Highlights
-
•
L.fermentum from buffalo milk grows efficiently without animal-derived medium components.
-
•
Highest viable biomass titers can be reached after only 8h improving productivity.
-
•
L. fermentum is suitable for large scale production: complete biotech approach.
-
•
L. fermentum demonstrates 60% cell survival after spray drying.
-
•
L. fermentum from buffalo milk displaces H. pylori in a gastric epithelial cell model.
Keywords: Limosilactobacillus fermentum, Helicobacter pylori, Probiotic, Scale up, Gastric model, Immunomodulation
Abstract
Probiotics are living microorganisms that give beneficial health effects while consumed, and each strain possesses diverse and unique properties and also different technological characteristics that affect its ability to be produced at large scale. Limosilactobacillus fermentum is a widely studied member of probiotics, however, few data are available on the development of fermentation and downstream processes for the production of viable biomasses for potential industrial applications.
In the present study a novel L. fermentum strain was isolated from buffalo milk and used as test example for biotechnological process development. The strain was able to produce up to 109 CFU/mL on a (glucose based) semi-defined medium deprived of animal-derived raw materials up to the pilot scale (150 L), demonstrating improved results compared to commonly used, although industrially not suitable, media rich of casein and beef extract. The study of strain behavior in batch experiments indicated that the highest concentration of viable cells was reached after only 8 h of growth, greatly shortening the process. Moreover, initial concentrations of glucose in the medium above 30 g/L, if not supported by higher nitrogen concentrations, reduced the yield of biomass and increased production of heterolactic fermentation by-products. Biomass concentration via microfiltration on hollow fibers, and subsequent spray-drying allowed to recover about 5.7 × 1010CFU/gpowder of viable cells, indicating strain resistance to harsh processing conditions.
Overall, these data demonstrate the possibility to obtain and maintain adequate levels of viable L. fermentum cells by using a simple approach that is potentially suitable for industrial development. Moreover, since often exopolysaccharides produced by lactobacilli contribute to the strain's functionality, a partial characterization of the EPS produced by the newly identified L. fermentum strain was carried out.
Finally, the effect of L. fermentum versus H. pylori in a gastric epithelial cell model was evaluated demonstrating its ability to stimulate the response of the immune system and displace the infective agent.
1. Introduction
Probiotics are live microorganisms that are intended to provide health benefits to their host [16], and every strain possesses different and often unique properties that justify the search for new probiotics. Lactic acid bacteria (LAB) are a group of probiotics with specific nutritional requirements (e.g. vitamins, amino acids etc.) and low redox potential for growth. They are often used as starter cultures for food fermentation due to the production of lactic acid (LA), bacteriocins (small proteins with antibacterial activity) and exopolysaccharides (EPS) as all these metabolites can compete with pathogenic microorganisms and prevent food decomposition [3]. Limosilactobacillus fermentum, an heterofermentative member of the LAB family (previously addressed as Lactobacillus fermentum and recently reclassified as Limosilactobacillus [62]), can be found mainly in vegetable and cereal-based fermented foods [17, 49]. Furthermore, it was also identified in the oral cavity [40], in human breast milk and in the vagina [27, 38]. Recently, L. fermentum was found in the fecal microbiota of Koreans from a village that is famous for the longevity of its inhabitants[46].
This microorganism was studied for several interesting applications and uses in recent years [43]. Different strains showed anti-infectious and immunomodulatory properties [61], cholesterol-reduction [53] on both mice [45] and human volunteers (strain ME3) [34], prevention of lactational mastitis in women during breastfeeding [31] and wound healing induction in animal models [5]. Moreover, it was also evaluated as antimicrobial agent against infections caused by Candida spp. [12, 18], Salmonella typhimurium [55] and Staphylococcus aureus [47]. Therefore, the industrial production of L. fermentum-based probiotic products is of great interest. However, very few scientific publications are available on the development of fermentation processes for the large-scale production [2, 19]. L. fermentum was demonstrated to grow on renewable feedstocks such as starch [11], whey [4], cereal-based substrates [13] and even anaerobic digester sludge [32]. Nevertheless, most studies and patents still use De Man, Rogosa and Sharpe (MRS) medium for bioreactor experiments [28, 33, 52] even if this medium is not well suited for commercial food or pharmaceutical production processes due to presence of animal derived components (beef extract and peptone). In fact, although several medicinal commonly used products are of animal origin, or rely on raw materials of animal origin, research is moving towards the identification of animal substitutes and the elimination of animal (and human) derived contaminants. Indeed, not only issues related to moral or religious commitments but also those regarding the risks due to the presence of prionic or viral particles, address biopharmaceutical product manufacturing to the avoidance of animal-derived supplements (https://www.ivtnetwork.com/article/animal-derived-ingredients-fda-and-regulations).
The aim of this study was the identification of a semi-defined medium deprived of ingredients of animal origin that could support growth of the newly identified L. fermentum strain isolated from buffalo milk, and to demonstrate scalability up to the pre-pilot scale. A simple and short fermentation process followed by biomass concentration and spray drying were also evaluated to implement a complete and industrially applicable biotechnological approach providing highly viable L. fermentum cells.
Since the EPS have been demonstrated to participate to beneficial effects of Lactobacilli on the gastric mucosa [26], a partial characterization of the EPS produced by the newly identified L. fermentum strain was carried out.
Moreover, due to the high prevalence rates of H. pylori infections in the adult population in industrialized countries, as well as in developing ones [29], and considering the ability of certain L. fermentum strains to improve immune response and resistance against infections caused by this pathogen [26], the ability of the newly isolated strain (produced with the developed process) to reduce the inflammation in adenocarcinoma gastric cells infected with H. pylori, was investigated in different experimental conditions.
2. Materials and methods
2.1. Bacterial strain and media
L. fermentum was isolated from buffalo milk and identified by molecular analysis and amplification of specific sequences [51] and stored at −80 °C in MRS broth. Twenty% v/v glycerol stock suspensions prepared with exponentially growing cells were stored at − 80 °C.
Before use for cell culture experiments, the strain was grown in MRS anaerobically at 37 °C for 24–48 h, centrifuged at 4.000 × g for 10 min, at 4 °C, washed twice with saline, resuspended at the concentration of 0.5 OD600 in DMEM without antibiotics and added to cultures.
Helicobacter pylori ATCC® 43,629™ was cultivated on Trypticase Soy Agar (Oxoid; Unipath, Basingstoke, UK). All medium components and salts were supplied by Sigma-Aldrich (St. Louis, MO, USA). Yeast extract was furnished by Organotechnie (La Corneuve, France), while sulphuric acid was purchased by Biochem s.r.l. (Turin, Italy). The different semi-defined media used for growth experiments are listed in table 1. Glucose to a final concentration of about 30 g/L, CaCl2 and Na2S were filter sterilised and added to all semi-defined media after autoclaving.
Table 1.
Composition of the semi-defined media used in 100 mL bottle experiments. a: Casein peptone; b: Bactocasitone; c: Soya peptone. d: Beef extract.
| Component (g/L) | MRS | Medium 1 | Medium 2 | Medium 3 | Medium 4 | Medium 5 |
|---|---|---|---|---|---|---|
| Yeast extract | 5 | 5 | 10 | 10 | 2.5 | 5 |
| Peptone | b*10 d*10 |
b* 10 | c* 10 | a* 7 | a* 5 c* 5 |
c* 10 |
| K2HPO4 | 2 | 2 | / | 2 | / | 3 |
| MgSO₄ * 7 H₂O | 0.1 | 0.1 | 0.25 | 0.2 | 0.1 | / |
| MnSO₄ * H₂O | 0.05 | 0.05 | 0.05 | 0.05 | / | / |
| Yeast nitrogen base | / | 5 | / | / | 5 | / |
| Na3C6H5O7 | / | 5 | 2 | / | / | / |
| Tween80 (mL/L) | 1 | 1 | 0.453 | / | / | / |
| C6H8O7 • 2NH3 | 2 | 2 | / | / | / | / |
| L-ascorbic acid | / | / | 0.5 | / | / | / |
| Sodium acetate | 5 | / | / | / | / | / |
| Di-sodium glycerophosphate | / | / | / | / | 19 | / |
| Ammonium sulfate | / | / | / | / | 2 | |
| CaCl2 * H2O | / | / | / | / | / | 0.2 |
| MgCl2 * 6 H2O | / | / | / | / | / | 0.2 |
| NaCl | / | / | 0.2 | / | / | 2 |
| Na2S *9H2O | / | / | / | / | / | 0.001 |
2.2. Bottle experiments
All experiments were performed in 100 mL screw-cap bottles with a working volume of 90 mL. Bottles were incubated at 37 °C and 150 rpm in a rotary shaker incubator (model Minitron, Infors, Bottmingen, Switzerland) for up to 24 h. Bottle inhibition tests were conducted on medium 2 supplemented with 20 g/L of glucose as carbon source. After sterilization, l-Lactic acid was added to reach a final concentration of 20, 40, 60, 80 and 100 g/L in the medium; before strain inoculation media were buffered with NaOH 20 M to a pH of 6.4. Samples were withdrawn every hour for 7 times to analyze optical density (600 nm) carbon source consumption, and acid and ethanol production. All bottle experiments were performed at least in duplicate.
