Abstract
Somatic embryogenesis (SE) is of great significance in Lilium bulb production, germplasm preservation, and genetic improvement. miRNAs are important regulators of plant growth and development at the transcriptional level. Previous research by our group has shown that lpu-miR171 and its target gene SCARECROW-LIKE 6 (SCL6) play an important regulatory role in lily SE, and we predicted and identified that endogenous target mimics (eTMs) can regulate lpu-miR171. However, the associated mechanism and internal regulatory network are not yet clear. In the present study, lpu-miR171 was used as an entry point to explore the regulatory network between its upstream eTMs and its downstream target gene LpSCL6, as well as to identify the mechanism of this regulatory network in Lilium SE. Tobacco transient transformation confirmed that miRNA171 significantly inhibited the expression of LpSCL6. On this basis, the Lilium stable genetic transformation system was used to demonstrate that silencing lpu-miR171a and lpu-miR171b and overexpressing LpSCL6-II and LpSCL6-I promoted starch accumulation in calli and the expression of key cell cycle genes, thus providing energy to meet preconditions for SE and accelerate the formation and development of Lilium somatic embryos. LpSCL6-II and LpSCL6-I are nuclear proteins with self-activation activity in yeast cells. In addition, we confirmed in Lilium that lpu-eTM171 is the eTM of lpu-miR171 that binds lpu-miR171 to prevent cleavage of the target gene LpSCL6, thereby promoting SE. Therefore, the present study established a new mechanism whereby the eTM-miR171-SCL6 module regulates SE in Lilium pumilum and provides new insights clarifying the mechanism of SE.
Introduction
Lilium species are among the most important ornamental plants and economic crops worldwide. This genus includes some of the most important cut flower species, potted plants, bedding plants, and garden crops, and the bulbs of this genus may be edible or medicinal. The improvement of genetic traits and the cultivation of high-quality bulbs are key, difficult problems in the field of international Lilium production and biotechnology. Somatic embryogenesis (SE) in plants, which presents the advantages of universal occurrence, genetic stability, a low mutation rate, and a high reproduction coefficient, is vital for bulb propagation, germplasm preservation, genetic trait improvement, mutant creation, and breeding by genetic engineering in Lilium [1]. However, due to the high specificity and poor synchronization of somatic embryo induction in different genotypes, the requirements for mass breeding and genetic engineering research in Lilium cannot be met, mainly because the mechanism of SE is not yet clear.
MicroRNAs (miRNAs) are a class of endogenous, small non-coding RNAs ~19–24 nucleotides in length. To date, 38 589 miRNAs have been published in the miRBase database (Release 22.1) [2]. Additionally, a large number of miRNA target genes have been identified by bioinformatics, omics, and immunoprecipitation, as well as other methods. As regulatory factors of endogenous gene expression, miRNAs have been widely recognized to play an important role in SE in a variety of plants, such as rice [3], citrus [4, 5], larch [6–8], cotton [9], maize [10], and longan [11, 12]. Specifically, miR156-SPL modules play a vital role during the initial phases of SE induction [5]. Overexpression of csi-miR156a or suppression of one of the two target genes, CsSPL3 and CsSPL14, can enhance the SE competence of citrus callus. miR159/LaMYB33 is involved in the maintenance of embryogenic ornon-embryogenic potential and somatic embryo maturation in larch [7]. miR166/ATHB15 regulates early somatic embryogenesis and miR167a/ARF8 regulates globular embryo formation and subsequent conversion to cotyledonary embryos in longan [11]. Similar stages of SE in different plants or different stages of SE in the same plants are regulated by different miRNAs or multiple miRNAs simultaneously. The molecular mechanism of miRNA regulation of SE needs further analysis.
Our previous study found that miR171 family members are differentially expressed in torpedo embryos and cotyledonary embryos compared with globular embryos in Lilium pumilum. Subsequently, we cloned miRNA171 and its target gene and preliminarily confirmed that miRNA171 plays an important role in torpedo embryo development in Lilium pumilum [13]. However, the molecular mechanism by which miR171 regulates Lilium SE has not been elucidated. The members of the miR171 family are 21 nucleotides long and are highly conserved in angiosperms [14]. miR171 regulates various plant growth and development processes by targeting members of the GRAS domain transcription factor family, which are essential for the maintenance of shoot apical meristems and axillary meristems [15]. A series of studies have shown that miR171 participates in the regulation of SE by modulating the SCL6 target gene. miRNA171 is one of the key miRNAs in the process of SE, as observed in larch [6], radish [16], citrus [4], and longan [12], and it has been shown to play an important regulatory role in the different stages of SE. In Larix kaempferi SE, 11 conserved miRNA families were detected by quantitative reverse transcription–PCR (qRT–PCR), among which miR171a/b might have proembryogenic functions, while miR171c acts in the induction process of larch SE [6]. In larch, the cleavage site of miR171 and the SCL6 target gene was identified through 5′ RACE (rapid amplification of cDNA ends) and through differential expression analysis, and it was shown that the posttranscriptional regulation of the SCL6 target gene by miR171 is related to the maintenance of embryonic potential [8]. In Citrus sinensis, the expression patterns of miR171 and the SCL6 target gene at different stages of somatic embryo development were analyzed, and the authors found that these patterns are closely related to the formation of somatic embryos [4]. In a study of Raphanus sativus L., miR171 accumulated in the early stage of SE, indicating that miR171 and the SCL6 target gene play a key regulatory role in the early stage of SE [16]. The above studies show that miR171 and the SCL6 target gene have an important regulatory function in SE; however, there are certain differences in their regulatory roles in the somatic embryos of different species. Their regulatory mechanism in SE has not been thoroughly analyzed, and there is little research on SE in lily. Current research directions have not included a deep analysis of the mechanism of miR171 in somatic embryos.
Since Franco-Zorrilla et al. first reported that the targeting activity of Arabidopsis miR399 is inhibited by endogenous target mimics (eTMs) [17], researchers have successively identified eTMs that regulate the activities of miR160, miR166, miRX27, and miR167 in plants such as Arabidopsis [18], rice [19], tomato [20], and longan [11]. The regulatory effect of eTMs on miRNAs has been verified in the processes of soybean lipid metabolism [21], tomato yellow leaf curl virus defense [22], and tobacco nicotine synthesis [23]. In addition, eTMs participate in a variety of plant development processes by regulating miRNAs. Regarding the regulation of SE, Lin et al. revealed the regulatory effects of miR160 and miR167 on auxin signaling in SE in longan by studying the differential expression levels of the miR160 and miR167 eTMs and the ARF target gene [11, 24]. However, the mechanism by which the eTM-miRNA module regulates SE has not been fully resolved.
In the early stage of our research, we used somatic embryos at different developmental stages as materials to construct the expression profiles of key miRNAs in SE. Although there are 13 members in the lily miR171 family, only miR171a and miR171b were differentially expressed during the somatic embryogenesis of L. pumilum, and the expression of the remaining mir171 members did not change significantly, with the expression level being extremely low [25]. We found that miR171a and miR171b are significantly differentially expressed during SE and that their expression peaks in embryogenic calli and during anaphase of SE [13]. RNA ligase-mediated (RLM) 5′ RACE has been used to verify the sites of action of lpu-miR171a and lpu-miR171b and the target genes, and a correlation analysis of the expression of lpu-miR171a and lpu-miR171b and the target genes in different stages of somatic embryo development has revealed that lpu-miR171a and lpu-miR171b have negative regulatory effects on LpSCL6-II and LpSCL6-I. To further clarify the mechanism by which lpu-miR171a and lpu-miR171b regulate SE, preliminary screening combined with bioinformatics methods was performed within the scope of the transcriptome to obtain standard eTMs. An expression correlation analysis of lpu-miR171a and lpu-miR171b and their corresponding eTMs revealed a negative correlation of the expression trends of lpu-miR171a and lpu-miR171b and their eTMs during SE, indicating that lpu-miR171 regulates target genes and is also regulated by the eTMs of upstream regulators [26]. Based on these previous studies, the present study explored the functions of lpu-miR171 and the LpSCL6 target gene in the process of SE in L. pumilum through stable overexpression and the use of artificial miRNA silencing technology to elucidate the regulatory relationship of lpu-miR171 with the LpSCL6 target gene. Overexpression and point mutation analyses confirmed the regulatory effect of eTM171 on lpu-miR171. Finally, the regulatory mode of eTM-miR171-SCL6 in the process of Lilium SE was clarified, laying a foundation for the comprehensive elucidation of the mechanism of Lilium SE.
