Significance
RNA molecules encode proteins and play numerous regulatory roles in cells. Targeting RNA with small molecules, as is routine with proteins, would create broad opportunities for modulating biology and creating new drugs. However, this opportunity has been difficult to realize because creating novel small molecules that bind RNA, especially using modest resources, is challenging. This study integrates two widely used technologies, SHAPE chemical probing of RNA and fragment-based ligand discovery, to craft an innovative strategy for creating small molecules that bind to and modulate the activity of a structured RNA. The anticipated impact is high because the methods are simple, can be implemented in diverse research and discovery contexts, and lead to realistic druglike molecules.
Keywords: RNA-targeted ligand discovery, SHAPE-MaP, cooperativity, fragment linking
Abstract
The transcriptome represents an attractive but underused set of targets for small-molecule ligands. Here, we devise a technology that leverages fragment-based screening and SHAPE-MaP RNA structure probing to discover small-molecule fragments that bind an RNA structure of interest. We identified fragments and cooperatively binding fragment pairs that bind to the thiamine pyrophosphate (TPP) riboswitch with millimolar to micromolar affinities. We then used structure-activity relationship information to efficiently design a linked-fragment ligand, with no resemblance to the native ligand, with high ligand efficiency and druglikeness, that binds to the TPP thiM riboswitch with high nanomolar affinity and that modulates RNA conformation during cotranscriptional folding. Principles from this work are broadly applicable, leveraging cooperativity and multisite binding, for developing high-quality ligands for diverse RNA targets.
The vast majority of small-molecule ligands developed to manipulate biological systems target proteins. This focus reflects the fact that proteins have a complex three-dimensional structure critical to function, including clefts and pockets into which small-molecule ligands can bind (1, 2). The transcriptome—the set of all RNA molecules produced in an organism—also includes promising targets for studying and manipulating biological systems. RNA can adopt three-dimensional structures of complexity rivaling that of proteins (3), a key feature needed for the development of highly selective ligands (4), and RNAs play pervasive roles in governing the behavior of biological systems (5). Small-molecule ligands that bind to messenger RNAs (mRNAs) have the potential to up- or downregulate transcription and translation efficiency, thus tuning protein expression in cells (6, 7). Noncoding RNAs also represent a rich pool of targets (8–10). There are challenges involved in identification of small-molecule RNA ligands not faced when targeting proteins (4, 11, 12), including the lack of enzymatic activity, rarity of hydrophobic pockets, conformational flexibility, and high overall molecular charge. These differences challenge conventional ligand screening methods, and new approaches have the potential to accelerate the discovery of small-molecule ligands that target RNA.
In the current study, we combine two principal strategies, fragment-based ligand discovery and nucleotide-resolution conformation-based high-throughput screening, to identify small-molecule RNA binders and to develop a highly specific RNA-binding compound. In fragment-based ligand discovery, one or more small-molecule “fragments” of low to moderate affinity are identified that bind a target of interest, and these fragments are then either elaborated or linked to create more potent ligands (13, 14). Fragments typically have molecular masses less than 250 Da and, in order to bind detectably at all, need to make substantial high-quality contacts with a target.
Fragment-based ligand discovery has been successfully employed to identify initial hit compounds that bind RNA (15–20). To date, fragment-based methods have not been used to create a high-affinity RNA-targeting compound de novo. Identification of multiple fragments that bind the same RNA would make it possible to take advantage of potential additive and cooperative interactions between fragments within the binding pocket (21, 22). We have recently shown that many RNAs bind their ligands via multiple subsites, which are regions of a binding pocket that contact a ligand in an independent or cooperative manner (23). We also showed that high-affinity RNA binding can occur even when subsite binding shows only modest cooperative effects and when the linking coefficient is unfavorable. These features bode well for the effectiveness of fragment-based ligand discovery as applied to RNA targets.
Inspired by these ideas, we first developed a technology to efficiently find fragments that bind an RNA of interest: here, the thiamine pyrophosphate (TPP) riboswitch. Our strategy melds fragment-based ligand discovery (13, 14) with SHAPE-MaP (selective 2'-hydroxyl acylation analyzed by primer extension and mutational profiling) RNA structure probing (24, 25), which was used both to identify RNA-binding fragments and to establish the individual sites of fragment binding at roughly nucleotide resolution. We identified second-site fragments that bound near the site of an initial fragment hit. The ligand ultimately created by linking two fragments has no resemblance to the native riboswitch ligand, binds the structurally complex TPP riboswitch RNA with high affinity, and modulates riboswitch conformation during cotranscriptional folding.
Results
Development and Validation of a Flexible SHAPE-Based Fragment Screening Technology.
Fragment-based ligand discovery has proven to be an effective approach for identifying compounds that form substantial intimate contacts with macromolecules, including RNA (13, 14, 17). A critical prerequisite for success of this discovery strategy is an adaptable, high-quality biophysical assay to detect ligand binding. To detect ligand binding, we used SHAPE RNA structure probing (24–26), which measures local nucleotide flexibility as the relative reactivity of the ribose 2'-hydroxyl group toward electrophilic reagents. SHAPE can be used on any RNA and provides data on virtually all nucleotides in the RNA in a single experiment, yielding per-nucleotide structural information in addition to simply detecting binding. SHAPE reactivity signals are largest in non-base-paired regions, which are critical elements in essentially all small-molecule RNA-binding sites. SHAPE-MaP (24, 25)—which melds SHAPE with a readout by high-throughput sequencing—enables, in principle, multiplexing and efficient high-throughput analysis of many thousands of samples.
We designed a modular RNA screening construct to implement SHAPE as a high-throughput assay for readout of ligand binding (Fig. 1A). The construct was designed to contain two target motifs: in this study, these were a pseudoknot from the 5′-UTR (untranslated region) of the dengue virus genome that reduces viral fitness when its structure is disrupted (27) and the TPP riboswitch aptamer domain (28–30). Including two distinct structural motifs in a single construct allowed each to serve as an internal specificity control for the other. Fragments that bound to both RNA structures were easily identified as nonspecific binders. These two structures were connected by a six-nucleotide linker, designed to be single stranded, to allow the two RNA structures to remain structurally independent. Flanking the structural core of the construct are structure cassettes (26); these stem-loop-forming regions encode primer-binding sites for steps required in the screening workflow and were designed not to interact with other structures in the construct (SI Appendix, Fig. S1).
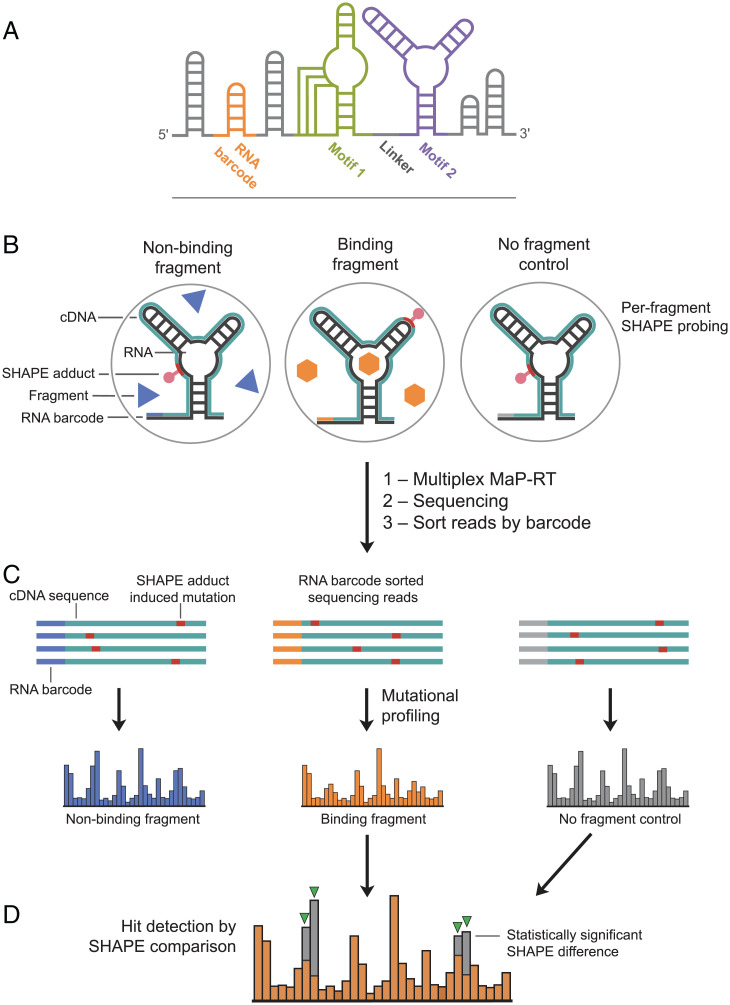
Fig. 1.
Schemes for RNA construct and fragment screening workflow. (A) RNA motifs 1 and 2 and the barcode helix are shown in colors; structure cassette helices are gray. (B) RNA is probed using SHAPE in the presence or absence of a small-molecule fragment. (C) Chemical modifications corresponding to ligand-dependent structural information are read out by multiplexed MaP sequencing. (D) Fragment hits are identified as multiple, statistically significant differences in SHAPE reactivities.
