Abstract
Arthropod-borne chikungunya virus (CHIKV) infection can cause a debilitating arthritic disease in human. However, there are no specific antiviral drugs and effective licensed vaccines against CHIKV available for clinical use. Here, we developed an mRNA-lipid nanoparticle (mRNA-LNP) vaccine expressing CHIKV E2-E1 antigen, and compared its immunogenicity with soluble recombinant protein sE2-E1 antigen expressed in S2 cells. For comparison, we first showed that recombinant protein antigens mixed with aluminum adjuvant elicit strong antigen-specific humoral immune response and a moderate cellular immune response in C57BL/6 mice. Moreover, sE2-E1 vaccine stimulated 12–23 folds more neutralizing antibodies than sE1 vaccine and sE2 vaccine. Significantly, when E2-E1 gene was delivered by an mRNA-LNP vaccine, not only the better magnitude of neutralizing antibody responses was induced, but also greater cellular immune responses were generated, especially for CD8+ T cell responses. Moreover, E2-E1-LNP induced CD8+ T cells can perform cytotoxic effect in vivo. Considering its better immunogenicity and convenience of preparation, we suggest that more attention should be placed to develop CHIKV E2-E1-LNP mRNA vaccine.
Keywords: Chikungunya virus (CHIKV), mRNA vaccine, Neutralizing antibody, Cytotoxic T-lymphocytes (CTL)
Highlights
-
•
Heterodimer sE2-E1 is a more promising antigen than sE1 or sE2 monomer.
-
•
CHIKV E2-E1-LNP mRNA vaccine is superior to subunit vaccine sE2-E1.
-
•
mRNA vaccine elicits robust CTL response but modest CD4+ T cell response.
1. Introduction
Chikungunya virus (CHIKV) is an arthropod-borne alphavirus transmitted by Aedes aegypti. It was first recognized as a human pathogen in 1952 (Robinson, 1955), and then caused sporadic human outbreaks in Africa and Southeast Asia in the next half century (Ann M. Powers, 2000). More recently, CHIKV has further spread to other places including islands around the India Ocean and Latin America (ECDC, 2020; PAHO, 2021). Global cumulative cases of CHIKV infection is estimated to have reached 10 million (Suhrbier, 2019). Along with the viral transmission, new genotypes such as the Asian and the India Ocean Lineage (IOL) had appeared, originated from the East/Central/South African (ECSA), associated with more urban transmission; while the original West Africa genotype was limited to sylvatic cycle (Volk et al., 2010). Of note, an adaptive mutation in the structure protein E1(A226V) has increased its viral fitness in Aedes albopictus (Tsetsarkin et al. 2007, 2009), allowing more efficient CHIKV transmission and making it a potential biosecurity threat.
The clinical manifestations of patients with CHIKV infection are divided into acute and chronic phases. In the acute phase (1–14 days), fever, headache, multiple joint pain, rash, muscle pain, back pain, fatigue and high viremia are the main clinical manifestations (Thiberville et al., 2013). In the chronic phase, persistent arthritis and arthralgia are the most common symptoms (Borgherini et al., 2008; Chopra et al., 2011). Vaccination is the most economic route to prevent CHIKV infection, however, there is no licensed CHIKV vaccine yet. Several vaccine strategies are under development, including inactivated vaccine (Tiwari et al., 2009; Kumar et al., 2012; DeZure et al., 2016), live-attenuated vaccine (LAV) (Edelman et al., 2000; Kim et al., 2011; Hallengard et al., 2014a), DNA vaccine (Tretyakova et al., 2014; Muthumani et al., 2016), chimeric vaccine (Brandler et al., 2013; Doel et al., 2014), virus-like particles (VLP) vaccine (Akahata et al., 2010; Metza et al., 2013; Chang et al., 2014) and subunit vaccine (Khan et al., 2012; Kumar et al., 2012; Metza et al., 2013). The antigen of choice is relatively straightforward. CHIKV envelope protein E1 and E2 form heterodimers which then further trimerize to form 80 spikes displayed on viral membrane (Li et al., 2010). E1 plays an important role during membrane fusion (Li et al., 2010) and E2 is responsible for receptor binding (Fox et al., 2015; Smith et al., 2015; Weaver et al., 2017). Therefore, E1 and E2 proteins are often selected as immunogens in vaccines. The crystal structure of CHIKV E2-E1 complex expressed in S2 cells can fit the spikes on the available cyto-EM structure of the alphavirus surface (Voss et al., 2010), indicating that CHIKV E2-E1 have attractive antigenic potential.
Humoral immunity plays an important role in virus clearance. The viremia can be rapidly cleared in wild type C57BL/6 mice when infected with an attenuated CHIKV strain (181/25), but not in B cell deficient (μMT) mice (Lum et al., 2013), indicating the directly cleanup effect of virus specific antibodies. Moreover, passive transfer of IgG or immune serum from CHIKV vaccine immunized animals to naïve mice can provide protection to recipients against lethal challenge (Akahata et al., 2010; Chu et al., 2013).
Recent studies suggest that T cells affect the outcomes of CHIKV infection. In experimental models, IFN-γ-producing CD4+ T cells are associated with pathology, as their depletion reduced joint swelling of CHIKV infected mice (Teo et al., 2013). However, CD4+ T cells can assist CD8+ T cells to better protect CHIKV infection. The depletion of CD8+ T cells alone can reduce the efficacy of cytotoxic T-lymphocytes (CTL)-based vaccine, but the depletion of both CD8+ and CD4+ T cells decreases the protective role of the vaccine further (Broeckel et al., 2019). In addition, the protective role of CD8+ T cells to control viral infection was also reported in other alphaviruses including Ross River virus (Burrack et al., 2015) and Sindbis virus (Gwendolyn K. Binder, 2001). Therefore, an ideal CHIKV vaccine need to induce protective antibodies and appropriate T cell responses. However, the ongoing researches of CHIKV vaccines can't meet these requirements completely, and there is still an urgent need to develop an effective and appropriate CHIKV vaccine.
Recently, mRNA vaccine platform has received widespread public attention due in part to its successful application to making vaccines against SARS-CoV-2 (Polack et al., 2020; Sahin et al., 2020). Scientific research on mRNA vaccines, however, had already shown its promise in the areas of human immunodeficiency virus (Pollard et al., 2013; Bahl et al., 2017), Zika virus (Richner et al., 2017), dengue virus (Zhang et al., 2020) and influenza virus (Vogel et al., 2018). The theoretical advantages of mRNA vaccine are to induce robust cellular immune response, especially CD8+ T cell response, and capable of inducing potent humoral immune responses without the addition of adjuvants.
Here, we first established a recombinant CHIKV subunit vaccine system for comparator and then chose the most promising antigen for construction of an mRNA vaccine. We show that an mRNA vaccine expressing the E2-E1 protein elicited better antibody responses and greater cellular immune responses than the recombinant protein in a murine model.
2. Materials and methods
2.1. Cell culture and viruses
HEK293T and 293A cells were cultured in DMEM (Gibco) containing 10% FBS (Gibco) and 1% Penicillin-Streptomycin; S2 cells were cultured in Schneiderís Drosophila Medium (SDM) (Gibco) containing 10% FBS and 1% Penicillin-Streptomycin or Express Five™ SFM (Gibco) containing 1% Penicillin-Streptomycin while expressing proteins.
2.2. Plasmid construction
Genes encoding chikungunya structure proteins were based on a CHIKV-Asian strain (GenBank accession No: CAX63317.3) and synthesized by GenScript (Nanjing, China). For protein expression, sequence of E3-E2-E1, E3-E2 and E1 were constructed into pMT/BiP/V5-His A plasmid (Invitrogen), respectively, which is the expression vector of the Drosophila expression system. Among these plasmids, transmembrane domain of E3-E2 and E1 sequence were deleted to obtain soluble E2 and E1 protein, and E3-E2-E1 sequence was designed according to the strategy of producing crystal E3-E2-E1 that had been reported previously (Voss et al., 2010). Briefly, transmembrane domain of E2, E1 and the sequence of 6k were deleted and replaced with a linker sequence of (GGGGS)4. His tag was added at E1 C-terminal. For the production of chikungunya pseudovirus, the chikungunya structure genes were constructed into the envelope vector pCMV-3tag-9 to obtain pCMV-CHIKV-E3-E1. For mRNA construction, sequence of E3-E2-linker-E1 was constructed into mRNA expression vector pok12-5′UTR-ORF-3′UTR (Zhang et al., 2020).
