Abstract
Unicellular marine cyanobacteria are ubiquitous in both coastal and oligotrophic regimes. The contribution of these organisms to primary production and nutrient cycling is substantial on a global scale. Natural populations of marine Synechococcus strains include multiple genetic lineages, but the link, if any, between unique phenotypic traits and specific genetic groups is still not understood. We studied the genetic diversity (as determined by the DNA-dependent RNA polymerase rpoC1 gene sequence) of a set of marine Synechococcus isolates that are able to swim. Our results show that these isolates form a monophyletic group. This finding represents the first example of correspondence between a physiological trait and a phylogenetic group in marine Synechococcus. In contrast, the phycourobilin (PUB)/phycoerythrobilin (PEB) pigment ratios of members of the motile clade varied considerably. An isolate obtained from the California Current (strain CC9703) displayed a pigment signature identical to that of nonmotile strain WH7803, which is considered a model for low-PUB/PEB-ratio strains, whereas several motile strains had higher PUB/PEB ratios than strain WH8103, which is considered a model for high-PUB/PEB-ratio strains. These findings indicate that the PUB/PEB pigment ratio is not a useful characteristic for defining phylogenetic groups of marine Synechococcus strains.
Unicellular marine cyanobacteria are abundant in both coastal and oligotrophic environments (6, 19, 21), where they contribute substantially to primary production (15). Phylogenetic analyses of isolates have shown that the Synechococcus group comprises multiple genetic lineages (9, 25, 30, 31, 39). These lineages have also been identified in situ, as demonstrated with shotgun-cloned rpoC1 libraries (11, 23). The relationship, if any, between specific genetic lineages and unique physiological adaptations is not currently understood.
Marine Synechococcus strains differ from each other physiologically in a number of ways. The differences include differences in the responses to nitrogen depletion (14), in preferences for nitrate or urea for growth (8, 26, 33), in cell size and growth rate (26, 33), and in cell cycle behavior (1).
There is also physiological diversity in the Synechococcus light-harvesting apparatus. Synechococcus cells harvest light with a structure known as the phycobilisome (13). In open-ocean Synechococcus strains phycoerythrin (PE) is the primary light-harvesting protein in the phycobilisome. There are two PE proteins encoded by different genes in marine Synechococcus strains, and they absorb light with the covalently attached chromophores phycourobilin (PUB) (maximum absorption at 495 nm) and phycoerythrobilin (PEB) (maximum absorption at 545 nm) (22). The in vivo absorption properties of cells are due mostly to the PUB and PEB chromophores and to chlorophyll a. It is thought that the presence of these two PE proteins in marine strains reflects evolutionary chromatic adaptation since these proteins enhance light harvesting at the blue wavelengths which penetrate the water column in oceanic waters (37).
Cultivated marine Synechococcus strains exhibit different PUB/PEB chromophore ratios (33), and the strain-specific differences can be relatively constant under different growth conditions (21, 38). Based on this observation, when multiple pigment signatures have been observed in the environment by using flow cytometry or fluorescence spectra, it has been proposed that they result from genetically distinct populations (17, 20, 21, 38, 40).
Another example of physiological diversity in the genus Synechococcus is the ability of some marine strains to swim by means of a unique type of motility that is characterized by the absence of flagella or any other visible motility organelle (35). It is likely that motility is an ecologically relevant trait, especially in the open ocean. Motile strains exhibit chemotactic responses to nitrogenous compounds, such as ammonia, nitrate, urea, glycine, and β-alanine, at concentrations relevant in the oligotrophic oceans (36). Hence, as an adaptive trait, motility may result in an enhanced ability to find and take up nitrogen, often a likely limiting resource in marine systems, as well as access to unique ecological niches, such as the microenvironments around particles and larger phytoplankton cells (36).
The motile marine Synechococcus strains currently in culture display a range of PUB/PEB chromophore ratios (33). In characterizing motile isolates from the California Current, we realized that this range was even broader and encompassed ratios exhibited by some nonmotile strains. As part of ongoing efforts to understand the spatial and temporal structure of the California Current cyanobacterial community, we investigated the link between two physiological characteristics, motility and pigment complement, and phylogeny. To do this, the phylogenetic diversity of motile Synechococcus strains and members of other unicellular cyanobacterial lineages was determined by using DNA-dependent RNA polymerase gene sequences. Furthermore, the PUB/PEB ratios of motile strains, including the ratios of several new isolates, were determined. We show here that motile marine Synechococcus strains form a monophyletic group whose members exhibit a wide range of PUB/PEB ratios; our data indicate that motility is a better marker of genetic affiliation than pigment ratios are.