A crucial component of the screening construct is the RNA barcode; barcoding enables multiplexing that substantially reduces the downstream workload. Each well in a 96-well plate used for screening a fragment library contains an RNA with a unique barcode in the context of an otherwise identical construct; the barcode sequence thus identifies the well position and the fragment (or fragments) present post multiplexing (Fig. 1A). The RNA barcode region was designed to fold into a self-contained structure that does not interact with any other part of the construct. The barcode structure is a seven-base-pair helix capped with a stable tetraloop and anchored with a G-C base pair to maintain hairpin stability (SI Appendix, Fig. S1). Barcodes (96) were designed such that any individual barcode must undergo two or more mutations to be misconstrued as another barcode.
This construct affords flexibility in choosing RNA structures to screen for ligand binding and supports a simple, straightforward screening experiment (Fig. 1B). Each well in a 96-well plate, containing an otherwise identical RNA construct with a unique RNA barcode, is incubated with one or a few small-molecule fragments or a no-fragment control (solvent vehicle) and then exposed to the SHAPE reagent. The resulting SHAPE adducts chemically encode per-nucleotide structural information. Post SHAPE probing, the information needed to determine fragment identity (RNA barcode) and fragment binding (SHAPE adduct pattern) is permanently encoded internally in each RNA strand. RNAs from the 96 wells of a plate can be pooled into a single sample. The fragment screening experiment is processed in a manner very similar to a standard MaP structure-probing workflow (25). Briefly, a specialized relaxed fidelity reverse transcription reaction is used to make complementary DNAs (cDNAs) that contain nontemplate-encoded sequence changes at the positions of any SHAPE adducts on the RNA (31) (Fig. 1 B and C). These cDNAs are then used to prepare a DNA library for high-throughput sequencing. Multiple plates of experiments can be barcoded at the DNA library level (25) to allow collection of data on thousands of compounds in a single sequencing run. The resulting sequencing data contain millions of individual reads, each corresponding to specific RNA strands. These reads are sorted by barcode to allow analysis of data for each small-molecule fragment or combination of fragments (Fig. 1C).
Detecting bound fragment signatures from per-nucleotide SHAPE-MaP mutation rates involves multiple steps to normalize data across a large experimental screen and to ensure statistical rigor (Fig. 1 C and D; SI Appendix, Supporting Methods). Key features of our SHAPE-based hit analysis strategy include 1) comparison of each fragment-exposed RNA, or “experimental sample,” to five negative, no-fragment-exposed, control samples to account for plate-to-plate and well-to-well variability; 2) hit detection performed independently for each of the two structural motifs in the construct, in this study, the pseudoknot and TPP riboswitch; 3) masking of individual nucleotides with low reactivities across all samples as these nucleotides are unlikely to show fragment-induced changes; and 4) calculation of per-nucleotide differences in mutation rates between the fragment-exposed experimental sample and the no-fragment-exposed negative control sample. Those nucleotides with a 20% or greater difference in SHAPE reactivity rate between one of the motifs and the no-fragment controls were selected for Z-score analysis. A fragment was determined to have significantly altered the SHAPE reactivity pattern if three or more nucleotides in one of the two motifs had Z-values greater than 2.7 [as determined by comparison of the Poisson counts for the two motifs (32); see SI Appendix, Supporting Methods]. Our primary screen included 1,500 fragment test compounds, 613 negative control samples, and 96 positive control samples. The 1,500 test fragments comprised a subset of the Maybridge Ro3 diversity library and were chosen to cover a large amount of chemical space while generally adhering to “rule of three” characteristics for fragments (33).
Primary Screening.
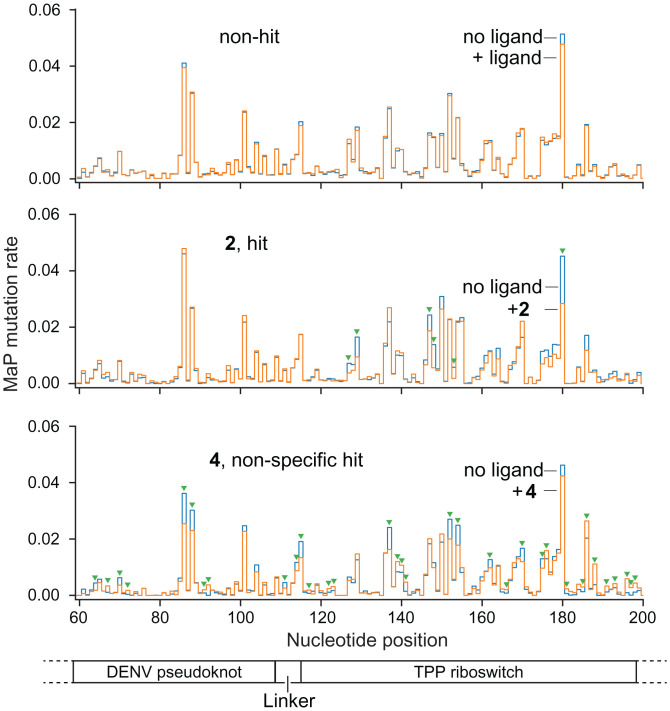
Out of the 1,500 tested fragments, 41 fragments were detected as hits, for an initial hit rate of 2.7%. Hit validation was performed via triplicate SHAPE analysis (Fig. 2 and SI Appendix, Fig. S2), and a compound was accepted as a true hit only if it was detected as a binder in all three replicates. These replicated hit compounds were then analyzed by isothermal titration calorimetry (ITC) to determine binding affinities for an RNA corresponding just to the target motif (omitting flanking sequences in the screening construct). Of these initial hits, eight were validated by replicate analysis and Kd values could be measured for six using ITC (Fig. 3A). Seven of the hits bound the TPP riboswitch specifically, based on their mutation signatures localizing mostly or entirely within the TPP riboswitch region of the test construct. The remaining hit [4] was nonspecific, as this fragment affected nucleotides across all portions of the RNA construct. No compounds were detected that specifically bound the dengue pseudoknot region of the test construct.
Fig. 2.
Representative SHAPE mutation rate comparisons for fragment hits and nonhits. Normalized mutation rates for fragment-exposed samples (orange) are compared to no-ligand traces (blue). Statistically significant changes in mutation rate are denoted with green triangles (see SI Appendix, Fig. S2 for SHAPE confirmation data). (Top) Mutation rate comparison for a representative fragment that does not bind the test construct. (Middle) Fragment hit to the TPP riboswitch region of the RNA. (Bottom) Nonspecific hit that induces reactivity changes across the entire test construct. RNA motif 1 and 2 landmarks are shown below SHAPE profiles.
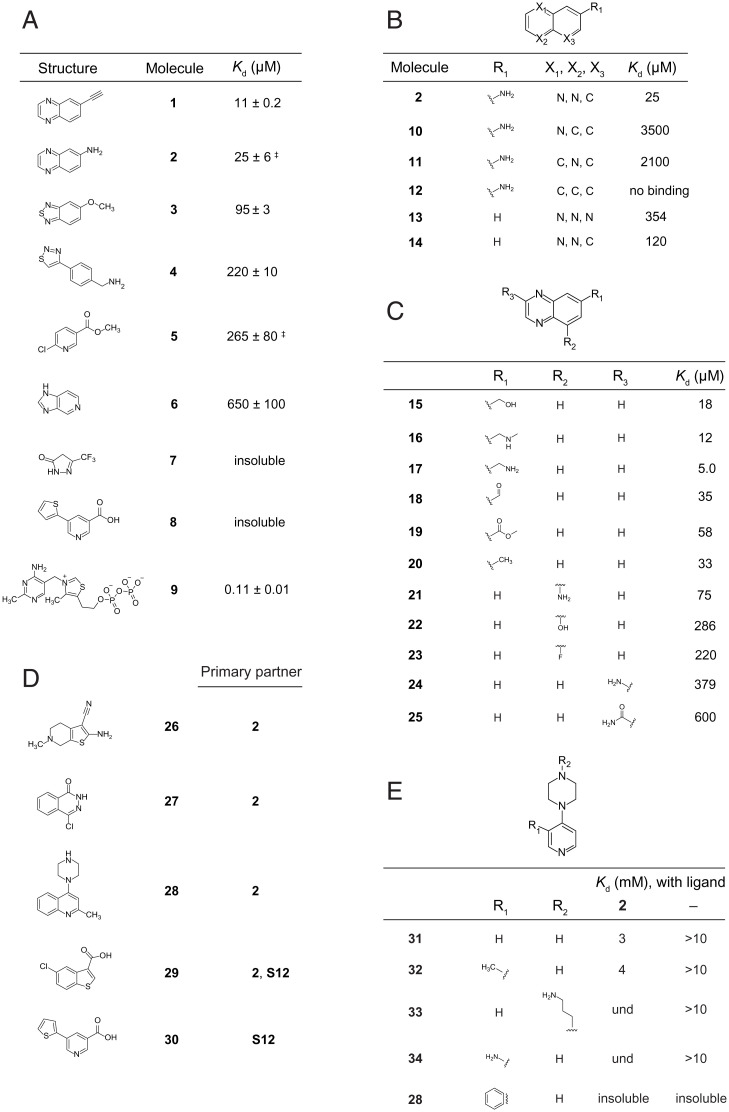
Fig. 3.