Viability was evaluated by serially diluting the samples and plating on MRS-agar medium. Plates were incubated at 37 °C for 36 h before counting viable cells. Each sample was analysed in triplicate.
Viability data on the different semi-defined media and on medium 2 supplemented with different carbon sources were analysed by one-way Anova and following post-hoc Tukey comparisons.
2.3. Bioreactor experiments
Medium 2 was used for all bioreactor processes. Fermentation experiments were performed in a Biostat CT plus (Sartorius Stedim, Gottingen, Germany) bioreactor with a working volume of about 2.2 L. Temperature was controlled at 37 °C, pH at 6.1 and agitation was fixed at 150 rpm. Glucose at a concentration of about 10 to 50 g/L was used as carbon source. Before each experiment, a concentrated stock solution was inoculated in 0.45 L of medium 2 at 37 °C and 150 rpm and grown for 8 h. The pre-culture was then transferred to the bioreactor with a peristaltic pump (model 313 U, Watson-Marlow, England) to reach up to 10% (v/v) of the working volume inside the fermenter. The airflow was kept constant at 0.44 vvm and stirring was set to 150 rpm.Experiments lasted up to 24 h. The batch process was scaled on a Biostat D100 (Sartorius Stedim, Gottingen, Germany) with a working volume of 60 L. Process parameters were those previously described. The scale-up strategy was based on maintaining constant the power input and tip speed used on the 3 L scale. In particular, the equations used were:
| (1) |
| (2) |
Where P0(W) is the ungassed power; Np is the ungassed power number of the impeller (rushtone in this case); ρ is the broth density; N is the number of impellers and DI is the diameter of the impellers. Samples were withdrawn during the experiments to analyze optical density (600 nm), cell viability, carbon source consumption, and acid and ethanol production. Viability was evaluated as previously described.
2.4. HPLC quantification of sugars, organic acids, and ethanol
Samples of about 10 mL were withdrawn during the experiments. The broth was centrifuged at 10,000 × g to separate the biomass and recover the supernatant. One mL was then UF/DF on 3 kDa centrifugal filter devices (Centricon, Amicon, Sigma-Aldrich) at 10,000 × g and the permeate was analysed for the determination of the concentrations of carbon source, acids and ethanol by HPLC (UHPLC Dionex Ultimate 3000; Thermofisher) on a Alltech IOA-2000 column (500 mm × 6.5 mm ID) as previously reported [15].
2.5. Downstream process
The broth collected at the end of the batch process performed on the 150 L bioreactor was treated on a 0.22 µm hollow fiber module. Eleven liters of culture were filtered on 1.65 m2 (with 1250 PES fibers, 1 mm thick) to separate biomass and supernatant recirculating for about 1 h. The concentrated biomass was spray-dried, as described in the following paragraph. The permeate was ultrafiltrated on 10 kDa cut-off membranes with a filtering area of 0.1 m2 (Sartorius Stedim, Gottingen, Germany). Tangential flow filtration was performed on a Sartoflow alpha (Sartorius Stedim, Gottingen, Germany) system connected with a thermostatic bath that kept a constant temperature of about 20–25 °C. After the concentration step, 3 vol of diafiltration (VDF) with milliQ water were performed to remove low molecular weight molecules still attached to the membrane, and salts. The retentate was then precipitated with 2 vol of 1:1 ethanol/acetone solution and dried in vacuum oven at 40 °C over-night. The obtained powder was then resuspended in milliQ water and treated with 2% p/v activated charcoal. Precipitation was repeated as described above to obtain a powder for EPSs characterization.
2.6. Phenol sulfuric acid assay for exopolysaccharides quantification
Quantification of exopolysaccharides was performed by phenol sulfuric acid test [21], a colorimetric assay used for the determination of total carbohydrates in a sample. Pentoses through hydrolysis are dehydrated to furfural, and hexoses to hydroxymethylfurfural producing a yellow-gold color in presence of phenol. The calibration curve was obtained with standard solutions of D (+) glucose at concentrations ranging from 0.01 to 0.1 mg/mL. Briefly, 200 μL of standards were placed in a reaction tube with 200 μL of aqueous solution of phenol 5% w/v. Then, 1 mL of concentrated sulfuric acid (98% w/w) was added, and the reaction tube was quickly closed and after vigorous stirring, the reaction was carried out for 30 min at 30 °C. Sample absorbance was read at 490 nm using distilled H2O as blank.
2.7. Spray dryer
After microfiltration the concentrated biomass (about 1.7 L) was diafiltered with 2 vol (3.4 L) of sterile phosphate buffer solution (PBS) and after addition of trehalose and sucrose it was spray-dried on a Mobile minor™ (Gea Process Engineering, Düsseldorf, Germany). Based on the theoretical wet to dry biomass ratio, a 1:1 ratio between dry biomass and total sugars was maintained. The sample was fed to the spray dryer at 2.4 L/min, with a peristaltic pump (model 730 U, Watson-Marlow, England). The inlet temperature was set at 165 °C, outlet temperature was 85 °C, while atomizer pressure was set to 1 bar.
2.8. Cell cultures
Human gastric adenocarcinoma cell-line AGS cells ATCC® CRL-1739™ (American Type Culture Collection, Rockville, MD, USA), were routinely cultured in Ham's F-12 K medium (Gibco, Waltham, Massachusetts,USA) supplemented with 1% (v/v) Penstrep, 1% (v/v) glutamine and 10% (v/v) fetal calf serum (Gibco) at 37 °C and 5% CO2.
2.9. Cell infection with H. pylori and/or L. fermentum
The ability of L. fermentum to reduce the inflammation in AGS cells after infection with H. pylori, was investigated in three different experimental types: (i) Competitive assay, in which AGS cells (105) were incubated simultaneously for with L. fermentum (108 CFU/mL) and H. pylori (108 CFU/mL) for 2 h; (ii) Inhibition assay, in which AGS cells (105 cells) were preincubated with L. fermentum (108 CFU/mL) for 1.5 h and then H. pylori (108 CFU/mL) was added and incubated for 2 h; (iii) Displacement assay in which AGS cells were pre-incubated with H. pylori (108 CFU/mL) for 2 h and then L. fermentum (108 CFU/mL) was added and further incubated for 1.5 h.
2.10. Evaluation of proinflammatory genes expression
At the end of cell culture experiments, to evaluate the expression of pro- and anti-inflammatory cytokines, the cells were washed three times with sterile PBS, and the total RNA was extracted using High Pure RNA Isolation Kit (Roche Diagnostics, Monza, Italy). Two hundred nanograms of total cellular RNA were reverse-transcribed (Expand Reverse Transcriptase, Roche Diagnostics) into complementary DNA (cDNA) using random hexamer primers (Random hexamers, Roche Diagnostics) at 42 °C for 45 min, according to the manufacturer's instructions. Real time PCR for IL-6, IL-8, TNF-α, IL-1α, IL-1β, TGF-β and HBD-2 was carried out with the LC Fast Start DNA Master SYBR Green kit using 2 µL of cDNA, corresponding to 10 ng of total RNA in a 20 mL final volume, 3 mM MgCl2 and 0.5 mM sense and antisense primers (Table 2). After amplification, melting curve analysis was performed by heating to 95 °C for 15 s with a temperature transition rate of 20 °C/s, cooling to 60 °C for 15 s with a temperature transition rate of 20 °C/s, and then heating the sample at 0.1 °C/s to 95 °C. The results were then analysed using LightCycler software (Roche Diagnostics). The standard curve of each primer pair was established with serial dilutions of cDNA. All PCR reactions were run in triplicate. The specificity of the amplification products was verified by electrophoresis on a 2% (w/v) agarose gel and visualization by ethidium bromide staining. Significant differences among groups were assessed by two-way ANOVA using GraphPad Prism 6.0, and the comparison between the means was calculated by t-student test. The data are expressed as means ± standard deviation (SD) of three independent experiments.
