Significance
Genome-wide association studies have identified two major risk loci for age-related macular degeneration (AMD) on chromosome (Chr) 1 and Chr10. Here, we use proteomics to analyze submacular stromal tissue punches from older eye donors without AMD, comparing tissue from donors who were homozygous for high-risk alleles at Chr1 or Chr10 with tissue from donors with low risk at these two loci. A common change found in Chr1/Chr10–risk eyes was increased mast cell proteases, and immunohistochemistry confirmed the presence of increased mast cell numbers. This study, therefore, provides a unifying mechanistic link between Chr1 and Chr10 risk and suggests that mast cell infiltration of the choroid and degranulation are early events in AMD pathogenesis.
Keywords: age-related macular degeneration, proteomics, mast cells
Abstract
Age-related macular degeneration (AMD) is a leading cause of visual loss. It has a strong genetic basis, and common haplotypes on chromosome (Chr) 1 (CFH Y402H variant) and on Chr10 (near HTRA1/ARMS2) contribute the most risk. Little is known about the early molecular and cellular processes in AMD, and we hypothesized that analyzing submacular tissue from older donors with genetic risk but without clinical features of AMD would provide biological insights. Therefore, we used mass spectrometry–based quantitative proteomics to compare the proteins in human submacular stromal tissue punches from donors who were homozygous for high-risk alleles at either Chr1 or Chr10 with those from donors who had protective haplotypes at these loci, all without clinical features of AMD. Additional comparisons were made with tissue from donors who were homozygous for high-risk Chr1 alleles and had early AMD. The Chr1 and Chr10 risk groups shared common changes compared with the low-risk group, particularly increased levels of mast cell–specific proteases, including tryptase, chymase, and carboxypeptidase A3. Histological analyses of submacular tissue from donors with genetic risk of AMD but without clinical features of AMD and from donors with Chr1 risk and AMD demonstrated increased mast cells, particularly the tryptase-positive/chymase-negative cells variety, along with increased levels of denatured collagen compared with tissue from low–genetic risk donors. We conclude that increased mast cell infiltration of the inner choroid, degranulation, and subsequent extracellular matrix remodeling are early events in AMD pathogenesis and represent a unifying mechanistic link between Chr1- and Chr10-mediated AMD.
Age-related macular degeneration (AMD) is a leading cause of visual loss; in 2015, 8.4 million people globally were estimated to have moderate to severe sight loss from AMD (1). Early AMD is characterized by the formation of drusen and pigmentary changes involving the retinal pigment epithelium (RPE). It can then progress to late-stage disease, which has two forms: geographic atrophy (the end stage of the underlying disease) and macular neovascularization (a late-stage complication). Genetic factors are very important contributors to AMD risk (2). Most of the genetic risk is attributed to two loci, one at chromosome 10q26 (Chr10q26) around the ARMS2 and HTRA1 genes and the other at Chr 1q32 around the CFH (complement factor H) gene (3) (hereafter referred to as Chr10 and Chr1 risk loci, respectively). The rs10490924 single-nucleotide polymorphism (SNP) is associated with Chr10 risk; it is present in 22% of individuals of European ancestry, and people who are homozygous for this change have a 10-fold increase in risk of late AMD (4). A haplotype including the rs1061170 SNP is associated with Chr1 risk, and this change corresponds to a coding variant (Y402H) in the CFH gene; it is present in ∼36% of Europeans, and homozygous individuals have a 7.4-fold increased risk of developing AMD (5). The association of this and other complement genes, including CFI, CFB, and C3, with AMD (3) highlights the central role of complement in the pathogenesis of this disorder, but the mechanism underpinning the Chr10 risk remains poorly understood. Apart from age, other nongenetic risk factors for AMD include smoking and high body mass index (2).
While in late-stage AMD, there is damage to all macular retinal layers and underlying choroid, the location and mechanisms of disease initiation and early progression are poorly understood. Soft drusen are an early clinical feature of the disease, and these form within Bruch’s membrane between the RPE basal lamina and the inner collagenous layer of Bruch’s membrane (6). Older histological research demonstrated that choriocapillary degeneration occurs in aging and is accentuated in AMD (7), and these findings have been supported by more recent studies including investigational clinical imaging (8–11). Studies have shown that increasing age and high-risk CFH (Chr1) genotypes are associated with increased levels of terminal complement complex (C5b9, membrane attack complex; formed as a result of complement activation) in Bruch’s membrane and the stroma of the choriocapillaris surrounding the capillaries, including the intercapillary septa (9, 12, 13). Therefore, we investigated the association between genetic risk of AMD and biochemical and cellular changes in the Bruch’s membrane/inner choroid.
Quantitative proteomics techniques, which compare the relative amounts of multiple proteins between different samples in an untargeted manner, are valuable for elucidating disease mechanisms. In this study, we applied this technique to analyze the composition of submacular stromal tissue punches from genotyped donor human eyes. Our aim was to determine whether genetic risk at Chr1 or Chr10 altered the proteome of the tissue as compared with tissue from donors without the common risk haplotypes at these loci. On discovering increased mast cell proteases in tissue from both Chr1 and Chr10 genetic high-risk groups, we used histology/immunohistochemistry and detected raised levels of inner choroidal mast cells and extracellular matrix damage in eyes with genetic risk of AMD both without and with early AMD.
Results
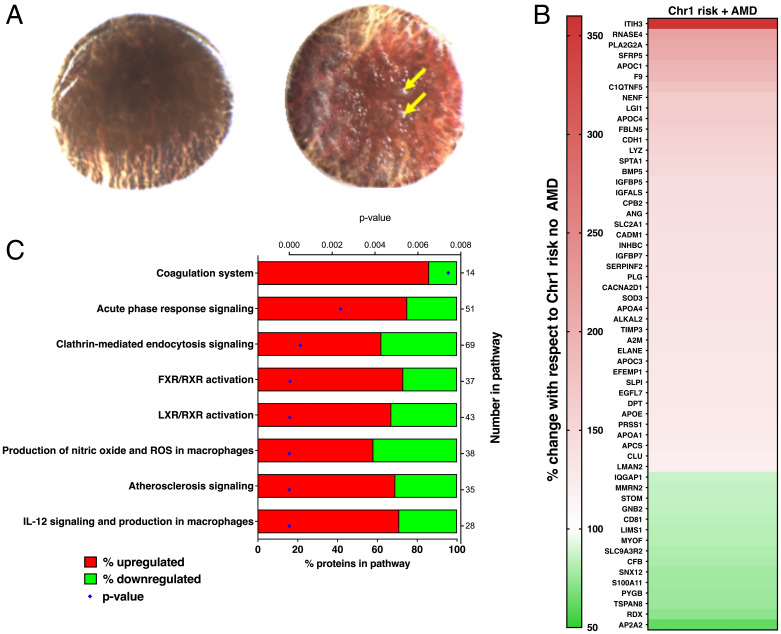
Over a 2-y period (2014 to 2016), 632 pairs of human donor eyes were processed and genotyped (including testing for five SNPs that were used to interrogate for Chr1 and Chr10 risk) (SI Appendix, Table S1). Submacular stromal tissue punches from 30 of these donors were subsequently used for relative quantitation proteomics; these samples were selected based upon genotype, presence or absence of AMD, and donor age (SI Appendix, Table S2). Submacular stromal tissue punches from donors without AMD (phenotyping for the presence or absence of AMD is in Methods) were compared from donors who were 1) homozygous for high-risk alleles at Chr1 and homozygous for low-risk alleles at Chr10 (“Chr1 risk group,” n = 8); 2) homozygous for high-risk alleles at Chr10 and homozygous for low-risk alleles at Chr1 (“Chr10 risk group,” n = 8); or 3) homozygous for low-risk alleles at both Chr1 and Chr10 (“low-risk group,” n = 8). A further subgroup that was homozygous for high-risk alleles at Chr1 and homozygous for low-risk alleles at Chr10 but with early AMD changes (“Chr1 risk group with AMD,” n = 6) (Fig. 1A shows representative images of macular drusen) was analyzed. Donors were matched as closely as possible considering sex, age, and tissue postmortem time. The mean ages of the groups were low risk, 77.3 y; Chr1 risk, 72 y; Chr10 risk, 69.1 y; and Chr1 risk with AMD, 82.3 y. ANOVA post hoc analysis revealed statistical differences in mean age between the low-risk and Chr10 risk groups, the Chr1 risk and Chr1 with AMD groups, and the Chr10 and Chr1 with AMD groups (SI Appendix, Table S3). This was unavoidable given the small numbers of donors with the specific genotypes analyzed.