Ligands and affinities for fragments that bind the TPP riboswitch. (A) Fragments that bind as detected in the initial round of SHAPE-enabled screening. Dissociation constants were determined by ITC; ‡ SE is derived from ≥3 replicates; other error estimates are calculated based on 95% confidence intervals for the least-squares regression of the binding curve. The native TPP ligand [9] is included for comparison. (B and C) Structure-activity relationships for analogs of fragment 2. Modifications to the (B) quinoxaline core and (C) pendant groups. (D) Fragments that bind in the presence of a prebound primary partner, as detected by SHAPE in round two screening. Hits were validated by replicate SHAPE analysis. (E) Structure-activity relationships for analogs of fragment 28 binding to the TPP riboswitch RNA, in the presence and absence of prebound fragment 2. und, undetermined due to inability to fit ITC binding curve; insoluble, compound insoluble at concentrations required for ITC.
The fragments that bound selectively to the TPP riboswitch have diverse chemotypes and are chemically dissimilar to the native TPP ligand (Fig. 3A). Overall, heteroaromatic nitrogen-containing rings predominate; these likely participate in hydrogen bonding interactions. Three compounds have pyridine rings, and two have pyrazine rings. An azole ring is present in three compounds: two thiadiazols and an imidazole. There is a thiazole ring in the native TPP ligand, but this moiety does not participate in binding interactions with the RNA (28, 29, 30, 34). Additionally, the identified fragments contain primary amines, esters and ethers, and fluorinated groups that could serve as hydrogen bond acceptors or donors. Overall, the fragments are diverse and, critically, are not overrepresented in primary amines or planar multiring systems that tend to interact with RNA nonspecifically.
Structure-Activity Relationships of Riboswitch-Binding Fragments.
We next examined analogs of our initial hits with goals of increasing binding affinity and identifying positions at which fragment hits could be modified with a linker without hindering binding. We focused on analogs of compounds 2 and 5, as these two fragments are structurally distinct, and relatively diverse analogs were commercially available. Analog-RNA binding was evaluated by ITC.
We tested 16 analogs of 2. Altering the core quinoxaline structure of 2 by removing one or both ring nitrogen atoms significantly decreased binding (Fig. 3B). Modest improvements in binding affinity resulted from introduction of a methylene-linked hydrogen bond donor or acceptor (Fig. 3C, compounds 16 and 17). Varying substituents at other positions on the quinoxaline ring core were generally detrimental. Compound 2 emerged as a good candidate for further development based on the high degree of flexibility and even improvement in binding, observed upon modification of the substituent at the C-6 position.
Our examination of 18 analogs of 5 suggested that this was a “flagpole” hit and not a promising candidate for further elaboration. The core pyridine functionality of the molecule appears to be crucial for binding, as changing the ring nitrogen position or adding or removing a ring nitrogen all reduced or abrogated binding (SI Appendix, Table S1A). Modifications to ring substituents generally proved to be detrimental to binding (SI Appendix, Table S1B). The only affinity-increasing analog featured a chlorine at the C-4 position, S12, yielding a compound that had approximately threefold-higher affinity than did 5.
Identification of Fragments That Bind to a Second Site on the TPP Riboswitch.
Second-round screens were employed to identify fragments that bound to the TPP riboswitch region of the screening construct prebound to compounds 2 or S12. In this screen, we expected to identify fragments that preferentially interacted with the TPP riboswitch when 2 or S12 was already bound, either due to cooperative effects or because new modes of binding become available due to structural changes that occur upon primary ligand binding (Fig. 4). Of the 1,500 fragments screened, five were validated to bind simultaneously with either 2 or S12 (Fig. 3D). One second-screen hit, 29, induced a very robust change in the SHAPE reactivity signal and appeared to alter the RNA structure, including unfolding of the P1 helix, consistent with nonspecific interactions, and was not considered further. Fragment 28 was insoluble at the concentrations needed for ITC analysis, so related analogs containing a pyridine instead of a quinoline ring were examined by ITC (Fig. 3E). These compounds bound with weak affinities; nonetheless, 31 and 32 showed clear, but modest, binding cooperativity with 2 (Fig. 3E).
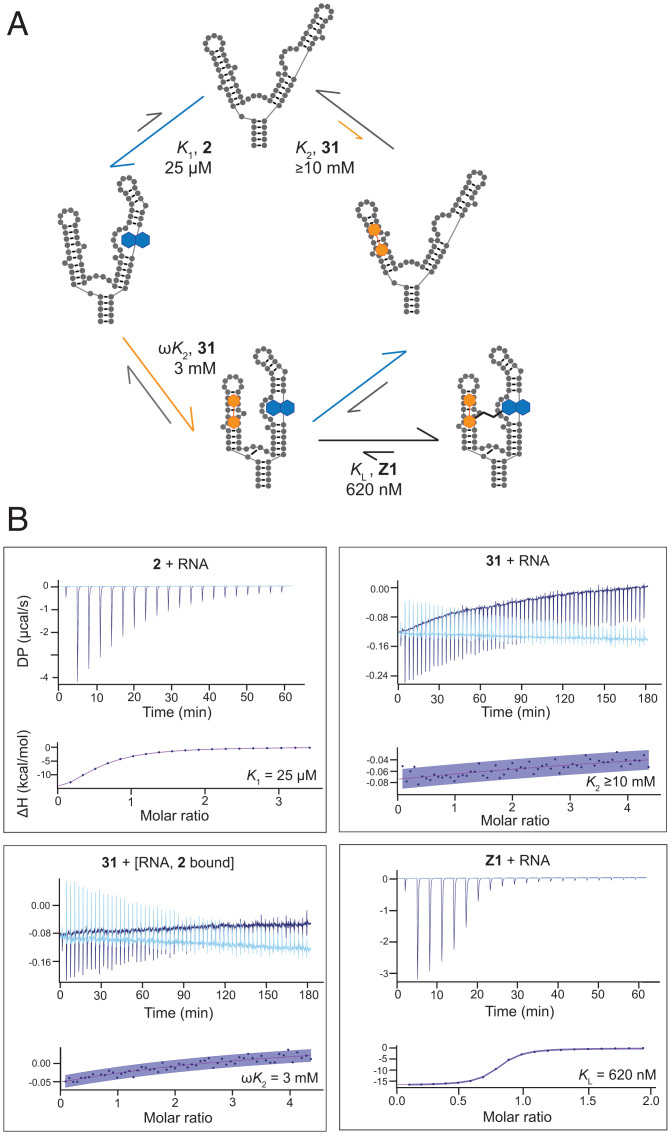
Fig. 4.
Thermodynamic cycle and stepwise ligand binding affinities for fragments 2 and 31. (A) Summary of binding by 2 (blue, K1) and 31 (orange, K2) fragments and the linked compound Z1. Kd values determined by ITC. (B) Representative ITC data showing single-compound and cooperative binding by 2 and 31. Linking the two fragments shows an additive effect in binding energies, resulting in a submicromolar ligand, Z1 (KL). ITC traces are shown with experimental traces in dark blue and background traces (ligand titrated into buffer) as light blue. Confidence intervals (95%) are in purple shading.
Cooperativity and Fragment Linking.
Cooperative binding interactions between 2 and 31 were quantified by ITC. Individually, 2 bound with a Kd of 25 µM and 31 with a much weaker Kd of ≥10 mM. We also examined the affinity of 31 when 2 was prebound to the TPP riboswitch RNA, forming a 2-RNA complex. Under these conditions, 31 bound to the 2-TPP RNA complex with a Kd of ∼3 mM (Fig. 4). This experiment also confirmed that when binding by 2 is saturated, 31 binds to the TPP RNA, implying that these two fragments do not bind in the same location. As 2 and 31 bound with excellent and reasonable affinity, respectively, to distinct regions of the TPP RNA, we explored linking of the two fragments with the goal of creating a high-affinity ligand.
