Significance
Retinal degenerative diseases affect specific regions of the retinal pigment epithelium (RPE), suggesting the presence of functionally different RPE subpopulations. To identify these subpopulations in human eyes, we generated the first complete morphometric map of the RPE at single-cell resolution using artificial intelligence–based software. We identified five concentric RPE subpopulations, including a ring of RPE cells with cell area similar to macula in the periphery of the eye. Moreover, we found that specific RPE subpopulations are differentially susceptible to monogenic and polygenic retinal diseases. The results obtained here will allow study of molecular and functional RPE differences responsible for regional retinal diseases and will help develop precise cell and gene therapies for specific degenerative eye diseases.
Keywords: cell morphometry, retinal degeneration, artificial intelligence, data science, AMD
Abstract
Regional phenotypic and functional differences in the retinal pigment epithelium (RPE) monolayer have been suggested to account for regional susceptibility in ocular diseases such as age-related macular degeneration (AMD), late-onset retinal degeneration (L-ORD), and choroideremia (CHM). However, a comprehensive description of human topographical RPE diversity is not yet available, thus limiting the understanding of regional RPE diversity and degenerative disease sensitivity in the eye. To develop a complete morphometric RPE map of the human eye, artificial intelligence–based software was trained to recognize, segment, and analyze RPE borders. Five statistically different, concentric RPE subpopulations (P1 to P5) were identified using cell area as a parameter, including a subpopulation (P4) with cell area comparable to that of macular cells in the far periphery of the eye. This work provides a complete reference map of human RPE subpopulations and their location in the eye. In addition, the analysis of cadaver non-AMD and AMD eyes and ultra-widefield fundus images of patients revealed differential vulnerability of the five RPE subpopulations to different retinal diseases.
The retinal pigment epithelium (RPE) is a monolayer of cells located between the photoreceptors and choroid that regulates both photoreceptor and choroidal health and function throughout life (1). With age, RPE cells undergo metabolic changes that can lead to degeneration of the choroid and functional defects of the overlying photoreceptors, causing vision loss (2). In addition, the RPE monolayer is not impacted uniformly across all types of RPE-associated retinal degenerations (RDs). For example, age-related macular degeneration (AMD) primarily affects macular RPE (2), while late-onset retinal degeneration (L-ORD) and choroideremia (CHM) affect the midperipheral RPE monolayer (3–8). High-resolution regional differences of RPE have not been mapped, despite evidence that such differences may contribute to disease phenotype (9).
Molecular and phenotypic differences between macular and peripheral RPE cells have been identified by several groups. For instance, macular RPE cells were shown to have higher activity of acid phosphatase and cathepsin D enzymes compared to peripheral RPE cells, suggesting differential lysosomal activity (10, 11). Na/K adenosine triphosphatase (ATPase) pumps were found to be 40 to 60% more expressed in peripheral RPE regions, reflecting regional differences in ionic regulation (12, 13). The elastic lamina of the Bruch’s membrane was demonstrated to be three to six times thinner in the macula in comparison to the peripheral RPE, suggesting different molecular composition of the Bruch’s membrane at different retinal locations (14, 15). Consistently, elastin (ELN)- and collagen-related genes have been shown to be more highly expressed in peripheral RPE (15). Macular RPE cells have been shown to express higher levels of genes involved in lipid synthesis, lipid metabolism, angiogenesis, and inflammation, while peripheral RPE cells express more monocarboxylate, α-ketoglutarate, sugar, leucine, proline, and choline transporters and have a higher expression of genes associated with extracellular matrix (16–18). Lastly, macular RPE cell density is higher than in the periphery; cell morphometry shows that macular cells are more regular, less elongated, and smaller in size compared to peripheral RPE cells (19, 20). However, the exact location of these geographic differences within the eye and how these differences relate RPE vulnerability to aging and retinal degenerative diseases are still unclear. Critical questions such as if there are RPE population subtypes, how many subpopulations there are, and which specific structural, molecular, or physiological properties make different RPE subpopulations more susceptible to different retinal diseases cannot be currently answered. We hypothesize that one approach to identify different RPE subpopulations is to analyze morphometric differences across the entire human eye. By developing a complete morphometric reference map of human RPE, we can begin to identify subpopulation-specific molecular pathways that lead to various retinal degenerative diseases and thus novel approaches to combat these diseases.
An artificial intelligence–based software was used to analyze cell morphometry from fluorescently labeled images of entire human eye RPE monolayers. We generated a comprehensive single-cell–resolution map of adult human RPE cells. Using cell area, we defined five statistically different RPE subpopulations and described their retinal location. We also discovered an RPE subpopulation with cell area similar to that of macula in the periphery of the eye. In addition, by analyzing the location of RPE lesions in human cadaver RPE monolayers and in patient retinal images, we discovered that specific RPE subpopulations are differentially susceptible to aging and to different types of retinal degenerative diseases.
Results
A comprehensive single-cell–resolution morphometric map of adult human RPE.
To generate a statistically robust and complete single-cell–resolution map of the human RPE, we analyzed 17 RPE flatmounts from the eye globes of nine donors with no significant existing eye conditions (SI Appendix, Table S1). RPE cell borders were stained with phalloidin-iFluor 647 (Fig. 1 A and B). Each RPE flatmount (about 23 mm in radius) was imaged in ∼3,000 tiled panels at 20× magnification; cell borders were recognized and segmented using a machine learning–based software, REShAPE (Fig. 1C; see SI Appendix for details). An average of 2.8 million RPE cells were detected and segmented in each flatmount (SI Appendix, Table S2). The resultant binary image of RPE cell borders was used to calculate cell morphometry features for every single cell in the entire human eye (Fig. 1 D and E and SI Appendix, Tables S3 and S4). We focused on four distinct morphometric features that define compact packing of epithelial cells in a sheet—cell area, aspect ratio (AR), hexagonality, and number of neighbors (21). Fig. 1F describes these four features analyzed by REShAPE and the range of measurements seen in adult human RPE samples as a heatmap. REShAPE produced single-cell–resolution heatmap images of the entire epithelium to visually display the quantification for these four shape metrics (Fig. 1 G, I, K, and M; a summary statistic for each RPE flatmount for these four metrics is shown in SI Appendix, Table S2). In the cell area heatmap, small RPE cells appear in dark blue and large RPE cells in red (Fig. 1G; Inset showing higher magnification in Fig. 1H). The fovea and the parafovea can be identified as a 3-mm-wide dark blue spot (arrowhead) in the center of the flatmount, next to the optic nerve (*) (Fig. 1G). RPE cell area increases gradually with retinal eccentricity up to 14 mm from the flatmount. We discovered a previously unreported ring of small RPE cells (dark blue; arrow) between radius 14 and 17 mm from the center of the flatmount. Outside of the ring of small RPE cells (dark blue; arrow), the cell area increased dramatically (green/yellow/red) (Fig. 1G). The flatmounts end at the start of the ora serrata. AR represents cell regularity and elongation and ranges in arbitrary score from 1 to 3. AR of 1 is assigned to a regular cell that has the same length of major and minor axes, while AR above 1 suggests an elongated cell with unequal sides, with AR of 3 being the most elongated cell. AR shows a different distribution compared to cell area. RPE cells in the macula and around the macula (10-mm radius) have an AR closer to 1; it increased between 10 and 14 mm to 1.23 and dropped again closer to 1.0 in the newly discovered ring of cells between 14 and 17 mm (Fig. 1I, arrow). This suggests compact cells in the central region and, in the peripheral, a newly discovered ring of cells (arrow), compared to central and farthest peripheries that contain more elongated cells (Fig. 1 I and J). The hexagonality score describes how close the cell shape is to a regular hexagon (equilateral and equiangular), with an arbitrary score ranging from 0 to 10, where 10 corresponds to a perfect hexagon. Similar to AR, RPE cells in the central region (0 to 10 mm) have a hexagonality score of 9.3, cells from 10 to 14 mm have a lower hexagonality score, and the ring of smaller cells in the far-periphery (Fig. 1K, arrow) again has a higher hexagonality score similar to RPE of the central region (Fig. 1 K and L). The number of neighbors for a given cell can vary from 1 to 14. Across the entire flatmount, 79% of cells have between five and seven neighbors per cell, consistent with high hexagonality scores in these regions of the eye. This trend is not followed by cells on the edges of the flatmount that only have five neighbors per cell (Fig. 1 M and N). Overall, an average adult human RPE cell has an area of 220.97 ± 15.34 µm2, an AR of 1.23 ± 0.02 (out of 3.0), a hexagonality score of 9.13 ± 0.06 (out of 10), and 5.47 ± 0.36 number of neighbors (SI Appendix, Table S2). Interestingly, the fovea and the parafovea are only distinguishable by eye in cell area heatmaps.