Table 2.
Primer sequences and amplification programs used for gene expression studies.
| Gene | Primers sequence | Conditions | Product size (bp) |
|---|---|---|---|
| IL-6 | 5′-ATGAACTCCTTCTCCACAAGCGC-3′ 5′-GAAGAGCCCTCAGGCTGGACTG-3′ |
5′’at 95 °C, 13″ at 56 °C, 25′’at 72 °C for 40 cycles | 628 |
| IL-8 | 5′-ATGACTTCCAAGCTGGCCGTG-3′ 5′-TGAATTCTCAGCCCTCTTCAAAAACTTCTC-3′ |
5′’at 94 °C, 6″ at 55 °C, 12′’at 72 °C for 40 cycles | 297 |
| IL-1β | 5′-GCATCCAGCTACGAATCTCC-3′ 5′-CCACATTCAGCACAGGACTC-3′ |
5′’at 95 °C, 14″ at 58 °C, 28′’at 72 °C for 40 cycles | 708 |
| TGF-β | 5′-CCGACTACTACGCCAAGGAGGTCAC-3′ 5′-AGGCCGGTTCATGCCATGAATGGTG-3′ |
5′’at 94 °C, 9″ at 60 °C, 18′’at 72 °C for 40 cycles | 439 |
| IL-1α | 5′- CATGTCAAATTTCACTGCTTCATCC-3′ 5′- GTCTCTGAATCAGAAATCCTTCTATC −3′ |
5″at 95 °C, 8″at 55 °C, 17″at 72 °C for 45 cycles | 421 |
| HBD-2 | 5′-GGATCCATGGGTATAGGCGATCCTGTTA-3′ 5′-AAGCTTCTCTGATGAGGGAGCCCTTTCT-3′ |
5″at 94 °C, 6″at 63 °C, 10″at 72 °C for 50 cycles | 198 |
| TNF-α | 5′- CAGAGGGAAGAGTTCCCCAG −3′ 5′- CCTTGGTCTGGTAGGAGACG −3′ |
5′’at 95 °C, 6″ at 57 °C, 13′’at 72 °C for 40 cycles | 324 |
2.11. ELISA assay
Supernatants of AGS cells infected as described aboveat the end of the experiment were harvested and the presence of cytokines IL-6, IL-8, IL-1α and HBD-2 was analysed by enzyme-linked immunosorbent assay (ELISA; ThermoFischer Scientific Inc., Waltham, Massachusetts,USA; Phoenix Pharmaceuticals, Burlingame, USA). Statistical analyses were performed as described in the previous paragraph.
2.12. Scanning electron microscope analysis
AGS cells (105) were infected with H. pylori or L. fermentum alone, both at 108 CFU/mL for 2 h and after incubation fixed in 4% v/v paraformaldehyde in PBS. Samples were then dehydrated by washing in increasing ethanol concentrations (30% to 95% for 10 min, and 100% 3 times for 15 min). Immediately afterwards the samples were further dried in the EMITECH K850 critical point dryer, then stutter coated in Denton Vacuum DESKV sputter coater with platinum – palladium target, at 77 mAmps for 120 s. The samples thus prepared were observed using a Fe-SEM Supra 40 field-emission scanning electron microscope, Zeiss, Germany (EHT = 5.00 kV, WD = 22 mm, Inlens detector).
2.13. Monosaccharide analysis and permethylation of EPS
Monosaccharide compositional analysis (acetylated methyl glycosides) was performed as reported elsewhere [3]. Monosaccharide derivatives were recognized based on their GC–MS spectrum fragmentation pattern and by comparison of their retention time with that of authentic standards.
A sample (1 mg) of the EPS was permethylated using methyl iodide and sodium hydroxide [14] as already reported [25]. After hydrolysis with 2 M TFA (1 h, 120 °C), the partially methylated monosaccharides were reduced with NaBD4. After the usual work-up, the sample was submitted to acetylation with acetic anhydride and pyridine 1:1 (30 min, 100 °C), and the mixture of partially methylated alditol acetates was analysed by GC–MS.
2.14. NMR spectroscopy
1H and 2D NMR spectra of the EPS from L. fermentum were recorded in deuterated water (D2O) using a Bruker 600 MHz instrument equipped with a cryoprobe at 298 K.
3. Results
3.1. Medium optimization and acid inhibition test
Initial bottle experiments were performed with 90 mL of each medium to evaluate viability and optical density. Results are showed in Fig. 1.
Fig. 1.
Small scale experiments performed in 100 mL bottles at 37 °C and 150 rpm. a) Optical density and concentration of viable cells obtained on different semi-defined media compared to MRS; b) Optical density and concentration of viable cells obtained on medium 2 with different carbon sources. Viability data were analysed by Anova and post hoc Tukey comparison; data significance was indicated as *p<0.05, **p<0.01. ***p<0.001.
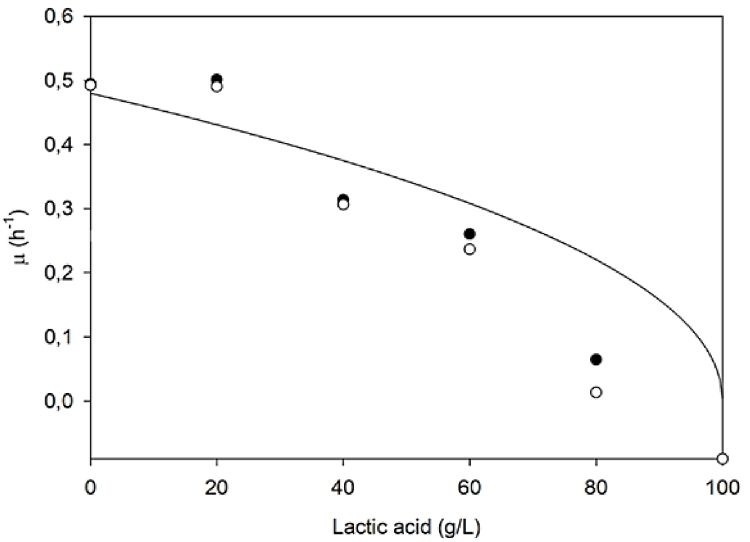
Different media were tested to identify those that better supported biomass production. Various nitrogen sources and their combinations were tested together with different C:N ratios. Glucose at a fixed concentration of about 30 g/L was chosen for these trials. Viability data were analysed by one-way Anova with post-hoc Tukey comparisons showing significantly higher concentrations of viable cells on medium 2 (p<0.001) compared to results obtained on the control medium (MRS). A maximum viability of about 9.3 ± 1.3 × 108 CFU/mL was reached after 16 h of growth, that corresponded to about 1.35±0.01 g/L of dry cell biomass and an optical density of about 4.5 ± 0.1 OD600 (Fig. 1a). This medium was then chosen for further experiments, and it was supplemented with different carbon sources at the same initial concentration (Fig 1b). L. fermentum reached the same final OD on media containing maltose and glucose however, the latter showed a 38% higher number of viable cells after 16 h of growth and was therefore selected for bioreactor experiments. Finally, bottle experiments on the same medium supplemented with increasing concentrations of lactic acid, were also performed. Growth was followed during the first 7 h and the specific growth rate (μ) was calculated as linear regression of OD measurements.
Luong-model [39] exponential inhibition Eq. (3) showed the best fit to experimental data:
| (3) |
where μmax is the maximum specific growth rate calculated in the absence of lactic acid, P is the concentration of product (lactic acid), Pmax is the critical product concentration (when P = Pmax, μ = 0) and np is the inhibition constant (for non-competitive inhibition np > 0). We found a Pmax of 100 g/L and a μmax equal to 0.59 h − 1 determined experimentally (Fig. 2). A regression coefficient (R2) equal to 0.93 was found analysing data.