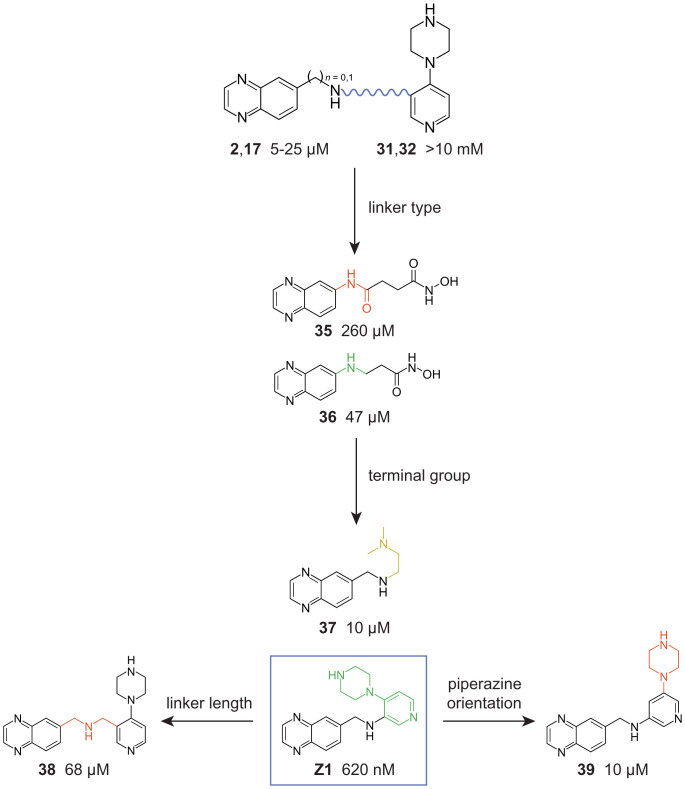
Based on our structure-activity relationship (SAR) analyses of fragment hits 2 (Fig. 3C) and 28 (Fig. 3E), we synthesized linked analogs of the most promising SAR fragments, focusing on the arylamine or aminomethyl position of 2 and 17 and two sites in the pyridine ring of 31 and 32 (Fig. 5 and SI Appendix, Fig. S3). First, we compared affinities of fragments conjugated with an amide or amine linker. A compound with a flexible amine linker [36] had fivefold-higher binding affinity than the amide-linked version (35, Fig. 5). These linkages were introduced in the context of a hydroxamic acid which might chelate a magnesium ion (35), as occurs with the pyrophosphate moiety of the native TPP ligand (28, 29). However, the amine-linked hydroxamic acid compound 36 bound more weakly than the parent fragment 17, suggesting that the hydroxamic acid moiety does not chelate an ion. The linked compound, called Z1, binds with a 620-nM affinity, showing that—with the right approximation—linking two fragments of modest affinity can achieve a high-nanomolar binder. Replacing the fragment 31 entity with a tertiary amine (compound 37) reduced affinity relative to Z1, suggesting the interaction of the 31/32 fragment with the RNA reflects more than just an electrostatic charge effect. Finally, changing the linkage between the 17 and 31 moieties by length [38] or pyridine ring linkage site [39] reduces affinity relative to Z1 (Fig. 5). Ultimately, by linking compounds that bound individually to the TPP riboswitch affinities of 5 µM [17] and ≥10 mM [31], we created a compound (Z1) that binds the RNA with a Kd of 620 nM.
Fig. 5.
Covalent linking of 17 and 31 as a function of linker type and length, terminal group chemotype, and terminal group orientation. Modifications that increase RNA binding affinity are colored green; negative and neutral modifications are colored red and yellow, respectively. Dissociation constants determined by ITC.
RNA-Ligand Structures and Modeling of Interactions between Z1 and RNA.
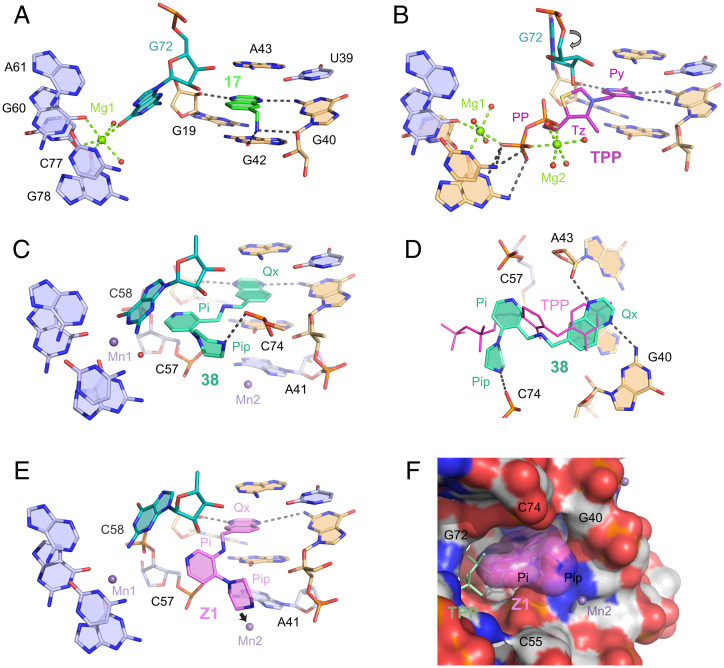
We attempted to obtain crystal structures of both the initial fragments and the final linked compounds and ultimately obtained structures for compounds 16, 17, 37, and 38 (SI Appendix, Table S2). Compounds 16 and 17 bind the TPP riboswitch in the same subsite pocket as the aminopyrimidine ring of the TPP ligand, stacking between G42 and A43 in the J3/2 junction (Fig. 6A and SI Appendix, Fig. S4) (28, 29). Compound 17 forms four hydrogen bonds with the RNA: one each to the ribose and sugar edge of G40, one to the ribose of G19, and one to G42. There is a significant change in local RNA structure relative to the RNA in complex with the native TPP ligand (Fig. 6B). G72 moves into the binding site where the pyrophosphate moiety of the TPP ligand would reside, consistent with prior work that visualized fragments bound in the thiamine subsite of the riboswitch binding pocket (17, 36).
Fig. 6.
Structures of the TPP riboswitch bound by compounds identified in this study and by the native TPP ligand. Nucleotides in direct contact with the organic ligands are beige. Mg2+ and Mn2+ cations and water molecules are depicted as green, lavender, and red spheres, respectively; spheres do not represent physical sizes of ions and water molecules. Intermolecular hydrogen and metal ion coordination bonds are shown as gray and green dashed lines, respectively. Crystal structures of riboswitch bound by (A) compound 17, (B) TPP, and (C) compound 38. Arrow in B shows rotation of G72 in the 17-bound structure. (D) Relative positions of 38 and TPP ligands. (E) Model of Z1 bound to riboswitch. Arrow shows postulated displacement of Mn2+ atom (Mn2). (F) Surface view for model of Z1 bound to RNA. TPP (light green color and thin sticks) is shown as a reference; for clarity, a few nucleotides at the front of the structure were removed in this image. Py, pyrimidine; Tz, thiazole; PP, pyrophosphate moieties of TPP; Ox, quinoxaline; Pi, pyridine; Pip, piperazine of 38. TPP-bound structure is from 2GDI (28).
Linked compounds 37 and 38 (SI Appendix, Fig. S4C and Fig. 6C) have the quinoxaline moiety flipped with respect to 17 such that the linker is oriented toward the interior of the riboswitch. Since the quinoxaline headgroup is symmetrical, hydrogen bonding is preserved. However, the linker orientation now places the pyridine moiety in an open space capped by G72 and surrounded by backbone atoms of C57, C58, and C74 (Fig. 6D). In the TPP-bound structure, this void is partially filled by the thiazole group and the linkage to pyrophosphate. The piperazine ring of 38 is oriented perpendicular to the TPP linker, points toward a new opening in the RNA, and ultimately adopts an L-shaped conformation (Fig. 6D). The elongated and weak electron density for the ethylene diamine and piperazine groups, respectively, in compounds 37 and 38 (SI Appendix, Fig. S4) suggests local orientational freedom that explains, in part, the low binding affinity in these complexes.
We were unable to crystallize the riboswitch in complex with the most potent molecule, linked compound Z1, and therefore used the structure of 38 as a framework for model building. Steric constraints imposed by the shorter linker and permissible orientations of the pyridine and piperazine rings reduced possible conformations and yielded a plausible solution for Z1 bound to the RNA (Fig. 6E). The pyridine and piperazine rings are positioned in the same void as in 38 but shift slightly away from G72. As a result, the piperazine amine protrudes into the binding site of a metal cation (Mn2+) (Fig. 6E, arrow). This metal ion, observed in all crystal structures for this riboswitch, stabilizes the local RNA structure and is likely essential for crystallization. In the modeled conformation, Z1 and 38 have similar buried surface areas and interfaces with the RNA; the reduced conformational freedom and snug fit in the binding pocket are consistent with the stronger RNA binding of Z1 (Fig. 6F).
Monitoring Riboswitch Activity with Z1 in a Cotranscriptional Context.
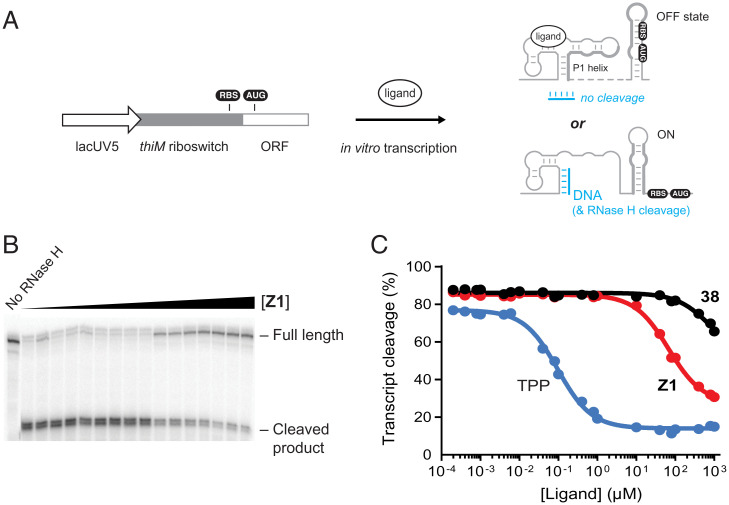
We investigated the ability of Z1 to modulate riboswitch conformation in a cotranscriptional structure switching assay (37). The DNA template contained the E. coli lacUV5 promoter, fused to the thiM riboswitch and the first 10 codons of the thiM open reading frame (Fig. 7A). We performed in vitro transcription reactions using E. coli RNA polymerase as a function of varying ligand concentration. The conformation of the nascent thiM riboswitch transcript was determined as the extent to which a structure resistant to RNase H cleavage formed (37, 38) (Fig. 7 A and B). Rapid and stable binding by a ligand favors the “OFF” state in which sequences downstream of the ligand-binding domain sequester the translation initiation site (RBS-AUG) region (Fig. 7A). In contrast, the absence of stable ligand engagement favors an alternative RNA fold, the “ON” state, that exposes the translation initiation site to allow ribosome binding.
