Abstract
The so-called ‘missing heritability problem’ is often characterized by behavior geneticists as a numerical discrepancy between alternative kinds of heritability. For example, while ‘traditional heritability’ derived from twin and family studies indicates that approximately ~50% of variation in intelligence is attributable to genetics, ‘SNP heritability’ derived from genome-wide association studies indicates that only ~10% of variation in intelligence is attributable to genetics. This 40% gap in variance accounted for by alternative kinds of heritability is frequently referred to as what’s “missing.” Philosophers have picked up on this reading, suggesting that “dissolving” the missing heritability problem is merely a matter of closing the numerical gap between traditional and molecular kinds of heritability. We argue that this framing of the problem undervalues the severity of the many challenges to scientific understanding of the “heritability” of human behavior. On our view, resolving the numerical discrepancies between alternative kinds of heritability will do little to advance scientific explanation and understanding of behavior genetics. Thus, we propose a new conceptual framework of the missing heritability problem that comprises three independent methodological and explanatory challenges: the numerical gap, the prediction gap, and the mechanism gap.
1. Introduction
First coined by Maher (2008), the so-called “missing heritability problem” (MHP) appears to refer to a conflict between two different ways of calculating heritability for any given phenotype. Traditional ‘quantitative’ genetic heritability, which is derived from twin and family studies, is usually significantly higher than its far more recent molecular counterparts, which are derived from genome-wide association studies (GWAS). Consider, for example, heritability of the Intelligence Quotient (IQ). Dating back nearly a century of human behavior genetics, heritability of IQ derived from twin and family studies ranges from .5 to .7, meaning that at least 50% of variance in IQ scores among related individuals is statistically associated with genetic differences between them. In stark contrast, however, cutting-edge GWAS have recently estimated that only 10% of variance in IQ is statistically associated with differences in DNA between unrelated individuals (Plomin & von Stumm, 2018; Selzam et al., 2016). This “missing” variance between traditional and molecular heritability – a 40% gap for IQ – is characteristic of the MHP. Importantly, there is not a single complex behavioral phenotype today for which there is no missing heritability.
Given the (unwise) tendency of researchers and the general public to interpret heritability as a measure of genetic influence on a trait, the MHP poses an interesting conceptual conflict. On the one hand, a century of traditional genetics suggests that genetic differences exhibit large influences on behavioral differences between individuals – typically 50% to 70% for any given behavioral trait. On the other hand, GWAS, which tracks actual differences in DNA between individuals, suggest that genes exhibit minimal influence on behavioral differences between individuals – typically 1% to 5% for any given behavioral trait. So what gives? Where is the missing heritability?
Although it has seen minimal attention from philosophers, we believe prospective explanations and resolutions to the MHP have implications for philosophy of biology. If, for example, one takes a monistic philosophical perspective of science, then the MHP means that something’s got to go: either traditional estimates of heritability derived from twin and family studies are approximately correct and molecular estimates are misconstrued, or vice versa – it can’t be that the heritability of IQ is both 10% and 50%. If this monistic perspective of science were accurate, then resolution of the MHP would have profound implications for philosophical debates about genetic causation (DiFrisco & Jaeger, 2020; Lynch, 2017; Lynch & Bourrat, 2016; Lynch & Kemp, 2014; Weber, 2005). In effect, resolution of the MHP in this case would mean that genetics exert either (a) little to no causal influence on human behavior or, conversely, (b) the lion’s share of causal influence on human behavior.
Alternatively, on a pluralistic philosophical perspective of behavior genetics – Helen Longino’s (2013), for example– explanations for the MHP would look quite different. From a pluralistic perspective, perhaps the MHP is no problem at all: traditional and molecular heritability are fundamentally different kinds of scientific estimates that are not necessarily in conflict. Eslwehere we argue, for example, that the MHP stands to impact philosophical literature regarding different kinds of scientific pluralism, such as integrative versus ineliminative pluralism (Matthews & Turkheimer, 2019). Plenty of philosophical literature hinges on the nature of the MHP and its prospective resolutions.
In what follows, we introduce and categorize the most prominent explanations and responses to the MHP. We will highlight a pervasive tendency among geneticists and philosophers of biology to assume that resolving the MHP is exclusively a numerical problem. That is, all existing literature on the MHP to date assumes that when the day comes that geneticists can make traditional heritability and DNA-based molecular heritability match, the MHP will be “dissolved.” To return to the IQ example, most researchers today assume that when the numerical value associated with the molecular heritability of IQ (currently .1) is not so dissimilar from the numerical value associated with traditional heritability of IQ (currently .5), then the profound mystery of heritability of IQ will finally be resolved! Importantly, most writing on the topic expresses optimism that this day will soon come as researchers collect larger datasets and develop more sophisticated statistical genetic models of heritability. Plomin & Stumm (2021), for example, highlight that the “missing heritability gap will be narrowed with bigger and better genome-wide association studies and with whole-genome sequencing that assesses all DNA differences in the genome rather than several hundred thousand SNPs assessed in current studies” (pp. 5–7).
Here we argue that framing the missing heritability problem in this way – as a relatively straightforward quantitative challenge of reconciling conflicting kinds of heritability – underappreciates the severe explanatory and methodological problems impeding scientific examination and understanding of heritability. We are not optimistic that the reconciliation of conflicting heritability estimates will confer any profound or meaningful scientific understanding of the heritability of human behavior. In the interest of promoting scientific humility about the many problems facing human behavior geneticists today, we propose a new framing of the MHP as one that comprises three independent scientific challenges, each of which vary in severity.
We refer to each independent challenge of the MHP as a kind of “gap,” as each is representative of some hole in scientific understanding of heritability that ought to be closed. On our proposal, the pervasive characterization of the MHP as a quantitative challenge is just the first and most tractable leg of the problem, which we call the numerical gap.1 Even if the numerical gap is closed, we argue that behavior geneticists face a second, greater problem of missing heritability: the prediction gap, which regards the challenge of making accurate and reliable prediction from DNA to behavior. Finally, the third and least tractable leg of the MHP regards the challenge of elucidating meaningful causal-mechanical stories that explain how and why genetic differences between individuals causes behavioral differences between individuals – we call this the mechanism gap. Each of the three legs of the MHP presents its own unique challenges and prospective solutions. We are pessimistic that the MHP will ever be “dissolved” for complex behavioral phenotypes (intelligence, for example). Contrary to popular belief, the MHP certainly isn’t going anywhere any time soon.
2. Formal Heritability Estimation
Broadly construed, ‘heritability’ is an estimate of the proportion of phenotypic variance that is statistically associated with genetic differences (Downes & Matthews, 2020). Historically, the paradigm of heritability estimation in humans is derived from Fisher’s (1918) seminal unification of discrete Mendelian inheritance with continuous (“biometric”) phenotypes (Downes & Turkheimer, in press). Fisher’s ideas were then adapted by the early twin researchers to estimate ratios of genetic to phenotypic variance (Newman et al., 1937). Lush (1935) refined the concept of heritability and developed it as a tool in artificial selection of plants and animals. Notably, traditional heritability today is derived primarily from quantitative analyses of pairs of monozygotic (MZ) ‘identical’ twins, who share 100% of their DNA, and dizygotic (DZ) ‘fraternal’ twins, who share roughly 50% of their DNA. We’ll refer to any kind of formal heritability estimate derived from twin and family studies as ‘twin heritability,’ or h2Twin.2 As a general rule of thumb, h2Twin ranges from .30 to .70 for most behavioral traits (Turkheimer, 2000).
