Abstract
Purpose
Healthcare professionals need a clear understanding of information about gene-drug interactions in order to make optimal use of pharmacogenetic (PGx) testing. In this report, we compare PGx information in the US Food and Drug Administration (FDA) Table of Pharmacogenetic Associations with information presented in Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines.
Summary
Information from CPIC guidelines and the FDA Table of Pharmacogenetic Associations do not have a high level of concordance. Many drugs mentioned in CPIC guidelines are not listed in the FDA table and vice versa, and the same gene-drug association and dosing recommendation was reported for only 5 of the 126 drugs included in either source. Furthermore, classification of drugs in specific sections of the FDA table does not correlate well with CPIC-assigned or provisionally assigned clinical actionability levels. The Pharmacogenomics Knowledge Base (PharmGKB) clinical annotation levels are generally high for drugs mentioned in CPIC guidelines. PharmGKB clinical annotation levels are often unassigned or are lower level for drugs listed on the FDA table but not in CPIC guidelines. These differences may be due in part to FDA having access to PGx information that is unavailable in published literature and/or because PGx classifications are based on criteria other than clinical actionability.
Conclusion
There are important differences between the PGx information presented in the FDA Table of Pharmacogenetic Associations and in CPIC guidelines. FDA and CPIC have different perspectives when evaluating PGx associations and use different approaches and information resources when considering clinical validity related to specific medicines. Understanding how information sources developed by each group differ and can be used together to form a holistic view of PGx may be helpful in increasing adoption of these information sources in practice.
Keywords: CPIC guidelines, Food and Drug Administration Table of Pharmacogenetic Associations, gene-drug associations, pharmacogenetics guidance
Pharmacogenetics (PGx) is a cornerstone of personalized medicine, providing a way to guide medication-based treatment and prevention strategies in accordance with each patient’s genetic characteristics. The use of PGx tests to detect genetic variants that influence response to associated drugs plays an important role in identifying patients at risk for therapy failure, avoiding adverse events, and optimizing drug choices and dosing. PGx tests have been clinically available for more than 15 years, and studies have demonstrated that PGx-guided therapy decisions for certain drugs can improve clinical outcomes.1 Most of the population has one or more actionable pharmacogenetic variants.2 Given this fact and the high prescription rates of many drugs with pharmacogenetic relevance, a significant percentage of patients would likely benefit from PGx testing and genotype-guided prescribing.2-5
However, PGx testing is not yet widely adopted in medical practice. One of numerous barriers is a general lack of knowledge and education among healthcare professionals about how to apply PGx information to clinical practice.6 In many cases, the evidence supporting PGx utilization varies depending on the source of information, causing confusion among implementers.7 Major sources of PGx guidance include clinical practice guidelines from medical organizations, guidelines published by the Clinical Pharmacogenetics Implementation Consortium (CPIC) and other international groups, the Pharmacogenomics Knowledge Base (PharmGKB), and the US Food and Drug Administration’s (FDA’s) drug labeling and Table of Pharmacogenetic Associations.8-13 In this report, we compare the FDA Table of Pharmacogenetic Associations12 (hereafter referred to as the FDA Table) with CPIC guidelines.8
Origins and purpose of the FDA Table.
For many FDA-approved medications (299 drugs as of December 31, 2020), PGx information is included in the drug labeling, although most labels do not provide specific recommendations regarding dosing or drug selection.14
The FDA Table was released in February of 2020. As stated by FDA, “The table contains information about gene-drug interactions for which FDA believes there is sufficient scientific evidence to support the described associations between certain genetic variants or genetic variant-inferred phenotypes and altered drug metabolism, and, in certain cases, differential therapeutic effects, including differences in risks of adverse events.” 12 The table categorizes gene-drug interactions into 3 sections of PGx associations: those for which the data support therapeutic management recommendations; those for which the data indicate a potential impact on medication safety or response; and those for which the data demonstrate a potential impact on pharmacokinetic properties only. Inclusion of a particular gene-drug interaction in the table does not necessarily mean FDA recommends that a PGx test be conducted before prescribing the corresponding medication (unless the test is an indicated FDA-approved companion diagnostic as described in the drug label).12 The criteria for which gene-drug pairs are included in the table have not been made clear. FDA characterizes the table as a “work in progress,” recognizing that “various other pharmacogenetic associations exist that are not included” and stating that the table will be “updated periodically with additional pharmacogenetic associations supported by sufficient evidence.” 12
Origins and purpose of CPIC clinical practice guidelines.
Launched in 2009 as a shared project between the National Institutes of Health (NIH)–funded PharmGKB and the Pharmacogenomics Research Network (PGRN), CPIC is an international consortium of volunteer PGx experts with a small, dedicated staff.15 CPIC has developed clinical practice guidelines (“CPIC guidelines” hereafter) for associated gene-drug interactions for which there is high-level evidence to support clinical actionability with respect to prescribing decisions. The guidelines are designed to assist healthcare providers in translating PGx test results into actionable prescribing decisions. CPIC uses evidence curated by PharmGKB and deep review of the available literature to develop each guideline. CPIC guidelines follow standardized formats, including systematic grading of evidence and clinical recommendations. The guidelines are peer reviewed and regularly updated. CPIC guidelines are used by healthcare practitioners, laboratories, and test manufacturers. They also influence reimbursement protocols and the development of clinical decision support software.
Interpretations and therapy recommendations in CPIC guidelines often differ from the information included in FDA-approved drug labels. These differences have led to a lack of clarity among providers, test developers, and clinical software developers regarding the appropriate use of PGx tests. In 2016, authors from FDA’s Center for Drug Evaluation and Research published a review of the information on gene-drug associations available in US drug labeling, practice guidelines, and recommendations.16 Comparisons of PGx information between various international clinical guidelines and some major regulatory bodies, including FDA, has shown that there was a lack of consensus regarding actionable pharmacogenomic labeling.17-19 A recent publication by Kisor et al20 pointed out differences between the FDA Table and CPIC guidelines focused on examples of drugs with pharmacokinetic implications well established by CPIC but without therapeutic recommendations by FDA. The report stated that as of January 2020, CPIC provided 23 PGx guidelines covering 47 drugs, of which 30 were included on the FDA Table.20
Objective.
