Significance
The mammalian target of rapamycin complex 1 (mTORC1) signaling pathway is frequently elevated in human disease, including cancer, type 2 diabetes, metabolic disorders, and neurodegeneration. We identify SNAT7 as an important regulator of mTORC1. We believe this research will provide valuable insight about mTORC1 biology and may uncover novel therapeutic targets for patients.
Keywords: mTOR, SNAT7, macropinocytosis
Abstract
Mammalian target of rapamycin complex 1 (mTORC1) senses amino acids to control cell growth, metabolism, and autophagy. Some amino acids signal to mTORC1 through the Rag GTPase, whereas glutamine and asparagine activate mTORC1 through a Rag GTPase-independent pathway. Here, we show that the lysosomal glutamine and asparagine transporter SNAT7 activates mTORC1 after extracellular protein, such as albumin, is macropinocytosed. The N terminus of SNAT7 forms nutrient-sensitive interaction with mTORC1 and regulates mTORC1 activation independently of the Rag GTPases. Depletion of SNAT7 inhibits albumin-induced mTORC1 lysosomal localization and subsequent activation. Moreover, SNAT7 is essential to sustain KRAS-driven pancreatic cancer cell growth through mTORC1. Thus, SNAT7 links glutamine and asparagine signaling from extracellular protein to mTORC1 independently of the Rag GTPases and is required for macropinocytosis-mediated mTORC1 activation and pancreatic cancer cell growth.
Cells have an inherent capability to sense nutrient availability and when this sensing mechanism is disrupted, the results can be catastrophic to the organism. Nutrient sensing is imperative to sustain normal cell growth and proliferation (1–4). One of the most significant players in this process is the mammalian target of rapamycin (mTOR), a conserved Ser/Thr protein kinase that is a key component of a multisubunit protein complex called mTOR complex 1 (mTORC1). This complex is responsible for integrating nutrients, such as amino acids and growth factors to regulate cell growth, autophagy, and other mTORC1-mediated events. Importantly, hyperactivated mTORC1 is frequently observed in many human diseases including cancer, obesity, type 2 diabetes, neurodegeneration, and metabolic disorders.
Multiple stimuli control mTORC1, like growth factors, amino acids, energy status, and stress. Amino acids are the most potent and vital stimuli for mTORC1 activation. For example, growth factor signaling cannot effectively activate mTORC1 without amino acids (5). Elevated amino acids promote mTORC1 lysosomal localization. Once mTORC1 is at the lysosome, it is activated by the small G protein Ras homolog enriched in brain (Rheb), which signals downstream of growth factors. The C-terminal CAAX (C = cysteine, A = aliphatic, X = terminal amino acid) box of Rheb anchors it to the lysosome surface (6, 7). Guanosine triphosphate (GTP)-bound Rheb binds to mTORC1, causing a conformational change in the mTOR active site and allosteric activation of the kinase (8). Growth factor signaling activates mTORC1 via Rheb by promoting tuberous sclerosis complex (TSC) dissociation from the lysosome and Rheb (6). TSC acts as a GTPase activating protein (GAP) for Rheb, promoting the inactive form of Rheb bound to guanosine diphosphate (GDP) (9–11). Thus, amino acid and growth factor signaling at the lysosome unite to achieve optimal mTORC1 activation.
The discovery of the Rag GTPases was the first critical link that coupled amino acid signaling to mTORC1 activation at the lysosome (5, 12). RagA or RagB forms a heterodimer with RagC or RagD, and this dimerization is important for mTORC1 activation and Rag GTPase protein stability (13). There are four Rag genes in mammals: RagA and RagB are high in sequence similarity and functionally redundant; likewise, RagC and RagD are also highly related in sequence and functionally redundant. RagA or RagB GTP-bound interacts with the mTORC1 component Raptor at the lysosome. RagC or RagD GDP-bound forms a heterodimer with the GTP-bound RagA or RagB. Following the identification of the Rag GTPases, multiple components were identified in the well-characterized Rag GTPase signaling pathway to mTORC1 (7, 14–18).
Several amino acids have been implicated in mTORC1 regulation such as leucine (Leu) (5, 19, 20), arginine (Arg) (19, 21, 22), methionine (Met) (23), glutamine (Gln) (13, 20, 24, 25), and asparagine (Asn) (26, 27). Leu, Arg, and Met require the Rag GTPases for mTORC1 activation (13, 20, 22–25, 28–31). Recently, the lysosomal solute like carrier (SLC) transporter SNAT9 (also referred to as SLC38A9) was identified as a Rag-dependent Arg and essential amino acid transporter required for mTORC1 activation (32–35). In addition to SNAT9, other SLC amino acid transporters such as SLC1A5, SLC7A5, SLC3A2, and SLC36A1 regulate mTORC1 activity through the Rag GTPases (13, 15, 20). SLC transporters comprise a family of ∼400 proteins that transport a wide array of substrates across biological membranes (36, 37). Importantly, SLC transporters are promising drug targets, with several classes of marketed drugs already approved. In contrast to the well-characterized Rag GTPase pathway, we previously discovered a signaling pathway independent of the Rag GTPases, whereby Gln and Asn signal to mTORC1 in a manner that requires ADP ribosylation factor-1 (Arf1) (13, 26). Consistently, at least some of this Gln signaling pathway to TOR is conserved in yeast (38, 39). However, SLCs and additional players in the Rag-independent pathway in mammals have yet to be identified.
Here, we show that the SLC Gln and Asn transporter SNAT7 regulates mTORC1 in a Rag-independent manner. SNAT7 localizes to the lysosome and exports Gln and Asn out of the lysosome into the cytoplasm after extracellular protein, such as albumin, is macropinocytosed (40–42). Macropinocytosis is a regulated form of endocytosis that mediates the nonselective uptake of solutes, nutrients, and antigens (43, 44). Highly oncogenic Ras family small GTPases enhance macropinocytosis through signaling cascades (45–50). Studies in Ras-activated cancer cells reveal that when Gln is limiting, extracellular proteins are internalized as a source of Gln to fuel cell growth and proliferation (40, 48, 49, 51). We show that SNAT7 is required for mTORC1 lysosomal localization, mTORC1 activation, and the growth of pancreatic cancer cells from macropinocytosed extracellular protein. In addition, deletion of SNAT7 in pancreatic cancer cells significantly hindered macropinocytosis-mediated activation of mTORC1, cell size, and cell proliferation. We anticipate that the results of this work will lead to a better understanding of the molecular mechanisms by which Gln and Asn signal to mTORC1.
Results
Lysosomal Localized SNAT7 Is Up-Regulated in the Absence of the Rag GTPases.
To study the Rag-independent pathway by which Gln and Asn regulate mTORC1, we previously generated and characterized RagA/B knockout (KO) cell lines (13, 26). RagA/B KO cells have low levels of RagC/RagD, because RagC/RagD protein levels are stabilized by the RagA/B-RagC/D heterodimerization. Thus, RagA/B KO cells do not have intact Rag GTPase complexes. As shown in previous studies, stimulation of both wild-type and RagA/B KO cells with Gln and Asn increase mTORC1 signaling as measured by protein immunoblotting using phospho-specific antibodies for the well-defined mTORC1 substrates: phosphorylation of S6K at Thr389 (pS6K), phosphorylation of 4EBP1 at Thr37 and Thr46 or the mobility shift of 4EBP1, and ULK1 at Ser758 (pULK1) (SI Appendix, Fig. S1 A and B) (13, 26). S6K and 4EBP1 phosphorylation promote protein synthesis, and ULK1 phosphorylation inhibits autophagy (1, 52). Like Gln, Asn signaling to mTORC1 also requires ADP ribosylation factor 1 (Arf1) (SI Appendix, Fig. S1 C and D). Thus, both Gln and Asn can signal to mTORC1 in the absence of the Rag GTPases.
