Significance
The mitochondrial genomes of land plants encode genes for cellular energy production and agriculturally important traits, but modification of the genomes is still difficult. Targeted base editing is one of the best ways to modify genes and intergenic regions and thus understand their functions, without drastically changing genome structure. In this study, we succeeded in creating plantlets of the model plant Arabidopsis thaliana, in which all of the many copies of the mitochondrial genomes in each cell had a targeted C:G base pair converted to a T:A pair. Introduced mutations were stably inherited by the next generation. This method will help to unravel the mysteries of plant mitochondrial genomes and may also serve as a basis for increasing crop yields.
Keywords: genome editing, plant mitochondria, RNA editing, mitochondrial genome, base editing
Abstract
Beyond their well-known role in respiration, mitochondria of land plants contain biologically essential and/or agriculturally important genes whose function and regulation are not fully understood. Until recently, it has been difficult to analyze these genes or, in the case of crops, to improve their functions, due to a lack of methods for stably modifying plant mitochondrial genomes. In rice, rapeseed, and Arabidopsis thaliana, mitochondria-targeting transcription activator-like effector nucleases (mitoTALENs) have recently been used to disrupt targeted genes in an inheritable and stable manner. However, this technique can also induce large deletions around the targeted sites, as well as cause ectopic homologous recombinations, which can change the sequences and gene order of mitochondrial genomes. Here, we used mitochondria-targeting TALEN-based cytidine deaminase to successfully substitute targeted C:G pairs with T:A pairs in the mitochondrial genomes of plantlets of A. thaliana without causing deletions or changes in genome structure. Expression vectors of the base editor genes were stably introduced into the nuclear genome by the easy-to-use floral dipping method. Some T1 plants had apparent homoplasmic substitutions that were stably inherited by seed progenies, independently of the inheritance of nuclear-introduced genes. As a demonstration of the method, we used it to restore the growth of an organelle transcript processing 87 (otp87) mutant that is defective in the editing of RNA transcripts of the mitochondrial atp1 gene and to identify bases in atp1 that affect the efficiency of RNA editing by OTP87.
Plant mitochondrial genomes not only encode genes involved in the electron transport chain, adenosine triphosphate synthesis, and translation of mitochondrial genes, but also many open reading frames (ORFs) of unknown function. The paucity of attempts to modify and characterize plant mitochondrial genomes is partly due to the limited availability of tools for modifying plant mitochondrial genomes and the associated difficulty in identifying single nucleotide polymorphisms (SNPs) in the genomes that affect agronomic traits. Bombardment methods have previously been used to stably introduce genes into the mitochondrial genomes of two unicellular organisms, Chlamydomonas (1) and yeast (2, 3), but so far they have not been successfully used to stably transform the mitochondrial genomes of land plants. We recently succeeded in knocking out mitochondrial genes of three land plants, rice, oilseed rape, and Arabidopsis thaliana (4, 5), by using mitochondria-targeting transcription activator-like effector nucleases (mitoTALENs) to introduce double-strand DNA breaks (DSBs) in targeted genes. A problem with transcription activator-like effector nuclease (TALEN)–induced DSBs is that they can cause deletions of up to several thousand bases around the cleavage site as well as cause changes in genome structure due to ectopic homologous or homeologous (i.e., partly homologous) recombination (4, 5). There is thus a need for a method that produces more precise changes in order to more accurately analyze gene functions and more safely conduct crop breeding. Recently, Mok et al. (6) substituted targeted C:G pairs to T:A pairs in mitochondrial genomes of human cultured cells by transiently expressing the halves of a bisected cytidine deaminase (CD) gene of Burkholderia cenocepacia DddA protein, each of which was fused to a uracil glycosylase inhibitor (UGI) and the DNA-binding domain of a TALEN. UGI was used to inhibit the removal of uracil bases, which were generated from cytosines by CD (6). Such substitutions occurred in up to 50% of mitochondrial genomes. Kang et al. (7) applied this technique to substitute targeted base pairs (C:G to T:A) in mitochondrial genomes of lettuce and rapeseed calli. They showed that the edit frequencies were as high as 25%, although inheritance of the introduced mutations by the next generation was not confirmed. Using the method of Mok et al. (6), we recently succeeded in substituting a targeted single nucleotide in the plastid genome of A. thaliana plantlets with high efficiency (up to 100% of the plastid genomes were edited) and confirmed the inheritance of these homoplasmic substitutions (8). In this study, we applied this method to targeted base editing in the mitochondrial genomes of A. thaliana plantlets via nuclear transformation.
In addition to our goal of introducing targeted changes in mitochondrial genomes, we wanted to see if the method could be used as a tool for better understanding of RNA editing, a process that has been revealed to have great importance in the expression of mitochondrial genes (9, 10). The mitochondrial genome of Arabidopsis, for example, has more than 400 RNA editing sites (11, 12). In RNA editing, PLS-class pentatricopeptide repeat (PPR) proteins edit targeted cytidines to uridines on RNA transcripts by recognizing a sequence upstream of each target site (13, 14). For this study, we selected a PPR protein, ORGANELLE TRANSCRIPT PROCESSING 87 (OTP87), and its target RNA editing site in the mitochondrial gene ATPase subunit 1 (atp1) as targets for analyses. First, to verify the feasibility of the method, we substituted a targeted C:G pair of atp1 of wild-type (WT) plants to a T:A pair, confirmed inheritance of the introduced mutations to the next generation, and investigated off-target effects of our base editing system on mitochondrial genomes. Second, we attempted to restore the growth of an otp87 retarded growth mutant (15) by base editing the atp1 gene, thus bypassing the need for RNA editing of the transcripts by OTP87. Third, to explore the usefulness of the method for understanding the process of target recognition in RNA editing, we used it to introduce other base changes in atp1 to analyze which nucleotides in the predicted OTP87 binding sequence (16) were important for RNA editing. Our results suggest that our method will be a useful tool for targeted editing of mitochondrial genes as well as a tool for exploring the role of RNA editing in mitochondrial genomes.
Results
Generation of Homoplasmic Mutations in atp1.
