Abstract
Environmental contamination with compounds containing oxyanions of chlorine, such as perchlorate or chlorate [(per)chlorate] or chlorine dioxide, has been a constantly growing problem over the last 100 years. Although the fact that microbes reduce these compounds has been recognized for more than 50 years, only six organisms which can obtain energy for growth by this metabolic process have been described. As part of a study to investigate the diversity and ubiquity of microorganisms involved in the microbial reduction of (per)chlorate, we enumerated the (per)chlorate-reducing bacteria (ClRB) in very diverse environments, including pristine and hydrocarbon-contaminated soils, aquatic sediments, paper mill waste sludges, and farm animal waste lagoons. In all of the environments tested, the acetate-oxidizing ClRB represented a significant population, whose size ranged from 2.31 × 103 to 2.4 × 106 cells per g of sample. In addition, we isolated 13 ClRB from these environments. All of these organisms could grow anaerobically by coupling complete oxidation of acetate to reduction of (per)chlorate. Chloride was the sole end product of this reductive metabolism. All of the isolates could also use oxygen as a sole electron acceptor, and most, but not all, could use nitrate. The alternative electron donors included simple volatile fatty acids, such as propionate, butyrate, or valerate, as well as simple organic acids, such as lactate or pyruvate. Oxidized-minus-reduced difference spectra of washed whole-cell suspensions of the isolates had absorbance maxima close to 425, 525, and 550 nm, which are characteristic of type c cytochromes. In addition, washed cell suspensions of all of the ClRB isolates could dismutate chlorite, an intermediate in the reductive metabolism of (per)chlorate, into chloride and molecular oxygen. Chlorite dismutation was a result of the activity of a single enzyme which in pure form had a specific activity of approximately 1,928 μmol of chlorite per mg of protein per min. Analyses of the 16S ribosomal DNA sequences of the organisms indicated that they all belonged to the alpha, beta, or gamma subclass of the Proteobacteria. Several were closely related to members of previously described genera that are not recognized for the ability to reduce (per)chlorate, such as the genera Pseudomonas and Azospirllum. However, many were not closely related to any previously described organism and represented new genera within the Proteobacteria. The results of this study significantly increase the limited number of microbial isolates that are known to be capable of dissimilatory (per)chlorate reduction and demonstrate the hitherto unrecognized phylogenetic diversity and ubiquity of the microorganisms that exhibit this type of metabolism.
Environmental contamination with compounds containing oxyanions of chlorine, such as perchlorate (ClO4−) or chlorate (ClO3−) [(per)chlorate], chlorite (ClO2−), and chlorine dioxide (ClO2), has been a constantly growing problem over the last 100 years (45, 46). In general, these compounds are not formed naturally and have been introduced into the environment in large quantities in the form of disinfectants, bleaching agents, and herbicides (1, 17, 38). Historical legal discharge of perchlorate-containing waste streams from munitions manufacturing and handling facilities has recently been identified as the predominant source of the perchlorate found in major drinking water supplies in the United States (32, 46). Chlorate is often a by-product of disproportionation reactions and photodecomposition of chlorine dioxide, chlorine, and chlorite used by drinking water suppliers and paper industries (4, 17, 39). In addition, the United States military, due to downsizing and periodic replacement of active military inventory having a limited shelf life, is expected to have more than 164 million pounds of perchlorate-containing rocket propellant that will require disposal over the next decade (49).
Perchlorate has been shown to affect iodide accumulation in the thyroid gland (40), while chlorate is toxic to brown algae at concentrations greater than 20 μg/liter. Both chlorate and chlorite have been shown to cause hemolytic anemia in laboratory animals (14, 15, 42). In 1992, the U.S. Environmental Protection Agency reviewed the health effects of perchlorate administered to patients with hyperthyroidism and found that doses of 6 μg per kg per day or more over a 2-month period resulted in fatal bone marrow changes (45). In 1998, using these data, workers at the California Department of Health Services calculated an action level of 18 μg/liter for drinking water supplies, which if exceeded required that water usage had to be stopped and remediation efforts had to begin (8, 45). This action level has now been increased by the U.S. Environmental Protection Agency to 32 μg/liter (33). Most perchlorate contamination found in the environment is the result of discharge of unregulated ammonium perchlorate-containing waste streams from rocket fuel-manufacturing plants and from the periodic servicing and maintenance of military inventories (45, 46). Perchlorate has been found in surface water and groundwater in Texas, Arkansas, Maryland, New York, California, Utah, and Nevada (46). In 1997, following the development of a highly sensitive analytical technique for determining perchlorate contents, monitoring studies revealed that perchlorate was a contaminant of major drinking water sources in the southwestern United States (32, 46). Perchlorate contamination has significantly affected California, Utah, and Nevada.
Although it has been recognized for more than 50 years that microbial reduction of chlorine oxyanions under anaerobic conditions is possible (5–7, 9, 19, 20, 23, 28, 29, 34, 41, 43, 50), relatively little known is known about the microorganisms involved in this type of respiratory metabolism. Generally, it is assumed that these organisms use either chlorate or perchlorate as a terminal electron acceptor (24), although this has been demonstrated in only a few isolated cases (5, 41, 50). The end product of the reductive metabolic process is innocuous chloride (5, 34, 50). Early studies revealed that microorganisms rapidly reduced chlorate as a competitive reaction for the nitrate reductase pathway (19, 20, 43). Chlorite was the end product, and growth was not associated with this reaction (16, 36). Until recently, only six microorganisms which can grow by dissimilatory (per)chlorate reduction had been described (5, 29, 34, 37, 41, 50). Only four of these six organisms, strain CKB (5), strain GR-1 (34), Ideonella dechloratans (29), and Wolinella succinogenes HAP-1 (50), have been studied in detail.
In order to determine the ubiquity and diversity of organisms capable of dissimilatory (per)chlorate reduction, we enumerated (per)chlorate-reducing bacteria (ClRB) in a broad spectrum of environments. We isolated 13 new ClRB from these environments. Several of the isolates obtained represent new genera in the class Proteobacteria. Our results demonstrate that dissimilatory reduction of (per)chlorate is a much more ubiquitous and diverse metabolic process than was thought previously.
MATERIALS AND METHODS
Sources of soils and sediments.