Results
Verification of the effects of lpu-miR171a on LpSCL6-II and lpu-miR171b on LpSCL6-I
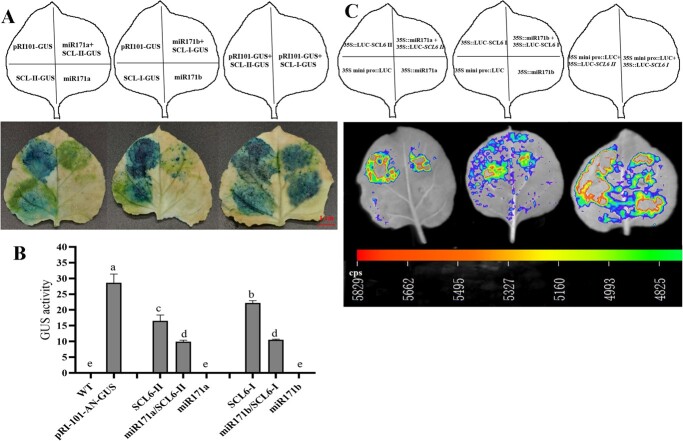
The overexpression vectors of lpu-miR171a, lpu-miR171b, LpSCL6-II, and LpSCL6-I were transiently transfected into tobacco leaves, and pRI101-ON-GUS was used as a control. GUS histochemical staining was performed 48 h after transfection. Fig. 1a shows that tobacco leaves containing the LpSCL6-II and LpSCL6-I vectors and the pRI101-ON-GUS empty vector were stained dark blue. No blue color was observed in Nicotiana benthamiana leaves transformed with the lpu-miR171a and lpu-miR171b vectors without the GUS reporter gene or the tobacco leaves of the wild-type control group. Tobacco leaves cotransformed with lpu-miR171a and lpu-miR171b and the LpSCL6-II and LpSCL6-I target genes showed less intense staining. Measurement of GUS activity in the tobacco leaves demonstrated that GUS enzyme activity was higher in tobacco leaves containing the LpSCL6-II and LpSCL6-I vectors and the empty vectors. GUS enzyme activity was not detected in tobacco leaves containing the lpu-miR171a and lpu-miR171b vectors without the GUS reporter gene or the wild-type control group, while GUS enzyme activity was significantly reduced in tobacco leaves cotransformed with lpu-miR171a + LpSCL6-II and lpu-miR171b + LpSCL6-I (Fig. 1b). The above results indicated that lpu-miR171a and lpu-miR171b have an inhibitory effect on LpSCL6-II and LpSCL6-I, thereby preventing the expression of the GUS gene. The LUC luminescence signal results are shown in Fig. 1c. No LUC luminescence signal was observed from the vectors containing the lpu-miR171a and lpu-miR171b precursors or the empty pRI-mini35S-LUC vectors. 35S::LUC-SCL6 I and 35S::LUC-SCL6 II transfected alone produced a stronger LUC luminescence signal, while the LUC luminescence signal of cotransformed tobacco leaves was weaker. These results showed that lpu-miR171a and lpu-miR171b have an inhibitory effect on LpSCL6-II and LpSCL6-I, thereby reducing the LUC luminescence signal. Based on the above test results, we demonstrated that the lpu-miR171a and lpu-miR171b candidates have negative regulatory effects on LpSCL6-II and LpSCL6-I.
Figure 1.
Interaction verification between lpu-miR171a and lpu-miR171b and the LpSCL6-II and LpSCL6-I target genes. a GUS histochemical staining. pRI101-GUS refers to the empty vectors. miR171a is a vector containing mature miR171a. miR171b is a vector containing mature miR171b. SCL-I-GUS is a vector containing LpSCL6-I. SCL-II-GUS is a vector containing LpSCL6-II. Scale bar = 1 cm. b GUS enzyme activity. c Verification results of lpu-miR171a and lpu-miR171b and the LpSCL6-II and LpSCL6-I target genes. 35S mini pro::LUC refers to the empty vectors. 35S::miR171a is a vector containing mature miR171a. 35S::SCL6 II is a vector containing LpSCL6-II. 35S::miR171a + 35S::SCL6 II refers to the observation result of the mixed injection of a vector containing mature miR171a and a vector containing LpSCL6-II. 35S::miR171b is a vector containing mature miR171b. 35S::SCL6 I is a vector containing LpSCL6-I. 35S::miR171b + 35S::SCL6 I refers to the observation result of the mixed injection of a vector containing mature miR171b and a vector containing LpSCL6-I. 35S mini pro::LUC + 35S::SCL6 I refers to the observation result of the mixed injection of empty vectors and a vector containing LpSCL6-I. 35S mini pro::LUC + 35S::SCL6 II refers to the observation result of the mixed injection of empty vectors and a vector containing LpSCL6-II.
LpSCL6-II and LpSCL6-I subcellular localization and transcriptional activation analysis
pRI-SCL6-I-GFP and pRI-SCL6-II-GFP were introduced into tobacco leaves to determine the subcellular localization of LpSCL6-I and LpSCL6-II. As shown in Fig. 2a, the pRI-SCL6-I-GFP and pRI-SCL6-II-GFP fusion proteins were localized in the nucleus and overlapped with the nuclear localization signal from DAPI, but the GFP (green fluorescent protein) signal of the positive control was distributed throughout the entire cell. Colocalization of DAPI with SCL-GFP indicated the localization of SCL in the nucleus. These results suggested that both the LpSCL6-I and LpSCL6-II proteins are nuclear proteins.
Figure 2.
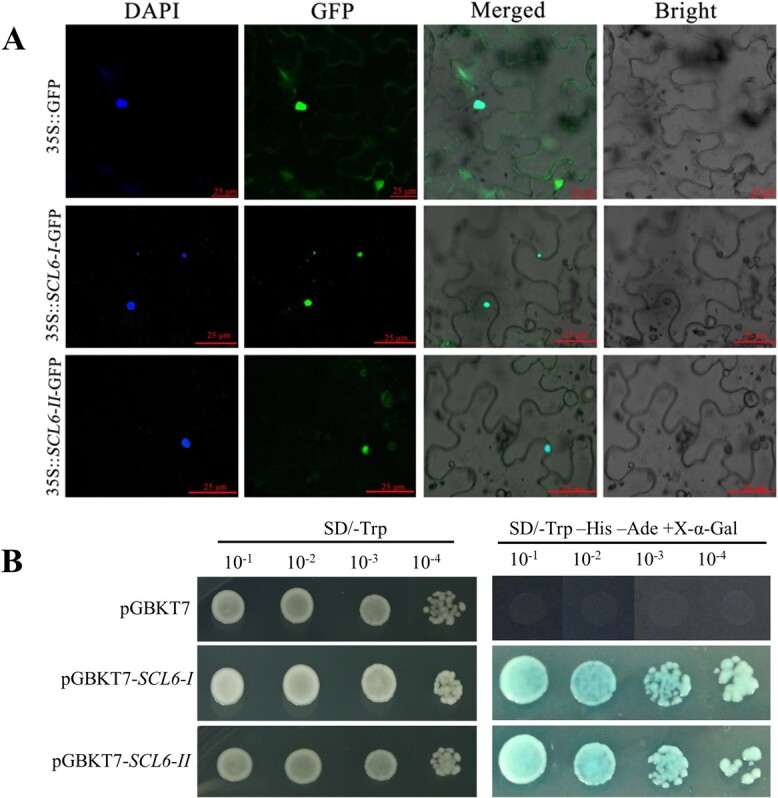
LpSCL6-I and LpSCL6-II subcellular localization and yeast transcriptional activity analysis. a LpSCL6-I and LpSCL6-II subcellular localization. DAPI, positive control for nuclear localization; GFP, green fluorescence channel; Merged, mixed field (superimposed photos); Bright, bright field. b LpSCL6-I and LpSCL6-II yeast transcriptional activity analysis. The left picture shows the result on a deficient medium (SD/−Trp) after the transformation of yeast. The right picture shows the result on triple-deficient medium (SD/−Ade/−His/−Trp) after the transformation of yeast.
