Abstract
A mercury-resistant bacterial strain which is able to reduce ionic mercury to metallic mercury was used to remediate in laboratory columns mercury-containing wastewater produced during electrolytic production of chlorine. Factory effluents from several chloralkali plants in Europe were analyzed, and these effluents contained total mercury concentrations between 1.6 and 7.6 mg/liter and high chloride concentrations (up to 25 g/liter) and had pH values which were either acidic (pH 2.4) or alkaline (pH 13.0). A mercury-resistant bacterial strain, Pseudomonas putida Spi3, was isolated from polluted river sediments. Biofilms of P. putida Spi3 were grown on porous carrier material in laboratory column bioreactors. The bioreactors were continuously fed with sterile synthetic model wastewater or nonsterile, neutralized, aerated chloralkali wastewater. We found that sodium chloride concentrations up to 24 g/liter did not inhibit microbial mercury retention and that mercury concentrations up to 7 mg/liter could be treated with the bacterial biofilm with no loss of activity. When wastewater samples from three different chloralkali plants in Europe were used, levels of mercury retention efficiency between 90 and 98% were obtained. Thus, microbial mercury removal is a potential biological treatment for chloralkali electrolysis wastewater.
Industrial use of mercury, a highly toxic metal, has led to significant mercury pollution of the environment (4, 16). Cleanup technologies which are capable of treating large volumes of soil, water, or sediment contaminated with relatively low levels of mercury in a cost-effective way are urgently needed (14). The potential of the microbial mer operon-based resistance mechanism, which functions by active enzymatic reduction of mercury ions to water-insoluble metallic mercury (5, 12), has been recognized for a long time because of its high levels of efficacy and specificity (3, 14). Metallic mercury produced by microbial reduction diffuses out of cells and accumulates in pure form in the medium. Since the microbial biomass acts as a catalyst, a process based on this principle can be run continuously without the production of large volumes of mercury-loaded biomass and with much greater efficacy than passive adsorption and immobilization treatments in which biomass is used. However, to our knowledge, until now microbial mercury reduction has not been used for treatment of industrial waste.
Chloralkali plants in which the amalgam process is used are the second largest users of mercury in Germany after electrical engineering (10). In the past, wastewater produced in the amalgam process was discharged into rivers and lakes, where mercury was detected long after the actual pollution had stopped (15) and is still a risk to humans since it accumulates in the food chain. Today, stringent legislation in Europe requires expensive treatment of wastewater in order to fulfill the discharge limit requirements. Therefore, we examined treatment of chloralkali wastewater with mercury-resistant microorganisms in an effort to develop an environmentally friendly, cost-effective, integrated, end-of-pipe remediation technology.
Fixed-bed reactors were chosen as most appropriate reactor design because of their robustness and relative ease of scale-up. It has been demonstrated previously that mercury is retained in laboratory columns containing immobilized merA gene-containing bacteria in a continuous process running for 3 months with high efficiency (3). Here, we examined whether actual chloralkali factory effluents could be treated with a mercury-resistant strain by using a stepwise approach. First, we determined the composition of chloralkali wastewater from several plants in Europe so that we could tailor the microbiological system to on site conditions. Since NaCl was the most significant copollutant and is known to interfere with mercuric reductase activity (2), we next studied, using defined mercury chloride solutions (model wastewater), the effect of inflow mercury and NaCl concentrations on the retention efficiencies of model reactors. Finally, original wastewater samples from three chloralkali plants in Europe were treated to determine the mercury retention efficiency of the microbial detoxification system for chloralkali wastewater.
MATERIALS AND METHODS
Strains.
