Abstract
The field of animal science, and especially animal nutrition, relies heavily on modeling to accomplish its day-to-day objectives. New data streams (“big data”) and the exponential increase in computing power have allowed the appearance of “new” modeling methodologies, under the umbrella of artificial intelligence (AI). However, many of these modeling methodologies have been around for decades. According to Gartner, technological innovation follows five distinct phases: technology trigger, peak of inflated expectations, trough of disillusionment, slope of enlightenment, and plateau of productivity. The appearance of AI certainly elicited much hype within agriculture leading to overpromised plug-and-play solutions in a field heavily dependent on custom solutions. The threat of failure can become real when advertising a disruptive innovation as sustainable. This does not mean that we need to abandon AI models. What is most necessary is to demystify the field and place a lesser emphasis on the technology and more on business application. As AI becomes increasingly more powerful and applications start to diverge, new research fields are introduced, and opportunities arise to combine “old” and “new” modeling technologies into hybrids. However, sustainable application is still many years away, and companies and universities alike do well to remain at the forefront. This requires investment in hardware, software, and analytical talent. It also requires a strong connection to the outside world to test, that which does, and does not work in practice and a close view of when the field of agriculture is ready to take its next big steps. Other research fields, such as engineering and automotive, have shown that the application power of AI can be far reaching but only if a realistic view of models as whole is maintained. In this review, we share our view on the current and future limitations of modeling and potential next steps for modelers in the animal sciences. First, we discuss the inherent dependencies and limitations of modeling as a human process. Then, we highlight how models, fueled by AI, can play an enhanced sustainable role in the animal sciences ecosystem. Lastly, we provide recommendations for future animal scientists on how to support themselves, the farmers, and their field, considering the opportunities and challenges the technological innovation brings.
Keywords: data science, education, modeling, precision livestock farming, simulation, smart livestock farming
The hype of artificial intelligence is at an end, revealing to a larger audience the inherent dependencies and limitations of modeling as a human process. Technology is good, but data and humans are essential in enabling a more sustainable role for models in the animal sciences ecosystem.
Introduction
An individual’s perception of what a model is depends on the scientific field that individual is working in. For mathematicians, a model is made up of formulas (Grimm et al., 2020), but for a social scientist, a (mental) model can be made entirely out of text (Johnson-Laird and Byrne, 1993). Where these models find commonality across disciplines is that they both share an intention to discover, describe, predict, and mimic worldly processes. By using mathematics, statistics, and content knowledge, scientists create models to make sense of physical and biological phenomena (Alber et al., 2019). Herein we discuss primarily on the formula-based models (as opposed to text-based or conceptual models), as it is the most applicable to the animal sciences.
The field of animal science, and especially animal nutrition, relies heavily on modeling to accomplish its day-to-day objectives. Models are often used to estimate how animals are projected to grow and determine the animal weight at slaughter, and for how long they should be fed to reach that objective in both group-based (van Milgen et al., 2008; NRC, 2012; NASEM, 2016) or individual-based management strategies (Tedeschi et al., 2004). Models can estimate milk production, fetal development, and the amount of nutrients needed to maximize estimated performance or decrease production costs (Dourmad et al., 2008; Weiss, 2021). Models have been demonstrated as tools to decrease the environmental impact of feeding animals by reducing nutrient excretion excess (Tedeschi et al., 2004; Dumas et al., 2010; Pomar et al., 2019b). Animal production practices can be modeled to estimate outcomes and provide eco-friendly solutions (Cadéro et al., 2020; Menendez and Tedeschi, 2020).
Mathematical modeling (MM), or systems biology, uses equations to represent complex biological phenomena and predict outcomes (France, 2008). But the model itself can be a combination of different classes or model types—mechanistic and empirical models, static and dynamic models, and deterministic and stochastic models (Thornley and France, 1984). To define these terms, a static model predicts a single time point, whereas a dynamic model considers changes over time (typically via a series of integrated differential equations or time step loops). A deterministic model typically considers the “average” animal; however, individual animals can be simulated with great reductions in modeling efficiency; further reduced by the implementation of discrete-time events. Whereas a stochastic model is composed of probabilistic and models variation. Then we have empirical models that describe correlations in the data (e.g., y = mx + b), vs. mechanistic models, which aim to reflect underlying causal pathways. To conceptualize the latter, if a cow is represented by level i, then the organs may be represented by level i − 1, cells as level i − 2, and the herd by level i + 1, and a mechanistic model may predict level i outcomes with a mathematical description of level i − 1 (always a level lower) attributes (Thornley and France, 1984).
In practice, the distinction of a model being a specific “type” is blurry at best as a mechanistic model may contain empirical elements, and a deterministic model can have a strategic variation or stochastic attributes introduced (e.g., inputs), but not throughout. Many of the most advanced models are a hybridization of approaches, for example, the new INRA feeding system for ruminants (Noziere et al., 2018).
Models are helpful in guiding the animal scientist to make informed decisions, but they require adaptation to the nature of the underlying data being captured (Petrie and Watson, 2013; Tedeschi and Fox, 2020). Traditional animal science models are mechanistic by nature, and work in a retrospective manner, meaning they use data from past populations to predict future outcomes of similar populations. In the new era of smart-livestock farming (SLF) or precision livestock farming (PLF; Tedeschi et al., 2021) such models might not be as successful mainly because these models were not developed to handle massive amounts or specific types of data such as images and audio files (Tedeschi, 2019). It is here that the field of artificial intelligence (AI), and more specifically machine learning (ML) and deep learning (DL), provide added benefit. Designed for iterating through a multitude of possible solutions in unstructured datasets, these models update themselves.
If traditional approaches keep modeling in a customary manner without including new technologies, they may reach a certain level of stagnation (Tedeschi, 2019; Ellis et al., 2020). They often represent an average response, in a typical condition, at a certain point in time (Pomar et al., 2003; France, 2008). In addition, they can take quite a long time to develop as they evolve via experimental design, and by the discovery of new biological facts that alter the initial representation (Tedeschi, 2006; Dumas et al., 2008; Pomar and Remus, 2021). Their strength in capturing biology may perhaps also be their biggest dependency.
The “new” availability of ML models (Ellis et al., 2020) has given rise to many types of empirical model categorizations to consider. ML models may be categorized as using supervised vs. unsupervised learning methods, and to work on data that is either continuous or discrete. Supervised learning is more familiar to modelers in animal science—whereby the inputs and outputs are known, and the goal is for the ML algorithm to learn a function that approximates the relationship between input and output data (e.g., linear regression). Unsupervised learning, on the other hand, has no labeled outputs—the goal is to infer the natural structure present within a set of data points (e.g., principal component analysis).
In terms of data type, these models may be developed on continuous data (individual numeric data points) or discrete (categorical data). The latter is perhaps where real innovation has evolved—partnering ML models with nonnumerical data types (including images, audios, videos, etc.). However, audiovisual data are made of pixels, and each pixel contains numerical values. As a result, the data is transformed into a matrix of integers requiring linear algebra to reach the proper form before being included in DL models. A classification summary of the model, including ML and DL models, can be found in Ellis et al. (2020).
The appearance of these “new” modeling methodologies has certainly elicited much hype within agriculture and well beyond. However, many of these modeling methodologies have been around for decades (Fradkov, 2020)—it is the new data streams (“big data”) with an exponential increase in computing power that have allowed new use of these methodologies to solve new problems (Wu et al., 2016).
One way to examine the emergence of big data, and the extensive marketing around the potential of ML models to address a suite of problems, is to take a close look at the Gartner Hype Cycle (Figure 1). The Hype Cycle tracks technological developments across time with five distinct phases: “Technology Trigger,” “Peak of Inflated Expectations,” “Trough of Disillusionment,” “Slope of Enlightenment,” and “Plateau of Productivity” (Linden and Fenn, 2003). There is even a specific cycle for AI (Dedehayir and Steinert, 2016). Each phase of the Hype Cycle has its own role to play. The “Innovation Trigger” is when the public becomes aware of the innovation. Based on the scientific breakthrough or applicability of the innovation, this may result in a hype. Investors and early adopters become very much interested which will lead to a “Peak of Inflated Expectations.” New technology is being trialed and proof-of-concepts emerge. This is when the phrase “do not miss the train” is frequently being used. More specifically, this is also where data science has resided for the past 5 yr. When the excitement subsides, the “Trough of Disillusionment” sets in. Interest wanes as the applicability of the innovation is not as far-reaching as believed, and its potential is much more dependent on the status of the (commercial) world. This stage is necessary to reach the “Slope of Enlightenment.” Here, early adopters begin to realize more fully how sustainable adoption of the technology looks like, subsequently leading to the “Plateau of Productivity.” This last stage is far less than what the first two stages ever promised, but it is here that the innovation has found its niche. Although the Hype Cycle sounds very appealing and intuitive, it is not without its incongruences and inconsistencies (Dedehayir and Steinert, 2016).
Figure 1.
Gartner Hype Cycle showing technological evolvement. There are five distinct phases: “Technology Trigger,” “Peak of Inflated Expectations,” “Trough of Disillusionment,” “Slope of Enlightenment,” and “Plateau of Productivity.” Adapted from Gartner and the Gartner Hype Cycle. https://www.gartner.com/en/research/methodologies/gartner-hype-cycle.
At the time of writing this review, both ML and DL (the latter referring to neural networks with more than one hidden layer; Dargan et al., 2020) have just passed the “Peak of Inflated Expectations” moving straight ahead for the “Trough of Disillusionment” (Chen and Asch, 2017; Lokhorst et al., 2019). According to the Hype Cycle, this is inevitable “healthy” technological development and “what goes up must come down” (Fenn and LeHong, 2011). And after 10 yr of exponential growth in computing power and model development, the hype of what AI could theoretically accomplish is being caught up by the actual situation of where the overall business is and most companies are, technologically. As a result, most data science projects never make it into production meaning that the model never sees the outside world (Weiner, 2020). This does not mean that we need to abandon ML/DL (Chen and Asch, 2017). What is most necessary is to demystify the field (Aho et al., 2020) and place a lesser emphasis on technology and more on business application.
Not every technology makes it to the production phase and technologies can be placed back in the curve (Dedehayir and Steinert, 2016). The problem with the often-cited Hype Cycle is that it showcases “old” technology and rebrands it as something new. For instance, a linear regression model that estimates its parameters via Maximum Likelihood, and not Ordinary Least Squares, is a model that acquires its solution through an iterative process. ML models require iterations (Vartak et al., 2015), but the method itself is much older than the term “machine learning” (Rosenblatt, 1958). As such, many labels in Hype Cycle are simply rebrands or the use of semantics—renaming and reframing of old concepts (Dedehayir and Steinert, 2016).
The aim of this review article is to share one view on the limitations and potential next steps for modeling and data analytics in the animal sciences. We discuss the inherent dependencies and limitations of modeling as a human process. Then, we highlight how models, including AI, can play a more sustainable role in the animal sciences ecosystem. Lastly, we make recommendations toward future animal scientists on how to support themselves, the farmers, and their field considering the opportunities and challenges technological innovation brings.
