Abstract
Cardiometabolic disease is an increasing cause of morbidity and death in society. While M1-like macrophages contribute to metabolic inflammation and insulin resistance, those polarized to an M2-like phenotype exert protective properties. Building on our observations reporting M2-like macrophage exosomes in atherosclerosis control, we tested whether they could serve to control inflammation in the liver and adipose tissue of obese mice. In thinking of clinical translation, we studied human THP-1 macrophages exposed to interleukin (IL)-4 as a source of exosomes (THP1-IL4-exo). Our findings show that THP1-IL4-exo polarized primary macrophages to an anti-inflammatory phenotype and reprogramed their energy metabolism by increasing levels of microRNA-21/99a/146b/378a (miR-21/99a/146b/378a) while reducing miR-33. This increased lipophagy, mitochondrial activity, and oxidative phosphorylation (OXPHOS). THP1-IL4-exo exerted a similar regulation of these miRs in cultured 3T3-L1 adipocytes. This enhanced insulin-dependent glucose uptake through increased peroxisome proliferator activated receptor gamma (PPARγ)-driven expression of GLUT4. It also increased levels of UCP1 and OXPHOS activity, which promoted lipophagy, mitochondrial activity, and beiging of 3T3-L1 adipocytes. Intraperitoneal infusions of THP1-IL4-exo into obese wild-type and Ldlr−/− mice fed a Western high-fat diet reduced hematopoiesis and myelopoiesis, and favorably reprogramed inflammatory signaling and metabolism in circulating Ly6Chi monocytes. This also reduced leukocyte numbers and inflammatory activity in the circulation, aorta, adipose tissue, and the liver. Such treatments reduced hepatic steatosis and increased the beiging of white adipose tissue as revealed by increased UCP1 expression and OXPHOS activity that normalized blood insulin levels and improved glucose tolerance. Our findings support THP1-IL4-exo as a therapeutic approach to control cardiometabolic disease and diabetes in obesity.
Keywords: exosomes, macrophage, adipocyte, microRNA-33, PPARγ, mitochondrial respiration, cardiometabolic inflammation, type II diabetes, obesity, beiging
Graphical abstract

Exosomes produced by IL-4-polarized macrophages carry a microRNA cluster that drives metabolic reprogramming, mitochondrial activity, and fatty acid catabolism in recipient cells, including macrophages and adipocytes. When infused into obese mice, these exosomes control cardiometabolic inflammation and enhance white adipose tissue beiging that together control features of type II diabetes.
Introduction
Cardiometabolic inflammatory disease and its associated complications are the leading causes of morbidity and mortality due to the increasing prevalence of diabetes.1 Risk factors contributing to its pathogenesis include obesity, insulin resistance, dyslipidemia, and hypertension.2 Recent findings point to chronic, unresolved inflammation as a major contributor to the onset and progression of cardiometabolic disease and its complications.2,3 A hallmark of this inflammatory response includes an accumulation of pro-inflammatory M1-like macrophages in the liver, as well as in adipose and vascular tissues.4, 5, 6, 7, 8 A number of studies have identified the release of pro-inflammatory cytokines, including tumor necrosis factor alpha (TNF-α) and interleukin-1-beta (IL-1β), from tissue-associated M1-like macrophages as factors that further exacerbates local inflammation and inhibit insulin sensitivity. In contrast, anti-inflammatory M2-like macrophages have been shown to exert protective properties in the liver, as well as in adipose and vascular tissues.9, 10, 11, 12, 13 While polarizing adipose tissue macrophages to an M2-like phenotype protects high-fat-diet-fed mice against obesity-induced insulin resistance and induces beiging in white adipose tissue (WAT),11,12 key signaling factors that could be exploited to drive this beneficial phenomenon remain elusive.
Extracellular vesicles (EVs), including exosomes, have recently been recognized as sources of intercellular communication in numerous disease states, including inflammation and metabolic disease. EVs, including exosomes, are increasingly reported to serve in modulating obesity beyond cytokines and other signaling factors.11,14, 15, 16 Specifically, exosomes produced by macrophages have been shown to control numerous disease states through the delivery of cargo, including microRNA.15, 16, 17, 18 Macrophages derived from obese adipose tissue and bone marrow (bone marrow-derived macrophages [BMDMs]) exposed to lipopolysaccharide (LPS) have both been reported to display features of inflammatory M1 macrophages.15 Exosomes released by such M1-like macrophages are enriched in microRNA-155 (miR-155), which can be communicated to suppress insulin sensitivity in cultured adipocytes and adipose tissue when injected into obese mice fed a high-fat diet.15 Recent findings from our study that examined exosomes derived from BMDM cultured with elevated glucose levels revealed the production of exosomes with altered microRNA cargo, including high levels of miR-486-5p, which downregulate the expression of Abca1 in macrophages.18 Furthermore, such “high-glucose” macrophage exosomes were found to drive hematopoiesis and accelerate atherosclerosis when infused into Apoe−/− mice.18 Collectively, results of these studies support exosomes produced by macrophages exposed to inflammatory cytokines and metabolic stress as contributors to cardiometabolic disease.
In contrast, increasing evidence points to exosomes produced by alternatively activated M2-like macrophages as contributors to inflammation control. Indeed, results of our study that examined exosomes produced by M2-like BMDMs exposed to IL-4 revealed their capacity to control hematopoiesis, inflammation, and atherosclerosis in Apoe−/− mice.17 Our findings showed that they did so in part through the delivery of a cluster of microRNA cargo including miR-99a/146b/378a that controlled nuclear factor (NF)-κB inflammatory activity.17 Results of a more recent study of M2-like BMDM exosomes further revealed their ability to improve insulin sensitivity and glucose tolerance in adipose tissue of obese mice through the delivery of miR-690.16 Taken together, these findings support M2-like macrophage exosomes as promising therapeutics for the control of cardiometabolic inflammatory disease. This study aimed at developing translational opportunities for M2-like macrophage exosomes by investigating properties conferred by those derived from IL-4-exposed human THP-1 macrophages.
Results
Functional studies of exosomes produced by human THP-1 macrophages
The human monocytic cell line THP-1 was differentiated to macrophages and exposed to recombinant human IL-4 for 24 h in RPMI medium depleted of exosomes according to the flow diagram (Figure S1A). Exosomes secreted by IL-4-exposed and naive THP-1 macrophages were purified using cushioned-density gradient ultracentrifugation (C-DGUC), a method that allows for a gentle concentration and purification of exosomes from conditioned culture medium and biofluids reported in our prior studies.17, 18, 19, 20, 21 Nanoparticle tracking analysis revealed similar particle concentrations of 4.7 × 1010 and 4.1 × 1010 particles/mL and average mode size of 100 and 90.5 nm for naive THP-1 exosomes (THP1-WT-exo) and IL-4-stimulated THP-1 exosomes (THP1-IL4-exo), respectively (Figures 1A–1C). Our data show that naive or IL-4-stimulated THP-1 macrophages generally secreted the same quantity of exosomes in a 24-h period, averaging 6 × 109 secreted particles per million cells for both conditions (Figure S1B). Morphological assessment of THP1-WT-exo and THP1-IL4-exo using transmission electron microscopy revealed an expected cup-shaped morphology and size averaging 100 nm (Figure 1D). Such isolates also showed similar average protein concentrations of 50 and 53 μg/mL for THP1-WT-exo and THP1-IL4-exo, respectively (Figure S1C). Western blot analysis showed the presence of exosomal proteins, including CD9, CD63, and CD81, and the absence of cell-associated proteins Calnexin and GM130 in a quantity of 1.5 × 109 particles (Figure 1E) or 1.5 μg of proteins of both THP1-IL4-exo and THP1-WT-exo (Figure S1D).
Figure 1.
Biophysical parameters of THP-1 macrophage exosomes
(A) Representative concentration and size distributions of THP1-WT-exo and THP1-IL-4-exo purified from THP-1 cell culture supernatants after a 24-h period of culture as determined using nanoparticle tracking analysis.
(B and C) Average concentration of purified exosomes in particles per milliliter (B) and mode of particle diameter in nanometers (C) (n = 4 samples per group).
(D) Electron micrograph of purified exosomes from THP-1 cells. Scale bar: 100 nm.
(E) Western blot analysis of Calnexin, GM130, CD9, CD63, and CD81 in exosome-free medium (EFM), cell lysate, and 1.5 × 109 particles of THP-1-derived exosomes (representative of three independent experiments). Data are represented as mean ± SEM.
THP1-IL4-exo induce immune and metabolic reprogramming in recipient macrophages
To assess in vitro signaling properties of THP-1 macrophage exosomes, we treated naive THP-1 macrophages with increasing concentrations of THP1-IL4-exo (2 × 109 or 4 × 109 particles/mL) or phosphate buffered saline (PBS) for 24 h and noted a dose response when measuring their impact on gene expression levels using qRT-PCR analysis. While recipient cells treated with 2 × 109 particles/mL of THP1-IL4-exo displayed a downregulation in M1-associated genes (Tnf, Il1b, Cd86, and Cd80) and upregulation in M2-associated genes (Cd206 and Cd163), a treatment using 4 × 109 particles/mL caused more profound effects (Figure S2A). Furthermore, the control of gene expression exerted by 4 × 109 particles/mL of THP1-IL4-exo in naive THP-1 macrophages was similar to what was observed when treating the cells directly with 20 ng/mL of IL-4 cytokine for 24 h (Figure S2B). Interestingly, while THP1-WT-exo also displayed an ability to suppress the expression of pro-inflammatory markers in recipient THP-1 macrophages, their capacity to do so was 2- to 3-fold less potent than what we observed in cells treated with THP1-IL4-exo or directly with IL-4 (Figures S2A and S2B).
In line with our prior findings with IL-4-stimulated BMDM exosomes,17 treatment with THP1-IL4-exo also upregulated the expression of anti-inflammatory M2 markers (Cd206 and Cd163), which was not observed with THP1-WT-exo or PBS treatment, although the effect was 2-fold less compared with a direct treatment with IL-4 at a concentration of 20 ng/mL (Figures S2A and S2B). Interestingly, we also found that exosomes derived from THP-1 macrophages treated with 100 ng/mL LPS and 20 ng/mL IFN-γ for 24 h upregulated the expression of pro-inflammatory cytokines (Tnf and Il1b) and M1 markers (Cd86 and Cd80) (Figure S2C). These results demonstrate the capacity for THP-1 macrophage exosomes to communicate pro- and anti-inflammatory signals depending on the polarization state of their parental cells.
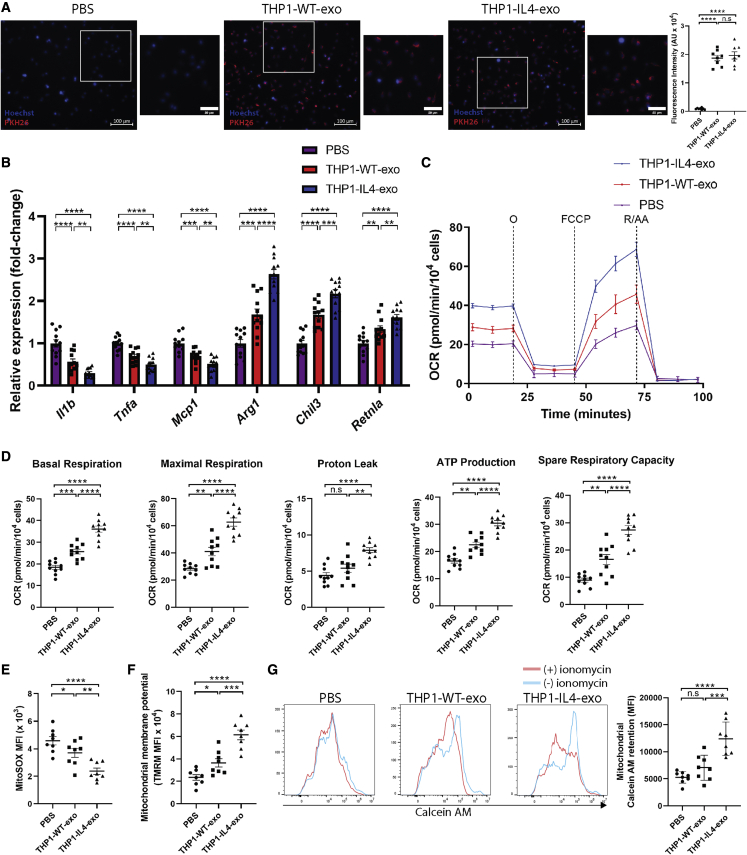
In testing their capacity to control metabolic pathways as a source of their anti-inflammatory signaling, we exposed THP1-IL4-exo to cultured primary BMDMs. Fluorescence intensity detection of PKH26-labeled THP-1 exosomes showed similar internalization efficiencies by recipient BMDMs (Figure 2A) and capacity to drive anti-inflammatory gene expression (Figure 2B) to those observed in recipient THP-1 macrophages (Figures S2A–S2C). We next tested the capacity of THP1-IL4-exo to drive metabolic reprogramming required to fuel macrophage polarization and their effector functions.22,23 We did so by measuring the oxygen consumption rate (OCR) using the Seahorse Mito Stress assay. Our data show that BMDMs treated with THP1-IL4-exo displayed enhanced oxidative phosphorylation (OXPHOS) as seen by elevated basal and maximal respiration associated with a higher proton leak and ATP production compared with BMDM treated with PBS or THP1-WT-exo (Figures 2C and 2D). To further document their metabolic reprogramming properties, we measured mitochondrial activity in BMDM treated with THP1-IL4-exo, THP1-WT-exo, or PBS alone for 24 h. Our findings revealed a reduced accumulation of superoxide in mitochondria as detected by flow cytometric analysis of BMDM stained with MitoSOX, with more potent effects observed in THP1-IL4-exo treatments (Figure 2E). Next, measurements of mitochondrial membrane potentials (ΔΨm) using flow cytometric analysis of tetramethylrhodamine (TMRM) staining further support improved mitochondrial activity in BMDM that had been treated with THP1-WT-exo and THP1-IL4-exo, with more profound effects observed when exposing cells to THP1-IL4-exo (Figure 2F). Furthermore, THP1-IL4-exo were found to potently control mitochondrial permeability by modulating the dynamic opening of the transition pores in BMDMs, resulting in improved mitochondrial calcein AM retention as detected by flow cytometry (Figure 2G). Together, our data demonstrate the ability for THP1-IL4-exo to confer robust immune and metabolic reprogramming properties in recipient macrophages.
Figure 2.
THP1-IL4-exo modulate inflammation and energy expenditure in recipient macrophages by inducing mitochondrial respiration
(A) Merged images showing quantification of the internalization of PKH26-labeled THP-1-derived exosomes by naive primary BMDMs counterstained with Hoechst (blue). BMDMs were co-incubated with 2 × 109 PKH26-labeled exosomes for 2 h at 37°C and washed repeatedly to remove unbound exosomes. All images were acquired using a Zeiss Axio microscope system with a 20× objective (n = 8 samples per group, pooled from two independent experiments). Scale bar: 100 μm and 50 μm (micrograph).