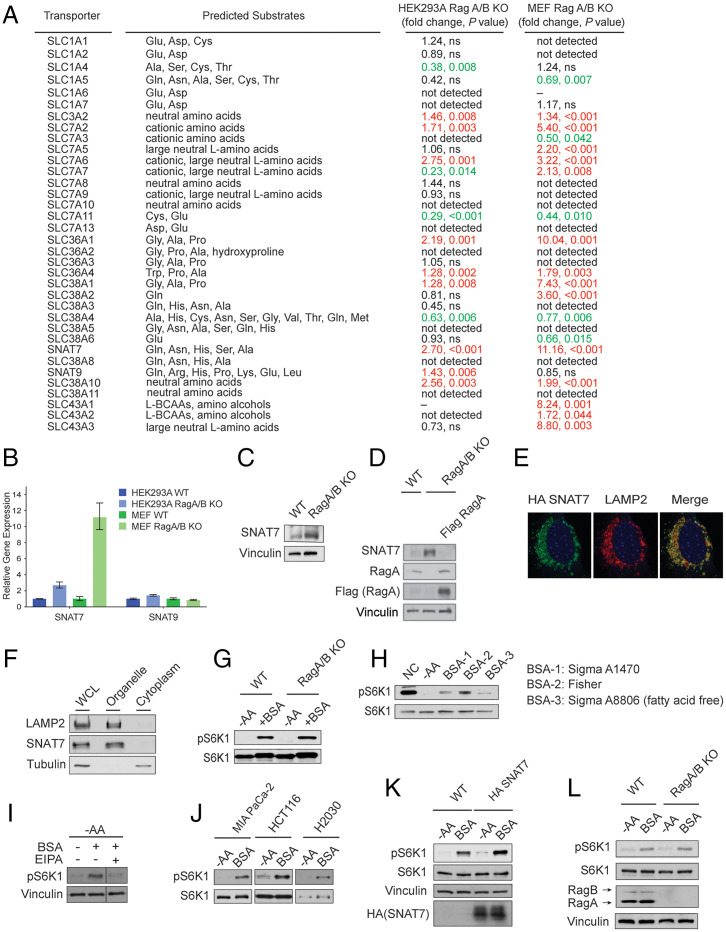
To identify new players in the Rag-independent pathway, we hypothesized that a SLC or multiple SLCs involved in transmitting Gln and Asn availability to mTORC1 may be up-regulated in RagA/B KO cells to compensate for a decrease in mTORC1 activity (∼30% decrease in mTORC1 activity in Rag-deficient cells) (13). We analyzed the expression of 35 SLCs, which are either annotated as or experimentally demonstrated to be amino acid transporters, in wild-type and Rag A/B KO MEF or HEK293A cells (Fig. 1A and SI Appendix, Fig. S2A) (36, 37). There was an approximate twofold and an approximate elevenfold increase in the mRNA expression of SNAT7 (also known as SLC38A7) in RagA/B KO HEK293A and MEF cells, respectively (Fig. 1 A and B). SNAT7 protein levels were also increased in the absence of the Rag GTPases (Fig. 1C). Consistently, the reconstitution of Flag-tagged RagA in RagA/B KO cells decreased SNAT7 protein level (Fig. 1D). Interestingly, overexpression of SNAT7, but not other tested SLCs, increased mTORC1 activity in amino acid depleted conditions (SI Appendix, Fig. S2B). mRNA levels of the Rag-dependent Arg and essential amino acid transporter SNAT9 (SLC38A9) and other SLC38 family transporters (Fig. 1 A and B and SI Appendix, Fig. S2A) were not significantly (greater than twofold) up-regulated in both RagA/B KO MEF and HEK293A cells. Thus, we focused on the role of SNAT7 in mTORC1 signaling.
Fig. 1.
SNAT7 is elevated and promotes mTORC1 signaling in the absence of the Rag GTPases. (A) Table summarizing mRNA levels of solute carrier transporters in wild-type (WT) and RagA/B knockout (KO) human embryonic kidney 293A (HEK293A) and mouse embryonic fibroblast (MEF) cells. P values were determined by Student’s t tests. P values greater than 0.05 were labeled as not significant (ns). Red text denotes increase in expression, whereas green text denotes a decrease in expression. mRNA levels that were not determined due to bad performance of primers were labeled as “–”. (B) Expression of SNAT7 and SNAT9 in WT and RagA/B KO HEK293A and MEF cells. mRNA levels of SNAT7 and SNAT9 were determined in WT or RagA/B KO MEF or HEK293A cells by real-time quantitative PCR. See (A) for P values. (C) SNAT7 expression is elevated in Rag A/B KO MEF cells. Protein levels of SNAT7 in WT or RagA/B KO MEFs were determined by Western blotting. Vinculin was used as a loading control. (D) Protein levels of SNAT7 in WT, Rag A/B KO MEFs, or Rag A/B KO MEFs stably expressing Flag-tagged Rag A were determined by Western blotting. RagA and Flag-tagged RagA were blotted for as controls. Vinculin was used as a loading control. (E) SNAT7 localizes to the lysosome, shown by immunofluorescence analysis depicting HA-tagged SNAT7 (green) and the lysosomal marker LAMP2 (red). (F) Subcellular fractionation experiments in HEK293A cells were performed separating the organelle and cytoplasmic fraction. SNAT7, LAMP2 (lysosomal marker), and Tubulin (cytoplasmic marker) were immunoblotted. (G) Bovine serum albumin (BSA) activates mTORC1 in the absence of the Rag GTPases. WT or RagA/B KO MEF cells were starved of amino acids (−AA) for 2 h and then stimulated with 3% BSA for 2 h. mTORC1 activity was analyzed by immunoblotting for the phosphorylation status of S6K1 (pS6K1) at threonine 389. S6K, actin, and vinculin were used as loading controls. (H) mTORC1 is activated by BSA. MIA PaCa-2 cells were starved of amino acids (−AA) for 2 h and then stimulated with different BSA (3%) for 4 h. mTORC1 activity was analyzed as described in (G). (I) BSA activates mTORC1 through macropinocytosis. MIA PaCa-2 cells were starved of amino acids (−AA) for 2 h and then stimulated with BSA (3%) for 4 h. Macropinocytosis inhibitor EIPA (25 μM) or DMSO as a control was maintained for the whole period of the starvation and stimulation process. mTORC1 activity was analyzed as described in (G). (J) BSA activates mTORC1 in pancreatic, colon, and lung cancer cell lines. Pancreatic (MIA PaCa-2), colon (HCT116), or lung (H2030) cancer Ras transformed cells were starved for amino acids (−AA) for 2 h followed by 3% BSA stimulation for 2 h (in HCT116) or 4 h (in MIA PaCa-2 and H2030). mTORC1 activity was analyzed as described in (G). (K) Expression of SNAT7 enhances BSA-induced mTORC1 activation. WT or HA-tagged SNAT7 stably expressed MIA PaCa-2 cells were starved of amino acids (−AA) for 2 h and then stimulated with 3% BSA for 4 h. mTORC1 activity was analyzed as described in (G). S6K1 and vinculin were used as loading controls. HA-tagged SNAT7 overexpression was confirmed by Western blotting. (L) Albumin signals to mTORC1 in the absence of the Rag GTPases. MIA PaCa-2 cells were starved of amino acids (−AA) for 2 h and then stimulated with 3% BSA for 4 h. mTORC1 activity was analyzed as described in (G). RagA/B levels were confirmed using Western blotting. Vinculin and S6K were used as loading controls.
SNAT7 is a lysosomal localized transporter which exports Gln and Asn from the lysosomal lumen into the cytoplasm (41). As expected, expression of HA-tagged SNAT7 (green) colocalized with LAMP2-positive lysosomal membranes (red) in Rag GTPase KO MEF cells (Fig. 1E) (40). Consistently, subcellular fractionation data revealed SNAT7 to be in the organelle fraction of cells containing lysosomes (Fig. 1F). Amino acids can accumulate within the lysosome through different routes, one being macropinocytosis where protein (such as albumin) is engulfed and subsequently proteolyzed (43, 44). Oncogenic Ras mutation in cancer cells enhances macropinocytosis, increases mitogen-activated protein kinases (MAPK) signaling, and promotes tumorigenesis (43–50, 53). Recent studies in Ras-activated cancer cells have revealed that when Gln is limiting, extracellular proteins, such as albumin, are internalized for a source of Gln to fuel cell growth and proliferation (40, 48, 49, 51). We starved wild-type and RagA/B KO MEF cells of amino acids, stimulated them with 3% bovine serum albumin (BSA), and observed mTORC1 activation (Fig. 1 G and H). Treatment of cells with the macropinocytosis inhibitor ethylisopropylamiloride (EIPA), blocked BSA-induced mTORC1 activation (Fig. 1I) (48). Human albumin concentrations in serum range from 2 to 5% (49), and increased mTORC1 activation was particularly observed in pancreatic MIA PaCa-2 (KRASG12C), colorectal HCT116 (KRASG13D), and lung NCI-H2030 (KRASG12C) carcinoma cell lines upon BSA treatment (Fig. 1J) (40, 48). We observed a modest increase in BSA-stimulated mTORC1 activation when HA-tagged SNAT7 was overexpressed in MIA PaCa-2 cells (Fig. 1K). Elevated SNAT7 expression did not alter mTORC1 activity under nutrient rich conditions (SI Appendix, Fig. S3 A and B). Moreover, BSA can also activate mTORC1 in Rag A/B KO cells (Fig. 1 G and L). Taken together, BSA stimulates mTORC1 signaling in the absence of the Rag GTPases, possibly through lysosomal export of Gln and Asn by SNAT7 after extracellular protein proteolysis via macropinocytosis.
The N Terminus of SNAT7 Interacts with mTORC1.