Fig. 2.
Product inhibition. Evaluation of lactic acid inhibition on the growth of L. fermentum in bottle experiments with an initial glucose concentration of 30 g/L, pH 6.5 and temperature of 37 °C. The experiment was run in duplicate.
3.2. Fermentation experiments
All fermentations were performed on medium 2 under controlled conditions of temperature, pH and aeration rate. Batch processes were initially performed with and without air sparging (0.44 vvm), and slightly better results were obtained in the presence of air (data not shown), therefore this condition was used in all further experiments. Initial batch processes with glucose at different concentrations (10, 30 and 50 g/L) were tested and summarized in table 3. During the first 8 h growth rate, glucose consumption and lactic acid production were evaluated. Batch processes with 30 g/L of initial glucose demonstrated the highest CFU/mL of about 1 × 109 ± 0.14 after 8 h of growth. This value did not change after 16 h, however it decreased to 7.4 × 109 after 24 h of growth (supplementary Fig 1).
Table 3.
Fermentation experiments performed on Biostat CT plus (3 L) and Biostat 100 (150 L) bioreactors.
| Process | OD max (600 nm) | Viability (CFU/mL) | Dry weight (g/L) | LA (g/L) | Et (g/L) | Yx/s (g/g) | YLA/x (g/g) | YEt/x (g/g) | EPS (glucose equivalents) (mg/mL) |
|---|---|---|---|---|---|---|---|---|---|
| Batch | |||||||||
| 10 g/L | 5.1 ± 0.4 | 3.6 ± 0.6 × 108 | 1.7 ± 0.1 | 6.5 ± 0.7 | 3.0 ± 0.6 | 0.17±0.04 | 3.8 ± 0.1 | 1.7 ± 0.2 | n.d |
| 30 g/L | 6.3 ± 0.3 | 1.0 ± 0.1 × 109 | 2.4 ± 0.3 | 19.5 ± 1.3 | 6.7 ± 1.4 | 0.09±0.01 | 8.3 ± 1.3 | 2.8 ± 0.7 | 0.450±0.040 |
| 50 g/L | 7.7 ± 1.5 | 6.9 ± 1.7 × 108 | 2.7 ± 0.5 | 26.8 ± 1.5 | 9.1 ± 1.1 | 0.07±0.001 | 9.8 ± 1.1 | 3.4 ± 0.2 | 0.630±0.006 |
| Batch_P | 7.3 ± 0.2 | 6.1 ± 0.9 × 108 | 2.7 ± 0.3 | 34.5 ± 0.7 | 9.6 ± 0.8 | 0.05±0.006 | 12.8 ± 1.7 | 3.6 ± 0.7 | 0.400±0.004 |
| Scale up | |||||||||
| Batch 150L | 6.8 | 8.0 × 108 | 2.6 | 21.4 | 8.2 | 0.10 | 8.2 | 3.1 | 0.47 |
EPS, exopolysaccharides; Batch_P, batch with pulse; LA, lactic acid; Et, ethanol.
Table 3 summarises bioreactor experiments in batch with pulse and batch mode. The highest concentration of viable cells was obtained during batch experiments with an initial concentration of glucose equal to 30 g/L. An Yx/s of about 0.17 g/g was obtained in the batch experiment with a starting concentration of about 10 g/L of glucose and it gradually decreased with 30 and 50 g/L of glucose initially in the medium, and when about 50±5 g/L of glucose were provided by adding a concentrated pulse after the first 8 h of growth (batch with pulse, Table 3). On the other hand, production of LA and ethanol proportionally increased with higher glucose concentrations in the medium, as indicated by higher YLA/x in the different experiments.
Batch processes with 30 g/L of glucose and an almost doubled amount of nitrogen sources (20 g/L yeast extract and 15 g/L soy peptone) led to a final concentration of viable cells of about 1.52±0.26 × 109 and an Yx/s of about 0.13±0.02 g/g (, ODmax 9.8 ± 1.0; LA 18.0 ± 2.0 g/L; Et 5.6 ± 1.5 g/L).
The batch process on medium 2 containing 30 g/L of glucose was then run on a Biostat D100 Sartorius bioreactor with 60 L of working volume, according to the scale-up strategy described in the materials and methods section. As shown in Table 3 all data were in line with those obtained on the 2 L scale. A slightly lower viability was obtained on the 150 L scale at the end of the fermentation (16 h), however, the value was within the variability interval observed on the 3 L scale.
The EPSs produced by L. fermentum and secreted in the broth were quantified at the end of all fermentation processes and results are reported in table 3. Higher concentrations of EPSs were obtained in batch processes with 50 g/L of glucose, whereas no polysaccharide could be detected when only 10 g/L of carbon source were present the medium.
3.3. Concentration and spray drying of probiotic biomass
Eleven L of broth were microfiltered and diafiltered with PBS in about 2 h. The initial flux was 40 LHM and it decreased by about 4-fold at the end of the process due to cake formation. Overall, the microfiltration and diafiltration steps yielded a 6.5-fold concentrated bacterial retentate, containing about 30 g of dry biomass. After addition of trehalose and sucrose the concentrated biomass was spray dried resulting in a survival of about 60% after drying. The recovered biomass showed a viability of 5.7 ± 1.2 × 1010 CFU/g of powder.
3.4. Chemical characterization and NMR study of EPSs
The glycosyl analysis of the recovered and partially purified EPSs indicated that the polysaccharides contained, as main constituents, mannose, galactose, glucose, and arabinose, as revealed by the comparison between the GC–MS chromatogram of standards with that of the isolated polymer (Fig. 3).
Fig. 3.
GC–MS chromatograms of acetylated methyl glycosides of EPS from L. fermentum (a), and standard sugars (b-e). Ara, arabinose; Man, mannose; Gal, galactose; Glc, glucose; Fuc, fucose; Rha, rhamnose; All, allose; Rib, ribose; GalA, galacturonic acid; Xyl, xylose; GlcA, glucuronic acid.
In addition, 1H and 2D 1H, 13C DEPT-HSQC NMR experiments were obtained (Fig. 4).
Fig. 4.
a) 1H NMR spectrum of L. fermentum EPS. b) DEPT-HSQC NMR experiment of L. fermentum EPS. The spectra were recorded in D2O at 298 K, at 600 MHz.
In the anomeric region of both spectra, many signals appeared, suggesting a mixture of exopolysaccharides. However, a more accurate analysis of the DEPT-HSQC experiment revealed some signals suggesting a mixture of at least two polysaccharides, one of which is a mannan whereas the other could be identified as a heteropolysaccharide (HePS). The structure of the last could be very similar to the neutral polysaccharide isolated from Lactobacillus delbrueckii ssp. bulgaricus LBB.B26 [50]. In fact, the carbon signal at δ 110 ppm correlating in the DEPT-HSQC spectrum with the anomeric proton at δ 5.18 ppm, together with signals of carbon nuclei in the range of δ 80–85 ppm could be assigned to galactofuranose units, due to the typical downfield shift of these signals [8]. A methylation analysis on the EPSs mixture, indicated terminal glucose, terminal galactose, terminal mannose, terminal arabinose, 4-substituted glucose, 6-substituted mannose, and 3,6-substituted galactose units. Some of these units could belong to the HePS, thus confirming the previous hypothesis. Finally, albeit the hexose units are frequent in lactobacilli EPS [12], the presence of arabinose has been detected and associated to bacterial stress resistance [44].
3.5. Effect of L. fermentum on H. pylori
The data obtained (Fig. 5a, b and c) show that L. fermentum can almost completely inhibit the inflammatory state strongly induced by H. pylori in the displacement assay. In fact, the genes encoding proinflammatory cytokines IL-1α, IL-6 and IL-8 are strongly downregulated in the presence of L. fermentum compared to infection with H. pylori alone. IL-1β, TGF-β and TNF-α were unmodulated. Therefore, the probiotic, which by itself can reduce the basal expression of proinflammatory molecules and increase antimicrobial defenses by inducing the expression of HBD-2, may improve the conditions of the gastric mucosa when damaged by H. pylori.
