Abstract
Exosomes are nanosized “off-the-shelf” lipid vesicles released by almost all cell types and play a significant role in cell–cell communication. Exosomes have already been proven to carry cell-specific cargos of proteins, lipids, miRNA, and noncoding RNA (ribonucleic acid). These vesicles can be selectively taken up by the neighboring cell and can regulate cellular functions. Herein, we have discussed three different roles of exosomes in neuroscience. First, we have discussed how exosomes play the role of a pathogenic agent as a part of cell–cell communication and transmit pathogens such as amyloid-beta (Aβ), further helping in the propagation of neurodegenerative and other neurological diseases. In the next section, the review talks about the role of exosomes in biomarker discovery in neurological disorders. Toward the end, we have reviewed how exosomes can be harnessed and engineered for therapeutic purposes in different brain diseases. This review is based on the current knowledge generated in this field and our comprehension of this domain.
Keywords: exosome, disease biomarker, cell-cell communication, neurotherapeutics, exosome engineering, pathogenic agent
1 Introduction
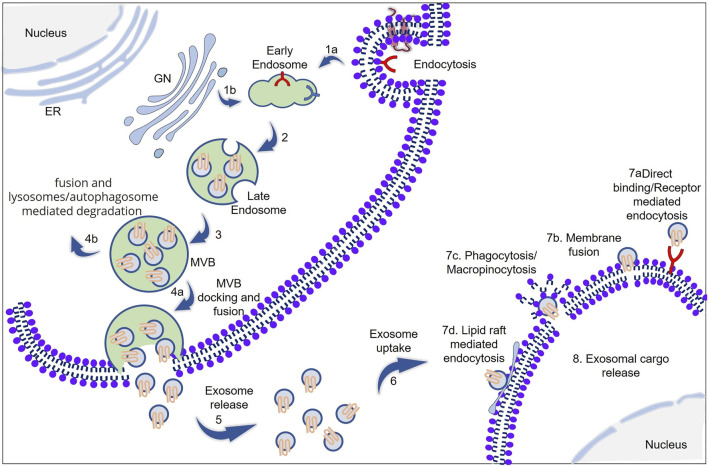
Exosomes, saucer-shaped vesicles of approximately 30–100 nm diameter (Théry et al., 2002b), are one of the different types of “extracellular vesicle” (EV) that are delimited by a lipid bilayer and are naturally released from the cell and play major roles in cell–cell communications. These vesicles are endosomal origin, cannot replicate, i.e., do not contain a functional nucleus, and float at a density of 1.13–1.19 g ml−1 in sucrose gradients (Théry et al., 2018). The process of exosome release can be divided into three steps: exosome biogenesis, intracellular movement of multivesicular bodies (MVBs), and MVB fusion with the plasma membrane. In the first step of exosome biogenesis (Figure 1), early endosomes are formed by inward invagination of the plasma membrane or from the trans-Golgi network. These early endosomes mature to form late endosomes. Invagination of the endosomal membrane into the lumen leads to the formation of intraluminal vesicles (ILVs), which finally leads to the generation of MVBs. Lastly, the generated MVBs will fuse with the plasma membrane or alternatively with lysosomes or autophagosomes. The former results in the release of the exosome, and the latter results in the degradation of MVBs. Several molecules are involved in this complicated process, and the details are summarized elsewhere (Teng and Fussenegger, 2021).
FIGURE 1.
Schematic representation of exosome biogenesis, cargo packing, and cellular uptake. (1a,b) Early endosomes are formed from the inward budding of the plasma membrane or Golgi network (GN). (2,3) Early endosomes mature to form late endosomes and subsequently to multivesicular bodies (MVB). (4a) MVB docking into the plasma membrane via SNAREs and SNAP23, and subsequent fusion with the plasma membrane results in exosome release (5). Alternatively, MVBs can fuse with lysosome and subsequent degradation (4b). (6) The released exosome will be taken up by the neighboring cell. The cellular uptake of exosomes can be (7a) direct binding and receptor-mediated endocytosis, (7b) membrane fusion, (7c) phagocytosis/micropinocytosis, or (7d) lipid raft-mediated endocytosis. The ESCRT machinery plays a key role in protein sorting, particularly for ubiquitinated cargos. During the process of exosome biogenesis, various proteins, including RNA-binding proteins (RBPs), are selectively sequestered into exosomes; these RBPs help in RNA cargo packing into the exosome. (ER: endoplasmic reticulum; GN: Golgi network; MVB: multivesicular bodies; ESCRT: endosomal sorting complex required for transport).
Exosomes are not merely lipid vesicle; they also contain membrane-associated proteins, transmembrane proteins, mRNA, noncoding RNAs, and other cell-specific cargos. Exosomes are equipped with endosomal sorting complex required for transport (ESCRT), Alix, TSG101, HSC70, CD63, CD81, and HSP90β protein that help in the formation and release of exosomes. CD63 and CD81 (Theos et al., 2006; Stuffers et al., 2009) are tetraspanin family proteins that are thought to help in exosome formation and release by an ESCRT-independent mechanism. These proteins are enriched in exosomes compared to the cell lysate, and these are termed “exosomal marker proteins,” which can be identified using Western blot with appropriate antibodies (Thompson et al., 2016; Doyle and Wang, 2019). ExoCarta database hosts about 41,860 proteins, >7540 RNA, and 1,116 lipid molecules from more than 286 exosomal studies; this can help us to have an idea about the diversity of exosomal content from different cell types, current cell state (e.g., transformed, differentiated, stimulated, and stressed), and culture conditions (Keerthikumar et al., 2016). A different method of isolation of exosomes from condition media has been introduced, from the most commonly used methods ultra-centrifugation, sucrose gradient centrifugation, different kit-based to microfluidics-based methods (Supplementary Table S1) (Théry et al., 2006; Wu et al., 2017). The choice of exosome isolation method greatly impacts the exosome quality and quantity (Patel et al., 2019; Brennan et al., 2020). The isolated exosomes can be characterized using nanoparticle tracking analysis (NTA) for size distribution and surface charge, transmission electron microscopy for size distribution and morphology (TEM), atomic force microscopy (AFM) for size distribution and surface morphology, and Western blotting to check the presence and absence of protein markers (Wu et al., 2015; Chopra et al., 2019). Experimental evidence and live-cell imaging (Sung et al., 2020) have already shown that exosomes are secreted by all cell types (Peters et al., 1989; Raposo et al., 1996; Théry et al., 2002a; Morelli et al., 2004) and brain cells like neurons, astrocytes, microglia, and oligodendrocytes are not the exception (Potolicchio et al., 2005; Fauré et al., 2006; Krämer-Albers et al., 2007; Taylor et al., 2007). In the late 1980s, when exosome was first discovered, it was thought to be cellular waste resulting from cell damage or by-products of cell homeostasis and had no significant impact on neighboring cells (Johnstone et al., 1987). But currently, it is crystal clear that exosomes and their cargo can play a major role in cellular processes like in immune response (Greening et al., 2015), signal transduction (Gangoda et al., 2015), antigen presentation (Mittelbrunn et al., 2011) as well as in disease state like chronic inflammation (Lässer et al., 2016), cardiovascular and renal diseases (Gonzalez-Calero et al., 2014), neurodegenerative diseases (Howitt and Hill, 2016), lipid metabolic diseases (Record et al., 2014), traumatic brain injury (Zhang et al., 2021), mental disorder (Saeedi et al., 2019), and tumors (Salem et al., 2016). It is now a well-established fact that exosomes can be found in almost all biological fluids like blood (Hornung et al., 2020), urine (Street et al., 2017), saliva (Machida et al., 2015), breast milk (Qin et al., 2016), cerebrospinal fluid (Yagi et al., 2017), semen (Madison et al., 2015), and amniotic fluid (Keller et al., 2007). Exosomes isolated from these fluids will reflect the cellular origin and its physiological state as a “fingerprint” or “signature” of the donor cell. From this, it is very clear that exosomes can be a potential target for biomarker discovery and early detection of many diseases. Apart from the role of exosomes in signal transduction and biomarker discovery, they can also be harnessed to be used as therapeutics in many diseases (Cooper et al., 2014; Zhuang et al., 2011). The ability to cross blood–brain barrier (BBB), non-immunogenicity, the option of surface engineering, and selective cargo packaging make exosomes emerge as a blockbuster therapeutic option in many diseases (Ghosh et al., 2020; Zhan et al., 2020; Mishra et al., 2021). The probable roles that can be played by exosomes are schematically summarized in Figure 2.
FIGURE 2.
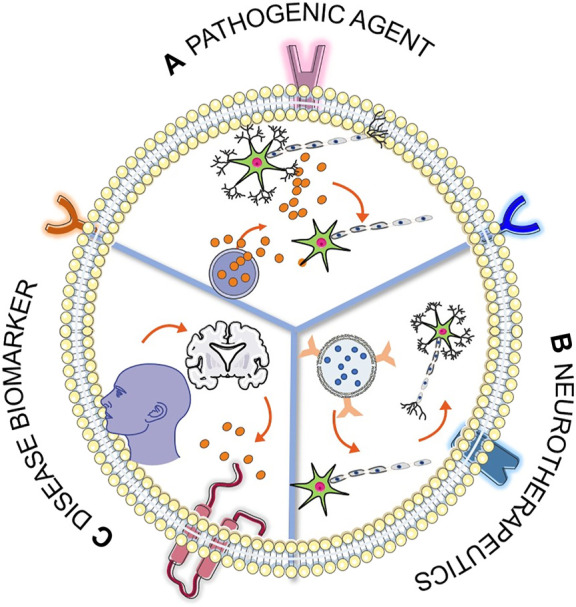
Schematic representation of roles of exosomes in three different fields. (A) The blue sphere represents MVB, and the red sphere represents the exosome; this section of the figure represents the release of pathogenic cargo from the exosome and subsequent disease transmission. (B) Therapeutic potential of surface engineered cargo loaded exosome and subsequent recovery from disease condition. (C) Exosomes released from the disease-affected cell can cross the BBB and can be found in blood circulation, which can be utilized in noninvasive biomarker discovery. [Some component of the figure is adapted from Servier Medical Art; Servier is licensed under a creative commons attribution 3.0 unported license (https://smart.servier.com/)].
2 Exosome as a Pathogenic Agent in Neurological Diseases
The cells in the central nervous system (CNS) communicate between themselves by intercellular and extracellular interactions. The former is mediated by ions and can be transduced and sensed by the cell through ion channels and neurotransmitter receptors present in neurons and glial cells (Yamazaki et al., 2007; Debanne and Rama, 2011). The latter could consist of either wiring transmission, which is primarily dependent on synapses or volume transmission, mediated by exosome for major vesicular carrier or by exocytosis of neurotransmitters (Trueta and De-Miguel, 2012; Borroto-Escuela et al., 2015). The secretion of exosomes from CNS cells was first demonstrated in cultured embryonic cortical neurons, and it can be released either presynaptically at the neuromuscular junction or postsynaptically by cortical neurons upon activation of synaptic NMDA receptors, which will then bind presynaptically to hippocampal neurons (Fauré et al., 2006; Zhang and Yang, 2018). Experimental evidence has also shown that neuron-derived exosomes are 50 times more abundant in soma and dendrites than axons in both peripheral nervous system (PNS) and CNS (Von Bartheld and Altick, 2011). In the brain, the exosomes act as local or distant messengers and communicators and can play a significant role in neural homeostasis, modulation of synaptic plasticity, synaptic transduction, modifying the cell surface properties of target cells, auto-protective mechanism for neurons, sequestering “toxic” (pathogenic) proteins, and promoting regeneration and neuroprotection both in the CNS and the PNS (Korkut et al., 2013; Lopez-Verrilli et al., 2013; Chivet et al., 2014; Kalani et al., 2014; Hornung et al., 2020). In addition to interneuronal communication, exosomes from neuronal culture when added to astrocyte culture have shown to have an effect in extracellular glutamate levels and modulation of synaptic activation (Morel et al., 2013). In the opposite case, when glial cell-derived exosomes are added to the neuronal culture, they significantly increase the firing rate of neurons and has a neuroprotective role under oxidative stress and starvation conditions (Smalheiser, 2007; Morel et al., 2013; Fröhlich et al., 2014). Apart from normal brain function, it is already a proven fact that exosomes have a role in disease progression and can act as a pathogen delivery agent (Hornung et al., 2020; Zhang et al., 2021). The different roles of exosomes in disease are as follows and are schematically represented in Figure 3.
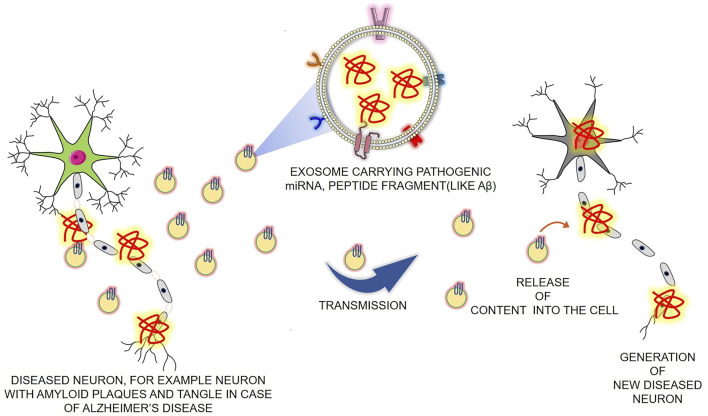
FIGURE 3.
Schematic representation of the role of exosomes as the pathogenic carrier in neurodegenerative disease. Pathogenic cargo-loaded exosomes are released from the diseased cell, and the cells that take up the exosomes get a similar kind of disease. This picture represents a similar phenomenon, taking an example of how exosomes can carry Aβ pathogenic peptides in their lumen and cause the propagation of Alzheimer’s disease. (Aβ: amyloid-beta) (Some component of the figure is adapted from Servier Medical Art; Servier is licensed under a creative commons attribution 3.0 unported license (https://smart.servier.com/).
