Highlights
-
•
ESKAPE pathogens display multidrug resistance (MDR) through various mechanisms.
-
•
Mechanisms adapted by ESKAPE pathogens to counteract antibiotics have been described.
-
•
Selected natural compounds with bactericidal effect towards ESKAPE pathogens have been listed.
Keywords: ESKAPE, Multidrug resistance, Natural compounds, Secondary metabolites
Abstract
The microorganisms that have developed resistance to available therapeutic agents are threatening the globe and multidrug resistance among the bacterial pathogens is becoming a major concern of public health worldwide. Bacteria develop protective mechanisms to counteract the deleterious effects of antibiotics, which may eventually result in loss of growth-inhibitory potential of antibiotics. ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) pathogens display multidrug resistance and virulence through various mechanisms and it is the need of the hour to discover or design new antibiotics against ESKAPE pathogens. In this article, we have discussed the mechanisms acquired by ESKAPE pathogens to counteract the effect of antibiotics and elaborated on recently discovered secondary metabolites derived from bacteria and plant sources that are endowed with good antibacterial activity towards pathogenic bacteria in general, ESKAPE organisms in particular. Abyssomicin C, allicin, anthracimycin, berberine, biochanin A, caffeic acid, daptomycin, kibdelomycin, piperine, platensimycin, plazomicin, taxifolin, teixobactin, and thymol are the major metabolites whose antibacterial potential have been discussed in this article.
1. Introduction
From 1940 to 1965 is the golden era in the field of antibiotics as many antimicrobials were discovered and introduced into modern medicine. The discovery of new antibiotics revolutionized modern medicine as these small molecules have been widely implemented in the treatment of bacterial diseases. Unfortunately, discontinuous or improper, or irregular intake and excessive usage of antibiotics have led to the emergence of antibiotic-resistant microorganisms. Antibiotic resistance is increasing at an alarming pace and has become one of the major health concerns of the globe due to various reasons including the availability of a limited number of antibiotics. According to the antibiotic resistance threats report (2019) of the Centers for Disease Control and Prevention (CDC, United States), 2.8 million people are infected with antibiotic-resistant infection and more than 35,000 people die annually in the United States alone. The existence of drug-resistant pathogens came into recognition only in the last two decades. Lack of development of new antimicrobials is one of the key factors for the crisis in the medical field that has been denoted elsewhere [1]. The Center for Disease Dynamics, Economics, and Policy (CDDEP) have well-documented the global status of antibiotics policy to understand the resistance and need for newer antimicrobials [2]. It is a matter of serious concern that, significant progress has not been made in the discovery and development of new antibiotics in the previous three decades. Many pharmaceutical companies have committed towards the development of drugs for the treatment of non-communicable diseases which are of substantial economic interest [3]. On one hand, pathogens are gaining resistance gradually towards existing therapeutics and on the other hand, a feeble amount of efforts have been made in the discovery of new antibiotics. Therefore, it is the need of the hour to discover new antimicrobials which are potent against a broad spectrum of microorganisms including drug-resistant pathogens.
Microorganisms develop an adaptive mechanism to survive in the favorable/adverse atmosphere and as a part of this, they subsequently develop several protective mechanisms to reduce their antibiotic susceptibility. This eventually leads to the loss of antibiotic potency and thus, making them ineffective for the treatment of bacterial infections. A group of microorganisms that possess multidrug resistance and virulence include Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. causing nosocomial infections and contributing a lion's share in hospital-acquired infections [4]. Louis B Rice gave the acronym for these pathogens as ESKAPE organisms and the following section discusses about the antibacterial resistance mechanisms of these pathogens [5].
2. ESKAPE pathogens
Hospital-acquired (nosocomial) infections can be caused by various microbes such as bacteria, fungi, viruses, parasites, and other agents. The transmission of these infections could be due to interpersonal communications between patients or with healthcare providers, through contaminated equipment, lack of proper sterilization measures, and many more [6]. Hospital-acquired infections may contribute to lengthier hospital stays, increased treatment-related expenses, and a high mortality rate [7]. The Infectious Diseases Society of America has grouped the bacterial pathogens responsible for hospital-acquired infections and referred to as “ESKAPE” pathogens. E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter spp. are the organisms that are grouped under ESKAPE pathogens and they can escape from the biocidal activity of antibacterial drugs. These organisms display a characteristic of multidrug resistance (MDR) and may pose a potentially fatal condition for immunocompromised individuals or patients with serious illness [6]. The World Health Organization (WHO) has recently included ESKAPE pathogens in the inventory of twelve bacteria which demands the development of new antibiotics urgently [8]. In a critical analysis, the multicriteria decision analysis method was used to prioritize antibiotic-resistant bacteria and classified bacteria into three categories (critical priority, high priority, and medium priority) depending on the need for the development of new antibiotics against antibiotic-resistant bacteria [8]. Among ESKAPE organisms, carbapenem-resistant A. baumannii and carbapenem-resistant P. aeruginosa were listed under critical priority, and vancomycin-resistant E. faecium (VREF) and methicillin/vancomycin resistant S. aureus (MRSA/VRSA) were categorised under high priority groups [8]. They also suggested the need for the development of new antibacterial agents against multidrug resistance (MDR)-tuberculosis, Gram-negative bacteria, antibiotic-resistant bacteria responsible for community-acquired infections such as Salmonella spp, Campylobacter spp, Neisseria gonorrhoeae, and Helicobacter pylori. The bacteria may develop MDR through various mechanisms including chemical alteration or breakdown of the drug, interference with influx and efflux of antibiotics, modification of drug target, and many more. In the below section, we have briefly discussed major mechanisms of drug resistance shown by ESKAPE organisms to escape from the antibiotics.
3. Major mechanisms of drug resistance acquired by ESKAPE organisms
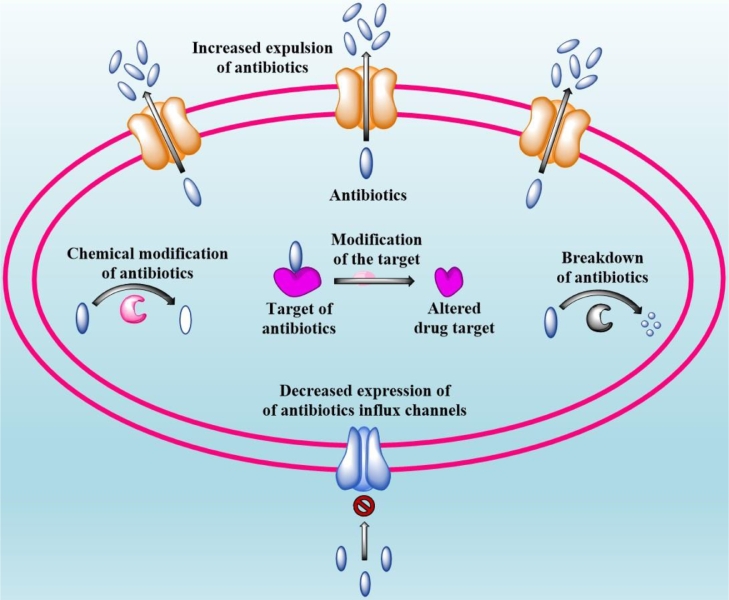
MDR bacteria have acquired various defense mechanisms to protect themselves from the bactericidal activity of antibiotics and some of the major drug-resistant mechanisms developed by ESKAPE organisms have been discussed in this section and graphical representation is provided in Fig. 1.
Fig. 1.
Bacteria develop resistance against antibiotics through chemical modification or breakdown of the drug, or preventing antibiotic influx, or antibiotic expulsion through efflux pumps or modification of antibiotic targets.
3.1. Chemical modification or breakdown of the drug
Bacteria express some crucial enzymes (such as β-lactamases) which offers them a survival benefit by chemically modifying or destroying the antibiotics. β-lactamases are some of the widely-studied enzymes in relevance to antibiotic resistance and they are characterized by their ability to cleave the amide bond of the four-membered β-lactam ring present in penicillins, cephalosporins, cephamycins, monobactams, and carbapenems [9]. Penicillinase, cephalosporinase, broad-spectrum β-lactamases, metallo-β-lactamases,extended-spectrum β-lactamases (ESBL), and carbapenemases are the major types of lactamases responsible for drug destruction which makes the antibiotic inactive and more than 2600 different β-lactamases have been identified so far [10]. The catalytic activity of some of these enzymes can be inhibited by compounds such as clavulanate, tazobactam, sulbactam, and ticarcillin and therefore administered them to patients along with β-lactam antibiotics [11]. β-lactamases based on molecular structure are classified into four classes according to Ambler scheme such as class A, B, C, and D. Class A family enzymes comprise the largest cluster of β- lactamases known to bear serine residue in their active site and act against various β-lactam antibiotics such as penicillins, early cephalosporins, third-generation oxyimino-cephalosporins, monobactams, cephamycins, and carbapenems [12]. Majority of the S. aureus of clinical relevance and few Enterococcus spp. produce blaZ–encoded penicillinases that belong to the class A family [13]. CTX-M (Cefotaximase from Munich), PER (Pseudomonas extended resistant), GES (Guiana extended-spectrum β-lactamase), and VEB (Vietnam extended-spectrum β-lactamase) families belong to class A ESBL enzymes and were found to be produced by all Gram-negative ESKAPE pathogens [14]. The majority of the class A ESBL enzymes can be inhibited by clavulanate except TEM-30 (Temoniera), SHV-10 (Sulphydral reagent variable), and TEM-50 [15]. Alarmingly, some strains of K. pneumoniae were reported to produce inhibitor-resistant β-lactamases (resistant to many known inhibitors of β-lactamase) such as Klebsiella pneumoniae carbapenemase (KPC) enzymes which degrade all β-lactams, including carbapenems [16]. Nevertheless, a combination of imipenem-cilastatin-relebactam or meropenem-vaborbactam was found to be effective against KPC-producing organisms [17]. The Ambler group B constitutes metallo-β–lactamases (MBLs) which contain Zn+ion in their active sites and are prominently found in Gram-negative ESKAPE pathogens [18]. IMP (Imipenemase)- and VIM (Verona integron-encoded MBL)-type MBLs are found to be present in ESKAPE pathogens such as P. aeruginosa, K. pneumoniae, Enterobacter cloacae complex isolates, and Acinetobacter spp [19,20]. NDM (New Delhi metallo-β–lactamase)-type MBLs are seen in most of the Gram-negative ESKAPE pathogens and they are of particular concern because they are incorporated into the mobile genetic elements that also encode determinants for resistance to other antibiotic classes [21]. The Amber Group C family contains cephalosporinases such as AmpC identified in the Enterobacter spp., P. aeruginosa, and Acinetobacter spp., which act against most narrow- to intermediate-spectrum cephalosporins plus aztreonam. Lastly, the Amber Group D family includes oxacillin-hydrolyzing enzymes which were reported to be present in Acinetobacter spp. and many Enterobacterales species [22]. Similarly, macrolide esterases hydrolyze the lactone ring of macrolides such as azithromycin and erythromycin.
Beside, some bacterial enzymes have the capability of chemically modifying the antibiotics (such as acetylation, phosphorylation, and adenylation) to inactivate them and to acquire antibiotic resistance. The major categories of antibiotic modifying enzymes are aminoglycoside-modifying enzymes (AMEs), rifamycin-modifying enzymes, macrolide phosphotransferases, chloramphenicol acetyltransferases, phosphomycin-modifying enzymes, and flavin-dependent monooxygenases. Rifamycin inhibits prokaryotic transcription by interacting with the β-subunit of the RNA polymerase, whereas rifamycin modifying enzymes modify the hydroxyl group which abolishes the potency of the antibiotic [23].
The aminoglycosides are broad-spectrum antibiotics derived from actinomycetes which inhibit protein synthesis by binding to ribosomal subunits. The resistance against these antibiotics majorly occurs through chemical modification by aminoglycoside-modifying enzymes (AMEs) [24]. The genes coding for these enzymes are generally present in the transmissible elements and have rarely been found in genome of some bacteria. The presence of AME activity in resistant bacterial strains has been documented previously 25, 26, 27. The members of the AME family are classified into aminoglycoside acetyltransferases (AACs), aminoglycoside phosphotransferases (APHs), and aminoglycoside nucleotidyltransferases (ANTs) based on the nature of chemical modifications they catalyze [28]. The chemical modifications catalyzed by these enzymes alter the binding affinity of aminoglycosides to ribosomal subunits. Aminoglycoside acetyltransferases are the largest class of AMEs which are further subdivided into four subclasses and are involved in the acetylation of amino groups present in the aminoglycoside antibiotics. AAC(1) and AAC(3) subclasses are involved in the acetylation of amino groups present in the 1st and 3rd position of the 2-deoxystreptamine moiety of aminoglycosides respectively, whereas AAC(2′) and AAC(6′) subclasses acetylate 2′ and 6′ amino group positions of 2,6-dideoxy-2,6-diamino-glucose moiety [24]. Recent epidemiological investigations in the United States, Europe, and Asia revealed that AAC(3) and AAC(6′) enzymes are most frequently observed in Gram-negative ESKAPE pathogens which confer resistance against gentamicin, tobramycin, and amikacin 29, 30, 31.
APHs are the second largest class of AMEs which are further subdivided into seven subclasses. These enzymes are involved in the phosphorylation of the hydroxyl group present in the aminoglycosides which diminish the binding affinity due to reduced hydrogen bonding interaction between the antibiotic and target. Out of seven subclasses, APH(3′) was reported to be found in S. aureus and Enterococcus spp. which confer resistance against amikacin [29].
ANTs are the third-largest class of AMEs which are involved in the addition of nucleotide monophosphate to hydroxyl groups of the aminoglycosides at 2″, 3″, 4′, 6, and 9 positions [24]. Out of five subclasses, ANT(2″) and ANT(4′) were reported to be produced by K. pneumoniae and S. aureus respectively. ANT(2″) confers resistance against 4,6-di-substituted aminoglycosides whereas ANT(4′) acts against kanamycin A/B/C, gentamicin A, amikacin, tobramycin, and neomycin B/C [32].
3.2. Prevention of antibiotic influx
The bactericidal efficacy of an antibiotic depends on the amount of drug taken into the bacteria and expelled out. Many antibacterial agents use intracellular targets to impart their growth-inhibitory activity and the maximum efficiency is expected only when a sufficient amount of antibiotic molecule reaches the inner compartments of the cell. Many bacteria have evolved with an adaptive mechanism in which they alter the expression levels of channels and efflux pumps. The outer membrane of Gram-negative bacteria has proteins termed porins that are involved in the formation of channels to enable the movement of antibiotics into the cell. A. baumannii which displayed the conspicuous loss of outer membrane protein (OMP, 29-kDa) was insensitive to imipenem activity [33]. Experimental evidence was provided to demonstrate the role of the OprD channel in the influx of panipenem/imipenem. OprD is present in the outer membrane of P. aeruginosa and a decrease in OprD levels enabled the bacteria to acquire resistance against imipenem [34].
3.3. Antibiotic expulsion through efflux pumps
Efflux pumps are responsible for the outflux of antibiotics from the bacterial cell and overexpression of various types of efflux pumps is observed in MDR bacteria. Drug expulsion could be either due to increased expression of efflux pump proteins or base substitution mutation in the gene that codes for the efflux pump protein which may result in the replacement of an amino acid that potentiates the activity of the efflux pump [35]. The genes that encode for efflux pumps can be present either in the bacterial genome or mobile genetic elements such as plasmids. Six families of bacterial drug efflux pumps have been described so far, they are ATP-binding cassette (ABC) family, the multidrug and toxin extrusion (MATE) family, major facilitator superfamily (MFS), the resistance-nodulation-cell division (RND) superfamily, the small multidrug resistance (SMR) family, and the proteobacterial antimicrobial compound efflux (PACE) family [36]. For an instance, P. aeruginosa strains are relatively less sensitive to tigecycline and the loss of MexXY (OprM) efflux pumps resulted in the increased sensitivity of microbe towards tigecycline [37]. The members of the RND superfamily such as MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexXY, and MuxBC—OpmB are involved in the expulsion of various antibiotics including β-lactams, aminoglycosides, chloramphenicol, fluoroquinolones, macrolides, quinolones, tetracycline, and novobiocin in P. aeruginosa [38]. It can be concluded that all resistant bacteria express their own set of efflux pumps to combat the harmful effects of antibiotics on them.