In this study, we cross-referenced the FDA Table and CPIC guidelines. Our analysis provides a comprehensive comparison of the 2 sources across multiple dimensions, including the drugs listed, gene-drug associations, dosing and usage recommendations, and the different categories of their respective gene-drug associations as described in the FDA Table (ie, in sections 1-3) and the assignment of CPIC “levels” for genes/drugs (ie, levels A-D).21 We also determined the percentages of drugs in CPIC guidelines, the FDA Table, or both sources that had each PharmGKB annotation level of evidence (ie, 1A-4).22,23 The analyses highlight areas where CPIC guidelines and FDA information about gene-drug associations match and where they differ. The findings may help inform community engagement among PGx test developers, users, and policymakers as they discuss how different sources of PGx information can be used together, and ultimately how healthcare providers also use these sources together for clinical decision-making.
Study methodology
Comparison of drugs listed in FDA Table and CPIC guidelines.
The drugs listed in the FDA Table were compared to the list of drugs mentioned in CPIC guidelines as of June 15, 2021. The drugs listed within these sources were categorized into 3 groups:
Listed in CPIC guidelines only
Listed in both CPIC guidelines and the FDA Table
Listed in FDA Table only
Two drugs appear twice in the FDA Table (carbamazepine and codeine, in sections 1 and 2). For the purpose of this analysis, these drugs were each counted once. Gene-drug associations that are mentioned in CPIC guidelines but have no actionable therapeutic recommendations were excluded from this analysis.
For each drug, the particular gene-drug associations and usage or dosing recommendations, as noted in CPIC guidelines and the FDA Table, were also compared. We created a table (see appendix) noting the following information for each drug: gene(s), FDA PGx section, CPIC guideline classification of recommendation, CPIC level, PharmGKB level, PGx information on FDA label, and differences in use recommendations between CPIC guidelines and the FDA Table. Gene-drug pairs with differences in specific prescribing recommendations based on genotypes or phenotypes and/or how the genotypes or phenotypes affect the drugs’ outcomes were noted. PGx information was considered concordant if the exact same dosing and/or use recommendations were made for the same genotypes or phenotypes (eg, both sources recommend avoiding abacavir in patients positive for the HLA-B*57:01 allele or considering an antiplatelet alternative to clopidogrel in CYP2C19 intermediate and poor metabolizers). PGx information was considered nonconcordant if dosing guidances or drug avoidance recommendations were different for any given genotype or phenotype and/or if the gene(s) related to the given drug were not the same within each source. Most differences were due to the presence of dosing recommendations by CPIC but a lack of dosing recommendations by FDA. The appendix describes these differences in detail.
Breakdown of drugs listed in FDA Table and/or CPIC guidelines by gene-drug association category.
FDA, CPIC, and PharmGKB use different grouping systems to classify PGx associations based on the information they use to determine the impact of genetic variation on drug response. FDA categorizes the drugs listed in its table into 3 sections based on the gene-drug information the agency has reviewed. However, the agency notes that most of the associations have not been thoroughly evaluated in terms of the impact of genetic testing on clinical outcomes.14 CPIC categorizes drugs based on gene-drug association clinical actionability levels A through D. PharmGKB categorizes drugs based on gene variant/drug association clinical annotation levels 1A through 4. Descriptions of the FDA Table sections and the CPIC and PharmGKB categories are shown in eTable 1.
We broke down the list of drugs in the FDA Table and CPIC guidelines by the gene-drug association categories used by the FDA and CPIC categorization systems. Drugs listed in CPIC guidelines have been thoroughly reviewed by CPIC and, by definition, are categorized at level A or B (clinically actionable). However, drugs included in the FDA Table that are not listed in CPIC guidelines have not necessarily been thoroughly reviewed by CPIC, so for these, we referred to CPIC provisional levels as indicated in the CPIC gene-drug table.24
We also performed a “cross-walk” of the FDA Table, section by section, and CPIC levels of evidence and determined the number of drugs with recommendations in CPIC guidelines that are not included in section 1 of the FDA Table.
We categorized the list of drugs in CPIC guidelines and in the FDA Table by PharmGKB clinical annotation level of evidence. PharmGKB clinical annotations summarize curated peer-reviewed literature reporting evidence of PGx associations between genetic variants and drug response, including but not limited to clinical utility. PharmGKB assesses its clinical annotation levels based on criteria including the number of studies finding positive versus negative results, P values, and study sizes.9,10 It is important to note that many drugs that have not been assigned a clinical annotation level of evidence by PharmGKB have simply not yet been assessed, and these drugs may be assigned annotation levels in the future. After completion of this analysis, PharmGKB created a new scoring system to help automate level of evidence assignment to clinical annotations with the goal of increasing transparency, consistency, and reproducibility.25
Results
Comparison of drugs listed in FDA Table and CPIC guidelines.
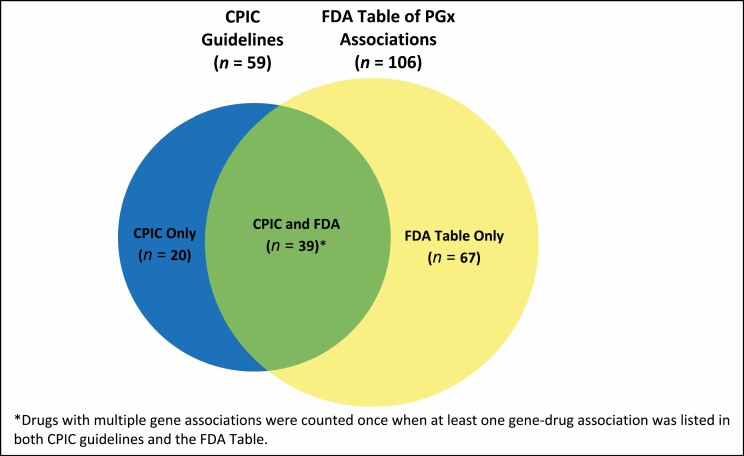
As of June 2021, a total of 126 drugs were listed in the FDA Table and/or CPIC guidelines. Of the 59 drugs listed in CPIC guidelines, 39 were listed in the FDA Table while 20 were not. A total of 106 drugs were listed in the FDA Table, including 39 that were in CPIC guidelines and 67 that were not (Figure 1).
