Abstract
The Getah virus (GETV), a mosquito-borne RNA virus, is widely distributed in Oceania and Asia. GETV is not the only pathogenic to horses, pigs, cattle, foxes and boars, but it can also cause fever in humans. Since its first reported case in Chinese mainland in 2017, the number of GETV-affected provinces has increased to seventeen till now. Therefore, we performed an epidemiologic investigation of GETV in the Xinjiang region, located in northwestern China, during the period of 2017–2020. ELISA was used to analyze 3299 serum samples collected from thoroughbred horse, local horse, sheep, goat, cattle, and pigs, with thoroughbred horse (74.8%), local horse (67.3%), goat (11.7%), sheep (10.0%), cattle (25.1%) and pigs (51.1%) being positive for anti-GETV antibodies. Interestingly, the neutralizing antibody titer in horses was much higher than in other species. Four samples from horses and pigs were positive for GETV according to RT-PCR. Furthermore, from the serum of a local horse, we isolated GETV which was designated as strain XJ-2019-07, and determined its complete genome sequence. From the phylogenetic relationships, it belongs to the Group III lineage. This is the first evidence of GETV associated to domestic animals in Xinjiang. Overall, GETV is prevalent in Xinjiang and probably has been for several years. Since no vaccine against GETV is available in China, detection and monitoring strategies should be improved in horses and pigs, especially imported and farmed, in order to prevent economic losses.
Keywords: Getah virus (GETV), Zoonoses, Arbovirus, Seroprevalence, Xinjiang Uygur Autonomous Region
Highlights
-
•
We firstly reported the epidemiology and isolation of GETV infection in livestock in Xinjiang.
-
•
GETV is prevalent in Xinjiang and probably has been for several years.
-
•
Since no vaccine against GETV is available in China, detection and monitoring strategies should be improved in horses and pigs, especially imported and farmed, in order to prevent economic losses.
1. Introduction
The re-emergence and growing burden of mosquito-borne virus infections, e.g., Zika virus (ZIKV) and Dengue virus (DENV), continue to threaten public health (Burt et al., 2017; Gutierrez-Bugallo et al., 2019; Messina et al., 2014). As a member of arbovirus, Getah virus (GETV) belongs to the genus Alphavirus in the family Togaviridae, along with Chikungunya virus (CHIKV) and Venezuelan equine encephalitis virus (VEEV). GETV was first isolated from mosquitoes in Malaysia in 1955 (Morita and Igarashi, 1984). At present, GETV has spread to 12 countries across Eurasia and the Pan-Pacific: China, Korea, Japan, Russia, Thailand, Mongolia, India, Australia, Philippines, Sri Lanka, Cambodia and Vietnam (Brown and Timoney, 1998; Bryant et al., 2005; Fukunaga et al., 2000; Ksiazek et al., 1981; Li et al., 2017; Lu et al., 2019; L'Vov et al., 2000; Nemoto et al., 2015; Turell et al., 2003).
Since GETV is known to be pathogenic to horses, pigs and cattle, it has caused large economic losses (Liu et al., 2019; Nemoto et al., 2015; Xing et al., 2020). The clinical manifestations of GETV infection in horses often include fever, rash on the body, hind limb oedema and lymph node enlargement. Nasal discharge has only been observed in experimentally inoculated horses, but not in cases of natural infection (Kamada et al., 1991). Notably, infection in pigs is more severe than in horses. Infected piglets exhibit depression, tremors, diarrhoea, hind limb paralysis, high mortality rates and aborted labour in infected sows (Xing et al., 2020). GETV was isolated from febrile sick cattle in China in 2019, which proved that it was capable of infecting cattle. GETV can also infect wild animals such as foxes, where it can cause fever, anorexia, depression, neurological symptoms and even death and wild boars which may experience fever, anorexia and depression (Kuwata et al., 2018; Shi et al., 2019).
More recent studies have shown that the virus has a broad spectrum of infection (Liu et al., 2019; Nemoto et al., 2015; Xing et al., 2020; Kuwata et al., 2018; Shi et al., 2019). Under experimental conditions, GETV is observed in blood of infected rabbits, monkeys, orangutans, mice, hamsters and guinea pigs, causing muscle inflammation, growth retardation and even fatal encephalitis (Kamada et al., 1991). Also, antibodies for GETV have been detected in serum of chickens, ducks, goats, kangaroos, birds and reptiles (Li et al., 1992, 2019; Lu et al., 2019). More importantly, neutralizing antibodies against GETV have been identified in human sera in Malaysia, Australia and China (Li et al., 1992; Lu et al., 2019). The specific antibody titer in people with fever is significantly higher than in healthy subjects, indicating that GETV infection is associated with disease in humans.
In China, the epidemiological status of GETV has become increasingly problematic and poses a threat to animal and public health. GETV infection of pigs in Chinese mainland was first observed in Hunan Province in 2017, originating from Taiwan in 2002. After that, GETV has appeared in eight other provinces (Ren et al., 2020; Xing et al., 2020; Lu et al., 2019). Xinjiang Uygur Autonomous Region is the largest province in China with major animal husbandry. It is located in northwestern China and borders eight countries including Mongolia, the Russian Federation, Kazakhstan, Kyrgyzstan, Tajikistan, Afghanistan, Pakistan, and India. To our knowledge, there are only a few reports about GETV prevalence in domestic animals in China (Li et al., 2019; Lu et al., 2020; Xing et al., 2020), and no information on the epidemiology of GETV has been reported in domestic animals in Xinjiang. Therefore, to understand the risk of GETV infection in the breeding industry, we conducted a surveillance of GETV infection among domestic animals in Xinjiang, China.