Twin heritability is ‘coarse-grained’ in the sense that it tracks large approximations of differences in genetic similarity (100% for MZ twins, 50% for DZ twins, 25% for cousins, etc.). In contrast, heritability estimates obtained using molecular (genomic) methods track ‘fine-grained’ genetic differences: single nucleotide polymorphisms (SNPs), which are the smallest units of genetic variation between individuals (i.e., A, C, G, and T). To the extent GWAS are conducted in efforts to estimate the amount of phenotypic variance attributable to molecular genetic differences, they have been said to provide ‘GWAS heritability’, or ‘h2GWAS.’ Broadly construed, h2GWAS sums the total effect size of each individual SNP that meets the genome-wide significance threshold (P = 5 × 10−8) in a GWAS. Because the individual effect sizes of SNPs are miniscule (r2<.05%), h2GWAS estimates are systematically smaller than h2Twin.
More recently, a new technique has been introduced that increases the amount of phenotypic variance accounted by molecular genetic differences derived from GWAS. Initially implemented in a software package called genome-wide complex trait analysis (GCTA), but now comprising a family of similar approaches, ‘SNP heritability’ (h2SNP) tracks all the genomic differences between SNP chips of unrelated individuals (Yang et al., 2010, 2011). That is Although h2GWAS is derived by summing the effects of only SNPs that meet the genome-wide P-value significance threshold for the phenotype in question, h2SNP is unrestricted, and analyzes all SNPs simultaneously. Roughly construed, h2SNP is estimated by comparing the overall SNP similarity of unrelated individuals to their phenotypic similarity, for any given trait. Thus, h2SNP is in part derived from ignoring effect sizes and statistical significance, in favor overall genomic similarity. For most traits, SNP heritability is significantly higher than GWAS heritability; yet still significantly lower that twin heritability: h2GWAS < h2SNP < h2Twin.3
3. Explanations and Solutions for Missing Heritability
GWAS proponents often respond to the MHP by downplaying it, either denying that heritability is actually missing or expressing optimism that discovery of the missing heritability is nigh. While introducing their technique for calculating molecular heritability (GCTA), Yang et al. (2010), for example, claim that “most of the heritability is not missing but has not previously been detected because the individual effects are too small to pass stringent significance tests” (p. 565). Also, elaborating techniques geared toward calculating molecular heritability, Lee et al. (2011) claim to “show that a good proportion of the heritability is not missing” (p. 303). As sample sizes increase, more SNPs are identified, more variance is explained, and, consequently, the numerical gap gets smaller. Although optimism about future research is characteristic of most literature on the subject, the remainder of this section outlines what we believe to be the five primary categories of explanations or prospective solutions to the MHP.
3.1. Missed Variants:
a common explanation for missing heritability appeals to the existence of low-penetrance, common genetic variants whose effects are too small to be detected by GWAS. GWAS are conducted by analyzing SNP microarrays or ‘SNP chips,’ which include only common genetic variants occurring at a minor allele frequency (MAF) >1% (Weedon et al., 2021). To achieve sufficient statistical power, however, effect sizes need to be proportional to sample size: the smaller the effect, the larger the sample required to power an analysis. Thus, given miniscule molecular genetic effect sizes, GWAS have been historically underpowered to detect the effects of common SNPs. Maher (2008), for example, notes “the possibility that there are many more frequent variants that have such a low penetrance that GWAS can’t statistically link them to a disease” (p. 19). Conversely, others have proposed high-penetrance, rare variants as a possible source of missing heritability. Because most GWAS to date consider SNP chips composed of common variants, some propose that rare variants occurring at frequencies <.5% may be a source of missing heritability (Zuk et al., 2014). The solution to the rare variant problem requires either whole genome sequencing or conducting GWAS with SNP chips that include low-frequency alleles.
Thus, in both cases – low-penetrance, common variants and high-penetrance, rare variants – contemporary methods of genomic investigation inadequately detect existing genetic effects. Moreover, although the reasons for the inadequate detection are different, both cases may be roughly characterized as a ‘missed variants’ explanation for missing heritability: some of the missing heritability is attributable to variants whose effects have been overlooked by current molecular techniques.4 Acknowledging that there may be subtle differences between the methods and kinds of variants inadequately captured by genomic methods, we believe the missed variants explanation is of one of the most prominent explanations for missing heritability.
3.2. Copy Number Variants:
(CNVs) are another source of genetic variation not detected by GWAS. A CNV is a sequence of nucleotide bases which does not vary in content, but rather in quantity of repetitions. The age of onset of Huntington’s disease, for example, is probabilistically determined by the quantity of repetitions of the Huntingtin gene (Htt): the more repetitions, the earlier the onset (Vittori et al., 2014). A variety of CNV repetitions have been associated with schizophrenia (Marshall et al., 2017). This explanation for missing heritability traces back to Maher (2008) who noted that CNVs “could begin to explain missing heritability in disorders such as schizophrenia and autism, for which GWAS have turned up almost nothing” (p. 20).5 Prospective solutions to CNVs as sources of missing heritability may include whole genome sequencing, as well as characterizing sequence content and structural arrangement (Eichler et al., 2010). We categorize any explanation or prospective solution to the MHP that appeals to copy number variants as the CNV explanation for missing heritability.
3.3. Epistasis:
The epistasis explanation appeals to non-additive ways in which genetic differences tracked by GWAS are interacting in ways that make heritability difficult to interpret (i.e., GxG interaction). Zuk et al. (2012) claim that “current estimates of missing heritability are not meaningful, because they ignore genetic interactions” (p. 1194). Paradigmatic cases of epistasis regard modifier genes or variants that amplify or diminish effects of other genes or variants, which cannot be tracked by traditional GWAS. To properly understand the effects of epistasis, studies would require assessments of populations of individuals who contain the relevant combinations of SNPs. Note, however, that these epistatic effects are potentially far more complicated than described above, likely involving hundreds or thousands of interacting variants. Thus, the prospective solution to the epistasis explanation for missing heritability is the development of unique methods for identifying SNPs subject to GxG interaction (e.g., Song et al., 2010, general epistatic model). Despite persistent efforts, little progress has been made in statistical modelling of epistasis in human populations. Notably, although easily detectable in non-human animal models, there is minimal evidence that statistical epistasis detectable in human populations (Sackton & Hartl, 2016; Wei et al., 2014).
3.4. Epigenetics:
The epigenesis explanation for missing heritability appeals to a broad array of ways in which genetic variants interact with cellular or environmental factors, which are not detected by traditional GWAS. Many researchers have listed or mentioned epigenesis as a possible explanation for missing heritability, but do little to flesh it out (Bloom et al., 2013; Clarke & Cooper, 2010; Liu et al., 2008; McCarthy & Hirschhorn, 2008). Others describe formal and theoretical challenges of accommodating how epigenetic factors influence molecular heritability estimates. Slatkin (2009), for example, offers a formal model that permits exploration of transgenerational epigenetic inheritance risk and recurrence of complex disease. Petronis (2010) offers a detailed analysis of epigenetic effects on heritability in the context of the MHP, including discussion of specific mechanisms through which environmental influences may lead to epigenetic inheritance of both sporadic and familial diseases. Furrow et al. (2011) construct a formal model of standard epigenetic and environment-sensitive epigenetic inheritance as factors contributing to the MHP.
