Evolutionary history of any living organism is as fascinating as it is complex. The causative agent of plague, the bacterium Yersinia pestis, is no exception. Having diverged from the enteropathogen Yersinia pseudotuberculosis, ancestral strains of Y. pestis spread all over Late-Neolithic Eurasia. In their study, Andrades Valtueña et al. (1) present a tour de force by reporting 17 new prehistoric Y. pestis genomes from Eurasian human burials (adding to 13 previously published) (1–7). Furthermore, their work, together with previously published data, lays the foundations for a new classification of Y. pestis strains and broadens our insight into the dynamics of emergence and spread of Y. pestis in prehistoric Eurasia.
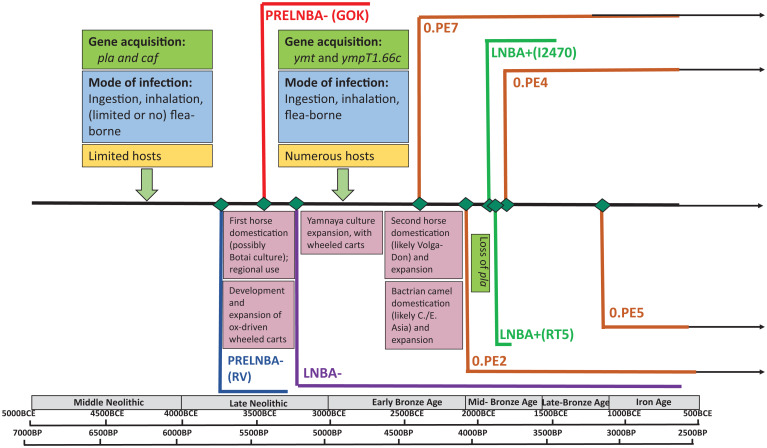
Of the ancient genomes, the authors classify the two oldest (previously published) genomes as the “preLNBA–” (pre-Late Neolithic/Early Bronze Age) lineage and 26 others as the “LNBA–” lineage (1). PreLNBA– genomes are from Latvia and Sweden, dated, respectively, to 5,300 to 5,050 and 5,040 to 4,867 y B.P. The low number of reported preLNBA– genomes suggests that these lineages may have died out several centuries after their rise. In contrast to preLNBA– lineages, LNBA– lineages were present over a wide geographic area (from Lake Baikal to central Europe) and existed for at least 2,500 y—from ca. 5,100 y B.P. until 2,736 to 2,457 y B.P. (the date of the latest genome).
The data reported by Andrades Valtueña et al. (1) indicate that the LNBA– lineage had a different evolutionary story compared to later lineages, such as “Branch 1” causing the second and third plague pandemics, characterized by phylogenetic and global focal diversity (8–10). Conversely, the LNBA– lineage is characterized by a genetic monophyly, lacking distinct subbranches, containing many evolutionary dead ends, and showing no correlation between genetic and geographic distance (1). Interestingly, the two earliest LNBA– genomes from central Siberia and North Caucasus are both chronologically concurrent (ca. 4,836 to 4,622 y B.P.) and phylogenetically positioned together. Given the 4,600 km that separate these genomes, it appears that the LNBA– lineage displayed a fast and extensive spread, presumably facilitated first by ox-hauled carts and, later, horse and camel domestication (11–13). Overall, LNBA– strains may have 1) all evolved from a single source deme, 2) spread with a high geographic mobility, and 3) had a limited reservoir.
In addition to new “LNBA–” genomes, the authors report a genome from El Sotillo (Spain), dated 3,361 to 3,181 y B.P. This genome, together with the previously published genome from the Volga region (Russia), dated 3,868 to 3,704 y B.P., represent two separate lineages that emerged at the beginning of the second millennium BCE and designated as “LNBA+” because they acquired ymt, which expands the range of mammalian hosts that sustain flea-borne plague (14). Interestingly, the LNBA+ and LNBA– temporally coexisted among themselves and with other early lineages (0.PE7, 0.PE2, 0.PE4, and 0.PE5), emerging successively in the later third and the second millennia BCE and also harboring ymt (Fig. 1) (1).
Fig. 1.
Early evolutionary history of Y. pestis based on the study and refs. 14–17 and 21–24 and its historical contexts, based on refs. 11–13.