2. Materials and methods
2.1. Collection of animal sera
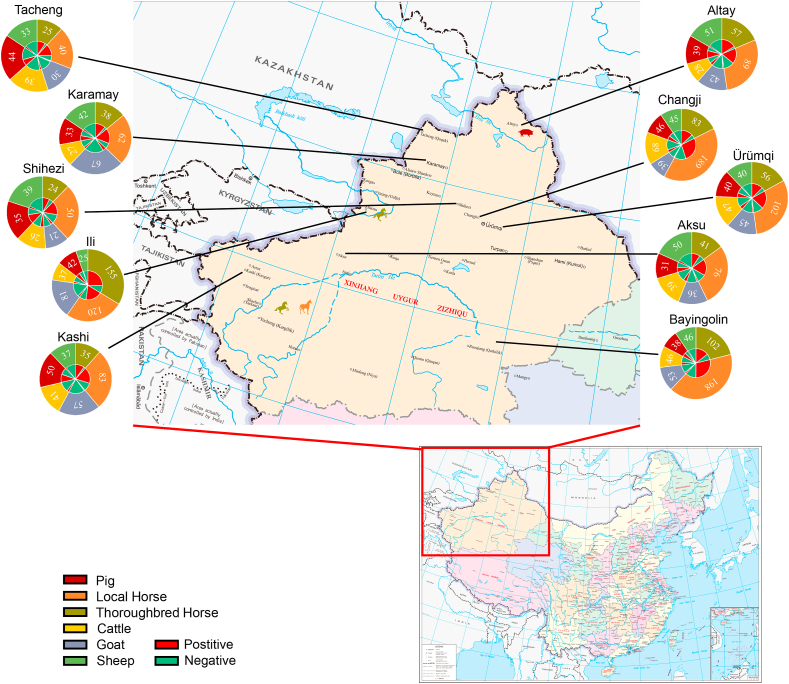
We conducted our study in ten cities of Xinjiang (Urumqi, Changji, Tacheng, Karamay, Altay, Shihezi, Ili, Aksu, Bayingolin and Kashi) during 2017–2020 (Fig. 1). A total of 3299 serum specimens were collected from six species: thoroughbred horse (616), local horse (1009), sheep (408), goat (471), cattle (398) and pig (397). Samples were originally received for animal health check analyses. All serum samples were recorded and testing results were classified by location, age and holding.
Fig. 1.
Molecular and serological surveillance of GETV. Different colors in the outer ring of the filled circles represent samples collected from individual species. Colors of the inner ring of the filled circle represent positive and negative rates of neutralizing antibody. Figures represent the number of samples.
2.2. Detection of GETV RNA
RNA was extracted from randomly selected 868 serum samples using a QIAamp Viral RNA Mini Kit (Qiagen). Quantitative RT-PCR was performed using RNA from all serum samples as described elsewhere (Shi et al., 2018).
2.3. Isolation of GETV
The positives for GETV, based on quantitative RT-PCR, were passed through 0.22-μm filters (Corning) and the filtrates were inoculated onto monolayers of three types of cultured cells: mammalian cells Vero-E6 and BHK-21 and mosquito cell C6/36. The cytopathogenic effect was observed within 96 h. When no CPE was evident, cells were blind-passaged five times.
2.4. Determination and phylogenetic analysis of GETV genome sequences
To determine the full genome sequences of isolated GETV, we used conventional RT-PCR (Li et al., 2017). To analyze the phylogenetic relationships of the complete genome of GETV, available sequences of the GETV strains were obtained from the National Center for Biotechnology Information (NCBI) (Supplementary Table S1). Establishment of a matrix alignment for almost and analysis of the aligned matrix data was performed using MEGA 7 software. Evolutionary history was inferred using the maximum likelihood method with the Tamura-Nei model and gamma-distributed rate heterogeneity in MEGA 7. The percentage of replicates in which the associated virus clustered together in the bootstrap test (1000 replicates) is shown next to the branch in each tree.
2.5. Enzyme-linked immunosorbent assay and virus neutralization test
To investigate the seroprevalence of GETV infection among domestic animals in Xinjiang, detection of anti-GETV antibodies was performed with ELISA (Bannai et al., 2019). The neutralizing antibody test was used to determine the serum neutralizing antibody titer with positive ELISA results (Li et al., 2019). Briefly, after inactivation at 56 °C for 30 min, serum specimens were diluted from 1:8 to 1:2048 and mixed with an equal volume containing 200 plaque-forming units (pfu) of GETV (Strain: SD17/09, Genbank: MH106780). After incubation at 37 °C for 1 h, the mixture was added to individual wells in a six-well plate containing a Vero-E6 cell monolayer, and incubated for 1 h. Subsequently, 1.3% methyl cellulose-DMEM medium containing 2% FBS was added (5 mL per well) and the cells were cultured for 3–4 more days. The plaques were stained with crystal violet and counted. The neutralizing antibody titer was taken as the highest dilution that reduced plaque formation by 90%.
3. Results
3.1. Molecular surveillance
The RT-PCR results of randomly selected 868 serum samples showed that four serum samples from horses and pigs collected from Altay, Ili and Kashi were positive for GETV (Fig. 1, Table 1). Total nucleic acid positive rates were 1.0% (2/200), 0.4% (1/210), 0.7% (1/128) in thoroughbred horse, local horse and pig, respectively. No viral RNA was detected in sheep (120), goat (100) and cattle samples (110).
Table 1.
Information on the four GETV positive samples in this study.
| Species | Total (%) | Year (%) |
City (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 2017 | 2018 | 2019 | 2020 | Altay | Ili | Kashi | Others | ||
| Thoroughbred horse | 2/200(1)a,b | 0/50(0) | 1/50(2) | 1/50(2) | 0/50(0) | 0/20(0) | 1/20(5)a | 1/20(5)b | 0/140(0) |
| Local horse | 1/210(0.4)c | 0/60(0) | 0/50(0) | 1/50(2)# | 0/50(0) | 0/20(0) | 0/20(0) | 1/20(5)c | 0/150(0) |
| Goat | 0/100(0) | 0/25(0) | 0/25(0) | 0/25(0) | 0/25(0) | 0/10(0) | 0/10(0) | 0/10(0) | 0/70(0) |
| Sheep | 0/120(0) | 0/30(0) | 0/30(0) | 0/30(0) | 0/30(0) | 0/10(0) | 0/10(0) | 0/10(0) | 0/90(0) |
| Cattle | 0/110(0) | 0/30(0) | 0/30(0) | 0/25(0) | 0/25(0) | 0/10(0) | 0/10(0) | 0/10(0) | 0/80(0) |
| Pig | 1/128(0.7)d | 1/38(0.3)d | 0/30(0) | 0/30(0) | 0/30(0) | 1/10(10)d | 0/10(0) | 0/10(0) | 0/98(0) |
XJ-2018-06 strain.