Fig. 1.
(A) Representative submacular tissue punches after removal of neurosensory retina from donors with no AMD (Left) and early AMD (Right); yellow arrows indicate drusen that had a diameter of greater than 63 µm. (B) Proteins with significantly changed levels when comparing submacular stromal tissue punches from Chr1 high-risk donors with and without AMD changes; red indicates increased with AMD, and green indicates decreased with AMD. (C) Ingenuity pathways analysis showing altered canonical pathways in submacular stromal tissue punches from Chr1 risk donors comparing with and without AMD. FXR, farnesoid X receptor; LXR, liver X receptor; ROS, reactive oxygen species.
Comparison of Submacular Stromal Tissue Punch Proteomes.
After exclusion of proteins identified with two or fewer spectra, the isobaric tag for relative and absolute quantitation (iTRAQ) mass spectrometry (MS) experiment identified ∼1,750 total proteins (Dataset S1). The study aimed to investigate extracellular proteins, so cells were removed as far as possible from the tissue; then, the residual cells were lysed by washing the tissue with distilled water during sample preparation. However, a systematic search of all detected proteins, using the GeneCards Human Gene Database as a reference, identified residual intracellular proteins. As the focus of this study was to analyze extracellular proteins, any proteins whose localization according to GeneCards was purely intracellular were removed prior to further analysis, but proteins that localized to plasma membranes were retained along with the extracellular proteins, leaving ∼800 proteins (Table 1); these are listed in Dataset S2. Bayesian mixed effects modeling was used to identify differential protein expression between the respective groups applying a global false discovery rate (gFDR) of <0.05. In samples without macroscopic AMD changes, when the Chr1 risk group was compared with the low-risk group, 20 proteins (2.5%) were significantly different (17 up-regulated and 3 down-regulated); when the Chr10 risk group was compared with the low-risk group, 16 proteins (2%) were significantly different (11 up-regulated and 5 down-regulated). These data are shown in Table 2 and Dataset S3. Strikingly, seven of the differentially expressed proteins were common to both the Chr1 and Chr10 risk groups, and three of these, which were also among the most highly differentially expressed proteins, were the mast cell–specific proteases chymase (CMA1), carboxypeptidase A3 (CPA3), and tryptase (TPSAB1/TPSB2). The serine protease cathepsin G (CTSG), which can be synthesized by various cell types including mast cells, was also significantly up-regulated. Adaptor Related Protein Complex 2 Subunit Alpha 2 (AP2A2), a subunit of the larger AP2 adaptor involved in endocytosis, and endothelial lipase (LIPG) were both significantly up-regulated in both the Chr1 and Chr10 risk groups, while hyaluronan and proteoglycan link protein 1 (HAPLN1) was significantly down-regulated in both groups.
Table 1.
Total numbers in each group analyzed by Bayesian mixed effects modeling and numbers of significantly up-regulated and down-regulated proteins
| Chr1 risk vs. low risk | Chr10 risk vs. low risk | Chr1 risk ± AMD | |
|---|---|---|---|
| Total proteins | 798 | 798 | 769 |
| Up-regulated (gFDR < 0.05) | 17 | 11 | 43 |
| Down-regulated (gFDR < 0.05) | 3 | 5 | 13 |
Table 2.
Proteins with significantly altered levels when comparisons were made between the Chr1 or Chr10 risk groups and the low-risk group
| Protein (gene identification) | Chr1 risk fold change vs. low risk | Chr10 risk fold change vs. low risk |
|---|---|---|
| CMA1* | 1.70 † | 1.51 † |
| CPA3* | 1.74 † | 1.59 † |
| CTSG* | 1.46 † | 1.50 † |
| TPSAB1/TPSB2* | 1.33 † | 1.70 † |
| LIPG | 1.33 † | 1.35 † |
| AP2A2 | 1.39 † | 1.38 † |
| HAPLN1 | 0.59 ‡ | 0.72 ‡ |
| BPIFA1 | 1.90 † | 1.25 |
| FCGRT | 1.48 † | 1.40 |
| IGKV3-20 | 1.47 † | 1.24 |
| DEFA1 | 1.34 † | 1.26 |
| PCOLCE | 1.29 † | 1.20 |
| ANPEP | 1.28 † | 1.14 |
| S100A9 | 1.26 † | 0.97 |
| LIMS1 | 1.24 † | 1.16 |
| NRP1 | 1.24 † | 1.16 |
| CFB | 1.22 † | 0.95 |
| COL15A1 | 1.20 † | 1.08 |
| SORBS1 | 0.86 ‡ | 0.96 |
| LMAN2 | 0.81 ‡ | 0.96 |
| MMP28 | 1.35 | 1.55 † |
| ADIRF | 1.10 | 1.45 † |
| FBLN5 | 1.13 | 1.31 † |
| C9orf139 | 1.20 | 1.33 † |
| PRKACA | 0.97 | 1.23 † |
| VCAM1 | 0.93 | 0.84 ‡ |
| CD47 | 0.94 | 0.82 ‡ |
| COL10A1 | 0.77 | 0.69 ‡ |
| TIMP1 | 0.76 | 0.68 ‡ |
Numbers are fold change and where significant these are in bold.
*Mast cell specific proteases.
†Significantly up-regulated in AMD risk group compared to low-risk group.
‡Significantly down-regulated in AMD risk group compared to low-risk group.
Several other proteins showed significant changes in just the Chr1 or Chr10 risk groups compared with the low-risk group, and a number of these were matrix related (Table 2). In the Chr1 risk group, these were procollagen C-endopeptidase enhancer (PCOLCE; increased) and collagen type XV alpha 1 (COL15A1; increased). In the Chr10 risk group, these were matrix metalloprotease 28 (MMP28; increased), fibulin-5 (FBLN5; increased), collagen type X alpha 1 chain (COL10A1; decreased), and tissue inhibitor of metalloproteinases 1 (TIMP1; decreased). Other changes of note in the Chr1 risk group included increased CFB, and the proangiogenic proteins neuropilin 1 (NRP1) and aminopeptidase N (ANPEP) were also both increased.
Further analysis was undertaken comparing the Chr1 risk groups with and without AMD to see if there were proteomic changes associated with progression to early AMD. There was no significant difference in the levels of tryptase, chymase, and CPA3, indicating that the raised levels of mast cell proteases persist in AMD. However, comparison of low risk vs. Chr1 risk with AMD showed significantly raised tryptase but not chymase or CPA3 (Dataset S3). Taken together, these data suggest that high tryptase levels are maintained as Chr1 risk eyes progress to AMD, but the effect is less clear cut for chymase and CPA3. Overall comparison between Chr1 risk samples with and without AMD demonstrated 56 (7.3%) differentially expressed proteins of the 769 detected, with 43 being up-regulated and 13 being down-regulated in submacular stromal tissue punches from maculae with macroscopic AMD changes (Table 1); a heat map shows these changes (Fig. 1B). Of the 56 differentially expressed proteins, there was a striking increase in interalpha-trypsin inhibitor heavy chain 3 (ITIH3) in AMD (3.6-fold increase). Among the proteins with significantly changed levels were proteins implicated in AMD, including APOE (increased) and CFB (decreased). Interestingly, there were significantly increased levels of proteins encoded by genes implicated in other macular conditions, including C1QTNF5 (late-onset retinal degeneration), EFEMP1 (dominant drusen), and TIMP3 (Sorsby fundus dystrophy). There were increased levels of neutrophil-secreted proteinase neutrophil elastase, which is specific to neutrophils and macrophages, and alpha-2-macroglobulin, which is secreted by macrophages, although it is not a specific marker for these cells. Ingenuity pathway analysis identified "interleukin (IL)-12 signaling and production in macrophages," "production of nitric oxide and ROS (reactive oxygen species) in macrophages," and "atherosclerosis signaling" with 5 of 20, 5 of 22, and 6 of 34 protein changes, respectively, mapping to these pathways (P = 0.000531, P = 0.00086, and P = 0.001, respectively, as determined by the Fisher’s exact test) (Fig. 1C). It is of note that "atherosclerosis signaling" was identified by ingenuity pathway analysis in this study, which is consistent with the findings of the large genome-wide association study undertaken by The AMD Gene Consortium (14).