Both LNBA– and LNBA+ acquired the loci (pla and caf) important to produce different “modern” forms of plague with high incidence: pneumonic plague transmitted by aerosol between humans and bubonic plague transmitted by fleas (15, 16). Also, LNBA+ would have evolved further to become more virulent in mammals and fleas, notably through the acquisition of the YPMT1.66c and ymt genes (14, 17) (Fig. 1).
Some argued that the early clinical manifestation of plague was the pneumonic form (2, 5). This rarest form of plague requires a permanent close contact between humans in densely settled environments to be maintained over space and time (18), which hardly characterizes late Neolithic and Bronze Age Eurasia. While some “proto-urban” sites in Eurasia, such as Cucuteni-Trypillia (northeastern Romania, Moldova, and western Ukraine) or Altyndepe (Turkmenistan) had high population densities (19, 20), most regions, especially the steppe, were sparsely populated. Therefore, one may wonder whether LNBA “–” and “+” lineages would have been more likely transmitted by fleas (or even lice) rather than inhalation of contaminated aerosols.
There are two models of flea-borne plague transmission. According to the “early-phase transmission” model, the transmission occurs within a few hours or days after an infectious blood meal and then quickly fades away if uninfected meals are subsequently ingested (21). According to the “blocked-flea model,” transmission requires an extrinsic incubation period of at least 4 to 5 d and occurs even after the ingestion of uninfected meals over 1 mo (22, 23). The mechanism of early-phase transmission is unclear. By contrast, we know that the “blocked-flea transmission” results from Y. pestis’ ability to form a solid mass in the foregut. This mass obstructs the ingestion of blood into the gut during a feeding attempt and increases the fleabite rate (due to the starving) and, therefore, favors the regurgitation of Y. pestis into the mammalian host. The transmission by “blocked” fleas sustains the long-term persistence of the flea–mammal–flea cycle, which may not be possible with early-phase transmission.
Both LNBA– and LNBA+ could, in theory, have been transmitted by fleas. However, fundamental genetic differences between the two lineages imply that the chances of transmission of the LNBA– by fleas is rather low and most likely limited to early-phase transmission. Indeed, LNBA– lineage has retained 1) a ureolytic activity killing >40% of fleas within the first day of infection and 2) functional ancestral genes that prevent the bacteria from producing a carbohydrate polymer important for flea blockage (23, 24). Furthermore, it lacks ymt that expands the range of mammalian hosts that sustains flea-borne plague (14). This genetic pattern may explain the particular LNBA– phylogeny, characterized by a low level of genetic diversification and numerous evolutionary dead ends, and evidently an eventual extinction of this lineage, judging by its absence from modern-day reservoirs.
The putative extinction of nonadapted flea LNBA– lineage occurred at least 2,500 y after its rise. How did this lineage survive and spread for such a long time? Is it possible that it spread via the ingestion of contaminated food, considering it emerged from an enteropathogen? In the Late Neolithic and Early Bronze Age, the Eurasian steppe between the Black Sea and western China was settled primarily by nomads, dwelling in proximity to a wider natural world in which sylvatic rodents abounded. While there is, at present, no paleodiet data on human consumption of rodents, there is archaeological evidence of their hunting and skinning, as well as using their teeth and bones for tools and crafts in late Neolithic Central Eurasia (5, 25, 26). However, as evidence from 20th-century Central Asia shows, primary outbreak of gastroenteritic plague in humans is confined to local communities, rather than spread across regions (27). Moreover, to maintain such a route of transmission would require constant contacts between infected rodents and humans all over Eurasia—hardly a feasible option. Finally, small migratory birds could be incriminated for the dissemination of LNBA– strains, given that a limited number of birds can be infected by Y. pestis and some may spread Y. pestis-infected fleas (28).
But what about flea-adapted LNBA+ lineages which, despite their theoretical capability to seed multiple reservoirs and become genetically diversified, became extinct, judging by their absence from modern-day reservoirs—in contrast to 0.PE branches, thriving today in different Asian foci (11, 29, 30)? All documented 0.PE2, 0.PE4, 0.PE5, and 0.PE7 genomes (>100 in total) come from sparsely populated regions in Asia. Could it be that local ecological and sociodemographic landscapes of 0.PE branches, marked by nomadic pastoralism, meant that plague was confined primarily to rodents, attacking humans only sporadically, and having less potential to burn itself out than LNBA+, spreading over a much wider geographic range, which included sedentary and more densely populated landscapes of western Eurasia, and circulating more intensively in humans? To appreciate why and when LNBA+ branches, in contrast with 0.PE, eventually died out, more genomes from various late Bronze and Iron Age and contexts are needed.