3.4. Modification of antibiotic target
Bacteria protect themselves from the action of antibiotics through modification of the drug target. The modifications include mutations in the gene that encode target protein, chemical alteration of the interaction site, and substitution of the original target [39]. Many antibiotics impart their antibacterial action by targeting ribosomal subunits (the 50S and 30S). Aminoglycoside antibiotics interact with a 16S rRNA of the 30S subunit and hamper the interaction of aminoacyl-tRNA to the anticodon which ultimately results in the abrogation of translation and bactericidal effects. 16S rRNA methyltransferases either methylate N7 of guanine (1405) or N1 of adenine (1408) of 16S rRNA to confer resistance against the majority of aminoglycosides [28]. Some rRNAs (5S and 23S rRNA) and ribosomal proteins of the 50S subunit are targeted by macrolides, ketolides, lincosamides, and streptogramin. Like 16S rRNA methyltransferases, 23S rRNA methyltransferases methylate the antibiotic binding site resulting in the abolishment of antibiotic activity [40]. Similarly, bacterial targets of antibiotics are modified by phosphoethanolamine transferases and peptidoglycan modifying enzymes.
4. Antibacterial agents derived from the Mother Nature
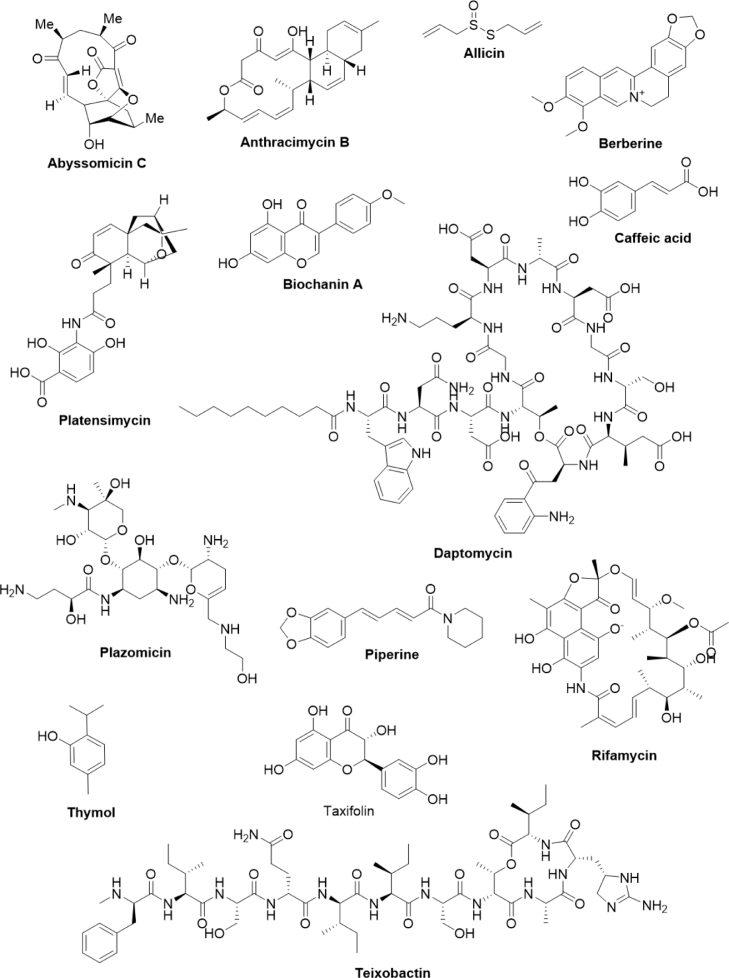
Mother Nature has been serving as an inexhaustible source of bioactive secondary metabolites which are endowed with a wide range of pharmacological activities including antibacterial, anticancer, antifungal, antihypertensive, antidepressants, antiviral, and many more 41, 42, 43, 44, 45, 46. The structural complexity of these compounds makes the synthetic process impractical. Thousands of secondary metabolites have been isolated from bacteria, plants, fungi, endophytes, and marine invertebrates and some of them have been approved as therapeutic agents for the treatment of several human ailments 47, 48, 49. Therefore, these biological sources are true treasure houses of secondary metabolites and drugs. In the following sections, we have discussed some of the antibacterial agents derived from bacteria (Fig. 2 and Table 1) and plants (Table 2).
Fig. 2.
Chemical structure of secondary metabolites derived from bacteria and plants.
Table 1.
Secondary metabolites derived from the bacterial origin with antibacterial activity towards ESKAPE pathogens.
| Sl. No. | Antibacterial compound | Source | Activity against | Mode of action | Reference |
|---|---|---|---|---|---|
| 1. | Lysobactin (Depsipeptide) | Several gram-negative gliding bacteria from soil | S. aureus (MIC=0.75 μg/mL), MRSA (MIC= 0.39 μg/mL), VRE (MIC= 0.78 μg/mL) | Interfere with peptidoglycan biosynthesis by binding to lipid I, Lipid II, Lipid III cell wall precursors | [164,165] |
| 2. | Salinamide F (Depsipeptide) | Streptomyces sp., strain CNB-091 | E. faecalis (MIC50 =12.5 μg/ml), S. aureus (MIC50 =100 μg /ml), H. influenzae, (MIC50 =12.5 μg/ml), Neisseria gonorrhoeae (MIC50 = 25 μg/ml), Enterobacter cloacae (MIC50 = 50 μg/ml) | Inhibition of RNA polymerase | [166] |
| 3. | Siamycin-I (Tricyclic peptide) | Streptomyces sp. | VRE (MIC = 7.4 μM), S. aureus (MIC = 3.7 μM), MRSA (MIC = 7.4 μM), multidrug resistant S. aureus (MIC = 14.8 μM) | Interferes in peptidoglycan biosynthesis by binding to lipid II. | [167] |
| 4. | Capistruin (Lasso peptide) | Burkholderia thailandensis E264 | E. coli 363 (MIC = 25 μM), P. aeruginosa AT27853 (MIC = 50 μM) | Inhibition of RNA polymerase | [168,169] |
| 5. | Mitomycin C (7-Amino-9alpha-methoxymitosane) | Streptomyces caespitosus | A. baumannii (MIC = 7 µg/mL) | DNA intercalation | [170,171] |
| 6. | Marthiapeptide A (Polythiazole cyclic peptide) | Marinactinospora thermotolerans SCSIO 00,652 |
M. luteus (2.0 µg/mL) S. aureus (8 µg/mL), |
ND | [172] |
| 7. | Lobophorin F | Streptomyces sp. (SCSIO 01,127) | S. aureus, E. faecalis (MIC for both = 8 μg/mL) | ND | [173] |
| 8. | Desotamide (Cyclic hexapeptide) | Streptomyces scopuliridis SCSIO ZJ46 | S. pnuemoniae (MIC = 12.5 μg/mL),, S. aureus (MIC = 16 μg/mL), methicillin-resistant S. epidermidis (MIC = 32 μg/mL) | ND | [174] |
| 9. | Abyssomicin C | Verrucosispora sp. AB-18–032 | MRSA (MIC =4 μg/mL), VRSA (MIC =13 μg/mL) | Inhibition of 4-amino-4-deoxychorismate (ADC) synthase | [50,51] |
| 10. | Ficellomycin | Streptomyces ficellus | Multidrug resistant strains of S. aureus | Interferes in DNA replication | [175] |
| 11. | Etamycin | Streptomyces sp | MRSA (MIC =1–2 mg/L) | Possible role in inhibition of protein synthesis | [176] |
| 12. | Thiomarinol A | Pseudoalteromonas sp. SANK 73390 | MRSA (MIC = 0.002 - 0.50 μM) | Inhibition of isoleucyl-tRNA synthetase | [177] |
| 13. | Mithramycin | Streptomyces plicatus, Streptomyces argillaceus | MRSA and MSSA strains (MIC = 0.125 – 0.25 μg/mL) vancomycin resistant enterococci and vancomycin sensitive enterococci (MIC = 1–16 µg/mL) | Inhibition of transcription | [178] |
| 14. | Marinopyrrole A | Streptomyces sp. CNQ-418 | S. aureus (MIC = 0.5–1 µg/mL), MRSA (MIC = 0.188–0.375 µg/mL), E. faecalis (MIC = 1 µg/mL) | ND | [179] |
| 15. | Bottromycin A2 (Macrocyclic peptide) | Streptomyces sp. KM-9459. | S. aureus (MIC = 1 µg/mL), MRSA (MIC = 1 µg/mL), VRE (MIC = 1 µg/mL) | Inhibition of protein synthesis | [180] |
| 16. | Tirandamycin C | Streptomyces sp. 307–9 | VREF | Inhibition of RNA polymerase | [181] |
| 17. | Anthracimycin B | Streptomyces cyaneofuscatus M-169, | MRSA, MSSA, E. faecium | ND | [59] |
| 18. | Fijimycin A (Cyclic depsipeptide) | Streptomyces sp. CNS-575 | MRSA (MIC = 4–16 µg/mL) | ND | [182] |
| 19. | Ecteinamycin | Actinomadura sp. WMMB499 | MRSA (MIC = 0.125 µg/mL), MSSA (MIC = 0.125 µg/mL), P. aeruginosa (MIC = 8 µg/mL) Clostridium difficile NAP1/B1/027 (MIC = 59 ng/µL) | Acts as an ionophore and is involved in membrane depolarization and dysregulation of potassium cation homeostasis. | [183] |
| 20. | Lajollamycin | S. nodosus NPS007994, | S. pneumoniae (MIC = 1.5 µg/mL), MRSA (MIC = 5 µg/mL) | ND | [184] |
ND: Not determined.
Table 2.
Secondary metabolites derived from plant origin with antibacterial activity towards ESKAPE pathogens.
| Sl. No. | Antibacterial compound | Source | Activity against | Mode of action | Reference |
|---|---|---|---|---|---|
| 1. | Resveratrol | Grapevines (Vitis vinifera), Peanuts (Arachis hypogea), Japanese knotweed (Polygonum cuspidatum) |
S. aureus, E. faecalis, and Streptococcus pyogenes (MICs = 100–200 μg/mL) M. smegmatis (MIC = 64 μg/mL) H. pylori (MIC = 25–50 μg/mL) N. gonorrhoeae (MIC = 75 μg/mL) M. tuberculosis (100 μg/mL) |
Partial inhibition of ATP hydrolysis and synthesis functions of ATP synthase complex. Possible role in the inhibition of FtsZ-mediated septum formation and cell division Cell membrane leakage. |
[185] |
| 2. | Glabrol | Licorice (Glycyrrhiza glabra L.) | MRSA T144 (MIC=2 μg/mL), MSSA 1518 (MIC=1 μg/mL) | Disruption of membrane permeability Dissipation of proton motive force |
[186] |
| 3. | Apigenin | Parsley (Petroselinum crispum), Chamomile (Matricaria chamomilla L.), Celery (Apium graveolens L.), Vine-spinach (Basella rubra L.), Artichokes (Cynara scolymus L.), and Oregano (Origanum vulgare L.), |
A. baumannii (RSKK 02026 Strain MIC=2 μg/mL, clinical isolate=64 μg/mL) E. aerogenes (MIC=64 μg/mL) |
Inhibition of D-alanine-D-alanine ligase and DNA gyrase enzyme | 187, 188, 189 |
| 4. | Baicalin | Baikal skullcap (Scutellaria baicalensis), Blue skullcap (Scutellaria lateriflora) | Staphylococcus saprophyticus, Streptococcus mutans | Inhibition of biofilm formation, Inhibition of efflux pump | [190] |
| 5. | Carvacrol | Conehead thyme (Thymus capitatus) Thyme (Thymus vulgaris) |
E. coli (MIC=8 μg/mL) | Disruption of membrane integrity | [191] |
| 6. | Sanguinarine | Celandine (Chelidonium majus L.) Blood Root (Sanguinaria canadensis L.), Fumitory (Fumaria officinalis L.) Plume poppy (Bocconia frutescens L.) |
S. aureus (MIC 1.9 mg/L) MRSA (3.12 μg/mL) MRSA (MIC=1.56 μg/mL) |
Release of autolytic enzymes and cell lysis. DNA intercalation leads to inhibition of replication and transcription |
[192] |
| 7. | Reserpine | Sarpagandha or Indian snake root (Rauvolfia serpentina (L.) Benth. ex Kurz) |
Staphylococcus sp., Streptococcus sp. Micrococcus sp. |
Efflux pump inhibitory activity | [193,194] |
| 8. | Tomatidine | Tomato (Solanum lycopersicum) | S. aureus, L. monocytogenes, Bacillus ssp. | Inhibition of ATP synthase complex | [195] |
| 9. | Sakuranetin | Asteraceae species (Polymnia fruticosa, Baccharis retusa), Himalayan cherry (Prunus puddum) | H. pylori (MIC= 87.3 μM) | Inhibition of HpFabZ enzyme | [196] |
| 10. | Curcumin (Diferuloylmethane) | Turmeric (Curcuma longa) | E. faecalis, S. aureus, P. aeruginosa | Membrane leakage | [197] |
| 11. | Agasyllin | Finocchiazzo (Ferulago campestris) | S. Typhi, E. aerogenes, E.cloacae, S. aureus (MIC=32 μg/mL) | Inhibition of DNA gyrase enzyme | [198,199] |
| 12. | Conessine | Kutaja Kurchi (Holarrhena antidysenterica), Kelinda (Holarrhena mitis), False rubber tree (Holarrhena floribunda) | P. aeruginosa, Bacillus spp. | Inhibition of MexAB-OprM efflux pump. | 200, 201, 202 |
| 13. | Shikimic acid | All plants | S. aureus (2.5 mg/mL) | Alteration of membrane potential and leakage of cellular contents, Inhibition of biofilm formation | [203,204] |
| 14. | Ferulic acid | All plants | E. coli (100 μg/mL), P.aeruginosa (100 μg/mL), S. aureus (1100 μg/mL), L. monocytogenes (1250 μg/mL) | Membrane rupture and release of intracellular contents | [205] |
| 15. | Kaempferol | Abudant in Tea (Camellia sinensis), Broccoli (Brassica oleracea), Apple (Malus domestica), Strawberry (Fragaria x ananassa). | S. aureus | Efflux pump inhibition, Inhibition of PriA helicase enzyme, Inhibition of biofilm formation, Inhibition of Sortase A activity | 206, 207, 208, 209 |
| 16. | Kurarinol | Shrubby sophora (Sophora flavescens) | S. aureus (219 μM), B. subtilis (219 μM), S. typhimurium (219 μM), P. vulgaris (109 μM) | Inhibition of Sortase A enzyme | [210] |
| 17. | Morusin | Mulberry (Morus alba) | S. aureus (14.9 μmol/L) | Disruption of membrane integrity, Reduction gene expression of acc C, Fab D, Fab F, Fab G, Fab H, Fab I, Fab Z, Pls X, Pls Y, Pls C (phosphatidic acid biosynthesis-associated genes) | [211] |
| 18. | Lonicerin | Japanse honeysuckle (Lonicera japonica) | P. aeruginosa | Inhibition of biofilm formation, Inhibition of alginate secretion protein (AlgE) | [212] |
| 19. | Plumbagin | Indian leadwort (Plumbago rosea), Ceylon leadwort (Plumbago zeylanica) | S. aureus (5 μg/mL), MRSA (4–8 μg/mL) | Possible role in the inhibition of DNA gyrase | 213, 214, 215 |
| 20. | Naringenin | Present in many citrus fruits such as Grapefruit (Citrus paradisi), Sour orange (Citrus aurantium), tart cherries (Prunus cerasus), tomatoes (Solanum lycopersicum), and greek oregano (Origanum vulgare L.). | E. faecalis (256 μg/mL), VERF (512 μg/mL) | Inhibition of β -Ketoacyl acyl carrier protein synthase (KAS) III (through in silico approach) | [140,216] |
4.1. Abyssomicin C
Abyssomicin C is one of the promising compounds belonging to the family of abyssomicin with antibacterial activity against many Gram-positive bacteria. Abyssomicin C along with its atrop isomer (atrop-abyssomicin C) and other inactive derivatives such as abyssomicins B and D were isolated from an actinomycete strain Verrucosispora AB-18-032. The actinomycete strain was isolated from the sediments of the deep Sea of Japan at a depth of 289 m [50]. Abyssomicin C has been reported to possess antibacterial activity against MRSA and VRSA with MIC values of 4 and 13 μg/mL, respectively [51]. The studies of Freundlich et al. demonstrated the antibacterial activity of Abyssomicin C and atrop-abyssomicin C against Mycobacterium tuberculosis H37Rv with MIC values of 3.6 and 7.2 μM, respectively [52]. Abyssomycin C and atrop-abyssomicin C are the first natural compounds reported to inhibit the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway in Gram-positive bacteria [53,54]. ADC synthase (4-amino-4-deoxy chorismic acid synthase) and ADC lyase (4-amino-4-deoxy chorismic acid lyase) are the two important enzymes involved in the biosynthesis of p-aminobenzoic acid from chorismic acid. ADC synthase has two subunits (namely PabA and PabB) which are involved in the conversion of chorismic acid and glutamine to 4-amino-4-deoxy chorismic acid and glutamate, respectively. ADC lyase converts 4-amino-4-deoxy chorismic acid into p-aminobenzoic acid which further participates in the folate biosynthesis. Abyssomycin C and atrop-abyssomicin C independently inhibit the activity of ADC synthase by covalently binding to the side chain of Cys263 of the PabB subunit of ADC synthase [55]. Monjas et al. synthesized truncated derivatives of abyssomycin C and studied their structure-activity relationship towards ADC synthase and these derivatives were found to have lower antibacterial activity compared to abyssomycin C [56].