Immunolocalization of Mast Cells in Submacular Choroid.
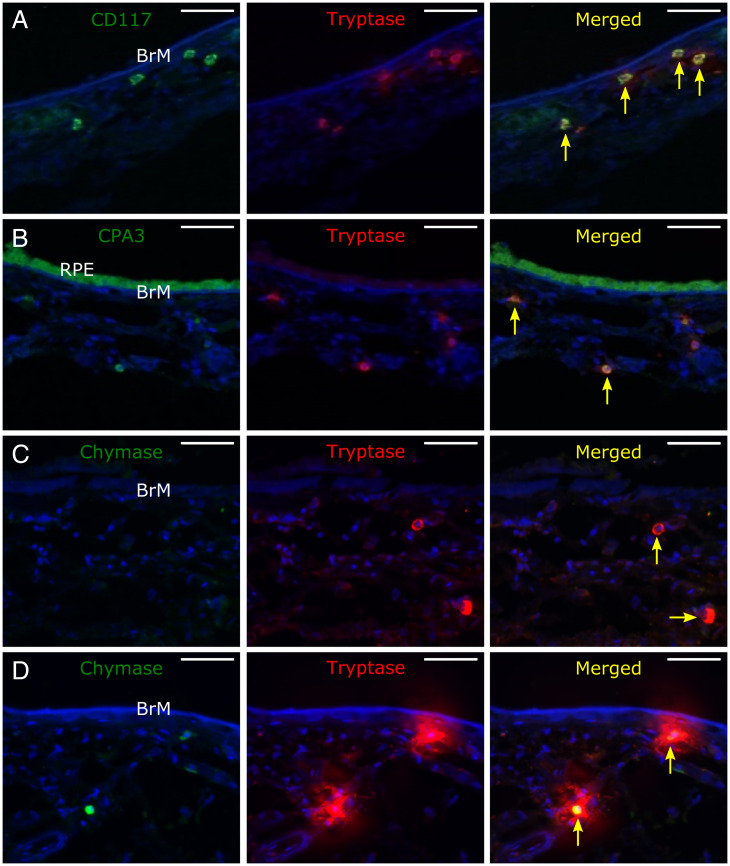
Since a key observation from the proteomic analysis was the identification of increased mast cell proteases both in donors who had high genetic risk of AMD but no disease and in donors with early AMD, we next sought to characterize mast cell populations in these groups. Submacular sections were probed with mast cell markers, including antibodies against tryptase, CD117, CPA3, and chymase. Tryptase-positive cells were observed throughout the choroid in submacular cross-sections, although they tended to be more concentrated in the inner choroid (Fig. 2 A–C), and these colocalized with CD117 (Fig. 2A), CPA3 (Fig. 2B), and chymase (Fig. 2D). Both tryptase- and chymase-positive cells (MCTC) and tryptase-positive/chymase-negative cells (MCT) were present, but MCT (Fig. 2C) predominated in the choroid of high–genetic risk donors. Strongly tryptase-positive cells with diffuse tryptase staining in the surrounding matrix were seen in high–genetic risk donors (Fig. 2D) but rarely in submacular choroid from donors with a low genetic risk.
Fig. 2.
Representative confocal micrograph z-stack projections of immunolabeled submacular sections labeled for (A) CD117 (green) and tryptase (red), (B) CPA3 (green; in this section, the RPE is present, which is autofluorescent, especially in the green channel) and tryptase (red), and (C and D) chymase (green) and tryptase (red) in C, showing MCT-type mast cells and in D, showing the halo of tryptase labeling surrounding degranulating MCTC-type mast cells. Sections were counterstained with Hoescht nuclear stain (blue), which stained Bruch’s membrane (BrM) and stroma of the inner choroid as well as cell nuclei. Yellow arrows point to mast cells. (Scale bars: 50 µM.)
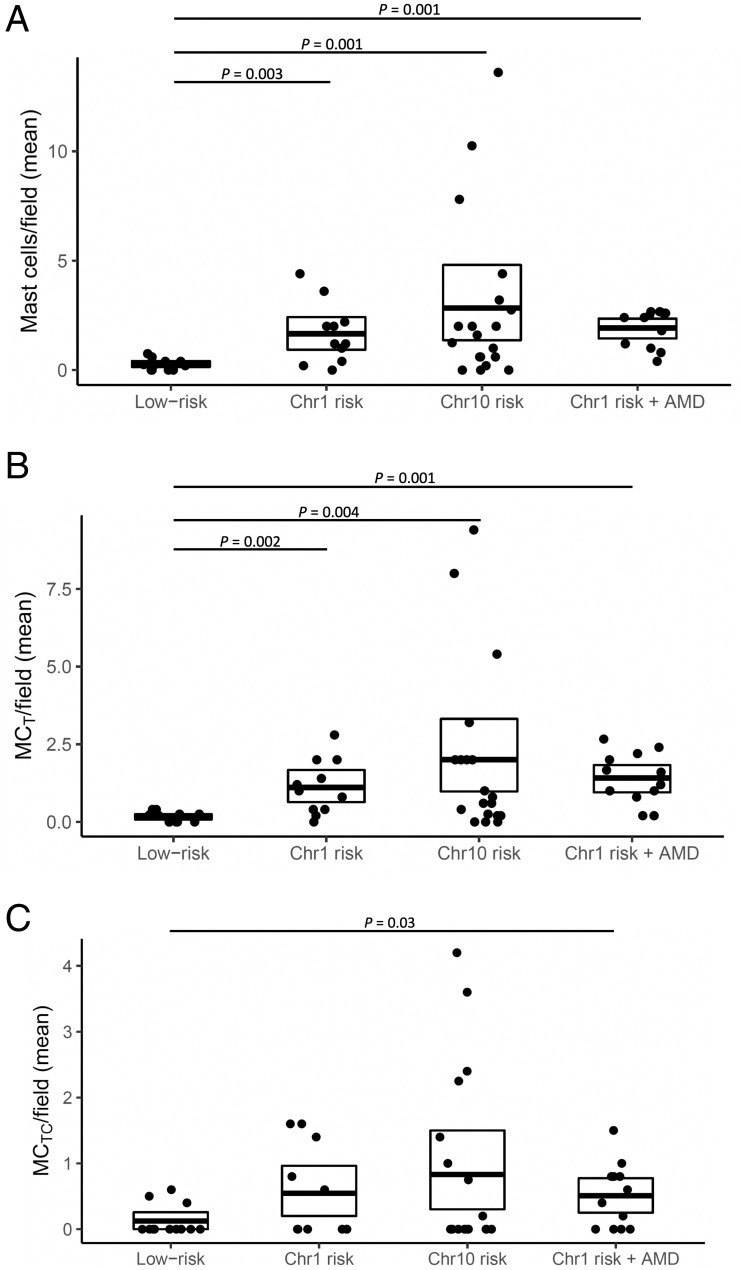
To determine whether the higher levels of mast cell proteases in submacular tissue from genetically high-risk maculae were associated with increased mast cell numbers, mast cells were quantitated in submacular cross-sections from donors with the different genetic risk profiles. CellProfiler was used as an automated means of quantifying cells that were labeled by immunohistochemistry for tryptase alone or tryptase and chymase (i.e., MCT or MCTC). Parameters were set so that weakly stained cells or staining where the localization could not be determined as intracellular or extracellular were excluded. Compared with low-risk submacular sections (0.23 mast cell counts per field of view, n = 13), those with high genetic risk all demonstrated significantly increased total mast cell counts (combined MCT and MCTC) per field of view, despite the absence of AMD. The Chr1 risk group had 1.5 mast cell counts per field of view (P = 0.003, n = 13), and the Chr10 risk group had 2.5 mast cell counts per field of view (P = 0.001, n = 16). The Chr1 risk with AMD group had 1.9 mast cell counts per field of view (P = 0.001, n = 11) (Fig. 3). Analysis of mast cell subtypes (Fig. 3) revealed that MCT were the more common mast cell subtype in high-risk maculae, accounting for 68.4% of all mast cells in Chr1 risk maculae and 62.6% in Chr10 risk maculae. MCT numbers were proportionally higher still when AMD changes were present (at 79% in Chr1 risk maculae with AMD); in low-risk maculae, there are comparable levels of both mast cell subtypes (∼50%). There were significantly increased levels of MCT in all high-risk subtypes, without and with AMD, as compared with low risk (Chr1 risk: P = 0.002, n = 13; Chr10 risk: P = 0.004, n = 16; Chr1 risk with AMD: P < 0.001, n = 11). However, MCTC numbers were only significantly increased in Chr1 risk with AMD compared with the low-risk group (P = 0.030, n = 11).