3.5. Twin Heritability Overestimation:
Another common response to the MHP appeals to various ways in which twin heritability may be inflated or overestimated (Maher, 2008; Manolio et al., 2009; Yang et al., 2015). The numerical gap arises, in part, because twin estimates are very high, for a number of reasons. Notably, twin heritability includes epistasis, epigenesis, and gene-environment correlations, as these non-linear or non-additive factors cannot be disentangled using traditional twin methods. Some authors have questioned the validity of the equal environments assumption of MZ twins, which may inflate twin heritability (Borkenau et al., 2002; Horwitz et al., 2003; Kendler et al., 1993; Scarr & Carter-Saltzman, 1979). Some commentators have noted that assortative mating may result in twin heritability overestimation as well.
4. Three Legs of the Missing Heritability Problem
In a boldly titled manuscript, “Dissolving the Missing Heritability Problem,” Bourrat & Lu (2017) claim “the missing heritability problem can largely be dissolved” (p. 1055). Although it remains a matter of fact that the numerical gap between twin heritability and SNP or GWAS heritability still exists for all behavioral phenotypes, Bourrat and Lu present a two-part explanation for missing heritability. Their account combines some of the aforementioned responses to suggest that dissolving the MHP is a matter of (1) decreasing twin heritability (because it is overestimated) and (2) increasing GWAS heritability (by modelling non-additive and non-linear molecular genetic interactions). Effectively, Bourrat and Lu’s explanation combines the twin heritability overestimation explanation for missing heritability with epigenesis and epistasis explanations.
Bourrat and Lu’s dissolution aligns with an underlying assumption of all the explanations and solutions: the missing heritability problem is a statistical problem. That is, each prospective solution to the MHP includes suggestions for how mathematical or statistical techniques might be tweaked or developed to close the numerical gap between heritability derived from either traditional twin and family studies, or molecular genetics (Génin, 2019). Although framing the issue numerically provides a quantifiable (and, therefore, potentially dissolvable) characterization of the MHP, our goal here is to bring attention to what we believe to be the more important and difficult challenges facing scientific efforts to elucidate the genetic underpinnings of human behavior – to fill the ‘gaps’ of what’s missing in current scientific and philosophical understanding of heritability.
The heritability of human behavior is a mystery machine. Honest geneticists will admit that although there are many leads, and genetical science has made wonderful leaps of technological progress in recent years, the relationship between genetics and behaviors remains one of the greatest challenges of modern biology. It is true: traditional genetics has revealed that all human behavioral traits are heritable, which is the first law of behavior genetics (Turkheimer 2000). It is true: hundreds of thousands of SNPs are statistically correlated with behavioral differences, which is the fourth law of behavior genetics (Chabris et al., 2015). It is also true, however: molecular genotypes are only weakly predictive of behavioral phenotypes. Importantly, it is also true: explanatory mechanisms that link molecular genotypes to behavioral phenotypes remain to be discovered. Our proposal is to reframe the missing heritability problem in a way that articulates these major methodological, practical, and explanatory impediments to understanding the heritability of human behavior. On our view, there are three primary aspects missing from even the most promising scientific accounts of heritability.
4.1. The Numerical Gap:
Conflicting results between alternative kinds of heritability is just one leg of the MHP, which we call the numerical gap (Figure 1, A). The challenge is ‘numerical’ in the sense that it is not grounded in alternative theories of heritability; bur rather, alternative numerical values derived from different methods of estimating heritability for the same phenotype. Thus, closing the numerical gap is a matter of finding ways to make traditional heritability (h2Twin) and molecular heritability (h2SNP) more similar. Recall that the current numerical gap regarding the MHP of IQ is approximately 40% (h2Twin = 50% versus h2SNP = 10%).
Figure 1: The Three Legs of the Missing Heritability Problem.
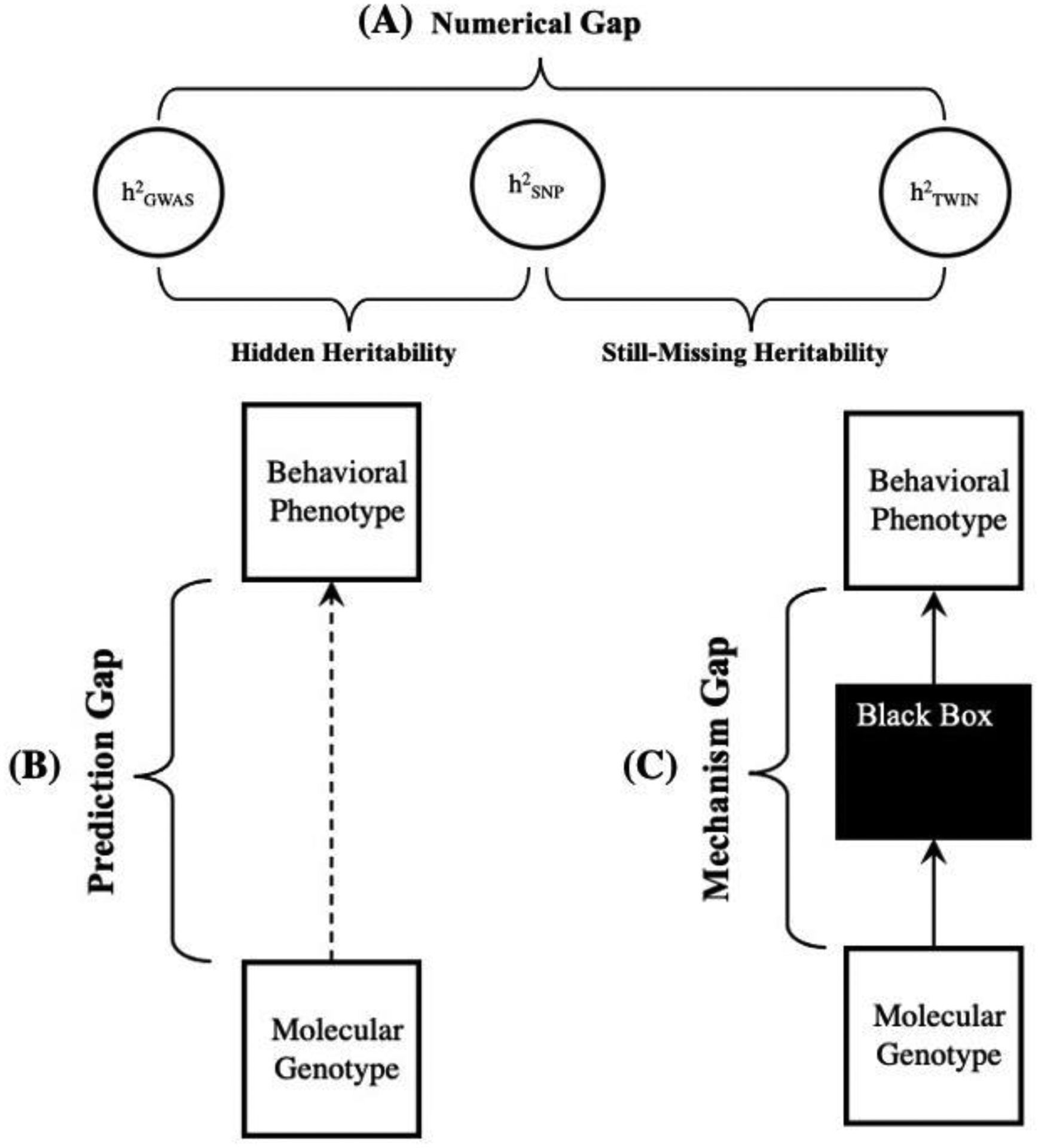
(A) The numerical gap regards the quantitative discrepancies that arise when different kinds of heritability are estimated for the same phenotype. GWAS Heritability is systematically lower than SNP heritability, which is systematically lower than traditional quantitative genetic heritability. For any given phenotype, the numerical gap would be closed if heritability derived from genomic data (h2GWAS or h2SNP) were approximately similar to heritability derived from twin and family studies (h2TWIN). (B) The prediction gap regards the practical challenge of making accurate and reliable prediction of behavioral phenotypes from molecular genotypes (i.e., DNA). The dotted line represents a predictive relationship. For any given phenotype, the prediction gap would be closed if it were possible to make useful individual-level DNA-based prediction across most populations and environments. (C) The mechanism gap regards the methodological and explanatory challenges of identifying and detailing a causal-mechanical pathway from genotype to behavioral phenotype. The solid arrows represent causal relationships and the black box represents undiscovered mechanism(s). For any given phenotype, the mechanism gap would be closed if mechanism(s) or mechanistic information were identified and detailed for the genotype-phenotype pathway of investigative interest.