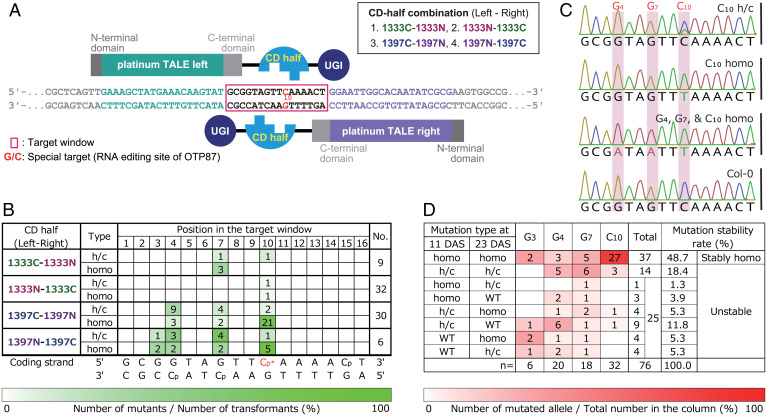
As a target for base editing, we selected a base pair corresponding to an RNA editing site in mitochondrial atp1, atp1-1178C. The cytidine in the atp1 gene should be converted into U after transcription. Therefore, we could eliminate possible negative influences of the C:G to T:A substitution to the plants when we evaluated the efficiency of the base editing and its inheritance. To achieve the targeted base editing in the mitochondrial atp1 gene, we constructed four vectors containing the C-terminal CD domain of DddA toxin of B. cenocepacia [1,427 aa (6)]. As in previous studies (6–8, 17), the coding sequence of the CD domain was split at the nucleotides immediately following the codons for Gly-1333 or Gly-1397. Each sequence of the split CD halves (N- and C-terminal halves) was fused to the 3′ side of the sequence of the DNA-binding domain of the platinum TALEN (hereafter referred to as pTALE), which recognizes a DNA sequence of up to 21 base pairs (18). The pTALE-CD sequences were fused to the 5′ side of the sequence of UGI to prevent removal of the uracil generated from cytosine [(6, 19); pTALE-CD-UGI]. The nucleotide sequences of CD and UGI were the same as in the previous study (8), whose codons were optimized for A. thaliana. The sequence of the mitochondrial targeting signal of the Arabidopsis adenosine triphosphatase (ATPase) delta prime subunit (5) was linked to the 5′ side of the pTALE-CD-UGI (mitoTALECD; SI Appendix, Fig. S1). A pair of mitoTALECDs, each under control of the Arabidopsis RPS5A promoter, were placed in tandem in a single binary vector (SI Appendix, Fig. S1). The RPS5A promoter has been used for highly efficient genome editing in A. thaliana (5, 8, 20). Four binary vectors (1333CN, 1333NC, 1397CN, and 1397NC) were constructed (Fig. 1A). As an example of this notation, 1333CN means that the C-terminal half of the CD domain split at Gly-1333 (1333C) is fused to the left TALE domain and that the N-terminal half (1333N) is fused to the right TALE domain. Each vector was transformed into the nuclear genome of A. thaliana by floral dipping (21) to substitute the targeted C:G pair to a T:A pair in the mitochondrial genomes. In addition to the targeted C:G pair [1178C of atp1 gene, C at position 10 (Fig. 1A)], other C:G pairs in the target window, which is the region between the left and right TALE-binding sequences, might also be substituted, as was observed in previous studies (8, 17).
Fig. 1.
Attempt to introduce a homoplasmic mutation at the target base in atp1. (A) A pair of pTALECD proteins, a target base, and a target window. The sequences that the left and right TALEs bind to are shown in green and purple, respectively. CD was split after Gly-1333 or Gly-1397, and the N- and C-terminal CD halves were fused to the left and right (or the right and left) TALEs, respectively. The sketches of TALEs, CD halves, and UGIs are based on Nakazato et al. (8). (B) Numbers of plants with edited bases of each type (h/c or homo) at each position in the target window at 11 DAS. Editing efficiencies are shown by the intensity of green color. Cp, C at the 3′ side of T; Cp*, special target C; homo, homoplasmically substituted; No., the number of total T1 plants. (C) Four representative Sanger sequences of PCR products amplified with primers specifically targeting mitochondrial DNA (not NUMT; SI Appendix, Table S2 provides detail). Positions at which base editing occurred in at least one of the four sequences are shaded in pink. Positions of the substituted base and substitution types are shown above each sequence. (D) Numbers of substituted bases in the target window categorized by substitution type at 11 and 23 DAS. Intensity of red shading indicates the numbers of substituted bases. Mutation stability rates were calculated by dividing the number of bases of each mutation type by the total number of substituted bases. “Unstable” means that the mutation type differed between 11 and 23 DAS.
The targeted region in the mitochondrial genomes was sequenced to determine whether the targeted C:G pair was converted to a T:A pair. Total DNAs from the leaves of the T1 transformants were amplified by PCR, and their amplicons were sequenced by the Sanger method. Of 78 T1 transformants that were examined, 36 (representing all four vectors) had C:G to T:A mutations in the target window (SI Appendix, Fig. S2 and Table S1). Plant nuclear genomes often acquire large fragments of mitochondrial DNA, called nuclear mitochondrial DNAs or NUMTs (22, 23). During genotyping, we noticed that we also amplified a nuclear sequence (At2g07698), almost identical to atp1, that was part of a NUMT in chromosome 2 of A. thaliana Columbia-0 (Col-0) (22). Therefore, we designed primers that specifically amplify the mitochondrial sequences and used them in subsequent analyses (SI Appendix, Table S2) to prevent unwanted amplification of the NUMT sequences. T1 plants in which mutations were detected in the first genotyping were regenotyped with the redesigned primers (SI Appendix, Table S3). Many of the transformants appeared to have homoplasmic substitutions in the target window (Fig. 1 B and C and SI Appendix, Table S3). In addition to mutating the targeted C at position 10, Gs at positions 3, 4, and 7 in the target window were mutated in some T1 plants. Most of the converted Cs were on the 3′ side of the T or A (Fig. 1B), as reported previously (8). The base-editing activities and preferences of the position of edited bases in the target window were different among the four vectors, and the C in the target window that was most frequently homoplasmically substituted was the C at position 10 when the vector was 1397C-1397N (1397CN; Fig. 1B). As a result, we obtained five mitochondrial mutants in which only the special target base (C at position 10) was substituted in the target window at both 11 and 23 d after stratification (DAS), that is, after the seeds were cold treated to induce germination (SI Appendix, Table S3).