Fig. 3.
CellProfiler quantitation of (A) total mast cell, (B) MCT, and (C) MCTC counts per field of maculae from donors who were genetically low risk at the Chr1 and Chr10 loci, high risk at the Chr1 without AMD, high risk at the Chr10 locus without AMD, and high risk at the Chr1 with early AMD. Scatterplots include the mean ± SD. Significantly different P values are shown, where P < 0.05 as determined using Poisson generalized linear mixed effects models.
Localization and Quantification of Denatured Collagen in Submacular Sections.
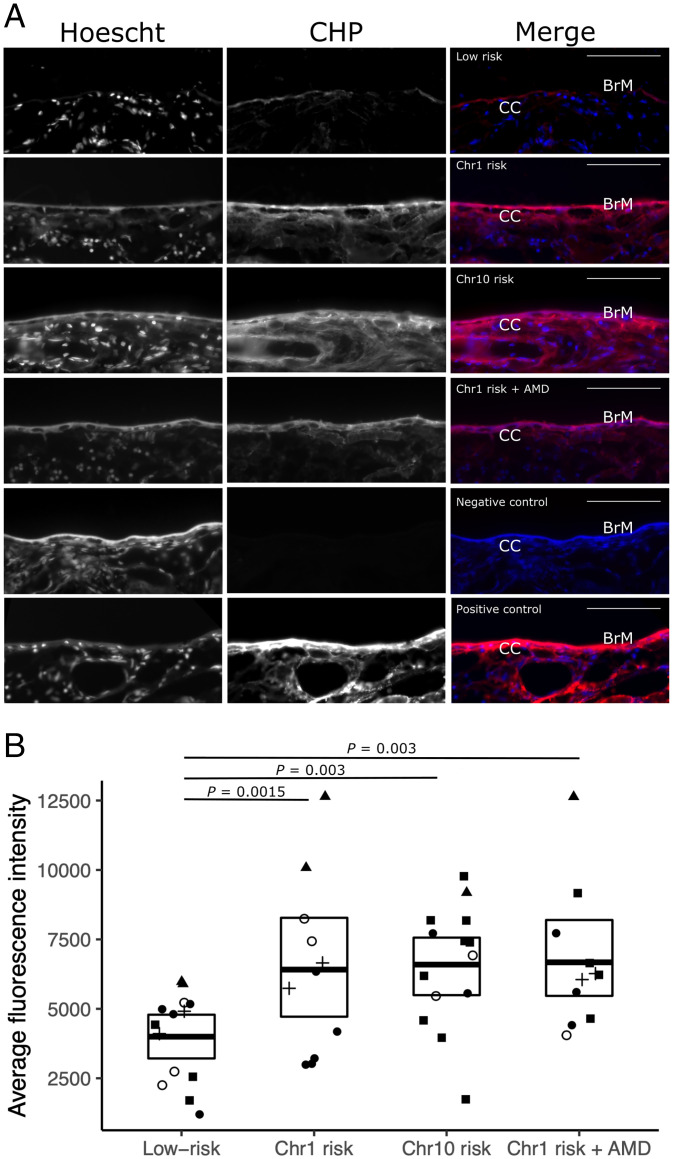
Mast cell proteases are able to degrade extracellular matrix directly or through activation of metalloproteinases (15). Given their increased levels in submacular stromal tissue punches from donors with a high genetic risk of AMD, we investigated whether there were changes in the amounts of denatured collagen, an indicator of matrix destruction, in submacular sections. These were treated with 15 µM collagen-hybridizing peptide (CHP), which specifically hybridizes to denatured collagen (16). Streptavidin-Cy5 was then used to fluorescently label the biotin-tagged CHP.
Submacular sections from donors with low AMD risk had low levels of CHP binding throughout Bruch’s membrane and the underlying choroid, indicative of largely intact collagen. However, both Chr1 and Chr10 risk and Chr1 risk with AMD submacular sections demonstrated increased CHP binding in the choroidal stroma and Bruch’s membrane (Fig. 4A). The intensity of CHP labeling of Bruch’s membrane was measured and found to be significantly increased in all groups with genetic high risk as compared with the low-risk group (Chr1 risk: 1.5-fold increase, P = 0.015, n = 11; Chr10 risk: 2-fold increase, P = 0.003, n = 14; Chr1 risk with AMD: 1.8-fold increase, P = 0.030, n = 11) (Fig. 4B).
Fig. 4.
(A) Submacular sections labeled with denatured CHP linked to Cy5 (CHP) and Hoescht nuclear stain (which labeled the Bruch’s membrane [BrM] and the choroidal stroma as well as cell nuclei) showing representative sections of genetically low risk, Chr1 risk, Chr10 risk, and Chr1 risk with AMD along with negative and positive controls. In merged images, CHP is red, and Hoescht nuclear stain blue; BrM and the choriocapillaris (CC) are labeled. (Scale bars: 50 µm.) (B) Quantification of denatured CHP binding to BrM, where the fold change of fluorescence intensity within a 5 µM width line over BrM was analyzed comparing low risk with Chr1 risk, Chr10 risk, and Chr1 risk with AMD. Significantly different P values are shown, where P < 0.05 as determined using a Poisson generalized linear mixed effects models; different experiments are indicated by different symbols.
Discussion
It is now well established that genetics plays a central role in determining AMD risk. Furthermore, tissue-based studies have implicated multiple cellular and biochemical abnormalities in established AMD. However, the mechanisms underpinning the early stages of the disease, particularly prior to the onset of clinical features, and the links between genetic predisposition and macular/submacular pathology are only partly understood. We hypothesized that analyzing submacular tissue from older donors who had genetic risk of AMD but did not have clinical features (defined by the presence of intermediate or large drusen) might shed light on these links, reasoning that multiple biochemical pathways may be altered once AMD is established. Most of the genetic risk is at two loci, one on Chr1 (CFH) and the other on Chr10 (ARMS2/HTRA1); therefore, we analyzed tissue from older donors who were homozygous for high-risk alleles at Chr1 (with low-risk alleles at Chr10) or homozygous for high-risk alleles at Chr10 (with low-risk alleles at Chr1) and compared them with tissue from donors who were homozygous for low-risk alleles at both loci. In addition, we were able to analyze tissue from donors who were homozygous for high-risk alleles at Chr1 (with low-risk alleles at Chr10) but who had early AMD (i.e., as defined by the presence of drusen with a diameter greater than 63 µm), allowing us to investigate changes associated with disease progression. We elected to use iTRAQ MS proteomics to make comparisons between these groups as this technique provides a powerful, untargeted approach for evaluating global differences in proteomes.
The most striking finding from our proteomics study was that submacular stromal tissue punches from donors with Chr1 risk or Chr10 risk but without macroscopic AMD changes both contained increased levels of mast cell–specific proteases, including tryptase, chymase, and CPA3, compared with those from eyes with low genetic risk at these loci. CTSG was also increased, which is produced by mast cells but is not specific to them. Subsequent immunohistochemical experiments demonstrated elevated numbers of mast cells in the choroid of both Chr1 risk and Chr10 risk maculae.
