Abstract
We have found that the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1 produces an extracellular chitinase. The gene encoding the chitinase (chiA) was cloned and sequenced. The chiA gene was found to be composed of 3,645 nucleotides, encoding a protein (1,215 amino acids) with a molecular mass of 134,259 Da, which is the largest among known chitinases. Sequence analysis indicates that ChiA is divided into two distinct regions with respective active sites. The N-terminal and C-terminal regions show sequence similarity with chitinase A1 from Bacillus circulans WL-12 and chitinase from Streptomyces erythraeus (ATCC 11635), respectively. Furthermore, ChiA possesses unique chitin binding domains (CBDs) (CBD1, CBD2, and CBD3) which show sequence similarity with cellulose binding domains of various cellulases. CBD1 was classified into the group of family V type cellulose binding domains. In contrast, CBD2 and CBD3 were classified into that of the family II type. chiA was expressed in Escherichia coli cells, and the recombinant protein was purified to homogeneity. The optimal temperature and pH for chitinase activity were found to be 85°C and 5.0, respectively. Results of thin-layer chromatography analysis and activity measurements with fluorescent substrates suggest that the enzyme is an endo-type enzyme which produces a chitobiose as a major end product. Various deletion mutants were constructed, and analyses of their enzyme characteristics revealed that both the N-terminal and C-terminal halves are independently functional as chitinases and that CBDs play an important role in insoluble chitin binding and hydrolysis. Deletion mutants which contain the C-terminal half showed higher thermostability than did N-terminal-half mutants and wild-type ChiA.
Chitins are a large family of glycans which are β-1,4-linked, insoluble linear polymers of N-acetylglucosamine (GlcNAc). They are present in the walls of higher fungi, in the exoskeletons of insects, arachnids, and many other groups of invertebrates, and as an extracellular polymer of some bacteria. Chitin is the second most abundant organic compound (after cellulose). It has been estimated that the annual formation rate and steady-state amount of chitin is on the order of 1010 to 1011 tons (11). Therefore, application of thermostable chitin-hydrolyzing enzymes (chitinases) is expected for effective utilization of this abundant biomass. Various chitinases and their genes from eucarya and bacteria have been investigated. However, studies on archaeal chitinases have been limited except for the enzyme from the hyperthermophilic archaeon Thermococcus chitonophagus (15). The strain produces a chitinase and utilizes chitin as a carbon source; however, sequence and structural information for archaeal chitinases have not yet been reported.
Recently, we reported that an archaeal strain, Pyrococcus kodakaraensis KOD1, possesses a chitinase homologue in its genome (9). As there has been no structural information concerning archaeal chitinases, we have pursued analysis of the gene and its protein product. This initial work should greatly contribute to basic studies (structure-function analyses) and industrial applications (recycling of biomass) of thermostable chitinases. Furthermore, it is intriguing that a hyperthermophilic archaeon which possesses many properties of a primitive form of life produces a hydrolytic enzyme of chitins, which are mainly found in the exoskeletons and cell walls of highly evolved organisms.
In the present study, the entire unique chitinase gene from strain KOD1 was cloned and sequenced. The cloned gene was expressed in Escherichia coli cells, and the recombinant protein was purified to homogeneity for biochemical analysis.
MATERIALS AND METHODS
Bacterial strains, plasmids, phages, and media.
E. coli JM109 and TG-1 were used as hosts for plasmid DNA and for preparing subclones for nucleotide sequencing. E. coli XL1-Blue MRA (P2) (Stratagene, La Jolla, Calif.) was used as a host strain for infecting lambda phages. E. coli BL21 (DE3) was used as a host for plasmids pET-25b(+) and pET-8c (Novagen, Madison, Wis.) to overexpress chiA and deletion derivatives. Plasmids pUC18 and pUC19 were used as vectors for DNA subcloning. Phage lambda EMBL4 (Stratagene) was used for construction of a genomic library of P. kodakaraensis KOD1. KOD1 was grown anaerobically for 17 h at 85°C in 1-liter screw-cap bottles, as described by Morikawa et al. (21). E. coli strains were cultivated in Luria-Bertani medium (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl in 1 liter of deionized water [DW], pH 7.0) at 37°C. NZCYM medium (10 g of NZ amine, 5 g of yeast extract, 1 g of Casamino Acids, 5 g of NaCl, and 2 g of MgSO4 · 7H2O in 1 liter of DW, pH 7.0) was used for cultivation of BL21 (DE3). Ampicillin, when added, was used at a final concentration of 50 μg/ml.
Detection of chitinase activity on plate medium.
In order to detect chitinase activity from P. kodakaraensis KOD1, plate cultivation was performed on medium supplemented with colloidal chitin (0.5%) and gellan gum (1%). The mixture (40 ml), in a 250-ml screw-cap bottle, was autoclaved, and then 80 μl of sulfur solution (10 g of Na2S · 9H2O and 3 g of elemental sulfur in 15 ml of DW) was added and solidified on the inside wall of the bottle. This bottle was immediately put in an anaerobic atmosphere and kept over night for deoxidation. One hundred microliters of diluted cell suspension (10−5 or 10−6) was inoculated and incubated at 85°C for 15 h. Colloidal chitin hydrolysis around the colonies was visualized by the Congo red-polysaccharide interaction method (29).
DNA manipulations.
DNA manipulations were performed by standard methods, as described by Sambrook et al. (26). Restriction enzymes and other modifying enzymes were purchased from Takara Shuzo (Kyoto, Japan) or Toyobo (Osaka, Japan). Small-scale preparation of E. coli plasmid DNA was performed with the Wizard Miniprep DNA purification kit (Promega, Madison, Wis.), and large-scale plasmid DNA preparation was performed with the Qiagen plasmid Maxi kit (Qiagen, Hilden, Germany).
Construction of a genomic library in lambda phage, screening, and subcloning.