Models have Limitations
Model transparency is key
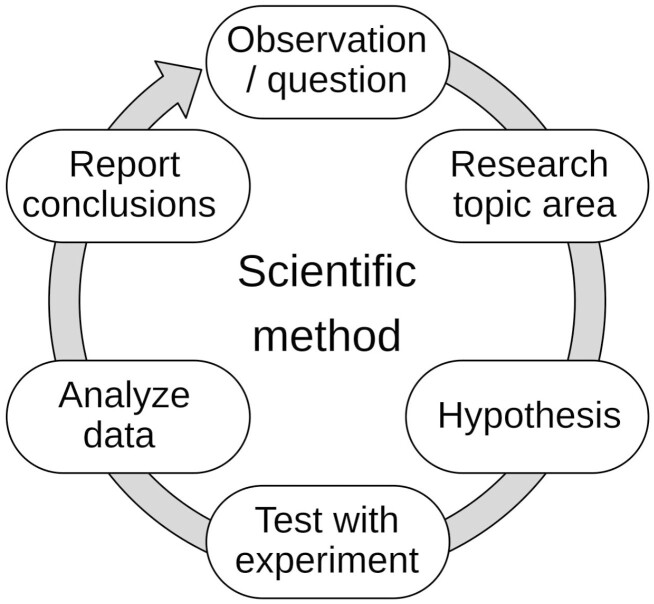
The scientific cycle (or “scientific method” or “research cycle”), describes a circular process in which a research question leads to a hypothesis, which leads to an experiment, which leads to an analysis, and from which then a conclusion can be derived (Figure 2; Bunge, 2001). Since the endeavor of answering research questions often leads to more questions, science represents a perpetual circular process of increasing knowledge.
Figure 2.
The scientific cycle describes a circular process in which a research question leads to a hypothesis, which leads to an experiment, followed by analysis, and from which then a conclusion can be derived.
The process of learning and discovery can be mathematically considered as Bayesian analysis. Contrary to the Frequentist approach of accepting or rejecting a null hypothesis (Mayo and Cox, 2006), Bayesian inference supports uncertainty and the fluidity of evidence. Here, models play a pivotal role in connecting new information (the likelihood) to previous knowledge (prior knowledge; Wagenmakers et al., 2008). The result is a new “understanding of the world” which is labeled “posterior knowledge” (Figure 3). Mathematically, posterior knowledge is literally the product of previous knowledge (the prior) and contemporary data (the likelihood). The Bayes’ theorem postulates that the probability of an outcome is a weighted combination of both old and new information. As a result, new information is placed in context and all probabilities are conditional on the past. Hence, to Bayesian theorists, the parameter estimates of a model, or the model itself, have (has) an expiration date that ends when a new dataset emerges.
Figure 3.
Bayesian analysis of calculating the posterior probability based on prior information and contemporary data (the likelihood).
The Bayesian framework integrates past and current knowledge via models, modelers, and actionable cycles. To allow a model to learn continuously, a tremendous amount of work on both data and model architecture is required using many iterations of the scientific cycle. A recent article showed that when given the exact same dataset, 49 researchers-modelers reached completely different solutions (Schweinsberg et al., 2021). With such a finding, how can anyone ever proclaim to have found the correct answer to a research question (Rosen, 2016)?
Likewise, an experiment is a contemporary product of the science known, the data available, and the capabilities and assumptions of the researcher. Even if you design the optimal study, randomize it perfectly, block for influential variables, and blind yourself throughout the experiment all the way up to the analysis phase, you will still end up (unintentionally) influencing the results of the study via unconscious choices. This does not mean that the experiment has been done incorrectly, or that the findings cannot be trusted. It only means that science is a human process (Hull, 2010). To acknowledge this, the intended experimental design needs to be placed in a protocol that should be (ideally) published and reviewed before conducting the study (Al-Jundi and SAkkA, 2016). Such a protocol should also describe how data are to be handled and for which purpose. We already see this in medicine (Tetzlaff et al., 2012), when researchers set out to do a meta-analysis (Higgins et al., 2019), but also in general when grant proposals are reviewed. After an experiment has been analyzed, deviations from the protocol should be highlighted and reasons provided (Rosen, 2016).
Another example can be found in the use of pre-developed algorithms. The development of the time-series algorithm “prophet” by Facebook has been described in full (Taylor and Letham, 2018), and the same can be said for the random forest (Breiman, 2001) and gradient boosting model family (Friedman, 2002). However, when included in a study to predict a particular outcome, the model is often just referenced as being applied. This is not enough detail or transparency, though, as the same model can be used in a multitude of different ways. Instead, descriptions should be provided explaining the choices modelers made when feeding a prophet or gradient boosting model, and how hyperparameters were tweaked to reach their results (Martinez-Moyano, 2012). A detailed and clear description of the model, its mathematical settings (e.g., hyperparameters), as well as the space, time, and sample size information regarding the analysis should be made available for verification of the reproducibility of an ML model (Eddy et al., 2012).
To share accurately and completely what was done, and to detect biases, nuances, and specifics, modelers will also need to become more comfortable with sharing data and code toward an open-science provision (Muñoz-Tamayo et al., 2022). Sharing via publications alone makes it near impossible to see how the final model came to be and accurately replicate it as assumptions and choices made are rarely described. These modeling decisions introduce bias and variability not reflected by standard errors.
To address error and bias, a technique called specification curve analysis (SCA) has been proposed (Simonsohn et al., 2020). The aim of the SCA procedure is to visualize the entire model response surface. This method analyzes and shows the changes in descriptive and inferential statistics when looping through all reasonable specifications (Cosme, 2020). For example, if the user has included five variables in your model, the SCA will run and visualize all possible model combinations, main effects, and up to five-factor interactions. In addition, you can specify how you dealt with data, such as missing, outliers, and the need to transform. As a result, the matrix of possibilities can become quite large, quite fast, and SCA will show you how results differ across combinations of choices. In a sense, an SCA is an extreme sensitivity analysis. As of now, the technique is still in development, and not widely applied, though it may be anticipated to mature rapidly and service for more sophisticated models.
Model validation needs to happen externally and continuously
A mathematical model cannot be truly validated but only allowed to demonstrate that it works within acceptable limits in a determined situation (Tedeschi, 2006). Although contradictory by nature, we define validation as a demonstration that a model has acceptable predictive accuracy for the end it was developed (Thornley and France, 2007). Models are often “validated” by testing them internally in datasets (Morota et al., 2018), and this process of internal validation can yield decent estimates when the data is sufficient and of good quality. To safeguard from overfitting, the datasets are split, and the model is repeatedly tested and adjusted via cross-validation. The true litmus test of any model is the use of an external “validation” set obtained via in vivo experiments (Remus et al., 2020a; Menendez et al., 2022).
Model validation, or calibration, is often conducted once but can be extended toward an internal cycle of repeated validation called cross-validation. Singular or repeated, internal validation happens mostly via statistical measures. A “validated” model is then shown to a customer and/or published but does not necessarily encompass their visions and/or wishes. At least not from the start.
This might not be the most appropriate way to validate models, and especially not for a real-time nutrition model. Here, the important question is: can this model correctly predict nutrient requirements in a timely manner for a real animal or animal group? It is important to keep in mind that, with the increased use of sensors, data models need to be able to quickly process data that varies within 1 d (and one animal). Data might also be incomplete, for instance, because of mechanical or health problems that resulted in missed information. Additionally, some of the assumptions made by the modeler, based on average responses presented in the literature, might not reflect the individual response observed in vivo (Daniel et al., 2017; Remus et al., 2020b, 2021). This is especially valid if the model in question has a mechanistic component.
Modeling animals is not easy as the animals themselves, and their requirements, change over time and among themselves (Hauschild et al., 2010; Muñoz-Tamayo et al., 2018; Remus et al., 2020c). This means that real-time precision feeding and nutrition models need to be calibrated for both individual and group predictions, ensuring enough flexibility to incorporate these variations. It also means that validation needs to be continuously monitored, together with potential end-users. In the data science field, continuous validation, monitoring, and updating of models are placed under the umbrella term ML operations (Heck et al., 2021).
Considering the rapid changes described above, it also means that conducting experiments on a regular basis becomes almost impossible to achieve as they are time-consuming and costly. It is not unfair to believe that they might even be outdated before they begin.
The need for a data-centric view
PLF involves sensors that capture large amounts of real-time information at the farm, herd, or animal level, which are later processed to inform the system (Wathes et al., 2008; Pomar et al., 2019b). The successful use of precision nutrition requires measuring individual key features (e.g., body weight, feed intake, backfat thickness, litter size, etc.) dependent on species (Pomar and Remus, 2019a, 2021; Gaillard et al., 2020). Using measuring devices (e.g., automated scales, automatic feeders, and cameras) decreases the workload and increases the efficiency with which data are collected and stored. In addition, it improves the model prediction for nutrient requirements, thereby decreasing economic and environmental costs linked to animal production (Andretta et al., 2016; Pomar et al., 2019b; Menendez et al., 2022). Although the main success is linked to the data processing (model and decision support tool), such sensor technologies allow a farmer to obtain data that seemed unattainable a few years ago.
With the greater popularization of sensors in the last decade (Banhazi et al., 2012; Halachmi et al., 2019; Tedeschi et al., 2021), data became more abundant and new types of data were introduced. It also aided in getting away from silo-data sets. This highlighted the importance of calculation methods, the second component required within PLF to implement this technology in the field successfully (Pomar et al., 2019b). Sensors will record different types of data, including images and audio files that cannot be integrated into MM, as they were not necessarily designed to handle this type of data and would require additional development to predict by the second/minute/hour. This makes real-time analysis infeasible by traditional MM (Parsons et al., 2007). In addition, MM are criticized for their complexity and, sometimes, impractical inputs required to simulate actual conditions (Wathes et al., 2008; Ellis et al., 2019; Pomar and Remus, 2019a), or even for being slow (Parsons et al., 2007). By contrast, a data-driven model predicts based on correlation and therefore encloses from linear and nonlinear regression to random forests and neural networks (Ellis et al., 2020), which solves the complexity and speed problem.
Still, the essential function of a MM is to provide understanding and guidance on the decision-making process. Data-driven (DD) models provide the “fish, but do not teach how to fish.” In other words, this type of model can yield good predictions, but rarely new knowledge or understanding of a process (Ribeiro et al., 2016). Whereas MM models allow the study of individual nutrients, metabolites, and their interactions, this advantage is lost in the simplicity of the DD models (Pomar et al., 2019b). Despite their differences, MM and DD models need to be calibrated to gather individual animal growth potential to operate in PLF systems (Pomar et al., 2015). In this case, a difficulty associated with MM is finding the correct reference population, highlighting the advantage of using DD flexible recursive technologies to obtain a range of possible parameters (Wathes et al., 2008; Pomar et al., 2015, 2019b).
A common mistake still being made is saying that models just need more data to work with. What models need is not necessarily more data, but the “right type” of data containing enough variation and granularity (Cirillo et al., 2021). It is not easy to predict what type of data a model needs to “become better,” and the search for useful data may very well extend beyond several scientific research cycles. In fact, data collection may by itself profit from several iterations of a dedicated scientific cycle in which we hypothesize and test which types have merit, look for combinations of data, and see if this makes a difference (Cortes-Ciriano et al., 2015).
However, if data is truly the new gold, the field of modeling should be made more familiar with the subject it wishes to mine. In fact, the often-used “data = gold” comparison is limited by itself, as data is made up of many sources and types. Hence, we are not just mining gold (numerical values), but also silver (images), diamonds (audio), emeralds (text), sapphires (videos), and titanium (management; human-dimensions). These seldom coexist, and all require a different approach and strategy (Greener et al., 2022).
For instance, the increase in image data has led to the development and use of DL models, which are extremely sophisticated neural networks sometimes containing up to trillions of connections. From a model-centric perspective, the increased usability of image data depends on the innovation of models. However, a smarter way of developing a DL model is to work on the images itself. By approaching the problem via a data-centric approach, images can be cropped, twisted, augmented, aggregated (pixel Dots Per Inch), and labeled (Hinterstoisser et al., 2018). Especially, the latter development has seen a sharp increase in the effectiveness of DL models. Simply a human telling a model where to look and what object to look for. Hence, we believe that an increasing effort in fundamental data research (a data-centric paradigm) will also help bring the fields of modeling and AI to a new level in animal sciences. This is already happening in other fields, such as automotive (Greener et al., 2022).
