Abstract
Background
Evidence has suggested that cytokine storms may be associated with T cell exhaustion (TEX) in COVID-19. However, the interaction mechanism between cytokine storms and TEX remains unclear.
Methods
With the aim of dissecting the molecular relationship of cytokine storms and TEX through single-cell RNA sequencing data analysis, we identified 14 cell types from bronchoalveolar lavage fluid of COVID-19 patients and healthy people. We observed a novel subset of severely exhausted CD8 T cells (Exh T_CD8) that co-expressed multiple inhibitory receptors, and two macrophage subclasses that were the main source of cytokine storms in bronchoalveolar.
Results
Correlation analysis between cytokine storm level and TEX level suggested that cytokine storms likely promoted TEX in severe COVID-19. Cell–cell communication analysis indicated that cytokines (e.g. CXCL10, CXCL11, CXCL2, CCL2, and CCL3) released by macrophages acted as ligands and significantly interacted with inhibitory receptors (e.g. CXCR3, DPP4, CCR1, CCR2, and CCR5) expressed by Exh T_CD8. These interactions formed the cytokine–receptor axes, which were also verified to be significantly correlated with cytokine storms and TEX in lung squamous cell carcinoma.
Conclusions
Cytokine storms may promote TEX through cytokine-receptor axes and be associated with poor prognosis in COVID-19. Blocking cytokine-receptor axes may reverse TEX. Our finding provides novel insights into TEX in COVID-19 and new clues for cytokine-targeted immunotherapy development.
Keywords: COVID-19, immune exhaustion, cytokine storm, single-cell sequencing data analysis, T cell, immune checkpoint
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has caused >500 million infections and 6.2 million deaths by 27th April 2022 (WHO data at https://www.who.int/emergencies/diseases/novel-coronavirus-2019). The over-production of pro-inflammatory cytokines known as a cytokine storm resulted in immune system disorders including constitutional symptoms, systemic inflammation, and multiorgan dysfunction that increase the risk of death.1,2 It is recognized that cytokine storms may be associated with CD8 T cell exhaustion (TEX) in severe COVID-19.3–5 Maraviroc, a C-C chemokine receptor 5 (CCR5) antagonist, which may reverse lymphoid exhaustion and alter the cell trafficking of inflammatory cells, has been tested in clinical trials for severe COVID-19 (ClinicalTrials.gov Identifier: NCT04435522, NCT04441385, and NCT04475991). CCR5 inhibition could also reduce cytokine storms, increase CD8 T cells, and decrease SARS-CoV-2 RNA abundance in severe COVID-19.6 However, the interplay mechanism between cytokine storms and TEX remains unclear. For example, how do cytokine storms affect TEX and further impact clinical outcomes? Answers to these questions are urgently needed for the development of new drugs and therapies that act by decreasing cytokine storms and reactivating exhausted T cells.
TEX also occurs in many chronic viral infections, including human immunodeficiency virus (HIV),7,8 hepatitis C virus (HCV),9 and hepatitis B virus (HBV).10 Similarly, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can potentially destroy the function of CD4 T cells and promote the excessive activation and possible subsequent exhaustion of CD8 T cells.11 Patients with severe COVID-19 might have cytokine storm syndrome or hyperinflammatory syndrome characterized by fulminant and fatal hypercytokinaemia with multiorgan failure.1 Recent reports also indicated that the increasing pro-inflammatory cytokine or chemokine responses damaged the homeostasis of the immune system, resulting in cytokine storm syndromes in patients with severe SARS-CoV-2 infection.1 Systemically elevated cytokines are cardiotoxic and possibly lead to myocardial injury. A minority of patients have the cardiovascular manifestations of COVID-19 disease, such as viral myocarditis, associated with a high risk of mortality.12
In this study, we aimed to dissect the molecular relationship between cytokine storms and TEX, and to determine the prognostic significance of cytokine storms. We first profiled and characterized immune cell types from bronchoalveolar lavage fluid (BALF) of COVID-19 patients and healthy people through single-cell RNA sequencing data analysis. We then focused the analysis on a subset of severely exhausted CD8 T cells that co-expressed several inhibitory receptors and significantly interacted with macrophages through the cytokines released by macrophages. Further, we found that cytokine–receptor interaction axes (e.g. CCL2-CCR2, CCL3-CCR1, CCL3-CCR5, CCL4-CCR5, CCL4L2-VSIR, and CCL3L1-CCR1) observed in severe COVID-19 were significantly correlated with TEX. Our study tried to provide new insights into the interplay mechanism between cytokine storms and TEX, and to suggest potential cytokine-targeted drug development for COVID-19.
Materials and methods
Source data retrieval
The raw count matrices of single-cell sequencing data for BALF (GSE145926)13 and peripheral blood mononuclear cells (PBMC) (GSE150728)14 were downloaded from NCBI Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). A total of 12 BALF samples were collected from 6 severe COVID-19 patients, 3 moderate COVID-19 patient samples, and 3 healthy controls. The 13 PBMC samples included 3 severe COVID-19 patient samples, 4 moderate COVID-19 patient samples, and 6 healthy controls. The clinical information of the patient samples and the definition of disease severity are summarized in Table 1.13,14 For the PBMC and BALF samples, most patients belonging to the severe group were males or >60 years old. Detailed information of the source data is listed in Supplementary Table 1, see online supplementary material. The tools used in this study are summarized in Supplementary Table 2, see online supplemetary material.
Table 1.
Clinical information of the patient samples used in this study.