Soil samples were collected from the top 6 cm of an uncontaminated soil in Thompson Woods on the Carbondale campus of Southern Illinois University and also from a hydrocarbon-contaminated soil at Tulsa Tape Incorporated in Carbondale, Ill. In addition, sediment samples were collected from campus lake and farm swine lagoons at Southern Illinois University in Carbondale, Ill.; from the Potomac River (Pohic Bay) in Virginia; from the Mississippi River in Chester, Ill.; from gold mine drainage sediment in Hotsprings, S.D.; and from swamp lands in Reston, Fla. All samples were freshly collected and transported directly to the lab, where they were immediately assayed to determine whether ClRB were present.
Medium and culture conditions.
Standard anaerobic culture techniques were used throughout this study (2, 22, 30). The medium was boiled under N2-CO2 (80:20) to remove dissolved O2 and then dispensed into anaerobic pressure tubes or serum bottles under N2-CO2; the tubes and bottles were closed with thick butyl rubber stoppers and sterilized by autoclaving. The basal medium used was the bicarbonate-buffered freshwater medium that was used previously to grow strain CKB (5). Unless otherwise noted, sodium salts of acetate and chlorate (10 mM each) were used as the electron donor and acceptor, respectively, and were added from sterile anoxic stock solutions.
Alternative electron donors were added from sterile anoxic aqueous stock solutions. Pure aromatic hydrocarbons (benzene, hexadecane, and toluene) were added directly (1 μl per 10 ml of medium). Electron acceptors were also added from anoxic aqueous stock solutions. Soluble Fe(III) was supplied as Fe(III) chelated with nitrilotriacetic acid (10 mM) (35). Mn(IV) was supplied as synthetic MnO2 that was prepared as previously described (26), and the final concentrations were 10 to 30 mM. Sulfur was supplied as a polysulfide solution prepared as described previously (51). All other electron acceptors were prepared as anoxic aqueous stock solutions of sodium salts, and the final concentration of each was 10 mM.
Isolation of ClRB.
(Per)chlorate-reducing enrichment cultures were established by transferring 1-g subsamples from each of the freshly collected soil and sediment samples into 9 ml of prepared anoxic medium under an N2-CO2 gas stream. Acetate (10 mM) was the electron donor, and chlorate (10 mM) was the electron acceptor. The preparations were incubated at 30°C in the dark. Positive enrichment cultures were identified on the basis of an increase in optical density (determined visually) and by microscopic examination. Once a positive enrichment culture was established, the (per)chlorate-reducing culture was transferred (10% inoculum) into 9 ml of fresh anoxic medium. Isolated colonies were obtained from transfers of positive enrichment cultures by the standard agar shake tube technique described previously (5, 52); acetate was the sole electron donor, and ClO3− (10 mM) was the sole electron acceptor.
MPN. counts.
The numbers of dissimilatory ClRB in soil and sediment samples were determined by three-tube most-probable-number (MPN) counting performed with 10 mM acetate as the electron donor. The results were expressed as the number of ClRB per gram (wet weight) of sample. The medium contained (per liter) 0.25 g of NH4Cl, 1.03 g of NaClO3, 1.36 g of CH3COONa, 0.60 g of NaH2PO4, 0.1 g of KCl, and 2.5 g of NaHCO3. Vitamins (10 ml/liter) and trace metals (10 ml/liter) were added from stock solutions prepared as previously described (5). MPN series preparations were incubated at room temperature in the dark for 60 days prior to analysis. Positive results in the MPN analysis were identified on the basis of an increase in optical density (determined visually) and also by microscopic examination.
Chlorite dismutase purification and determination of activity.
Washed cell suspensions of each of the ClRB isolates were examined for chlorite dismutase activity by using a Clark O2 electrode as previously described (5, 10). In addition, chlorite dismutase was purified to homogeneity from the soluble fraction of lysed cell preparations of the previously described (5) (per)chlorate-reducing organism strain CKB. Each lysed cell preparation was prepared from a washed pellet (10 g, wet weight) of strain CKB cells grown with chlorate and acetate as the electron acceptor and electron donor, respectively. The cells were harvested by centrifugation (10,000 × g, 10 min, 4°C). The resulting cell pellet was resuspended in 14 mM phosphate buffer (pH 7.2) supplemented with 0.5 mM phenylmethylsulfonyl fluoride. The cells were broken by three passes through a French pressure cell at 20,000 lb/in2 and were treated for 60 min at room temperature with a DNase solution (10 mg/ml of homogenate; 0.1% DNase in 50 mM MgCl2 buffer). The lysed cells were centrifuged at 26,000 × g for 5 min to remove the cell debris, and the resulting supernatant was fractionated into soluble and membrane-bound protein portions by ultracentrifugation (110,000 × g, 1 h, 4°C). The pink cell extract was stored at 4°C until it was analyzed.
The enzyme was purified from the cell extract by using sequential chromatography. At each step of the purification protocol, the fractions were examined for chlorite dismutation specific activity by microassay. A 30-ml sample of cell extract was loaded onto a column (2.5 by 10 cm) packed with Q-Sepharose Fast Flow (Amersham Pharmacia Biotech, Piscataway, N.J.) medium, and the column was developed with a 0 to 300 mM KCl gradient in 50 mM Tris-HCl (pH 7.5). Fractions with chlorite dismutase activity were identified and pooled. The pooled active fractions were loaded onto a column (2.5 by 20 cm) that was packed with hydroxyapatite (Bio-Rad Laboratories, Richmond, Calif.) and was developed with a potassium phosphate buffer gradient (10 to 250 mM; pH 7.2). The resulting fractions with chlorite dismutase activity were pooled and supplemented with ammonium chloride (final concentration, 2 M) before they were loaded onto a Phenyl Sepharose high-performance column (1 by 1 cm). The Phenyl Sepharose column was developed with a descending ammonium chloride gradient (2 to 0 M) in 50 mM Tris-HCl (pH 7.5). Chlorite dismutase eluted as a pale pink fraction, which was concentrated by ultrafiltration (molecular weight cutoff, 30,000). The resulting concentrated fraction was passed through a column (1.6 by 60 cm) that was packed with Superdex 200 medium (Amersham Pharmacia Biotech) and was developed with 150 mM NaCl in 50 mM potassium phosphate buffer (pH 7.2). The pure chlorite dismutase was collected and stored at 4°C until it was analyzed.
SDS-PAGE.