To quantify the base editing efficiency, the sequence including the target window was amplified by PCR using mitochondria-specific primers (SI Appendix, Table S2), and the PCR product was deep-sequenced (SI Appendix, Fig. S3 A and B). This analysis was performed on 18 of 36 T1 plants in which the bases in the target window were substituted as detected with the Sanger method (SI Appendix, Table S3). Of the 51 bases in the 18 plants that appeared to be homoplasmically substituted (SI Appendix, Table S3), 44 (86.3%) of them were substituted at rates ≥99.00% (SI Appendix, Fig. S3C). These results confirmed that the introduced mutations in the target window were nearly homoplasmic, even in the T1 generation.
To see if the type of introduced mutations changed during plant development, the type of mutation in each transformant was checked by Sanger sequencing of PCR fragments from total DNAs of different leaves at 11 and 23 DAS. A total of 76 mutated bases were detected on at least one of these days (Fig. 1D). Of these, 14 bases were heteroplasmically and/or chimerically (h/c; i.e., not homoplasmically) substituted on both days and 25 bases were substituted in different manners on the 2 d (see Fig. 1D for the numbers and percentages of each type of substitution). The substitution frequencies of some bases were very different on the two days (G3 and G4 in 1397CN 9, G4 and G7 in 1397CN 12, and G3 and G7 in 1397CN26; SI Appendix, Table S3 and Fig. S3D). The remaining 37 bases, which accounted for about half of the mutated bases that were detected, were homoplasmically substituted on both days [48.7% (37/76); Fig. 1D]. This tendency was confirmed by deep sequencing (i.e., bases that were substituted at rates ≥99.00% on both days were observed in many of the examined plants; SI Appendix, Fig. S3A). These results indicate that C:G pairs in the target window were efficiently substituted to T:A pairs by mitoTALECD and that some of the mutations were stably homoplasmic because they were detected in two leaves at two different time points, even in the T1 generation.
Inheritance of Mutations by Seed Progeny.
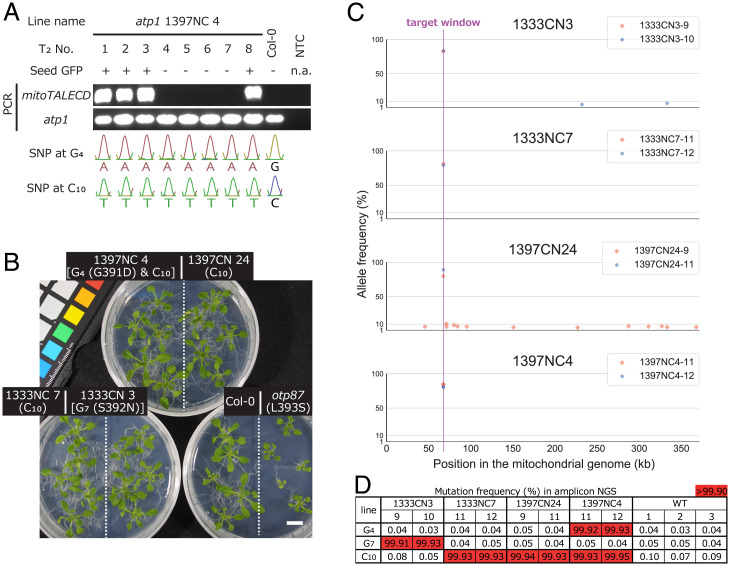
To see if the introduced mutations were inherited by seed progenies, we genotyped 13 T2 progenies of each of four T1 plants whose C:G pairs in the target window were homoplasmically substituted. All examined plantlets inherited the parental homoplasmic mutations, regardless of whether they had mitoTALECD genes in their nuclei (Fig. 2A and SI Appendix, Fig. S4 and Table S4). This indicates that homoplasmic mutations in the mitochondrial genomes introduced by mitoTALECD were stably inherited by the seed progenies. For each of the four lines, the progenies that did not have mitoTALECD genes grew as well as WT plants. This was even the case for plants with two distinct additional mutations that caused amino acid substitutions (G391D and S392N; Fig. 2B). Some bases that were mutated in a heteroplasmic and/or chimeric manner in the T1 generation were observed to have a uniform genotype in the T2 generation (SI Appendix, Fig. S4 and Table S4).
Fig. 2.
Genotypes and phenotypes of T2 plants. (A) Genotypes of eight T2 progenies of atp1 1397NC 4. Seed-specific GFP expression from the T-DNA (SI Appendix, Fig. S1) was confirmed by its fluorescence. Positive signals of mitoTALECD amplification show inheritance of nuclear-introduced mitoTALECD genes. atp1 was used as a positive control for the PCR amplification of mitoTALECD. Sanger data of two bases (G4 and C10) in the target window, where the parent had mutations, are shown at the bottom. NTC, no template control. n.a., not applicable. (B) Phenotypes of T2 progenies of four lines (atp1 1397NC 4, 1397CN 24, 1333NC 7, and 1333CN 3), Col-0, and otp87 at 20 DAS. All of the T2 plants did not contain the nuclear mitoTALECD gene but inherited mitochondrial homoplasmic mutations. They all grew as well as Col-0 and grew better than otp87. The dashed lines separate distinct lineage of plants in one plate. Scale bar, 1 cm. (C) On- and off-target SNPs in mitochondrial genomes in eight representative T2 plants (two progenies of four T1 lines). None of these plants contained mitoTALECD genes. The x- and y-axes show the positions and frequencies of the mutated SNPs that differed by at least 5% from the reference genome BK010421.1. AFs were calculated as AFmu − AFWT, where AFmu is an AF of a SNP of each mutant and AFWT is the average of the AFs of the same SNP of three WT plants. (D) Results of amplicon NGS for the same samples as those subjected to whole-genome sequencing. AFs of mutated bases in the target window are shown, and those more than 99.90% are highlighted in red. Read depth varied from about 41,000 to 161,000.
Off-Target Mutations in the Mitochondrial Genome.
To investigate the off-target effects of mitoTALECD in the mitochondrial genomes, we determined SNP frequencies in T2 plants that had already been confirmed to inherit parental homoplasmic mutations in the target window (SI Appendix, Table S4). The locations and frequencies of line-specific mutated SNPs that were different from the reference sequence (BK010421.1) are shown by dots in Fig. 2C. These data show that none of the detected off-target sites were homoplasmic; that is, they were observed in fewer than 10% of the mitochondrial DNA copies in each plant (except for one site in 1397CN 24–9). Such off-target mutations did not occur within 2 kb of the target window or around sequences similar (≥70% identical) to the sequences recognized by the TALE domains.