| Donor | Gender | Age (years) | Severity | Disease | Tissue | GEO Sample ID (GSM) |
|---|---|---|---|---|---|---|
| M1 | Male | 36 | Moderate | COVID-19 | BALF | GSM4339769 |
| M2 | Female | 37 | Moderate | COVID-19 | BALF | GSM4339770 |
| M3 | Male | 35 | Moderate | COVID-19 | BALF | GSM4339772 |
| S1 | Male | 62 | Severe | COVID-19 | BALF | GSM4339773 |
| S2 | Male | 66 | Severe | COVID-19 | BALF | GSM4339771 |
| S3 | Male | 38 | Severe | COVID-19 | BALF | GSM4339774 |
| S4 | Female | 65 | Severe | COVID-19 | BALF | GSM4475051 |
| S5 | Female | 36 | Severe | COVID-19 | BALF | GSM4475052 |
| S6 | Male | 46 | Severe | COVID-19 | BALF | GSM4475053 |
| HC1 | Female | 38 | Healthy | COVID-19 | BALF | GSM4475048 |
| HC2 | Male | 24 | Healthy | COVID-19 | BALF | GSM4475049 |
| HC3 | Male | 22 | Healthy | COVID-19 | BALF | GSM4475050 |
| C1A | Male | 60–69 | Moderate | COVID-19 | PBMC | GSM4557327 |
| C1B | Male | 60–69 | Severe | COVID-19 | PBMC | GSM4557328 |
| C2 | Male | 40–49 | Moderate | COVID-19 | PBMC | GSM4557329 |
| C3 | Male | 60–69 | Severe | COVID-19 | PBMC | GSM4557330 |
| C4 | Male | 30–39 | Severe | COVID-19 | PBMC | GSM4557331 |
| C5 | Male | 50–59 | Moderate | COVID-19 | PBMC | GSM4557332 |
| C7 | Male | 20–29 | Moderate | COVID-19 | PBMC | GSM4557333 |
| H1 | Female | 40–49 | Healthy | COVID-19 | PBMC | GSM4557334 |
| H2 | Male | 40–49 | Healthy | COVID-19 | PBMC | GSM4557335 |
| H3 | Female | 30–39 | Healthy | COVID-19 | PBMC | GSM4557336 |
| H4 | Male | 40–49 | Healthy | COVID-19 | PBMC | GSM4557337 |
| H5 | Male | 40–49 | Healthy | COVID-19 | PBMC | GSM4557338 |
| H6 | Male | 30–39 | Healthy | COVID-19 | PBMC | GSM4557339 |
Quality control on single-cell RNA-seq data
We performed quality control for the raw count data of each single-cell RNA-seq sample using Seurat (v4). To ensure the consistency of the analysis, we applied the same criteria to all the samples of healthy, moderate, and severe groups. During acute infection, T cell metabolic activity is increased, possibly resulting in mitochondrial gene expression.15 Therefore, the mitochondrial gene percentage, a key parameter for filtering cells, was set at 20%. In addition, the cells that expressed <200 genes or >6000 genes were also discarded. For gene filtering, genes that were expressed in <10 cells were removed from the final count matrix. After the quality control, 84 114 cells from BALF and 48 507 PBMC were retained for further analysis.
Clustering, annotation, and identification of cell types
Due to the heterogeneity of BALF and PBMCs, we adopted two different strategies of clustering and analysis. For BALF, each dataset was normalized and identified with the top 2000 variable features by the “vst” method in Seurat (v4). Then, to correct batch effects among the samples, all datasets were integrated into a filtered gene-barcode matrix using the “FindIntegrationAnchors” and “IntegrateData” functions in Seurat. Dimensionality reduction was performed by principal component analysis (PCA). Then we used the JackStraw procedure16 wrapped in Seurat and the Elbow plot to evaluate how many principal components (PCs) should be chosen to perform further clustering analysis. According to the evaluation result of the PCs, we tried a series of PC numbers (40–60) (Supplementary Fig. 2A, see online supplementary material) to cluster cells and subsequently identified the best PC number based on the expression of the well-known cell type (T cell) markers by using the “VlnPlot” function. To our prior knowledge, T cells have similar expression patterns and should be clustered together during cell clustering. When the PCs number was set as 50, the T cell cluster was clearly observed in our cell clusters (Fig. 1C). Therefore, the top 50 PCs were used to conduct uniform manifold approximation and projection (UMAP) for the visualization of single-cell groups in 2D space. Meanwhile, the top 50 PCs were also used to construct a shared nearest-neighbour graph (SNN; FindNeighbors), followed by graphical clustering (FindClusters) with the graph-based modularity-optimization algorithm Louvain for community detection. The resolution was a flexible parameter varying with the total number of cells during cell clustering. Larger resolution makes more clusters. To cluster 84 114 cells of BALF, we tested different resolution values from 0.8 to 1.5 and then set 1.2 as the best resolution. Furthermore, specific markers in each cluster were identified by the “FindAllMarkers” function and each cluster was assigned to a known cell type using the combination of SingleR automatic annotation and manual annotation according to canonical genes from CellMarker.17 Then we again applied the “FindAllMarkers” function to obtain highly expressed genes of each cell type. Finally, to understand the molecular biological characteristics of exhausted CD8 T cells, we applied Metascape (https://metascape.org/) to perform a pathway enrichment analysis on all highly expressed genes with the average log2 (fold-change) of >0.25 and adjusted P value < 0.05.18
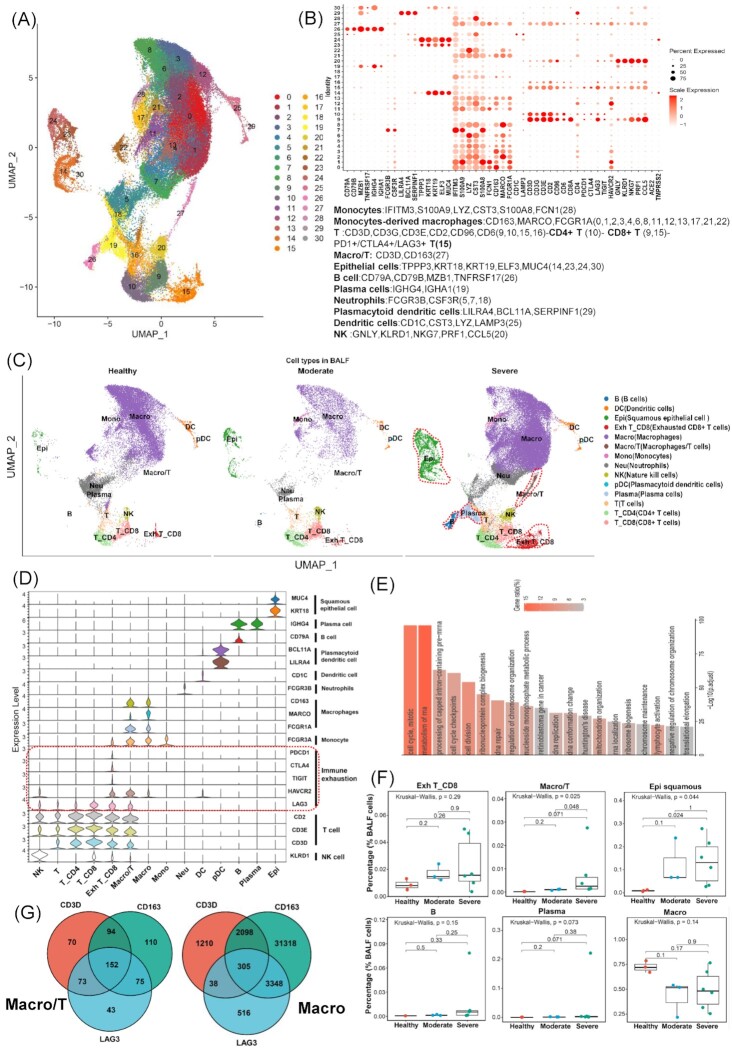
Figure 1.