Throughout the purification protocol, the purity of chlorite dismutase fractions was routinely determined by using sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). The 1.5-mm 15% polyacrylamide gels were prepared by using (per gel) 15 ml of a 30.8% acrylamide–2.7% bisacrylamide monomer solution, 7.5 ml of 1.5 M Tris-HCl (pH 8.8), 0.12 ml of 25% SDS, 7.0 ml of water, 0.15 ml of 10% ammonium persulfate, and 0.10 ml of N,N,N′,N′-tetramethylethylenediamine. The gel was poured vertically, and when it was set, a 4% acrylamide stacker gel was laid on top of it. The samples were treated with an equal volume of a buffer containing 0.125 M Tris-HCl (pH 6.8), 4% SDS, 20% (vol/vol) glycerol, 0.2 M dithiothreitol, and 0.02% bromophenol blue. The treated samples were boiled for 3 min and loaded immediately. The gels were electrophoresed at 10 mM for 16 h with a tank buffer containing 0.025 M Tris, 0.192 M glycine, and 0.1% SDS (pH 8.3). The gels were typically stained with Coomassie blue, and the final purity was confirmed by silver nitrate staining.
Molecular mass determination.
The molecular mass of chlorite dismutase was determined by gel filtration performed with a column packed with Superdex 200 (Amersham Pharmacia Biotech). The Superdex 200 column was calibrated by using the following molecular mass standards: beta-amylase (200,000 Da), alcohol dehydrogenase (150,000 Da), and bovine serum albumin (66,000 Da).
Cytochrome content.
In a preliminary investigation of the cytochrome contents of the (per)chlorate-reducing isolates, dithionite-reduced–versus–air-oxidized difference spectra were obtained for washed cell suspensions of acetate-chlorate-grown cells suspended in anoxic bicarbonate buffer (2.5 g/liter) that was sparged with N2-CO2 (80:20, vol:vol), as previously described (5, 11–13, 25).
The abilities of potential electron acceptors to oxidize type c cytochromes were determined as previously described (13). Briefly, cell suspensions (2 ml) were placed into two sealed glass cuvettes under N2-CO2. The suspensions were bubbled with H2-CO2 (80:20) for 2 min to reduce the cytochromes and then bubbled with N2-CO2 for 1 min. An aliquot (0.5 ml) of an anoxic 2.5 mM stock solution of a potential electron acceptor in bicarbonate buffer was added to one cuvette, and 0.5 ml of the anoxic bicarbonate buffer was added to the second cuvette. Difference absorbance spectra for the two treatments were recorded with a scanning spectrophotometer.
16S rRNA gene sequencing and analysis.
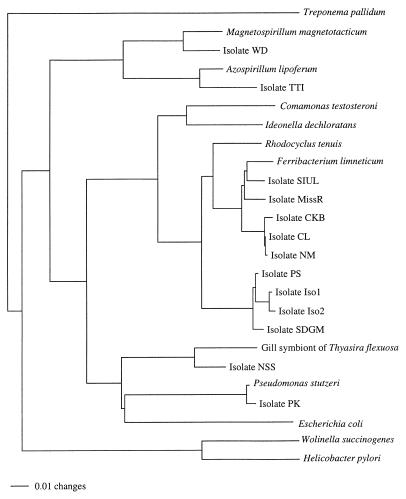
Cells from 2-ml cultures of ClRB were harvested by centrifugation, resuspended in 40 μl of sterile water, and lysed by adding 5 μl of chloroform and incubating the preparations for 10 min at 95°C. Primers specific for bacterial 16S ribosomal DNA (rDNA) (primer 8F [5′-AGAGTTTGATCCTGGCTCAG-3′] and primer 1525R [5′-AAGGAGGTGATCCAGCC-3′]) were used in 50-μl PCR mixtures that contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, 1.2 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 75 ng of each primer, 0.5 μl of Taq polymerase (Gibco/BRL), and 1 μl of lysed cells. Amplification was performed by using the following conditions: 94°C for 3 min, followed by 30 cycles consisting of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and a final step consisting of 10 min at 72°C. The amplification products were gel purified (GeneClean II; Bio 101) and cycle sequenced (ThermoSequenase; Amersham) by using internal primers. Some of the amplification products were cloned (TOPO TA cloning kit; Invitrogen) and then sequenced. Sequence entry and manipulation were performed with the MacVector 6.1 sequence analysis software program for the Macintosh (Oxford Molecular). Sequences of select 16S rRNAs were downloaded from the Ribosomal Database Project (27) and GenBank (3) into the computer program SeqApp (18). ClRB 16S rDNA sequences were manually added to the alignment by using secondary structure information for proper alignment. Distance, parsimony, and maximum-likelihood analyses of the aligned sequences were performed with a G3 computer by Power Macintosh using PAUP*, version 4.0d65 (44). A bootstrap analysis with 100 replications was conducted by using a heuristic search strategy to assess the confidence levels of various clades. The GenBank accession numbers for the sequences used to prepare Fig. 3 are as follows: Treponema pallidum, M88726; Magnetospirillum magnetotacticum, Y10110; strain WD, AF170352; Azospirillum lipoferum, X79730; strain TTI, AF170353; Comamomas testosteroni, M11224; I. dechloratans, X72724; Rhodocyclus tenuis, D16209; Ferribacterium limneticum, Y17060; strain SIUL, AF170356; strain MissR, AF170357; strain CKB, AF047462; strain CL, AF170354; strain NM, AF170355; strain PS, AF170348; strain Iso1, AF170350; strain Iso2, AF170351; strain SDGM, AF170349; gill symbiont of Thyasira flexosa, L01575; strain NSS, AF170359; Pseudomonas stutzeri, U26415; strain PK, AF170358; Escherichia coli, J01859; W. succinogenes ATCC 29543, M26636; and Helicobacter pylori, M88157.
FIG. 3.
Phylogenetic tree based on 16S rDNA sequence data resulting from a distance analysis performed with the Jukes-Cantor correction. The same topology was obtained by using either parsimony or maximum likelihood and was supported by bootstrap analysis.
Analytical techniques.
Acetate concentrations were determined by high-performance liquid chromatography with UV detection (Shimadzu model CDD-6A instrument) by using an HL-75H+ cation-exchange column (Hamilton model 79476). The eluent was 0.016 N H2SO4, and the flow rate was 0.4 ml per min. Perchlorate, chlorate, and chloride concentrations were determined by high-performance liquid chromatography with conductivity detection (Shimadzu model CDD-6A instrument) by using a PRP-X100 anion-exchange column (Hamilton model 79434). The eluent was 4 mM p-hydroxybenzoic acid in 2.5% methanol with the pH adjusted to 8.5, and the flow rate was 2.0 ml per min. Growth of cultures on soluble electron acceptors was determined by measuring the increase in optical density at 600 nm. The oxygen concentrations resulting from chlorite dismutation were determined with an O2 electrode (model 5300; Yellow Springs Instrument Co.). Chlorite dismutase enzyme activity was determined by performing a microassay with horseradish peroxidase (Sigma Chemical Co., St. Louis, Mo.) coupled to dianisidine as the electron donor. In the presence of chlorite a yellow-brown color was produced, which could be read spectrophotometrically at a wavelength of 450 nm (8a). Protein concentrations were determined colorimetrically at 595 nm by performing a Bradford assay.