4.2. Anthracimycin
Anthracimycin was isolated initially in 2013 from the Streptomyces strain CNH365 that was collected from the marine sediments of Santa Barbara, USA. Anthracimycin is a 14-membered macrolide that possesses a rare chemical structure that closely resembles the structure of chlorotonil A (antimalarial agent). In the initial studies, anthracimycin demonstrated potent antibacterial activity against the Gram-positive organisms such as Bacillus anthracis, S. aureus, Enterococcus faecalis, Streptococcus pneumoniae with MIC values of 0.031, 0.0625, 0.125, 0.25 μg/mL, respectively [57]. Later, anthracimycin was also demonstrated to possess antibacterial activity against the methicillin-susceptible/resistant S. aureus (MSSA/MRSA), and VRSA with MIC values less than 0.25 mg/L. In the in vivo studies, anthracimycin protected the mice from MRSA-induced mortality at 1 or 10 mg/kg. They also showed that anthracimycin inhibits the DNA and RNA synthesis in S. aureus and the inhibition is independent of DNA intercalation. Using genome mining strategy, Sirota et al., identified that Nocardiopsis kunsanensis produces an anthracimycin analog named anthracimycin BII-2619 [58]. Notably, both anthracimycin BII-2619 and anthracimycin displayed antibacterial activity against the MSSA and MRSA with MIC values ≤1 μM. In another study, Rodríguez et al., isolated the anthracimycin B along with anthracimycin from actinomycete Streptomyces cyaneofuscatus M-169 species obtained from gorgonian coral that was collected from Avilés submarine Canyon at 1500 m depth [59]. The anthracimycin B exhibited potent antibacterial activity against MSSA, MRSA, vancomycin sensitive E. faecium, and vancomycin sensitive E. faecalis. Interestingly, Davison et al. developed a method for the chemical synthesis of anthracimycin which opened a new avenue for the synthesis of potent anthracimycin analogs [60].
4.3. Daptomycin
Daptomycin is a cyclic lipopeptide antibiotic produced by Streptomyces roseosporus. It received FDA approval for the treatment of skin infections and S. aureus bacteremia. Daptomycin is the only lipopeptide antibiotic that is in clinical practice and has potent activity against Gram-positive bacterial infections, MRSA, and vancomycin-resistant Enterococci (VRE) [61]. Daptomycin is used as an alternative therapeutic agent to vancomycin for invasive MRSA or MSSA infections in patients who are allergic to β-lactams. It has been considered as a last-resort treatment for heterogeneous vancomycin-intermediate S. aureus (hVISA) and vancomycin-intermediate S. aureus (VISA) infections [62]. Although the precise mechanism of action of daptomycin is yet to be understood, a model proposes that daptomycin interacts with the plasma membrane resulting in its permeabilization and depolarization caused by a loss of cytoplasmic potassium ions [62]. Although daptomycin resistance is uncommon, failure of treatment up to 33% may be observed in invasive infections including bacteremia or osteomyelitis [63]. In one of the interesting studies, S. aureus was demonstrated to release phospholipids in response to daptomycin treatment to sequester the drug and to escape from its bactericidal effects [64]. In a similar study, E. faecalis and pathogenic streptococci were demonstrated to nullify the effect of daptomycin by shedding phospholipids [61]. Considering the therapeutic importance of daptomycin, a large number of studies are being carried out to potentiate its bactericidal action in combination with other antibacterial agents.
4.4. Kibdelomycin
Kibdelomycin is a novel class of antibiotics produced by Kibdelosporangium sp. (MA7385) [65]. S. aureus fitness test strategy was employed for the discovery of kibdelomycin which signifies the importance of a high-throughput chemical genetic approach in the screening of potential natural compounds. The kibdelomycin showed potent antibacterial activity against many Gram-positive organisms such as wild-type S. aureus, MRSA, S. pneumoniae, E. faecalis with MIC values 2, 0.5, 1, and 2 μg/mL, respectively and Haemophilus influenzae (Gram-negative organism) with a MIC value of 2 μg/mL. Kibdelomycin was found to impart their antibacterial activity by interrupting DNA replication through the inhibition of ATPase activity of DNA gyrase and topoisomerase IV of Escherichia coli and S. aureus [65]. Subsequently, kibdelomycin A, a congener of kibdelomycin was isolated from Kibdelosporangium sp. (MA7385) and its derivatives (kibdelomycin C-33 acetate and tetrahydro-bisdechloro derivative of kibdelomycin) were prepared [66]. Kibdelomycin A and its derivatives were reported to have inferior antibacterial activity compared with kibdelomycin against MSSA, MRSA, S. pneumoniae, E. faecalis, Bacillus subtilis, H. influenzae, E. coli, and Candida albicans [66]. Kibdelomycin A displayed relatively lesser inhibitory activity compared to kibdelomycin towards DNA gyrase and topoisomerase IV of S. aureus whereas both the compounds showed similar inhibitory activity towards DNA gyrase and topoisomerase IV of E. coli [66]. Interestingly, co-crystallization studies between kibdelomycin and gyrase B or topoisomerase IV of S. aureus revealed that kibdelomycin exhibits a “dual-arm” U-shaped binding mode with both enzymes [67]. Kibdelomycin is a wide-spectrum antibiotic without cross-resistance to known gyrase inhibitors, including clinically effective quinolones. The ‘dual arm’ U-shaped binding interaction of kibdelomycin is unique and not seen in other gyrase inhibitors [67]. This may be the reason to present kibdelomycin as an antibiotic without cross-resistance to other gyrase inhibitors. Beside, kibdelomycin was reported with potent antibacterial activity against Clostridium difficile, a causative agent of C. difficile-associated diarrhea, especially in elder people [68]. Kibdelomycin acted as a broad range bactericidal agent and found to be effective against the Gram-negative organisms such as Moraxella catarrhalis and A. baumannii with MIC90 values of 0.5 and 0.125 μg/mL, respectively. Taken together, kibdelomycin can be considered as the potential antimicrobial compound against many pathogenic bacteria [69].
4.5. Platensimycin
Platensimycin and platencin are produced by Streptomyces platensis that represents a unique structural class of non-toxic natural antibiotics [70]. Platensimycin has been identified as an effective inhibitor of bacterial fatty acid synthases by selectively targeting β-ketoacyl-(acyl-carrier-protein (ACP)) synthase I/II [71]. The discovery of platensimycin has provided a new direction to the discovery of antibiotics with structural diversity. MRSA biofilms in wounds are difficult to treat with conventional antibiotics. Platensimycin and sulfur-Michael derivatives suppressed the biofilm formation by S. aureus beyond 95% at 2 μg/mL [72]. Notably topical application of ointments having platensimycin was successful in treating MRSA infections in a BABL/c mouse burn wound model [72]. However, its clinical development was hindered due to its poor solubility and pharmacokinetic properties. To overcome these shortcomings, several efforts have been made including the preparation of platensimycin nanoformulations. In one such study, platensimycin-loaded nanoparticles substantially decreased residual bacteria relative to free platensimycin in the MRSA-infected macrophage cell model [73]. Notably, encapsulation of platensimycin with liposomes or micelles significantly enhanced its pharmacokinetic properties in Sprague–Dawley rats with an increase in the survival rate of MRSA-infected C57BL/6J mice [73]. These studies suggest that pitfalls of platensimycin may be overcome either through its chemical modifications or the utilization of better delivery systems.
4.6. Plazomicin
Plazomicin [6′-(hydroxylethyl)-1-(haba)-sisomicin] is a next-generation semisynthetic aminoglycoside antibiotic that originated from sisomicin which was isolated from Micromonopora inyoensis [74]. Plazomicin obtained approval from the US Food and Drug Administration for the treatment of complicated urinary tract infections caused by Gram-negative aerobic bacteria. Importantly, plazomicin is a relatively efficacious aminoglycoside towards carbapenem-resistant Enterobacteriaceae and ESBL-producing Enterobacteriaceae [75,76]. The positively charged plazomicin belongs to the class of aminoglycosides. These classes of antibiotics penetrate the periplasm of bacteria by interacting with the negatively charged cell membrane [77,78]. Further with the help of oxygen-nitrogen dependant electron chain machinery, they enter the aerobic bacteria [77]. The plazomicin is an inhibitor of protein synthesis and showed synergistic activity in combination with the cefepime, doripenem, imipenem, or piperacillin-tazobactam against P. aeruginosa. In addition, it also displayed synergistic antibacterial activity in combination with daptomycin or ceftobiprole against MRSA, hVISA, VISA, and VRSA [79]. The studies of Thwaites et al. showed that plazomicin displayed potent bactericidal activity against MDR E. coli, K. pneumoniae, and Enterobacter spp. with a minimum bactericidal concentration (MBC50/90) of 0.5/ 4 μg/mL respectively [80].
4.7. Teixobactin
Teixobactin is a new class of undecapeptide-containing antibiotic recently isolated using iChip (isolation Chip) technology from the uncultured bacteria Eleftheria terrae, which belongs to the new genus aquabacteria [81]. The iChip technology employs simultaneous isolation and growth of uncultured bacteria in its natural environment by providing suitable nutrients and growth factors [82]. Teixobactin exhibited excellent antibacterial activity against antibiotic-resistant microorganisms such as E. faecalis, E. faecium, MRSA, and many other Gram-positive organisms. The observed antibacterial effect of teixobactin was due to the inhibition of cell wall synthesis by interacting with lipid II (precursor of peptidoglycan) and lipid III (precursor of teichoic acid). The interaction of teixobactin with these precursors interferes with the precursor recycling and synthesis of peptidoglycans and teichoic acid [81]. Interestingly, teixobactin showed no hemolytic activity in human red blood cells and no toxicity against NIH/3T3 and HepG2 cells at 100 μg/mL. Subsequently, after the discovery of this antibiotic, many synthetic routes for the preparation of teixobactin and its analogs were developed using solid and solution phase peptide synthesis 83, 84, 85, 86.
Structurally, teixobactin contains four D-amino acids (D-phenylalanine [N-Me-D-Phe], D-glutamine, D-allo-isoleucine, D-threonine), and one rare amino acid (L-allo-enduracididine) out of 11 residues. In the initial approaches of preparation of teixobactin analogs, L-allo-enduracididine was replaced with various acidic, basic, and neutral amino acids followed by evaluation of their antibacterial potency against MRSA 87, 88, 89. The analogs in which L-allo-enduracididine is substituted with Ile, Leu, Val, and Met were found to have equal or higher antibacterial efficacy than teixobactin suggesting that L-allo-enduracididine is not critical for the antibacterial activity of teixobactin [90,91]. The replacement of N-Me-D-Phe with N-benzyl, N-decyl, N, N-dodecyl, N-decanoyl, N-morpholine, and tetramethyl-N-guanidinylated groups resulted in inactive compounds [92]. On the other hand, 4-phenyl and 4-chlorophenyl analogs of teixobactin were found to be more potent and Phe(4-benzoyl)1-teixobactin was almost three-fold more effective than the teixobactin. These studies suggest that the phenyl moiety of teixobactin plays a key role in imparting antibacterial activity. Concerning this, Zong et al. proposed a hydrophobic interaction between lipid II of the cell wall of the bacteria and the N-terminal of teixobactin analogs [93]. Recently, Gunjal et al. comprehensively reviewed different aspects of teixobactin including its antibiotic potential, the effect of replacement of amino acids on antibacterial activity, synthetic efforts for its preparation, and structural-activity-relationships [90]. Unconventionally and unlike other antibiotics, teixobactin targets lipids for bactericidal activity (rather than proteins) which minimizes the risk of development of drug resistance by mutations of drug targets.
5. Antibacterials from plant origin
5.1. Allicin
Allicin (diallylthiosulfinate) is an organosulfur compound present in the garlic (Allium sativum L.) which is endowed with good antibacterial activity against MRSA, Pseudomonas syringae, Salmonella typhimurium, and Vibrio cholerae [94]. Allicin is synthesized from the non-protein amino acid named alliin by the enzyme alliinase in the garlic [95]. Allicin is a volatile compound and it was used to treat some infections of the respiratory system in the pre-antibiotic era. The antimicrobial effect of the allicin is imparted through its inhibitory activity towards the thiol group-containing enzymes such as alcohol dehydrogenase, thioredoxin reductase, and RNA polymerase [96]. The electrophilic sulfur center of the allicin readily oxidizes thiol groups of the target enzymes to form S-allylmercapto adduct and thereby leads to inactivation of these enzymes. Allicin has also been shown to partially inhibit the synthesis of DNA and proteins. The possible reason for translation inhibition is due to the reaction of allicin with thiol groups of elongating amino acid chains on ribosomes [97]. Some investigations have revealed that allicin augments the antibacterial activity of some of the antibiotics such as cefoperazone, tobramycin, and ciprofloxacin, against P. aeruginosa [98]. Despite being a good antibacterial agent, it is unstable at room temperature and antimicrobial activity is lost within minutes upon heating to >80 °C. The thiosulfinate analogs of allicin such as dimethyl-, diethyl-, diallyl-, dipropyl-, and dibenzyl-thiosulfinates were reported to have variable antibacterial activity against E. coli K12, Pseudomonas fluorescens, and P. syringae pv. phaseolicola 4612, Micrococcus luteus, and were found to be much more thermally stable than allicin [99].
5.2. Berberine
Berberine is a phytogenous isoquinoline alkaloid present in the plants that belong to the family of Berberidaceae, Papaveraceae, and Ranunculaceae [100]. The antibacterial property of berberine is extensively reported in the literature and its age-old usage in traditional Chinese and Ayurvedic medicinal systems against bacterial infections is well-documented. The antibacterial activity of berberine is mediated through diverse mode-of-actions such as intercalation of DNA, inhibition of enzymes (such as RNA polymerase, gyrase, and topoisomerase IV), and blockage of cell division [101]. The antibacterial activity of berberine and in combination with the antibiotics such as ampicillin and oxacillin against the clinical isolates of MRSA was studied and berberine inhibited the growth of MRSA with MIC values ranging from 32 to 128 μg/mL [102]. It has also been identified that berberine decreases the MRSA adhesion and intracellular invasion to the cultured monolayers of human gingival fibroblasts. The antibacterial activity of berberine alone and in combination with ampicillin or oxacillin resulted in additive and synergistic effects against MRSA isolates respectively [102]. In another study, berberine (30–45 μg/mL) induced antibacterial activity and inhibited the biofilm formation in Staphylococcus epidermidis [103]. Berberine also inhibited the biofilm formation in other organisms such as C. albicans, S. typhimurium, and S. aureus 104, 105, 106. Chu et al. provided the mechanistic insight for the inhibition of biofilm formation in MRSA by berberine. It was revealed that berberine inhibits biofilm formation in a dose-dependent manner (1 to 64 μg/mL) by disrupting the aggregation of phenol-soluble modulins (PSMs) into amyloid fibrils [107]. PSMs are a family of amphipathic, α-helical peptides present in staphylococci and have drawn attention due to their involvement in the virulence of S. aureus. PSMs are involved in biofilm development which is thought to have a prominent role in staphylococcal colonization [108]. Berberine displayed a synergistic antibacterial effect with azithromycin against the clinical isolates of P. aeruginosa [109]. The combination of berberine and azithromycin significantly reduced the production of virulence factors such as alginate, LasA protease, LasB protease, pyoverdin, pyocyanin, chitinase, and extracellular DNA. In addition, the combination also inhibits the expression of quorum sensing-related genes such as lasI, lasR, rhlI, and rhlR. Recently, Li et al., demonstrated that berberine exhibit the synergistic effect with sodium new houttuyfonate against the MRSA and VISA, and the effect is likely to be mediated through interruption of the cell membrane [110]. Overall, these reports suggest that berberine has the potential to be developed as a drug that can be administered along with other clinically approved antibacterials.