Fig. 1.
Complete morphometric analysis of an entire human RPE monolayer. (A) Representative image of an entire human RPE flatmount (red: phalloidin). Approximately 2 to 3.5 million cells were captured in about 200,000 images, projected in two-dimensional space, and tiled together. (B) Representative higher magnification image of phalloidin (red)-stained RPE cells. (C) REShAPE, a U-net convolutional neural network (CNN), recognizes and segments RPE cell borders from fluorescent images and analyzes RPE cell morphometry for the entire human RPE flatmount. (Conv 3x3, 3x3 convolutions; ReLU, rectified line unit.) (D and E) REShAPE-generated image for the entire human RPE flatmount with cell borders segmented for each RPE cell (D) and a representative higher magnification image (E). (F) Schematic representation of four distinct cell morphometry parameters (cell area, AR, hexagonality score, and number of neighbors) used for RPE cell shape analysis, specific examples highlighted. (G, I, K, and M) REShAPE-segmented cadaver human RPE flatmount images that are color-coded for cell area, AR, hexagonality score, and number of neighbors illustrate shape metrics of individual RPE cells in human eyes. Heatmaps on the top left corner of each image show range of values used for these four morphometry parameters. Arrowhead, fovea; *, optic nerve; arrow, peripheral ring of small RPE cells. (H, J, L, and N) Zoomed-in color-coded images display RPE shape metrics at single-cell level.
REShAPE discovers five RPE subpopulations with a peripheral ring of small RPE cells.
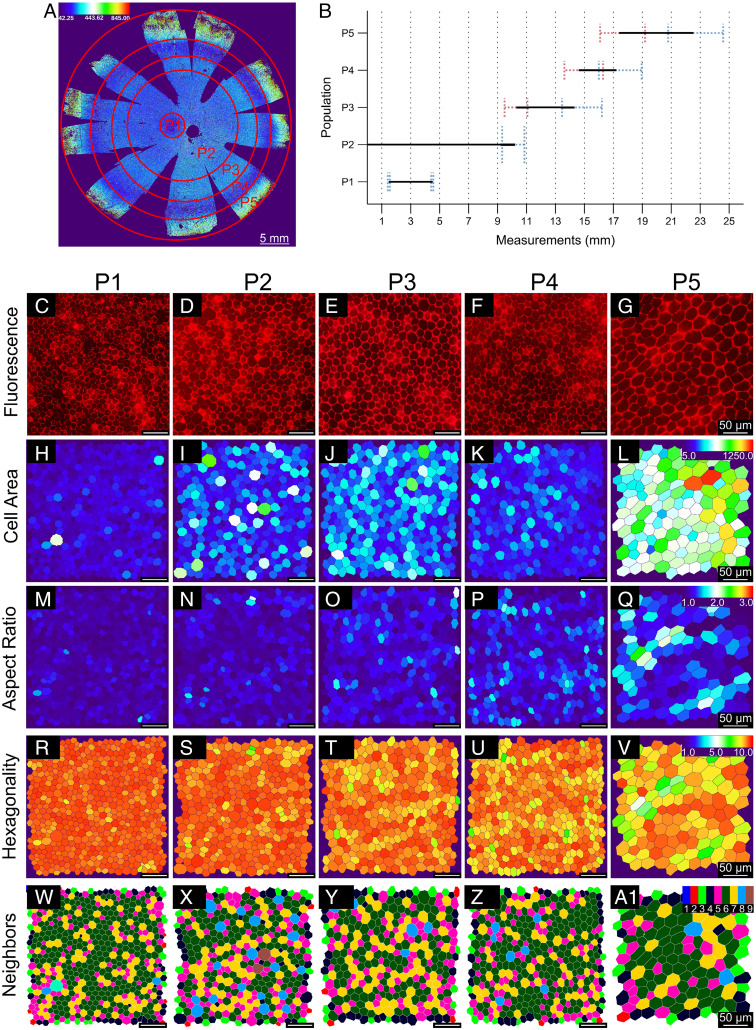
Cell area heatmaps revealed the presence of five RPE subpopulations organized in concentric circles, with distinct cell area distributions compared to the neighboring subpopulation (Fig. 2A). To isolate morphometric information from individual subpopulations, we developed another image analysis tool—REShAPE Selection (see Materials and Methods for details)—that isolated concentric circles representing the five RPE subpopulations and retrieved shape information for every cell in each subpopulation. Going outward from the fovea toward the periphery of the monolayer, we named these five RPE subpopulations P1 to P5 (morphometry features for each subpopulation are reported in SI Appendix, Tables S7–S9). The distance of each RPE subpopulation from the center of the flatmount was determined as the average length (in millimeters) of the radii of the inner and outer circles used for isolating a given subpopulation (Fig. 2B and SI Appendix, Table S5). P1, which roughly corresponds to fovea and parafovea, is a 3-mm-wide (1.5-mm radius) spot located ∼3 mm to the temporal side of the optic nerve. P1 cells have an average area of 147.24 ± 15.36 µm2, AR 1.15 ± 0.04, hexagonality 9.31 ± 0.11, and number of neighbors 5.55 ± 0.35 (Fig. 2 C, H, M, R, and W and SI Appendix, Table S6). P2 covers most of the center of the RPE monolayer, including the perifovea, up to a 10-mm radial distance. P2 cells have an average area of 201.74 ± 17.45 µm2, AR 1.18 ± 0.02, hexagonality 9.25 ± 0.05, and number of neighbors 5.47 ± 0.43 (Fig. 2 D, I, N, S, and X and SI Appendix, Table S6). P3 consists of a midperipheral ring of RPE cells located 10 to 14 mm from the center with an average cell area of 231.21 ± 18.08 µm2, AR 1.23 ± 0.03, hexagonality 9.12 ± 0.08, and number of neighbors 5.46 ± 0.63 (Fig. 2 E, J, O, T, and Y and SI Appendix, Table S6). REShAPE Selection Tool worked well on isolating shape metric information on all subpopulations except P4, the newly discovered small RPE cells of the periphery situated on average 14 to 17 mm from the center. Due to the peripheral location and the small width of P4, some cells from P3 and P5 were included with P4. Therefore, we manually isolated the darkest blue regions of P4 and found that they had an average cell area of 176.76 ± 18.68 µm2, AR 1.27 ± 0.04, hexagonality 9.00 ± 0.12, and number of neighbors 5.64 ± 0.25 (Fig. 2 F, K, P, U, and Z and SI Appendix, Table S6). P5 comprises far-peripheral RPE cells positioned 17 mm away from the center of the flatmount until the start of the ora serrata, at the very edge of the flatmount (22 to 25 mm from the center), with an average cell area of 331.87 ± 27.23 µm2, AR 1.33 ± 0.03, hexagonality 8.79 ± 0.11, and number of neighbors 5.04 ± 0.46 (Fig. 2 G, L, Q, V, and A' and SI Appendix, Table S6). All of the RPE subpopulations are posterior to the ora serrata. Comparative analysis of different RPE subpopulations revealed progressively increasing cell area from P1 to P3 regions. P5 had the largest average cell area, 2.3× larger than that of the P1 subpopulation (Figs. 2 and 3 A and B). Four distinct regions of the monolayer can be identified with statistically distinguishable ARs: a central region containing P1 and P2 with average AR of 1.17 and three peripheral subpopulations corresponding to P3, P4, and P5 with progressively increasing ARs (1.23, 1.27, and 1.33, respectively) (Figs. 2 and 3 C and D; P value comparisons between P1/P2 versus P3 versus P4 versus P5). The same four regions can also be identified as significantly different using the hexagonality score. P1 had the highest mean hexagonality score (9.31 ± 0.11), while P5 had the lowest hexagonality score (8.79 ± 0.11) (Figs. 2 and 3 E and F; P value comparisons between P1/P2 versus P3 versus P4 versus P5). The number of RPE cell neighbors does not reflect the same trend as the other shape metrics. P1 and P4 had the highest mean number of neighbors (5.56 ± 0.35 and 5.64 ± 0.25, respectively), while P5 had the lowest mean number of neighbors (5.04 ± 0.46). P5 was statistically different from all the other subpopulations for number of neighbors.