Fig. 7.
Cotranscriptional riboswitch ligand-binding assays. (A) DNA template for cotranscriptional assay with the thiM riboswitch. Ligand binding is detected as the presence, or not, of an RNA conformation that protects the final transcript from RNase H cleavage by a DNA oligonucleotide that binds the P1 helix. (B) Representative cleavage assay, performed in the presence of Z1. (C) Kswitch determination for TPP, Z1, and 38. Compounds Z1 and 38 differ by a single –CH2– group (Fig. 5). Data were fit to a single-site binding equation. Data shown are representative of experiments performed in triplicate.
As expected, the thiM riboswitch construct was responsive to the native TPP ligand. Cleavage efficiency by RNase H decreased with increasing TPP concentration. The TPP concentration required for half-maximal protection (Kswitch) was 96 ± 7 nM (Fig. 7C). The Kswitch value is comparable to the directly measured Kd value (Fig. 4A) (37, 39, 40). When cotranscriptional engagement was measured for Z1 and the weak-binding 38 analog, Kswitch values of 68 ± 4 and 820 ± 360 µM, respectively, were obtained (Fig. 7 B and C). Thus, Z1 binds selectively to nascent thiM riboswitches and stabilizes the OFF state in a functional, cotranscriptional context. The Kswitch value is ∼100-fold higher than the measured Kd value for binding to the aptamer domain alone suggesting, that for Z1, the thiM riboswitch operates under a kinetic regime where the off rate for ligand binding allows an RNA structural rearrangement during transcription (40).
Discussion
Both coding (mRNA) and noncoding RNAs can potentially be manipulated to alter the course of cellular regulation and disease, and to this end, we sought to develop an efficient strategy to identify small-molecule ligands for structured RNAs. Our study demonstrates the promise of using a SHAPE screening readout to detect ligand binding to RNA melded with a fragment-based strategy. We devised a ligand, compound Z1, that is completely unrelated in structure to the native TPP riboswitch ligand, binds the aptamer domain with a Kd of 620 nM, and has a (QED; ref. 41) druglikeness of 0.77. Druglikeness is comparable to that of the very best RNA-targeted ligands reported to date (4). Our structural data provide a basis for further optimization of Z1. This study focused on finding high-quality ligands for a riboswitch, an RNA evolved to recognize a small-molecule ligand. We did not identify ligands for the dengue virus pseudoknot, also present during screening. Further work will be required to devise the best strategy for liganding arbitrary sites in other RNAs, not known to bind a defined ligand.
Z1 was identified based on a biophysical emphasis on RNA-binding affinity and, opportunistically, also functions in cotranscriptional riboswitch structure switching, albeit with lower efficiency than the natural TPP ligand. Distinct from most riboswitches, the TPP ligand binds the riboswitch outside of the central multihelix junction region and instead bridges two RNA stems at a distance from the central junction. The resulting relative orientation of the stems likely stabilizes the junction and facilitates formation of the regulatory (P1) helix emerging from the junction. In contrast to the pyrophosphate moiety of TPP, Z1 does not form extensive interactions with the pyrophosphate-sensing stem of the riboswitch, consistent with requiring higher compound concentrations for switching in the cotranscriptional assay. The TPP riboswitch is widespread in bacteria (42) and often present in several instances that control multiple operons in a given cell (42, 43). Focusing on TPP riboswitches may, therefore, create opportunities to target these organisms with reduced propensity to develop resistance.
The melded SHAPE and fragment-based screening approach is generic with respect to both the RNA structure that can be targeted and the ligand chemotypes that can be developed. The strategy is well suited to finding ligands specifically for RNAs capable of forming complex three-dimensional pockets, which is probably essential for identifying specific RNA-ligand interactions (4). The MaP readout enables multiple levels of multiplexing through both RNA and DNA barcoding. The effort required to screen a 1,000-plus member fragment library is thus dramatically streamlined; the primary screen described here was accomplished by a single person in roughly 3 weeks. The steps required to set up a screen—involving construct design, barcode selection, and synthesis of the required set of barcoded RNAs—can be accomplished by a single person in 3 to 4 weeks, enabling efficient screening of diverse targets.
Many of the ligands that we obtained were similar to those reported previously for a single-round screen also performed against the TPP riboswitch (15–17). Our hits in the primary screen appeared to be biased toward higher-affinity ligands, relative to this prior work, such that most ligands detected by SHAPE bound in the 10- to 300-µM range. Our hit detection strategy likely favors both higher-affinity fragment binders and binders that induce substantial changes in SHAPE reactivity. We think this bias toward tight-binding fragments is an advantage overall. We did not identify fragments that bound the dengue pseudoknot, also present in our screening construct, that reached the affinity and specificity required to meet our screening criteria. The dengue pseudoknot RNA is highly structured, and the likelihood that a fragment can perturb the SHAPE reactivity of this structure might be low. Another possibility is that this pseudoknot might not contain a ligandable pocket.
Our fragment-pair identification strategy, in which a fragment hit from the primary screen was prebound to the RNA and screened for additional fragment binding partners, specifically leveraged the per-nucleotide information obtainable by SHAPE and was successfully used here to discover induced-fit fragment pairs (Fig. 4). One tenet of fragment-based ligand development is that cooperativity between two fragments can be achieved through proximal binding, and this additive binding can be exploited by linking fragments together with a minimally invasive covalent linker (21, 22, 44, 45) . Development of the linked compound Z1 from primary and secondary fragment hits shows that fragment-based ligand discovery can be efficiently applied to RNA targets. There is a modest degree of cooperativity between 2 and 31: binding by compound 31 was stronger by 3- to 10-fold when 2 was prebound to the RNA. Upon linking these two fragments, we observed modest additivity in their binding energies: Z1 had an affinity of 620 nM. Notably, Z1 was identified efficiently after synthesizing and testing only a handful of linked fragments. We did not observe a superadditive effect (44) of linking fragments 2 and 31, likely because we have not achieved perfect positioning of the fragments. Small changes in the length or geometry of the linker resulted in large changes in affinity for the linked ligand (Fig. 5), implying that precise orientation of the linker is necessary to optimally position the two fragments. Successful development of compound Z1 reinforces a key observation based on analysis of TPP-like ligands (23): it is not necessary to achieve perfection in either the degree of cooperativity between fragments or construction of the covalent linker joining them to efficiently devise a submicromolar ligand.
Crystallographic analysis shows that compounds from the primary screen bind RNA in the same subpocket (site 1) bound by the pyrimidine moiety of the natural TPP ligand; in contrast, site 2 is a side opening in the RNA not used by bound TPP (Fig. 6F). This site can likely accommodate other extended groups, and their interactions with RNA could be improved by shifting their position within site 2 via different linkers, as demonstrated by differences in binding of Z1 and 38. These ligand variations could also maximize interactions with the pyrophosphate-sensing stem to improve the switching ability of the riboswitch. Our screening strategy ultimately identified binders of the riboswitch and revealed a ligand binding site not targeted by a natural ligand of the riboswitch. Z1 is not a synthetic mimetic of the natural riboswitch ligand, should not bind TPP-dependent human enzymes, and thus provides an entry to using this riboswitch as an RNA-specific genetic control element (46, 47).