A lambda phage library of P. kodakaraensis KOD1 was prepared by ligating genomic DNA partially digested with EcoRI into EcoRI-digested arms of lambda EMBL4. The phage clone (no. 3-1) which carries β-glycosidase and chitinase genes was screened by plaque hybridization, as we reported previously (9).
Nucleotide sequence determination.
DNA sequencing was performed by the dideoxy chain termination method with fluorescent primers (A.L.F. DNA sequencer; Amersham Pharmacia Biotech Inc., Uppsala, Sweden). Nucleotide and amino acid sequence analyses were performed with DNASIS software (Hitachi Software Engineering Co. Ltd., Yokohama, Japan).
Construction of expression plasmids.
The expression plasmid pET-ChiA was constructed by two steps. First, the 5′ part of chiA was amplified by PCR with primers F1 and R1 (sequences are given below) with lambda phage clone DNA (no. 3-1) as a template. The amplified DNA (1,571 bp) was digested by NcoI and SacI, and the resulting NcoI-SacI fragment (288 bp) was inserted between the NcoI and SacI sites of pET-25b(+). The constructed plasmid was designated pET-NS1. Second, a 3.5-kbp SacI fragment, including the 3′ part of chiA from the phage DNA, was inserted into the SacI site of pET-NS1. The resulting plasmid was designated pET-ChiA. The expression plasmids for various deletion mutants, shown in Fig. 2, were constructed by PCR as described below. Six oligonucleotides (F1, 5′-TTCCATGGCGGAGAGCGTAAGCCTGAGCGG-3′; F2, 5′-CATATGCCGAGCAACATCACCGTTCC-3′; F3, 5′-CTTCCGGAGCACTTCTTCGCCCC-3′; R1, 5′-GCAGATCTCAGCCGAGGTGCTGGAGAACAGTATC-3′; R2, 5′-GGAAGCTTCAACCTGGGCCTGCTGGAACGTACGG-3′; R3, 5′-TGGGATCCAGCTGAAGAACTGGCTG-3′ [italic type indicates an NcoI site in F1, an NdeI site in F2, a BglII site in R1, a HindIII site in R2, and a BamHI site in R3]) were utilized as primers for PCR. Lambda phage DNA (no. 3-1) was used as a template for DNA amplification. For ChiAΔ1 construction, primers F1 and R2 were used for PCR. The amplified DNA (2,660 bp) was digested with NcoI and HindIII and then inserted into the NcoI and HindIII sites of plasmid pET-25b(+). The resulting plasmid was designated pET-ChiAΔ1. For ChiAΔ2, primers F2 and R3 were utilized for PCR. The amplified fragment (2,326 bp) inserted into the SmaI site of pUC18 was digested with NdeI and BamHI and then inserted into the NdeI and BamHI sites of pET-25b(+). The construct was designated pET-ChiAΔ2. For ChiAΔ3 construction, primers F1 and R1 were utilized. The amplified DNA (1,571 bp) was digested with NcoI and BglII and then inserted into the NcoI and BamHI sites of pET-25b(+). The resulting plasmid was designated pET-ChiAΔ3. For ChiAΔ4 construction, primers F3 and R3 were utilized for PCR. Plasmid pET-8c was digested with NcoI, treated with T4 DNA polymerase to fill in the cohesive ends, and digested with BamHI. The amplified fragment (1,282 bp) was digested with BamHI, phosphorylated with T4 kinase, and ligated into the prepared plasmid. The resulting plasmid was designated pET-ChiAΔ4.
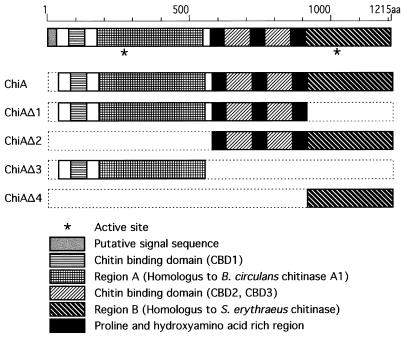
FIG. 2.
Structural features of the chitinase from P. kodakaraensis (ChiA) and schematic drawings of deletion mutants. A putative signal sequence, regions A and B, three CBDs, and three proline- and hydroxyamino acid residue-rich regions are shown as indicated.
Purification of recombinant ChiA and deletion mutants.
E. coli cells harboring pET-ChiA were induced for overexpression with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at the mid-exponential growth phase and incubated for 4 h at 37°C. Cells were harvested by centrifugation (7,000 × g for 10 min at 4°C), washed with buffer A (50 mM Tris-HCl [pH 7.5], 1 mM EDTA), and then resuspended in buffer A. The cells were disrupted by sonication, and the supernatant was obtained by centrifugation (24,000 × g for 20 min at 4°C). The supernatant was incubated at 70°C for 15 min and centrifuged (15,000 × g for 15 min at 4°C) to obtain heat-stable crude extract. The crude supernatant was brought to 40% saturation with solid ammonium sulfate, followed by overnight stirring at 4°C. The suspension was centrifuged (12,000 × g for 30 min at 4°C), and the resulting precipitate was dissolved in buffer A. The protein solution was dialyzed overnight against the same buffer and then centrifuged again to remove insoluble proteins formed during dialysis. A Resource Q column (Amersham Pharmacia Biotech) was equilibrated with buffer A, and the obtained supernatant was applied to the column. ChiA was eluted by a linear gradient of NaCl (0 to 0.5 M) with a protein purification apparatus (ÄKTA Explorer 10S; Amersham Pharmacia Biotech). The peak fractions were concentrated by using Centricon-100 (Millipore, Bedford, Mass.), and the sample was applied to a Superdex-200 HR 10/30 column (Amersham Pharmacia Biotech) equilibrated with buffer B (50 mM Tris-HCl [pH 7.5], 150 mM NaCl). The active fractions were collected and stored at 4°C. Purifications for deletion mutants were conducted with slight modifications. The heat-stable crude extract of ChiAΔ4 was brought to 50% saturation with solid ammonium sulfate. For concentration before gel filtration, Centricon-50 (for ChiAΔ1, ChiAΔ2, and ChiAΔ3) or Centricon-30 (for ChiAΔ4) was used. A Superdex-75 HR 10/30 column (Amersham Pharmacia Biotech) instead of Superdex-200 HR 10/30 was used for ChiAΔ4. The protein concentration was determined by the Bio-Rad protein assay system (Bio-Rad, Hercules, Calif.) with bovine serum albumin as a standard.