Fig. 2.
Analysis of cell area revealed the presence of five morphometrically different RPE subpopulations in human cadaver eyes. (A) A custom-made software called REShAPE Selection Tool divided segmented color-coded RPE flatmount images into five subpopulations arranged in concentric rings based on differences in cell area, from the center toward the periphery. (B) The graph describes the distance of each RPE subpopulation from the center of the eye in millimeters. Line boundaries represent the averages and the SDs calculated from the radii of the cropped circles (red corresponds to the SD of measurements of inner radii and blue to measurements of outer radii). (C–A1) The panel shows zoomed-in representative images of cells for each RPE subpopulation (P1 to P5). (C–G) Correspond to phalloidin (red)-stained images. (H–A1) Illustrate border-segmented images color coded for cell area (H–L), AR; (M–Q), hexagonality (R–V), and neighbors (W–A1) for all five RPE subpopulations. Heatmap scale for cell area (range, 50 to 1,250 µm2) is shown on top-right corner of L, for AR (range 1 to 3 arbitrary units) on top-right corner of Q, for hexagonality (range, 1 to 10 arbitrary units) on top-right corner of V, and for neighbors (range, 1 to 9) on top-right corner of A1.
Fig. 3.
Graphs showing morphometric differences between RPE subpopulations. (A–H) Morphometric data for each individual RPE cell in each of the five RPE subpopulations (P1 to P5) were collected using the REShAPE Selection Tool and quantified for cell area in square micrometers.(A), AR in arbitrary units 1 to 3 (C), hexagonality in arbitrary units 1 to 10 (E), and neighbors 1 to 14 (G) (box limits represent the first and third quartile, the central line shows the median and the whiskers indicate the 5th and 95th percentile). (B, D, F, and H) Corresponding statistical analysis of each RPE subpopulation and shape metric is presented as pairwise comparisons tables performed using Tukey test. (A linear mixed-effects model and a Tukey test for multiple comparisons were performed, n = 17; ***P < 0.001, **P < 0.01, *P < 0.05; SI Appendix, Tables S6–S9). (I–P) Shape-metric comparison of P4 with the three known macular populations—fovea, parafovea, and perifovea—and P2 for cell area (I), AR (K), hexagonality (M), and neighbors (O) (box limits represent the first and third quartile, the central line shows the median and the whiskers indicate the 5th and 95th percentile. ***P < 0.001). (J, L, N, and P) Corresponding statistical analysis of each RPE subpopulation and shape metric is presented as pairwise comparisons tables performed using Tukey test. (A linear mixed-effects model and a Tukey test for multiple comparisons were performed, n = 17; ***P < 0.001, **P < 0.01, *P < 0.05; SI Appendix, Tables S10 and S11). HSD, honest significant difference; ns, not significant.
In our measurements, P1 corresponds to fovea and parafovea, while P2 includes the perifovea and RPE of the posterior pole up to 10-mm radius from the center of the flatmount. To better understand the newly discovered P4 ring of small RPE cells, we compared it with the three known macular populations—fovea, parafovea, and perifovea—and P2. We isolated morphometric features specifically of the fovea (1.5-mm diameter in the center of the flatmount), the parafovea (1.5- to 3-mm diameter concentric ring), and the perifovea (3- to 5-mm diameter concentric ring; SI Appendix, Tables S10 and S11). Foveal cells had an average area of 131.93 ± 14.96 µm2, AR 1.15 ± 0.04, hexagonality 9.30 ± 0.10, and number of neighbors 5.58 ± 0.36 (Fig. 3 I, K, M, and O). RPE cells in the parafovea had an average area of 159.71 ± 18.47 µm2, AR 1.15 ± 0.05, hexagonality 9.32 ± 0.12, and number of neighbors 5.53 ± 0.40 (Fig. 3 I, K, M, and O). RPE cells in the perifovea had an average area of 177.56 ± 15.69 µm2, AR 1.16 ± 0.04, hexagonality 9.30 ± 0.09, and number of neighbors 5.42 ± 0.40 (Fig. 3 I, K, M, and O). Comparative analysis of P4 with these three regions revealed that P4 cells had the same dimension as perifoveal RPE cells (176.76 ± 18.68 µm2 versus 177.56 ± 15.69 µm2) (Fig. 3 I and J) and a similar number of neighbors to foveal, parafoveal, and perifoveal RPE cells and P2 [5.64 ± 0.25 versus 5.58 ± 0.36 (fovea), 5.53 ± 0.40 (parafovea), and 5.42 ± 0.40 (perifovea) and 5.47 ± 0.43 (P2)] (Fig. 3 O and P). However, P4 RPE cells had a statistically larger value of AR and lower hexagonality (1.27 ± 0.04 and 9.00 ± 0.12, respectively) (Fig. 3 K–N).
To determine if the isolated RPE subpopulations also display molecular differences, we stained non-AMD RPE flatmounts for ELN and myocilin (MYOC) proteins, previously shown to have higher expression in midperipheral RPE regions compared to the macula (14, 15, 18). Antibodies against ELN and MYOC were validated on cryosections of non-AMD eyes (SI Appendix, Fig. S1 A and B). We found that both proteins had higher expression in P2 and P3 compared to the other subpopulations (SI Appendix, Fig. S1 C–E). Specifically, ELN intensity was about threefold higher in P2 compared to P1, P4, and P5 (median: P2 = 1.9, P1 = 0.5, P4 = 0.5, and P5 = 0.7); MYOC intensity was 2.5 to 4 times higher in P2 compared to P1, P4, and P5 (median: P2 = 1.6, P1 = 0.6, P4 = 0.4, and P5 = 0.6). Interestingly, P1, P4, and P5 showed comparable levels of ELN and MYOC expression, suggesting similarity between macular and far-peripheral regions. Overall, our result is consistent with the literature, demonstrating lower expression of ELN and MYOC in the macula compared to the midperiphery. In addition, we showed similar expression of the two molecular markers between macula and far-peripheral regions.
Taken together, these results indicate that the RPE cells of the newly discovered subpopulation P4 have an area similar to perifoveal RPE, a number of neighbors similar to central and midperipheral RPE subpopulations, similar expression of ELN and MYOC to P1 and P5, and values of AR and hexagonality between P3 and P5.
Geographic correlation between RPE subpopulations and photoreceptor cells.