As outlined in Section 3, all the explanations and solutions to the MHP are concerned with the numerical gap. Four of the five common responses to the MHP – missed variants, CNVs, epistasis, and epigenesis – for example, describe genetic effects that may account for missing heritability, were they amenable to investigation by GWAS or related methods. Relatedly, the twin heritability overestimation explanation suggests that the numerical value conferred by traditional statistical genetic methods is simply inaccurate. Thus, resolving the numerical gap would be a methodological challenge for statistical geneticists interested in finding ways to make traditional and molecular heritability values more similar. For example, if both the twin and molecular heritability of IQ were approximately .5, then the numerical gap of the MHP would be resolved.
The numerical gap is the most tractable leg of the MHP. This is the case, in part, because geneticists have already had some success closing the gap for some basic physical phenotypes, such as height and weight (Wainschtein et al., 2019). More importantly, however, the numerical gap is the most tractable leg of the MHP because its resolution requires the least demand of researchers working to resolve the problem. Resolution of the numerical gap does not require that researchers can explain or understand the causal relationships between the genotype and phenotype in question. Nor does resolution of the numerical gap require that researchers can demonstrate any practical applications of heritability, such as genetic prediction – h2 is a strictly theoretical construct. Finally, resolution of the numerical gap requires no experimental intervention or biological assays involving the genotype-phenotype relationship in question; heritability works from a black box, independent of knowledge of underlying causes. Although resolution of the numerical gap of the MHP is an important scientific challenge, we also believe that there are far greater and more fruitful challenges to investigating, understanding, and explaining the heritability of human behavior.
4.2. The Prediction Gap:
Distinct from the challenge of reconciling numerical heritability discrepancies is a problem of genetic prediction (Turkheimer, 2015). At least one useful step toward explaining the overwhelmingly complex relationships between genes and behavior is found in the capacity to predict the latter from the former. On our framing of the missing heritability problem, the prediction gap regards the challenge of making useful and reliable predictions of complex human behavior from genomic data. The prediction gap for the MHP of IQ, for example, would be resolved on the day that it were possible to accurately predict any individual’s IQ from a sample of DNA across wide range of populations and environments. As we will discuss in further detail in Section 5, polygenic scores are currently the best tools in this respect, as they permit prediction of behavioral phenotypes from genomic data.
As of today, however, the prediction gap remains daunting. Although our best methods of polygenic prediction have increased SNP-to-behavior prediction in recent years, this predictive validity remains practically useless for complex behavioral traits. As of the writing of this manuscript, the most tractable prediction gap for a complex behavioral phenotype regards educational attainment. A polygenic score for educational attainment (EA-PGS) accounts for approximately 15% of variance in EA (J. J. Lee et al., 2018; Selzam et al., 2016; Stumm et al., 2020).
Among behavioral geneticist, that predictive value is astounding – especially given the fact that the predictive value of any given SNP is so small. From a practical perspective, however, 15% variance is useless. It is practically useless, in part, because of the minimal amount of variance accounted for: on average, populations of individuals with higher EA-PGS are likely to complete more years of schooling in their lifetime than populations of individuals with lower EA-PGS. The flip side of this meager population-level statistic, however, is that any single individual’s EA-PGS does not provide useful information about that particular individual’s likelihood of doing well in educational settings. With only 15% variance explained, there will inevitably be many high EA-PGS individuals who accrue below-average years of schooling and many low EA-PGS individuals who accrue above-average years of schooling.
Aside from the group-to-individual problem of genetic prediction, polygenic scores are currently subject to a severe problem of portability (Bitarello & Mathieson, 2020; Coop, 2019; Majara et al., 2021; Manrai et al., 2016; Martin et al., 2017; Martschenko & Matthews, 2021; Matthews, 2022; Mostafavi et al., 2019; Schultz et al., 2021; Scutari et al., 2016). The problem of portability is grounded in the diminishing predictive validity of polygenic scores in populations that are different than the original GWAS population sample from which the PGS is derived. Problems of portability have been demonstrated for a variety of population characteristics including genetic ancestry, sex, age, and socioeconomic status (Mostafavi et al., 2019). Most notably, PGS derived from European-ancestry GWAS are far less predictive in individuals of African ancestry than individuals of European ancestry. This particular challenge of portability has been most profoundly exemplified in a study demonstrating that PGS for height genetically predicts individuals of African ancestry to be shorter than all Europeans and only minimally taller than individuals of Asian ancestry – a demonstrably false prediction (Martin et al., 2017).
The prediction gap is the second-most tractable leg of the MHP. Again, this is in part because resolution of any given prediction gap does not demand that scientists explain or understand a putatively causal genotype-phenotype relationship – prediction does not entail explanation, nor does explanation entail prediction. The prediction gap is a practical problem, which is demonstrated by the manner in which geneticists have worked to improve the predictive validity of PGS at the cost of statistical and explanatory scientific rigor. In efforts to improve PGS predictive validity, behavior geneticists routinely include SNPs that do not meet the criteria for statistical significance, for example. Regardless of whether any given SNP is putatively causal or statistically significant, it is included in the construction of a PGS as long as it improves the predictive validity of the PGS (Plomin & Stumm, 2021).
4.3. The Mechanism Gap:
Heritability values and polygenic prediction say nothing about the underlying biological etiology of human behavior. Like most behavior geneticists, we believe that there is at least some causal-mechanical relationship between SNPs and behavioral differences between individuals, although the specifics of those relationships and their magnitude remain unknown. The mechanism gap is the formidable scientific and philosophical challenge of identifying and elucidating a meaningful causal-mechanical, biological pathway from genotype to phenotype. To dissolve the mechanism gap of the MHP for IQ, for example, would be grounded in the capacity to describe a mechanism that helps explain how and why individuals with high-percentile IQ-PGS perform better on IQ tests than individuals with low-percentile IQ-PGS.
We defer to the growing body of philosophical literature on the so-called new philosophy of mechanisms in regards to the details of what it might look like to close any given mechanism gap (Craver & Tabery, 2016; Machamer et al., 2000). In most cases we anticipate that the mechanism gap will be closed if scientists have thoroughly elucidated at least one organized set of entities/parts that give rise to differences in behavior between individuals. Huntington’s disease (HD) provides an uncommon example of a behavioral phenotype for which the mechanism gap is mostly resolved. Individuals with HD share a specific genotype that results in mutation of the Huntingtin (Htt) protein, which causes damage to neurological cells and manifests phenotypically as hyperkinetic movement (chorea), lack of coordination, and dementia. Robust details of the underlying biological etiology aside, the important point is that there is minimal mechanism gap with respect to the genotype-phenotype HD causal pathway.