The transcriptional activity test results are shown in Fig. 2b. Yeast containing the pGBKT7 empty vector grew only on SD/−Trp medium, while yeast containing pGBKT7-LpSCL6-II and pGBKT7-LpSCL6-I fusion vectors grew normally on both SD/−Trp medium and SD/−Trp−His−Ade medium. After adding X-α-Gal, the cells appeared blue, indicating α-galactosidase activity. These results indicated that LpSCL6-I and LpSCL6-II are transcriptional activators.
Generation and expression analysis of lpu-miR171a, lpu-miR171b, LpSCL6-II, and LpSCL6-I transgenic lines
qRT–PCR was used to detect target gene expression levels in plants with lpu-miR171a and lpu-miR171b overexpression or silencing to screen transgenic lines. The same method was used to screen LpSCL6-II and LpSCL6-I overexpression lines. The identification of precursors is a prerequisite for the identification of mature miRNAs. Therefore, the expression levels of lpu-miR171a and lpu-miR171b precursor and mature forms were analyzed. The results showed that the expression of miR171a and miR171b in the lpu-miR171a- and lpu-miR171b-overexpressing transgenic lines was significantly increased (Fig. 3a and b), whereas the expression of miR171a and miR171b in the silenced transgenic lines was significantly inhibited (Fig. 3c and d). The expression trends of precursor miR171a and miR171b and mature miR171a and miR171b were consistent. The lpu-miR171a- and lpu-miR171b-overexpressing and lpu-miR171a- and lpu-miR171b-silenced transgenic lines were next evaluated to determine the expression levels of LpSCL6-II and LpSCL6-I. The relative expression levels of LpSCL6-II and LpSCL6-I in the lpu-miR171a- and lpu-miR171b-overexpressing transgenic lines were significantly suppressed (Fig. 3a and b), while the relative expression levels of the target genes in the silenced transgenic lines were significantly upregulated (Fig. 3c and d).
Figure 3.
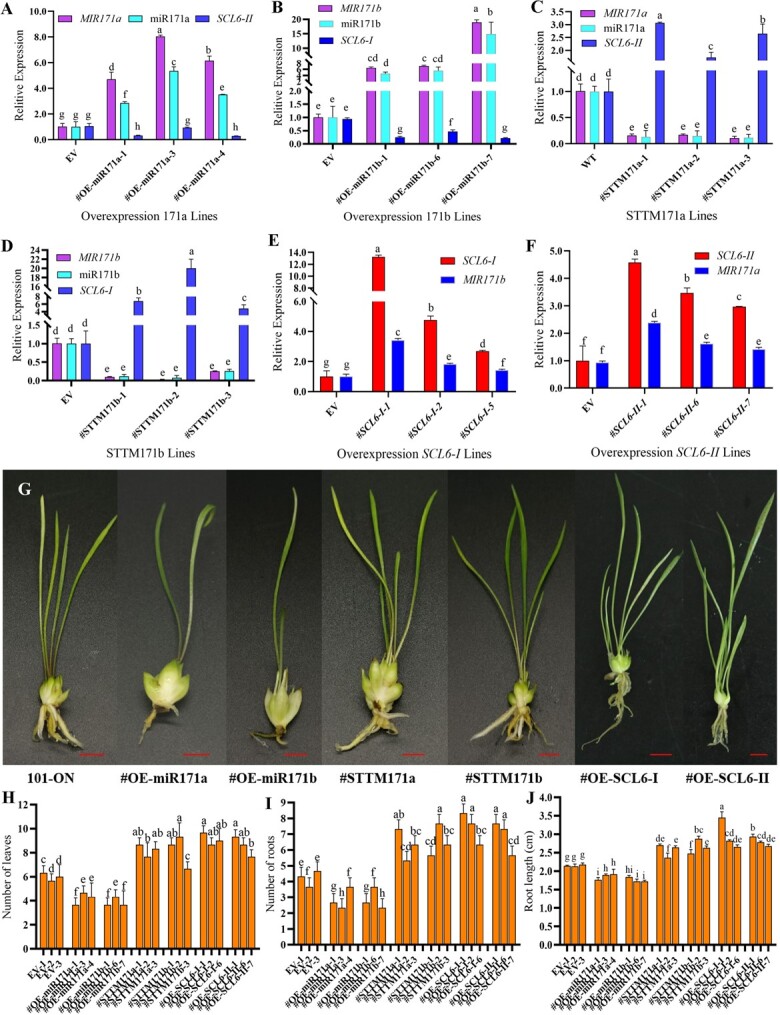
Expression detection, phenotypic observation and growth index determination in transgenic lines. a Expression levels of MIR171a, miR171a, and the target gene LpSCL6-II in miR171a-overexpressing lines. b Expression levels of MIR171b, miR171b, and the target gene LpSCL6-I in miR171b-overexpressing lines. c Expression levels of miR171a and the target gene LpSCL6-II in STTM171a-silenced transgenic lines. d Expression levels of miR171b and the target gene LpSCL6-I in STTM171b-silenced transgenic lines. e Expression level of LpSCL6-I and precursor MIR171b in LpSCL6-I-overexpressing lines. f Expression level of LpSCL6-II and precursor MIR171a in LpSCL6-II-overexpressing lines. g Phenotypic characteristics of different transgenic plants. Scale bars = 1 cm. h Leaf numbers of different transgenic plants. i Root numbers of different transgenic plants. j Root lengths of different transgenic plants.
In the LpSCL6-II- and LpSCL6-I-overexpressing transgenic lines, the expression levels of LpSCL6-II and LpSCL6-I were significantly increased (Fig. 3e and f). Subsequently, the expression levels of miR171a and miR171b in LpSCL6-II- and LpSCL6-I-overexpressing lines were evaluated. Upregulation of LpSCL6-II and LpSCL6-I caused the accumulation of lpu-miR171a and lpu-miR171b, which was significantly upregulated relative to the control (Fig. 3e and f). These results indicated that overexpression of LpSCL6-II and LpSCL6-I affects the transcription of lpu-miR171a and lpu-miR171b. In addition, these findings indicated that in Lilium lpu-miR171a and lpu-miR171b cleave the target genes LpSCL6-II and LpSCL6-I, thereby inhibiting their expression, and that a positive feedback regulation relationship exists between lpu-miR171a and lpu-miR171b and the target genes LpSCL6-II and LpSCL6-I.
The lpu-miR171a- and lpu-miR171b-overexpressing transgenic lines generally showed blockage of root system development and a markedly reduced number of leaves. In contrast, the lpu-miR171a- and lpu-miR171b-silenced and LpSCL6-II- and LpSCL6-I-overexpressing lines showed strong roots and many leaves (Fig. 3g). One month after the transgenic lines were transferred, the numbers of leaves and roots and root system length were quantified. Nine plants from each transgenic line were counted. The number of leaves, the root coefficient, and root system length were significantly different between the transgenic plants and the control (Fig. 3h–j).
Lpu-miR171a and lpu-miR171b silencing or LpSCL6-II and LpSCL6-I overexpression promotes the formation of Lilium somatic embryos
Somatic embryo induction was performed in different transgenic lines. At 28 days, we observed that, in contrast to the control, the lpu-miR171a- and lpu-miR1711b-overexpressing scale surfaces did not produce obvious calli. When viewed under a microscope, obvious globular embryo protrusions were observed around the vascular tissue of the scales. However, the scales of STTM171a- and STTM171b-silenced and LpSCL6-II- and LpSCL6-I-overexpressing lines formed obvious calli, and globular embryos were observed on the surface. In addition, many somatic embryos germinated and formed obvious cotyledon-shaped embryos. At 50 days, relative to the control, lpu-miR171a- and lpu-miR171b-overexpressing scales produced a small number of somatic embryos, and no somatic embryo germination leading to the formation of cotyledon-shaped embryos was observed. However, the scales of the STTM171a- and STTM171b-silenced and LpSCL6-II- and LpSCL6-I-overexpressing lines formed abundant calli and produced more somatic embryos, and more of the somatic embryos germinated and became seedlings (Fig. 4a). The results of the statistical analysis of the SE efficiency of different transgenic lines were consistent with the observation results, as shown in Supplementary Data Table S1. These results indicated that lpu-miR171a and lpu-miR171b overexpression inhibits the formation and development of somatic embryos and reduces the efficiency of SE. In contrast, these results indicated that silencing lpu-miR171a and lpu-miR171b or overexpressing LpSCL6-II and LpSCL6-I accelerates the formation of somatic embryos, increases the rate of cotyledon-type embryo formation, and significantly shortens the development time of somatic embryos.