Identification of cell types in BALF and cell–cell interaction of exhausted CD8 T cells. (A) Overview of the cell clusters in BALFs (n = 12) derived from across healthy people (n = 3), moderate (n = 3), and severe (n = 6) COVID-19 patients. (B) Dot plots show the expression level of canonical markers representing cell types and genes related to SARS-CoV-2 entry into cells. Cell types and their corresponding genes are listed at the bottom. Each dot is colored according to the scale expression and sized by the percentage of cells in a cluster (row) which expresses a given gene (column). (C) UMAP plots of BALF colored by cell types across three disease states including healthy (left panel), moderate (middle panel), and severe (right panel). (D) Violin plots show the expression of canonical maker genes (rows) which represent the distinct cell types (columns) and multiple inhibitory receptors indicating TEX. (E) Top 20 pathway/process enrichment analysis of all highly expressed genes in exhausted CD8 T cells; adjusted P value < 0.0001 (hypergeometric test, adjusted P-values obtained by the Benjamini–Hochberg procedure). (F) The cell percentages of Exh T_CD8, Macro/T, Epi squamous, B, Plasma, and Macro cells among healthy controls (n = 3), moderate patients (n = 3), and severe patients (n = 6). The Kruskal–Wallis H test was used for comparing the three disease states. (G) Venn diagrams display the numbers of cells concurrently expressing CD3D, CD163, and LAG3 in Macro/T (left panel) and Macro (right), respectively. B, B cells. DC, dendritic cells. Epi, squamous epithelial cells. Exh T_CD8, exhausted CD8 T cells. Macro, macrophages. Macro/T, a mixture of macrophages and T cells. Mono, monocytes. Neu, neutrophils. NK, nature kill cells. pDC, plasmacytoid dendritic cells. Plasma, plasma cells. T, T cells. T_CD4, CD4 T cells. T_CD8, CD8 T cells.
For the PBMC data analysis, all filtered datasets were first merged into a gene-barcode matrix with the “merge” function in Seurat. The gene-barcode matrix was first normalized by using the ‘LogNormalize’ method in Seurat (v4) with default parameters. The top 2000 variable genes were then identified using the ‘vst’ method in the Seurat “FindVariableFeatures” function. Using the same method in the BALF data analysis, we tested a series of PC numbers (15–20) (Supplementary Fig. 3A, see online supplementary material) and finally determined 18 as the best PC number for cell clustering. So, dimension reduction was performed based on the top 2000 variable genes with PCA and then UMAP was performed on the top 18 PCs. We also tested the various values of resolution ranging from 0.5 to 1.0, and found 0.7 made the finest clustering for the PBMC data. Finally, we assigned each cluster to a known cell type in the same manner for BALF.
Additionally, we also reviewed and tried other deep-learning-based methods19–24 to cluster the single cell RNAseq data to validate our results of cell clustering. To identify the cell types enriched in BALF and PBMC of the severe COVID-19 patient group, we applied the Kruskal–Wallis test to compare the percentages of each cell type among severe, moderate, and healthy groups.
Cell–cell interaction analysis
To establish a cell–cell interaction network via ligand–receptor interactions, we applied CellPhoneDB25 to the single cell raw count matrices of BALF and PBMC respectively in severe COVID-19 patients. The ligand–receptor pairs that were expressed in <10% of cells were discarded. To identify the significant cell–cell interactions, we performed permutation tests between two cell types mediated by a specific ligand–receptor pair. The permutation tests were based on the mean gene expression of the ligand from one cell type and the corresponding receptor from another cell type. According to the protocol of CellPhoneDB, the smaller the P-value, the more reliable the resulting ligand-receptor complexes. Therefore, to reduce the false positives, P-value < 0.01 was considered statistically significant. Finally, we constructed a cell–cell crosstalk network based on the number of cell–cell interactions.
Association analysis of cell types related to strong cytokine storm and immune exhaustion scores for BALF
To explore cytokine storm-related cell types, we collected the cytokine storm genes (CXCL10, CCL3, CCL2, IL2, IL7, CSF3, TNF, and IL6) from a previous study1 and defined cytokine storm scores using the “AddModuleScore” function in Seurat (v4). We then used the sigmoid function to normalize cytokine storm scores to a range from 0 to 1 and plotted the normalized scores in a UMAP plot. We defined the immune exhaustion score based on six inhibitory receptors (PDCD1, CTLA4, LAG3, BTLA, TIGIT, and HAVCR2) through the same definition method as for cytokine storm score. We performed the Mann–Whitney rank test for each cell type's score versus all other types’ score, and the Kruskal–Wallis test for every cell type score comparison among severe, moderate, and healthy groups. Pearson correlation was applied to correlate the median cytokine storm scores of macrophages with the median immune exhaustion scores of (exhausted) CD8 T cells across three groups for BALF.
Re-clustering the macrophages of BALF to identify subclasses related to cytokine storm
To investigate which macrophage subclasses were the main source of cytokines, we separated macrophages from all the cell types of BALF by the “SplitObject” function. Then we tested a series of resolution values (0.1–0.9) to re-cluster macrophages and combined the expression of cytokine storm genes in the identified clusters to determine the best resolution value. Then we re-clustered these cells into 7 subclasses by using the “FindClusters” function with a resolution of 0.2. We tried UMAP andt-distributed stochastic neighbor embedding (tSNE) to visualize the 7 subclasses of macrophages and found tSNE made more clear clustering 2D space. Following the same approach for determining the previously mentioned PC parameter, we set 45 as the best PC number to cluster. We performed tSNE on the top 45 PCs to visualize the distribution of macrophage subclasses among severe, moderate, and healthy groups. Then we calculated the expression level of cytokine storm-related genes across 7 macrophage subclasses and identified differently expressed genes in each subclass using the “FindAllMarkers” function. We compared the percentages of 7 subclasses among three disease states using the Kruskal–Wallis test. We conducted the pathway and process enrichment analyses based on the top 100 highly expressed genes in subclasses enriched in the severe group.
Statistical analysis
Statistical discrete analyses were performed in R (version 3.5.1, http://www.r-project.org). The Kaplan–Meier estimate and log-rank testing were used to perform survival analysis. We used the Wilcoxon rank-sum test to analyse the correlation between continuous variables and categorical variables. We used the ‘corr.test’ function wrapped in psych (https://CRAN.R-project.org/package = psych) for Pearson analysis and P-value adjustment.
Results
Identifying severe CD8 TEX and relevant cell types in BALF of COVID-19 patients via single-cell transcriptomic analysis
To explore TEX in the bronchoalveolar immune microenvironment, we conducted an analysis on the scRNA-seq data derived from BALF cells of patients with moderate and severe COVID-19, and of healthy controls. The original sequencing data and the clinical features are available from the publicly reported study by Liao et al.13 Graph-based clustering identified 30 clusters (Fig. 1A and B). Based on the expression with at least two canonical genes from multiple COVID-19 single-cell studies,13,17,26 we assigned each cluster to 14 cell types (Fig. 1B and C). In particular, we defined the exhausted CD8 T cell (Exh T_CD8) cluster based on the elevated expression of multiple inhibitory receptors (TIGIT, LAG3, PDCD1, CTLA4, and HAVCR2) (Fig. 1D and Supplementary Table 3, see online supplementary material), and this cluster has not been reported by previous studies.13 Gene annotation and enrichment analysis of all differently expressed genes in the exhausted CD8 T cell cluster versus other cell clusters (Supplementary Table 3) suggested that these T cells might have enhanced the metabolism of RNA and cell division (Fig. 1E). Notable differences of cell type distribution could be observed based on the UMAP among the different infection states (healthy, moderate, and severe) (Fig. 1C). Exh T_CD8, squamous epithelial cells (Epi), and the mixture of macrophage and T cells (Macro/T) were visually enriched in the severe COVID-19 group (Fig. 1C). In addition, we used several deep-learning-based methods19–24 to cluster the cells and found scvi-tools produced a comparable result. Specifically, 45 clear clusters were identified (Supplementary Fig. 1A). When applying the cell annotation from the Seurat result to these clusters, most of the cells for each cell type were clustered together in UMAP (Supplementary Fig. 1B).
