Significance
Dietary fibers contain complex mixtures of biomolecules, making it difficult to develop/test hypotheses about how different fiber-types impact different components of the human gut microbiome and how microbiome changes that they produce are linked to human physiology. Here, we analyze microbiome and plasma proteome responses to consumption of two fiber-enriched snacks in two human studies. We use a variety of computational methods to correlate their effects on gut microbiome genes encoding enzymes that degrade complex fiber-associated polysaccharides, the microbial products of polysaccharide degradation, and plasma proteins representing diverse physiological processes. This approach can be used to guide the design of fiber-containing snacks that more precisely manipulate microbiome features in ways that improve nutritional and health status.
Keywords: gut microbiome-directed foods, carbohydrate-active enzymes, fiber-glycan metabolism, microbiome-plasma proteome relationships
Abstract
Increases in snack consumption associated with Westernized lifestyles provide an opportunity to introduce nutritious foods into poor diets. We describe two 10-wk-long open label, single group assignment human studies that measured the effects of two snack prototypes containing fiber preparations from two sustainable and scalable sources; the byproducts remaining after isolation of protein from the endosperm of peas and the vesicular pulp remaining after processing oranges for the manufacture of juices. The normal diets of study participants were supplemented with either a pea- or orange fiber-containing snack. We focused our analysis on quantifying the abundances of genes encoding carbohydrate-active enzymes (CAZymes) (glycoside hydrolases and polysaccharide lyases) in the fecal microbiome, mass spectrometric measurements of glycan structures (glycosidic linkages) in feces, plus aptamer-based assessment of levels of 1,300 plasma proteins reflecting a broad range of physiological functions. Computational methods for feature selection identified treatment-discriminatory changes in CAZyme genes that correlated with alterations in levels of fiber-associated glycosidic linkages; these changes in turn correlated with levels of plasma proteins representing diverse biological functions, including transforming growth factor type β/bone morphogenetic protein-mediated fibrosis, vascular endothelial growth factor-related angiogenesis, P38/MAPK-associated immune cell signaling, and obesity-associated hormonal regulators. The approach used represents a way to connect changes in consumer microbiomes produced by specific fiber types with host responses in the context of varying background diets.
Advances in our understanding of the role of the gut microbiome in regulating many aspects of human physiology hold the promise of evolving our view of human nutrition by establishing mechanistic connections between the foods we consume and how they affect health status. One manifestation of this effort is a series of studies, performed on well-phenotyped cohorts, that seek to relate features of gut microbial community composition (organisms, genes), dietary practices, and pre- and postprandial cardiometabolic responses to test meals (1–4). A key question raised by these initiatives relates to the nature of the “bioactive” components of foods. Specifically, what are the nutrients utilized by various gut community members or microbiome-encoded metabolic pathways? What products are produced by biotransformation of these nutrients? How are these products linked to specific host physiologic (or pathophysiologic) processes?
Plant-derived dietary fibers represent a “poster child” for these efforts and illustrate the formidable challenges faced. The health benefits of dietary fibers are widely known, as is their inadequate representation in Western diets. However, natural fibers are structurally complex and highly diverse. They contain numerous, typically undefined polysaccharide structures and largely unspecified protein, lipid, and small molecule constituents. Their composition varies as a function of their origin (food staple and cultivar), the different methods employed to recover them from these sources, as well as the different techniques used to incorporate them into processed foods with acceptable organoleptic properties (5). Moreover, analyzing the host effects of metabolism of different fibers is confounded by the fact that there is substantial intra- and interpersonal variation in microbiome configuration (6, 7).
Snacking is becoming an ever more dominant feature of daily life worldwide and thus provides an opportunity to introduce nutritious ingredients, such as fibers, into diets. However, obtaining structure-activity relationships for specific fiber types and their corresponding targets in the gut community is foundational for designing snack foods that evoke and/or reinforce microbiome responses that are beneficial to the host.
Degradation of dietary polysaccharides is a function primarily performed by bacterial carbohydrate-active enzymes (CAZymes). The gut microbiome harbors tens of thousands of CAZyme genes belonging to at least 136 glycoside hydrolase (GH) and 29 polysaccharide lyase (PL) families [extrapolated and updated from El Kaoutari et al. (8)]. In contrast, the human genome only contains 98 GH and no PL genes (9), of which <20% contribute to the processing of dietary glycans.
In the current study, we test the effects of dietary supplementation with two snack food prototypes, one containing pea fiber and the other orange fiber, in two pilot studies of overweight and obese individuals consuming their normal, unrestricted diets. Our strategy was to focus on fiber-associated changes in the abundances of microbial GH and PL genes to determine whether responses to the pea or orange fiber prototypes in the gut microbiome and host are decipherable against a background of varying dietary practices and starting microbiome configurations. Higher order singular value decomposition (10) was utilized as a feature selection tool to identify treatment-discriminating changes in GH and PL gene representation. Mass spectrometric assays of the levels of fecal glycan structures (glycosidic linkages) were subsequently performed and the results were correlated with changes in the abundances of treatment-discriminating GH and PL genes with known or predicted substrate specificities. Our analysis concluded by measuring changes in levels of 1,305 plasma proteins in each study participant as a function of fiber treatment and applying computational tools to identify links between these microbiome and plasma proteome changes in response to fiber consumption. Our results provide an approach, using pilot human studies, for selecting specific fiber preparations, plus informative microbiome and host biomarkers, that can be advanced to proof-of-concept clinical trials which assess their capacity for precise manipulation of microbiome and host features.
Results
Study Design.
The snack prototypes used in this study incorporated plant fibers from one of two sustainable and scalable sources: (i) the byproducts remaining after isolation of protein from the endosperm of peas, and (ii) the vesicular pulp recovered after processing oranges for the manufacture of juice. Arabinose and galacturonic acid are the most abundant monosaccharides in both fiber preparations; however, arabinan is more abundant in pea fiber (22.4% arabinose vs. 13.9% in orange fiber) while galacturonan (in the form of homogalacturonan and rhamnogalacturonan I) predominates in orange fiber (42.9% galacturonic acid vs. 13.9% in pea fiber). Other glycans found in these two fiber preparations include xylans (defined by the presence of 4-linked xylose), galactans (containing 4-linked galactose), arabinogalactans (3-linked galactose), as well as amylopectins, amyloses, xyloglucans, and cellulose which all possess 4-linked glucose and 4,6-linked glucose (Dataset S1A) (11–13).
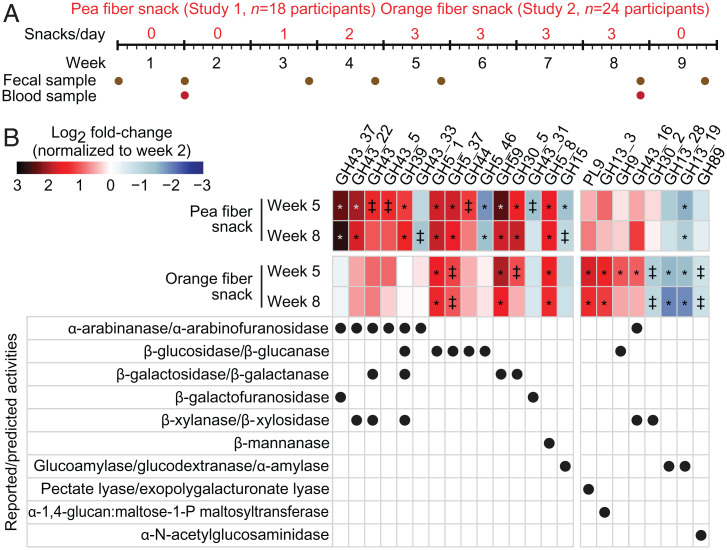
We first performed an exploratory 10-wk open label, single group assignment study of the effects of snack prototypes containing 6.7–8.1 g of pea fiber per 35 g serving on the fecal microbiomes of nine pairs of adult dizygotic twins (36.6 ± 2.9 y [mean ± SD]) recruited from the Missouri Adolescent Female Twin Study (MOAFTS) cohort (14) (Dataset S1B). The study design is summarized in Fig. 1A. In the 2-wk-long preintervention phase, participants consumed their normal diets. Starting at week three, they supplemented their diets with a single pea fiber snack serving a day for 1 wk (either in the form of a biscuit [6.7 g pea fiber] or bar [8.1 g extruded pea fiber]; note that alternate product formats of the pea fiber snack were offered to accommodate personal preferences and ensure compliance). The “dose” was increased to two snack servings a day during the following week and then to three servings per day for the next 4 wk (weeks 5–8) (see Dataset S1A for nutritional analysis of snacks).