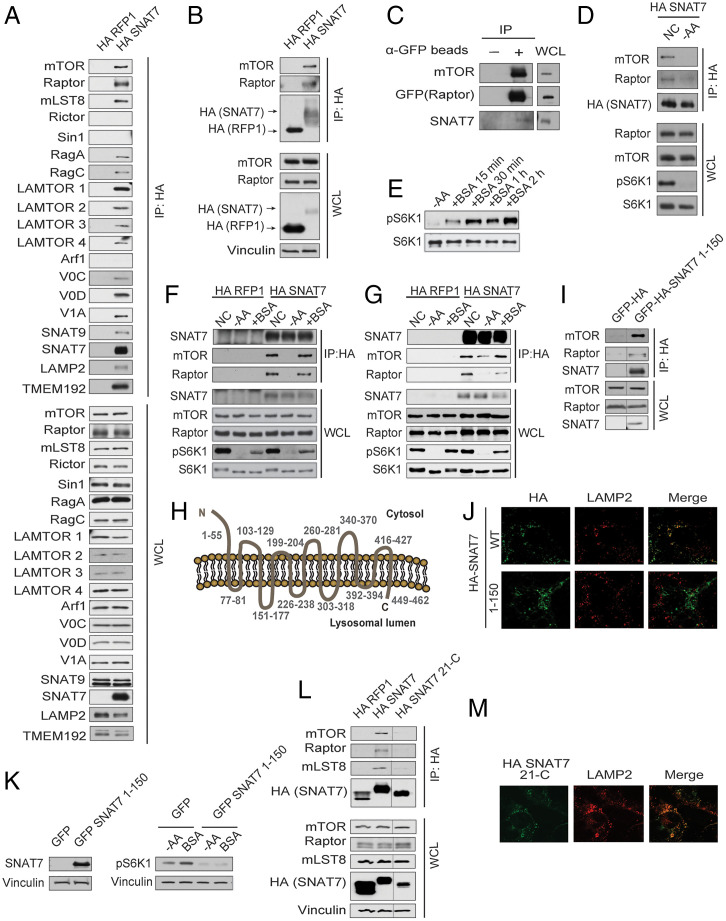
Sequence alignment analysis of SNAT9 and SNAT7 revealed that SNAT7 is lacking a large part of the N-terminal domain of SNAT9, including critical residues required for interaction with the Rag GTPases (32–34). Previous immunoprecipitation and subsequent mass spectrometry studies using 1% Nonidet P-40 lysis buffer revealed that SNAT7 does not appear to interact with the Rag GTPases or other components involved in the Rag-dependent signaling pathway (33). Immunoprecipitation studies using 0.3% CHAPS in wild-type cells show that HA-tagged SNAT7 can pull down multiple proteins localized to the lysosome including the Rag GTPases (Fig. 2A). CHAPS (0.3%) is typically used when performing immunoprecipitation experiments to preserve the mTORC1 component interaction (54). When performing HA-tagged SNAT7 immunoprecipitations using 1% Triton X-100, SNAT7 did not interact with the Rag GTPases (SI Appendix, Fig. S4). In both conditions, (0.3% CHAPs and 1% Triton X-100) SNAT7 interacts with mTORC1. SNAT7 and mTORC1 both localize to the lysosome; however, it is currently unclear if the SNAT7-mTORC1 interaction is direct or indirect. Interestingly, HA-tagged SNAT7 interacts with mTORC1 in the absence of Rag GTPases (Fig. 2B). Moreover, SNAT7 does not bind to mTORC2 indicating specificity in the SNAT7-mTORC1 interaction (Fig. 2A). Arf1 did not bind to SNAT7 or mTORC1, and inhibition of Arf1 activity does not alter macropinocytosis (Fig. 2A and SI Appendix, Fig. S5 A and B). Analysis of anti-GFP immunoprecipitates from RagA/B KO haploid cell line (HAP1) with Raptor endogenously labeled with GFP also revealed that there is an SNAT7-mTORC1 complex (Fig. 2C) (55). In the presence of amino acids, anti-Raptor-GFP or HA-SNAT7 immunoprecipitates contained SNAT7, suggesting that the SNAT7-mTORC1 interaction is at the lysosome (Fig. 2 C and D). Because BSA activates mTORC1 and SNAT7 binds to mTORC1 (Figs. 1 G–L and 2 A–D), we tested if the SNAT7-mTORC1 interaction corresponds with the time frame in which BSA induces mTORC1 activation. MIA PaCa-2 cells were starved of amino acids and treated with BSA for different time points (Fig. 2E). BSA activated mTORC1 and promoted SNAT7-mTORC1 binding as early as 15 min (Fig. 2F), and the SNAT7-mTORC1 interaction was sustained for at least 4 h (Fig. 2G). Thus, SNAT7 and mTORC1 interact under amino acid rich conditions or BSA stimulation regardless of Rag GTPases.
Fig. 2.
SNAT7 interacts with mTORC1 at the lysosome. (A) Overexpressed SNAT7 interacts with mTORC1. MIA PaCa-2 cells overexpressing HA-tagged SNAT7 or HA-tagged RFP (control) were immunoprecipitated with anti-HA beads, and then analyzed for indicated proteins (mTOR, Raptor, mLST8, Rictor, Sin 1, RagA, RagC, LAMTOR1-4, Arf1, V0C, V0D, V1A, SNAT9, SNAT7, LAMP2, and TMEM 192). (B) Overexpressed SNAT7 interacts with mTORC1 in the absence of the Rag GTPases. RagA/B knockout (KO) human embryonic kidney 293A (HEK293A) cells were transfected with HA-tagged RFP1 or HA-tagged SNAT7 for 24 h. Under normal culturing conditions (with amino acids present), anti-HA lysates were analyzed for mTORC1 components (mTOR, Raptor, and mLST8) via Western blot analysis. HA-tagged SNAT7, HA-tagged RFP1, and vinculin were probed for as controls. IP, immunoprecipitation; WCL, whole cell lysate. (C) Endogenous SNAT7 interacts with mTORC1. RagA/B KO HAP1 cells with an endogenously tagged GFP on Raptor were used to detect an endogenous interaction between mTORC1 and SNAT7 under normal culturing conditions. Anti-GFP immunoprecipitates were probed for GFP-tagged Raptor, mTOR and SNAT7 by Western blot analysis. (D) SNAT7 forms a nutrient sensitive complex with mTORC1. HA-tagged SNAT7 stably overexpressed RagA/B KO HEK293A cells were starved of amino acids (−AA) for 4 h or not starved (NC), and anti-HA lysates were analyzed for mTORC1 components (mTOR and Raptor). Phosphorylation status of S6K1 (pS6K1) at threonine 389 was assessed for mTORC1 activity by Western blot. S6K were probed for as loading controls. NC, normal culturing conditions. (E) BSA activates mTORC1. MIA PaCa-2 cells were starved of amino acids (−AA) for 2 h and then stimulated with BSA for 15 mins, 30 mins, 1 h, or 2 h. mTORC1 activity was analyzed as in (D). (F) Albumin regulates SNAT7-mTORC1 binding. HA-tagged SNAT7 stably overexpressed in MIA PaCa-2 cells were starved for amino acids (−AA) for 2 h followed by 3% bovine serum albumin (BSA) stimulation for 15 mins or (G) 4 h, and anti-HA lysates were analyzed for mTORC1 components (mTOR and Raptor). mTORC1 activity was analyzed as in (D). (H) A schematic of the 11-transmembrane protein SNAT7 on lysosomal membrane. “N” and “C” denote N terminus and C terminus, respectively. Number ranges indicate amino acid sequences of corresponding regions of this protein. (I) The N terminus of SNAT7 interacts with mTORC1. RagA/B KO HEK293A cells were transfected with GFP-tagged HA, or GFP-HA-tagged SNAT7 1–150 for 24 h. Proteins were immunoprecipitated with HA beads, and probed for mTOR, Raptor, mLST8, and SNAT7. (J) HA-tagged SNAT7 or HA-tagged SNAT7 1–150 (green) was overexpressed in HEK293A RagA/B KO cells for 24 h. Colocalization of HA-tagged SNAT7 or HA-tagged SNAT7 1–150 and the lysosomal protein (LAMP2, red) was analyzed via confocal microscopy. (K) Overexpression of the N terminus of SNAT7 inhibits mTORC1 signaling. RagA/B KO HEK293A cells were transfected with GFP-HA or GFP-HA-tagged SNAT7 1–150 for 24 h. (Left) Overexpression of HA-tagged SNAT7 1–150 truncation was confirmed by Western blot and Vinculin was probed for as control. (Right) Cells were then starved of amino acids (−AA) and stimulated with 3% BSA for 4 h. mTORC1 activity was determined as described in (D). Vinculin was probed for as a loading control. (L) The N terminus (amino acids 1–20) of SNAT7 is required to interact with mTORC1. RagA/B KO HEK293A cells were transfected with HA-tagged RFP1, HA-tagged SNAT7, and HA-tagged SNAT7 missing amino acids 1–20 (HA-tagged SNAT7 21-C) for 24 h. Proteins were immunoprecipitated with HA beads, and probed for mTOR, Raptor, mLST8, and HA. WCL were probed for mTOR, Raptor, mLST8, HA, and vinculin as controls. (M) Deletion of the N terminus (amino acids 1–20) does not alter SNAT7 lysosomal localization. HA-tagged SNAT7 21-C (green) was overexpressed in HEK293A RagA/B KO cells for 24 h. Colocalization of HA-tagged SNAT7 21-C and the lysosomal protein (LAMP2, red) was analyzed via confocal microscopy.