(B) qRT-PCR analysis of Il1b, Tnf, Mcp1, Arg1, Chil3, and Retnla mRNA expression in BMDMs treated with PBS (control), THP1-WT-exo, or THP1-IL-4-exo for 24 h. Results were normalized to B2m and Gapdh mRNA and are presented relative to control (n = 12 per group, pooled from three independent experiments).
(C) Graph showing representative Seahorse mitochondrial stress tests. O, oligomycin (1 μM); F, FCCP (2 μM); R/AA, rotenone/antimycin A (0.5 μM). One representative experiment out of two experiments is shown; n = 5 per group.
(D) Graphs showing quantified cell-normalized mitochondrial OCR from stress tests; n = 10 per group, pooled from two independent experiments.
(E–G) Graphs showing MFI of MitoSOX (E), TMRM (F), and calcein AM (G) signals in BMDMs treated with 4 × 109 particles/mL of THP1-WT-exo, THP1-IL4-exo, or PBS (control) as measured by flow cytometric analysis; n = 8 in each group, pooled from two independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 as determined using one-way ANOVA and Holm-Sidak post-test. Data are represented as mean ± SEM.
THP1-IL4-exo induce lipophagy and modulate microRNA levels that control metabolism in recipient macrophages
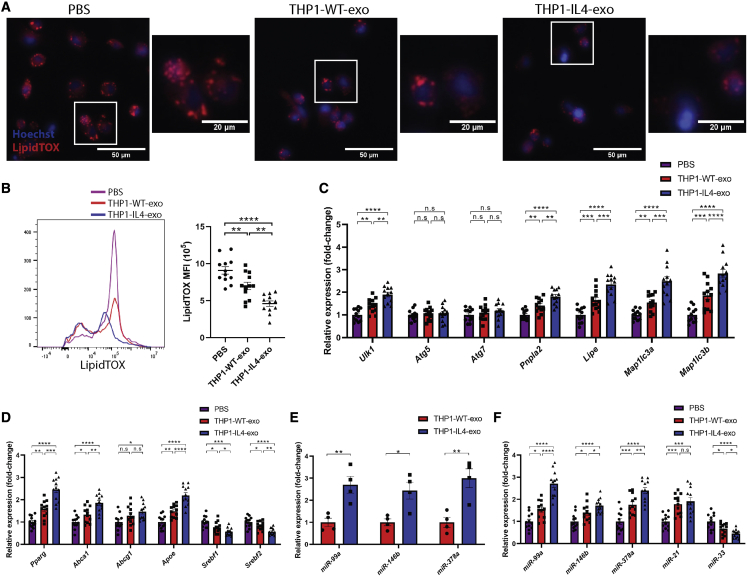
To further examine the extent to which THP1-IL4-exo reprogram cellular metabolism, we assessed their impact on lipid droplet sizes and densities in recipient BMDMs stained with LipidTOX using fluorescent microscopy and flow cytometry. Our findings show that THP1-IL4-exo treatments reduced neutral lipid accumulation in BMDMs (Figures 3A and 3B). Furthermore, gene expression analysis of RNA extracted from these cells revealed substantial increases in the expression of genes associated with lipophagy that included Ulk1, Pnpla2, Lipe, Map1lc3a, and Map1lc3b (Figure 3C). We also found a robust increase in the expression of peroxisome proliferator activated receptor gamma (PPARγ), Pparg, along with its target genes involved in cholesterol efflux that included Abca1 and Apoe (Figure 3D). Remarkably, THP1-IL4-exo exerted opposite effects in controlling the expression of genes responsible for cellular lipid synthesis and uptake including the sterol regulatory element-binding protein 1 and 2 (SREBP-1 and SREBP-2), Srebf1 and Srebf2 (Figure 3D).
Figure 3.
THP1-IL4-exo regulate lipid homeostasis, induce lipophagy, and control the expression of distinct microRNAs in recipient macrophages
(A) Merged images showing LipidTOX staining of neutral lipids in naive cultured BMDMs counterstained with Hoechst (blue). BMDM were co-incubated with 4 × 109 particles/mL of THP1-WT-exo, THP1-IL4-exo, or PBS for 24 h at 37°C and subsequently stained with LipidTOX for 30 min. All images were acquired using a Zeiss microscope system with a 20× objective. Scale bars: 50 μm and 20 μm (micrograph).
(B) Representative flow cytometric plot and graph showing average mean fluorescent intensity (MFI) of LipidTOX signals in BMDMs treated with 4 × 109 particles/mL of THP1-WT-exo, THP1-IL4-exo, or PBS for 24 h at 37°C; n = 12 per group, pooled from three independent experiments.
(C) qRT-PCR analysis of Ulk1, Atg5, Atg7, Pnpla2, Lipe, Map1lc3a, and Map1lc3b mRNA expression in BMDMs treated with PBS (control), THP1-WT-exo, or THP1-IL-4-exo for 24 h.
(D) qRT-PCR analysis of Pparg, Abca1, Abcg1, Apoe, Srebf1, and Srebf2 expression in BMDMs treated with PBS (control), THP1-WT-exo, or THP1-IL-4-exo for 24 h. Results were normalized to B2m and Gapdh mRNA and are presented relative to control (n = 12 per group, pooled from three independent experiments).
(E) qRT-PCR analysis of miR-99a-5p, miR-146b-5p, and miR-378a-3p microRNA expression in THP1-WT-exo and THP1-IL-4-exo. Results were normalized to U6 snRNA and miR-16-5p expression, with UniSp6 used as a spike-in control. Data are presented relative to THP1-WT-exo (n = 4 per group).
(F) qRT-PCR analysis of miR-99a-5p, miR-146b-5p, miR-378a-3p, miR-21-5p, and miR-33-5p microRNA expression in BMDMs treated with PBS (control), THP1-WT-exo, or THP1-IL-4-exo for 24 h. Results were normalized to U6 snRNA and miR-16-5p expression, with UniSp6 used as a spike-in control. Data are presented relative to control (n = 12 per group, pooled from three independent experiments). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 as determined using either unpaired Student’s t test (for two-group comparison) or one-way ANOVA followed by Holm-Sidak post-test (for multiple-group comparison). Data are represented as mean ± SEM.
Findings from our laboratory and those of others recently revealed that M1- and M2-polarized primary BMDMs release select sets of microRNAs into exosomes that can communicate inflammatory and metabolic signaling to recipient macrophages,17,18 hematopoietic stem/progenitor cells (HSPCs),17,18 and adipocytes.15,16 In testing whether THP-1 macrophages can release similar sets of microRNAs as those we reported to be highly enriched in IL-4-treated BMDM exosomes,17 we measured levels of microRNA-99a/146b/378a. Our findings show an enrichment of all three of these microRNAs in THP1-IL4-exo compared with THP1-WT-exo (Figure 3E). Interestingly, naive THP-1 macrophages and murine BMDMs treated with THP1-IL4-exo for 24 h also displayed increased levels of these microRNAs compared with cells treated with either THP1-WT-exo or PBS (Figures S2D and 3F), supporting their communication from exosomes as we reported.17 Stemming from their potent capacity to polarize macrophages and control their lipid metabolism, we tested the impact of THP1-IL4-exo in modulating cellular levels of miR-21-5p, a central regulator of macrophage polarization,24 along with miR-33-5p, which is recognized as a regulatory hub for energy metabolism in macrophages.25,26 While we noted increased expression of miR-21-5p in THP-1 macrophages and BMDMs treated with THP1-IL4-exo, we uncovered opposite effects when looking at miR-33-5p. Indeed, we noted a 3-fold reduction in levels of miR-33-5p in cells treated with THP1-IL4-exo (Figures S2D and 3F). This finding is in line with our data documenting substantial reduction in Srebf2 mRNA (Figure 3D), which serves as its host.27 We also noted no significant difference in miR-21-5p and miR-33-5p cargo in THP1-IL4-exo compared with THP1-WT-exo (Figure S2E). Taken together, these results support a capacity for THP1-IL4-exo to communicate anti-inflammatory and metabolic control in macrophages by modulating levels of cellular microRNA recognized to play critical roles in regulating lipophagy, energy metabolism, and inflammatory signaling in these cells.
THP1-IL4-exo promote beiging in recipient 3T3-L1 adipocytes by modulating cellular microRNA
While studies have shown that exosomes produced by primary mouse macrophages can control insulin sensitivity and metabolism in mouse adipocyte,15,16 whether those produced by human THP-1 cells can exert similar properties is not known. To test this possibility, we treated fully differentiated 3T3-L1 adipocytes with THP1-IL4-exo or control for 24 h. Fluorescence intensity detection of PKH26-labeled exosomes produced by THP-1 cells exposed to IL-4 or control showed similar internalization efficiencies in recipient 3T3-L1 adipocytes (Figure 4A). THP1-IL4-exo treatments increased expression in Pparg and its target gene Slc2a4 (GLUT4),15,28 as well as Ucp1 (Figure 4B). We also noted a concomitant reduction in Srebf1 and Srebf2 (Figure 4B). Western blot analysis confirmed increased protein levels for the gene products of PPARγ, GLUT4, and UCP1 in cell lysates (Figures 4C and 4D). Having observed pro-thermogenic properties conferred by THP-1 macrophage exosomes to recipient adipocytes, we next wondered whether they could also enhance mitochondrial respiration in these cells by testing the OCR using the Seahorse Mito Stress assay. Data shown in Figures 4E and 4F demonstrate the capacity for THP1-IL4-exo to profoundly increase OXPHOS activity in 3T3-L1 adipocytes compared with treatments with THP1-WT-exo or PBS alone.
Figure 4.
THP1-IL4-exo induce energy expenditure and control expression of distinct microRNAs in recipient 3T3-L1 adipocytes
(A) Merged images showing and quantification of the internalization of PKH26-labeled THP-1-derived exosomes by 3T3-L1 adipocytes counterstained with Hoechst (blue). 3T3-L1 adipocytes were co-incubated with 2 × 109 PKH26-labeled exosomes for 2 h at 37°C and washed repeatedly to remove unbound exosomes. All images were acquired using a Zeiss Axio microscope system with a 20× objective (n = 8 samples per group, pooled from two independent experiments). Scale bar: 500 μm and 250 μm (micrograph).
(B) qRT-PCR analysis of Pparg, Slc2a4, Srebf1, Srebf2, and Ucp1 mRNA expression in 3T3-L1 adipocytes treated with PBS (control), THP1-WT-exo, or THP1-IL-4-exo for 24 h. Results were normalized to B2m and Gapdh mRNA and are presented relative to control (n = 12 per group, pooled from three independent experiments).
(C and D) Western blot analysis (C) and quantification (D) of GLUT4, PPARγ, and UCP1 protein levels in cell lysates of 3T3-L1 adipocytes treated with PBS (control), THP1-WT-exo, or THP1-IL-4-exo for 24 h. Quantification was performed using ImageJ and data normalized to loading controls β-Actin or GAPDH (n = 6 per group, pooled from two independent experiments).
(E) Graph showing representative Seahorse mitochondrial stress tests. O, oligomycin (1 μM); F, FCCP (0.25 μM); R/AA, rotenone/antimycin A (0.5 μM). One representative experiment out of two experiments is shown; n = 6 per group.
(F) Graphs showing quantified cell-normalized mitochondrial OCR from stress tests; n = 12 in each group, pooled from two independent experiments. qRT-PCR analysis of miR-99a-5p, miR-146b-5p, miR-378a-3p, miR-21-5p, and miR-33-5p expression levels in 3T3-L1 adipocytes treated with PBS (control), THP1-WT-exo, or THP1-IL-4-exo for 24 h. Results were normalized to U6 snRNA and miR-16-5p expression, with UniSp6 used as a spike-in control. Data are presented relative to control (n = 12 per group, pooled from three independent experiments). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 as determined using one-way ANOVA and Holm-Sidak post-test. Data are represented as mean ± SEM.
To further assess the mechanism through which THP1-IL4-exo induce metabolic activity in 3T3-L1 adipocytes, we detected the expression levels of microRNAs found to be altered in BMDM and THP-1 cells treated with THP-1 exosomes (Figures S2D and 3F). In doing so, we detected an enrichment of miR-99a-5p, miR-146b-5p, and miR-378a-3p in 3T3-L1 adipocytes treated with THP1-IL4-exo compared with those treated with either THP1-WT-exo or PBS (Figure 4G). We next measured the expression of microRNAs central to adipocyte beiging and homeostasis, including miR-21-5p and miR-33-5p,29,30 which we found to show opposing sensitivity to THP1-IL4-exo in cultured macrophages (Figures S2D and 3F). While we noted a nearly 2-fold increase in miR-21-5p levels (Figure 4G), we detected a sharp 2-fold reduction in miR-33-5p levels in 3T3-L1 adipocytes treated with THP1-IL4-exo (Figure 4G). Together, our findings provide compelling evidence supporting THP1-IL4-exo as a source of beiging and OXPHOS in recipient adipocytes by modulating microRNA that parallels those seen in recipient BMDM.
THP1-IL4-exo induce lipophagy and mitochondrial activity during the differentiation of 3T3-L1 adipocytes
We next examined metabolic activities controlled by THP1-IL4-exo during 3T3-L1 preadipocyte differentiation performed in basic Dulbecco’s Modified Eagle’s Medium (DMEM) medium with 1 μg/mL bovine insulin, 0.5 mM methylisobutylxanthine (IBMX), and 1.0 mM dexamethasone (DEX) for 2 days as widely reported.31 Cells were treated with THP1-IL4-exo every 2 days at a concentration of 4 × 109 particles/mL along with a fresh change of DMEM supplemented with 1 μg/mL bovine insulin until fully differentiated on day 15. Monitoring cellular neutral lipids using LipidTOX staining revealed that THP1-IL4-exo treatments reduced neutral lipid droplet accumulation during the differentiation of 3T3-L1 preadipocytes. In contrast to adipocytes treated with PBS that displayed large lipid vacuoles, those treated repeatedly with THP1-exo displayed far fewer and smaller lipid vacuoles that were even further reduced in adipocytes treated with THP1-IL4-exo (Figure 5A). Quantification of neutral lipid staining by flow cytometric analysis (Figure 5B) confirmed the microscopy data in Figure 5A. Cells exposed to THP1-IL4-exo displayed lower mean fluorescent intensity of LipidTOX compared with cells treated with either THP1-WT-exo or PBS alone (Figure 5B). Quantifying oil red O (ORO) extracted from sets of cells treated in a similar manner further confirmed benefits that THP1-IL4-exo exerted to reduce neutral lipid accumulation in differentiated 3T3-L1 adipocytes (Figures S3A and S3B).
Figure 5.