A data-centric focus will help us remove models from a pedestal and place them in a more symbiotic relationship with humans called Human-Centered AI (Shneiderman, 2022). Very much still in the “Innovation Trigger” phase, Human-Centered AI is a natural evolvement from the heavy AI focus of the past 10 yr. Instead of human labor being replaced by AI, this new field is all about integration, synchronization, and even symbiosis. By letting humans help models, models are better able to fit the need of a human. However, to enable these developments, data is needed on human–AI interaction. For instance, if a model gives advice, does the farmer accept that advice, discard it, or act in between? What is the underlying reason for the action of the farmer? To assess model acceptance, specific data needs to be collected, but to even reach the advice phase both modelers and models need data as well (Tedeschi and Boston, 2011).
Hence, perhaps the most fundamental shift in the scientific cycle would be achieved by sharing data. Model results are openly published, but data is rarely if ever shared (Tenopir et al., 2011). Although uncomfortable, sharing data and code are required for science to prosper (Fecher et al., 2015). By restricting knowledge to be obtained mostly via scientific articles, it remains unclear how “good” the data is, and the extraction itself can be tedious if not impossible. In fact, this process can introduce new forms of bias, or at least introduce a new set of assumptions. For instance, a meta-analysis containing individual (animal) data is preferred over a meta-analysis containing only aggregated summaries because it allows for less bias (Stewart and Parmar, 1993), and much greater flexibility and transparency in the form of meta-regression (Thompson and Higgins, 2002).
For a research area to move on faster, collected data need to be “open and free.” In addition, societal pressure is asking for less experimentation on animals or at the very least to include the least number of animals possible within a single experiment (Ibrahim, 2006; Daneshian et al., 2011; DeMello, 2021). This means that experiments will become fragile, and their findings highly dependent on the results of other experiments. It also means that the sharing of data will become more and more expected, which we are seeing now during Covid-19 (Cosgriff et al., 2020).
The need to protect sensitive information, however, cannot be completely discarded. Companies and individuals need to acquire a taste for sharing, and the first steps should be beneficial to all. Here, two technological advances will most likely play a key role in the years to come: 1) federated learning and 2) synthetic data. Federated learning (known as collaborative learning) is a technique in which an algorithm is trained across different data sources (Li et al., 2020). Hence, the model is trained, but the data is not exchanged which means that a modeler can access models’ results coming from different data sources but not the data itself. Multiple ways of training a federated model exist, but perhaps the most straightforward way of visualizing it is by imagining 10 different data sources being used to train 10 different models. In the end, the models are pooled. The technique sounds easier than it is, and many caveats exist, but it is an exciting field in which data privacy and security remain safeguarded.
Another technological advance is the creation of synthetic data (Reiter, 2002; Surendra and Mohan, 2017). This kind of data is created via algorithms that have stored the key relationships of an actual existing dataset. Many data generation methods exist, and their possible application extends far beyond data privacy and security. By using the variance-covariance matrix of a real dataset, synthetic data can be created and further augmented to help trained models deal with new and unforeseen information (Varga and Bunke, 2003). Experiments can be created (Bolón-Canedo et al., 2013), and it is not so hard to imagine how multiple synthetic datasets can be used to form extensive digital twins (San, 2021). In fact, synthetic data can be created in parallel with real-time “real” data to conduct a synthetic stress test.
Models cannot prove causality
Pearl describes the ladder of causality, which is made up of three rungs: association (seeing), intervention (doing), and causality (imagining; Pearl and Mackenzie, 2018). Most of us know association via its synonym “correlation” and statistical models are true masters of the craft. It means that as A shifts, B must shift as well, but they do not have to shift because of each other.
The second rung, “intervention,” is more difficult to achieve and requires the type of experimental design most researchers are familiar with (imposing A and watching if B happens). Here, a change in A must lead to a change in B, whereas a change in B cannot lead to a change in A. In fact, you want B to only change because of A. For an intervention to actually intervene, its action must lead to a singular result, and most statistical models are trained to pick up the inference if it occurs. However, they do require help in the form of a good experimental design, and the field of “statistical significance” is not without its critics and rightfully so (French, 1988; Ioannidis, 2005).
The most fascinating rung is that of causality itself, or “imagining.” Pearl describes imagining as the ability to conjure up a parallel universe in your head and play through fictitious scenarios. Whereas an intervention is more difficult to prove than an association, it goes without saying that a parallel universe can never be “proven.” Models, however, are regularly abused to do just that. For instance, imagine a model predicting a suboptimal growth curve in a batch of animals, leading to a dietary intervention of the farmer. In the end, the curve improved yet never reached its optimal scenario. So, in terms of data, we now have an observed growth curve, a predicted growth curve, an intervention, and the difference between predicted and observed.
Could we proclaim that the intervention has led to the improvement in growth? We know an intervention was conducted prior to the improvement (rung two), and we have a parallel universe provided by the model showing how the curve would have most likely transpired without the intervention (rung three). However, what was predicted to transpire before the intervention happened, deviated noticeably from what happened. In addition, this was not an experimental setting in which all known and unknown factors where kept constant, and model predictions are inherently uncertain. Furthermore, there are no replications conducted, and perhaps not even possible anymore due to a changing environment.
The key questions to answer now are: 1) what lead to the improvement of the growth curve and 2) why did we see this specific curve? Was it the action of the farmer, which deviated from the advice given? Was it the genetic make-up of the animal? Its surroundings? Why did one animal adapt better than another? And, most importantly, what would have happened if the farmer had done nothing? This is when the scientific cycle continues, and more research is needed.
A problem here is that models should be created to solve a problem, not a system. Describing a problem in such a way that it can be adequately modeled is already hard enough, let alone an entire system. Hence, perhaps the most dangerous thing a modeler can do is proclaim that a model can do more than it can. Instead, they should focus on model limitations as models in themselves represent a hypothesis of how as system is regulated—if it is proved wrong, we learned more. We all have models that fail, reveal holes in our knowledge, we do experimental work, and redevelop that portion of the model. Accepting this will help us to better accept models for what they really are, which is a mirror of our contemporary knowledge (Hobbs and Hooten, 2015).
Models need adoption to have an impact
Despite the challenges of model development and implementation, models are a necessary component in the PLF development. This system aims to obtain and process data continuously allowing the automated management and feeding of animals (Pomar et al., 2019b). Meaning that the farmer can obtain detailed information on animal’s health and growth performance in real-time through the system control. Such information can allow for early disease detection based on changes on feed intake (Colin et al., 2021; Thomas et al., 2021), or determining optimal slaughter weight. Data on feed intake and body weight allow to make decisions in terms of right amount and right feed composition to be provided to an individual or to a group of animals (Pomar et al., 2019b). Depending on the production objectives, the controller can be programmed to maximize growth rate, to minimize feed cost, to minimize nutrient excretion, or to meet another objective (O’Grady and O’Hare, 2017; Pomar et al., 2019b).
Once a model is found adequate for a PLF application it must be implemented by the user group. Typically, sensor products come with turn-key solutions, including models. The turn-key package implies that there is confidence in the model’s ability to work with the sensor and provide the necessary information. The acceptance of a model is a psychological process (Briggs, 2016). To some, the model needs to be accurate and precise in its predictions. To others, the model needs to be (fully) explainable to justify model output (e.g., enteric emission footprint) or to gather insights (learning-based outcomes). For the majority, their needs are somewhere in between. Whereas “prediction” is straightforward in its description (i.e., to foretell what has yet to come), the definition of “explanation” is more contemporary, and they share overlap (Briggs, 2016). For instance, a model that can predict the milk yield of a cow within acceptable limits but does so by using the texture of the skin, is deemed predictable but not explainable; “the mathematical/statistically relationship exists but not the understanding of biological process that relate to milk yield and skin texture.” To some farmers, a well-predicting unexplainable model might not get accepted. To others, it does not really matter what is inside the model if important technical or financial key performance indicators are met. Here, end-users do not question the ability of the model to explain but rather gain confidence through repeatability of gains in performance. However, open-source models and more rigorous documentation is shifting scientific model development, transparency, and how end results are interpreted.
The field of data science has led to an explosion of models in which a common distinction is made between descriptive, predictive, and prescriptive models (Roy et al., 2022). Descriptive models describe past and current processes, predictive models aim to predict or forecast the future, and prescriptive models provide a recommendation that they authoritatively bring forward (Frazzetto et al., 2019). One could say that the first type is made for an explanation, whereas the other two are future-oriented via prediction. If a farmer would use the milk yield model previously described, and deploy it as prescriptive, part of the farm would be run based on the skin texture of the cows. The choice of model depends on the level of understanding of both the modeler and the potential user, and the scenario of deployment. Often, we see variables included that a specialist would never include, because biologically it makes no sense. When complex and nonmeasurable variables are included, or spawned inside the model via dimension reduction, the model loses its ability to explain. In addition, some of these variables are also impossible to measure in real-life, as they are “latent” or invisible.
Most modelers will go for a model that has both an acceptable level of prediction and explanation. However, most algorithms are not designed to do both, and with the rise of “data science” the focus has clearly shifted toward prediction (Lundberg and Lee, 2017). By using large quantities of data, ML/DL models are used to detect unknown cross-correlations with predictive power (Dobson, 2013). As a result, these models might have no biological precedence. Being able to explain why a highly accurate prediction has become so in the first place is a daunting and often overlooked task by novice modelers.
AI: From Hype Toward Sustainable Adoption
Synonyms for “hype” include “advertising,” “plugging,” and “promotion.” Although hype may aid in getting a technological innovation started, it adds little to establishing a sustainable environment in which it can be nurtured, molded, and further developed (Christensen, 2013). Perhaps AI would prosper more if the focus was not on selling it as an end-solution, but rather as a building block amongst other, less technological, building blocks. Instead of seeing it as an automatic solution toward previously unsolvable problems, we could perhaps benefit more if we would approach it as an enabler toward possibilities such as environmental sustainability via multiscaling (Favino et al., 2016; Khamis et al., 2019), synthetic data, and federated learning. Although enabling processes should act like a conductor, they require by themselves a lot of hard work, time, and money.
The process of accepting and adopting technological development is largely dependent on humans. On the far left of the bell-shaped distribution called the “Rogers” Adoption Curve” (Rogers, 2003) we have “Innovators” who like about any technological innovation they can find. Next to the “Innovators” are the “Early Adopters.” Then there are the “Early Majority,” the “Late Majority,” and finally, the “Laggards.” Although it would be great to only have a population of “Innovators,” technology also needs to be adopted by the greater target audience population if it is to move from hype to normalization. Adoption does not happen overnight nor is it guaranteed.
The hype of collecting large amounts of data via sensors and using AI may overwhelm livestock producers if the process is not directed. In fact, they might not even want these changes to happen, or at the very least do not see a benefit (Kelly et al., 2015). For the animal scientist of the future, these are feelings and cognitions that need to be acknowledge and explored. Just using expensive equipment to collect a lot of data paired with sophisticated models for the sake of technological progress is not a recipe for sustainable adoption.
Dealing with variation
Feeding animals is the biggest environmental and economic cost for nonruminants (Andretta et al., 2021b). One of the main problems linked to nutritional programs is their gap in understanding of animal variability. As animals use and retain nutrients differently, it affects the environmental and economic performance of a system (Pomar and Remus, 2019b; Pomar et al., 2021).