SNAT7 is predicted to have 11 transmembrane domains, with a cytosolic N-terminal region of 55 amino acids (Fig. 2H). Because the N terminus of SNAT7 is exposed to the cytoplasm where mTORC1 resides, we tested whether the N terminus is involved in the SNAT7-mTORC1 interaction. Overexpression of a cytosolic N-terminal segment (residues 1–150) of SNAT7 was sufficient to bind to mTORC1 (Fig. 2 I and J), and potently inhibited mTORC1 signaling, possibly by disrupting the endogenous SNAT7-mTORC1 interaction (Fig. 2K). Moreover, removal of the first 20 amino acids of SNAT7 prevented SNAT7-mTORC1 interaction (Fig. 2 L and M). Thus, the N terminus of SNAT7 is necessary for the SNAT7-mTORC1 interaction.
SNAT7 Regulates Albumin-Induced mTORC1 Lysosomal Localization and Activation.
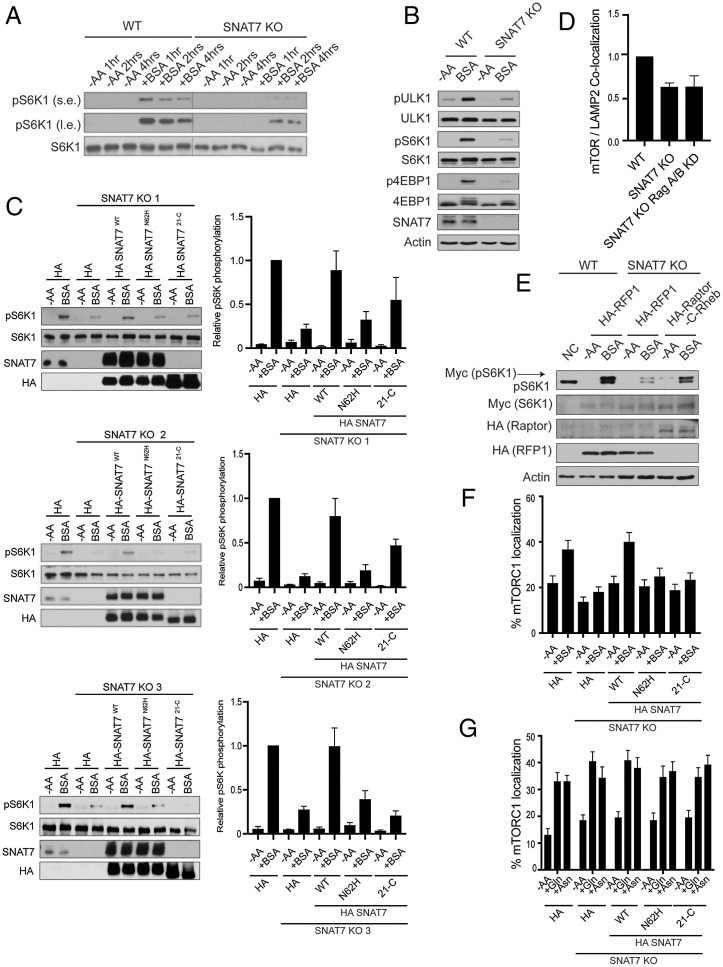
To further investigate the role of SNAT7 in macropinocytosis mediated mTORC1 activation, we generated SNAT7 KO MIA PaCa-2 cells via CRISPR-Cas9 genome editing. We starved wild-type and SNAT7 KO cells of amino acids, stimulated them with BSA, and analyzed mTORC1 activity (Fig. 3 A and B). Remarkably, loss of SNAT7 significantly reduced albumin-induced mTORC1 activation. However, depletion of SNAT7 had no effect on Gln- or Asn-induced mTORC1 activation, suggesting that SNAT7 exports Gln and Asn out of the lysosome to activate mTORC1 following macropinocytosis, and is dispensable for free Gln and Asn signaling to mTORC1 (SI Appendix, Fig. S6 A–C). Expression of SNAT7 can slightly increase Gln and Asn signaling to mTORC1, perhaps by increasing the export of lysosomal Gln/Asn further increasing mTORC1 activity (SI Appendix, Fig. S6D). Because a previous study showed that SNAT7 can export Gln and Asn out of the lysosome in vitro using biochemical assays with Gln and Asn esters (40), we tested if Gln and Asn esters could activate mTORC1 in cells (SI Appendix, Fig. S6E). Gln and Asn esters were able to activate mTORC1 in both wild-type and SNAT7 KO cells. It is thought that amino acid esters freely diffuse across membranes at neutral pH due to the low pKa of the amino group, where they are hydrolyzed by esterases into native amino acids within the lysosome (56). Then the free amino acid cannot diffuse back out of the lysosome due to its high polarity, becomes trapped, and accumulates to high concentrations within the lysosome (56). However, we were unable to see the accumulation of Gln and Asn esters in lysosomes after the isolation of lysosomes (SI Appendix, Fig. S6 F–H). SNAT7 protein levels did not change under different nutrient conditions (SI Appendix, Fig. S7). Moreover, depletion of SNAT7 did not alter lysosomal function (SI Appendix, Fig. S8A) or macropinocytosis (SI Appendix, Fig. S8 B and C). To confirm that the loss of mTORC1 activation by albumin is due to the deletion of SNAT7, we stably overexpressed HA-tagged SNAT7 in three different SNAT7 KO MIA PaCa-2 cell lines and found that HA-tagged SNAT7 in SNAT7 KO cells can rescue mTORC1 activation when compared to wild-type cells (Fig. 3C). However, overexpression of the transporter-defective HA-tagged SNAT7 mutant (Asn-62 mutated to Histidine 62, N62H) was not able to restore mTORC1 activation by albumin. SNAT7 Asn-62 is crucial for amino acid transport and the SNAT7 N62H mutant prevents Gln and Asn export out of the lysosome (40, 57, 58). Overexpression of the SNAT7 with the deleted cytosolic N-terminal segment (HA-tagged SNAT721-C) of SNAT7 was also unable to fully restore mTORC1 activity. Collectively, these results demonstrate that SNAT7 controls albumin-induced mTORC1 activation via macropinocytosis.
Fig. 3.
SNAT7 is required for albumin-induced mTORC1 activation. (A) SNAT7 deletion impairs mTORC1 activation by macropinocytosis at multiple time points. WT or SNAT7 KO MIA PaCa-2 cells (SNAT7 KO) were starved of amino acids for 2 h and then maintained in amino acid starvation media (−AA) or stimulated with 3% bovine serum albumin (+BSA) for the indicated times. mTORC1 activity was analyzed by immunoblotting for the phosphorylation status of S6K1 (pS6K1) at threonine 389. s.e., short exposure; l.e., long exposure. S6K1 was probed for as a loading control. (B) SNAT7 is required for macropinocytosis mediated mTORC1 activation of multiple downstream substrates. SNAT7 KO MIA PaCa-2 cells were starved of serum and amino acids (−AA) and stimulated with 3% BSA for 4 h. mTORC1 activity was analyzed by immunoblotting for the phosphorylation status of ULK1 at serine 758 (pULK1), S6K1 at threonine 389 (pS6K1), 4EBP1 at serine 65 (p4EBP1), or by mobility shift of the total 4EBP1 protein. ULK1, S6K1, SNAT7, and actin were probed for as controls. (C) SNAT7 is required for BSA-induced mTORC1 activation. WT MIA PaCa-2 cells, SNAT7 KO (3 different clones as indicated), or SNAT7 KO cells overexpressing HA-tagged SNAT7WT, HA-tagged SNAT7N62H, or HA-tagged SNAT721-C were starved of amino acids (−AA) for 2 h and then stimulated with BSA. (Left) mTORC1 activity was analyzed by the phosphorylation of S6K. SNAT7 levels were confirmed using Western blotting. S6K and HA are loading controls. (Right) Quantification of pS6K levels and statistical analysis. Clone 1: WT + BSA vs. SNAT7 KO + BSA, P < 0.001; WT + BSA vs. HA-SNAT7N62H + BSA, P < 0.001; WT + BSA vs. HA-SNAT721-C + BSA, P > 0.05; and SNAT7 KO + BSA vs. HA-SNAT7WT + BSA, P < 0.05. Clone 2: WT + BSA vs. SNAT7 KO + BSA, P < 0.0001; WT + BSA vs. HA-SNAT7N62H + BSA, P < 0.001; WT + BSA vs. HA-SNAT721-C + BSA, P < 0.01; and SNAT7 KO + BSA vs. HA-SNAT7WT + BSA, P < 0.05. Clone 3: WT + BSA vs. SNAT7 KO + BSA, P < 0.0001; WT + BSA vs. HA-SNAT7N62H + BSA, P < 0.01; WT + BSA vs. HA-SNAT721-C + BSA, P < 0.001; and SNAT7 KO + BSA vs. HA-SNAT7WT + BSA, P < 0.05. (D) SNAT7 regulates mTORC1 lysosomal localization. WT, SNAT7 KO, or SNAT7 KO with RagA/B knocked down (KD) MIA PaCa-2 cells were assessed for mTORC1 lysosomal localization. Images were quantified for mTOR/LAMP2 colocalization. Significance (P value): WT vs. SNAT7 KO, P < 0.01; WT vs. SNAT7 KO Rag A/B KD, P < 0.05; and SNAT7 KO vs. SNAT7 KO Rag A/B KD, P = 0.984. (E) mTORC1 activation deficiency in SNAT7 KO cells can be rescued by artificially targeting mTORC1 to the lysosome. WT or KO MIA PaCa-2 cells were cotransfected with HA-tagged RFP1, Raptor, or Raptor fused with the C terminus of Rheb (Raptor-C-Rheb), together with Myc-tagged S6K1. Cells were starved of amino acids (−AA) for 2 h and stimulated with 3% BSA for 4 h. mTORC1 activity was assessed as in (A). Myc, HA, and actin were probed for as controls. (F) SNAT7 is required for BSA-induced mTORC1 lysosomal localization. WT, SNAT7 KO, or SNAT7 KO cells overexpressing HA-tagged SNAT7WT, HA-tagged SNAT7N62H, or HA-tagged SNAT721-C were starved of amino acids (−AA) for 2 h and then stimulated with 3% BSA for 4 h. Colocalization of mTOR and the lysosomal protein (LAMP2) was analyzed via confocal microscopy. P values: WT -AA vs. WT + BSA, P < 0.05; HA-SNAT7WT −AA vs. HA-SNAT7WT + BSA, P < 0.01; WT + BSA vs. SNAT7 KO + BSA, P < 0.001; SNAT7 KO + BSA vs. HA-SNAT7WT + BSA, P < 0.001; WT + BSA vs. HA-SNAT7N62H + BSA, P < 0.05; WT + BSA vs. HA-SNAT721-C + BSA, P < 0.05; SNAT7WT + BSA vs. HA-SNAT7N62H + BSA, P < 0.05; and SNAT7WT + BSA vs. HA-SNAT721-C + BSA, P < 0.01. (G) SNAT7 is not required for glutamine and asparagine induced mTORC1 lysosomal localization. WT, SNAT7 knockout (KO), or SNAT7 KO MIA PaCa-2 cells overexpressing HA-tagged SNAT7WT, HA-tagged SNAT7N62H, or HA-tagged SNAT721-C were starved of amino acids (−AA) for 2 h and then stimulated with 4 mM glutamine (+Gln) or asparagine (+Asn) for 2 h. mTOR and lysosomal protein (LAMP2) colocalization was analyzed with confocal microscopy. P values: WT −AA vs. WT + Gln, P < 0.001; WT −AA vs. WT + Asn, P < 0.001; SNAT7 KO −AA vs. SNAT7 KO + Gln, P < 0.001; SNAT7 KO −AA vs. SNAT7 KO +Asn, P < 0.01; SNAT7WT −AA vs. SNAT7WT + Gln, P < 0.001; SNAT7WT −AA vs. SNAT7WT + Asn, P < 0.001; HA-SNAT7N62H −AA vs. HA-SNAT7N62H + Gln, P < 0.01; HA-SNAT7N62H −AA vs. HA-SNAT7N62H + Asn, P < 0.01; HA-SNAT721-C −AA vs. HA-SNAT721-C + Gln, P < 0.01; and HA-SNAT721-C −AA vs. HA-SNAT721-C + Asn, P < 0.001.
Amino acids promote mTORC1 lysosomal localization and subsequent activation (5). Because SNAT7 interacts with mTORC1 at the lysosome under amino acid sufficient and BSA stimulated conditions (Fig. 2 A–D and F–G), we tested if SNAT7 may regulate mTORC1 lysosomal translocation. We examined the colocalization of mTOR and LAMP2 in SNAT7 KO cells. Interestingly, SNAT7 depletion significantly decreased (∼36%) mTORC1 lysosomal localization with or without the Rag GTPases (Fig. 3D). Based on these results, we reasoned that SNAT7 may be important for mTORC1 lysosomal localization and subsequent activation by Rheb. Forcing mTORC1 to the lysosome near Rheb could perhaps rescue BSA-induced mTORC1 activation when SNAT7 is depleted. Indeed, artificially targeting of mTORC1 to the lysosomal surface by the addition of the C-terminal lysosomal targeting motif of Rheb (last 15 amino acids including the CAAX box) to Raptor (7, 13), rescued BSA-induced mTORC1 activation even when SNAT7 was depleted (Fig. 3E). BSA stimulates the translocation of mTORC1 to the lysosome (49), and SNAT7 depletion inhibited BSA-induced mTORC1 lysosomal translocation (Fig. 3F). HA-tagged SNAT7 overexpression in SNAT7 KO MIA PaCa-2 rescued BSA-induced mTORC1 lysosomal localization when compared to wild-type cells, whereas HA-tagged SNAT7 N62H and HA-SNAT7 21-C did not fully rescue mTORC1 lysosomal localization. In contrast, SNAT7 depletion had no effect on Gln- or Asn-induced mTORC1 lysosomal localization (Fig. 3G). Thus, SNAT7 plays a role in the recruitment of mTORC1 to the lysosome and subsequent activation.
SNAT7 Is Essential for Pancreatic Cancer Cell Growth through mTORC1.
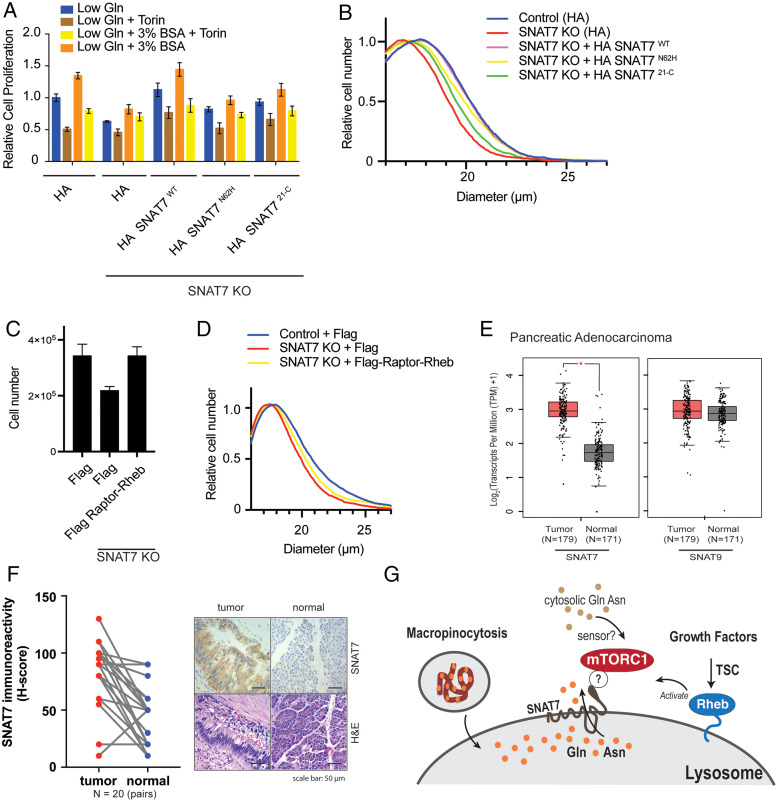
SNAT7 is widely expressed in multiple cell lines (59) and is altered (mutated or amplified) in several human cancers (60, 61). Because oncogenic Ras mutations enhance macropinocytosis and are present in PDAC, we overexpressed two different KRAS mutants (KRAS G12D or KRAS G12V) and assessed SNAT7 protein levels (SI Appendix, Fig. S9A). SNAT7 protein levels were not increased with the expression of KRAS mutants, and SNAT7 mRNA levels are not up-regulated in other cancers with KRAS mutations (lung adenocarcinoma) (SI Appendix, Fig. S9B). Cancer cells require high levels of nutrients, especially Gln, for cell growth and proliferation (51, 62, 63). When extracellular Gln is limiting, Ras transformed cells initiate the uptake of extracellular proteins, which are proteolyzed in the lysosome and supply the cell with amino acids (40, 48, 49, 51). Because SNAT7 is a lysosomal transporter, we investigated whether SNAT7 was required for the extracellular protein-dependent proliferation of pancreatic cancer cells. We and others observe that MIA PaCa-2 cells rely on free extracellular Gln, and the supplement of albumin under low Gln conditions can increase MIA PaCa-2 cell growth (Fig. 4A) (40, 48). The mTOR inhibitor Torin 1 prevented albumin-induced MIA PaCa-2 cell growth under low Gln conditions. In contrast, in MIA PaCa-2 SNAT7 KO cells, albumin was unable to significantly increase cell proliferation in low Gln conditions. HA-tagged SNAT7 overexpression in SNAT7 KO MIA PaCa-2 cells rescued BSA-induced cell proliferation under low Gln conditions. In contrast, HA-tagged SNAT7 N62H and HA-tagged SNAT7 21-C could not rescue cell proliferation to the level of wild-type cells. Moreover, depletion of SNAT7 significantly impaired other mTORC1-mediated biology, such as cell size (Fig. 4B). Reconstitution of HA-tagged SNAT7 rescued cell size in SNAT7 KO cells compared to wild-type, whereas HA-tagged SNAT7 N62H and HA-tagged SNAT7 21-C only partially rescued. Overexpression of Raptor fused to the C terminus of Rheb (Flag Raptor-C-Rheb) increased cell proliferation in SNAT7 KO cells (Fig. 4C), and partially rescued cell size in SNAT7 KO cells (Fig. 4D). SNAT7 did not appear to alter Gln starvation induced mTORC1 reactivation through autophagy (SI Appendix, Fig. S10 A and B). Importantly, RNA sequencing data indicate that SNAT7 is significantly up-regulated in pancreatic adenocarcinoma (PDAC), whereas SNAT9 is not (Fig. 4E). We assessed the protein levels of SNAT7 in a cohort of paired PDAC patient samples by immunohistochemistry (IHC) staining. SNAT7 protein levels are significantly elevated in PDAC tumor samples when compared to benign control tissues (Fig. 4F). Taken together, SNAT7 is essential for mTORC1 activation and the growth of pancreatic cancer cells from macropinocytosed extracellular protein.
