2017; Ketter et al., 2018). In these models, intestinal inflammation was not specifically examined. However, our apical inside-out organoid model revealed that certain strains of A. calcoaceticus stimulated pro-inflammatory cytokines. This data can be interpreted in several ways. First, it is possible that in the setting of a complex gut microbiota in vivo, other microbes may dampen pro-inflammatory signatures associated with Acinetobacter. Second, the existing in vivo studies used A. baumannii and this study focused on A. calcoaceticus. It is therefore possible that differences may exist between the species. Third, our data suggests that pro-inflammatory responses are strain dependent and these strain differences may also exist for A. baumannii strains. Future studies using mouse models are warranted to fully dissect the colonization capacity and epithelial crosstalk with A. calcoaceticus.
Elevated levels of Acinetobacter in the setting of IBD has been observed in several studies (Gophna et al., 2006; Lucke et al., 2006; Leung et al., 2014; Kevans et al., 2015; Tang et al., 2015; Sjoberg et al., 2017; Sekido et al., 2020; He et al., 2021; Qi et al., 2022). Two studies found high abundance of Acinetobacter in pediatric patients with newly diagnosed ulcerative colitis (Kevans et al., 2015; Sjoberg et al., 2017); providing potential evidence that Acinetobacter species could be contributing to the onset of intestinal inflammation in genetically susceptible patients. Another study found that Acinetobacter was enriched in the mucosa-associated bacteria during active colitis in ulcerative colitis patients (Tang et al., 2015). This study found that Acinetobacter levels significantly correlated with microbial pathways in actively inflamed colitis tissue, suggesting a potential causal relationship between Acinetobacter and intestinal inflammation. Another study identified Acinetobacter in the CD14+CD11c + CD163low subset macrophages in the lamina propia of ulcerative colitis patients (Sekido et al., 2020), which indicates these microbes were able to bypass the epithelial barrier. In our intestinal organoid model, we found that several A. calcoaceticus strains stimulated pro-inflammatory cytokine expression and suppressed mucin production. These findings mirror what has been observed in ulcerative colitis patients (Trabucchi et al., 1986; Raouf et al., 1992; Pullan et al., 1994; Tytgat et al., 1996; Hanski et al., 1999; Heazlewood et al., 2008; Zhao et al., 2010; Larsson et al., 2011; Antoni et al., 2014; Johansson et al., 2014; Wenzel et al., 2014). Our findings provide further evidence for the potential link between Acinetobacter, intestinal inflammation and IBD. In the future, we plan to dissect the mechanisms of how A. calcoaceticus initiates inflammation in more depth.
Another observation from our organoid model was that not all the clinical isolates stimulated pro-inflammation cytokine expression. Gram-negative bacteria such as Acinetobacter can activate TLR4 on host cells via the cell wall component lipopolysaccharide (LPS) (Pelletier et al., 2013). LPS is comprised of lipid A, the core oligosaccharide, and the O-specific antigen. Lipid A is considered the bioactive component of LPS and is responsible for activating immune responses (Pelletier et al., 2013). A. baumannii has been shown to modify their lipid A with the addition of positively charged residues including ethanolamine, phosphoethanolamine, aminoarabinose, and glucosamine (Moskowitz et al., 2004; Arroyo et al., 2011; Basheer et al., 2011; Beceiro et al., 2011; Llobet et al., 2011; Pelletier et al., 2013). These modifications enhance the resistance of A. baumannii to the antibiotic colistin and suppresses their immunostimulatory capacity (Pelletier et al., 2013). In addition to modifying LPS, some clinical strains of A. baumannii have been identified with loss-of-function mutations in genes in the LPS biosynthetic pathway (Moffatt et al., 2010; Nagy et al., 2019); resulting in strains which completely lack LPS. Although A. baumannii can survive in the absence of LPS, these microbes have distinct morphological defects and growth alterations under laboratory conditions (Nagy et al., 2019; Beceiro et al., 2011; Bojkovic et al., 2015; Powers et al., 2018; Boll et al., 2016). All of our A. calcoaceticus strains were resistant to colistin (data not shown) suggesting that some LPS modification might have occurred in these microbes. Since we didn’t observe significant morphological or growth differences between our strains, we think that all our stains harbor LPS, but we speculate that their may be different modifications between our A. calcoaceticus strains which could account for the variability in cytokine stimulation. We plan in future studies to examine the LPS structures of our A. calcoaceticus strains.