The mechanism gap is, by far, the greatest challenge of the three legs of the MHP. This is the case, in part, because historically the mechanism gap has seen the fewest resolutions in practice. Human behavioral traits for which there is a well-established genotype-phenotype mechanistic pathway are extremely rare. Rather, the norm of behavioral genetics is that all behavioral phenotypes are correlated with genetic differences, but only putative mechanistic explanations – at best – are on offer. Although many in the field believe that genetic mechanisms play a key role in IQ, for example, the mechanism gap is poorly understood (Haier, 2017; Plomin & von Stumm, 2018; Ritchie, 2015). The reasoning goes, perhaps, that specific differences in genotype produce proteins that influence synaptic connectivity or grey matter in such a way that ultimately results in differences in how well individuals score on IQ tests. Unlike HD, however, robust biological mechanisms that explain IQ variation have not been discovered. Rather, in most cases researchers note some relationship between the phenotype in question and the kind of cell in which it is expressed. Regarding SNPs associated with EA, for example, researchers note that, “relative to other genes, genes near our lead SNPs were overwhelmingly enriched for expression in the central nervous system” (Lee et al. 2018, p. 1114).
5. Discussion of Interrelations Among Heritability, Prediction, and Mechanism
Our tripartite characterization of the MHP lends itself to analyses of the interrelations between each of the three legs: the numerical gap, the prediction gap, and the mechanism gap. A key aspect of our characterization is that each of the three legs of the MHP present independent scientific, explanatory, and methodological challenges: progress in any single leg of the problem does not entail progress in any of the other legs. In this section we explore some of these interrelationships between the three legs of the MHP.
5.1. Heritability ←→ Prediction:
There are two ingredients to successful genetic prediction: substantial heritability, which is relatively easy to come by, and the availability of a close genetic replicate to predict from, which is not. The allure of twin studies can be understood in these terms, because they have both heritable traits and the extraordinary fact of identical twinning. The magic of twin studies – the satisfying specification of r = 1.0 for MZ pairs and r = .5 for DZ, the remarkable similarities of separated twins who have never met – occurs because of the conjunction of these two factors. Heritability does not necessitate predictability, however. Suppose there were no identical twins, so quantitative geneticists had to base their investigations on the similarity of half-siblings and cousins. Given a few simplifying assumptions, half-sibling studies would yield the same heritabilities as twin studies, but strong prediction would not be possible. The heritability of height may be quite high but predicting your height from that of your second cousin is never going to work very well.
This point is important for understanding the predictive limitations of SNP heritability. It is tempting to think that if a matrix of SNP-based genetic similarities produces an estimate of the heritability of IQ of .5, then it ought to be possible to predict an individual’s IQ score at r = .5, but such is not the case. The problem, as above, is that in a SNP heritability matrix there is by design no close replicate from which to predict. The prediction protocol corresponding to a SNP heritability is a polygenic score (PGS); SNP heritability provides an upper limit on the validity of a PGS. The difference between a SNP heritability and a polygenic score is the difference between all information about similarity that can be extracted from a SNP chip and that which is comprised by a linear sum. Twins extract all the available information from a genetic sequence by actually growing an organism from it, which is why twins are the most effective means of generating genetic predictions (Turkheimer, 2015).
5.2. Mechanism ←→ Prediction:
On a simplified characterization, resolution of what we call the mechanism gap requires knowledge of mechanism. Again, we defer to the burgeoning philosophical literature on mechanisms and mechanistic explanation to cash out in precise terms what it means to have knowledge of mechanism (e.g., Craver & Tabery, 2016). The important point is that for any putatively causal genotype-phenotype pathway – from SNPs statistically associated with IQ, to genes, to brain development, to actual IQ differences between individuals, for example – the presumption of most behavior geneticists today is that there is some robust set of biological mechanisms to be discovered and detailed. Definitional details of what mechanisms are and how they interact aside, simply “knowledge of mechanism” suits the purposes of our analysis.
So how does the mechanism gap relate to the prediction gap? Knowledge of mechanism and validity of prediction are inversely related, for two main reasons. The first is that prediction requires variation. Rare events are difficult to predict, and it is difficult to predict even common outcomes from rare events. Consider Down’s Syndrome, which has a large and relatively well-understood effect on IQ of about 30 points. Given such a large effect, shouldn’t we be able to predict people’s IQs by noting whether or not they are afflicted with Down’s Syndrome? The answer is no. Although Down’s syndrome has a large causal effect, its relative rarity means that it has very little variance in the general population. Knowing that a given individual does not have Down’s Syndrome tells us next to nothing about his or her IQ because the same can be said of practically everyone. To be a good predictor a mechanism would have to vary in the general population, which is exactly what doesn’t exist for complex polygenic traits. The second reason mechanism and prediction are inversely related is that statistical prediction works best in a black box, without the constraints of explanation. As we have noted, polygenic prediction is optimized when all SNPs are included in the predictor, regardless of their statistical significance, never mind their biological relevance.
5.3. Heritability ←→ Mechanism:
Closing the numerical gap would be a matter of mitigating discrepancies between alternative kinds of heritability; namely, traditional heritability versus any kind of heritability derived from molecular genetics. The development of SNP heritability was the first step toward closing this gap, as it provides a method for bringing heritability estimates informed by molecular genetics closer to those estimates derived from twin studies. Importantly, though, there is no strong or informative relationship between heritability and mechanism discovery. Increasing SNP heritability does nothing to unravel biological etiology and vice versa. One does not generally speak of the heritability of genetic conditions such as Huntington’s disease for which the mechanism is known. The reason for this is because knowledge of mechanism underlying any given genotype-phenotype pathway is the eye of the geneticist’s prize. Heritability is at best a biologically vacuous, rough approximation of genetic influence.
Moreover, heritability takes a back seat to mechanism when both are known. The concept of numerical heritability applies much more directly to common traits that vary according to the fourth Law of behavior genetics, as the binomial combination of an uncountable number of loci.6 Such traits have reliably high heritabilities, yet underlying mechanisms remain unknown. There is evidence that the most heritable cancers are common and polygenic, as opposed to rare and therefore relatively invariant cancers that are the result of a single variant or mutation (Risch, 2001). The former are heritable in the usual sense but lack mechanisms; the latter have mechanisms but lack statistical heritabilities because there is little reason in practice to estimate the heritability of a phenotype for which the mechanism has already known (Claus et al., 1996). The reason for this is because heritability becomes an afterthought in the presence of a robustly elucidated causal mechanism for the phenomenon in question.
We see here for the first time an important theme: opposition between mechanistic accounts that are well-specified causally, and statistical models that can “account for” variability while remaining black boxes. This inverse relationship is no accident. The less attention one is compelled to pay to the causal meaning of genetic variance, the more of it can be put in the numerator of a heritability ratio. Identical twins comprise all genetic similarity and all the ways genes can combine. Twins could thus be used to estimate large heritabilities long before DNA was discovered. Genetic variation based on candidate genes obviously requires knowledge of the specific gene or SNP in question, and as a consequence the “heritabilities” based on candidate genes are much smaller. Combining individual SNPs into a polygenic score sacrifices knowledge of the action of the individual SNPs for the added variance one can “explain” by combining them. Finally, SNP heritability abandons any interest in mechanisms, using the SNPs only to estimate pairwise genetic similarities in the same causally agnostic sense as twin studies, and therefore produces larger heritabilities. In many ways, this places the field back where it started.