Fig. 4.
SNAT7 regulates pancreatic cancer cell growth through mTORC1. (A) SNAT7 depletion inhibits BSA-induced mTORC1 activation and cell proliferation in pancreatic cancer cell lines. Relative cell proliferation was determined for WT MIA PaCa-2 cells, SNAT7 KO, SNAT7 KO MIA PaCa-2 cells expressing HA-tagged SNAT7WT, HA-tagged SNAT7N62H, or HA-tagged SNAT721-C in low glutamine (Gln) with or without Torin, or low Gln + 3% BSA with or without Torin for 24 h. P values: WT Gln vs. WT Gln + BSA, P < 0.001; WT Gln vs. SNAT7 KO Gln, P < 0.001; WT Gln vs. SNAT7 KO Gln + BSA, P < 0.05; WT Gln vs. HA-SNAT7WT Gln, not significant; WT Gln vs. HA- SNAT7WT Gln + BSA, P < 0.01; WT Gln vs. HA-SNAT7N62H Gln, P < 0.01; WT Gln vs. HA-SNAT7N62H Gln + BSA, not significant; WT Gln vs. HA-SNAT721-C Gln, not significant; WT Gln vs. HA-SNAT721-C Gln + BSA, not significant; WT Gln + BSA vs. SNAT7 KO Gln + BSA, P < 0.001; WT Gln + BSA vs. HA- SNAT7WT Gln + BSA, not significant; WT Gln + BSA vs. HA-SNAT7N62H Gln + BSA, P < 0.001; WT Gln + BSA vs. HA-SNAT721-C Gln + BSA, P < 0.05; SNAT7 KO Gln vs. HA-SNAT7WT Gln, P < 0.001; SNAT7 KO Gln vs. HA-SNAT7N62H Gln, P < 0.01; SNAT7 KO Gln vs. HA-SNAT721-C Gln, P < 0.001; SNAT7 KO Gln + BSA vs. HA-SNAT7WT Gln + BSA, P < 0.001; SNAT7 KO Gln + BSA vs. HA-SNAT7N62H Gln + BSA, not significant; and SNAT7 KO Gln + BSA vs. HA-SNAT721-C Gln + BSA, P < 0.01. (B) SNAT7 depletion inhibits cell growth in pancreatic cancer cell lines. The cell size of control, SNAT7 KO, SNAT7 KO expressing HA-tagged SNAT7WT, SNAT7 KO expressing HA-tagged SNAT7N62H, or SNAT7 KO expressing HA-tagged SNAT721-C MIA PaCa-2 cells were analyzed after 48 h in low glutamine with 3% BSA condition. P values: control vs. SNAT7 KO, P < 0.001; control vs. HA-SNAT7WT, not significant; control vs. HA-SNAT7N62H, P < 0.01; control vs. SNAT7 KO, P < 0.01; SNAT7 KO vs. HA- SNAT7WT P < 0.001; SNAT7 KO vs. HA-SNAT7N62H, P < 0.001; and SNAT7 KO vs. HA-SNAT721-C, P < 0.01. (C) Targeting mTORC1 to the lysosome rescues cell proliferation when SNAT7 is depleted. Cell proliferation was analyzed in WT MIA PaCa-2 cells (control), SNAT7 KO, or SNAT7 KO cells expressing Flag-tagged Raptor-Rheb. P values: control vs. SNAT7 KO, P < 0.01 and SNAT7 KO vs. SNAT7 KO expressing Flag-tagged Raptor-Rheb, P < 0.01. (D) Targeting mTORC1 to the lysosome partially rescues cell size when SNAT7 is depleted. Cell size was measured for WT MIA PaCa-2 cells (control), SNAT7 KO, or SNAT7 KO cells expressing Flag-tagged Raptor-Rheb 48 h after seeding. P values: control vs. SNAT7 KO, P < 0.01 and SNAT7 KO vs. SNAT7 + Flag-Raptor-Rheb, P < 0.05. (E) Expression of SNAT7 is elevated in pancreatic adenocarcinomas. mRNA expression of SNAT7 or SNAT9 in pancreatic adenocarcinomas was analyzed at the GEPIA platform based on TCGA and GTEx dataset. TPM, transcripts per million. *q < 0.001, ANOVA analysis followed by Benjamini and Hochberg false discovery rate correction. (F) SNAT7 protein expression is up-regulated in pancreatic ductal adenocarcinoma tissues. SNAT7 protein expression was analyzed by immunohistochemistry (IHC) staining in 20 pairs of pancreatic ductal adenocarcinoma tissues. (Left) Immunoreactivity scores (H-score) was calculated based on percentage and intensity of staining (see Materials and Methods for details). Statistical analysis was performed by paired Student’s t test. P < 0.0001. (Right) Representative staining images of IHC and corresponding hematoxylin and eosin staining are also shown for tumor and normal samples as indicated. (G) Working model of glutamine (Gln) and asparagine (Asn) signaling pathway to mTORC1. Macropinocytosis engulfs proteins, such as albumin where it is targeted to the lysosome and degraded by proteolysis. Then lysosomal amino acids, specifically Gln and Asn, are exported out of the lysosome into the cytoplasm by SNAT7. Cytosolic Gln and Asn (from macropinocytosis or intracellular/extracellular stores) can then activate mTORC1 through an unknown sensor(s). Growth factors activate mTORC1 through tuberous sclerosis complex (TSC)-Rheb signaling axis. ? denotes that another protein may bridge the SNAT7-mTORC1 interaction.
Discussion
Amino acid sensing by mTORC1 is crucial in regulating cell growth, metabolism, and other mTORC1-mediated biology. Like Gln, Asn signals to mTORC1 in a Rag GTPase-independent pathway and requires Arf1 (26). We identified a poorly characterized SLC, referred to as SNAT7, that links Gln and Asn sensing by mTORC1 at the lysosome via macropinocytosis (Fig. 4G). This pathway is important in Ras transformed cells, where macropinocytosis fuels cells with extracellular nutrients resulting in mTORC1 activation and pancreatic cancer cell growth. SNAT7 exports Gln and Asn out of the lysosome (40), and our results suggest that Gln and Asn exported out of the lysosome by SNAT7 activate mTORC1 and fuel pancreatic cancer cell growth. Cytosolic Gln and Asn do not require SNAT7 to activate mTORC1. The precise mechanisms by which Gln and Asn signal to and activate mTORC1, and whether Gln and Asn sensors exist, await discovery.
Many mechanistic details have been revealed for how Leu, Arg, and Met can regulate mTORC1 activity through the Rag GTPases (5, 19–23). For example, protein sensors have been identified for Leu and Arg as Sestrin2 and CASTOR1, respectively (22, 28–31). CASTOR1 has ACT domains, evolutionarily conserved ancient domains that function as small molecule binding modules for diverse ligands, including amino acids and nucleotides (64–66). Only a few ACT-containing proteins to date have been identified and characterized in mammals and other amino acid sensors involved in the mTOR pathway may contain these ACT domains (65–70). GlnD is a bifunctional uridylyltransferase/uridylyl-removing enzyme that senses Gln levels through its C-terminal ACT domains in Escherichia coli (71); however, there is no clear eukaryotic homolog. In addition, a recent study reveals aminoacyl-tRNA synthetases (ARSs) transfer amino acids to other proteins (72). Leu can modify RagA at lysine 142 and promote mTORC1 activity. Intriguingly, these findings suggest protein modification via amino acids may regulate mTORC1 signaling. Gln and Asn sensors, or proteins modified by Gln and Asn involved in mTORC1 signaling have yet to be discovered. Given the structural similarity between Gln and Asn, it is possible that they have the same sensor.
Amino acid transporters play a major role in mTORC1 regulation. Plasma membrane localized SLCs like SLC3A2, SLC7A5, SLC1A5, SLC38A2, and SLC38A5 modulate mTORC1 by facilitating amino acid transport across the plasma membrane (20, 73, 74). Lysosomal localized SLCs, such as SLC36A1, also modulate mTORC1 activity by releasing lysosomal amino acids into the cytoplasm (75). Furthermore, the lysosomal localized transporter SNAT9 is an Arg and essential amino acid transporter required for mTORC1 activation in a Rag-dependent manner (32–34). Recently, SNAT9 has been shown to transport several essential amino acids in an Arg-regulated fashion (35). Moreover, depletion of SNAT9 in Ras-transformed pancreatic cancer cells lines decreased macropinocytosis-induced pancreatic tumor growth. Our results show that the Gln and Asn transporter SNAT7 is essential for mTORC1 activation independently of the Rag GTPases and is crucial for the growth of pancreatic cancer cells from macropinocytosed extracellular protein. Because depletion of SNAT7 in pancreatic cancer cells maintained a small degree of mTORC1 activation, we suspect that both SNAT9 and SNAT7 play a role in albumin-induced pancreatic cancer cell growth (Fig. 3A). Other SLCs involved in macropinocytosis and mTORC1 regulation have yet to be discovered.