Fig. 1.
Characterizing fecal microbiome responses in participants consuming the fiber-snack prototypes based on changes in the representation of CAZyme genes. (A) Design of human study 1 (pea fiber) and study 2 (orange fiber). (B) Heatmap summarizing the log2 fold change in the representation of glycoside hydrolase (GH) and polysaccharide lyase (PL) genes with statistically significant changes in their abundances in response to pea or orange fiber-snack consumption compared to baseline. The reported or predicted activities of these CAZymes and the magnitude of the changes in the abundance of their genes are shown (mean values for each study cohort). Linear mixed-effects model (false discovery rate-corrected): ‡q < 0.1, *q < 0.05; n = 18 and 24 participants for study 1 and study 2, respectively, n = 120 fecal samples analyzed.
We subsequently performed a second human study with the same design (Fig. 1A). However, instead of a pea fiber-containing snack, the diets of participants were supplemented with an escalating dose of 1, 2, and finally 3 servings/d of a 35 g snack bar containing 10.2 g of extruded orange fiber. This study involved 12 dizygotic twin pairs from the MOAFTS cohort (37 ± 2.9 y [mean± SD]), including nine pairs who had participated in the pea fiber study. For these 18 participants, the interval between cessation of pea fiber snack consumption and initiation of orange fiber consumption ranged from 50 to 106 d (84 ± 26 d [mean± SD]). Because we did not sample the fecal microbiota or collect diet histories during this interval, we treated the pea and orange fiber supplementation studies as independent (although we cannot formally rule out residual effects from study 1 carrying over to study 2). Blood samples were obtained for clinical chemistry (Dataset S1C) and plasma proteomic analyses, while fecal samples were collected at the time points shown in Fig. 1A for the microbiome and carbohydrate analyses described below. Dataset S1 D–G documents participants’ self-reported weekly dietary histories and any gastrointestinal symptoms.
The choice of twins for these two studies was not related to the primary outcome measures, but rather because substantial clinical data were available for this long-studied cohort together with preclinical data generated from transplantation of their microbiomes into gnotobiotic mice (12, 14). Twenty-two of the 24 members of the twin cohort who were enrolled in the two studies were either overweight or obese (body mass index [BMI] ≥25 kg/m2). No statistically significant differences in BMI were observed over the period of intervention for either study, nor were there significant effects on levels of C-reactive protein or on fasting levels of insulin, total cholesterol, low-density lipoprotein, or triglycerides. High-density lipoprotein levels were marginally decreased after orange fiber treatment (see Dataset S1C for results and statistical tests).
Identifying CAZyme Genes Responsive to the Different Fiber Snack Prototypes.
Pea fiber.
We performed shotgun sequencing of DNA isolated from fecal samples collected from all 18 participants while they were consuming their normal unsupplemented diet (week 2) and during supplementation with three pea fiber snacks a day (weeks 5 and 8) (Dataset S2A). CAZymes were annotated by their family, and where appropriate by subfamily, as defined in the CAZy database (15) (see Dataset S2B for the reported or predicted functions of all CAZymes in the resulting dataset). Using the procedure for higher-order singular value decomposition (HOSVD) illustrated in SI Appendix, Fig. S1A, we constructed a tensor composed of three matrices, one for each fecal collection timepoint (weeks 2, 5, and 8), where rows represent the 18 participants and columns represent the log2 fold-change in CAZyme gene representation compared to week 2 (preintervention). We then employed a numeric approximation method known as canonical-polyadic alternating least squares (CP-ALS) to define the dimensions of a diagonal “core tensor” that relates all components of the tensor. We subsequently generated a “randomized tensor” by shuffling the rows (each participant), columns (each CAZyme) and z axis (timepoints) of the original tensor; this allowed us to identify a reduced number of tensor components that could be used to characterize changes in microbiome configuration in response to pea fiber snack supplementation. GH and PL genes whose abundances were statistically significantly different after pea fiber snack consumption were defined as having a q-value <0.1 (linear-mixed effects model) and positioned at the tails (α <0.1) of the distribution of CAZymes along tensor component 1 (TC1) (Dataset S2C).
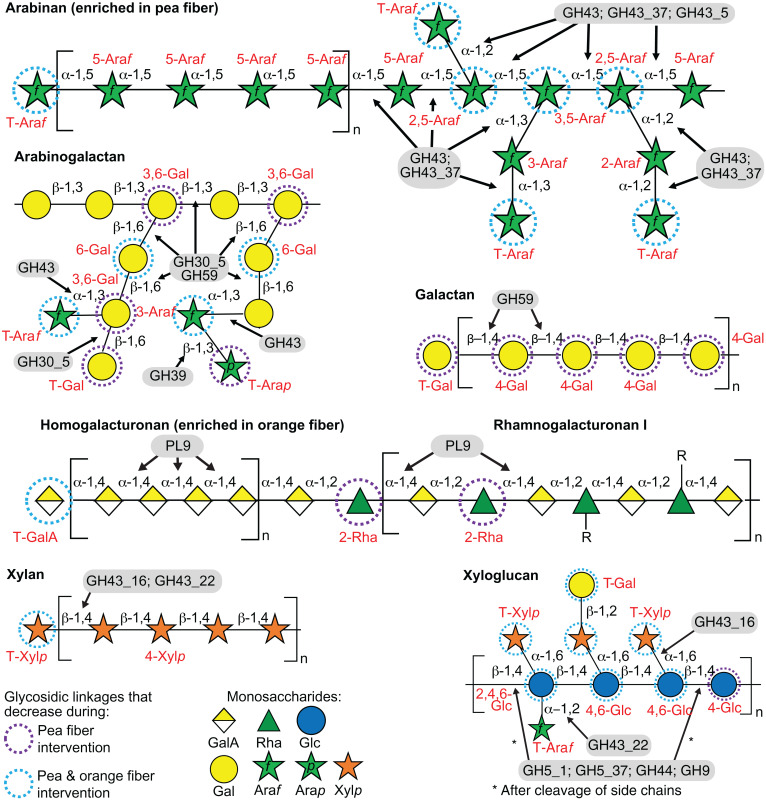
The top two rows of Fig. 1B integrate data from all 18 participants and include GH and PL genes with statistically significant mean log2 fold increases in their abundances at weeks 5 and 8 (i.e., after 1 and 4 wk of consumption of three pea fiber snacks per day, respectively) compared to preintervention. These increases can be related to the composition of the pea fiber preparation; they include genes encoding CAZymes that (i) have α-L-arabinanase and α-L-arabinofuranosidase activity (GH43_37, GH43_22, GH43, and GH43_5) (16, 17), (ii) metabolize galactans and arabinogalactans (GH59 [β-galactosidase] and GH30_5 [β-galactanase]) (18, 19), (iii) metabolize xylans (GH43_22 [β-xylanase]) (19), (iv) process cellulose and xyloglucans (GH5_1, GH5_37, and GH44 [β-glucanases and β-glucosidases]) (20, 21), and (v) belong to the multifunctional family GH39 with α-L-arabinofuranosidase, β-glucosidase, β-galactosidase, β-xylosidase activities (22, 23) (see Fig. 2 for a schematic representation of the interaction between these CAZymes and glycan structures, and SI Appendix, Fig. S2A for changes in the abundances of these CAZyme genes after treatment in each of the participants).
Fig. 2.