THP1-IL4-exo enhance mitochondrial activity, induce lipophagy, and promote beiging during 3T3-L1 adipocyte differentiation
(A) Merged images showing LipidTOX staining of 3T3-L1 adipocytes counterstained with Hoechst (blue). 3T3-L1 adipocytes were treated with 4 × 109 particles/mL of THP1-WT-exo, THP1-IL4-exo, or PBS every 2 days following a 2-day induction period by IBMX, DEX, and bovine insulin. By day 15, cells were stained with LipidTOX for 30 min. All images were acquired using a Zeiss microscope system with a 20× objective. Scale bars: 50 μm and 50 μm (micrograph).
(B) Representative flow cytometric plot and graph showing average MFI of LipidTOX signals in 3T3-L1 adipocytes treated with 4 × 109 particles/mL of THP1-WT-exo, THP1-IL4-exo, or PBS every 2 days following the induction period; n = 12 per group, pooled from three independent experiments.
(C–E) Graphs showing MFI of MitoSOX (C), TMRM (D), and calcein AM (E) signals in 3T3-L1 adipocytes treated with 4 × 109 particles/mL of THP1-WT-exo, THP1-IL4-exo, or PBS every 2 days following the induction period; n = 8 per group, pooled from two independent experiments.
(F) qRT-PCR analysis of Ucp1, Ppargc1a, Tbx1, Dio2, Zfp516, Prdm16, and Slc25a25 mRNA expression in 3T3-L1 adipocytes treated with 4 × 109 particles/mL of THP1-WT-exo, THP1-IL4-exo, or PBS every 2 days following the induction period.
(G) qRT-PCR analysis of Ulk1, Atg5, Atg7, Pnpla2, Lipe, Map1lc3a, and Map1lc3b mRNA expression in 3T3-L1 adipocytes treated with 4 × 109 particles/mL of THP1-WT-exo, THP1-IL4-exo, or PBS every 2 days following the induction period.
(H) qRT-PCR analysis of Adipoq and Lep in 3T3-L1 adipocytes treated with 4 × 109 particles/mL of THP1-WT-exo, THP1-IL4-exo, or PBS every 2 days following the induction period. Results were normalized to B2m and Gapdh mRNA and are presented relative to control (n = 12 per group, pooled from three independent experiments). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 as determined using one-way ANOVA and Holm-Sidak post-test. Data are represented as mean ± SEM.
To further document evidence of beiging in 3T3-L1 cells exposed to THP-1 macrophage exosomes, we used flow cytometric analyses to monitor three established functional parameters to assess mitochondrial activity. In detecting levels of MitoSOX dye fluorescence, we noted a marked reduction of mitochondrial superoxide in 3T3-L1 adipocytes treated with THP1-IL4-exo that was less prominent in those treated with THP1-WT-exo (Figure 5C). Next, measurements of TMRM levels revealed increased mitochondrial membrane potential ΔΨm in 3T3-L1 adipocytes repeatedly treated with THP-1 exosomes, with more profound effects observed when using THP1-IL4-exo treatments (Figure 5D). Finally, we observed increased mitochondrial calcein AM retention in THP1-IL4-exo-treated 3T3-L1 adipocytes (Figure 5E), demonstrating a similar level of control over mitochondrial permeability as observed in BMDMs (Figure 2G). Together, these data demonstrate the capacity for THP1-IL4-exo signaling to improve mitochondrial health and functions in recipient 3T3-L1 adipocytes during differentiation, a hallmark of beiging in white adipocytes.31, 32, 33, 34
In assessing pathways associated with beiging in 3T3-L1 adipocytes treated with THP1-IL4-exo, we measured the expression levels of genes associated with thermogenesis. Our findings revealed a concerted upregulation of canonical markers of adipose tissue beiging that included Ucp1, Ppargc1a, Tbx1, Dio2, Zfp516, Prdm16, and Slc25a25. Importantly, their level of upregulation was increased when adipocytes were treated with THP1-IL4-exo rather than THP1-WT-exo (Figure 5F). In agreement with our findings seen in recipient macrophages, such treatments also led to an upregulated expression of genes associated with lipophagy during adipocyte beiging, namely Ulk1, Pnpla2, Lipe, Map1lc3a, and Map1lc3b34 (Figure 5G). As with other tested parameters, the modulation of these genes was more potent in adipocytes treated with THP1-IL4-exo. Last, we documented an increased expression of adiponectin (Adipoq) with a concomitant decrease in the expression of leptin (Lep) (Figure 5H). Taken together, our findings reveal that THP1-IL4-exo can serve to drive beiging during the differentiation of 3T3-L1 preadipocyte into adipocytes by inducing lipophagy and improving mitochondrial activity that together increase the expression of adipokines central to the regulation of obesity and diabetes.
THP1-IL4-exo resolve cardiometabolic inflammation in mice with diet-induced hyperlipidemia, obesity, and insulin resistance
In light of provocative evidence pointing to THP1-IL4-exo as potent mediators of cellular metabolic reprogramming in cultured macrophages and adipocytes, we next explored their capacity to control cardiometabolic tissue inflammation in mice. We did so by repeatedly treating obese mice with intraperitoneal (i.p.) infusions of THP-1 macrophage exosomes and monitored parameters of inflammation as recently reported in our studies of Apoe−/− mice treated with exosomes derived from IL-4-exposed BMDMs.17 To avoid complications associated with a loss of apoE expression on adiposity and inflammation,35, 36, 37, 38 we chose to study hypomorphic Apoeh/h Ldlr−/− mice that we previously reported as a model of spontaneous hyperlipidemia and atherosclerosis in the setting of reduced cellular Apoe expression that results in plasma apoE- and apoB-lipoprotein accumulation due to impaired clearance mechanisms in the liver.39 For the purpose of this study, 20-week-old Apoeh/h Ldlr−/− mice were fed a diet rich in saturated fat and sucrose (Western diet) for 10 weeks to induce obesity and insulin resistance. Owing to reduced Apoe expression levels in all tissues, Apoeh/h Ldlr−/− mice were more sensitive to Western diet feeding than Ldlr−/− mice, and therefore displayed greater plasma cholesterol and triglyceride levels, averaging 2,000 mg/dL and 1,300 mg/dL, respectively (Figures S4A and S4B).
To assess their biodistribution, THP-1 macrophage exosomes were labeled with 1,1′-dioctadecyl-3,3,3′,3′ tetramethylindotricarbocyanine iodide (DiR [DiIC18(7)]) and infused i.p. into 20-week-old Apoeh/h Ldlr−/− mice that had been fed the Western diet for 10 weeks. Six hours post infusion, the presence of DiR-positive exosomes was detected in the circulation (Figure 6A), as well as the epididymal WAT (eWAT), lung, liver, brain, kidney, aorta, spleen, bone, and intestine (Figure 6B). Data shown in Figure 6B also demonstrate a similar pattern of biodistribution between THP1-WT-exo and THP1-IL4-exo that included the eWAT, liver, bone marrow, and spleen.
Figure 6.
THP1-IL4-exo resolve systemic inflammation and hematopoiesis in mice with diet-induced obesity and hyperlipidemia
(A and B) Images of DiR fluorescence in blood (A) and organs (B) 6 h post injection from 20-week-old Western-diet-fed Apoeh/hLdlr−/− mice injected i.p. with PBS as control or 1 × 1010 particles of THP1-WT-exo or THP1-IL4-exo.
(C) Schematic diagram detailing the duration of Western diet feeding and injection strategy in 26-week-old Apoeh/hLdlr−/− mice.
(D) Quantification of circulating CD11b+ cells, neutrophils, Ly6Chi monocytes, and Ly6Clo monocytes in mice injected with PBS or THP1-IL4-exo by flow cytometry; n = 10 per group, pooled from three independent experiments.
(E) Quantification of splenic CD11b+ cells, neutrophils, Ly6Chi monocytes, and Ly6Clo monocytes in mice injected with PBS or THP1-IL4-exo by flow cytometry; n = 10 per group, pooled from three independent experiments.
(F) Quantification of bone marrow LSK, MPP1, MPP2, MPP3, MPP4, CMP, GMP, and MEP cell populations by flow cytometry; n = 8 per group, pooled from two independent experiments.
(G) Quantification of splenic LSK, MPP1, MPP2, MPP3, MPP4, CMP, GMP, and MEP cell populations by flow cytometry; n = 8 per group, pooled from two independent experiments.
(H) qRT-PCR analysis of Tnf, Il1b, and Mcp1 mRNA expression in circulating Ly6Chi monocytes.
(I) qRT-PCR analysis of Pparg, Abca1, Abcg1, Srebf1, and Srebf2 mRNA expression in circulating Ly6Chi monocytes.
(J) qRT-PCR analysis of miR-99a-5p, miR-146b-5p, miR-378a-3p, miR-21-5p, and miR-33-5p microRNA expression levels in circulating Ly6Chi monocytes isolated by FACS. Results were normalized to B2m and Gapdh for mRNA analysis and U6 snRNA, miR-16-5p, and UniSp6 (spike-in control) for miRNA analysis. Data are presented relative to control (n = 8 per group, pooled from two independent experiments).
(K) Multiplex immunoassay analysis of plasma TNF-α, IFN-γ, IL-6, and IL-1β; n = 10 per group, pooled from three independent experiments. Data are taken in 26-week-old Apoeh/hLdlr−/− mice fed with a Western diet and injected with PBS or THP1-IL-4-exo (1 × 1010 particles/mouse every 2 days for 6 weeks while on Western diet). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 as determined using either unpaired Student’s t test (for two-group comparison) or one-way ANOVA followed by Holm-Sidak post-test (for multiple-group comparison). Data are represented as mean ± SEM.
Based on the similar pattern of biodistribution displayed by the two forms of THP-1 macrophage exosome preparations, we opted to study the in vivo properties of THP1-IL4-exo due to their more potent capacity to control inflammation and metabolic activities observed through our in vitro studies in Figures 2, 3, 4, and 5. Cohorts of 20-week-old Western-diet-fed Apoeh/h Ldlr−/− mice were treated with either 1 × 1010 particles of THP1-IL4-exo, a dose that we determined represents approximately 2%–5% of total exosomes in normal mouse plasma,18 or PBS three times a week (every 2 days) for 6 weeks while the mice were kept on Western diet. At the end of the exosome infusion period, all mice displayed similar obesity with an average weight of 42 g for both THP1-IL4-exo-infused and PBS-infused groups (Figure S4C). Moreover, all mice displayed a similar eWAT fat-to-body ratio of 0.05 (Figure S4D). A diagram of experimental parameters is shown in Figure 6C. In agreement with our recent studies of BMDM exosomes, the infusion of THP1-IL4-exo into mice provoked changes in hematopoiesis and leukocyte numbers in the circulation.17,18 Indeed, mice infused with THP1-IL4-exo displayed a profound reduction in circulating CD45+ CD11b+ myeloid cells (Figures S4E and 6D). We also found significantly less inflammatory Ly6Chi monocytes and neutrophils in the circulation of mice infused with THP1-IL4-exo (Figures S4E and 6D). In addition, we observed a reduced accumulation of splenic CD11b+ Ly6G− monocytes and CD11b+ Ly6G+ neutrophils in mice infused with THP1-IL4-exo (Figures S4F and 6E). THP1-IL4-exo-treated mice also displayed reduced numbers of Ly6Chi and increased Ly6Clo subpopulations of splenic CD11b+ Ly6G− monocytes (Figures S4F and 6E). The reduction of circulating and splenic myeloid cells paralleled an observed reduction of HSPCs in the bone marrow and spleen. This included a reduction of Lin− Sca-1+ c-Kit+ (LSK) cells, multipotent progenitor cells (MPP1-4), common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs), in the bone marrow and spleen (Figures S4G, S4H, 6F, and 6G).
In seeking to uncover mechanisms through which THP1-IL4-exo controlled hematopoiesis when infused into Western-diet-fed Apoeh/h Ldlr−/− mice, we noted their capacity to reprogram metabolic and inflammatory gene expression in peritoneal macrophages and circulating Ly6Chi monocytes isolated by fluorescence-activated cell sorting (FACS) (Figures S4I, 6H, and 6I). This included by decreasing the expression of pro-inflammatory cytokines Tnf, Il1b, and Mcp1 (Figures S4I and 6H). In contrast, THP1-IL4-exo upregulated the expression of Pparg, Abca1, and Abcg1, and decreased levels of miR-33-5p and its host gene Srebf1 and Srebf2 (Figures 6I and 6J). A mechanistic basis to explain this level of cellular gene expression control likely stems from increases in levels of miR-99a-5p/146b-5p/378a-3p and miR-21-5p in Ly6Chi monocytes (Figure 6J). Finally, the control of leukocyte activation via THP1-IL4-exo also led to a profound reduction in the levels of plasma cytokine that included TNF-α, IFN-γ, IL-6, and IL-1β in mice infused with THP1-IL4-exo as determined using a multiplex immunoassay analysis (Figure 6K). Together, our results support a role for THP1-IL4-exo in contributing to actively reprogram cellular metabolism and inflammatory function in hematopoietic compartments of mice continually maintained on a Western diet.
Infusions of THP1-IL4-exo suppress leukocyte expansion in aortic, hepatic, and adipose tissues of hyperlipidemic and obese mice
To determine whether benefits of controlled hematopoietic cell expansion and activation could extend to solid organs, we examined leukocyte subsets in the aorta, liver, and eWAT of mice infused with IL4-THP1-exo. Enzymatic digestion of these tissues followed by flow cytometric detection of leukocytes revealed a marked reduction in CD45+ leukocytes in all three organs (Figures 7A–7D and 7G). Subpopulation analyses further revealed lower numbers of total CD11b+ cells and CD11b+ F4/80+ macrophages in the aorta (Figures 7A and 7B). Moreover, within the aortic CD11b+ population, we observed an expansion of Ly6Clo monocytes and contraction of Ly6Chi monocyte and neutrophil subpopulations (Figures 7A and 7B). We also observed a similar reduction in CD45+ leukocytes in livers of mice infused with THP1-IL4-exo (Figures 7C and 7D). While there was no difference in the number of liver-associated CD11blo F4/80hi Kupffer cells, there was a significant reduction in infiltrating CD11bhi F4/80lo macrophages in the livers of mice infused with THP1-IL4-exo (Figures 7C and 7D). Furthermore, we detected lower numbers of total CD11b+ cells in the liver and, within this population, lower numbers of Ly6Chi and neutrophils subpopulations in THP1-IL4-exo-infused mice (Figures 7C and 7D). An analysis of gene expression levels from RNA extracted from the livers of these mice also showed a decrease in the expression of Adgre1 (F4/80) and inflammatory markers associated with M1 macrophages (Tnf, Il1b, Mcp1, and Nos2) (Figure 7F). In contrast, we noted an increased expression of M2-associated markers (Arg1, Chil3, and Retnla) (Figure 7E). Last, histological assessments of the liver revealed reduced hepatocyte enlargement and ORO retention in liver sections taken from Western-diet-fed Apoeh/h Ldlr−/− mice treated with THP1-IL4-exo (Figures S5A and S5B). Together, these data support the benefits of THP-IL4-exo in controlling the onset of diet-induced steatohepatitis.