2.3. Recombinant proteins production and purification
pMT-sE2-E1, pMT-sE2, or pMT-sE1 were co-transfected with pCoblast plasmid into Drosophila melanogaster Schneider 2 (S2) cells, respectively, using a Calcium Phosphate Transfection kit (Invitrogen). The transfected S2 cells were selected by SDM with 25 μg/mL blasticidin and 10% FBS for four weeks. 10 μmol/L CdCl2 was then added into to the survived S2 cells to induce protein expression. After verified by Western blot, the selected S2 cells were scale-up cultured in SFM (Expressive Five™ SFM) with 10 μg/mL blasticidin and produced proteins under the induction of 10 μmol/L CdCl2 condition. After 7 days post induction, the supernatants were collected and proteins were purified following the protocol of Ni-NTA purification system. Briefly, concentrated supernatants were mixed with Ni-NTA agarose (Invitrogen) in native binding buffer and rotated at 4 °C for 4 h, followed by washing twice with native washing buffer. Proteins were eluted by native purification buffer with concentration gradient of imidazole. The purity of protein was verified by SDS-PAGE. Finally, purified proteins were dialyzed in PBS for 3 cycles and stocked in −80 °C freezer.
2.4. SDS-PAGE and western blot
Purified CHIKV recombinant proteins were verified by SDS-PAGE for molecular size and Western blot assay for specificity. In denaturing condition, proteins with SDS-PAGE sample loading buffer (Beyotime) were incubated at 100 °C for 10 min and separated on a 10% SDS-PAGE gel. In non-denaturing condition, proteins with native gel sample loading buffer (Beyotime) and separated on a 10% SDS-PAGE gel. The gel was stained with Coomassie brilliant blue G-250 (Thermo Scientific) to check out the purity of recombinant proteins or transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore), followed by being recognized with anti-chikungunya polyclonal antibody (pAb) (IBT bioservices) and a secondary anti-mouse IgG antibody (Promega) and finally colorized using the BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime). For detection of mRNA mediated protein expression, cell culture supernatant and cell lysate samples were incubated at 100 °C for 10 min and separated on a 10% SDS-PAGE gel and transferred onto a PVDF membrane (Millipore). The specific protein was recognized by his-tag monoclonal antibody (proteintech) and a secondary goat anti-mouse IgG-HRP antibody (Sounthern Biotech) and finally colorized by chemiluminescence imaging (Tanon).
2.5. mRNA and lipid nanoparticle (LNP) production
The production of mRNA-LNP was described previously (Zhang et al., 2020). Briefly, RNA was produced using T7 RNA polymerase on linearized plasmids (pok12-5′UTR-CHIKV-E2-E1-3′UTR) encoding CHIKV E2-E1 protein. The UTP was substituted with 1 mψ (1-methylpseudouridine-5′-triphosphate) (TriLink BioTechnologies) to improve protein expression. Then the transcriptional RNA was added with Cap1 and poly(A) tails by using the Vaccinia Capping System and E. coli poly(A) polymerase (New England Biolabs), respectively. The mRNA was stored at −80 °C freezer.
For LNP production, D-Lin-MC3-DMA (MedChemExpress), DSPC (Avanti Polar Lipids), cholesterol (Sigma), and PEG-lipid (Avanti Polar Lipids) were solubilized in ethanol respectively and mixed with a molar ratio of 50:10:38.5:1.5. The lipid mixture was then mixed with an aqueous buffer (50 mmol/L citrate buffer [pH 4.0]), reaching the final lipid concentration of 7.37 mg/mL. Then the lipid mixture and mRNA were added into the NanoAssemblr™ with the volume ratio of 1:3 to obtain mRNA-LNP. Then the diluted LNP-encapsulated mRNA samples were concentrated to appropriate volume and passed through 0.22 μm filters. The produced mRNA-LNP was stored at 4 °C until use. The encapsulation efficiency was measured with the Quant-iT RiboGreen RNA Assay Kit (Life Technologies) using a microplate reader.
2.6. Mouse immunization
For subunit vaccine immune strategy, immunogens including protein sE2-E1, sE2 and sE1 were diluted in PBS to 200 μg/mL respectively and mixed with aluminum adjuvant in PBS to the same volume. Twenty-four 6-week-old female C57BL/6 mice were divided into four groups and were immunized with 100 μL of the mixed solution, equivalently 10 μg protein each mouse. Three injections were operated at weeks 0, 2 and 4 respectively. Sera were collected at weeks 4 and 6. For mRNA vaccine immune strategy, eleven mice were divided into two groups and immunized with 10 μg E2-E1 LNP (six mice) or Empty-LNP (five mice) at weeks 0, 4, and 8. Serum samples were collected at two weeks after immunization. For mRNA vaccine immune strategy of dosage optimization, twenty-nine C57BL/6 mice were divided into five groups and were immunized with 0.2 μg, 1 μg, 5 μg or 10 μg E2-E1 LNP (six mice per group) or Empty-LNP (five mice) in a same volume (50 μL) at weeks 0, 4, and 8, respectively. Sera and splenocytes were harvested at 6 weeks after the last immunization for measurement of antibody responses and T cell responses.
2.7. ELISA
For binding antibody measurement in subunit vaccine immunized mice, V5-tag antibody (GenScript) was diluted to 1 μg/mL in PBS and coated in 96 well high binding plate (Corning, USA) overnight at 4 °C. After three times of wash with PBS containing 0.05% Tween-20 (PBST), the plates were blocked with PBST containing 3% BSA at 37 °C for 2 h and then washed 5 times with PBST. Then, protein sE1 or sE2 was diluted in PBST containing 1% BSA to 5 μg/mL and added into plate and incubated at 37 °C for 1 h. Mouse serum samples were incubated at 56 °C for 30 min and 2-fold diluted in PBST containing 1% BSA, then were incubated in plates at 37 °C for 1 h. After five times of washes, secondary HRP-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology, USA) was diluted in PBST with 1% BSA with the ratio of 1:5000 and incubated at 37 °C for 1 h. Excessive antibody was washed off and TMB substrate (Life technology, USA) was added to the plates for chromogenic reaction, which was stopped with 2 mol/L HCl. The absorbance was measured at 450 nm using a microplate reader. For binding antibody measurement in mRNA vaccine immunized mice, protein sE2-E1 was diluted to 1.5 μg/mL in PBS and then was directly coated in 96 well high binding plate overnight at 4 °C. After blocking and washing, plates were incubated with diluted serum, and the following steps were the same as above.
2.8. Production of CHIKV pseudotyped virus
Production of CHIKV pseudovirus was similar to that of influenza pseudovirus which has been described previously (Ren et al., 2016). Briefly, 14 μg plasmid pHIV-luc, 14 μg pCMV-Δ8.9 and 14 μg pCMV-CHIKV-E3-E1 in opti-MEM were mixed with 84 μg polyetherimide in opti-MEM, and incubated in room temperature for 30 min. Then the mixture was co-transfected to HEK293T cells cultured in DMEM in 10 cm2 dish. Four hours later, added 3 mL DMEM with 10% FBS and cultured at 37 °C overnight, followed by changing the medium with 10 mL DMEM containing 10% FBS and 100 μmol/L sodium butyrate. 8 h later, the medium was replaced with 10 mL fresh DMEM containing 10% FBS, and cells were cultured at 37 °C for 48 h. Chikungunya pseudovirus-containning supernatants were collected after centrifuging at 6000 rpm for 20 min and stocked in −80 °C freezer.
2.9. Pseudovirus neutralizing assay
9 × 104 293A cells each well were seeded in 48-well-plate (Nunc) and cultured at 37 °C overnight. Mouse serum samples were incubated at 56 °C for 30 min and 3-fold diluted in DMEM containing 3% FBS, and then were incubated with CHIKV pseudoviruses at room temperature for 1 h. The mixture of CHIKV pseoduviruses and mouse serum samples were added into 293A cells. After 18 h, the medium was replaced with fresh DMEM with 10% FBS. 72 h later, the medium was removed and 70 μL 1 × Glo lysis buffer was added into each well. Then the lysate was transferred into 96-well plate and added with 30 μL Bright-Glo luciferase substrates (Promega, USA). The luminescence was measured using a multimode plate-reader.