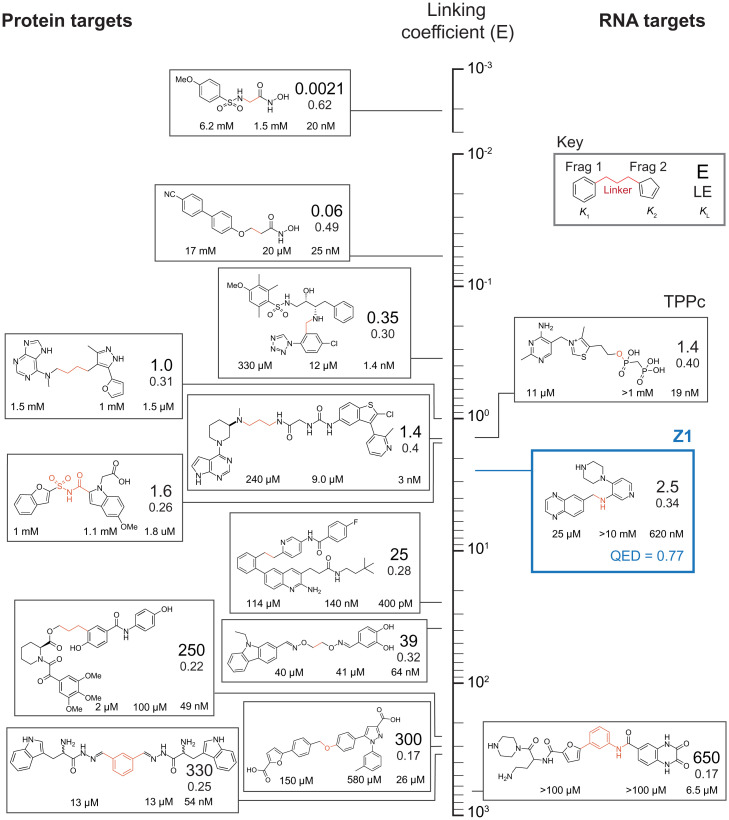
There have been many efforts designed to exploit cooperativity between fragments to obtain tight-binding ligands that target proteins; in contrast, targeting RNA is in its infancy. This study fully applies a fragment approach to elaborate a fragment hit and create a potent, druglike ligand that binds RNA. We explored how well our SHAPE-based screening strategy, melded with fragment linking, compared with prior (protein-focused) efforts. We ranked compounds discovered previously using fragment-based strategies according to their linking coefficients (E), a measure of how well the entire system functions together when linked (22, 48) (Fig. 8; expanded in SI Appendix, Table S3). In the absence of positive or negative contributing factors, the binding energies of the two fragments are exactly additive, the linker is inert, and E is equal to 1.0. Cooperative effects or favorable linker interactions decrease E. E values can vary by orders of magnitude in protein systems. The linking coefficient for Z1 is 2.5, slightly above average for linked (protein-targeted) ligands in the academic literature. Z1 has a ligand efficiency (LE), the free energy of binding divided by the number of nonhydrogen atoms, that compares favorably to examples of linked fragment ligands targeting proteins (Fig. 8). By these metrics, Z1 performs nearly as well TPPc—a ligand closely related to the native TPP riboswitch ligand, whose fragment-like properties we investigated recently (23)—and is actually superior to TPP, the natural ligand of the riboswitch. We infer that fragment-based ligand discovery, especially as efficiently implemented by SHAPE-enabled multiplexed screening, holds significant promise to enable rapid development of unique small molecules that target the vast world of ligandable RNA structures.
Fig. 8.
Comparison of fragment-linker-fragment ligands, ordered by their linking coefficient (E). Cooperative (more efficient) linking corresponds to smaller E values (top of vertical axis). Values are shown on a logarithmic axis. Z1 is highlighted in blue. Dissociation constants for individual fragments and linked ligand are denoted below component fragments; E-value and LE are shown (see Key). Covalent linkage introduced between fragments is highlighted in red. Structures for component fragments are detailed in SI Appendix, Table S3, and values for Z1 are reported relative to initial fragment hits.
Materials and Methods
Compounds.
The fragment screening library was obtained from Maybridge as a subset of their Ro3 diversity fragment library and contained 1,500 compounds dissolved in DMSO at 50 mM. Most of these compounds adhere to the “rule of three” for fragment compounds; having a molecular mass <300 Da, containing ≤3 hydrogen bond donors and ≤3 hydrogen bond acceptors, and having a ClogP ≤3.0. All compounds used for ITC, with the exception of those listed in the Chemical Synthesis section, were purchased from Millipore-Sigma and used without further purification.
Construct Design.
The screening construct was designed to allow incorporation of a wide variety of one or more internal target RNA motifs. For this study, two motifs were present in the construct: the TPP riboswitch domain (28) and a pseudoknot from the 5′-UTR of the dengue virus (27). The design for the complete construct sequence, including structure cassettes, the RNA barcode helix, and the two test RNA structures (separated by a six-nucleotide linker), was evaluated using RNAstructure (49). To reduce the likelihood of the two test structures interacting, a small number of sequence alterations were made to discourage misfolded structures predicted by RNAstructure while retaining the native fold (SI Appendix, Fig. S1). The structure of the final construct was confirmed by SHAPE-MaP.
RNA barcodes were designed to fold into self-contained hairpins (SI Appendix, Fig. S1). All possible permutations of RNA barcodes were computed and folded in the context of the full construct sequence, and any barcodes that had the potential to interact with another part of the RNA construct were removed from the set. Barcoded constructs were probed by SHAPE-MaP using the “no ligand” protocol and folded using RNAstructure with SHAPE reactivity constraints to confirm that barcode helices folded into the desired self-contained hairpins.
Preparation of RNA.
DNA templates (Integrated DNA Technologies) for in vitro transcription encoded the target construct sequence (containing the dengue pseudoknot sequence, single-stranded linker, and TPP riboswitch sequence) and flanking structure cassettes (26): 5′-GTGGG CACTT CGGTG TCCAC ACGCG AAGGA AACCG CGTGT CAACT GTGCA ACAGC TGACA AAGAG ATTCC TAAAA CTCAG TACTC GGGGT GCCCT TCTGC GTGAA GGCTG AGAAA TACCC GTATC ACCTG ATCTG GATAA TGCCA GCGTA GGGAA GTGCT GGATC CGGTT CGCCG GATCA ATCGG GCTTC GGTCC GGTTC-3′; primer binding sites are underlined. Forward PCR primers containing unique RNA barcodes and the T7 promoter sequence were used to individually add RNA barcodes to each of 96 constructs in individual PCR reactions. A sample forward primer sequence, with barcode nucleotides in bold and the primer binding site underlined, is 5′-GAAAT TACGA CTCAC TATAG GTCGC GAGTA ATCGC GACCG GCGCT AGAGA TAGTG CCGTG GGCAC TTCGG TGTC-3′.
DNA was amplified by PCR using 200 µM dNTP mix (New England Biolabs), 500 nM forward primer, 500 nM reverse primer, 1 ng DNA template, 20% (vol/vol) Q5 reaction buffer, and 0.02 U/µL Q5 hot-start high-fidelity polymerase (New England Biolabs) to create templates for in vitro transcription. DNA was purified (PureLink Pro 96 PCR Purification Kit; Invitrogen) and quantified (Quant-iT dsDNA high sensitivity assay kit; Invitrogen) on a Tecan Infinite M1000 Pro microplate reader.
In vitro transcription was carried out in 96-well plate format with each well containing 100 µL total reaction volume. Each well contained 5 mM NTPs (New England Biolabs), 0.02 U/µL inorganic pyrophosphatase (yeast, New England Biolabs), 0.05 mg/mL T7 polymerase in 25 mM MgCl2, 40 mM Tris, pH 8.0, 2.5 mM spermidine, 0.01% Triton, 10 mM dithiothreitol (DTT), and 200 to 800 nM of a uniquely barcoded DNA template (generated by PCR). Reactions were incubated at 37 °C for 4 h, then treated with TurboDNase (RNase-free, Invitrogen) at a final concentration of 0.04 U/µL and incubated at 37 °C for 30 min, followed by a second DNase addition to a total final concentration of 0.08 U/µL and an additional 30-min incubation at 37 °C. Enzymatic reactions were halted by the addition of EDTA to a final concentration of 50 mM and placed on ice. RNA was purified (Agencourt RNAclean XP magnetic beads; Beckman Coulter) in a 96-well format and resuspended in 10 mM Tris pH 8.0, 1 mM EDTA. RNA concentrations were quantified (Quant-iT RNA broad range assay kit; Invitrogen) on a Tecan Infinite M1000 Pro microplate reader, and RNAs in each well were individually diluted to 1 pmol/µL. RNA was stored at −80 °C.
Chemical Modification and Screening.
Screening experiments were carried out in 25 µL in 96-well plate format on a Tecan Freedom Evo-150 liquid handler equipped with an 8-channel air displacement pipetting arm, disposable filter tips, robotic manipulator arm, and an EchoTherm RIC20 remote controlled heating/cooling dry bath (Torrey Pines Scientific).
For the first fragment-ligand screen, 5 pmol RNA per well were diluted to 19.6 µL in RNase-free water on a 4 °C cooling block. The plate was heated at 95 °C for 2 min, immediately followed by snap cooling at 4 °C for 5 min. To each well was added 19.6 µL of 2× folding buffer (final concentrations: 50 mM Hepes pH 8.0, 200 mM potassium acetate, and 10 mM MgCl2), and plates were incubated at 37 °C for 30 min. For the second fragment-ligand screen, 24.3 µL folded RNA per well were added to 2.7 µL of primary binding fragment in DMSO to a final concentration of 10× the Kd of the fragment, and samples were incubated at 37 °C for 10 min. To combine the target RNA with fragment, 24.3 µL RNA solution or RNA plus primary binding fragment were added to wells containing 2.7 µL 10× screening fragments (in DMSO to yield a final fragment concentration of 1 mM). Solutions were mixed thoroughly by pipetting and incubated for 10 min at 37 °C. For SHAPE probing, 22.5 µL RNA-fragment solution from each well of the screening plate were added to 2.5 µL 10× SHAPE reagent in DMSO on a 37 °C heating block and rapidly mixed by pipetting to achieve homogenous distribution of the SHAPE reagent with the RNA. After the appropriate reaction time, samples were placed on ice. For the first fragment screen, 1-methyl-7-nitroisatoic anhydride was used as the SHAPE reagent at a final concentration of 10 mM with reaction for 5 min. For the second fragment screen, 5-nitroisatoic anhydride (50) was used as the SHAPE reagent at a final concentration of 25 mM with reaction for 15 min. Excess fragments, solvent, and hydrolyzed SHAPE reagent were removed using AutoScreen-A 96-well plates (GE Healthcare Life Sciences), and 5 µL modified RNA from each well of a 96-well plate were pooled into a single sample per plate for sequencing library preparation.