We compared the percentage of each cell type among three disease states, and found that Exh_T CD8, Macro/T, and Epi cells were enriched in the severe group and the other cell types were not enriched in any group (Fig. 1F and Supplementary Fig. 2B). Specifically, Exh T_CD8 did not show significant statistical significance, but the median percentage of Exh T_CD8 tended to be higher in the severe COVID-19 group (Fig. 1F). Notably, the Macro/T cell type was a unique cluster, which was observed in the severe group only (Fig. 1C and F) and expressed the canonical genes of both macrophages (CD163, MARCO, and FCGR1A) and T cells (CD2, CD3E, and CD3D) (Fig. 1D). Epi were significantly enriched in both the moderate and severe groups with SARS-CoV-2 infection, and the median percentage in the severe group was higher than that in the moderate group (Fig. 1F).
Cytokine storm promoting TEX in severe COVID-19 via cytokine-receptor interaction axes
Recent studies suggested that cytokine storm was strongly associated with COVID-19 severity, and macrophages may contribute to cytokine storms in BALF.13,26–29 Therefore, we analysed the single-cell sequencing data of BALF samples infected by SARS-CoV-2, and interrogated the potential relationship between cytokine storm and TEX. We computed the cytokine storm scores based on the expression of eight reported pro-inflammatory cytokines (IL2, IL7, CSF3, CXCL10, CCL2, CCL3, TNF, and IL6) detected in patients with cytokine storms,1 as well as the immune exhaustion scores based on the expression of six inhibitory receptors (PDCD1, CTLA4, LAG3, BTLA, TIGIT, and HAVCR2). As expected, the macrophages in the severe group had the highest cytokine storm scores among healthy, moderate, and severe groups (Fig. 2A and B), and the cytokine storm scores of macrophages gradually increased across the three groups (Fig. 2B). Surprisingly, the mixture of macrophages and T cells (Macro/T) had both high cytokine storm scores (Fig. 2B top panel) and high immune exhaustion scores (Fig. 2B bottom panel), suggesting that cytokine storm likely promoted the interaction between macrophages and T cells. On the other hand, all Exh T_CD8, T_CD4, and T cells exhibited gradually increasing exhaustion level across healthy, moderate, and severe groups (Fig. 2B bottom panel), indicating that TEX occurred in the moderate group and was more serious in the severe group. Pearson correlation analysis illustrated that with increasing cytokine storm scores of macrophages, TEX level increased as well (Fig. 2C and D). Overall, these results indicated that cytokine storms produced by macrophages promoted TEX in the bronchoalveolar immune microenvironment of severe COVID-19.
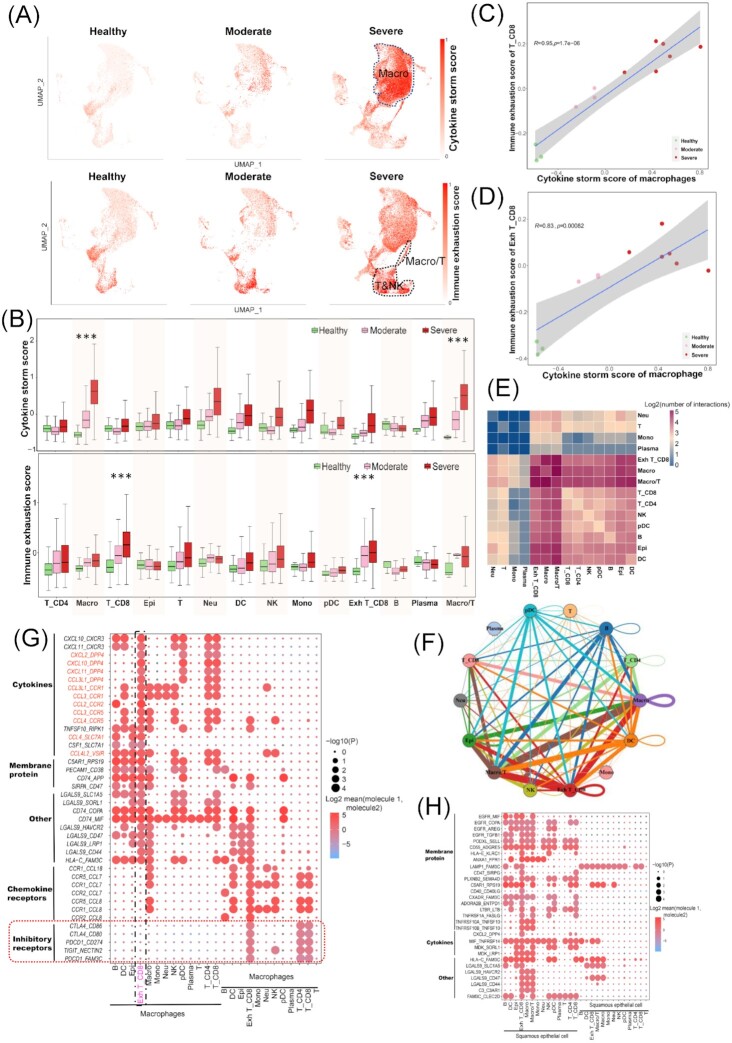
Figure 2.
Impacts of cytokine storms derived from macrophages on CD8 T cell exhaustion. (A) UMAP plots of BALF cells colored by cytokine storm (top panel) and immune exhaustion (bottom panel) scores across healthy controls (n = 3), moderate patients (n = 3), and severe patients (n = 6). The scores were scaled as 0–1. (B) Cytokine storms (top panel) and immune exhaustion (bottom panel) scores. (C and D) Pearson correlation between immune exhaustion score of CD8 T (T_CD8 and Exh T_CD8 respectively) and cytokine storm score of macrophages. (E) Heatmap shows the log2 (transformed number) of interactions between different cell types. (F) The cell–cell interaction network shows interaction frequencies between different cell types. Colorful nodes represent cell types, edges stand for cell–cell interaction, and their sizes indicate interaction numbers. (G and H) Dot plots show the selected ligand–receptor interactions mediating cell–cell communications: (G) is for macrophages and other cell types and (H) for squamous epithelial cell and other cell types. The circle size indicates P value. The means of the average expression levels of interacting molecule 1 in cluster 1 and interacting molecule 2 in cluster 2 are indicated by colour.