Mast cells are normally present in the choroid but not in the retina (17). Increased numbers of mast cells have previously been reported in the choroid of individuals with established AMD (18). In a further study, mast cell tryptase was identified in the choroidal matrix and Bruch’s membrane of eyes with geographic atrophy, a form of late-stage AMD (19). Importantly, our study builds upon these observations by showing that there are increased mast cells in the choroid of donors with genetic risk of AMD but without established AMD, suggesting that this is an early event in the disease process.
Mast cells originate from hematopoietic progenitors, and the control of migration of mast cell progenitors into tissues is locally regulated in a tissue-specific manner. Once within tissues, maturation is controlled by stem cell factor and other growth factors and cytokines. As well as their involvement in immunoglobulin E (IgE)-mediated hypersensitivity reactions, IgE-independent activation through a variety of mechanisms, including the complement system, can cause the release of a wide variety of biologically active mediators. This can be through mast cell degranulation, which releases proteases, vasoactive amines, heparin, cytokines, and growth factors, or through the secretion of proinflammatory lipid mediators (prostaglandins and leukotrienes), cytokines, chemokines, and growth factors (20). Two major types of mast cells have been described: MCTC, which secrete tryptase, chymase, CPA3, and cathepsin, and MCT, which just secrete tryptase. May (17) showed that most of the mast cells in the normal human choroid are of the MCTC variety, whereas McLeod et al. (19) found that MCT were the predominant variety in the choroid in aged eyes with and without AMD. We demonstrated similar proportions of MCTC and MCT in the choroid of eyes with low genetic risk, but MCT in particular were increased in the choroid of donors with Chr1 risk (with or without AMD changes) and Chr10 risk. However, all of the proteases secreted by MCTC were increased in submacular stromal tissue punches from donors with Chr1 and Chr10 risk, suggesting that while numbers of MCTCs were not significantly increased in high–genetic risk donors without AMD, there was increased degranulation to account for raised protease levels.
One potential action of the proteases released by mast cell degranulation is disruption of extracellular matrix. These effects can be direct, such as chymase cleaving laminin and fibronectin and tryptase cleaving aggrecan. In addition, they can have indirect effects by cleaving and thereby, activating metalloproteinases. Tryptase can activate MMP13 and MMP3 by cleaving the prometalloproteinases, which in turn, can activate MMP1 (latent collagenase); chymase can cleave and thereby, activate pro-MMP1, pro-MMP2, and pro-MMP9 (15). Therefore, we investigated whether the increased mast cells and their proteases were associated with matrix damage using a histological probe that specifically binds to denatured collagen (CHP). Increased staining for denatured collagen was evident throughout the choroid of Chr1 and Chr10 risk tissue donors compared with tissue sections from genetically low-risk samples, suggesting that mast cell–induced damage to choroidal extracellular matrix occurs early in AMD pathogenesis. Quantitative analysis of the labeling of submacular Bruch’s membrane with CHP revealed significantly increased binding of CHP in all genetic risk groups, including those without AMD. Matrix remodeling could account for the choroidal thinning observed when comparing donor eyes with Chr1 risk with low risk (12). A previous quantitative proteomics study (21) comparing macular Bruch’s membrane/choroid complex from AMD and normal eyes identified weakened extracellular matrix integrity as a feature of early-stage/midstage AMD, thereby supporting these findings. Mast cell protease-mediated degradation of the choriocapillaris basement membrane could lead to endothelial damage and vascular dropout. Damage to the integrity of Bruch’s membrane could alter its permeability, allowing large blood-derived proteins to access the space between the RPE basal lamina and inner collagenous layer and thereby, contributing toward drusen formation (22), along with RPE instability and damage. In addition, degradation of Bruch’s membrane could predispose to choroidal neovascularization.
The biochemical and cellular consequences of Chr10 risk are poorly understood, so the reasons for the association with increased choroidal mast cells and mast cell degranulation are unclear. Chr1 risk results in increased complement activation in the inner choroid (12, 13), and complement proteins may be derived from the circulation (23, 24) or expressed locally (25, 26). Complement activation will result in increased production of the anaphylatoxins C3a and C5a; these have been shown to act as chemoattractants for mast cells and to stimulate mast cell degranulation (27). Mast cells may also contribute to complement activation as they can secrete C3 and C5, and both tryptase and chymase can cleave C3 to produce C3a (28). In addition, heparin released during mast cell degranulation could dislodge factor H and FHL-1 bound to the extracellular matrix of the choroid and Bruch’s membrane, thereby promoting complement activation (29, 30).
Mast cells stimulate inflammation through a variety of mechanisms. Through the production of tumor necrosis factor-α, they can promote the recruitment of macrophages and neutrophils. Vascular permeability is also increased through the secretion of histamine, vascular endothelial growth factor, IL-6, and IL-8. It is of note that when comparing Chr1 risk eyes with and without AMD, neutrophil elastase (secreted by neutrophils and macrophages) was increased. Also, the strongest signals in pathway analysis were for “IL-12 signaling and production of macrophages” and for “production of nitric oxide and reactive oxygen species in macrophages.” The analysis also highlighted retinoid X receptor (RXR) activation, and this pathway is an important regulator of macrophage function, regulating immune function, uptake, and lipid handling (31). As there are increased submacular choroidal macrophages in eyes with AMD (32), these data suggest that mast cells could stimulate macrophage recruitment as AMD becomes established.
Parallels can be drawn between our observations and rat models, where ocular mast cell degranulation is induced using compound 48/80. When compound 48/80 was injected subconjunctivally, the resultant mast cell degranulation resulted in inflammation, increased permeability of choroidal vessels, and macrophage infiltration (33). In a modified model with a slower release of compound 48/80 delivered subconjunctivally, choroidal (and retinal) thinning, macrophage activation, and RPE degeneration were observed, and these pathological changes were prevented by either the inhibition of mast cell degranulation or tryptase activity (34). Taken together, these models suggest that genetically driven mast cell infiltration of the choroid and degranulation could initiate AMD.
There were a number of limitations to the study. The donor eye tissue was processed up to 49 h postmortem, and it is possible that some protein degradation could have occurred during that time. It is conceivable that the increased collagen denaturation observed in the Chr1 and Chr10 risk donor tissue occurred postmortem due to higher levels of proteases. However, as all sample groups had similar postmortem times, this would not be expected to introduce bias. Furthermore, because the proteins were identified by MS following tryptic digestion, limited degradation would not necessarily prevent detection. A further limitation was that differences between protein levels detected by MS were not verified using other technologies. Because of postmortem opacification of the neurosensory retina, the diagnosis of AMD was made by examination of macular tissue punches after removal of the neurosensory retina and determining whether drusen of greater than 63 µm in diameter were present or absent. We did not attempt to determine whether clinical features of AMD, including pigmentation associated with the RPE and subretinal drusenoid deposits, were present, as we deemed that we could not reliably detect them. In addition, other features of AMD, such as basal laminar or basal linear deposits, were not detected using this methodology (6, 35). Tissue punches where debris of uncertain origin was observed were excluded from the study. Nonetheless, the presence and severity of AMD in the samples could have been underestimated in the tissue punches analyzed.
In conclusion, we have identified increased mast cell proteases and choroidal mast cells in eyes from older donors with genetic risk of AMD at both Chr1 and Chr10 but without macroscopic changes of AMD. This signal persists as early AMD develops, at least in Chr1-associated disease. These findings provide insights into the early stages of AMD pathogenesis and establish a common mechanistic pathway for Chr1 and Chr10 risk. The increased levels of mast cells in the choroid may lead to matrix destruction, recruitment of other immune cells, and inflammation as the disease progresses. Therefore, targeting mast cell recruitment and degranulation could represent a therapeutic approach for treating AMD at an early stage.
Methods
Collection and Dissection of Donor Ocular Tissue.