2.1 Alzheimer’s Disease
Extracellular deposition of polymerized amyloid-β (Aβ) protein, also called plaques, and intracellular filamentous inclusions of hyperphosphorylated tau protein, known as neurofibrillary tangles, are the two major neuropathology involved in Alzheimer’s disease (AD) (Yamaguchi et al., 1989; Lee et al., 1991). Several lines of evidence pointed toward the fact that the exosomes have a role in amyloid pathology in AD. Scientists have shown that in HeLa and N2a cells, after beta-cleavage of amyloid precursor protein (APP) in early endosomes, a minute fraction of Aβ peptides can be secreted from the cells in association with exosomes; not only that but also exosomal proteins have also been found to accumulate in the plaques of AD patients’ brains (Rajendran et al., 2006). Intraperitoneal injection of GW4869, the moiety responsible for inhibition of neutral sphingomyelinase 2 (nSMase2) in the 5XFAD mouse, shows reduced levels of brain and serum exosomes, brain ceramide, and also reduces Aβ1–42 plaque load. This result suggests that exosomes are involved in the generation of Aβ plaques (Dinkins et al., 2014). In recent years, it has been experimentally proven that exosomes isolated from AD brains contain elevated levels of amyloid-beta oligomers; these exosomes can act as vehicles for the neuron-to-neuron transfer of such toxic species. Inhibition of the formation, secretion, or uptake of exosomes has been found to reduce both the spread of oligomers and the related toxicity (Sinha et al., 2018). Recent experimental evidence indicates that soluble pre-fibrillar Aβ species are more toxic than insoluble fibrils (Ladiwala et al., 2012). Scientists have shown that the exosomes of microglial origin are strikingly high in AD patients and in subjects with mild cognitive impairment and are toxic for cultured neurons. Studies have also found that the neurotoxicity of these exosomes is due to the capability of exosome lipids to promote the formation of soluble Aβ species and from the trafficking of neurotoxic Aβ via exosomes after Aβ got internalized into microglia (Joshi et al., 2014). Apart from Aβ, exosomes are also involved in tauopathy. A recent study shows much of the tau phosphorylated at Thr-181 is secreted by M1C cells and occurs via exosomal release (Saman et al., 2012). A group of scientists developed an adeno-associated virus-based model exhibiting rapid tau propagation from the entorhinal cortex to the dentate gyrus and has shown that microglia spread tau via exosomes secretion. Inhibiting exosome synthesis significantly reduces tau propagation in-vitro and in-vivo (Asai et al., 2015). Another group of scientists has discovered cambinol, an inhibitor of the neutral sphingomyelinase 2 (nSMase2) enzyme, and shown that cambinol works in a dose-dependent manner and suppresses extracellular vesicle (EV) production, which in turn reduce tau seed propagation (Bilousova et al., 2018). Apolipoprotein E (apoE) and bridging integrator-1(Bin1), the genetic risk factors for late-onset AD (LOAD), are involved in exosome biogenesis and cargo sorting (Cohn et al., 2021). Experiments have shown that overexpression of BIN1 in PS19 mice promotes the release of Tau via extracellular vesicles (Crotti et al., 2019). On the other hand, the apolipoprotein E4 genotype is involved in the downregulation of exosome biosynthesis and release; this will lead to decreased elimination of materials from the endo-lysosomal system. The failure of the endo-lysosomal system will contribute to amyloidogenic amyloid-β precursor protein processing, compromise trophic signaling and synaptic function, and interfere with a neuron’s ability to degrade material, all of which will result in neuronal vulnerability and a higher risk of AD development (Peng et al., 2019).
2.2 Parkinson's Disease
Parkinson’s disease (PD) is the second most common neurodegenerative disease (Lebouvier et al., 2009), which is characterized by the degenerative death of dopaminergic (DA) neurons in the substantia nigra, a significant decrease in striatal DA content, and the appearance of Lewy bodies due to the accumulation and aggregation of α-synuclein (α-syn) in the cytoplasm of residual nigrostriatal neurons (Spillantini et al., 1998). Experimental evidence has shown that α-syn is packaged into exosomes via the endosome pathway, and it can fuse with the plasma membrane for secretion as exosomal cargo with the assistance of VPS4 and SUMO proteins (Cabin et al., 2002; Lee et al., 2005). Recent studies suggest that exosomes provide favorable conditions for α-syn aggregate formation; this, in turn, promotes the propagation of PD pathology (Grey et al., 2015). Scientists have already discovered the presence of oligomeric α-syn in the exosome, which is readily taken up by the neighboring cell and is more toxic as compared to free α-syn (Danzer et al., 2012). In an experiment with an SH-SY5Y cell line, overexpressing α-syn has shown to have α-syn in isolated exosomes, and this can get transferred to normal SH-SY5Y cells (Alvarez-Erviti et al., 2011a). Later, it was also shown that the exosome-packed α-syn could promote the cell death of recipient neuronal cells. These experiments provide support to the hypothesis of exosome-mediated α-syn propagation between neurons and facilitate PD progression (Emmanouilidou et al., 2010). Recent research by a group of scientists suggested that the presence of α-syn oligomers in CD11b + exosomes of microglia origin can induce α-syn aggregation within neurons (Guo et al., 2020). Overall, it is very clear that exosomes can act as a pathogenic agent in PD propagation.
2.3 Frontotemporal Dementia and Amyotrophic Lateral Sclerosis
FTD involves progressive deficits in behavior, executive function, or language (Bang et al., 2015). Transactive response DNA-binding protein (TDP-43), its aggregation, and cytoplasmic translocation are thought to represent significant steps in the pathogenesis of FTD or ALS (Hu and Grossman, 2009). ALS is a distinct neurodegenerative disease affecting motor neurons in the brain and spinal cord; SOD1 was the first gene discovered to cause familial ALS and was the most studied cause of ALS (Sheng et al., 2012). FTD and ALS appear to be on a spectrum, and some patients display mixed phenotypes of both diseases (Kawakami et al., 2019). The TDP-43 gene is involved in the pathogenesis of both the disease, but SOD1 is only related to ALS but not FTD (Hornung et al., 2020; Jo et al., 2020). Using mouse motor neuron-like NSC-34 cells overexpressing human wild-type or mutant SOD1, scientists have shown that exosomes derived from NSC-34 cells contain SOD1; this gave the evidence of secretion and cell-to-cell transmission of SOD1 (Gomes et al., 2007). In the similar way, TDP-43 can also get transmitted from cell to cell (Nonaka et al., 2013; Iguchi et al., 2016).
2.4 Traumatic Brain Injury
TBI occurs due to the sudden external force in the brain that leads to temporary or permanent neurological deficits. TBI pathogenesis is a complex process due to primary and secondary injuries. The primary deficit occurs immediately, and the secondary injury can occur from minutes to days after the primary impact and consists of a molecular, chemical, and inflammatory cascade responsible for further cerebral damage. The injury involves depolarization of the neurons and release of excitatory neurotransmitters such as glutamate and aspartate that lead to increased intracellular calcium levels, which in turn activates caspases and free radicals that result in the degradation of cells either directly or indirectly through an apoptotic process. These cell deaths result in an inflammatory response that further damages neuronal cells and the blood–brain barrier (BBB) and promotes cerebral edema. The secondary injury phase is followed by the recovery period that involves reorganization at an anatomical, molecular, and functional level. The brain parenchyma, cerebrospinal fluid, and blood make up the volume of the intracranial compartment. An increase in intracranial volume via mass effect from blood, both cytotoxic and vasogenic edema, and venous congestion is also a hallmark of TBI. This would lead to pathological brain compression and, finally, death (Galgano et al., 2017). Exosomes are actively participating in traumatic brain injury pathogenesis; in a case study involving military personnel with mild TBIs (mTBI) and chronic symptoms, it is found that there is a higher level of tau, amyloid-beta 42, and IL-10 in neuron-derived exosomes (NDEs) (Gill et al., 2018). A group of scientists postulated that exosomes could mediate the induction of chronic traumatic encephalopathy (CTE) from mTBI. They have hypothesized a pathway to show how exosomes can mediate pathogenesis from normal to mild deterioration after one mTBI to advanced CTE pathology after the repeated occurrence of mTBIs. According to their hypothesis, initial mTBI leads to the production of NDEs that contains pathogenic complexes of PRPc-Abo-Fyn, SNGY3 + P-tau, and IL-6-sIL-6R; this leads to damage in the donor neurons and other neurons that receive the neurotoxic NDEs. Apart from the neurons, microglia (MG) and astrocytes (AG) will also produce microglia-derived exosomes (MDEs) and astrocytes-derived exosomes (ADEs), respectively, carrying elevated levels of APP, BACE-1, and IL-6, which will further cause neurotoxic damage to neurons. With subsequent episodes of mTBI, this series of processes will increase and cause neuronal apoptosis, which will subsequently lead to the induction of CTE (Goetzl et al., 2019). From these studies, it is believable that the NDE from TBI patients contains neurotoxic cargo, and this NDE causes neuronal damage in proximal or distal cells that receive it.
2.5 Glioblastoma
Apart from its role as a mediator of neurodegenerative disease, exosomes also play a significant role as a pathogenic agent in brain malignancies. The most frequent intrinsic tumors of the central nervous system are glioma. It encompasses two principal subgroups: (World Health Organisation) WHO grade I or “nondiffuse gliomas,” showing a more circumscribed growth pattern, and WHO grades II–IV or “diffusely infiltrating gliomas,” arising from glial cells or glial precursors (Wesseling and Capper, 2018). Scientifically accepted hallmark of cancers includes sustaining proliferative signaling, resisting cell death, evading growth suppression, activating invasion and metastasis, enabling replicative immortality, and inducing angiogenesis (Hanahan and Weinberg, 2011). Exosomes play a significant role as a pathogenic agent in all the six hallmark scenarios (Choi et al., 2018; Oushy et al., 2018; Bian et al., 2019; Hallal et al., 2019; Gao et al., 2020; Lucero et al., 2020). Apart from these roles, studies have shown that glioblastoma-derived exosomes can promote the immunosuppressive properties of microglia when they are taken up by tumor-associated microglia (Abels et al., 2019). Recent studies have shown that exosomes have active participation in the acquisition of resistance to therapy in glioblastomas (Yekula et al., 2021). From the aforementioned evidence, it is clear that exosomes play a major role as a pathogenic agent in maintaining the tumor microenvironment and further metastasis of tumors.
3 Exosome in Biomarker Discovery for Neurological Diseases
The blood–brain barrier (BBB) is a complex physical barrier between the brain and the peripheral circulation that regulates the influx and efflux of molecules to the brain to preserve CNS homeostasis and maintains the stable local ionic microenvironment necessary for neuronal function (Armulik et al., 2010; Kheirandish-Gozal et al., 2017). This barrier makes it difficult for biomolecules to pass from the brain side to the peripheral circulation and remains the main obstacle in the discovery of biomarkers from peripheral blood or serum for brain-related diseases. After the discovery that exosomes can cross BBB and the exosomal content remains active, the interest in exosome-based biomarker discovery in neurological disorder has increased (Saeedi et al., 2019). Exosomal content can help us gain insights into early disease detection, disease state, and disease severity (Figure 4). The ability to compare the biomarkers in exosomes originating from different cell types gives an added advantage to biomarker analysis in CNS-derived blood exosomes as compared to CSF (Hornung et al., 2020). Different biofluids and their exosomal content for biomarker discovery in neurological diseases are listed in Supplementary Table S2.
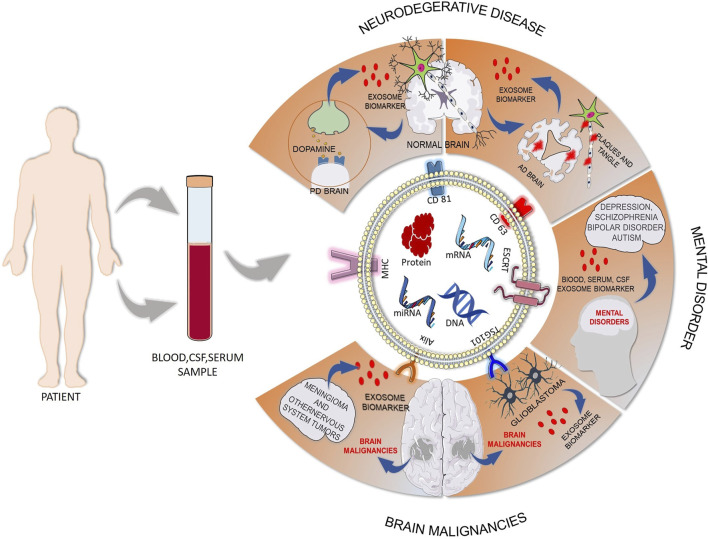
FIGURE 4.
Exosome-based biomarker discovery in neurological disorders from biofluids. Blood, cerebrospinal fluid, plasma, or serum sample can be collected from patients, and the content of the isolated exosomes from that sample can give us the opportunity in early detection of disease to gain knowledge about disease state and disease severity. This figure gives a snapshot of exosome release from disease states like neurogenerative disease, mental disorder, and brain malignancies and the biomarker potential of exosomes. (AD: Alzheimer’s disease; PD: Parkinson's disease) [Some component of the figure is adapted from Servier Medical Art; Servier is licensed under a creative commons attribution 3.0 unported license (https://smart.servier.com/)].
4 Exosome as Neurotherapeutics
In the previous section, we briefly introduced the role of exosomes as a biomarker and pathogenic agent. In this section, we will be focusing on the recent progress surrounding exosomal surface engineering and engineering exosome for packaging cargo of interest. We will also discuss how exosome engineering can increase the value of exosomes as therapeutics in different neurological disorders.
4.1 Engineered Exosome
The main issue with exosome therapeutics is it does not have targeting ability. There are two types of exosome engineering: surface engineering and packaging of cargo of interest. The former endows the exosomes with targetability, and the latter makes the exosomes a better delivery agent and increases therapeutic value.
4.1.1 Methods of Exosome Engineering
Methods of exosome engineering involve two strategies: i) surface engineering strategy (Richardson and Ejima, 2019) and ii) exosome packaging strategy (Donoso-Quezada et al., 2020).
4.1.1.1 Surface Engineering Strategy
Currently available methods for exosome surface functionalization can be classified into two main approaches: 1) genetic engineering and 2) chemical modification. The former method is effective for displaying genetically engineered proteins on the surface of exosomes, but it is only limited to genetically encodable peptides and proteins. But the latter chemical modification method can be used to functionalize exosomes with a wide range of molecules by utilizing noncovalent or covalent interactions. This method remains challenging because of the membrane complexity and because of the various issues with the purification steps necessary to separate the unreacted chemicals from the exosomes (Richardson and Ejima, 2019).