Interestingly we noted differences between our commercially available strains and clinical isolates in many of our results. For example, the clinical isolates were more adapted at survival in high concentrations of ethanol and NaCl than the lab adapted strains. The growth of the clinical isolates was also not as robust as the ATCC 23055 strain in utilizing many of the carbon sources, such as glucose, L-arbinose, D-trehalose, GluNAc, galactose, mannose and fructose. In our organoid model, we found that lab adapted ATCC 23055 and CB1 and one clinical isolate T82482 stimulate multiple pro-inflammatory cytokines, while the other strains had minimal stimulation of cytokines. A number of groups have begun to question the adequacy of laboratory-adapted reference strains to represent “real world” pathogenesis (Fux et al., 2005). Some laboratory strains have been sub-cultured for years, which may result in the loss of important pathophysiological characteristics or the dependence on lab-specific media components. This limitation can be overcome by including multiple strains, including clinical isolates, and examining their collective behavior. While we did note several differences, in general we found that all strains were fairly resistant to environmental stressors (pH, osmolarity, ethanol, and hydrogen peroxide) and we found several common nutrient sources across strains. Based on these studies, we believe that A. calcoaceticus species are well adapted to colonize the gastrointestinal tract and can consume a variety of nutritional sources. We also believe this work highlights the benefits of incorporating clinical isolates into future work.
There are several strengths in our study. To the best of our knowledge, this is the only study that has examined the ability of A. calcoaceticus to withstand conditions of the gastrointestinal tract and the first show that A. calcoaceticus can colonize human stool communities and modulate the gut epithelium. We incorporated several clinical isolates, which has allowed us to identify some global attributes of A. calcoaceticus. However, there are also several limitations. This work was all done in vitro and in vivo studies are necessary to truly identify the colonization capacity of A. calcoaceticus. Although we hypothesize that A. calcoaceticus activates TLR4 on the gut epithelium to drive inflammatory signatures, this work did not identify a specific mechanism and more work is needed to delineate how A. calcoaceticus modulates the intestinal epithelium and immune cells.
In summary, we demonstrate that A. calcoaceticus strains can withstand intestinal conditions and thrive with several dietary sources. We believe these attributes make Acinetobacter spp. ecologically fit for colonizing the gut. This information is clinically important since the gut likely serves as a reservoir for secondary Acinetobacter spp. infections. Thus, it might be possible to prevent secondary infections, like blood stream infections or pneumonia, by inhibiting Acinetobacter gut colonization and we believe this is an exciting area for future studies.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author Contributions
Concept and design JG and ME; intellectual contribution JG, BB, TT, AE, and ME; data acquisition JG, BB, TT, AE, and ME; data analysis, statistics, and interpretation JG, BB, and ME; drafting manuscript JG; funding AE and ME.