Based on morphometric similarity between P4 and perifoveal RPE cells, we asked if P4 RPE cells interact with the same ratio of rods and cones as perifoveal RPE cells. Furthermore, a 1-mm-wide cone-rich rim of photoreceptors has been previously described at the very edge of the retina, adjacent to the ora serrata (22–25). Given that the width of the P4 RPE cell ring (2 mm) is similar to the cone-rich rim (1 mm), we also asked if the cone rim overlapped with P4 RPE cells. To test these hypotheses, photoreceptors and RPE densities were calculated across a piece of nasal retina from three different donors (SI Appendix, Fig. S2A), where the cone-enriched peripheral rim has previously been described as most pronounced (24). Seventeen regions of interest evenly distributed across the nasal retina were selected to quantify cell density (SI Appendix, Fig. S2 A–K). Consistent with published work (25), a high density of cone photoreceptors (∼7,000 cones/mm2) was detected at the very edge of the retina, but this region corresponded to RPE subpopulation P5 and not to P4 (SI Appendix, Fig. S2L).
We divided the rod and cone ratio by the RPE density across the three pieces of nasal retina analyzed and found similarity between this ratio in P4 and the start (1 to 2 mm from the optic nerve) of P2, an area with photoreceptor composition similar to that of perifovea (SI Appendix, Fig. S2M). Both areas had a ratio close to 0.006 (see Discussion for more explanation about P4).
Variable RPE morphometry within subpopulations in AMD compared to non-AMD flatmounts.
The discovery of five distinct RPE subpopulations prompted the idea that different subpopulations are differentially sensitive to different retinal degenerative diseases. It is well known that AMD lesions occur predominantly in the macular and submacular regions (P1 and P2 subpopulations) (26). To determine if the morphometric similarity between P4 and perifovea also resulted in a physiological similarity, we asked if P4 was similarly affected in AMD eyes (SI Appendix, Table S12). We used non-AMD eye maps as a reference and compared morphometric differences between corresponding RPE subpopulations for each shape metric in AMD eyes. Five AMD eyes were analyzed using the same parameters used to analyze non-AMD eyes (SI Appendix, Table S13, representative flatmount shown in SI Appendix, Fig. S3 A–E). The entire RPE/choroid flatmounts were imaged; cell borders were segmented, and morphometric parameters were analyzed using REShAPE (SI Appendix, Tables S14–S19). Due to extensive RPE-dropout lesions in AMD eyes, an average of 1.6 million cells were analyzed per AMD eye, compared to 2.8 million cells in non-AMD eyes (compare SI Appendix, Tables S13 and S2, respectively). In all AMD flatmounts, P1 and up to the centermost part of P2 RPE cells were lost; large areas of geographic atrophy were also visible at the far periphery of the flatmounts in P4 and P5 regions and often extended into the P3 region (SI Appendix, Fig. S3 A and B). Higher magnification representative images of RPE stained with phalloidin-iFluor 647 and corresponding heatmaps revealed higher variability in cell area within each subpopulation (compare SI Appendix, Fig. S3 F–M to Fig. 2 C–L). A similar trend of increased variability within each subpopulation was seen for other shape metrics as well (compare SI Appendix, Fig. S3 N–Y to Fig. 2 M–A′).
To better understand increased variability in RPE shape metrics in AMD eyes, we quantified all four metrics across P2 to P5 subpopulations (P1 was missing in most AMD eyes). Statistical analysis revealed large spread of the whiskers in the boxplots (Fig. 4 A, C, E and G). The difference in RPE cell area seen in non-AMD eyes (Fig. 3 A and B) between P2, P3, and P4 faded, while P5 still remained the subpopulation with significantly larger average cell area (279.91 ± 38.52 µm2) (SI Appendix, Fig. S3 J–M and Fig. 4 A and B). Furthermore, there was no statistical difference among subpopulations for any of the other three shape metrics except P2 versus P5 for AR (Fig. 4 C–H). This suggests that in AMD eyes RPE cells lose their epithelial phenotype, which may also result in loss of RPE functionality. These data further underscore physiological similarities between the P4 subpopulation and the perifovea beyond just the morphological aspects.
Fig. 4.
Graphs showing morphometric differences between RPE subpopulations in AMD and non-AMD donor eyes. (A, C, E, and G) Quantification of each RPE subpopulation for cell area (square micrometers) (A); AR (C); hexagonality (E); and number of neighbors (G) presented as boxplots (box limits represent the first and third quartile, the central line shows the median and the whiskers indicate the 5th and 95th percentile. **P < 0.01, *P < 0.05). (B, D, F, and H) Corresponding statistical analysis table for cell area (B), AR (D), hexagonality (F), and number of neighbors (H) presented for pairwise comparisons with Tukey test. (A linear mixed-effects model and a Tukey test for multiple comparisons were performed, n = 5; **P < 0.01, *P < 0.05; SI Appendix, Tables S16–S18). (I, K, M, and O) Quantification of each RPE subpopulation for cell area (square micrometers) (I), AR (K), hexagonality (M), and number of neighbors (O) for AMD (red) and non-AMD donors (blue) are presented side by side to show the differences (the error bars indicate the 5th and 95th percentile). (J, L, N, and P) Corresponding tables of statistical analysis for cell area (J), AR (L), hexagonality (N), and number of neighbors (P) where AMD and non-AMD measurements for each RPE subpopulation were compared with t test; *P < 0.05. HSD, honest significant difference.
To better understand how the entire RPE morphometric map changes at a single-cell level, we next performed comparative analysis of morphological features between AMD and non-AMD eyes (Fig. 4 I–P). Compared to the cell area of non-AMD donors (blue), the average cell area of P3 and P5 in AMD (red) eyes was smaller but showed higher variability (P3: 214.42 ± 40.57 µm2 AMD versus 231.21 ± 18.08 µm2 non-AMD; P5: 279.91 ± 38.52 µm2 AMD versus 331.87 ± 27.23 µm2 non-AMD) (Fig. 4 I and J). Similar to non-AMD eyes, RPE cells were more compact in the center of AMD flatmounts (P2) and became more elongated toward the periphery, but only P2 and P5 differed statistically (P2: 1.26 ± 0.7; P5: 1.38 ± 0.04) (SI Appendix, Fig. S3 N–Q, Fig. 4 C and D, and SI Appendix, Table S16). In comparison with non-AMD donors, RPE cells were 7 to 8% more elongated in each subpopulation, and the SD was 75% larger in AMD donor eyes (Fig. 4 K and L and SI Appendix, Tables S6 and S16). The higher variability of AMD subpopulations is also reflected by a 15 to 30% larger interquartile range of sample distribution compared to non-AMD eyes. Similar to non-AMD eyes, RPE cells tended to be more hexagonal in the central compared to peripheral subpopulations (SI Appendix, Table S16), but the difference among subpopulations was not statistically significant in AMD eyes (SI Appendix, Fig. S3 R–U and Fig. 4 E and F). In each RPE subpopulation, the average value of hexagonality score was 3 to 4% lower in AMD eyes than the value for non-AMD eyes, and the SD was 30 to 70% larger than non-AMD eyes (Fig. 4 M and N and SI Appendix, Tables S6 and S16). For hexagonality score, AMD subpopulations had a 15 to 40% larger interquartile range of sample distribution compared to non-AMD eyes. No significant difference was found between RPE subpopulations in the number of RPE cell neighbors in AMD flatmounts (SI Appendix, Fig. S3 V–Y and Fig. 4 G and H). Compared to non-AMD eyes, the number of neighbors was reduced by 8 to 15%, and the SD was up to 70% larger in AMD eyes (Fig. 4 O and P and SI Appendix, Tables S6 and S16). Overall, in AMD flatmounts compared to non-AMD flatmounts, there was much larger variability within RPE subpopulations for each morphologic feature, while the shape metric differences among RPE subpopulations were reduced.