Genetic engineering-based surface engineering includes designing plasmids, transfecting cells with the designed plasmid, and exosomes isolation which itself is a challenging and expensive task. In many works, lactadherin that localizes to exosomes via binding of its C1C2 domain to exosome lipids has been utilized for the generation of chimeric protein and exosome functionalization (Delcayre et al., 2005). The protein of interest is cloned to the C1C2 domain of the lactadherin, which results in chimeric proteins being trafficked to the exosomes, and the N-terminal region was displayed outward on the exosome surface. Different scientific groups have utilized lactadherin to display GLuc reporter protein, carcinoembryonic antigen, and HER2 and anti-HER2 antibodies on the exosome surface (Hartman et al., 2011; Takahashi et al., 2013; Wang et al., 2018b). In another set, scientists have utilized exosomal membrane protein, Lamp2b, and fused targeting peptides, e.g., RVG and RGD peptides to the N terminus of the protein for exosome surface functionalization and targeted delivery to neurons (Kumar et al., 2007) and breast cancer cell (Tian et al., 2014). Yim et al. fused CIBN to the N-terminus of EGFP tagged CD9 (CIBN-EGFP-CD9) for the vector preparation and fused the cargo proteins with CRY2 (cargo protein-CRY2). This system helped to immobilize proteins to the inner surface of exosomes and loaded cargo proteins into the newly generated exosomes (Yim et al., 2016a). Lai et al. (2014) have generated a lentivirus vector encoding the transmembrane domain of PDGFR, BAP domain, and GLuc, and this construct generates surface-engineered exosomes with Gluc and BAP domain which gives an opportunity for in vivo multimodal imaging to monitor tissue distribution, blood levels, and clearance dynamics of the EV. Though the genetic engineering-based methods have advantages, this method still has risks of the engineered biomolecules appearing on the internal exosome surface rather than the desired external surface.
Chemical modification involves covalent or noncovalent interaction for exosome surface functionalization. Alkyl chains can be utilized to anchor molecules into the lipid bilayer membranes of exosomes through hydrophobic interactions. Using this strategy, PEG-lipid conjugates were inserted into exosomal membranes to increase blood circulation time (Kooijmans et al., 2016). Wan et al. (2018) utilized lipids to modify cell membranes for the formation of exosome-mimetic vesicles. They conjugated nucleolin-targeting aptamer AS1411 to cholesterol-PEG and generated surface-functionalized exosomes for in vivo anticancer drug delivery. Functionalized exosomes can also be generated by fusing liposomes, consisting of DOPS and PEG-DSPE with exosomes via freeze−thaw cycles. This method also justifies the efficient packing of liposomal cargo into the newly generated liposome fused exosome (Sato et al., 2016). Wan et al. (2017) have demonstrated the hybridization-mediated assembly of DNA on the exosomal surface for the generation of targeted exosomes. Apart from noncovalent modifications, exosomes can be covalently modified; these modifications are less prone to dissociate away from the exosomes, unlike noncovalent modifications. Smyth et al. (2014) modified the exosomes with alkyne-containing 4-pentynoic acid used carbodiimide coupling onto amines in the exosomal membrane. Then, they used these functionalized exosomes to conjugated azide-tagged fluorophores via azide−alkyne Huisgen cycloaddition or click chemistry. Wang et al. (2015) used metabolic engineering to introduce azide groups on the surface of exosomes; they have also used azide−alkyne Huisgen cycloaddition reaction to covalently introduce small molecules and proteins onto the exosomal surface. Many other works also used similar chemical-based approaches for surface engineering of exosomes (Qi et al., 2016; Kumar et al., 2018; Tian et al., 2018).
4.1.1.2 Exosome Packaging Strategy
Several studies have explored the natural properties of exosomes as nanocarriers, and in recent years, numerous techniques have been developed to improve the immunogenicity, drug loading efficiency, or targeting ability of exosomes. In this section, we will discuss the state-of-the-art packaging strategies to load a cargo of interest into the exosome. Packaging strategies include
Passive loading of hydrophobic compounds: The lipidic nature of the exosome membrane enables several hydrophobic compounds to be passively loaded into the exosomes by co-incubation. Using this strategy, curcumin (Sun et al., 2010), doxorubicin, and paclitaxel (Yang et al., 2015) have been successfully incorporated into different cell-derived exosomes. Apart from therapeutic molecules, large protein, e.g., the tetrameric protein catalase, has efficiently been loaded into Raw 264.7-derived exosomes by simple diffusion (Haney et al., 2015).
Physical methods for molecule loading: The previously discussed passive loading method is not efficient for packaging hydrophobic molecules like DNA or RNA. In these scenarios, physical methods like electroporation, sonication, and extrusion-based methods are used. Momen-Heravi et al. (2014) loaded miRNA-155 into the exosomes using electroporation; they concluded that higher voltages (between 0.14 and 0.2 kV) and a total exosomal protein concentration between 500 and 1,000 μg/ml resulted in better loading yields. Wahlgren et al. (2012) used a similar method to load mitogen-activated protein kinase-1 siRNA (MAPK1-siRNA), and they have found that the optimum electroporation voltage was between 0.150 and 0.200 kV and the exosomal protein concentration was between 250 L and 1,000 μg/ml. Larger nucleic acids, like double-stranded DNA, have also been successfully packaged into exosomes by electroporation; it was found that the loading efficiency of dsDNA significantly decreases for sizes above 750 bp (Lamichhane et al., 2015). By using membrane-permeabilizing agents, the issue of aggregation and fusion of exosomes after electroporation can be resolved (Hood et al., 2014). Another physical method is sonication, and it is reported that sonication can successfully incorporate doxorubicin and paclitaxel into exosomes with more efficiency than other physical methods (Kim et al., 2016). Though this technique is more efficient as it is the most damaging technique for exosomal membrane, sonication is very rarely used for exosomal cargo packaging. Another less explored method for exosome cargo loading is cell extrusion, where cells are extruded through 100–400 nm pore size membrane filters to break up the cell and then cells reform the cell membrane to generate exosome-mimics. Using this method, exosome-mimics have been generated from MCF10A cells and loaded with siRNA by electroporation (Yang et al., 2016). Similarly, catalase was loaded into Raw 264.7-derived exosomes by extruding the catalase mixture with exosomes (Haney et al., 2015).
Hydrophobic modification of nucleic acids: To avoid the problem of aggregation, vesicle fusion, and variations of surface zeta potential associated with electroporation-based siRNA, miRNA cargo loading, hydrophobic modification of nucleic acids has evolved as a strategy to pack cargo into the exosomes. Didiot et al. have modified the siRNA by adding a cholesterol moiety conjugated to the 3’ end of the passenger strand and successfully loaded the modified cargo into U87 glioblastoma cell-derived exosomes (Didiot et al., 2016).
Labeling of target proteins for loading into exosomes: This method gives the opportunity to utilize the protein that plays a major role in exosomal cargo packaging; one such protein is ESCRT which specifically shortens the ubiquitinated proteins in the exosomes. In many studies, scientists are leveling cargo protein with a peptide that can selectively interact with ESCRT, which increases the probability of cargo protein getting packed into the exosomes (Villarroya-Beltri et al., 2014). Cheng and Schorey (2016) fused ubiquitin to the C-terminal region of enhanced green fluorescent protein (EGFP), tumor antigenic protein nHer2, and Mycobacterium tuberculosis proteins Ag58B and ESAT6; ubiquitin labeling increases the loading of all the protein into the exosomes. Other than ESCRT, late-domains (L-domains), which recognizes the WW tag in the protein of interest, also give similar opportunity in cargo loading (Sterzenbach et al., 2017).
Light-induced exosome loading: optically reversible protein–protein interaction (EXPLORE) can be used to load proteins into exosomes. This process involves endogenous biogenesis processes and the delivery of cargo proteins into the cytosol by light-mediated signal (Yim et al., 2016b). Scientists have explored exosomal CD9-CIBN-CRY2-based systems to pack many cargos into the exosomes (Yim et al., 2016a; Huang et al., 2019).
4.2 Applications of Cell-Derived and Surface-Engineered Novel Cargo-Loaded Exosome as Neurotherapeutics
Natural exosomes have various potentials; their clinical application is associated with some inherent limitations of targetability, immunogenicity, and less efficient cargo delivery. Recently, to overcome these limitations, exosome engineering and the development of designer exosomes are coming into the picture. In this section, we will first discuss the role of natural exosomes as neurotherapeutics, and toward the end, we will discuss the role of surface engineered novel cargo-loaded exosomes as neurotherapeutics (Figure 5). Mesenchymal stem cell (MSC) is already extensively studied for regenerative medicine, cell therapy, and tissue engineering (Ankrum and Karp, 2010). Accordingly, research based on exosomes derived from MSCs (MSC-exosomes) has great value as this has the advantage of exosomes and also the characteristics of MSCs (Ghosh et al., 2020). Numerous studies have demonstrated the therapeutic value of MSC-exosomes in tumors, neurodegenerative diseases, cardiovascular and cerebrovascular diseases, wound repair, etc (Cui et al., 2018; Zhao et al., 2018; Moon et al., 2019; Rao et al., 2019; Riazifar et al., 2019). MSC-derived exosomes can exert their therapeutic effect by removing or inhibiting pathological processes or by promoting regenerative mechanisms. In the former case, it is known to reduce amyloid-beta (Aβ) aggregate in AD, rescue dopaminergic neurons from 6-OHDA-induced apoptosis in PD, reduce demyelination in multiple sclerosis (MS), and inhibit apoptosis, inflammation, and promotes angiogenesis in TBI and stroke (Jarmalavičiūtė et al., 2015; Huang et al., 2017; Yang et al., 2017b; Ding et al., 2018; Elia et al., 2019). In the latter case of regeneration, MSC-derived exosomes exert their effect by neuroprotection, neurogenesis, neuromodulation, and angiogenesis in many disease conditions like AD, TBI, and stroke (Doeppner et al., 2015; Zhang et al., 2015; Elia et al., 2019; Otero-Ortega et al., 2020). Other than the aforementioned process, MSC-exosomes show their effect in reducing oxidative stress, restoring the integrity of the BBB, inhibiting tumor growth, and improving behavioral and biochemical deficits in mental disorders like schizophrenia (Jarmalavičiūtė et al., 2015; Chen et al., 2016; Li et al., 2019; Tsivion-Visbord et al., 2020; Luo et al., 2021). Apart from MSC-exosomes, other cell-derived exosomes are explored for a similar application. Sharma et al. (2019) have shown exosomes released by neural cultures can rescue deficits in neuronal proliferation, differentiation, synaptogenesis, and synchronized firing in MECP2-knockdown human primary neural cultures, a model for Rett syndrome. Chen et al. (2020b) have shown exosomes derived from astrocytes promoted the recovery of damaged neurons by downregulation of the apoptosis rate and upregulating mitochondrial function. Lopez-Verrilli et al. (2013) have shown Schwann cells (SC)-derived exosomes increase axonal regeneration in-vitro and increase regeneration after sciatic nerve injury in vivo. Webb et al. (2018) demonstrated that human neural stem cell-derived EVs improve behavior and mobility by removing intracranial hemorrhage, reducing the volume of the cerebral lesions and brain swelling, which eventually leads to recovery from ischemic stroke in a pig model. From the previous discussion, it is clear that natural exosomes have a potential therapeutic effect; many attempts have been made to improve the therapeutic potential and load cargo of interest. Alvarez-Erviti et al. (2011b) have generated engineered dendritic cells that express Lamp2b fused to the neuron-specific RVG peptide and isolated exosomes from the engineered cell. They have packed the exosomes with exogenous siRNA by electroporation and demonstrated the targeted delivery of cargo specifically to neurons, microglia, and oligodendrocytes in the brain, resulting in a specific gene knockdown and subsequent therapeutic effect in AD (Alvarez-Erviti et al., 2011b). Using a similar approach, Yang et al. (2017a) have successfully delivered miR-124 to the infarct site, which leads to amplification of adult neurogenesis in ischemia. Tian et al. (2018) have developed c (RGDyK)-conjugated curcumin-loaded exosomes (cRGD-Exo), which can target the lesion region and show strong suppression of the inflammatory response and cellular apoptosis in the lesion region of the ischemic brain after intravenous administration. A group of scientists developed superparamagnetic iron oxide nanoparticles (SPIONs) and curcumin (Cur)-loaded exosomes. By click chemistry, they have conjugated the exosome membrane with neuropilin-1-targeted peptide (RGERPPR, RGE); these engineered exosomes have the ability to target glioma cells and also have imaging and therapeutic functions (Jia et al., 2018). Ye et al. (2018) have developed methotrexate (MTX)-loaded EVs functionalized with therapeutic [Lys-Leu-Ala (KLA)] and targeted [low-density lipoprotein (LDL)] peptides which show targetability toward brain tumors and show therapeutic effects. Wang and Han (2019) have modified the exosomes and loaded a plasmid expressing B-cell lymphoma-2 (Bcl-2) and Bcl-2-associated X-protein (Bax) short hairpin RNA (shRNA); these exosomes show therapeutic effect in apoptosis and neural functions after TBI. Zhang et al. (2019) have developed c (RGDyK) peptide conjugated, cholesterol-modified miR-210 engineered exosomes, which show upregulated expressions of integrin β3, vascular endothelial growth factor (VEGF), and CD34 and subsequent angiogenesis after middle cerebral artery occlusion (MCAO). In addition to different scientific groups, many companies are developing exosome-based neurotherapeutics; one such company is Evox Therapeutics. Evox Therapeutics uses a biotechnological-based approach to generate drug-loaded brain and central nervous system targeted engineered exosomes (https://www.evoxtherapeutics.com/). Aruna Bio is working on pharmaceutical exosomes for drug delivery to the brain and neurons (https://www.arunabio.com/). Some major disadvantages of cell therapy (induced pluripotent stem cell, iPSC), such as necrosis or abnormal cell differentiation, tumorigenesis, immune rejection caused by cell transplantation, and microvascular embolism, can be overcome by exosome-based therapies (Ghosh et al., 2020). The main advantages of exosome therapies are as followed. First, exosomes mediate stem cell paracrine action, participate in cell–cell communication and are already proven as the main mechanism of disease treatment mediated by cell-based therapies. Second, exosomes can be engineered and can be packed with a cargo of interest like existing, newly developed compositions, and can work as drug delivery vehicles. Third, in some cases, exosomes have autonomous targeting capabilities which make exosome-specific tissue or cell-targeted drug carriers (Mathieu et al., 2019; Wei et al., 2021). From the aforementioned studies, we can have an idea that exosome-based neurotherapeutics have made huge progress in recent years and with the development of new technologies, more progress will follow in upcoming days and can be an alternative to cell-based therapies, like iPSC therapies.
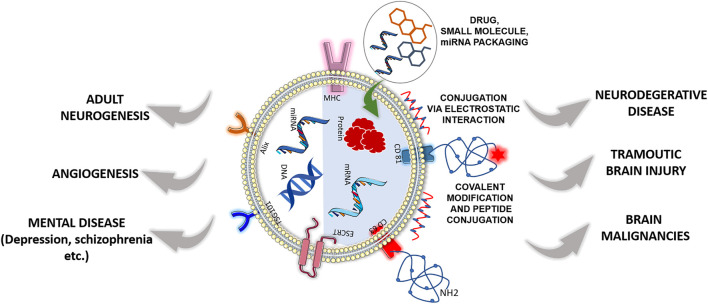
FIGURE 5.