Figure 4.
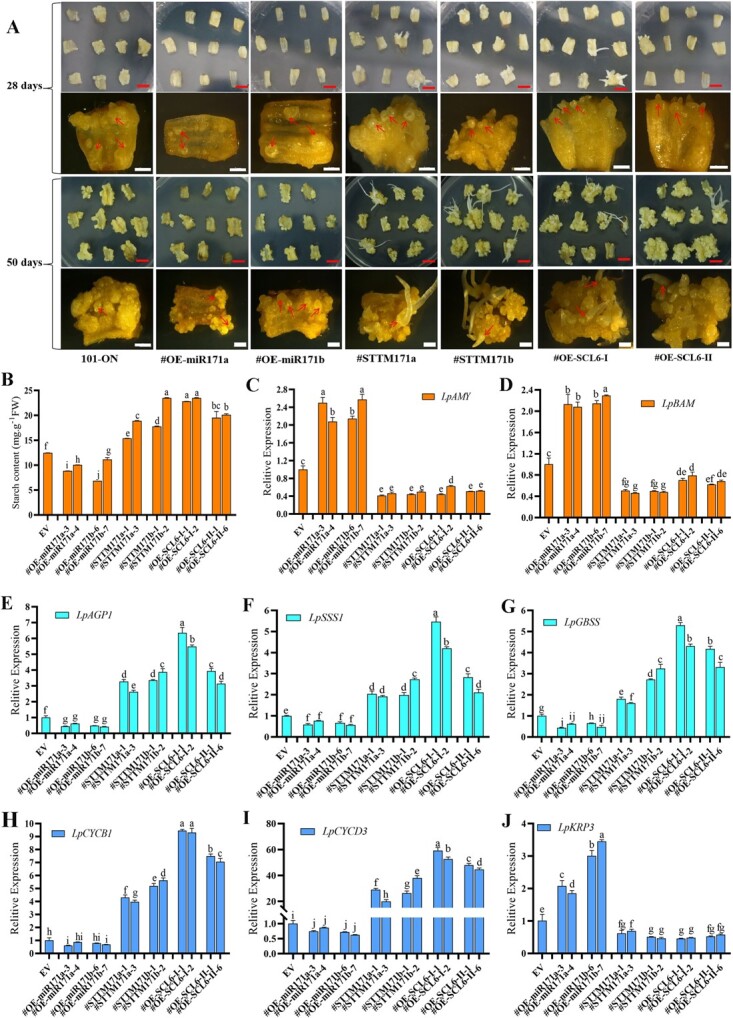
Comparison of somatic embryo induction, tissue morphology observations, starch content determination, and related gene expression. a Growth status of scales at 28 and 50 days of induction. The top images are photographs, and the bottom images are micrographs. Red scale bars = 0.5 cm; white scale bars = 2 mm. Red arrows point to somatic embryos. b Determination of starch contents in different transgenic plants. c and d Expression levels of LpAMY and LpBAM, encoding key starch metabolism enzymes. e Expression level of LpAGP1, a key starch synthesis gene. f Expression level of LpSSS1, a key starch synthesis gene. g Starch synthase LpGBSS expression. h and i Expression of the key cell cycle genes LpCYCB1 and LpCYCD3. j Expression of the cell cycle inhibitor gene LpKRP3.
According to histomorphological observations, the number of embryonic cells in lpu-miR171a- and lpu-miR171b-overexpressing were lower than those in the controls at 28 days and 50 days, and few starch granules were observed. However, obvious embryonic cells were observed in the cells of the lpu-miR171a- and lpu-miR171b-silenced and LpSCL6-II- and LpSCL6-I-overexpressing lines. In addition, much larger and more abundant amyloplasts accumulated (Supplementary Data Fig. S2). Starch content measurements showed that the starch contents of the lpu-miR171a- and lpu-miR171b-overexpressing lines were lower than that of the control, and the starch content was significantly increased after lpu-miR171a and lpu-miR171b silencing and LpSCL6-II and LpSCL6-I overexpression (Fig. 4b). The expression levels of the key starch synthase genes LpAGP1, LpSSS1, and LpGBSS as well as the starch metabolism enzyme genes LpAMY and LpBAM were determined. The results showed that LpAMY and LpBAM were upregulated in lpu-miR171a- and lpu-miR171ab-overexpressing lines and significantly downregulated in lpu-miR171a- and lpu-miR171b-silenced and LpSCL6-II- and LpSCL6-I-overexpressing lines (Fig. 4c and d). After lpu-miR171a and lpu-miR171b overexpression, LpAGP, LpSSS, and LpGBSS were downregulated, whereas their expression levels were significantly upregulated after lpu-miR171a and lpu-miR171b silencing and LpSCL6-II and LpSCL6-I overexpression (Fig. 4e–g). These changes are conducive to maintaining the starch content in accordance with the results of the tissue morphological observation.
Since the somatic embryo formation cycles of the different transgenic lines were different, we evaluated the cell cycle-related gene expression of somatic embryos of different lines. The results showed that lpu-miR171a and lpu-miR171b silencing and LpSCL6-II and LpSCL6-I overexpression were accompanied by significant upregulation of the key cell cycle genes LpCYCB1 and LpCYCD3. In particular, LpCYCD3 was upregulated 40-fold in the somatic embryos of lpu-miR171b-silenced lines relative to the control, and LpCYCD3 was upregulated 60-fold in the somatic embryos of LpSCL6-I-overexpressing lines relative to the control. However, the levels of LpCYCB1 and LpCYCD3 in the lpu-miR171a- and lpu-miR171b-overexpressing lines were slightly decreased relative to the control (Fig. 4h and i). The cell cycle suppressor gene LpKRP3 was significantly upregulated in the lpu-miR171a- and lpu-miR171b-overexpressing lines and significantly downregulated in the lpu-miR171a- and lpu-miR171b-silenced and LpSCL6-II- and LpSCL6-I-overexpressing lines (Fig. 4j). These results further confirmed that lpu-miR171a and lpu-miR171b silencing and LpSCL6-II and LpSCL6-I overexpression accelerate the formation of somatic embryos and significantly shorten the developmental cycle of somatic embryos.
eTM171 acts as an miRNA decoy to suppress the expression of lpu-miR171
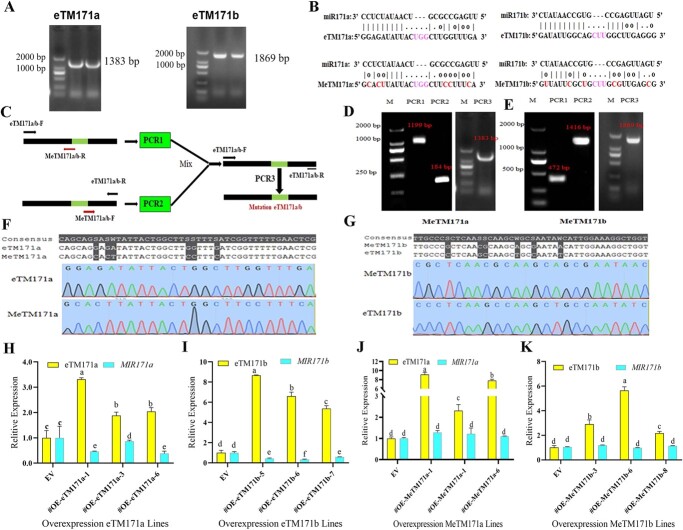
According to the full-length cDNA sequence information of the candidate unigene40819 and unigene40253, specific fragments of 1383 bp (unigene40819) and 1869 bp (unigene40253) were amplified (Fig. 5a). To indicate their relationships with miR171a and miR171b, the cloned unigene40819 and unigene40253 were renamed lpu-eTM171a and lpu-eTM171b, respectively. Overlapping PCR was used to introduce synonymous point mutations at six bases in the lpu-eTM171a and lpu-eTM171b sequences paired with lpu-miR171a and lpu-miR171b (Fig. 5b). After three rounds of PCR (Fig. 5c), the sequences of lpu-MeTM171a and lpu-MeTM171b were amplified (Fig. 5d and e). The sequencing results showed that six bases were mutated in the lpu-eTM171a and lpu-eTM171b sequences (Fig. 5f and g). The lpu-eTM171a, lpu-eTM171b, lpu-MeTM171a, and lpu-MeTM171b overexpression vectors were constructed and transformed into Lilium, and overexpression lines with significantly up-regulated expression were selected by qRT–PCR (Fig. 5h–k).