RESULTS
MPN studies.
The MPN counts obtained when chlorate was the electron acceptor indicated that acetate-oxidizing ClRB are present in many diverse environments. The (per)chlorate-reducing microbial community was significant in all of the environments tested (Table 1). The numbers of ClRB ranged from 2.31 × 103 ± 1.33 × 103 to 2.40 × 106 ± 1.74 × 106 cells per g. The highest MPN counts obtained were the counts obtained for swine waste lagoons.
TABLE 1.
MPN counts of acetate-oxidizing (per)chlorate reducers in different environments
| Environment | Counts (cells per g) |
|---|---|
| Swine waste lagoon | (2.40 ± 1.74) × 106 |
| Pristine aquatic sediment | (4.62 ± 1.75) × 103 |
| Mississippi river sediment | (4.27 ± 2.14) × 103 |
| Pristine soil | (2.31 ± 1.33) × 103 |
| Gold mine drainage sediment | (4.27 ± 2.14) × 103 |
| Petroleum-contaminated soil | (9.33 ± 4.17) × 103 |
| Pohic Bay | (1.49 ± 0.60) × 104 |
| Florida swamp | (2.31 ± 1.33) × 104 |
ClRB isolates.
After 2 weeks of incubation good growth was observed in the primary enrichment cultures prepared from all of the environments sampled. Enrichment cultures were transferred into fresh basal medium (10% inoculum). Good growth was observed in the resulting preparations after 24 h, as determined by increases in optical density and by microscopic examination. Highly enriched (per)chlorate-reducing cultures were obtained after sequential transfers during the following week prior to serial dilution into agar tubes. Small colonies with consistent morphology were apparent in the higher-dilution agar tubes obtained from each enrichment culture after 1 week of incubation. The colonies were generally pink, wet, doomed, entire, smooth, and small (diameter, 1 to 4 mm). Several of the colonies were selected from each of the enrichment series cultures, and (per)chlorate-reducing isolates were obtained from all of the environments sampled.
Phenotypic characteristics.
All of the ClRB isolates were completely oxidizing, gram-negative, nonfermenting facultative anearobes. Morphologically, most of the isolates were short motile rods that were 0.5 μm in diameter and 2 μm long. However, some of the isolates, such as strain WD, which was a spirillum whose cells were 0.2 by 7 μm, had different morphologies. Spores were not visible in wet mounts of any of the isolates when phase-contrast microscopy was used, and no growth was observed in fresh acetate-chlorate medium after pasteurization at 80°C for 3 min. All of the isolates could grow aerobically on L-broth, and colonies on L-broth agar plates were generally white, smooth, and approximately 0.5 mm in diameter.
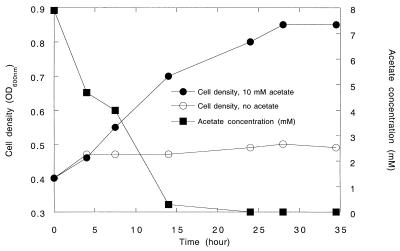
All of the ClRB isolates were strict respirers and could not grow on anoxic basal media amended with glucose (10 mM), yeast extract (10 g/liter), and Casamino Acids (10 g/liter) in the absence of a suitable electron acceptor. All of the ClRB isolates could couple the complete oxidation of acetate to the reduction of chlorate in defined basal medium (Fig. 1). The increases in cell numbers coincided with the oxidation of acetate and the production of chloride (Fig. 1). Chlorate was reduced to innocuous chloride by all of the isolates tested (strains PK, WD, NSS, and PS). Oxidation of 7.9 mM acetate resulted in reduction of 9.3 mM chlorate, giving a stoichiometry of 1.18, a value which, when assimilation into biomass was considered, was in close agreement with the theoretical value according to the following equation:
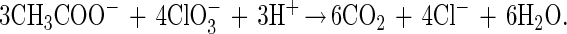 |
Chlorite, the potential intermediate in chlorate reduction, was not detected in the culture broth. In addition to acetate, the ClRB isolates tested used short-chain volatile fatty acids and simple dicarboxylic acids as alternative electron donors (Table 2). None of the ClRB isolates could use H2 or hydrocarbons as alternative electron donors, although some could use inorganic electron donors, such as Fe(II) (Table 2). The ClRB isolates were relatively limited in the range of electron acceptors used. In addition to perchlorate, chlorate, and O2, some of the isolates, but not all of them, could use nitrate for anaerobic growth (Table 3). A broad range of alternative electron acceptors were not used by the ClRB isolates (Table 3).
FIG. 1.
Growth curve for (per)chlorate-reducing strain PK with when acetate was the electron donor and chlorate (10 mM) was the sole electron acceptor. The data are averages based on triplicate determinations. OD600nm, optical density at 600 nm.
TABLE 2.
Compounds used as electron donors by (per)chlorate-reducing isolates when chlorate (10 mM) is the electron acceptor
| Electron donor | Concn | (Per)chlorate-reducing isolates
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| NM | CL | MissR | PS | Iso1 | Iso2 | SDGM | WD | ||
| Acetate | 10 mM | +a | + | + | + | + | + | + | + |
| Propionate | 10 mM | + | + | + | + | + | + | + | + |
| Isobutyrate | 10 mM | + | + | + | + | + | + | + | + |
| Butyrate | 10 mM | + | + | + | + | + | + | + | + |
| Valerate | 10 mM | + | + | + | + | + | + | + | + |
| Formate | 10 mM | − | − | + | − | − | − | − | − |
| Methanol | 5 mM | − | − | − | − | − | − | − | − |
| Ethanol | 5 mM | − | − | + | + | + | + | + | − |
| Catechol | 1 mM | − | − | − | − | − | − | − | − |
| Glycerol | 5 mM | − | − | − | − | − | − | − | − |
| Benzoate | 0.5 mM | − | − | − | − | − | − | − | − |
| Pyruvate | 5 mM | + | ND | ND | + | ND | ND | ND | ND |
| Citrate | 5 mM | − | − | − | − | − | − | − | − |
| Succinate | 1 mm | + | + | − | + | − | − | − | + |
| Lactate | 10 mM | + | + | + | + | + | + | + | + |
| Glucose | 10 mM | − | − | − | − | − | − | − | − |
| Casamino Acids | 1 g/liter | + | − | + | + | + | + | + | + |
| Fumarate | 5 mM | + | + | + | + | + | + | + | + |
| Malate | 1 mM | + | + | + | + | + | + | + | + |
| Hydrogen | 101 kPa | − | − | − | − | − | − | − | − |
| Fe(II) | 5 mM | ND | ND | ND | + | ND | ND | ND | + |
+, electron donor utilized; −, electron donor not utilized; ND, not determined.