SNAT7 regulates mTORC1 through macropinocytosis. However, SNAT7 does not appear to alter autophagy-dependent reactivation of mTORC1 following glutamine starvation (SI Appendix, Fig. S10). This is somewhat surprising because macropinocytosis and autophagy are both thought to result in the release of amino acids (such as glutamine) from the lysosome. SNAT9 has previously been shown to play a role in autophagy (35). Glutamine starvation may trigger autophagy where Arg or other amino acids are exported out of the lysosome by SNAT9 resulting in mTORC1 activation. Perhaps macropinocytosis results in higher export of Gln from the lysosome, than that of autophagy. Alternatively, long-term glutamine starvation may initiate de novo glutamine synthesis leading to mTORC1 reactivation independently of SNAT7. Studies have previous showed that glutamine starvation can promote de novo glutamine synthesis (76, 77). Last, glutamine starvation (to some extent) could result in activation of GCN2-ATF4 pathway promoting extracellular amino acid uptake and subsequent mTORC1 reactivation (78).
Both Gln and Asn are implicated in tumorigenesis and cancer. Multiple tumor cell lines display increased rates of Gln consumption to support macromolecular biosynthesis and cell proliferation (51, 62, 63). For example, Gln drives the tricarboxylic acid cycle, activates mTORC1, and contributes to the synthesis of lipids, nucleotides, and nonessential amino acids. Interestingly, previous studies show Asn is capable of rescuing cancer cell growth in the absence of Gln (79). Asn is important for tumor growth, due to the effectiveness of treating low-ASNS expressing leukemia with asparaginase (80). Asparaginase is effective as a therapeutic for cancers that obtain most of their Asn from their extracellular environment. Asn bioavailability also contributes to metastatic breast cancer, and asparaginase treatment reduces this metastasis (81). We anticipate that the findings presented here will have a significant impact on understanding mTORC1 activation in cancer. Furthermore, SNAT7 may represent a new target for Gln- and Asn-related anticancer therapies.
Materials and Methods
Amino Acid Starvation and Amino Acid or BSA Stimulation of Cells.
Amino acid-free medium was made following the Sigma (#D5796) high-glucose DMEM recipe with the exception that all amino acids were omitted. All experiments with amino acid starvation and amino acid or BSA stimulation contained 10% dialyzed FBS (#F0392 from Sigma) instead of regular FBS (#F2442 from Sigma) unless otherwise indicated. Amino acid starvation was performed by replacing regular medium with amino acid-free medium for 2 h prior to amino acid or BSA stimulation unless otherwise indicated. For all amino acid stimulation experiments, amino acids were used with indicated concentration and time points, and “+AA” denotes replacement of regular media containing amino acids. For BSA stimulation, cells were starved of amino acids for 2 h, followed by replacement of starvation media containing 3% BSA for the indicated times.
Immunofluorescent Microscopy.
Cells were seeded in 24-well plates on coverslips (VWR #89015–725) one day prior to experimentation [similar procedure as previous reported (82)]. Coverslips were pretreated with 5 μg/mL of fibronectin (#F4759 from Sigma) at 37 °C for 1 h, with a quick phosphate-buffered saline (PBS) wash prior to cell seeding. The following steps were performed at room temperature: cells were briefly washed with PBS and fixed with 4% paraformaldehyde (#15710 from Electron Microscopy Sciences) in PBS for 20 min, followed by washing with PBS three times for 5 min each. Cells were permeabilized with 0.01% saponin in PBS (for MEF cells) or 0.2% Triton-X100 in PBS (for HEK293A and MIA PaCa-2 cells) for 10 min. Cells on coverslips were blocked in 2% BSA in TBS-Tween (TBST) for 1 h, followed by washing with TBST three times (5 min each). Cells were incubated for 1 h with primary antibodies diluted in PBS, followed by washing with PBS three times (5 min each). Cells were incubated for 1 h with secondary antibodies diluted in PBS, followed by washing with PBS twice (5 min each), then washed with double distilled water twice (5 min each).
For LysoTracker experiments, cells were treated with 100 nM LysoTracker Red DND-99 1 h prior to fixation. For dextran-TMR experiments, cells were treated with 1 mg/mL dextran-TMR 30 min prior to fixation.
Coverslips were mounted onto microscope slides (#12-550-15 from Thermo Fisher Scientific) using ProLong Gold Antifade Reagent with DAPI (#P36931 from Thermo Fisher Scientific). Images were captured with a Zeiss LSM 800 confocal microscope. Images depicted in figures were exported from Zeiss ZEN imaging software. Individual channels were pseudocolored using ImageJ prior to assembling figures. For Fig. 3 D, F, and G, the colocalization of mTOR and LAMP2 was analyzed using the Squassh segmentation and analysis algorithm in ImageJ (83).
Lysosomal-Immunoprecipitation and Metabolite Extraction for Liquid Chromatography-Mass Spectrometry.
Wild-type (WT) and SNAT7 KO MIA PaCa-2 cells stably expressing Flag-tagged or HA-tagged TMEM192 were plated in 15-cm plates for the following conditions: amino acid starvation (−AA), glutamine/asparagine (2 mM) stimulation, and glutamine ester/asparagine ester (2 mM) stimulation. Cells were then starved for 2 h or stimulated with either +glutamine/asparagine or +glutamine/asparagine esters. Samples were processed separately on ice or 4 °C. Cells were rinsed with ice cold 1× PBS and then scraped in 1 mL KPBS (136 mM KCL, 10 mM KH2PO4, and pH −7.25 was adjusted with KOH). Further processing of samples was done according to (35).
Cell Proliferation.
A total of 5 × 104 MIA PaCa-2 cells were seeded in triplicate in 12-well plates. After 48 h, cells were trypsinized for the counting. Cells were placed on the counting slide (Bio-Rad, #145-0011) and counted using a Bio-Rad TC20 Automated Cell Counter (Bio-Rad, #1450102). For SNAT7 WT, SNAT7 KO, HA-SNAT7WT, HA-SNAT7N62H, and HA-SNAT721-C cell proliferation, 5 × 104 cells were plated in triplicate in 12-well plates. After 24 h, cells were stimulated in 0.2 mM glutamine (Sigma, #G3126) medium (Thermo Fisher Scientific, #11960069) with or without 3% BSA (Sigma, #A1470) and 500 nM Torin (Sigma, #475991) and were incubated for 48 h. Then, cells were washed three times with 1× PBS and stained with 0.5% crystal violet (Sigma, #C6158) in 20% methanol. For measuring absorbance at 590 nm, stained cells were resolved in ethanol. Cells incubated for 24 h in low glutamine were used as a basal level.
Cell Size.
A total of 5 × 104 WT or SNAT7 KO MIA PaCa-2 cells, HA-SNAT7WT, HA-SNAT7N62H, or HA-SNAT721-C were seeded into 12-well plates and were cultured in low glutamine medium (0.2 mM glutamine). After 48 h, cells were lifted by 0.25% Trypsin-EDTA solution (Sigma, #5642) and then resuspended with 1× PBS. Cell size was measured by using a Z2 Coulter Particle Count and Size Analyzer (Beckman Coulter) and processed by Z2 Accucomp software (Beckman Coulter).
Forty-eight hours later, the cells were harvested by trypsinization and resuspended in 1 mL PBS, diluted 1:50 with counting solution (isoton, Beckman Coulter), and cell diameters determined using a particle size counter (Coulter Z2, Beckman Coulter) with Coulter Z2 AccuComp software.
Statistical Analysis.
Statistical analyses were conducted using Prism 7 (GraphPad). Student’s t tests were used for comparison between two groups and P values less than 0.05 were considered as statistically significant. Paired t test was performed for immunohistochemistry scores in paired patient samples.