Thus, an important aspect of our tripartite take on the MHP is that each of the three legs represent distinct and independent practical, scientific, and explanatory challenges. Reconciling conflicting heritability estimates (i.e., closing the numerical gap) does not entail genetic prediction. The MHP of height, for example, presents an elegant example of this. In large part, the numerical gap for the MHP of height has been closed: traditional and molecular estimates of heritability of height are approximately similar. Despite that impressive scientific achievement, genetic prediction of height has yet to demonstrate clinical utility in comparison to alternative predictors, such as mid-parental height (Lu et al., 2021).
Similarly, closing the prediction gap for any given phenotype does not entail any resolution of the mechanism gap for the same trait. As the predictive accuracy of PGS improve with time, there may come a day when it is possible to accurately and reliably predict an individual’s height from a sample of their DNA. As described previously, however, PGS are not explanatory – quite the opposite actually. One may be capable of explaining a genotype-phenotype pathway by describing a mechanism without being able to make accurate predictions about that pathway, and vice versa.
6. Concluding remarks: on the particularly formidable case of human behavior
Although it may be tempting to characterize the missing heritability problem as no more than a numerical issue of conflicting estimates of heritability (h2GWAS < h2SNP < h2Twin), here we encourage a more sobering perspective of the challenges facing scientific efforts to account for the “heritability” of human behavior. We are motivated by a tendency among geneticists and philosophers to maintain that the longstanding mysteries regarding the genetic bases of complex human behaviors, such as IQ, will soon be resolved. Importantly, we are not optimistic that, perhaps with the advent of more sophisticated statistical models and bigger data, the capacity to match traditional and GWAS or SNP-based estimates of heritability will drastically advance scientific understanding of the genetics of human behavior. It won’t.
Rather, the gap between alternative methods of heritability estimates – what we call the numerical gap – is an issue independent of a greater scientific challenge: making accurate individual-level genetic prediction of behavioral phenotypes. Polygenic scores have made large gains in this respect, but, realistically, they’re not even close to resolving what we refer to as the prediction gap of the missing heritability problem. Finally, an old lesson from the philosophy of science: prediction does not entail explanation. We take it to be a crucial goal of scientific practice to develop meaningful explanations of why genetic differences between individuals correlate to behavioral differences between individuals. Although there is ample optimism in the genomics community, GWAS and PGS have also failed to deliver on this front. As of the writing of this manuscript, over 1,200 SNPs are statistically associated with the number of years of schooling a person completes in their lifetime (i.e., educational attainment), yet the putatively causal relationship between those SNPs and differences in educational outcomes is entirely opaque, other than the very general assertion that many of the SNPs are close to genes that are expressed in neural tissue. Until scientists have identified, described, and substantiated causal-mechanical etiologies that would explain why countless SNPs are correlated with behavioral outcomes like IQ and educational attainment, then what we call the mechanism gap of the missing heritability problem remains a daunting and persistent scientific challenge.
Thus, on our view the MHP is far from dissolved, nor will it be any time soon. The many challenges related to the heritability of human behavior are quite alive and well. Moreover, it is not that there is just one MHP that may someday be solved in one fell swoop of elegant science. Rather, there is a different MHP for each phenotype of interest to genetic researchers. For example, the numerical gap for height is drastically smaller than the numerical gap for educational attainment. Because each of the three legs of any given MHP are independent, so are problems of missing heritability independent from one another: resolving the MHP of height does not entail resolution of the MHP of educational attainment, or any other phenotype for that matter. There’s a different missing heritability problem for literally every single phenotype of scientific interest.
Moreover, just as each of the three legs of the MHP vary in difficulty and tractability – the numerical gap being the most tractable and the mechanism gap being the least so – some cases of missing heritability are more problematic than others. As a general rule of thumb: the more complex the phenotype, there more difficult the MHP. Highly complex human behavioral traits and outcomes such as intelligence and educational attainment are farthest from dissolution: the numerical gaps, predictions gaps, and mechanism gaps for these cases may never be resolved. At least one rung down the ladder of MHP difficulty would be basic, human physical phenotypes, such as height, weight, and cardiovascular disease. Although alarmingly complex in their own ways, basic physical phenotypes are much more investigatively tractable. As previously mentioned, the numerical gap for the MHP of height is all but closed – and we would not be surprised if one day it were possible to accurately and reliably predict an individual’s height from their DNA.
The elucidation of meaningful biological etiologies that underly phenotypes of scientific interest – i.e., closing mechanism gaps – however, will inevitably remain a persistent challenge for human genetics. The reason why this is the case speaks to the severity of the MHP beyond human phenotypes to the genetics of non-human animals more broadly: the ethics of experimentation precludes the elucidation of mechanistic pathways for most human phenotypes. The MHP of non-human animals is far less severe and far more tractable than it is for any human phenotype precisely because a wide array of experimental interventions and assays are on the table. Although scientists may poke and prod nematodes to their heart’s desire – conducting gene knockouts to elucidate genotype-phenotype mechanistic pathways, for example – there will never be a day when it is ethically permissible to conduct the kinds of experiments in human that would be required to close any given mechanism gap.7 The missing heritability problem isn’t going anywhere any time soon.
Credit Statement:
This research was conducted at the University of Virginia Center for Genetics & Human Agency, the NIH-funded, Columbia University Center for Research on Ethical, Legal & Social Implications of Psychiatric, Neurologic & Behavioral Genetics (5RM1HG007257-07), and The Hastings Center.
Footnotes
In an earlier iteration of this this idea, we briefly referred to this numerical gap, instead, as a “statistical gap” (Matthews & Turkheimer 2019).
Sometimes referred to as ‘twin study heritability’ (h2TS), ‘family heritability (h2family)’, or just ‘traditional heritability.’
Witte et al. (2014) and Tropf et al. (2017) offer a similar characterization and distinguish ‘still-missing heritability’ (as the gap between h2SNP and h2Twin) from ‘hidden heritability’ (as the gap between h2GWAS and h2SNP).
Elsewhere referred to the ‘Common vs. Rare Allele Hypothesis for Complex Diseases’ (El-Fishawy, 2013) or the ‘rare variants hypothesis’ (Yang et al. 2010).
It’s worth noting that applications of GWAS to schizophrenia and autism have been more successful since Maher’s statement in 2008. While in 2009 GWAS accounted for 3% variance, polygenic scores today predict 6% of the liability variance for schizophrenia (Pardiñas et al., 2018).
Recall that the fourth law of behavior genetics is that a typical human behavioral trait is associated with very many genetic variants, each of which accounts for a very small percentage of the behavioral variability (Chabris et al., 2015).
See (Matthews, 2017) for a more detailed account of the role of mechanistic reasoning and experimental intervention in scientific efforts to elucidate genotype-phenotype pathways in non-human animals.
Contributor Information
Lucas J. Matthews, Columbia University, The Hastings Center
Eric Turkheimer, University of Virginia.