Then we explored the potential molecular mechanism via which cytokine storms promoted TEX. Of 719 cells belonging to Macro/T, 152 (21.14%) showed expression of LAG3, which is a typical inhibitory receptor representing TEX (Fig. 1G left). We suspected that this cell type may be a technical artefact known as “doublets” during single-cell RNA sequencing, as it was observed in the severe group only. The percentage of macrophages in the severe group was lower than that in the healthy group, indicating that the doublets may have no association with the high cell mass of macrophages. All of this evidence suggested an underlying interaction of T cells and macrophages, which possibly resulted in the occurrence of the Macro/T “doublets”. Therefore, we performed a cell–cell communication analysis through CellPhoneDB25 based on the combined expression of multi-subunit ligand–receptor complexes for the severe group cells. We found that Exh CD8_T cells had higher numbers of interactions with macrophages (Macro) and Macro/T cells (Fig. 2E). Specifically, cytokines (CXCL10, CXCL11, CXCL2, CCL2, CCL3, and so on) expressed in macrophages acted as ligands and significantly interacted with receptors (CXCR3, DPP4, CCR1, CCR2, CCR5, and so on) expressed in exhausted CD8 T cells (Exh T_CD8), forming the cytokine–receptor interaction axes (Fig. 2G). Multiple inhibitory receptors (CTLA4, PDCD1, TIGIT and HAVCR2) expressed in exhausted CD8 T cells significantly interacted with membrane protein genes (CD86, CD80, CD274/PD-L1, and NECTIN2). Cell–cell interaction analysis indicated that macrophages interacted with Exh T_CD8 cells via the cytokine–receptor axes in the severe group (Fig. 2F). Meanwhile, Epi cells had a high number of interactions with Macro, Macro/T, and Exh T_CD8 cells (Fig. 2F), and many membrane proteins expressed in Epi cells interacted with membrane proteins, cytokines and secretory proteins expressed in Exh T_CD8 cells (Fig. 2H).
Two macrophage subtypes were the main source of cytokine storms
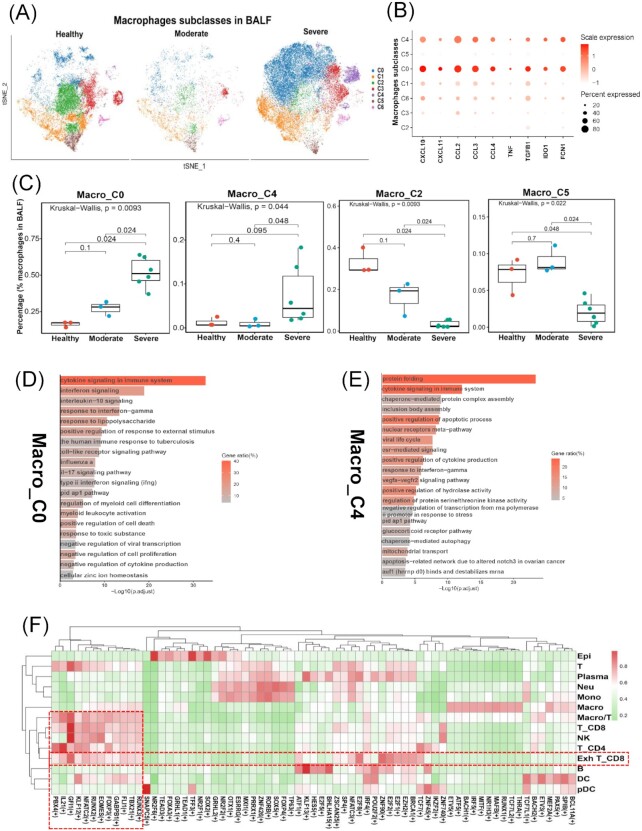
To further decompose the macrophage heterogeneity and identify macrophage subsets that contributed to cytokine storms, we re-clustered these macrophage cells into six subclasses (Macro_C0 to Macro_C5) (Fig. 3A). Macro_C0 and Macro_C4 were enriched in the severe group (Fig. 3C) and had higher expression levels of cytokine-storm genes (CXCL10, CXCL11, CCL2, CCL3, CCL4, TNF, and TGFB1) than other subclasses (Fig. 3B). Macro_C2 and Macro_C5 were enriched in the healthy control (Fig. 3C) but lacked expression of cytokine-storm genes (Fig. 3B). This evidence suggested that Macro_C0 and Macro_C4 macrophage subclasses were the main sources of cytokine storms in the bronchoalveolar immune environment. Pathway and process enrichment analysis of the top 100 differently expressed genes (Supplementary Table 4, see online supplementary material) in Macro_C0 and Macro_C4 respectively illustrated that these genes played vital roles in cytokine signalling in the immune system, interferon signalling, and interleukin-10 signalling (Fig. 3D and E).
Figure 3.
Identification of macrophage subclasses related to cytokine storms. (A) tSNE map plot of the macrophages colored by heterogeneous subclasses and split into the three disease states (healthy, moderate, and severe). (B) Dot plots show the expression levels of cytokine storms-related genes across 7 macrophage subclasses. (C) Boxplots show the cell percentages of 4 macrophage subclasses across three disease states. (D and E) The top 20 enriched pathways/processes of the top 100 differently expressed genes in Macro_C0 (D) and Macro_C4 (E) (hypergeometric test, adjusted P-values obtained by the Benjamini–Hochberg procedure). (F) Heatmap shows the regulon specificity score (RSS) for transcriptional factors (columns) corresponding to cell types (rows).
Identification of transcription factors related to TEX and cytokine storms
The biogenesis of TEX and cytokine storm relies on the intracellular transcriptional state driven by transcription factors.30,31 The transcriptional state of a cell emerges from an underlying gene regulatory network (GRN) in which a limited number of transcription factors (TFs) and cofactors regulate each other and their downstream target genes.32 Using pySCENIC, we reconstructed regulons based on the raw count matrix of the severe group and assessed the enrichment score for each regulon's target genes (AUCell). We identified master regulons that were specific to cell types via the regulon specificity score (RSS) (Supplementary Table 5, see online supplementary material). Finally, we predicted that transcription factor genes TBX21, NFATC2, and EOMES, which were involved in the transcriptional regulatory network for the development of TEX,33 were the master regulons with higher RSS for exhausted T cells (Fig. 3F). GFI1, ZNF90, E2F8, E2F2, and EZH2 were also specific to exhausted T cells and may play a vital role in TEX in severe COVID-19. In terms of macrophages, interferon regulatory factor 5 (IRF5) and NR1H3 involved in the inflammation-related transcriptional programs of macrophages34 were predicted as master regulons that might regulate cytokine-related gene expression (Fig. 3F).