Interactions between CAZymes and glycan structures within pea and orange fiber preparations. Illustration of the predominant polysaccharide structures enriched in pea or orange fiber and their respective glycosidic linkages. From Top, arabinan, type II arabinogalactan, galactan, homogalacturonan, and rhamnogalacturonan I, plus other polysaccharides found in each fiber preparation including hemicelluloses such as xylan and xyloglucan (Bottom). Red font indicates the glycosidic-linked sugars that were recovered after acid hydrolysis/derivatization of polysaccharides in feces and analyzed by UHPLC-dMRM mass-spectrometry in our study. Fiber-responsive CAZymes are shown within gray ellipses and their predicted cleavage sites (linkages) marked with black arrows. The Inset indicates fecal glycosidic linkages that significantly decrease after consumption of the respective fiber type (dashed circles). Monosaccharide abbreviations: galacturonic acid (GalA), rhamnose (Rha), arabinofuranose (Araf), galactose (Gal), xylose (Xyl), glucose (Glc), mannose (Man), terminal (T), and R-group (R).
Orange fiber.
We performed HOSVD analysis in the same way as for the pea fiber study. For each of the 24 participants in this study, changes in the abundances of genes encoding CAZymes were computed as the log2 fold-change from their levels during the preintervention period (week 2) (Dataset S2A). The third and fourth rows in Fig. 1B describe GH and PL genes with statistically significant changes in their abundances (q-value <0.1 [linear-mixed effects model]; positioning at the tails [α <0.1] of the distribution along TC1) (Dataset S2C). CAZyme genes whose abundances increased encode enzymes with reported (i) pectate lyase/exopolygalacturonate lyase activity (PL9) (24) that can process galacturonan, a major component of the orange fiber preparation, (ii) β-galactosidase (GH59) (18) and β-galactanase (GH30_5) (19) activities that degrade galactans and arabinogalactans, (iii) α-L-arabinofuranosidase and β-xylosidase activities (GH43_16) (25) that metabolize arabinan and arabinoxylan, plus (iv) a reported β-mannanase (GH5_8) (20), as well as β-glucanases (GH5_1, GH5_37, and GH9) (20, 26) that can process cellulose and xyloglucans (Fig. 2) (see SI Appendix, Fig. S2B for the responses in individual participants).
Changes in the Abundances of Bacterial Taxa.
SI Appendix, Fig. S3A shows changes in the relative abundances of bacterial taxa (amplicon sequence variants [ASVs]) in subjects consuming the maximum dose (3 snack servings/d) of pea fiber or orange fiber for 1 and 4 wk, compared to their abundances prior to initiating the intervention. Statistically significant changes were defined by q-value <0.1, (linear mixed-effects model) and, using HOSVD, by their positioning at the tails (α <0.1) of the distribution along TC1. A notable feature of the microbiota response was the significant increase in the representation of Ruminococcaceae and Lachnospiraceae, including members of the butyrate-producing genera Lachnospira, Ruminococcus, and Faecalibacterium (Dataset S3 A–C). A recent study has described reductions in the relative abundances of these genera associated with low fiber intake (27). We also observed increases in the abundances of ASVs corresponding to two pectin-degrading specialists, Monoglobus pectinilyticus (ASV71, pea fiber) and Lachnospira pectinoschiza (ASV87, ASV167, orange fiber) (28, 29). This latter finding is consistent with the polysaccharide composition of pea fiber (rich in arabinan) and orange fiber (rich in galacturonan). Surprisingly, members of the Bacteroides were not prominently represented among the ASVs with statistically significant increases in their relative abundances. Taxa that decreased significantly included Collinsella aerofaciens (reported to be negatively associated with dietary fiber intake in overweight and obese women) (30) and Ruminococcus bromii (a primary degrader of resistant starch) (31).
Gas chromatography-mass spectrometry revealed no statistically significant increases in fecal levels of acetate, butyrate, propionate, lactate, or succinate in any participant in either study; the only statistically significant change was a reduction in acetate levels in those consuming the orange fiber snack prototype (Dataset S3D). Comparing the log2 fold-change in the levels of these short chain fatty acids and ASVs whose abundances changed in a statistically significant fashion between baseline and week 8 in each study disclosed statistically significant positive correlations (Spearman’s ρ >0.5; P < 0.05) between acetate and Monoglobus pectinilyticus (ASV71), the Lachnospiraceae NK4A136 group (ASV148), and Marvinbryantia sp. (ASV212) in participants consuming pea fiber, and a negative correlation (Spearman’s ρ <−0.5; P = 0.005) between acetate and Lachnospira pectinoschiza (ASV87) in participants consuming the orange fiber snack (Dataset S3E). The biological significance of these latter findings remains to be defined. Notably, in both studies, there was a substantially greater degree of interpersonal variation in the pattern of change in the abundances of ASVs compared to genes encoding CAZymes (P < 0.0001, Dunn’s Kruskal–Wallis test of Bray–Curtis dissimilarity distances) (SI Appendix, Fig. S3 B and C and Dataset S3F).
Correlations between CAZymes, Fecal Glycans and the Plasma Proteome.
CAZymes and glycosidic linkages.
We used ultrahigh-performance liquid chromatography triple quadrupole mass spectrometry to quantify the levels of 50 glycosidic linkages in fecal samples collected during the preintervention phase and after 1 and 4 wk of consuming the maximum dose of each fiber snack. Levels of these linkages were determined after their liberation from fecal glycans by acid hydrolysis (see Materials and Methods and Dataset S4 A and B). Among the 37 linkages that exhibited statistically significant changes after consumption of the pea fiber snack, 35 decreased. Similarly, a majority of the linkages that changed significantly after orange fiber supplementation decreased (22 of 28) (SI Appendix, Fig. S4A). We focused on linkages whose levels were negatively correlated with the abundances of genes encoding CAZymes known or reported to have substrate specificities for glycans containing these linkages. We did so based on the supposition that increased levels of the CAZyme and decreased levels of the related glycosidic linkage could provide prima facie evidence for microbial consumption of the parent glycan (see Discussion for caveats).
Our approach yielded 11 pea fiber-discriminatory CAZyme genes and nine orange fiber-discriminatory CAZyme genes whose increases were significantly correlated with glycosidic linkages that decreased after treatment (Spearman’s ρ values |≥ 0.45|) (SI Appendix, Fig. S4 B and C and Dataset S5 A and B). For example, in the pea fiber study, the significant negative correlations between (i) fecal levels of GH43 and GH43_37 (α-L-arabinofuranosidase and α-L-arabinanase activities) and fecal levels of 3-Araf and 2-Araf, and (ii) levels of GH43_22 (β-xylanase and α-L-arabinofuranosidase activities) and levels of 2,4,6-Glc and 3,5-Araf/3,4-Xylp, are consistent with the reported activities of these enzymes (note that the removal of α-1,2-Araf side chains by GH43_22 enables cleavage of the β-1,4 glucose bonds of the xyloglucan backbone by GH5_1, GH5_37, GH44, and GH9) (Fig. 2). As noted above, homo- and to a lesser extent rhamno-galacturonan are the major glycan components of orange fiber. The abundance of PL9, which encodes a pectate lyase/exopolygalacturonate lyase that can process galacturonan, increased significantly when the orange fiber snack was consumed but did not change significantly in the pea fiber study. The increases in PL9 were negatively correlated with levels of several pectin components; for example, the decrease in T-GalA linkages is consistent with microbial consumption of homogalacturonan, while decreases in 3,5-Araf/3,4-Xylp, T-Ara, and 2,3Araf linkages are consistent with consumption of arabinan side chains (e.g., in rhamnogalacturonans) (Fig. 2). Finally, several correlations were shared between the pea and orange fiber studies: e.g., the abundances of genes encoding GH5_1 and GH5_37 β-glucanases were negatively correlated with levels of 4,6-Glc (a component of xyloglucan, Fig. 2) in both studies.
CAZymes and plasma proteins.