Fig. 5.
Real-Time PCR (a) and ELISA (b and c) show the expression levels of proinflammatory cytokines and HBD-2 in AGS cells infected with H. pylori and/or L. fermentum. Data are expressed as relative mRNAs expression (A) and protein concentration (C) in each group and are representative of three different experiments ± SD. Significant differences are indicated by *p<0.05, **p<0.01, ***p<0.001.
3.6. Effects of H. pylori and L. fermentum on the gastric epithelium morphology
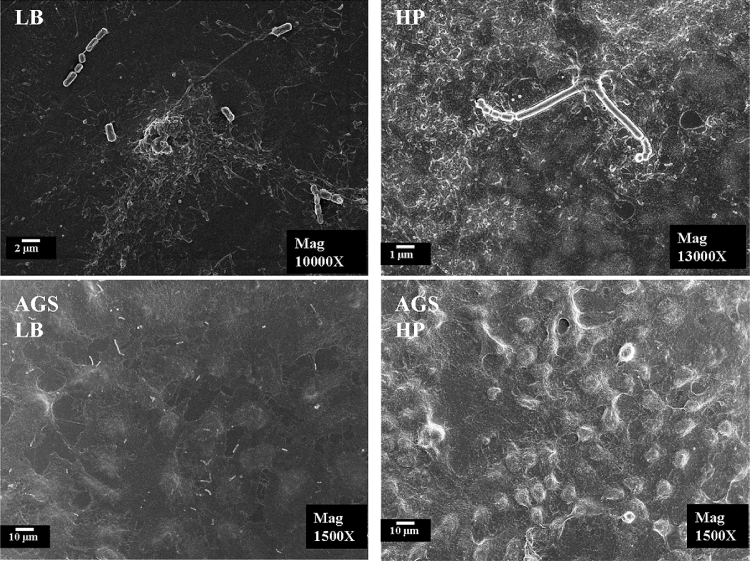
Scanning electron microscope observation at high magnification (10–13 KX) revealed the typical rod-like shape of L. fermentum (Fig. 6 LB panel), while H. pylori shows the characteristic gull-wing shape (Fig. 6 HP panel). Low magnification images (1500X) demonstrate different cell morphologies: L. fermentum doesn't alter the flattened physiological aspect of the cell monolayer, while the presence of the pathogen causes cell surface ruffles and loss of flattened shape.
Fig. 6.
Microorganism and co-culture micrograph obtained by scanning electron microscopy. LB, L. fermentum; HP, H. pylori; AGS and LB or HP indicates co-culture. Different magnifications highlight bacteria or cells morphology.
4. Discussion
L. fermentum is a widely studied probiotic, however, distinct strains often differ in terms of physiology and biological properties [6, 11, [35], [36], [37], 42, 48, 54, 57, 60]. The aim of this study was to develop an improved fermentation process and demonstrate its suitability for scale-up and subsequent concentration, including the drying process, for the potential industrial production of viable biomass by using a L. fermentum strain, that was newly isolated from buffalo milk, as test example. The work focused on the identification of an alternative to the commonly used MRS medium, that could support growth up to the pilot scale, despite the absence of complex animal derived constituents. In fact, regulatory (safety), ethical and religious issues increasingly limit the use of ingredients of animal origin and steer product manufacturing towards the search for safer solutions.
A very critical point regarding the potential commercial use of probiotics is the delivery of an adequate number of viable cells in the target area. This is affected by the survival rate during the manufacturing process, and during transit in the gastrointestinal tract. Therefore, it is quite important to obtain high densities of viable cells during fermentation processes and further processing.
Media with different composition were tested in small scale bottle experiments. The elimination of beef extract and the replacement of bactocasitone or casein, which are typical but animal derived complex nitrogen sources, with soy peptone in medium 2 were demonstrated to be superior with respect to achieved biomass concentration as well as viability. Higher concentration of YE, as well as lower C:N ratios, and the presence of additional elements for growth (eg. MgSO4, NaCl, ascorbic acid, sodium citrate) (Wayah and Philip. 2018) in this medium compared to medium 1, 4 and 5 greatly improved fermentation performance. This not only shows that it is possible to eliminate and/or replace animal derived media components, but that these traditionally used media are clearly not optimal for commercial manufacturing processes of L. fermentum and most likely also of other LAB. Among all carbon sources tested glucose and maltose mostly improved cell density, however the latter yielded a significantly lower concentration of viable cells (two tailed non homoscedastic, t-student p<0.00001, Anova p<0.05). In a different study [10, 57] L. fermentum Ogi E1 also showed similar cell densities on these two substrates, whereas compared to the results presented in this study L. fermentum GA715 showed lower biomass concentrations. In general, both strains showed a different ranking for the same carbon sources, which together with our results indicate a high diversity on the efficiency for using different carbon sources within the same species.
Fermentation process duration is a key aspect of industrial applications. Preliminary 24 h bottle and bioreactor experiments demonstrated that the maximum concentration of viable cells was obtained after 8 h of growth (it remained constant until 16 h, whereas it greatly decreased after 24 h of growth). These data indicate the possibility to shorten the process, thereby greatly improving the overall productivity.
Therefore, different initial concentrations of glucose (10, 30, 50 g/L) were next evaluated in 3 L bioreactors, in batch mode, with 8 h processes. With up to 30 g/L of glucose in the medium viability increased, whereas no further improvement was observed with higher substrate concentrations. Interestingly, a decreasing Yx/s trend and an increase of YLA/x and YEt/x, were observed with higher initial glucose concentrations (10, 30, 50 g/L). Even splitting substrate feeding (25 g/L initially in the medium and 25 g/L at the end of the exponential phase) did not improve cell viability, suggesting a major carbon flux towards LA and ethanol instead of biomass. Considering an average bacterial biomass elemental composition equal to CH1.66N0.20O0.27−—CH2N0.24 O0.33 [7], and nitrogen content in medium 2, the moles of N needed to support the production of biomass in the presence of 30 and 50 g/L of glucose might have been insufficient, generating a nitrogen limitation condition. For this reason, batch processes with 30 g/L of glucose and an 80% higher total nitrogen concentration were performed. Results showed an improvement of the number of viable cells of about 50% (two tailed non homoscedastic t-student, p<0.005) and a 39% improvement of the Yx/s, probably indicating the need of additional N source to further support biomass production and slightly decrease lactic acid secretion. These data are also in accordance with results obtained with L. fermentum Ogi E1 on starch and yeast extract, since the authors found that increasing the concentration of YE in the medium above 20 g/L (Cg:Ng equal to 1:1) did not improve biomass production [11]. Since, as indicated for the first time in this study, L. fermentum tolerates high LA concentrations (Pm=100 g/L) as other LAB, strategies that allow administration of higher amounts of carbon source should be investigated to maximize biomass production.
One of the necessary technological characteristics of probiotics is their “ability to be produced at large scale” [23]. Therefore, the development of biotechnological processes up to the pilot scale and the use of a suitable drying process are important steps to industrial application. Considering the lack of literature on this aspect, the batch process was scaled to the 150 L reactor. Considering the different reactor geometry of the 3 and 150 L fermenter, a constant power input and tip speed were maintained in the two reactor set-ups. This approach allowed to obtain very similar results to that observed on lab scale experiments. Further downstream processing was performed to simulate a complete manufacturing cycle. Biomass concentration on hollow fibers was the most critical point of the downstream process [4]. Clogging of the membrane, either due to production of EPS, or to the lumen of the hollow fibers, allowed processing of about 11 L of broth. The low volume caused a partial loss of dried sample in the spray-drier chamber thereby reducing the final yield of recovered powder. However, this problem can be avoided on industrial scale by increasing the surface area of filtration modules and the volume of spray dried sample. Among the available drying techniques, spray-drying is one of the most predominant in the dairy industry since it guarantees lower energy costs and higher sustainability [30]. However, due to the high inlet and outlet temperatures, and considering the sensitivity of probiotics, variable tolerance among different strains and operating conditions is observed [30]. Notwithstanding the stressful microfiltration treatment, the L. fermentum strain isolated in this work showed 60% survival after spray-drying, demonstrating a good resistance to harsh conditions that prevail during the process.