Therapeutic effect of designer exosomes or cell-derived natural exosomes in neurological disorders. The left half of the exosome represents cell-derived natural exosome, and the right half represents surface-functionalized engineered exosome. Both cell-derived and engineered exosomes have a therapeutic effect in neurological disorders with advantages and disadvantages. [Some component of the figure is adapted from Servier Medical Art; Servier is licensed under a creative commons attribution 3.0 unported license (https://smart.servier.com/)].
5 Discussion and Future Perspective
Exosomes are a rising star and a complete package in the era of advanced medical science due to their multiple roles in cell–cell communication, biomarker discovery, disease progression, and therapeutics. The following are some of the benefits that exosomes have: 1) they can pass the blood–brain barrier, and are less invasive, non-tumorigenic, and non-immunogenic, 2) their shelf life and half-life in patients are longer, allowing for long-term storage without loss of function, and 3) they do not reproduce or cause a microvascular embolism (Ghosh et al., 2020). These advantages make the exosomes a superior tool for biomarker discovery and therapeutic development. Apart from advantages, the main challenges for bringing exosomes into the clinical practice include the following: First, the urgent need for standard, efficient, and sensitive methods with a low biofluid volume requirement and high purity and yield for classification and extraction of exosomes from different body fluids and cells. Second, the identification of specific subtypes of EVs is urgently needed, as different vesicles may exert various biological effects. Current methods of exosome isolation are too diverse to confirm the purity of the product. Therefore, it is necessary to standardize the protocols and identification methods when attempting to use exosomes widely in clinical testing. Additionally, more reliable biomarkers should be confirmed. Although many molecules carried by exosomes have been documented to serve as potential biomarkers, few of them are qualified for application. Documented biomarkers need to be validated on a larger scale to create a standard hallmark for diseases. Third, for exosome-based therapeutics development, the targetability of exosomes needs to be checked, as different culture conditions change exosomal cargo. A standardized protocol needs to be developed for large production of exosomes from the cell; cell banks need to be developed. A specific purification method and sensitive method for specific exosomal marker identification need to be developed to avoid ambiguity. Last but not least, the biological safety, targeted efficacy, and adverse effects of exosomes must be confirmed before clinical use. In recent years, to endow the exosomes with targetability and to make exosomes more potent delivery and therapeutic agents, exosome engineering is coming into the picture, which will resolve many issues that cell-derived exosomes have. In conclusion, if the abovementioned lags are resolved and guidelines prescribed by the International Society for Extracellular Vesicles (Théry et al., 2018) are followed, then exosomes can be in the spotlight of clinical practice as biomarkers and therapeutics in the near future.
Author Contributions
SaG and SuG discussed this area for exploration of the role of exosomes as pathogenic agents, disease biomarkers, and neurotherapeutics and jointly wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.878058/full#supplementary-material
References
- Abels E. R., Maas S. L. N., Nieland L., Wei Z., Cheah P. S., Tai E., et al. (2019). Glioblastoma-associated Microglia Reprogramming Is Mediated by Functional Transfer of Extracellular miR-21. Cell Rep. 28 (12), 3105–e7. 10.1016/j.celrep.2019.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akers J. C., Ramakrishnan V., Kim R., Skog J., Nakano I., Pingle S., et al. (2013). MiR-21 in the Extracellular Vesicles (EVs) of Cerebrospinal Fluid (CSF): a Platform for Glioblastoma Biomarker Development. PLoS One 8 (10), e78115. 10.1371/journal.pone.0078115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L., Seow Y., Schapira A. H., Gardiner C., Sargent I. L., Wood M. J., et al. (2011a). Lysosomal Dysfunction Increases Exosome-Mediated Alpha-Synuclein Release and Transmission. Neurobiol. Dis. 42 (3), 360–367. 10.1016/j.nbd.2011.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M. J. (2011b). Delivery of siRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat. Biotechnol. 29 (4), 341–345. 10.1038/nbt.1807 [DOI] [PubMed] [Google Scholar]
- Ankrum J., Karp J. M. (2010). Mesenchymal Stem Cell Therapy: Two Steps Forward, One Step Back. Trends Mol. Med. 16 (5), 203–209. 10.1016/j.molmed.2010.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik A., Genové G., Mäe M., Nisancioglu M. H., Wallgard E., Niaudet C., et al. (2010). Pericytes Regulate the Blood-Brain Barrier. Nature 468 (7323), 557–561. 10.1038/nature09522 [DOI] [PubMed] [Google Scholar]
- Asai H., Ikezu S., Tsunoda S., Medalla M., Luebke J., Haydar T., et al. (2015). Depletion of Microglia and Inhibition of Exosome Synthesis Halt Tau Propagation. Nat. Neurosci. 18 (11), 1584–1593. 10.1038/nn.4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banack S. A., Dunlop R. A., Cox P. A. (2020). An miRNA Fingerprint Using Neural-Enriched Extracellular Vesicles from Blood Plasma: towards a Biomarker for Amyotrophic Lateral Sclerosis/motor Neuron Disease. Open Biol. 10 (6), 200116. 10.1098/rsob.200116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang J., Spina S., Miller B. L. (2015). Frontotemporal Dementia. Lancet 386 (10004), 1672–1682. 10.1016/S0140-6736(15)00461-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian E. B., Chen E. F., Xu Y. D., Yang Z. H., Tang F., Ma C. C., et al. (2019). Exosomal lncRNA-ATB A-ctivates A-strocytes that P-romote G-lioma C-ell I-nvasion. Int. J. Oncol. 54 (2), 713–721. 10.3892/ijo.2018.4644 [DOI] [PubMed] [Google Scholar]
- Bilousova T., Elias C., Miyoshi E., Alam M. P., Zhu C., Campagna J., et al. (2018). Suppression of Tau Propagation Using an Inhibitor that Targets the DK-Switch of nSMase2. Biochem. Biophys. Res. Commun. 499 (4), 751–757. 10.1016/j.bbrc.2018.03.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroto-Escuela D. O., Agnati L. F., Bechter K., Jansson A., Tarakanov A. O., Fuxe K. (2015). The Role of Transmitter Diffusion and Flow versus Extracellular Vesicles in Volume Transmission in the Brain Neural-Glial Networks. Philos. Trans. R. Soc. Lond B Biol. Sci. 370 (1672), 20140183. 10.1098/rstb.2014.0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan K., Martin K., FitzGerald S. P., O'Sullivan J., Wu Y., Blanco A., et al. (2020). A Comparison of Methods for the Isolation and Separation of Extracellular Vesicles from Protein and Lipid Particles in Human Serum. Sci. Rep. 10 (1), 1–13. 10.1038/s41598-020-57497-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabin D. E., Shimazu K., Murphy D., Cole N. B., Gottschalk W., McIlwain K. L., et al. (2002). Synaptic Vesicle Depletion Correlates with Attenuated Synaptic Responses to Prolonged Repetitive Stimulation in Mice Lacking Alpha-Synuclein. J. Neurosci. 22 (20), 8797–8807. 10.1523/JNEUROSCI.22-20-08797.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. Y., Lu J. M., Zhao Z. Q., Li M. C., Lu T., An X. S., et al. (2017). MicroRNA Biomarkers of Parkinson's Disease in Serum Exosome-like Microvesicles. Neurosci. Lett. 644, 94–99. 10.1016/j.neulet.2017.02.045 [DOI] [PubMed] [Google Scholar]
- Ceylan D., Tufekci K. U., Keskinoglu P., Genc S., Özerdem A. (2020). Circulating Exosomal microRNAs in Bipolar Disorder. J. Affect. Disord. 262, 99–107. 10.1016/j.jad.2019.10.038 [DOI] [PubMed] [Google Scholar]
- Cha D. J., Mengel D., Mustapic M., Liu W., Selkoe D. J., Kapogiannis D., et al. (2019). miR-212 and miR-132 Are Downregulated in Neurally Derived Plasma Exosomes of Alzheimer's Patients. Front. Neurosci. 13, 1208. 10.3389/fnins.2019.01208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanteloup G., Cordonnier M., Moreno-Ramos T., Pytel V., Matías-Guiu J., Gobbo J., et al. (2019). Exosomal HSP70 for Monitoring of Frontotemporal Dementia and Alzheimer's Disease: Clinical and FDG-PET Correlation. J. Alzheimers Dis. 71 (4), 1263–1269. 10.3233/JAD-190545 [DOI] [PubMed] [Google Scholar]
- Chen C., Skog J., Hsu C. H., Lessard R. T., Balaj L., Wurdinger T., et al. (2010). Microfluidic Isolation and Transcriptome Analysis of Serum Microvesicles. Lab. Chip 10 (4), 505–511. 10.1039/B916199F [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. H., Chen C. H., Wallace C. G., Yuen C. M., Kao G. S., Chen Y. L., et al. (2016). Intravenous Administration of Xenogenic Adipose-Derived Mesenchymal Stem Cells (ADMSC) and ADMSC-Derived Exosomes Markedly Reduced Brain Infarct Volume and Preserved Neurological Function in Rat after Acute Ischemic Stroke. Oncotarget 7 (46), 74537–74556. 10.18632/oncotarget.12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. C., Wu D., Hu C. J., Chen H. Y., Hsieh Y. C., Huang C. C. (2020a). Exosomal TAR DNA-Binding Protein-43 and Neurofilaments in Plasma of Amyotrophic Lateral Sclerosis Patients: A Longitudinal Follow-Up Study. J. Neurol. Sci. 418, 117070. 10.1016/j.jns.2020.117070 [DOI] [PubMed] [Google Scholar]
- Chen W., Zheng P., Hong T., Wang Y., Liu N., He B., et al. (2020b). Astrocytes-derived Exosomes Induce Neuronal Recovery after Traumatic Brain Injury via Delivering Gap Junction Alpha 1-20 K. J. Tissue Eng. Regen. Med. 14 (3), 412–423. 10.1002/term.3002 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Schorey J. S. (2016). Targeting Soluble Proteins to Exosomes Using a Ubiquitin Tag. Biotechnol. Bioeng. 113 (6), 1315–1324. 10.1002/bit.25884 [DOI] [PubMed] [Google Scholar]
- Cheng L., Doecke J. D., Sharples R. A., Villemagne V. L., Fowler C. J., Rembach A., et al. (2015). Prognostic Serum miRNA Biomarkers Associated with Alzheimer's Disease Shows Concordance with Neuropsychological and Neuroimaging Assessment. Mol. Psychiatry 20 (10), 1188–1196. 10.1038/mp.2014.127 [DOI] [PubMed] [Google Scholar]
- Chivet M., Javalet C., Laulagnier K., Blot B., Hemming F. J., Sadoul R. (2014). Exosomes Secreted by Cortical Neurons upon Glutamatergic Synapse Activation Specifically Interact with Neurons. J. Extracell. Vesicles 3 (1), 24722. 10.3402/jev.v3.24722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D., Montermini L., Kim D. K., Meehan B., Roth F. P., Rak J. (2018). The Impact of Oncogenic EGFRvIII on the Proteome of Extracellular Vesicles Released from Glioblastoma Cells. Mol. Cell Proteomics 17 (10), 1948–1964. 10.1074/mcp.RA118.000644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra N., Dutt Arya B., Jain N., Yadav P., Wajid S., Singh S. P., et al. (2019). Biophysical Characterization and Drug Delivery Potential of Exosomes from Human Wharton's Jelly-Derived Mesenchymal Stem Cells. ACS omega 4 (8), 13143–13152. 10.1021/acsomega.9b01180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn S. W., Melnik M., Huang C., Teter B., Chandra S., Mcintire L. B., et al. (2021). Multi-omics Analysis of Microglial Extracellular Vesicles from Human Alzheimer's Disease Brain Reveals Disease-Associated Signatures. Front. Pharmacol. 12, 3078. 10.3389/fphar.2021.766082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. M., Wiklander P. B., Nordin J. Z., Al-Shawi R., Wood M. J., Vithlani M., et al. (2014). Systemic Exosomal siRNA Delivery Reduced Alpha-Synuclein Aggregates in Brains of Transgenic Mice. Mov. Disord. 29 (12), 1476–1485. 10.1002/mds.25978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti A., Sait H. R., McAvoy K. M., Estrada K., Ergun A., Szak S., et al. (2019). BIN1 Favors the Spreading of Tau via Extracellular Vesicles. Sci. Rep. 9 (1), 1–20. 10.1038/s41598-019-45676-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G. H., Wu J., Mou F. F., Xie W. H., Wang F. B., Wang Q. L., et al. (2018). Exosomes Derived from Hypoxia-Preconditioned Mesenchymal Stromal Cells Ameliorate Cognitive Decline by Rescuing Synaptic Dysfunction and Regulating Inflammatory Responses in APP/PS1 Mice. FASEB J. 32 (2), 654–668. 10.1096/fj.201700600R [DOI] [PubMed] [Google Scholar]
- Cumba Garcia L. M., Peterson T. E., Cepeda M. A., Johnson A. J., Parney I. F. (2019). Isolation and Analysis of Plasma-Derived Exosomes in Patients with Glioma. Front. Oncol. 9, 651. 10.3389/fonc.2019.00651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzer K. M., Kranich L. R., Ruf W. P., Cagsal-Getkin O., Winslow A. R., Zhu L., et al. (2012). Exosomal Cell-To-Cell Transmission of Alpha Synuclein Oligomers. Mol. Neurodegener. 7 (1), 42–18. 10.1186/1750-1326-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debanne D., Rama S. (2011). Astrocytes Shape Axonal Signaling. Sci. Signal. 4 (162), pe11. 10.1126/scisignal.2001884 [DOI] [PubMed] [Google Scholar]