TABLE 3.
Compounds used as electron acceptors by (per)chlorate-reducing isolates when acetate (10 mM) is the electron donor
| Electron acceptor | Concn | (Per)chlorate-reducing isolates
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| NM | CL | MissR | PS | Iso1 | Iso2 | SDGM | WD | ||
| Chlorate | 10 mM | +a | + | + | + | + | + | + | + |
| Nitrate | 5 mM | − | − | + | + | + | + | + | + |
| Sulfate | 10 mM | − | − | − | − | − | − | − | − |
| Selenate | 2 mM | − | − | − | − | − | − | − | − |
| Fumarate | 25 mM | − | − | − | − | − | − | − | − |
| Malate | 5 mM | − | − | − | − | − | − | − | − |
| Mn(IV) | 2 mM | − | − | − | − | − | − | − | − |
| Fe(III) | 10 mM | − | − | − | − | − | − | − | − |
| O2 | 101 kPa | + | + | + | + | + | + | + | + |
| AQDSb | 5 mM | − | − | − | − | − | − | − | − |
| Fermentation | − | − | − | − | − | − | − | − | |
+, electron acceptor utilized; −, electron acceptor not utilized.
AQDS, 2,6-anthraquinone disulfonate.
Cytochrome content and oxidation by potential electron acceptors.
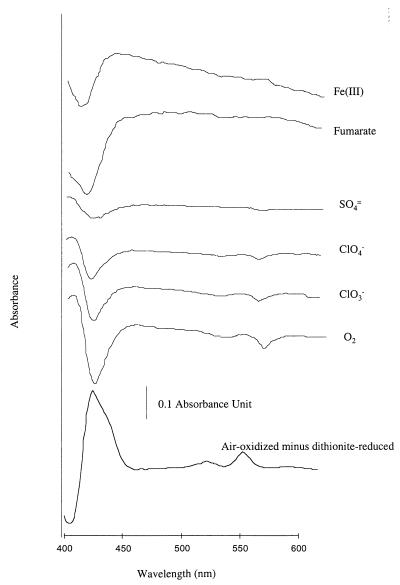
Air-oxidized–minus–dithionite-reduced spectra of washed whole-cell suspensions of all of the ClRB isolates grown with chlorate as an electron acceptor had absorbance maxima close to 425, 525, and 552 nm, which are indicative of type c cytochrome(s) (Fig. 2). A hydrogen-reduced type c cytochrome(s) in anoxic washed cell suspensions of one of the previously described isolates (strain CKB) (5) was reoxidized by compounds that are known to act as electron acceptors for this organism, such as chlorate or perchlorate (Fig. 2). A hydrogen-reduced cytochrome(s) was not reoxidized by compounds such as sulfate, Fe(III), and fumarate, which are not used by this organism as electron acceptors for anaerobic growth (Fig. 2).
FIG. 2.
Difference absorbance spectra of H2-reduced washed whole-cell suspensions of (per)chlorate-reducing strain CKB in the presence of various potential electron acceptors.
Phylogeny of the ClRB.
Analyses of the 16S rDNA sequences indicated that all of the isolates were members of the class Proteobacteria of the Bacteria (Fig. 3). The new ClRB isolates belonged to three subgroups (the alpha, beta, and gamma subclasses) of the Proteobacteria, which demonstrated that this type of metabolism is widespread in the class (Fig. 3). Some of the isolates, such as strain PK, were closely related to members of previously described genera that are not known for the potential to grow by dissimilatory (per)chlorate reduction, while others, such as strain NSS, had no close relatives and represented novel genera in the Proteobacteria (Fig. 3). The majority of the isolates obtained were closely related to each other and to the phototrophic Rhodocyclus species in the beta subclass of the Proteobacteria.
Chlorite dismutase.
Washed whole-cell suspensions of all of the ClRB isolates could dismutate chlorite to chloride and molecular oxygen. At room temperature, O2 evolution was rapid, linear, and proportional to the chlorite concentration (data not shown). No O2 production was observed in the absence of cells or if the ClRB was heat killed. At a higher chlorite concentration (10 mM), O2 evolution was so extensive that a copious amount of froth was observed in each cell suspension.
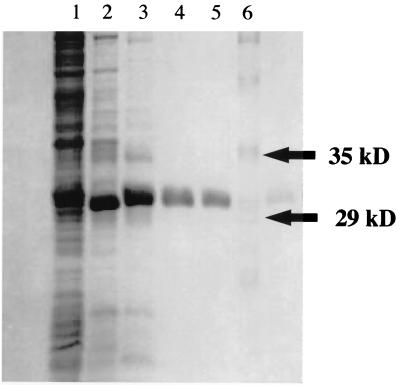
A single enzyme with chlorite dismutase activity was purified to homogeneity from a previously characterized strain (strain CKB) (5) (Fig. 4 and Table 4). The specific activity of the purified enzyme was 1,928 μmol of chlorite per mg of protein per min (Table 4). A comparison with molecular mass standards in SDS-PAGE denaturing gels indicated that the molecular mass of the denatured protein was 32 kDa, while size exclusion chromatography indicated that the molecular mass of the native protein was 120 kDa. These data suggest that the enzyme is a homotetramer with a molecular mass of approximately 120 kDa.
FIG. 4.
SDS-PAGE gel containing the chlorite dismutase active fractions from strain CKB. Lane 1, cell lysate fraction; lane 2, chlorite dismutase active pool from the Q-Sepharose column; lane 3, active pool from the hydroxyapatite column; lane 4, chlorite dismutase active fraction from the Phenyl Sepharose column; lane 5, purified chlorite dismutase from the Superdex 200 column; lane 6, molecular mass standard.