Figure 1.
Number of drugs with pharmacogenetics (PGx) associations as listed in the FDA Table of Pharmacogenetic Associations and Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines.
Table 1 lists the individual drugs in the FDA Table only (by section), CPIC guidelines only, or both. There is a lack of concordance between many of the drugs listed in the FDA Table and those listed in CPIC guidelines. Many of the overlapping drugs listed by both sources have additional or different gene-drug associations and/or different dosing or use recommendations. Regarding the 39 drugs included in both sources, FDA and CPIC report on the same gene association and the same dosing recommendation for only 5, while 5 others have different gene-drug associations and 29 have different dosing or use recommendations.
Table 1.
Drugs Listed in CPIC Guidelines Only, Both CPIC Guidelines and FDA Table, and FDA Table Only
| CPIC Guidelines Only (n = 20) | CPIC Guidelines and FDA Tablea (n = 39) | FDA Table Only (n = 67) |
|---|---|---|
| Data Support Therapeutic Management Recommendations | ||
| Atazanavir | Abacavir | Amifampridine |
| Desflurane | Atomoxetine | Amifampridine phosphate |
| Enflurane | Azathioprine | Amphetamine |
| Fosphenytoin | Capecitabine | Aripiprazole |
| Halothane | Carbamazepine | Aripiprazole lauroxil |
| Hydrocodone | Celecoxib | Belinostat |
| Isoflurane | Citalopram | Brexpiprazole |
| Ivacaftor | Clopidogrel | Brivaracetam |
| Lansoprazole | Codeine | Clobazam |
| Lornoxicam | Fluorouracil | Clozapine |
| Methoxyflurane | Flurbiprofen | Deutetrabenazine |
| Ondansetron | Meloxicam | Dronabinol |
| Peginterferon alfa-2a | Mercaptopurine | Eliglustat |
| Peginterferon alfa-2b | Pantoprazole | Erdafitinib |
| Phenytoin | Piroxicam | Flibanserin |
| Rasburicase | Succinylcholine | Gefitinib |
| Sertraline | Tacrolimus | Iloperidone |
| Sevoflurane | Thioguanine | Irinotecan |
| Tenoxicam | Tramadol | Lofexidine |
| Tropisetron | Warfarin | Meclizine |
| Metoclopramide | ||
| Mivacurium | ||
| Oliceridine | ||
| Pimozide | ||
| Pitolisant | ||
| Propafenone | ||
| Sacituzumab | ||
| Siponimod | ||
| Tetrabenazine | ||
| Thioridazine | ||
| Valbenazine | ||
| Venlafaxine | ||
| Vortioxetine | ||
| Potential Impact on Safety/Response | ||
| Allopurinol | Carvedilol | |
| Carbamazepineb | Cevimeline | |
| Codeineb | Isoniazid | |
| Efavirenz | Lapatinib | |
| Oxcarbazepine | Nilotinib | |
| Simvastatin | Pazopanib | |
| Voriconazole | Perphenazine | |
| Procainamide | ||
| Sulfamethoxazole/ trimethoprim | ||
| Sulfasalazine | ||
| Tolterodine | ||
| Impact on PK Only | ||
| Amitriptyline | Amoxapine | |
| Clomipramine | Atorvastatin | |
| Desipramine | Avatrombopag | |
| Dexlansoprazole | Carisoprodol | |
| Doxepin | Darifenacin | |
| Escitalopram | Diazepam | |
| Fluvoxamine | Dolutegravir | |
| Ibuprofen | Donepezil | |
| Imipramine | Elagolix | |
| Nortriptyline | Esomeprazole | |
| Omeprazole | Fesoterodine | |
| Paroxetine | Galantamine | |
| Tamoxifen | Hydralazine | |
| Trimipramine | Metoprolol | |
| Mirabegron | ||
| Nebivolol | ||
| Propranolol | ||
| Protriptyline | ||
| Raltegravir | ||
| Rabeprazole | ||
| Risperidone | ||
| Rosuvastatin | ||
| Tamsulosin |
Abbreviations: CPIC, Clinical Pharmacogenetics Implementation Consortium; FDA, Food and Drug Administration; PK, pharmacokinetics.
aFor drugs in both CPIC guidelines and the FDA Table of Pharmacogenetic Associations, many had different dosing or use recommendations (these drugs are indicated by ochre color) or additional or different gene/drug associations mentioned in the CPIC and FDA sources (indicated by red).
bCodeine and carbamazepine are listed in two FDA sections but counted only once each in the overall count.
Detailed comparisons of differing gene-drug associations or drug use recommendations are shown in the appendix. For many of the drugs included in both sources, there are differences between the FDA Table and CPIC guidelines in use or dosage recommendations of various types and strengths. Several examples help illustrate the nature of these differences. For carbamazepine, CPIC guidelines include “strong/optional” recommendations for HLA-A and HLA-B, whereas the FDA Table makes a recommendation (in section 1) for only HLA-B, while HLA-A is listed as having a potential impact (section 2). For another gene-drug pair (CYP2C19 and voriconazole), CPIC recommends an alternative medication for ultrarapid, rapid, and poor CYP2C19 metabolizers, whereas the FDA Table does not mention ultrarapid or rapid metabolizers and does not include therapy recommendations for poor metabolizers. For capecitabine and fluorouracil (both with DPYD associations), the FDA Table indicates a higher adverse reaction risk in intermediate and poor DPYD metabolizers, which aligns with the CPIC guideline, but states there is insufficient data available to recommend a dosage in intermediate metabolizers. CPIC specifies a reduced recommended dosage for intermediate metabolizers depending on the variant identified. eTable 2 may be helpful in informing potential PGx community discussions aimed at harmonizing clinical recommendations.