MATERIALS AND METHODS
Strain isolation and growth conditions.
The strains used in this work and some of their relevant properties are shown in Table 1. Strains CC9702 and CC9703 were isolated from the oligotrophic edge of the California Current at stations 93.120 (30 m) and 77.100 (70 m), respectively, of the CalCOFI (California Cooperative Oceanic Fisheries Investigations, cruise 9709) grid pattern. The isolation procedure used was the procedure described by Toledo and Palenik (30), with the following modification: the original inoculum was either filtered through a 1.2-μm-pore-size filter or not filtered. The unfiltered treatment resulted in enrichment cultures containing eukaryotic cells and unicellular cyanobacteria. After dense cyanobacterial growth was observed, the samples were filtered through a 1.2-μm-pore-size filter to remove large eukaryotic cells and to obtain a cyanobacterial enrichment culture; this was followed by plating. Clonal isolates were obtained by two rounds of pour plating (3) and isolation of single colonies. All strains were grown in SN medium (34) at a light intensity of 0.15 × 1016 quanta m−2 s−1 under cool white fluorescent tubes at 23°C. Freshwater Synechococcus sp. strain E. Lake was grown in BG11 medium (7) containing 1.7 mM NaNO3.
TABLE 1.
Synechococcus strains used in this study and some of their characteristics
| Strain | Geographic region of isolation | PUB/PEB ratioa | Source | Motility |
|---|---|---|---|---|
| E. Lake | Freshwater (Yosemite National Park) | No PUB | B. Palenikb | − |
| WH7803 | Atlantic Ocean | 0.4 | J. Waterburyc | − |
| WH7805 | Atlantic Ocean | No PUB | J. Waterburyc | − |
| WH8011 | Atlantic Ocean | 0.8 | J. Waterburyc | + |
| WH8102 | Atlantic Ocean | 2.0 | J. Waterburyc | + |
| WH8103 | Atlantic Ocean | 1.2 | J. Waterburyc | + |
| WH8112 | Atlantic Ocean | 1.1 | J. Waterburyc | + |
| WH8113 | Atlantic Ocean | 1.0 | J. Waterburyc | + |
| CC9301d | California Current | 2.0 | B. Brahamsha | + |
| CC9702 | California Current | 1.6 | This study | + |
| CC9703 | California Current | 0.4 | This study | + |
| C129 | Red Sea | 0.9 | A. Post and D. Lindell | + |
Motility was assessed by phase-contrast microscopy by using wet mounts of logarithmic- and stationary-phase cultures. Motile isolates also exhibited diffuse colony morphology in pour plates, as reported previously by Brahamsha (3).
DNA extraction and PCR amplification.
Genomic DNA was extracted as described by Brahamsha (3) and was purified further with a Gene Clean kit (Bio 101). Alternatively, DNA was extracted from cells by using a High Pure PCR template preparation kit (Boehringer Mannheim) as recommended by the manufacturer. The PCR primers used and the method used to amplify the 612-bp rpoC1 gene fragment have been described previously (24, 30). PCR performed with DNA from motile strains sometimes resulted in low yields of the 612-bp fragment and often a smaller PCR product. Based on its sequence, the latter product is not related to rpoC1 (data not shown). The amplified band whose size corresponded to the size of rpoC1 was then excised from the agarose gel, purified, cloned, and sequenced as described below. DNA was purified from the agarose gel with a Gene Clean glassmilk purification kit (Bio 101).
Cloning and sequencing.
The products of PCR were cloned either directly or after gel purification into the pCR2.1 cloning vector (TA cloning kit or TA-topo kit; Invitrogen) by following the manufacturer’s instructions. The nucleotide sequences of both strands of the cloned 612-bp rpoC1 fragment were determined by automated sequencing of plasmid templates performed with a PRISM Ready Reaction DyeDeoxy terminator cyle sequencing kit and a model 373 DNA sequencer (Applied Biosystems).
Phylogenetic analysis.