Each screen consisted of 19 fragment test plates, 2 plates containing a distribution of positive (fragment 2, final concentration 1 mM) and negative (solvent, DMSO) controls, and 1 negative SHAPE control plate treated with solvent (DMSO) instead of SHAPE reagent. For hit validation experiments, well locations of each hit fragment were changed to control for well location and RNA barcode effects.
Library Preparation and Sequencing.
Reverse transcription was performed on pooled, modified RNA in a 100-µL volume. To 71 µL of pooled RNA was added 6 µL reverse transcription primer to achieve a final concentration of 150 nM primer, and the sample was incubated at 65 °C for 5 min and then placed on ice. To this solution, 6 µL 10× first-strand buffer (500 mM Tris pH 8.0, 750 mM KCl), 4 µL 0.4 M DTT, 8 µL dNTP mix (10 mM each), and 15 µL 500 mM MnCl2 were added, and the solution was incubated at 42 °C for 2 min before adding 8 µL SuperScript II Reverse Transcriptase (Invitrogen). The reaction was incubated at 42 °C for 3 h, followed by a 70 °C heat inactivation for 10 min before being placed on ice. The resulting cDNA product was purified (Agencourt RNAclean magnetic beads; Beckman Coulter), eluted into RNase-free water, and stored at −20 °C. The sequence of the reverse transcription primer was 5′- CGGGC TTCGG TCCGG TTC-3′.
DNA libraries were prepared for sequencing using a two-step PCR to amplify the DNA and to add the necessary TruSeq adapters (25). DNA was amplified by PCR using 200 µM dNTP mix (New England Biolabs), 500 nM forward primer, 500 nM reverse primer, 1 ng cDNA or double-stranded DNA template, 20% (vol/vol) Q5 reaction buffer (New England Biolabs), and 0.02 U/µL Q5 hot-start high-fidelity polymerase (New England Biolabs). Excess unincorporated dNTPs and primers were removed by affinity purification (Agencourt AmpureXP magnetic beads; Beckman Coulter; at a 0.7:1 sample-to-bead ratio). DNA libraries were quantified (Qubit dsDNA High Sensitivity assay kit; Invitrogen) on a Qubit fluorometer (Invitrogen), checked for quality (Bioanalyzer 2100 on-chip electrophoresis instrument; Agilent), and sequenced on an Illumina NextSeq 550 high-throughput sequencer.
The SHAPE-MaP library preparation amplicon-specific forward primer was 5′-CCCTA CACGA CGCTC TTCCG ATCTN NNNNG GCCTT CGGGC CAAGG A-3′. The SHAPE-MaP library preparation amplicon-specific reverse primer was 5′-GACTG GAGTT CAGAC GTGTG CTCTT CCGAT CTNNN NNTTG AACCG GACCG AAGCC CGATT T-3′. The sequences overlapping the RNA screening construct are underlined.
ITC.
ITC experiments were performed using a Microcal PEAQ-ITC automated instrument (Malvern Analytical) under RNase-free conditions (51). In vitro transcribed RNA was exchanged into folding buffer containing 100 mM CHES, pH 8.0, 200 mM potassium acetate, and 3 mM MgCl2 using centrifugal concentration (Amicon Ultra centrifugal filters, 10K MWCO, Millipore-Sigma). Ligands were dissolved into the same buffer (to minimize heat of mixing upon addition of ligand to RNA) at a concentration 10 to 20 times the desired experimental concentration of RNA. RNA concentration was quantified (Nanodrop UV-VIS spectrometer; ThermoFisher Scientific) and diluted to 1 to 10 times the expected Kd in buffer, and the diluted RNA was requantified to confirm the final experimental RNA concentration. The RNA, diluted in folding buffer, was heated at 65 °C for 5 min, placed on ice for 5 min, and allowed to fold at 37 °C for 15 min. If needed, the primary binding ligand (e.g., 2) was prebound to the RNA by adding 0.1 volume at 10 times the desired final concentration of the bound ligand, followed by incubation at room temperature for 10 min.
Each ITC experiment involved two runs: one in which the ligand was titrated into RNA (the experimental trace) and one in which the same ligand was titrated into buffer (the control trace). ITC experiments were performed using the following parameters: 25 °C cell temperature, 8 µCal/sec reference power, 750 RPM stirring speed, high feedback mode, and 0.2 µL initial injection, followed by 19 injections of 2 µL. Each injection required 4 s to complete, and there was a 180-s spacing between injections.
ITC data were analyzed using MicroCal PEAQ-ITC Analysis Software (Malvern Analytical). First, the baseline for each injection peak was manually adjusted to resolve any incorrectly selected injection endpoints. Second, the control trace was subtracted from the experimental trace by point-to-point subtraction. Third, a least-squares regression line was fit to the data using the Levenberg-Marquardt algorithm. Confidence intervals (95%) were typically less than ±30% of the fit value; floating N values were generally 1.0 ± 0.5 for compounds binding with Kd < 300 μM. In the case of weakly binding ligands (Kd > 500 µM), N was set to 1.0 to enable fitting of low c-value curves.
Linking Parameter and LE.
E is the affinity of the linked compound (L) relative to coupled binding by the constituent ligands (A and B) and was calculated as E = KL/(KAKB). LE is the binding energy per (nonhydrogen) heavy atom (HA) of a ligand to its binding partner and is calculated as LE = –RT ln KL/HA (in units of kcal/mol).
Chemical Synthesis.
Reaction schemes for all synthesized ligands are detailed in SI Appendix, Fig. S3.
X-Ray Crystallography.
Compounds 16 and 17 were crystallized using TPP riboswitch RNA prepared by in vitro transcription as described (28). TPP riboswitch RNA (0.2 mM) and 16 or 17 (2 mM) were heated in a buffer containing 50 mM potassium acetate (pH 6.8) and 3 (compound 16) or 5 (compound 17) mM MgCl2 at 60 °C for 3 min, snap cooled in crushed ice, and incubated at 4 °C for 30 min prior to crystallization. For crystallization, 1.0 μL of the RNA-ligand complex was mixed with 1.0 μL reservoir solution containing 0.1 M sodium acetate (pH 4.8), 0.35 M ammonium acetate, and 28% (wt/vol) PEG4000. Cocrystals with 16 appeared in 2 mo, while crystals with 17 grew in 2 wk. Compounds 37 and 38 were crystallized using an RNA described previously (29). The RNA (0.15 mM) was incubated in a buffer containing 5 mM Tris⋅HCl, pH 8.0, 3 mM MgCl2, 10 mM NaCl, 0.1 M KCl, and 0.5 mM spermine with 1.0 mM compound 37 or 0.7 mM compound 38 at 37 °C for 30 min, followed by at 4 °C for 60 min prior to crystallization. For crystallization, 1.5 μL of the RNA-ligand complex was mixed with 0.75 μL reservoir solution. For 37, reservoir solution was 50 mM Bis-Tris, pH 6.5, 0.5 M ammonium chloride, 15 mm MnCl2, and 28% (wt/vol) PEG2000. For 38, reservoir solution was 50 mM Bis-Tris, pH 6.5, 0.45 M ammonium chloride, 10 mm MnCl2, and 30% (wt/vol) PEG2000. Crystals grew in 1 wk. Crystallization was performed at 291 K by hanging drop vapor diffusion. The crystals were cryoprotected in mother liquor supplemented with 15% of glycerol and small-molecule compound at the concentration used for preparing the complexes prior to snap freezing in liquid nitrogen. Data were processed with HKL2000 (52). The structure was solved by molecular replacement using Phenix (53) and the 2GDI or 2HOJ riboswitch RNA structures (28, 29). The structures were refined in Phenix. Organic ligands, water molecules, and ions were added based on Fo-Fc and 2Fo-Fc electron density maps and verified with simulated annealing omit maps.
Cotranscriptional Ligand Binding and Conformation Switching Assays.
Assays were performed as reported (37, 38). Briefly, the DNA template, including an added promoter (lowercase), was created by PCR from E. coli genomic DNA (primers: forward, 5′-gggca cccca ggctt tacac tttat gcttc cggct cgtat aatgt gtggC TGCGA TTTAT CATCG CAACC AAAC; reverse, 5′-TTGCG CTGAA CCCAG CAGGT CGACT). In vitro transcription assays were performed in 20 mM Tris⋅HCl (pH 8.0), 20 mM MgCl2, 20 mM NaCl, 14 mM 2-mercaptoethanol, and 0.1 mM EDTA. The DNA template (3 pmol), sigma70 factor (12 pmol), and E. coli RNA polymerase (6 pmol) were incubated at 37 °C for 5 min. The CUGC tetranucleotide (100 μM), ATP and GTP nucleotides (25 μM) and [α-32P]-UTP were added, and the reaction incubated at 37 °C for 10 min. Samples were passed through G50 columns to remove free nucleotides. Reactions were completed by adding all four nucleotides (25 μM final concentration) with heparin (6 mg/μL) to create single-round transcription conditions. Tested ligands were added during this step to final concentrations of 0.2 nM to 1 mM. Aliquots (8 µL) of the transcription reactions were then mixed with DNA probe (2 µL), final concentration 20 μM (5′-CCGAGTCGTT), for 5 min at 37 °C. Following this incubation, RNase H cleavage assays were performed by adding RNase H (0.12 U) in 5 mM Tris⋅HCl (pH 8.0), 20 mM MgCl2, 100 mM KCl, 50 μM EDTA, and 10 mM 2-mercaptoethanol (total volume of each RNase H reaction was 10 µL) at 37 °C for 5 min. Reactions were stopped by adding 20 μL stop solution (95% formamide, 20 mM EDTA, and 0.4% SDS). Samples were resolved by (10%) denaturing polyacrylamide gel electrophoresis; experiments were performed in triplicate.