Traditional models have for years tried to explain the nutrient utilization of an average individual. For instance, conventional MM estimates the average population responses using historical population information. Many models have a matrix of variance (e.g., stochasticity) aiming to simulate different animals of the population (e.g., 90th percentile and 50th percentile). What they do not consider is that a 90th percentile animal (e.g., greater feed efficiency) might have a greater performance because it presents differences in the regulation of biological processes compared to the 50th percentile animal (e.g., median feed efficiency; Vigors et al., 2019; Hu et al., 2022). Instead of explaining variability, these models assume that differences among animals are random. Therefore, important limitations of these models are the assumption that all the individuals of the population have the same response to a given nutrient provision and that they have not been developed for real-time estimations using up-to-date available information (Pomar and Remus, 2019a, 2021; Pomar et al., 2019a).
With the increasing information on individual performance, the rather large variability in metabolic response to nutrient intake has highlighted the weakness of MM applied for PLF (Pomar and Remus, 2019a, 2021). Real-time nutrition models have focused on decreasing nutrient excess, to decrease environmental and production costs (Gauthier et al., 2019; Pomar and Remus, 2019b; Gaillard et al., 2020). This type of approach makes the technology adoption more attractive to producers once they can see the return on investment.
With the collection of individual growth, and drinking and feeding patterns, milk yield and composition or individual behavior can be recorded throughout the day (Pomar et al., 2015, 2019b; Ellis et al., 2020). Therefore, MM must be developed specifically for PLF and operate in real-time at the individual or small group level, considering the between- and within-animal variation. Growth patterns, nutrient utilization, and behavior vary among animals and herds. There are opportunities to combine DD AI with knowledge-driven MM to control more complex PLF components. AI thrives in large complex datasets, where establishing connections can be otherwise difficult due to data complexity, volume, and flexibility to process real-time data from individuals (Tedeschi et al., 2021; Thomas et al., 2021). In contrast, knowledge-driven MM can simplify complex biological systems based on well-established concepts and information (Ellis et al., 2020). In both cases, PLF models must be flexible enough to consider changes over time for the same animal or herd and among animals and herds, acknowledging the method limitation while using its strength, handling big and complex data. Therefore, real-time models are essential in this type of system.
For more complex responses, such as the amount of nutrients needed for fetal development, maintenance, and/or growth gray-box models (Roush et al., 2006), there seems to be a better chance of success when integrating empirical and DD models (Hauschild et al., 2012; Tedeschi, 2019; Gaillard et al., 2020).
Gray-box models are the new black
Mechanistic models are often labeled “white-box” models because they are fully transparent and manually built from the bottom-up based on biological principles. This is contrary to “black-box” models of which the input and outputs are known, but the “inside function” of the model is not (Estrada-Flores et al., 2006; Pomar et al., 2015). Despite some authors trying to give biological meaning to statistical model coefficients (Remus et al., 2014; Sauvant and Nozière, 2016), the true relationship remains purely mathematical. Gray-box models (Figure 4) are integrations of black- and white-box models (Estrada-Flores et al., 2006; Roush et al., 2006; Pomar et al., 2015) and are known as hybrid models (Estrada-Flores et al., 2006; Lo-Thong et al., 2020).
Figure 4.
Example of a gray-box or hybrid model use within precision (digital) livestock farming systems, adapted from Remus et al. (2021).
Although the colors have intuitive appeal, they are not the best descriptors considering that all models are derived from “white-box” mathematical algorithms. In fact, black boxes are the direct result of combining known algorithms with extreme computing power allowing billions of neural network layers to iterate toward a solution via trillions of tries. Because of these developments, the 2020 Gartner Hype Cycle for Emerging Technologies has started to include eXplainable AI (XAI; Pereira, 2020). For now, the field of XAI is mostly focused on visualizing the inner workings of models (Samek et al., 2019). For instance, by showing how the different parts of a DL model are built up across iterations, the user can get an immediate “feeling” of how the model achieved its result. We expect more algorithmic explanatory support in future developments of XAI (Samek et al., 2019).
The need to explain the insights of a model has brought increased attention to gray-box models (Tedeschi, 2019; Ellis et al., 2020). The term hybrid intelligent mechanist model (HIMM) is proposed for models that combine AI with mechanistic models (Tedeschi et al., 2021). The term gray-box also describes a model aiming to understand and explain ML models (Pintelas et al., 2020). Although these models have received increased attention over the last few years, they are not new. In fact, the practice of combining models such as neural networks (Roush et al., 2006), a revised simplex algorithm (Parsons et al., 2007), weighted time-series (Hauschild et al., 2012) to mechanistic models have been proposed in the nonruminant field for almost two decades.
As previously discussed, modeling mimics the scientific cycle, and likely due to the need for farm modernization, availability of new measuring devices, and a new generation thirsty for technology, the new tendency is HIMM which will likely focus on combining sophisticated machine and DL algorithms to obtain data needed for MM.
Although the use of such models seems tangible for academics, their adoption and popularization have been limited due to the availability of measuring and controlling systems allowing their adoption (Pomar and Remus, 2021). Surprising enough, factor limiting the adoption of PLF and SLF itself was the availability of useful decision support tools (Pomar et al., 2019b; Tedeschi et al., 2021). One may assume that data scientists, animal scientists, and engineers working together are what it will take to make PLF a reality in the field.
Combining white- and black-box approaches allows taking advantage of their main strengths. The DD models can be used to classify groups, predict outputs where the understanding of that prediction is not necessary, and reduce dimensional problems (Ellis et al., 2020). Thus, the black-box approach (e.g., artificial neural network, random forest, and time-series) is helpful to estimate the unknown parameters that vary among individuals based on real-time measured variables and estimates obtained from previously developed algorithms (Pomar et al., 2019b). Mechanistic models should be used to provide an understanding of a prediction, such as determining nutrient requirements, estimating the potential environmental and growth impact of nutritional additives, the impact of feed composition, and feedstuff changes on growth composition, among others (Ellis et al., 2020; Pomar and Remus, 2021). Therefore, the gray-box approach allows combining the advantages of these two approaches using each method based on its strength: data-driven to obtain short-term predictions of an input variable and MM can be used as a knowledge-driven prediction based on the input offered by the DD.
The modeling enabled precision nutrition approach within PLF offers possibilities for decreasing nitrogen, phosphorus, greenhouse gas emissions, and production costs (Cadéro et al., 2020; Andretta et al., 2021a). Still, the advance of this approach depends on a better understanding of biological animal variability and the ability to identify this difference among animals (Pomar and Remus, 2019b, 2021; Remus et al., 2020b). Here, applying DL models on top of vision technology (Fernandes et al., 2019; Samperio et al., 2021) and biosensors (Neethirajan, 2017), might help contribute greatly to this development. For instance, the use of simple surveillance cameras allows for counting animals (smaRt Counting, Conception Ro-main inc, Saint-Lambert-de-Lauzon, Québec, Canada), or detecting sow’s ovulation (Labrecque and Rivest, 2018). Cameras could potentially be used to estimate body composition through DL algorithms (Fernandes et al., 2019). Sound recording and video images are being used to detect behavior anomalies (Wurtz et al., 2019), and replace the use of eartags with automatic recognition (Tassinari et al., 2021). From the need to detect and intervene due to an undesired behavior or for early disease detection, DL algorithms have been developed (Cowton et al., 2018; Liu et al., 2020). What all these technologies have in common is the need for certain information to be used by a model aiming to identify, feed, or treat an individual animal with tailored measures. Hence the need to consider which model or combination of models (e.g., HIMM) are required to achieve a specific purpose.
Virtual experimentation
Models need time to learn new situations. To allow sustainable adoption, models should be allowed to forecast situations and incidences that have never occurred before, and multiple ways exist to teach models how to train for such situations (Peng et al., 2021). For one, they could be allowed to run what-if scenario’s outside of regular conventional bounds using enhanced theory and causal hypotheses about the known farming system (Rotz, 2018). Another more advanced way would be to simulate noisy data via Generative Adversarial Networks and create huge volumes of “outside the known parameter” data (Creswell et al., 2018). As a result, models can be trained to deal with potentially unknown future events and provide critical insight in their behavior.
Computer simulation has existed for some time, but the field is becoming increasingly more powerful to train models on what-has-never-occurred scenarios and on helping models train other models (i.e., meta modeling; Sutton and Barto, 2018; Tedeschi and Fox, 2020). Hence, model training is shifting, and the trend now is not solely to rely on collecting data from the real world, but to venture into the “metaverse” or “virtual world” and create and develop various “what-if scenarios” (Owens et al., 2011). Thereby converting the virtual digital world into trillions of terabytes of data for a meager cost.
A successful model deployment, here, would most likely relate to a model being able to make choices on its own based on the goals and boundaries provided. For instance, these prescriptive models could be deployed to oversee the development of optimal growth curves and adjust the feed of the animal once deviations are predicted (Risbey et al., 1999; Pomar and Remus, 2021; Zuidhof, 2021). This would require real-time tracking of feed and growth curves, among many other parameters. By being allowed to make direct real-time adjustments, the model is also able to assess its own impact and retrain itself by estimating the impact of its “choices.”
The evaluation process can be repeated many times over, both internally when building the model (e.g., cross-validation) as well as externally (via Bayesian Inference and a test set). The internal validation mimics faster and smaller iterations of the Scientific Cycle, as all model aspects undergo a “stress test” (Browne, 2000). During such a test, the response of the model is tested under extreme conditions to ensure that it is obeying the laws of physics. The benefit of this exercise is that the modeler will acquire additional information on the strengths and weaknesses of the model, its dependency on the training data, and thus its likely ability to deal with new and unforeseen scenarios. Of course, not all questions may necessarily be answered in a single build, and so each model needs to be rebuilt several times using new tests sets; an iterative process (e.g., experiments; Browne, 2000; Sterman, 2000).
The most exhaustive stress test for a model may be commercial deployment. It is here that the model is asked to mimic the real-life scenario of the farm and show that it can simulate an actual environment. If the same type of data on which the model was built also flows within the farm, the model should be able to create a digital copy within a few iterations. However, this is seldom the case, and most farms only have in part the data a model needs to provide acceptable performance. As a result, models (regardless of type) can severely underperform compared with training scenarios. Modelers should expect this to happen frequently, and internally stress their models to different levels of digital maturity. This will help them offer solutions more quickly or communicate to the customer what is best attainable given the limitations of the farm, creating clear expectations for data collection requirements.
These situations also lend themselves to opportunities if the modeler and the potential user are willing. Despite not having shown an acceptable performance, the model should still be allowed to continue learning from the new situation and to see how far it can go with the limited data at hand. Comparing model performance across different scenarios can lead to fundamental insights in the necessities of the model. In addition, it can show the consumer the importance of collecting more (granular) data.
Since a model can theoretically act as a digital twin (Smith, 2018), it should be allowed to work through hypothetical scenarios. These scenarios can be univariate, changing one parameter at a time, or multivariate in which multiple interacting variables are altered. To validate different model scenarios the changes must be conducted according to a plan, and the results must be benchmarked to the predicted results (Koketsu et al., 2010). This is an excellent result of external evaluation. The downside of this approach however is that it can take quite some time before benchmark results are available. If something goes amiss during that period, or if influential changes are made, the model needs to be rerun to take these changes into account. Predictions up to the rerun can be verified, but the use of the model is quite ad hoc or has at the very least considerable lag time in response. This is because models are often run once, prior to a new batch, and then verified after the batch has ended. There is often no intermittent verification nor auto-adjustment when the variables that flow in the model show unexpected patterns that could heavily impact the (predicted) results. Consequently, there is a need for real-time models to have built in warning systems to identify extreme outliers which indicate a true anomaly from the animal or a sensor error.