To test if REShAPE was able to detect morphometric changes occurring with age and disease status, we divided non-AMD donors in age categories (<65, 65 to 79, and 80 to 100 y) and measured age-dependent changes within each RPE subpopulation. All AMD donor eyes were in the 80- to 100-y age category. Strikingly, we saw a significant increase in AR and a decrease in hexagonality and number of neighbors in the P1 subpopulation of 80- to 100-y non-AMD donors compared to younger non-AMD donors (<65, 65 to 79 y) (SI Appendix, Fig. S4 B–D). In addition, REShAPE detected changes in AR, hexagonality, and number of neighbors between age-matched non-AMD and AMD donors, especially in P2 (SI Appendix, Fig. S4 B–D). This indicates that changes of RPE morphology are exacerbated in AMD. Overall, these results suggest that REShAPE can detect changes of cell morphology prior to the development of visibly apparent atrophic areas. Moreover, age-related morphometric changes may appear in P1 earlier than in other subpopulations. Future development of adaptive optics image resolution could help predict changes in RPE health.
Lastly, we compared the morphometry of embryonic stem cell- and induced pluripotent stem cell-derived RPE cells (PSC-RPE) from published literature (27–38) with non-AMD (SI Appendix, Fig. S5 A–D) and AMD (SI Appendix, Fig. S5 E–H) donor eyes. We found that PSC-RPE cell area is similar to subpopulation P1 of non-AMD donors (SI Appendix, Fig. S5A), while AR and hexagonality are more similar to subpopulation P5 (SI Appendix, Fig. S5 B and C). The number of neighbors in PSC-RPE was comparable to all RPE subpopulations (SI Appendix, Fig. S5D). The result was similar when PSC-RPE cells were compared to AMD donors (SI Appendix, Fig. S5 E–H). The high variability within the PSC-RPE group across all shape metrics is likely due to a highly variable source of different cell lines and differentiation protocols used by different laboratories.
Far-peripheral subpopulations are more vulnerable than midperipheral subpopulations in AMD and non-AMD flatmounts.
We noticed micro RPE lesions that were seen more frequently in several subpopulations in non-AMD flatmounts than in the P1-P2 macular/perimacular regions (Fig. 5 A and B). RPE cell borders in these lesions were poorly segmented by the REShAPE software (Fig. 5B). To better understand these lesions and further determine the differential vulnerability of diverse RPE subpopulations to degeneration, we generated dual RPE/retina flatmounts from three non-AMD donors. The dual RPE/retina flatmounts were optically cleared with BABB (a 1:2 mixture of benzyl alcohol and benzyl benzoate) and were stained with phalloidin to label actin structures and with peanut agglutinin (PNA) to label cone inner and outer segments. All the lesions analyzed had the same characteristics: Micro lesions were present in all RPE subpopulations, RPE cells were still present in the lesioned areas but were raised by underlying deposits (Fig. 5C, indicated by *, and Movie S1), and RPE cells were irregular in shape in these lesions. Photoreceptors located above these lesions appeared displaced toward the sides by the deposits (Fig. 5C and Movie S1). To determine if any subpopulation had a higher prevalence of lesions, we quantified how many lesions were present in each RPE subpopulation. In non-AMD flatmounts, far-peripheral RPE subpopulation P5 contained significantly more lesions than midperipheral subpopulation P3 (median value P5: 24% versus P3: 7%, P value = 0.02; Fig. 5D). In AMD flatmounts, we documented the same trend, although not statistically different between subpopulations (Fig. 5E). Overall, far-peripheral RPE subpopulation P5 seems to be more prone to develop micro lesions than midperipheral RPE subpopulation P3. Interestingly, P5 RPE cells overlapped with the rim of high-density cone photoreceptors.
Fig. 5.
Far-peripheral RPE subpopulation P5 contains sub-RPE deposits reminiscent of macular RPE P1. (A and B) Low- and high-magnification (Inset) images of P4 RPE subpopulation stained with phalloidin (white) dye (A) and RPE cell areas color coded (B) reveal lesions in non-AMD donor RPE flatmounts. (C) Three-dimensional imaging of a piece of retina and RPE over the area of RPE lesion stained with phalloidin (white) and PNA (magenta) dyes reveals intact RPE and photoreceptors in the area of lesions. Asterisks mark sub-RPE deposits in the lesion area (n = 3 donors). (D and E) The quantity of lesioned (lifted) RPE for each population was calculated as a percentage of the area of not-segmented cells for both non-AMD (D) and AMD donor (E) eyes. Far-peripheral subpopulation P5 contains more lesions than midperipheral population P3 in non-AMD eyes (n = 17). The same tendency is observed in AMD eyes, although the variability between each donor is greater (n = 5) (box limits represent the first and third quartile, the central line shows the median and the whiskers indicate the 5th and 95th percentile. One-way ANOVA and Tukey test for multiple comparisons were performed; *P < 0.05). Note that for AMD eyes our quantification does not distinguish between geographic atrophies and lesions due to sub-RPE deposits. ONL, outer nuclear layer.
We also examined if the prevalence of RPE lesions in each subpopulation correlated with choroidal endothelial cell dropout. Using published protocols (39), we stained cryosections from age-matched non-AMD and AMD donors with UEA-1 lectin to detect the presence of ghost vessels and correlated their density within RPE subpopulations (SI Appendix, Fig. S6 A and B). In accordance with data on RPE lesions (Fig. 5D), we found that midperipheral subpopulation P3 had the least density of ghost vessels in non-AMD donor (median: 0.1/mm) (SI Appendix, Fig. S6C). Ghost vessel density increased gradually toward the center of the eye (P1, P2) and toward the periphery (P4, P5), with subpopulations P1 and P5 having the highest concentration (median: P1 = 1.5/mm, P5 = 1.0/mm) (SI Appendix, Fig. S6C). A similar trend was observed in the AMD donor, with P1 having the highest concentration (median: 7.8/mm), although ghost vessel density in P5 was likely underestimated because of distortions of the choroid in that region (SI Appendix, Fig. S6D). When comparing ghost vessel density between non-AMD and AMD, we found a fivefold increase in P1 (median: 1.5/mm non-AMD versus 7.8/mm AMD) and P2 (median: 0.4/mm non-AMD versus 1.9/mm AMD) in AMD compared to the age-matched non-AMD donor (SI Appendix, Fig. S6E). These data support the idea that midperipheral RPE subpopulation P3 is more resistant to the development of RPE/choroid damage compared to central and far-peripheral subpopulations.
RPE subpopulations are differentially sensitive to different types of retinal diseases.