- Delcayre A., Estelles A., Sperinde J., Roulon T., Paz P., Aguilar B., et al. (2005). Exosome Display Technology: Applications to the Development of New Diagnostics and Therapeutics. Blood Cells Mol. Dis. 35 (2), 158–168. 10.1016/j.bcmd.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Denk J., Oberhauser F., Kornhuber J., Wiltfang J., Fassbender K., Schroeter M. L., et al. (2018). Specific Serum and CSF microRNA Profiles Distinguish Sporadic Behavioural Variant of Frontotemporal Dementia Compared with Alzheimer Patients and Cognitively Healthy Controls. PloS one 13 (5), e0197329. 10.1371/journal.pone.0197329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot M. C., Hall L. M., Coles A. H., Haraszti R. A., Godinho B. M., Chase K., et al. (2016). Exosome-mediated Delivery of Hydrophobically Modified siRNA for Huntingtin mRNA Silencing. Mol. Ther. 24 (10), 1836–1847. 10.1038/mt.2016.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M., Shen Y., Wang P., Xie Z., Xu S., Zhu Z., et al. (2018). Exosomes Isolated from Human Umbilical Cord Mesenchymal Stem Cells Alleviate Neuroinflammation and Reduce Amyloid-Beta Deposition by Modulating Microglial Activation in Alzheimer's Disease. Neurochem. Res. 43 (11), 2165–2177. 10.1007/s11064-018-2641-5 [DOI] [PubMed] [Google Scholar]
- Dinkins M. B., Dasgupta S., Wang G., Zhu G., Bieberich E. (2014). Exosome Reduction In Vivo Is Associated with Lower Amyloid Plaque Load in the 5XFAD Mouse Model of Alzheimer's Disease. Neurobiol. Aging 35 (8), 1792–1800. 10.1016/j.neurobiolaging.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doeppner T. R., Herz J., Görgens A., Schlechter J., Ludwig A. K., Radtke S., et al. (2015). Extracellular Vesicles Improve Post-Stroke Neuroregeneration and Prevent Postischemic Immunosuppression. Stem Cells Transl. Med. 4 (10), 1131–1143. 10.5966/sctm.2015-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Zieren R. C., Horie K., Kim C. J., Mallick E., Jing Y., et al. (2020). Comprehensive Evaluation of Methods for Small Extracellular Vesicles Separation from Human Plasma, Urine and Cell Culture Medium. J. Extracell. Vesicles 10 (2), e12044. 10.1002/jev2.12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Gu H., Guo Q., Liang S., Xue J., Yao F., et al. (2021). Profiling of Serum Exosome MiRNA Reveals the Potential of a MiRNA Panel as Diagnostic Biomarker for Alzheimer's Disease. Mol. Neurobiol., 1–11. 10.1007/s12035-021-02323-y [DOI] [PubMed] [Google Scholar]
- Donoso-Quezada J., Ayala-Mar S., González-Valdez J. (2020). State-of-the-art Exosome Loading and Functionalization Techniques for Enhanced Therapeutics: a Review. Crit. Rev. Biotechnol. 40 (6), 804–820. 10.1080/07388551.2020.1785385 [DOI] [PubMed] [Google Scholar]
- Doyle L. M., Wang M. Z. (2019). Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 8 (7), 727. 10.3390/cells8070727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Yu Y., Hu Y., Li X. W., Wei Z. X., Pan R. Y., et al. (2019). Genome-wide, Integrative Analysis Implicates Exosome-Derived microRNA Dysregulation in Schizophrenia. Schizophr. Bull. 45 (6), 1257–1266. 10.1093/schbul/sby191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia C. A., Tamborini M., Rasile M., Desiato G., Marchetti S., Swuec P., et al. (2019). Intracerebral Injection of Extracellular Vesicles from Mesenchymal Stem Cells Exerts Reduced Aβ Plaque Burden in Early Stages of a Preclinical Model of Alzheimer's Disease. Cells 8 (9), 1059. 10.3390/cells8091059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanouilidou E., Melachroinou K., Roumeliotis T., Garbis S. D., Ntzouni M., Margaritis L. H., et al. (2010). Cell-produced Alpha-Synuclein Is Secreted in a Calcium-dependent Manner by Exosomes and Impacts Neuronal Survival. J. Neurosci. 30 (20), 6838–6851. 10.1523/JNEUROSCI.5699-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauré J., Lachenal G., Court M., Hirrlinger J., Chatellard-Causse C., Blot B., et al. (2006). Exosomes Are Released by Cultured Cortical Neurones. Mol. Cell. Neurosci. 31 (4), 642–648. 10.1016/j.mcn.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Feneberg E., Steinacker P., Lehnert S., Schneider A., Walther P., Thal D. R., et al. (2014). Limited Role of Free TDP-43 as a Diagnostic Tool in Neurodegenerative Diseases. Amyotroph. Lateral Scler. Front. Degener. 15 (5-6), 351–356. 10.3109/21678421.2014.905606 [DOI] [PubMed] [Google Scholar]
- Fiandaca M. S., Kapogiannis D., Mapstone M., Boxer A., Eitan E., Schwartz J. B., et al. (2015). Identification of Preclinical Alzheimer's Disease by a Profile of Pathogenic Proteins in Neurally Derived Blood Exosomes: A Case-Control Study. Alzheimers Dement. 11 (6), 600–e1. 10.1016/j.jalz.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich D., Kuo W. P., Frühbeis C., Sun J.-J., Zehendner C. M., Luhmann H. J., et al. (2014). Multifaceted Effects of Oligodendroglial Exosomes on Neurons: Impact on Neuronal Firing Rate, Signal Transduction and Gene Regulation. Phil. Trans. R. Soc. B 369 (1652), 20130510. 10.1098/rstb.2013.0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgano M., Toshkezi G., Qiu X., Russell T., Chin L., Zhao L. R. (2017). Traumatic Brain Injury: Current Treatment Strategies and Future Endeavors. Cell Transpl. 26 (7), 1118–1130. 10.1177/0963689717714102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangoda L., Boukouris S., Liem M., Kalra H., Mathivanan S. (2015). Extracellular Vesicles Including Exosomes Are Mediators of Signal Transduction: Are They Protective or Pathogenic? Proteomics 15 (2-3), 260–271. 10.1002/pmic.201400234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Zhang Z., Mashimo T., Shen B., Nyagilo J., Wang H., et al. (2020). Gliomas Interact with Non-glioma Brain Cells via Extracellular Vesicles. Cell Rep. 30 (8), 2489–e5. 10.1016/j.celrep.2020.01.089 [DOI] [PubMed] [Google Scholar]
- Gelle T., Samey R. A., Plansont B., Bessette B., Jauberteau-Marchan M. O., Lalloué F., et al. (2021). BDNF and Pro-BDNF in Serum and Exosomes in Major Depression: Evolution after Antidepressant Treatment. Prog. Neuropsychopharmacol. Biol. Psychiatry 109, 110229. 10.1016/j.pnpbp.2020.110229 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Garg S., Ghosh S. (2020). Cell-derived Exosome Therapy: a Novel Approach to Treat Post-traumatic Brain Injury Mediated Neural Injury. ACS Chem. Neurosci. 11 (14), 2045–2047. 10.1021/acschemneuro.0c00368 [DOI] [PubMed] [Google Scholar]
- Gill J., Mustapic M., Diaz-Arrastia R., Lange R., Gulyani S., Diehl T., et al. (2018). Higher Exosomal Tau, Amyloid-Beta 42 and IL-10 Are Associated with Mild TBIs and Chronic Symptoms in Military Personnel. Brain Inj. 32 (11), 1277–1284. 10.1080/02699052.2018.1471738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl E. J., Boxer A., Schwartz J. B., Abner E. L., Petersen R. C., Miller B. L., et al. (2015). Altered Lysosomal Proteins in Neural-Derived Plasma Exosomes in Preclinical Alzheimer Disease. Neurology 85 (1), 40–47. 10.1212/WNL.0000000000001702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl E. J., Ledreux A., Granholm A. C., Elahi F. M., Goetzl L., Hiramoto J., et al. (2019). Neuron-Derived Exosome Proteins May Contribute to Progression from Repetitive Mild Traumatic Brain Injuries to Chronic Traumatic Encephalopathy. Front. Neurosci. 13, 452. 10.3389/fnins.2019.00452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl E. J., Mustapic M., Kapogiannis D., Eitan E., Lobach I. V., Goetzl L., et al. (2016). Cargo Proteins of Plasma Astrocyte-Derived Exosomes in Alzheimer's Disease. FASEB J. 30 (11), 3853–3859. 10.1096/fj.201600756R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C., Keller S., Altevogt P., Costa J. (2007). Evidence for Secretion of Cu,Zn Superoxide Dismutase via Exosomes from a Cell Model of Amyotrophic Lateral Sclerosis. Neurosci. Lett. 428 (1), 43–46. 10.1016/j.neulet.2007.09.024 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Calero L., Martin-Lorenzo M., Alvarez-Llamas G. (2014). Exosomes: a Potential Key Target in Cardio-Renal Syndrome. Front. Immunol. 5, 465. 10.3389/fimmu.2014.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening D. W., Gopal S. K., Xu R., Simpson R. J., Chen W. (2015). Exosomes and Their Roles in Immune Regulation and Cancer. Semin. Cell Dev. Biol. 40, 72–81. 10.1016/j.semcdb.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Grey M., Dunning C. J., Gaspar R., Grey C., Brundin P., Sparr E., et al. (2015). Acceleration of α-synuclein Aggregation by Exosomes. J. Biol. Chem. 290 (5), 2969–2982. 10.1074/jbc.M114.585703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes V. A., Kenney K., Shahim P., Qu B. X., Lai C., Devoto C., et al. CENC Multisite Observational Study Investigators (2020). Exosomal Neurofilament Light: A Prognostic Biomarker for Remote Symptoms after Mild Traumatic Brain Injury? Neurology 94 (23), e2412–e2423. 10.1212/WNL.0000000000009577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Wang J., Zhao Y., Feng Y., Han S., Dong Q., et al. (2020). Microglial Exosomes Facilitate α-synuclein Transmission in Parkinson's Disease. Brain 143 (5), 1476–1497. 10.1093/brain/awaa090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallal S., Mallawaaratchy D. M., Wei H., Ebrahimkhani S., Stringer B. W., Day B. W., et al. (2019). Extracellular Vesicles Released by Glioblastoma Cells Stimulate Normal Astrocytes to Acquire a Tumor-Supportive Phenotype via P53 and MYC Signaling Pathways. Mol. Neurobiol. 56 (6), 4566–4581. 10.1007/s12035-018-1385-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Peng C., Yi J., Zhang D., Xiang X., Peng X., et al. (2021). Highly Efficient Exosome Purification from Human Plasma by Tangential Flow Filtration Based Microfluidic Chip. Sensors Actuators B Chem. 333, 129563. 10.1016/j.snb.2021.129563 [DOI] [Google Scholar]
- Hanahan D., Weinberg R. A. (2011). Hallmarks of Cancer: the Next Generation. cell 144 (5), 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Haney M. J., Klyachko N. L., Zhao Y., Gupta R., Plotnikova E. G., He Z., et al. (2015). Exosomes as Drug Delivery Vehicles for Parkinson's Disease Therapy. J. Control. Release 207, 18–30. 10.1016/j.jconrel.2015.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman Z. C., Wei J., Glass O. K., Guo H., Lei G., Yang X. Y., et al. (2011). Increasing Vaccine Potency through Exosome Antigen Targeting. Vaccine 29 (50), 9361–9367. 10.1016/j.vaccine.2011.09.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi N., Doi H., Kurata Y., Kagawa H., Atobe Y., Funakoshi K., et al. (2020). Proteomic Analysis of Exosome-Enriched Fractions Derived from Cerebrospinal Fluid of Amyotrophic Lateral Sclerosis Patients. Neurosci. Res. 160, 43–49. 10.1016/j.neures.2019.10.010 [DOI] [PubMed] [Google Scholar]
- Hermann S., Buschmann D., Kirchner B., Borrmann M., Brandes F., Kotschote S., et al. (2019). Transcriptomic Profiling of Cell-free and Vesicular microRNAs from Matched Arterial and Venous Sera. J. Extracell. Vesicles 8 (1), 1670935. 10.1080/20013078.2019.1670935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisey C. L., Dorayappan K. D. P., Cohn D. E., Selvendiran K., Hansford D. J. (2018). Microfluidic Affinity Separation Chip for Selective Capture and Release of Label-free Ovarian Cancer Exosomes. Lab. Chip 18 (20), 3144–3153. 10.1039/C8LC00834E [DOI] [PubMed] [Google Scholar]
- Holcar M., Kandušer M., Lenassi M. (2021). Blood Nanoparticles - Influence on Extracellular Vesicle Isolation and Characterization. Front. Pharmacol. 12. 10.3389/fphar.2021.773844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood J. L., Scott M. J., Wickline S. A. (2014). Maximizing Exosome Colloidal Stability Following Electroporation. Anal. Biochem. 448, 41–49. 10.1016/j.ab.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung S., Dutta S., Bitan G. (2020). CNS-derived Blood Exosomes as a Promising Source of Biomarkers: Opportunities and Challenges. Front. Mol. Neurosci. 13, 38. 10.3389/fnmol.2020.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt J., Hill A. F. (2016). Exosomes in the Pathology of Neurodegenerative Diseases. J. Biol. Chem. 291 (52), 26589–26597. 10.1074/jbc.R116.757955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. T., Grossman M. (2009). TDP-43 and Frontotemporal Dementia. Curr. Neurol. Neurosci. Rep. 9 (5), 353–358. 10.1007/s11910-009-0052-3 [DOI] [PubMed] [Google Scholar]
- Huang J. H., Yin X. M., Xu Y., Xu C. C., Lin X., Ye F. B., et al. (2017). Systemic Administration of Exosomes Released from Mesenchymal Stromal Cells Attenuates Apoptosis, Inflammation, and Promotes Angiogenesis after Spinal Cord Injury in Rats. J. Neurotrauma 34 (24), 3388–3396. 10.1089/neu.2017.5063 [DOI] [PubMed] [Google Scholar]
- Huang K., Fang C., Yi K., Liu X., Qi H., Tan Y., et al. (2018). The Role of PTRF/Cavin1 as a Biomarker in Both Glioma and Serum Exosomes. Theranostics 8 (6), 1540–1557. 10.7150/thno.22952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Gu N., Zhang X.-E., Wang D.-B. (2019). Light-Inducible Exosome-Based Vehicle for Endogenous RNA Loading and Delivery to Leukemia Cells. Adv. Funct. Mat. 29 (9), 1807189. 10.1002/adfm.201807189 [DOI] [Google Scholar]