Figure 7.
Infusions of THP1-IL4-exo suppress aortic, hepatic, and adipose tissue inflammation in obese hyperlipidemic mice
(A and B) Representative flow cytometry plots of leukocyte subsets from aorta (A) and quantification of aortic CD45+ cells, macrophages, CD11b+ cells, neutrophils, Ly6Chi monocytes, and Ly6Clo monocytes (B).
(C and D) Representative flow cytometry plots of leukocyte subsets from livers (C) and quantification of hepatic CD45+ cells, infiltrating macrophages/monocytes, Kupffer cells, neutrophils, and Ly6Chi monocytes (D).
(E) qRT-PCR analysis of Adgre1, Tnf, Il1b, Mcp1, Nos2, Arg1, Chil3, and Retnla mRNA expression in livers. Results were normalized to B2m and Gapdh mRNA and are presented relative to control. Data are pooled from three independent experiments, n = 10 per group.
(F) Representative images and quantification of F4/80+ cells that formed crown-like structures (CLSs) in eWAT; n = 8 per group, pooled from two independent experiments. Scale bar: 200 μm.
(G) Quantification of eWAT CD45+ cells, macrophages, CD11b+ cells, neutrophils, and Ly6Chi monocytes measured by flow cytometry.
(H) qRT-PCR analysis of Adgre1, Tnf, Il1b, Mcp1, Nos2, Arg1, Chil3, and Retnla mRNA expression in eWAT. Results were normalized to B2m and Gapdh mRNA and are presented relative to control. Data are pooled from three independent experiments, n = 10 per group. All data are taken from 26-week-old Apoeh/hLdlr−/− mice fed with a Western diet and injected with PBS or THP1-IL-4-exo (1 × 1010 particles/mouse every 2 days for 6 weeks while on Western diet). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 as determined using either unpaired Student’s t test (for two-group comparison) or one-way ANOVA followed by Holm-Sidak post-test (for multiple-group comparison). Data are represented as mean ± SEM.
In spite of a lack of significant changes in weight gains (Figures S4C and S4D), our data show that THP1-IL4-exo infusions profoundly suppressed eWAT inflammation. Immunohistochemical analysis revealed reduced numbers of F4/80+ crown-like structures (CLSs) in eWAT derived from mice treated with THP1-IL4-exo compared with those treated with PBS (Figure 7F). Flow cytometric analysis of cellular digests from eWAT collected from Western-diet-fed Apoeh/h Ldlr−/− mice showed a reduction in CD45+ leukocytes in mice infused with THP1-IL4-exo (Figures S6 and 7G). Subpopulation analyses further showed reduced numbers of total CD11b+ cells and CD11b+ F4/80+ macrophages in eWAT of these mice (Figures S6 and 7G). Among all eWAT-associated CD11b+ cells, we detected lower numbers of Ly6Chi monocytes and neutrophils (Figures S6 and 7G). Similar to our findings of liver tissue taken from these mice (Figure 7E), gene expression analysis of eWAT RNA extracts showed a marked decrease in the expression of Adgre1 (F4/80) and M1 markers/pro-inflammatory cytokines (Tnf, Il1b, Mcp1, and Nos2) with a concomitant increase in M2/anti-inflammatory markers (Arg1, Chil3, and Retnla) (Figure 7H).
To confirm the efficacy of THP1-IL4-exo in controlling cardiometabolic inflammation in a second model system, we repeatedly infused them into 20-week-old Western-diet-fed wild-type mice for 6 weeks while keeping the mice on the diet. These mice displayed significantly lower plasma cholesterol and triglycerides than Apoeh/h Ldlr−/− mice, averaging 300 mg/dL and 100 mg/dL respectively (Figures S7A and S7B). Despite lower blood lipids, the Western-diet-fed wild-type mice displayed similar total body weights and eWAT fat-to-body ratio similar to Western-diet-fed Apoeh/h Ldlr−/− mice, averaging 43 g and 0.057 and 0.051 fat-to-body ratio for PBS-treated and THP1-IL4-exo-treated groups, respectively (Figures S7C and S7D). As observed with the Apoeh/h Ldlr−/− model, repeated infusions of THP1-IL4-exo suppressed the accumulations of CD45+ leukocytes, CD11bhi F4/80lo macrophages, neutrophils, and Ly6Chi CD11b+ cells in the livers of wild-type mice fed this diet (Figures S7E and S7F). We also detected a similar reduction of CD45+ leukocytes, macrophages, neutrophils, and Ly6Chi CD11b+ cells in eWAT compared with mice infused with PBS (Figures S7G and S7H). Collectively, our data demonstrate the efficacy of repeated THP1-IL4-exo infusions in exerting a profound control over chronic vascular, hepatic, and adipose tissue inflammation that develops in response to Western diet feeding in wild-type mice and low-density lipoprotein receptor-deficient mice. Our findings also support this treatment to drive an expansion of protective M2-like macrophages in these tissues.
THP1-IL4-exo drive the beiging of WAT that improves glucose disposal and insulin resistance in obese mice
Our in vivo findings so far provide compelling evidence demonstrating the efficacy of THP1-IL4-exo in suppressing inflammation in cardiometabolic tissue including eWAT. Based on our data demonstrating their capacity to drive the beiging of cultured adipocytes, we wondered whether THP1-IL4-exo could protect hyperlipidemic Apoeh/h Ldlr−/− mice against obesity-induced insulin resistance and impaired glucose disposal. We noted that 6 weeks of tri-weekly i.p. infusions substantially reduced fasting glucose and insulin levels in these mice (Figure 8A). Furthermore, such treatments profoundly enhanced glucose disposal as measured by a glucose tolerance test (GTT) (Figures 8B and 8C). A correction of all of these metabolic parameters related to type II diabetes was reproduced in our second mouse model using Western-diet-fed wild-type mice infused with a similar course of THP1-IL4-exo (Figures S8A–S8C).
Figure 8.
Infusions of THP1-IL4-exo enhance energy expenditure and induce eWAT beiging, which improves glucose disposal and normalizes fasting blood glucose and insulin levels in hyperlipidemic and obese mice
(A) Fasting glucose and insulin levels measured from mouse plasma; insulin was measured by ELISA.
(B and C) Plot showing blood glucose levels in mice during GTT (B) and values of area under the curve in GTT (C). Data are pooled from two independent experiments, n = 8 per group.
(D) Detection of 2-DG uptake in differentiated 3T3-L1 adipocytes treated with 4 × 109 particles/mL of THP1-IL4-exo, THP1-WT-exo, or PBS for 24 h; n = 10 per group, pooled from two independent experiments.
(E) Graph showing representative Seahorse mitochondrial stress tests. O, oligomycin (10 μM); F, FCCP (10 μM); R/AA, rotenone/antimycin A (5 μM). One representative experiment out of two experiments is shown; n = 4 per group.
(F) Graphs showing quantified protein-normalized mitochondrial OCR from stress tests; n = 8 in each group, pooled from two independent experiments.
(G) Representative images of hematoxylin and eosin staining and quantification of adipocyte sizes in eWAT; n = 8 per group, pooled from two independent experiments. Scale bar: 300 μm.
(H) qRT-PCR analysis of Pparg and Slc2a4 mRNA expression in eWAT; n = 10 per group, pooled from three independent experiments.
(I) Western blot analysis and quantification of GLUT4 protein levels in eWAT tissue lysates. Quantification was performed using ImageJ and data normalized to loading controls Vinculin (n = 8 per group, pooled from two independent experiments).
(J) qRT-PCR analysis of Ucp1, Ppargc1a, Tbx1, Dio2, Zfp516, Prdm16, and Slc25a25 mRNA expression in eWAT; n = 10 per group, pooled from three independent experiments.
(K) Images of UCP1 staining in eWAT; two representative images are shown from n = 8 per group. Scale bar: 500 μm.
(L) qRT-PCR analysis of Ulk1, Atg5, Atg7, Pnpla2, Lipe, Map1lc3a, and Map1lc3b mRNA expression in eWAT; n = 10 per group, pooled from three independent experiments.
(M) qRT-PCR analysis of Adipoq and Lep mRNA expression in eWAT; n = 10 per group, pooled from three independent experiments.
(N) Adiponectin:leptin ratio as measured by ELISA from plasma of fasted mice; n = 8 per group, pooled from three independent experiments. Data are taken from 26-week-old Apoeh/hLdlr−/− mice fed with a Western diet and injected with PBS or THP1-IL-4-exo (1 × 1010 particles/mouse every 2 days for 6 weeks while on Western diet), or 26-week-old chow-fed wild-type C57BL/6 mice. Results were normalized to B2m and Gapdh mRNA and are presented relative to control. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 as determined using either unpaired Student’s t test (for two-group comparison) or one-way ANOVA followed by Holm-Sidak post-test (for multiple-group comparison). Data are represented as mean ± SEM.
To investigate mechanisms responsible for improved glucose disposal observed in obese mice treated with THP1-IL4-exo, we examined levels of 2-deoxyglucose (2-DG) uptake in cultured 3T3-L1 adipocytes. Using this approach, we noted increased 2-DG uptake in 3T3-L1 adipocytes treated with insulin and THP1-IL4-exo (Figure 8D). Interestingly, the use of THP1-WT-exo also increased 2-DG uptake in 3T3-L1 cells upon insulin stimulation, although to a lesser extent compared with THP1-IL4-exo treatment (Figure 8D). To further interrogate the metabolic benefits that THP1-IL4-exo contribute to adipose tissue, we measured the OCR in whole eWAT taken from Western-diet-fed Apoeh/h Ldlr−/− mice using the Seahorse Mito Stress assay. Paralleling the observed control of tissue inflammation, eWAT taken from THP1-IL4-exo-infused mice displayed a profound increase in OXPHOS activity. This was revealed by an elevated basal and maximal respiration associated with a higher proton leak and ATP production compared with PBS-infused mice (Figures 8E and 8F). Remarkably, eWAT from Western-diet-fed mice infused with THP1-IL4-exo displayed greater OXPHOS activity than eWAT from control chow-fed mice (Figures 8E and 8F), demonstrating the capacity of these exosomes to profoundly upregulate metabolic activity in eWAT even when challenged by a high-fat, high-sucrose diet. Furthermore, the benefits that THP1-IL4-exo infusions conferred to improve energy metabolism and WAT homeostasis mitigated adipocyte hypertrophy in eWAT of Western-diet-fed Apoeh/h Ldlr−/− mice (Figure 8G). These findings support the benefit of M2 macrophage expansion in restoring WAT homeostasis in obese mice.11
Last, we measured the expression levels of genes responsible for energy metabolism in eWAT tissue extracts. Consistent with our in vitro studies, we observed an increase in the mRNA expression of Pparg, as well as the mRNA and protein products of its target gene, Slc2a4 (GLUT4) in eWAT extracts of THP1-IL4-exo-infused mice (Figures 8H and 8I). We also detected an upregulation of genes involved in eWAT beiging and thermogenesis that included Ucp1, Ppargc1a, Tbx1, Dio2, Zfp516, Prdm16, and Slc25a25 (Figure 8J). Immunohistochemical analysis revealed increased levels of UCP1 in eWAT of mice treated with THP1-IL4-exo compared with PBS-treated mice (Figure 8K). The altered expression pattern of these genes and protein products supports the observed increase in OXPHOS activity in eWAT derived from THP1-IL4-exo-infused mice in Figures 8E and 8F. Remarkably, we further observed an upregulated expression in genes associated with lipophagy (Ulk1, Pnpla2, Lipe, Map1lc3a, and Map1lc3b)34 in eWAT of THP1-IL4-exo-infused mice (Figure 8L). Together, these findings are consistent with our in vitro data from cultured 3T3-L1 adipocytes in Figures 5F and 5G. In line with data from our in vitro studies from cultured 3T3-L1 adipocytes in Figure 5H, we also recorded increased gene expression of adiponectin (Adipoq) and reduced expression of leptin (Lep) in eWAT of mice treated with THP1-IL4-exo (Figure 8M). These findings parallel a nearly 2-fold increase in the plasma adiponectin:leptin ratio detected in Western-diet-fed mice infused with THP1-IL4-exo (Figure 8N), supporting improved adipose tissue function40 in response to this treatment.
Discussion
Findings from our study reveal that exosomes produced by the human monocytic cell line THP-1 are effective in controlling cardiometabolic inflammation along with the onset of diabetes in mice fed a Western high-fat diet. We also show that an exposure to IL-4 is critical for THP-1 cells to produce exosomes capable of controlling inflammation in the liver, adipose tissue, and aorta of obese diabetic mice. While benefits of the exosome treatments likely included their potent control of hematopoiesis in the bone marrow and spleen as we recently reported with studies of M2-like murine macrophage exosomes,17 they also resulted from profound shifts in cellular energy metabolism that led to increased mitochondrial respiration observed both in ex vivo adipose tissue as well as in cultured adipocytes and macrophages. A common signaling cascade observed in both cultured cell types and circulating Ly6Chi monocytes was an upregulation of PPARγ expression and reduced levels of miR-33-5p that together increased the expression of genes associated with lipophagy and cellular lipid elimination. Augmented aerobic bioenergetic activity in adipose tissue of mice treated with THP1-IL4-exo was accompanied by the beiging of WAT that improved glucose clearance and normalized resting blood glucose and insulin levels in obese mice. Together, our findings highlight cell-signaling properties through which THP1-IL4-exo rewire energy metabolism in target cells to drive mitochondrial respiration and introduce these exosomes as possible treatments for cardiometabolic inflammation and type II diabetes in obesity.
Exosomes produced by macrophages are increasingly recognized as mediators of intercellular signaling in cardiovascular inflammation17,18 and diabetes.15,16 Furthermore, accumulating evidence points to the activation status of macrophages as a source of microRNA cargo diversity in secreted exosomes that contribute to cellular signaling in numerous pathologies.15,18 Indeed, macrophages exposed to inflammatory cytokines and metabolic stressors, including LPS, oxidized low-density lipoprotein, and high glucose, have been documented to produce exosomes enriched with microRNAs that contribute to cardiovascular inflammation,18,41 insulin resistance,15 and atherosclerosis acceleration.18 In contrast, macrophages exposed to anti-inflammatory cytokines enriched exosomes with microRNAs that resolved systemic inflammation and atherosclerosis in mice with hyperlipidemia17 and restored insulin sensitivity in obese mice.16 Findings from our study of THP1-IL4-exo support the value of M2-like macrophage exosomes to control chronic inflammatory disorders caused by excessive energy storage. Furthermore, our findings introduce the capacity for THP1-IL4-exo to modulate mitochondrial bioenergetics in macrophages and adipocytes to drive beiging in WAT, leading to the resolution of cardiometabolic inflammation in obese diabetic mice. Our findings thus provide translational relevance for exosomes produced by a well-defined human macrophage cell line that could be suitable for human therapy.