Pseudomonas putida Spi3 was isolated from sediments of the Spittelwasser River, a tributary of the Elbe River, by directly plating sediment serial dilutions onto 0.1× Luria-Bertani agar (10 g of tryptone per liter, 0.5 g of yeast extract per liter, 1 g of NaCl per liter) containing 50 μg of Hg(II) per liter. The Spittelwasser River was subjected to massive industrial pollution, including pollution with inorganic and organic mercury compounds, up to 1989. The isolate was identified as P. putida strain on the basis of 16S ribosomal DNA sequencing data (level of similarity, 99.8%) and analyses performed at the German Culture Collection of Microorganisms and Cell Cultures, including fatty acid methyl ester analysis, phenotypic and physiological tests, and ribotyping. The maximum concentrations of HgCl2 transformed by P. putida Spi3 were 70 mg/liter on solid medium and 10 mg/liter in liquid medium (0.1× Luria-Bertani medium). The presence of the merA and merB genes was confirmed by performing specific PCR with primers based on the alignment of merA sequences in the GenBank database (15a).
Determination of wastewater composition.
Standard kits (Aquanal; Riedel-de Haen, Seelze, Germany) were used to determine hardness and phosphate ammonia, nitrate, and nitrite concentrations. Sulfate concentrations were determined by using Aquaquant (Merck, Darmstadt, Germany). To determine chloride concentrations, chemical oxygen demand, and sulfite concentrations, we used kits obtained from Dr. Lange (Düsseldorf, Germany). Free and total chlorine contents were measured with a kit obtained from Hach (Loveland, Colo.). Oxygen contents, pH, and conductivity were determined by using electrodes (models oxi 96-A, LF 96-A, and pH330; wtw, Weilheim, Germany).
Mercury measurements.
Mercury contents were determined by flameless cold vapor adsorption spectroscopy by using a flow injection system (model FIAS 200; Perkin-Elmer, Überlingen, Germany) which was linked to an atomic adsorption spectrophotometer (model AAS 2100; Perkin-Elmer). To determine soluble mercury contents, samples (5 ml) were routinely oxidized by adding 0.01 volume of 65% HNO3. Ionic mercury was then reduced with NaBH4 (4 g/liter) to metallic mercury, which was volatilized by the carrier gas argon and detected at 253.7 nm by the atomic adsorption spectrophotometer. If necessary, samples were diluted so that they contained less than 100 μg of Hg per liter. To determine total mercury concentrations, 7-ml samples were pretreated by oxidizing them with 3 ml of 65% HNO3 for 2 h at 140°C. Samples were obtained with an autosampler once per hour (Fig. 1 and 2) or a 24-h period (Fig. 3B); by taking subsamples which were pooled over a 48-h period (Fig. 3A); or by sampling at the indicated intervals (Fig. 3C). The reactor effluents were collected in vials containing 1% of the final volume of HNO3 (65%) to stabilize the dissolved mercury.
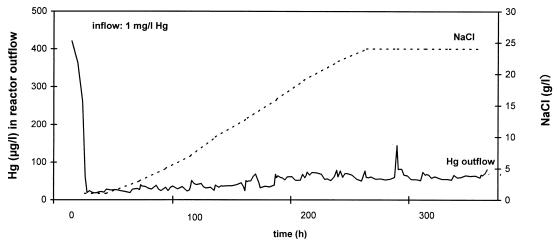
FIG. 1.
Removal of mercury from model wastewater (containing 1 mg of Hg per liter) in the presence of different concentrations of NaCl by a P. putida biofilm immobilized on Siran beads in an upflow column reactor. l, liter.
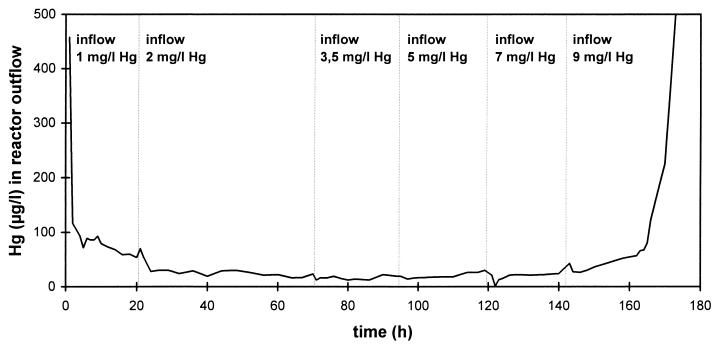
FIG. 2.
Removal of different concentrations of mercury from model wastewater (containing 10 g of NaCl per liter) by a P. putida Spi3 biofilm immobilized on Siran beads in an upflow column reactor. l, liter.