In both the pea and orange fiber studies, plasma samples were obtained from participants during the preintervention phase and at week 8. We employed the procedure described in SI Appendix, Fig. S1B to identify significant correlations between changes in treatment-responsive GH and PL genes and the abundances of plasma protein biomarkers and mediators of a range of physiologic functions. A cross-correlation (CC) matrix was created for each study where columns are levels of 1,305 plasma proteins that we had quantified using an aptamer-based platform (Dataset S6 A–C) and rows are the GH and PL genes with statistically significant increases in their abundances in response to consumption of one or the other snack prototype (Fig. 1B). Singular value decomposition (SVD) was performed on this matrix (32). We focused on the first singular vector (SV1) because it explained the highest percentage of the cross-correlation variance for protein responses to each of the snacks. Using a histogram of protein projections along SV1, we selected proteins in the 10th and 90th percentiles of the distribution (α <0.1) (Fig. 3A and Dataset S6D). Proteins falling within the 10th and the 90th percentile groups were analyzed separately with the comprehensive multiomics platform for biological interpretation (CompBio) software package (33, 34). CompBio uses contextual language processing and a biological language dictionary that is not restricted to fixed pathway and ontology knowledge bases, such as KEGG (35, 36) or Gene Ontology (37), to extract “knowledge” from all PubMed abstracts that reference entities of interest (in this case proteins). As described previously (12), CompBio then applies a conditional probability analysis to compute the statistical enrichment of biological “concepts” (e.g., pathways comprised of proteins represented in the 10th or 90th percentile bins) over the probability of these concepts being generated from similarly sized, randomly generated lists of all known proteins in UniProt. Concepts are further clustered into higher-level “themes” based on the co-occurrence of concepts in PubMed abstracts. Themes represented in the 10th and 90th percentiles are ranked by their normalized enrichment score (NES) that utilizes an empirical P value derived from several thousand random lists of proteins, each containing a similar number of proteins to those in the 10th and 90th percentile groups (12). Normalized enrichment scores for concepts are first calculated and these scores are then used to compute the NES score for their associated themes (12):
Fig. 3.
Plasma proteomic biological themes correlated with changes in CAZyme gene abundances after consumption of the fiber-snack prototypes. (A) Schematic illustrating the distribution of plasma protein projections along singular vector 1 (SV1) from a CC-SVD analysis of the abundance of CAZyme genes versus changes in levels of plasma proteins after fiber snack consumption. Proteins with SV1 projections along the tails (α = 0.1) of the distribution are highlighted (green and red); these proteins, belonging to the 10th and 90th percentile groups, were analyzed using CompBio to identify biological themes enriched in each of the two percentile groups for each treatment. (B) Diagram illustrating pathways in which microbiome CAZyme-correlated plasma proteins (boldfaced) are involved in three interrelated biological themes (TGF-β/BMP-mediated fibrosis, P38/MAPK-associated immune biomarkers, and VEGF-mediated angiogenesis). (C, D) Network of biological themes identified from the CC-SVD and CompBio analysis. Themes are depicted as spheres; the number in each sphere corresponds to the theme enriched after pea or orange fiber treatment that is listed in SI Appendix, Dataset S6D. The size of a sphere is proportional to the CompBio enrichment score of its theme. The thickness of the lines connecting themes is proportional to the number of proteins shared between them. Purple spheres represent themes related to TGF-β-BMP signaling (fibrosis), p38/MAPK immune biomarkers, and VEGF-mediated angiogenesis that were enriched in the plasma proteomes of pea and orange fiber study participants (see Table 1).
Themes generated by CompBio incorporate proteins in a given pathway or process, independent of whether they have a positive or negative function.
By applying CompBio to plasma proteomic datasets generated before the intervention and after consuming 3 servings/d of either the pea or orange fiber snacks for 4 wk, we identified a total of 106 biological themes that satisfied our threshold cutoff for statistically significant enrichment (log2 NES ≥15). These included themes that were treatment-specific (37 for pea fiber and 35 for orange fiber) and those that were shared between the two treatments (n = 17) (Dataset S6D).
Table 1 and Fig. 3 B–D highlight three groups of themes related to vascular health that were significantly correlated with CAZyme changes identified in participants consuming pea or orange fiber snacks; transforming growth factor type β/bone morphogenetic protein (TGF-β/BMP)-mediated fibrotic responses, vascular endothelial growth factor (VEGF)-related angiogenic responses, and proteins involved in P38/MAPK signaling in immune cells. Note that treatment-responsive CAZyme genes are positively correlated with proteins in the 90th percentile bin and negatively correlated with proteins in the 10th percentile bin (see Dataset S6 A and B for the abundances of all measured plasma proteins in all participants before and after pea and orange fiber treatment, respectively).
Table 1.
CAZyme-associated plasma proteomic themes enriched after consumption of the fiber snacks
| Biological themes | Pea fiber snack | Orange fiber snack | |||
|---|---|---|---|---|---|
| Log2 (ES) | No. of proteins | Log2 (ES) | No. of proteins | ||
| 10th percentile | |||||
| TGF-β/BMP signaling (fibrosis) | FGF signaling (fibroblast/ligands) | 17.1 | 65 | 15.6 | 62 |
| PAK kinase signaling | 16.8 | 44 | 16.4 | 44 | |
| TGF-β signaling-SMADs | 16.7 | 86 | 16.8 | 76 | |
| Actin cytoskeleton reorganization | 16.6 | 62 | 15.9 | 29 | |
| Metalloproteinases (MMP1) | 15.8 | 52 | |||
| VEGF-mediated angiogenesis | VEGF-mediated angiogenesis/lymphangiogenesis | 16.3 | 69 | 16.6 | 73 |
| PDGF-receptor activity | 16.0 | 70 | |||
| Blood coagulation, fibrin clot formation (platelets/serpine) | 15.1 | 39 | 15.3 | 39 | |
| Vascular abnormalities | 15.4 | 26 | |||
| P38/MAPK-associated immune cell biomarkers | p38/MAPK/JNK activation | 17.9 | 129 | 17.7 | 120 |
| IFN stimulated genes | 16.6 | 26 | 16.7 | 42 | |
| MyD88-dependent toll-like receptor signaling | 16.2 | 76 | 15.3 | 56 | |
| MAPK signaling | 16.0 | 35 | |||
| RTKs | 15.4 | 96 | |||
| Macrophage-alveolar | 15.3 | 40 | |||
| GM-CSF-megakaryocytes | 16.3 | 73 | |||
| CLEC-Fcγ-related | 15.9 | 34 | |||
| 90th percentile | |||||
| TGF-β/BMP signaling (fibrosis) | FGF signaling (FGF1,9,18) | 15.8 | 16 | ||
| BMP signaling | 15.4 | 21 | |||
| TGF-β receptor signaling- SMADs (inhba,gdfs) | 15.6 | 44 | |||
| ECM-metalloproteinases (MMP7) | 15.4 | 57 | |||
| VEGF-mediated angiogenesis | VEGFD-mediated angiogenesis-immune infiltration | 15.4 | 30 | 16.6 | 58 |
| VEGF-activated signaling involved in axon guidance | 15.4 | 28 | |||
| Blood coagulation, fibrin clot formation (fibrinogen) | 15.1 | 13 | |||
| P38/MAPK-associated immune cell biomarkers | Receptor tyrosine kinase (ROR) signaling | 15.0 | 21 | ||
| Lymphocyte/dendritic cell markers | 15.1 | 18 | 15.1 | 43 | |
| Signaling events regulated by Ret tyrosine kinase | 15.9 | 30 | |||
| FCGR-neutralization | 15.2 | 28 | |||
SI Appendix, Fig. S5 A and D show the cross-correlation between levels of proteins in these three themes and the abundances of CAZyme genes that exhibited statistically significant increases in the microbiomes of participants consuming the pea fiber or orange fiber snacks. These CAZymes are ordered based on their projections along SV1 in the U matrix described in step 2 of the cross-correlation singular value decomposition (CC-SVD) analysis outlined in SI Appendix, Fig. S1B. In the case of pea fiber, for example, the increase in the abundance of GH39 (a family reported to include α-L-arabinofuranosidase, β-glucosidase, β-galactosidase, and β-xylosidase activities) was positively correlated with plasma levels of BMP1, BMPR2, PAK3, ANG, and VEGFD (Spearman’s ρ values ranging from 0.28–0.50), and negatively correlated with levels of PAK6, PDGFA, PDGFB, VEGFC, and ANGPT1 (angiopoietin 1) (Spearman’s ρ from −0.30 to −0.48). GH43_22 (α-L-arabinofuranosidase and β-xylanase) as well as a member of GH44 with endo-β-glucanase and xyloglucanase activities shared a similar pattern of positive and negative correlations with these proteins as GH39. Conversely, CAZymes with distributions on the other end of SV1 (e.g., GH5_1 [β-glucanase], GH59 [β-galactosidase], and GH5_8 [β-mannanase]) exhibited an opposite pattern of correlation with these proteins. Increased levels of PDGFA and PDGFB are associated with atherosclerosis (38), while VEGFD and VEGFC are paralogs that interact with both VEGFR-2 and VEGFR-3 during angiogenesis and lymphangiogenesis. The development of white adipose tissue (WAT) is closely linked to angiogenesis; in early onset obesity, ANGPT1 expression in WAT is correlated with both adipocyte size and standardized BMI (39).