Human donor eye tissue was obtained from the Manchester Eye Tissue Repository, an ethically approved Research Tissue Bank (UK NHS Health Research Authority reference no. 15/NW/0932). Eye tissue was acquired after the corneas had been removed for transplantation and explicit consent had been obtained to use the remaining tissue for research. Guidelines established in the Human Tissue Act of 2004 (United Kingdom) and the tenets of the Declaration of Helsinki were adhered to. All ocular tissue was processed within 49 h postmortem (SI Appendix, Table S2).
Pairs of donor eyes were flat mounted after using a scalpel to make four equally spaced incisions into the globe from the anterior to posterior poles (ensuring that the central macular region was not cut). Flat-mounted donor eyes were imaged using a Stereozoom microscope GXMXTL3T at ×7 and ×10 magnification with an iLUMos high-intensity dual gooseneck light emitting diode illuminator. Images were then captured using a GXCAM-5 camera (all GT-vision Ltd.). The lens and vitreous were discarded, but samples of iris/ciliary body were retained and stored at −80 °C for subsequent genotyping. Using the yellow coloration of the macula lutea as a guide, a 6-mm macula tissue punch (Oncall Medical Supplies) was taken from the flat-mounted eye tissue, which incorporated all tissue layers through to the sclera. The overlying neurosensory retina was then removed, and the underlying RPE was imaged at a range of magnifications from ×20 to ×35. The submacular tissue punches were then processed for proteomic or histological analysis (relevant Methods sections).
Phenotyping for the Presence or Absence of AMD.
Images of macular tissue punches were subsequently analyzed to look for AMD changes, including drusen, pigmentary changes, atrophy, and scarring. When drusen were observed, the images were analyzed using GXCapture software (GT-vision Ltd.), and the diameter of the drusen was measured. In practice, it was concluded that trying to detect AMD changes, including pigmentary changes, involving the RPE, subretinal drusenoid deposits, and atrophy was unreliable. Therefore, the diagnosis of AMD was made on the basis of donors who had one or more macular drusen with a diameter greater than 63 µm. In addition, some eyes were diagnosed with disciform scars. Donor tissue with disciform scars; cases where the phenotypic classification of AMD was unclear, including cases where there were deposits of uncertain origin; or cases where there was other pathology were excluded from this study.
Isolation of Genomic DNA for Genotyping.
DNA was isolated from frozen iris/ciliary body tissue excised from postmortem human eyes at the point of dissection using the Qiagen BioRobot EZ1 and the EZ1 Advanced XL DNA Tissue Card (Qiagen) as per the manufacturer’s instructions. The quality of all DNA samples was ensured by independent quantification, and all samples were normalized to a concentration of 20 ng/µL.
Genotyping and Sanger Sequencing.
A Sequenom assay was designed to analyze five SNPs that identified the common risk haplotypes on Chr1 (containing the Y402H variant) and Chr10 (near ARMS2/HTRA1). Primers were designed using Sequenom SNP Assay Design software version 3.0 for iPLEX reactions. The protocol and reaction conditions were in accordance with the manufacturer’s instructions. The panel analyzed three SNPs that identified the common Chr1 risk haplotype and two SNPs that identified the common Chr10 risk haplotype (SI Appendix, Table S1); more than one SNP was analyzed for each of these common risk loci to ensure that the genotyping of samples was correct. Genotype identification was performed using the MassARRAY system from Sequenom. The matrix-assisted laser desorption/ionization time-of-flight MS spectra from the Sequenom MassARRAY Analyzer 4 instrument were analyzed using Typer 4.0.20 software (Sequenom). Automated fluorescent cycle DNA sequencing was performed using an Applied Biosystems model 3730 DNA analyzer. STADEN (36) package Pregap4 1.4b1 software version 1.8b1 (Medical Research Council, Laboratory of Molecular Biology) was used for analysis of sequencing data.
Preparation of Submacular Stromal Tissue Punches for Proteomic Analysis.
This procedure has been described previously (37). The submacular tissue punches were washed in phosphate-buffered saline (PBS); then, using a cell scraper, the overlying RPE was gently removed while taking care to not rip the underlying tissue. The remaining Bruch’s membrane and choroid complex were then peeled away from the underlying sclera, and the tissue was turned over so that the choroid was exposed and then, gently scraped away. The tissue was then washed extensively in ultrapure water to remove residual blood, and the process then repeated until the tissue appeared translucent. Previously published histology following this technique demonstrated that the remaining tissue consisted of Bruch’s membrane and attached stroma from the inner choroid, primarily the intercapillary septa (37). These samples, referred to as submacular stromal tissue punches, were stored at −80 °C prior to proteomics analysis.
Sample Preparation and Proteome Comparison by iTRAQ Labeling and Liquid Chromatography–MS.
Using a scalpel, each submacular stromal tissue punch was cut in to quarters and then, transferred into Covaris microtubes-50 (Covaris Ltd.) containing 50 µL of lysis buffer (0.5 M triethylammonium bicarbonate [TEAB], 5 mM dithiothreitol [DTT], 0.05% sodium dodecyl sulfate). Samples were then sonicated using an S220 focused ultrasonicator (Covaris Ltd.) with the following settings: 20% duty cycle, 450 cycles/burst, and an intensity of eight with an overall peak power of ∼64 W for a total of 12 min while the sample was kept chilled in a water bath at 4 °C. All samples were then centrifuged at 13,000 rpm for 5 min, and protein concentrations were determined using Bradford assay reagent as per the manufacturer’s instructions (Bio-Rad) and a SpectraMax M5 plate reader (Molecular Devices).
Sample preparation for iTRAQ MS analysis was performed using previously published methods (38, 39) with the following minor modifications; a volume equivalent to 100 µg of protein was taken from each sample, and the final volume was adjusted to 40 µL using 100 mM TEAB. Samples were then reduced using 5 mM DTT for 30 min at 60 °C followed by alkylation using 15 mM iodoacetamide for 15 min at room temperature in the dark. Proteins were then digested using 5 µg modified porcine trypsin (Promega) in 100 mM TEAB overnight at 37 °C. The following day, samples were dried in a SpeedVac concentrator and then, resuspended in 25 µL 1 M TEAB. iTRAQ labeling was performed as per the manufacturer’s instructions using the eight-plex iTRAQ kit (AB Sciex). In brief, iTRAQ reagent vials were briefly centrifuged, and 70 µL isopropanol was added to each vial. Reagent vials were then added to the appropriate sample and incubated at room temperature for 2 h. Samples volume was then reduced to ∼30 µL using the SpeedVac concentrator at room temperature. Each eight-plex kit contained two samples from each of the four risk groups. These batches of eight samples were then pooled and dried in the SpeedVac concentrator. High-pH reverse-phase chromatographic peptide fractionation and low-pH reverse-phase liquid chromatography–MS data acquisition were performed as previously described (39). Briefly, pooled iTRAQ-labeled peptides were fractionated by high-pH reverse-phase chromatography into 78 peptide-containing fractions using a ZORBAX 300Extend-C18 4.6- × 150-mm, 3.5-μm column (Agilent). Fractions were dried by centrifugal evaporation and resuspended in 27 µL 3% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid for liquid chromatography–MS/MS analysis. Nine microliters of sample was loaded onto a nanoACQUITY 2G-V/M Trap Sym C18 5 µm 180 µm × 20 mm on a nanoAcquity UPLC system (Waters), and peptides were separated using a nanoACQUITY BEH300 C18 1.7-μm 75-μm × 250-mm column (Waters) at 300 nL/min using a 90-min gradient from 3 to 40% acetonitrile with 0.1% (vol/vol) formic acid. Eluted peptides were analyzed on a QStar Elite Q-ToF mass spectrometer (Sciex) running in information dependent acquisition mode with four precursors selected per cycle.
Proteomic Analysis and Bayesian Modeling.
For the identification of proteins from their peptide spectra, the data were analyzed using ProteinPilot v4.5 (AB SCIEX) using “thorough” search settings; the “biological modifications” option was selected for identification focus. To perform false discovery rate (FDR) analysis on the protein identification, the search database (all human sequences from SWISS-PROT, release 2017_04) was reversed, concatenated with the forward database, and used as the search database within ProteinPilot (40,370 proteins in total). FDR was determined by calculating the number of reverse “hits” as a proportion of forward hits using the dedicated worksheet exported from the search software.