Figure 5.
eTM171 acts as an endogenous target mimic to inhibit the expression of lpu-miR171. a PCR amplification product of lpu-eTM171a and lpu-eTM171b full-length cDNA (1383 and 1869 bp). b The mutated base sequences introduced by lpu-miR171a and lpu-miR171b. c The overlapping PCR method. d and e lpu-MeTM171a and lpu-MeTM171b: two rounds of PCR amplification products. f and g Sequence alignment of lpu-eTM171a and lpu-eTM171b and of lpu-MeTM171a and lpu-MeTM171b. h and j Expression level of the MIR171a precursor in lines with lpu-eTM171a overexpression and point mutation overexpression. i and k Expression level of the MIR171b precursor in lines with lpu-eTM171b overexpression and point mutation overexpression.
To verify whether lpu-eTM171a and lpu-eTM171b affect Lilium SE, somatic embryo induction was performed on different transgenic lines. The results showed that at 28 days the somatic embryo induction rate and somatic embryo growth status of the lpu-eTM171a- and lpu-eTM171b-overexpressing lines and the lpu-MeTM171a and lpu-MeTM171b point mutation-overexpressing lines were not significantly different from those of the control. At 50 days, only a small number of somatic embryos germinated and formed cotyledon-shaped embryos among the somatic embryos of the lpu-eTM171b-overexpressing lines (Supplementary Data Table S1).
The expression levels of lpu-miR171a and lpu-miR171b in lpu-eTM171a and lpu-eTM171b and point mutation-overexpressing lines were detected. The results showed that the expression of miR171a and miR171b in the lpu-eTM171a- and lpu-eTM171b-overexpressing lines was significantly downregulated by ~50% (Fig. 5h and i). However, the point mutation-overexpressing (OE-MeTM171a and OE-MeTM171b) lines did not show changes in the expression of miR171a and miR171b (Fig. 5j and k). The above results demonstrated that high lpu-eTM171a and lpu-eTM171b expression has a negative regulatory effect on the corresponding lpu-miR171a and lpu-miR171b targets. However, high expression of the lpu-MeTM171a and lpu-MeTM171b point mutants had no negative regulatory effect on lpu-miR171a and lpu-miR171b, indicating that the sequence structure that binds to miRNA inhibits miRNA accumulation.
Mode of eTM171-miR171-SCL6 involvement in Lilium somatic embryogenesis
We examined the expression of lpu-miR171 and LpSCL6 during SE in lpu-eTM171a and lpu-eTM171b and point mutation-overexpressing lines. The results showed that after 28 and 50 days of somatic embryo induction the expression of lpu-miR171a and lpu-miR171b in the OE-eTM171a and OE-eTM171b lines was downregulated, and the expression of the LpSCL6-II and LpSCL6-I target genes was significantly increased. The expression of lpu-miR171a and lpu-miR171b and the LpSCL6-II and LpSCL6-I target genes in the OE-MeTM171a and OE-MeTM171b lines was not changed significantly relative to the control (Fig. 6a–d). These results demonstrated that lpu-eTM171 acts as a decoy for lpu-miR171 and binds lpu-miR171 to prevent cleavage of the LpSCL6 target gene.
Figure 6.
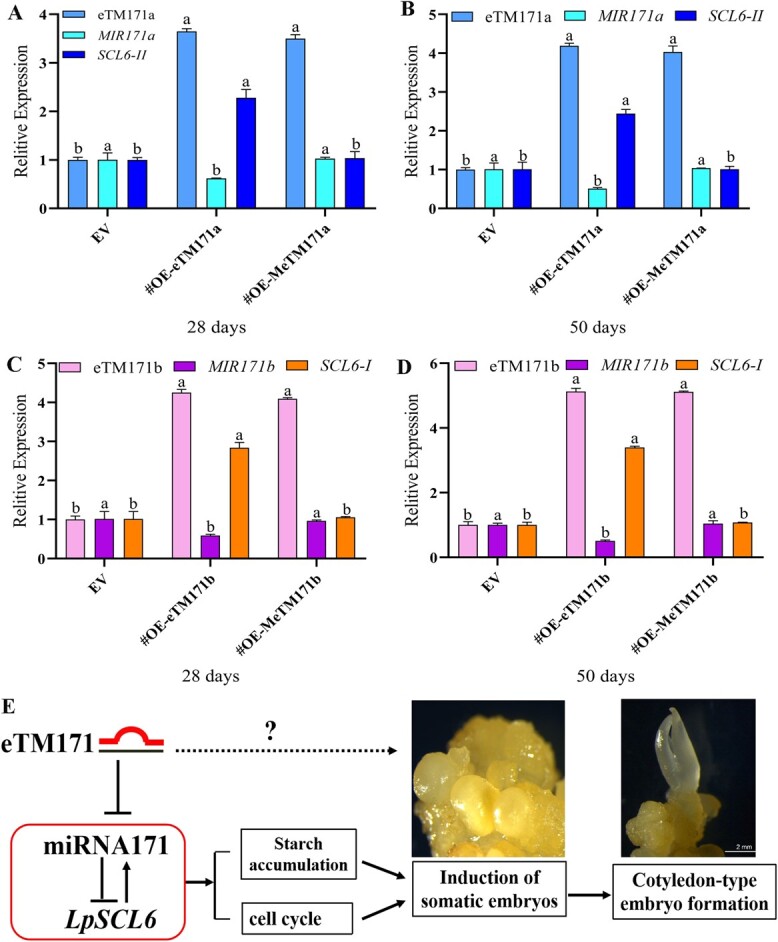
Hypothetical model of the eTM-miR171-SCL6 module regulating SE in L. pumilum. a and b Expression levels of MIR171a and LpSCL6-II on the 28th and 50th days of lpu-eTM171a overexpression and point mutant line somatic embryo induction. c and d Expression levels of MIR171b and LpSCL6-I on the 28th and 50th days of lpu-eTM171b overexpression and point mutant line somatic embryo induction. Different lower-case letters in a-d indicate significant differences (tested by ANOVA) with respect to the same gene in different transgenic lines as a group. e Hypothetical model of the eTM-miR171-SCL6 module regulating SE in L. pumilum. The solid and dotted arrows indicate positive regulation, and the inverted T-shape indicates negative regulation. The solid lines are the results obtained from the verification of this research, and the dotted lines are the results of speculation based on model plant research but not verified.
Discussion
Because miRNA has no coding function, it only functions by cleaving target genes or inhibiting target gene translation. In plants, RLM 5′ RACE technology is mainly used to verify the interactions between miRNAs and target genes [33], but this method cannot intuitively reflect the interaction between miRNAs and target genes. Agrobacterium-mediated tobacco transformation results in high transient expression efficiency and a long expression time, making this approach suitable for the study of gene interactions in plants [34]. This method is mainly used to verify the interaction between miRNAs and target genes by detecting the expression of the GUS gene in tobacco after transiently expressing miRNAs and target genes [35, 36]. In the present study, before verifying the function of lpu-miR171a and lpu-miR171b and their target genes, we used the tobacco transient transformation method to verify the interaction between lpu-miR171a and lpu-miR171b and the LpSCL6-II and LpSCL6-I target genes. GUS histochemical staining and enzyme activity test results showed that the corresponding miRNA inhibited its target gene, but the inhibition efficiency was not 100%. There may be two reasons for this result. First, there is a balanced relationship between miRNA cleavage and target gene transcription, and this relationship is strongly affected by miRNA expression levels [37]. Second, only part of the target mRNA is cleaved and degraded by miRNA, while the remaining target mRNA is separated from the cleavage system and transcribed normally [38].