Detailed material and methods for cell lines, tissue culture, antibodies, chemicals, plasmids, cDNA transfection, RNA interference, GFP nanobody purification, cell lysis, cell immunoprecipitation, subcellular fractionation, Western blot, real-time PCR, generation of stable cell lines, generation of KO cells using CRISPR/Cas9 genome editing, patient issue samples, immunohistochemistry, and immunohistochemistry scoring are available in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to Kun-Liang Guan, Melanie Cobb, Vincent Tagliabracci, Kim Orth, Beth Levine, and the members of the Jewell laboratory for insightful discussions. We thank Ting-Sung Hsieh, Huyen Nguyen, Teresa Yoon, Anderson Frank, Thu Nguyen, and Linda Zhong for suggestions and technical help. This work was supported by grants from Cancer Prevention Research Institute of Texas (CPRIT) Scholar Recruitment of First-Time, Tenure-Track Faculty Member (RR150032), Cancer Prevention Research Institute of Texas (CPRIT) High-Impact/High-Risk Research Award (RP160713), The Welch Foundation (I-1927-20170325 and I-1927-20200401), 2017 UT Southwestern President’s Research Council Distinguished Researcher Award, American Cancer Society Research Scholar Grant (133894-RSG-19-162-01-TBE), and National Institutes of Health (R01GM129097-01) to J.L.J. R.J.D. is supported by grants from CPRIT (RP180778), the National Cancer Institute (R35CA22044901), and The Welch Foundation (I-1733). C.H.M. is supported by National Institutes of Health (T32GM008203).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123261119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Jewell J. L., Guan K. L., Nutrient signaling to mTOR and cell growth. Trends Biochem. Sci. 38, 233–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Palm W., Thompson C. B., Nutrient acquisition strategies of mammalian cells. Nature 546, 234–242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomes A. P., Blenis J., A nexus for cellular homeostasis: The interplay between metabolic and signal transduction pathways. Curr. Opin. Biotechnol. 34, 110–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sancak Y., et al. , The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menon S., et al. , Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sancak Y., et al. , Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H., et al. , Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552, 368–373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoki K., Li Y., Xu T., Guan K. L., Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., et al. , Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5, 578–581 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J., Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13, 1259–1268 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L., Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jewell J. L., et al. , Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 347, 194–198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bar-Peled L., Schweitzer L. D., Zoncu R., Sabatini D. M., Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoncu R., et al. , mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334, 678–683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bar-Peled L., et al. , A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsun Z. Y., et al. , The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 52, 495–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfson R. L., et al. , KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara K., et al. , Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273, 14484–14494 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Nicklin P., et al. , Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136, 521–534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauchart-Thevret C., Cui L., Wu G., Burrin D. G., Arginine-induced stimulation of protein synthesis and survival in IPEC-J2 cells is mediated by mTOR but not nitric oxide. Am. J. Physiol. Endocrinol. Metab. 299, E899–E909 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Chantranupong L., et al. , The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165, 153–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu X., et al. , SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durán R. V., et al. , Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell 47, 349–358 (2012). [DOI] [PubMed] [Google Scholar]
- 25.van der Vos K. E., et al. , Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat. Cell Biol. 14, 829–837 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Meng D., et al. , Glutamine and asparagine activate mTORC1 independently of Rag GTPases. J. Biol. Chem. 295, 2890–2899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krall A. S., Xu S., Graeber T. G., Braas D., Christofk H. R., Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun. 7, 11457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfson R. L., et al. , Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saxton R. A., et al. , Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 351, 53–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxton R. A., Chantranupong L., Knockenhauer K. E., Schwartz T. U., Sabatini D. M., Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 536, 229–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chantranupong L., et al. , The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 9, 1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung J., Genau H. M., Behrends C., Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol. Cell. Biol. 35, 2479–2494 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rebsamen M., et al. , SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 519, 477–481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S., et al. , Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyant G. A., et al. , mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell 171, 642–654.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin L., Yee S. W., Kim R. B., Giacomini K. M., SLC transporters as therapeutic targets: Emerging opportunities. Nat. Rev. Drug Discov. 14, 543–560 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.César-Razquin A., et al. , A call for systematic research on solute carriers. Cell 162, 478–487 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Stracka D., Jozefczuk S., Rudroff F., Sauer U., Hall M. N., Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J. Biol. Chem. 289, 25010–25020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanigawa M., Maeda T., An In Vitro TORC1 kinase assay that recapitulates the gtr-independent glutamine-responsive TORC1 activation mechanism on yeast vacuoles. Mol. Cell. Biol. 37, e00075-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdon Q., et al. , SNAT7 is the primary lysosomal glutamine exporter required for extracellular protein-dependent growth of cancer cells. Proc. Natl. Acad. Sci. U.S.A. 114, E3602–E3611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chapel A., et al. , An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol. Cell. Proteomics 12, 1572–1588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jadot M., et al. , Accounting for protein subcellular localization: A compartmental map of the rat liver proteome. Mol. Cell. Proteomics 16, 194–212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanson J. A., Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 9, 639–649 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayor S., Pagano R. E., Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 8, 603–612 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Bar-Sagi D., Feramisco J. R., Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by Ras proteins. Science 233, 1061–1068 (1986). [DOI] [PubMed] [Google Scholar]
- 46.Porat-Shliom N., Kloog Y., Donaldson J. G., A unique platform for H-Ras signaling involving clathrin-independent endocytosis. Mol. Biol. Cell 19, 765–775 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walsh A. B., Bar-Sagi D., Differential activation of the Rac pathway by Ha-Ras and K-Ras. J. Biol. Chem. 276, 15609–15615 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Commisso C., et al. , Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palm W., et al. , The utilization of extracellular proteins as nutrients is suppressed by mTORC1. Cell 162, 259–270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramirez C., Hauser A. D., Vucic E. A., Bar-Sagi D., Plasma membrane V-ATPase controls oncogenic RAS-induced macropinocytosis. Nature 576, 477–481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pavlova N. N., Thompson C. B., The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jewell J. L., Russell R. C., Guan K. L., Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14, 133–139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox A. D., Fesik S. W., Kimmelman A. C., Luo J., Der C. J., Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim D. H., et al. , mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Manifava M., et al. , Dynamics of mTORC1 activation in response to amino acids. eLife 5, e19960 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reeves J. P., Accumulation of amino acids by lysosomes incubated with amino acid methyl esters. J. Biol. Chem. 254, 8914–8921 (1979). [PubMed] [Google Scholar]
- 57.Bröer S., Amino acid transport across mammalian intestinal and renal epithelia. Physiol. Rev. 88, 249–286 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Zhang Z., Gameiro A., Grewer C., Highly conserved asparagine 82 controls the interaction of Na+ with the sodium-coupled neutral amino acid transporter SNAT2. J. Biol. Chem. 283, 12284–12292 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uhlén M., et al. , Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hensley C. T., Wasti A. T., DeBerardinis R. J., Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J. Clin. Invest. 123, 3678–3684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wise D. R., Thompson C. B., Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chipman D. M., Shaanan B., The ACT domain family. Curr. Opin. Struct. Biol. 11, 694–700 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Grant G. A., The ACT domain: A small molecule binding domain and its role as a common regulatory element. J. Biol. Chem. 281, 33825–33829 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Lang E. J., Cross P. J., Mittelstädt G., Jameson G. B., Parker E. J., Allosteric ACTion: The varied ACT domains regulating enzymes of amino-acid metabolism. Curr. Opin. Struct. Biol. 29, 102–111 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Aravind L., Koonin E. V., Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 287, 1023–1040 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Carluccio C., Fraternali F., Salvatore F., Fornili A., Zagari A., Structural features of the regulatory ACT domain of phenylalanine hydroxylase. PLoS One 8, e79482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kobe B., et al. , Structural basis of autoregulation of phenylalanine hydroxylase. Nat. Struct. Biol. 6, 442–448 (1999). [DOI] [PubMed] [Google Scholar]
- 70.Siltberg-Liberles J., Martinez A., Searching distant homologs of the regulatory ACT domain in phenylalanine hydroxylase. Amino Acids 36, 235–249 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y., Pohlmann E. L., Serate J., Conrad M. C., Roberts G. P., Mutagenesis and functional characterization of the four domains of GlnD, a bifunctional nitrogen sensor protein. J. Bacteriol. 192, 2711–2721 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He X. D., et al. , Sensing and transmitting intracellular amino acid signals through reversible lysine aminoacylations. Cell Metab. 27, 151–166.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Pinilla J., et al. , SNAT2 transceptor signalling via mTOR: A role in cell growth and proliferation? Front. Biosci. (Elite Ed.) 3, 1289–1299 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Kim J., et al. , Amino acid transporter Slc38a5 controls glucagon receptor inhibition-induced pancreatic α cell hyperplasia in mice. Cell Metab. 25, 1348–1361.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ögmundsdóttir M. H., et al. , Proton-assisted amino acid transporter PAT1 complexes with Rag GTPases and activates TORC1 on late endosomal and lysosomal membranes. PLoS One 7, e36616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tajan M., et al. , A role for p53 in the adaptation to glutamine starvation through the expression of SLC1A3. Cell Metab. 28, 721–736.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tardito S., et al. , Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat. Cell Biol. 17, 1556–1568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Staschke K. A., Wek R. C., Adapting to cell stress from inside and out. Nat. Cell Biol. 21, 799–800 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Pavlova N. N., et al. , As extracellular glutamine levels decline, asparagine becomes an essential amino acid. Cell Metab. 27, 428–438.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pieters R., et al. , L-asparaginase treatment in acute lymphoblastic leukemia: A focus on Erwinia asparaginase. Cancer 117, 238–249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Knott S. R. V., et al. , Asparagine bioavailability governs metastasis in a model of breast cancer. Nature 554, 378–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meng D., et al. , ArfGAP1 inhibits mTORC1 lysosomal localization and activation. EMBO J. 40, e106412 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rizk A., et al. , Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat. Protoc. 9, 586–596 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.