The question of the context in which Y. pestis lineages emerged, spread, evolved, and disappear is as important. Establishing the origins of the early lineages is, at present, not feasible, but a hypothesis may be offered. Given that the two earliest LNBA– genomes (RISE509 from central Siberia and RK1001 from North Caucasus) are both chronologically concurrent and phylogenetically positioned together, despite being situated some 4,600 km apart, may indicate that their common source was somewhere in between: in Central Asia, a home to both several Bronze-Age 0.PE lineages and all Iron-Age and medieval 0.ANT branches. If the preLNBA–, LNBA–, and LNBA+ branches arose in Central Asia, then the local climatic context of their chronologies is to be considered. The successive emergence of the three earliest known lineages between ca. 5,600 and 5,100 y B.P. as well as the 0.PE2, the two LNBA+ lineages and 0.PE4 between ca. 4,000 and 3,800 y B.P. (Fig. 1) occurred in the context of excessively wet episodes in Central Asia (31–34). Could it be that excessive rainfall, creating abundant grass biomass and facilitating population growth of rodent hosts, provided the optimal conditions for intense bacterial activity leading to divergence events?
Overall, Andrades Valtueña et al.’s (1) work opens the door to some exciting big questions such as when, where, how, and why ancestral strains have emerged, evolved, spread, and sometimes counterselected to extinction, and how they got transmitted from wildlife reservoirs to human populations. The work also invites the question of where we draw a border between attributes defining Y. pestis and its ancestor Y. pseudotuberculosis. Future collaborative synergistic research will undoubtedly advance our understanding of these fascinating questions.
Footnotes
The authors declare no competing interest.
See companion article, “Stone Age Yersinia pestis genomes shed light on the early evolution, diversity, and ecology of plague,” 10.1073/pnas.2116722119.
Change History
May 18, 2022: The title has been updated to correct a typographical error.
References
- 1.Andrades Valtueña A., et al. , Stone Age Yersinia pestis genomes shed light on the early evolution, diversity, and ecology of plague. Proc. Natl. Acad. Sci. U.S.A. 119, 10.1073/pnas.2116722119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimbler D. L., Schroeder J. A., Eddy J. L., Lathem W. W., Early emergence of Yersinia pestis as a severe respiratory pathogen. Nat. Commun. 6, 7487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasmussen S., et al. , Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell 163, 571–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrades Valtueña A., et al. , The Stone Age plague and its persistence in Eurasia. Curr. Biol. 27, 3683–3691.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Rascovan N., et al. , Emergence and spread of basal lineages of Yersinia pestis during the Neolithic Decline. Cell 176, 295–305.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Yu H., et al. , Paleolithic to Bronze Age Siberians reveal connections with First Americans and across Eurasia. Cell 181, 1232–1245.e20 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Susat J., et al. , A 5,000-year-old hunter-gatherer already plagued by Yersinia pestis. Cell Rep. 35, 109278 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Spyrou M. A., et al. , Phylogeography of the second plague pandemic revealed through analysis of historical Yersinia pestis genomes. Nat. Commun. 10, 4470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slavin P., Out of the West: Formation of a permanent plague reservoir in South-Central Germany (1349–1356) and its implications. Past Present 252, 3–51 (2021). [Google Scholar]
- 10.Cui Y., et al. , Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc. Natl. Acad. Sci. U.S.A. 110, 577–582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klimscha F., Transforming technical know-how in time and space. Using the Digital Atlas of Innovations to understand the innovation process of animal traction and the wheel. eTopoi 6, 16–63 (2017). [Google Scholar]