XJ-2019-06 strain.
XJ-2019-07 strain.
XJ-2017-08 strain.
3.2. Isolation of GETV
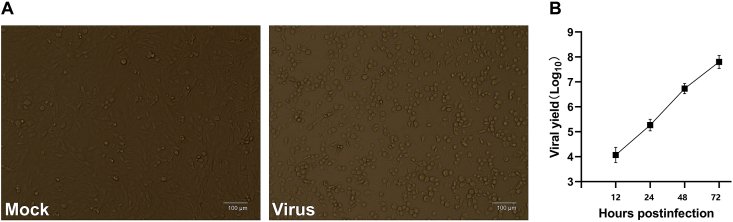
GETV was isolated from a sero-positive local horse and was named as XJ-2019-07. CPEs produced by infection of Vero-E6 cell monolayers included cell aggregation, shrinking, followed by detachment, with > 95% of cells. Peak titers reached 107.8 TCID50/mL by 72 h (Fig. 2).
Fig. 2.
Characteristics of isolated GETV. A CPE of strain XJ-2019-07 at 60 h. B The viral titers (TCID50/mL) were determined at 12, 24, 48, and 72 h. The graph shows the mean of three different experiments.
3.3. Complete genome sequencing and mutation analysis of GETV XJ-2019-07
A nearly complete genome of this strain (GenBank accession no. MZ388464) was obtained from the first generation virus isolate using conventional RT-PCR (Kanamitsu et al., 1979). The nucleotide identity of the genome between XJ-2019-07 and other GETV strains was calculated (Table 2). The results showed that XJ-2019-07 had a nucleotide identity of 95.1%–99.1% with other GETV strains at the genome level, and had the lowest homology (95.1%) with the original strain MM2021 and the highest homology (99.1%) with strain SC266, which was recently isolated from pig in China. In addition, XJ-2019-07 had a nucleotide identity of 98.2%–98.5% with other horse-derived GETV strains at the genome level, with the highest homology (98.5%) with 14-I-605-C1, 15-I-752, 16-I-599, which was detected in Japan in 2014–2016, and the lowest homology (98.2%) with strain GZ201808, which were detected in China in 2018. It has no special amino acid sites mutation. Like most strains, strain XJ-2019-07 revealed amino acid substitutions S27F, T90V and D262N in the E2 protein, which is different from the original strain MM2021.
Table 2.
Nucleotide/amino acid identity of the genome and nonstructural and structural polyprotein coding sequences between XJ-2019-07 and other GETV strains.
| No. | GenBank accession No. | Strain | Complete genome |
Non-structural polyprotein |
Structural polyprotein |
||
|---|---|---|---|---|---|---|---|
| nt | nt | aa | nt | aa | |||
| 1 | AB032553.1 | Sagiyama virus | 97.0 | 97.2 | 98.9 | 96.9 | 98.5 |