Bayesian protein-level differential expression was performed using version v1.0.0 of the in house–developed software “BayesProt” (https://github.com/biospi/bayesprot/releases/tag/v1.0.0) as presented in Xu et al. (39). This method combines ProteinPilot (AB Sciex) sample normalization (“bias correction”) with a Bayesian linear mixed effects model implemented with the MCMCglmm R package (40). The Bayesian model performs both protein-level aggregation and infers differential expression simultaneously by modeling fixed effects between the baseline risk group and the other three plus a separate variance component for each risk group. As the study is balanced and blocked (each eight-plex containing two samples from each group), these modeling constraints also enable the inference of normalization fixed effects across the eight-plexes using the samples themselves as implicit “common controls” (41). For each of the comparisons, using the inferred posterior distribution of the condition fixed effect, for each protein we performed a one-sided significance test on the posterior probability that the mean fold change was at least 5% above or below zero. The lower of these two statistics defines the local FDR. For each comparison, a gFDR-controlled significant set of proteins (i.e., controlled for multiple hypothesis testing) was then defined as the largest set of proteins with an average local FDR < 5% (42).
Tissue Section Preparation.
The 6-mm submacular tissue punches from the contralateral eyes to those used in the proteomics experiment and additional genotyped tissue from other donors collected after the proteomic analysis were fixed in 4% paraformaldehyde (Merck Millipore) and buffered at pH 7.4 at room temperature for 1 to 2 h. The tissue punches were then embedded directly in full-strength optimal cutting temperature cryoprotectant compound (RA Lamb), and 10-µm tissue sections were cut using a Leica CM1950 cryostat and mounted on X-tra adhesive slides (both from Leica Biosystems). Slides were then stored at −80 °C.
Immunofluorescence.
Prior to staining, all slides were air-dried for 1 h at room temperature. Slides were then treated with antigen retrieval reagent universal (CTS015; R&D Systems) at 95 °C for 3 min, with a further 10 min in the retrieval solution at room temperature. Slides were then gently washed with PBS containing 0.05% Tween-20 (PBST) and fixed in ice-cold acetone (all from Merck Millipore) for 20 s before further washing in PBST. Next, a hydrophobic pen (Vector Laboratories) was used to draw around sections, which were then treated with blocking buffer (10% goat serum [Merck Millipore] diluted in PBST [vol/vol]) for 1 h at room temperature. Next, tissue sections were treated with primary antibodies (SI Appendix, Table S4) diluted in blocking buffer overnight at 4 °C (control sections were treated with blocking buffer alone). Then, the sections were extensively washed in PBST prior to treating with either biotinylated goat anti-rabbit IgG or biotinylated goat anti-mouse IgG (both Vector Laboratories) diluted 1:250 in blocking buffer for 1 h at room temperature. For costaining of submacular sections, after incubation with combined primary antibodies, sections were incubated with biotinylated goat anti-mouse IgG and goat anti-rabbit AlexaFlour plus 555 (Thermo Fisher Scientific). The slides were washed thoroughly in PBST followed by incubation with streptavidin-Cy5 conjugate (Thermo Fisher Scientific) diluted 1:250 in blocking buffer for 30 min at room temperature. Slides were washed, treated with Hoescht nuclear stain (Merck Millipore) at 1 µg/mL for 2 min, and mounted in Vectashield antifade (Vector Laboratories).
Denatured Collagen Staining.
A biotin-conjugated CHP (Echelon Biosciences), which specifically hybridizes to denatured collagen chains, was applied to submacular tissue sections as per the manufacturer’s instructions. In brief, reconstituted CHP was diluted to a 15 µM stock, which was then heated in a water bath at 80 °C for 5 min followed by immersion in an ice-water bath for 60 s. Submacular tissue sections, which had been air-dried for 1 h, were then incubated in 100 µL of 15 µM CHP overnight at 4 °C. Control sections were treated with PBS alone, and positive control sections were incubated in PBS heated to 96 °C for 3 min (to denature collagen) prior to incubation with CHP. The following day, sections were washed three times in PBST for 5 min. Sections were then incubated with streptavidin-Cy5 conjugate (Thermo Fisher Scientific) diluted 1:250 in PBS for 30 min at room temperature. Slides were washed, treated with Hoescht nuclear stain (Merck Millipore) at 1 µg/mL for 2 min, and then, mounted in Vectashield antifade (Vector Laboratories).
Imaging and Image Processing.
For immunofluorescence and imaging denatured collagen, images were collected on a Zeiss Axioimager M2 upright microscope using a 20× objective and a 0.5–numerical aperture enhanced contrast Plan-Neofluar objective and captured using a Coolsnap HQ2 camera (Photometrics) through Micromanager software v1.4.23. To prevent bleed through from one channel to the next for immunofluorescence-specific band-pass filter sets for DAPI (4',6-diamidino-2-phenylindole), Cy3 and Cy5 were used; for imaging denatured collagen, DAPI and Cy5 were used. Images were then processed and analyzed using Fiji ImageJ (https://imagej.net/Fiji/Downloads).
An analysis pipeline was developed for the quantification of mast cells using CellProfiler software 3.1.9 (https://cellprofiler.org/) to automate the quantitation of the number of MCT- or MCTC-stained cells in submacular sections. In brief, red tryptase-positive objects were identified within a diameter range of 20 to 80 pixels and a fluorescence intensity threshold greater than 0.1. Clumped objects were distinguished on the basis of shape, and objects within 20 pixels of one another were excluded (so that larger positively stained cells could not be counted more than once). All objects that fell within these criteria were applied as a primary object mask to tryptase- and chymase-costained images, where average staining intensity was then calculated per pixel squared. Chymase staining with an intensity less than 0.1 was omitted. Data were analyzed with a Poisson generalized linear mixed effects model with donor as a random effect using R package “afex” version 0.28-1, function “mixed.” P values were computed based on 1,000 samples of parametric bootstrap.
CHP binding to Bruch’s membrane was quantified. Using ImageJ, a 5-µm-thick line was drawn along the entire length of Bruch’s membrane on the image (SI Appendix, Fig. S1). Bruch’s membrane was analyzed where the RPE had been artifactually detached, except in areas where it was folded or had autofluorescent debris attached, and the average fluorescence intensity per unit of cross-sectional area of Bruch’s membrane measured from four to five randomly acquired images per donor. Data were analyzed with a linear mixed effects model with random effects for donor and experiment using afex mixed. P values were computed with Kenwood–Roger approximation of the degrees of freedom.
Confocal microscopy was used to acquire exemplar images of the mast cells shown in Fig. 2. Images were acquired on an upright Leica TCS SP8 using either a 20× objective, 0.5–numerical aperture Plan Fluotar or a 63× objective, 1.4–numerical aperture Plan Apo (oil:Ph3) objective. The SP8 system’s tunable white light laser, acoustic optical beam splitter, and spectral detectors provide a completely filter-free detection system. Images were collected using photomultiplier tube (PMT) 1 and hybrid detection (HyD) 4 detectors with the following detection mirror settings: DAPI 410 to 493 nm and Cy5 651 to 780 nm using the 405-nm (11%) and 647-nm (35%) laser lines, respectively. For confocal imaging of chymase- and tryptase-costained macular sections, the settings are as follows: pinhole one airy unit, scan speed 400 Hz unidirectional, and format 1,024 × 1,024. Images were collected using PMT 1 and HyD 2 and 4 detectors with the following detection mirror settings: DAPI 410 to 493 nm, Cy3 560 to 628 nm, and Cy5 651 to 780 nm using the 405-nm (25%), 553-nm (40%), and 647-nm (35%) laser lines, respectively. When it was not possible to eliminate bleed through between channels, the images were collected sequentially.