Funding
T32GM132055-01 (JG), T32DK124191-01A1 (TT), K01DK121869 (AE), and K01K123195-01 (ME).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.880024/full#supplementary-material
References
- Agusti C., Pujol M., Argerich M. J., Ayats J., Badia M., Dominguez M. A., et al. (2002). Short-term Effect of the Application of Selective Decontamination of the Digestive Tract on Different Body Site Reservoir ICU Patients Colonized by Multi-Resistant Acinetobacter Baumannii. J. Antimicrob. Chemother. 49, 205–208. 10.1093/jac/49.1.205 [DOI] [PubMed] [Google Scholar]
- Aljindan R., Bukharie H., Alomar A., Abdalhamid B. (2015). Prevalence of Digestive Tract Colonization of Carbapenem-Resistant Acinetobacter Baumannii in Hospitals in Saudi Arabia. J. Med. Microbiol. 64, 400–406. 10.1099/jmm.0.000033 [DOI] [PubMed] [Google Scholar]
- Antoni L., Nuding S., Wehkamp J., Stange E. F. (2014). Intestinal Barrier in Inflammatory Bowel Disease. Wjg 20, 1165–1179. 10.3748/wjg.v20.i5.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo L. A., Herrera C. M., Fernandez L., Hankins J. V., Trent M. S., Hancock R. E. W. (2011). The pmrCAB Operon Mediates Polymyxin Resistance in Acinetobacter Baumannii ATCC 17978 and Clinical Isolates through Phosphoethanolamine Modification of Lipid A. Antimicrob. Agents Chemother. 55, 3743–3751. 10.1128/aac.00256-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayats J., Corbella X., Ardanuy C., Domínguez M. A., Ricart A., Ariza J., et al. (1997). Epidemiological Significance of Cutaneous, Pharyngeal, and Digestive Tract Colonization by Multiresistant Acinetobacter Baumannii in ICU Patients. J. Hosp. Infect. 37, 287–295. 10.1016/s0195-6701(97)90145-6 [DOI] [PubMed] [Google Scholar]
- Basheer S. M., Guiso N., Tirsoaga A., Caroff M., Novikov A. (2011). Structural Modifications Occurring in Lipid A of Bordetella Bronchiseptica Clinical Isolates as Demonstrated by Matrix-Assisted Laser Desorption/ionization Time-Of-Flight Mass Spectrometry. Rapid Commun. Mass Spectrom. 25, 1075–1081. 10.1002/rcm.4960 [DOI] [PubMed] [Google Scholar]
- Beceiro A., Llobet E., Aranda J., Bengoechea J. A., Doumith M., Hornsey M., et al. (2011). Phosphoethanolamine Modification of Lipid A in Colistin-Resistant Variants of Acinetobacter Baumannii Mediated by the pmrAB Two-Component Regulatory System. Antimicrob. Agents Chemother. 55, 3370–3379. 10.1128/aac.00079-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkovic J., Richie D. L., Six D. A., Rath C. M., Sawyer W. S., Hu Q., et al. (2015). Characterization of an Acinetobacter baumannii lptD Deletion Strain: Permeability Defects and Response to Inhibition of Lipopolysaccharide and Fatty Acid Biosynthesis. J. Bacteriol. 198, 731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll J. M., Crofts A. A., Peters K., Cattoir V., Vollmer W., Davies B. W., et al. (2016). A Penicillin-Binding Protein Inhibits Selection of Colistin-Resistant, Lipooligosaccharide-Deficient Acinetobacter baumannii . Proc Natl Acad Sci U S A 113, E6228–e6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Di Segni A., Benshoshan M., Asaf R., Squires J. E., Farage Barhom S., et al. (2017). Fecal Microbial Characterization of Hospitalized Patients with Suspected Infectious Diarrhea Shows Significant Dysbiosis. Sci. Rep. 7, 1088. 10.1038/s41598-017-01217-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V. C. C., Chen J. H. K., So S. Y. C., Wong S. C. Y., Yan M. K., Chau P. H., et al. (2015). Use of Fluoroquinolones Is the Single Most Important Risk Factor for the High Bacterial Load in Patients with Nasal and Gastrointestinal Colonization by Multidrug-Resistant Acinetobacter Baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 34, 2359–2366. 10.1007/s10096-015-2489-4 [DOI] [PubMed] [Google Scholar]