| 2 | KY434327.1 | YN12031 | 96.1 | 96.3 | 98.5 | 97.3 | 98.5 |
| 3 | EF631998.1 | LEIV 16275 Mag | 97.3 | 97.5 | 99.1 | 97.1 | 99.3 |
| 4 | AB859822.1 | Kochi/01/2005 | 97.6 | 97.8 | 99.2 | 98.3 | 99.4 |
| 5 | AY702913.1 | South Korea | 98.6 | 98.7 | 99.4 | 98.4 | 99.7 |
| 6 | EF011023.1 | Alphavirus M1 | 97.7 | 97.9 | 98.9 | 97.5 | 98.9 |
| 7 | EF631999.1 | LEIV 17741 MPR | 98.3 | 98.3 | 99.1 | 98.2 | 99.8 |
| 8 | EU015062.1 | HB0234 | 98.5 | 98.7 | 99.2 | 98.5 | 99.4 |
| 9 | EU015063.1 | YN0540 | 98.3 | 98.4 | 99.3 | 98.4 | 99.7 |
| 10 | KY363862.1 | HNJZ-S1 | 98.5 | 98.6 | 99.2 | 98.6 | 99.8 |
| 11 | KY399029.1 | GETV-V1 | 98.4 | 98.5 | 99.4 | 98.8 | 99.5 |
| 12 | KY450683.1 | YN12042 | 97.9 | 98.2 | 99.3 | 98.2 | 99.6 |
| 13 | LC079086.1 | MI-110-C1 | 98.3 | 98.4 | 99.4 | 98.9 | 99.7 |
| 14 | LC079088.1 | 14-I-605-C1 | 98.5 | 98.6 | 99.4 | 98.8 | 99.7 |
| 15 | LC107870.1 | SC1210 | 98.2 | 98.3 | 99.3 | 98.3 | 99.5 |
| 16 | LC152056.1 | 12IH26 | 98.6 | 98.6 | 99.4 | 98.5 | 99.7 |
| 17 | LC212972.1 | 15-I-752 | 98.5 | 98.6 | 99.3 | 98.6 | 99.7 |
| 18 | LC223130.1 | 16-I-599 | 98.5 | 98.5 | 99.2 | 98.1 | 99.6 |
| 19 | MF741771.1 | HuN1 | 97.4 | 97.4 | 99.1 | 97.9 | 99.3 |
| 20 | MG865965.1 | AH9192 | 98.1 | 98.3 | 99.2 | 98.1 | 99.2 |
| 21 | MG865966.1 | HNNY-1 | 98.5 | 98.6 | 99.3 | 98.0 | 99.8 |
| 22 | MG865968.1 | HNPDS-1 | 98.5 | 98.6 | 99.4 | 98.5 | 99.8 |
| 23 | MH106780.1 | SD17/09 | 97.5 | 97.8 | 99.2 | 98.3 | 99.2 |
| 24 | MH722255.1 | JL1707 | 98.4 | 98.6 | 99.3 | 98.2 | 99.4 |
| 25 | MH722256.1 | JL1808 | 97.6 | 97.8 | 99.3 | 97.9 | 99.4 |
| 26 | MK487997.1 | GZ201808 | 98.2 | 98.4 | 99.6 | 98.8 | 99.2 |
| 27 | MK693225.1 | SC201807 | 99.0 | 99.3 | 99.3 | 98.5 | 99.8 |
| 28 | NC 006558.1 | Getah virus | 98.6 | 99.2 | 99.4 | 98.5 | 99.7 |
| 29 | MN849355.1 | MM2021 | 95.1 | 95.4 | 98.6 | 97.1 | 98.2 |
| 30 | MT269657.1 | GX201808 | 97.1 | 97.3 | 99.0 | 97.9 | 99.0 |
| 31 | MN478486.1 | SC483 | 99.0 | 99.3 | 99.4 | 98.9 | 99.6 |
| 32 | MN478487.1 | SC266 | 99.1 | 99.4 | 99.6 | 99.0 | 99.6 |
| 33 | MT086508.1 | GDFS2-2018 | 98.5 | 98.5 | 99.2 | 98.6 | 99.4 |
| 34 | LC534253.1 | SW_boar_Thailand | 96.0 | 96.3 | 98.7 | 97.2 | 98.3 |
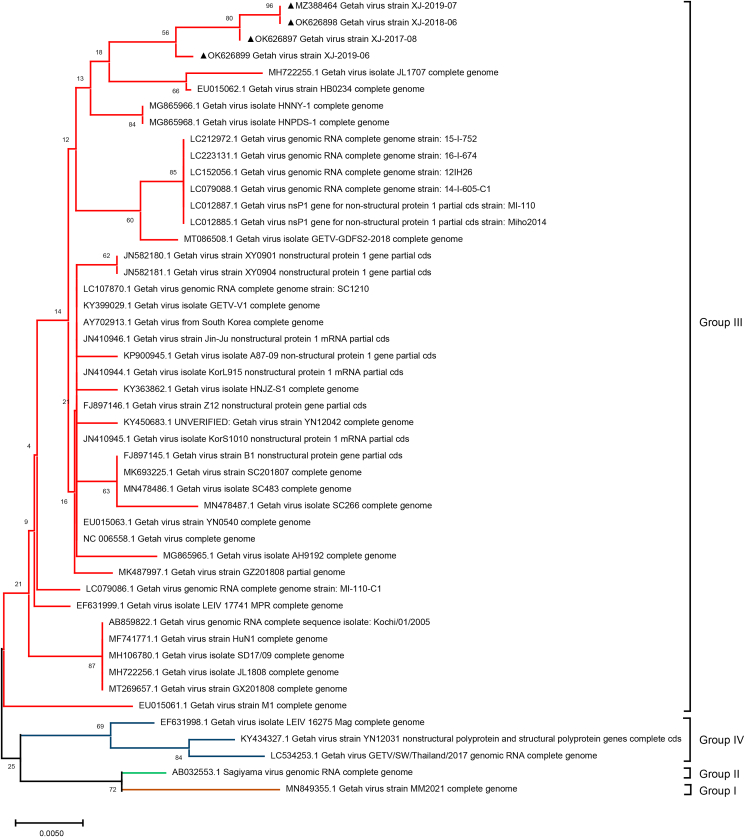