Breakdown of drugs listed in FDA Table and/or CPIC guidelines by gene-drug association category.
Table 2 summarizes the number of drugs in the FDA Table and in CPIC guidelines broken down by gene-drug association categories assigned by the FDA and CPIC grouping systems, as described in Table 1. The 67 drugs listed in the FDA Table that are not included in CPIC guidelines have not been thoroughly reviewed by the organization; however, CPIC has assigned provisional classifications to most of the drugs, which are also broken down in Table 2.
Table 2.
Gene-Drug Association Categories (FDA Table Sections and CPIC Clinical Actionability Levels) for Drugs Listed in CPIC Guidelines and FDA Table of Pharmacogenetic Associations
| No. (%) of Drugs | |||
|---|---|---|---|
| CPIC Guideline Only | CPIC/FDA Overlap | FDA Table Only | |
| FDA Table section | |||
| 1. Data support therapeutic management recommendations | 0 (0) | 20 (49) | 33 (49) |
| 2. Potential impact on safety/response | 0 (0) | 7 (17)b | 11 (16) |
| 3. Impact on PK only | 0 (0) | 14 (34) | 23 (35) |
| Not in FDA Table | 20 (100) | 0 (0) | 0 (0) |
| CPIC level | |||
| A | 18 (90) | 32 (82) | 3 (5)a |
| B (A/B) | 2 (10) | 7 (18) | 14 (20)a |
| C (B/C) | 0 (0) | 0 (0) | 48 (72)a |
| D | 0 (0) | 0 (0) | 0 (0) |
| No CPIC level assigned | 2 (3) |
Abbreviations: CPIC, Clinical Pharmacogenetics Implementation Consortium; FDA, Food and Drug Administration; PK, pharmacokinetics.
aReflects CPIC provisional levels.
bCodeine and carbamazepine are listed in 2 FDA sections for different gene-drug associations.
We note that although the 5 drugs with concordant use recommendations specified in the FDA Table and CPIC guidelines are found in FDA section 1, the inclusion of a drug in a particular section of the FDA Table does not predict a higher probability it will have a CPIC guideline. The distribution of drugs listed in FDA Table sections 1 through 3 is similar whether they are in CPIC guidelines or in the FDA Table only. About half of all of the drugs listed in the FDA Table that are not listed in a CPIC guideline are in section 1. Similarly, of the drugs in the FDA Table that are also listed in CPIC guidelines, about half are in section 1. Additionally, whereas all of the drugs listed in CPIC guidelines are classified by CPIC as level A or B (ie, prescribing action is recommended), CPIC classifies most drugs (73%) that are only listed in the FDA Table as level C (ie, no prescribing actions are recommended).
A cross-walk comparison of the drugs in each section of the FDA Table against CPIC level determinations (eTable 2) found 52 drugs listed in section 1, 20 listed in section 2, and 36 listed in section 3 of the FDA Table.
We note that many drugs (n = 39) determined by CPIC to be clinically actionable (with CPIC guideline therapy recommendations) are not in FDA Table section 1. Conversely, section 1 of the FDA Table includes 20 drugs that CPIC categorizes provisionally as level B/C or lower, and 1 drug does not have a CPIC categorization. There are 13 drugs in FDA section 3 determined by CPIC to be clinically actionable (with CPIC guideline therapy recommendations).
The percentage of drugs in CPIC guidelines only, the CPIC/FDA overlap group, and the FDA Table only was determined with respect to PharmGKB annotation levels (eFigure 1). All the drugs listed in CPIC guidelines have an assigned PharmGKB level. However, of the 67 drugs listed in the FDA Table only, 51% were not assigned a PharmGKB level at the time of our study. (This analysis was performed prior to the July 2021 implementation by PharmGKB of a new quantitative system for assigning levels of evidence to clinical annotations.25,26) Many of the drugs that were not assigned a PharmGKB annotation level have not yet been assessed by the organization.
We note that PharmGKB clinical annotation levels are generally high for drugs listed in CPIC guidelines, including those drugs also listed within the FDA Table. This would be expected given that CPIC uses PharmGKB clinical annotation levels 1A through 2B as one reason for assigning CPIC levels to genes/drugs in its prioritization approach for guideline development. PharmGKB has not assigned annotation levels to many of the drugs listed in the FDA Table that are not mentioned in CPIC guidelines, and those drugs in this group that do have clinical annotations have lower assigned levels (on average) than those included in CPIC guidelines. This implies that FDA may have based the PGx section assignments on evidence other than what is available to PharmGKB in published literature and/or on criteria other than clinical actionability.
Discussion
Inconsistencies between pharmacogenomics resources, including CPIC guidelines, the FDA Table, and medical organization clinical practice guidelines, create confusion for PGx implementers, test developers, physicians, and pharmacists regarding applying PGx to medication therapy decisions. Our analysis confirms that CPIC and FDA present information that is not concordant for most drugs.
CPIC and FDA reported on the same gene-drug association and/or the same dosing recommendation for only 5 of the 126 drugs included in either source. In many cases, the differences involved phenotype subgroups missing from the FDA Table for which therapy changes are recommended in CPIC guidelines. Although all 5 of the drugs that had concordant CPIC and FDA recommendations were in section 1 of the FDA Table, a substantial number of drugs with recommendations in CPIC guidelines (n = 39) are not listed in section 1 of the FDA Table. Conversely, CPIC guidelines have been issued for fewer than 40% of drugs in section 1. Also noteworthy, there are CPIC guidelines for 36% of the drugs in Section 3 of the FDA Table that are considered by FDA to have associations with a potential impact on pharmacokinetics only. When considering PharmGKB annotation evidence levels, those drugs listed in the FDA Table for which there are no CPIC guidelines have either not yet been considered for assignment of a clinical annotation or have lower annotation levels on average, regardless of the FDA Table section in which they are listed.