Neighbor-joining trees were constructed with Jukes-Cantor corrected distances. A bootstrap analysis was performed with 100 subsamples. All of the programs used for analysis were obtained from the PHYLIP package, version 3.572 (10). Strains CC9616, CC9617, CC9701, CC9605, and SS9401 are nonmotile marine Synechococcus spp. strains that were isolated from either the California Current (CC9616, CC9617, CC9701, and CC9605) or the Sargasso Sea (SS9401) and will be described elsewhere (26).
Pigment analysis.
In vivo absorption spectra for strains WH7803, WH8103, CC9702, and CC9703 were obtained by using cultures in the logarithmic growth phase. The spectral absorption coefficients of cell suspensions were determined in 1-cm quartz cuvettes with a dual-beam spectrophotometer (model Cary 100E; Varian) equipped with an integrating sphere (model DRA-CA-30; Labsphere). Measurements were made in the spectral region from 400 to 750 nm at a resolution of 1 nm, and sterile SN medium was used to determine the instrument baseline and as the reference. Individual spectra were smoothed with a 5-nm moving average, and the spectral absorption coefficient at 750 nm was subtracted from the entire spectrum to correct for scattering. The mean cellular absorption cross-section, ς̄a (28), was calculated by normalizing the spectral absorption coefficient to the number of cells per unit of suspension volume. Cells were enumerated with a FACSort flow cytometer (Becton Dickinson) equipped with an air-cooled laser providing 15 mW at 488 nm. A 5-μl aliquot of each culture was diluted in 990 μl of sterile phosphate-buffered saline (pH 7.5), and 5 μl of green fluorescent microbeads (diameter, 0.9 μm; Duke Scientific) was added as an internal reference. Cell counts were standardized to the volume of the sample analyzed by the instrument, as estimated from the running time.
Fluorescence excitation spectra were obtained with whole-cell suspensions by using a Fluoromax 2 spectrofluorometer (ISA Instruments). Emission was measured at 570 nm. The excitation slit was 1 nm, while the emission slit was 5 nm. Spectra were obtained in the ratio mode. The PUB/PEB ratio was calculated by determining the ratio of relative fluorescence from the PUB peak (excitation around 492 nm) to relative fluorescence from the PEB peak (excitation around 545 nm). The actual peaks were used rather than specific wavelengths. This ratio estimated the PUB/PEB ratio in PE since presumably excitation of PUB or PEB chromophores (sometimes part of phycocyanin) would result in fluorescence emission from the terminal energy acceptor in phycocyanin, a phycocyanobilin chromophore (29). This would occur at 644 nm, a wavelength significantly longer than 570 nm. When no PUB peak was present (strains WH7805 and E. Lake), the result was indicated as No PUB.
Nucleotide sequence accession numbers.
DNA sequences of the amplified 612-bp rpoC1 fragment from Synechococcus spp. motile strains CC9301, WH8112, WH8011, WH8113, WH8102, WH9702, WH9703, and C129 have been deposited in the GenBank database under accession no. AF153332 to AF153339, respectively. The nucleotide sequence accession numbers for strains CC9605, CC9616, CC9617, and SS9401 are AF154560 to AF154563, respectively, while the nucleotide sequence accession number for strain CC9701 is AF155131. Accession numbers for the other strains used in this study have been published previously (24, 25, 30).
RESULTS
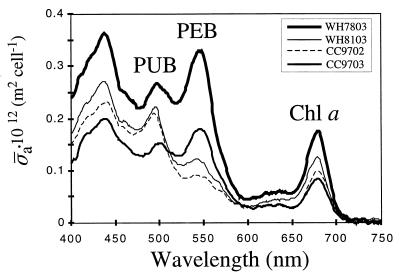
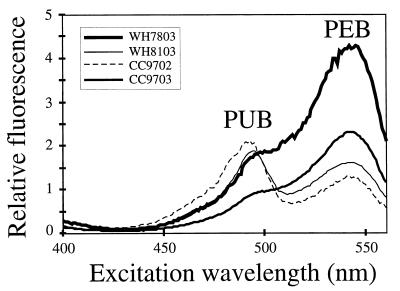
Two new motile Synechococcus strains, designated strains CC9702 and CC9703, were isolated from the California Current. During our characterization of these isolates, we observed that they had noticeably different pigment characteristics; cultures of CC9702 were brown, while cultures of CC9703 were pink (data not shown), suggesting that the PUB/PEB ratios of the two strains were different. This was confirmed by comparing the in vivo absorption spectra of the strains (Fig. 1), as well as their fluorescence excitation spectra (Fig. 2). The absorption spectrum of strain CC9703 was remarkably similar to that of nonmotile isolate WH7803 (Fig. 1), a model representative of strains that have low PUB/PEB ratios (group II [33]), while the absorption spectrum of CC9702 was more similar to that of motile strain WH8103, which is considered a model organism for the high-PUB/PEB-ratio group (group V [33]) (Fig. 1).