5.3. Biochanin A
Biochanin A (5,7-dihydroxy-4′‑methoxy isoflavone) is majorly present in the soybean, chickpea, peanuts, Indian rosewood, etc. It was initially isolated from red clover (Trifolium pretense L.), which belongs to the Fabaceae family [111]. Biochanin A showed a synergistic effect in combination with ciprofloxacin against clinical isolates of S. aureus and also reduced the NorA gene expression in MRSA indicating that biochanin A may suppress the efflux of antibiotics [112,113]. In an interesting study, Lechner et al. examined the antibacterial effect of several plant phenolic compounds (such as baicalein, baicalin, biochanin A, daidzein, formononetin, genistein, luteolin, myricetin, resveratrol, chlorpromazine, reserpine, and verapamil) through ethidium bromide efflux assay in Mycobacterium smegmatis. Out of all these compounds, only biochanin A potentiated the effects of ethidium bromide and demonstrated efflux inhibition comparable to standard efflux pump inhibitors [114]. Biochanin A displayed the selective growth inhibitory effect towards Clostridium spp. over Bifidobacterium spp. [115]. The antibacterial activity of biochanin A was reported against pathogenic organisms such as Leishmania chagasi (EC50= 18.96 μg/mL), and Chlamydia pneumoniae (IC50 =12 μM) [116,117]. An inclusion complex of biochanin A with (2-hydroxypropyl)-β-cyclodextrin was prepared to enhance its aqueous solubility and its antibacterial examination against E. coli and K. pneumoniae did not display significant activity over pure biochanin A [118]. Recently, the pharmacological effects of biochanin A have been thoroughly reviewed by Sarfraz et al. [111]. Only a limited number of studies have reported the antimicrobial activity towards ESKAPE organisms. Many more studies are essential to comprehensively demonstrate the mode-of-action of this compound.
5.4. Caffeic acid and its derivatives
Caffeic acid is a phenolic compound present in many plant commodities such as olives, cinnamon, nutmeg, blueberries, apple, star anise, etc. [119]. Chemically, caffeic acid is a 3, 4-dihydroxy cinnamic acid-containing phenylpropanoid with hydroxyl groups at the third and fourth position of the aromatic ring [120]. The plant system contains monomeric, dimeric, and trimeric forms of caffeic acid such as organic acid esters, sugar esters, amides, glycosides, etc. The caffeic acid is endowed with diverse pharmacological properties such as antibacterial, antiviral, anti-inflammatory, antioxidant, antiatherosclerotic, immune-stimulatory, antidiabetic, cardioprotective, antiproliferative, hepatoprotective, anticancer, and many other activities. The poor aqueous solubility and lack of stability are the major limitations of caffeic acid and some of its derivatives. They get degraded rapidly in vitro and in vivo setup and hence attempts were made to encapsulate caffeic acid and its derivatives into nanoparticles to increase the water solubility. Caffeic acid imparted growth inhibitory activity against S. aureus and the observed antibacterial activity of caffeic acid is due to the presence of hydroxyl groups in the phenolic ring [121]. The different derivatives of caffeic acid amides showed considerable antibacterial activity against B. subtilis with MIC values ranging between 1.18 and 15.5 µg/mL. The caffeic acid conjugated with chitosan exhibited a synergistic antibacterial effect in combination with antibiotics such as tetracycline, erythromycin, and lincomycin against S. aureus, S. epidermidis, P. aeruginosa, Propionibacterium acnes [122]. The observed growth inhibitory effects of the conjugates could be due to the alteration of bacterial membrane permeability. Caffeic acid-coated polymeric fibrous materials such as poly(3-hydroxybutyrate) and poly(ethylene glycol) showed antibacterial activity against S. aureus and E. coli and they were proposed to be useful in controlling wound infections [123]. To address the solubility and stability issues of caffeic acid, several nano-formulations of caffeic acid were made and their antibacterial activity was studied. The caffeic acid immobilized on zinc oxide nanoparticles showed strong antimicrobial activity against S. aureus, MRSA, and E. coli [124]. The encapsulation of caffeic acid with cyclodextrins improved the solubility of caffeic acid and was found to have enhanced antibacterial activity against the K. pneumoniae and S. aureus [125].
The caffeic acid phenethyl ester (CAPE) exhibited antibacterial activity against both Gram-positive and Gram-negative organisms such as S. aureus, Streptococcus mutans, Streptococcus mitis, Bacillus megaterium, B. subtilis, Lactobacillus acidophilus, etc. [126]. In another study, CAPE displayed remarkable antimicrobial activity against common oral cariogenic bacteria (S. mutans, Streptococcus sobrinus, Actinomyces viscosus, and L. acidophilus) [127]. Additionally, CAPE inhibited the formation of S. mutans biofilms and their metabolic activity in mature biofilms. CAPE has been reported to induce antibacterial effects through various mechanisms including permeabilization of the cell membrane, alteration of membrane transport, membrane depolarization, and inhibition of bacterial RNA polymerase [128]. The caffeic acid entrapped in silica gel matrix presented good antibacterial activity against E. faecalis and E. coli. In view of all these studies, caffeic acid and its derivatives are emerging as potential antibacterial agents against a wide variety of bacteria.
Chlorogenic acid is one of the naturally occurring derivatives of caffeic acid seen in coffee, tea, apples, pears, carrots, tomatoes, sweet potatoes, grapes, etc. Chlorogenic acid is a major constituent of some traditional Chinese medicine preparations and it is endowed with many medicinal properties including antibacterial activity against many Gram-positive and Gram-negative organisms [129,130]. A significant synergetic bactericidal and antibiofilm effect was produced against S. aureus by the combination of ultrasound and chlorogenic acid treatment which was mediated through increasing the cell membrane permeability, release of ATP, and nucleic acids, and decreasing the exopolysaccharide contents in the biofilm [131]. Similarly, chlorogenic acid presented strong bactericidal and antibiofilm activity in the clinical isolates of Stenotrophomonas maltophilia including the trimethoprim/sulfamethoxazole resistant strain [132]. The antimicrobial roles of caffeic acid, its combinational uses, and synthetic strategies have been thoroughly reviewed recently [119]. These reports suggest that the caffeic acid derivatives or structural analogs could serve as potent chemicals against bacteria with pathological relevance.
5.5. Piperine
Piperine is one of the primary alkaloids present in the black pepper (Piper nigrum L.) and long pepper (Piper longum L.). The piperine was first isolated by Hans Christian Ørsted in 1819 from the extracts of pepper as a yellow crystalline compound. The IUPAC name of piperine is 1-(5-[1,3-benzodioxol5-yl]-1-oxo-2,4-pentadienyl)piperidine and it is present in four isomeric forms such as piperine, isopiperine, chavicine, and isochavicine [133]. Some of the piperine analogs were reported as the inhibitors of NorA efflux pumps in S. aureus [134]. Multidrug efflux pumps are the leading player for bacterial antibiotic resistance due to their involvement in the expulsion of antibiotics out of the cell. NorA multidrug-resistant efflux pump contributes to the reduced susceptibility of S. aureus to antibiotics [135]. Notably, inhibition of efflux pumps using natural and synthetic compounds has emerged as an important strategy to counteract bacterial multidrug resistance [136]. Piperine potentiated the effect of ciprofloxacin against S. aureus which resulted in a two-fold reduction of the MIC value of ciprofloxacin [137]. The nanoliposomes with co-loaded piperine and gentamicin displayed good antibacterial activity with almost a 32-fold reduction of MIC value compared to free gentamycin against the MRSA [138] and these activities are proposed to be imparted through the inhibition of efflux pumps [134]. These reports emphasize the efflux pump inhibitory activity of piperine and the need for its evaluation with other antibiotics against drug-resistant organisms.
5.6. Taxifolin
Taxifolin is chemically known as 3,5,7,3,4-pentahydroxy flavanone or dihydroquercetin, a polyphenolic flavonoid, which is majorly present in plants such as Silybum marianum, Allium cepa, Pseudotsuga taxifolia, and Pinus pinaster. Taxifolin possesses diverse medicinal properties such as anti-inflammatory, anticancer, antiangiogenic, hepatoprotective, etc. Taxifolin was found to have antibacterial activity against S. sobrinus, a pathogen responsible for dental caries, with an IC50 value of 21.8 ± 1.7 μg/mL [139]. It also showed good inhibition towards glucosyltransferase derived from S. sobrinus with an IC50 value of 53.0 ± 0.7 μg/mL [139]. The inhibition of glucosyltransferase is one of the strategies in the prevention of dental caries and this report suggests that taxifolin can be developed as a potent agent for preventing dental caries. Taxifolin inhibited the growth of E. faecalis and VREF with MIC values of 128 and 512 μg/mL, respectively [140]. Based on the docking studies, the authors demonstrated that β-ketoacyl acyl carrier protein synthase III, a key enzyme in bacterial fatty acid biosynthesis, is the potential target of taxifolin with a binding affinity of 6.76 × 107 M−1 [140]. In another study, the antibacterial activity of taxifolin was evaluated at 0.5, 1.0, 2.0, and 5.0% concentrations against E. coli, S. epidermidis, P. aeruginosa, and M. luteus and out of which S. epidermidis displayed high sensitivity to taxifolin at 5% concentration [141]. DNA gyrase and aminoacyl-tRNA synthetases are the bacterial enzymes crucial for DNA replication and protein synthesis, respectively. Eight flavonoids were investigated for their inhibitory activity towards DNA gyrase and isoleucyl-tRNA synthetase on a molecular docking platform and taxifolin was identified as a lead inhibitor [142]. They also reported that taxifolin interacts with a good number of amino acids at the active sites of DNA gyrase and aminoacyl-tRNA synthetase of M. tuberculosis based on the in silico molecular docking and dynamics simulation approach. Additionally, taxifolin displayed antimycobacterial activity against M. tuberculosis with a MIC value of ≤ 12.5 μg/mL [142]. In a recent investigation, taxifolin was reported as the inhibitor of sortase A of MRSA, an enzyme essential for bacterial colonization in the host tissues [143]. Taxifolin is also believed to be effective against many Gram-positive organisms which demand further investigation [143].
5.7. Thymol
Thymol (also known as 2-isopropyl-5-methylphenol) is a primary monoterpene present in the essential oils of plants belonging to the genera of Thymus, Ocimum, Origanum, Satureja, Thymbra, and Monarda. The usage of essential oils of these plants as a flavoring and preservative agent is an age-old practice in the food industry. The FDA has listed the thymol and other components present in these essential oils as ‘generally recognized as safe’ (GRAS) for their supplementation in food products [144]. Many studies reported the antibacterial activity of essential oils that are rich in thymol against various Gram-positive and Gram-negative organisms. Thymol presented antibacterial activity against clinical isolates and standard cultures of MRSA and S. epidermidis and the results showed that all isolates were susceptible to thymol with MIC values ranging from 0.03 to 0.06% (v/v) [145]. Thymol displayed antibacterial activity against S. aureus and E. coli with a MIC of 0.31 and 5.0 mg/mL respectively and mode-of-action analysis revealed that thymol influences the permeability of the lipid membrane of the bacteria which results in leakage of cellular contents [146]. Xu et al. showed that thymol induces bactericidal activity against E. coli through the permeabilization and depolarization of the bacterial cytoplasmic membrane. They showed the potential use of flow cytometry as a tool for the investigation of the mode of antibacterial action of essential oil components [147]. The chemical derivatives of thymol were examined for antibacterial effect against S. mutans, S. aureus, B. subtilis, S. epidermidis, and E. coli. Thymyl acetate (2-isopropyl-5-methylphenyl acetate), thymyl propionate (2-isopropyl-5-methylphenyl propionate), and thymyl isobutyrate (2-isopropyl-5-methylphenyl isobutyrate) displayed consistent activity against all the Gram-positive strains [148]. The effect of thymol against Salmonella spp. biofilm on polypropylene was examined. Thymol suppressed bacterial counts about 1–2 log during biofilm formation, reduction of about 1–5 log in the established biofilms [149]. These results suggest the antibiofilm potential of thymol against Salmonella spp. which can be examined in other bacterial systems. In another study, thymol imparted antibacterial activity towards S. aureus, S. epidermidis, B. cereus, E. coli, P. aeruginosa, E. feacalis, Vibrio paraheamolyticus, Vibrio alginolyticus, and Salmonella enteritidis with MIC values ranging between 32 and 64 μg/mL [150]. Additionally, thymol reduced the MIC values of tetracycline and benzalkonium chloride towards these microorganisms. Thymol induced the intracellular accumulation of ethidium bromide in a dosage-dependent manner indicating that thymol may sensitize bacteria to antibiotics through the inhibition of efflux pumps [150]. Marchese et al. thoroughly reviewed the antibacterial and antifungal effects of thymol and its derivatives [144]. Taken together, thymol or its derivatives may be developed as antibacterials towards drug-sensitive/-resistant microbes.
6. Conclusion and future perspectives
ESKAPE organisms are posing a life-threatening condition in the global context and the discovery of new potent antibiotics against these organisms is the need of the hour. ESKAPE organisms are heterogenous at the genetic level, but share common resistance mechanisms such as the chemical modification or breakdown of the drug, prevention of antibiotic influx, antibiotic expulsion through efflux pumps, modification of antibiotic target, etc. It is a global challenge for the discovery of novel potent bactericidal agents with favorable toxicity profiles. Many pathogenic bacteria are developing various strategies to counteract the bactericidal effect of most effective antibiotics through different mechanisms which pose an alarm for the need for newer antibiotics. It is important to note that, a large number of secondary metabolites have been isolated from bacteria, fungi, plants, and marine invertebrates and have been developed as potent drugs against various human ailments 151, 152, 153, 154, 155. Over 13,000 natural compounds have been isolated from bacterial sources indicating that these microorganisms remain as a treasure house of bioactive compounds [156,157]. Beside, about 70% of these natural compounds have been extracted from actinomycetes (a group of Gram-positive mycelial bacteria) and merely 1% of actinomycetes have been cultured so far indicating the abundance of hidden bioactive compounds in the bacterial kingdom. It is highly essential to speed up the discovery of bioactive compounds from unexplored bacterial and plant sources 158, 159, 160, 161. Antibiotic discovery faces several constraints. Since from 1960s, very few classes of antibiotics were discovered such as quinolones, lincosamides, oxazolidinones, and cyclic lipopeptides which still have a major share in the world's global antibiotic market [162]. Beside, novel antibiotics that were discovered later on were all majorly derived from established antibiotic classes and they are used as last-resort antibiotics which further discouraged the investment in antibiotic drug discovery research by many pharmaceutical industries [163]. In addition, the expensive operational cost of clinical trials poses a significant constraint in the R & D of antimicrobial drug design. Despite these constraints, several alternate strategies are employed in the current antibiotic research to counter the antimicrobial resistance of ESKAPE organisms. Combinatorial drug therapy is one such strategy where adjuvants are used along with the antibiotics to improve the patient response. To combat against the threatening antimicrobial resistance of ESKAPE organisms, other various novel strategies utilized nowadays are repurposing of drugs, monoclonal antibody therapy, vaccine development, bacteriophage therapy, etc. Along with these various strategies, concerted efforts are needed in the global and regional platforms in policymaking, funding, guidelines for appropriate use of antibiotics, diagnostics and treatment, and employment strategies applied against ESKAPE pathogens to keep the antimicrobial resistance mechanisms of these organisms under surveillance and control.
Declaration of Competing Interest
I, Prof Rangappa on behalf of all the co-authors declare that there is no conflict of interest associated with this study. I also declare that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. I also declare that the manuscript has been solely submitted to Biotechnology Reports and it is not under consideration in any other journal for publication.
Contributor Information
Chakrabhavi Dhananjaya Mohan, Email: cd.mohan@yahoo.com.
Kanchugarakoppal S. Rangappa, Email: rangappaks@gmail.com.