Previous studies have also highlighted differences in recommendations between PGx organizations, regulatory bodies, and professional societies. A recent analysis demonstrated that nearly one half of clinical PGx recommendations from FDA drug labels, CPIC, and major US-based professional medical organizations have inconsistencies.7 A comparison between guidelines published by CPIC and the Dutch Pharmacogenetics Working Group (DPWG) revealed several differences resulting from variable guideline development methods.27 Previous additional studies individually highlighted the number of PharmGKB level 1A clinical annotations that had CPIC, DPWG, or FDA drug label support, differences in PGx information between drug labels from various international regulatory bodies, and high discordance among product labeling and professional societies, which may be due to the way in which biomarkers are evaluated and the intent of practice guidelines.1,17,28 Our study focuses on all germline PGx recommendations from the newest set of FDA tables and CPIC guidelines and provides a comprehensive, yet granular, comparison of use recommendations and categories of drug-gene associations.
FDA and CPIC use different approaches and information resources and have different perspectives and objectives when evaluating PGx gene associations related to specific medicines. The FDA Table groups drugs within 3 sections based on the gene-drug information that the agency has reviewed. When reviewing new drugs, FDA examines drug applications with consideration of any evidence of PGx associations affecting a drug’s safe and effective use. According to FDA, “Drug labels may contain information on genomic biomarkers and can describe: drug exposure and clinical response variability, risk for adverse events, genotype-specific dosing, mechanisms of drug action, polymorphic drug target and disposition genes, and/or trial design features.” 14 This labeled information can include data that are known to the FDA but not necessarily reported in peer-reviewed literature. Drug labels are sometimes, but not often, updated with additional PGx information that may emerge after approval.29 In the absence of a regular process for FDA to consider emerging gene-drug association data post approval, the PGx information in drug labels for older medications may not reflect the current state of the science. CPIC assigns levels of gene-drug associations based on clinical actionability. CPIC guidelines are developed for gene-drug associations that are determined to be actionable (CPIC level A or B).8 CPIC guidelines are based on primary published information and include a process for review and updates as additional evidence emerges. All supporting evidence is cited or listed in the guideline publications. Understanding the basis for these differences and how information sources developed by different expert groups can be used together may be helpful to increase the adoption of PGx in practice.
Data supporting CPIC guidelines and PharmGKB clinical annotation levels is from peer-reviewed published literature. FDA likely had access to additional information when developing the Table of Pharmacogenetic Associations, including unpublished information from new drug submissions from drug manufacturers. Some discrepancies regarding clinical recommendations made by the 2 groups may be due to the differing data sources and evaluation processes involved. Importantly, each group uses different definitions and sources to assign sections, actionability, or annotation levels, and definitions may evolve over time. All lists will evolve as the PGx evidence base from studies continues to grow.
A consistent and standardized process for interpreting PGx information may help ensure patient access to accurate, clinically actionable PGx test results. Evidentiary standards should account for the different perspectives of FDA and CPIC and take into consideration additional sources for evidence evaluation, including clinical annotations made by PharmGKB. Resources do not necessarily need to align, but it is important that the PGx community recognize there are major differences between FDA and CPIC in recommendations, evidence/classification levels, and sources of data. It is also important to recognize that professional medical organization guidelines greatly influence the adoption of new practices across therapeutic areas. Most practitioners (outside of PGx implementers) are currently unaware of CPIC guidelines and don’t use PGx information in FDA labeling.30 There should be a concerted effort from professional medical organizations to evaluate PGx information using sources from CPIC, PharmGKB, and FDA to inform clinical practice recommendations.
Pharmacogenetic and policy leaders have been calling for improved collaboration between FDA, CPIC, clinical guideline developers, professional medical and pathology organizations, NIH, and the private sector. A collaborative approach will minimize duplicative efforts and harmonize solutions for an immediate impact on public health. FDA has formally recognized and joined a new pharmacogenomics collaborative community called Standardizing Laboratory Practices in Pharmacogenomics (STRIPE), which aims to bring diverse stakeholders together to address challenges and opportunities for PGx testing.31 The Pharmacogenomic Clinical Annotation Tool (PharmCAT) is being developed through a collaboration between the PharmGKB and the former PGRN Statistical Analysis Resource, with input from other groups, to extract guideline variants from a genetic dataset, interpret the variant alleles, and make prescribing recommendations that can be used to inform treatment decisions.32,33
Conclusion
The information in this report may help with future updates to the FDA Table and to inform STRIPE participants, PharmCAT developers, and the larger PGx community as they facilitate consensus approaches to help overcome challenges in PGx and clarify the appropriate clinical use of PGx in practice. This, in turn, may help advance personalized medicine by improving the ability of laboratories, clinicians, and health systems to provide patients with access to clinical PGx testing.
Supplementary Material
Acknowledgments
The authors acknowledge the important input of Annette Taylor, PhD towards the development of this report. The authors also acknowledge members of the Personalized Medicine Coalition Pharmacogenetics Working Group for external review, and members of CPIC, including Michelle Whirl-Carrillo, PhD, Teri Klein, PhD, and Kelly Caudle, PharmD, PhD, for providing helpful feedback for early versions of the report.