In these eight plants, the coverage patterns over the whole mitochondrial genome were very similar to the coverage pattern in the WT plants (SI Appendix, Fig. S5). The coverage showed no evidence of deletions or changes in mitochondrial genome structure, such as sequence rearrangements or creations of new repeat sequences, such as those that were observed in our previous reports of mitoTALENs (4, 5).
About 20% of the reads at the position of targeted SNPs in the target window did not have mutated bases (Fig. 2C). However, the sequences of the mitochondrial atp1 PCR products from these eight plants had homoplasmic substitutions of such C:G pairs to T:A pairs (SI Appendix, Fig. S4 and Table S4), while the sequences of the nuclear atp1-like (At2g07698) PCR products showed no substitutions in the sequence corresponding to the target window (SI Appendix, Fig. S6). Moreover, deep sequencing of PCR products of the mitochondrial target window in these eight plants revealed that the bases in the target window were substituted at rates >99.9% (Fig. 2D). These results support the idea that WT C:G SNPs detected in whole-genome sequencing were derived from the atp1-like sequences in the nucleus. In addition, this sequence showed virtually no edited bases (SI Appendix, Fig. S6), and the minor off-target mutations in the sequence (1397CN 24–10 and 12; SI Appendix, Fig. S6) can be removed by cross breeding. In any case, no major off-target mutations were detected either in the mitochondrial genomes (Fig. 2C) or in nuclear DNA sequences resembling the target window (SI Appendix, Fig. S6).
Phenotypic Complementation of a ppr Mutant, otp87, by mitoTALECD.
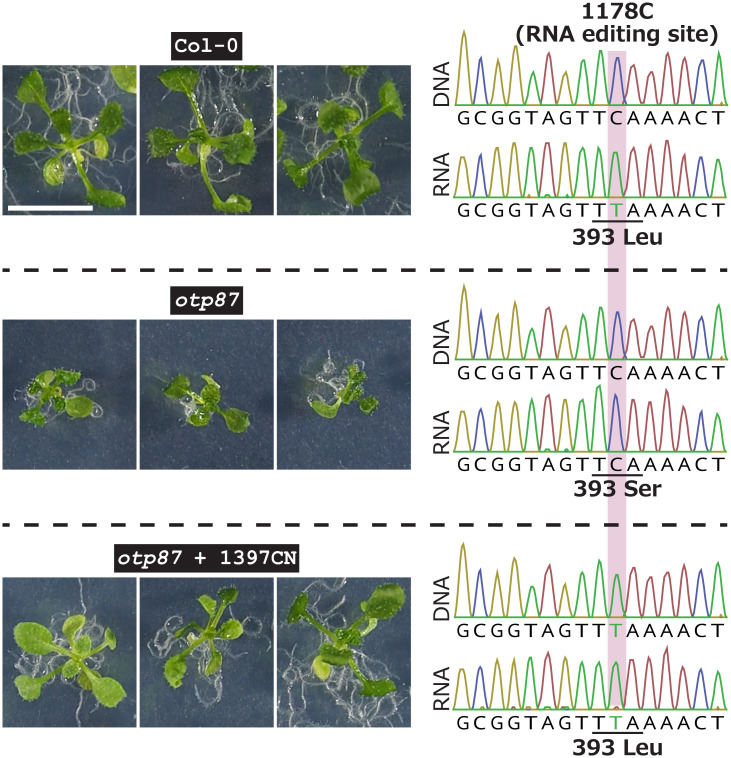
RNA editing is a characteristic feature of terrestrial plant mitochondrial (and plastid) genomes, in which specific C-to-U changes are made in RNA molecules after transcription. They are mediated by nuclear-encoded mitochondria-targeted PPR proteins (9). To test the usefulness of mitoTALECD for molecular analysis of mitochondrial genomes, two experiments related to RNA editing were performed (described in this and the following sections). First, we investigated the otp87 mutant, which has a retarded growth phenotype. In WT plants, the PPR protein OTP87 converts 1178C (C10 in the target window; Fig. 1A) of atp1 transcripts and 24C in nad7 transcripts to U (15). Since only the former RNA editing event causes an amino acid substitution (S393L), the failure of this process has been proposed to be responsible for the retarded growth of otp87. We investigated whether the deficiency in RNA editing, and thus retarded growth, could be corrected by substituting the 1178C of atp1 to T at the DNA level by mitoTALECD. One of the mitoTALECD expression vectors, 1397CN (Fig. 1B), was introduced into the nuclear genome of otp87 mutants. Among 14 T1 plants that were examined, seven plants grew as well as WT plants (Fig. 3 and SI Appendix, Fig. S7A). These seven plants had a homoplasmic substitution at 1178C (C10) to T (or U) at both DNA and RNA levels in a true leaf (Fig. 3 and SI Appendix, Fig. S7B). These results indicate that the inability to edit the 1178C of atp1 transcripts is responsible for the retarded growth of otp87 mutants.
Fig. 3.
Repair of mitochondrial atp1 RNA in otp87 mutant by mitoTALECD. Left, images of representative plants of Col-0, otp87 mutant, and otp87 at 13 DAS in which atp1 was modified by mitoTALECD. Right, representative DNA and RNA sequences around 393Leu of atp1. Top, the C in the 393Leu codon is normally changed to T by RNA editing by OTP87. In the otp87 mutant (Middle), this change is not made, resulting in a Leu > Ser substitution and stunted growth. To restore the mutant to normal growth, we made a C-to-T substitution in atp1 using mitoTALECD (Bottom) so that RNA editing by OTP87 was not needed. This substitution restored growth to that of the WT. Images and genotypes of all other examined plants are shown in SI Appendix, Fig. S7 A and B. Scale bar, 1 cm.
Recognition of atp1 RNA by OTP87.