The molecular source of signaling through which THP1-IL4-exo altered metabolic properties of target cells, while not directly examined in this study, likely derived in part from a delivery of microRNA cargo that includes miR-99a-5p/146b-5p/378a-3p as reported in our recent study of mouse primary macrophages.17 Indeed, our findings support functional transfer of these microRNAs as they were enriched in THP1-IL4-exo and all recipient cells tested. Furthermore, we noted miR-21-5p to be highly enriched in both the exosomes and their target cells, consistent with our data reporting it as the most abundant microRNA in exosomes produced by macrophages irrespective of their stimulation with IL-4.17 Future studies will be required to validate functional transfer of microRNA and other bioactive cargo enriched in THP1-IL4-exo that could include long non-coding RNA and even metabolites of mitochondrial respiration as sources of cellular bioenergetic regulation. Regardless of how THP1-IL4-exo provoked an upregulation of all four of these microRNAs in their target cells, their functional significance was striking with respect to our observed rewiring of energy metabolism in cultured cells and cardiometabolic tissue in obese mice. The mechanistic basis for these benefits likely derived from the targeted control of mRNA gene expression by the exosomes.
Based on our prior findings and those of others, increased cellular miR-99a-5p likely contributed to control TNF-α mRNA levels to suppress M1-like macrophage marker genes that included iNOS, which plays a central role in promoting insulin resistance and atherosclerosis.17,42 Furthermore, exosomal miR-99a-5p is likely beneficial in improving glucose tolerance, insulin sensitivity, and preserved liver function by preventing cellular oxidative stress via NOX4 mRNA control as reported in models of obesity and fatty liver disease.43 Similarly, increased cellular miR-378a-3p levels likely exerted numerous metabolic benefits, including the control of macrophage proliferation44 and obesity in our cohorts of Western-diet-fed mice. While not directly examined in our study, exosomal miR-378a-3p could have increased lipolysis and the expansion and thermogenic properties of brown adipose tissue by downregulating Scd1 and Pde1b45,46 in adipocytes and WAT of obese mice (Figures S3C and S9A), contributing to the normalization of their blood glucose and insulin levels. Furthermore, based on a recent report of miR-378a-3p activity in WAT,47 its exosomal delivery to this tissue likely served a central role in promoting beiging and improved glucose and insulin signaling in obese mice injected with THP1-IL4-exo. Favorable metabolic reprograming in target tissues likely also arose from an upregulated expression of miR-146b-5p. Indeed, we recently reported the exosomal delivery of this microRNA in controlling inflammatory activity by reducing TLR/NF-κB signaling in macrophages and HSPCs that attenuated atherosclerosis progression.17 Interestingly, the control of macrophage activation via miR-146a has been reported to control metabolic dysfunction in diet-induced obesity,48 a pathway that could be shared by its homolog miR-146b-5p, which will require further investigation.
Above all microRNAs found to be upregulated in BMDM and 3T3-L1 adipocytes treated with THP-1 exosomes, miR-21-5p likely served to substantially improve cardiometabolic inflammation and attenuate atherosclerosis in our cohorts of obese mice. Indeed, miR-21-5p is well recognized as a central regulator of macrophage metabolic reprograming49,50 and cellular activity in atherosclerosis control.51 MiR-21-5p also protects against insulin resistance and improves glucose uptake in adipocytes by modulating the PTEN-AKT pathway, resulting in increased mobilization of GLUT4 upon insulin stimulation.52 Furthermore, recent findings have reported its potent capacity to improve metabolic activity, WAT beiging, and brown adipose tissue (BAT) induction in obese mice through the modulation of vascular endothelial growth factor A (VEGF-A), p53, and TGFβ1 signaling pathways.30 Taken together, our data point to the cellular enrichment of microRNA carried by THP1-IL4-exo as a key element to explain their control of cardiometabolic inflammation in obese mice.
Central to the metabolic signaling properties exerted by THP1-IL4-exo was the profound reduction in miR-33-5p that the exosomes induced in cultured BMDMs, 3T3-L1 adipocytes, and circulating Ly6Chi monocytes. Reduced levels of miR-33-5p can in part be explained by the downregulation of its host, Srebf2, in addition to the homolog, Srebf1, which are both repressed by PPARγ.53, 54, 55 Our findings therefore support the importance of PPARγ induction in response to THP1-IL4-exo treatment as a major driver of metabolic reprogramming in target cells by downregulating SREBP-1/2 gene expression and reducing levels of miR-33-5p, which is a critical hub for bioenergetic metabolism.26,29,56 An induced expression of PPARγ likely arose by a functional transfer and increased cellular levels of miR-99a-5p and miR-378a-3p, which have both been reported to drive the gene expression of PPARγ.42,46,57,58 Taken together, our findings reveal novel insights into molecular properties through which M2-like macrophage exosomes control the reprogramming of cellular metabolism via the upregulation of microRNAs that drive the expression of PPARγ and subsequent reduction of miR-33-5p.
While our findings support those from a recent study demonstrating benefits of antagonizing miR-33-5p to improve thermogenesis and adipose tissue beiging,29 they also introduce M2-like macrophage exosomes as effectors of these metabolic processes. In testing the effects of miR-33-5p downregulation conferred by THP1-IL4-exo, we observed a marked increase in its target genes involved in adipocyte beiging in both cultured 3T3-L1 adipocytes and WAT of Western-diet-fed obese mice. In particular, we found a profound increase in the transcriptional regulators and coregulators Zfp516, Ppargc1a, and Prdm16, as well as the enzyme type II iodothyronine deiodinase Dio2, and the calcium-binding mitochondrial carrier protein involved in mitochondrial function Slc25a25.29 The enhanced expression of these genes likely contributed to our observations of increased uncoupling protein 1 (UCP1) gene and protein expression, which is recognized as a key regulator in thermogenesis and adipocyte beiging.29 This pattern of gene and protein expression likely led to an observed increase in mitochondrial respiration and subsequent OXPHOS activity in both cultured adipocytes and WAT, a critical process during thermogenesis and adipocyte beiging.29,31 The improvements in mitochondrial function caused by the cellular uptake of THP1-IL4-exo likely also stemmed from their ability to reduce oxidative stress (Figures 2E and 5C), resulting in the control of mitochondrial permeability by restricting the opening of the transition pore.
Such functional benefits in the adipose tissue were likely also driven from our observed upregulation of genes associated with lipophagy, lipid catabolism, and cholesterol efflux, all of which are controlled by miR-33-5p.26,29,56 In particular, we found an increase in expression of genes associated with lipophagy, including Ulk1, Pnpla2, Lipe, Map1lc3a, and Map1lc3b, in BMDMs, 3T3-L1 adipocytes, and eWAT of mice treated with THP1-IL4-exo. Among these genes, Ulk1 (Unc-51 like autophagy activating kinase 1), Map1lc3a (microtubule-associated protein 1 light chain 3 alpha), and Map1lc3b (microtubule-associated protein 1 light chain 3 beta) are known inducers of autophagy.34 In addition, Pnpla2 (adipose triglyceride lipase) and Lipe (hormone-sensitive lipase) are known to induce fatty acid and cholesterol catabolism in macrophages and adipocytes during cardiovascular and metabolic inflammation.59, 60, 61
Beyond serving as a checkpoint for miR-33-5p expression, PPARγ induction by THP1-IL4-exo exposure increased the expression of lipid mobilizing and efflux genes including Abca1 and Apoe, as well as the insulin-dependent glucose transporter GLUT4 (Slc2a4), in cells cultured in vitro and those tested in mice. Indeed, an upregulation of Abca1 observed in BMDMs and circulating Ly6Chi monocytes could have directly derived from a reduction of miR-33-5p, resulting in a dual mode of transcriptional regulation.29,62 Interestingly, the expression of Abcg1 was not as highly upregulated as Abca1, despite both being targets of miR-33-5p, suggesting that Abcg1 may be subject to post-transcriptional regulation in macrophages and monocytes.62 Regardless, the profound decrease in neutral lipids detected in both BMDM and 3T3-L1 adipocytes treated with THP1-IL4-exo support a functional relevance to our observed upregulation of genes involved in lipid catabolism that likely serve as biofuels to satisfy demands of increased mitochondrial respiration and efflux of free cholesterol that would otherwise be deleterious.63,64 Indeed, enhanced cellular cholesterol elimination observed in macrophages and monocytes has long been noted to control monocytosis and hematopoiesis, including in diabetic mice during atherosclerosis regression in response to miR-33-5p inhibition.62 Together, our observations demonstrate that THP1-IL4-exo are effective in activating pathways critical for the catabolism and efflux of fatty acids and cholesterol in macrophages and adipocytes that are controlled by miR-33-5p.26,29,56
A critical element for improving metabolic health in the treatment of obesity is a restoration of balance in adipokine expression, particularly in maintaining the adiponectin:leptin ratio at a level of 1.0 or higher.40 Such pattern of adipokine expression enhances cold-induced beiging of WAT, fosters M2 macrophage proliferation,65 and restores white adipocyte homeostasis.11 Remarkably, THP1-IL4-exo treatments induced this beneficial effect by upregulating adiponectin (Adipoq), while concomitantly downregulating leptin (Lep) in adipocytes in cultured cells and in vivo in adipose tissues of obese Western-diet-fed mice infused with THP1-IL4-exo. This profound shift in endocrine function likely contributed to normalize levels of insulin and glucose in obese mice fed a Western high-fat diet.
Our findings are in line with those that revealed the utility of miRNA delivery in diabetes control.16,30,42, 43, 44, 45, 46, 47,52 Indeed, miR-99a-5p/378a-3p/21-5p that have all been reported to improve cellular aspects of metabolism in diabetes are all enriched in THP1-IL4-exo and in their target cells. Furthermore, while our data generally support beneficial properties reported for M2-like BMDM exosomes in improving insulin sensitivity in obese high-fat-diet-fed mice,16 the mechanisms to explain such benefits differ. Indeed, BMDM exposed to a combination of IL-4 and IL-13 led to exosomes enriched with miR-690 that restored insulin sensitivity in obese mice via the control of a Nadk-relevant pathway involved in macrophage inflammatory response and insulin signaling.16 As miR-690 is a mouse-specific microRNA not carried by THP1-IL4-exo, it is not surprising that our data of BMDMs and 3T3-L1 adipocytes treated with THP1-IL4-exo did not reveal changes in the expression of Nadk (Figure S9C). These results suggest that THP1-IL4-exo exert protective properties that do not overlap with the mechanism identified by the study of exosomes derived from IL-4-/IL-13-treated primary mouse macrophages in controlling insulin resistance in obese mice.16
In conclusion, our findings unveil a plethora of benefits exerted by IL-4-stimulated THP-1 macrophage exosomes that improve adipokine homeostasis and rewire energy metabolism in target cells, including macrophages and adipocytes, to control the onset of cardiometabolic tissue inflammation during obesity-driven diabetes. Together, the results of our study provide compelling evidence for the broad and potent capacity of THP-IL4-exo to resolve inflammation and improve energy homeostasis in adipose tissue and hematopoietic compartments. Our data therefore support future studies of THP1-IL4-exo in large mammals to determine their suitability for the treatment of obesity-driven cardiometabolic disease and type II diabetes.
Study limitations
Interpretations of results from this study are limited by the targeted analysis of microRNA using qRT-PCR. Other microRNAs enriched in THP1-IL4-exo likely contribute to their overall beneficial signaling capacity. While future unbiased RNA-seq studies will be needed to fully assess the microRNA cargo in THP1-IL4-exo, the cluster of microRNAs identified in the current study is of value owing to their established roles in modulating inflammation and metabolic activity. Furthermore, while THP1-IL4-exo display a potent capacity to suppress inflammation, there remains limited knowledge as to their entire functional cargo. Exploring the contribution of long non-coding RNA, proteins, and even metabolites of the tricarboxylic acid (TCA) cycle as signaling cargo could add to the understanding of their therapeutic potential.
Materials and methods
Animal studies
In vivo studies were conducted using hypomorphic Apoeh/h Ldlr−/− mouse strain by breeding Ldlr−/− mice on a C57BL/6J background (Jackson Laboratories, USA) to hypomorphic (Apoeh/h) mice.66 Ten-week-old male Apoeh/h Ldlr−/− or C57BL/6 wild-type mice were fed a Western diet (Research Diets, USA) for 10 weeks before being randomly assigned to be infused with THP1-IL4-exo or PBS as control for 6 weeks while remaining on the Western diet for the duration of the study (n = 8–10 for each treatment group, pooled from two to three cohorts of mice). Data collection and analyses were conducted in a blinded fashion. All mice were housed and bred in specific pathogen-free conditions in the Animal Research Facility at the San Francisco Veterans Affairs Medical Center. All animal experiments were approved by the Institutional Animal Care and Use Committee at the San Francisco Veterans Affairs Medical Center.
Cell culture
The human monocytic cell line (THP-1) was purchased from the University of California San Francisco (UCSF) Cell and Genome Engineering Core (CGEC) as an authenticated stock. Cells were cultured in RPMI 1640 medium (Corning, USA) supplemented with 10% fetal bovine serum (GIBCO, USA), 1% GlutaMAX (Gibco, USA), and 1% penicillin-streptomycin (Gibco, USA). THP-1 cells were grown and expanded in suspension in a T-75 flask (Fisher Scientific, USA) until reaching a density of 1 × 106 cells/mL. Cells were then seeded to 10-cm plates (Corning, USA) at a density of 4 × 106 cells/plate and differentiated into macrophages by culturing in 25 ng/mL phorbol 12-myristate 13-acetate (PMA) (Fisher Scientific, USA) for 48 h. Cells were then cultured in PMA-free medium for an additional 48 h and subsequently exosome-free medium with or without recombinant human IL-4 (Peprotech, USA) for exosome production.
For in vitro experiments, THP-1 cells were seeded to a 12-well plate (Corning, USA) at a density of 4 × 105 cells/well and differentiated into macrophages by culturing in 25 ng/mL PMA (Fisher Scientific, USA) for 48 h. Cells were then cultured in PMA-free medium for an additional 48 h. Fresh culture medium was then added with 2 × 109 or 4 × 109 particles/mL of exosomes or equal volume of PBS (Corning, USA) as control and cultured for 24 h prior to analysis.