Cytokine storms showed no association with TEX in PBMC
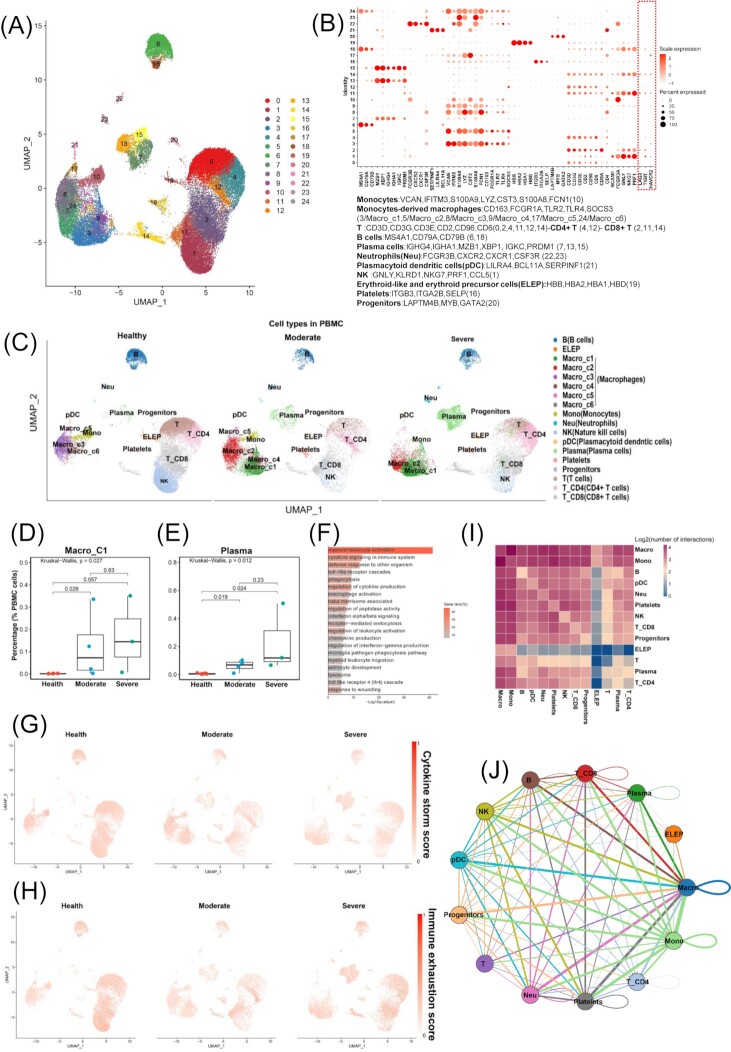
To investigate how cytokine storms impact TEX in peripheral blood of patients with COVID-19, we performed scRNA-Seq data analysis of PBMC samples of moderate COVID-19, severe COVID-19, and healthy control. A total of 24 cell clusters were identified, including harboured monocyte, monocytesderived macrophage, T, CD4 T, CD8 T, B, plasma, and NK cells (Fig. 4A and B). We compared the percentage of each cell type and observed that the plasma cells and a subclass of macrophages (Macro_C1) were significantly enriched in PBMC of severe COVID-19 (Fig. 4D and E, and Supplementary Fig. 3B). Pathway and process enrichment analysis of the top 100 highly expressed genes in Macro_C1 showed that these genes were also associated with cytokine-related pathways and processes (Fig. 4F). With respect to TEX, compared with the exhausted T cell cluster which expressed multiple inhibitory receptors in BALF, only a subset of T cells had the expression of TIGIT, LAG3, or HAVCR2 in PBMC (Fig. 4B). We then calculated immune exhaustion scores and cytokine storm scores of PBMC. We found a minority of cells with high immune exhaustion scores or cytokine storm scores, but these cells were not significantly enriched in each of the groups or cell types (Fig. 4G and H). Cell–cell interaction analysis illustrated that macrophages did not have high frequency of interactions with T cells in PBMC of severe COVID-19 (Fig. 4I and J). These results indicated that although a macrophage subclass related to cytokine production occurred in PBMC of severe COVID-19, the macrophages did not cause cytokine storms in PBMC.
Figure 4.
Single-cell RNA-seq analysis to investigate the impact of cytokine storms on TEX in PBMC. (A) Overview of the cell clusters in PBMCs (n = 13) derived from across healthy people (n = 6), moderate (n = 4), and severe (n = 3) COVID-19 patients. (B) Dotplots show the expression level of canonical markers representing cell types in PBMC. Cell types and their corresponding genes are listed at the bottom. (C) UMAP plots of PBMC colored by cell type among three disease states (healthy control, moderate, and severe). Cell percentage comparisons of Macro_C1 (D) and plasma (E) in PBMC across healthy controls, moderate, and severe patients. Kruskal–Wallis H test. (F) Pathway and process enrichment analysis of the top 100 highly expressed genes in a subclass of macrophages (Macro_C1). UMAP plots of PBMCs colored by cytokine storm (G) and immune exhaustion (H) scores across three disease states. The scores are scaled to 0–1. (I) Log2-transformed frequency of interactions between cell types of PBMC. (J) Cell–cell interaction network shows the interaction frequencies between cell types of PBMC.