- Corbella X., Pujol M., Ayats J., Sendra M., Ardanuy C., Dominguez M. A., et al. (1996). Relevance of Digestive Tract Colonization in the Epidemiology of Nosocomial Infections Due to Multiresistant Acinetobacter Baumannii. Clin. Infect. Dis. 23, 329–334. 10.1093/clinids/23.2.329 [DOI] [PubMed] [Google Scholar]
- Coron N., Pavlickova S., Godefroy A., Pailhoriès H., Kempf M., Cassisa V., et al. (2017). Mouse Model of Colonization of the Digestive Tract with Acinetobacter Baumannii and Subsequent Pneumonia. Future Microbiol. 12, 707–719. 10.2217/fmb-2016-0203 [DOI] [PubMed] [Google Scholar]
- Dijkshoorn L., Van Aken E., Shunburne L., Van Der Reijden T. J. K., Bernards A. T., Nemec A., et al. (2005). Prevalence of Acinetobacter Baumannii and Other Acinetobacter Spp. In Faecal Samples from Non-hospitalised Individuals. Clin. Microbiol. Infect. 11, 329–332. 10.1111/j.1469-0691.2005.01093.x [DOI] [PubMed] [Google Scholar]
- Engevik M. A., Versalovic J. (2017). Biochemical Features of Beneficial Microbes: Foundations for Therapeutic Microbiology. Microbiol. Spectr. 1, 3-47. 10.1128/microbiolspec.BAD-0012-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engevik M. A., Aihara E., Montrose M. H., Shull G. E., Hassett D. J., Worrell R. T. (2013). Loss of NHE3 Alters Gut Microbiota Composition and influencesBacteroides Thetaiotaomicrongrowth. Am. J. Physiology-Gastrointestinal Liver Physiology 305, G697–G711. 10.1152/ajpgi.00184.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engevik M. A., Danhof H. A., Auchtung J., Endres B. T., Ruan W., Bassères E., et al. (2021). Fusobacterium Nucleatum Adheres to Clostridioides Difficile via the RadD Adhesin to Enhance Biofilm Formation in Intestinal Mucus. Gastroenterology 160, 1301–1314. e1308. 10.1053/j.gastro.2020.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fux C. A., Shirtliff M., Stoodley P., Costerton J. W. (2005). Can Laboratory Reference Strains Mirror 'real-World' Pathogenesis? Trends Microbiol. 13, 58–63. 10.1016/j.tim.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Glew R. H., Moellering R. C., Jr., Kunz L. J. (1977). Infections with Acinetobacter Calcoaceticus (Herellea Vaginicola). Medicine 56, 79–98. 10.1097/00005792-197703000-00001 [DOI] [PubMed] [Google Scholar]
- Gophna U., Sommerfeld K., Gophna S., Doolittle W. F., Veldhuyzen Van Zanten S. J. O. (2006). Differences between Tissue-Associated Intestinal Microfloras of Patients with Crohn's Disease and Ulcerative Colitis. J. Clin. Microbiol. 44, 4136–4141. 10.1128/jcm.01004-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanski C., Born M., Foss H. D., Marowski B., Mansmann U., Arasteh K., et al. (1999). Defective Post-transcriptional Processing of MUC2 Mucin in Ulcerative Colitis and in Crohn's Disease Increases Detectability of the MUC2 Protein Core. J. Pathol. 188, 304–311. [DOI] [PubMed] [Google Scholar]
- He X.-X., Li Y.-H., Yan P.-G., Meng X.-C., Chen C.-Y., Li K.-M., et al. (2021). Relationship between Clinical Features and Intestinal Microbiota in Chinese Patients with Ulcerative Colitis. Wjg 27, 4722–4737. 10.3748/wjg.v27.i28.4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood C. K., Cook M. C., Eri R., Price G. R., Tauro S. B., Taupin D., et al. (2008). Aberrant Mucin Assembly in Mice Causes Endoplasmic Reticulum Stress and Spontaneous Inflammation Resembling Ulcerative Colitis. PLoS Med. 5, e54. 10.1371/journal.pmed.0050054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. E. V., Gustafsson J. K., Holmén-Larsson J., Jabbar K. S., Xia L., Xu H., et al. (2014). Bacteria Penetrate the Normally Impenetrable Inner Colon Mucus Layer in Both Murine Colitis Models and Patients with Ulcerative Colitis. Gut 63, 281–291. 