TABLE 4.
Purification and specific activity of chlorite dismutase from (per)chlorate-reducing strain CKB
| Prepn | Vol (ml) | Protein concn (mg/ml) | Total protein (mg) | Sp act (U/mg)a | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 30.0 | 27.72 | 831.6 | 35.0 | 1 |
| Q-Sepharose | 120.0 | 0.777 | 93.24 | 398.0 | 11.4 |
| Hydroxyapatite | 144.0 | 0.200 | 28.80 | 640.0 | 18.3 |
| Phenyl-Sepharose | 48.0 | 0.080 | 3.84 | 1,102.0 | 32.0 |
| Superdex 200 | 10.0 | 0.307 | 3.07 | 1,928.0 | 55.0 |
One unit was defined as 1 μmol of sodium chlorite dismutated per min.
DISCUSSION
The results of this study demonstrate the hitherto unrecognized ubiquity of microbial (per)chlorate reduction and the broad phylogenetic diversity of the organisms capable of this type of metabolism. Contamination of drinking water, groundwater, and surface water by oxyanions of chlorine, especially chlorate and perchlorate, has only recently been recognized as a potentially serious health risk (32, 45, 46). Although the fact that microbial reduction of (per)chlorate occurs has been recognized for the last 50 years and although microbial reduction of (per)chlorate has been identified as a potentially important metabolic process for the treatment of perchlorate and chlorate contamination in the environment (45, 46, 49), very little is known about the microorganisms involved in (per)chlorate reduction. Several organisms, including Proteus mirabilis (16), Rhodobacter capsulatus, and Rhodobacter sphaeroides (36), have been shown to be capable of reducing chlorate to chlorite. However, no growth is associated with this type of metabolism, and the chlorite end product is generally toxic to the organisms. The MPN counts obtained in this study indicate that significant levels of C1RB occur in very diverse environments, many of which have not been exposed to chlorine oxyanions. This finding supports and expands the observations made in a previous investigation (48), in which it was shown that chlorate reduction was prevalent in several diverse environments. This is, however, unexpected as there are no known natural sources of these compounds (21) and they have been introduced into the environment only in the last 100 years due to human activity (45). Early studies suggested that microbial (per)chlorate reduction may simply be a competitive reaction for the nitrate reductase system of denitrifying bacteria in the environment (19, 20, 43), but this suggestion does not explain the presence of chlorate reductase enzymes, such as the chlorate reductase C purified from P. mirabilis, which can only use chlorate as a substrate (31).
Only six dissimilatory (per)chlorate-reducing organisms have been identified previously, and only four of these have been studied in detail. Thus, the true diversity of microbial (per)chlorate reduction is still not known. Our findings significantly increase the number of (per)chlorate-reducing isolates that have been described. All of the isolates which we obtained are members of the Proteobacteria, and they represent three of the five subclasses of this class. Although in this study we did not obtain any ClRB isolates which are members of the epsilon or delta subclass of the Proteobacteria, a previously described ClRB, strain HAP-1 (50), was identified as a new strain of W. succinogenes, which is a member of the epsilon subclass. Thus, the ability to reduce (per)chlorate is widespread in the Proteobacteria. The broad phylogenetic diversity of organisms observed in this study which are capable of this type of metabolism has some interesting evolutionary implications due to the relatively short time in which (per)chlorate reduction could have evolved.
Several of the isolates which we obtained were representatives of previously defined genera not recognized for the ability to reduce (per)chlorate. When some of the previously described close relatives of the ClRB isolates, such as P. stutzeri, the closest known relative of strain PK (99.4% similarity, as determined by 16S rDNA sequence analysis), were tested for (per)chlorate reduction, no growth was observed, and none of the organisms could dismutate chlorite. This result is similar to previous observations made with the ClRB W. succinogenes HAP-1, which is 99.3% similar (as determined by 16S rDNA sequence analysis) to the type strain of W. succinogenes (strain ATCC 29543), which cannot grow by (per)chlorate reduction (50).
Comparison with other ClRB.
Many of the ClRB isolates obtained in this study were not closely related to any previously described organism and represented new genera in the Proteobacteria. All of the ClRB isolates contained type c cytochromes, which in the case of the previously described ClRB (5) strain CKB were reoxidized in the presence of physiological electron acceptors used by this organism. Other compounds, such as sulfate, fumarate, and Fe(III), which were not reduced by this organism in anaerobic culture, also did not reoxidize the reduced cytochrome(s). Although not conclusive, this data suggests that type c cytochromes may be involved in the transport of electrons to perchlorate or chlorate.
Only four of the previously described dissimilatory (per)chlorate reducers, strain GR-1 (34), I. dechloratans (29), W. succinogenes HAP-1 (50), and strain CKB (5), have been well characterized. Strains CKB (5) and GR-1 (34) and I. dechloratans (29) are members of the beta subclass of the Proteobacteria. I. dechloratans is phylogenetically distinct from any of the ClRB isolates obtained in this study. Similar comparisons with strain GR-1 could not be made as the 16S rDNA sequence of this isolate is not available. W. succinogenes HAP-1 is a member of the epsilon subclass of the Proteobacteria and as such is very distantly related to the ClRB isolates which we obtained (50). All of the ClRB isolates are similar to strain GR-1 (34) and I. dechloratans (29) in terms of their ability to couple growth to the oxidation of acetate when chlorate is the sole electron acceptor. As previously observed with other dissimilatory (per)chlorate reducers (29, 34, 50), chlorate is completely reduced to chloride.
Similar to strain CKB (5) and in contrast to previously described (per)chlorate reducers (29, 34, 41, 50), the new ClRB isolates were relatively limited in terms of the range of electron donors or acceptors used. None of the isolates utilized carbohydrates, which are used by I. dechloratans (29). In addition, none of the new ClRB isolates could oxidize H2, an important end product of fermentation which serves as an electron donor for (per)chlorate reduction by W. succinogenes HAP-1 (50). The only electron acceptors tested that were utilized by the ClRB isolates were O2, perchlorate, chlorate, and (in some cases) nitrate. The ability of the new isolates to grow aerobically is similar to the ability of the previously described ClRB, and this finding suggests that all ClRB are facultative anaerobes. This conclusion is supported by the fact that all of the ClRB isolates obtained dismutate chlorite into chloride and O2, which would be toxic to strict anaerobes. Although it was originally suggested that W. succinogenes HAP-1 is a strict anaerobe (50), a recent study indicates that this organism is in fact a microaerophile (49). The fact that not all of the ClRB isolates can use nitrate also supports the hypothesis that the chlorate reduction pathway and the nitrate reduction pathway are unrelated pathways and is in contrast to suggestions made in previous studies (19, 20, 43).