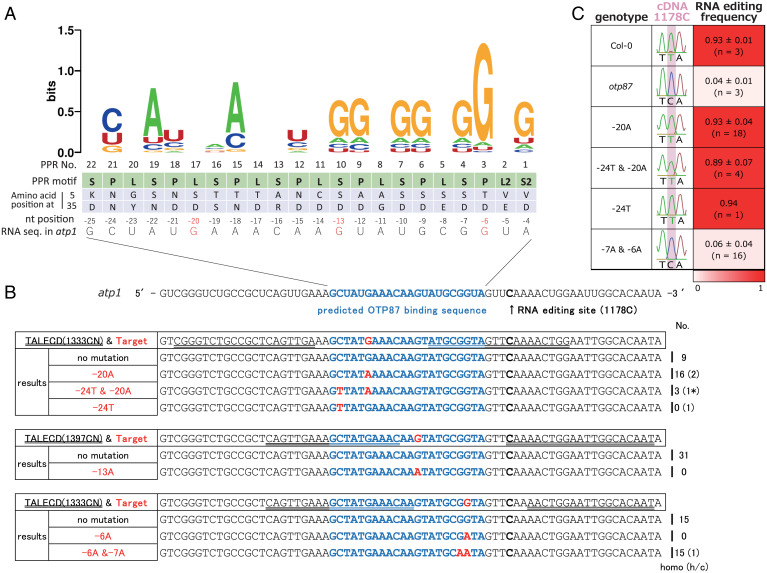
In a second experiment, we tested the atp1 sequence to which OTP87 is predicted to bind [(16); Fig. 4A and SI Appendix, Fig. S8A]. The putative nucleotides bound by the PLS-type PPR protein, OTP87, and their probabilities are shown as a nucleotide logo at the top of Fig. 4A. They were predicted according to the combination of the two key amino acid residues at positions 5 and 35 of each PPR motif [e.g., P, L, and S (16, 24–26)]. The actual atp1 sequence upstream of the RNA editing site, to which OTP87 is predicted to bind, is shown at the bottom of Fig. 4A. In this experiment, we tried to mutate some C:G pairs in this sequence to see whether this sequence is required for RNA editing and, if it is, which bases are involved. Three mitoTALECD expression vectors were constructed to substitute each of three Gs located 20, 13, and 6 bases upstream of 1178C to A (denoted -20G, -13G, and -6G; Fig. 4A and SI Appendix, Fig. S8A). Thirty four T1 seeds of each line (Col-0 background) were sown, and DNA and RNA sequences of the seedlings were analyzed to see the patterns of DNA mutation by the mitoTALECDs and their effects on RNA editing efficiency at 1178C. Although -13G was not successfully substituted in this study, mitochondrial genome mutants of the following four allele patterns were obtained at the predicted binding sequence of OTP87: (i) -24C was substituted to T; (ii) -20G was substituted to A; (iii) -24C and -20G were substituted to T and A, respectively; and (iv) both -7G and -6G were substituted to As (Fig. 4B). RNA editing efficiency, expressed as Sanger sequencing data of RT-PCR products of atp1 transcripts, was decreased only in the fourth allele pattern (Fig. 4 B and C and SI Appendix, Fig. S8 B–D). These results indicate that at least one base of the predicted binding sequence of OTP87 actually affects the efficiency of RNA editing and that the -7G and/or -6G is needed to edit 1178C and probably to recognize and bind atp1 transcripts. On the other hand, -24C and -20G can be substituted to U and A, respectively, without disrupting these activities.
Fig. 4.
Effects of mutations in the predicted binding sequence of OTP87 in atp1 on its RNA editing. (A) RNA sequence logo showing the bases that each of the 22 PPR motifs of OTP87 is predicted to bind to. The PPR motifs are numbered starting from the C terminus. The binding is predicted by the amino acid residues at positions 5 and 35 of each PPR motif. The actual RNA sequence corresponding to the predicted bound site, which is located upstream of the RNA editing site by OTP87 in atp1, is shown at the bottom. The C-terminal S2 domain and the N-terminal S domain correspond to nucleotides 4 and 25 bases upstream of the editing site, respectively (-4A and -25G). Targeted bases of mitoTALECD (see the legend of B) are shown in red. nt, nucleotide. (B) The results of targeted base editing at the predicted binding sequence of OTP87 in atp1. Shown at Top is a portion of the atp1 messenger RNA sequence that is predicted to be involved in OTP87 binding. The binding region is shown in blue, and the RNA editing site is shown in bold. Each of the Gs at positions -20, -13, and -6 of the sequence was targeted for substitution to A by specific pairs of mitoTALECDs. All alleles that were obtained and were intended to be obtained and the number of plants of each allele are shown under the RNA sequence. TALE-binding sequences are underlined. homo, homoplasmically substituted. * means that -20G was homoplasmically substituted while -24C was heteroplasmically or chimerically substituted. (C) Representative RNA (complementary DNA) sequences around the RNA editing site of the obtained alleles. Right column shows RNA editing efficiencies (mean ± SD) estimated by EditR (Materials and Methods). The efficiencies are shown by the intensity of red color. Images of plant Nos. 1 to 15 for each line and genotype of all examined plants are shown in SI Appendix, Fig. S8 B–D.
Discussion
C:G Pairs of A. thaliana Mitochondrial atp1 Gene Were Homoplasmically Substituted to T:A Pairs by the mitoTALECD Method.
Targeted base editing of mitochondrial genomes was first achieved in human cultured cells (6) and subsequently used in mice (17) and protoplasts and calli of lettuce and rapeseed (7). These experiments achieved heteroplasmic and/or chimeric substitutions of specific C:G pairs to T:A pairs (6, 7, 17). Inheritance of the introduced mutations was confirmed in mice (17) but not in plants (7). In this study, C:G pairs in the target window appeared to be homoplasmically substituted to T:A pairs, even in the T1 generation (Fig. 1B and SI Appendix, Fig. S3 and Table S3), and the introduced mutations were inherited by the T2 generation (Fig. 2 A and D and SI Appendix, Fig. S4 and Table S4). The reason for the homoplasmy is not clear, but it might be partly due to expressing mitoTALECDs with the Arabidopsis RPS5A promoter, which strongly drives gene expression in Arabidopsis (27). This promoter was successfully used for genome editing in A. thaliana (5, 8, 20). Another possible contributor to homoplasmy is the use of TALE repeats of efficient platinum TALEN, whose amino acid residues differ from those of previous TALENs (18).
Characteristics of Cytosines That Were Converted to Thymines.