FIG. 1.
In vivo spectral absorption cross-section for Synechococcus sp. strains WH7803, WH8103, CC9702, and CC9703. The peaks corresponding to PUB, PEB, and chlorophyll a (Chl a) are indicated.
FIG. 2.
In vivo fluorescence excitation spectra of Synechococcus sp. strains WH7803, WH8103, CC9702, and CC9703. Emission was measured at 570 nm. The peaks corresponding to PUB and PEB are indicated.
In order to examine pigment diversity in the motile strains, we obtained the fluorescence excitation spectra of nine motile strains and determined the PUB/PEB ratios (Table 1). We obtained a wide range of PUB/PEB chromophore ratios for the strains analyzed. It appears that the PUB/PEB ratios of the motile strains that we examined are diverse and do not fit into discrete high- and low-ratio groups. The range encompasses the values obtained for WH8103 and for WH7803.
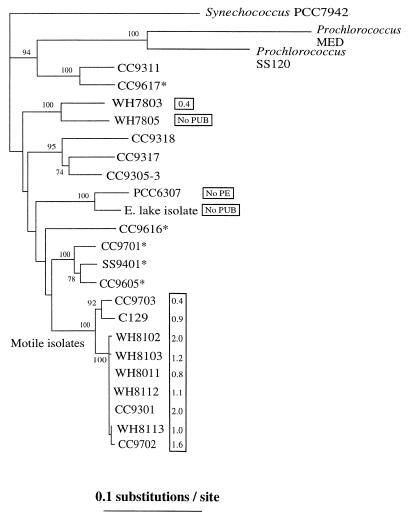
rpoC1 gene fragments were amplified from nine motile strains (Table 1). A phylogenetic analysis of the rpoC1 gene sequences of motile Synechococcus strains showed that these organisms belong to a single lineage that is independent of all other marine or freshwater groups (Fig. 3). Our phylogenetic analyses were robust, as shown by the high bootstrap values at each node based on 100 replicates. There was some genetic diversity within the group, as all WH motile isolates and strains CC9702 and CC9301 were closely related (99% identity at the nucleotide level), while strains CC9703 and C129 were more divergent (Fig. 3) and exhibited 96% identity at the nucleotide level with the members of the former group. A comparison with 40 environmental cloned sequences detected in shotgun clone libraries revealed that there are at least two closely related clones (MLG5 and MLG30) obtained from the California Current (11) that have not been cultivated yet but potentially have a motile phenotype. These environmental clones and all motile strains contain two unique amino acid residues that are potentially diagnostic among Synechococcus spp., a valine and a serine at positions 199 and 202 of the 204-amino-acid fragment (data not shown).
FIG. 3.
Neighbor-joining tree constructed with Jukes-Cantor corrected distances by using a 612-bp fragment of the DNA-dependent RNA polymerase (rpoC1) gene sequence of different Synechococcus-like isolates. Bootstrap values based on 100 replicates are shown at each node. Scale bar = 0.1 nucleotide substitution per site. The values in boxes are PUB/PEB fluorescence ratios. Asterisks indicate strains whose sequences were obtained from reference 26.
DISCUSSION
Natural populations of marine Synechococcus include multiple genetic lineages, but the link, if any, between unique physiological traits and genetic groups is not well understood. In this work, we found that marine Synechococcus strains that share the unique property of nonflagellar swimming form a monophyletic group despite differences in pigment characteristics. Although we do not understand the mechanism of swimming in Synechococcus spp. yet, it, like diverse other forms of prokaryotic motility (16, 18), is likely to require numerous genes and gene products (2, 7). Such putative complexity may account for the fact that nonflagellar swimming appears to have arisen only once in marine Synechococcus spp.