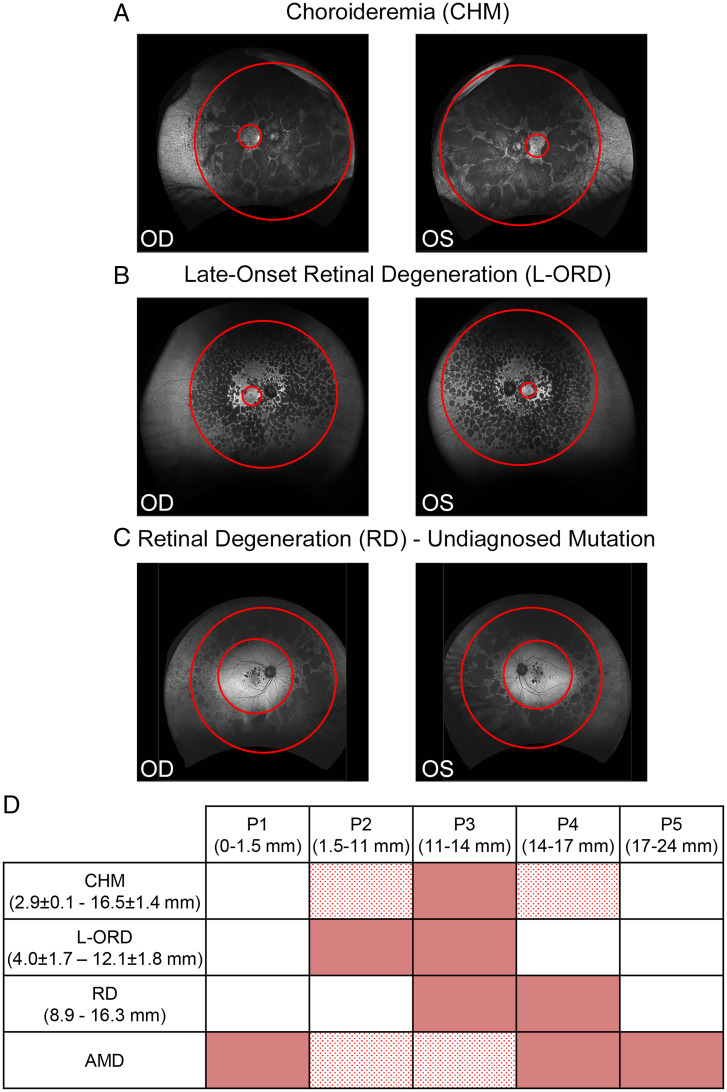
To further test the hypothesis that different RDs affect specific RPE subpopulations, we analyzed ultra-widefield fundus autofluorescence images from patients affected by CHM (two patients) (Fig. 6A) and L-ORD (three patients) (Fig. 6B) and an RD patient with no identified molecular cause (one patient) (Fig. 6C). We measured the horizontal and vertical radii of the inner and outer boundaries of the areas of discrete hypoautofluorescence and correlated these measurements to RPE subpopulations. We found that the fovea/parafovea (P1) was relatively spared in these diseases at the stages examined, while midperipheral RPE subpopulations contained areas of RPE atrophy. Specifically, the lesions of CHM patients spanned from part of P2 to part of P4 (Fig. 6D, red and dotted red boxes). The lesions of L-ORD patients occupied the entirety of P2 and P3 (Fig. 6D, red boxes). The atrophy of the RD patient geographically corresponded to the entirety of P3 and P4 (Fig. 6D, red boxes). A comparative analysis of RPE regions affected in AMD eyes, analyzed from the RPE flatmounts, shows that P2 and the midperipheral population P3 were relatively spared compared to the other populations (Fig. 6D, dotted red boxes). The analysis suggests that different RPE subpopulations are vulnerable to different types of retinal degenerative diseases. It is critical to note that these images come from one point in time, while these conditions are inherently progressing. Imaging these diseases at early stages is fundamental in order to speculate about which RPE subpopulation is involved in the etiology of the disease.
Fig. 6.
Different retinal diseases affect different RPE subpopulations. (A–C) Right (OD) and Left (OS) eye fundus images of patients affected by CHM (A) and L-ORD (B) and a patient with RD with an undiagnosed mutation (C) with RPE degeneration in different regions of the eye. Red circles higlight the inner and outer boundaries of the areas of retinal degeneration. (D) Table summarizes defects in RPE subpopulations in different forms of RDs [mean ± SD of radii (in millimeters) of inner and outer limits of atrophic regions]. Red boxes correspond to fully affected RPE subpopulations, dotted red boxes correspond to partially affected subpopulations, while white boxes indicate unaffected subpopulations.
Discussion
In this study, we provide a complete single-cell–resolution map of adult human RPE cells from non-AMD and AMD donors. We were able to identify five morphometrically distinct RPE subpopulations, including a subpopulation with cell area similar to macular cells in the far-periphery of the eye. This work provides a reference standard for further molecular and physiological analysis of different RPE subpopulations in the human eye. By analyzing cadaver AMD eyes and ultra-widefield fundus images of patients with monogenic retinal degenerative diseases, we identified differential disease vulnerability in these RPE subpopulations. Lesions identified in far-peripheral RPE subpopulations in non-AMD cadaver eyes underscore the importance of imaging peripheral RPE regions to better understand disease progression.
The accuracy of REShAPE was validated previously using RPE cells (40). The machine learning algorithm for REShAPE was trained using images of 12,750 hand-segmented RPE cells, leading to an accuracy of 90% for extracting epithelial cell morphometric features. Here, we demonstrate the utility of REShAPE in analyzing cellular morphometry features in ocular tissues. REShAPE is able to complete the analysis of large images (150,000 × 150,000 pixels) containing 3 to 4 million cells within 4 h. The code for REShAPE (https://github.com/nih-nei/REShAPE) takes in microscopy images and outputs a table of morphological features. We propose broad application of REShAPE for segmentation and analysis of cell borders of other epithelia, for instance, to study phenomena such as planar cell polarity in Drosophila wing epithelial cells (41).
Our morphometry data are consistent with previous work on RPE cell density and shape (19, 20). We found a similar trend of increasing cell size and elongation and decreasing hexagonality and number of neighbors with outward retinal locations (19, 20). Notably, we report the presence of a ring of small RPE cells in the periphery (P4), similar in cell area to perifovea, that was not described previously. We observed differences in cell area and hexagonality between macular and midperipheral cells, while Bhatia et al. (19) did not. This discrepancy likely stems from the choice of retinal locations that were averaged in Bhatia et al. (19). REShAPE provides considerable advancement compared to previous studies (19, 20). While Bhatia et al. (19) and Watzke et al. (20) only analyzed a few regions on the temporal side of the eye, we developed a single-cell–resolution map of the entire human eye. This approach is meant to provide a standard by defining human RPE subpopulations at single-cell resolution and their exact location within the human eye. Researchers in the field could use the geographic references provided here (Fig. 2B and SI Appendix, Table S5) to isolate specific RPE subpopulations for further molecular and physiological analysis. It is important to note that the variability observed in the location of the RPE subpopulations may stem from the individual variability in eye dimension. Nevertheless, the parameters provided in this work can help standardize sampling of RPE subpopulations from different locations.
The concentric circles of RPE cells with different sizes raise a question about the correlation between cell size and cell function. In fact, this is one of the longest debated questions in biology and is relevant at all scales of biological organization (42). Larger organisms have a lower size-normalized metabolic rate, a phenomenon known as allometric scaling of metabolism (43). This property applies to individual cells as well (44). A few studies have indeed shown that there is a decline in metabolic activity with increasing cell size (45, 46). Whether cell size regulates cellular metabolism or vice versa, a connection between cell size and physiology appears to be more and more evident (47). The presence of RPE subpopulations of different cell size suggests that these subpopulations are physiologically different. Evidence of regional RPE heterogeneity between central and peripheral areas of the monolayer has been described previously by gene expression (15, 17, 18, 48), lysosomal enzyme activity (11, 49, 50), Na/K ATPases expression (12), and Bruch’s membrane thickness (14). In this study, we propose that different RPE subpopulations are differentially sensitive to different retinal diseases. In AMD eyes, P1, P4, and P5 appear to be mostly affected, while midperipheral RPE (P3) seems to degenerate first in L-ORD, CHM, and RD patients (Fig. 6D). Molecular and functional differences between RPE subpopulations are the likely cause of differential RPE sensitivity to different diseases. In this paper, we report variation of expression of two molecular markers, ELN and MYOC, across RPE subpopulations (SI Appendix, Fig. S1). In future studies, it would be important to characterize the molecular profile of each RPE subpopulation to find correlations with regional RPE sensitivity.