FIG. 3.
Removal of mercury from neutralized, aerated chloralkali electrolysis wastewater samples obtained from three factories (Table 1) by P. putida Spi3 biofilms immobilized on Siran beads (●) and on cellulose fibers (□) in an upflow column reactor. (A) Wastewater A. (B) Wastewater B. (C) Wastewater C. bv, bed volume. l, liter; d, day.
Bioreactor setup and experimental design.
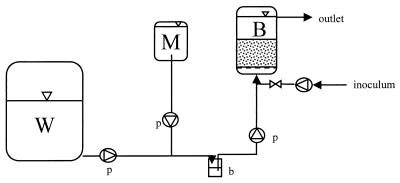
The experimental design is shown in Fig. 4. Sterile model wastewater or nonsterile, neutralized, aerated chloralkali wastewater was pumped into laboratory column bioreactors in the upflow mode by using a peristaltic pump. Sterile medium was added through a second peristaltic pump. The total volume entering the column was 20 ml/h unless indicated otherwise; this 20 ml/h consisted of 2 ml of medium (containing 1 g of yeast extract per liter) per h and 18 ml of wastewater per h. A bubble trap was used to prevent contamination of the medium and also to prevent the transport of gas bubbles into the columns. For inoculation, a bypass was used. Experiments in which model wastewater and chloralkali wastewaters A and B were used (Fig. 1, 2, 3A, and 3B) were performed with chromatographic columns (type XK16; length, 19 cm; inside diameter, 1.6 cm; Pharmacia, Erlangen, Germany). For experiments in which chloralkali wastewater C was used (Fig. 3C), glass columns (length, 16 cm; inside diameter, 2.2 cm) were used. The glass columns were filled with glass beads (Schott Glaswerke, Mainz, Germany) whose diameters decreased from the bottom to the top (from 5 mm at the bottom to 1 to 3 mm at the top; total volume, approximately 20 cm3) to obtain good distribution of the wastewater inflow. Carrier material (20 cm3) was placed above the glass beads. The chromatographic columns each contained a grid at the bottom for even inflow distribution, and the columns were filled with only carrier material (approximately 20 cm3). The carrier material was Siran beads (SiO2; diameter, 0.4 to 1.0 mm; density, 0.49 g/cm3; porosity, 55 to 60%; pore size, <120 μm; Schott Glaswerke). In addition, chloralkali wastewater C was also remediated in glass columns containing two other types of carrier material, lignocell (wood; diameter, 2.0 to 2.5 mm; density, 0.27 to 0.33 g/cm3; type HBK 1500-3000; J. Rettenmaier & Söhne GmbH & Co., Holzmühle, Germany) and arbocell (cellulose; fiber length, 700 μm; fiber diameter, 20 μm; density, 0.03 to 0.045 g/cm3; type BC 1000; J. Rettenmaier & Söhne GmbH & Co.). The columns and tubing were sterilized by autoclaving them at 121°C for 20 min or by flushing them for 24 h with 70% ethanol. All experiments were conducted at room temperature (approximately 25°C).
FIG. 4.
Design of model bioreactor columns used to remove mercury from industrial effluents. W, wastewater container (1-m3 container for chloralkali wastewater; 1-liter bottle for model wastewater); M, medium (1-liter bottle); B, bioreactor (bed volume, 20 ml); b, bubble trap; p, peristaltic pump. See text for explanation.
Inoculation.
To prepare the inoculum, one colony of P. putida Spi3 was picked from a plate, suspended in 500 ml (Fig. 3C), 800 ml (Fig. 1 and 2), or 1,000 ml (Fig. 3A and B) of liquid growth medium [10 g of yeast extract, per liter, 10 g of NaCl per liter, 1 mg of Hg(II) per liter] and grown at 30°C on a rotary shaker for 2 days. Portions (400 to 500 ml) of the inoculum were run through the sterilized bioreactors at a rate of 20 ml/h for approximately 1 day. Subsequently, fresh sterilized medium was run through the columns for 2 days (Fig. 1 and 2), 5 days (Fig. 3A and B), or 10 days (Fig. 3C) so that a biofilm could grow on the carrier material. Then the medium was changed to a medium containing 1 g of yeast extract per liter, which was added to the columns at a rate of 2 ml/h. Model wastewater or chloralkali wastewater was added at a rate of 18 ml/h by using a second peristaltic pump, which resulted in a total inflow rate of 20 ml/h. Chloralkali wastewater was neutralized with NaOH or HCl and aerated until the oxygen saturation level was 100% prior to biological treatment.