- 12.Librado P., et al. , The origins and spread of domestic horses from the Western Eurasian steppes. Nature 598, 634–640 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ming L., et al. , Whole-genome sequencing of 128 camels across Asia reveals origin and migration of domestic Bactrian camels. Commun. Biol. 3, 1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bland D. M., Miarinjara A., Bosio C. F., Calarco J., Hinnebusch B. J., Acquisition of yersinia murine toxin enabled Yersinia pestis to expand the range of mammalian hosts that sustain flea-borne plague. PLoS Pathog. 17, e1009995 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sebbane F., Jarrett C. O., Gardner D., Long D., Hinnebusch B. J., Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. U.S.A. 103, 5526–5530 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebbane F., Jarrett C., Gardner D., Long D., Hinnebusch B. J., The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect. Immun. 77, 1222–1229 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pradel E., et al. , New insights into how Yersinia pestis adapts to its mammalian host during bubonic plague. PLoS Pathog. 10, e1004029 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. K. Cohn, Jr, Epidemiology of the Black Death and successive waves of plague. Med. Hist. Suppl. 27, 74–100 (2008). [PMC free article] [PubMed] [Google Scholar]
- 19.Rassmann K., et al. , High precision tripolye settlement plans, demographic estimations and settlement organization. J. Neolithic Archaeol. 16, 96–134 (2014). [Google Scholar]
- 20.Masson V. M., Altyn-Depe (University of Pennsylvania Press, Philadelphia, PA, 1988). [Google Scholar]
- 21.Eisen R. J., Dennis D. T., Gage K. L., The role of early-phase transmission in the spread of Yersinia pestis. J. Med. Entomol. 52, 1183–1192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dewitte A., et al. , A refined model of how Yersinia pestis produces a transmissible infection in its flea vector. PLoS Pathog. 16, e1008440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y. C., Jarrett C. O., Bosio C. F., Hinnebusch B. J., Retracing the evolutionary path that led to flea-borne transmission of Yersinia pestis. Cell Host Microbe 15, 578–586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chouikha I., Hinnebusch B. J., Silencing urease: A key evolutionary step that facilitated the adaptation of Yersinia pestis to the flea-borne transmission route. Proc. Natl. Acad. Sci. U.S.A. 111, 18709–18714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen S., Bradley B., Maki D., Outram A., “Community organisation among Copper Age sedentary horse pastoralists of Kazakhstan” in Beyond the Steppe and the Sown: Proceedings of the 2002 University of Chicago Conference on Eurasian Archaeology, Peterson D. L., Popova L. M., Smith A. T., Eds. (Brill, Leiden, 2006), pp. 89–111.
- 26.Macāne A., Nordqvist K., Kostyleva E., Marmot incisors and bear tooth pendants in Volosovo hunter-gatherer burials. New radiocarbon and stable isotope data from the Sakhtysh complex, Upper-Volga region. J. Archaeol. Sci. Rep. 26, 101908 (2019). [Google Scholar]
- 27.Rivkus Y., Blyummer A. G., Endemiya Chumy v Pustynyakh Srednei Azii i Kazakhstana (Voronezh, 2016). [Google Scholar]
- 28.Mahmoudi A., et al. , Plague reservoir species throughout the world. Integr. Zool. 16, 820–833 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Kutyrev V. V., et al. , Phylogeny and classification of Yersinia pestis through the lens of strains from the plague foci of Commonwealth of Independent States. Front. Microbiol. 9, 1106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eroshenko G. A., et al. , Yersinia pestis strains of ancient phylogenetic branch 0.ANT are widely spread in the high-mountain plague foci of Kyrgyzstan. PLoS One 12, e0187230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathis M., Sorrel P., Klotz S., Huang X., Oberhänsli H., Regional vegetation patterns at lake Son Kul reveal Holocene climatic variability in central Tien Shan (Kyrgyzstan, Central Asia). Quat. Sci. Rev. 89, 169–185 (2014). [Google Scholar]
- 32.Wolff C., et al. , Precipitation evolution of Central Asia during the last 5000 years. Holocene 27, 142–154 (2016). [Google Scholar]
- 33.Zhang D., Feng Z., Holocene climate variations in the Altai Mountains and the surrounding areas: A synthesis of pollen records. Earth Sci. Rev. 185, 847–869 (2018). [Google Scholar]
- 34.Leroy S., Ricketts R. D., Rasmussen K. A., Climatic and limnological changes 12,750 to 3600 years ago in the Issyk-Kul catchment, Tien Shan, based on palynology and stable isotopes. Quat. Sci. Rev. 259, 106897 (2021). [Google Scholar]