This study may provide useful information for designing target windows. The CD of B. cenocepacia DddA protein was previously found to mainly cause TC > TT substitutions in mammalian mitochondrial genomes (6, 17). In this report, in addition to TC > TT substitutions (Fig. 1B), we observed some AC > AT substitutions (Fig. 1B), as was the case in a previous study of plastid genome base editing (8). Furthermore, a GC > GT substitution was observed at -24C in Fig. 4B. No CC > CT substitution was observed, but CC > TT substitutions were observed in two cases (ACC > ATT; Figs. 1B and 4B). In these cases, consecutive base editing (ACC > ATC > ATT) might have occurred. These results suggest that, in the case of base editing in plant mitochondrial genomes with this method, bases located on the 5′ side of the converted C are more flexible, so more Cs in the genome can be candidates of targets for substitution than in the cases of base editing of mammalian mitochondrial DNA (6, 17).
Some Characteristics of the mitoTALECD Method.
mitoTALEN methods for modifying mitochondrial genes (4, 5) reported large changes in the mitochondrial genome, including deletions around the cleavage sites, rearrangement of gene order, and generation of new repeat sequences, probably as a result of ectopic homologous or homeologous recombination. In contrast, no such changes were observed with mitoTALECDs (SI Appendix, Fig. S5). Also, the method is well suited for use with A. thaliana because its nuclear genome can be easily modified through floral dipping (21). Our method can also more cleanly knock out mitochondrial genes by creating a premature stop codon in the reading frame. This is not so easy to do with the mitoTALENs methods because of their tendency to cause other changes in the genome.
During the course of revision of this manuscript, another group reported an editing method that introduces homoplasmic and inheritable point mutations in tobacco mitochondrial genomes using TALEN and chemical mutagens (28). This method differed from our method in several respects. It 1) caused C:G > T:A, C:G > A:T, T:A > C:G, and T:A > A:T substitutions at the TALE-binding sequence, while our method caused only C:G > T:A substitutions; 2) achieved substitutions in about 5% of the plants that were genotyped, while our substitution frequencies ranged from 0 to 100% depending on the vector; 3) caused substitutions at random locations within the TALE-binding sequence, whereas our method has a greater ability to target the position of substitution; and 4) obtained mutants after one or more rounds of tissue culture of the T1 plants, while our method could generate mutants in the T1 generation. Which of the two methods is best will depend on the type of base substitution that is desired.
A problem with our method is that we occasionally observed some bystander errors, in which bases near the target base (C10) and in the target window were mutated (SI Appendix, Tables S3 and S4 and Fig. S4). Such errors might be avoided by changing the location and length of the target window or by changing the length of the linker sequence (29).
mitoTALECD Is a Useful Tool to Characterize RNA Editing, Analyze Mitochondrial ORFs of Unknown Function, and Conduct Future Crop Breeding.
The two experiments regarding RNA editing demonstrate the potential of the mitoTALECD method. In the first experiment, transforming otp87 plants with a mitoTALECD expression vector restored the growth of seven of the T1 plants (Fig. 3 and SI Appendix, Fig. S7). All of the seven plants had a substitution of the 1178C to T (U) at both DNA and RNA levels (Fig. 3 and SI Appendix, Fig. S7), which suggests that the growth retardation of otp87 is caused by its inability to edit 1178C of atp1 transcripts (15). However, some of the T1 plants were as small as otp87, even though the substitutions were homoplasmic (SI Appendix, Fig. S7 A and B). The reason is unclear, but it may be due to effects other than targeted base editing because some T1 plants with a Col-0 background also grew slowly (cf. phenotype columns in SI Appendix, Table S3 and Fig. S9).
So far, over 100 PLS-class PPR proteins have been identified as site recognition factors of RNA editing in plant mitochondria (10). Mutant plants of RNA editing factors often showed severe phenotypes, indicating that RNA editing events they participate in are essential for mitochondrial function (15, 30, 31). However, since about half of the PPR proteins target multiple sites, it has been difficult to distinguish the effect of each RNA editing event on plant phenotypes. mitoTALECD is thus useful for characterizing the impact of each RNA editing event on phenotype as demonstrated in the first RNA editing experiment (Fig. 3 and SI Appendix, Fig. S7).
The second RNA editing experiment showed that specific bases in atp1 were important for recognition by OTP87 (Fig. 4 and SI Appendix, Fig. S8). Although the PPR code seems to be a basic mechanism for recognition of the target RNA editing site (16, 25, 26), how each PPR protein specifically recognizes its target sites in vivo remains unclear. The contribution of each PPR motif–nucleotide interaction to recognizing the target site seems to be different depending on the intensity of the interaction and the type of the nucleotide, as well as the distance between the nucleotide and the target cytidines (16, 32). Furthermore, the C-terminal E1, E2, and DYW deaminase domains of PLS class PPR proteins (33–36) and other types of RNA editing factors such as MORF, ORRM, and NUWA proteins are likely to affect target recognition (37–40). mitoTALECD is useful for characterizing bases that are critical for RNA editing as demonstrated in the second RNA editing experiment (Fig. 4 and SI Appendix, Fig. S8). Moreover, this approach can also be potentially applied to analysis of other types of proteins that bind to specific RNA or DNA sequences and are involved in various gene expression processes in plant mitochondria.
Finally, we want to point out two ways in which mitoTALECD single-base substitution can be useful in plant studies. First, it can be used to study mitochondrial SNPs. SNPs in mitochondrial genomes should affect the phenotype, but few SNPs associated with a given phenotype have been identified. So mitoTALECD single-base substitution can be a valuable tool for identifying which of the SNPs are important. Second, mitoTALECD can be used to clarify the roles of mitochondrial ORFs of unknown function, including candidates for the genes underlying cytoplasmic male sterility, which is a useful trait for hybrid breeding (41, 42). The method described in this study can potentially substitute an amino acid, create a premature stop codon, and mutate cis elements in mitochondrial genes. We expect that through such analyses, this method will help to clarify the functions of mitochondrial genes as well as accelerate the search for SNPs in mitochondrial genomes that can be useful for crop breeding.
Materials and Methods
Plant Materials, Growth Conditions, Agrobacterium-Mediated Plant Transformation, and Screening Transformants.