Supplementary Material
Acknowledgments
This research was supported by Fight for Sight UK Grant 1517/18 and the Macular Society (United Kingdom). Development and application of the Bayesian models were funded by Medical Research Council UK Grant MR/N028457/1 (to A.W.D.). The Bioimaging Facility microscopes used in this study were purchased with grants from the Biotechnology and Biological Sciences Research Council (United Kingdom), the Wellcome Trust (United Kingdom), and the University of Manchester Strategic Fund. We thank Peter March and Roger Meadows for their help with the microscopy and Peter Walker at the University of Manchester Histology Core Facility for his support.
Footnotes
Competing interest statement: A.J.D. is a founder and shareholder of Link Biologics, which is developing a protein biologic (unrelated to these studies) for eye indications. S.J.C., R.D.U., and P.N.B. are inventors named in patent applications that describe the use of complement inhibitors for therapeutic purposes and the use of circulating complement protein measurements for patient stratification, and they are cofounders of and shareholders in Complement Therapeutics, a company that focuses on the development of complement targeted therapeutics, including for AMD.
This article is a PNAS Direct Submission. C.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118510119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Flaxman S. R., et al. ; Vision Loss Expert Group of the Global Burden of Disease Study, Global causes of blindness and distance vision impairment 1990-2020: A systematic review and meta-analysis. Lancet Glob. Health 5, e1221–e1234 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Seddon J. M., Silver R. E., Kwong M., Rosner B., Risk prediction for progression of macular degeneration: 10 common and rare genetic variants, demographic, environmental, and macular covariates. Invest. Ophthalmol. Vis. Sci. 56, 2192–2202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritsche L. G., et al. , A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 48, 134–143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakravarthy U., et al. , ARMS2 increases the risk of early and late age-related macular degeneration in the European Eye Study. Ophthalmology 120, 342–348 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Klein R. J., et al. , Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L., Messinger J. D., Kar D., Duncan J. L., Curcio C.A., Biometrics, impact, and significance of basal linear deposit and subretinal drusenoid deposit in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 62, 33 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramrattan R. S., et al. , Morphometric analysis of Bruch’s membrane, the choriocapillaris, and the choroid in aging. Invest. Ophthalmol. Vis. Sci. 35, 2857–2864 (1994). [PubMed] [Google Scholar]
- 8.Mullins R. F., Johnson M. N., Faidley E. A., Skeie J. M., Huang J., Choriocapillaris vascular dropout related to density of drusen in human eyes with early age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 52, 1606–1612 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitmore S. S., et al. , Complement activation and choriocapillaris loss in early AMD: Implications for pathophysiology and therapy. Prog. Retin. Eye Res. 45, 1–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohn E. H., et al. , Choriocapillaris degeneration in geographic atrophy. Am. J. Pathol. 189, 1473–1480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutty G. A., McLeod D. S., Bhutto I. A., Edwards M. M., Seddon J. M., Choriocapillaris dropout in early age-related macular degeneration. Exp. Eye Res. 192, 107939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullins R. F., et al. , The membrane attack complex in aging human choriocapillaris: Relationship to macular degeneration and choroidal thinning. Am. J. Pathol. 184, 3142–3153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keenan T. D., et al. , Assessment of proteins associated with complement activation and inflammation in maculae of human donors homozygous risk at chromosome 1 CFH-to-F13B. Invest. Ophthalmol. Vis. Sci. 56, 4870–4879 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Fritsche L. G., et al. ; AMD Gene Consortium, Seven new loci associated with age-related macular degeneration. Nat. Genet. 45, 433–439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douaiher J., et al. , Development of mast cells and importance of their tryptase and chymase serine proteases in inflammation and wound healing. Adv. Immunol. 122, 211–252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang J., et al. , In situ imaging of tissue remodeling with collagen hybridizing peptides. ACS Nano 11, 9825–9835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.May C. A., Mast cell heterogeneity in the human uvea. Histochem. Cell Biol. 112, 381–386 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Bhutto I. A., et al. , Increased choroidal mast cells and their degranulation in age-related macular degeneration. Br. J. Ophthalmol. 100, 720–726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLeod D. S., et al. , Mast cell-derived tryptase in geographic atrophy. Invest. Ophthalmol. Vis. Sci. 58, 5887–5896 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wernersson S., Pejler G., Mast cell secretory granules: Armed for battle. Nat. Rev. Immunol. 14, 478–494 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Yuan X., et al. , Quantitative proteomics: Comparison of the macular Bruch membrane/choroid complex from age-related macular degeneration and normal eyes. Mol. Cell. Proteomics 9, 1031–1046 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergen A. A., et al. , On the origin of proteins in human drusen: The meet, greet and stick hypothesis. Prog. Retin. Eye Res. 70, 55–84 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Cipriani V., et al. , Beyond factor H: The impact of genetic-risk variants for age-related macular degeneration on circulating factor-H-like 1 and factor-H-related protein concentrations. Am. J. Hum. Genet. 108, 1385–1400 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cipriani V., et al. , Increased circulating levels of Factor H-Related Protein 4 are strongly associated with age-related macular degeneration. Nat. Commun. 11, 778 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voigt A. P., et al. , Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 116, 24100–24107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulfaul K., et al. , Local Factor H production by human choroidal endothelial cells mitigates complement deposition: Implications for macular degeneration. J. Pathol. 257, 29–38 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elieh Ali Komi D., Shafaghat F., Kovanen P. T., Meri S., Mast cells and complement system: Ancient interactions between components of innate immunity. Allergy 75, 2818–2828 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Lubbers R., van Essen M. F., van Kooten C., Trouw L. A., Production of complement components by cells of the immune system. Clin. Exp. Immunol. 188, 183–194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark S. J., et al. , Identification of factor H-like protein 1 as the predominant complement regulator in Bruch’s membrane: Implications for age-related macular degeneration. J. Immunol. 193, 4962–4970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark S. J., et al. , Impaired binding of the age-related macular degeneration-associated complement factor H 402H allotype to Bruch’s membrane in human retina. J. Biol. Chem. 285, 30192–30202 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rőszer T., Menéndez-Gutiérrez M. P., Cedenilla M., Ricote M., Retinoid X receptors in macrophage biology. Trends Endocrinol. Metab. 24, 460–468 (2013). [DOI] [PubMed] [Google Scholar]
- 32.McLeod D. S., et al. , Distribution and quantification of choroidal macrophages in human eyes with age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 57, 5843–5855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bousquet E., et al. , Choroidal mast cells in retinal pathology: A potential target for intervention. Am. J. Pathol. 185, 2083–2095 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Ogura S., et al. , A role for mast cells in geographic atrophy. FASEB J. 34, 10117–10131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sura A. A., et al. , Measuring the contributions of basal laminar deposit and Bruch's membrane in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 61, 19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staden R., Beal K. F., Bonfield J. K., The Staden package, 1998. Methods Mol. Biol. 132, 115–130 (2000). [DOI] [PubMed] [Google Scholar]
- 37.McHarg S., Brace N., Bishop P. N., Clark S. J., Enrichment of Bruch’s membrane from human donor eyes. J. Vis. Exp. 105, 53382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unwin R. D., Griffiths J. R., Whetton A. D., Simultaneous analysis of relative protein expression levels across multiple samples using iTRAQ isobaric tags with 2D nano LC-MS/MS. Nat. Protoc. 5, 1574–1582 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Xu J., et al. , Regional protein expression in human Alzheimer’s brain correlates with disease severity. Commun. Biol. 2, 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hadfield J. D., MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R Package. J. Stat. Softw. 33, 1–22 (2010).20808728 [Google Scholar]
- 41.Phillips A., Unwin R. D., Hubbard S., Dowsey A. W., “Uncertainty aware protein-level quantification and differential expression analysis of proteomics data with seaMass” in Statistical Analysis of Proteomic Data: Methods and Tools, Burger T., Ed. (Humana, New York, NY, 2022). [DOI] [PubMed] [Google Scholar]
- 42.Morris J. S., Brown P. J., Herrick R. C., Baggerly K. A., Coombes K. R., Bayesian analysis of mass spectrometry proteomic data using wavelet-based functional mixed models. Biometrics 64, 479–489 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.