2.10. ELISpot assay
ELISpot assays were performed according to the manufacturer's protocol (Mabtech) to detect cytokines-producing T cells in the splenocytes. Briefly, ELISpot plates were coated with cytokine specific coating antibody at 4 °C overnight. After removing excess antibody and 5 times of washes with PBS, plates were blocked with RPMI 1640 medium containing 10% FBS for 30 min at room temperature. Then we removed the medium and added 2 μg protein sE2-E1 or 6 μg CD4+ T cell-restricted peptide E2EP3 or 6 μg CD8+ T cell-restricted peptide E1328-345 each well as the stimulation. Then splenocytes isolated from immunized mice were added into the plate and cultured at 37 °C for 48 h. Next, the plates were washed with PBS for 5 times and incubated with the detection antibody for 2 h at room temperature. After the wash and incubation with streptavidin-HRP for 1 h, immune spots were developed by using substrate solution TMB and counted with the ImmunoSpot Analyzer.
2.11. Flow cytometry
Splenocytes were harvested and suspended in RPMI 1640 medium containing 10% FBS to 2 × 107 cells/mL. And 100 μL of each sample was added into 96U well plate. These splenocytes were then stimulated with PBS, sE2-E1 protein, E2EP3 peptide, E1328-345 peptide or PMA/Inomycin, respectively. After 1 h incubation at 37 °C, Golgi plug were added into samples to prevent IFN-γ secretion. Five hours later, cells were transferred in FCS tube, and then incubated with AF488 anti-mouse CD3 antibody, APC anti-mouse CD8a antibody and BV785 anti-mouse CD4 antibody (Biolegend) at 4 °C for 30 min. After permeabilization and fixation, cells were washed with PBS twice. Then cells were incubated with PE anti-mouse IFN-γ antibody in fixation solution for 30 min and washed with PBS twice. After resuspending with 300 μL PBS, cells were analyzed using BD LSRFortessa flow cytometry.
2.12. In vivo killing
Splenocytes from 6 to 8 weeks-old naïve C57BL/6 mice were used as target cells and incubated with 50 μg/mL CHIKV E1328-345 peptide or control peptide (Ovalbumin (OVA) CD8+ T cell restricted peptide) at 37 °C for 4 h, and then labelled with either 1 μmol/L carboxy fluorescein succinimidyl ester (CFSE) for cells pulsed with CHIKV peptide, or 0.1 μmol/L CFSE for cells pulsed with control OVA peptide. Subsequently, these two groups of target cells were mixed equally, and transferred intravenously (i.v.) at 1.5 × 107 cells/mouse into CHIKV mRNA vaccine immunized mice at 6 weeks after the 3rd immunization. 16 h later, the recipient mice were sacrificed, and the splenocytes were isolated and analyzed with BD LSRFortessa flow cytometer for CFSE staining on target cells. The ratios of in vivo killing in immunized mice were calculated with the formula: R (R: killing ratio; F: frequency of cells, e.g. means frequency of CFSE higher cells in CHIKV mRNA immunized mice).
2.13. Statistical analysis
All data were analyzed with GraphPad Prism 7 software. Neutralization EC50 was analyzed by SPSS. Statistical difference was calculated by t-test and reported as follows, ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; ∗∗∗∗, P < 0.0001.
3. Results
3.1. sE2-E1 heterodimer subunit vaccine elicits potent humoral immune responses
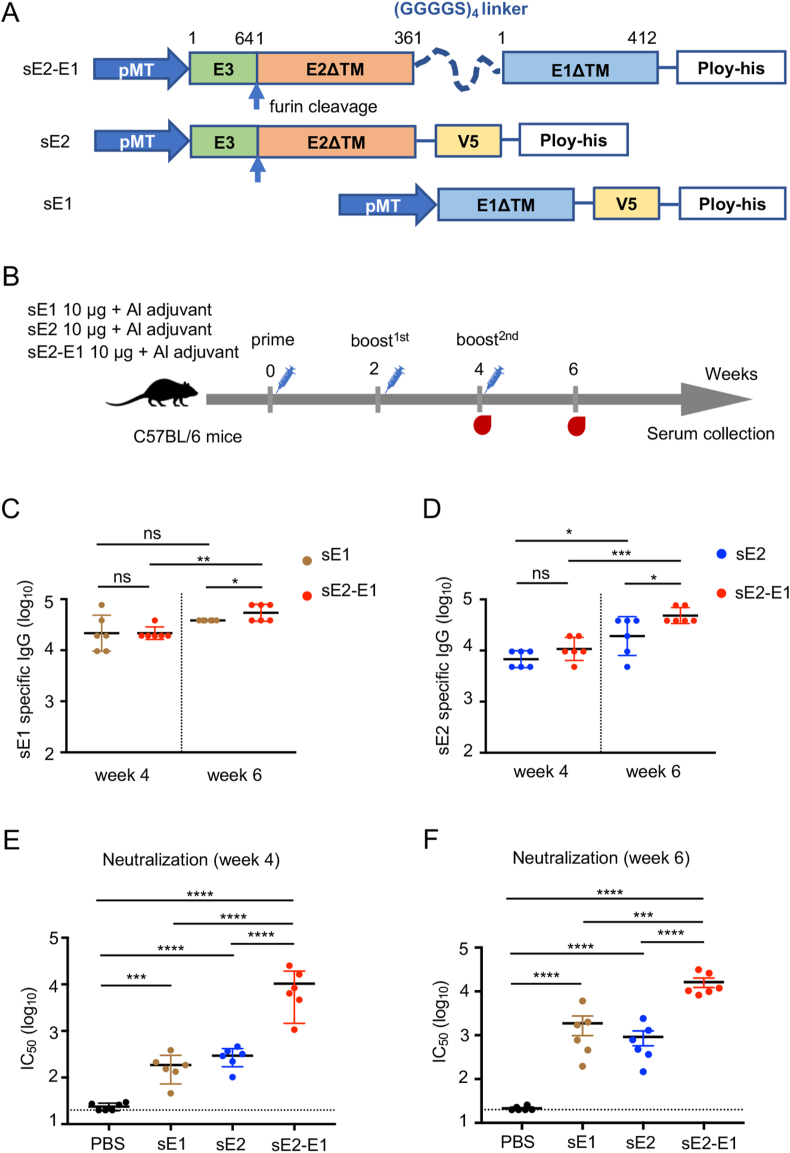
To establish a reference based on which the novel mRNA vaccine could be compared, we first prepared two types of subunit protein vaccines, one type is monomer protein including soluble E1 and soluble E2 proteins (sE1, sE2), another is soluble E2-E1 heterodimer (sE2-E1), which connects E1 and E2 using four repeated GGGGS as linker. The transmembrane domains of the proteins were deleted to increase the protein secretion (Fig. 1A). The proteins were expressed in Drosophila S2 cells, and then purified with Ni-NTA chelate agarose. Purified CHIKV proteins were identified by SDS-PAGE for size and Western blot for specificity. And these purified proteins are mostly in the form of monomers (Supplementary Figure S1).
Fig. 1.
sE2-E1 heterodimer subunit vaccine elicits potent humoral immune responses. A Schematic diagram of CHIKV subunit vaccine candidates. Soluble E2-E1 heterodimer consists of structure protein E1 and E2 with a (GGGGS)4 liner. Transmembrane domains were deleted to improve protein secretion. B Immune strategy of CHIKV subunit vaccines, 6–8 weeks-old C57BL/6 mice were divided into four groups and immunized with vaccine candidates (10 μg, n = 6) or PBS with aluminium adjuvant at weeks 0, 2, 4 via subcutaneous injection, and sera were collected at weeks 4 and 6. (C–D) CHIKV subunit vaccines induced antigen specific IgG titers were detected by ELISA, and antibodies were captured with sE1 protein (C) or sE2 protein (D) using V5-pAb pre-coating ELISA plate. E–F CHIKV neutralizing antibodies were detected by neutralizing assay based on CHIKV pseudovirus at two weeks after the 2nd immunization (E) and the 3rd immunization (F). Statistical differences were determined using t-test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001).