Next Steps for Animal Scientists
Content experts are often paired with statisticians or data scientists in a data-heavy project. Despite the intuitive appeal of joining complementary minds, pairing also requires solid communication and project management. The mind of one researcher needs to be “transferred” to the other, and vice versa, and commercially it means having to pay for two salaries instead of one. There is little evidence that pairing will lead to better results. Scientists are trained to complete the scientific cycle by themselves, and a big part of this cycle is the design, conduct, and analysis of experiments. Although curricula for “design of experiments” differs from that of “systems modeling,” “data science,” or “biostatistics” there is an unmistakable overlap.
Despite the explosion of easy-to-use software applications, knowing how to code in multiple programming languages is a fundamental skill that requires honing and shaping and continual practice. In a sense, the field of computer science will merge with every other research field to date, and primary schools (K-12), universities and companies need to support their students and workforce in achieving desirable levels of competency; programming is the new “second language” of the digital world. As a result, some universities have started to offer minor tracks in data science and modeling that accompany the core base of the animal scientist. It is good to focus but even better to integrate.
Future scientists should embrace being asked to handle bigger, more granular data like audio or image files. Not only will these experiences add additional capabilities to the resume of the researcher, but it will also mean acquiring a more holistic and agnostic view of models.
Learn how to program and get dirty
Admittedly, animal scientists do not always wish to become computer scientists. Driven by time constraints and a lack of mathematical knowledge, most researchers will turn to fully functioning and documented libraries instead of building algorithms from scratch(Menendez et al., 2022). Although such behavior is understandable, it is also concerning as the fields of modeling and statistics are heavily dependent on assumptions (mathematical and statistical principles; Von Bertalanffy, 2010). There is, of course, merit in not having to recreate past work, but it remains the responsibility of the researcher to know what is and what is not possible and what kind of assumptions underlie each model. Besides scientific principles, statistical principles need to be adopted as well, and every model deployed by the researcher needs to be understood and documented at the level where they can be explained and shared with others.
A great animal scientist does not need to be a great mathematician. For them, the integration of modeling and statistics needs to be as close as possible to their field of interest. For instance, an animal scientist that wants to specialize on how piglets develop needs to understand which data types are collected during that phase, and how to best apply the appropriate type models on them, interpret results (analysis), and generate inferences (synthesis). By connecting the techniques as close as possible to the interest of the scientist, any additional requested effort fits the greater good.
Regardless of your level of interest in models or data, you will need to learn how to program. Although there is certainly a role to be played in primary or high school, it is not until university that most animal scientists get to explore the scientific cycle. This is also when an understanding of data and modeling becomes more important. To the interested student, there is already a lot of material available in the form of online courses which exist to teach almost every topic related to basic science, modeling, and statistics. From algebra and calculus to full-fledged courses on systems modeling and machine or DL. Many of these courses are provided via high-ranking universities, and for a minor fee and a passed test the online student can obtain a certification of completion. The ability to now be trained via any remote place in the world by a prestigious university is truly the marvel of our time.
However, courses do not constitute work experience, and the transition from university to a first job can feel like stepping into another world. Although they help you build technological prowess, on-line courses cannot show you how to deal with situations that you and your model have never faced before. To build experience, you need to fail, and an on-line exam is not the same as a disappointed customer who no longer trusts your nutritional or management recommendations. It is in those moments where most of us develop grit, expand key principles of programming and modeling to achieve custom solutions, and gain a more realistic view of models and their potential use.
Universities would do well to offer students these useful, and sometimes painful experiences, by pairing them to the real-life issues livestock producers face. Not only will these assignments shape the modeling base of the student, but it will also highlight what truly matters to a livestock producer. Including the expectations they have from a modeling solution and the perquisites for implementing one. Most importantly, future animal scientists can see for themselves how a changing world also impacts requirements.
The role of industry
The greatest asset any company has is its (future) workforce, especially in a science-heavy field such as agriculture. Considering the increased use of sensors for automatic data collection and the ever-present promise of AI, it is no surprise that many have invested in “data science.” Unfortunately, companies believed that by hiring one or two data scientists, or by building a multidisciplinary team, dormant data could now be automatically transformed into sustainable revenue. As a result, ML now finds itself in the “Trough of Disillusionment.”
For a company to utilize models and data analytics and thus embark on data science, it needs to become a data science company. Not only are basic investments in modeling and statistics needed, but models also need a technical basis on which to be deployed. Building these data lakes and accompanying data pipelines requires continued investment in a non-sexy topic that has no immediate return of investment. Without these fundamental basic steps, models cannot support further automation, and as result of misplaced investments and unreasonable expectations, most companies have either disbanded or outsourced their data science teams.
To adopt and harness the potential of digitalization and AI, the industry needs to join forces. Large companies need to work with smaller start-ups and scale-ups to combine money and a large customer base with the technological prowess and more agility (Menendez et al., 2022). To remain successful, companies need to hire analytical savvy people and support universities in training them; not only by funding research but also by offering real-world cases that could profit all parties involved. Here, companies can direct universities in extending their curricula by showing the needs of the industry whereas universities can showcase cutting-edge technology. The biggest benefactor of the symbiosis will be the student—the future employee.
Conclusions and Perspectives
Technological adoption can be a bumpy road. The scientific cycle can be long and requires many iterations to build knowledge. The same holds true for data science. As AI becomes increasingly more powerful and applications start to diverge, new research fields are being created. Sustainable application is still many years away and companies and universities alike do well to remain at the forefront. This requires investments in hardware, software, and analytical talent. It also requires a solid connection to the outside world to test what does and does not work in practice and to critically determine when the field of agriculture is ready to take its next big steps. Other research fields, such as engineering and automotive, have shown that the application power of AI can be far-reaching. This is because there is a uniformity and consistency from the production-consumer-and-regulatory level making. As agriculture, specifically, animal science and livestock production strive to be far reaching, it must determine what and where models and precision technology will be the most successful in both the short and long term.
Acknowledgments
Based on multiple presentations given at the ASAS-NANP Symposium: “Mathematical Modeling in Animal Nutrition: Training the Future Generation in Data and Predictive Analytics for a Sustainable Development—Basic Training” at the 2021 Annual Meeting of the American Society of Animal Science held in Louisville, KY, July 14–17, with publications sponsored by the Journal of Animal Science and the American Society of Animal Science. This work was partially supported by the Data Science for Food and Agricultural Systems (DSFAS) program (2021-67021-33776) from the United States Department of Agriculture (USDA), National Institute of Food and Agriculture (NIFA), the National Research Support Project #9 from the National Animal Nutrition Program (https://animalnutrition.org/), and the Next Level Animal Sciences (NLAS) program of the Wageningen University & Research, The Netherlands.
Glossary
Abbreviations
- AI
artificial intelligence
- DD
data-driven (model)
- DL
deep learning
- HIMM
hybrid intelligent mechanistic model
- KPI
key performance indicator
- ML
machine learning
- MM
mathematical modeling
- PCA
principal component analysis
- PLF
precision livestock farming
- SCA
specification curve analysis
- SLF
smart-livestock farming
- XAI
eXplainable AI
Contributor Information
Marc Jacobs, FR Analytics B.V., 7642 AP Wierden, The Netherlands.
Aline Remus, Sherbrooke Research and Development Centre, Sherbrooke, QC J1M 1Z3, Canada.
Charlotte Gaillard, Institut Agro, PEGASE, INRAE, 35590 Saint Gilles, France.
Hector M Menendez, III, Department of Animal Science, South Dakota State University, Rapid City, SD 57702, USA.
Luis O Tedeschi, Department of Animal Science, Texas A&M University, College Station, TX 77843-2471, USA.
Suresh Neethirajan, Farmworx, Adaptation Physiology, Animal Sciences Group, Wageningen University, 6700 AH, The Netherlands.
Jennifer L Ellis, Department of Animal Biosciences, University of Guelph, Guelph, ON N1G 2W1, Canada.
Conflict of interest statement
The authors declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
LITERATURE CITED
- Aho, T., Sievi-Korte O., Kilamo T., Yaman S., and Mikkonen T.. . 2020. Demystifying data science projects: a look on the people and process of data science today. In: Morisio, M., Torchiano M., and Jedlitschka A., editors. International Conference on product-focused software process improvement. Cham: Springer International Publishing Switzerland; p. 153–167. doi: 10.1007/978-3-030-64148-1_10 [DOI] [Google Scholar]
- Al-Jundi, A., and SAkkA S.. . 2016. Protocol writing in clinical research. J. Clin. Diagn. Res. 10:ZE10. doi: 10.7860/JCDR/2016/21426.8865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber, M., Tepole A. B., Cannon W. R., De S., Dura-Bernal S., Garikipati K., Karniadakis G., Lytton W. W., Perdikaris P., and Petzold L.. . 2019. Integrating machine learning and multiscale modeling—perspectives, challenges, and opportunities in the biological, biomedical, and behavioral sciences. NPJ Digit. Med. 2:1–11. doi: 10.1038/s41746-019-0193-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andretta, I., Hickmann F. M. W., Remus A., Franceschi C. H., Mariani A. B., Orso C., Kipper M., Létourneau-Montminy M. -P., and Pomar C.. . 2021a. Environmental impacts of pig and poultry production: insights from a systematic review. Front. Vet. Sci. 8:733–750. doi: 10.3389/fvets.2021.750733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andretta, I., Pomar C., Rivest J., Pomar J., and Radünz J.. . 2016. Precision feeding can significantly reduce lysine intake and nitrogen excretion without compromising the performance of growing pigs. Animal 10(7):1137–1147. doi: 10.1017/S1751731115003067. Epub 2016 Jan 13. PMID: 26759074. [DOI] [PubMed] [Google Scholar]
- Andretta, I., Remus A., Franceschi C. H., Orso C., and Kipper M.. . 2021b. Chapter 3 - Environmental impacts of feeding crops to poultry and pigs. In: Galanakis, C. M. editor. Environmental impact of agro-food industry and food consumption. Cambridge, Massachusetts: Academic Press; p. 59–79. [Google Scholar]
- Banhazi, T. M., Lehr H., Black J., Crabtree H., Schofield P., Tscharke M., and Berckmans D.. . 2012. Precision livestock farming: an international review of scientific and commercial aspects. Int. J. Agric. Biol. Eng. 5:1–9. doi: 10.3965/j.ijabe.20120503.001 [DOI] [Google Scholar]
- Bolón-Canedo, V., Sánchez-Maroño N., and Alonso-Betanzos A.. . 2013. A review of feature selection methods on synthetic data. Knowl. Inf. Syst. 34:483–519. doi: 10.1007/S10115-012-0487-8 [DOI] [Google Scholar]
- Breiman, L. 2001. Random forests. Mach. Learn. 45:5–32. doi: 10.1023/A:1010933404324 [DOI] [Google Scholar]
- Briggs, W. 2016. Uncertainty: the soul of modeling, probability & statistics. Cham: Springer International Publishing Switzerland. [Google Scholar]
- Browne, M. W. 2000. Cross-validation methods. J. Math. Psychol. 44:108–132. doi: 10.1006/jmps.1999.1279 [DOI] [PubMed] [Google Scholar]
- Bunge, M. A. 2001. Scientific realism: selected essays of Mario Bunge. Amherst, New York: Prometheus Books. [Google Scholar]
- Cadéro, A., Aubry A., Dourmad J. Y., Salaün Y., and Garcia-Launay F.. . 2020. Effects of interactions between feeding practices, animal health and farm infrastructure on technical, economic and environmental performances of a pig-fattening unit. Animal 14(Suppl_2):s348–s359. ISSN: 1751-7311. doi: 10.1017/S1751731120000300 [DOI] [PubMed] [Google Scholar]
- Chen, J. H., and Asch S. M.. . 2017. Machine learning and prediction in medicine—beyond the peak of inflated expectations. N Engl. J. Med. 376:2507. doi: 10.1056/NEJMp1702071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C. M. 2013. The innovator’s dilemma: when new technologies cause great firms to fail. Brighton, Massachusetts: Harvard Business Review Press. [Google Scholar]
- Cirillo, D., Núñez-Carpintero I., and Valencia A.. . 2021. Artificial intelligence in cancer research: learning at different levels of data granularity. Mol. Oncol. 15:817–829. doi: 10.1002/1878-0261.12920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin, B., Germain S., and Pomar C.. . 2021. Early detection of individual growing pigs’ sanitary challenges using functional data analysis of real-time feed intake patterns. Commun. Stat. Case Stud. Data Anal. Appl. 1–21. doi: 10.1080/23737484.2021.1991855 [DOI] [Google Scholar]
- Cortes-Ciriano, I., Bender A., and Malliavin T. E.. . 2015. Comparing the influence of simulated experimental errors on 12 machine learning algorithms in bioactivity modeling using 12 diverse data sets. J. Chem. Inf. Model. 55:1413–1425. doi: 10.1021/acs.jcim.5b00101 [DOI] [PubMed] [Google Scholar]
- Cosgriff, C. V., Ebner D. K., and Celi L. A.. . 2020. Data sharing in the era of COVID-19. Lancet Digit. Heal. 2:e224. doi: 10.1016/S2589-7500(20)30082-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosme, D. 2020. Specification curve analysis a practical guide. [accessed February 08, 2022].https://dcosme.github.io/specification-curves/SCA_tutorial_inferential_presentation#1.
