To the Editor,
Human rhinovirus (HRV) is the most frequent cause of the common cold and acute viral rhinosinusitis.1 Traditionally, HRV identification was performed using culture-based, biochemical, and serologic methods and limited to HRV-A and HRV-B species. The advent of PCR-based sequencing allowed for the rapid and sensitive diagnosis of HRV, and the discovery of a novel HRV-C species in 2006.2 Researchers using advanced molecular diagnostic techniques have determined that HRV infections, in particular HRV-A and HRV-C species, are the major viral trigger for childhood and adult exacerbations of asthma and chronic obstructive pulmonary disease.3 However, the role of HRV infections in chronic rhinosinusitis (CRS) is not well understood.4 Given that different HRV species have been shown to exhibit varying degrees of disease severity in chronic airway disorders, scores we performed a study to (a) detect HRV species and associated viral symptoms in adults with CRS and (b) test the differential epithelial response after RV-A16 and RV-C15 infections in air-liquid interface (ALI) cultures derived from their surgical specimens.
We performed a retrospective study of 218 adults who were diagnosed with CRS after obtaining IRB approval and written consent (Table S1). At each visit, viral nasal swabs and Wisconsin Upper Respiratory Symptom Surveys (WURSS-11) were collected. A total of 412 successfully screened viral swabs and WURSS-11 questionnaires were collected over the course of 2 years. Some patients were tested multiple times as a part of their routine office visits. We used a linear mixed model to account for the repeated sampling of patients. Of those patients whom tested negative for HRV at their first visit, 40% were asymptomatic, and among the remaining 60%, their median WURSS score was 11. Of those patients whom tested positive for HRV, a higher percentage was identified in the symptomatic (WURSS > 0) compared to the asymptomatic (WURSS = 0) group. We focused on HRV-A and HRV-C species differences based on the small number of those with HRV-B, or multi-HRV infections (Figure 1A). We identified patients with positive HRV infections, and their WURSS score was log-transformed before analysis to reduce skewness and heteroscedasticity. In a linear regression, HRV-C infections (22 ± 12.4), as compared to HRV-A infections (12 ± 6.8), and female gender resulted in significantly higher symptom scores (P = .01) in symptomatic participants with CRS. This trend was not significant when including all participants (WURSS ≥ 0) (Figure 1B, Tables S2 and S3).
FIGURE 1.
Human rhinovirus (HRV) species and their relation to WURSS-11 symptom severity. A, 11% of all viral swabs were positive for HRV infection (n = 17) from all asymptomatic patients (WURSS-11 = 0; n = 161). HRV-positive/asymptomatic swabs were divided by HRV-A (35%, n = 6), HRV-B (12%, n = 2), HRV-C (47%, n = 8), and multi-HRV (6%, n = 1). 17% of all viral swabs were positive (n = 43) from all symptomatic patients (WURSS-11 > 0; n = 251). HRV-positive/symptomatic swabs were divided by HRV-A (40%, n = 17), HRV-B (5%, n = 2), HRV-C (44%, n = 19), and multi-HRV (12%, n = 5). B, Linear regression of the log of WURSS scores in those with positive HRV infection was performed in all patients (WURSS ≥ 0) and in symptomatic patients (WURSS > 0). A mixed model was used to account for repeated measures among patients. There were no significant factors associated with symptom severity in all patients. In patients who were symptomatic, HRV-C species (P = .01) compared to HRV-A species, and female gender (P = .01) compared to male gender resulted in more severe symptom scores
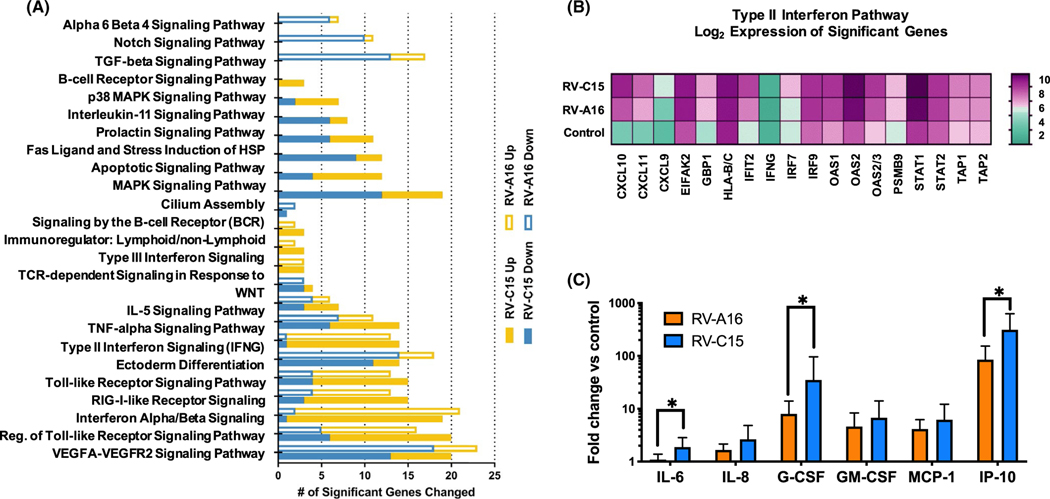