Preparation of colloidal chitin.
Ten grams of chitin (Wako Pure Chemical Industries Ltd., Osaka, Japan) was mixed with 500 ml of 85% phosphoric acid and stirred for 24 h at 4°C. The suspension was poured into 5 liters of DW and centrifuged (12,000 × g for 10 min). The resulting precipitate was washed with DW until the pH reached 5.0 and then neutralized by addition of 6 N NaOH. The suspension was centrifuged (12,000 × g for 10 min) and washed with 3 liters of DW for desalting. The resulting precipitate was suspended with DW. The chitin content in the suspension was determined by drying a sample (final concentration, 1%).
Enzyme assay.
Chitinase was assayed by a modification of the Schales procedure (16) with colloidal chitin as the substrate (final concentration, 0.17%). The standard assay was performed at 80°C in 50 mM sodium acetate buffer (pH 5.0) for 10 min. The reaction was terminated by cooling samples in an ice-cold bath, and the amount of reducing sugar generated was measured. One unit of chitinase activity was defined as the amount of enzyme which produces 1 μmol of reducing sugar per min. The pH dependence of chitinase activity was determined at 80°C with the following buffers: pH 2.5 to 4.0, 50 mM disodium hydrogen citrate-HCl; pH 4.0 to 5.5, 50 mM sodium acetate; pH 5.5 to 7.0, 50 mM morpholinoethanesulfonic acid-NaOH; pH 7.0 to 9.0, 50 mM Tris-HCl; pH 9.0 to 10.5; 50 mM glycine-NaOH. The optimal temperature for chitinase activity was examined at 50 to 100°C with colloidal chitin as a substrate in 50 mM sodium acetate buffer (pH 5.0).
Chitinase activity was also measured by a fluorometric assay with 4-methylumbelliferyl N-acetyl-β-d-glucosaminide (GlcNAc-4MU), 4-methylumbelliferyl β-d-N,N′-diacetyl chitobioside (GlcNAc2-4MU), and 4-methylumbelliferyl β-d-N,N′,N"-triacetyl chitotrioside (GlcNAc3-4MU) (Sigma, St. Louis, Mo.). The fluorescence of liberated 4-methylumbelliferone (4MU) was measured (350-nm excitation, 440-nm emission) in fluorescence spectrophotometer F-2000 (Hitachi Ltd., Tokyo, Japan).
TLC.
The reaction products for various N-acetyl-chitooligosaccharides and colloidal chitin were analyzed by silica gel thin-layer chromatography (TLC). Aliquots (1 μl) of the reaction mixtures were chromatographed on a silica gel plate (Kieselgel 60; Merck Co., Berlin, Germany) with n-butanol–methanol–25% ammonia solution–water (5:4:2:1 [vol/vol/vol/vol]), and the products were detected by spraying the plate with aniline-diphenylamine reagent (4 ml of aniline, 4 g of diphenylamine, 200 ml of acetone, and 30 ml of 85% phosphoric acid) and baking it at 180°C for 3 min.
Chitin binding assay.
The chitin binding assay was based on the procedure for the cellulose binding assay, with slight modifications (10). The standard assay mixture contained 35 μl of enzyme extract (ChiAΔ2 and ChiAΔ4 in 50 mM sodium phosphate buffer [pH 7.0]) and 35 μl of 1% colloidal chitin. After the mixture was incubated for 1 h at room temperature with occasional stirring, the supernatant containing unadsorbed protein was separated from colloidal chitin by centrifugation (15,000 × g for 10 min at 4°C). The precipitate was washed twice with 50 mM sodium phosphate buffer (pH 7.0). The supernatant and precipitate were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the objective band was detected.
Nucleotide sequence accession number.
The nucleotide sequence of chiA is available in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession no. AB02740.
RESULTS
Detection of chitinase activity from P. kodakaraensis KOD1.
P. kodakaraensis KOD1 cells were able to colonize on the plate medium containing colloidal chitin. Clear zones due to chitin hydrolysis were detected around the colonies by Congo red staining (data not shown). This result indicated that P. kodakaraensis KOD1 produced and secreted thermostable chitinase.
Cloning and sequence analysis of the P. kodakaraensis chitinase gene.
A genomic library was constructed from P. kodakaraensis KOD1 in the lambda phage EMBL4 vector system (9). Previously, we obtained a phage clone harboring the complete β-glycosidase gene in a 14-kb EcoRI fragment. Sequence analysis revealed that the β-glycosidase gene was clustered with an incomplete open reading frame (ORF) that shows high similarity with chitinases (9). The nucleotide sequence of the entire ORF was determined (Fig. 1), revealing that the ORF starts from the initiation codon GTG, located 7 bases downstream of the putative ribosomal binding sequence (GGGGTG). The GTG codon is known as a translational initiation codon of archaea (2, 6, 8). The ORF encodes a protein of 1,215 amino acids, and the deduced molecular mass is 134,259 Da, which is the largest among known chitinases. The ORF has been designated the chiA gene.
FIG. 1.
Nucleotide and deduced amino acid sequences of the chiA gene. The putative Shine-Dalgarno (SD) sequence is double underlined. The translation termination codon is designated by an asterisk. A putative signal sequence is underlined. CBDs are boxed. Proline- and hydroxyamino acid residue-rich regions are shown by dotted lines.
Structural features of ChiA.