References
- Bitarello BD, & Mathieson I (2020). Polygenic Scores for Height in Admixed Populations. G3: Genes, Genomes, Genetics, 10(11), 4027–4036. 10.1534/g3.120.401658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JS, Ehrenreich IM, Loo WT, Lite T-LV, & Kruglyak L (2013). Finding the sources of missing heritability in a yeast cross. Nature, 494(7436), 234–237. 10.1038/nature11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkenau P, Riemann R, Angleitner A, & Spinath FM (2002). Similarity of childhood experiences and personality resemblance in monozygotic and dizygotic twins: A test of the equal environments assumption. Personality and Individual Differences, 33(2), 261–269. 10.1016/S0191-8869(01)00150-7 [DOI] [Google Scholar]
- Bourrat P, & Lu Q (2017). Dissolving the Missing Heritability Problem. Philosophy of Science, 84(5), 1055–1067. 10.1086/694007 [DOI] [Google Scholar]
- Chabris C, Lee J, Cesarini D, Benjamin D, & Laibson D (2015). The Fourth Law of Behavior Genetics. Current Directions in Psychological Science, 24(4), 304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AJ, & Cooper DN (2010). GWAS: Heritability missing in action? European Journal of Human Genetics, 18(8), 859–861. 10.1038/ejhg.2010.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus EB, Schildkraut JM, Thompson WD, & Risch NJ (1996). The genetic attributable risk of breast and ovarian cancer. Cancer, 77(11), 2318–2324. [DOI] [PubMed] [Google Scholar]
- Coop G (2019). Reading tea leaves? Polygenic scores and differences in traits among groups. ArXiv:1909.00892 [q-Bio]. http://arxiv.org/abs/1909.00892
- Craver C, & Tabery JG (2016). Mechanisms in Science. In Zalta EN (Ed.), The Stanford Encyclopedia of Philosophy (Fall 2016). [Google Scholar]
- DiFrisco J, & Jaeger J (2020). Genetic Causation in Complex Regulatory Systems: An Integrative Dynamic Perspective. BioEssays, 42(6), 1900226. 10.1002/bies.201900226 [DOI] [PubMed] [Google Scholar]
- Downes SM, & Matthews LJ (2020). Heritability. The Stanford Encyclopedia of Philosophy. <https://plato.stanford.edu/archives/spr2020/entries/heredity/> [Google Scholar]
- Downes SM, & Turkheimer E (in press). An Early History of the Heritability Coefficient Applied to Humans (1918–1960). Biological Theory. [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, & Nadeau JH (2010). Missing heritability and strategies for finding the underlying causes of complex disease. Nature Reviews Genetics, 11(6), 446–450. 10.1038/nrg2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Fishawy P (2013). Common Disease-Rare Variant Hypothesis. In Encyclopedia of Autism Spectrum Disorders (pp. 720–722). Springer, New York, NY. 10.1007/978-1-4419-1698-3_1997 [DOI] [Google Scholar]
- Fisher RA (1918). The Correlation Between Relatives on the Supposition of Mendelian Inheritance. Transactions of the Royal Society of Edinburgh, 52, 399–433. [Google Scholar]
- Furrow RE, Christiansen FB, & Feldman MW (2011). Environment-Sensitive Epigenetics and the Heritability of Complex Diseases. Genetics, 189(4), 1377–1387. 10.1534/genetics.111.131912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Génin E (2019). Missing heritability of complex diseases: Case solved? Human Genetics. 10.1007/s00439-019-02034-4 [DOI] [PubMed] [Google Scholar]
- Haier RJ (2017). The Neuroscience of Intelligence. Cambridge University Press. [Google Scholar]
- Horwitz AV, Videon TM, Schmitz MF, & Davis D (2003). Rethinking Twins and Environments: Possible Social Sources for Assumed Genetic Influences in Twin Research. Journal of Health and Social Behavior, 44(2), 111–129. 10.2307/1519802 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Kessler RC, Heath AC, & Eaves LJ (1993). A test of the equal-environment assumption in twin studies of psychiatric illness. Behavior Genetics, 23(1), 21–27. 10.1007/BF01067551 [DOI] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, Nguyen-Viet TA, Bowers P, Sidorenko J, Linnér RK, Fontana MA, Kundu T, Lee C, Li H, Li R, Royer R, Timshel PN, Walters RK, Willoughby EA, … Cesarini D (2018). Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nature Genetics, 50(8), 1112. 10.1038/s41588-018-0147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR, Goddard ME, & Visscher PM (2011). Estimating Missing Heritability for Disease from Genome-wide Association Studies. The American Journal of Human Genetics, 88(3), 294–305. 10.1016/j.ajhg.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li Y, & Tollefsbol TO (2008). Gene-Environment Interactions and Epigenetic Basis of Human Diseases. Current Issues in Molecular Biology, 10(1–2), 25–36. [PMC free article] [PubMed] [Google Scholar]
- Lu T, Forgetta V, Wu H, Perry JRB, Ong KK, Greenwood CMT, Timpson NJ, Manousaki D, & Richards JB (2021). A Polygenic Risk Score to Predict Future Adult Short Stature Among Children. The Journal of Clinical Endocrinology & Metabolism, 106(7), 1918–1928. 10.1210/clinem/dgab215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lush JL (1935). Progeny Test and Individual Performance as Indicators of an Animal’s Breeding Value. Journal of Dairy Science, 18(1), 1–19. 10.3168/jds.S0022-0302(35)93109-5 [DOI] [Google Scholar]
- Lynch KE (2017). Heritability and causal reasoning. Biology & Philosophy, 32(1), 25–49. 10.1007/s10539-016-9535-1 [DOI] [Google Scholar]
- Lynch KE, & Bourrat P (2016). Interpreting Heritability Causally. Philosophy of Science. 10.1086/688933 [DOI] [Google Scholar]
- Lynch KE, & Kemp DJ (2014). Nature-via-nurture and unravelling causality in evolutionary genetics. Trends in Ecology & Evolution, 29(1), 2–4. 10.1016/j.tree.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Machamer P, Darden L, & Craver CF (2000). Thinking about Mechanisms. Philosophy of Science, 67(1), 1–25. [Google Scholar]
- Maher B (2008). The case of the missing heritability. Nature, 456, 18–21. [DOI] [PubMed] [Google Scholar]
- Majara L, Kalungi A, Koen N, Zar H, Stein DJ, Kinyanda E, Atkinson EG, & Martin AR (2021). Low generalizability of polygenic scores in African populations due to genetic and environmental diversity. BioRxiv, 2021.01.12.426453. 10.1101/2021.01.12.426453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, … Visscher PM (2009). Finding the missing heritability of complex diseases. Nature, 461(7265), 747–753. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, & Kohane IS (2016). Genetic Misdiagnoses and the Potential for Health Disparities. New England Journal of Medicine, 375(7), 655–665. 10.1056/NEJMsa1507092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D, Gujral M, Brandler WM, Malhotra D, Wang Z, Fajarado KVF, Maile MS, Ripke S, Agartz I, Albus M, … Sebat J (2017). Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nature Genetics, 49(1), 27–35. 10.1038/ng.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, & Kenny EE (2017). Human demographic history impacts genetic risk prediction across diverse populations. The American Journal of Human Genetics, 100(4), 635–649. 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martschenko D, & Matthews LJ (2021). Genomics, Behavior, and Social Outcomes. Hastings Center Bioethics Briefings. https://www.thehastingscenter.org/publications-resources/hastings-center-bioethics-briefings/ [Google Scholar]
- Matthews LJ (2017). On mechanistic reasoning in unexpected places: The case of population genetics. Biology & Philosophy, 32(6), 999–1018. 10.1007/s10539-017-9588-9 [DOI] [Google Scholar]