10.1136/gutjnl-2012-303207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketter P. M., Yu J. J., Guentzel M. N., May H. C., Gupta R., Eppinger M., et al. (2018). Acinetobacter Baumannii Gastrointestinal Colonization Is Facilitated by Secretory IgA Which Is Reductively Dissociated by Bacterial Thioredoxin A. mBio 9, e01298. 10.1128/mBio.01298-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevans D., Tyler A. D., Holm K., Jørgensen K. K., Vatn M. H., Karlsen T. H., et al. (2015). Characterization of Intestinal Microbiota in Ulcerative Colitis Patients with and without Primary Sclerosing Cholangitis. Eccojc 10, 330–337. 10.1093/ecco-jcc/jjv204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J. M. H., Karlsson H., Crespo J. G., Johansson M. E. V., Eklund L., Sjövall H., et al. (2011). Altered O-Glycosylation Profile of MUC2 Mucin Occurs in Active Ulcerative Colitis and Is Associated with Increased Inflammation. Inflamm. Bowel Dis. 17, 2299–2307. 10.1002/ibd.21625 [DOI] [PubMed] [Google Scholar]
- Leung J. M., Davenport M., Wolff M. J., Wiens K. E., Abidi W. M., Poles M. A., et al. (2014). IL-22-producing CD4+ Cells Are Depleted in Actively Inflamed Colitis Tissue. Mucosal Immunol. 7, 124–133. 10.1038/mi.2013.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Yang M., Zhou K., Zhang L., Tian L., Lv S., et al. (2015). Diversity of Duodenal and Rectal Microbiota in Biopsy Tissues and Luminal Contents in Healthy Volunteers. J. Microbiol. Biotechnol. 25, 1136–1145. 10.4014/jmb.1412.12047 [DOI] [PubMed] [Google Scholar]
- Li S., Duan X., Peng Y., Rui Y. (2019). Molecular Characteristics of Carbapenem-Resistant Acinetobacter Spp. From Clinical Infection Samples and Fecal Survey Samples in Southern China. BMC Infect. Dis. 19, 900. 10.1186/s12879-019-4423-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. J., Cheng A. C., Kennon J., Spelman D., Hale D., Melican G., et al. (2014). Prevalence of Multidrug-Resistant Organisms and Risk Factors for Carriage in Long-Term Care Facilities: a Nested Case-Control Study. J. Antimicrob. Chemother. 69, 1972–1980. 10.1093/jac/dku077 [DOI] [PubMed] [Google Scholar]
- Llobet E., Campos M. A., Giménez P., Moranta D., Bengoechea J. A. (2011). Analysis of the Networks Controlling the Antimicrobial-peptide-dependent Induction of Klebsiella pneumoniae Virulence Factors. Infect. Immun. 79, 3718–3732. 10.1128/iai.05226-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucke K., Miehlke S., Jacobs E., Schuppler M. (2006). Prevalence of Bacteroides and Prevotella Spp. In Ulcerative Colitis. J. Med. Microbiol. 55, 617–624. 10.1099/jmm.0.46198-0 [DOI] [PubMed] [Google Scholar]
- Madar Johansson M., Azzouz M., Häggendal B., Säljö K., Malmi H., Zaviolov A., et al. (2020). Glycosphingolipids Recognized by Acinetobacter Baumannii. Microorganisms 8, 612. 10.3390/microorganisms8040612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancilla-Rojano J., Ochoa S. A., Reyes-Grajeda J. P., Flores V., Medina-Contreras O., Espinosa-Mazariego K., et al. (2020). Molecular Epidemiology of Acinetobacter calcoaceticus-Acinetobacter baumannii Complex Isolated From Children at the Hospital Infantil de México Federico Gómez. Front. Microbiol. 11, 576673. 10.3389/fmicb.2020.576673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Presentado J. C., Seija V., Vignoli R., Pontet J., Robino L., Cordeiro N. F., et al. (2013). Polyclonal Endemicity of Acinetobacter Baumannii in Ventilated Patients in an Intensive Care Unit in Uruguay. Int. J. Infect. Dis. 17, e422–e427. 10.1016/j.ijid.2012.12.025 [DOI] [PubMed] [Google Scholar]