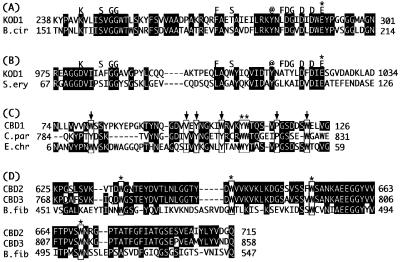
Figure 2 shows a schematic drawing of ChiA. In the N-terminal portion, a hydrophobic core region which might be functional as a signal sequence was found. The Ala-X-Ala sequence is frequently observed at signal peptide processing sites of secretory proteins from eucarya and bacteria (33). Archaeal signal peptides have also been shown to be processed at the same site (19, 28). Interestingly, two putative active sites and three putative chitin binding domains (CBDs) were found in ChiA. The N-terminal region is similar to Bacillus circulans WL-21 chitinase A1 (36% identity, 403 amino acids) (35), and the C-terminal region is similar to Streptomyces erythraeus chitinase (30% identity, 283 amino acids) (18). The N-terminal and C-terminal regions are tentatively termed region A and region B, respectively. Both chitinase regions are classified in family 18 of glycosyl hydrolases based on amino acid sequence similarities (12). Family 18 bacterial chitinases contain consensus residues, as indicated in Fig. 3A and B (27). Conserved glutamate residues are considered to be involved in the catalytic reaction as proton donors (24, 34). A unique conserved region which is known as a cellulose binding domain is found in cellulases, xylanases, and chitinases. ChiA was found to possess three conserved regions (CBD1, CBD2, and CBD3). One is in front of region A, and the other two are between regions A and B. These three regions are considered to be functional for chitin binding. The first, CBD1, shows sequence similarity with the CBD of Clostridium paraputrificum chitinase B (22) and the cellulose binding domain of Erwinia chrysanthemi endoglucanase Z (EGZ) (4) (Fig. 3C). They are classified into family V of cellulose binding domains (30, 31). It has been already reported that the CBD of C. paraputrificum chitinase B binds to chitin. The tertiary structure of the cellulose binding domain of EGZ has been solved and investigated in detail by nuclear magnetic resonance (4). The cellulose binding domain of EGZ contains several hydrophobic core residues and two solvent-exposed aromatic residues thought to stack directly against the pyranose rings of cellulose (4). These residues were found to be well conserved in CBD1 of ChiA. The other two CBDs (CBD2 and CBD3), which possess almost identical sequences (Fig. 3D), show sequence similarity with the Butyrivibrio fibrisolvens H17c endoglucanase cellulose binding domain (3). They contain four strictly conserved tryptophan residues (7). CBD2 and CBD3 were classified into family II of cellulose binding domains (bacterial type) according to similarities in primary structure (30, 31).
FIG. 3.
(A and B) Two putative active sites of ChiA. Amino acid sequences of two putative active sites of regions A and B are aligned with respective similar amino acid sequences of the active sites of B. circulans (B.cir) chitinase A1 (A) and S. erythraeus (S.ery) chitinase (B). Consensus residues among family 18 bacterial chitinases are indicated above the alignments. Conserved aromatic residues are indicated by “@.” Glutamate residues that probably act as proton donors are indicated by asterisks. (C) Comparison of amino acid sequences among ChiA CBD1, C. paraputrificum (C.par) chitinase B chitin binding domain, and E. chrysanthemi (E.chr) EGZ cellulose binding domain. Identical amino acids are shown against a black background. Two solvent-exposed aromatic residues in the cellulose binding domain of EGZ are indicated by squares and asterisks. Hydrophobic core residues in the cellulose binding domain of EGZ are squared with arrows. (D) Comparison of amino acid sequences among ChiA CBD2, CBD3, and B. fibrisolvens (B.fib) H17c endoglucanase cellulose binding domain. Identical amino acids are shown against a black background. Four strictly conserved tryptophan residues of family II cellulose binding domains are squared with asterisks.
The region occupied by many proline and hydroxyamino acid residues such as serine and threonine is known to be functional as a domain linker (31). Proline- and hydroxyamino acid-rich regions were observed around CBD2 and CBD3, as shown in Fig. 1. These regions of ChiA might function as domain linkers to connect the two catalytic and chitin binding domains.
Overexpression and purification of the recombinant chitinase.
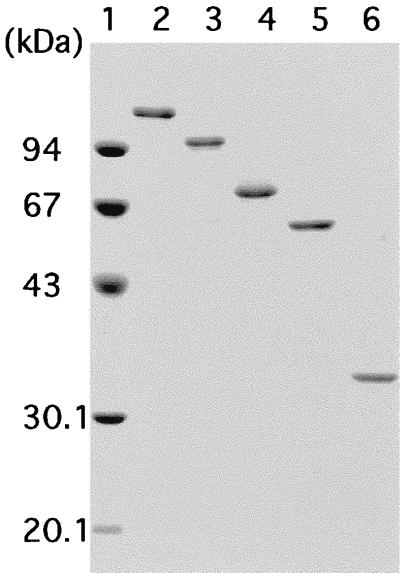
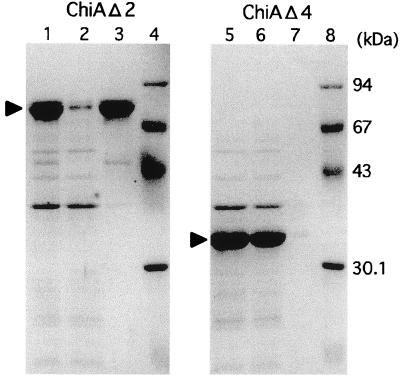
The chiA gene was expressed by using an expression plasmid pET-25b(+) which possesses a secretion signal to the periplasmic space of E. coli cells because efficient expression was not achieved by the recombinant plasmid harboring the entire signal region of chiA (data not shown). Hence, the putative signal region of ChiA was replaced with a bacterial signal sequence for efficient periplasm secretion. Significant chitinase activity was detected from cells harboring the recombinant plasmid pET-ChiA after induction by IPTG. The cells were disrupted by sonication, and the protein was purified to homogeneity by heat treatment and column chromatography with ResourceQ and Superdex-200, as described in Materials and Methods. The molecular mass (131,235 Da) calculated from the amino acid sequence without the signal peptide is in reasonable agreement with the 130 and 117 kDa assessed by SDS-PAGE (Fig. 4, lane 2) and gel filtration, respectively, indicating that ChiA is a monomeric enzyme. Purified ChiA was used for further enzymatic characterization.