The miR171-SCL6 module is involved in branch formation [39], meristem maintenance [15], root growth, leaf formation [40], and SE [8]. The miR171 miRNA family is highly conserved. In Arabidopsis, three MIR171 genes (a, b, and c) are predicted to regulate three SCL6 genes (SCLII, SCLIII, and SCLIV) [41–43]. The expression domains of miR171 family members overlap with the mRNA sequences of SCL6-II and SCL6-III, indicating that both miRNAs and target mRNAs have redundant functions [44]. In Arabidopsis, plants overexpressing miR171c (OE171c) and triple scl6 mutants show similar pleiotropic phenotypes, including reduced branch numbers, plant height, chlorophyll accumulation, shortened main roots, and changes in flower structure and leaf shape. These results indicate that miR171 mainly suppresses the expression of the SCL6 gene to control extensive developmental processes during bud development [39]. However, the functions of Lilium miR171s are unclear.
miR171s play diverse roles in different plant species [14]. In rice, osa-miR171c accumulated substantially under abiotic stress, and transgenic lines overexpressing osa-miR171c displayed increased sensitivity to salt stress [45]. Both knockout of mdm-miR171i and overexpression of MsSCL26.1 improved drought stress tolerance in the cultivated apple line ‘GL-3’ [46]. In this study, after lpu-miR171a and lpu-miR171b overexpression in L. pumilum, the plants generally showed hindered root development and fewer leaves. In contrast, the silenced plants showed strong roots and many leaves, similar to the phenotype observed in Arabidopsis [39] and tomato plants [20]. The expression analysis of the LpSCL6-II and LpSCL6-I target genes in transgenic lines with lpu-miR171a and lpu-miR171b overexpression and STTM silencing showed that, after lpu-miR171a and lpu-miR171b overexpression, the expression of the target genes was significantly downregulated. However, the silencing of lpu-miR171a and lpu-miR171b relieved the inhibitory effect on the target genes and significantly increased the expression of LpSCL6-II and LpSCL6-I. These findings further demonstrated that miR171a and miR171b regulate the growth and development of Lilium through the targeted regulation of the SCL6 target gene.
Our previous studies have shown that miR171a and miR171b accumulate in large amounts in the early and late stages of somatic embryogenesis [13]. In this study, we found that lpu-miR171a and lpu-miR171b overexpression inhibited the formation and development of somatic embryos, and the efficiency of SE was low. In contrast, lpu-miR171a and lpu-miR171b silencing accelerated the formation of somatic embryos and significantly shortened the development of somatic embryos. The above research results indicate that in lily plants the accumulation of miR171 has a negative regulatory effect on plant growth and somatic embryo formation. miR171 was induced by abiotic stress in embryogenic callus tissues from Larix leptolepis [47]. In this study, plant growth and somatic embryo development were blocked after miR171 overexpression, which may be related to abiotic stress, but the relationship between miR171 and somatic embryo development under abiotic stress needs further study. The functions of miR171a and miR171b are determined by the functions of the corresponding target genes SCL6-II and SCL6-I. We found that there were 12 SCL6 members in the lily SE transcriptome database, but only LpSCL6-II and LpSCL6-I were the target genes corresponding to miR171a and miR171b [13]. In this study, some of the functions of LpSCL6-II and LpSCL6-I were similar, while others were different. Both genes play important roles in the early and late stages of somatic embryogenesis; however, LpSCL6-I has a more obvious effect than LpSCL6-II.
CYCB and CYCD are key genes in the G2/M transition that are often used as markers of cell division [48]. Cell cycle repressor kip-related proteins (KRPs) mainly inhibit cell cycle proteins and negatively regulate cell proliferation [49]. In this study, the expression levels of LpCYCB1 and LpCYCD3 were significantly upregulated after LpSCL6 overexpression, while the expression of the LpKRP gene was inhibited, indicating that LpSCL6 overexpression promoted the proliferation of Lilium embryogenic cells and was beneficial to the formation of somatic embryos. Studies have shown that the increase in the contents of embryogenic calli, especially amyloplasts, can provide an energy and material basis for the formation of somatic embryos [5]. In this study, histomorphological observations showed that few amyloplasts accumulated in embryonic cells overexpressing lpu-miR171a and lpu-miR171b compared with the control lines. However, lpu-miR171a and lpu-miR171b silencing and LpSCL6-II and LpSCL6-I overexpression resulted in the accumulation of much larger and more abundant amyloplasts. In Carica papaya, increased sucrose and starch content is a key point in somatic embryo transformation, indicating that carbohydrates such as starch play a key role in the morphogenesis of somatic embryos [50]. Among the key enzymes in starch synthesis, adenosine 5′-diphosphate glucose pyrophosphorylase (AGPase) is considered to be the key enzyme required for the first step of plant starch synthesis [51]. Starch synthase (SS) includes granule-bound starch synthase (GBSS) and soluble starch synthase (SSS). GBSS mainly catalyzes the synthesis of amylose, and SSS mainly catalyzes the synthesis of amylopectin [52]. α-Amylase (AMY) and β-amylase (BAM) are key enzymes in starch degradation [53]. In this study, overexpression of LpSCL6 promoted the expression of starch synthase genes LpAGP, LpSSS and LpGBSS, and inhibited the expression of starch degradation genes LpAMY and LpBAM. These results were consistent with the previous conclusion that overexpression of LpSCL6 promoted the accumulation of starch.
Tang et al. [28] developed STTM technology based on TMs, which is an effective tool for studying miRNA functions in plants. The overexpression of STTMs has been used to identify miRNA functions in a variety of plants, such as Arabidopsis and wheat [54, 55]. To further explore the function of lpu-miR171a and lpu-miR171b candidates, we used STTM technology to silence endogenous miRNAs based on stable transformation. The results showed that lpu-miR171a and lpu-miR171b silencing effectively shortened the SE cycle of L. pumilum, thereby revealing a new function of miR171 in the regulation of Lilium and plant growth and development.
The present study confirmed the function of lpu-miR171 and the LpSCL6 target gene in the SE of L. pumilum and elucidated the regulatory effect of lpu-miR171 on the LpSCL6 target gene. In summary, a hypothetical model whereby the eTM-miR171-SCL6 module regulates SE in L. pumilum is proposed based on combining our results and previous reports involving model plants (Fig. 6e). Our research provides promising candidate genes for enhancing the SE ability of Lilium and lays a good foundation for studying the mechanism of SE in this genus.
Materials and methods
Plant materials
Embryogenic calli and somatic embryos of L. pumilum DC. Fisch. were obtained according to a previously described method [27]. Tobacco seedlings aged 28 days were also used.
Gene cloning
Genomic DNA was extracted via the CTAB method as described previously [26]. According to the predicted sequence information on the lpu-eTM171 candidate, Primer 5.0 software was used to design primers (shown in Supplementary Data Table S2), and the DNA was used as a template for PCR amplification. The PCR products were inserted into the pMD18-T vector and sequenced. Sequence alignment was performed using DNAMAN8.
Plasmid construction and Lilium transformation
The precursor fragments of lpu-miR171a and lpu-miR171b and the coding sequence (CDS) regions of LpSCL6-II and LpSCL6-I and lpu-eTM171a and lpu-eTM171b were amplified by PCR. According to the seamless cloning principle of the In-Fusion HD Cloning Kit (TaKaRa, Dalian, China), these products were cloned into the plasmid pRI101-ON. A short tandem target mimic (STTM) sequence (Supplementary Data Table S2) that specifically inhibited lpu-miR171a and lpu-miR171b was designed and synthesized according to the method of Tang et al. [28] and cloned into the pRI101-ON plasmid. A schematic diagram of the vector structure is shown in Supplementary Data Fig. S1. All of the above vectors were used to transform the embryogenic calli of L. pumilum. For genetic transformation, the method that we reported previously was used [29]. The precursor fragments of lpu-miR171a and lpu-miR171b and the CDS regions of LpSCL6-II and LpSCL6-I were cloned into the pRI101-ON-GUS and pRI-ON-LUC plasmids for the transient transformation of tobacco leaves.