The 3T3-L1 preadipocyte cell line was purchased from the UCSF CGEC as authenticated stock. Cells were cultured in DMEM (Corning, USA) supplemented with 10% fetal bovine serum (Gibco, USA), 1% GlutaMAX (Gibco, USA), and 1% penicillin-streptomycin (Gibco, USA). To differentiate into adipocytes, 3T3-L1 cells were seeded into a 12-well culture plate (Corning, USA) at a density of 4 × 104 cells/well and allowed to grow until fully confluent. The cells were then incubated as a confluent culture for 48 h and subsequently cultured in differentiation medium containing DMEM supplemented with 10% fetal bovine serum (Gibco, USA), 1.0 μM dexamethasone (Sigma-Aldrich, USA), 0.5 mM methylisobutylxanthine (IBMX) (Sigma-Aldrich, USA), 1.0 μg/mL bovine insulin (Sigma-Aldrich, USA), 1% GlutaMAX (Gibco, USA), and 1% penicillin-streptomycin (Gibco, USA) for an additional 48 h. The cells were then changed into adipocyte maintenance medium containing DMEM supplemented with 10% fetal bovine serum (Gibco, USA), 1.0 μg/mL bovine insulin (Sigma-Aldrich, USA), 1% GlutaMAX (Gibco, USA), and 1% penicillin-streptomycin (Gibco, USA). The adipocyte maintenance medium was changed every 48–72 h and cells were fully differentiated by 15 days of induction. Differentiated adipocytes were then cultured with 4 × 106 particles/mL of exosomes or an equal volume of PBS (Corning, USA) as control for 24 h before being collected for analysis. In some experiments, 4 × 106 particles/mL of exosomes or equal volume of PBS (Corning, USA) were given every 2 days after the cells were induced in differentiation medium.
Murine BMDMs were obtained as previously described.17,18 Briefly, bone marrow cells were flushed from the tibia and femurs of 6- to 12-week-old male C57BL/6J mice. Cells were cultured in DMEM (Corning, USA) supplemented with 10% fetal bovine serum (Gibco, USA), 1% GlutaMax (Gibco, USA), and 1% penicillin-streptomycin (Gibco, USA) and differentiated with 25 ng/mL mouse macrophage colony-stimulating factor (M-CSF,Peprotech, USA) for 6 days. BMDMs were seeded into 12-well culture plates (Corning, USA) at a concentration of 3 × 105 cells/well and stimulated with 2 × 109 or 4 × 109 particles/mL of exosomes or an equal volume of PBS (Corning, USA) as control for 24 h before collected for analysis.
THP-1 macrophage exosome isolation and nanoparticle tracking analysis
Our exosome isolation and characterizations were performed in adherence to the MISEV2018 guidelines.67 THP-1 cells were seeded into 10-cm plates (Corning, USA) at a density of 4 × 106 cells/plate as described above. PMA-free medium was discarded after 48 h of incubation. The cells were then washed once with PBS (Corning, USA) and cultured in exosome-depleted medium prepared by ultracentrifugation for 18 h at 100,000 × g (Type 45 Ti rotor, Beckman Coulter, USA) and filtration (0.2 μm) supplemented with or without 20 ng/mL human IL-4 (Peprotech, USA). After 24 h of incubation, the conditioned medium was collected. Exosomes were isolated from conditioned medium using C-DGUC as previously described.17, 18, 19, 20 Briefly, the conditioned medium was centrifuged at 400 × g for 10 min at 4°C to pellet dead cells and debris followed by centrifugation at 2000 × g for 20 min at 4°C to eliminate debris and large vesicles. The supernatant was then filtered (0.2 μm) and centrifuged on a 60% iodixanol cushion (Sigma-Aldrich, USA) at 100,000 × g for 3 h (Type 45 Ti, Beckman Coulter, USA). OptiPrep density gradient (5%, 10%, 20% w/v iodixanol) was employed to further purify exosomes at 100,000 × g for 18 h at 4°C (SW 40 Ti rotor, Beckman Coulter, USA). Afterward, twelve 1-mL fractions were collected starting from the top of the tube. Fraction 7 of the gradient was dialyzed in PBS with the Slide-A-Lyzer MINI Dialysis Device (Thermo Fisher Scientific, USA) and used for subsequent experiments and analyses. A flow diagram depicting the production and purification steps of THP-1 macrophage exosomes is shown in Figure S1A.
Particles in fraction 7 were subjected to size and concentration measurement by NanoSight LM14 (Malvern Instruments, Westborough, USA) at a 488-nm detection wavelength. The analysis settings were optimized and kept identical for each sample, with a detection threshold set at 3, three videos of 1 min each were analyzed to give the mean, mode, median, and estimated concentration for each particle size. Samples were diluted in 1:100 or 1:200 PBS and measured in triplicate. Data were analyzed with the NTA 3.2 software. All exosome samples were store at 4°C and used within 1 month after isolation. Details relevant to exosome isolation and physical characterization data have been submitted to the EV-TRACK knowledgebase (EV-TRACK: EV200042).
Labeling and in vitro/in vivo tracking of THP-1 macrophage exosomes
Fluorescently detectable THP-1 exosomes were generated using PKH26 (Sigma-Aldrich, USA) or DiR (DiIC18(7); Invitrogen, USA). The dye was added to the 3-mL iodixanol cushion layer containing exosome or to 3 mL of PBS to achieve a final concentration of 3.5 mM for PKH26 or 1 μM for DiR and incubated for 20 min at room temperature. Labeled exosomes and control were loaded below an iodixanol step gradient as described above in the section “THP-1 macrophage exosome isolation and nanoparticle tracking analysis.” Free dye and nonspecific protein-associated dye were eliminated from labeled exosomes or from PBS control during this separation step. For in vitro experiments, BMDMs or 3T3-L1 adipocytes were exposed to 4 × 109 PKH26-labeled exosome for 2 h, washed three times with PBS, and imaged using a Zeiss Observer microscope. Fluorescence intensity of the PKH26-positive cells was measured by using ImageJ. For in vivo experiments, 20-week-old Western-diet-fed Apoeh/h Ldlr−/− mice were infused i.p. with PBS or 1 × 1010 DiR-labeled exosomes for 6 h. The mice were then extensively perfused with PBS. Blood, aortas, hearts, livers, eWAT, bones, spleen, lungs, brains, intestines, and kidneys were collected, imaged, and quantified for DiR fluorescence signal using the Odyssey Infrared Imaging System and Image Studio software.
Transmission electron microscopy
An assessment of exosome morphology was assessed by electron microscopy by loading 7 × 108 exosomes onto a glow discharged 400-mesh Formvar-coated copper grid (Electron Microscopy Sciences, USA). The nanoparticles were left to settle for 2 min, and the grids were washed four times with 1% uranyl acetate. Excess uranyl acetate was blotted off with filter paper. Grids were then allowed to dry and subsequently imaged at 120 kV using a Tecnai 12 transmission electron microscope (FEI, USA).
Protein extraction and immunoblotting
Each fraction of the C-DGUC purified exosomes (37.5-μL sample) was mixed with 12.5 mL of 4× Laemmli buffer (Bio-Rad, USA). For cell lysates, cells were lysed in radioimmunoprecipitation assay (RIPA) Buffer (Cell Signaling, USA) containing cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail (Roche, Switzerland) and 1 mM PMSF (Cell Signaling, USA). For tissue lysates, 25 mg of eWAT were placed in RIPA Buffer (Cell Signaling, USA) containing cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail (Roche, Switzerland) and 1 mM PMSF (Cell Signaling, USA) and homogenized with a Tissue-Tearor. Protein concentrations were measured using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, USA). A total of 15 μg of proteins was diluted with PBS to 37.5 μL, then mixed with 12.5 μL 4× Laemmli buffer (Bio-Rad, USA). Samples were subsequently heated at 95C for 5 min. Samples were then loaded on a 10% SDS-PAGE gel and transferred onto a polyvinylidene fluoride (PVDF) membrane (Bio-Rad, USA). The membranes were blocked with 5% non-fat milk dissolved in PBS for 1 h and then incubated with primary antibodies overnight at 4°C. Primary antibodies for exosomes markers include anti-CD9 (1:100, BD Biosciences, USA), anti-CD63 (1:100, BD Biosciences, USA), and anti-CD81 (1:100, Santa Cruz, USA). Primary antibodies for cell lysate markers include anti-Calnexin (1:500, Abcam, USA) and anti-GM130 (1:250, BD Biosciences, USA). Primary antibodies for 3T3-L1 adipocytes and eWAT tissues include anti-PPARγ (1:200, Santa Cruz, USA), anti-GLUT4 (1:200, Santa Cruz, USA), anti-UCP1 (1:200, Santa Cruz, USA), anti-b-actin (1:1,000, Santa Cruz, USA), anti-HSP70 (1:1,000, Santa Cruz, USA), and anti-GAPDH (1:1,000, Cell Signaling, USA), and anti-Vinculin (1:1,000, Santa Cruz, USA). After four washes in PBS containing 0.1% Tween (PBST), membranes were incubated with corresponding HRP-conjugated secondary antibodies: anti-mouse IgG-HRP (1:1,000, Santa Cruz, USA) or anti-rabbit IgG-HRP (1:1,000, Thermo Fisher Scientific, USA) for 1 h and washed in PBST. Signals were visualized after incubation with Amersham ECL Prime substrate and imaged using an ImageQuant LAS 4000. Quantification was analyzed using ImageJ. A list of antibodies used is available in Table S1.
RNA extraction and gene expression analysis using qRT-PCR
Exosomes were treated with 0.4 mg/mL of RNase A/T1 Mix (Thermo Fisher Scientific, USA) for 20 min at 37°C before RNA extraction. For RNA extraction from tissues, 100 mg of adipose tissue or 25 mg of liver tissue was placed into Qiazol Lysis Buffer (QIAGEN, Germany), and the tissue was then homogenized using a Tissue-Tearor and spun at 12,000 × g at 4°C for 10 min. The supernatant was then collected for RNA extraction. Circulating Ly6Chi monocytes were stained with the antibody cocktail indicated in the circulating and tissue-associated leukocyte detection using flow cytometry section below and FACS sorted using a BD FACSAria II (BD Biosciences, USA) into Qiazol Lysis Buffer (QIAGEN, Germany).
Total RNA isolated from exosomes, cells, and tissues was extracted using Qiazol Lysis Buffer and purified using the RNeasy Mini Kit (QIAGEN, Germany) according to the manufacturer’s protocol. RNA from liver and fat tissues was purified using on-column DNase digestion (QIAGEN, Germany) according to the manufacturer’s protocol. RNA was quantified using Nanodrop or Quant-iT RiboGreen RNA Assay Kit (Thermo Fisher Scientific, USA) and reverse transcribed using the iScript Reverse Transcription Supermix (Bio-Rad, USA) for mRNA or the miRCURY LNA RT Kit (QIAGEN, Germany) for microRNA analysis. qPCR reactions were performed using the Fast SYBR Green Master Mix (Applied Biosystems, USA) for mRNA or the miRCURY LNA SYBR Green PCR Kit (QIAGEN, Germany) for microRNA and processed using a QuantStudio 7 Flex Real-Time PCR System. Cycle threshold (Ct) values were normalized to the housekeeping genes Gapdh and B2m for mouse and Gapdh and Actb for human mRNA. For microRNA expression, UniSp6 was used as a spike-in control while U6 small nuclear RNA (snRNA), miR-16-5p, and miR-21-5p (QIAGEN, Germany) were used as reference genes. All reactions were done in triplicate. Primers used for qRT-PCR are listed in Table S2.
Glucose uptake assay in cultured 3T3-L1 adipocytes
3T3-L1 cells were seeded at a density of 2,000 cells/well in a 96-well culture plate and differentiated into mature adipocytes as described above. Adipocytes were then treated with 6 × 109 particles/mL PBS, THP1-WT-exo, or THP1-IL4-exo for 24 h. Cells were then starved in serum-free medium overnight. The next day, adipocytes were preincubated with Krebs-Ringer-Phosphate-HEPES (KRPH) buffer containing 2% BSA, 20 mM HEPES, 5 mM KH2PO4, 1 mM MgSO4, 1 mM CaCl2, 136 mM NaCl, and 4.7 mM KCl, pH 7.4 (all from Sigma-Aldrich, USA) for 40 min. Cells were then stimulated with 1 μM human insulin (Sigma-Aldrich, USA) for 20 min. Subsequently, 10 μ L/well of 10 mM 2-DG was added and incubated for 20 min. Next, cells were washed three times with PBS to remove exogenous 2-DG. Adipocytes were then lysed and 2-DG uptake was processed using a Glucose Uptake Assay Kit (Abcam, USA) according to the manufacturer’s protocol. Absorbance reading was measured at optical density (OD) 412 nm on a microplate reader (Molecular Devices, USA).
Detection of lipid vacuoles and mitochondria health in macrophages and 3T3-L1 adipocytes
For analysis of lipid vacuoles, BMDMs or differentiated 3T3-L1 adipocytes were stained with LipidTOX (Invitrogen, USA) (1:250) and Hoechst (1:1,000) in Live Cell Imaging Solution (Invitrogen, USA) for 30 min at room temperature and imaged using a Zeiss Observer microscope. Alternatively, cells were stained with LipidTOX (Invitrogen, USA) (1:250) for 30 min, dissociated with trypsin-EDTA, and analyzed using a CytoFLEX S cytometer (Beckman, USA). For analysis of mitochondrial activity, cells were stained with MitoSOX or tetramethylrhodamine at final concentrations of 5 μM and 0.1 μM, respectively. The cells were then incubated at 37°C for 30 min. Cells were then dissociated with trypsin-EDTA and analyzed using a CytoFLEX S cytometer (Beckman, USA). To measure mitochondrial transition pore opening, BMDMs or 3T3-L1 adipocytes were analyzed using the MitoProbe Transition Pore Assay Kit (Invitrogen, USA) according to the manufacturer’s protocol. Briefly, cell suspensions were mixed with 2 μM calcein AM and 160 μM CoCl2. For negative control, cells were also mixed with 0.2 μM ionomycin. Cells were then analyzed for mitochondrial calcein AM retention using the CytoFLEX S cytometer (Beckman, USA).
Circulating and tissue-associated leukocyte detection using flow cytometry
Mice were anesthetized with isoflurane (Forane, Baxter, USA) and peripheral blood was collected by retro-orbital bleeding with heparinized micro-hematocrit capillary (Fisher Scientific, USA) in tubes containing 0.5 M EDTA. Red blood cells were lysed in red blood cell (RBC) Lysis Buffer (BioLegend, USA). Nonspecific binding was blocked with TruStain FcX Ab (BioLegend, USA) for 10 min at 4°C in FACS buffer (Ca2+/Mg2+-free PBS with 2% fetal bovine serum and 0.5 mM EDTA) before staining with appropriate antibodies: CD11b (clone M1/70), Ly-6C (clone HK1.4), CD115 (clone AFS98), and CD45 (clone 30-F11) (all BioLegend, USA) for 30 min at 4°C. The antibody dilutions ranged from 1:200 to 1:100. Splenocytes were isolated using mechanical dissociation. Briefly, spleens were mashed using the bottom of a 3-mL syringe (BD Biosciences, USA). The cells were then passed through a 70-μm cell strainer and incubated in RBC Lysis Buffer (BioLegend, USA). Nonspecific binding was blocked with TruStain FcX Ab (BioLegend, USA) for 10 min at 4°C in FACS buffer before staining with appropriate antibodies: CD11b (clone M1/70), Ly-6C (clone HK1.4), Ly-6G (clone 1A8), and CD11c (clone N418). The antibody dilutions ranged from 1:200 to 1:100.