Other notable relationships between CAZyme gene levels and plasma proteins were evident in the orange fiber study. SERPINE1, CXCL11, CTSA, NXPH1, and CD47 were among proteins in the 10th percentile bin that had largest negative projections on SV1; these proteins were negatively correlated with CAZymes whose levels increased after orange fiber snack supplementation (Dataset S6D). Blood levels of SERPINE1 (plasminogen activator inhibitor 1) and CTSA (cathepsin A) have both been linked to BMI in three independent studies that included over 4,600 participants (40). CXCL11 is a chemokine chemotactic for interleukin-activated T cells; it is elevated in several inflammatory conditions, including bacterial flagellin-induced intestinal inflammation (41). NXPH1 (neurexophilin 1) is a secreted protein originally identified as being involved in regulating neuronal function, although genome-wide association studies have identified several SNPs in NXPH1 that are associated with type 2 diabetes, and plasma levels of cholesterol, triglycerides and C-reactive protein (42). Mice deficient in CD47 are resistant to diet-induced obesity and exhibit improved insulin sensitivity compared to their wild type counterparts (43). Taken together, these data provide further evidence for a link between the consumption of specific fiber glycans, bacterial CAZyme genes involved in their utilization and features of the plasma proteome associated with metabolic health.
Plasma proteins, CAZymes, and glycosidic linkages.
Correlation analyses between levels of plasma proteins, and fecal CAZymes and glycosidic linkages can be integrated and used to compare responses to consumption of a specific fiber. For example, SI Appendix, Fig. S5 B and E and Dataset S7 A and B show the linkages in the pea fiber and orange fiber studies, respectively, that have strong correlations (Spearman’s ρ values |≥0.35|) with changes in levels of proteins with high enrichment scores within concepts related to vascular biology (Dataset S6D). Ordering the linkages identified in each study based on the absolute values of their Spearman correlations with the corresponding proteins revealed very distinctive groups of glycosidic linkage-plasma protein relationships. These groups can be used to define interpersonal differences in responses to fiber snack consumption. For example, SI Appendix, Fig. S5C considers pea fiber-induced changes in GH43 and GH43_37, 3-Araf, and three plasma proteins (SMAD2, MAPK14, and MAPK2) involved in TGF-β/BMP1 and MAPK signaling pathways related to fibrosis. White adipose tissue fibrosis is associated with inflammation, insulin resistance, and reduced adipocyte size, and is a characteristic feature of obesity (44). With the exception of three individuals, there is an inverse relationship between levels of these α-L-arabinofuranosidase and α-L-arabinanase activities, fecal 3-Araf, and these fibrosis-associated plasma proteins. Similarly, the relationship between levels of T-GalA and three proteins components of fibrosis-associated TGF-β/BMP1 and MAPK signaling pathways (SMAD2, SMAD3, and MAPK1), which were all negatively correlated with PL9 levels, allows interpersonal variations in the responses of orange fiber study participants to be categorized (SI Appendix, Fig. S5F) (also see SI Appendix, Fig. S6).
A comparable series of analyses was performed of the relationship between snack fiber discriminatory CAZymes, glycosidic linkages and regulators of various facets of energy homeostasis (pro-opiomelanocortin [POMC], leptin and its receptor [LEP and LEPR], pancreatic polypeptide [PPY], insulin [INS], adiponectin [ADIPOQ], and atrial natriuretic peptide [NPPA]) (Fig. 4). Hypothalamic leptin signaling leads to activation of the MC4 receptor pathway, resulting in increased energy expenditure and satiety (45). The increase in levels of POMC that occurs in consumers of the pea fiber snack was positively correlated with levels of GH39 (Spearman’s ρ, 0.68), and strongly negatively correlated with 2,4,6-Glc levels (Spearman’s ρ, −0.53), suggesting a possible relationship between xyloglucan metabolism and levels of this protein (Fig. 4 A–C and G). Mutations in LEPR are associated with hyperphagia and early-onset obesity (46). LEPR deficiency is also associated with obesity (47). LEPR levels were positively correlated with increases in PL9, one of the distinguishing features of the microbiome response to the orange fiber snack, and negatively correlated with levels of T-GalA, consistent with a relationship to galacturonan breakdown (Fig. 4 D–F and H). In addition, a strong positive correlation (Spearman’s ρ, 0.64) was documented between levels of LEP and GH9 (β-glucanase), suggesting a relationship that involves degradation of the xyloglucan constituents of orange fiber (Fig. 2).
Fig. 4.
Correlating changes in abundances of CAZyme genes and plasma proteins involved in energy homeostasis with fiber snack consumption. Heatmaps plotting Spearman’s ρ values for correlations between changes in the abundances of plasma proteins and CAZyme genes after pea and orange fiber consumption (A, D) and correlations between plasma proteins and fecal glycosidic linkages (B, E). Color denotes the direction of the correlation. The size of each circle and its color intensity represent the strength of the correlation. (C, F) Bar/dot plots showing, for individual participants, log2 fold-changes (week 8 versus baseline) in the fecal abundance of the GH39 gene and 2,4,6-Glc containing glycans, as well as changes in levels of plasma POMC in participants consuming the pea fiber snack prototype (C), and analogous changes in PL9, T-GalA, and LEPR for those consuming the orange fiber snack prototype (F). (G, H) Heatmaps plotting the log2-fold change in the abundances of plasma proteins after consumption of the pea fiber snack prototype (G), and the orange fiber snack prototype (H), from the preintervention period to week 8. Participants are grouped based on hierarchical clustering (Euclidean distances) of their plasma protein profiles. Symbols with matching colors adjacent to the participant IDs denote members of the same twin pair; circles represent individuals who were obese while pentagons indicate non-obese individuals. 18 participants provided samples for the analyses in study 1 (pea fiber) and 20 for study 2 (orange fiber).
Discussion
This report and its antecedents (11, 12) illustrate a multistep approach for identifying food ingredients that can reshape the gut microbiome in ways that could enhance beneficial community and host functions. The current study focuses on fecal CAZyme gene abundances and targeted mass spectrometric measurements of fecal levels of glycosidic linkages because we wanted to deliberately assess how consumption of a class of foods, in this case plant fibers, impacts the representation of genes involved in their metabolism. In principle, the approach we use can be generalized to measurements of other classes of microbiome genes with other enzymatic functions of interest and their products. We show how a suite of computational tools enable fiber types, whose effects on the human microbiome were first defined in preclinical models, to be evaluated in pilot human studies involving participants with varied dietary practices and starting microbiome configurations. These tools include HOSVD, cross-correlation singular value decomposition, and a software package (CompBio) that mines all PubMed abstracts to compute statistical enrichments for biological “themes” in the plasma proteome that are affected by fiber consumption—specifically themes that are correlated with changes in fecal levels of fiber-discriminatory CAZyme genes.
We demonstrate the feasibility and value of adding results from mass spectrometric analysis of fecal glycosidic linkages to analyses of microbiome CAZyme gene-plasma proteome correlations. The analytic method, which relies on ultrahigh-performance liquid chromatography coupled to a triple quadrupole mass spectrometer, and employing multiple reaction monitoring (48, 49), enables measurement of glycosidic linkages with sufficiently robust quantitation and sensitivity to detect signals above background in the highly complex matrix of feces. The analysis is rapid (performed in minutes) and conducted in a 96-well plate format making its application to larger proof-of-concept studies feasible. Correlations between glycosidic linkage content and CAZyme gene abundances provide a ‘first pass’ assessment of the changes that occur in community saccharolytic activity associated with consumption of fiber-enriched snacks. This method for quantifying linkages could provide one way of overcoming a confounder often encountered in studies that attempt to link dietary interventions to gut microbiome responses; namely, that by having to record to dietary intake and/or body weight on a weekly basis, study participants change their dietary habits making it important to determine the (glycan) content of the diets that they consume.