Supplementary Material
Acknowledgments
This work was supported by NIH grants to K.M.W. (R01-EUREKA GM098662 and R01 AI068462) and A.S. (R01 GM112940) and by a Canadian Institutes of Health Research grant to D.A.L. K.L. was supported by the UNC Lineberger Comprehensive Cancer Center. D.H. was supported by an RNA Innovation Fellowship. ITC experiments were performed at the UNC Macromolecular Interactions Facility (NCI P30CA016086). This work used NE-CAT beamlines (GM124165), a Pilatus detector (RR029205), and a Eiger detector (OD021527) at the Advanced Photon Source (DE-AC02-06CH11357) of the Argonne National Laboratory and FMX (17ID-2) and AMX (17ID-1) beamlines at NSLS-II (Brookhaven National Laboratory), supported by the NIH NIGMS (1P30GM133893) and BER-BO 070. NSLS-II is supported by DOE, BES-FWP-PS001.
Footnotes
Competing interest statement: The authors declare a competing interest. K.M.W. is an advisor to and holds equity in Ribometrix and MeiraGTx. Patents have been filed on compounds described herein.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122660119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. The atomic coordinates for the complex of the TPP riboswitch with fragment and linked ligands have been deposited in the Protein Data Bank (PDB) under accession codes 7TZR [16], 7TZS [17], 7TZT [37], and 7TZU [38].
References
- 1.Hajduk P. J., Huth J. R., Tse C., Predicting protein druggability. Drug Discov. Today 10, 1675–1682 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Vukovic S., Huggins D. J., Quantitative metrics for drug-target ligandability. Drug Discov. Today 23, 1258–1266 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Batey R. T., Rambo R. P., Doudna J. A., Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed. Engl. 38, 2326–2343 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Warner K. D., Hajdin C. E., Weeks K. M., Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 17, 547–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharp P. A., The centrality of RNA. Cell 136, 577–580 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Kozak M., Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361, 13–37 (2005). [DOI] [PubMed] [Google Scholar]
- 7.McCown P. J., Corbino K. A., Stav S., Sherlock M. E., Breaker R. R., Riboswitch diversity and distribution. RNA 23, 995–1011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cech T. R., Steitz J. A., The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Adams B. D., Parsons C., Walker L., Zhang W. C., Slack F. J., Targeting noncoding RNAs in disease. J. Clin. Invest. 127, 761–771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsui M., Corey D. R., Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 16, 167–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan L., Disney M. D., Recent advances in developing small molecules targeting RNA. ACS Chem. Biol. 7, 73–86 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Connelly C. M., Moon M. H., J. S. Schneekloth, Jr, The emerging role of RNA as a therapeutic target for small molecules. Cell Chem. Biol. 23, 1077–1090 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray C. W., Rees D. C., The rise of fragment-based drug discovery. Nat. Chem. 1, 187–192 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Doak B. C., Norton R. S., Scanlon M. J., The ways and means of fragment-based drug design. Pharmacol. Ther. 167, 28–37 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Chen L., Cressina E., Leeper F. J., Smith A. G., Abell C., A fragment-based approach to identifying ligands for riboswitches. ACS Chem. Biol. 5, 355–358 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Cressina E., Chen L., Abell C., Leeper F. J., Smith A. G., Fragment screening against the thiamine pyrophosphate riboswitch thiM. Chem. Sci. (Camb.) 2, 157–165 (2011). [Google Scholar]
- 17.Warner K. D., et al. , Validating fragment-based drug discovery for biological RNAs: Lead fragments bind and remodel the TPP riboswitch specifically. Chem. Biol. 21, 591–595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeiger M., et al. , Fragment based search for small molecule inhibitors of HIV-1 Tat-TAR. Bioorg. Med. Chem. Lett. 24, 5576–5580 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Bottini A., et al. , Targeting influenza A virus RNA promoter. Chem. Biol. Drug Des. 86, 663–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sreeramulu S., et al. , Exploring the druggability of conserved RNA regulatory elements in the SARS-CoV-2 genome. Angew. Chem. Int. Ed. Engl. 60, 19191–19200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter C. A., Anderson H. L., What is cooperativity? Angew. Chem. Int. Ed. Engl. 48, 7488–7499 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Ichihara O., Barker J., Law R. J., Whittaker M., Compound design by fragment-linking. Mol. Inform. 30, 298–306 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Zeller M. J., et al. , Subsite ligand recognition and cooperativity in the TPP riboswitch: Implications for fragment-linking in RNA ligand discovery. ACS Chem. Biol. 17, 438–448 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siegfried N. A., Busan S., Rice G. M., Nelson J. A. E., Weeks K. M., RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smola M. J., Rice G. M., Busan S., Siegfried N. A., Weeks K. M., Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 10, 1643–1669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merino E. J., Wilkinson K. A., Coughlan J. L., Weeks K. M., RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Liu Z.-Y., et al. , Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization. J. Virol. 87, 6804–6818 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serganov A., Polonskaia A., Phan A. T., Breaker R. R., Patel D. J., Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 441, 1167–1171 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards T. E., Ferré-D’Amaré A. R., Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure 14, 1459–1468 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Thore S., Frick C., Ban N., Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. J. Am. Chem. Soc. 130, 8116–8117 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Busan S., Weeks K. M., Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA 24, 143–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woolson R., Statistical Methods for the Analysis of Biomedical Data (John Wiley & Sons, 1987). [Google Scholar]
- 33.Jhoti H., Williams G., Rees D. C., Murray C. W., The ‘rule of three’ for fragment-based drug discovery: Where are we now? Nat. Rev. Drug Discov. 12, 644–645 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Chen L., et al. , Probing riboswitch-ligand interactions using thiamine pyrophosphate analogues. Org. Biomol. Chem. 10, 5924–5931 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Codd R., Traversing the coordination chemistry and chemical biology of hydroxamic acids. Coordination Chemistry Reviews 252, 1387–1408 (2008). [Google Scholar]
- 36.Warner K. D., Ferré-D’Amaré A. R., Crystallographic analysis of TPP riboswitch binding by small-molecule ligands discovered through fragment-based drug discovery approaches. Methods Enzymol. 549, 221–233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chauvier A., et al. , Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation. Nat. Commun. 8, 13892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chauvier A., et al. , Monitoring RNA dynamics in native transcriptional complexes. Proc. Natl. Acad. Sci. U.S.A. 118, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastet L., et al. , Translational control and Rho-dependent transcription termination are intimately linked in riboswitch regulation. Nucleic Acids Res. 45, 7474–7486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickiser J. K., Winkler W. C., Breaker R. R., Crothers D. M., The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 18, 49–60 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Bickerton G. R., Paolini G. V., Besnard J., Muresan S., Hopkins A. L., Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blount K. F., Breaker R. R., Riboswitches as antibacterial drug targets. Nat. Biotechnol. 24, 1558–1564 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Barrick J. E., Breaker R. R., The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8, R239 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jencks W. P., On the attribution and additivity of binding energies. Proc. Natl. Acad. Sci. U.S.A. 78, 4046–4050 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olejniczak E. T., et al. , Stromelysin inhibitors designed from weakly bound fragments: Effects of linking and cooperativity. J. Am. Chem. Soc. 119, 5828–5832 (1997). [Google Scholar]
- 46.Etzel M., Mörl M., Synthetic riboswitches: From plug and pray toward plug and play. Biochemistry 56, 1181–1198 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Tickner Z. J., Farzan M., Riboswitches for controlled expression of therapeutic transgenes delivered by adeno-associated viral vectors. Pharmaceuticals 14, 1–29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borsi V., Calderone V., Fragai M., Luchinat C., Sarti N., Entropic contribution to the linking coefficient in fragment based drug design: A case study. J. Med. Chem. 53, 4285–4289 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Reuter J. S., Mathews D. H., RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11, 129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Busan S., Weidmann C. A., Sengupta A., Weeks K. M., Guidelines for SHAPE reagent choice and detection strategy for RNA structure probing studies. Biochemistry 58, 2655–2664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilbert S. D., Batey R. T., Monitoring RNA-ligand interactions using isothermal titration calorimetry. Methods Mol. Biol. 540, 97–114 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 53.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. The atomic coordinates for the complex of the TPP riboswitch with fragment and linked ligands have been deposited in the Protein Data Bank (PDB) under accession codes 7TZR [16], 7TZS [17], 7TZT [37], and 7TZU [38].