All of the ClRB isolates exhibit chlorite dismutase activity. Transformation of chlorite by these isolates, like transformation of chlorite by strain GR-1 (34), is not dependent on the presence of acetate. As in strain GR-1, a single enzyme is responsible for the dismutation activity in strain CKB. The purified enzyme has a high specific activity, which is the same order of magnitude as the specific activity of the enzyme isolated from strain GR-1 (47).
Environmental significance.
The role of ClRB in environments that have not been exposed to chlorine oxyanions has yet to be determined. Although a few dissimilatory (per)chlorate reducers have been described (5, 29, 34, 41, 50), all of the isolates were obtained from contaminated sediments or wastewater treatment sludges. This study is the first study in which it was demonstrated that organisms with (per)chlorate-reducing ability can be readily isolated from pristine environments.
Although (per)chlorate reduction has been recognized for more than 50 years, the presence of oxyanions of chlorine in the environment is the result of human activity over the last 100 years. Thus, the evolution of a phylogenetically diverse group of organisms with the ability to couple growth to the reduction of (per)chlorate was not expected. This metabolic ability appears to be centered around the unique ability of the organisms to dismutate chlorite into chloride and oxygen. Although chlorite dismutation has not been demonstrated yet for all of the known dissimilatory ClRB, it was shown previously for strain GR-1 (34) and strain CKB (5, 9, 10) and in this study for 13 ClRB isolates, suggesting that it is a characteristic of all ClRB. The fact that the purified chlorite dismutase enzymes of strains GR-1 and CKB are similar in terms of general structure, molecular mass, and specific activity suggests that a single gene coding for this enzyme may be conserved in these (per)chlorate-reducing organisms and that (per)chlorate reduction may be the result of horizontal gene transfer events.
ACKNOWLEDGMENTS
This research was supported in part by grant DE-FG02-98ER62689 from the Department of Energy to J.D.C. and L.A.A. and by the 1998 Oak Ridge Associated Universities Junior Faculty award to J.D.C.
REFERENCES
- 1.Agaev R, Danilov V, Khachaturov V, Kasymov B, Tishabaev B. The toxicity to warm-blooded animals and fish of new defoliants based on sodium and magnesium chlorates. Uzb Biol Zh. 1986;1:40–43. [Google Scholar]
- 2.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergnor E, Germgard U, Kolar J, Lindgren O. Formation of chlorate in chlorine dioxide bleaching. Cellul Chem Technol. 1987;21:307–314. [Google Scholar]
- 5.Bruce R A, Achenbach L A, Coates J D. Dechlorimonas agitatus strain CKB gen. nov., sp. nov., a novel dissimilatory chlorate-reducer from a paper mill. Environ Microbiol. 1999;1:319–331. doi: 10.1046/j.1462-2920.1999.00042.x. [DOI] [PubMed] [Google Scholar]
- 6.Bryan E H. Application of the chlorate BOD procedure to routine measurement of wastewater strength. J Water Pollut Control Fed. 1966;38:1350–1362. [PubMed] [Google Scholar]
- 7.Bryan E H, Rohlich G A. Biological reduction of sodium chlorate as applied to measurement of sewage BOD. Sewage Ind Wastes. 1954;26:1315–1324. [Google Scholar]
- 8.California Department of Health Services. 23 September 1997, posting date. Perchlorate in California drinking water. [Online.] http://www.dhs.cahwnet.gov/ps/ddwem/chemicals/perchl/perchlindex.htm. [12 October 1999, last date accessed.]
- 8a.Coates, J. D. Unpublished data.
- 9.Coates J D, Bruce R A, Haddock J D. Anoxic bioremediation of hydrocarbons. Nature. 1998;396:730. doi: 10.1038/25470. [DOI] [PubMed] [Google Scholar]
- 10.Coates, J. D., R. A. Bruce, J. A. Patrick, and L. A. Achenbach. Hydrocarbon bioremediative potential of (per)chlorate-reducing bacteria. Bioremediat. J., in press.
- 11.Coates J D, Lonergan D J, Lovley D R. Desulfuromonas palmitatis sp. nov., a long-chain fatty acid oxidizing Fe(III) reducer from marine sediments. Arch Microbiol. 1995;164:406–413. [PubMed] [Google Scholar]
- 12.Coates J D, Ellis D J, Blunt Harris E L, Gaw C V, Roden E R, Lovley D R. Recovery of humic-reducing bacteria from a diversity of environments. Appl Environ Microbiol. 1998;64:1504–1509. doi: 10.1128/aem.64.4.1504-1509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coates J D, Phillips E J P, Lonergan D J, Jenter H, Lovley D R. Isolation of Geobacter species from a variety of sedimentary environments. Appl Environ Microbiol. 1996;62:1531–1536. doi: 10.1128/aem.62.5.1531-1536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Condie L. Toxicological problems associated with chlorine dioxide. J Am Water Wks Assoc. 1986;78:73–78. [Google Scholar]
- 15.Daniel F, Condie L, Robinson M, Stober J, York R, Olson G, Wang S. Comparative subchronic toxicity studies of three disinfectants. J Am Water Works Assoc. 1990;82:61–69. [Google Scholar]
- 16.de Groot G N, Stouthamer A H. Regulation of reductase formation in Proteus mirabilis. I. Formation of reductases and enzymes of the formic hydrogenlyase complex in the wild type and in chlorate resistant mutants. Arch Microbiol. 1969;66:220–233. [PubMed] [Google Scholar]
- 17.Germgard U, Teder A, Tormund D. Chlorate formation during chlorine dioxide bleaching of softwood kraft pulp. Paperi ja Puu. 1981;3:127–133. [Google Scholar]
- 18.Gilbert D G. SeqApp, version 1.9a157. 1993. Biocomputing Office, Biology Department, Indiana University, Bloomington. [Google Scholar]
- 19.Hackenthal E. Die Reduktion von Perchlorat durch Bacterien. II. Die Identitat der Nitratreduktase und des Perchlorat Reduzierenden Enzyms aus B. cereus. Biochem Pharmacol. 1965;14:1313–1324. doi: 10.1016/0006-2952(65)90118-8. [DOI] [PubMed] [Google Scholar]
- 20.Hackenthal E, Mannheim W, Hackenthal R, Becher R. Die Reduktion von Perchlorat durch Bakterien. I. Untersuchungen an intakten Zellen. Biochem Pharmacol. 1964;13:195–206. doi: 10.1016/0006-2952(64)90137-6. [DOI] [PubMed] [Google Scholar]
- 21.Herman D, Frankenberger W T. Microbial-mediated reduction of perchlorate in groundwater. J Environ Qual. 1998;27:750–754. [Google Scholar]
- 22.Hungate R E. A roll tube method for cultivation of strict anaerobes. Methods Microbiol. 1969;3B:117–132. [Google Scholar]
- 23.Korenkov, V., V. Romanenko, S. Kuznetsov, and J. Voronov. March 1976. Process for purification of industrial waste waters from perchlorates and chlorates. U.S. patent 3,943,055.