References
- 1.Walsh C. American Society for Microbiology (ASM); 2003. Antibiotics: Actions, Origins, Resistance. [Google Scholar]
- 2.Gelband H., Molly Miller P., Pant S., Gandra S., Levinson J., Barter D., White A., Laxminarayan R. The state of the world's antibiotics 2015. Wound Heal. South. Afr. 2015;8(2):30–34. [Google Scholar]
- 3.Coates A., Hu Y., Bax R., Page C. The future challenges facing the development of new antimicrobial drugs. Nat. Rev. Drug Discov. 2002;1(11):895–910. doi: 10.1038/nrd940. [DOI] [PubMed] [Google Scholar]
- 4.Mukhopadhyay S., Bharath Prasad A., Mehta C.H., Nayak U.Y. Antimicrobial peptide polymers: no escape to ESKAPE pathogens—a review. World J. Microbiol. Biotechnol. 2020;36(9):1–14. doi: 10.1007/s11274-020-02907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice L.B. vol. 197. The University of Chicago Press; 2008. (Federal Funding For the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE). [DOI] [PubMed] [Google Scholar]
- 6.Santajit S., Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016;2016 doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watson D., Spaulding A.B., Dreyfus J. Risk-set matching to assess the impact of hospital-acquired bloodstream infections. Am. J. Epidemiol. 2018;188(2):461–466. doi: 10.1093/aje/kwy252. [DOI] [PubMed] [Google Scholar]
- 8.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 9.Bush K., Bradford P.A. Interplay between β-lactamases and new β-lactamase inhibitors. Nat. Rev. Microbiol. 2019;17(5):295–306. doi: 10.1038/s41579-019-0159-8. [DOI] [PubMed] [Google Scholar]
- 10.Naas T., Oueslati S., Bonnin R.A., Dabos M.L., Zavala A., Dortet L., Retailleau P., Iorga B.I. Beta-lactamase database (BLDB)–structure and function. J. Enzym. Inhib. Med. Chem. 2017;32(1):917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tooke C.L., Hinchliffe P., Bragginton E.C., Colenso C.K., Hirvonen V.H.A., Takebayashi Y., Spencer J. β-Lactamases and β-Lactamase inhibitors in the 21st century. J. Mol. Biol. 2019;431(18):3472–3500. doi: 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bush K., Jacoby G.A. Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 2010;54(3):969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter S., Doern G., Heilmann K., Miner S., Tendolkar S., Riahi F., Diekema D. Detection and prevalence of penicillin-susceptible Staphylococcus aureus in the United States in 2013. J. Clin. Microbiol. 2016;54(3):812–814. doi: 10.1128/JCM.03109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Oliveira D.M., Forde B.M., Kidd T.J., Harris P.N., Schembri M.A., Beatson S.A., Paterson D.L., Walker M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020;33(3):e00119–e00181. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canton R., Morosini M., Martin O., De la Maza S., De La Pedrosa E.G.G. IRT and CMT β-lactamases and inhibitor resistance. Clin. Microbiol. Infect. 2008;14:53–62. doi: 10.1111/j.1469-0691.2007.01849.x. [DOI] [PubMed] [Google Scholar]
- 16.Nordmann P., Cuzon G., Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 2009;9(4):228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 17.Vena A., Castaldo N., Bassetti M. The role of new β-lactamase inhibitors in Gram-negative infections. Curr. Opin. Infect. Dis. 2019;32(6):638–646. doi: 10.1097/QCO.0000000000000600. [DOI] [PubMed] [Google Scholar]
- 18.Lauretti L., Riccio M.L., Mazzariol A., Cornaglia G., Amicosante G., Fontana R., Rossolini G.M. Cloning and characterization of bla VIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999;43(7):1584–1590. doi: 10.1128/aac.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumura Y., Peirano G., Devinney R., Bradford P.A., Motyl M.R., Adams M.D., Chen L., Kreiswirth B., Pitout J.D. Genomic epidemiology of global VIM-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2017;72(8):2249–2258. doi: 10.1093/jac/dkx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peirano G., Matsumura Y., Adams M.D., Bradford P., Motyl M., Chen L., Kreiswirth B.N., Pitout J.D. Genomic epidemiology of global carbapenemase-producing Enterobacter spp., 2008–2014. Emerg. Infect. Dis. 2018;24(6):1010. doi: 10.3201/eid2406.171648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jovcic B., Lepsanovic Z., Suljagic V., Rackov G., Begovic J., Topisirovic L., Kojic M. Emergence of NDM-1 metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob. Agents Chemother. 2011;55(8):3929–3931. doi: 10.1128/AAC.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitout J.D., Peirano G., Kock M.M., Strydom K.A., Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin. Microbiol. Rev. 2019;33(1):e00102–e00119. doi: 10.1128/CMR.00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Pascale G., Wright G.D. Antibiotic resistance by enzyme inactivation: from mechanisms to solutions. ChemBioChem. 2010;11(10):1325–1334. doi: 10.1002/cbic.201000067. [DOI] [PubMed] [Google Scholar]
- 24.Krause K.M., Serio A.W., Kane T.R., Connolly L.E. Aminoglycosides: an overview. Cold Spring Harb. Perspect. Med. 2016;6(6) doi: 10.1101/cshperspect.a027029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holbrook S.Y., Garneau-Tsodikova S. Evaluation of aminoglycoside and carbapenem resistance in a collection of drug-resistant Pseudomonas aeruginosa clinical isolates. Microb. Drug Resist. 2018;24(7):1020–1030. doi: 10.1089/mdr.2017.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costa B.O., Cardoso M.H., Franco O.L. Development of peptides that inhibit aminoglycoside-modifying enzymes and β-lactamases for control of resistant bacteria. Curr. Protein Pept. Sci. 2020;21(10):1011–1026. doi: 10.2174/1389203721666200915113630. [DOI] [PubMed] [Google Scholar]
- 27.Shi W.F., Jiang J.P., Mi Z.H. Relationship between antimicrobial resistance and aminoglycoside-modifying enzyme gene expressions in Acinetobacter baumannii. Chin. Med. J. 2005;118(2):141–145. [PubMed] [Google Scholar]
- 28.Egorov A., Ulyashova M., Rubtsova M.Y. Bacterial enzymes and antibiotic resistance. Acta Nat. 2018;4(39):10. (англоязычная версия) [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez M.S., Tolmasky M.E. Aminoglycoside modifying enzymes. Drug Resist. Updates. 2010;13(6):151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castanheira M., Davis A.P., Serio A.W., Krause K.M., Mendes R.E. In vitro activity of plazomicin against Enterobacteriaceae isolates carrying genes encoding aminoglycoside-modifying enzymes most common in US Census divisions. Diagn. Microbiol. Infect. Dis. 2019;94(1):73–77. doi: 10.1016/j.diagmicrobio.2018.10.023. [DOI] [PubMed] [Google Scholar]
- 31.Costello S.E., Deshpande L.M., Davis A.P., Mendes R.E., Castanheira M. Aminoglycoside-modifying enzyme and 16S ribosomal RNA methyltransferase genes among a global collection of Gram-negative isolates. J. Glob. Antimicrob. Resist. 2019;16:278–285. doi: 10.1016/j.jgar.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 32.Benveniste R., Davies J. R-factor mediated gentamicin resistance: a new enzyme which modifies aminoglycoside antibiotics. FEBS Lett. 1971;14(5):293–296. doi: 10.1016/0014-5793(71)80282-x. [DOI] [PubMed] [Google Scholar]
- 33.Limansky A.S., Mussi M.A., Viale A.M. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 2002;40(12):4776–4778. doi: 10.1128/JCM.40.12.4776-4778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuoka T., Ohya S., Narita T., Katsuta M., Iijima M., Masuda N., Yasuda H., Trias J., Nikaido H. Activity of the carbapenem panipenem and role of the OprD (D2) protein in its diffusion through the Pseudomonas aeruginosa outer membrane. Antimicrob. Agents Chemother. 1993;37(2):322–327. doi: 10.1128/AAC.37.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piddock L.J.V. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006;19(2):382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du D., Wang-Kan X., Neuberger A., van Veen H.W., Pos K.M., Piddock L.J.V., Luisi B.F. Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 2018;16(9):523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 37.Dean C.R., Visalli M.A., Projan S.J., Sum P.E., Bradford P.A. Efflux-mediated resistance to tigecycline (GAR-936) in pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 2003;47(3):972–978. doi: 10.1128/AAC.47.3.972-978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernando D.M., Kumar A. Resistance-nodulation-division multidrug efflux pumps in gram-negative bacteria: role in virulence. Antibiotics. 2013;2(1):163–181. doi: 10.3390/antibiotics2010163. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munita J.M., Arias C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016;4(2) doi: 10.1128/microbiolspec.VMBF-0016-2015. 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fyfe C., Grossman T.H., Kerstein K., Sutcliffe J. Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb. Perspect. Med. 2016;6(10) doi: 10.1101/cshperspect.a025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohan C.D., Bharathkumar H., Bulusu K.C., Pandey V., Rangappa S., Fuchs J.E., Shanmugam M.K., Dai X., Li F., Deivasigamani A., Hui K.M., Kumar A.P., Lobie P.E., Bender A., Basappa S., Sethi G., Rangappa K.S. Development of a novel azaspirane that targets the janus kinase-signal transducer and activator of transcription (STAT) pathway in hepatocellular carcinoma in vitro and in vivo*. J. Biol. Chem. 2014;289(49):34296–34307. doi: 10.1074/jbc.M114.601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baburajeev C.P., Dhananjaya Mohan C., Ananda H., Rangappa S., Fuchs J.E., Jagadish S., Sivaraman Siveen K., Chinnathambi A., Ali Alharbi S., Zayed M.E., Zhang J., Li F., Sethi G., Girish K.S., Bender A., Basappa, Rangappa K.S. Development of novel triazolo-thiadiazoles from heterogeneous “green” catalysis as protein tyrosine phosphatase 1B inhibitors. Sci. Rep. 2015;5(1):14195. doi: 10.1038/srep14195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey V., Wang B., Mohan C.D., Raquib A.R., Rangappa S., Srinivasa V., Fuchs J.E., Girish K.S., Zhu T., Bender A., Ma L., Yin Z., Basappa Rangappa KS, Lobie P.E. Discovery of a small-molecule inhibitor of specific serine residue BAD phosphorylation. Proc. Natl. Acad. Sci. 2018;115(44):E10505–E10514. doi: 10.1073/pnas.1804897115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J.H., Mohan C.D., Deivasigamani A., Jung Y.Y., Rangappa S., Basappa S., Chinnathambi A., Alahmadi T.A., Alharbi S.A., Garg M., Lin Z.X., Rangappa K.S., Sethi G., Hui K.M., Ahn K.S. Brusatol suppresses STAT3-driven metastasis by downregulating epithelial-mesenchymal transition in hepatocellular carcinoma. J. Adv. Res. 2020;26:83–94. doi: 10.1016/j.jare.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohan C.D., Liew Y.Y., Jung Y.Y., Rangappa S., Preetham H.D., Chinnathambi A., Alahmadi T.A., Alharbi S.A., Lin Z.X., Rangappa K.S., Ahn K.S. Brucein D modulates MAPK signaling cascade to exert multi-faceted anti-neoplastic actions against breast cancer cells. Biochimie. 2021;182:140–151. doi: 10.1016/j.biochi.2021.01.009. [DOI] [PubMed] [Google Scholar]
- 46.Mohan C.D., Yang M.H., Rangappa S., Chinnathambi A., Alharbi S.A., Alahmadi T.A., Deivasigamani A., Hui K.M., Sethi G., Rangappa K.S., Ahn K.S. 3-formylchromone counteracts STAT3 signaling pathway by elevating SHP-2 expression in hepatocellular carcinoma. Biology. 2022;11(1):29. doi: 10.3390/biology11010029. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manu K.A., Shanmugam M.K., Ramachandran L., Li F., Fong C.W., Kumar A.P., Tan P., Sethi G. First evidence that γ-tocotrienol inhibits the growth of human gastric cancer and chemosensitizes it to capecitabine in a xenograft mouse model through the modulation of NF-κB pathway. Clin. Cancer Res. 2012;18(8):2220–2229. doi: 10.1158/1078-0432.ccr-11-2470. [DOI] [PubMed] [Google Scholar]
- 48.Ahn K.S., Sethi G., Chaturvedi M.M., Aggarwal B.B. Simvastatin, 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor, suppresses osteoclastogenesis induced by receptor activator of nuclear factor-kappaB ligand through modulation of NF-kappaB pathway. Int. J. Cancer. 2008;123(8):1733–1740. doi: 10.1002/ijc.23745. [DOI] [PubMed] [Google Scholar]
- 49.Ramachandran L., Manu K.A., Shanmugam M.K., Li F., Siveen K.S., Vali S., Kapoor S., Abbasi T., Surana R., Smoot D.T., Ashktorab H., Tan P., Ahn K.S., Yap C.W., Kumar A.P., Sethi G. Isorhamnetin inhibits proliferation and invasion and induces apoptosis through the modulation of peroxisome proliferator-activated receptor γ activation pathway in gastric cancer. J. Biol. Chem. 2012;287(45):38028–38040. doi: 10.1074/jbc.M112.388702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keller S., Nicholson G., Drahl C., Sorensen E., Fiedler H.P., Süssmuth R.D. Abyssomicins G and H and atrop-abyssomicin C from the marine Verrucosispora strain AB-18-032. J. Antibiot. 2007;60(6):391–394. doi: 10.1038/ja.2007.54. [DOI] [PubMed] [Google Scholar]
- 51.Riedlinger J., Reicke A., Zähner H., Krismer B., Bull A.T., Maldonado L.A., Ward A.C., Goodfellow M., Bister B., Bischoff D. Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J. Antibiot. 2004;57(4):271–279. doi: 10.7164/antibiotics.57.271. [DOI] [PubMed] [Google Scholar]
- 52.Freundlich J.S., Lalgondar M., Wei J.R., Swanson S., Sorensen E.J., Rubin E.J., Sacchettini J.C. The abyssomicin C family as in vitro inhibitors of Mycobacterium tuberculosis. Tuberculosis. 2010;90(5):298–300. doi: 10.1016/j.tube.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicolaou K., Harrison S.T., Chen J.S. Discoveries from the abyss: the abyssomicins and their total synthesis. Synthesis. 2009;2009(01):33–42. doi: 10.1055/s-0028-1083259. (Mass) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.KeC Nicolaou, Chen J.S., Edmonds D.J., Estrada A.A. Recent advances in the chemistry and biology of naturally occurring antibiotics. Angew. Chem. Int. Ed. 2009;48(4):660–719. doi: 10.1002/anie.200801695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keller S., Schadt H.S., Ortel I., Süssmuth R.D. Action of atrop-abyssomicin C as an inhibitor of 4-Amino-4-deoxychorismate synthase PabB. Angew. Chem. Int. Ed. 2007;46(43):8284–8286. doi: 10.1002/anie.200701836. [DOI] [PubMed] [Google Scholar]