For aorta digestion, single-cell suspension from an aorta segment including the aortic arch and thoracic aorta was prepared by incubation with an enzyme mixture containing 400 U/mL collagenase I, 120 U/mL collagenase XI, 60 U/mL hyaluronidase, and 60 U/mL DNase I (all from Sigma-Aldrich, USA) in Hank’s balanced salt solution for 50 min at 37°C. Cells were then passed through a 70-μm cell strainer and spun down at 300 × g for 5 min at 4°C. The resulting cell pellet was blocked with TruStain FcX antibody (BioLegend, USA) for 10 min at 4°C in FACS buffer before staining with appropriate antibodies: CD11b (clone M1/70), Ly-6C (clone HK1.4), Ly-6G (clone 1A8), CD45 (clone 30-F11), and F4/80 (clone BM8). The antibody dilutions ranged from 1:200 to 1:100.
For adipose tissue and liver digestion, 1 g of eWAT or 1 g of liver was put in an enzyme mix from the Adipose Tissue or Liver Dissociation Kit (Miltenyi, Germany) and digested using the gentleMACS Dissociator (Miltenyi, Germany) according to the manufacturer’s protocol. Cells were then passed through a 100-μm cell strainer and centrifuged at 300 × g for 10 min. Cellular debris was cleared by mixing the cell suspension with Debris Removal Solution (Miltenyi, Germany) and performing density centrifugation according to the manufacturer’s protocol. The resulting cell pellet was incubated in RBC Lysis Buffer (BioLegend, USA) and blocked with TruStain FcX Ab (BioLegend, USA) for 10 min at 4°C in FACS buffer before staining with appropriate antibodies: CD11b (clone M1/70), Ly-6C (clone HK1.4), Ly-6G (clone 1A8), CD45 (clone 30-F11), and F4/80 (clone BM8). The antibody dilutions ranged from 1:200 to 1:100. All euthanized mice were fully perfused with PBS before the tissues were harvested. All flow cytometric analyses were conducted using a CytoFLEX S cytometer (Beckman, USA). Sorting of circulating Ly6Chi monocytes was performed using a FACSAria II Cell Sorter (BD Biosciences, USA). A list of antibodies used is available in Table S1.
Histological assessments in liver and eWAT
Frozen liver tissues were cryosectioned at 10 μm, with two sections per mouse per slide. Neutral lipid accumulation was analyzed by staining with ORO (Sigma-Aldrich, USA) and counterstaining with modified Mayer’s hematoxylin (Thermo Fisher Scientific, USA). ORO-positive areas were captured by a Zeiss Observer microscope and quantified using ImageJ. The average between the two sections is shown for each mouse. Paraffinized liver and eWAT tissues were deparaffinized and cut at 10 μm, with two sections per mouse per slide. The slides were then stained with modified Mayer’s hematoxylin (Thermo Fisher Scientific, USA) and Eosin Y (Sigma-Aldrich, USA) and captured by a Zeiss Observer microscope. Steatotic areas in the liver were measured by ImageJ, and the average between the two sections is shown for each mouse. Average adipocyte sizes were also measured by ImageJ.
For F4/80 or UCP1 staining, deparaffinized eWAT sections underwent heat-induced antigen unmasking in 10 mM citrate buffer with 0.5% Tween 20. Sections were then blocked with 10% normal goat serum and labeled with a primary rabbit anti-mouse F4/80 or UCP1 (Cell Signaling, USA). Endogenous peroxidase activity was then blocked by incubating with 3% H2O2 (Sigma-Aldrich, USA). Sections were then incubated with SignalStain Boost IHC Detection Reagent anti-rabbit (Cell Signaling, USA) and visualized for peroxidase activity using SignalStain DAB Substrate Kit (Cell Signaling, USA). Sections were counterstained with Vector Methyl Green (Vector Laboratories, USA). Images were captured using a Zeiss Observer microscope. F4/80-positive CLSs were enumerated, and the average between the two sections is shown for each mouse.
Measurement of inflammatory cytokines and metabolic markers in mice
Peripheral blood was collected as indicated above and spun at 1,500 × g for 30 min at 4°C to collect the plasma. Cholesterol and triglycerides levels were measured from plasma using the Cholesterol E Assay Kit or L-Type Triglyceride M Assay Kit (Wako Diagnostics, Fujifilm, Japan). Insulin level was measured in the plasma using a Mouse Insulin ELISA Kit (Abcam, USA). Plasma adiponectin and leptin were measured using a Mouse Adiponectin or Leptin ELISA Kit (Abcam, USA). Plasma cytokines (TNF-α, IFN-γ, IL-6, and IL-1β) were measured using the V-Plex Mouse Custom Cytokine Kit (Meso Scale Discovery, USA). For GTT, mice received one dose of glucose (2 g/kg body weight) via i.p. injection after 18 h of fasting. Blood glucose levels were measured before glucose injection and 30 min, 60 min, 90 min, and 120 min after injection.
Seahorse extracellular flux analysis
For BMDMs, cells were plated at 60,000 cells/well into XFe24 cell culture microplates (Agilent, USA) and incubated overnight at 37°C and 5% CO2. The following day, BMDMs were stimulated with PBS, THP1-WT-exo, or THP1-IL4-exo for 24 h. Before analysis, cells were washed with Seahorse XF DMEM assay buffer (Agilent, USA) supplemented with 10 mM glucose (Agilent, USA), 1 mM pyruvate (Agilent, USA), and 2 mM glutamine (Agilent, USA) and incubated for 1 h at 37°C without CO2. OCR and extracellular acidification rate (ECAR) were measured using the mitochondrial stress test kit (Agilent, USA) in response to 1 μM oligomycin, 2 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and 0.5 μM rotenone/antimycin A (R/AA) with the Seahorse XFe-24 Bioanalyzer (Agilent, USA). After OCR measurements, cells were incubated in Hoechst (1:1,000) diluted in Live Cell Imaging Solution (Invitrogen, USA) and imaged under a Zeiss Observer microscope. Total cell counts were measured using ImageJ.
For adipocytes, fully differentiated 3T3-L1 cells were plated at 30,000 cells/well into XFe24 cell culture microplates (Agilent, USA) and incubated overnight at 37°C and 5% CO2. The following day, cells were stimulated with PBS, THP1-WT-exo, or THP1-IL4-exo for 24 h. Before analysis, cells were washed with Seahorse XF DMEM assay buffer (Agilent, USA) supplemented with 10 mM glucose (Agilent, USA), 1 mM pyruvate (Agilent, USA), and 2 mM glutamine (Agilent, USA) and incubated for 1 h at 37°C without CO2. OCR and ECAR were measured using the mitochondrial stress test kit (Agilent, USA) in response to 1 μM oligomycin, 0.25 μM FCCP, and 0.5 μM R/AA with the Seahorse XFe-24 Bioanalyzer (Agilent, USA). After OCR measurements, cells were incubated in Hoechst (1:1,000) diluted in Live Cell Imaging Solution (Invitrogen, USA) and imaged under a Zeiss Observer microscope. Total cell counts were measured using ImageJ.
For ex vivo OCR measurements of eWAT, 10 mg/well of eWAT was washed in Seahorse XF DMEM assay buffer (Agilent, USA) supplemented with 25 mM HEPES (Sigma-Aldrich, USA) and placed in an XF24 Islet Capture Microplate (Agilent, USA). The tissue was then locked in a capture screen and incubated in Seahorse XF DMEM assay buffer (Agilent, USA) supplemented with 10 mM glucose (Agilent, USA), 1 mM pyruvate (Agilent, USA), and 2 mM glutamine (Agilent, USA) and incubated for 1 h at 37°C without CO2. OCR and ECAR were measured using the mitochondrial stress test kit (Agilent, USA) in response to 10 μM oligomycin, 10 μM FCCP, and 5 μM R/AA with the Seahorse XFe-24 Bioanalyzer (Agilent, USA). Proteins from eWAT were extracted using a Tissue-Tearor in RIPA Buffer (Cell Signaling, USA) containing cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail (Roche, Switzerland) and 1 mM PMSF (Cell Signaling, USA). Protein concentrations were measured using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, USA).
All OCR measurements from cells were normalized to cell number and used to calculate various parameters of mitochondrial activity, including basal mitochondrial OCR, proton leak, ATP synthesis, maximal OCR, and spare respiratory capacity. All OCR measurements from eWAT were normalized to total proteins and used to calculate parameters similar to those indicated above. Data were analyzed using XFe Wave software.
Statistical analysis
Statistical analysis was performed with GraphPad Prism v8, using the unpaired, two-tailed, Student’s t test (two groups) and one-way or two-way analysis of variance (ANOVA) with post-tests, Holm-Sidak, as indicated in figure legends for multiple groups. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 ∗∗∗∗p < 0.0001. Normality test was performed using the Shapiro-Wilk test on GraphPad Prism v8, with p > 0.05 indicating normal distribution. All error bars represent the mean ± the standard error of the mean (SEM unless stated). All experiments were repeated at least twice or performed with independent samples.
Acknowledgments
We thank the staff at the University of California, Berkeley, Electron Microscope Laboratory for advice and assistance with electron microscopy sample preparation and imaging. We also thank the Diabetes Research Center at the University of California, San Francisco, for a Pilot & Feasibility Award and assistance with the instruments at the Parnassus Flow Cytometry Core, which are supported by grant P30 DK063720 from the National Institutes of Health. Funding: Department of Veterans Affairs Merit grant I01BX003928 (R.L.R); Department of Veterans Affairs Merit grant I01BX003928 supplement (R.L.R); Department of Veterans Affairs Research Career Scientist Award grant IK6BX005692 (R.L.R); National Institutes of Health grant R01HL133575 (R.L.R); and National Institutes of Health grant UG3CA241703 (R.L.R).
Author contributions
Conceptualization, T.A.P. and R.L.R.; methodology, T.A.P., L.B., and R.L.R.; investigation, T.A.P., M.N., N.K.V., and R.L.R.; visualization, T.A.P. and N.K.V.; funding acquisition, R.L.R.; project administration, R.L.R.; supervision, R.L.R.; writing – original draft, T.A.P. and R.L.R.; writing – review & editing, T.A.P. and R.L.R.
Declaration of interests
T.A.P., M.N., N.K.V., L.B., and R.L.R. have filed an invention disclosure related to some aspects of this work with the University of California, San Francisco, and the US Department of Veterans Affairs.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.03.008.
Supplemental information
Data and materials availability
All data presented in the main text or the supplementary materials are available upon request.
References
- 1.Leon B.M., Maddox T.M. Diabetes and cardiovascular disease: epidemiology, biological mechanisms, treatment recommendations and future research. World J. Diabetes. 2015;6:1246–1258. doi: 10.4239/wjd.v6.i13.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirk E.P., Klein S. Pathogenesis and pathophysiology of the cardiometabolic syndrome. J. Clin. Hypertens. 2009;11:761–765. doi: 10.1111/j.1559-4572.2009.00054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donath M.Y., Meier D.T., Boni-Schnetzler M. Inflammation in the pathophysiology and therapy of cardiometabolic disease. Endocr. Rev. 2019;40:1080–1091. doi: 10.1210/er.2019-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lumeng C.N., Deyoung S.M., Bodzin J.L., Saltiel A.R. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes. 2007;56:16–23. doi: 10.2337/db06-1076. [DOI] [PubMed] [Google Scholar]
- 5.Lumeng C.N., DelProposto J.B., Westcott D.J., Saltiel A.R. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57:3239–3246. doi: 10.2337/db08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morinaga H., Mayoral R., Heinrichsdorff J., Osborn O., Franck N., Hah N., Walenta E., Bandyopadhyay G., Pessentheiner A.R., Chi T.J., et al. Characterization of distinct subpopulations of hepatic macrophages in HFD/obese mice. Diabetes. 2015;64:1120–1130. doi: 10.2337/db14-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh D.Y., Morinaga H., Talukdar S., Bae E.J., Olefsky J.M. Increased macrophage migration into adipose tissue in obese mice. Diabetes. 2012;61:346–354. doi: 10.2337/db11-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glass C.K., Witztum J.L. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 9.Back M., Yurdagul A., Jr., Tabas I., Oorni K., Kovanen P.T. Inflammation and its resolution in atherosclerosis: mediators and therapeutic opportunities. Nat. Rev. Cardiol. 2019;16:389–406. doi: 10.1038/s41569-019-0169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman K., Fisher E.A. Insights from pre-clinical and clinical studies on the role of innate inflammation in atherosclerosis regression. Front. Cardiovasc. Med. 2018;5:32. doi: 10.3389/fcvm.2018.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H., Shang Q., Pan Z., Bai Y., Li Z., Zhang H., Zhang Q., Guo C., Zhang L., Wang Q. Exosomes from adipose-derived stem cells attenuate adipose inflammation and obesity through polarizing M2 macrophages and beiging in white adipose tissue. Diabetes. 2018;67:235–247. doi: 10.2337/db17-0356. [DOI] [PubMed] [Google Scholar]