To examine the immunogenicity of these vaccine candidates, 6-week-old female C57BL/6 mice were immunized subcutaneously with sE2-E1 (10 μg), or sE1 (10 μg), or sE2 (10 μg) mixed in aluminum adjuvant at week 0, 2 and 4 (Fig. 1B). Sera were collected at week 4 and 6, and used for measurement of antibody responses by ELISA and neutralization assay. After two immunizations, all three immunogens showed induction of CHIKV specific binding antibodies (Fig. 1C and D). A third immunization further boosted the antibody titers, the mean titer of anti-sE1 specific antibody IgG reached 38,400 (sE1 group) and 57,600 (sE2-E1 group) (Fig. 1C), and anti-sE2 specific antibody IgG reached 24,800 (sE2 group) and 51,200 (sE2-E1 group) (Fig. 1D). sE2-E1 elicited higher antigen specific binding antibody titers than sE1 (1.5 folds) and sE2 (2 folds) at week six post immunization (Fig. 1C and D). Significantly, all three groups had neutralizing antibodies to pseudotyped CHIKV after two immunizations, and sE2-E1 elicited the greatest antibody level, reaching mean IC50 titers to 6480 (Fig. 1E). Moreover, neutralizing antibody levels were all boosted after the third immunization, reaching mean IC50 titers to 1219 (sE1 group), 626 (sE2 group), and 14,903 (sE2-E1 group), respectively (Fig. 1F). These results indicated that sE2-E1 is a superior antigen to elicit neutralizing antibody.
3.2. sE2-E1 heterodimer elicits moderate cellular immune response
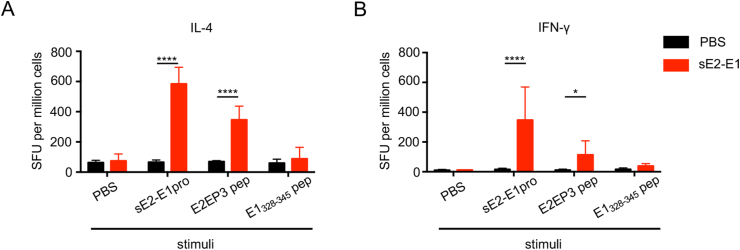
Besides antibody responses, we further assessed the vaccine induced T cell responses in mice. Splenocytes were harvested at two weeks after the 3rd immunization and used to detect IL-4 or IFN-γ production by antigen-specific T cells through ELISpot assay. sE2-E1 protein, or an identified CD4+ T cell restricted-peptide E2EP3 (Teo et al., 2017), or a CD8+ T cell restricted-peptide E1328-345 (Broeckel et al., 2019) were used as stimulators. Results showed that high level of IL-4+ T cells was induced with sE2-E1 protein or E2EP3 peptide stimulation, but not with E1328-345 peptide (Fig. 2A); and modest level of IFN-γ+ T cells was also induced with sE2-E1 protein or E2EP3 peptide stimulation, but not with E1328-345 peptide (Fig. 2B). These results indicated that sE2-E1 subunit vaccine elicited primarily CD4+ T cell responses of both Th1 and Th2 types, but not CD8+ T cell response.
Fig. 2.
sE2-E1 heterodimer elicits moderate cellular immune responses. A–B Splenocytes were harvested at weeks 6 and used to quantify antigen specific IL-4-producing T cells (A) or IFN-γ-producing T cells (B) by ELISpot assay. Protein sE2-E1, CD4+ T cell restricted peptide E2EP3 (STKDNFNVYKATRPYLAH) and CD8+ T cell restricted peptide E1328-345 (CAVHSMTNAVTIREAEIE) were used as stimulators. Statistical differences were determined using t-test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001).
3.3. CHIKV E2-E1 mRNA vaccine elicits more promising antibody responses than the sE2-E1 heterodimer recombinant protein vaccine
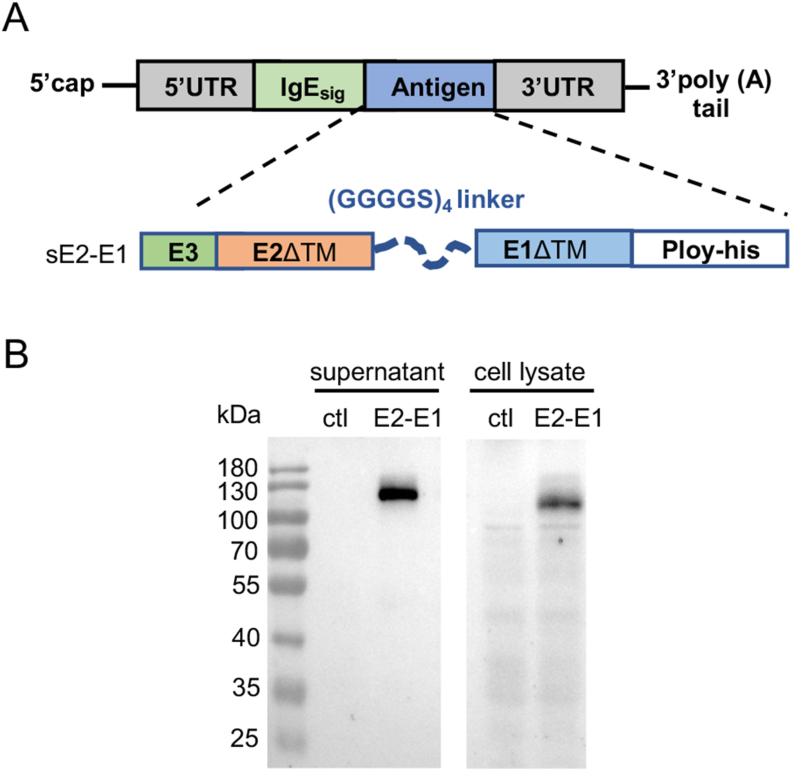
To test the mRNA platform in the development of CHIKV vaccine, we constructed an mRNA vaccine expressing the E2-E1 heterodimer protein (mRNA-E2-E1) (Fig. 3A), and first examined the protein expression by transfecting mRNA-E2-E1 using HEK293T cells. Western blot showed the E2-E1 protein could be expressed in mRNA-E2-E1 transfected cells and secreted into culture supernatant (Fig. 3B).
Fig. 3.
The design and expression of CHIKV mRNA antigen in vitro. A Schematic illustration of CHIKV mRNA consisting of a 5′ cap, a 5′ UTR, a signal peptide IgE, an antigen sE2-E1, a 3′ UTR, and a 3′ poly(A) tail. B Protein expression in mRNA-transfected HEK293T cells. Protein was detected by Western blot with his-tag monoclonal antibody.
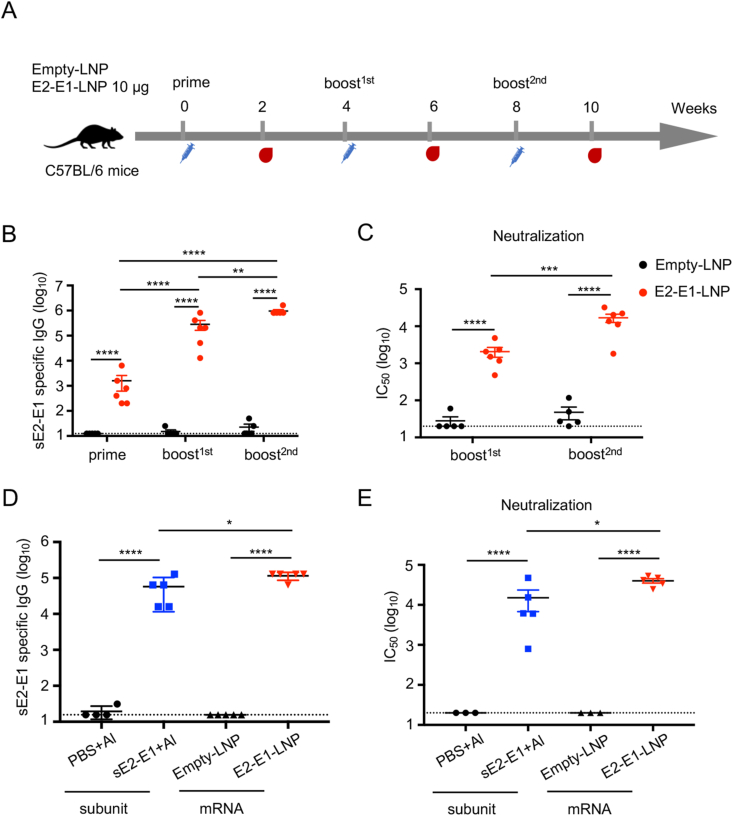
To prepare a vaccine for immunization, the mRNA-E2-E1 was packaged into LNP (E2-E1-LNP) as the method previously described (Zhang et al., 2020), and was used to inoculate 6-week-old female C57BL/6 mice intramuscularly, with three times of 10 μg E2-E1-LNP or Empty-LNP at four weeks interval. Serum samples were collected at two weeks after each immunization for antibody evaluation (Fig. 4A). Results showed that E2-E1-LNP elicited antigen specific binding antibodies after the first immunization, which was boosted after each subsequent immunization and reached 106 after the 3rd immunization (Fig. 4B). The neutralizing antibodies as measured by mean IC50 were elevated after each boosting and reached 16,686 after the 3rd immunization (Fig. 4C). For comparison, three doses of E2-E1-LNP elicited 2 folds higher binding antibody levels and 2.6 folds higher neutralizing antibody levels than three doses of sE2-E1 recombinant protein (Fig. 4D and E).