- 56.Monjas L., Fodran P., Kollback J., Cassani C., Olsson T., Genheden M., Larsson D.J., Wallentin C.J. Synthesis and biological evaluation of truncated derivatives of abyssomicin C as antibacterial agents. Beilstein J. Org. Chem. 2019;15(1):1468–1474. doi: 10.3762/bjoc.15.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jang K.H., Nam S.J., Locke J.B., Kauffman C.A., Beatty D.S., Paul L.A., Fenical W. Anthracimycin, a potent anthrax antibiotic from a marine-derived actinomycete. Angew. Chem. 2013;125(30):7976–7978. doi: 10.1002/anie.201302749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sirota F.L., Goh F., Low K.N., Yang L.K., Crasta S.C., Eisenhaber B., Eisenhaber F., Kanagasundaram Y., Ng S.B. Isolation and identification of an Anthracimycin analogue from Nocardiopsis kunsanensis, a halophile from a Saltern, by genomic mining strategy. J Genom. 2018;6:63. doi: 10.7150/jgen.24368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodríguez V., Martín J., Sarmiento-Vizcaíno A., De la Cruz M., García L.A., Blanco G., Reyes F. Anthracimycin B, a potent antibiotic against gram-positive bacteria isolated from cultures of the deep-sea actinomycete Streptomyces cyaneofuscatus M-169. Mar. Drugs. 2018;16(11):406. doi: 10.3390/md16110406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davison E.K., Freeman J.L., Zhang W., Wuest W.M., Furkert D.P., Brimble M.A. Asymmetric Total Synthesis of the Naturally Occurring Antibiotic Anthracimycin. Org. Lett. 2020;22(14):5550–5554. doi: 10.1021/acs.orglett.0c01913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ledger E.V.K., Pader V., Edwards A.M. Enterococcus faecalis and pathogenic streptococci inactivate daptomycin by releasing phospholipids. Microbiology. 2017;163(10):1502–1508. doi: 10.1099/mic.0.000529. (Reading, Engl.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cafiso V., Stracquadanio S., Lo Verde F., De Guidi I., Zega A., Pigola G., Stefani S. Genomic and long-term transcriptomic imprints related to the daptomycin mechanism of action occurring in daptomycin- and methicillin-resistant staphylococcus aureus under daptomycin exposure. Front. Microbiol. 2020;11:1893. doi: 10.3389/fmicb.2020.01893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seaton R.A., Menichetti F., Dalekos G., Beiras-Fernandez A., Nacinovich F., Pathan R., Hamed K. Evaluation of effectiveness and safety of high-dose daptomycin: results from patients included in the European cubicin® outcomes registry and experience. Adv. Ther. 2015;32(12):1192–1205. doi: 10.1007/s12325-015-0267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pader V., Hakim S., Painter K.L., Wigneshweraraj S., Clarke T.B., Edwards A.M. Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat. Microbiol. 2016;2:16194. doi: 10.1038/nmicrobiol.2016.194. [DOI] [PubMed] [Google Scholar]
- 65.Phillips J.W., Goetz M.A., Smith S.K., Zink D.L., Polishook J., Onishi R., Salowe S., Wiltsie J., Allocco J., Sigmund J. Discovery of kibdelomycin, a potent new class of bacterial type II topoisomerase inhibitor by chemical-genetic profiling in Staphylococcus aureus. Chem. Biol. 2011;18(8):955–965. doi: 10.1016/j.chembiol.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 66.Singh S.B., Goetz M.A., Smith S.K., Zink D.L., Polishook J., Onishi R., Salowe S., Wiltsie J., Allocco J., Sigmund J. Kibdelomycin A, a congener of kibdelomycin, derivatives and their antibacterial activities. Bioorg. Med. Chem. Lett. 2012;22(23):7127–7130. doi: 10.1016/j.bmcl.2012.09.071. [DOI] [PubMed] [Google Scholar]
- 67.Lu J., Patel S., Sharma N., Soisson S.M., Kishii R., Takei M., Fukuda Y., Lumb K.J., Singh S.B. Structures of kibdelomycin bound to Staphylococcus aureus GyrB and ParE showed a novel U-shaped binding mode. ACS Chem. Biol. 2014;9(9):2023–2031. doi: 10.1021/cb5001197. [DOI] [PubMed] [Google Scholar]
- 68.Miesel L., Hecht D.W., Osmolski J.R., Gerding D., Flattery A., Li F., Lan J., Lipari P., Polishook J.D., Liang L. Kibdelomycin is a potent and selective agent against toxigenic Clostridium difficile. Antimicrob. Agents Chemother. 2014;58(4):2387–2392. doi: 10.1128/AAC.00021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh S.B., Dayananth P., Balibar C.J., Garlisi C.G., Lu J., Kishii R., Takei M., Fukuda Y., Ha S., Young K. Kibdelomycin is a bactericidal broad-spectrum aerobic antibacterial agent. Antimicrob. Agents Chemother. 2015;59(6):3474–3481. doi: 10.1128/AAC.00382-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martens E., Demain A.L. Platensimycin and platencin: promising antibiotics for future application in human medicine. J. Antibiot. 2011;64(11):705–710. doi: 10.1038/ja.2011.80. [DOI] [PubMed] [Google Scholar]
- 71.Wang J., Soisson S.M., Young K., Shoop W., Kodali S., Galgoci A., Painter R., Parthasarathy G., Tang Y.S., Cummings R., Ha S., Dorso K., Motyl M., Jayasuriya H., Ondeyka J., Herath K., Zhang C., Hernandez L., Allocco J., Basilio A., Tormo J.R., Genilloud O., Vicente F., Pelaez F., Colwell L., Lee S.H., Michael B., Felcetto T., Gill C., Silver L.L., Hermes J.D., Bartizal K., Barrett J., Schmatz D., Becker J.W., Cully D., Singh S.B. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441(7091):358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 72.Su M., Qiu L., Deng Y., Ruiz C.H., Rudolf J.D., Dong L.B., Feng X., Cameron M.D., Shen B., Duan Y., Huang Y. Evaluation of platensimycin and platensimycin-inspired thioether analogues against methicillin-resistant staphylococcus aureus in topical and systemic infection mouse models. Mol. Pharm. 2019;16(7):3065–3071. doi: 10.1021/acs.molpharmaceut.9b00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deng Y., Weng X., Li Y., Su M., Wen Z., Ji X., Ren N., Shen B. (2019) Late-stage functionalization of platensimycin leading to multiple analogues with improved antibacterial activity in vitro and in vivo. 62 (14):6682–6693. 10.1021/acs.jmedchem.9b00616. [DOI] [PMC free article] [PubMed]
- 74.Boumehira A.Z., El-Enshasy H.A., Hacène H., Elsayed E.A., Aziz R., Park E.Y. Recent progress on the development of antibiotics from the genus Micromonospora. Biotechnol. Bioprocess Eng. 2016;21(2):199–223. [Google Scholar]
- 75.Dozzo P., Moser H.E. New aminoglycoside antibiotics. Expert Opin. Ther. Pat. 2010;20(10):1321–1341. doi: 10.1517/13543776.2010.506189. [DOI] [PubMed] [Google Scholar]
- 76.Hover B.M., Kim S.H., Katz M., Charlop-Powers Z., Owen J.G., Ternei M.A., Maniko J., Estrela A.B., Molina H., Park S., Perlin D.S., Brady S.F. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 2018;3(4):415–422. doi: 10.1038/s41564-018-0110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Citron D., Tyrrell K., Leoncio E., Serio A., Krause K., Goldstein E. (2017) In vitro activity of plazomicin (PLZ) and comparators against facultative anaerobes incubated aerobically and anaerobically and obligate anaerobes (abstract no. 333 plus poster). 2nd American Society for Microbiology Microbe, New Orleans.
- 78.Denervaud-Tendon V., Poirel L., Connolly L.E., Krause K.M., Nordmann P. Plazomicin activity against polymyxin-resistant Enterobacteriaceae, including MCR-1-producing isolates. J. Antimicrob. Chemother. 2017;72(10):2787–2791. doi: 10.1093/jac/dkx239. [DOI] [PubMed] [Google Scholar]
- 79.Ramirez M.S., Tolmasky M.E. Amikacin: uses, resistance, and prospects for inhibition. Molecules. 2017;22(12):2267. doi: 10.3390/molecules22122267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thwaites M., Hall D., Shinabarger D., Serio A., Krause K., Marra A., Pillar C. Evaluation of the bactericidal activity of plazomicin and comparators against multidrug-resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2018;62(8):e00218–e00236. doi: 10.1128/AAC.00236-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P., Mueller A., Schäberle T.F., Hughes D.E., Epstein S. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517(7535):455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nichols D., Cahoon N., Trakhtenberg E., Pham L., Mehta A., Belanger A., Kanigan T., Lewis K., Epstein S. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010;76(8):2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin K., Sam I.H., Po K.H.L., Zadeh E.H.G., Chen S., Yuan Y., Li X. Total synthesis of teixobactin. Nat. Commun. 2016;7(1):1–6. doi: 10.1038/ncomms12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giltrap A. Total Synthesis of Natural Products With Antimicrobial Activity. Springer Singapore; Singapore: 2018. Total synthesis of teixobactin; pp. 33–69. [DOI] [Google Scholar]
- 85.Liu L., Wu S., Wang Q., Zhang M., Wang B., He G., Chen G. Total synthesis of teixobactin and its stereoisomers. Org. Chem. Front. 2018;5(9):1431–1435. [Google Scholar]
- 86.Yang H., Chen K.H., Nowick J.S. Elucidation of the teixobactin pharmacophore. ACS Chem. Biol. 2016;11(7):1823–1826. doi: 10.1021/acschembio.6b00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parmar A., Iyer A., Vincent C.S., Van Lysebetten D., Prior S.H., Madder A., Taylor E.J., Singh I. Efficient total syntheses and biological activities of two teixobactin analogues. Chem. Commun. 2016;52(36):6060–6063. doi: 10.1039/C5CC10249A. [DOI] [PubMed] [Google Scholar]
- 88.Parmar A., Iyer A., Lloyd D.G., Vincent C.S., Prior S.H., Madder A., Taylor E.J., Singh I. Syntheses of potent teixobactin analogues against methicillin-resistant Staphylococcus aureus (MRSA) through the replacement of l-allo-enduracididine with its isosteres. Chem. Commun. 2017;53(55):7788–7791. doi: 10.1039/C7CC04021K. [DOI] [PubMed] [Google Scholar]
- 89.Jad Y.E., Acosta G.A., Naicker T., Ramtahal M., El-Faham A., Govender T., Kruger H.G., de la Torre B.G., Albericio F. Synthesis and biological evaluation of a teixobactin analogue. Org. Lett. 2015;17(24):6182–6185. doi: 10.1021/acs.orglett.5b03176. [DOI] [PubMed] [Google Scholar]
- 90.Gunjal V.B., Thakare R., Chopra S., Reddy D.S. Teixobactin: a paving stone toward a new class of antibiotics? J. Med. Chem. 2020;63(21):12171–12195. doi: 10.1021/acs.jmedchem.0c00173. [DOI] [PubMed] [Google Scholar]
- 91.Jin K., Po K.H.L., Kong W.Y., Lo C.H., Lo C.W., Lam H.Y., Sirinimal A., Reuven J.A., Chen S., Li X. Synthesis and antibacterial studies of teixobactin analogues with non-isostere substitution of enduracididine. Bioorg. Med. Chem. 2018;26(5):1062–1068. doi: 10.1016/j.bmc.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 92.Jin K., Po K.H.L., Wang S., Reuven J.A., Wai C.N., Lau H.T., Chan T.H., Chen S., Li X. Synthesis and structure-activity relationship of teixobactin analogues via convergent Ser ligation. Bioorg. Med. Chem. 2017;25(18):4990–4995. doi: 10.1016/j.bmc.2017.04.039. [DOI] [PubMed] [Google Scholar]
- 93.Zong Y., Sun X., Gao H., Meyer K.J., Lewis K., Rao Y. Developing equipotent teixobactin analogues against drug-resistant bacteria and discovering a hydrophobic interaction between lipid II and TEIXOBACTIN. J. Med. Chem. 2018;61(8):3409–3421. doi: 10.1021/acs.jmedchem.7b01241. [DOI] [PubMed] [Google Scholar]
- 94.Cutler R., Wilson P. Antibacterial activity of a new, stable, aqueous extract of allicin against methicillin-resistant Staphylococcus aureus. Br. J. Biomed. Sci. 2004;61(2):71–74. doi: 10.1080/09674845.2004.11732646. [DOI] [PubMed] [Google Scholar]
- 95.Ilić D.P., Nikolić V.D., Nikolić L.B., Stanković M.Z., Stanojević L.P., Cakić M.D. Allicin and related compounds: biosynthesis, synthesis and pharmacological activity. Facta universitatis-series: physics. Chem. Technol. 2011;9(1):9–20. [Google Scholar]
- 96.El-Saber Batiha G., Magdy Beshbishy A., G Wasef L., Elewa Y.H., A Al-Sagan A., El-Hack A., Mohamed E., Taha A.E., M Abd-Elhakim Y., Prasad Devkota H. Chemical constituents and pharmacological activities of garlic (Allium sativum L.): a review. Nutrients. 2020;12(3):872. doi: 10.3390/nu12030872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Borlinghaus J., Albrecht F., Gruhlke M.C., Nwachukwu I.D., Slusarenko A.J. Allicin: chemistry and biological properties. Molecules. 2014;19(8):12591–12618. doi: 10.3390/molecules190812591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cai Y., Wang R., An M.M., Liang B.B., Fang Y. In vitro bactericidal activity of allicin combined with cefoperazone, tobramycin and ciprofloxacin. Int. J. Antimicrob. Agents. 2008;31(2):179–180. doi: 10.1016/j.ijantimicag.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 99.Leontiev R., Hohaus N., Jacob C., Gruhlke M.C., Slusarenko A.J. A comparison of the antibacterial and antifungal activities of thiosulfinate analogues of allicin. Sci. Rep. 2018;8(1):1–19. doi: 10.1038/s41598-018-25154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Singh N., Sharma B. Toxicological effects of berberine and sanguinarine. Front. Mol. Biosci. 2018;5:21. doi: 10.3389/fmolb.2018.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peng L., Kang S., Yin Z., Jia R., Song X., Li L., Li Z., Zou Y., Liang X., Li L. Antibacterial activity and mechanism of berberine against Streptococcus agalactiae. Int. J. Clin. Exp. Pathol. 2015;8(5):5217. [PMC free article] [PubMed] [Google Scholar]
- 102.Yu H.H., Kim K.J., Cha J.D., Kim H.K., Lee Y.E., Choi N.Y., You Y.O. Antimicrobial activity of berberine alone and in combination with ampicillin or oxacillin against methicillin-resistant Staphylococcus aureus. J. Med. Food. 2005;8(4):454–461. doi: 10.1089/jmf.2005.8.454. [DOI] [PubMed] [Google Scholar]
- 103.Wang X., Yao X., Tang T., Dai K., Sadovskaya I., Flahaut S., Jabbouri S. Effect of berberine on Staphylococcus epidermidis biofilm formation. Int. J. Antimicrob. Agents. 2009;34(1):60–66. doi: 10.1016/j.ijantimicag.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 104.Wei G.X., Xu X., Wu C.D. In vitro synergism between berberine and miconazole against planktonic and biofilm Candida cultures. Arch. Oral Biol. 2011;56(6):565–572. doi: 10.1016/j.archoralbio.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 105.Liu Q., Niu H., Zhang W., Mu H., Sun C., Duan J. Synergy among thymol, eugenol, berberine, cinnamaldehyde and streptomycin against planktonic and biofilm-associated food-borne pathogens. Lett. Appl. Microbiol. 2015;60(5):421–430. doi: 10.1111/lam.12401. [DOI] [PubMed] [Google Scholar]
- 106.Guo N., Zhao X., Li W., Shi C., Meng R., Liu Z., Yu L. The synergy of berberine chloride and totarol against Staphylococcus aureus grown in planktonic and biofilm cultures. J. Med. Microbiol. 2015;64(8):891–900. doi: 10.1099/jmm.0.000106. [DOI] [PubMed] [Google Scholar]
- 107.Chu M., Zhang M.B., Liu Y.C., Kang J.R., Chu Z.Y., Yin K.L., Ding L.Y., Ding R., Xiao R.X., Yin Y.N. Role of berberine in the treatment of methicillin-resistant Staphylococcus aureus infections. Sci. Rep. 2016;6(1):1–9. doi: 10.1038/srep24748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Otto M. Phenol-soluble modulins. Int. J. Med. Microbiol. 2014;304(2):164–169. doi: 10.1016/j.ijmm.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li Y., Huang J., Li L., Liu L. Synergistic activity of berberine with azithromycin against Pseudomonas aeruginosa isolated from patients with cystic fibrosis of lung in vitro and in vivo. Cell. Physiol. Biochem. 2017;42(4):1657–1669. doi: 10.1159/000479411. [DOI] [PubMed] [Google Scholar]
- 110.Li X., Wang P., Hu X., Zhang Y., Lu X., Li C., Nie T., Li G., Wang X., Pang J. The combined antibacterial effects of sodium new houttuyfonate and berberine chloride against growing and persistent methicillin-resistant and vancomycin-intermediate Staphylococcus aureus. BMC Microbiol. 2020;20(1):1–11. doi: 10.1186/s12866-020-02003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sarfraz A., Javeed M., Shah M.A., Hussain G., Shafiq N., Sarfraz I., Riaz A., Sadiqa A., Zara R., Zafar S. Biochanin A: a novel bioactive multifunctional compound from nature. Sci. Total Environ. 2020;722 doi: 10.1016/j.scitotenv.2020.137907. [DOI] [PubMed] [Google Scholar]