Gene expression of cytokine–receptor interaction axes correlated with poor prognosis in lung squamous cell carcinoma
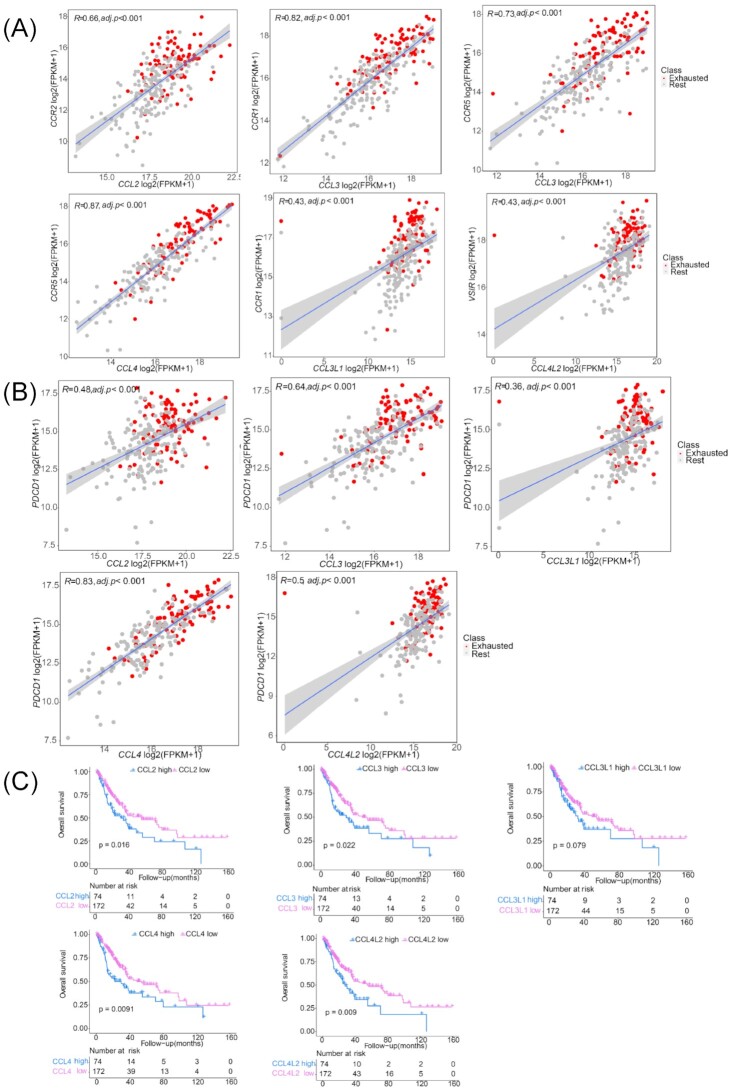
Cytokine storm is a common clinical feature in both severe COVID-19 and lung squamous cell carcinoma (LUSC).35 Exhausted T cells are widely present in the tumour microenvironment, leading to immune evasion.36 To validate cytokine storm-related axes that impact TEX, we performed expression analysis in a TCGA LUSC cohort to explore the co-expression of cytokines and receptors that formed the interaction axes in severe COVID-19. The paired genes ofCCL2-CCR2 (R = 0.66, adjusted P < 0.001), CCL3-CCR1 (R = 0.82, adjusted P < 0.001), CCL3-CCR5 (R = 0.73, adjusted P < 0.001), CCL4-CCR5 (R = 0.87, adjusted P < 0.001), CCL4L2-VSIR (R = 0.43, adjusted P < 0.001), andCCL3L1-CCR1 (R = 0.43, adjusted P < 0.001) axes were co-expressed in LUSC (Fig. 5A), suggesting that cytokines may also interact with the receptors of T cells and promote TEX in LUSC. Of these cytokine–receptor axes, the CCL2-CCR2 axis has been reported to be involved in immune evasion through PD-1 signalling in esophageal carcinogenesis.37,38 In our study, the cytokine genes (CCL2, CCL3, CCL4, CCL3L1, and CCL4L2) showed correlations with PD-1 (PDCD1) expression (Fig. 5B). Survival analysis based on the grouping of cytokine gene expression levels indicated that the high expression of CCL2, CCL3, CCL4, CCL3L1, and CCL4L2 was significantly associated with poor prognosis in LUSC (Fig. 5C). In brief, these evidences demonstrated that similar cytokine storms promoted TEX in COVID-19 and LUSC, and were associated with poor clinical outcomes. The cytokine-receptor axes may be a potential targets of immunotherapy. The targeted drugs of the cytokine–receptor axes are summarized in Table 2.
Figure 5.
Expression correlation analysis of cytokines and receptors in a LUSC cohort and survival analysis based on cytokine expression level. (A) Correlation of expression between the paired genesCCL4-CCR2, CCL3-CCR1, CCL3-CCR5, CCL4-CCR5, CCL4L2-VSIR, and CCL3L1-CCR1. (B) Pearson correlation analyses indicate the strong association of cytokines with PDCD1 expression. (C) High expression of CCL2, CCL3, CCL4, CCL3L1, and CCL4L2 is associated with poor overall survival of LUSC patients.
Table 2.
Cytokine–receptor axes identified in this study.
| Axis | Cytokine/receptor | Description | Correlation among LUSC cohort | Drug |
|---|---|---|---|---|
| CCL3L1_CCR1 | CCL3L1 | C-C motif chemokine ligand 3 like 1 | R = 0.43, P = 1.1e−12 | No record |
| CCR1 | C-C motif chemokine receptor 1 | DAPTA, BX 471, CCL3 ,CCL4, J 113863 | ||
| CCL3_CCR1 | CCL3 | C-C motif chemokine ligand 3 | R = 0.82, P < 2.2e−16 | ROX-888 |
| CCR1 | C-C motif chemokine receptor 1 | DAPTA, BX 471, CCL3, CCL4, J 113863 | ||
| CCL2_CCR2 | CCL2 | C-C motif chemokine ligand 2 | R = 0.66, P < 2.2e−16 | Danazol, Atorvastatin, Simvastatin, Chondroitin sulfate, Risperidone |
| CCR2 | C-C motif chemokine receptor 2 | INCB3284, CCX915, Plozalizumab, MK-0812, DAPTA | ||
| CCL3_CCR5 | CCL3 | C-C motif chemokine ligand 3 | R = 0.73, P < 2.2e−16 | ROX-888 |
| CCR5 | C-C motif chemokine receptor 5 | Maraviroc, Dexamethasone, Efavirenz, Etravirine, Ritonavir | ||
| CCL4_CCR5 | CCL4 | C-C motif chemokine ligand 4 | R = 0.87, P < 2.2e−16 | No record |
| CCR5 | C-C motif chemokine receptor 5 | Maraviroc, Dexamethasone, Efavirenz, Etravirine, Ritonavir | ||
| CCL4L2_VSIR | CCL4L2 | C-C motif chemokine ligand 4 like 2 | R = 0.43, P = 2.1e−12 | No record |
| VSIR | V-set immunoregulatory receptor | No record |
Discussion
Cytokine storms involve elevated levels of pro-inflammatory cytokine and immune cell hyperactivation, leading to coagulopathy, multiple organ failure and even death.1,35 It has been reported that cytokine storms are associated with TEX.4,5 However, knowledge about how cytokine storms impact TEX remains lacking. In this study, we revealed that cytokine storms released by macrophages promoted TEX via CCL2-CCR2, CCL3-CCR1, CCL3-CCR5, CCL4-CCR5, CCL4L2-VSIR, and CCL3L1-CCR1 axes, that are associatied with poor prognosis of COVID-19.
To investigate the interaction mechanism between cytokine storms and TEX, we first analysed single-cell sequencing data of BALF samples of COVID-19 to identify and define 14 cell-type clusters. Of them, the severely exhausted CD8 T cell cluster expressed multiple distinct inhibitory receptors and was enriched in BALF of severe COVID-19. The co-expression of multiple inhibitory receptors was associated with higher exhaustion levels of CD8 T cells and more severe infection during viral infection.39 This severe exhausted CD8 T cell type has not been reported by previous studies.13,26 Because severe exhausted T cells displayed accumulation of mitochondria,40 the low mitochondrial gene percentage parameter setting in the single-cell quality control led to the removal of these cells in previous studies. Consistent with previous studies,13,26 Epi cells accounted for a significantly higher cell proportion of BALF in severe COVID-19 compared with that in moderate COVID-19 or healthy controls. Interestingly, the Macro/T cell type which may be “doublets” of macrophages and T cells and expressed exhaustion markers such as LAG3 and HAVCR2, only existed in BALF of severe COVID-19.