Examples (such as the one presented here) in which a phylogenetically coherent group of cyanobacteria can be defined by a physiological characteristic are rare. Recently, Garcia-Pichel et al. (12) showed that a group of extremely halotolerant cyanobacteria, the Halothece cluster, is monophyletic. Within this group there is a great deal of morphological and physiological diversity. Garcia-Pichel et al. suggested that gaining access to the hypersaline niche allowed the ancestor of the Halothece group to evolve adaptations suited to more specific niches within that environment. Similarly, the physiological diversity in pigment types among motile Synechococcus strains may stem from adaptations of motile populations to environmental niches with different light intensities, spectral compositions, or nutrient (especially nitrogen) regimes, which resulted in selection for different PUB/PEB ratios. It has been proposed that PE plays a role in nitrogen storage (41), and it is possible that in addition to affecting light-harvesting properties, chromophore diversity may be linked to differences in nitrogen metabolism and storage.
Motile marine Synechococcus strains are not usually found in nutrient-rich coastal regions, but they are found in oligotrophic regions of the world’s oceans where nitrogen is often limiting (20, 33). While motile Synechococcus strains appear to exhibit neither phototactic nor photophobic responses, Willey and Waterbury (36) have shown that at least one motile Synechococcus strain (strain WH8113) is chemotactic toward a variety of nitrogenous compounds at ecologically relevant concentrations. These researchers suggested that for organisms living in oligotrophic environments, the ability to detect and seek out patches of nutrient enrichment, such as larger phytoplankton cells or microaggregates (marine snow), may provide an ecological advantage. Hence, chemotaxis toward particles or other cells exuding nitrogenous compounds may provide a unique niche for motile Synechococcus strains compared to nonmotile strains that share the same environment and have the same pigment composition.
In order to determine whether motility is correlated with particular physical and chemical properties of the water column, such as nitrogen content or aggregate abundance, it will be necessary to understand the distribution of motile Synechococcus strains. On the basis of the number of isolates obtained from enrichment cultures (20, 33), motile Synechococcus strains have been reported to be numerous, especially in the open ocean. However, very little is known about the in situ abundance of these organisms relative to the abundance of nonmotile strains. An examination of environmental libraries has indicated that clones that group with motile strains do not appear to be well-represented (11). It is possible that this is due to the poor efficiency of rpoC1 amplification from motile strains with the standard primers.
The approaches used for in situ enumeration of cyanobacteria have included immunofluorescence, which has been used successfully to enumerate certain PE-containing Synechococcus serogroups in field samples (5), and, more recently, fluorescence in situ hybridization (27). A direct assay for motile strains currently being developed utilizes an antibody specific for SwmA (2), an abundant cell surface polypeptide that is required for swimming in Synechococcus spp. This antiserum reacts with all of the motile strains examined to date but not with nonmotile strains (4) and hence should be useful for assessing the distribution of motile strains in field samples.
Finally, the data presented here indicate that pigment characteristics, particularly PUB/PEB ratios, are not useful for delineating Synechococcus phylogenetic groups. One of the principal characteristics used to define marine Synechococcus groups has been the PUB/PEB ratio (33). While some phylogenetic analyses have demonstrated that there is a correspondence between pigment groups and specific lineages (9, 30, 38), these analyses were based on a limited number of isolates. We show here that there is pigment diversity within a unique lineage, the motile Synechococcus lineage. The PUB/PEB ratios of nine motile strains were very diverse, spanning the known range of values obtained for marine Synechococcus strains. Such pigment diversity was also observed with two nonmotile Synechococcus clusters, a cluster consisting of marine strains WH7803 and WH7805 and a cluster consisting of freshwater strains PCC6307 (32) and E. Lake (Fig. 3). We may eventually learn that only some clades exhibit pigment diversity or that all clades exhibit pigment diversity.
ACKNOWLEDGMENTS
This research was supported by NSF grant OCE9633111. G.T. was partially supported by CONACYT grant 92755.
We thank Rick Reynolds for help with the cellular absorption cross-section analysis; Greg Mitchell and Vic Vacquier for the use of the integrating sphere spectrophotometer and the spectrofluorometer, respectively; and Anton Post and Debbie Lindell for providing Synechococcus sp. strain C129.