Fig. 4.
CHIKV E2-E1-LNP mRNA vaccine elicits potent antibody responses better than the sE2-E1 heterodimer subunit vaccine. A Immune strategy of CHIKV E2-E1-LNP. 6-8 weeks-old female C57BL/6 mice were immunized with 10 μg E2-E1-LNP or Empty-LNP (n = 6) at weeks 0, 4 and 8 through intramuscular injection. Antibody responses were measured at weeks 2, 6 and 10. B CHIKV specific binding antibodies were detected by ELISA coating with sE2-E1 heterodimer protein. C CHIKV specific neutralizing antibodies were detected by neutralization assay based on CHIKV pseudoviral system. D Antigen specific binding antibodies of the subunit vaccine or mRNA vaccine immunized mice after the 3rd immunization were captured with sE2-E1 protein using V5-pAb pre-coating ELISA plate. E CHIKV neutralizing antibodies detection of serum samples from sE2-E1 protein or E2-E1-LNP immunized mice after the 3rd immunization. Statistical differences were determined using t-test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001).
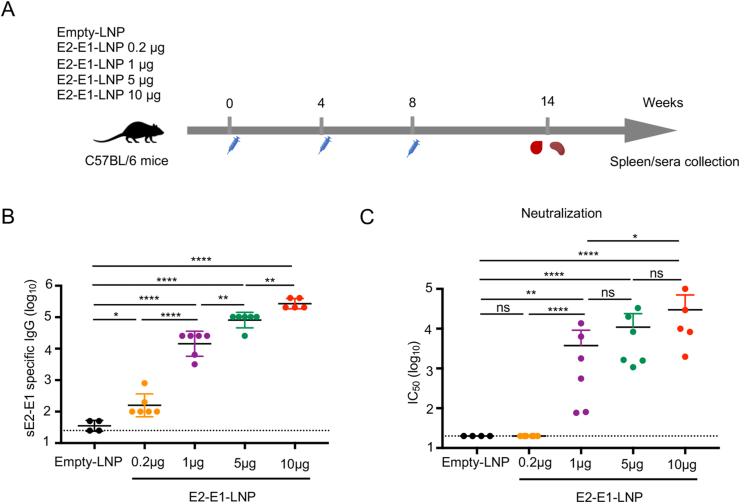
3.4. CHIKV specific antibody induced by E2-E1-LNP is dose dependent
To optimize the vaccination dosage of mRNA-E2-E1 vaccine, we immunized mice with 0.2 μg, 1 μg, 5 μg or 10 μg E2-E1-LNP via intramuscular injection at weeks 0, 4 and 8 and sacrificed mice at week 14 (6 weeks after the last immunization), when serum samples were collected for antibody measurement and splenocytes were harvested for T cell evaluation (Fig. 5A). The results showed that all E2-E1-LNP immunized groups elicited antigen-specific binding antibody in a dose dependent manner (Fig. 5B). Moreover, all but the lowest dose (0.2 μg) E2-E1-LNP immunized groups elicited neutralizing antibody showing a similar dose dependent trend (Fig. 5C).
Fig. 5.
CHIKV specific antibody induced by E2-E1-LNP is dose dependent. A 6-8 weeks-old female C57BL/6 mice were immunized with 0.2 μg, 1 μg, 5 μg, 10 μg E2-E1-LNP or Empty-LNP (n = 4–6) at weeks 0, 4 and 8 through intramuscular injection. Sera were collected at 6 weeks after the last immunization. B Antigen specific binding antibodies were determined by ELISA. C Antigen specific neutralizing antibody were determined by pseudoviral neutralization assay. Statistical differences were determined using t-test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001).
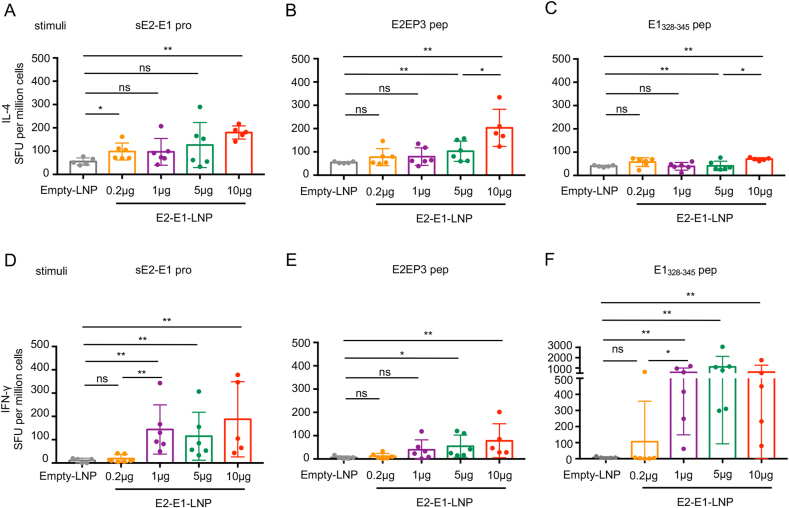
3.5. CHIKV mRNA-E2-E1 vaccine induces potent CD8+ T cell responses
T cell responses induced by E2-E1-LNP were then measured at 6 weeks after the last immunization by ELISpot assay. The results showed that IL-4+ T cells were moderately activated by the stimulation with sE2-E1 protein or the E2EP3 peptide in the high dose group (Fig. 6A and B), but not with the E1328-345 peptide (Fig. 6C). IFN-γ-producing T cells was moderately induced by stimulation with sE2-E1 protein, but strongly induced by the stimulation with the CD8+ T cell restricted peptide (Fig. 6D and F). Moreover, except for the lowest dose (0.2 μg), there is no difference in the T cell responses induced among the dose groups. Meanwhile, CD4+ T cell restricted peptide E2EP3 did not significantly activate IFN-γ-producing T cells (Fig. 6E).
Fig. 6.
CHIKV E2-E1-LNP mRNA vaccine induces antigen specific T cell responses. A–F Mice (n = 5–6 each group) were sacrificed and splenocytes were harvested at 6 weeks after the last immunization and used to quantify antigen specific IL-4-producing T cells (A–C) or IFN-γ-producing T cells (D–F) by ELISpot assay. Protein sE2-E1, CD4+ T cell restricted peptide E2EP3 and CD8+ T cell restricted peptide E1328-345 were used as stimulations. Statistical differences were determined using t-test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001).
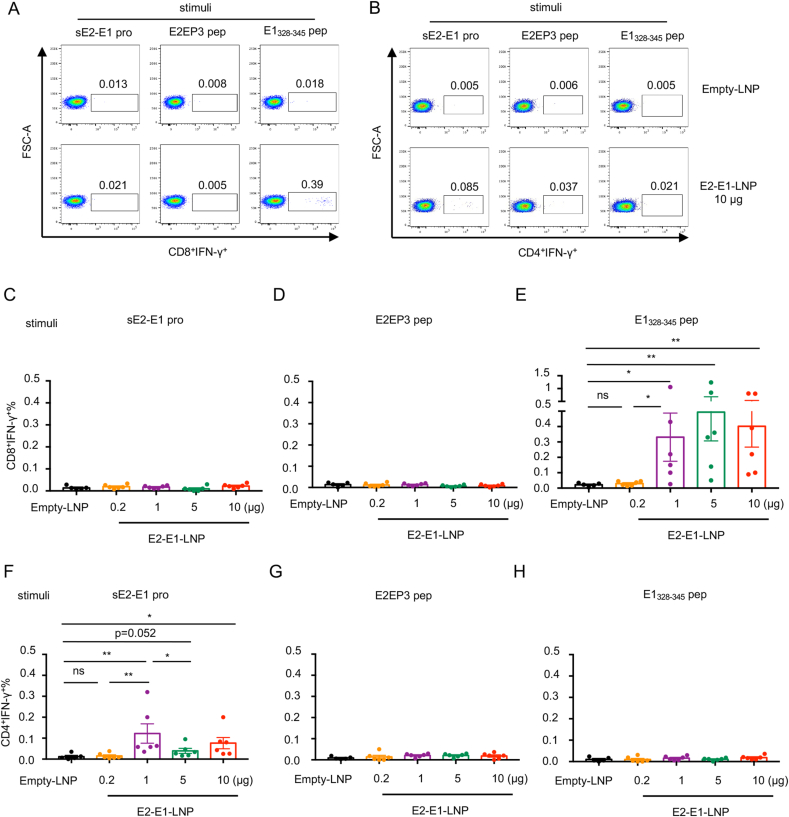
To further verify the phenotype of responding T cells, we performed flow cytometry assay. Representative data showed that E1328-345 peptide dominantly activated IFN-γ-producing CD8+ T cells, while IFN-γ-producing CD4+ T cells were mainly stimulated by sE2-E1 protein (Fig. 7A and B). Consistent with results of ELISpot assay, there is no dose dependency for IFN-γ response in either CD8+ T cells or CD4+ T cells (Fig. 7C–H). These results indicated that 1 μg dosage of CHIKV mRNA vaccine is enough to induce a suitable CD8+ T cell response in mice.