FIG. 4.
SDS-PAGE of purified ChiA and deletion mutants. Lane 1, molecular mass standards, namely, rabbit muscle phosphorylase b (94 kDa), bovine serum albumin (67 kDa), egg white ovalbumin (43 kDa), bovine erythrocyte carbonic anhydrase (30.1 kDa), and soybean trypsin inhibitor (20.1 kDa); lane 2, purified ChiA (131,235 Da); lane 3, purified ChiAΔ1 (97,553 Da); lane 4, purified ChiAΔ2 (70,567 Da); lane 5, purified ChiAΔ3 (58,871 Da); lane 6, purified ChiAΔ4 (33,832 Da).
Characterization of the purified chitinase.
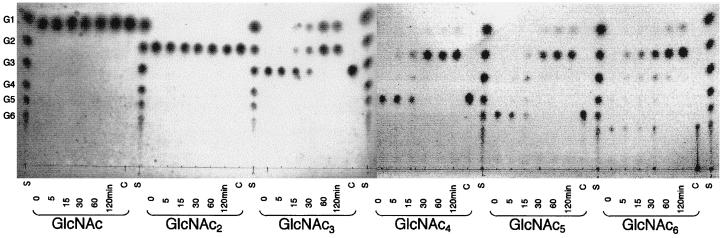
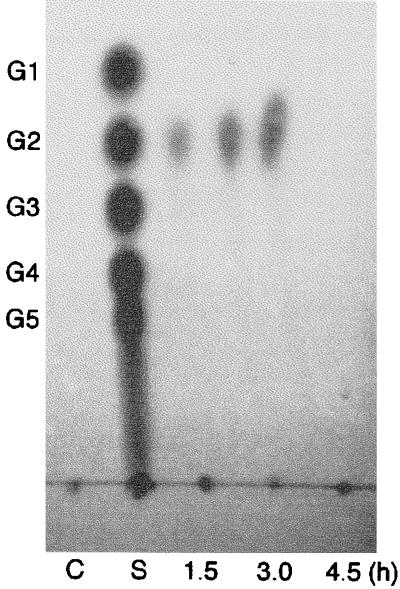
The optimal temperature and pH of ChiA for colloidal chitin were found to be 85°C and 5.0, respectively. Reaction products produced by ChiA were analyzed by TLC. When oligosaccharides (GlcNAc4–6) were used as a substrate, the major product was chitobiose (GlcNAc2), along with a small amount of GlcNAc. When GlcNAc3 was used as a substrate, the products were GlcNAc2 and GlcNAc. However, ChiA did not hydrolyze GlcNAc2 (Fig. 5). When colloidal chitin was used as a substrate, GlcNAc2 was detected by TLC at about 1.5 h after initiation of the reaction (Fig. 6). These results indicate that the major reaction product of digestion of ChiA is chitobiose (GlcNAc2).
FIG. 5.
TLC of restriction products by ChiA from various N-acetyl-chitooligosaccharides. The reaction mixture (50 μl) containing 0.7 mg of substrates in 70 mM sodium acetate buffer (pH 5.0) was incubated with enzyme (GlcNAc1–3, 0.45 μg; GlcNAc4–6, 0.9 μg) at 80°C. The reaction products at 0, 5, 15, 30, 60, and 120 min were analyzed. Lanes C, negative control incubated without enzyme at 80°C for 120 min; lanes S, standard N-acetyl-chitooligosaccharides ranging from GlcNAc (G1) to GlcNAc6 (G6).
FIG. 6.
TLC of restriction products by ChiA from colloidal chitin. The reaction mixture (1 ml) containing 0.16 mg of colloidal chitin in 50 mM sodium acetate buffer (pH 5.0) was incubated with enzyme (0.6 μg) at 80°C for 1.5, 3.0, and 4.5 h. The reaction products (250 μl each) were centrifuged, and the supernatants were concentrated and analyzed. Lane C, negative control incubated without enzyme at 80°C for 4.5 h; lane S, standard N-acetyl-chitooligosaccharides ranging from GlcNAc (G1) to GlcNAc5 (G5).
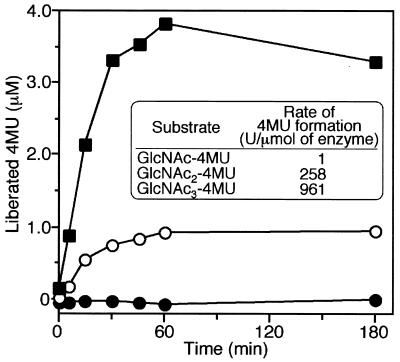
The reaction profile was further characterized by using fluorescently labeled substrates (Fig. 7). Three kinds of substrates, GlcNAc-4MU, GlcNAc2-4MU, and GlcNAc3-4MU, were used, and reaction velocities were compared based on the amount of free 4-MU released from the substrates and detected by fluorescence at 440 nm. It has been reported that the digestion pattern of chitinase can be examined by comparing activity for GlcNAc2-4MU with that for GlcNAc3-4MU (25). A typical endo-type enzyme is known to show higher activity for GlcNAc3-4MU than for GlcNAc2-4MU. The experimental results shown in Fig. 7 clearly indicate that ChiA does not hydrolyze GlcNAc-4MU and that the enzyme reaction velocity is higher for GlcNAc3-4MU than for GlcNAc2-4MU, suggesting that ChiA is an endo-type enzyme. In addition, the TLC pattern for reaction products of GlcNAc6 does not show major GlcNAc4 spots in the course of the enzyme reaction, indicating that ChiA is not an exo-type enzyme.