RESULTS
Chloralkali wastewater composition.
Table 1 shows the compositions of the wastewater samples obtained from three chloralkali plants in Europe. The mercury concentrations ranged from 1.6 to 7.6 mg/liter. The difference between the total mercury and the soluble mercury was the mercury which was detected only after a sample was pretreated with hot salpetric acid; this mercury was designated the bound mercury. Mercury is easily attached to suspended solids or complexed by components of wastewater (e.g., chloride anions). Bound mercury may not have been available to the microbial detoxification system used. Due to reducing agents present in the water, a small fraction of the mercury occurred as Hg0. This fraction was almost zero for wastewater C, which was oxidized at the factory with Cl2 in order to allow mercury removal by ion-exchange columns. The wastewaters were alkaline (pH 12.0 to 13.0) or acidic (pH 2.4) depending on the process conditions. Therefore, the wastewater had to be neutralized prior to microbiological treatment. Chlorine was detected in very small amounts in wastewater C but not in wastewaters A and B; it is generally present in chloralkali wastewater or is even added in some plants (wastewater C) to completely oxidize the mercury to Hg(II), but it is subsequently removed by adding Na2SO3. Chlorine is a sterilizing agent and thus must not be present for microbiological treatment. All of the chloralkali wastewaters contained chloride, and the concentrations ranged from 6.7 to 25.0 g/liter. Since it is known that NaCl interferes with the activity of the mercuric reductase, probably due to the formation of mercuro-chlorocomplexes (2), it was necessary to determine the efficiency of microbial mercury removal in the presence of different salt concentrations. Oxygen was always present, and the temperature of the water was either the same as the temperature of process water taken from a river (wastewater B) or between 35 and 37°C (wastewaters A and C), depending on the sampling point at the factory. Accompanying ions (NH4, NO2, NO3, PO4, SO4) occurred at levels which were not considered inhibitory to microbiological treatment. The chemical oxygen demand, which correlated roughly with the load of suspended solids, was low (1.3 mg/liter in wastewater B).
TABLE 1.
Characteristics of chloralkali effluents from three plants in Europe
| Wastewater | Hg concn (μg/liter)
|
pH | Concn (mg/liter) of:
|
Chemical oxygen demand (mg/liter) | Temp (°C) | Conductivity (mS/cm) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Solved | Hg0 | Cl− | Total Cl2 | Free Cl2 | NH4+ | NO2− | NO32− | PO43− | SO42− | O2 | |||||
| A | 1,560 | 1,345 | 36 | 12.0 | 25,000 | 0 | 0 | 0 | 0.02 | 10 | <3 | >300 | 6.8 | NDa | 37 | 58.1 |
| B | 1,693 | 1,020 | 15 | 13.0 | 6,700 | 0 | 0 | 0.8 | 0.04 | 150 | 0.42 | 650 | 9.0 | 1.3 | 9 | 28.4 |
| C | 7,610 | 6,960 | 0.4 | 2.4 | 10,000 | 0.3 | 0 | 0.2 | 0 | <5 | <3 | ND | 9.1 | ND | 35 | 26.4 |
ND, not determined.
Effects of different salt concentrations on mercury retention.
Figure 1 shows the effects of different salt concentrations on the efficiency of mercury removal in a model reactor. At NaCl concentrations up to 24 g/liter, representing up to 14.6 g of Cl− per liter, the efficiencies of removal by the biofilm were practically the same. The outflow concentrations of Hg ranged from 20 to 80 μg/liter and thus were at the theoretical lower limit for this type of reactor operation, which is defined by the solubility of Hg0 in water. It is 60 μg/liter for distilled water at room temperature and decreases as the salt concentration increases (10). For an inflow concentration of mercury of 1 mg/liter the efficiency of removal by the reactor was 92 to 98%.