- Cowton, J., Kyriazakis I., Plötz T., and Bacardit J.. . 2018. A combined deep learning GRU-autoencoder for the early detection of respiratory disease in pigs using multiple environmental sensors. Sensors 18:2521. doi: 10.3390/s18082521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creswell, A., White T., Dumoulin V., Arulkumaran K., Sengupta B., and Bharath A.. . 2018. Generative adversarial networks: an overview. IEEE Signal Process. Mag. 35:53–65. doi: 10.1109/MSP.2017.2765202 [DOI] [Google Scholar]
- Daneshian, M., Akbarsha M. A., Blaauboer B., Caloni F., Cosson P., Curren R., Goldberg A., Gruber F., Ohl F., and Pfaller W.. . 2011. A framework program for the teaching of alternative methods (replacement, reduction, refinement) to animal experimentation. ALTEX-Altern. Anim. Ex. 28:341–352. doi: 10.14573/altex.2011.4.341 [DOI] [PubMed] [Google Scholar]
- Daniel, J. -B., Friggens N., Van Laar H., Ferris C., and Sauvant D.. . 2017. A method to estimate cow potential and subsequent responses to energy and protein supply according to stage of lactation. J. Dairy Sci. 100:3641–3657. doi: 10.3168/jds.2016-11938 [DOI] [PubMed] [Google Scholar]
- Dargan, S., Kumar M., Ayyagari M. R., and Kumar G.. . 2020. A survey of deep learning and its applications: a new paradigm to machine learning. Arch. Comput. Methods Eng. 27:1071–1092. doi: 10.1007/s11831-019-09344-w [DOI] [Google Scholar]
- Dedehayir, O., and Steinert M.. . 2016. The hype cycle model: a review and future directions. Technol. Forecast. Soc. Change 108:28–41. doi: 10.1016/j.techfore.2016.04.005 [DOI] [Google Scholar]
- DeMello, M. 2021. Chapter 9 Animals and science, animals and society: an introduction to human-animal studies. New York City: Columbia University Press; p. 204–232. [Google Scholar]
- Dobson, A. J. 2013. Introduction to statistical modelling. Cham: Springer International Publishing Switzerland. [Google Scholar]
- Dourmad, J. -Y., Étienne M., Valancogne A., Dubois S., van Milgen J., and Noblet J.. . 2008. InraPorc: a model and decision support tool for the nutrition of sows. Anim. Feed Sci. Technol. 143:372–386. doi: 10.1016/j.anifeedsci.2007.05.019 [DOI] [Google Scholar]
- Dumas, A., Dijkstra J., and France J.. . 2008. Mathematical modelling in animal nutrition: a centenary review. J. Agric. Sci. 146:123–142. doi: 10.1017/S0021859608007703 [DOI] [Google Scholar]
- Dumas, A., France J., and Bureau D.. . 2010. Modelling growth and body composition in fish nutrition: where have we been and where are we going? Aquacult. Res. 41:161–181. doi: 10.1111/j.1365-2109.2009.02323.x [DOI] [Google Scholar]
- Eddy, D. M., Hollingworth W., Caro J. J., Tsevat J., McDonald K. M., and Wong J. B.. . 2012. Model transparency and validation: a report of the ISPOR-SMDM modeling good research practices task Force–7. Med. Decis. Making 32:733–743. doi: 10.1177/0272989X12454579 [DOI] [PubMed] [Google Scholar]
- Ellis, J., Jacobs M., Dijkstra J., van Laar H., Cant J., Tulpan D., and Ferguso N.. . 2019. The role of mechanistic models in the era of big data and intelligent computing. Animal 10:286. [DOI] [PubMed] [Google Scholar]
- Ellis, J. L., Jacobs M., Dijkstra J., van Laar H., Cant J. P., Tulpan D., and Ferguson N.. . 2020. Review: synergy between mechanistic modelling and data-driven models for modern animal production systems in the era of big data. Animal 14:s223–s237. doi: 10.1017/S1751731120000312 [DOI] [PubMed] [Google Scholar]
- Estrada-Flores, S., Merts I., De Ketelaere B., and Lammertyn J.. . 2006. Development and validation of “grey-box” models for refrigeration applications: a review of key concepts. Int. J. Refrig. 29:931–946. doi: 10.1016/j.ijrefrig.2006.03.018 [DOI] [Google Scholar]
- Favino, M., Quaglino A., Pozzi S., Krause R., and Pivkin I.. . 2016. Multiscale modeling, discretization, and algorithms: a survey in biomechanics. arXiv preprint arXiv 1609.07719. doi: 10.48550/arXiv.1609.07719 [DOI] [Google Scholar]
- Fecher, B., Friesike S., and Hebing M.. . 2015. What drives academic data sharing? PLoS One 10:e0118053. doi: 10.1371/journal.pone.0118053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn, J., and LeHong H.. . 2011. Hype cycle for emerging technologies, 2011. Gartner. [Google Scholar]
- Fernandes, A. F. A., Dórea J. R. R., Fitzgerald R., Herring W., and Rosa G. J. M.. . 2019. A novel automated system to acquire biometric and morphological measurements and predict body weight of pigs via 3D computer vision. J. Anim. Sci. 97:496–508. doi: 10.1093/jas/sky418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradkov, A. L. 2020. Early history of machine learning. IFAC-PapersOnLine 53:1385–1390. doi: 10.1016/j.ifacol.2020.12.1888 [DOI] [Google Scholar]
- France, J. 2008. Modelling animal systems. J. Agric. Sci. 146:121. doi: 10.1017/S0021859607007666 [DOI] [Google Scholar]
- Frazzetto, D., Nielsen T. D., Pedersen T. B., and Šikšnys L.. . 2019. Prescriptive analytics: a survey of emerging trends and technologies. VLDB J. 28:575–595. doi: 10.1007/s00778-019-00539-y [DOI] [Google Scholar]
- French, S. 1988. How significant is statistical significance? a critique of the use of statistics in research. Physiotherapy 74:266–268. doi: 10.1016/S0031-9406(10)63189-6 [DOI] [Google Scholar]
- Friedman, J. H. 2002. Stochastic gradient boosting. Comput. Stat. Data Anal. 38:367–378. doi: 10.1016/s0167-9473(01)00065-2 [DOI] [Google Scholar]
- Gaillard, C., Quiniou N., Gauthier R., Cloutier L., and Dourmad J. -Y.. . 2020. Evaluation of a decision support system for precision feeding of gestating sows. J. Anim. Sci. 98:1–12. doi: 10.1093/jas/skaa255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier, R., Largouët C., Gaillard C., Cloutier L., Guay F., and Dourmad J. Y.. . 2019. Dynamic modeling of nutrient use and individual requirements of lactating sows. J. Anim. Sci. 97:2822–2836. doi: 10.1093/jas/skz167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greener, J. G., Kandathil S. M., Moffat L., and Jones D. T.. . 2022. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 23:40–55. doi: 10.1038/s41580-021-00407-0 [DOI] [PubMed] [Google Scholar]
- Grimm, V., Railsback S. F., Vincenot C. E., Berger U., Gallagher C., DeAngelis D. L., Edmonds B., Ge J., Giske J., and Groeneveld J.. . 2020. The ODD protocol for describing agent-based and other simulation models: a second update to improve clarity, replication, and structural realism. J. Artif. Soc. Soc. Simul. 23(2):7. doi: 10.18564/jasss.425933204215 [DOI] [Google Scholar]
- Halachmi, I., Guarino M., Bewley J., and Pastell M.. . 2019. Smart animal agriculture: application of real-time sensors to improve animal well-being and production. Annu. Rev. Anim. Biosci. 7:403–425. doi: 10.1146/annurev-animal-020518-114851 [DOI] [PubMed] [Google Scholar]
- Hauschild, L., Lovatto P. A., Pomar J., and Pomar C.. . 2012. Development of sustainable precision farming systems for swine: estimating real-time individual amino acid requirements in growing-finishing pigs. J. Anim. Sci. 90:2255–2263. doi: 10.2527/jas.2011-4252 [DOI] [PubMed] [Google Scholar]
- Hauschild, L., Pomar C., and Lovatto P. A.. . 2010. Systematic comparison of the empirical and factorial methods used to estimate the nutrient requirements of growing pigs. Animal 4:714–723. doi: 10.1017/S1751731109991546 [DOI] [PubMed] [Google Scholar]
- Heck, P., Schouten G., and Cruz L.. . 2021. A software engineering perspective on building production-ready machine learning systems. In Handbook of research on applied data science and artificial intelligence in business and industry. Hershey, Pennsylvania: IGI Global; p. 23–54. [Google Scholar]
- Higgins, J. P., Thomas J., Chandler J., Cumpston M., Li T., Page M. J., and Welch V. A.. . 2019. Cochrane handbook for systematic reviews of interventions. Hoboken, New Jersey: John Wiley & Sons. [Google Scholar]
- Hinterstoisser, S., Lepetit V., Wohlhart P., and Konolige K.. . 2018. On pre-trained image features and synthetic images for deep learning. In: Proceedings of the European Conference on Computer Vision (ECCV) Workshops. Cham: Springer International Publishing Switzerland. [Google Scholar]
- Hobbs, N. T., and Hooten M.. . 2015. Bayesian models: a statistical primer for ecologists. William Street, Princeton, New Jersey: Princeton University Press. [Google Scholar]
- Hu, G., Do D. N., Davoudi P., and Miar Y.. . 2022. Emerging roles of non-coding RNAs in the feed efficiency of livestock species. Genes 13:297. doi: 10.3390/genes13020297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, D. L. 2010. Science as a process: an evolutionary account of the social and conceptual development of science. Chicago: University of Chicago Press. [Google Scholar]
- Ibrahim, D. M. 2006. Reduce, refine, replace: the failure of the three R’s and the future of animal experimentation. University of Chicago Legal Forum, 2006, Arizona Legal Studies Discussion Paper No. 06-17, Available at SSRN: https://ssrn.com/abstract=888206
- Ioannidis, J. P. A. 2005. Why most published research findings are false. PLoS Med. 2:e124. doi: 10.1371/journal.pmed.0020124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Laird, P. N., and Byrne R. M.. . 1993. Mental models or formal rules? Behav. Brain Sci. 16:368–380. doi: 10.1017/S0140525X0003065X [DOI] [Google Scholar]
- Kelly, E., Heanue K., Buckley C., and O’Gorman C.. . 2015. Proven science versus farmer perception. In: 2015 Conference International Association of Agricultural Economists; August 9–14. 2015; Milan, Italy.