We introduced a six-base synonymous point mutation into the lpu-eTM171a and lpu-eTM171b sequences paired with lpu-miR171a and lpu-miR171b by referring to the reported method of lncRNA23468 point mutation in tomato [30]. The lpu-eTM171a and lpu-eTM171b mutations were generated by overlapping PCR, which involved the use of lpu-eTM171a and lpu-eTM171b as templates for amplification and mutagenesis. Three rounds of PCR were performed to amplify the lpu-MeTM171a and lpu-MeTM171b mutants (the procedure is illustrated in Fig. 5c). Point mutation overexpression vectors were constructed (named pRI101-MeTM171a and pRI101-MeTM171b) and transformed into embryogenic calli of Lilium (the primers used are shown in Supplementary Data Table S2).
β-Glucuronidase histochemical staining and luciferase reporter system detection
β-Glucuronidase (GUS) histochemical staining and enzyme activity detection were performed as described previously [26]. The pRI-mini35S-LUC, 35S::LUC-SCL6 I, 35S::LUC-SCL6 II, pRI-101-miR171a, and pRI-101-miR171b luciferase (LUC) reporter gene vectors as well as the coexpression vectors (35S::miR171a + 35S::LUC-SCL6 II and 35S::miR171b + 35S::LUC-SCL6 I) were transferred into tobacco leaves and observed with a digital in vivo luminescence imager after 48 h.
Subcellular localization and transcriptional activation analysis
The full-length LpSCL6-II and LpSCL6-I CDSs without the termination codon were amplified in the pRI101-GFP vector. The primers used for this purpose are shown in Supplementary Data Table S2. Using the DAPI (4′,6-diamidino-2-phenylindole) nuclear localization dye signal as a positive control and pRI-101-GFP as a negative control, the vectors were injected into Nicotiana benthamiana leaves. After transformation for 2 days, the leaves were visualized using a laser confocal fluorescence microscope (TCSSP8-SE, Leica, Wetzlar, Germany).
The transactivation assay was performed using the Matchmaker GAL4 Two-Hybrid System 3 (Clontech, Palo Alto, CA, USA). The coding regions of LpSCL6-II and LpSCL6-I were inserted into the pGBKT7 vector. The pGBKT7-LpSCL6-I and pGBKT7-LpSCL6-II constructs as well as the pGBKT7 negative control were transformed into the Y2H Gold yeast strain. The transformed strains were grown on SD/−Trp medium and selected on SD/−Trp−His−Ade medium.
Gene expression analysis
Leaves and calli of the control and transgenic lilies were collected separately and stored at −80°C. The HiPure HP Plant RNA Kit (Magen, Shanghai, China) was used to extract total RNA, and the FastKing cDNA Kit (Tiangen, Beijing, China) was used to reverse-transcribe the RNA into cDNA. Referring to our previous report [31], a QuantStudio 3 Real-Time PCR System was used for qRT–PCR analysis. The qRT–PCR primers are listed in Supplementary Data Table S2. Gene expression levels were calculated using the 2–ΔΔCt method. Each sample included three biological replicates and three technical replicates.
Comparison of somatic embryo inducibility
In the SE system, somatic embryo induction medium was inoculated with different transgenic lines of Lilium. Transgenic lines carrying empty vectors were used as controls to compare the SE and regeneration efficiency of the transgenic lines. The process of somatic embryogenesis was observed, and the somatic embryogenesis induction rate and cotyledon-shaped embryo induction rate were calculated at 28 and 50 days. Each experiment included 30 explants and was carried out three times. The SE induction rate (%) was calculated as 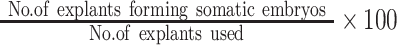 . The cotyledon-shaped embryo induction rate (%) was calculated as
. The cotyledon-shaped embryo induction rate (%) was calculated as 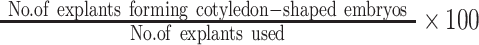 .
.
Histomorphology and determination of starch content
Morphological observations of the somatic embryos induced from different transgenic lines were performed at different developmental stages. The samples were fixed in formalin–acetic acid–alcohol (FAA; 90% ethanol, 5% formaldehyde, and 5% acetic acid; v/v/v) for 24 h at 4°C, and paraffin sections were prepared according to the method of Fu et al. [32]. After dehydration-transparency embedding, the embedded wax blocks were sliced into 6-μm sections using a microtome (RM 2245, Leica, Germany). After staining with 0.01% toluidine blue, the sections were observed and photographed under an optical microscope (DM3000, Leica, Germany). The morphological structures of somatic embryos at different stages, somatic embryo contents, and starch granule contents were observed and compared. The starch content was determined according to the instructions of the starch determination kit (Suzhou Keming Biotechnology Co., Ltd., Suzhou, China).
Statistical analysis
The data were analyzed with IBM SPSS Statistics 23.0 (SPSS Inc., Chicago, USA) for one-way ANOVA and Duncan’s test. The effects were considered significant at P < .05.
Acknowledgements
This work was supported by the National Key R&D Program of China (2018YFD1000407), the National Natural Science Foundation of China (Grant Nos. 31672179 and 31872150) and the LiaoNing Revitalization Talents Program (XLYC2002052).
Author contributions
R.Y. and H.M.S. conceived and designed the experiments. R.Y. and S.L.S. performed the experiments and analyzed the data. S.L.S. and H.Y.L. provided the experimental methods and participated in the discussion. R.Y. and H.M.S. wrote and revised the manuscript. R.Y. and S.L.S. contributed equally to this work. All authors read and approved the final manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of interest
The authors declare that they have no conflicts of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
Supplementary Material
Contributor Information
Rui Yan, Key Laboratory of Protected Horticulture of Education Ministry, College of Horticulture, Shenyang Agricultural University, National and Local Joint Engineering Research Center of Northern Horticultural Facilities Design and Application Technology, Shenyang 110866, China; School of Agriculture, Ningxia University, Yinchuan, Ningxia 750021, China.
Shengli Song, Key Laboratory of Protected Horticulture of Education Ministry, College of Horticulture, Shenyang Agricultural University, National and Local Joint Engineering Research Center of Northern Horticultural Facilities Design and Application Technology, Shenyang 110866, China.
Hongyu Li, College of Life Science and Bioengineering, Shenyang University, Shenyang 110866, China.
Hongmei Sun, Key Laboratory of Protected Horticulture of Education Ministry, College of Horticulture, Shenyang Agricultural University, National and Local Joint Engineering Research Center of Northern Horticultural Facilities Design and Application Technology, Shenyang 110866, China.