The partially purified EPSs isolated from the supernatant of L. fermentum cultures were subjected to a partial characterization by GC–MS and NMR spectroscopy. Results indicated that the EPS fractions contained a mixture of a homopolysaccharide, such as a mannan, and a heteropolysaccharide. The last resulted to be very similar to that already characterised from another strain of Lactobacillus [50], due to the distinguishable signals in the NMR experiments of the galactofuranose units and to the identification of the same attachment points reported for the neutral polysaccharide from L. delbrueckii in the methylation analysis. The detection of mannan could be due to the presence of residues of medium components (yeast extract) as often previously found [9, 24], whereas the arabinose content is quite rare in lactobacilli [44].
In healthy individuals, the gastric epithelium, owing to the shape and polarization of its cells and to cell-cell and cell-matrix adhesions, represents the first barrier of defense against pathogens. H. pylori, colonizing the gastric mucus, manages to disassemble this epithelial barrier and induce an inflammatory state that can sometimes lead to the onset of neoplastic changes [58]. Clinical treatment consists in the standard triple therapy (lansoprazole, clarithromycin, and metronidazole), which may cause serious side effects and increased antibiotic resistance [1, 56]. The use of Lactobacillus spp. instead of antibiotics could avoid this drawback [22] and L. fermentum is interesting in this respect. In fact, recent studies demonstrated the ability of L. fermentum UCO-979C to improve immune response and resistance against infections caused by Helicobacter pylori [26]. The L. fermentum strain newly isolated in this work was evaluated during H. pylori infection to demonstrate whether it could improve the inflammatory state of the intestinal epithelium.
For this purpose, competition, inhibition, and displacement assays in which AGS cells were infected with L. fermentum or H. pylori alone, or co-infected at different times, were conducted. The expression of proinflammatory cytokines, soluble mediators of natural immunity and the immune response [41], and of Human β-defensin-2 (HBD-2), inducible antimicrobial peptide active against Gram-positive and Gram-negative bacteria, fungi, and the envelope of some viruses, and involved in the innate immune response [20], was evaluated.
Results showed that L. fermentum by itself can increase the production of HBD-2 by gastric epithelial cells, which is of great importance as it induces the enhancement of antimicrobial defenses and mechanisms of innate immunity; it was also shown to have a strong anti-inflammatory effect by downregulating the expression of proinflammatory cytokines in the displacement assay, therefore following infection with H. pylori.
The neoplastic changes induced by H. pylori are caused mainly by a rearrangement of the actin filaments of the cytoskeleton, following the formation of protrusions and massive stress fibers in gastric epithelial cell cultures that destroy cell-cell junctions, altering cell morphology [59].
In a second set of experiments, it was verified that L. fermentum, unlike H. pylori, does not induce any morphological changes in the structure of the gastric epithelium as shown by SEM analysis.
Differently from H. pylori, L. fermentum manages to interact with the gastric epithelial cells, even at the level of the cell-cell junctions, preserving their structure.
5. Conclusions
This work presents a comprehensive approach towards the development of a potential probiotic production process demonstrating the suitability of the newly isolated L. fermentum strain to upstream and downstream process development. Exopolysaccharides have been partially purified showing peculiar structural features as found by NMR analyses.
Moreover, the biological activity of the newly isolated L. fermentum strain was evaluated in a gastric epithelial cell model demonstrating defensin upregulation and H. pylori inhibition, also modulating inflammatory cytokines.
Author contributions
DC, SD and AF drafted the manuscript; SD, MV and DC conducted fermentation experiments; SD conducted downstream processing; AD, SD and MV performed cell viability counts; AF conducted biological assays; AC and MMC characterized the EPSs and drafted the related manuscript sections; MC performed SEM experiments; DC, CS and GD conceived the study.
Supplementary files
Supplementary figure 1- Viability monitoring during growth in bottle and batch experiments at different time points.
Declaration of Competing Interest
The authors declare non conflict of interest.
Acknowledgements
The work was funded by the “Ministero delle Infrastrutture e dello Sviluppo Economico” project “Integratori Innovativi per l'infiammazione” (Incube).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.btre.2022.e00732.
Appendix. Supplementary materials
References
- 1.Adachi T., Matsui S., Watanabe T., Okamoto K., Okamoto A., Kono M., Yamada M., Nagai T., Komeda Y., Minaga K., Kamata K., Yamao K., Takenaka M., Asakuma Y., Sakurai T., Nishida N., Kashida H., Kudo M. Comparative study of clarithromycin- versus metronidazole-based triple therapy as first-line eradication for helicobacter pylori. Oncology. 2017;93:15–19. doi: 10.1159/000481224. [DOI] [PubMed] [Google Scholar]
- 2.Alfano A., Donnarumma G., Cimini D., Fusco A., Marzaioli I., De Rosa M., Schiraldi C. Lactobacillus plantarum: microfiltration experiments for the production of probiotic biomass to be used in food and nutraceutical preparations. Biotechnol. Prog. 2015;2:325–333. doi: 10.1002/btpr.2037. [DOI] [PubMed] [Google Scholar]
- 3.Alfano A., Perillo F., Fusco A., Savio V., Corsaro M.M., Donnarumma G., Schiraldi C., Cimini D. Lactobacillus brevis CD2: fermentation strategies and extracellular metabolites characterization. Probiotics Antimicrob. Proteins. 2020;12:1542–1554. doi: 10.1007/s12602-020-09651-w. [DOI] [PubMed] [Google Scholar]
- 4.Aragón-Rojas S., Quintanilla-Carvajal M.X., Hernández-Sánchez H. Multifunctional role of the whey culture medium in the spray drying microencapsulation of lactic acid bacteria. Food Technol. Biotechnol. 2018;3:381–397. doi: 10.17113/ftb.56.03.18.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashoori, Y., Mohkam, M., Heidari, R., Abootalebi, S.N., Mousavi, S.M., Hashemi, S.A., Golkar, N., Gholami, A. (2020) Development and in vivo characterization of probiotic lysate-treated chitosan nanogel as a novel biocompatible formulation for wound healing. BioMed Res. Int. 8868618. 10.1155/2020/8868618. [DOI] [PMC free article] [PubMed]
- 6.Aziz K., Haseeb Zaidi A., Fatima H.N., Tariq M. Lactobacillus fermentum strains of dairy-product origin adhere to mucin and survive digestive juices. J. Med. Microbiol. 2019;68:1771–1786. doi: 10.1099/jmm.0.001090. [DOI] [PubMed] [Google Scholar]
- 7.Bailey J.E., Ollis D.F. McGraw-Hill Chemical Engineering Series. 2nd ed. McGraw-Hill; New York, NY, USA: 1986. Biochemical engineering fundamentals. ISBN 9780070032125. [Google Scholar]
- 8.Bock K., Pedersen C. Carbon-13 nuclear magnetic resonance spectroscopy of monosaccharides. Adv. Carbohydr. Chem. Bio-chem. 1983;41:27–66. [Google Scholar]
- 9.Butorac K., Novak J., Bellich B., Terán L.C., Banić M., Leboš Pavunc A., Zjalić S., Cescutti P., Šušković J., Kos B. Lyophilized alginate-based microspheres containing Lactobacillus fermentum D12, an exopolysaccharides producer, contribute to the strain's functionality in vitro. Microb. Cell. Fact. 2021;20(1):85–101. doi: 10.1186/s12934-021-01575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calderon Santoyo M., Loiseau G., Rodriguez Sanoja R., Guyot J.P. Study of starch fermentation at low pH by Lactobacillus fermentum Ogi E1 reveals uncoupling between growth and alpha-amylase production at pH 4.0. Int. J. Food Microbiol. 2003;1:77–87. doi: 10.1016/s0168-1605(02)00140-x. [DOI] [PubMed] [Google Scholar]