- Khamis, A., Li H., Prestes E., and Haidegger T.. . 2019. AI: a key enabler of sustainable development goals, part 1 [industry activities]. IEEE Robot. Autom. Mag. 26(3):95–102. doi: 10.1109/MRA.2019.2928738 [DOI] [Google Scholar]
- Koketsu, Y., Sasaki Y., Ichikawa H., and Kaneko M.. . 2010. Benchmarking in animal agriculture: concepts and applications. J. Vet. Epidemiol. 14:105–117. doi: 10.2743/jve.14.105 [DOI] [Google Scholar]
- Labrecque, J., and Rivest J.. . 2018. A real-time sow behavior analysis system to predict an optimal timing for insemination. In: 10th International Livestock Environment Symposium (ILES X); p. 1. doi: 10.13031/iles.18-023 [DOI] [Google Scholar]
- Li, T., Sahu A. K., Talwalkar A., and Smith V.. . 2020. Federated learning: challenges, methods, and future directions. IEEE Signal Process. Mag. 37:50–60. doi: 10.48550/arXiv.1908.07873 [DOI] [Google Scholar]
- Linden, A., and Fenn J.. . 2003. Understanding Gartner’s hype cycles. Strategic analysis report Nº R-20-1971. Gartner, Inc 88:1423. [Google Scholar]
- Liu, D., Oczak M., Maschat K., Baumgartner J., Pletzer B., He D., and Norton T.. . 2020. A computer vision-based method for spatial-temporal action recognition of tail-biting behaviour in group-housed pigs. Biosyst. Eng. 195:27–41. doi: 10.1016/j.biosystemseng.2020.04.007 [DOI] [Google Scholar]
- Lokhorst, C., De Mol M. R., and Kamphuis C.. . 2019. Invited review: big data in precision dairy farming. Animal 13(7):1519–1528. doi: 10.1017/S1751731118003439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo-Thong, O., Charton P., Cadet X. F., Grondin-Perez B., Saavedra E., Damour C., and Cadet F.. . 2020. Identification of flux checkpoints in a metabolic pathway through white-box, grey-box and black-box modeling approaches. Sci. Rep. 10:13446. doi: 10.1038/s41598-020-70295-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg, S. M., and Lee S.-I.. . 2017. A unified approach to interpreting model predictions. In: Proceedings of the 31st international conference on neural information processing systems (NIPS 2017), Long Beach, CA, USA; p. 4768–4777. doi: 10.48550/arXiv.1705.07874 [DOI] [Google Scholar]
- Martinez-Moyano, I. J. 2012. Documentation for model transparency. Syst. Dyn. Rev. 28:199–208. doi: 10.1002/sdr.1471 [DOI] [Google Scholar]
- Mayo, D. G., and Cox D. R.. . 2006. Frequentist statistics as a theory of inductive inference, Optimality. Institute of Mathematical Statistics; p. 77–97. [Google Scholar]
- Menendez, H. M., Brennan J. R., Neethirajan S., Gaillard C., Remus A., Jacobs M., Ehlert K., and Tedeschi L. O.. . 2022. ASAS-NANP symposium: mathematical modeling in animal nutrition: opportunities and challenges of confined and extensive precision livestock production. J. Anim. Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez, H. M., and Tedeschi L. O.. . 2020. The characterization of the cow-calf, stocker and feedlot cattle industry water footprint to assess the impact of livestock water use sustainability. J. Agric. Sci. 158:416–430. doi: 10.1017/S0021859620000672 [DOI] [Google Scholar]
- van Milgen, J., Valancogne A., Dubois S., Dourmad J. -Y., Sève B., and Noblet J.. . 2008. InraPorc: a model and decision support tool for the nutrition of growing pigs. Anim. Feed Sci. Technol. 143:387–405. doi: 10.1016/j.anifeedsci.2007.05.020 [DOI] [Google Scholar]
- Morota, G., Ventura R. V., Silva F. F., Koyama M., and Fernando S. C.. . 2018. Big data analytics and precision animal agriculture symposium: machine learning and data mining advance predictive big data analysis in precision animal agriculture. J. Anim. Sci. 96:1540–1550. doi: 10.1093/jas/sky014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Tamayo, R., Nielsen B. L., Gagaoua M., Gondret F., Krause E. T., Morgavi D. P., Olsson I. A. S., Pastell M., Taghipoor M., Tedeschi L., . et al. 2022. Seven steps to enhance open science practices in animal science. Zenodo version 1. doi: 10.5281/zenodo.5891771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Tamayo, R., Puillet L., Daniel J. -B., Sauvant D., Martin O., Taghipoor M., and Blavy P.. . 2018. To be or not to be an identifiable model. Is this a relevant question in animal science modelling? Animal 12:701–712. doi: 10.1017/S1751731117002774 [DOI] [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine. 2016. Nutrient requirements of beef cattle: Eighth Revised Edition. Washington, DC: The; National Academies of Sciences, Engineering, and Medicine. doi: 10.17226/19014 [DOI] [Google Scholar]
- Neethirajan, S. 2017. Recent advances in wearable sensors for animal health management. Sens. Bio-Sens. Res. 12:15–29. doi: 10.1016/j.sbsr.2016.11.004 [DOI] [Google Scholar]
- Noziere, P., Sauvant, D., & Delaby, L.. 2018. INRA feeding system for ruminants. Wageningen (The Netherlands): Academic Publishers. [Google Scholar]
- NRC. 2012. Nutrient requirements of swine, 11th rev ed, 12th ed. Washington (DC): The National Academies Press. [Google Scholar]
- O’Grady, M. J., and O’Hare G. M.. . 2017. Modelling the smart farm. Inf. Process. Agric. 4:179–187. doi: 10.1016/j.inpa.2017.05.001 [DOI] [Google Scholar]
- Owens, D., Mitchell A., Khazanchi D., and Zigurs I.. . 2011. An empirical investigation of virtual world projects and metaverse technology capabilities. ACM SIGMIS Database 42:74–101. doi: 10.1145/1952712.1952717 [DOI] [Google Scholar]
- Parsons, D. J., Green D. M., Schofield C. P., and Whittemore C. T.. . 2007. Real-time control of pig growth through an integrated management system. Biosyst. Eng. 96:257–266. doi: 10.1016/j.biosystemseng.2006.10.013 [DOI] [Google Scholar]
- Pearl, J., and Mackenzie D.. . 2018. The book of why: the new science of cause and effect. New York: Basic books. [Google Scholar]
- Peng, G. C. Y., Alber M., Tepole A. B., Cannon W. R., De S., Dura-Bernal S., Garikipati K., Karniadakis G., Lytton W. W., Perdikaris P., . et al. 2021. Multiscale modeling meets machine learning: what can we learn? Arch. Comput. Methods Eng. 28:1017–1037. doi: 10.1007/s11831-020-09405-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, D. 2020. Gartner 2020 hype cycle for emerging technologies. What’s in it for AI leaders? [accessed January 15, 2022].
- Petrie, A., and Watson P.. . 2013. Statistics for veterinary and animal science. Hoboken, New Jersey: John Wiley & Sons. [Google Scholar]
- Pintelas, E., Livieris I. E., and Pintelas P.. . 2020. A grey-box ensemble model exploiting black-box accuracy and white-box intrinsic interpretability. Algorithms 13:17. doi: 10.3390/a13010017 [DOI] [Google Scholar]
- Pomar, C., Andretta I., and Remus A.. . 2021. Feeding strategies to reduce nutrient losses and improve the sustainability of growing pigs. Front. Vet. Sci. 8:742220. doi: 10.3389/fvets.2021.742220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomar, C., Kyriazakis I., Emmans G. C., and Knap P. W.. . 2003. Modeling stochasticity: dealing with populations rather than individual pigs. J. Anim. Sci. 81(14 suppl 2):E178–E186. doi: 10.2527/2003.8114_suppl_2E178x [DOI] [Google Scholar]
- Pomar, C., van Milgen J., and Remus A.. . 2019a. Precision livestock feeding, principle and practice. Wageningen, Netherlands: Wageningen Academic Publishers. [Google Scholar]
- Pomar, C., van Milgen J., and Remus A.. . 2019b. Precision livestock feeding, principle and practice . In: Poultry and pig nutrition challenges of the 21st century. Wageningen, Netherlands: Wageningen Academic Publishers; p. 397–418. doi: 10.3920/978-90-8686-884-1_18. [DOI] [Google Scholar]
- Pomar, C., Pomar J., Rivest J., Cloutier L., Letourneau-Montminy M. P., Andretta I., and Hauschild L.. . 2015. Estimating real-time individual amino acid requirements in growing-finishing. In: Sakomura, N. K., Gous R. M., Kyriazakis, I. and Hauschild L., editors. Nutritional modelling for pigs and poultry. Wallingford (UK): CABI Publishing; p. 157–174. doi: 10.1079/9781780644110.0157 [DOI] [Google Scholar]
- Pomar, C., and Remus A.. . 2019a. Are actual animal growth models adequate to predict growth and estimate amino acid and other nutrient requirements? In: Teixeira, I. A. M. A. , Biagioli B., Hauschild, L. and Sakomura, N. editors. Proceedings of the 9th Workshop on Modelling Nutrient Digestion and Utilization in Farm Animals (MODNUT) No. 10. Itamambuca (Brazil): Cambridge University Press, p. 294. [Google Scholar]
- Pomar, C., and Remus A.. . 2019b. Precision pig feeding: a breakthrough toward sustainability. Anim. Front. 9:52–59. doi: 10.1093/af/vfz006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomar, C., and Remus A.. . 2021. Basic elements, difficulties and pitfalls on the development and application of precision nutrition techniques for smart pig farming Animal Nutrition Conference of Canada 2021; p 138–152. https://animalnutritionconference.ca/, Virtual event
- Reiter, J. P. 2002. Satisfying disclosure restrictions with synthetic data sets. J. Off. Stat. 18:531. [Google Scholar]
- Remus, A., del Castillo J. R. E., and Pomar C.. . 2020a. Improving the estimation of amino acid requirements to maximize nitrogen retention in precision feeding for growing-finishing pigs. Animal 14:2032–2041. doi: 10.1017/S1751731120000798 [DOI] [PubMed] [Google Scholar]
- Remus, A., Hauschild L., Andretta I., Kipper M., Lehnen C. R., and Sakomura N. K.. . 2014. A meta-analysis of the feed intake and growth performance of broiler chickens challenged by bacteria. Poult. Sci. 93:1149–1158. doi: 10.3382/ps.2013-03540 [DOI] [PubMed] [Google Scholar]
- Remus, A., Hauschild L., Létourneau-Montminy M. -P., and Pomar C.. . 2021. Estimating amino acid requirements in real-time for precision-fed pigs: the challenge of variability among individuals. Animals 11:3354. doi: 10.3390/ani11123354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus, A., Hauschild L., Methot S., and Pomar C.. . 2020b. Precision livestock farming: real-time estimation of daily protein deposition in growing–finishing pigs. Animal 14:s360–s370. doi: 10.1017/S1751731120001469 [DOI] [PubMed] [Google Scholar]