In order to determine what factors may contribute to this difference in severity between HRV-A and HRV-C in symptomatic patients, we used differentiated air-liquid interface (ALI) cultures derived from the middle turbinate epithelia of patients with CRS. ALI cultures from each donor (n = 4) were infected with a mock control, RV-A16 or RV-C15. No significant differences in viral binding after 4 hours and replication at 48 hours post-infection were identified between the two viruses (data not shown). Affymetrix gene expression analysis was performed on mRNA derived from whole-cell lysates (Figure 2A). The α6/β4, Notch, and TGFβ signaling pathways were uniquely downregulated with RV-A16 infection, while a set of pro-apoptotic and stress response pathways were uniquely deregulated in response to RV-C15. The “Type II Interferon Pathway” (F test P < .0001) specifically the induction of CXCL10 (interferon gamma-inducible protein 10; IP-10) was significantly upregulated after RV-C15 infection (52-fold increase) and RV-A16 infection (22-fold increase) when compared to control (P < .0001) (Figure 2B). Based on our gene expression array data, we looked specifically at the secretion of a subset of inflammatory cytokines associated with host-viral immune responses. Basolateral media were collected from the same infected ALI samples discussed above at the 48-hour post-infection time point. With the exception of IL-1ra and IL-4, all measured cytokines showed increased expression following RV-A16 or RV-C15 infection when compared to the control treated samples (Figure 2C), and all showed a larger magnitude of induction with RV-C. IL-6, G-CSF, and IP-10 (CXCL10) in particular were significantly more upregulated by RV-C15 infection than RV-A16.
FIGURE 2.
Differentially expressed genes and changes in cytokine production after RV-A16 and RV-C15 infection in sinus ALI cultures from patients with CRS. A, Sinonasal epithelial cells from 4 CRS patients were differentiated at ALI. Cultures were infected for 48 h with mock (control), HRV-A16, or HRV-C15 in duplicate. Pathway analysis of Affymetrix mRNA following HRV infection. Significant changes in response to RV-A16 infection are shown as solid boxes, and those due to RV-C15 are shown as empty bars; # upregulated genes = yellow, # downregulated genes = blue. B, Heatmap of significantly changed genes in the Type II Interferon signaling pathway (pathway significance F-test P-value < .00001). Log2 averaged gene expression values are shown. C, Cytokine secretion after RV-A16 and RV-C15 compared to control. Values shown are the average of two biologic replicates and three technical replicates, with fold change of infected versus uninfected controls. T test P-values *: IL-6 = .0033, G-CSF (CSF3) = .0131, and IP-10 (CXCL10) = 8.72 × 10−9
To date, this is the largest study assessing the prevalence of HRV in adults with CRS and the first to characterize HRV-C infections in adult CRS subjects. McCulloch et al5 identified all hospitalized adult patients with positive RV infections and by genotyping found that RV-A (54%) and RV-C (35%) were the predominant species. Similar rates of RV species prevalence have been found in non-hospitalized adults.6 Lee et al7 screened 111 adult CRS patients and were unable to detect any HRV-C infections in their nasal samples. However, they excluded any subjects who reported symptoms of upper respiratory infections over the past 4 weeks; therefore, active symptomatic RV-C infections would have been missed. A limitation of our study was its retrospective nature which did not allow for longitudinal assessments of patients before and after HRV infection as well as associated clinical disease measures.