FIG. 7.
Reaction profiles with various 4-methylumbelliferyl saccharides (GlcNAc1–3-4MU, 350-nm excitation, 440-nm emission). The enzyme reaction mixture containing 10 μl of 1 mM GlcNAc1–3-4MU, 990 μl of 100 mM acetate buffer, and 20 μl of enzyme solution with 18 ng of ChiA was incubated at 80°C. The reaction mixture (100 μl) was added to 900 μl of ice-cold 100 mM glycine-NaOH (pH 11) to terminate the reaction. Liberated 4MU was detected at a wavelength of 440 nm. The rate of 4MU liberation was calculated from the initial reaction velocity. ●, GlcNAc-4MU; ○, GlcNAc2-4MU; ■, GlcNAc3-4MU.
Construction and characterization of deletion derivatives.
As mentioned above, ChiA possesses unique structural features with an unusually large molecular mass (ca. 130 kDa without the signal sequence). In order to examine whether regions A and B are independently functional as a chitinase, several deletion derivatives were constructed. A series of deletion mutants of the chitinase gene were constructed as shown in Fig. 2. ChiAΔ1 and ChiAΔ2 lack region B and region A with CBD1, respectively. ChiAΔ3 lacks region B, CBD2, and CBD3. ChiAΔ4 lacks all CBDs and region A. Deletion mutants were expressed in E. coli cells and purified to homogeneity as described in Materials and Methods (Fig. 4). All deletion mutants showed enzyme activity toward colloidal chitin, indicating that the respective active sites in region A and region B are independently functional (Table 1). The specific activity of ChiA was the highest among those of the enzymes examined. The sum of the specific activities of mutants containing region A (ChiAΔ3) or region B (ChiAΔ2) was nearly the same as ChiA activity, suggesting that the hydrolytic effects of region A and B were additive and not synergistic in this case. The optimal pH and temperature of these deletion mutants were similar to those of the wild-type enzyme (pH 5.0 and 85°C). The thermostabilities of ChiA and the four deletion mutants were examined by monitoring the remaining activity after heat treatment. When heat treatment was performed at 90°C, relative thermostabilities were as follows: ChiAΔ2 > ChiAΔ4 > ChiAΔ1 > ChiA > ChiAΔ3. In the case of heat treatment at 100°C, ChiA, ChiAΔ1, and ChiAΔ3 were inactivated within 3 min; however, ChiAΔ2 and ChiAΔ4 retained more than 70% activity for 3 h. The C-terminal half of ChiA was clearly indicated to be much more thermostable than the N-terminal half. Removal of the N-terminal region might induce a more thermostable conformation.
TABLE 1.
Specific activities of ChiA and its deletion mutants toward colloidal chitin
| Enzyme | Sp acta (U/nmol of enzyme) | % Activityb |
|---|---|---|
| ChiA | 2.42 | 100.0 |
| ChiAΔ1 | 1.23 | 50.9 |
| ChiAΔ2 | 1.42 | 58.4 |
| ChiAΔ3 | 0.94 | 38.7 |
| ChiAΔ4 | 1.02 | 42.1 |
Chitinase assays were performed under standard conditions.
Values represent activities relative to that of ChiA.
In order to confirm that CBD2 and CBD3 are functional as CBDs, in vitro binding experiments were performed. It was revealed that ChiAΔ2 exhibits binding to colloidal chitin but that ChiAΔ4 does not (Fig. 8). Therefore, CBD2 and/or CBD3 is functional as a chitin binding domain(s). In addition, ChiAΔ1 and ChiAΔ2, harboring CBD2 and CBD3, were found to show slightly higher activities toward colloidal chitin than do the corresponding mutants lacking CBD2 and CBD3 (Table 1). These results indicate that CBD2 and/or CBD3 plays an important role for both region A and region B in hydrolysis of the insoluble substrate.
FIG. 8.
Chitin binding assay of ChiAΔ2 and ChiAΔ4. Lanes 1 and 5, enzyme extract used for chitin binding experiment; lanes 2 and 6, unbound fraction to colloidal chitin; lanes 3 and 7, bound fraction to colloidal chitin; lanes 4 and 8, molecular mass standards, namely, rabbit muscle phosphorylase b (94 kDa), bovine serum albumin (67 kDa), egg white ovalbumin (43 kDa), and bovine erythrocyte carbonic anhydrase (30.1 kDa).
DISCUSSION
Many chitinases and their genes from eucarya and bacteria have been investigated. However, there have been no reports on chitinases from archaea except for one from the hyperthermophilic archaeon T. chitonophagus. T. chitonophagus produces a chitinase and utilizes chitin as a carbon and energy source (15). Strain KOD1 is the second example of a hyperthermophilic archaeon which produces chitinase, and this is the first report to describe unique structural and biochemical features of archaeal chitinase.
Many insoluble polysaccharide hydrolases, such as cellulases, xylanases, and chitinases, are composed of a catalytic domain joined to one or more substrate binding domains (cellulose binding domain, etc.) or modules (fibronectin type III module, etc.). Frequently they are linked by one or several recognizable linker sequences (31). Some of them have two catalytic domains, such as Anaerocellum thermophilum CelA, with separate glycosyl hydrolase family 9 and 48 catalytic domains (37), and Clostridium thermocellum CelJ, which is the largest catalytic component of the cellulosome (1). However, a chitinase with two catalytic domains has not yet been found. The structure of ChiA is considered to be unique among known bacterial and eucaryal chitinases.
At present, glycosyl hydrolases are classified into 57 families, based on amino acid sequence similarities (12–14). Family 18 includes chitinases from bacteria, fungi, viruses, animals, and some plants (classes III and V). On the other hand, family 19 includes almost exclusively plant chitinases (classes I, II, and IV) except for one bacterial chitinase from Streptomyces griseus HUT 6037 (23). Both catalytic regions of ChiA are classified into family 18 of glycosyl hydrolases.