A. thaliana Col-0, otp87 (homozygous transfer DNA [T-DNA] insertion line GK-073C06-011724), and transgenic plants were grown under a long-day condition (16 h light and 8 h dark) at 22 °C. Col-0 seeds were sown on half-strength Murashige & Skoog medium containing agar (8). Two- to three-week-old plantlets were transferred to Jiffy-7 (Jiffy Products International) and thereafter subjected to Agrobacterium transfection. Mature Col-0 and otp87 plants were transformed by floral dipping (21). Obtained T1 seeds were selected by their seed-specific fluorescence of green fluorescent protein (GFP) (8, 43). These T1 seeds were sown on the medium described above containing 125 mgL−1 of claforan. T1 plants were transplanted to Jiffy-7 at 23 DAS. otp87 seeds (GABI_073C06) were obtained from ABRC Stock Center. Homozygous T-DNA insertions in OTP87 in plants were checked by PCR (15).
Designing the TALE-Binding Sequence and Vector Constructions.
TALE-binding sequences are listed in Figs. 1A and 4B. The TALE-recognized bases were adjacent to the 3′ side of thymine and set about 20 bp in length. The length of the target window (16 bp) and the position of special target cytosine (C10) were set by estimation from the successes in a previous study (8). Binary vectors that express mitoTALECD were constructed by using the Platinum Gate TALEN system (18) and multisite Gateway (Thermo Fisher Scientific), almost as well as the same previous study (8). Some exceptions were a destination vector and an entry vector used for multi-LR reaction; in this study, we used ones that have a mitochondria-localization signal instead of a plastid-targeted signal [for the destination vector, we used a vector that was made from the one (Addgene ID 163145) by adding an Oleosin-GFP expression cassette; for the entry vector, Addgene ID 163146 (5)].
Genotyping T1 and T2 Plants.
PCRs for Sanger sequencing (for Figs. 1 and 2 and SI Appendix, Figs. S2–S4 and S6 and Tables S1 and S3) were examined using KOD One PCR Master Mix (TOYOBO) with roughly extracted DNA from an emerging leaf or cotyledon with standard protocols. Nucleic acid templates of PCRs for Sanger sequencing (for Figs. 3 and 4 and SI Appendix, Figs. S7 and S8) were extracted with Maxwell RSC Plant RNA Kit (Promega) not using included deoxyribonuclease. RNA templates of RT-PCRs were prepared by degrading DNA in the extracted nucleic acid with Deoxyribonuclease (RT Grade) for Heat Stop (Nippon Gene). RT-PCRs were performed using PrimeScript II High Fidelity One Step RT-PCR Kit (TaKaRa). A part of a mitoTALECD reading frame was amplified with primers (SI Appendix, Table S2) to identify transgenic plants. Mitochondrial DNA and complementary DNA sequences around the target window and its homologous sequences in nuclear DNA were amplified with primers shown in SI Appendix, Table S2. Purified PCR products were read by Sanger sequencing, and the data were analyzed with Geneious Prime (v. 2021. 2.2).
Total DNAs for whole-genome sequencing were extracted from mature leaves with DNeasy Plant Pro Kit (QIAGEN). Paired-end libraries for 11 samples using a VAHTS Universal Pro DNA Library Prep Kit for Illumina (Vazyme) and sequencing of 5Gbase/sample using the Illumina NovaSEq. 6000 platform were carried out by GENEWIZ Japan. We obtained whole-genome sequence data to perform SNP calling for three WT samples and eight T2 samples (two samples each for four lines). As preprocess for analysis, low-quality and adaptor sequences in the reads were trimmed using PEAT [v1.2.4 (44)]. Paired-end reads of each strain were mapped to the reference sequences (mitochondrial genome BK010421.1 and chloroplast genome AP000423.1) using BWA (v 0.7.12) in single-ended mode (45). We filtered out inadequate mapped reads with sequence identity ≤97% or alignment cover rate ≤80%. SNPs were then called using samtools mpileup command (-uf -d 50000 -L 2000) and bcftools call command [-m -A -P 0.1 (46)]. According to allele frequency (AF) calculated by bcftools, we finally detected SNPs with (AF of the T2 sample) − (average AF of three WT strains) ≥ 0.05 as off-target SNP candidates to remove the many artificial SNPs derived from NUMTs and plastid genome sequences similar to the sequences in the mitochondrial genome (Fig. 2C).
To evaluate whether closeness to target sites or similarity to TALE sequences influenced the locations of off-target mutations, we tallied off-target mutations that were either within 2,000 bp of the target site or within 10 to 20 bp of the region with homology (identity ≥70% and alignment cover rate ≥70%) to a sequence recognized by either TALE left or TALE right.
Templates for PCRs for amplicon next-generation sequencing (NGS) were the same as those used for PCRs for Sanger sequencing or whole-genome sequencing. PCRs were performed using primers that had sample-specific tags at the head (SI Appendix, Table S2). Pair-end libraries were prepared by using a VAHTS Universal Pro DNA Library Prep Kit for Illumina (Vazyme). Sequencing was carried out by GENEWIZ Japan using Illumina NovaSEq. 6000 platform. The allele frequencies of each base on the target sequence (5′-GCGGTAGTTCAAAACT-3′) were calculated based on the paired-reads that satisfied the following three conditions: 1) When paired-end reads were mapped to the target sequence, the 5′ ends of both reads were soft-clipped 6 bp each and the clipped sequence matched the sample-specific tag sequence (SI Appendix, Table S2); 2) the tag sequence of the read1 was identical to that of the read2; and 3) the average quality score of the target sequence was ≥36.
PPR Binding Sequence Prediction.
To predict the OTP87 binding site in atp1, the PPR code (16, 24) was used. In this code, the combination of two key amino acid residues at positions 5 and 35 of each PPR repeat was used to calculate the probabilities of which nucleotide each individual PPR repeat might recognize. Binding probabilities for each motif were depicted by weblogo (http://weblogo.berkeley.edu/), as shown in Fig. 4A.
Estimating RNA Editing Efficiency.
To estimate RNA editing efficiency at 1178C in Fig. 4C, we used EditR (http://baseeditr.com/), an algorithm that predicts base editing efficiency from raw data of Sanger sequencing (i.e., .ab1 files). This algorithm was designed to evaluate the editing efficiency of the CRISPR-Cas–based base editing system (47). Instead of using the guide RNA sequence, we used the sequence from the first base in the atp1 sequence to which OTP87 is predicted to bind, to 1178C (5′-GCTATGAAACAAGTATGCGGTAGTTC-3′). To filter out low-quality regions, we trimmed about 50 bases at both ends of the data before estimating base editing efficiency.