References
- 1. Bakhshaie M, Babalar M, Mirmasoumi M et al. Somatic embryogenesis and plant regeneration of Lilium ledebourii (Baker) Boiss., an endangered species. Plant Cell Tissue Organ Cult. 2010;102:229–35. [Google Scholar]
- 2. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo Y, Zhou H, Li Y et al. Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 2006;580:5111–6. [DOI] [PubMed] [Google Scholar]
- 4. Wu X, Kou SJ, Liu YL et al. Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA-and siRNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol J. 2015;13:383–94. [DOI] [PubMed] [Google Scholar]
- 5. Long JM, Liu C-Y, Feng M-Q et al. miR156-SPL modules regulate induction of somatic embryogenesis in citrus callus. J Exp Bot. 2018;69:2979–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang J, Zhang S, Han S et al. Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta. 2012;236:647–57. [DOI] [PubMed] [Google Scholar]
- 7. Li W, Zhang S-G, Han S-Y et al. Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.) Carr. Plant Cell Tissue Organ Cult. 2013;113:131–6. [Google Scholar]
- 8. Li W, Zhang S-G, Han S-Y et al. The post-transcriptional regulation of LaSCL6 by miR171 during maintenance of embryogenic potential in Larix kaempferi (Lamb.) Carr. Tree Genet Genomes. 2014;10:223–9. [Google Scholar]
- 9. Yang X, Wang L, Yuan D et al. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J Exp Bot. 2013;64:1521–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chavez-Hernandez EC, Alejandri-Ramirez ND, Juarez-Gonzalez VT et al. Maize miRNA and target regulation in response to hormone depletion and light exposure during somatic embryogenesis. Front Plant Sci. 2015;6:555–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin Y, Lai Z-X, Lin L et al. Endogenous target mimics, microRNA167, and its targets ARF6 and ARF8 during somatic embryo development in Dimocarpus longan Lour. Mol Breed. 2015;35:1–15. [Google Scholar]
- 12. Lin Y, Lai Z. Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan Lour.). PLoS One. 2013;8:e60337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H, Zhang J, Yang Y et al. miR171 and its target gene SCL6 contribute to embryogenic callus induction and torpedo-shaped embryo formation during somatic embryogenesis in two lily species. Plant Cell Tissue Organ Cult. 2017;130:591–600. [Google Scholar]
- 14. Zhu X, Leng X, Sun X et al. Discovery of conservation and diversification of miR171 genes by phylogenetic analysis based on global genomes. Plant Genome. 2015;8. [DOI] [PubMed] [Google Scholar]
- 15. Fan T, Li X, Yang W et al. Rice Osa-miR171c mediates phase change from vegetative to reproductive development and shoot apical meristem maintenance by repressing four OsHAM transcription factors. PLoS One. 2015;10:e0125833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhai L, Xu L, Wang Y et al. Genome-wide identification of embryogenesis-associated microRNAs in radish (Raphanus sativus L.) by high-throughput sequencing. Plant Mol Biol Rep. 2014;32:900–15. [Google Scholar]
- 17. Franco-Zorrilla JM, Valli A, Todesco M et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–7. [DOI] [PubMed] [Google Scholar]
- 18. Wu HJ, Wang ZM, Wang M et al. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013;161:1875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meng Y, Shao C, Wang H et al. Target mimics: an embedded layer of microRNA-involved gene regulatory networks in plants. BMC Genomics. 2012;13:197–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kravchik M, Stav R, Belausov E et al. Functional characterization of microRNA171 family in tomato. Plants. 2019;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye C, Xu H, Shen E et al. Genome-wide identification of non-coding RNAs interacted with microRNAs in soybean. Front Plant Sci. 2014;5:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang J, Yu W, Yang Y et al. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci Rep. 2015;5:16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li F, Wang W, Zhao N et al. Regulation of nicotine biosynthesis by an endogenous target mimicry of microRNA in tobacco. Plant Physiol. 2015;169:1062–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin Y, Lai Z, Tian Q et al. Endogenous target mimics down-regulate miR160 mediation of ARF10, -16, and -17 cleavage during somatic embryogenesis in Dimocarpus longan Lour. Front Plant Sci. 2015;6:956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang J, Xue B, Gai M et al. Small RNA and transcriptome sequencing reveal a potential miRNA-mediated interaction network that functions during somatic embryogenesis in Lilium pumilum DC. Fisch. Front Plant Sci. 2017;8:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Wang J, Yan R et al. Functional characterization of the MiR171a promoter and endogenous target mimics identification in Lilium pumilum DC. Fisch. during somatic embryogenesis. Plant Cell Tissue Organ Cult. 2020;144:345–57. [Google Scholar]
- 27. Zhang J, Gai M, Li X et al. Somatic embryogenesis and direct as well as indirect organogenesis in Lilium pumilum DC. Fisch., an endangered ornamental and medicinal plant. Biosci Biotechnol Biochem. 2016;80:1898–906. [DOI] [PubMed] [Google Scholar]
- 28. Tang G, Yan J, Gu Y et al. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods. 2012;58:118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan R, Wang Z, Ren Y et al. Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven. Int J Mol Sci. 2019;20:2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang N, Cui J, Shi Y et al. Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato–Phytophthora infestans interaction. Hortic Res. 2019;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang J, Gai MZ, Xue BY et al. The use of miRNAs as reference genes for miRNA expression normalization during Lilium somatic embryogenesis by real-time reverse transcription PCR analysis. Plant Cell Tissue Organ Cult. 2017;129:105–18. [Google Scholar]
- 32. Fu L, Zhu Y, Li M et al. Autopolyploid induction via somatic embryogenesis in Lilium distichum Nakai and Lilium cernuum Komar. Plant Cell Tissue Organ Cult. 2019;139:237–48. [Google Scholar]
- 33. Llave C, Xie Z, Kasschau KD et al. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–6. [DOI] [PubMed] [Google Scholar]
- 34. Yin J, Wong W, Jang I et al. Co-expression of peppermint geranyl diphosphate synthase small subunit enhances monoterpene production in transgenic tobacco plants. New Phytol. 2017;213:1133–44. [DOI] [PubMed] [Google Scholar]
- 35. Feng H, Zhang Q, Wang Q et al. Target of tae-miR408, a chemocyanin-like protein gene (TaCLP1), plays positive roles in wheat response to high-salinity, heavy cupric stress and stripe rust. Plant Mol Biol. 2013;83:433–43. [DOI] [PubMed] [Google Scholar]
- 36. Han L, Weng K, Ma H et al. Identification and characterization of Erysiphe necator-responsive microRNAs in Chinese wild Vitis pseudoreticulata by high-throughput sequencing. Front Plant Sci. 2016;7:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nikovics K, Blein T, Peaucelle A et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adam H, Marguerettaz M, Qadri R et al. Divergent expression patterns of miR164 and CUP-SHAPED COTYLEDON genes in palms and other monocots: implication for the evolution of meristem function in angiosperms. Mol Biol Evol. 2011;28:1439–54. [DOI] [PubMed] [Google Scholar]
- 39. Wang L, Mai Y, Zhang Y et al. MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant. 2010;3:794–806. [DOI] [PubMed] [Google Scholar]
- 40. Hai BZ, Qiu Z, He Y et al. Characterization and primary functional analysis of Pinus densata miR171. Biol Plant. 2018;62:318–24. [Google Scholar]
- 41. Engstrom EM, Andersen CM, Gumulak-Smith J et al. Arabidopsis homologs of the petunia hairy meristem gene are required for maintenance of shoot and root indeterminacy. Plant Physiol. 2011;155:735–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reinhart BJ, Weinstein EG, Rhoades MW et al. MicroRNAs in plants. Genes Dev. 2002;16:1616–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schulze S, Schafer BN, Parizotto EA et al. Lost meristems genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J. 2010;64:668–78. [DOI] [PubMed] [Google Scholar]
- 44. Parizotto EA, Dunoyer P, Rahm N et al. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18:2237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang W, Fan T, Hu X et al. Overexpressing Osa-miR171c decreases salt stress tolerance in rice. J Plant Biol. 2017;60:485–92. [Google Scholar]
- 46. Wang Y, Feng C, Zhai Z et al. The apple microR171i-SCARECROW-LIKE PROTEINS26.1 module enhances drought stress tolerance by integrating ascorbic acid metabolism. Plant Physiol. 2020;184:194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang S, Zhou J, Han S et al. Four abiotic stress-induced miRNA families differentially regulated in the embryogenic and non-embryogenic callus tissues of Larix leptolepis. Biochem Biophys Res Commun. 2010;398:355–60. [DOI] [PubMed] [Google Scholar]
- 48. Scofield S, Jones A, Murray JAH. The plant cell cycle in context. J Exp Bot. 2014;65:2557–62. [DOI] [PubMed] [Google Scholar]
- 49. Jun SE, Okushima Y, Nam J et al. Kip-related protein 3 is required for control of endoreduplication in the shoot apical meristem and leaves of Arabidopsis. Mol Cells. 2013;35:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vale EM, Reis RS, Passamani LZ et al. Morphological analyses and variation in carbohydrate content during the maturation of somatic embryos of Carica papaya. Physiol Mol Biol Plants. 2018;24:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang Y, Gao T, Xu M et al. Functional analysis of a wheat AGPase plastidial small subunit with a truncated transit peptide. Molecules. 2017;22:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang H, Jang S-G, Lar SM et al. Genome-wide identification and genetic variations of the starch synthase gene family in rice. Plants. 2021;10:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fulton DC, Stettler M, Mettler T et al. Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell. 2008;20:1040–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jia X, Ding N, Fan W et al. Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci. 2015;233:11–21. [DOI] [PubMed] [Google Scholar]
- 55. Jiao J, Wang Y, Selvaraj JN et al. Barley stripe mosaic virus (BSMV) induced microRNA silencing in common wheat (Triticum aestivum L.). PLoS One. 2015;10:e0126621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.