- Moffatt J. H., Harper M., Harrison P., Hale J. D. F., Vinogradov E., Seemann T., et al. (2010). Colistin Resistance in Acinetobacter Baumannii Is Mediated by Complete Loss of Lipopolysaccharide Production. Antimicrob. Agents Chemother. 54, 4971–4977. 10.1128/aac.00834-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz S. M., Ernst R. K., Miller S. I. (2004). PmrAB, a Two-Component Regulatory System of Pseudomonas aeruginosa that Modulates Resistance to Cationic Antimicrobial Peptides and Addition of Aminoarabinose to Lipid A. J. Bacteriol. 186, 575–579. 10.1128/jb.186.2.575-579.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E., Losick R., Kahne D. (2019). Robust Suppression of Lipopolysaccharide Deficiency in Acinetobacter Baumannii by Growth in Minimal Medium. J. Bacteriol. 201, e00420. 10.1128/JB.00420-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemec A., Krizova L., Maixnerova M., Sedo O., Brisse S., Higgins P. G. (2015). Acinetobacter Seifertii Sp. nov., a Member of the Acinetobacter Calcoaceticus-Acinetobacter Baumannii Complex Isolated from Human Clinical Specimens. Int. J. Syst. Evol. Microbiol. 65, 934–942. 10.1099/ijs.0.000043 [DOI] [PubMed] [Google Scholar]
- Overduin J., Tylee T. S., Frayo R. S., Cummings D. E. (2014). Hyperosmolarity in the Small Intestine Contributes to Postprandial Ghrelin Suppression. Am. J. Physiology-Gastrointestinal Liver Physiology 306, G1108–G1116. 10.1152/ajpgi.00072.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P. K., Verma P., Kumar H., Bavdekar A., Patole M. S., Shouche Y. S. (2012). Comparative Analysis of Fecal Microflora of Healthy Full-Term Indian Infants Born with Different Methods of Delivery (Vaginal vs Cesarean): Acinetobacter Sp. Prevalence in Vaginally Born Infants. J. Biosci. 37, 989–998. 10.1007/s12038-012-9268-5 [DOI] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter Baumannii : Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 21, 538–582. 10.1128/cmr.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M. R., Casella L. G., Jones J. W., Adams M. D., Zurawski D. V., Hazlett K. R. O., et al. (2013). Unique Structural Modifications Are Present in the Lipopolysaccharide from Colistin-Resistant Strains of Acinetobacter Baumannii. Antimicrob. Agents Chemother. 57, 4831–4840. 10.1128/aac.00865-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers M. J., Trent M. S. (2018). Expanding the Paradigm for the Outer Membrane: Acinetobacter baumannii in the Absence of Endotoxin. Mol. Microbiol. 107, 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullan R. D., Thomas G. A., Rhodes M., Newcombe R. G., Williams G. T., Allen A., et al. (1994). Thickness of Adherent Mucus Gel on Colonic Mucosa in Humans and its Relevance to Colitis. Gut 35, 353–359. 10.1136/gut.35.3.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q., Liu Y.-N., Lv S.-Y., Wu H.-G., Zhang L.-S., Cao Z., et al. (2022). Gut Microbiome Alterations in Colitis Rats after Moxibustion at Bilateral Tianshu Acupoints. BMC Gastroenterol. 22, 62. 10.1186/s12876-022-02115-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf A. H., Tsai H. H., Parker N., Hoffman J., Walker R. J., Rhodes J. M. (1992). Sulphation of Colonic and Rectal Mucin in Inflammatory Bowel Disease: Reduced Sulphation of Rectal Mucus in Ulcerative Colitis. Clin. Sci. (Lond) 83, 623–626. 10.1042/cs0830623 [DOI] [PubMed] [Google Scholar]
- Raplee I., Walker L., Xu L., Surathu A., Chockalingam A., Stewart S., et al. (2021). Emergence of Nosocomial Associated Opportunistic Pathogens in the Gut Microbiome after Antibiotic Treatment. Antimicrob. Resist Infect. Control 10, 36. 10.1186/s13756-021-00903-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Viswanathan R., Singh A., Das P., Basu S. (2010). Gut Colonization by Multidrug-Resistant and Carbapenem-Resistant Acinetobacter Baumannii in Neonates. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1495–1500. 10.1007/s10096-010-1030-z [DOI] [PubMed] [Google Scholar]