- Iguchi Y., Eid L., Parent M., Soucy G., Bareil C., Riku Y., et al. (2016). Exosome Secretion Is a Key Pathway for Clearance of Pathological TDP-43. Brain 139 (12), 3187–3201. 10.1093/brain/aww237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmalavičiūtė A., Tunaitis V., Pivoraitė U., Venalis A., Pivoriūnas A. (2015). Exosomes from Dental Pulp Stem Cells Rescue Human Dopaminergic Neurons from 6-Hydroxy-Dopamine-Induced Apoptosis. Cytotherapy 17 (7), 932–939. 10.1016/j.jcyt.2014.07.013 [DOI] [PubMed] [Google Scholar]
- Jia G., Han Y., An Y., Ding Y., He C., Wang X., et al. (2018). NRP-1 Targeted and Cargo-Loaded Exosomes Facilitate Simultaneous Imaging and Therapy of Glioma In Vitro and In Vivo . Biomaterials 178, 302–316. 10.1016/j.biomaterials.2018.06.029 [DOI] [PubMed] [Google Scholar]
- Jia L., Qiu Q., Zhang H., Chu L., Du Y., Zhang J., et al. (2019). Concordance between the Assessment of Aβ42, T-Tau, and P-T181-Tau in Peripheral Blood Neuronal-Derived Exosomes and Cerebrospinal Fluid. Alzheimers Dement. 15 (8), 1071–1080. 10.1016/j.jalz.2019.05.002 [DOI] [PubMed] [Google Scholar]
- Jia L., Zhu M., Kong C., Pang Y., Zhang H., Qiu Q., et al. (2021). Blood Neuro-Exosomal Synaptic Proteins Predict Alzheimer's Disease at the Asymptomatic Stage. Alzheimers Dement. 17 (1), 49–60. 10.1002/alz.12166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Qian J., Yang J., Yan X., Xue X., Chang Q. (2018). Advances in Exosome-Related Biomarkers for Glioblastoma: Basic Research and Clinical Application. Glioma 1 (5), 159. 10.4103/glioma.glioma_35_1 [DOI] [Google Scholar]
- Jo M., Lee S., Jeon Y. M., Kim S., Kwon Y., Kim H. J. (2020). The Role of TDP-43 Propagation in Neurodegenerative Diseases: Integrating Insights from Clinical and Experimental Studies. Exp. Mol. Med. 52 (10), 1652–1662. 10.1038/s12276-020-00513-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone R. M., Adam M., Hammond J. R., Orr L., Turbide C. (1987). Vesicle Formation during Reticulocyte Maturation. Association of Plasma Membrane Activities with Released Vesicles (Exosomes). J. Biol. Chem. 262 (19), 9412–9420. 10.1016/S0021-9258(18)48095-7 [DOI] [PubMed] [Google Scholar]
- Joshi P., Turola E., Ruiz A., Bergami A., Libera D. D., Benussi L., et al. (2014). Microglia Convert Aggregated Amyloid-β into Neurotoxic Forms through the Shedding of Microvesicles. Cell Death Differ. 21 (4), 582–593. 10.1038/cdd.2013.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalani A., Tyagi A., Tyagi N. (2014). Exosomes: Mediators of Neurodegeneration, Neuroprotection and Therapeutics. Mol. Neurobiol. 49 (1), 590–600. 10.1007/s12035-013-8544-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami I., Arai T., Hasegawa M. (2019). The Basis of Clinicopathological Heterogeneity in TDP-43 Proteinopathy. Acta Neuropathol. 138 (5), 751–770. 10.1007/s00401-019-02077-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., et al. (2016). ExoCarta: a Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 428 (4), 688–692. 10.1016/j.jmb.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S., Rupp C., Stoeck A., Runz S., Fogel M., Lugert S., et al. (2007). CD24 Is a Marker of Exosomes Secreted into Urine and Amniotic Fluid. Kidney Int. 72 (9), 1095–1102. 10.1038/sj.ki.5002486 [DOI] [PubMed] [Google Scholar]
- Kheirandish-Gozal L., Khalyfa A., Gozal D. (2017). Exosomes, Blood-Brain Barrier, and Cognitive Dysfunction in Pediatric Sleep Apnea. Sleep. Biol. Rhythms 15 (4), 261–267. 10.1007/s41105-017-0108-8 [DOI] [Google Scholar]
- Kim M. S., Haney M. J., Zhao Y., Mahajan V., Deygen I., Klyachko N. L., et al. (2016). Development of Exosome-Encapsulated Paclitaxel to Overcome MDR in Cancer Cells. Nanomedicine 12 (3), 655–664. 10.1016/j.nano.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijmans S. A. A., Fliervoet L. A. L., Van Der Meel R., Fens M. H. A. M., Heijnen H. F. G., van Bergen En Henegouwen P. M. P., et al. (2016). PEGylated and Targeted Extracellular Vesicles Display Enhanced Cell Specificity and Circulation Time. J. Control. Release 224, 77–85. 10.1016/j.jconrel.2016.01.009 [DOI] [PubMed] [Google Scholar]
- Korkut C., Li Y., Koles K., Brewer C., Ashley J., Yoshihara M., et al. (2013). Regulation of Postsynaptic Retrograde Signaling by Presynaptic Exosome Release. Neuron 77 (6), 1039–1046. 10.1016/j.neuron.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer-Albers E. M., Bretz N., Tenzer S., Winterstein C., Möbius W., Berger H., et al. (2007). Oligodendrocytes Secrete Exosomes Containing Major Myelin and Stress-Protective Proteins: Trophic Support for Axons? Proteomics Clin. Appl. 1 (11), 1446–1461. 10.1002/prca.200700522 [DOI] [PubMed] [Google Scholar]
- Kumar P., Wu H., McBride J. L., Jung K. E., Kim M. H., Davidson B. L., et al. (2007). Transvascular Delivery of Small Interfering RNA to the Central Nervous System. Nature 448 (7149), 39–43. 10.1038/nature05901 [DOI] [PubMed] [Google Scholar]
- Kumar S., Michael I. J., Park J., Granick S., Cho Y. K. (2018). Cloaked Exosomes: Biocompatible, Durable, and Degradable Encapsulation. Small 14 (34), e1802052. 10.1002/smll.201802052 [DOI] [PubMed] [Google Scholar]
- Ladiwala A. R., Litt J., Kane R. S., Aucoin D. S., Smith S. O., Ranjan S., et al. (2012). Conformational Differences between Two Amyloid β Oligomers of Similar Size and Dissimilar Toxicity. J. Biol. Chem. 287 (29), 24765–24773. 10.1074/jbc.M111.329763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. P., Mardini O., Ericsson M., Prabhakar S., Maguire C., Chen J. W., et al. (2014). Dynamic Biodistribution of Extracellular Vesicles In Vivo Using a Multimodal Imaging Reporter. ACS Nano 8 (1), 483–494. 10.1021/nn404945r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane T. N., Raiker R. S., Jay S. M. (2015). Exogenous DNA Loading into Extracellular Vesicles via Electroporation Is Size-dependent and Enables Limited Gene Delivery. Mol. Pharm. 12 (10), 3650–3657. 10.1021/acs.molpharmaceut.5b00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lässer C., O’Neil S. E., Shelke G. V., Sihlbom C., Hansson S. F., Gho Y. S., et al. (2016). Exosomes in the Nose Induce Immune Cell Trafficking and Harbour an Altered Protein Cargo in Chronic Airway Inflammation. J. Transl. Med. 14 (1), 1–14. 10.1186/s12967-016-0927-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebouvier T., Chaumette T., Paillusson S., Duyckaerts C., Bruley des Varannes S., Neunlist M., et al. (2009). The Second Brain and Parkinson's Disease. Eur. J. Neurosci. 30 (5), 735–741. 10.1111/j.1460-9568.2009.06873.x [DOI] [PubMed] [Google Scholar]
- Lee H. J., Patel S., Lee S. J. (2005). Intravesicular Localization and Exocytosis of Alpha-Synuclein and its Aggregates. J. Neurosci. 25 (25), 6016–6024. 10.1523/JNEUROSCI.0692-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V. M., Balin B. J., Otvos L., Trojanowski J. Q. (1991). A68: a Major Subunit of Paired Helical Filaments and Derivatized Forms of Normal Tau. Science 251 (4994), 675–678. 10.1126/science.1899488 [DOI] [PubMed] [Google Scholar]
- Li C., Fei K., Tian F., Gao C., Yang S. (2019). Adipose-derived Mesenchymal Stem Cells Attenuate Ischemic Brain Injuries in Rats by Modulating miR-21-3p/MAT2B Signaling Transduction. Croat. Med. J. 60 (5), 439–448. 10.3325/cmj.2019.60.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Huang L., Chen J., Ni F., Zhang Y., Liu F. (2021). Isolation of Exosome Nanoparticles from Human Cerebrospinal Fluid for Proteomic Analysis. ACS Appl. Nano Mat. 4 (4), 3351–3359. 10.1021/acsanm.0c02622 [DOI] [Google Scholar]
- Liu C. G., Song J., Zhang Y. Q., Wang P. C. (2014). MicroRNA-193b Is a Regulator of Amyloid Precursor Protein in the Blood and Cerebrospinal Fluid Derived Exosomal microRNA-193b Is a Biomarker of Alzheimer's Disease. Mol. Med. Rep. 10 (5), 2395–2400. 10.3892/mmr.2014.2484 [DOI] [PubMed] [Google Scholar]
- Lopez-Verrilli M. A., Picou F., Court F. A. (2013). Schwann Cell-Derived Exosomes Enhance Axonal Regeneration in the Peripheral Nervous System. Glia 61 (11), 1795–1806. 10.1002/glia.22558 [DOI] [PubMed] [Google Scholar]
- Lucero R., Zappulli V., Sammarco A., Murillo O. D., Cheah P. S., Srinivasan S., et al. (2020). Glioma-derived miRNA-Containing Extracellular Vesicles Induce Angiogenesis by Reprogramming Brain Endothelial Cells. Cell Rep. 30 (7), 2065–e4. 10.1016/j.celrep.2020.01.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Xian P., Wang T., Wu S., Sun T., Wang W., et al. (2021). Antioxidant Activity of Mesenchymal Stem Cell-Derived Extracellular Vesicles Restores Hippocampal Neurons Following Seizure Damage. Theranostics 11 (12), 5986–6005. 10.7150/thno.58632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida T., Tomofuji T., Ekuni D., Maruyama T., Yoneda T., Kawabata Y., et al. (2015). MicroRNAs in Salivary Exosome as Potential Biomarkers of Aging. Int. J. Mol. Sci. 16 (9), 21294–21309. 10.3390/ijms160921294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison M. N., Jones P. H., Okeoma C. M. (2015). Exosomes in Human Semen Restrict HIV-1 Transmission by Vaginal Cells and Block Intravaginal Replication of LP-BM5 Murine AIDS Virus Complex. Virology 482, 189–201. 10.1016/j.virol.2015.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M., Martin-Jaular L., Lavieu G., Théry C. (2019). Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-To-Cell Communication. Nat. Cell Biol. 21 (1), 9–17. 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- McKeever P. M., Schneider R., Taghdiri F., Weichert A., Multani N., Brown R. A., et al. (2018). MicroRNA Expression Levels Are Altered in the Cerebrospinal Fluid of Patients with Young-Onset Alzheimer's Disease. Mol. Neurobiol. 55 (12), 8826–8841. 10.1007/s12035-018-1032-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Singh P., Qayoom I., Prasad A., Kumar A. (2021). Current Strategies in Tailoring Methods for Engineered Exosomes and Future Avenues in Biomedical Applications. J. Mat. Chem. B 9 (32), 6281–6309. 10.1039/D1TB01088C [DOI] [PubMed] [Google Scholar]
- Mittelbrunn M., Gutiérrez-Vázquez C., Villarroya-Beltri C., González S., Sánchez-Cabo F., González M. Á., et al. (2011). Unidirectional Transfer of microRNA-Loaded Exosomes from T Cells to Antigen-Presenting Cells. Nat. Commun. 2 (1), 1–10. 10.1038/ncomms1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen-Heravi F., Bala S., Bukong T., Szabo G. (2014). Exosome-mediated Delivery of Functionally Active miRNA-155 Inhibitor to Macrophages. Nanomedicine 10 (7), 1517–1527. 10.1016/j.nano.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondello S., Guedes V. A., Lai C., Czeiter E., Amrein K., Kobeissy F., et al. (2020). Circulating Brain Injury Exosomal Proteins Following Moderate-To-Severe Traumatic Brain Injury: Temporal Profile, Outcome Prediction and Therapy Implications. Cells 9 (4), 977. 10.3390/cells9040977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon G. J., Sung J. H., Kim D. H., Kim E. H., Cho Y. H., Son J. P., et al. (2019). Application of Mesenchymal Stem Cell-Derived Extracellular Vesicles for Stroke: Biodistribution and microRNA Study. Transl. Stroke Res. 10 (5), 509–521. 10.1007/s12975-018-0668-1 [DOI] [PubMed] [Google Scholar]
- Morel L., Regan M., Higashimori H., Ng S. K., Esau C., Vidensky S., et al. (2013). Neuronal Exosomal miRNA-dependent Translational Regulation of Astroglial Glutamate Transporter GLT1. J. Biol. Chem. 288 (10), 7105–7116. 10.1074/jbc.M112.410944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli A. E., Larregina A. T., Shufesky W. J., Sullivan M. L., Stolz D. B., Papworth G. D., et al. (2004). Endocytosis, Intracellular Sorting, and Processing of Exosomes by Dendritic Cells. Blood 104 (10), 3257–3266. 10.1182/blood-2004-03-0824 [DOI] [PubMed] [Google Scholar]
- Muraoka S., Jedrychowski M. P., Tatebe H., DeLeo A. M., Ikezu S., Tokuda T., et al. (2019). Proteomic Profiling of Extracellular Vesicles Isolated from Cerebrospinal Fluid of Former National Football League Players at Risk for Chronic Traumatic Encephalopathy. Front. Neurosci. 13, 1059. 10.3389/fnins.2019.01059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka S., Jedrychowski M. P., Yanamandra K., Ikezu S., Gygi S. P., Ikezu T. (2020). Proteomic Profiling of Extracellular Vesicles Derived from Cerebrospinal Fluid of Alzheimer's Disease Patients: A Pilot Study. Cells 9 (9), 1959. 10.3390/cells9091959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasca C., Dobbin J., Bigio B., Watson K., de Angelis P., Kautz M., et al. (2021). Insulin Receptor Substrate in Brain-Enriched Exosomes in Subjects with Major Depression: on the Path of Creation of Biosignatures of Central Insulin Resistance. Mol. Psychiatry 26 (9), 5140–5149. 10.1038/s41380-020-0804-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neerukonda S. N., Egan N. A., Patria J., Assakhi I., Tavlarides-Hontz P., Modla S., et al. (2020). A Comparison of Exosome Purification Methods Using Serum of Marek's Disease Virus (MDV)-vaccinated and -Tumor-Bearing Chickens. Heliyon 6 (12), e05669. 10.1016/j.heliyon.2020.e05669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T., Masuda-Suzukake M., Arai T., Hasegawa Y., Akatsu H., Obi T., et al. (2013). Prion-like Properties of Pathological TDP-43 Aggregates from Diseased Brains. Cell Rep. 4 (1), 124–134. 10.1016/j.celrep.2013.06.007 [DOI] [PubMed] [Google Scholar]