Several studies have compared the epithelial response after infecting ALI cultures with different RV species and determined that RV-A and RV-C infections resulted in increased cellular damage and cytokine responses compared to RV-B infections.8 We also identified upregulation of genes associated with apoptotic pathways in response to RV-C15 infection and increased cytokine levels induced by RV-C15 compared to RV-A16 infection that could reflect host antiviral response or disease severity. In particular, CXCL10 expression levels and corresponding IP-10 protein levels showed significantly higher levels after RV-C infection when compared to RV-A infections and uninfected controls. CXCL10 is a cytokine secreted by epithelial cells in response to rhinovirus replication, and IP-10 protein levels have been shown to be significantly increased after RV infections in asthma both in vivo and in vitro and are a specific biomarker for acute virus-induced airway disease.9
In summary, our clinical and in vitro molecular data suggest that HRV-C infections are common and stimulate increased epithelial inflammatory responses compared to HRV-A infections. Because we used single clinical isolates of RV species for our infections (RV-A16, RV-C15) in CRS tissues, these findings may not be generalized to all RV species strains. Future longitudinal studies detailing host-virome interactions to specific RV strains and their associated clinical characteristics are required to determine their role in the pathophysiology of CRS.
Supplementary Material
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
CONFLICTS OF INTEREST
Drs. Chang, Billheimer, and Martinez report grants from NIH (R01 HL132523) during the conduct of the study. The remaining authors (AW, JC, JC, AK, RL, ET, and CL) have nothing to disclose. AW, FM, and EC designed the study; AW, JC, JC, AK, and RL performed the experiments; ET and DB performed the statistical analyses; and CL and EC provided the clinical data and analysis. All authors have reviewed and approved the manuscript.
REFERENCES
- 1.Pitkäranta A, Arruda E, Malmberg H, Hayden FG. Detection of rhinovirus in sinus brushings of patients with acute community-acquired sinusitis by reverse transcription-PCR. J Clin Microbiol 1997;35(7):1791–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominguez SR, Briese T, Palacios G, et al. Multiplex MassTag-PCR for respiratory pathogens in pediatric nasopharyngeal washes negative by conventional diagnostic testing shows a high prevalence of viruses belonging to a newly recognized rhinovirus clade. J Clin Virol. 2008;43(2):219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gern JE. The ABCs of rhinoviruses, wheezing, and asthma. J Virol. 2010;84(15):7418–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basharat U, Aiche MM, Kim MM, Sohal M, Chang EH. Are rhinoviruses implicated in the pathogenesis of sinusitis and chronic rhinosinusitis exacerbations? A comprehensive review. Int Forum Allergy Rhinol. 2019;9(10):1159–1188. [DOI] [PubMed] [Google Scholar]
- 5.McCulloch DJ, Sears MH, Jacob JT, et al. Severity of rhinovirus infection in hospitalized adults is unrelated to genotype. Am J Clin Pathol 2014;142(2):165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe A, Carraro E, Kamikawa J, Leal E, Granato C, Bellei N. Rhinovirus species and their clinical presentation among different risk groups of non-hospitalized patients. J Med Virol. 2010;82(12):2110–2115. [DOI] [PubMed] [Google Scholar]
- 7.Lee SB, Yi JS, Lee BJ, et al. Human rhinovirus serotypes in the nasal washes and mucosa of patients with chronic rhinosinusitis. Int Forum Allergy Rhinol. 2015;5(3):197–203. [DOI] [PubMed] [Google Scholar]
- 8.Nakagome K, Bochkov YA, Ashraf S, et al. Effects of rhinovirus species on viral replication and cytokine production. J Allergy Clin Immunol. 2014;134(2):332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wark PA, Bucchieri F, Johnston SL, et al. IFN-gamma-induced protein 10 is a novel biomarker of rhinovirus-induced asthma exacerbations. J Allergy Clin Immunol. 2007;120(3):586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.