Fig. 7.
Antigen specific T cell response elicited by E2-E1-LNP mRNA vaccine is CD8+ T cell dominant. Splenocytes from each group (n = 5–6) were harvested and used for quantifying antigen specific CD8+IFN-γ+ T cells or CD4+IFN-γ+ T cells by flow cytometry. A Representative results of mRNA vaccine induced CD8+IFN-γ+ T cells. B Representative results of mRNA vaccine induced CD4+IFN-γ+ T cells. C–E Percentage of CD8+IFN-γ+ T cells stimulated with sE2-E1 protein (C), E2EP3 peptide (D) and E1328-345 peptide (E). F–H Percentage of CD4+IFN-γ+ T cells stimulated with sE2-E1 protein (F), E2EP3 peptide (G) and E1328-345 peptide (H). Statistical differences were determined using t-test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001).
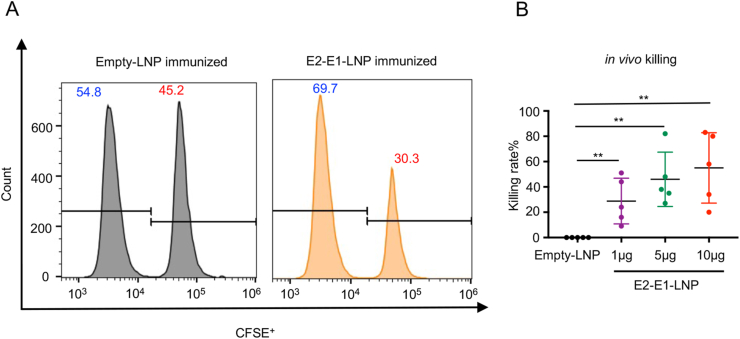
Finally, we examined the function of CHIKV antigen specific CD8+ T cells using an in vivo killing assay, in which different concentration CFSE labelled naïve splenocytes that had been incubated with CHIKV E1328-345 peptide or a control OVA peptide were transferred into CHIKV E2-E1-LNP mRNA vaccine immunized mice. Results showed that E1328-345 peptide labelled cells significantly reduced in immunized but not control mice, indicating that CD8+ T cells induced by E2-E1-LNP vaccine can kill target cells in vivo (Fig. 8A and B).
Fig. 8.
CHIKV E2-E1-LNP mRNA vaccine induced antigen specific CD8+ T cells perform cytotoxic effect in vivo. Naïve splenocytes pulsed with control peptide were labelled with 0.1 μmol/L CFSE and naïve splenocytes pulsed with E1328-345 peptide were labelled with 1 μmol/L CFSE. Then the CFSEhigh cells and CFSElow cells were mixed at 1:1 and transferred into mRNA vaccine immunized mice (n = 5) at 6 weeks after the last immunization. 16 h after adoptive transfer of naïve splenocytes, immunized mice were sacrificed. Percentage of specific cytotoxic killing was analyzed by flow cytometry. A Representative results of mRNA vaccine induced CD8+ T cells mediated cytotoxic effect. B Killing rate of each dose groups. Statistical differences were determined using t-test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001).
4. Discussion
Many efforts had been made to develop a safe and effective CHIKV vaccine. Here, we demonstrate a novel mRNA vaccine E2-E1-LNP shows greater immunogenicity compared to the recombinant protein CHIKV vaccine candidates expressing various forms of the viral envelope protein. Specifically, recombinant protein sE2-E1 can induce better neutralizing antibody responses than recombinant sE1 and sE2; but sE2-E1 in aluminum adjuvant only induces negligible CD8+ T cell responses. In comparison, mRNA vaccine expressing E2-E1 antigen can not only elicit stronger neutralizing antibody responses than recombinant protein sE2-E1, but also better protective CD8+ T cell response. These results suggest that CHIKV E2-E1-LNP vaccine is a promising vaccine candidate warrants rapid development.
It is interesting that E2-E1 heterodimer, and E1 and E2 monomers have significantly different immunogenicity in our study, despite both antigens are located on the virion. Epitopes recognized by the monoclonal neutralizing antibodies isolated from infected people were reported to locate on both E1 and E2 proteins, indicating that both E1 and E2 can induce neutralizing antibodies, which may neutralize virus through various processes, such as receptor binding, viral entry, membrane fusion and virus release (Masrinoul et al., 2014; Fox et al., 2015). Therefore, both E1 and E2 are often selected as immunogens in various vaccine platforms (Mallilankaraman et al., 2011; Khan et al., 2012; Kumar et al., 2012; Brandler et al., 2013). Previous studies show that recombinant E1 and E2 proteins often induce only low levels of antibody response(Khan et al., 2012; Metza et al., 2013); but mixing E1 and E2 into a cocktail can induce higher level of neutralizing antibodies than using E1 or E2 alone (Khan et al., 2012). The crystallographic study of E2-E1 complex expressed in S2 cells revealed a heterodimeric structure, which is very similar to those formed within spikes structure on virion surface (Voss et al., 2010). Moreover, the alphavirus receptor mxra8 binding to CHIKV E proteins involves both E1 and E2 protein (Basore et al., 2019; Song et al., 2019). These may help to explain why our recombinant sE2-E1 has an outstanding neutralization potency. Thus, we chose E2-E1 as the antigen of mRNA vaccine.
Our mRNA vaccine expressing E2-E1 is of high immunogenicity, which elicits not only high level of neutralizing antibody, but also strong CTL immune response. Previous study indicates that antigen-specific IgG titer exceeds 104 was correlated with significant protection (Hallengard et al., 2014b). In this study, the IgG titers in mRNA vaccinated mice after the 3rd immunization was close to 106, which far exceeded 104, suggesting the mRNA vaccine may be able to confer protection. Moreover, cellular immune responses also affect the outcomes of CHIKV infection. Our results showed both mRNA and recombinant protein vaccines activated CD4+ T cells, only E2-E1-LNP elicit potent CD8+ T cell responses, whereas the recombinant protein vaccines did not. Because CD8+ T cells generally protect against viral infections including CHIKV, and CD4+ T cells contribute to the development of joint swelling in mouse model (Teo et al. 2013, 2017), we think a vaccine that activates CD8+ T cells but not very much CD4+ T cells should be a better one for CHIKV. Theoretically, the endogenous expressed E2-E1 protein can promote the efficiency of processing and presentation of CD8+ T cell epitopes compared to recombinant protein vaccine (Sahin et al., 2014). Therefore, mRNA CHIKV vaccine is superior to recombinant protein vaccine in this regard.
5. Conclusions
In conclusion, we developed a novel CHIKV E2-E1-LNP mRNA vaccine that can induce potent neutralizing antibodies, modest level of CD4+ T cells, and functional CD8+ T cells. In comparison, recombinant protein vaccine expressing the same antigen can only elicit antibody responses and CD4+ T cell responses, but not CD8+ T cells. Thus, mRNA vaccine as a new vaccine form for CHIKV should be developed with increase priority. In future we will perform animal challenge experiments to observe efficacy of CHIKV E2-E1-LNP mRNA vaccine, compare the protective efficacy of the mRNA vaccine and the sE2-E1 subunit vaccine, and explore the relationship between the vaccine induced immune responses and the protective efficacy.
Data availability
The authors declare that the data supporting the conclusions during the current study are included in this article.
Ethics statement
All animal experiments were conducted strictly according to the instructions of SPF containments in Shanghai Public Health Clinical Center. The protocol and animal ethics were approved by the Shanghai Public Health Clinical Center Laboratory Animal Welfare and Ethics Committee (Approval number: 2020-A009-01).