Effects of different mercury concentrations on microbial mercury retention.
To simulate chloralkali wastewater, we used model wastewater which contained 10 g of NaCl per liter and different concentrations of mercuric chloride. Figure 2 shows that the outflow concentration for the model reactor was not affected by inflow concentrations of mercury up to 7 mg/liter. The outflow concentrations were less than 60 μg/liter, which represents the theoretical optimum which can be achieved (see above). Thus, the removal efficiency increased from 95% (inflow Hg concentration, 1 mg/liter) to 99.2% (inflow Hg concentration, 7 mg/liter). The relatively high NaCl concentration in the model wastewater did not decrease the removal efficiency of the reactor. When the Hg concentration in the inflow was 9 mg/liter, breakthrough occurred. Thus, the upper Hg concentration limit for microbial removal of mercury from chloralkali wastewater was between 7 and 9 mg/liter.
Microbial removal of mercury from chloralkali wastewater.
Wastewater samples from three chloralkali plants in Europe, whose compositions corresponded to the compositions of wastewaters A, B, and C in Table 1, were treated in model reactors. Figure 3A shows the removal of mercury from wastewater A (Table 1). In this experiment outflow concentrations of Hg around 50 μg/liter were obtained with neutralized, oxygen-saturated chloralkali wastewater; this value corresponded to the theoretical optimum and was similar to the value obtained with model wastewater. The efficiency of removal by the bioreactor for chloralkali wastewater A was 97%. An increase in the volumetric load to 60 ml/h, corresponding to 3 bed volumes per h, decreased the efficiency of mercury removal, probably because of a lack of adaptation of the microbial biofilm.
Figure 3B shows the results obtained for wastewater B. The maximum performance was obtained with model wastewater (inflow mercury concentration, 3 mg/liter; outflow Hg concentrations, 50 to 60 μg/liter). When chloralkali wastewater B was fed into the bioreactor, the outflow Hg concentration increased to 180 μg/liter, although the mercury concentration of the wastewater was only 1.7 mg/liter (compared to 3 mg of Hg per liter for the model wastewater). When the inflow water was changed back to model wastewater, the outflow concentrations returned to less than <50 μg/liter. A mercury concentration of around 50 μg/liter was also detected in the reactor effluent if no mercury was present in the inflow, illustrating the fact that it is the equilibrium between dissolved mercury and metallic mercury which is the reason for the residual effluent concentration. The maximum removal efficiencies of the bioreactor were therefore 98.4% for model wastewater and 90% for chloralkali wastewater B.
Figure 3C shows the results obtained for wastewater C with two different carrier materials, Siran beads and cellulose fibers, in separate bioreactors which were run in parallel. When model wastewater with a mercury concentration of 3 mg/liter was used, maximum mercury retention was observed (outflow Hg concentration, <20 μg/liter). Then, 67% chloralkali wastewater was added to the columns (inflow mercury concentration, 5 mg/liter). For the bioreactor filled with Siran beads, the outflow Hg concentrations increased to approximately 200 μg/liter and remained at that level during the following shift to 100% wastewater (inflow Hg concentration, 7.6 mg/liter). When a different batch of wastewater with a mercury concentration of 5.5 mg/liter was added to the column, the outflow Hg concentrations were also approximately 200 μg/liter. For the bioreactor filled with cellulose fibers, the mercury retention was slightly better; the outflow Hg concentrations were of approximately 110 μg/liter for both batches of wastewater C. When model wastewater was again added to the bioreactors with a mercury concentration identical to that in the previous batch of chloralkali wastewater (5.5 mg/liter), the outflow concentrations decreased to around 60 μg/liter and thus returned to the theoretical optimum. The maximum removal efficiencies for the bioreactors were 99% for model wastewater and between 97.3% (Siran) and 98.5% (cellulose fibers) for chloralkali wastewater C.
DISCUSSION
Effect of chloride on mercury removal.