While the methodologic approach can be used to facilitate the design of fiber-containing snacks/supplements that selectively target microbiome features, there are a number of caveats and limitations of the current study that need to be considered. It is important to be cautious when interpreting CAZyme-glycan-proteome correlations. For example, alterations in glycosidic linkage levels may reflect metabolism of glycans that are nonsnack components of participant’s diets or of their gut habitats, which we did not attempt to control for here. Furthermore, CAZyme gene abundances are not measures of CAZyme expression. In addition, the substrate specificities of many CAZymes have notbeen directly validated in biochemical assays. These considerations highlight the need to develop in vivo assays of how microbial communities process food components (in this case, measurements of the degradation of glycan and nonglycan components of a fiber-enriched snack). One approach that has been validated in gnotobiotic mice colonized with human gut communities is to orally administer collections of microscopic paramagnetic glass beads where different bead types have different glycans covalently bound to their surfaces and different surface-bound identifying fluorescent tags. These artificial food particles can be retrieved based on their magnetism, sorted based on their fluorophore, and the degradation of bound polysaccharide quantified by comparing the amount of glycan on input beads to the amount remaining after recovery of beads from feces/cecal contents (11, 50). While the safety and utility of this type of analytic approach remains to be determined for humans, using these “microbiota functional activity biosensors” represents one way of identifying substrates utilized by a microbiome as a function of variables such as host diet, diet supplementation, or various other perturbations. The knowledge gained could guide decisions related to bioequivalence of fibers from different sources, food processing methods used to manufacture a fiber snack, and the amount of fiber required to produce the intended microbiome/host effects. This latter consideration is particularly important given that the amount of fiber present has marked effects on the organoleptic properties of a food product and hence its consumer acceptability.
Identifying changes in the plasma proteome that correlate with changes in the abundances of treatment discriminatory CAZymes represents one strategy for connecting fiber snack-induced changes in the microbiome with physiologic responses. The current studies were insufficiently powered to draw conclusions about the effects of BMI (or twin pair membership) on the magnitude or direction of these responses. Previous studies involving hundreds to thousands of individuals have demonstrated that dietary fiber promotes weight loss in the context of caloric restriction (51, 52) but they have generally not taken into account the specific and distinct effects of different fiber types on features of the microbiome such as CAZymes. Therefore, additional clinical trials are needed to confirm and precisely quantify differences in disease-relevant proteomic biomarkers identified through the type of wide scale proteomics approach described here. The substantial number of microbiome and plasma proteomic biomarkers revealed by CC-SVD and CompBio from our pilot studies provide an opportunity to select endpoints based on exemplary fiber-discriminatory changes in the microbiome and plasma proteome and use these endpoints as a basis for powering a larger, longer proof-of-concept study. Given the interpersonal variation in microbiome and host responses we document, this type of larger study is a critical next step in determining whether there is robust evidence for the beneficial effects of specific fiber components in microbiome-directed foods. This effort would include assessments of the generalizability, safety, durability, and dose-dependency of their clinical effects.
Materials and Methods
Production of Fiber Snacks.
Pea fiber and orange fiber enriched snacks with organoleptic properties designed to satisfy USA consumer preferences were prepared by Mondelēz Global LLC and tested to confirm the absence of microbial contamination. The nutritional composition of these prototypes is described in Dataset S1A.
Human Study Design.
The pea and orange fiber studies were separate open-label, single group assignment studies that enrolled members of the MOAFTS cohort (14) aged 31–45 y. The first (pea fiber) and second (orange fiber) studies were performed between April and August 2017, and August and December 2017, respectively. The studies were approved by the Washington University Institutional Review Board (IRB ID #201611122). All participants provided written informed consent (ClinicalTrials.gov NCT03078283).
Nine adult female dizygotic twin pairs were recruited for study 1 (Dataset S1B). Individuals allergic to tree nuts, peanuts, dairy, eggs, fish, crustacean shellfish, sesame seeds, wheat, gluten, celery, soybeans, or mustard, and those with inflammatory bowel disease, gastrointestinal cancer, hepatitis, HIV, or renal failure were excluded from the study, along with those who were pregnant or trying to get pregnant. Blood (10 mL) was collected after a 12-h overnight fast between study days 10–14 (week 2) and between days 52–56 (end of week 8), and plasma was prepared. Self-recorded weights were collected on a weekly basis, and participants received weekly shipments of the snack prototypes. The diets of participants were not adjusted in any way other than by adding a fiber snack supplement. For study 2, twelve adult female dizygotic twin pairs concordant or discordant for obesity in the MOAFTS cohort were recruited for testing the orange fiber snack prototype; nine of these twin pairs had also participated in study 1 (Dataset S1B). The same inclusion/exclusion criteria and design features were used for both studies. Participants completed questionnaires related to their diets, any antibiotics taken, and measures of abdominal discomfort (Dataset S1 D–G). Weekly phone calls were scheduled by the study coordinator to monitor compliance. The primary outcome measures were the effects of the respective fiber snack prototypes on gut microbial community structure and function.
Fecal samples collected by participants in small medically approved collection containers were frozen immediately (−20 °C) in dedicated freezers that were provided prior to initiation of each study. Samples were shipped on a regular basis in a frozen state to a biospecimen repository located at Washington University in St. Louis and stored at −80 °C until the time of processing. Blood samples were obtained after an overnight fast in the Clinical Translational Research Unit (CTRU) at Washington University, or during home visits by licensed phlebotomists (PCM TRIALS, CO). Blood was aliquoted into EDTA-K2 treated tubes and centrifuged at 2,000 × g for 10 min at 4 °C. Following centrifugation, plasma was immediately transferred into cryo-resistant polypropylene tubes (0.5 mL aliquots). Plasma samples collected during home visits were stored for 1–3 d at −20 °C and then shipped in a frozen state to the study biospecimen repository. Plasma prepared at the CTRU was immediately stored at −80 °C until analyzed.
Fecal DNA Sequencing.
Each fecal sample was pulverized with a mortar and pestle while immersed in liquid nitrogen. DNA was then extracted from an aliquot of the homogenized sample (∼50–100 mg) by bead-beating (BioSpec Minibeadbeater-96) for 4 min in the presence of 250 µL of 0.1 mm-diameter zirconium oxide beads, a 3.97 mm-diameter steel ball, 500 µL of buffer A (200 mM NaCl, 200 mM Trizma base, 20 mM EDTA), 210 µL of 20% SDS, and 500 µL of phenol:chloroform:isoamyl alcohol (25:24:1). After centrifugation (3,220 × g for 4 min), DNA was purified (QiaQuick 96 purification kit; Qiagen) and quantified (Quant-iT dsDNA broad range kit; Invitrogen).
16S rDNA Analysis.
This analysis followed experimental and analytic methods described in a previous publication (12). Polymerase chain reaction (PCR) of purified DNA samples (1 ng/µL) was performed using barcoded primers targeting variable region 4 of the bacterial 16S ribosomal RNA gene (53). 16S ribosomal DNA (rDNA) amplicons, labeled with sample-specific barcodes were quantified, pooled, and sequenced using an Illumina MiSeq instrument (paired-end 250 nt reads). Following demultiplexing, paired-end reads were trimmed to 200 nt, merged, and chimeric sequences were removed (DADA2 v. 1.13.0) (54). Amplicon sequence variants (ASVs) were generated using DADA2 and aligned against the GreenGenes 2016 (v. 13.8) database to 97% sequence identity. Taxonomic assignments were made using the Ribosomal Database Project (RDP) (release 11.5) and SILVA (v. 128). Only ASVs with a relative abundance ≥0.1% in at least five samples were retained for further analyses.
Shotgun Sequencing.