- 11.Calderon M., Loiseau G., Guyot J.P. Nutritional requirements and simplified cultivation medium to study growth and energetics of a sourdough lactic acid bacterium Lactobacillus fermentum Ogi E1 during heterolactic fermentation of starch. J. Appl Microbiol. 2001;90:508–516. doi: 10.1046/j.1365-2672.2001.01272.x. [DOI] [PubMed] [Google Scholar]
- 12.Castro-Bravo N., Wells J.M., Margolles A., Ruas-Madiedo P. Interactions of surface exopolysaccharides from bifidobacterium and lactobacillus within the intestinal environment. Front. Microbiol. 2018;9:24–26. doi: 10.3389/fmicb.2018.02426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charalampopoulos D., Pandiella S.S., Webb C. Growth studies of potentially probiotic lactic acid bacteria in cereal-based substrates. J. Appl. Microbiol. 2002;92:851–859. doi: 10.1046/j.1365-2672.2002.01592.x. [DOI] [PubMed] [Google Scholar]
- 14.Ciucanu I., Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohyd. Res. 1984;131:209–217. doi: 10.1016/0008-6215(84)85242-8. [DOI] [Google Scholar]
- 15.D'ambrosio S., Alfano A., Cassese E., Restaino O.F., Barbuto Ferraiuolo S., Finamore R., Cammarota M., Schiraldi C., Cimini D. Production and purification of higher molecular weight chondroitin by metabolically engineered Escherichia coli K4 strains. Sci. Rep. 2020;10:13–20. doi: 10.1038/s41598-020-70027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Degnan F.H. The US food and drug administration and probiotics: regulatory categorization. Clin. Infect. Dis. 2008;46:S133–S151. doi: 10.1086/523324. [DOI] [PubMed] [Google Scholar]
- 17.Di Cagno R., Surico R.F., Siragusa S., De Angelis M., Paradiso A., Minervini F., De Gara L., Gobbetti M. Selection and use of autochthonous mixed starter for lactic acid fermentation of carrots, French beans or marrows. Int. J. Food Microbiol. 2008;127:220–228. doi: 10.1016/j.ijfoodmicro.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 18.do Carmo M.S., Noronha F.M., Arruda M.O., Costa Ê.P., Bomfim M.R., Monteiro A.S., Ferro T.A., Fernandes E.S., Girón J.A., Monteiro-Neto V. Lactobacillus fermentum ATCC 23271 displays in vitro inhibitory activities against candida spp. Front. Microbiol. 2016;7:17–22. doi: 10.3389/fmicb.2016.01722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donnarumma G., Molinaro A., Cimini D., De Castro C., Valli V., De Gregorio V., De Rosa M., Schiraldi C. Lactobacillus crispatus L1: high cell density cultivation and exopolysaccharide structure characterization to highlight potentially beneficial effects against vaginal pathogens. BMC Microbiol. 2014;14:137–148. doi: 10.1186/1471-2180-14-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnarumma G., Paoletti I., Fusco A., Perfetto B., Buommino E., de Gregorio V., Baroni A. β-Defensins: work in progress. Adv. Exp. Med. Biol. 2016;901:59–76. doi: 10.1007/5584_2015_5016. [DOI] [PubMed] [Google Scholar]
- 21.Dubois M., Gilles K.A., Hamilton J.K., Rebers P.A., Smith F. Colorimetric method for determination of sugars and related substance. Anal. Chem. 1956;28(3):350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- 22.Fekete T. ACP journal club. Review: probiotics reduce clostridium difficile-associated diarrhea in patients receiving antibiotics. Ann. Intern. Med. 2013;10:158–167. doi: 10.7326/0003-4819-158-10-201305210-02010. [DOI] [PubMed] [Google Scholar]
- 23.Forssten S.D., Sindelar C.W., Ouwehand A.C. Probiotics from an industrial perspective. Anaerobe. 2011;17(6):410–413. doi: 10.1016/j.anaerobe.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Fraunhofer M.E., Geissler A.J., Wefers D., Bunzel M., Jakob F., Vogel R.F. Characterization of β-glucan formation by Lactobacillus brevis TMW 1.2112 isolated from slimy spoiled beer. Int. J. Biol. Macromol. 2018;107(Pt A):874–881. doi: 10.1016/j.ijbiomac.2017.09.063. [DOI] [PubMed] [Google Scholar]
- 25.Fresno, S., Jiménez, N., Canals, R., Merino, S., Corsaro, M.M., Lanzetta, R., Parrilli, M., Pieretti, G., Regué, M., Tomás. J.M. (2007) A second galacturonic acid transferase is required for core lipopolysaccharide biosynthesis and complete capsule association with the cell surface in klebsiella pneumoniae. J. Bacteriol. 189:1128–1137. [DOI] [PMC free article] [PubMed]
- 26.Garcia-Castillo V., Marcial G., Albarracín L., Tomokiyo M., Clua P., Takahashi H., Kitazawa H., Garcia-Cancino A., Villena J. The Exopolysaccharide of lactobacillus fermentum UCO-979C is partially involved in its immunomodulatory effect and its ability to improve the resistance against Helicobacter Pylori Infection. Microorganisms. 2020;8:479. doi: 10.3390/microorganisms8040479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardiner G.E., Heinemann C., Bruce A.W., Beuerman D., Reid G. Persistence of lactobacillus fermentum RC-14 and lactobacillus rhamnosus GR-1 but not L. rhamnosus GG in the human vagina as demonstrated by randomly amplified polymorphic DNA. Clin. Diagn. Lab. Immunol. 2002;9:92–96. doi: 10.1128/cdli.9.1.92-96.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jing, W., Baolong, S., Baotao, R., Huapeng, Z., Yanting, Z., Jinli, Z., Hong, L. (2021) Lactobacillus fermentum, Culture of Lactobacillus fermentum and Preparation Method of Culture. CN112940968A.
- 29.Hooi J.K.Y., Lai W.Y., Ng W.K., Suen M.M.Y., Underwood F.E., Tanyingoh D., Malfertheiner P., Graham D.Y., Wong V.W.S., Wu J.C.Y., Chan F.K.L., Sung J.J.Y., Kaplan G.G., Ng S.C. Global prevalence of helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153(2):420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 30.Huang S., Vignolles M.R., Dong Chen X., Le Loir Y., Jan G., Schuck P., Jeantet R. Spray drying of probiotics and other food-grade bacteria: a review. Trends Food Sci. Technol. 2017;63:1–17. doi: 10.1016/j.tifs.2017.02.007. [DOI] [Google Scholar]
- 31.Hurtado J.A., Maldonado-Lobón J.A., Díaz-Ropero M.P., Flores-Rojas K., Uberos J., Leante J.L., Affumicato L., Couce M.L., Garrido J.M., Olivares M., Fonollá J. Oral administration to nursing women of lactobacillus fermentum CECT5716 prevents lactational mastitis development: a randomized controlled trial. Breastfeed. Med. 2017;12:202–209. doi: 10.1089/bfm.2016.0173. [DOI] [Google Scholar]
- 32.Kim D.H., Lim W.T., Lee M.K., Kim M.S. Effect of temperature on continuous fermentative lactic acid (LA) production and bacterial community, and development of LA-producing UASB reactor. Bioresour. Techonl. 2012;119:355–361. doi: 10.1016/j.biortech.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 33.Knox K.W., Campbell L.K., Broady K.W., Wicken A.J. Serological studies on chemostat-grown cultures of Lactobacillus fermentum and Lactobacillus plantarum. Infect. Immun. 1979;24:12–18. doi: 10.1128/iai.24.1.12-18.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kullisaar T., Zilmer K., Salum T., Rehema A., Zilmer M. The use of probiotic L. fermentum ME-3 containing Reg'Activ Cholesterol supplement for 4 weeks has a positive influence on blood lipoprotein profiles and inflammatory cytokines: an open-label preliminary study. Nutr. J. 2016;15:93. doi: 10.1186/s12937-016-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim S.M., Lee N.K., Kim K.T., Paik H.D. Probiotic Lactobacillus fermentum KU200060 isolated from watery kimchi and its application in probiotic yogurt for oral health. Microb. Pathog. 2020;147 doi: 10.1016/j.micpath.2020.104430. [DOI] [PubMed] [Google Scholar]