Author contributions
Ningning Ge: conceptualization, investigation, formal analysis, data curation, methodology, writing-original draft, writing-review & editing. Jin Sun: investigation, methodology, writing-review & editing. Zhihua Liu: funding acquisition, investigation, methodology, writing-review & editing. Jiayi Shu: writing-review & editing. Huimin Yan: methology. Zhihua Kou: conceptualization, supervision, writing-review & editing. Yu Wei: supervision, writing-review & editing. Xia Jin: conceptualization, supervision, project administration, funding acquisition, writing-review & editing.
Conflict of interest
All authors declare no conflicts of interest.
Acknowledgements
We thank Dr. Guiqing Wang (Institute Pasteur of Shanghai, Shanghai, China) for kindly providing vectors for pseudotyped virus. This work was supported by the following grants: the National Key R&D Program of China (Grant 2016YFC1201000 to X.J.) and the Institute Fund of Shanghai Public Health Clinical Center (Grant KY-GW-2021-17 to ZH.L.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.01.032.
Contributor Information
Zhihua Kou, Email: kouzhihua@shphc.org.cn.
Yu Wei, Email: yuwei@ips.ac.cn.
Xia Jin, Email: jinxia@serum-china.com.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Akahata W., Yang Z.Y., Andersen H., Sun S., Holdaway H.A., Kong W.P., Lewis M.G., Higgs S., Rossmann M.G., Rao S., Nabel G.J. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat. Med. 2010;16:334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson O., Thompson J., Ribeiro A.M., Watson M., Zaks T., Ciaramella G. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basore K., Kim A.S., Nelson C.A., Zhang R., Smith B.K., Uranga C., Vang L., Cheng M., Gross M.L., Smith J., Diamond M.S., Fremont D.H. Cryo-EM structure of chikungunya virus in complex with the Mxra8 receptor. Cell. 2019;177:1725–1737. doi: 10.1016/j.cell.2019.04.006. e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgherini G., Poubeau P., Jossaume A., Gouix A., Cotte L., Michault A., Arvin-Berod C., Paganin F. Persistent arthralgia associated with chikungunya virus: a study of 88 adult patients on reunion island. Clin. Infect. Dis. 2008;47:469–475. doi: 10.1086/590003. [DOI] [PubMed] [Google Scholar]
- Brandler S., Ruffié C., Combredet C., Brault J.-B., Najburg V., Prevost M.-C., Habel A., Tauber E., Desprès P., Tangy F. A recombinant measles vaccine expressing chikungunya virus-like particles is strongly immunogenic and protects mice from lethal challenge with chikungunya virus. Vaccine. 2013;31:3718–3725. doi: 10.1016/j.vaccine.2013.05.086. [DOI] [PubMed] [Google Scholar]
- Broeckel R.M., Haese N., Ando T., Dmitriev I., Kreklywich C.N., Powers J., Denton M., Smith P., Morrison T.E., Heise M., DeFilippis V., Messaoudi I., Curiel D.T., Streblow D.N. Vaccine-induced skewing of T cell responses protects against chikungunya virus disease. Front. Immunol. 2019;10:2563. doi: 10.3389/fimmu.2019.02563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack K.S., Montgomery S.A., Homann D., Morrison T.E. CD8+T cells control Ross River virus infection in musculoskeletal tissues of infected mice. J. Immunol. 2015;194:678–689. doi: 10.4049/jimmunol.1401833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.-J., Dowd K.A., Mendoza F.H., Saunders J.G., Sitar S., Plummer S.H., Yamshchikov G., Sarwar U.N., Hu Z., Enama M.E., Bailer R.T., Koup R.A., Schwartz R.M., Akahata W., Nabel G.J., Mascola J.R., Pierson T.C., Graham B.S., Ledgerwood J.E., Team tVS. Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: a phase 1 dose-escalation trial. Lancet. 2014 doi: 10.1016/S0140-6736(14)61185-52046-2052. [DOI] [PubMed] [Google Scholar]
- Chopra A., Anuradha V., Ghorpade R., Saluja M. Acute Chikungunya and persistent musculoskeletal pain following the 2006 Indian epidemic: a 2-year prospective rural community study. Epidemiol. Infect. 2011;140:842–850. doi: 10.1017/S0950268811001300. [DOI] [PubMed] [Google Scholar]
- Chu H., Das S.C., Fuchs J.F., Suresh M., Weaver S.C., Stinchcomb D.T., Partidos C.D., Osorio J.E. Deciphering the protective role of adaptive immunity to CHIKV/IRES a novel candidate vaccine against Chikungunya in the A129 mouse model. Vaccine. 2013;31:3353–3360. doi: 10.1016/j.vaccine.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZure A.D., Berkowitz N.M., Graham B.S., Ledgerwood J.E. Whole-inactivated and virus-like particle vaccine strategies for chikungunya virus. JID (J. Infect. Dis.) 2016;214:S497–S499. doi: 10.1093/infdis/jiw352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC . Multistate (World). Monitoring global outbreaks; 2020. Communicable Disease Threats Report, 14- 20 June 2020, Week 25. Chikungunya and Dengue.https://www.ecdc.europa.eu/en/publications-data/communicable-disease-threats-report-14-20-june-2020-week-25 [Google Scholar]
- Edelman R., Tacket C.O., Wasserman S.S., Bodison S.A., Perry J.G., Mangiafico J.A. Phase II safety and immunogenicity study of live Chikungunya virus vaccine TSI-GSD-218. Am. J. Trop. Med. Hyg. 2000;62:681–685. doi: 10.4269/ajtmh.2000.62.681. [DOI] [PubMed] [Google Scholar]
- Fox J.M., Long F., Edeling M.A., Lin H., van Duijl-Richter M.K.S., Fong R.H., Kahle K.M., Smit J.M., Jin J., Simmons G., Doranz B.J., Crowe J.E., Fremont D.H., Rossmann M.G., Diamond M.S. Broadly neutralizing alphavirus antibodies bind an epitope on E2 and inhibit entry and egress. Cell. 2015;163:1095–1107. doi: 10.1016/j.cell.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwendolyn K., Binder D.E.G. Interferon-gamma-Mediated site-specific clearance of alphavirus from CNS neurons. Science. 2001;293:303–306. doi: 10.1126/science.1059742. [DOI] [PubMed] [Google Scholar]
- Hallengard D., Kakoulidou M., Lulla A., Kummerer B.M., Johansson D.X., Mutso M., Lulla V., Fazakerley J.K., Roques P., Le Grand R., Merits A., Liljestrom P. Novel attenuated Chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J. Virol. 2014;88:2858–2866. doi: 10.1128/JVI.03453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallengard D., Lum F.M., Kummerer B.M., Lulla A., Lulla V., Garcia-Arriaza J., Fazakerley J.K., Roques P., Le Grand R., Merits A., Ng L.F., Esteban M., Liljestrom P. Prime-boost immunization strategies against Chikungunya virus. J. Virol. 2014;88:13333–13343. doi: 10.1128/JVI.01926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Dhanwani R., Rao P.V., Parida M. Subunit vaccine formulations based on recombinant envelope proteins of Chikungunya virus elicit balanced Th1/Th2 response and virus-neutralizing antibodies in mice. Virus Res. 2012;167:236–246. doi: 10.1016/j.virusres.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Kim D.Y., Atasheva S., Foy N.J., Wang E., Frolova E.I., Weaver S., Frolov I. Design of chimeric alphaviruses with a programmed, attenuated, cell type-restricted phenotype. J. Virol. 2011;85:4363–4376. doi: 10.1128/JVI.00065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Sudeep A.B., Arankalle V.A. Evaluation of recombinant E2 protein-based and whole-virus inactivated candidate vaccines against chikungunya virus. Vaccine. 2012;30:6142–6149. doi: 10.1016/j.vaccine.2012.07.072. [DOI] [PubMed] [Google Scholar]