Using a protocol outlined in Delannoy-Bruno et al. (12), sequencing libraries were generated from each purified DNA sample (Nextera DNA Library Prep Kit, Illumina) (55), pooled, and sequenced (Illumina NextSeq 550 and HiSeq 3000 instruments; 10.7 ± 0.6 × 106 [mean ± SD] and 6.9 ± 1.1 × 106 [mean ± SD] 150 nt paired-end reads/sample for studies 1 and 2, respectively). Reads were demultiplexed (bcl2fastq, Illumina), adapter sequences were trimmed (cutadapt) (56) and the reads were quality filtered (Sickle) (57). Human DNA sequences were removed [Bowtie2 (58); hg19 build of the genome]. Filtered reads were assembled (IDBA-UD) (59) and annotated (prokka) (60). Paired-end reads from each sample were mapped to the corresponding assembled contigs in that sample to generate counts for each open reading frame (ORF). Duplicate reads (optical, PCR-generated) were removed from the mapped data (Picard MarkDuplicates tool v 2.9.3 – http://broadinstitute.github.io/picard/). Count data were generated from alignments (featureCounts; Subread v. 1.5.3 package) (61) for each ORF in each sample and normalized (TPM).
Microbiome Gene Annotation.
The procedure for annotating CAZyme genes is described elsewhere (12). Briefly, each predicted amino acid sequence was compared to full-length sequences stored in the CAZy database using Blastp (62). Query sequences were assigned to the same family/subfamily based on threshold cutoffs of 100% coverage, >50% amino acid sequence identity and an E-value ≤10−6 with a sequence in the database. A second, two-step similarity search was performed on remaining sequences: (i) comparison (Blastp) of the sequence against a library of individual (isolated) modules (i.e., where isolated modules [catalytic or ancillary] are considered as opposed to full-length sequences in CAZy which can contain several modules); and (ii) a HMMER3 (63) search against a curated collection of Hidden Markov Models based on each of the CAZy module families. Based on these two steps, sequences were assigned to the corresponding family/families (and subfamily/subfamilies) based on the following criteria: (i) Blastp E-value <10−4 and (ii) hmmhitstart ≤0.05 or hmmhitend ≥0.95. CAZyme gene family/subfamily abundance tables were generated by aggregating abundance data for each sample. As described in Delannoy-Bruno et al. (12), the abundances of genes annotated with multiple CAZyme families/subfamilies were propagated to each individual family/subfamily member; the abundances were then summed across all corresponding CAZyme families represented in each fecal sample.
Analysis of Blood Samples.
Levels of 1,305 proteins were quantified in a 50 μL aliquot of plasma using the SOMAscan 1.3K Proteomic Assay plasma/serum kit (SomaLogic, Boulder, CO). Procedures used for quality control filtering and analysis of differential protein abundances are described elsewhere (12, 32). The filtering step yielded 1,254 and 1,260 proteins that were used in downstream analyses for human studies 1 and 2, respectively. Conventional blood chemistry tests were performed by the Clinical Laboratory Improvement Amendments (CLIA)-certified Core Laboratory for Clinical Studies (CLCS) at Washington University School of Medicine.
HOSVD, CC-SVD, and CompBio Analyses.
For a description of HOSVD see Delannoy-Bruno et al. (12) and SI Appendix, Fig. S1A. CC-SVD is described in our previous publications (12, 32) and in schematic form in SI Appendix, Fig. S1B. CompBio V2.0 is a commercial software package available from PercayAI Inc. (St. Louis, MO) (https://www.percayai.com).
Mass Spectrometry-Based Analyses.
Glycosidic linkages.
Fecal samples (100-200 mg) were dried to completion by lyophilization and dry blended using 1.4 mm stainless steel beads. Stock solutions of fecal samples were prepared (10 mg/mL in water). To homogenize the samples further, stock solutions were bead-blended, heated at 100 °C for 1 h and then subjected to a second round of bead-blending. Three replicate 5 µL aliquots of each blended stock solution were incubated in saturated NaOH and iodomethane (in dimethyl sulfoxide [DMSO]) to achieve methylation of free hydroxyl groups. Excess NaOH and DMSO were removed (extraction with dichloromethane and water) and the permethylated samples were hydrolyzed (4 M trifluoroacetic acid for 2 h at 100 °C). Hydrolyzed samples were subsequently derivatized with 1-phenyl-3-methyl-5-pyrazolone (PMP) (incubation in 0.2 M PMP [prepared in methanol] and 28% NH4OH for 30 min at 70 °C). Derivatized glycosides were dried to completion (vacuum centrifuge) and reconstituted in 100 µL of 70% methanol. A 1-µL aliquot was analyzed using an Agilent 1290 infinity II ultrahigh-performance liquid chromatography (UHPLC) system coupled to an Agilent 6495A triple quadrupole mass spectrometer in dynamic multiple reaction monitoring (dMRM) mode. Glycosidic linkages present in samples were identified using a pool of oligosaccharide standards and a comprehensive linkage library (48, 49).
Short chain fatty acids.
The method used for quantifying short chain fatty acids is described in Cowardin et al. (64).
Supplementary Material
Acknowledgments
We are indebted to Denise Schmitz for assistance with coordination and oversight of the human studies, including collecting clinical meta-data and fecal samples; Su Deng, Justin Serugo, Kazi Ahsan, Julia Veitinger, Samantha Bale, Jessica Forman, and Sebastian Karlsson for archiving and processing human biospecimens; Marty Meier, Jessica Hoisington-López, and MariaLynn Crosby for fecal microbiome shotgun sequencing; Twyla Juehne, Andrew Lutz, and Jinsheng Yu (Genome Technology Access Center [GTAC] at Washington University in St. Louis) for generating SOMAscan datasets; and Chad Storer and Rich Head (GTAC) for their assistance with CompBio analyses. We also thank Gautier Cesbron Lavau and Monika Okoniewska (Mondelēz Global LLC) for preparing the snack prototypes used in the human studies and Laura Kyro for assistance with figure illustrations. This study was supported by the NIH (DK78669 and DK70977), and Mondelēz Global, LLC. as part of an academic-industrial collaboration. O.D.-B. received predoctoral stipend support from NIH (R25GM103757, T32GM007067, and T32HL130357). J.I.G. is the recipient of a Thought Leader Award from Agilent Technologies.
Footnotes
Reviewers: E.C., The University of Chicago; and E.E., Weizmann Institute of Science.
Competing interest statement: C.B.L. is a cofounder of Evolve Biosystems, InterVenn Biosciences, and BCD Bioscience, companies involved in the characterization of glycans and developing carbohydrate applications for human health. D.K.H., A.M., and S.V. are employees of Mondelēz Global LLC, a multi-national company engaged in production of snack foods. R.A.B. may receive royalty income based on the CompBio technology licensed by Washington University to PercayAI. The remaining authors declare that they have no competing financial interests. A patent application related to the fiber-snack formulations described in this report has been filed (WO 2021/016129).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123411119/-/DCSupplemental.
Data Code and Availability
Shotgun and 16S rDNA amplicon sequencing datasets generated from fecal DNA have been deposited at the European Nucleotide Archive (study accession PRJEB44020). SOMAscan aptamer-based proteomics data have been deposited at the European Genome-Phenome Archive (study accession EGAS00001005330). Fecal monosaccharide and linkage data are available in Glyopost (ID GPST000212). Code for HOSVD (CP-ALS plus randomization code) and CC-SVD is available via Zenodo (https://doi.org/10.5281/zenodo.4767887).