- Sekido Y., Nishimura J., Nakano K., Osu T., Chow C.-E. T., Matsuno H., et al. (2020). Some Gammaproteobacteria Are Enriched within CD14+ Macrophages from Intestinal Lamina Propria of Crohn's Disease Patients versus Mucus. Sci. Rep. 10, 2988. 10.1038/s41598-020-59937-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöberg F., Barkman C., Nookaew I., Östman S., Adlerberth I., Saalman R., et al. (2017). Low-complexity Microbiota in the Duodenum of Children with Newly Diagnosed Ulcerative Colitis. PLoS One 12, e0186178. 10.1371/journal.pone.0186178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M. S., Poles J., Leung J. M., Wolff M. J., Davenport M., Lee S. C., et al. (2015). Inferred Metagenomic Comparison of Mucosal and Fecal Microbiota from Individuals Undergoing Routine Screening Colonoscopy Reveals Similar Differences Observed during Active Inflammation. Gut Microbes 6, 48–56. 10.1080/19490976.2014.1000080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom K. A., Hsiao W. W. L., Harris A. D., Stine O. C., Rasko D. A., Johnson J. K. (2010). Patients with Acinetobacter Baumannii Bloodstream Infections Are Colonized in the Gastrointestinal Tract with Identical Strains. Am. J. Infect. Control 38, 751–753. 10.1016/j.ajic.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit J.-F., Garrait V., Misset B., Goldstein F. W., Renaud B., Carlet J., et al. (1993). The Digestive Tract Is a Major Site for Acinetobacter Baumannii Colonization in Intensive Care Unit Patients. J. Infect. Dis. 168, 1336–1337. 10.1093/infdis/168.5.1336 [DOI] [PubMed] [Google Scholar]
- Trabucchi E., Mukenge S., Baratti C., Colombo R., Fregoni F., Montorsi W. (1986). Differential Diagnosis of Crohn's Disease of the Colon from Ulcerative Colitis: Ultrastructure Study with the Scanning Electron Microscope. Int. J. Tissue React. 8, 79–84. [PubMed] [Google Scholar]
- Tytgat K. M. A. J., Van Der Wal J.-W. G., Einerhand A. W. C., Büller H. A., Dekker J. (1996). Quantitative Analysis of MUC2 Synthesis in Ulcerative Colitis. Biochem. Biophysical Res. Commun. 224, 397–405. 10.1006/bbrc.1996.1039 [DOI] [PubMed] [Google Scholar]
- Wenzel U. A., Magnusson M. K., Rydström A., Jonstrand C., Hengst J., Johansson M. E., et al. (2014). Spontaneous Colitis in Muc2-Deficient Mice Reflects Clinical and Cellular Features of Active Ulcerative Colitis. PLoS One 9, e100217. 10.1371/journal.pone.0100217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H., Edmond M. B., Pfaller M. A., Jones R. N., Wenzel R. P., Seifert H. (2000). Nosocomial Bloodstream Infections Caused by Acinetobacter Species in United States Hospitals: Clinical Features, Molecular Epidemiology, and Antimicrobial Susceptibility. Clin. Infect. Dis. 31, 690–697. 10.1086/314040 [DOI] [PubMed] [Google Scholar]
- Wood B., Katzberg R. (1978). Tween 80/diatrizoate Enemas in Bowel Obstruction. Am. J. Roentgenol. 130, 747–750. 10.2214/ajr.130.4.747 [DOI] [PubMed] [Google Scholar]
- Xavier C., Miquel P., Josefina A., Montserrat S., Carmen A., Dom X., et al. (1996). Relevance of Digestive Tract Colonization in the Epidemiology of Nosocomial Infections Due to Multiresistant Acinetobacter Baumannii. Clin. Infect. Dis. 23, 329–334. [DOI] [PubMed] [Google Scholar]
- Zhao F., Edwards R., Dizon D., Afrasiabi K., Mastroianni J. R., Geyfman M., et al. (2010). Disruption of Paneth and Goblet Cell Homeostasis and Increased Endoplasmic Reticulum Stress in Agr2−/− Mice. Dev. Biol. 338, 270–279. 10.1016/j.ydbio.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.