- 112.Liu G., Liang J.C., Wang X.L., Li Z.H., Wang W., Guo N., Wu X.P., Shen F.G., Xing M.X., Liu L.H. In vitro synergy of biochanin A and ciprofloxacin against clinical isolates of Staphylococcus aureus. Molecules. 2011;16(8):6656–6666. doi: 10.3390/molecules16086656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zou D., Xie K., Wang H., Chen Y., Xie M. Inhibitory effects of biochanin A on the efflux pump of methicillin-resistant Staphylococcus aureus (MRSA) Acta Microbiol. Sin. 2014;54(10):1204–1211. Wei sheng wu xue bao= [PubMed] [Google Scholar]
- 114.Lechner D., Gibbons S., Bucar F. Plant phenolic compounds as ethidium bromide efflux inhibitors in Mycobacterium smegmatis. J. Antimicrob. Chemother. 2008;62(2):345–348. doi: 10.1093/jac/dkn178. [DOI] [PubMed] [Google Scholar]
- 115.Sklenickova O., Flesar J., Kokoska L., Vlkova E., Halamova K., Malik J. Selective growth inhibitory effect of biochanin A against intestinal tract colonizing bacteria. Molecules. 2010;15(3):1270–1279. doi: 10.3390/molecules15031270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sartorelli P., Carvalho C.S., Reimão J.Q., Ferreira M.J.P., Tempone A.G. Antiparasitic activity of biochanin A, an isolated isoflavone from fruits of Cassia fistula (Leguminosae) Parasitol. Res. 2009;104(2):311–314. doi: 10.1007/s00436-008-1193-z. [DOI] [PubMed] [Google Scholar]
- 117.Hanski L., Genina N., Uvell H., Malinovskaja K., Gylfe Å., Laaksonen T., Kolakovic R., Mäkilä E., Salonen J., Hirvonen J. Inhibitory activity of the isoflavone biochanin A on intracellular bacteria of genus Chlamydia and initial development of a buccal formulation. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0115115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nikolic I.L., Savic I.M., Popsavin M.M., Rakic S.J., Mihajilov-Krstev T.M., Ristic I.S., Eric S.P., Savić-Gajic I.M. Preparation, characterization and antimicrobial activity of inclusion complex of biochanin A with (2-hydroxypropyl)-β-cyclodextrin. J. Pharm. Pharmacol. 2018;70(11):1485–1493. doi: 10.1111/jphp.13003. [DOI] [PubMed] [Google Scholar]
- 119.Khan F., Bamunuarachchi N.I., Tabassum N., Kim Y.M. Caffeic acid and its derivatives: antimicrobial drugs toward microbial pathogens. J. Agric. Food Chem. 2021;69(10):2979–3004. doi: 10.1021/acs.jafc.0c07579. [DOI] [PubMed] [Google Scholar]
- 120.Silva T., Oliveira C., Borges F. Caffeic acid derivatives, analogs and applications: a patent review (2009–2013) Expert Opin. Ther. Pat. 2014;24(11):1257–1270. doi: 10.1517/13543776.2014.959492. [DOI] [PubMed] [Google Scholar]
- 121.Stojković D., Petrović J., Soković M., Glamočlija J., Kukić-Marković J., Petrović S. In situ antioxidant and antimicrobial activities of naturally occurring caffeic acid, p-coumaric acid and rutin, using food systems. J. Sci. Food Agric. 2013;93(13):3205–3208. doi: 10.1002/jsfa.6156. [DOI] [PubMed] [Google Scholar]
- 122.Kim J.H., Yu D., Eom S.H., Kim S.H., Oh J., Jung W.K., Kim Y.M. Synergistic antibacterial effects of chitosan-caffeic acid conjugate against antibiotic-resistant acne-related bacteria. Mar. Drugs. 2017;15(6):167. doi: 10.3390/md15060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ignatova M., Manolova N., Rashkov I., Markova N. Quaternized chitosan/κ-carrageenan/caffeic acid–coated poly (3-hydroxybutyrate) fibrous materials: preparation, antibacterial and antioxidant activity. Int. J. Pharm. 2016;513(1–2):528–537. doi: 10.1016/j.ijpharm.2016.09.062. [DOI] [PubMed] [Google Scholar]
- 124.Choi K.H., Nam K.C., Lee S.Y., Cho G., Jung J.S., Kim H.J., Park B.J. Antioxidant potential and antibacterial efficiency of caffeic acid-functionalized ZnO nanoparticles. Nanomaterials. 2017;7(6):148. doi: 10.3390/nano7060148. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pinho E., Soares G., Henriques M. Evaluation of antibacterial activity of caffeic acid encapsulated by β-cyclodextrins. J. Microencapsul. 2015;32(8):804–810. doi: 10.3109/02652048.2015.1094531. [DOI] [PubMed] [Google Scholar]
- 126.Meyuhas S., Assali M., Huleihil M., Huleihel M. Antimicrobial activities of caffeic acid phenethyl ester. J. Mol. Biochem. 2015;4(2) [Google Scholar]
- 127.Niu Y., Wang K., Zheng S., Wang Y., Ren Q., Li H., Ding L., Li W., Zhang L. Antibacterial effect of caffeic acid phenethyl ester on cariogenic bacteria and streptococcus mutans biofilms. Antimicrob. Agents Chemother. 2020;64(9) doi: 10.1128/aac.00251-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Veloz J.J., Alvear M., Salazar L.A. Antimicrobial and antibiofilm activity against Streptococcus mutans of individual and mixtures of the main polyphenolic compounds found in Chilean propolis. BioMed. Res. Int. 2019 doi: 10.1155/2019/7602343. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li G., Wang X., Xu Y., Zhang B., Xia X. Antimicrobial effect and mode of action of chlorogenic acid on Staphylococcus aureus. Eur. Food Res. Technol. 2014;238(4):589–596. [Google Scholar]
- 130.Luís Â., Silva F., Sousa S., Duarte A.P., Domingues F. Antistaphylococcal and biofilm inhibitory activities of gallic, caffeic, and chlorogenic acids. Biofouling. 2014;30(1):69–79. doi: 10.1080/08927014.2013.845878. [DOI] [PubMed] [Google Scholar]
- 131.Sun J., Wang D., Sun Z., Liu F., Du L., Wang D. The combination of ultrasound and chlorogenic acid to inactivate Staphylococcus aureus under planktonic, biofilm, and food systems. Ultrason. Sonochem. 2021;80 doi: 10.1016/j.ultsonch.2021.105801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Karunanidhi A., Thomas R., van Belkum A., Neela V. In vitro antibacterial and antibiofilm activities of chlorogenic acid against clinical isolates of Stenotrophomonas maltophilia including the trimethoprim/sulfamethoxazole resistant strain. Biomed. Res. Int. 2013;2013 doi: 10.1155/2013/392058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gorgani L., Mohammadi M., Najafpour G.D., Nikzad M. Piperine—the bioactive compound of black pepper: from isolation to medicinal formulations. Compr. Rev. Food Sci. Food Saf. 2017;16(1):124–140. doi: 10.1111/1541-4337.12246. [DOI] [PubMed] [Google Scholar]
- 134.Nargotra A., Sharma S., Koul J.L., Sangwan P.L., Khan I.A., Kumar A., Taneja S.C., Koul S. Quantitative structure activity relationship (QSAR) of piperine analogsfor bacterial NorA efflux pump inhibitors. Eur. J. Med. Chem. 2009;44(10):4128–4135. doi: 10.1016/j.ejmech.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 135.Kumar A., Khan I.A., Koul S., Koul J.L., Taneja S.C., Ali I., Ali F., Sharma S., Mirza Z.M., Kumar M., Sangwan P.L., Gupta P., Thota N., Qazi G.N. Novel structural analogues of piperine as inhibitors of the NorA efflux pump of Staphylococcus aureus. J. Antimicrob. Chemother. 2008;61(6):1270–1276. doi: 10.1093/jac/dkn088. [DOI] [PubMed] [Google Scholar]
- 136.Zhang L., Ma S. Efflux pump inhibitors: a strategy to combat P-Glycoprotein and the NorA multidrug resistance pump. ChemMedChem. 2010;5(6):811–822. doi: 10.1002/cmdc.201000006. [DOI] [PubMed] [Google Scholar]
- 137.Khan I.A., Mirza Z.M., Kumar A., Verma V., Qazi G.N. Piperine, a phytochemical potentiator of ciprofloxacin against Staphylococcus aureus. Antimicrob. Agents Chemother. 2006;50(2):810–812. doi: 10.1128/AAC.50.2.810-812.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Khameneh B., Iranshahy M., Ghandadi M., Ghoochi Atashbeyk D., Fazly Bazzaz B.S., Iranshahi M. Investigation of the antibacterial activity and efflux pump inhibitory effect of co-loaded piperine and gentamicin nanoliposomes in methicillin-resistant Staphylococcus aureus. Drug Dev. Ind. Pharm. 2015;41(6):989–994. doi: 10.3109/03639045.2014.920025. [DOI] [PubMed] [Google Scholar]
- 139.Kuspradini H., Mitsunaga T., Ohashi H. Antimicrobial activity against Streptococcus sobrinus and glucosyltransferase inhibitory activity of taxifolin and some flavanonol rhamnosides from kempas (Koompassia malaccensis) extracts. J. Wood Sci. 2009;55(4):308–313. [Google Scholar]
- 140.Jeong K.W., Lee J.Y., Kang D.I., Lee J.U., Shin S.Y., Kim Y. Screening of flavonoids as candidate antibiotics against Enterococcus faecalis. J. Nat. Prod. 2009;72(4):719–724. doi: 10.1021/np800698d. [DOI] [PubMed] [Google Scholar]
- 141.Artem'Eva O., Pereselkova D., Fomichev Y.P. (2015) Dihydroquercetin, the bioactive substance, to be used against pathogenic microorganisms as an alternative to antibiotics. Сельскохозяйственная биология (4 (eng)).
- 142.Davis C.K., Nasla K., Anjana A., Rajanikant G. Taxifolin as dual inhibitor of Mtb DNA gyrase and isoleucyl-tRNA synthetase: in silico molecular docking, dynamics simulation and in vitro assays. In Silico Pharmacol. 2018;6(1):1–11. doi: 10.1007/s40203-018-0045-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang L., Wang G., Qu H., Wang K., Jing S., Guan S., Su L., Li Q., Wang D. Taxifolin, an Inhibitor of Sortase A, Interferes With the Adhesion of Methicillin-Resistant Staphylococcal aureus. Front Microbiol. 2021;12:1876. doi: 10.3389/fmicb.2021.686864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Marchese A., Orhan I.E., Daglia M., Barbieri R., Di Lorenzo A., Nabavi S.F., Gortzi O., Izadi M., Nabavi S.M. Antibacterial and antifungal activities of thymol: a brief review of the literature. Food Chem. 2016;210:402–414. doi: 10.1016/j.foodchem.2016.04.111. [DOI] [PubMed] [Google Scholar]
- 145.Nostro A., Blanco A.R., Cannatelli M.A., Enea V., Flamini G., Morelli I., Sudano Roccaro A., Alonzo V. Susceptibility of methicillin-resistant staphylococci to oregano essential oil, carvacrol and thymol. FEMS Microbiol. Lett. 2004;230(2):191–195. doi: 10.1016/S0378-1097(03)00890-5. [DOI] [PubMed] [Google Scholar]
- 146.Trombetta D., Castelli F., Sarpietro M.G., Venuti V., Cristani M., Daniele C., Saija A., Mazzanti G., Bisignano G. Mechanisms of antibacterial action of three monoterpenes. Antimicrob. Agents Chemother. 2005;49(6):2474–2478. doi: 10.1128/AAC.49.6.2474-2478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu J., Zhou F., Ji B.P., Pei R.S., Xu N. The antibacterial mechanism of carvacrol and thymol against Escherichia coli. Lett. Appl. Microbiol. 2008;47(3):174–179. doi: 10.1111/j.1472-765X.2008.02407.x. [DOI] [PubMed] [Google Scholar]
- 148.Mathela C.S., Singh K.K., Gupta V.K. Synthesis and in vitro antibacterial activity of thymol and carvacrol derivatives. Acta Pol. Pharm. 2010;67(4):375–380. [PubMed] [Google Scholar]
- 149.Amaral V.C., Santos P.R., da Silva A.F., dos Santos A.R., Machinski Jr M., Mikcha J.M.G. Effect of carvacrol and thymol on Salmonella spp. biofilms on polypropylene. Int. J. Food Sci. Technol. 2015;50(12):2639–2643. [Google Scholar]
- 150.Miladi H., Zmantar T., Chaabouni Y., Fedhila K., Bakhrouf A., Mahdouani K., Chaieb K. Antibacterial and efflux pump inhibitors of thymol and carvacrol against food-borne pathogens. Microb. Pathog. 2016;99:95–100. doi: 10.1016/j.micpath.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 151.Uzma F., Mohan C.D., Hashem A., Konappa N.M., Rangappa S., Kamath P.V., Singh B.P., Mudili V., Gupta V.K., Siddaiah C.N., Chowdappa S., Alqarawi A.A., Abd_Allah E.F. Endophytic fungi—alternative sources of cytotoxic compounds: a review. Front. Pharmacol. 2018;9(309) doi: 10.3389/fphar.2018.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mohan C.D., Hari S., Preetham H.D., Rangappa S., Barash U., Ilan N., Nayak S.C., Gupta V.K., Basappa Vlodavsky I, Rangappa K.S. Targeting heparanase in cancer: inhibition by synthetic, chemically modified, and natural compounds. iScience. 2019;15:360–390. doi: 10.1016/j.isci.2019.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mohan C.D., Rangappa S., Preetham H.D., Chandra Nayaka S., Gupta V.K., Basappa S., Sethi G., Rangappa K.S. Targeting STAT3 signaling pathway in cancer by agents derived from mother Nature. Semin. Cancer Biol. 2020 doi: 10.1016/j.semcancer.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 154.Shanmugam M.K., Manu K.A., Ong T.H., Ramachandran L., Surana R., Bist P., Lim L.H., Kumar A.P., Hui K.M., Sethi G. Inhibition of CXCR4/CXCL12 signaling axis by ursolic acid leads to suppression of metastasis in transgenic adenocarcinoma of mouse prostate model. Int. J. Cancer. 2011;129(7):1552–1563. doi: 10.1002/ijc.26120. [DOI] [PubMed] [Google Scholar]
- 155.Kashyap D., Tuli H.S., Yerer M.B., Sharma A., Sak K., Srivastava S., Pandey A., Garg V.K., Sethi G., Bishayee A. Natural product-based nanoformulations for cancer therapy: opportunities and challenges. Semin. Cancer Biol. 2021;69:5–23. doi: 10.1016/j.semcancer.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 156.Mohan C.D., Rangappa S., Nayak S.C., Jadimurthy R., Wang L., Sethi G., Garg M., Rangappa K.S. Bacteria as a treasure house of secondary metabolites with anticancer potential. Semin. Cancer Biol. 2021 doi: 10.1016/j.semcancer.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 157.Uzma F., Mohan C.D., Siddaiah C.N., Chowdappa S., Singh B.P. Advances in Endophytic Fungal Research: Present Status and Future Challenges. Springer International Publishing; Cham: 2019. Endophytic fungi: promising source of novel bioactive compounds; pp. 243–265. [DOI] [Google Scholar]
- 158.Ahn K.S., Sethi G., Chao T.H., Neuteboom S.T., Chaturvedi M.M., Palladino M.A., Younes A., Aggarwal B.B. Salinosporamide A (NPI-0052) potentiates apoptosis, suppresses osteoclastogenesis, and inhibits invasion through down-modulation of NF-kappaB regulated gene products. Blood. 2007;110(7):2286–2295. doi: 10.1182/blood-2007-04-084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lee J.H., Mohan C.D., Shanmugam M.K., Rangappa S., Sethi G., Siveen K.S., Chinnathambi A., Alahmadi T.A., Alharbi S.A., Basappa S., Rangappa K.S., Ahn K.S. Vitexin abrogates invasion and survival of hepatocellular carcinoma cells through targeting STAT3 signaling pathway. Biochimie. 2020;175:58–68. doi: 10.1016/j.biochi.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 160.Mohan C.D., Kim C., Siveen K.S., Manu K.A., Rangappa S., Chinnathambi A., Alharbi S.A., Rangappa K.S., Kumar A.P., Ahn K.S. Crocetin imparts antiproliferative activity via inhibiting STAT3 signaling in hepatocellular carcinoma. IUBMB Life. 2021;73(11):1348–1362. doi: 10.1002/iub.2555. [DOI] [PubMed] [Google Scholar]
- 161.Ramchandani S., Mohan C.D., Mistry J.R., Su Q., Naz I., Rangappa K.S., Ahn K.S. The multifaceted antineoplastic role of pyrimethamine against human malignancies. IUBMB Life. 2022;74(3):198–212. doi: 10.1002/iub.2590. [DOI] [PubMed] [Google Scholar]