Image Processing.
Plant images were taken with a digital camera (OLYMPUS OM-D E-M5) and processed with Adobe Photoshop 2021.
Supplementary Material
Acknowledgments
We thank Mrs. Yoshiko Tamura, Mrs. Reiko Masuda, and Mr. Hiroki Ayabe for their technical support. The otp87 seeds were kindly provided by the Arabidopsis Biological Resource Center. This work was supported by grants from the Japan Society for the Promotion of Science (Grant Nos. 16H06279 (PAGS), 19H02927, and 19KK0391 to S.-i.A. and 20H05680 to N.T.).
Footnotes
Competing interest statement: The authors declare a competing interest. A patent related to the method described in this paper is pending in Japan (No. 2021-9001) and is in preparation for submission to countries in the Patent Cooperation Treaty. The patent is held by the University of Tokyo.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121177119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. DNA data and plasmids have been deposited in GenBank (LC708159–LC708165) and Addgene (186198–186204).
References
- 1.Remacle C., Cardol P., Coosemans N., Gaisne M., Bonnefoy N., High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl. Acad. Sci. U.S.A. 103, 4771–4776 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox T. D., Sanford J. C., McMullin T. W., Plasmids can stably transform yeast mitochondria lacking endogenous mtDNA. Proc. Natl. Acad. Sci. U.S.A. 85, 7288–7292 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston S. A., Anziano P. Q., Shark K., Sanford J. C., Butow R. A., Mitochondrial transformation in yeast by bombardment with microprojectiles. Science 240, 1538–1541 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Kazama T., et al. , Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nat. Plants 5, 722–730 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Arimura S. I., et al. , Targeted gene disruption of ATP synthases 6-1 and 6-2 in the mitochondrial genome of Arabidopsis thaliana by mitoTALENs. Plant J. 104, 1459–1471 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Mok B. Y., et al. , A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang B.-C., et al. , Chloroplast and mitochondrial DNA editing in plants. Nat. Plants 7, 899–905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakazato I., et al. , Targeted base editing in the plastid genome of Arabidopsis thaliana. Nat. Plants 7, 906–913 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Small I. D., Schallenberg-Rüdinger M., Takenaka M., Mireau H., Ostersetzer-Biran O., Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 101, 1040–1056 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Takenaka M., Jörg A., Burger M., Haag S., RNA editing mutants as surrogates for mitochondrial SNP mutants. Plant Physiol. Biochem. 135, 310–321 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Bentolila S., Oh J., Hanson M. R., Bukowski R., Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 9, e1003584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giegé P., Brennicke A., RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. U.S.A. 96, 15324–15329 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zehrmann A., Verbitskiy D., van der Merwe J. A., Brennicke A., Takenaka M., A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21, 558–567 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotera E., Tasaka M., Shikanai T., A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Hammani K., et al. , The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. J. Biol. Chem. 286, 21361–21371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takenaka M., Zehrmann A., Brennicke A., Graichen K., Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One 8, e65343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H., et al. , Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nat. Commun. 12, 1–6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakuma T., et al. , Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci. Rep. 3, 1–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mol C. D., et al. , Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: Protein mimicry of DNA. Cell 82, 701–708 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Tsutsui H., Higashiyama T., pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol. 58, 46–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Noutsos C., Richly E., Leister D., Generation and evolutionary fate of insertions of organelle DNA in the nuclear genomes of flowering plants. Genome Res. 15, 616–628 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang G.-J., et al. , Nuclear integrants of organellar DNA contribute to genome structure and evolution in plants. Int. J. Mol. Sci. 21, 707 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan J., et al. , Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Res. 47, 3728–3738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barkan A., et al. , A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8, e1002910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T., Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One 8, e57286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weijers D., et al. , An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289–4299 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Forner J., et al. , Targeted introduction of heritable point mutations into the plant mitochondrial genome. Nat. Plants 8, 245–256 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan J., Zhang F., Karcher D., Bock R., Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat. Commun. 10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glass F., Härtel B., Zehrmann A., Verbitskiy D., Takenaka M., MEF13 requires MORF3 and MORF8 for RNA editing at eight targets in mitochondrial mRNAs in Arabidopsis thaliana. Mol. Plant 8, 1466–1477 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Sung T. Y., Tseng C. C., Hsieh M. H., The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. Plant J. 63, 499–511 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Oldenkott B., Yang Y., Lesch E., Knoop V., Schallenberg-Rüdinger M., Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2, 85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruwe H., Gutmann B., Schmitz-Linneweber C., Small I., Kindgren P., The E domain of CRR2 participates in sequence-specific recognition of RNA in plastids. New Phytol. 222, 218–229 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Cheng S., et al. , Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Takenaka M., et al. , DYW domain structures imply an unusual regulation principle in plant organellar RNA editing catalysis. Nat. Catal. 4, 510–522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ichinose M., Sugita M., The DYW domains of pentatricopeptide repeat RNA editing factors contribute to discriminate target and non-target editing sites. Plant Cell Physiol. 59, 1652–1659 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Takenaka M., et al. , Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5104–5109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bentolila S., et al. , RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. U.S.A. 109, E1453–E1461 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi X., Germain A., Hanson M. R., Bentolila S., RNA recognition motif-containing protein ORRM4 broadly affects mitochondrial RNA editing and impacts plant development and flowering. Plant Physiol. 170, 294–309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He S., et al. , A novel imprinted gene NUWA controls mitochondrial function in early seed development in Arabidopsis. PLoS Genet. 13, e1006553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kubo T., Newton K. J., Angiosperm mitochondrial genomes and mutations. Mitochondrion 8, 5–14 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Chen Z., et al. , Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit. Rev. Plant Sci. 36, 55–69 (2017). [Google Scholar]
- 43.Shimada T. L., Shimada T., Hara-Nishimura I., A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61, 519–528 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Li Y.-L., et al. , PEAT: An Intelligent and Efficient Paired-End Sequencing Adapter Trimming Algorithm in BMC Bioinformatics (BioMed Central, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., Durbin R., Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kluesner M. G., et al. , EditR: A method to quantify base editing from Sanger sequencing. CRISPR J. 1, 239–250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. DNA data and plasmids have been deposited in GenBank (LC708159–LC708165) and Addgene (186198–186204).