Cellulose binding domains can be classified into 13 families (30). Three-dimensional structures of families I, II, III, IV, and V have been solved for at least one member of each family (4, 17, 20, 32, 36). CBD2 and CBD3 of ChiA were classified into family II. Family II cellulose binding domains are comprised of domains from various cellulases and chitinases (30). As mentioned in Results, the nucleotide and deduced amino acid sequences of CBD2 and CBD3 are almost identical (Fig. 3D). It is unclear why almost identical sequences are arranged in tandem. One of the regions might have been duplicated in the course of evolution to acquire more efficient substrate binding. CBD1 is homologous to the cellulose binding domain of EGZ, which is a member of family V. It was recently reported that family V sequences of cellulose binding domains have been found in several chitinases. The cellulose binding domain of EGZ and some CBDs from chitinase may form a new family (4). Most cellulose binding domains seem to be functional for efficient binding of the catalytic domains to crystalline and amorphous portions of cellulose substrates, leading consequently to effective enzyme concentration on the substrate surface. In addition, it has been proposed that cellulose binding domains mediate nonhydrolytic disruption of the crystalline structure and thereby facilitate subsequent enzymatic hydrolysis by the catalytic domains (31). Our experimental results showing that CBD2 and/or CBD3 surely binds to colloidal chitin (Fig. 8) and is important for hydrolysis (Table 1) does not contradict the above hypothesis.
Chitinases can be classified into two major categories: endo-chitinases and exo-chitinases. Exochitinases can be divided into two subcategories: chitobiosidases, which catalyze the progressive release of GlcNAc2 starting at the nonreducing end of the chitin microfibril, and 1-4-β-N-acetylglucosaminidases, which cleave the oligomeric products of endochitinases and chitobiosidases, generating monomers of GlcNAc (5). It is suggested that ChiA is an endochitinase for the following reasons: (i) ChiA does not hydrolyze GlcNAc2 and GlcNAc-4MU, indicating that ChiA is not a 1-4-β-N-acetylglucosaminidase; (ii) ChiA hydrolyzes GlcNAc3-4MU faster than GlcNAc2-4MU; (iii) when GlcNAc6 is used as a substrate, ChiA does not produce GlcNAc4 as a major product in the course of the reaction; and (iv) when GlcNAc4 and GlcNAc5 are used as a substrate, GlcNAc3 and GlcNAc4, respectively, are detected. If ChiA were a chitobiosidase, the enzyme should not be able to hydrolyze GlcNAc4 and GlcNAc5 to GlcNAc3 and GlcNAc4, respectively. Reasons ii through iv indicate that ChiA is not a chitobiosidase.
The structure of ChiA, with two catalytic regions, is very interesting. We have indicated that these catalytic regions are functional independently as thermostable chitinases (Table 1). Further studies are required to explain how these two catalytic domains and three CBDs interact with each other and whether there are synergistic effects of these domains on hydrolysis of crystalline chitin. It is noteworthy that the deletion mutants which harbor the C-terminal half show much higher thermostability at extremely high temperatures. These thermostable chitinases would be applicable for industrial uses. Moreover, it will be very interesting to clarify why a hyperthermophile exhibiting many properties of primitive forms of life possesses a large hydrolase of chitins, which occur mainly in the exoskeletons and cell walls of various creatures living at moderate temperatures. It is speculated that bacterial genes of chitinases have been horizontally transferred from bacteria to strain KOD1 in the process of evolution. Experiments to collect more information about the biochemical and physiological characteristics of this enzyme are under way.
ACKNOWLEDGMENT
This work was supported by a grant from CREST (Core Research for Evolutional Science and Technology), Japan.
REFERENCES
- 1.Ahsan M M, Kimura T, Karita S, Sakka K, Ohmiya K. Cloning, DNA sequencing, and expression of the gene encoding Clostridium thermocellum cellulase CelJ, the largest catalytic component of the cellulosome. J Bacteriol. 1996;178:5732–5740. doi: 10.1128/jb.178.19.5732-5740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcari P, Russo A D, Ianniciello G, Gallo M, Bocchini V. Nucleotide sequence and molecular evolution of the gene coding for glyceraldehyde-3-phosphate dehydrogenase in the thermoacidophilic archaebacterium Sulfolobus solfataricus. Biochem Genet. 1993;31:241–251. doi: 10.1007/BF00557333. [DOI] [PubMed] [Google Scholar]
- 3.Berger E, Jones W A, Jones D T, Woods D R. Cloning and sequencing of an endoglucanase (end1) gene from Butyrivibrio fibrisolvens H17c. Mol Gen Genet. 1989;219:193–198. doi: 10.1007/BF00261176. [DOI] [PubMed] [Google Scholar]
- 4.Brun E, Moriaud F, Gans P, Blackledge M J, Barras F, Marion D. Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi. Biochemistry. 1997;36:16074–16086. doi: 10.1021/bi9718494. [DOI] [PubMed] [Google Scholar]
- 5.Cohen-Kupiec R, Chet I. The molecular biology of chitin digestion. Curr Opin Biotechnol. 1998;9:270–277. doi: 10.1016/s0958-1669(98)80058-x. [DOI] [PubMed] [Google Scholar]
- 6.Cubellis M V, Rozzo C, Nitti G, Arnone M I, Marino G, Sannia G. Cloning and sequencing of the gene coding for aspartate aminotransferase from the thermoacidophilic archaebacterium Sulfolobus solfataricus. Eur J Biochem. 1989;186:375–381. doi: 10.1111/j.1432-1033.1989.tb15219.x. [DOI] [PubMed] [Google Scholar]