- 162.Fischbach M.A., Walsh C.T. Antibiotics for emerging pathogens. Science. 2009;325(5944):1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Payne D.J., Gwynn M.N., Holmes D.J., Pompliano D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007;6(1):29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 164.Hou J., Robbel L., Marahiel M.A. Identification and characterization of the lysobactin biosynthetic gene cluster reveals mechanistic insights into an unusual termination module architecture. Chem. Biol. 2011;18(5):655–664. doi: 10.1016/j.chembiol.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 165.Lee W., Schaefer K., Qiao Y., Srisuknimit V., Steinmetz H., Müller R., Kahne D., Walker S. The mechanism of action of lysobactin. J. Am. Chem. Soc. 2016;138(1):100–103. doi: 10.1021/jacs.5b11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hassan H.M., Degen D., Jang K.H., Ebright R.H., Fenical W. Salinamide F, new depsipeptide antibiotic and inhibitor of bacterial RNA polymerase from a marine-derived Streptomyces sp. J. Antibiot. 2015;68(3):206–209. doi: 10.1038/ja.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Daniel-Ivad M., Hameed N., Tan S., Dhanjal R., Socko D., Pak P., Gverzdys T., Elliot M.A., Nodwell J.R. An engineered allele of afsQ1 facilitates the discovery and investigation of cryptic natural products. ACS Chem. Biol. 2017;12(3):628–634. doi: 10.1021/acschembio.6b01002. [DOI] [PubMed] [Google Scholar]
- 168.Knappe T.A., Linne U., Zirah S., Rebuffat S., Xie X., Marahiel M.A. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J. Am. Chem. Soc. 2008;130(34):11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 169.Kuznedelov K., Semenova E., Knappe T.A., Mukhamedyarov D., Srivastava A., Chatterjee S., Ebright R.H., Marahiel M.A., Severinov K. The antibacterial threaded-lasso peptide capistruin inhibits bacterial RNA polymerase. J. Mol. Biol. 2011;412(5):842–848. doi: 10.1016/j.jmb.2011.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cruz-Muñiz M.Y., López-Jacome L.E., Hernández-Durán M., Franco-Cendejas R., Licona-Limón P., Ramos-Balderas J.L., Martinéz-Vázquez M., Belmont-Díaz J.A., Wood T.K., García-Contreras R. Repurposing the anticancer drug mitomycin C for the treatment of persistent Acinetobacter baumannii infections. Int. J. Antimicrob. Agents. 2017;49(1):88–92. doi: 10.1016/j.ijantimicag.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 171.Avendaño C., Menéndez J. Medicinal Chemistry of Anticancer Drugs. Elsevier; 2015. Anticancer drugs that interact with the DNA minor groove; pp. 243–271. [Google Scholar]
- 172.Zhou X., Huang H., Chen Y., Tan J., Song Y., Zou J., Tian X., Hua Y., Ju J. Marthiapeptide A, an anti-infective and cytotoxic polythiazole cyclopeptide from a 60L scale fermentation of the deep sea-derived Marinactinospora thermotolerans SCSIO 00652. J. Nat. Prod. 2012;75(12):2251–2255. doi: 10.1021/np300554f. [DOI] [PubMed] [Google Scholar]
- 173.Niu S., Li S., Chen Y., Tian X., Zhang H., Zhang G., Zhang W., Yang X., Zhang S., Ju J. Lobophorins E and F, new spirotetronate antibiotics from a South China Sea-derived Streptomyces sp. SCSIO 01127. J. Antibiot. 2011;64(11):711–716. doi: 10.1038/ja.2011.78. [DOI] [PubMed] [Google Scholar]
- 174.Song Y., Li Q., Liu X., Chen Y., Zhang Y., Sun A., Zhang W., Zhang J., Ju J. Cyclic hexapeptides from the deep South China sea-derived streptomyces scopuliridis SCSIO ZJ46 active against pathogenic Gram-positive bacteria. J. Nat. Prod. 2014;77(8):1937–1941. doi: 10.1021/np500399v. [DOI] [PubMed] [Google Scholar]
- 175.He X., Li M., Song S., Wu X., Zhang J., Wu G., Yue R., Cui H., Song S., Ma C. Ficellomycin: an aziridine alkaloid antibiotic with potential therapeutic capacity. Appl. Microbiol. Biotechnol. 2018;102(10):4345–4354. doi: 10.1007/s00253-018-8934-4. [DOI] [PubMed] [Google Scholar]
- 176.Haste N.M., Perera V.R., Maloney K.N., Tran D.N., Jensen P., Fenical W., Nizet V., Hensler M.E. Activity of the streptogramin antibiotic etamycin against methicillin-resistant Staphylococcus aureus. J. Antibiot. 2010;63(5):219–224. doi: 10.1038/ja.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Johnson R.A., Chan A.N., Ward R.D., McGlade C.A., Hatfield B.M., Peters J.M., Li B. Inhibition of Isoleucyl-tRNA Synthetase by the hybrid antibiotic thiomarinol. J. Am. Chem. Soc. 2021;143(31):12003–12013. doi: 10.1021/jacs.1c02622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bhave S.V., Sawant A.V., Shanbhag P., Parab R.R., Ranadive P.V., Mishra P.D., Mahajan G.B. Fermentation, isolation of mithramycin from Streptomyces of Playa Region and its novel anti-MRSA and anti-VRE activity. SOJ Microbiol. Infect. Dis. 2015;3(2):1–8. [Google Scholar]
- 179.Haste N.M., Hughes C.C., Tran D.N., Fenical W., Jensen P.R., Nizet V., Hensler M.E. Pharmacological properties of the marine natural product marinopyrrole A against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011;55(7):3305–3312. doi: 10.1128/AAC.01211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kobayashi Y., Ichioka M., Hirose T., Nagai K., Matsumoto A., Matsui H., Hanaki H., Masuma R., Takahashi Y., Ōmura S. Bottromycin derivatives: efficient chemical modifications of the ester moiety and evaluation of anti-MRSA and anti-VRE activities. Bioorg. Med. Chem. Lett. 2010;20(20):6116–6120. doi: 10.1016/j.bmcl.2010.08.037. [DOI] [PubMed] [Google Scholar]
- 181.Carlson J.C., Li S., Burr D.A., Sherman D.H. Isolation and characterization of tirandamycins from a marine-derived Streptomyces sp. J. Nat. Prod. 2009;72(11):2076–2079. doi: 10.1021/np9005597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sun P., Maloney K.N., Nam S.J., Haste N.M., Raju R., Aalbersberg W., Jensen P.R., Nizet V., Hensler M.E., Fenical W. Fijimycins A–C, three antibacterial etamycin-class depsipeptides from a marine-derived Streptomyces sp. Bioorg. Med. Chem. 2011;19(22):6557–6562. doi: 10.1016/j.bmc.2011.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wyche T.P., Alvarenga R.F.R., Piotrowski J.S., Duster M.N., Warrack S.R., Cornilescu G., De Wolfe T.J., Hou Y., Braun D.R., Ellis G.A. Chemical genomics, structure elucidation, and in vivo studies of the marine-derived anticlostridial ecteinamycin. ACS Chem. Biol. 2017;12(9):2287–2295. doi: 10.1021/acschembio.7b00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Manam R.R., Teisan S., White D.J., Nicholson B., Grodberg J., Neuteboom S.T., Lam K.S., Mosca D.A., Lloyd G.K., Potts B.C. Lajollamycin, a Nitro-tetraene Spiro-β-lactone-γ-lactam Antibiotic from the Marine Actinomycete Streptomyces n odosus. J. Nat. Prod. 2005;68(2):240–243. doi: 10.1021/np049725x. [DOI] [PubMed] [Google Scholar]
- 185.Vestergaard M., Ingmer H. Antibacterial and antifungal properties of resveratrol. Int. J. Antimicrob. Agents. 2019;53(6):716–723. doi: 10.1016/j.ijantimicag.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 186.Wu S.C., Yang Z.Q., Liu F., Peng W.J., Qu S.Q., Li Q., Song X.B., Zhu K., Shen J.Z. Antibacterial effect and mode of action of flavonoids from licorice against methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2019;10:2489. doi: 10.3389/fmicb.2019.02489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Özçelik B., Kartal M., Orhan I. Cytotoxicity, antiviral and antimicrobial activities of alkaloids, flavonoids, and phenolic acids. Pharm. Biol. 2011;49(4):396–402. doi: 10.3109/13880209.2010.519390. [DOI] [PubMed] [Google Scholar]
- 188.Basile A., Sorbo S., Giordano S., Ricciardi L., Ferrara S., Montesano D., Cobianchi R.C., Vuotto M., Ferrara L. Antibacterial and allelopathic activity of extract from Castanea sativa leaves. Fitoterapia. 2000;71:S110–S116. doi: 10.1016/s0367-326x(00)00185-4. [DOI] [PubMed] [Google Scholar]
- 189.Górniak I., Bartoszewski R., Króliczewski J. Comprehensive review of antimicrobial activities of plant flavonoids. Phytochem. Rev. 2019;18(1):241–272. [Google Scholar]
- 190.Wang J., Jiao H., Meng J., Qiao M., Du H., He M., Ming K., Liu J., Wang D., Wu Y. Baicalin inhibits biofilm formation and the quorum-sensing system by regulating the MsrA drug efflux pump in Staphylococcus saprophyticus. Front. Microbiol. 2019;10:2800. doi: 10.3389/fmicb.2019.02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Althunibat O.Y., Qaralleh H., Al-Dalin S.Y.A., Abboud M., Khleifat K., Majali I.S., Aldal'in H., Rayyan W.A., Jaafraa A. Effect of thymol and carvacrol, the major components of Thymus capitatus on the growth of Pseudomonas aeruginosa. J. Pure Appl. Microbiol. 2016;10(1):367–374. [Google Scholar]
- 192.Obiang-Obounou B.W., Kang O.H., Choi J.G., Keum J.H., Kim S.B., Mun S.H., Shin D.W., Kim K.W., Park C.B., Kim Y.G. The mechanism of action of sanguinarine against methicillin-resistant Staphylococcus aureus. J. Toxicol. Sci. 2011;36(3):277–283. doi: 10.2131/jts.36.277. [DOI] [PubMed] [Google Scholar]
- 193.Sridevi D., Shankar C., Prakash P., Park J.H., Thamaraiselvi K. Inhibitory effects of reserpine against efflux pump activity of antibiotic resistance bacteria. Chem. Biol. Lett. 2017;4(2):69–72. [Google Scholar]
- 194.Negi J., Bisht V., Bhandari A., Bisht D., Singh P., Singh N. Quantification of reserpine content and antibacterial activity of Rauvolfia serpentina (L.) Benth. ex Kurz. Afr. J. Microbiol. Res. 2014;8(2):162–166. [Google Scholar]
- 195.Lamontagne Boulet M., Isabelle C., Guay I., Brouillette E., Langlois J.P., Jacques P.É., Rodrigue S., Brzezinski R., Beauregard P.B., Bouarab K. Tomatidine is a lead antibiotic molecule that targets Staphylococcus aureus ATP synthase subunit C. Antimicrob. Agents Chemother. 2018;62(6):e02117–e02197. doi: 10.1128/AAC.02197-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stompor M. A review on sources and pharmacological aspects of sakuranetin. Nutrients. 2020;12(2):513. doi: 10.3390/nu12020513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tyagi P., Singh M., Kumari H., Kumari A., Mukhopadhyay K. Bactericidal activity of curcumin I is associated with damaging of bacterial membrane. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0121313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Vaou N., Stavropoulou E., Voidarou C., Tsigalou C., Bezirtzoglou E. Towards advances in medicinal plant antimicrobial activity: a review study on challenges and future perspectives. Microorganisms. 2021;9(10):2041. doi: 10.3390/microorganisms9102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Basile A., Sorbo S., Spadaro V., Bruno M., Maggio A., Faraone N., Rosselli S. Antimicrobial and antioxidant activities of coumarins from the roots of Ferulago campestris (Apiaceae) Molecules. 2009;14(3):939–952. doi: 10.3390/molecules14030939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kumar N., Singh B., Bhandari P., Gupta A.P., Kaul V.K. Steroidal alkaloids from holarrhena antidysenterica (L.) W ALL. Chem. Pharm. Bull. 2007;55(6):912–914. doi: 10.1248/cpb.55.912. [DOI] [PubMed] [Google Scholar]
- 201.Siriyong T., Srimanote P., Chusri S., Yingyongnarongkul B.E., Suaisom C., Tipmanee V., Voravuthikunchai S.P. Conessine as a novel inhibitor of multidrug efflux pump systems in Pseudomonas aeruginosa. BMC Complement. Altern. Med. 2017;17(1):1–7. doi: 10.1186/s12906-017-1913-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Patrice K., Véronique P.B., David L., François-Xavier E. Antibacterial activities of the extracts and conessine from Holarrhena floribunda G. Don.(Apocynaceae) Afr. J. Tradit. Complement. Altern. Med. 2007;4(3):352–356. doi: 10.4314/ajtcam.v4i3.31229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bai J., Wu Y., Liu X., Zhong K., Huang Y., Gao H. Antibacterial activity of shikimic acid from pine needles of cedrus deodara against staphylococcus aureus through damage to cell membrane. Int. J. Mol. Sci. 2015;16(11):27145–27155. doi: 10.3390/ijms161126015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bai J.R., Zhong K., Wu Y.P., Elena G., Gao H. Antibiofilm activity of shikimic acid against staphylococcus aureus. Food Control. 2019;95:327–333. [Google Scholar]
- 205.Borges A., Ferreira C., Saavedra M.J., Simões M. Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb. Drug Resist. 2013;19(4):256–265. doi: 10.1089/mdr.2012.0244. [DOI] [PubMed] [Google Scholar]
- 206.Somerset S.M., Johannot L. Dietary flavonoid sources in Australian adults. Nutr. Cancer. 2008;60(4):442–449. doi: 10.1080/01635580802143836. [DOI] [PubMed] [Google Scholar]
- 207.Ming D., Wang D., Cao F., Xiang H., Mu D., Cao J., Li B., Zhong L., Dong X., Zhong X. Kaempferol inhibits the primary attachment phase of biofilm formation in Staphylococcus aureus. Front. Microbiol. 2017;8:2263. doi: 10.3389/fmicb.2017.02263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Brown A.R., Ettefagh K.A., Todd D., Cole P.S., Egan J.M., Foil D.H., Graf T.N., Schindler B.D., Kaatz G.W., Cech N.B. A mass spectrometry-based assay for improved quantitative measurements of efflux pump inhibition. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0124814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Huang Y.H., Huang C.C., Chen C.C., Yang K.J., Huang C.Y. Inhibition of staphylococcus aureus PriA helicase by flavonol kaempferol. Protein J. 2015;34(3):169–172. doi: 10.1007/s10930-015-9609-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Oh I., Yang W.Y., Chung S.C., Kim T.Y., Oh K.B., Shin J. In vitro sortase A inhibitory and antimicrobial activity of flavonoids isolated from the roots of Sophora flavescens. Arch. Pharm. Res. 2011;34(2):217–222. doi: 10.1007/s12272-011-0206-0. [DOI] [PubMed] [Google Scholar]
- 211.Pang D., Liao S., Wang W., Mu L., Li E., Shen W., Liu F., Zou Y. Destruction of the cell membrane and inhibition of cell phosphatidic acid biosynthesis in Staphylococcus aureus: an explanation for the antibacterial mechanism of morusin. Food Funct. 2019;10(10):6438–6446. doi: 10.1039/c9fo01233h. [DOI] [PubMed] [Google Scholar]
- 212.Xu Z., Li K., Pan T., Liu J., Li B., Li C., Wang S., Diao Y., Liu X. Lonicerin, an anti-algE flavonoid against Pseudomonas aeruginosa virulence screened from Shuanghuanglian formula by molecule docking based strategy. J. Ethnopharmacol. 2019;239 doi: 10.1016/j.jep.2019.111909. [DOI] [PubMed] [Google Scholar]
- 213.Nair S.V., Baranwal G., Chatterjee M., Sachu A., Vasudevan A.K., Bose C., Banerji A., Biswas R. Antimicrobial activity of plumbagin, a naturally occurring naphthoquinone from Plumbago rosea, against Staphylococcus aureus and Candida albicans. Int. J. Med. Microbiol. 2016;306(4):237–248. doi: 10.1016/j.ijmm.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 214.Periasamy H., Iswarya S., Pavithra N., Senthilnathan S., Gnanamani A. In vitro antibacterial activity of plumbagin isolated from Plumbago zeylanica L. against methicillin-resistant Staphylococcus aureus. Lett. Appl. Microbiol. 2019;69(1):41–49. doi: 10.1111/lam.13160. [DOI] [PubMed] [Google Scholar]
- 215.Dissanayake D., Perera D., Keerthirathna L., Heendeniya S., Anderson R.J., Williams D.E., Peiris L.D.C. Antimicrobial activity of Plumbago indica and ligand screening of plumbagin against methicillin-resistant Staphylococcus aureus. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1846622. [DOI] [PubMed] [Google Scholar]
- 216.Amin I., Majid S., Farooq A., Wani H.A., Noor F., Khan R., Shakeel S., Bhat S.A., Ahmad A., Madkhali H. Naringenin (4, 5, 7-trihydroxyflavanone) as a potent neuroprotective agent: from chemistry to medicine. Stud. Nat. Prod. Chem. 2020;65:271–300. [Google Scholar]