- Remus, A., Hauschild L., and Pomar C.. . 2020c. Simulated amino acid requirements of growing pigs differ between current factorial methods. Animal 14:725–730. doi: 10.1017/S1751731119002660 [DOI] [PubMed] [Google Scholar]
- Ribeiro, M. T., Singh S., and Guestrin C.. . 2016. Model-agnostic interpretability of machine learning. arXiv preprint arXiv 1606.05386. [Google Scholar]
- Risbey, J., Kandlikar M., Dowlatabadi H., and Graetz D.. . 1999. Scale, context, and decision making in agricultural adaptation to climate variability and change. Mitig. Adapt. Strateg. Glob. Chang 4:137–165. doi: 10.1023/A:1009636607038 [DOI] [Google Scholar]
- Rogers, E. 2003. Diffusion of innovations, 5th ed. New York (NY): The Free Press; p. 107, 111, 127. [Google Scholar]
- Rosen, J. 2016. Research protocols: a forest of hypotheses. Nature 536:239–241. doi: 10.1038/nj7615-239a [DOI] [PubMed] [Google Scholar]
- Rosenblatt, F. 1958. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol. Rev. 65:386. doi: 10.1037/h0042519 [DOI] [PubMed] [Google Scholar]
- Rotz, C. A. 2018. Modeling greenhouse gas emissions from dairy farms. J. Dairy Sci. 101:6675–6690. doi: 10.3168/jds.2017-13272 [DOI] [PubMed] [Google Scholar]
- Roush, W. B., Dozier W. A., and Branton S. L.. . 2006. Comparison of gompertz and neural network models of broiler growth. Poult. Sci. 85:794–797. doi: 10.1093/ps/85.4.794 [DOI] [PubMed] [Google Scholar]
- Roy, D., Srivastava R., Jat M., and Karaca M. S.. . 2022. A complete overview of analytics techniques: descriptive, predictive, and prescriptive. In: Decision intelligence analytics and the implementation of strategic business management. Cham: Springer International Publishing Switzerland; p. 15–30. doi: 10.1007/978-3-030-82763-2_2 [DOI] [Google Scholar]
- Samek, W., Montavon G., Vedaldi A., Hansen L. K., and Müller K.-R.. . 2019. Explainable AI: interpreting, explaining and visualizing deep learning. London, UK: Springer Nature. [Google Scholar]
- Samperio, E., Lidón I., Rebollar R., Castejón-Limas M., and Álvarez-Aparicio C.. . 2021. Lambs’ live weight estimation using 3D images. Animal 15:100212. doi: 10.1016/j.animal.2021.100212 [DOI] [PubMed] [Google Scholar]
- San, O. 2021. The digital twin revolution. Nat. Comput. Sci. 1:307–308. doi: 10.1038/s43588-021-00077-0 [DOI] [PubMed] [Google Scholar]
- Sauvant, D., and Nozière P.. . 2016. Quantification of the main digestive processes in ruminants: the equations involved in the renewed energy and protein feed evaluation systems. Animal 10:755–770. doi: 10.1017/S1751731115002670 [DOI] [PubMed] [Google Scholar]
- Schweinsberg, M., Feldman M., Staub N., van den Akker O. R., van Aert R. C., Van Assen M. A., Liu Y., Althoff T., Heer J., and Kale A.. . 2021. Same data, different conclusions: radical dispersion in empirical results when independent analysts operationalize and test the same hypothesis. Organ. Behav. Hum. Decis. Process. 165:228–249. ISSN 0749-5978. doi: 10.1016/j.obhdp.2021.02.003 [DOI] [Google Scholar]
- Shneiderman, B. 2022. Human-Centered AI. Oxford, England: Oxford University Press. [Google Scholar]
- Simonsohn, U., Simmons J. P., and Nelson L. D.. . 2020. Specification curve analysis. Nat. Hum. Behav. 4:1208–1214. doi: 10.1038/s41562-020-0912-z [DOI] [PubMed] [Google Scholar]
- Smith, M. J. 2018. Getting value from artificial intelligence in agriculture. Anim. Prod. Sci. 60:46–54. doi: 10.1071/AN18522 [DOI] [Google Scholar]
- Sterman, J. 2000. Business dynamics. New York City: McGraw-Hill, Inc. [Google Scholar]
- Stewart, L. A., and Parmar M. K.. . 1993. Meta-analysis of the literature or of individual patient data: is there a difference? Lancet 341:418–422. doi: 10.1016/0140-6736(93)93004-k [DOI] [PubMed] [Google Scholar]
- Surendra, H., and Mohan H.. . 2017. A review of synthetic data generation methods for privacy preserving data publishing. Int. J. Sci. Technol. Res. 6:95–101. [Google Scholar]
- Sutton, R. S., and Barto A. G.. . 2018. Reinforcement learning: an introduction. Cambridge, MA: MIT press. [Google Scholar]
- Tassinari, P., Bovo M., Benni S., Franzoni S., Poggi M., Mammi L. M. E., Mattoccia S., Di Stefano L., Bonora F., Barbaresi A., . et al. 2021. A computer vision approach based on deep learning for the detection of dairy cows in free stall barn. Comput. Electron. Agric. 182:106030. doi: 10.1016/j.compag.2021.106030 [DOI] [Google Scholar]
- Taylor, S. J., and Letham B.. . 2018. Forecasting at scale. Am. Stat. 72:37–45. [Google Scholar]
- Tedeschi, L. O. 2006. Assessment of the adequacy of mathematical models. Agric. Sys. 89:225–247. doi: 10.1016/j.agsy.2005.11.004 [DOI] [Google Scholar]
- Tedeschi, L. O. 2019. Mathematical modeling in ruminant nutrition: approaches and paradigms, extant models, and thoughts for upcoming predictive analytics. J. Anim. Sci. 97(5):1921–1944. doi: 10.1093/jas/skz092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi, L. O., and Boston R.. . 2011. Identifiability and accuracy: a closer look at contemporary contributions and changes in these vital areas of mathematical modelling. In: Modelling nutrient digestion and utilisation in farm animals; p. 91–99. doi: 10.3920/978-90-8686-712-7_10 [DOI] [Google Scholar]
- Tedeschi, L. O., and Fox D. G.. . 2020. The ruminant nutrition system: an applied model for predicting nutrient requirements and feed utilization in ruminants. XanEdu. Acton, MA. ISBN-13: 978-1506697758; ISBN-10: 1506697755. doi: 10.1017/S0021859617000296. [DOI] [Google Scholar]
- Tedeschi, L. O., Fox D. G., and Guiroy P. J.. . 2004. A decision support system to improve individual cattle management. 1. A mechanistic, dynamic model for animal growth. Agric. Sys. 79:171–204. doi: 10.1016/S0308-521X(03)00070-2 [DOI] [Google Scholar]
- Tedeschi, L. O., Greenwood P. L., and Halachmi I.. . 2021. Advancements in sensor technology and decision support intelligent tools to assist smart livestock farming. J. Anim. Sci. 99:skab038. doi: 10.1093/jas/skab038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenopir, C., Allard S., Douglass K., Aydinoglu A. U., Wu L., Read E., Manoff M., and Frame M.. . 2011. Data sharing by scientists: practices and perceptions. PLoS One 6:e21101. doi: 10.1371/journal.pone.0021101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff, J. M., Chan A. -W., Kitchen J., Sampson M., Tricco A. C., and Moher D.. . 2012. Guidelines for randomized clinical trial protocol content: a systematic review. Systematic reviews 1:1–11. doi: 10.1186/2046-4053-1-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J., Rousselière Y., Marcon M., and Hémonic A.. . 2021. Early detection of diarrhea in weaned piglets from individual feed, water and weighing data. Front. Anim. Sci. 2:688902. doi: 10.3389/fanim.2021.688902 [DOI] [Google Scholar]
- Thompson, S. G., and Higgins J. P.. . 2002. How should meta-regression analyses be undertaken and interpreted? Stat. Med. 21:1559–1573. doi: 10.1002/sim.1187 [DOI] [PubMed] [Google Scholar]
- Thornley, J. H. M., and France J.. . 1984. Role of modeling in animal production research and extension work. Oakland, California: University of California. [Google Scholar]
- Thornley, J. H., and France J.. . 2007. Mathematical models in agriculture. Quantitative methods for the plant, animal and ecological sciences, 2nd edn. Eds Thornley J. H. M. & France J.. 906 pp. Wallingford, UK: CABI. [Google Scholar]
- Varga, T., and Bunke H.. . 2003. Effects of training set expansion in handwriting recognition using synthetic data. In: Proc. 11th Conf. of the Int. Graphonomics Society; p 200–203. [Google Scholar]
- Vartak, M., Ortiz P., Siegel K., Subramanyam H., Madden S., and Zaharia M.. . 2015. Supporting fast iteration in model building. In: NIPS Workshop Learning Sys; p. 1–6. [Google Scholar]
- Vigors, S., O’Doherty J. V., Bryan K., and Sweeney T.. . 2019. A comparative analysis of the transcriptome profiles of liver and muscle tissue in pigs divergent for feed efficiency. BMC Genomics 20:461. doi: 10.1186/s12864-019-5740-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bertalanffy, L. 2010. General systems theory. In: The science of synthesis: exploring the social implications of general systems theory. Boulder, Colorado: University Press of Colorado; p. 103–143. [Google Scholar]
- Wagenmakers, E.-J., Lee M., Lodewyckx T., and Iverson G. J.. . 2008. Bayesian versus frequentist inference. In: Hoijtink, H., Klugkist I., Boelen P. A., editors. Bayesian evaluation of informative hypotheses. Statistics for social and behavioral sciences. New York, NY: Springer; p. 181; –207. doi: 10.1007/978-0-387-09612-4_9 [DOI] [Google Scholar]
- Wathes, C., Kristensen H. H., Aerts J. -M., and Berckmans D.. . 2008. Is precision livestock farming an engineer’s daydream or nightmare, an animal’s friend or foe, and a farmer’s panacea or pitfall? Comput. Electron. Agric. 64:2–10. doi: 10.1016/j.compag.2008.05.005 [DOI] [Google Scholar]
- Weiner, J. 2020. Why AI/data science projects fail: how to avoid project pitfalls. Synth. Lect. Comput. Anal. 1:i–77. doi: 10.2200/S01070ED1V01Y202012CAN001 [DOI] [Google Scholar]
- Weiss, W. 2021. Brief introduction to the NASEM (formerly known as NRC) of the nutrient requirements of dairy cattle. Cornell University Library. https://hdl.handle.net/1813/110230 [Google Scholar]
- Wu, C., Buyya R., and Ramamohanarao K.. . 2016. Big data analytics= machine learning+ cloud computing. arXiv preprint arXiv 1601.03115. doi: 10.48550/arXiv.1601.03115 [DOI] [Google Scholar]
- Wurtz, K., Camerlink I., D’Eath R. B., Fernández A. P., Norton T., Steibel J., and Siegford J.. . 2019. Recording behaviour of indoor-housed farm animals automatically using machine vision technology: a systematic review. PLoS One 14:e0226669. doi: 10.1371/journal.pone.0226669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuidhof, M. J. 2021. Precision Feeding of Poultry: Matching Nutrient Supply with the Nutrient Requirements of Individual birds 5th annual Animal Nutrition Conference of Canada (ANCC), Virtual event. Animal Nutrition Association of Canada. [Google Scholar]