- Otake K., Kamiguchi H., Hirozane Y. (2019). Identification of Biomarkers for Amyotrophic Lateral Sclerosis by Comprehensive Analysis of Exosomal mRNAs in Human Cerebrospinal Fluid. BMC Med. Genomics 12 (1), 7–11. 10.1186/s12920-019-0473-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero-Ortega L., Laso-García F., Frutos M. C. G., Diekhorst L., Martínez-Arroyo A., Alonso-López E., et al. (2020). Low Dose of Extracellular Vesicles Identified that Promote Recovery after Ischemic Stroke. Stem Cell Res. Ther. 11 (1), 70–15. 10.1186/s13287-020-01601-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oushy S., Hellwinkel J. E., Wang M., Nguyen G. J., Gunaydin D., Harland T. A., et al. (2018). Glioblastoma Multiforme-Derived Extracellular Vesicles Drive Normal Astrocytes towards a Tumour-Enhancing Phenotype. Philos. Trans. R. Soc. Lond B Biol. Sci. 373 (1737), 20160477. 10.1098/rstb.2016.0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G. K., Khan M. A., Zubair H., Srivastava S. K., Khushman M., Singh S., et al. (2019). Comparative Analysis of Exosome Isolation Methods Using Culture Supernatant for Optimum Yield, Purity and Downstream Applications. Sci. Rep. 9 (1), 1–10. 10.1038/s41598-019-41800-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K. Y., Pérez-González R., Alldred M. J., Goulbourne C. N., Morales-Corraliza J., Saito M., et al. (2019). Apolipoprotein E4 Genotype Compromises Brain Exosome Production. Brain 142 (1), 163–175. 10.1093/brain/awy289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters P. J., Geuze H. J., Van der Donk H. A., Slot J. W., Griffith J. M., Stam N. J., et al. (1989). Molecules Relevant for T Cell-Target Cell Interaction Are Present in Cytolytic Granules of Human T Lymphocytes. Eur. J. Immunol. 19 (8), 1469–1475. 10.1002/eji.1830190819 [DOI] [PubMed] [Google Scholar]
- Potolicchio I., Carven G. J., Xu X., Stipp C., Riese R. J., Stern L. J., et al. (2005). Proteomic Analysis of Microglia-Derived Exosomes: Metabolic Role of the Aminopeptidase CD13 in Neuropeptide Catabolism. J. Immunol. 175 (4), 2237–2243. 10.4049/jimmunol.175.4.2237 [DOI] [PubMed] [Google Scholar]
- Qi H., Liu C., Long L., Ren Y., Zhang S., Chang X., et al. (2016). Blood Exosomes Endowed with Magnetic and Targeting Properties for Cancer Therapy. ACS Nano 10 (3), 3323–3333. 10.1021/acsnano.5b06939 [DOI] [PubMed] [Google Scholar]
- Qin W., Tsukasaki Y., Dasgupta S., Mukhopadhyay N., Ikebe M., Sauter E. R. (2016). Exosomes in Human Breast Milk Promote EMT. Clin. Cancer Res. 22 (17), 4517–4524. 10.1158/1078-0432.CCR-16-0135 [DOI] [PubMed] [Google Scholar]
- Rajendran L., Honsho M., Zahn T. R., Keller P., Geiger K. D., Verkade P., et al. (2006). Alzheimer's Disease Beta-Amyloid Peptides Are Released in Association with Exosomes. Proc. Natl. Acad. Sci. U. S. A. 103 (30), 11172–11177. 10.1073/pnas.0603838103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F., Zhang D., Fang T., Lu C., Wang B., Ding X., et al. (2019). Exosomes from Human Gingiva-Derived Mesenchymal Stem Cells Combined with Biodegradable Chitin Conduits Promote Rat Sciatic Nerve Regeneration. Stem Cells Int. 2019, 2546367. 10.1155/2019/2546367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., et al. (1996). B Lymphocytes Secrete Antigen-Presenting Vesicles. J. Exp. Med. 183 (3), 1161–1172. 10.1084/jem.183.3.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M., Poirot M., Silvente-Poirot S. (2014). Emerging Concepts on the Role of Exosomes in Lipid Metabolic Diseases. Biochimie 96, 67–74. 10.1016/j.biochi.2013.06.016 [DOI] [PubMed] [Google Scholar]
- Rekker K., Saare M., Roost A. M., Kubo A. L., Zarovni N., Chiesi A., et al. (2014). Comparison of Serum Exosome Isolation Methods for microRNA Profiling. Clin. Biochem. 47 (1-2), 135–138. 10.1016/j.clinbiochem.2013.10.020 [DOI] [PubMed] [Google Scholar]
- Riazifar M., Mohammadi M. R., Pone E. J., Yeri A., Lässer C., Segaliny A. I., et al. (2019). Stem Cell-Derived Exosomes as Nanotherapeutics for Autoimmune and Neurodegenerative Disorders. ACS Nano 13 (6), 6670–6688. 10.1021/acsnano.9b01004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. J., Ejima H. (2019). Surface Engineering of Extracellular Vesicles through Chemical and Biological Strategies. Chem. Mat. 31 (7), 2191–2201. 10.1021/acs.chemmater.9b00050 [DOI] [Google Scholar]
- Saeedi S., Israel S., Nagy C., Turecki G. (2019). The Emerging Role of Exosomes in Mental Disorders. Transl. PsychiatryPsychiatry 9 (1), 1–11. 10.1038/s41398-019-0459-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem K. Z., Moschetta M., Sacco A., Imberti L., Rossi G., Ghobrial I. M., et al. (2016). Exosomes in Tumor Angiogenesis. Methods Mol. Biol. 1464, 25–34. 10.1007/978-1-4939-3999-2_3 [DOI] [PubMed] [Google Scholar]
- Saman S., Kim W., Raya M., Visnick Y., Miro S., Saman S., et al. (2012). Exosome-associated Tau Is Secreted in Tauopathy Models and Is Selectively Phosphorylated in Cerebrospinal Fluid in Early Alzheimer Disease. J. Biol. Chem. 287 (6), 3842–3849. 10.1074/jbc.M111.277061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo A., Imbrucè P., Gardenghi B., Belli L., Agushi R., Tamanini A., et al. (2018). A microRNA Signature from Serum Exosomes of Patients with Glioma as Complementary Diagnostic Biomarker. J. Neurooncol. 136 (1), 51–62. 10.1007/s11060-017-2639-x [DOI] [PubMed] [Google Scholar]
- Sardar Sinha M., Ansell-Schultz A., Civitelli L., Hildesjö C., Larsson M., Lannfelt L., et al. (2018). Alzheimer's Disease Pathology Propagation by Exosomes Containing Toxic Amyloid-Beta Oligomers. Acta Neuropathol. 136 (1), 41–56. 10.1007/s00401-018-1868-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y. T., Umezaki K., Sawada S., Mukai S. A., Sasaki Y., Harada N., et al. (2016). Engineering Hybrid Exosomes by Membrane Fusion with Liposomes. Sci. Rep. 6 (1), 1–11. 10.1038/srep21933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., Chung J., Balaj L., Charest A., Bigner D. D., Carter B. S., et al. (2012). Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat. Med. 18 (12), 1–11. 10.1038/nm.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Mesci P., Carromeu C., McClatchy D. R., Schiapparelli L., Yates J. R., et al. (2019). Exosomes Regulate Neurogenesis and Circuit Assembly. Proc. Natl. Acad. Sci. U. S. A. 116 (32), 16086–16094. 10.1073/pnas.1902513116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., Chattopadhyay M., Whitelegge J., Valentine J. S. (2012). SOD1 Aggregation and ALS: Role of Metallation States and Disulfide Status. Curr. Top. Med. Chem. 12 (22), 2560–2572. 10.2174/1568026611212220010 [DOI] [PubMed] [Google Scholar]
- Shi M., Kovac A., Korff A., Cook T. J., Ginghina C., Bullock K. M., et al. (2016). CNS Tau Efflux via Exosomes Is Likely Increased in Parkinson's Disease but Not in Alzheimer's Disease. Alzheimers Dement. 12 (11), 1125–1131. 10.1016/j.jalz.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Liu C., Cook T. J., Bullock K. M., Zhao Y., Ginghina C., et al. (2014). Plasma Exosomal α-synuclein Is Likely CNS-Derived and Increased in Parkinson's Disease. Acta Neuropathol. 128 (5), 639–650. 10.1007/s00401-014-1314-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtam T., Evtushenko V., Samsonov R., Zabrodskaya Y., Kamyshinsky R., Zabegina L., et al. (2020). Evaluation of Immune and Chemical Precipitation Methods for Plasma Exosome Isolation. Plos one 15 (11), e0242732. 10.1371/journal.pone.0242732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser N. R. (2007). Exosomal Transfer of Proteins and RNAs at Synapses in the Nervous System. Biol. Direct 2 (1), 35–15. 10.1186/1745-6150-2-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth T., Petrova K., Payton N. M., Persaud I., Redzic J. S., Graner M. W., et al. (2014). Surface Functionalization of Exosomes Using Click Chemistry. Bioconjug. Chem. 25 (10), 1777–1784. 10.1021/bc500291r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares Martins T., Catita J., Martins Rosa I., A B da Cruz E Silva O., Henriques A. G. (2018). Exosome Isolation from Distinct Biofluids Using Precipitation and Column-Based Approaches. PloS one 13 (6), e0198820. 10.1371/journal.pone.0198820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini M. G., Crowther R. A., Jakes R., Cairns N. J., Lantos P. L., Goedert M. (1998). Filamentous Alpha-Synuclein Inclusions Link Multiple System Atrophy with Parkinson's Disease and Dementia with Lewy Bodies. Neurosci. Lett. 251 (3), 205–208. 10.1016/S0304-3940(98)00504-7 [DOI] [PubMed] [Google Scholar]
- Sterzenbach U., Putz U., Low L. H., Silke J., Tan S. S., Howitt J. (2017). Engineered Exosomes as Vehicles for Biologically Active Proteins. Mol. Ther. 25 (6), 1269–1278. 10.1016/j.ymthe.2017.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street J. M., Barran P. E., Mackay C. L., Weidt S., Balmforth C., Walsh T. S., et al. (2012). Identification and Proteomic Profiling of Exosomes in Human Cerebrospinal Fluid. J. Transl. Med. 10 (1), 5–7. 10.1186/1479-5876-10-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street J. M., Koritzinsky E. H., Glispie D. M., Star R. A., Yuen P. S. (2017). Urine Exosomes: an Emerging Trove of Biomarkers. Adv. Clin. Chem. 78, 103–122. 10.1016/bs.acc.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Stuffers S., Sem Wegner C., Stenmark H., Brech A. (2009). Multivesicular Endosome Biogenesis in the Absence of ESCRTs. Traffic 10 (7), 925–937. 10.1111/j.1600-0854.2009.00920.x [DOI] [PubMed] [Google Scholar]
- Sun D., Zhuang X., Xiang X., Liu Y., Zhang S., Liu C., et al. (2010). A Novel Nanoparticle Drug Delivery System: the Anti-inflammatory Activity of Curcumin Is Enhanced when Encapsulated in Exosomes. Mol. Ther. 18 (9), 1606–1614. 10.1038/mt.2010.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung B. H., Von Lersner A., Guerrero J., Krystofiak E. S., Inman D., Pelletier R., et al. (2020). A Live Cell Reporter of Exosome Secretion and Uptake Reveals Pathfinding Behavior of Migrating Cells. Nat. Commun. 11 (1), 1–15. 10.1038/s41467-020-15747-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Nishikawa M., Shinotsuka H., Matsui Y., Ohara S., Imai T., et al. (2013). Visualization and In Vivo Tracking of the Exosomes of Murine Melanoma B16-BL6 Cells in Mice after Intravenous Injection. J. Biotechnol. 165 (2), 77–84. 10.1016/j.jbiotec.2013.03.013 [DOI] [PubMed] [Google Scholar]
- Tan Y. J., Wong B. Y. X., Vaidyanathan R., Sreejith S., Chia S. Y., Kandiah N., et al. (2021). Altered Cerebrospinal Fluid Exosomal microRNA Levels in Young-Onset Alzheimer's Disease and Frontotemporal Dementia. J. Alzheimers Dis. Rep. 5, 805–813. 10.3233/ADR-210311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. R., Robinson M. B., Gifondorwa D. J., Tytell M., Milligan C. E. (2007). Regulation of Heat Shock Protein 70 Release in Astrocytes: Role of Signaling Kinases. Dev. Neurobiol. 67 (13), 1815–1829. 10.1002/dneu.20559 [DOI] [PubMed] [Google Scholar]
- Teng F., Fussenegger M. (2021). Shedding Light on Extracellular Vesicle Biogenesis and Bioengineering. Adv. Sci. 8 (1), 2003505. 10.1002/advs.202003505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos A. C., Truschel S. T., Tenza D., Hurbain I., Harper D. C., Berson J. F., et al. (2006). A Lumenal Domain-dependent Pathway for Sorting to Intralumenal Vesicles of Multivesicular Endosomes Involved in Organelle Morphogenesis. Dev. Cell 10 (3), 343–354. 10.1016/j.devcel.2006.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Amigorena S., Raposo G., Clayton A. (2006). Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. Chapter 3 (1), Unit–22. 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- Théry C., Witwer K. W., Aikawa E., Alcaraz M. J., Anderson J. D., Andriantsitohaina R., et al. (2018). Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): a Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles. 7 (1), 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Duban L., Segura E., Véron P., Lantz O., Amigorena S. (2002a). Indirect Activation of Naïve CD4+ T Cells by Dendritic Cell-Derived Exosomes. Nat. Immunol. 3 (12), 1156–1162. 10.1038/ni854 [DOI] [PubMed] [Google Scholar]
- Théry C., Zitvogel L., Amigorena S. (2002b). Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2 (8), 569–579. 10.1038/nri855 [DOI] [PubMed] [Google Scholar]
- Thompson A. G., Gray E., Heman-Ackah S. M., Mäger I., Talbot K., Andaloussi S. E., et al. (2016). Extracellular Vesicles in Neurodegenerative Disease - Pathogenesis to Biomarkers. Nat. Rev. Neurol. 12 (6), 346–357. 10.1038/nrneurol.2016.68 [DOI] [PubMed] [Google Scholar]