The highly systematic arrangement of five RPE subpopulations with clear morphologically distinct features raises another question about the developmental and/or physiological role of this arrangement. We hypothesize that size differences are established during eye development and help maintain the correct photoreceptor/RPE ratio at different retinal locations, but it is not clear if this RPE map is instructive to photoreceptor development or vice versa. It has been suggested that in the fovea and the parafovea, RPE cells are smaller in area to meet the metabolic demand required to support a high cone density (51). However, even where rod density is high and cone density is low, such as in the posterior pole, RPE cells are still relatively small (P2). We think there could be a relation between RPE cell size and rod/cone photoreceptor ratio. For instance, because of the morphometric similarity between P4 and perifovea, we hypothesized that they may interact with a similar ratio of rod and cone photoreceptors. Given the similarity of our rod and cone density to data obtained by Curcio et al. (52), we used their data to obtain the average photoreceptor densities overlaying each RPE subpopulation, including the macula. P4 has an average density of 85,000 rods/mm2 and 5,000 cones/mm2, while perifovea has an average density of 120,000 rods/mm2 and 10,000 cones/mm2. We calculated the rod/cone ratio divided by RPE density for the two subpopulations and found that they have a similar ratio of 0.096 for P4 and 0.068 for perifovea. The ratio calculated for other RPE subpopulations is 0.002 (P1), 0.129 (P2), 0.087 (P3), and 0.026 (P5). Another potential explanation for the presence of P4 could be that the geometry of the eyeball requires RPE cells to be small around the equator of the eye. In mice, RPE cells at the equator appear to be different from neighboring cells—oriented circumferentially instead of radially (53). Another possibility is that P4 consists of a pool of RPE cells that retain the capacity to enter cell cycle to replenish dying RPE cells. In rats, peripheral RPE cells can reenter cell cycle and complete cellular division (54). Moreover, Ki67-positive cells were identified in equatorial/peripheral regions of the human RPE monolayer, although their exact location could not be determined (54). Salero et al. (55) also described a pool of human RPE cells with stem cell–like properties. Although the reason for having a smaller size of RPE cells in the P4 ring is unclear, the discovery of this peripheral RPE population is most intriguing.
The larger RPE cell size in P5 is also not easily understandable. Here, RPE cells could be larger because rod and cone densities in this region are not very high, but we cannot exclude that variability and larger cell size in P5 is caused by age-induced changes in the eyeball size (9, 53). All eyes in our study were from donors older than 50 y. In fact, an extreme effect of aging and degeneration is highlighted in the analysis of eyes from AMD patients. In AMD, RPE cells that are degenerating or undergoing apoptosis are eventually extruded from the monolayer. The space left by the dying cells is replenished by expansion of the neighboring cells (56). Thus, the RPE monolayer will contain unusually small cells undergoing an active apoptotic process (57–59) and unusually large cells that fill the gap of those apoptotic cells. This is the likely reason why the variability of RPE morphometry is up to 70% larger across all shape metrics in AMD eyes. The analysis of RPE monolayer from younger donors, as well as globes of varying axial length, could help clarify if P5 is established during development or if it is a consequence of aging.
The peripheral retina has mostly been neglected, perhaps because of limited far-peripheral imaging techniques in vivo and the natural focus on the high-acuity region in the center of the eye. The presence of a peripheral ring of small RPE cells and a peripheral rim enriched in cone photoreceptors (23, 24) suggests that we do not know enough about the function of the peripheral retina. These peripheral regions could have implications in our understanding of retinal diseases and of human vision itself.
In conclusion, we uncovered five morphometrically distinct human RPE subpopulations, which could be isolated for further molecular and physiological analysis. We discovered a peripheral RPE subpopulation (P4) that is similar in cell size to perifoveal RPE. We found that a peripheral rim of cone photoreceptors geographically overlaps with peripheral subpopulation P5. Lastly, we found that the five RPE subpopulations are differentially susceptible to different types of retinal diseases. Future studies should focus on the analysis of RPE morphometry in fetal eyes and young individuals to understand if RPE subpopulations change over time. In addition, this study provides a reference guide for a precision medicine–based approach whereby different RPE subpopulations derived from pluripotent stem cells will help develop disease-specific cell, gene, and small molecule–based therapies.
Materials and Methods
Seventeen non-AMD and five AMD cadaver eye globes from nine and three donors, respectively, were obtained from Advancing Sight Network (Birmingham, AL) and Lions Eye Institute (Tampa, FL). Only non-AMD eyes from donors with no history of RD were considered for this study. AMD donors were diagnosed with AMD before death. Eyes were collected from both sexes between the age of 46 and 100 y. For fundus imaging, all participants or guardians provided written, informed consent, and the study was approved by the institutional review board of the NIH and adhered to the tenets of the Declaration of Helsinki. The average death to preservation time for tissues used for RPE flatmount was 24 h; for all other experiments, it was 5 h. Detailed methods for tissue processing, flatmount staining and imaging, tissue clearing, statistics, and human subject imaging and analysis are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank the National Eye Institute (NEI) histology core for the use of the Zeiss Axio Scan.Z1; Jorge Ferrari Escoto for helping with the isolation of RPE subpopulations; and the donors, their families, the Advancing Sight Network, and the Lions Eye Institute for their generosity. We also thank Michael B. Gorin for the invaluable comments on the manuscript. This work was supported by NEI IRP funds (grant number ZIA EY000533-04).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117553119/-/DCSupplemental.
Data Availability
The REShAPE code is available on GitHub (https://github.com/nih-nei/REShAPE). All other study data are included in the article and/or supporting information.
References
- 1.Bharti K., Miller S. S., Arnheiter H., The new paradigm: Retinal pigment epithelium cells generated from embryonic or induced pluripotent stem cells. Pigment Cell Melanoma Res. 24, 21–34 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma R., Bose D., Maminishkis A., Bharti K., Retinal pigment epithelium replacement therapy for age-related macular degeneration: Are we there yet? Annu. Rev. Pharmacol. Toxicol. 60, 553–572 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuntz C. A., et al. , Sub-retinal pigment epithelial deposits in a dominant late-onset retinal degeneration. Invest. Ophthalmol. Vis. Sci. 37, 1772–1782 (1996). [PubMed] [Google Scholar]
- 4.Jacobson S. G., Cideciyan A. V., Wright E., Wright A. F., Phenotypic marker for early disease detection in dominant late-onset retinal degeneration. Invest. Ophthalmol. Vis. Sci. 42, 1882–1890 (2001). [PubMed] [Google Scholar]
- 5.Styles C. J., Dhillon B., Wright A. F., The diagnosis of autosomal dominant late-onset retinal degeneration in two sisters. Eye 17, 530–532 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Pennesi M. E., Birch D. G., Duncan J. L., Bennett J., Girach A., Choroideremia: Retinal degeneration with an unmet need. Retina 39, 2059–2069 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacDonald I. M., Russell L., Chan C.-C., Choroideremia: New findings from ocular pathology and review of recent literature. Surv. Ophthalmol. 54, 401–407 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coussa R. G., Traboulsi E. I., Choroideremia: A review of general findings and pathogenesis. Ophthalmic Genet. 33, 57–65 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Harman A. M., Fleming P. A., Hoskins R. V., Moore S. R., Development and aging of cell topography in the human retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 38, 2016–2026 (1997). [PubMed] [Google Scholar]
- 10.Boulton M., Moriarty P., Jarvis-Evans J., Marcyniuk B., Regional variation and age-related changes of lysosomal enzymes in the human retinal pigment epithelium. Br. J. Ophthalmol. 78, 125–129 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayasaka S., Shiono T., Hara S., Mizuno K., Regional distribution of lysosomal enzymes in the retina and choroid of human eyes. Albrecht Von Graefes Arch. Klin. Exp. Ophthalmol. 216, 269–273 (1981). [DOI] [PubMed] [Google Scholar]
- 12.Burke J. M., McKay B. S., Jaffe G. J., Retinal pigment epithelial cells of the posterior pole have fewer Na/K adenosine triphosphatase pumps than peripheral cells. Invest. Ophthalmol. Vis. Sci. 32, 2042–2046 (1991). [PubMed] [Google Scholar]