Appendix—Drug-Gene Associations, Clinical Recommendations, Evidence Levels, and Differences in Dosing or Use Recommendations for Drugs Listed in Both FDA Table of Pharmacogenetic Associations and CPIC Guidelinesa
| Drug (n = 39) | Gene(s) (n = 52) | FDA Table Section Classificationb | CPIC Guideline Classification of Recommendation | CPIC Level | PharmGKB Level | PGx Info on FDA Label | Differences in Use Recommendations in CPIC Guideline(s) and FDA Table |
|---|---|---|---|---|---|---|---|
| Abacavir | HLA-B | Recommendation | Strong | A | 1A | Testing required | None |
| Allopurinol | HLA-B | Potential Impact | Strong | A | 1A | NA | CPIC states contraindicated if HLA-B*5801 positive; FDA states higher risk of adverse reaction |
| Amitriptyline | CYP2C19 | Not included | Optional UM/RM, moderate PM | A | 1A | NA | CPIC recommends avoiding amitriptyline in UMs/RMs/PMs |
| CYP2D6 | PK Only | Strong UM/PM, moderate IM | A | 1A | Actionable PGx | CPIC lists additional gene (CYP2C19) and provides dose guidance in IMs and PMs and avoid in Ums; FDA states alters systemic concentrations | |
| Atomoxetine | CYP2D6 | Recommendation | Strong or moderate depending on AS and children vs adults | A | 1A | Actionable PGx | CPIC provides dose guidance for all phenotypes and indicates potential for adverse reactions and improved efficacy compared with non-PMs; FDA states adverse reaction risk in PMs |
| Azathioprine | TPMT | Recommendation | Strong IM/PM | A | 1A | Testing recommended | CPIC provides dose guidance for IMs and for malignancy in PMs, advises considering alternatives for other PMs; FDA indicates consider alternatives in PM and dose reduction in IM |
| NUDT15 | Recommendation | Strong | A | 1A | Testing recommended | ||
| Capecitabine | DPYD | Recommendation | Strong or moderate depending on AS | A | 1A | Actionable PGx | CPIC provides dose guidance for IMs; FDA indicates insufficient data available for dosing guidance for IMs |
| Carbamazepine | HLA-B | Recommendation | Strong/optional | A | 1A | Testing required | None |
| HLA-A | Potenial impact | Strong/optional | A | 1A | Actionable PGx | CPIC states avoid use if HLA-A*31:01 positive; FDA states consider risk and benefit; CPIC and FDA have different dosing recommendations | |
| Celecoxib | CYP2C9 | Recommendation | Moderate IM (AS of 1) and PM | A | 1A | Actionable PGx | None |
| Citalopram | CYP2C19 | Recommendation | Strong IM, moderate PM/UM | A | 1A | Actionable PGx | FDA recommends maximum dose of 20 mg in PMs; CPIC recommends 50% dose reduction in PMs and alternative therapy in UMs |
| Clomipramine | CYP2D6 | PK Only | Optional | B | 1A | Actionable PGx | CPIC lists additional gene (CYP2C19), provides dose guidance, and suggests alternative therapy in UMs and PMs; FDA states alters systemic concentrations only |
| CYP2C19 | Not included | Optional UM/RM/PM | B | 1A | NA | CPIC provides optional recommendation to avoid tertiary amines in UMs/RMs/PMs | |
| Clopidogrel | CYP2C19 | Recommendation | Strong PM, moderate IM | A | 1A | Actionable PGx | None |
| Codeine | CYP2D6 | Recommendation and potential impact | Strong UM, moderate IM, optional PM | A | 1A | Actionable PGx | CPIC recommends avoiding use in UMs and PMs; FDA states drug contraindicated in children under 12 years of age, regardless of phenotype |
| Desipramine | CYP2D6 | PK Only | Optional | B | 1A | Actionable PGx | CPIC provides dose guidance and suggests alternative therapy in UMs and PMs; FDA states alters systemic concentrations only |
| Dexlansoprazole | CYP2C19 | PK Only | Moderate for RM, NM, and PM | B | 1A | Actionable PGx | CPIC provides optional dosing recommendations for all phenotypes; FDA states IMs and PMs may have higher systemic concentrations |
| Doxepin | CYP2C19 | PK Only | Optional | B | 1A | Actionable PGx | CPIC provides dose guidance; FDA states alters systemic concentrations only |
| CYP2D6 | PK Only | Optional | B | 1A | Actionable PGx | CPIC states alternative therapy in UMs and provides dose guidance in PMs; FDA states alters systemic concentrations only | |
| Efavirenz | CYP2B6 | Potential Impact | Moderate PM/IM | A | 1A | Actionable PGx | CPIC provides dose guidance in IMs and PMs; FDA states higher concentrations and QT prolongation in PMs |
| Escitalopram | CYP2C19 | PK Only | Moderate PM/UM | A | 1A | Actionable PGx | CPIC recommends considering alternative for UMs and dose reduction for PMs; FDA states alters systemic concentrations (drug was 1 of 2 drugs that were subject of FDA warning letter to INOVA Health) |
| Fluorouracil | DPYD | Recommendation | Strong or moderate depending on AS | A | 1A | Actionable PGx | CPIC provides dose guidance for IMs; FDA indicates insufficient data available for dose guidance for IMs |
| Flurbiprofen | CYP2C9 | Recommendation | Moderate IM (AS of 1) and PM | A | 1A | Actionable PGx | None |
| Fluvoxamine | CYP2D6 | PK Only | Optional PM | B | 1A | Actionable PGx | CPIC recommends dose reduction in PMs; FDA states use with caution |
| Ibuprofen | CYP2C9 | PK Only | Moderate PM | A | 1A | CPIC recommends dose reduction in PMs; FDA states *3 allele carriers or PMs may have higher systemic concentrations | |
| Imipramine | CYP2C19 | Not included | Optional UM/RM/PM | B | 1A | NA | CPIC provides optional recommendation to avoid tertiary amines in UM/RM/PMs |
| CYP2D6 | PK Only | Optional | B | 1A | Actionable PGx | CPIC lists additional gene (CYP2C19), provides dose guidance, and states alternative therapy in UMs and PMs; FDA states alters systemic concentrations | |