3.4. Phylogenetic analysis of GETV XJ-2019-07
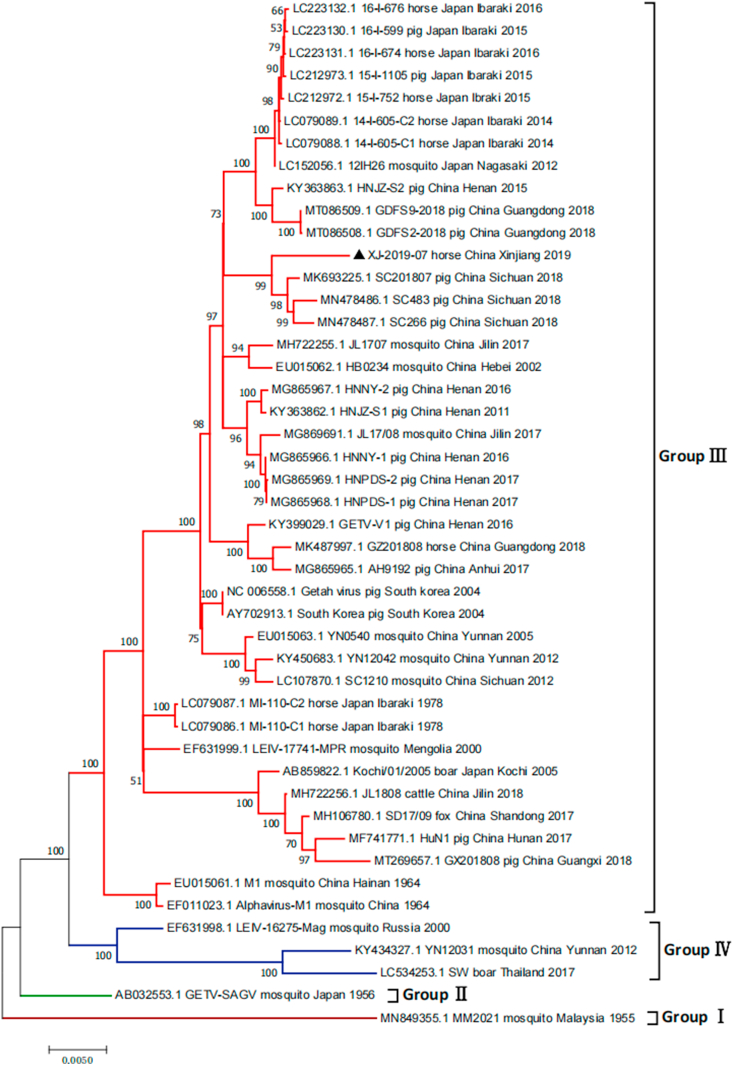
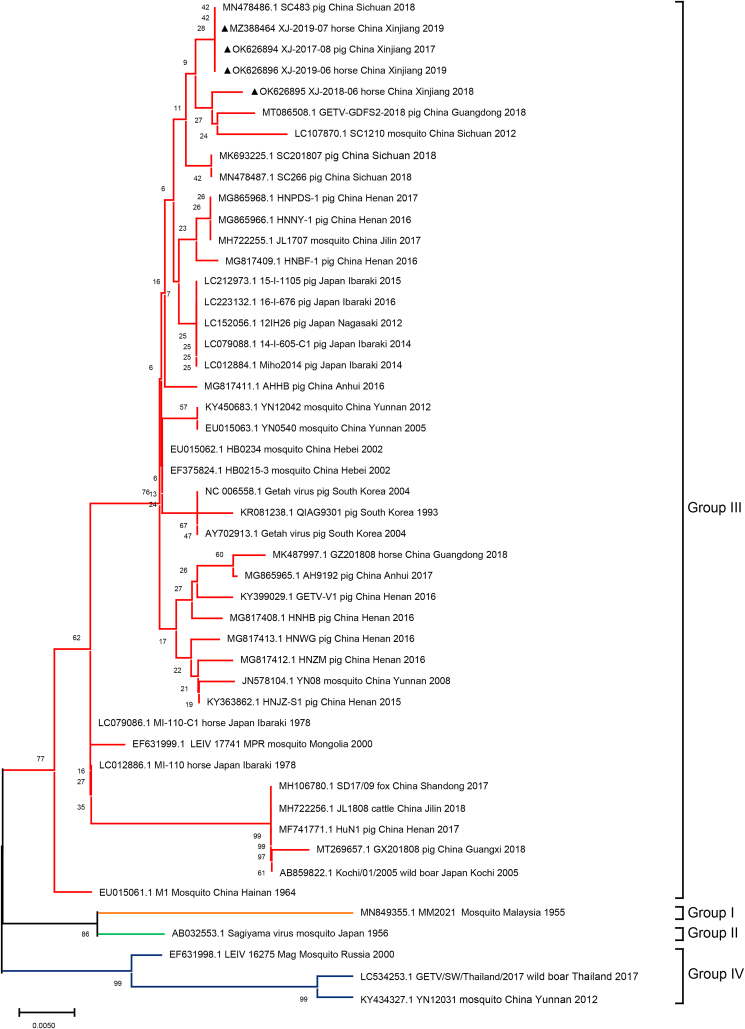
We analyzed the sequences of the NSP1 (381 bp) and capsid (552 bp) genes of four positive samples and the complete genome of XJ-2019-07. Phylogenetic analysis showed that GETVs were divided into four evolutionary groups (Fig. 3, Fig. 4, and Fig. 5). Group I only had the oldest GETV strain (MM2021) isolated in 1964. One strain isolated in Japan in 1956 formed Group II. Most of the GETV strains isolated from mosquitoes, pigs, horses, cattle, and other animals, including the strain XJ-2019-07 in this study were classified as Group III. Three GETV strains, YN12031, LEIV/16275/Mag and SW_boar_Thailand were classified as Group IV.
Fig. 3.
Phylogenetic analyses of complete genome of GETV. The strain isolated in this study is labeled by triangle. XJ-2019-07 GenBank accession no. MZ388464.
Fig. 4.
Phylogenetic analyses of Cap gene nucleotide sequences of GETV. The strain isolated in this study is labeled by triangle.
Fig. 5.
Phylogenetic analyses of Nsp1 gene nucleotide sequences of GETV. The strain isolated in this study is labeled by triangle.
3.5. Seroprevalence of GETV infection
According to the ELISA results, total positive rates of anti-GETV antibodies were 74.8% (461/616), 67.3% (679/1009), 11.7% (55/471), 10.0% (41/408), 25.1% (100/398) and 51.1% (203/397) in thoroughbred horse, local horse, goat, sheep, cattle and pig, respectively (Table 3). Geographically, these were detected in all ten cities in all species and with similar positive rates. From an age perspective, the positive rate in both thoroughbred and local horses increased with age from 0 to 80.2%. For pigs, the positive rate of pups was 85.0%, higher than for young (65.0%) or adults (43.8%). All samples from goat, sheep and cattle were from adults (11.7%, 10.0%, 25.1%, respectively). In thoroughbred and local horses, positive rate of farm animals (76.7%, 73.2%) were higher than in small holders (53.1%, 37.9%), whereas in cattle, sheep, goat and pigs these differences were not significant.
Table 3.
Seroprevalence of Getah virus (GETV) and the association of antibody occurrence in Xinjiang Uygur Autonomous Region.