- Tian T., Zhang H. X., He C. P., Fan S., Zhu Y. L., Qi C., et al. (2018). Surface Functionalized Exosomes as Targeted Drug Delivery Vehicles for Cerebral Ischemia Therapy. Biomaterials 150, 137–149. 10.1016/j.biomaterials.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Tian Y., Li S., Song J., Ji T., Zhu M., Anderson G. J., et al. (2014). A Doxorubicin Delivery Platform Using Engineered Natural Membrane Vesicle Exosomes for Targeted Tumor Therapy. Biomaterials 35 (7), 2383–2390. 10.1016/j.biomaterials.2013.11.083 [DOI] [PubMed] [Google Scholar]
- Trueta C., De-Miguel F. F. (2012). Extrasynaptic Exocytosis and its Mechanisms: a Source of Molecules Mediating Volume Transmission in the Nervous System. Front. Physiol. 3, 319. 10.3389/fphys.2012.00319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsivion-Visbord H., Perets N., Sofer T., Bikovski L., Goldshmit Y., Ruban A., et al. (2020). Mesenchymal Stem Cells Derived Extracellular Vesicles Improve Behavioral and Biochemical Deficits in a Phencyclidine Model of Schizophrenia. Transl. Psychiatry 10 (1), 305–310. 10.1038/s41398-020-00988-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C., Baixauli F., Gutiérrez-Vázquez C., Sánchez-Madrid F., Mittelbrunn M. (2014). Sorting it Out: Regulation of Exosome Loading. Seminars Cancer Biol. 28, 3–13. 10.1016/j.semcancer.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bartheld C. S., Altick A. L. (2011). Multivesicular Bodies in Neurons: Distribution, Protein Content, and Trafficking Functions. Prog. Neurobiol. 93 (3), 313–340. 10.1016/j.pneurobio.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlgren J., De L Karlson T., Brisslert M., Vaziri Sani F., Telemo E., Sunnerhagen P., et al. (2012). Plasma Exosomes Can Deliver Exogenous Short Interfering RNA to Monocytes and Lymphocytes. Nucleic Acids Res. 40 (17), e130. 10.1093/nar/gks463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S., Zhang L., Wang S., Liu Y., Wu C., Cui C., et al. (2017). Molecular Recognition-Based DNA Nanoassemblies on the Surfaces of Nanosized Exosomes. J. Am. Chem. Soc. 139 (15), 5289–5292. 10.1021/jacs.7b00319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Wang L., Zhu C., Zheng Q., Wang G., Tong J., et al. (2018). Aptamer-conjugated Extracellular Nanovesicles for Targeted Drug Delivery. Cancer Res. 78 (3), 798–808. 10.1158/0008-5472.CAN-17-2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Han S. (2019). Modified Exosomes Reduce Apoptosis and Ameliorate Neural Deficits Induced by Traumatic Brain Injury. Asaio J. 65 (3), 285–292. 10.1097/MAT.0000000000000810 [DOI] [PubMed] [Google Scholar]
- Wang G., Wen Y., Faleti O. D., Zhao Q., Liu J., Zhang G., et al. (2020). A Panel of Exosome-Derived Mirnas of Cerebrospinal Fluid for the Diagnosis of Moyamoya Disease. Front. Neurosci. 14, 548278. 10.3389/fnins.2020.548278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Atik A., Stewart T., Ginghina C., Aro P., Kerr K. F., et al. (2018a). Plasma α-synuclein and Cognitive Impairment in the Parkinson's Associated Risk Syndrome: A Pilot Study. Neurobiol. Dis. 116, 53–59. 10.1016/j.nbd.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. H., Forterre A. V., Zhao J., Frimannsson D. O., Delcayre A., Antes T. J., et al. (2018b). Anti-HER2 scFv-Directed Extracellular Vesicle-Mediated mRNA-Based Gene Delivery Inhibits Growth of HER2-Positive Human Breast Tumor Xenografts by Prodrug Activation. Mol. Cancer Ther. 17 (5), 1133–1142. 10.1158/1535-7163.MCT-17-0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Altinoglu S., Takeda Y. S., Xu Q. (2015). Integrating Protein Engineering and Bioorthogonal Click Conjugation for Extracellular Vesicle Modulation and Intracellular Delivery. PLoS One 10 (11), e0141860. 10.1371/journal.pone.0141860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Liu Z., Ye T., Mabrouk O. S., Maltbie T., Aasly J., et al. (2017). Elevated LRRK2 Autophosphorylation in Brain-Derived and Peripheral Exosomes in LRRK2 Mutation Carriers. Acta Neuropathol. Commun. 5 (1), 86–13. 10.1186/s40478-017-0492-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb R. L., Kaiser E. E., Jurgielewicz B. J., Spellicy S., Scoville S. L., Thompson T. A., et al. (2018). Human Neural Stem Cell Extracellular Vesicles Improve Recovery in a Porcine Model of Ischemic Stroke. Stroke 49 (5), 1248–1256. 10.1161/STROKEAHA.117.020353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Zhao L., Kong G., Liu X., Zhu S., Zhang S., et al. (2020). Combination of Size-Exclusion Chromatography and Ultracentrifugation Improves the Proteomic Profiling of Plasma-Derived Small Extracellular Vesicles. Biol. Proced. Online 22 (1), 12–11. 10.1186/s12575-020-00125-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Ao Q., Wang X., Cao Y., Liu Y., Zheng S. G., et al. (2021). Mesenchymal Stem Cell-Derived Exosomes: A Promising Biological Tool in Nanomedicine. Front. Pharmacol. 11. 10.3389/fphar.2020.590470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welton J. L., Loveless S., Stone T., von Ruhland C., Robertson N. P., Clayton A. (2017). Cerebrospinal Fluid Extracellular Vesicle Enrichment for Protein Biomarker Discovery in Neurological Disease; Multiple Sclerosis. J. Extracell. vesicles 6 (1), 1369805. 10.1080/20013078.2017.1369805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling P., Capper D. (2018). WHO 2016 Classification of Gliomas. Neuropathol. Appl. Neurobiol. 44 (2), 139–150. 10.1111/nan.12432 [DOI] [PubMed] [Google Scholar]
- Winston C. N., Goetzl E. J., Akers J. C., Carter B. S., Rockenstein E. M., Galasko D., et al. (2016). Prediction of Conversion from Mild Cognitive Impairment to Dementia with Neuronally Derived Blood Exosome Protein Profile. Alzheimers Dement. (Amst) 3, 63–72. 10.1016/j.dadm.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston C. N., Romero H. K., Ellisman M., Nauss S., Julovich D. A., Conger T., et al. (2019). Assessing Neuronal and Astrocyte Derived Exosomes from Individuals with Mild Traumatic Brain Injury for Markers of Neurodegeneration and Cytotoxic Activity. Front. Neurosci. 13, 1005. 10.3389/fnins.2019.01005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Ouyang Y., Wang Z., Zhang R., Huang P. H., Chen C., et al. (2017). Isolation of Exosomes from Whole Blood by Integrating Acoustics and Microfluidics. Proc. Natl. Acad. Sci. U. S. A. 114 (40), 10584–10589. 10.1073/pnas.1709210114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Deng W., Klinke D. J., II (2015). Exosomes: Improved Methods to Characterize Their Morphology, RNA Content, and Surface Protein Biomarkers. Analyst 140 (19), 6631–6642. 10.1039/C5AN00688K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Zhao Y., Zhou X., Luan J., Cui Y., Han J. (2018). Comparison of the Extraction and Determination of Serum Exosome and miRNA in Serum and the Detection of miR-27a-3p in Serum Exosome of ALS Patients. Intractable Rare Dis. Res. 7 (1), 13–18. 10.5582/irdr.2017.01091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Ohkubo T., Kawaji H., Machida A., Miyata H., Goda S., et al. (2017). Next-generation Sequencing-Based Small RNA Profiling of Cerebrospinal Fluid Exosomes. Neurosci. Lett. 636, 48–57. 10.1016/j.neulet.2016.10.042 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H., Hirai S., Shoji M., Harigaya Y., Okamoto Y., Nakazato Y. (1989). Alzheimer Type Dementia: Diffuse Type of Senile Plaques Demonstrated by Beta Protein Immunostaining. Prog. Clin. Biol. Res. 317, 467–474. 10.1007/BF00687420 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y., Hozumi Y., Kaneko K., Sugihara T., Fujii S., Goto K., et al. (2007). Modulatory Effects of Oligodendrocytes on the Conduction Velocity of Action Potentials along Axons in the Alveus of the Rat Hippocampal CA1 Region. Neuron Glia Biol. 3 (4), 325–334. 10.1017/S1740925X08000070 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhang X., Chen X., Wang L., Yang G. (2017a). Exosome Mediated Delivery of miR-124 Promotes Neurogenesis after Ischemia. Mol. Ther. Nucleic Acids 7, 278–287. 10.1016/j.omtn.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Martin P., Fogarty B., Brown A., Schurman K., Phipps R., et al. (2015). Exosome Delivered Anticancer Drugs across the Blood-Brain Barrier for Brain Cancer Therapy in Danio rerio . Pharm. Res. 32 (6), 2003–2014. 10.1007/s11095-014-1593-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. T., Liu C. G., Gao S. C., Zhang Y., Wang P. C. (2018). The Serum Exosome Derived MicroRNA-135a, -193b, and -384 Were Potential Alzheimer's Disease Biomarkers. Biomed. Environ. Sci. 31 (2), 87–96. 10.3967/bes2018.011 [DOI] [PubMed] [Google Scholar]
- Yang Y., Ye Y., Su X., He J., Bai W., He X. (2017b). MSCs-Derived Exosomes and Neuroinflammation, Neurogenesis and Therapy of Traumatic Brain Injury. Front. Cell. Neurosci. 11, 55. 10.3389/fncel.2017.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Xie J., Zhu J., Kang C., Chiang C., Wang X., et al. (2016). Functional Exosome-Mimic for Delivery of siRNA to Cancer: In Vitro and In Vivo Evaluation. J. Control. Release 243, 160–171. 10.1016/j.jconrel.2016.10.008 [DOI] [PubMed] [Google Scholar]
- Ye Z., Zhang T., He W., Jin H., Liu C., Yang Z., et al. (2018). Methotrexate-loaded Extracellular Vesicles Functionalized with Therapeutic and Targeted Peptides for the Treatment of Glioblastoma Multiforme. ACS Appl. Mat. Interfaces 10 (15), 12341–12350. 10.1021/acsami.7b18135 [DOI] [PubMed] [Google Scholar]
- Yekula A., Taylor A., Beecroft A., Kang K. M., Small J. L., Muralidharan K., et al. (2021). The Role of Extracellular Vesicles in Acquisition of Resistance to Therapy in Glioblastomas. Cdr 4 (1), 1–16. 10.20517/cdr.2020.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim N., Ryu S. W., Choi K., Lee K. R., Lee S., Choi H., et al. (2016a). Exosome Engineering for Efficient Intracellular Delivery of Soluble Proteins Using Optically Reversible Protein-Protein Interaction Module. Nat. Commun. 7 (1), 12277–12279. 10.1038/ncomms12277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim N., Ryu S.-W., Choi K., Lee K. R., Lee S., Choi H., et al. (2016b). “Abstract 2167: Efficient and Rapid Cellular Delivery of Bioactive Proteins Using EXPLOR: Exosomes Engineered for Protein Loading via Optically Reversible Protein-Protein Interaction,” in Proceedings of the 107th Annual Meeting of the American Association for Cancer Research. 10.1158/1538-7445.AM2016-2167 [DOI] [Google Scholar]
- Yoo C. E., Kim G., Kim M., Park D., Kang H. J., Lee M., et al. (2012). A Direct Extraction Method for microRNAs from Exosomes Captured by Immunoaffinity Beads. Anal. Biochem. 431 (2), 96–98. 10.1016/j.ab.2012.09.008 [DOI] [PubMed] [Google Scholar]
- Zhan Q., Yi K., Qi H., Li S., Li X., Wang Q., et al. (2020). Engineering Blood Exosomes for Tumor-Targeting Efficient Gene/chemo Combination Therapy. Theranostics 10 (17), 7889–7905. 10.7150/thno.45028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Yang P. (2018). A Novel Cell-Cell Communication Mechanism in the Nervous System: Exosomes. J. Neurosci. Res. 96 (1), 45–52. 10.1002/jnr.24113 [DOI] [PubMed] [Google Scholar]
- Zhang H., Wu J., Wu J., Fan Q., Zhou J., Wu J., et al. (2019). Exosome-mediated Targeted Delivery of miR-210 for Angiogenic Therapy after Cerebral Ischemia in Mice. J. Nanobiotechnology 17 (1), 29–13. 10.1186/s12951-019-0461-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chopp M., Meng Y., Katakowski M., Xin H., Mahmood A., et al. (2015). Effect of Exosomes Derived from Multipluripotent Mesenchymal Stromal Cells on Functional Recovery and Neurovascular Plasticity in Rats after Traumatic Brain Injury. J. Neurosurg. 122 (4), 856–867. 10.3171/2014.11.JNS14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang Y., Chopp M., Pang H., Zhang Z. G., Mahmood A., et al. (2021). MiR-17-92 Cluster-Enriched Exosomes Derived from Human Bone Marrow Mesenchymal Stromal Cells Improve Tissue and Functional Recovery in Rats after Traumatic Brain Injury. J. Neurotrauma 38 (11), 1535–1550. 10.1089/neu.2020.7575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wu X., Qian M., Song Y., Wu D., Zhang W. (2018). Knockdown of TGF-Β1 Expression in Human Umbilical Cord Mesenchymal Stem Cells Reverts Their Exosome-Mediated EMT Promoting Effect on Lung Cancer Cells. Cancer Lett. 428, 34–44. 10.1016/j.canlet.2018.04.026 [DOI] [PubMed] [Google Scholar]
- Zhao Z. H., Chen Z. T., Zhou R. L., Zhang X., Ye Q. Y., Wang Y. Z. (2018). Increased DJ-1 and α-Synuclein in Plasma Neural-Derived Exosomes as Potential Markers for Parkinson's Disease. Front. Aging Neurosci. 10, 438. 10.3389/fnagi.2018.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Xiang X., Grizzle W., Sun D., Zhang S., Axtell R. C., et al. (2011). Treatment of Brain Inflammatory Diseases by Delivering Exosome Encapsulated Anti-inflammatory Drugs from the Nasal Region to the Brain. Mol. Ther. 19 (10), 1769–1779. 10.1038/mt.2011.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Guo L., Wei B., Yu A., Yu B., Xu A. (2020). Long Noncoding RNA POU3F3 and α-Synuclein in Plasma L1CAM Exosomes Combined with β-Glucocerebrosidase Activity: Potential Predictors of Parkinson's Diseasee. Neurotherapeutics 17 (3), 1104–1119. 10.1007/s13311-020-00842-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.