| Meloxicam | CYP2C9 | Recommendation | Moderate IM (AS of 1) and PM | A | 1A | Actionable PGx | CPIC recommends dose reduction in IMs with AS of 1 and recommends alternative therapy in PMs; FDA states PMs or *3 allele carriers have higher systemic concentrations and advises considering dose reductions in PMs |
| Mercaptopurine | TPMT | Recommendation | Strong PM/IM | A | 1A | Testing recommended | CPIC recommends dose guidance in PMs and IMs; FDA provides dose guidance in PMs and states IMs may require dosage reduction based on tolerability |
| NUDT15 | Recommendation | Strong PM/IM | A | 2B | Testing recommended | ||
| Nortriptyline | CYP2D6 | PK Only | Strong UM/PM, moderate IM | A | 1A | Actionable PGx | CPIC provides dose guidance; FDA states alternative therapy in UMs and PMs or dose reduction in PMs |
| Omeprazole | CYP2C19 | PK Only | Moderate for RM, NM, and PM | A | 1A | Actionable PGx | CPIC provides dosing recommendations for all phenotypes; FDA states IM and PMs may have higher systemic concentrations only |
| Oxcarbazepine | HLA-A | Not included | Strong/optional | C | 3 | NA | CPIC recommends avoiding use if oxcarbazepine naïve and alternative agents are available; FDA does not mention HLA-A |
| HLA-B | Potential Impact | Strong/optional | A | 1A | Testing recommended | CPIC recommends avoiding use if oxcarbazepine naïve and alternative agents are available; FDA states higher adverse reaction risk only | |
| Pantoprazole | CYP2C19 | Recommendation | Moderate for RM, NM, and PM | A | 1A | Actionable PGx | CPIC provides dosing recommendations for all phenotypes; FDA states PMs may have higher systemic concentrations and advises considering dose reduction in children |
| Paroxetine | CYP2D6 | PK Only | Strong UM, optional PM | A | 1A | Informative PGx | CPIC recommends alternative for UMs and dose reduction for PMs; FDA states alters systemic concentrations only |
| Piroxicam | CYP2C9 | Recommendation | Moderate PM/IM | A | 1A | Actionable PGx | CPIC recommends avoiding use in IMs with AS of 1 and PMs; FDA states consider reducing dosage in PMs |
| Simvastatin | SLCO1B1 | Potential Impact | Strong | A | 1A | NA | CPIC provides dose guidance and states consider alternative statins as an option; FDA states risk of adverse reaction is higher at high doses only |
| Succinylcholine | BCHE | Recommendation | C/D | 3 | Actionable PGx | CPIC guidelines mention 2 genes (CACNA15 and RYR1); FDA recommendations are based on a different gene: BCHE | |
| CACNA15 | Not included | Strong (selected genes) | A | 1A | Actionable PGx | ||
| RYR1 | Not included | Strong | A | 1A | Actionable PGx | ||
| Tacrolimus | CYP3A5 | Recommendation | Strong NM/IM | A | 1A | NA | CPIC provides dose guidance; FDA states may result in lower systemic concentrations and adjust dose based on trough concentrations |
| Tamoxifen | CYP2D6 | PK Only | Strong or moderate depending on AS | A | 1A | Actionable PGx | CPIC provides dose and drug selection guidance; FDA states impact on efficacy not well established |
| Thioguanine | TPMT | Recommendation | Strong PM, moderate IM | A | 3 | Testing recommended | CPIC recommends dose guidance in PMs and IMs; FDA provides dose guidance in PMs and states IMs may require dosage reduction based on tolerability |
| NUDT15 | Recommendation | Strong PM, moderate IM | A | 3 | Testing recommended | ||
| Tramadol | CYP2D6 | Recommendation | Strong UM/PM | A | 1B | Actionable PGx | CPIC recommends avoiding use in UMs and PMs; FDA states risks in UMs and avoid use in children under 12 |
| Trimipramine | CYP2D6 | PK Only | Optional | B | 1A | Actionable PGx | CPIC lists additional gene (CYP2C19) and lists dose guidance or alternative therapy in UMs and PMs |
| CYP2C19 | Not included | Optional UM/RM/PM | B | 1A | NA | CPIC provides optional recommendation to avoid tertiary amines in UMs/RMs/PMs. Gene-drug pair not included in FDA table. | |
| Voriconazole | CYP2C19 | PK Only | Moderate UM/RM/PM | A | 1A | Actionable PGx | CPIC recommends alternative for UMs, RMs, and PMs; FDA states higher concentrations in IM and PMs |
| Warfarin | CYP2C9 | Recommendation | Strong or moderate | A | 1A | Actionable PGx | CPIC provides specific dose guidance and algorithm; FDA states may alter systemic concentrations and dosage requirements |
| CYP4F2 | Recommendation | Optional | A | 1A | NA | ||
| VKORC1 | Recommendation | Strong or moderate | A | 1A | Actionable PGx |
Abbreviations: AS, activity score; CPIC; Clinical Pharmacogenetics Implementation Consortium; FDA; Food and Drug Administration; IM, intermediate metabolizer; NA, not applicable; NM, normal metabolizer; PGx, pharmacogenetics; PM, poor metabolizer; RM, rapid metabolizer; UM, ultrarapid metabolizer.
aOchre color denotes drugs listed in both CPIC guidelines and the FDA Table for which dosing or use recommendations provided in the 2 sources differ; red denotes drugs for which additional or different gene-drug associations are mentioned.
b“Recommendations” denotes therapeutic management recommendations; “Potential Impact,” potential impact on safety or response; “PK Only,” impact on pharmacokinetics only.
Contributor Information
Daryl Pritchard, Personalized Medicine Coalition, Washington, DC, USA.
Jai N Patel, Department of Cancer Pharmacology and Pharmacogenomics, Levine Cancer Institute, Atrium Health, Charlotte, NC, USA.
Lindsay E Stephens, Personalized Medicine Coalition, Washington, DC, USA.