- 7.Din N, Forsythe I J, Burtnick L D, Gilkes N R, Miller R C, Jr, Warren R A, Kilburn D G. The cellulose-binding domain of endoglucanase A (CenA) from Cellulomonas fimi: evidence for the involvement of tryptophan residues in binding. Mol Microbiol. 1994;11:747–755. doi: 10.1111/j.1365-2958.1994.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 8.Dong G, Vieille C, Savchenko A, Zeikus J G. Cloning, sequencing, and expression of the gene encoding extracellular α-amylase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl Environ Microbiol. 1997;63:3569–3576. doi: 10.1128/aem.63.9.3569-3576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ezaki S, Miyaoku K, Nishi K, Tanaka T, Fujiwara S, Takagi M, Atomi H, Imanaka T. Gene analysis and enzymatic properties of thermostable β-glycosidase from Pyrococcus kodakaraensis KOD1. J Biosci Bioeng. 1999;88:130–135. doi: 10.1016/s1389-1723(99)80190-x. [DOI] [PubMed] [Google Scholar]
- 10.Gilkes N R, Warren R A J, Miller R C, Jr, Kilburn D G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J Biol Chem. 1988;263:10401–10407. [PubMed] [Google Scholar]
- 11.Gooday G W. Physiology of microbial degradation of chitin and chitosan. In: Ratledge C, editor. Biochemistry of microbial degradation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 279–312. [Google Scholar]
- 12.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber R, Stöhr J, Hohenhaus S, Rachel R, Burggraf S, Jannasch H W, Stetter K O. Thermococcus chitonophagus sp. nov., a novel, chitin-degrading, hyperthermophilic archaeum from a deep-sea hydrothermal vent environment. Arch Microbiol. 1995;164:255–264. [Google Scholar]
- 16.Imoto T, Yagishita K. A simple activity measurement of lysozyme. Agric Biol Chem. 1971;35:1154–1156. [Google Scholar]
- 17.Johnson P E, Joshi M D, Tomme P, Kilburn D G, McIntosh L P. Structure of the N-terminal cellulose-binding domain of Cellulomonas fimi CenC determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1996;35:14381–14394. doi: 10.1021/bi961612s. [DOI] [PubMed] [Google Scholar]
- 18.Kamei K, Yamamura Y, Hara S, Ikenaka T. Amino acid sequence of chitinase from Streptomyces erythraeus. J Biochem. 1989;105:979–985. doi: 10.1093/oxfordjournals.jbchem.a122791. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi T, Kanai H, Aono R, Horikoshi K, Kudo T. Cloning, expression, and nucleotide sequence of the α-amylase gene from the haloalkaliphilic archaeon Natronococcus sp. strain Ah-36. J Bacteriol. 1994;176:5131–5134. doi: 10.1128/jb.176.16.5131-5134.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraulis P J, Clore G M, Nilges M, Jones T A, Pettersson G, Knowles J, Gronenborn A M. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry. 1989;28:7241–7257. doi: 10.1021/bi00444a016. [DOI] [PubMed] [Google Scholar]
- 21.Morikawa M, Izawa Y, Rashid N, Hoaki T, Imanaka T. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl Environ Microbiol. 1994;60:4559–4566. doi: 10.1128/aem.60.12.4559-4566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J Bacteriol. 1997;179:7306–7314. doi: 10.1128/jb.179.23.7306-7314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohno T, Armand S, Hata T, Nikaidou N, Henrissat B, Mitsutomi M, Watanabe T. A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J Bacteriol. 1996;178:5065–5070. doi: 10.1128/jb.178.17.5065-5070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perrakis A, Tews I, Dauter Z, Oppenheim A B, Chet I, Wilson K S, Vorgias C E. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure. 1994;2:1169–1180. doi: 10.1016/s0969-2126(94)00119-7. [DOI] [PubMed] [Google Scholar]
- 25.Robbins P W, Albright C, Benfield B. Cloning and expression of a Streptomyces plicatus chitinase (chitinase-63) in Escherichia coli. J Biol Chem. 1988;263:443–447. [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Svitil A L, Kirchman D L. A chitin-binding domain in a marine bacterial chitinase and other microbial chitinases: implications for the ecology and evolution of 1,4-β-glycanases. Microbiology. 1998;144:1299–1308. doi: 10.1099/00221287-144-5-1299. [DOI] [PubMed] [Google Scholar]
- 28.Tachibana Y, Mendez M L, Fujiwara S, Takagi M, Imanaka T. Cloning and expression of the α-amylase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. J Ferment Bioeng. 1996;82:224–232. [Google Scholar]
- 29.Teather R M, Wood P J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomme P, Boraston A, McLean B, Kormos J, Creagh A L, Sturch K, Gilkes N R, Haynes C A, Warren R A J, Kilburn D G. Characterization and affinity applications of cellulose-binding domains. J Chromatogr B. 1998;715:283–296. doi: 10.1016/s0378-4347(98)00053-x. [DOI] [PubMed] [Google Scholar]
- 31.Tomme P, Warren R A J, Gilkes N R. Cellulose hydrolysis by bacteria and fungi. Adv Microb Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 32.Tormo J, Lamed R, Chirino A J, Morag E, Bayer E A, Shoham Y, Steitz T A. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 33.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe T, Kobori K, Miyashita K, Fujii T, Sakai H, Uchida M, Tanaka H. Identification of glutamic acid 204 and aspartic acid 200 in chitinase A1 of Bacillus circulans WL-12 as essential residues for chitinase activity. J Biol Chem. 1993;268:18567–18572. [PubMed] [Google Scholar]
- 35.Watanabe T, Suzuki K, Oyanagi W, Ohnishi K, Tanaka H. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J Biol Chem. 1990;265:15659–15665. [PubMed] [Google Scholar]
- 36.Xu G Y, Ong E, Gilkes N R, Kilburn D G, Muhandiram D R, Harris-Brandts M, Carver J P, Kay L E, Harvey T S. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:6993–7009. [PubMed] [Google Scholar]
- 37.Zverlov V, Mahr S, Riedel K, Bronnenmeier K. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile ‘Anaerocellum thermophilum’ with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology. 1998;144:457–465. doi: 10.1099/00221287-144-2-457. [DOI] [PubMed] [Google Scholar]