| Species | Total (%) | Year (%) |
Age (%) |
Holding (%) |
Neutralizing antibodies titer (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2017 | 2018 | 2019 | 2020 | Pups | Young | Adult | Open yard or small holder | Farm | <1:64 | 1:64–1:128 | 1:128–1:512 | >1:512 | ||
| Thoroughbred horse | 461/616 (74.8) | 115/150 (76.7) | 135/198 (68.1) | 136/178 (76.4) | 75/90 (83.3) | 0/23 (0) | 1/20 (2.5) | 460/573 (80.2) | 26/49 (53.1) | 435/567 (76.7) | 71/461 (15.4) | 80/461 (17.4) | 200/461 (43.3) | 110/461 (23.9) |
| Local horse | 679/1009 (67.3) | 163/289 (56.4) | 259/352 (73.6) | 168/279 (60.2) | 89/123 (72.4) | 0/21 (0) | 2/23 (8.7) | 677/965 (70.2) | 64/169 (37.9) | 615/840 (73.2) | 148/679 (21.8) | 149/679 (21.9) | 286/679 (42.1) | 96/679 (14.1) |
| Goat | 55/471 (11.7) | 17/103 (16.5) | 14/147 (9.5) | 19/150 (12.7) | 5/71 (7.0) | NT | NT | 55/471 (11.7) | 2/18 (11.1) | 53/453 (11.7) | 55/55 (100) | – | – | – |
| Sheep | 41/408 (10.0) | 8/110 (7.2) | 12/150 (8.0) | 15/103 (14.6) | 6/45 (13.3) | NT | NT | 41/408 (10.0) | 3/21 (14.3) | 38/387 (9.8) | 41/41 (100) | – | – | – |
| Cattle | 100/398 (25.1) | 32/110 (29.1) | 33/132 (25.0) | 26/103 (25.2) | 9/53 (17.0) | NT | NT | 100/398 (25.1) | 2/18 (11.1) | 98/380 (25.8) | 90/100 (90.0) | 10/100 (10.0) | – | – |
| Pig | 203/397 (51.1) | 80/150 (53.3) | 73/147 (49.7) | 23/60 (38.3) | 27/40 (67.5) | 34/40 (85.0) | 39/60 (65.0) | 130/297 (43.8) | 5/30 (16.7) | 198/367 (54.0) | 160/203 (78.8) | 30/203 (14.8) | 13/203 (6.4) | – |
NT: not tested because sample was not available. -: negative.
Differences in titer of neutralizing antibodies were compared among various animal specimens. Neutralizing antibody titers varied by specimen: low titers (<1:128) were detected in serum samples from sheep, goat and cattle, whereas titers of more than 1:512 were observed in thoroughbred horse and local horse. These accounted for 23.9% (110/461) and 14.1% (96/679) of the total positive specimens, respectively. The serum titers of pigs were lower than 1:512, most of them were in 1:64 and accounted for 6.4% (13/203) (Table 3).
4. Discussion
In the present study, we report for the first time the epidemiology of GETV infection among domestic animals in Xinjiang. We also report the first isolation of GETV in Xinjiang.
To find serum samples containing GETV, we performed RT-PCR using a universal primer and found four positive pig serum sample. From one serum of local horse, we successfully isolated XJ-2019-07 and determined the complete genome sequence. The phylogenetic tree shows that all strain are close to the strains from China and belong to the Group III lineage. It shows that GETV probably has been prevalent for several years.
ELISA results showed that positive rates of GETV antibodies varied from 10.0% to 74.8% in six domestic animals, with no significant species difference among cities. The time distribution analysis showed that all the species were positive during 2017–2020, with slightly different positive rates, although this may be related to the outbreak of COVID-19 and African swine fever. Given that infections have been observed in a wide range of vertebrates (Li et al., 2019; Liu et al., 2019; Nemoto et al., 2015; Shi et al., 2019; Yago et al., 1987; Yang et al., 2018), it is not surprising that GETV infects a variety of hosts in Xinjiang.
In our study, goat had low positive rates (11.7%), similar to Yunnan Province (Li et al., 2019). In contrast, the positive rates in pigs and horse were high, similar to results reported in Japan and Korea (Nemoto et al., 2015; Yago et al., 1987). Cattle presented low positive rates (25.1%) and this contrasts with results from Yunnan and Jilin provinces, possibly because differences in number of samples, climate, environment or ecology (Li et al., 2019; Liu et al., 2019). We also found the antibody in sheep, which suggests that a variety of domestic animals can potentially act as hosts for GETV.
In India and Korea, horse antibodies to GETV have been reported with an average of 17% (10%28%) and 37%–47%, respectively (Brown and Timoney, 1998; Kuwata et al., 2018; Rhee et al., 1986). In Japan the range is wider, from 7.5% to 93% (Bannai et al., 2017; Sugiyama et al., 2009). However, in our study, positive rates of GETV in thoroughbred and local horses were higher than in India and South Korea, but lower than in some regions of Japan. This suggests a high GETV transmission in Xinjiang, although the possibility that positive thoroughbred horses were imported from countries like Japan cannot be ruled out (Li et al., 2017). Indeed, Xinjiang is a major province of animal husbandry and borders on eight countries (Zhou et al., 2019), therefore constantly GETV monitoring should be strengthened. Thoroughbred and local horses differed in their rates of positivity for the antibody, as well as in their antibody titers, suggesting response to GETV infection may differ for subspecies.
The positive rate of both thoroughbred and local horses increased while with age, whereas in pigs the trend was the opposite. This may be related to differences in sampling time and growth cycle between these species. Specifically, a factor could be maternal antibodies; from foals, we collected samples after weaning for several months, whereas piglets had just been weaned. Also, horses can be bred for many years and may be repeatedly infected, whereas pigs are slaughtered early for meat. Finally, the pig breeding environment is much more intensive than for horses. The limited number of samples for some of the species (e.g., sheep, goat and cattle) may have also led to an underestimate of the seroprevalence level. Thus, a long-term serological survey with larger samples is needed.
Interestingly, there was a difference in prevalence in horses and pigs under different breeding regimes. Indeed, seroprevalence in farm-bred horses reached 76.7%, possibly because of the intensive breeding environment in large-scale farms which accelerate the spread of GETV in the population.
No viral RNA was detected in sheep, goat and cattle samples. We speculate that GETV might have been transferred to other regions through mosquitoes or migratory birds. Indeed, many strains have been isolated from mosquitoes (Li et al., 2019) and both herons and egrets have antibodies against “Sagiyama virus”, synonym of GETV (Scherer et al., 1962).
5. Conclusions
In summary, we show that GETV is epidemic in domestic animals in Xinjiang, and for the first time it has been found in sheep. Overall, GETV is prevalent in Xinjiang and probably has been for several years. Since no vaccine against GETV is available in China, detection and monitoring strategies should be improved in horses and pigs, especially imported and farmed, in order to prevent economic losses.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. The raw sequence data reported in this article have been deposited in NCBI (accession numbers: MZ388464, OK626894–OK626899).
Ethics statement
The protocol of this study was approved by the Animal Ethical and Welfare Committee, Northwest A&F University, China. All Serum samples for the study were collected with the animal owner's approval, and the experiments were conducted under the guidelines of Northwest A&F University for animal experiments.