REFERENCES
- 1.Binder B J, Chisholm S W. Cell cycle regulation in marine Synechococcus sp. strains. Appl Environ Microbiol. 1995;61:708–717. doi: 10.1128/aem.61.2.708-717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brahamsha B. An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc Natl Acad Sci USA. 1996;93:6504–6509. doi: 10.1073/pnas.93.13.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahamsha B. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl Environ Microbiol. 1996;62:1747–1751. doi: 10.1128/aem.62.5.1747-1751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahamsha, B. 1999. Unpublished data.
- 5.Campbell L. Identification of marine chroococcoid cyanobacteria by immunofluorescence. In: Yentsch C, Mague F C, Horan P K, editors. Lecture notes on coastal and estuarine studies. New York, N.Y: Springer-Verlag; 1988. pp. 208–229. [Google Scholar]
- 6.Campbell L, Vaulot D. Photosynthetic picoplankton community structure in the subtropical North Pacific Ocean near Hawaii (station ALOHA) Deep Sea Res I. 1993;40:2043–2060. [Google Scholar]
- 7.Castenholz R W. Culturing methods for cyanobacteria. Methods Enzymol. 1988;167:68–93. [Google Scholar]
- 8.Collier J L, Brahamsha B, Palenik B. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5.) to utilize urea as nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology. 1999;145:447–459. doi: 10.1099/13500872-145-2-447. [DOI] [PubMed] [Google Scholar]
- 9.Douglas S E, Carr N. Examination of genetic relatedness of marine Synechococcus spp. by using restriction fragment length polymorphisms. Appl Environ Microbiol. 1988;54:3071–3078. doi: 10.1128/aem.54.12.3071-3078.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5p. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 11.Ferris M, Palenik B. Niche adaptation in ocean cyanobacteria. Nature. 1998;396:226–228. [Google Scholar]
- 12.Garcia-Pichel F, Nübel U, Muyzer G. The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch Microbiol. 1998;169:469–482. doi: 10.1007/s002030050599. [DOI] [PubMed] [Google Scholar]
- 13.Glazer A. Light harvesting by phycobilisomes. Annu Rev Biophys Biophys Chem. 1985;14:47–77. doi: 10.1146/annurev.bb.14.060185.000403. [DOI] [PubMed] [Google Scholar]
- 14.Glibert P M, Kana T M, Olson R J, Kirchman D L, Alberte R S. Clonal responses of growth and photosynthetic responses to nitrogen availability in marine Synechococcus spp. J Exp Mar Biol Ecol. 1986;101:199–208. [Google Scholar]
- 15.Goericke R, Welschmeyer N A. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep Sea Res I. 1993;40:2283–2294. [Google Scholar]
- 16.Hartzell P L, Youderian P. Genetics of gliding motility and development in Myxococcus xanthus. Arch Microbiol. 1995;164:309–323. doi: 10.1007/BF02529977. [DOI] [PubMed] [Google Scholar]
- 17.Lantoine F, Neveux J. Spatial and seasonal variations in abundance and spectral characteristics of phycoerythrins in the tropical northeastern Atlantic Ocean. Deep Sea Res I. 1997;44:223–246. [Google Scholar]
- 18.Macnab R M. Flagella and motility. In: Neidhardt F C, editor. Escherichia coli and Salmonella. Washington, D.C.: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- 19.Olson R J, Chisholm S W, Zettler E R, Altabet M A, Dusenberry J A. Spatial and temporal distributions of prochlorophyte picoplankton in the North Atlantic Ocean. Deep Sea Res. 1990;37:1033–1051. [Google Scholar]
- 20.Olson R J, Chisholm S W, Zettler E R, Armbrust E V. Analysis of Synechococcus pigment types in the sea using single and dual beam flow cytometry. Deep Sea Res. 1988;35:425–440. [Google Scholar]
- 21.Olson R J, Chisholm S W, Zettler E R, Armbrust E V. Pigments, size, and distribution of Synechococcus in the North Atlantic and Pacific oceans. Limnol Oceanogr. 1990;35:45–58. [Google Scholar]
- 22.Ong L J, Glazer A N. Phycoerythrins of marine unicellular cyanobacteria. J Biol Chem. 1991;266:9515–9527. [PubMed] [Google Scholar]
- 23.Palenik B. Cyanobacterial community structure as seen from RNA polymerase gene sequence analysis. Appl Environ Microbiol. 1994;60:3212–3219. doi: 10.1128/aem.60.9.3212-3219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palenik B, Haselkorn R. Multiple evolutionary origins of prochlorophytes, the chlorophyll b-containing prokaryotes. Nature. 1992;355:265–267. doi: 10.1038/355265a0. [DOI] [PubMed] [Google Scholar]
- 25.Palenik B, Swift H. Cyanobacterial evolution and prochlorophyte diversity as seen in DNA-dependent RNA polymerase gene sequences. J Phycol. 1996;32:638–646. [Google Scholar]
- 26.Palenik, B., G. Toledo, R. Goericke, and B. Brahamsha. Unpublished data.