- Matthews LJ (2022). Half a century later and we’re back where we started: How the problem of locality turned in to the problem of portability. Studies in History and Philosophy of Science Part A, 91, 1–9. 10.1016/j.shpsa.2021.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews LJ, & Turkheimer E (2019). Across the great divide: Pluralism and the hunt for missing heritability. Synthese. 10.1007/s11229-019-02205-w [DOI] [Google Scholar]
- McCarthy MI, & Hirschhorn JN (2008). Genome-wide association studies: Potential next steps on a genetic journey. Human Molecular Genetics, 17(R2), R156–R165. 10.1093/hmg/ddn289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi H, Harpak A, Conley D, Pritchard JK, & Przeworski M (2019). Variable prediction accuracy of polygenic scores within an ancestry group. BioRxiv, 629949. 10.1101/629949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman HH, Freeman FN, & Holzinger KJ (1937). Twins: A study of heredity and environment (pp. xvi, 369). Univ. Chicago Press. [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, Han J, Hubbard L, Lynham A, Mantripragada K, Rees E, MacCabe JH, McCarroll SA, Baune BT, Breen G, … Walters JTR (2018). Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nature Genetics, 50(3), 381–389. 10.1038/s41588-018-0059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A (2010). Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature, 465(7299), 721–728. [DOI] [PubMed] [Google Scholar]
- Plomin R, & von Stumm S. (2021). Polygenic scores: Prediction versus explanation. PsyArXiv. 10.31234/osf.io/xbfmr [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, & von Stumm S (2018). The new genetics of intelligence. Nature Reviews Genetics, 19(3), 148–159. 10.1038/nrg.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N (2001). The Genetic Epidemiology of Cancer: Interpreting Family and Twin Studies and Their Implications for Molecular Genetic Approaches. Cancer Epidemiology and Prevention Biomarkers, 10(7), 733–741. [PubMed] [Google Scholar]
- Ritchie S (2015). Intelligence: All that matters. John Murray Learning. [Google Scholar]
- Sackton TB, & Hartl DL (2016). Genotypic Context and Epistasis in Individuals and Populations. Cell, 166(2), 279–287. 10.1016/j.cell.2016.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr S, & Carter-Saltzman L (1979). Twin method: Defense of a critical assumption. Behavior Genetics, 9(6), 527–542. 10.1007/BF01067349 [DOI] [PubMed] [Google Scholar]
- Schultz LM, Merikangas AK, Ruparel K, Jacquemont S, Glahn DC, Gur RE, Barzilay R, & Almasy L (2021). Stability of Polygenic Scores Across Discovery Genome-Wide Association Studies. BioRxiv, 2021.06.18.449060. 10.1101/2021.06.18.449060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutari M, Mackay I, & Balding D (2016). Using Genetic Distance to Infer the Accuracy of Genomic Prediction. PLOS Genetics, 12(9), e1006288. 10.1371/journal.pgen.1006288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzam S, Krapohl E, von Stumm S, O’Reilly PF, Rimfeld K, Kovas Y, Dale PS, Lee JJ, & Plomin R (2016). Predicting educational achievement from DNA. Molecular Psychiatry. 10.1038/mp.2016.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M (2009). Epigenetic Inheritance and the Missing Heritability Problem. Genetics, 182(3), 845–850. 10.1534/genetics.109.102798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YS, Wang F, & Slatkin M (2010). General Epistatic Models of the Risk of Complex Diseases. Genetics, 186(4), 1467–1473. 10.1534/genetics.110.119008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stumm S, Smith‐Woolley E, Ayorech Z, McMillan A, Rimfeld K, Dale PS, & Plomin R (2020). Predicting educational achievement from genomic measures and socioeconomic status. Developmental Science, 23(3), e12925. 10.1111/desc.12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropf FC, Lee SH, Verweij RM, Stulp G, van der Most PJ, de Vlaming R, Bakshi A, Briley DA, Rahal C, Hellpap R, Nyman A, Esko T, Metspalu A, Medland SE, Martin NG, Barban N, Snieder H, Robinson MR, & Mills MC (2017). Hidden heritability due to heterogeneity across seven populations. Nature Human Behaviour, 1(10), 757–765. 10.1038/s41562-017-0195-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkheimer E (2000). Three laws of behavior genetics and what they mean. Current Directions in Psychological Science, 9, 160–164. [Google Scholar]
- Turkheimer E (2015). Genetic Prediction. Hastings Center Report, 45(5), S32–S38. [DOI] [PubMed] [Google Scholar]
- Vittori A, Breda C, Repici M, Orth M, Roos RAC, Outeiro TF, Giorgini F, Hollox EJ, & REGISTRY investigators of the European Huntington’s Disease Network. (2014). Copy-number variation of the neuronal glucose transporter gene SLC2A3 and age of onset in Huntington’s disease. Human Molecular Genetics, 23(12), 3129–3137. 10.1093/hmg/ddu022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainschtein P, Jain DP, Yengo L, Zheng Z, TOPMed Anthropometry Working Group, T.-O. for P. M. C., Cupples LA, Shadyab AH, McKnight B, Shoemaker BM, Mitchell BD, Psaty BM, Kooperberg C, Roden D, Darbar D, Arnett DK, Regan EA, Boerwinkle E, Rotter JI, Allison MA, … Visscher PM (2019). Recovery of trait heritability from whole genome sequence data. BioRxiv, 588020. 10.1101/588020 [DOI] [Google Scholar]
- Weber M (2005). Genes, Causation and Intentionality. History and Philosophy of the Life Sciences, 27(3/4), 407–420. [PubMed] [Google Scholar]
- Weedon MN, Jackson L, Harrison JW, Ruth KS, Tyrrell J, Hattersley AT, & Wright CF (2021). Use of SNP chips to detect rare pathogenic variants: Retrospective, population based diagnostic evaluation. BMJ (Clinical Research Ed.), 372, n214. 10.1136/bmj.n214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W-H, Hemani G, & Haley CS (2014). Detecting epistasis in human complex traits. Nature Reviews. Genetics, 15(11), 722–733. 10.1038/nrg3747 [DOI] [PubMed] [Google Scholar]
- Witte JS, Visscher PM, & Wray NR (2014). The contribution of genetic variants to disease depends on the ruler. Nature Reviews. Genetics, 15(11), 765–776. 10.1038/nrg3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AAE, Lee SH, Robinson MR, Perry JRB, Nolte IM, van Vliet-Ostaptchouk JV, Snieder H, The LifeLines Cohort Study, Esko T, Milani L, Mägi R, Metspalu A, Hamsten A, Magnusson PKE, Pedersen NL, … Visscher PM (2015). Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nature Genetics, 47(10), 1114–1120. 10.1038/ng.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, & Visscher PM (2010). Common SNPs explain a large proportion of the heritability for human height. Nature Genetics, 42(7), 565–569. 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, & Visscher PM (2011). GCTA: A Tool for Genome-wide Complex Trait Analysis. The American Journal of Human Genetics, 88(1), 76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, & Lander ES (2012). The mystery of missing heritability: Genetic interactions create phantom heritability. Proceedings of the National Academy of Sciences, 109(4), 1193–1198. 10.1073/pnas.1119675109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, & Lander ES (2014). Searching for missing heritability: Designing rare variant association studies. Proceedings of the National Academy of Sciences, 111(4), E455–E464. 10.1073/pnas.1322563111 [DOI] [PMC free article] [PubMed] [Google Scholar]