References
- 1.Asnicar F., et al. , Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat. Med. 27, 321–332 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendes-Soares H., et al. , Model of personalized postprandial glycemic response to food developed for an Israeli cohort predicts responses in Midwestern American individuals. Am. J. Clin. Nutr. 110, 63–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D. D., et al. , The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat. Med. 27, 333–343 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeevi D., et al. , Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Ozyurt V. H., Ötles S., Effect of food processing on the physicochemical properties of dietary fibre. Acta Sci. Pol. Technol. Aliment. 15, 233–245 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Flint H. J., Duncan S. H., Scott K. P., Louis P., Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 74, 13–22 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Leshem A., Segal E., Elinav E., The gut microbiome and individual-specific responses to diet. mSystems 5, e00665-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Kaoutari A., Armougom F., Gordon J. I., Raoult D., Henrissat B., The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11, 497–504 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Drula E., et al. , The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 50, D571–D577 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martino C., et al. , Context-aware dimensionality reduction deconvolutes gut microbial community dynamics. Nat. Biotechnol. 39, 165–168 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patnode M. L., et al. , Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell 179, 59–73.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delannoy-Bruno O., et al. , Evaluating microbiome-directed fibre snacks in gnotobiotic mice and humans. Nature 595, 91–95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hisamatsu M., York W. S., Darvill A. G., Albersheim P., Characterization of seven xyloglucan oligosaccharides containing from seventeen to twenty glycosyl residues. Carbohydr. Res. 227, 45–71 (1992). [DOI] [PubMed] [Google Scholar]
- 14.Bucholz K. K., Heath A. C., Madden P. A., Transitions in drinking in adolescent females: Evidence from the Missouri adolescent female twin study. Alcohol. Clin. Exp. Res. 24, 914–923 (2000). [PubMed] [Google Scholar]
- 15.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B., The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones D. R., et al. , SACCHARIS: An automated pipeline to streamline discovery of carbohydrate active enzyme activities within polyspecific families and de novo sequence datasets. Biotechnol. Biofuels 11, 27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mewis K., Lenfant N., Lombard V., Henrissat B., Dividing the large glycoside hydrolase family 43 into subfamilies: A motivation for detailed enzyme characterization. Appl. Environ. Microbiol. 82, 1686–1692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar R., Henrissat B., Coutinho P. M., Intrinsic dynamic behavior of enzyme:substrate complexes govern the catalytic action of β-galactosidases across clan GH-A. Sci. Rep. 9, 10346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita K., et al. , Degradative enzymes for type II arabinogalactan side chains in Bifidobacterium longum subsp. longum. Appl. Microbiol. Biotechnol. 103, 1299–1310 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Aspeborg H., Coutinho P. M., Wang Y., Brumer H. III, Henrissat B., Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12, 186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warner C. D., Go R. M., García-Salinas C., Ford C., Reilly P. J., Kinetic characterization of a glycoside hydrolase family 44 xyloglucanase/endoglucanase from Ruminococcus flavefaciens FD-1. Enzyme Microb. Technol. 48, 27–32 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Morrison J. M., Elshahed M. S., Youssef N., A multifunctional GH39 glycoside hydrolase from the anaerobic gut fungus Orpinomyces sp. strain C1A. PeerJ 4, e2289 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki Y., et al. , Characterization of a novel 3-O-α-d-galactosyl-α-l-arabinofuranosidase for the assimilation of gum arabic AGP in Bifidobacterium longum subsp. longum. Appl. Environ. Microbiol. 87, e02690-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakraborty S., et al. , Role of pectinolytic enzymes identified in Clostridium thermocellum cellulosome. PLoS One 10, e0116787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orita T., Sakka M., Kimura T., Sakka K., Characterization of Ruminiclostridium josui arabinoxylan arabinofuranohydrolase, RjAxh43B, and RjAxh43B-containing xylanolytic complex. Enzyme Microb. Technol. 104, 37–43 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Foley M. H., et al. , A cell-surface GH9 endo-glucanase coordinates with surface glycan-binding proteins to mediate xyloglucan uptake in the gut symbiont bacteroides ovatus. J. Mol. Biol. 431, 981–995 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayerhofer C. C. K., et al. , Low fibre intake is associated with gut microbiota alterations in chronic heart failure. ESC Heart Fail. 7, 456–466 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim C. C., et al. , Genomic insights from Monoglobus pectinilyticus: A pectin-degrading specialist bacterium in the human colon. ISME J. 13, 1437–1456 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornick N. A., Jensen N. S., Stahl D. A., Hartman P. A., Allison M. J., Lachnospira pectinoschiza sp. nov., an anaerobic pectinophile from the pig intestine. Int. J. Syst. Bacteriol. 44, 87–93 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Gomez-Arango L. F., et al. , Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes 9, 189–201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ze X., Duncan S. H., Louis P., Flint H. J., Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6, 1535–1543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R. Y., et al. , Duodenal microbiota in stunted undernourished children with enteropathy. N. Engl. J. Med. 383, 321–333 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winkler E. S., et al. , Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell 184, 1804–1820.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou W., et al. , Ablation of fat cells in adult mice induces massive bone gain. Cell Metab. 32, 801–813.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M., New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The Gene Ontology Consortium, The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raines E. W., PDGF and cardiovascular disease. Cytokine Growth Factor Rev. 15, 237–254 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Gaebler N., et al. , Age- and BMI-associated expression of angiogenic factors in white adipose tissue of children. Int. J. Mol. Sci. 20, 5204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaghlool S. B., et al. , Revealing the role of the human blood plasma proteome in obesity using genetic drivers. Nat. Commun. 12, 1279 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z., et al. , Chemokine CXCL11 links microbial stimuli to intestinal inflammation. Clin. Exp. Immunol. 164, 396–406 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraja A. T., et al. , Genetic analysis of 16 NMR-lipoprotein fractions in humans, the GOLDN study. Lipids 48, 155–165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maimaitiyiming H., Norman H., Zhou Q., Wang S., CD47 deficiency protects mice from diet-induced obesity and improves whole body glucose tolerance and insulin sensitivity. Sci. Rep. 5, 8846 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Divoux A., et al. , Fibrosis in human adipose tissue: Composition, distribution, and link with lipid metabolism and fat mass loss. Diabetes 59, 2817–2825 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farooqi I. S., O’Rahilly S., Mutations in ligands and receptors of the leptin-melanocortin pathway that lead to obesity. Nat. Clin. Pract. Endocrinol. Metab. 4, 569–577 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Nunziata A., et al. , Functional and phenotypic characteristics of human leptin receptor mutations. J. Endocr. Soc. 3, 27–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleinendorst L., et al. , Leptin receptor deficiency: A systematic literature review and prevalence estimation based on population genetics. Eur. J. Endocrinol. 182, 47–56 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Galermo A. G., et al. , Liquid chromatography-tandem mass spectrometry approach for determining glycosidic linkages. Anal. Chem. 90, 13073–13080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galermo A. G., Nandita E., Castillo J. J., Amicucci M. J., Lebrilla C. B., Development of an extensive linkage library for characterization of carbohydrates. Anal. Chem. 91, 13022–13031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wesener D. A., et al. , Microbiota functional activity biosensors for characterizing nutrient metabolism in vivo. eLife 10, e64478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miketinas D. C., et al. , Fiber intake predicts weight loss and dietary adherence in adults consuming calorie-restricted diets: The POUNDS lost (preventing overweight using novel dietary strategies) study. J. Nutr. 149, 1742–1748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sylvetsky A. C., et al. Diabetes Prevention Program Research Group, A high-carbohydrate, high-fiber, low-fat diet results in weight loss among adults at high risk of type 2 diabetes. J. Nutr. 147, 2060–2066 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caporaso J. G., et al. , Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108 (suppl. 1), 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Callahan B. J., et al. , DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baym M., et al. , Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10, e0128036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011). [Google Scholar]
- 57.Joshi N., Fass J., Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files, Version 1.33. https://github.com/najoshi/sickle. Accessed 8 January 2022.
- 58.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng Y., Leung H. C. M., Yiu S. M., Chin F. Y. L., IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Camacho C., et al. , BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mistry J., Finn R. D., Eddy S. R., Bateman A., Punta M., Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41, e121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cowardin C. A., et al. , Mechanisms by which sialylated milk oligosaccharides impact bone biology in a gnotobiotic mouse model of infant undernutrition. Proc. Natl. Acad. Sci. U.S.A. 116, 11988–11996 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Shotgun and 16S rDNA amplicon sequencing datasets generated from fecal DNA have been deposited at the European Nucleotide Archive (study accession PRJEB44020). SOMAscan aptamer-based proteomics data have been deposited at the European Genome-Phenome Archive (study accession EGAS00001005330). Fecal monosaccharide and linkage data are available in Glyopost (ID GPST000212). Code for HOSVD (CP-ALS plus randomization code) and CC-SVD is available via Zenodo (https://doi.org/10.5281/zenodo.4767887).