Because of its high reactivity, mercury easily forms complexes with anions. It is assumed that the bioavailability of Hg(II) is reduced at high chloride concentrations due to the formation of mercuro-chloro complexes. Using a geochemical equilibrium speciation model, Barkay et al. (2) calculated the concentrations of different species of Hg(II) complexes in various assay solutions. In particular, the concentrations of HgCl3− and HgCl4− in the presence of different chloride concentrations were calculated. Using a mer lux HgCl4− reporter strain (13), Barkay et al. (2) were able to show that the light emitted by the reporter strain decreased as the concentration of NaCl increased from 1 to 100 mM, corresponding to significant increases in the concentration of mercuro-chloro complexes. However, no additional inhibition of reductase activity was detected in the presence of 200 mM NaCl, corresponding to 12 g of NaCl per liter. The effects of NaCl concentrations greater than 12 g/liter have not been studied with a defined system. In our experiments, biofilms grown on silica beads did not exhibit a reduction in mercury reduction activity when they were exposed to NaCl concentrations up to 24 g/liter. Our data cannot be compared directly to those of Barkay et al. for a number of reasons. The number of cells in the microbial biofilm could not be quantified. A potential decrease in the level of reductase activity per cell could have been compensated for by an increase in the number of actively reducing cells and thus may not have been evident in the overall reactor performance. Moreover, biofilm physiology is known to differ fundamentally from the physiology of bacterial cells living in suspension. In addition, the effect of chloride on mercury reductase activity might not be linear, as found by Barkay et al. (2) at NaCl concentrations of ≥6 g/liter. Finally, mercury reductase activity is strongly dependent on accompanying ions in the solution and therefore is hard to predict when complex wastewater is examined. With respect to the applicability of microbial removal of mercury to a technical process, we concluded from our experiments that salt concentrations which were in the range of the concentrations occurring in chloralkali wastewaters did not inhibit microbial removal of mercury in biofilms.
Upper limit for microbial mercury reduction.
The level of resistance of bacteria to mercury is known to be greater on plates than in liquid culture (11). Moreover, it is dependent on cell density (7). Mercury reductase activity levels are usually determined in buffered solutions containing mercaptoethanol (1, 3, 11), which stabilizes Hg(II) by complexation and results in much higher levels of resistance of the bacteria than the levels of resistance in solutions lacking mercaptoethanol. Consequently, the levels of resistance for mercury-resistant bacteria that have been reported previously vary greatly. Therefore, the upper limit for mercury concentrations that could be treated by a microbial biofilm in unbuffered industrial wastewater had to be determined experimentally. To do this, we used model wastewater which contained the NaCl concentration frequently encountered in chloralkali wastewater. Our experiment showed that mercury concentrations up to 7 mg/liter had no effect on the mercury removal activity of a biofilm. As reported previously (3), Hg(II) was completely transformed to Hg0, which was retained in the bioreactor. The upper limit of the mercury concentrations that could be treated by the biofilm investigated here was between 7 and 9 mg/liter. In older biofilms which develop during several months of reactor operation, the value may be even higher due to the buffering effect of dead biomass. Moreover, it might be possible to reconstitute biofilm activity by decreasing the mercury concentration, since some bacteria may survive in the reactor. Thus, microbial removal of mercury occurs at the concentrations regularly encountered in chloralkali wastewater. However, to protect the bacteria from toxic mercury shock (≥9 mg of Hg per liter), a continuously operating plant needs to have emergency valves controlled by the inflow mercury concentration.
Bioavailability and solubility of mercury.
In the model wastewater, the total mercury concentration equaled the solved mercury concentration. In actual chloralkali wastewater, a fraction of the total mercury was detected only after hot salpetric acid pretreatment. This mercury was designated bound mercury, and we assumed that it was not available for microbial transformation (Table 1). Therefore, solved mercury was used to determine the efficiency of microbial mercury removal. The lower limit of the outflow mercury concentration which can be obtained with the type of reactor used here is the concentration which corresponds to the water solubility of metallic mercury. At room temperature in distilled water, this value is 60 μg of Hg per liter (10), but the value strongly depends on the temperature and the accompanying ions (e.g., chloride). The fact that physical solution processes were responsible for the residual mercury concentration determined during optimum reactor performance was demonstrated by eluting approximately 60 μg of Hg per liter from columns fed inflow water containing no mercury (Fig. 3B); similar experiments have been performed previously (3).