- 27.Schönhuber W, Zarda B, Eix S, Rippka R, Herdman M, Ludwig W, Amman R. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1999;65:1259–1267. doi: 10.1128/aem.65.3.1259-1267.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stramski D, Shalapyonok A, Reynolds R A. Optical characterization of the oceanic unicellular cyanobacterium Synechococcus grown under a day-night cycle in natural irradiance. J Geophys Res. 1995;100:295–307. [Google Scholar]
- 29.Swanson R V, Ong L J, Wilbanks S M, Glazer A N. Phycoerythrins of marine unicellular cyanobacteria. II. Characterization of phycobiliproteins with unusually high phycourobilin content. J Biol Chem. 1991;266:9528–9534. [PubMed] [Google Scholar]
- 30.Toledo G, Palenik B. Synechococcus diversity in the California Current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl Environ Microbiol. 1997;63:4298–4303. doi: 10.1128/aem.63.11.4298-4303.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urbach E, Scanlan D J, Distel D L, Waterbury J B, Chisholm S W. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (cyanobacteria) J Mol Evol. 1998;46:188–201. doi: 10.1007/pl00006294. [DOI] [PubMed] [Google Scholar]
- 32.Waterbury J B, Rippka R. Subsection I. Order Chroococcales Wettstein 1924, emend. Rippka et al., 1979. In: Staley J T, Bryant M P, Pfennig N, Holt J B, editors. Bergey’s manual of systematic bacteriology. Vol. 3. Baltimore, Md: Williams and Wilkins; 1989. pp. 1728–1738. [Google Scholar]
- 33.Waterbury J B, Watson F W, Valois F W, Franks D G. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. In: Platt T, Li W K W, editors. Photosynthetic picoplankton. Ottawa, Ontario, Canada: Canadian Department of Fisheries and Oceans; 1986. pp. 71–120. [Google Scholar]
- 34.Waterbury J B, Willey J M. Isolation and growth of marine planktonic cyanobacteria. Methods Enzymol. 1988;167:100–105. [Google Scholar]
- 35.Waterbury J B, Willey J M, Franks D G, Valois F W, Watson S W. A cyanobacterium capable of swimming motility. Science. 1985;230:74–76. doi: 10.1126/science.230.4721.74. [DOI] [PubMed] [Google Scholar]
- 36.Willey J M, Waterbury J B. Chemotaxis toward nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl Environ Microbiol. 1989;55:1888–1894. doi: 10.1128/aem.55.8.1888-1894.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood A M. Adaptation of photosynthetic apparatus of marine ultraphytoplankton to natural light fields. Nature. 1985;316:253–255. [Google Scholar]
- 38.Wood A M, Horan P K, Muirhead K, Phinney D A, Yentsch C M, Waterbury J B. Discrimination between types of pigments in marine Synechococcus spp. by scanning spectroscopy, epifluorescence microscopy, and flow cytometry. Limnol Oceanogr. 1985;30:1303–1315. [Google Scholar]
- 39.Wood A M, Townsend D. DNA polymorphism within the WH7803 serogroup of marine Synechococcus spp. (cyanobacteria) J Phycol. 1990;26:576–585. [Google Scholar]
- 40.Wood A M, Phinney D A, Yentsch C S. Water column transparency and the distribution of spectrally distinct forms of phycoerythrin-containing organisms. Mar Ecol Prog Ser. 1998;162:25–31. [Google Scholar]
- 41.Wyman M, Gregory R P F, Carr N G. Novel role for phycoerythrin in a marine cyanobacterium, Synechococcus strain DC2. Science. 1985;230:818–820. doi: 10.1126/science.230.4727.818. [DOI] [PubMed] [Google Scholar]