- 12.Song M.Y., Kim S.H., Ryoo G.H., Kim M.K., Cha H.N., Park S.Y., Hwang H.P., Yu H.C., Bae E.J., Park B.H. Adipose sirtuin 6 drives macrophage polarization toward M2 through IL-4 production and maintains systemic insulin sensitivity in mice and humans. Exp. Mol. Med. 2019;51:1–10. doi: 10.1038/s12276-019-0256-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alisi A., Carpino G., Oliveira F.L., Panera N., Nobili V., Gaudio E. The role of tissue macrophage-mediated inflammation on NAFLD pathogenesis and its clinical implications. Mediators Inflamm. 2017;2017:8162421. doi: 10.1155/2017/8162421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J., Sun X., Zhang F.L., Jin H., Yan X.L., Huang S., Guo Z.N., Yang Y. Clinical potential of extracellular vesicles in type 2 diabetes. Front. Endocrinol. 2020;11:596811. doi: 10.3389/fendo.2020.596811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ying W., Riopel M., Bandyopadhyay G., Dong Y., Birmingham A., Seo J.B., Ofrecio J.M., Wollam J., Hernandez-Carretero A., Fu W., et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171:372–384.e312. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 16.Ying W., Gao H., Dos Reis F.C.G., Bandyopadhyay G., Ofrecio J.M., Luo Z., Ji Y., Jin Z., Ly C., Olefsky J.M. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 2021;33:781–790.e785. doi: 10.1016/j.cmet.2020.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouchareychas L., Duong P., Covarrubias S., Alsop E., Phu T.A., Chung A., Gomes M., Wong D., Meechoovet B., Capili A., et al. Macrophage exosomes resolve atherosclerosis by regulating hematopoiesis and inflammation via MicroRNA cargo. Cell Rep. 2020;32:107881. doi: 10.1016/j.celrep.2020.107881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouchareychas L., Duong P., Phu T.A., Alsop E., Meechoovet B., Reiman R., Ng M., Yamamoto R., Nakauchi H., Gasper W.J., et al. High glucose macrophage exosomes enhance atherosclerosis by driving cellular proliferation & hematopoiesis. iScience. 2021;24:102847. doi: 10.1016/j.isci.2021.102847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duong P., Chung A., Bouchareychas L., Raffai R.L. Cushioned-Density Gradient Ultracentrifugation (C-DGUC) improves the isolation efficiency of extracellular vesicles. PLoS One. 2019;14:e0215324. doi: 10.1371/journal.pone.0215324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorrentino T.A., Duong P., Bouchareychas L., Chen M., Chung A., Schaller M.S., Oskowitz A., Raffai R.L., Conte M.S. Circulating exosomes from patients with peripheral artery disease influence vascular cell migration and contain distinct microRNA cargo. JVS Vasc. Sci. 2020;1:28–41. doi: 10.1016/j.jvssci.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srinivasan S., Yeri A., Cheah P.S., Chung A., Danielson K., De Hoff P., Filant J., Laurent C.D., Laurent L.D., Magee R., et al. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell. 2019;177:446–462.e416. doi: 10.1016/j.cell.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galvan-Pena S., O'Neill L.A. Metabolic reprograming in macrophage polarization. Front. Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van den Bossche J., Baardman J., Otto N.A., van der Velden S., Neele A.E., van den Berg S.M., Luque-Martin R., Chen H.J., Boshuizen M.C., Ahmed M., et al. Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Rep. 2016;17:684–696. doi: 10.1016/j.celrep.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Cui H., He Y., Chen S., Zhang D., Yu Y., Fan C. Macrophage-derived miRNA-containing exosomes induce peritendinous fibrosis after tendon injury through the miR-21-5p/smad7 pathway. Mol. Ther. Nucleic Acids. 2019;14:114–130. doi: 10.1016/j.omtn.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouimet M., Ediriweera H.N., Gundra U.M., Sheedy F.J., Ramkhelawon B., Hutchison S.B., Rinehold K., van Solingen C., Fullerton M.D., Cecchini K., et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Invest. 2015;125:4334–4348. doi: 10.1172/JCI81676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouimet M., Ediriweera H., Afonso M.S., Ramkhelawon B., Singaravelu R., Liao X., Bandler R.C., Rahman K., Fisher E.A., Rayner K.J., et al. microRNA-33 regulates macrophage autophagy in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2017;37:1058–1067. doi: 10.1161/ATVBAHA.116.308916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerin I., Clerbaux L.A., Haumont O., Lanthier N., Das A.K., Burant C.F., Leclercq I.A., MacDougald O.A., Bommer G.T. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J. Biol. Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Z., Xie Y., Morrison R.F., Bucher N.L., Farmer S.R. PPARgamma induces the insulin-dependent glucose transporter GLUT4 in the absence of C/EBPalpha during the conversion of 3T3 fibroblasts into adipocytes. J. Clin. Invest. 1998;101:22–32. doi: 10.1172/JCI1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Afonso M.S., Verma N., van Solingen C., Cyr Y., Sharma M., Perie L., Corr E.M., Schlegel M., Shanley L.C., Peled D., et al. MicroRNA-33 inhibits adaptive thermogenesis and adipose tissue beiging. Arterioscler. Thromb. Vasc. Biol. 2021;41:1360–1373. doi: 10.1161/ATVBAHA.120.315798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lhamyani S., Gentile A.-M., Giráldez-Pérez R.M., Feijóo-Cuaresma M., Romero-Zerbo S.Y., Clemente-Postigo M., Zayed H., Olivera W.O., Bermúdez-Silva F.J., Salas J., et al. miR-21 mimic blocks obesity in mice: a novel therapeutic option. Mol. Ther. - Nucleic Acids. 2021;26:401–416. doi: 10.1016/j.omtn.2021.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan Y., Zhao W., Zhao D., Wang C., Yu N., An T., Mo F., Liu J., Miao J., Lv B., et al. Salvianolic acid B improves mitochondrial function in 3T3-L1 adipocytes through a pathway involving PPARgamma coactivator-1alpha (PGC-1alpha) Front. Pharmacol. 2018;9:671. doi: 10.3389/fphar.2018.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cedikova M., Kripnerova M., Dvorakova J., Pitule P., Grundmanova M., Babuska V., Mullerova D., Kuncova J. Mitochondria in white, Brown, and beige adipocytes. Stem Cells Int. 2016;2016:6067349. doi: 10.1155/2016/6067349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo C.Y., Jang J.E., Lee S.E., Koh E.H., Lee K.U. Mitochondrial dysfunction in adipocytes as a primary cause of adipose tissue inflammation. Diabetes Metab. J. 2019;43:247–256. doi: 10.4093/dmj.2018.0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ro S.H., Jang Y., Bae J., Kim I.M., Schaecher C., Shomo Z.D. Autophagy in adipocyte browning: emerging drug target for intervention in obesity. Front. Physiol. 2019;10:22. doi: 10.3389/fphys.2019.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espiritu D.J., Mazzone T. Oxidative stress regulates adipocyte apolipoprotein e and suppresses its expression in obesity. Diabetes. 2008;57:2992–2998. doi: 10.2337/db08-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Z.H., Gu D., Mazzone T. Role of adipocyte-derived apoE in modulating adipocyte size, lipid metabolism, and gene expression in vivo. Am. J. Physiol. Endocrinol. Metab. 2009;296:E1110–E1119. doi: 10.1152/ajpendo.90964.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonacina F., Coe D., Wang G., Longhi M.P., Baragetti A., Moregola A., Garlaschelli K., Uboldi P., Pellegatta F., Grigore L., et al. Myeloid apolipoprotein E controls dendritic cell antigen presentation and T cell activation. Nat. Commun. 2018;9:3083. doi: 10.1038/s41467-018-05322-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li K., Ching D., Luk F.S., Raffai R.L. Apolipoprotein E enhances microRNA-146a in monocytes and macrophages to suppress nuclear factor-kappaB-driven inflammation and atherosclerosis. Circ. Res. 2015;117:e1–e11. doi: 10.1161/CIRCRESAHA.117.305844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaudreault N., Kumar N., Posada J.M., Stephens K.B., Reyes de Mochel N.S., Eberle D., Olivas V.R., Kim R.Y., Harms M.J., Johnson S., et al. ApoE suppresses atherosclerosis by reducing lipid accumulation in circulating monocytes and the expression of inflammatory molecules on monocytes and vascular endothelium. Arterioscler. Thromb. Vasc. Biol. 2012;32:264–272. doi: 10.1161/ATVBAHA.111.238964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fruhbeck G., Catalan V., Rodriguez A., Ramirez B., Becerril S., Salvador J., Colina I., Gomez-Ambrosi J. Adiponectin-leptin ratio is a functional biomarker of adipose tissue inflammation. Nutrients. 2019;11:454. doi: 10.3390/nu11020454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen M.A., Karunakaran D., Geoffrion M., Cheng H.S., Tandoc K., Perisic Matic L., Hedin U., Maegdefessel L., Fish J.E., Rayner K.J. Extracellular vesicles secreted by atherogenic macrophages transfer MicroRNA to inhibit cell migration. Arterioscler. Thromb. Vasc. Biol. 2018;38:49–63. doi: 10.1161/ATVBAHA.117.309795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaiswal A., Reddy S.S., Maurya M., Maurya P., Barthwal M.K. MicroRNA-99a mimics inhibit M1 macrophage phenotype and adipose tissue inflammation by targeting TNFalpha. Cell Mol. Immunol. 2019;16:495–507. doi: 10.1038/s41423-018-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li P., Fan C., Cai Y., Fang S., Zeng Y., Zhang Y., Lin X., Zhang H., Xue Y., Guan M. Transplantation of brown adipose tissue up-regulates miR-99a to ameliorate liver metabolic disorders in diabetic mice by targeting NOX4. Adipocyte. 2020;9:57–67. doi: 10.1080/21623945.2020.1721970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruckerl D., Jenkins S.J., Laqtom N.N., Gallagher I.J., Sutherland T.E., Duncan S., Buck A.H., Allen J.E. Induction of IL-4Ralpha-dependent microRNAs identifies PI3K/Akt signaling as essential for IL-4-driven murine macrophage proliferation in vivo. Blood. 2012;120:2307–2316. doi: 10.1182/blood-2012-02-408252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Li C., Li H., Song Y., Zhao Y., Zhai L., Wang H., Zhong R., Tang H., Zhu D. miR-378 activates the pyruvate-PEP futile cycle and enhances lipolysis to ameliorate obesity in mice. EBioMedicine. 2016;5:93–104. doi: 10.1016/j.ebiom.2016.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan D., Mao C., Quattrochi B., Friedline R.H., Zhu L.J., Jung D.Y., Kim J.K., Lewis B., Wang Y.X. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat. Commun. 2014;5:4725. doi: 10.1038/ncomms5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du J., Zhang P., Luo J., Shen L., Zhang S., Gu H., He J., Wang L., Zhao X., Gan M., et al. Dietary betaine prevents obesity through gut microbiota-drived microRNA-378a family. Gut Microbes. 2021;13:1–19. doi: 10.1080/19490976.2020.1862612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Runtsch M.C., Nelson M.C., Lee S.H., Voth W., Alexander M., Hu R., Wallace J., Petersen C., Panic V., Villanueva C.J., et al. Anti-inflammatory microRNA-146a protects mice from diet-induced metabolic disease. PLOS Genet. 2019;15:e1007970. doi: 10.1371/journal.pgen.1007970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Usuelli V., Ben Nasr M., D'Addio F., Liu K., Vergani A., El Essawy B., Yang J., Assi E., Uehara M., Rossi C., et al. miR-21 antagonism reprograms macrophage metabolism and abrogates chronic allograft vasculopathy. Am. J. Transpl. 2021;21:3280–3295. doi: 10.1111/ajt.16581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheedy F.J. Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front. Immunol. 2015;6:19. doi: 10.3389/fimmu.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Canfran-Duque A., Rotllan N., Zhang X., Fernandez-Fuertes M., Ramirez-Hidalgo C., Araldi E., Daimiel L., Busto R., Fernandez-Hernando C., Suarez Y. Macrophage deficiency of miR-21 promotes apoptosis, plaque necrosis, and vascular inflammation during atherogenesis. EMBO Mol. Med. 2017;9:1244–1262. doi: 10.15252/emmm.201607492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ling H.Y., Hu B., Hu X.B., Zhong J., Feng S.D., Qin L., Liu G., Wen G.B., Liao D.F. MiRNA-21 reverses high glucose and high insulin induced insulin resistance in 3T3-L1 adipocytes through targeting phosphatase and tensin homologue. Exp. Clin. Endocrinol. Diabetes. 2012;120:553–559. doi: 10.1055/s-0032-1311644. [DOI] [PubMed] [Google Scholar]
- 53.Han T., Lv Y., Wang S., Hu T., Hong H., Fu Z. PPARgamma overexpression regulates cholesterol metabolism in human L02 hepatocytes. J. Pharmacol. Sci. 2019;139:1–8. doi: 10.1016/j.jphs.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 54.Klopotek A., Hirche F., Eder K. PPAR gamma ligand troglitazone lowers cholesterol synthesis in HepG2 and Caco-2 cells via a reduced concentration of nuclear SREBP-2. Exp. Biol. Med. 2006;231:1365–1372. doi: 10.1177/153537020623100810. [DOI] [PubMed] [Google Scholar]
- 55.Yoshikawa T., Ide T., Shimano H., Yahagi N., Amemiya-Kudo M., Matsuzaka T., Yatoh S., Kitamine T., Okazaki H., Tamura Y., et al. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. I. PPARs suppress sterol regulatory element binding protein-1c promoter through inhibition of LXR signaling. Mol. Endocrinol. 2003;17:1240–1254. doi: 10.1210/me.2002-0190. [DOI] [PubMed] [Google Scholar]
- 56.Ouimet M., Koster S., Sakowski E., Ramkhelawon B., van Solingen C., Oldebeken S., Karunakaran D., Portal-Celhay C., Sheedy F.J., Ray T.D., et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 2016;17:677–686. doi: 10.1038/ni.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krist B., Florczyk U., Pietraszek-Gremplewicz K., Jozkowicz A., Dulak J. The role of miR-378a in metabolism, angiogenesis, and muscle biology. Int. J. Endocrinol. 2015;2015:281756. doi: 10.1155/2015/281756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerin I., Bommer G.T., McCoin C.S., Sousa K.M., Krishnan V., MacDougald O.A. Roles for miRNA-378/378∗ in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 2010;299:E198–E206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Escary J.L., Choy H.A., Reue K., Schotz M.C. Hormone-sensitive lipase overexpression increases cholesteryl ester hydrolysis in macrophage foam cells. Arterioscler. Thromb. Vasc. Biol. 1998;18:991–998. doi: 10.1161/01.atv.18.6.991. [DOI] [PubMed] [Google Scholar]
- 60.Radovic B., Aflaki E., Kratky D. Adipose triglyceride lipase in immune response, inflammation, and atherosclerosis. Biol. Chem. 2012;393:1005–1011. doi: 10.1515/hsz-2012-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jha P., Claudel T., Baghdasaryan A., Mueller M., Halilbasic E., Das S.K., Lass A., Zimmermann R., Zechner R., Hoefler G., Trauner M. Role of adipose triglyceride lipase (PNPLA2) in protection from hepatic inflammation in mouse models of steatohepatitis and endotoxemia. Hepatology. 2014;59:858–869. doi: 10.1002/hep.26732. [DOI] [PubMed] [Google Scholar]
- 62.Distel E., Barrett T.J., Chung K., Girgis N.M., Parathath S., Essau C.C., Murphy A.J., Moore K.J., Fisher E.A. miR33 inhibition overcomes deleterious effects of diabetes mellitus on atherosclerosis plaque regression in mice. Circ. Res. 2014;115:759–769. doi: 10.1161/CIRCRESAHA.115.304164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yvan-Charvet L., Pagler T., Gautier E.M., Avagyan S., Siry R.L., Han S., Welch C.L., Wang N., Randolph G.J., Snoeck H.W., Tall A.R. ATP-binding cassette transporters and HDL suppress hematopoietic stem cell proliferation. Science. 2010;328:1689–1693. doi: 10.1126/science.1189731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tall A.R., Costet P., Wang N. Regulation and mechanisms of macrophage cholesterol efflux. J. Clin. Invest. 2002;110:899–904. doi: 10.1172/JCI16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hui X., Gu P., Zhang J., Nie T., Pan Y., Wu D., Feng T., Zhong C., Wang Y., Lam K.S., Xu A. Adiponectin enhances cold-induced browning of subcutaneous adipose tissue via promoting M2 macrophage proliferation. Cell Metab. 2015;22:279–290. doi: 10.1016/j.cmet.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 66.Raffai R.L., Weisgraber K.H. Hypomorphic apolipoprotein E mice: a new model of conditional gene repair to examine apolipoprotein E-mediated metabolism. J. Biol. Chem. 2002;277:11064–11068. doi: 10.1074/jbc.M111222200. [DOI] [PubMed] [Google Scholar]
- 67.Thery C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in the main text or the supplementary materials are available upon request.