Howard L McLeod, Geriatric Oncology Consortium, Tampa, FL, USA.
Disclosures
The authors have declared no potential conflicts of interest.
References
- 1. Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. Lancet. 2019;394(10197):521-532. PMID:31395440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heise CW, Gallo T, Curry SC, et al. Identification of populations likely to benefit from pharmacogenomics testing. Pharmacogenet Genomics. 2020;30:91-95. [DOI] [PubMed] [Google Scholar]
- 3. Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five United States medical centers. Annu Rev Pharmacol Toxicol. 2015;55:89-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hicks JK, Rouby NE, Ong HH, et al. Opportunity for genotype-guided prescribing among adult patients in 11 US health systems. Clin Pharmacol Ther. 2021;110(1):179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chanfreau-Coffinier C, Hill LE, Lunch JA, et al. Projected prevalence of actionable pharmacogenetic variants and level A drugs prescribed among US Veterans Health Administration pharmacy users. JAMA Netw Open. 2019;2(6):1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krebs K, Milani L. Translating pharmacogenomics into clinical decisions: do not let the perfect be the enemy of the good. Hum Genomics. 2019;13:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shugg T, Pasternak AL, London B, et al. Prevalence and types of inconsistencies in clinical pharmacogenetic recommendations among major U.S. sources. NPJ Genom Med. 2020; 5(48):1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical Pharmacogenetics Implementation Consortium. Clinical practice guidelines. Accessed February 22, 2021. https://cpicpgx.org/guidelines/
- 9. Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92(4):414-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. PharmGKB. Overview of the PharmGKB. Accessed November 17, 2020. https://www.pharmgkb.org/page/overview
- 11. US Food and Drug Administration. Drugs@FDA: FDA-Approved Drugs. Accessed February 22, 2021. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm
- 12. US Food and Drug Administration. Table of Pharmacogenetic Associations. Updated March 18, 2021. Accessed May 28, 2021. https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations
- 13. PharmGKB. DPWG: Dutch Pharmacogenetics Working Group. Accessed July 1, 2021. https://www.pharmgkb.org/page/dpwg
- 14. US Food and Drug Administration. Table of pharmacogenetic biomarkers in drug labeling. Published August 2020. Accessed December 3, 2020. https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling
- 15. Relling MV, Klein TE, Gammal RS, et al. The Clinical Pharmacogenetics Implementation Consortium: 10 years later. Clin Pharmacol Ther. 2020;107:171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Filipski KK, Pacanowski MA, Ramamoorthy A, et al. Dosing recommendations for pharmacogenetic interactions related to drug metabolism. Pharmacogenet Genomics. 2016;26(7):334-339. [DOI] [PubMed] [Google Scholar]
- 17. Shekhani R, Steinacher L, Swen JJ, et al. Evaluation of current regulation and guidelines of pharmacogenomic drug labels: opportunities for improvements. Clin Pharmacol Ther. 2020;107(5):1240-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagy M, Attya M, Patrinos GP. Unraveling heterogeneity of the clinical pharmacogenomic guidelines in oncology practice among major regulatory bodies. Pharmacogenomics. 2020;21(17):1247-1264. [DOI] [PubMed] [Google Scholar]
- 19. Yoon DY, Lee S, Ban MS, et al. Pharmacogenomic information from CPIC and DPWG guidelines and its application on drug labels. Transl Clin Pharmacol. 2020;28(4):189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kisor DF, Monte AA, Mueller DJ. Pharmacogenetic associations and evidence-based pharmacogenetics guidelines: supporting label and off-label use of drug-gene interaction data. Pharmacogenomics. 2020;21(7):427-430. [DOI] [PubMed] [Google Scholar]
- 21. Clinical Pharmacogenetics Implementation Consortium. Prioritization. Accessed November 17, 2020. http://www.cpicpgx.org/prioritization/#cpicLevels
- 22. PharmGKB. Guideline annotations. Accessed November 17, 2020. http://www.pharmgkb.org/guidelineAnnotations
- 23. PharmGKB. Clinical annotation levels of evidence. Accessed August 6, 2021. https://www.pharmgkb.org/page/clinAnnLevels
- 24. Clinical Pharmacogenetics Implementation Consortium. Genes-drugs. Accessed November 17, 2020. https://cpicpgx.org/genes-drugs/
- 25. Whirl-Carrillo M, Huddart R, Gong L, Sangkuhl K, et al. An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2021;110(3):563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whirl-Carrillo M. PharmCAT version 1.0 released. The PharmGKB blog. Published September 27, 2021. Accessed September 27, 2021. https://pharmgkb.blogspot.com/2021/09/pharmcat-version-10-released.html
- 27. Bank PCD, Caudle KE, Swen JJ, et al. Comparison of the guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin Pharmacol Ther. 2018;103(4):599-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Filipski KK, Pacanowski MA, Ramamoorthy A, Feero WG, Freedman AN. Dosing recommendations for pharmacogenetic interactions related to drug metabolism. Pharmacogenet Genomics. 2016;26(7):334-339. [DOI] [PubMed] [Google Scholar]
- 29. Drozda K, Pacanowski MA, Grimstein C, et al. Pharmacogenetic labeling of FDA-approved drugs: a regulatory retrospective. JACC Basic Transl Sci. 2018;3(4):545-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rahawi S, Naik H, Blake KV, et al. Knowledge and attitudes on pharmacogenetics among pediatricans. J Hum Genet. 2020;64:437-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. American Society of Pharmacovigilance. STRIPE Collaborative Community. Accessed May 2, 2021. https://www.stopadr.org/stripe
- 32. Pharmacogenomics Clinical Annotation Tool. Accessed August 6, 2021. http://pharmcat.org/
- 33. Sangkuhl K, Whirl-Carrillo M, Whaley RM, et al. Pharmacogenomics Clinical Annotation Tool (PharmCAT). Clin Pharmacol Ther. 2020;107(1):203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.