Author contributions
Ning Shi: investigation, data curation, formal analysis, writing-original draft. Xiangshu Qiu: investigation, validation. Xinyu Cao: investigation. Zhanhai Mai: resources, methodology. Xiangyu Zhu: software, supervision. Nan Li: validation, visualization. He Zhang: writing-review&editing. Jinyong Zhang: writing-review&editing. Jinyong Zhang: validation, writing-review&editing. Zhuoxin Li: validation, writing-review&editing. Nuerlan Shaya: resources, methodology. Huijun Lu: funding acquisition, methodology, conceptualization, methodology. Ningyi Jin: funding acquisition, methodology, conceptualization, methodology.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This work was supported by the National Program on Key Research Project of China (2018YFD0500104 and 2018YFD0500803), and Technologies for Prevention and Control of Virus Zoonoses, Chinese Academy of Medical Sciences (2020–12M-5-001).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.02.004.
Contributor Information
Huijun Lu, Email: huijun_lu@126.com.
Ningyi Jin, Email: ningyik@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bannai H., Nemoto M., Niwa H., Murakami S., Tsujimura K., Yamanaka T., Kondo T. Geospatial and temporal associations of Getah virus circulation among pigs and horses around the perimeter of outbreaks in Japanese racehorses in 2014 and 2015. BMC Vet. Res. 2017;13:187. doi: 10.1186/s12917-017-1112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H., Nemoto M., Tsujimura K., Yamanaka T., Kokado H. Development of an enzyme-linked immunosorbent assay for Getah virus infection in horses using recombinant E2 protein as an antigen. J. Virol. Methods. 2019;271:113681. doi: 10.1016/j.jviromet.2019.113681. [DOI] [PubMed] [Google Scholar]
- Brown C.M., Timoney P.J. Getah virus infection of Indian horses. Trop. Anim. Health Prod. 1998;30:241–252. doi: 10.1023/a:1005079229232. [DOI] [PubMed] [Google Scholar]
- Bryant J.E., Crabtree M.B., Nam V.S., Yen N.T., Duc H.M., Miller B.R. Isolation of arboviruses from mosquitoes collected in northern Vietnam. Am. J. Trop. Med. Hyg. 2005;73:470–473. [PubMed] [Google Scholar]
- Burt F.J., Chen W., Miner J.J., Lenschow D.J., Merits A., Schnettler E., Kohl A., Rudd P.A., Taylor A., Herrero L.J., Zaid A., Ng L., Mahalingam S. Chikungunya virus: an update on the biology and pathogenesis of this emerging pathogen. Lancet Infect. Dis. 2017;17:e107–e117. doi: 10.1016/S1473-3099(16)30385-1. [DOI] [PubMed] [Google Scholar]
- Fukunaga Y., Kumanomido T., Kamada M. Getah virus as an equine pathogen. Vet. Clin. N. Am. Equine Pract. 2000;16:605–617. doi: 10.1016/s0749-0739(17)30099-8. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Bugallo G., Piedra L.A., Rodriguez M., Bisset J.A., Lourenco-de-Oliveira R., Weaver S.C., Vasilakis N., Vega-Rua A. Vector-borne transmission and evolution of Zika virus. Nat. Ecol. Evol. 2019;3:561–569. doi: 10.1038/s41559-019-0836-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada M., Kumanomido T., Wada R., Fukunaga Y., Imagawa H., Sugiura T. Intranasal infection of Getah virus in experimental horses. J. Vet. Med. Sci. 1991;53:855–858. doi: 10.1292/jvms.53.855. [DOI] [PubMed] [Google Scholar]
- Kanamitsu M., Taniguchi K., Urasawa S., Ogata T., Wada Y., Wada Y., Saroso J.S. Geographic distribution of arbovirus antibodies in indigenous human populations in the Indo-Australian archipelago. Am. J. Trop. Med. Hyg. 1979;28:351–363. doi: 10.4269/ajtmh.1979.28.351. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Trosper J.H., Cross J.H., Basaca-Sevilla V. Isolation of Getah virus from Nueva Ecija province, republic of the Philippines. Trans. R. Soc. Trop. Med. Hyg. 1981;75:312–313. doi: 10.1016/0035-9203(81)90346-1. [DOI] [PubMed] [Google Scholar]
- Kuwata R., Shimoda H., Phichitraslip T., Prasertsincharoen N., Noguchi K., Yonemitsu K., Minami S., Supriyono, Tran N., Takano A., Suzuki K., Nemoto M., Bannai H., Yokoyama M., Takeda T., Jittapalapong S., Rerkamnuaychoke W., Maeda K. Getah virus epizootic among wild boars in Japan around 2012. Arch. Virol. 2018;163:2817–2821. doi: 10.1007/s00705-018-3897-4. [DOI] [PubMed] [Google Scholar]
- Li X.D., Qiu F.X., Yang H., Rao Y.N., Calisher C.H. Isolation of Getah virus from mosquitos collected on Hainan Island, China, and results of a serosurvey. Southeast Asian J. Trop. Med. Publ. Health. 1992;23:730–734. [PubMed] [Google Scholar]
- Li Y.Y., Liu H., Fu S.H., Li X.L., Guo X.F., Li M.H., Feng Y., Chen W.X., Wang L.H., Lei W.W., Gao X.Y., Lv Z., He Y., Wang H.Y., Zhou H.N., Wang G.Q., Liang G.D. From discovery to spread: the evolution and phylogeny of Getah virus. Infect. Genet. Evol. 2017;55:48–55. doi: 10.1016/j.meegid.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Li Y., Fu S., Guo X., Li X., Li M., Wang L., Gao X., Lei W., Cao L., Lu Z., He Y., Wang H., Zhou H., Liang G. Serological survey of Getah virus in domestic animals in Yunnan province, China. Vector Borne Zoonotic Dis. 2019;19:59–61. doi: 10.1089/vbz.2018.2273. [DOI] [PubMed] [Google Scholar]