- 24.Logan B. A review of chlorate- and perchlorate-respiring microorganisms. Bioremediat J. 1998;2:69–79. [Google Scholar]
- 25.Lovley D R, Giovannoni S J, White D C, Champine J E, Phillips E J P, Gorby Y A, Goodwin S. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159:336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- 26.Lovley D R, Phillips E J P. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol. 1988;54:1472–1480. doi: 10.1128/aem.54.6.1472-1480.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malmqvist A, Welander T, Gunnarsson L. Anaerobic growth of microorganisms with chlorate as an electron acceptor. Appl Environ Microbiol. 1991;57:2229–2232. doi: 10.1128/aem.57.8.2229-2232.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malmqvist A, Welander T, Moore E, Ternstrom A, Molin G, Stenstrom I-M. Ideonella dechloratans gen. nov., sp. nov., a new bacterium capable of growing anaerobically with chlorate as an electron acceptor. Syst Appl Microbiol. 1994;17:58–64. [Google Scholar]
- 30.Miller T L, Wolin M J. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl Microbiol. 1974;27:985–987. doi: 10.1128/am.27.5.985-987.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oltmann L F, Reifnders W N M, Stouthamer A H. Characterization of purified nitrate reductase A and chlorate reductase C from Proteus mirabilis. Arch Microbiol. 1976;111:25–35. doi: 10.1007/BF00446546. [DOI] [PubMed] [Google Scholar]
- 32.Renner R. Perchlorate-tainted wells spur government action. Environ Sci Technol News. 1998;32:210A. doi: 10.1021/es983489t. [DOI] [PubMed] [Google Scholar]
- 33.Renner R. EPA draft almost doubles safe dose of perchlorate in water. Environ Sci Technol News. 1999;33:110A–111A. doi: 10.1021/es992699i. [DOI] [PubMed] [Google Scholar]
- 34.Rikken G, Kroon A, van Ginkel C. Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl Microbiol Biotechnol. 1996;45:420–426. [Google Scholar]
- 35.Roden E E, Lovley D R. Dissimilatory Fe(III) reduction by the marine microorganism Desulfuromonas acetoxidans. Appl Environ Microbiol. 1993;59:734–742. doi: 10.1128/aem.59.3.734-742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roldan M D, Reyes F, Moreno-Vivian C, Castillo F. Chlorate and nitrate reduction in phototrophic bacteria Rhodobacter capsulatus and Rhodobacter sphaeroides. Curr Microbiol. 1994;29:241–245. [Google Scholar]
- 37.Romanenko V I, Korenkov V N, Kuznetsov S I. Bacterial decomposition of ammonium perchlorate. Mikrobiologiya. 1976;45:204–209. [PubMed] [Google Scholar]
- 38.Rosemarin A, Lehtinen K, Notini M. Effects of treated and untreated softwood pulp mill effluents on Baltic Sea algae and invertebrates in model ecosystems. Nord Pulp Pap Res J. 1990;2:83–87. [Google Scholar]
- 39.Siddiqui M. Chlorine-ozone interactions: formation of chlorate. Water Res. 1996;30:2160–2170. [Google Scholar]
- 40.Stanbury J B, Wyngaarden J B. Effect of perchlorate on the human thyroid gland. Metabolism. 1952;1:533–539. [PubMed] [Google Scholar]
- 41.Stepanyuk V, Smirnova G, Klyushnikova T, Kanyuk N, Panchenko L, Nogina T, Prima V. New species of the Acinetobacter genus Acinetobacter thermotoleranticus sp. nov. Mikrobiologiya. 1992;61:347–356. [Google Scholar]
- 42.Stettler R. Utilisation del’ozone dans le traitment de eaux de boisson. Gas Wass Abwass. 1977;57:81–95. [Google Scholar]
- 43.Stouthamer A. Nitrate reduction in Aerobacter aerogenes. I. Isolation properties of mutant strains blocked in nitrate assimilation and resistant against chlorate. Arch Microbiol. 1967;56:68–75. [PubMed] [Google Scholar]
- 44.Swofford D L. PAUP*: phylogenetic analysis using parsimony (and other methods), version 4.0. Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 45.Urbansky E T. Perchlorate chemistry: implications for analysis and remediation. Bioremediat J. 1998;2:81–95. [Google Scholar]
- 46.U.S. Environmental Protection Agency. Perchlorate. [Online.] wysiwgy://2/http://www.epa.gov/ncea/perch.htm. [12 October 1999, last date accessed.]
- 47.van Ginkel C, Rikken G, Kroon A, Kengen S. Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch Microbiol. 1996;166:321–326. doi: 10.1007/s002030050390. [DOI] [PubMed] [Google Scholar]
- 48.van Ginkel C G, Plugge C M, Stroo C A. Reduction of chlorate with various energy substrates and inocula under anaerobic conditions. Chemosphere. 1995;31:4057–4066. [Google Scholar]
- 49.Wallace W, Beshear S, Williams D, Hospadar S, Owens M. Perchlorate reduction by a mixed culture in an up-flow anaerobic fixed bed reactor. J Ind Microbiol Biotechnol. 1998;20:126–131. [Google Scholar]
- 50.Wallace W, Ward T, Breen A, Attaway H. Identification of an anaerobic bacterium which reduces perchlorate and chlorate as Wolinella succinogenes. J Ind Microbiol. 1996;16:68–72. [Google Scholar]
- 51.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 3356–3362. [Google Scholar]
- 52.Widdel F, Hansen T A. The dissimilatory sulfate- and sulfur-reducing bacteria. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 583–624. [Google Scholar]