Mercury removal from chloralkali wastewater.
With all columns, model wastewater was treated with the maximum efficiency possible for this type of reactor. In comparison, chloralkali wastewater was treated with the maximum possible efficiency for wastewater A, while for wastewaters B and C, the outflow mercury concentrations were greater than 60 μg/liter, indicating that microbial mercury retention was inhibited. This inhibition was completely reversible if the inflow was changed to model wastewater (containing mercury at concentrations that were the same as or higher than the concentrations in the chloralkali wastewater) and therefore must have been caused by components of the chloralkali wastewater. Moreover, since biofilm activity was restored instantly, the microbial biofilm was not permanently impaired.
Due to the complex ion content of chloralkali wastewater, mercury speciation cannot be predicted. Reduced transformation activity might be caused by the presence of mercury complexes which cannot be treated by the mer operon-encoded enzymes. Alternatively, components of the wastewater (e.g., surplus Na2SO3) or traces of chlorine might be slightly inhibitory to the microorganisms. For wastewater C, small differences in removal rates were observed with different carrier materials, indicating that characteristics of the fixed bed affected reactor performance. Further improvement in reactor performance should be obtained by optimizing reactor design and operation (e.g., the carrier material used to immobilize the microbial biofilm and the flow of the wastewater through the bioreactor). Moreover, the performance of mixed-community biofilms specifically adapted to chloralkali wastewater, as well as the performance of genetically engineered mercury-reducing bacteria (6, 8, 9, 11), should be examined.
Bioprocess development.
Using the data in Table 1, we identified characteristics which were shared by all of the chloralkali wastewaters and also significant differences between wastewaters from different plants (different Hg concentrations, pH values, chloride concentrations and temperatures). Thus, it should be possible to develop a general bioprocess for chloralkali wastewater which can be adapted to individual plants. With the exception of pH, none of the characteristics of the wastewater was critical for microbiological activity. Therefore, the only indispensable pretreatment was neutralization of the wastewater. However, some parameters definitely affected the performance of microorganisms; these included high mercury concentrations combined with high chloride concentrations, the wastewater temperature, and the concentration of dissolved oxygen. In our study, we adjusted the temperature and the oxygen concentration so that each of these parameters was the same in all experiments; this allowed us to compare the results obtained. Moreover, the experiments were performed with wastewater batches which were mixed so that they were homogeneous. A pilot plant operating on site at an electrolysis factory will be exposed to significant variations in some of the parameters mentioned above. Since the discharge limit for industrial wastewater is 50 μg of Hg per liter in the European Community, a final cleaning step (e.g., activated carbon filtration) needs to be included in any biotechnological process for mercury remediation in order to fulfill cleanup goals defined by current law.
It has been shown that high-efficiency removal of mercury from chloralkali wastewater by a biofilm of mercury-resistant bacteria in laboratory columns is feasible. The overall levels of mercury removal from chloralkali wastewater in our study were between 90 and 98%, which indicated that the microbial detoxification system for mercury was highly effective under in situ wastewater conditions. Scale-up of the process to a pilot plant will require engineering solutions in order to maintain a suitable environment for the microbial biofilm under all possible on site conditions and the development of a wastewater-adapted, safe inoculum.
ACKNOWLEDGMENTS
We thank Ulrich Becker for help during the initial phase of the project, especially during sampling of Spittelwasser River sediments and measurement of mercury concentrations. We acknowledge K.-H. Ujma, Preussag AG, Hannover, Germany, and R.-D. Wilken, ESWE Institute for Water Research, Wiesbaden, Germany, for help with establishing contacts in chloralkali electrolysis companies. We also thank all of the company employees who supported the work, particularly those who provided information and helped obtain samples.
This work was supported by grants LIFE97-ENV/D/000463 and BIO4-CT98-0168 from the European Community’s LIFE and BIOTECHNOLOGY programs, respectively.