- 36.Lin W.H., Yu B., Jang S.H., Tsen H.Y. Different probiotic properties for Lactobacillus fermentum strains isolated from swine and poultry. Anaerobe. 2007;13:107–113. doi: 10.1016/j.anaerobe.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Linninge C., Xu J., Bahl M.I., Ahrné S., Molin G. Lactobacillus fermentum and lactobacillus plantarum increased gut microbiota diversity and functionality, and mitigated Enterobacteriaceae, in a mouse model. Benef. Microbes. 2019;10:413–424. doi: 10.3920/BM2018.0074. [DOI] [PubMed] [Google Scholar]
- 38.López-Huertas E. Safety and efficacy of human breast milk Lactobacillus fermentum CECT 5716. A mini-review of studies with infant formulae. Benef. Microbes. 2015;6:219–224. doi: 10.3920/BM2014.0091. [DOI] [PubMed] [Google Scholar]
- 39.Luong J.H. Kinetics of ethanol inhibition in alcohol fermentation. Biotechnol. Bioeng. 1985;27:280–285. doi: 10.1002/bit.260270311. [DOI] [PubMed] [Google Scholar]
- 40.Mann S., Park M.S., Johnston T.V., Ji G.E., Hwang K.T., Ku S. Isolation, characterization and biosafety evaluation of lactobacillus fermentum ok with potential oral probiotic properties. Probiotics. Antimicrob. Proteins. 2021 doi: 10.1007/s12602-021-09761-z. [DOI] [PubMed] [Google Scholar]
- 41.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends. Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 42.Muhialdin B.J., Hassan Z., Sadon S. Antifungal activity of lactobacillus fermentum Te007, Pediococcus pentosaceus Te010, lactobacillus pentosus G004, and L. paracasi D5 on selected foods. J. Food Sci. 2011;76:493–499. doi: 10.1111/j.1750-3841.2011.02292.x. [DOI] [PubMed] [Google Scholar]
- 43.Naghmouchi K., Belguesmia Y., Bendali F., Spano G., Seal B.S., Drider D. Lactobacillus fermentum: a bacterial species with potential for food preservation and biomedical applications. Crit. Rev. Food Sci. Nutr. 2020;60:3387–3399. doi: 10.1080/10408398.2019.1688250. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen P.T., Nguyen T.T., Vo T.N.T., Nguyen T.T.X., Hoang Q.K., Nguyen H.T. Response of lactobacillus plantarum VAL6 to challenges of pH and sodium chloride stresses. Sci Rep. 2021;11:1301–1309. doi: 10.1038/s41598-020-80634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan D.D., Zeng X.Q., Yan Y.T. Characterisation of lactobacillus fermentum SM-7 isolated from koumiss, a potential probiotic bacterium with cholesterol-lowering effects. J. Sci. Food Agric. 2011;91:512–518. doi: 10.1002/jsfa.4214. [DOI] [PubMed] [Google Scholar]
- 46.Park J.S., Shin E., Hong H., Shin H.J., Cho Y.H., Ahn K.H., Paek K., Lee Y. Characterization of lactobacillus fermentum PL9988 isolated from healthy elderly Korean in a longevity village. J. Microbiol. Biotechnol. 2015;25:1510–1518. doi: 10.4014/jmb.1505.05015. [DOI] [PubMed] [Google Scholar]
- 47.Peng Z., Wei B., Huang T., Liu Z., Guan Q., Xie M., Li H., Xiong T. Screening, safety evaluation, and mechanism of two lactobacillus fermentum strains in reducing the translocation of staphylococcus aureus in the Caco-2 monolayer model. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.566473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodrigues J., Passos M.R., Silva de Macêdo Neres N., Almeida R.S., Pita L.S., Santos I.A., Santana Silveira P.H., Reis M.M., Santos I.P., de Oliveira Negrão Ricardo L., Lima B.O., D'Orleans Farias Marinho P., Soares A.B., Silva Bastos Andrade L.O., Brasileiro Pessoa S.M., Leles Silva M.M., Oliveira M.C., Pinheiro da Silva J., Moura M.A., Cruz M.P., Yatsuda R. Antimicrobial activity of lactobacillus fermentum TcUESC01 against streptococcus mutans UA159. Microb. Pathog. 2020;142:40–63. doi: 10.1016/j.micpath.2020.104063. [DOI] [PubMed] [Google Scholar]
- 49.Sánchez I., Palop L., Ballesteros C. Biochemical characterization of lactic acid bacteria isolated from spontaneous fermentation of 'Almagro' eggplants. Int. J. Food Microbiol. 2000;59:9–17. doi: 10.1016/s0168-1605(00)00256-7. [DOI] [PubMed] [Google Scholar]
- 50.Sa´nchez-Medina I., Gerwig G.J., Urshev Z.L., Kamerling J.P. Structure of a neutral exopolysaccharide produced by Lactobacillus delbrueckii ssp. bulgaricus LBB.B26. Carbohyd. Res. 2007;342:2430–2439. doi: 10.1016/j.carres.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 51.Settanni L., van Sinderen D., Rossi J., Corsetti A. Rapid differentiation and in situ detection of 16 sourdough lactobacillus species by multiplex PCR. Appl. Environ. Microbiol. 2005;71:3049–3059. doi: 10.1128/AEM.71.6.3049-3059.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song J., Peng S., Yang J., Zhou F., Suo H. Isolation and identification of novel antibacterial peptides produced by lactobacillus fermentum SHY10 in Chinese pickles. Food. Chem. 2021;348:90–97. doi: 10.1016/j.foodchem.2021.129097. [DOI] [PubMed] [Google Scholar]
- 53.Tomaro-Duchesneau C., Saha S., Malhotra M., Jones M.L., Rodes L., Prakash S. Lactobacillus fermentum NCIMB 5221 and NCIMB 2797 as cholesterol-lowering probiotic biotherapeutics: in vitro analysis. Benef. Microbes. 2015;6:861–869. doi: 10.3920/BM2015.0021. [DOI] [PubMed] [Google Scholar]
- 54.Toral M., Robles-Vera I., Romero M., de la Visitación N., Sánchez M., O'Valle F., Rodriguez-Nogales A., Gálvez J., Duarte J., Jiménez R. Lactobacillus fermentum CECT5716: a novel alternative for the prevention of vascular disorders in a mouse model of systemic lupus erythematosus. FASEB J. 2019;9:10005–10018. doi: 10.1096/fj.201900545RR. [DOI] [PubMed] [Google Scholar]
- 55.Truusalu K., Mikelsaar R.H., Naaber P., Karki T., Kullisaar T., Zilmer M., Mikelsaar M. Eradication of salmonella typhimurium infection in a murine model of typhoid fever with the combination of probiotic lactobacillus fermentum ME-3 and ofloxacin. BMC Microbiol. 2008;8:132. doi: 10.1186/1471-2180-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tshibangu-Kabamba, E., & Yamaoka, Y. (2021) Helicobacter pylori infection and antibiotic resistance - from biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 10.1038/s41575-021-00449-x. [DOI] [PubMed]
- 57.Wayah S.B., Philip K. Characterization, yield optimization, scale up and biopreservative potential of fermencin SA715, a novel bacteriocin from Lactobacillus fermentum GA715 of goat milk origin. Microb. Cell Factories. 2018;17:125. doi: 10.1186/s12934-018-0972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wessler S., Backert S. Molecular mechanisms of epithelial-barrier disruption by Helicobacter pylori. Trends Microbiol. 2008;16:397–405. doi: 10.1016/j.tim.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 59.Wessler S., Gimona M., Rieder G. Regulation of the actin cytoskeleton in Helicobacter pylori-induced migration and invasive growth of gastric epithelial cells. Cell Commun. Signal. 2011;9:27. doi: 10.1186/1478-811X-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yong C.C., Lim J., Kim B.K., Park D.J., Oh S. Suppressive effect of lactobacillus fermentum Lim2 on Clostridioides difficile 027 toxin production. Lett. Appl. Microbiol. 2019;68:386–393. doi: 10.1111/lam.13124. [DOI] [PubMed] [Google Scholar]
- 61.Zarłok K. Lactobacillus fermentum CECT5716 – Probiotic from human milk with interesting properties. Wiad. Lek. 2016;69:271–275. [PubMed] [Google Scholar]
- 62.Zheng J., Wittouck S., Salvetti E., Franz C.M.A.P., Harris H.M.B., Mattarelli P., O'Toole P.W., Pot B., Vandamme P., Walter J., Watanabe K., Wuyts S., Felis G.E., Gänzle M.G., Lebeer S. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus lactobacillus Beijerinck 1901, and union of lactobacillaceae and leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70(4):2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.