- 13.Miller S. S., Steinberg R. H., Oakley B. II, The electrogenic sodium pump of the frog retinal pigment epithelium. J. Membr. Biol. 44, 259–279 (1978). [DOI] [PubMed] [Google Scholar]
- 14.Chong N. H. V., et al. , Decreased thickness and integrity of the macular elastic layer of Bruch’s membrane correspond to the distribution of lesions associated with age-related macular degeneration. Am. J. Pathol. 166, 241–251 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Soest S. S., et al. , Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch’s membrane. Mol. Vis. 13, 1608–1617 (2007). [PubMed] [Google Scholar]
- 16.Li B., et al. , Metabolic features of mouse and human retinas: Rods versus cones, macula versus periphery, retina versus RPE. iScience 23, 101672 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitmore S. S., et al. , Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-seq. Exp. Eye Res. 129, 93–106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radeke M. J., Peterson K. E., Johnson L. V., Anderson D. H., Disease susceptibility of the human macula: Differential gene transcription in the retinal pigmented epithelium/choroid. Exp. Eye Res. 85, 366–380 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Bhatia S. K., et al. , Analysis of RPE morphometry in human eyes. Mol. Vis. 22, 898–916 (2016). [PMC free article] [PubMed] [Google Scholar]
- 20.Watzke R. C., Soldevilla J. D., Trune D. R., Morphometric analysis of human retinal pigment epithelium: Correlation with age and location. Curr. Eye Res. 12, 133–142 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Hannezo E., Prost J., Joanny J.-F., Theory of epithelial sheet morphology in three dimensions. Proc. Natl. Acad. Sci. U.S.A. 111, 27–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greeff R., “Mikroscopische anatomie der sehnerven und der netzhaut” in Handbuch Der Gesamten Augenheilkunde, von Graefe A., Saemisch T., Eds. (Springer, 1900), pp. 74–127. [Google Scholar]
- 23.Mollon J. D., Regan B. C., Bowmaker J. K., What is the function of the cone-rich rim of the retina? Eye (Lond.) 12 (Pt 3b), 548–552 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Williams R. W., The human retina has a cone-enriched rim. Vis. Neurosci. 6, 403–406 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Osterberg G. A., Topography of the layer of rods and cones in the human retina. Acta Ophthalmol. Suppl. 6, 1–97 (1935). [Google Scholar]
- 26.Lim L. S., Mitchell P., Seddon J. M., Holz F. G., Wong T. Y., Age-related macular degeneration. Lancet 379, 1728–1738 (2012). [DOI] [PubMed] [Google Scholar]
- 27.da Cruz L., et al. , Phase 1 clinical study of an embryonic stem cell-derived retinal pigment epithelium patch in age-related macular degeneration. Nat. Biotechnol. 36, 328–337 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Foltz L. P., Clegg D. O., Rapid, directed differentiation of retinal pigment epithelial cells from human embryonic or induced pluripotent stem cells. J. Vis. Exp. 128, e56274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galloway C. A., et al. , Drusen in patient-derived hiPSC-RPE models of macular dystrophies. Proc. Natl. Acad. Sci. U.S.A. 114, E8214–E8223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galloway C. A., et al. , Characterization of human iPSC-RPE on a prosthetic Bruch’s membrane manufactured from silk fibroin. Invest. Ophthalmol. Vis. Sci. 59, 2792–2800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamao H., et al. , Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Reports 2, 205–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leach L. L., et al. , Induced pluripotent stem cell-derived retinal pigmented epithelium: A comparative study between cell lines and differentiation methods. J. Ocul. Pharmacol. Ther. 32, 317–330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben M’Barek K., et al. , Human ESC-derived retinal epithelial cell sheets potentiate rescue of photoreceptor cell loss in rats with retinal degeneration. Sci. Transl. Med. 9, eaai7471 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Osakada F., et al. , In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J. Cell Sci. 122, 3169–3179 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Sharma R., et al. , Clinical-grade stem cell-derived retinal pigment epithelium patch rescues retinal degeneration in rodents and pigs. Sci. Transl. Med. 11, eaat5580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh R., et al. , iPS cell modeling of Best disease: Insights into the pathophysiology of an inherited macular degeneration. Hum. Mol. Genet. 22, 593–607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R., et al. , Functional analysis of serially expanded human iPS cell-derived RPE cultures. Invest. Ophthalmol. Vis. Sci. 54, 6767–6778 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pennington B. O., Clegg D. O., Melkoumian Z. K., Hikita S. T., Defined culture of human embryonic stem cells and xeno-free derivation of retinal pigmented epithelial cells on a novel, synthetic substrate. Stem Cells Transl. Med. 4, 165–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mullins R. F., Johnson M. N., Faidley E. A., Skeie J. M., Huang J., Choriocapillaris vascular dropout related to density of drusen in human eyes with early age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 52, 1606–1612 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaub N. J., et al. , Deep learning predicts function of live retinal pigment epithelium from quantitative microscopy. J. Clin. Invest. 130, 1010–1023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Classen A. K., Anderson K. I., Marois E., Eaton S., Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Dev. Cell 9, 805–817 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Marshall W. F., How cells measure length on subcellular scales. Trends Cell Biol. 25, 760–768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miettinen T. P., Björklund M., Mitochondrial function and cell size: An allometric relationship. Trends Cell Biol. 27, 393–402 (2017). [DOI] [PubMed] [Google Scholar]
- 44.West G. B., Woodruff W. H., Brown J. H., Allometric scaling of metabolic rate from molecules and mitochondria to cells and mammals. Proc. Natl. Acad. Sci. U.S.A. 99 (suppl. 1), 2473–2478 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miettinen T. P., Björklund M., Cellular allometry of mitochondrial functionality establishes the optimal cell size. Dev. Cell 39, 370–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savage V. M., et al. , Scaling of number, size, and metabolic rate of cells with body size in mammals. Proc. Natl. Acad. Sci. U.S.A. 104, 4718–4723 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ginzberg M. B., Kafri R., Kirschner M., Cell biology: On being the right (cell) size. Science 348, 1245075 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishibashi K., Tian J., Handa J. T., Similarity of mRNA phenotypes of morphologically normal macular and peripheral retinal pigment epithelial cells in older human eyes. Invest. Ophthalmol. Vis. Sci. 45, 3291–3301 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Burke J. M., Twining S. S., Regional comparisons of cathepsin D activity in bovine retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 29, 1789–1793 (1988). [PubMed] [Google Scholar]
- 50.Cabral L., et al. , Regional distribution of lysosomal enzymes in the canine retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 31, 670–676 (1990). [PubMed] [Google Scholar]
- 51.Snodderly D. M., Sandstrom M. M., Leung I. Y. F., Zucker C. L., Neuringer M., Retinal pigment epithelial cell distribution in central retina of rhesus monkeys. Invest. Ophthalmol. Vis. Sci. 43, 2815–2818 (2002). [PubMed] [Google Scholar]
- 52.Curcio C. A., Sloan K. R., Kalina R. E., Hendrickson A. E., Human photoreceptor topography. J. Comp. Neurol. 292, 497–523 (1990). [DOI] [PubMed] [Google Scholar]
- 53.Kim Y. K., et al. , Morphometric analysis of retinal pigment epithelial cells from C57BL/6J mice during aging. Invest. Ophthalmol. Vis. Sci. 62, 32–32 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Hussaini H., Kam J. H., Vugler A., Semo M., Jeffery G., Mature retinal pigment epithelium cells are retained in the cell cycle and proliferate in vivo. Mol. Vis. 14, 1784–1791 (2008). [PMC free article] [PubMed] [Google Scholar]
- 55.Salero E., et al. , Adult human RPE can be activated into a multipotent stem cell that produces mesenchymal derivatives. Cell Stem Cell 10, 88–95 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Zhang Q., et al. , Comparison of histologic findings in age-related macular degeneration with RPE flatmount images. Mol. Vis. 25, 70–78 (2019). [PMC free article] [PubMed] [Google Scholar]
- 57.Bortner C. D., Cidlowski J. A., Apoptotic volume decrease and the incredible shrinking cell. Cell Death Differ. 9, 1307–1310 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Elmore S., Apoptosis: A review of programmed cell death. Toxicol. Pathol. 35, 495–516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gómez-Angelats M., Cidlowski J. A., Cell volume control and signal transduction in apoptosis. Toxicol. Pathol. 30, 541–551 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The REShAPE code is available on GitHub (https://github.com/nih-nei/REShAPE). All other study data are included in the article and/or supporting information.