REFERENCES
- 1.Barbiere P, Bestetti G, Reniero D, Galli E. Mercury resistance in aromatic compound degrading Pseudomonas strains. FEMS Microbiol Ecol. 1996;20:185–194. [Google Scholar]
- 2.Barkay T, Gillman M, Turner R R. Effects of dissolved organic carbon and salinity on bioavailability of mercury. Appl Environ Microbiol. 1997;63:4267–4271. doi: 10.1128/aem.63.11.4267-4271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunke M, Deckwer W-D, Frischmuth A, Horn J M, Lünsdorf H, Rhode M, Röhricht M, Timmis K N, Weppen P. Microbial retention of mercury from waste streams in laboratory columns containing merA gene bacteria. FEMS Microbiol Rev. 1993;11:145–152. doi: 10.1111/j.1574-6976.1993.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 4.Bryan G W, Langston W J. Bioavailability, accumulation and effects of heavy metals in sediments with special reference to United Kingdom estuaries: a review. Environ Pollut. 1992;76:89–131. doi: 10.1016/0269-7491(92)90099-v. [DOI] [PubMed] [Google Scholar]
- 5.Cervantes C, Silver S. Metal resistance in pseudomonads: genes and mechanisms. In: Nakazawa T, et al., editors. Molecular biology of pseudomonads. Washington, D.C.: ASM Press; 1996. pp. 398–416. [Google Scholar]
- 6.Chang J-S, Chao Y-P, Law W-S. Repeated fed-batch operations for microbial detoxification of mercury using wild-type and recombinant mercury-resistant bacteria. J Biotechnol. 1998;64:219–230. doi: 10.1016/s0168-1656(98)00112-6. [DOI] [PubMed] [Google Scholar]
- 7.Chang J-S, Hong J. Estimation of kinetics of mercury detoxification from low-inoculum batch cultures of Pseudomonas aeruginosa PU21 (Rip64) J Biotechnol. 1995;42:85–90. doi: 10.1016/0168-1656(95)00032-l. [DOI] [PubMed] [Google Scholar]
- 8.Chen S, Wilson D B. Construction and characterization of Escherichia coli genetically engineered for bioremediation of Hg2+-contaminated environments. Appl Environ Microbiol. 1997;63:2442–2445. doi: 10.1128/aem.63.6.2442-2445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S-L, Kim E-K, Shuler M, Wilson D W. Hg2+ removal by genetically engineered Escherichia coli in a hollow fiber bioreactor. Biotechnol Prog. 1998;14:667–671. doi: 10.1021/bp980072i. [DOI] [PubMed] [Google Scholar]
- 10.Elvers B, Hawkins S, Schulz G, editors. Ullmann’s encyclopedia of industrial chemistry. 5th ed. A16. Weinheim, Germany: VCH; 1990. p. 270. [Google Scholar]
- 11.Horn J M, Brunke M, Deckwer W-D, Timmis K N. Pseudomonas putida strains which constitutively overexpress mercury resistance for biodetoxification of organomercurial pollutants. Appl Environ Microbiol. 1994;60:357–362. doi: 10.1128/aem.60.1.357-362.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osborn M, Bruce K D, Strike P, Ritchie D A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen L D, Turner R R, Barkay T. Cell-density-dependent sensitivity of a mer-lux bioassay. Appl Environ Microbiol. 1997;63:3291–3293. doi: 10.1128/aem.63.8.3291-3293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saouter E, Turner R, Barkay T. Mercury microbial transformations and their potential for the remediation of a mercury-contaminated site. In: Means J L, Hinchee R E, editors. Emerging technology for bioremediation of metals. Boca Raton, Fla: CRC Press; 1994. pp. 99–105. [Google Scholar]
- 15.Smith J N, Schafer C T. Sedimentation, bioturbation and Hg uptake in the sediments of the estuary and Gulf of St. Lawrence Limnol Oceanogr. 1999;44:207–219. [Google Scholar]
- 15a.von Canstein, H. Unpublished data.
- 16.Zilloux E J, Porcella D B, Benott J M. Mercury cycling and effects in freshwater wetland ecosystems. Environ Toxicol Chem. 1993;12:2245–2264. [Google Scholar]