- Li L., Jose J., Xiang Y., Kuhn R.J., Rossmann M.G. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum F.-M., Teo T.-H., Lee W.W.L., Kam Y.-W., Rénia L., Ng L.F.P. An essential role of antibodies in the control of chikungunya virus infection. J. Immunol. 2013;190:6295–6302. doi: 10.4049/jimmunol.1300304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K., Shedlock D.J., Bao H., Kawalekar O.U., Fagone P., Ramanathan A.A., Ferraro B., Stabenow J., Vijayachari P., Sundaram S.G., Muruganandam N., Sarangan G., Srikanth P., Khan A.S., Lewis M.G., Kim J.J., Sardesai N.Y., Muthumani K., Weiner D.B. A DNA vaccine against chikungunya virus is protective in mice and induces neutralizing antibodies in mice and nonhuman primates. PLoS Neglected Trop. Dis. 2011;5:e928. doi: 10.1371/journal.pntd.0000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masrinoul P., Puiprom O., Tanaka A., Kuwahara M., Chaichana P., Ikuta K., Ramasoota P., Okabayashi T. Monoclonal antibody targeting chikungunya virus envelope 1 protein inhibits virus release. Virology. 2014;464–465:111–117. doi: 10.1016/j.virol.2014.05.038. [DOI] [PubMed] [Google Scholar]
- Metza S.W., Martinab B.E., Pvd Doelb, Geertsemaa C., Osterhausb A.D., Vlaka J.M., Pijlman G.P. Chikungunya virus-like particles are more immunogenic in a lethal AG129 mouse model compared to glycoprotein E1 or E2 subunits. Vaccine. 2013 doi: 10.1016/j.vaccine.2013.09.0456092-6096. [DOI] [PubMed] [Google Scholar]
- Muthumani K., Block P., Flingai S., Muruganantham N., Chaaithanya I.K., Tingey C., Wise M., Reuschel E.L., Chung C., Muthumani A., Sarangan G., Srikanth P., Khan A.S., Vijayachari P., Sardesai N.Y., Kim J.J., Ugen K.E., Weiner D.B. Rapid and long-term immunity elicited by DNA-encoded antibody prophylaxis and DNA vaccination against chikungunya virus. JID (J. Infect. Dis.) 2016;214:369–378. doi: 10.1093/infdis/jiw111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAHO . 2021. Cases of Chikungunya Virus Disease by Contury or Territory Cumulative Cases.https://www3.paho.org/data/index.php/en/mnu-topics/chikv-en/550-chikv-weekly-en.html [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Tureci O., Nell H., Schaefer A., Unal S., Tresnan D.B., Mather S., Dormitzer P.R., Sahin U., Jansen K.U., Gruber W.C., Group C.C.T. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard C., Rejman J., De Haes W., Verrier B., Van Gulck E., Naessens T., De Smedt S., Bogaert P., Grooten J., Vanham G., De Koker S. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 2013;21:251–259. doi: 10.1038/mt.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers Acb Ann M., Tesh Robert B., Weaver Scott C. Re-emergence of chikungunya and o’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J. Gen. Virol. 2000;81:471–479. doi: 10.1099/0022-1317-81-2-471. [DOI] [PubMed] [Google Scholar]
- Doel P.V.D., Volz A., Roose J.M., Sewbalaksing V.D., Pijlman G.P., Iv Middelkoop, Duiverman V., Evd Wetering, Sutter G., Osterhaus A.D.M.E., Martina B.E.E. Recombinant modified vaccinia virus Ankara expressing glycoprotein E2 of Chikungunya virus protects AG129 mice against lethal challenge. PLoS Neglected Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Wang G., Wang S., Chen H., Chen Z., Hu H., Cheng G., Zhou P. Cross-protection of newly emerging HPAI H5 viruses by neutralizing human monoclonal antibodies: a viable alternative to oseltamivir. mAbs. 2016;8:1156–1166. doi: 10.1080/19420862.2016.1183083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., Ciaramella G., Diamond M.S. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;168:1114–1125 e1110. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.C. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. I. Clinical features. Trans. R. Soc. Trop. Med. Hyg. 1955;49:28–32. doi: 10.1016/0035-9203(55)90080-8. [DOI] [PubMed] [Google Scholar]
- Sahin U., Kariko K., Tureci O. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., Brachtendorf S., Lorks V., Sikorski J., Hilker R., Becker D., Eller A.K., Grutzner J., Boesler C., Rosenbaum C., Kuhnle M.C., Luxemburger U., Kemmer-Bruck A., Langer D., Bexon M., Bolte S., Kariko K., Palanche T., Fischer B., Schultz A., Shi P.Y., Fontes-Garfias C., Perez J.L., Swanson K.A., Loschko J., Scully I.L., Cutler M., Kalina W., Kyratsous C.A., Cooper D., Dormitzer P.R., Jansen K.U., Tureci O. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Smith S.A., Silva L.A., Fox J.M., Flyak A.I., Kose N., Sapparapu G., Khomandiak S., Ashbrook A.W., Kahle K.M., Fong R.H., Swayne S., Doranz B.J., McGee C.E., Heise M.T., Pal P., Brien J.D., Austin S.K., Diamond M.S., Dermody T.S., Crowe J.E., Jr. Isolation and characterization of broad and ultrapotent human monoclonal antibodies with therapeutic activity against chikungunya virus. Cell Host Microbe. 2015;18:86–95. doi: 10.1016/j.chom.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Zhao Z., Chai Y., Jin X., Li C., Yuan F., Liu S., Gao Z., Wang H., Song J., Vazquez L., Zhang Y., Tan S., Morel C.M., Yan J., Shi Y., Qi J., Gao F., Gao G.F. Molecular basis of arthritogenic alphavirus receptor MXRA8 binding to chikungunya virus envelope protein. Cell. 2019;177:1714–1724. doi: 10.1016/j.cell.2019.04.008. e1712. [DOI] [PubMed] [Google Scholar]
- Suhrbier A. Rheumatic manifestations of chikungunya: emerging concepts and interventions. Nat. Rev. Rheumatol. 2019;15:597–611. doi: 10.1038/s41584-019-0276-9. [DOI] [PubMed] [Google Scholar]
- Teo T.H., Lum F.M., Claser C., Lulla V., Lulla A., Merits A., Renia L., Ng L.F. A pathogenic role for CD4+ T cells during Chikungunya virus infection in mice. J. Immunol. 2013;190:259–269. doi: 10.4049/jimmunol.1202177. [DOI] [PubMed] [Google Scholar]
- Teo T.-H., Chan Y.-H., Lee W.W.L., Lum F.-M., Amrun S.N., Her Z., Rajarethinam R., Merits A., Rötzschke O., Rénia L., Ng L.F.P. Fingolimod treatment abrogates chikungunya virus–induced arthralgia. Sci. Transl. Med. 2017;9(375):eaal1333. doi: 10.1126/scitranslmed.aal1333. [DOI] [PubMed] [Google Scholar]
- Thiberville S.D., Moyen N., Dupuis-Maguiraga L., Nougairede A., Gould E.A., Roques P., de Lamballerie X. Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy. Antivir. Res. 2013;99:345–370. doi: 10.1016/j.antiviral.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari M., Parida M., Santhosh S.R., Khan M., Dash P.K., Rao P.V.L. Assessment of immunogenic potential of Vero adapted formalin inactivated vaccine derived from novel ECSA genotype of Chikungunya virus. Vaccine. 2009;27:2513–2522. doi: 10.1016/j.vaccine.2009.02.062. [DOI] [PubMed] [Google Scholar]
- Tretyakova I., Hearn J., Wang E., Weaver S., Pushko P. DNA vaccine initiates replication of live attenuated chikungunya virus in vitro and elicits protective immune response in mice. J. Infect. Dis. 2014;209:1882–1890. doi: 10.1093/infdis/jiu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K.A., Vanlandingham D.L., McGee C.E., Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3 doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K.A., McGee C.E., Volk S.M., Vanlandingham D.L., Weaver S.C., Higgs S. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H., Reece S.T., Sahin U., Tregoning J.S. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk S.M., Chen R., Tsetsarkin K.A., Adams A.P., Garcia T.I., Sall A.A., Nasar F., Schuh A.J., Holmes E.C., Higgs S., Maharaj P.D., Brault A.C., Weaver S.C. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J. Virol. 2010;84:6497–6504. doi: 10.1128/JVI.01603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss J.E., Vaney M.C., Duquerroy S., Vonrhein C., Girard-Blanc C., Crublet E., Thompson A., Bricogne G., Rey F.A. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- Weaver S.C., Weber C., Berberich E., von Rhein C., Henß L., Hildt E., Schnierle B.S. Identification of functional determinants in the chikungunya virus E2 protein. PLoS Neglected Trop. Dis. 2017;11:e0005318. doi: 10.1371/journal.pntd.0005318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Sun J., Li M., Jin X. Modified mRNA-LNP vaccines confer protection against experimental DENV-2 infection in mice. Molecular Therapy - Methods & Clinical Development. 2020;18:702–712. doi: 10.1016/j.omtm.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the conclusions during the current study are included in this article.