- Liu H., Zhang X., Li L.X., Shi N., Sun X.T., Liu Q., Jin N.Y., Si X.K. First isolation and characterization of Getah virus from cattle in northeastern China. BMC Vet. Res. 2019;15:320. doi: 10.1186/s12917-019-2061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Chen R., Shao R., Dong N., Liu W., Li S. Getah virus: an increasing threat in China. J. Infect. 2020;80:350–371. doi: 10.1016/j.jinf.2019.11.016. [DOI] [PubMed] [Google Scholar]
- Lu G., Ou J., Ji J., Ren Z., Hu X., Wang C., Li S. Emergence of Getah virus infection in horse with fever in China, 2018. Front. Microbiol. 2019;10:1416. doi: 10.3389/fmicb.2019.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Vov S D., Gromashevski'I V.L., Aristova V.A., Morozova T.N., Skvortsova T.M., Gushchina E.A., Petrova E.S., L'Vov D.K. Isolation of Getah virus (Togaviridae, Alfavirus) strains in North- Eastern Asia. Vopr. Virusol. 2000;45:14–18. (In Russian) [PubMed] [Google Scholar]
- Messina J.P., Brady O.J., Scott T.W., Zou C., Pigott D.M., Duda K.A., Bhatt S., Katzelnick L., Howes R.E., Battle K.E., Simmons C.P., Hay S.I. Global spread of dengue virus types: mapping the 70 year history. Trends Microbiol. 2014;22:138–146. doi: 10.1016/j.tim.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Igarashi A. Oligonucleotide fingerprint analysis of strains of Getah virus isolated in Japan and Malaysia. J. Gen. Virol. 1984;65(Pt 11):1899–1908. doi: 10.1099/0022-1317-65-11-1899. [DOI] [PubMed] [Google Scholar]
- Nemoto M., Bannai H., Tsujimura K., Kobayashi M., Kikuchi T., Yamanaka T., Kondo T. Getah virus infection among Racehorses, Japan, 2014. Emerg. Infect. Dis. 2015;21:883–885. doi: 10.3201/eid2105.141975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T., Mo Q., Wang Y., Wang H., Nong Z., Wang J., Niu C., Liu C., Chen Y., Ouyang K., Huang W., Wei Z. Emergence and phylogenetic analysis of a Getah virus isolated in Southern China. Front. Vet. Sci. 2020;7:552517. doi: 10.3389/fvets.2020.552517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee Y.O., Heo Y., Kim Y.H. Serological survey of horses in Korea for evidence of Getah virus infection. Korean J. Vet. Res. 1986;26:93–96. [Google Scholar]
- Scherer W.F., Funkenbusch M., Buescher E.L., Izumi T. Sagiyama virus, a new group A arthropod-borne virus from Japan. I. Isolation, immunologic classification, and ecologic observations. Am. J. Trop. Med. Hyg. 1962;11:255–268. doi: 10.4269/ajtmh.1962.11.255. [DOI] [PubMed] [Google Scholar]
- Shi N., Li L.X., Lu R.G., Yan X.J., Liu H. Highly pathogenic swine Getah virus in blue foxes, Eastern China, 2017. Emerg. Infect. Dis. 2019;25:1252–1254. doi: 10.3201/eid2506.181983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi N., Liu H., Li L.X., Hu B., Zhang L., Zhao C.F., Deng X.Y., Li X.T., Xue X.H., Bai X., Zhang H.L., Lu R.G., Lian S.Z., Wang Y., Yan M.H., Yan X.J. Development of a TaqMan probe-based quantitative reverse transcription PCR assay for detection of Getah virus RNA. Arch. Virol. 2018;163:2877–2881. doi: 10.1007/s00705-018-3927-2. [DOI] [PubMed] [Google Scholar]
- Sugiyama I., Shimizu E., Nogami S., Suzuki K., Miura Y., Sentsui H. Serological survey of arthropod-borne viruses among wild boars in Japan. J. Vet. Med. Sci. 2009;71:1059–1061. doi: 10.1292/jvms.71.1059. [DOI] [PubMed] [Google Scholar]
- Turell M.J., O'Guinn M.L., Wasieloski L.J., Dohm D.J., Lee W.J., Cho H.W., Kim H.C., Burkett D.A., Mores C.N., Coleman R.E., Klein T.A. Isolation of Japanese encephalitis and Getah viruses from mosquitoes (Diptera: Culicidae) collected near camp greaves, gyonggi province, Republic of Korea, 2000. J. Med. Entomol. 2003;40:580–584. doi: 10.1603/0022-2585-40.4.580. [DOI] [PubMed] [Google Scholar]
- Xing C., Jiang J., Lu Z., Mi S., He B., Tu C., Liu X., Gong W. Isolation and characterization of Getah virus from pigs in Guangdong province of China. Transbound Emerg. Dis. 2020;67:2249–2253. doi: 10.1111/tbed.13567. [DOI] [PubMed] [Google Scholar]
- Yago K., Hagiwara S., Kawamura H., Narita M. A fatal case in newborn piglets with Getah virus infection: isolation of the virus. Nihon Juigaku Zasshi. 1987;49:989–994. doi: 10.1292/jvms1939.49.989. [DOI] [PubMed] [Google Scholar]
- Yang T., Li R., Hu Y., Yang L., Zhao D., Du L., Li J., Ge M., Yu X. An outbreak of Getah virus infection among pigs in China, 2017. Transbound Emerg. Dis. 2018;65:632–637. doi: 10.1111/tbed.12867. [DOI] [PubMed] [Google Scholar]
- Zhou H., Ma Z., Hu T., Bi Y., Mamuti A., Yu R., Carr M.J., Shi M., Li J., Sharshov K., Gao G.F., Shi W. Tamdy virus in ixodid ticks infesting bactrian camels, Xinjiang, China, 2018. Emerg. Infect. Dis. 2019;25:2136–2138. doi: 10.3201/eid2511.190512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. The raw sequence data reported in this article have been deposited in NCBI (accession numbers: MZ388464, OK626894–OK626899).