We suspected that the interaction of macrophages and T cells caused the presence of Macro/T and was associated with TEX. Therefore, we then conducted cell–cell communication analyses which confirmed the presence of interaction between exhausted T cells and macrophages in severe COVID-19 via cytokine–receptor axes (CXCL2-DPP4, CXCL11-DPP4, CCL3L1-DPP4, CCL3L1-CCR1, CCL3-CCR1, CCL2-CCR2, CCL3-CCR5, CCL4-CCR5, and CCL4L2-VSIR). CXCL10, CXCL11, CCL2, CCL4 and CCL3 were involved in cytokine storms as soluble mediators.31 Of these cytokine–receptor axes, CCL2-CCR2 and CCL3-CCR5 have been reported to regulate T cell differentiation.41 Our result indicated that cytokine–receptor axes mediate the communication between macrophages and exhausted T cells, impacting TEX.
Further, as reported in previous studies,42 cytokine storm was not observed in the moderate group of COVID-19, but was more pronounced in the severe group. While TEX had been observed in moderate patients, subsequently more serious TEX was seen in the severe group. Correlation analysis indicated that cytokine storm level was significantly positively correlated with TEX. This evidence indicated that the presence of cytokine storm promoted TEX in severe COVID-19. Additional cell subtype clustering in macrophages suggested that macrophages might be a key source of cytokine storms in BALF of severe COVID-19.
Blockade of the cytokine–receptor axes mediating the communication between macrophages and exhausted T cells, such as CCL2-CCR2, CCL3-CCR5, and CCL4-CCR5, may attenuate TEX. Clinical trials (NCT04435522, NCT04441385, and NCT04475991) to test the efficacy of maraviroc, a CCR5 antagonist for severe COVID-19, are currently ongoing. Maraviroc may reverse TEX and reduce cytokine storms.6 Moreover, dexamethasone, a drug for CCR5 gene (GeneCards GCID: GC03P046383), was reported to be able to reduce 28-day mortality for severe COVID-19.43 These findings illustrated that the cytokine–receptor axes may be important therapeutic targets.
Cytokine storms showed no association with TEX in PBMC of COVID-19; however, in lung cancer, especially LUSC, an interleukin-6-related cytokine storm was observed.35 In our study, the cytokine storm-related genes and corresponding receptor genes found in severe COVID-19, were significantly correlated with each other in LUSC, and these cytokines had strongly positive correlation with PD-1 expression. This evidence confirmed that a similar cytokine storm was present in LUSC and positively correlated with severe TEX. The co-expression of cytokine and receptor genes (CCL2-CCR2,CCL3-CCR1, CCL3-CCR5,CCL4-CCR5, CCL4L2-VSIR, and CCL3L1-CCR1) confirmed that cytokine–receptor axes found in COVID-19 may also play a critical role in TEX of LUSC. Of these axes, a previous study demonstrated that the CCL2-CCR2 axis can induce immune evasion via PD-1 signalling and the blockade of this axis can enhance the antitumor efficacy of CD8 T cells in esophageal carcinogenesis.37 Anlotinib can induce CCL2 decreases and improve the survival of refractory advanced nonsmall cell lung cancer patients,44 suggesting that patients with high CCL2 expression level may benefit from this drug. High expression of cytokine storm-related genes (CCL2, CCL3, CCL4, CCL3L1, and CCL4L2) was significantly associated with poor prognosis, confirming the prognostic value of cytokine storms in LUSC.
In conclusion, we revealed that cytokine storms may promote TEX through cytokine–receptor axes associated with poor prognosis in COVID-19. Similar findings were verified in LUSC. Blocking the cytokine–receptor axes, such as CCL2-CCR2, CCL3-CCR1, CCL3-CCR5, CCL4-CCR5, CCL3L1-VSIR, and CCL3L1-CCR1, may be a novel strategy to reverse TEX and furhter to treat severe COVID-19 with cytokine storm-related immune exhaustion. Some cytokine–receptor axes (e.g. CCL2-CCR2 and CCL3-CCR2) have been confirmed to be involved in TEX-related immune evasion in other diseases though experiments. However, the molecular relationship between cytokine storms and TEX were not fully investigated experimentally. Through bioinformatics analysis during this study, we provided a new insight into the molecular relationship between cytokine storms and TEX, suggesting a clue to potential cytokine-targeted strategies for immunotherapy and indicating the prognostic significance of cytokine storms.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Key R&D Program of China (Grants No. 2021YFF1200900, 2021YFF1200903, 2016YFC0901604 & 2018YFC091040), the Natural Science Foundation of Guangdong Province (Grant No. 2021A1515012108), the Guangdong Project (Grant No. 2017GC010608), and the Support Scheme of Guangzhou for Leading Talents in Innovation and Entrepreneurship (Grant No. 2020007).
Contributor Information
Minglei Yang, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou 510080, China.
Chenghao Lin, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou 510080, China.
Yanni Wang, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou 510080, China.
Kang Chen, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou 510080, China.
Yutong Han, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou 510080, China.
Haiyue Zhang, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou 510080, China.
Weizhong Li, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou 510080, China; Center for Precision Medicine, Sun Yat-sen University, Guangzhou 510080, China; Key Laboratory of Tropical Disease Control of Ministry of Education, Sun Yat-Sen University, Guangzhou 510080, China.
Conflict of interest
The authors declare no conflicts of interest. In addition, as an Editorial Board Member of Precision Clinical Medicine, the corresponding author Weizhong Li was blinded from reviewing or making decisions on this manuscript.
References
- 1. Mehta P, Mcauley DF, Brown M, et al. . COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet North Am Ed. 2020;395:1033–4.. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mulchandani R, Lyngdoh T, Kakkar AK. Deciphering the COVID-19 cytokine storm: Systematic review and meta-analysis. Eur J Clin Invest. 2021;51:e13429. doi: 10.1111/eci.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mahmoudi S, Rezaei M, Mansouri N, et al. . Immunologic features in Coronavirus Disease 2019: functional exhaustion of T cells and cytokine storm. J Clin Immunol. 2020;40:974–6.. doi: 10.1007/s10875-020-00824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Biasi S, Meschiari M, Gibellini L, et al. . Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen J, Vitetta L. Increased PD-L1 expression may be associated with the cytokine storm and CD8+ T-cell exhaustion in severe COVID-19. J Infect Dis. 2021;223:1659–60.. doi: 10.1093/infdis/jiab061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patterson BK, Seethamraju H, Dhody K, et al. . CCR5 inhibition in critical COVID-19 patients decreases inflammatory cytokines, increases CD8 T-cells, and decreases SARS-CoV2 RNA in plasma by day 14. Int J Infect Dis. 2021;103:25–32.. doi: 10.1016/j.ijid.2020.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shankar P, Russo M, Harnisch B, et al. . Impaired function of circulating HIV-specific CD8(+) T cells in chronic human immunodeficiency virus infection. Blood. 2000;96(9):3094–101. [PubMed] [Google Scholar]
- 8. Day CL, Kaufmann DE, Kiepiela P, et al. . PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443(7109):350–4.. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 9. Gruener NH, Lechner F, Jung M-C, et al. . Sustained dysfunction of antiviral CD8(+) T lymphocytes after infection with hepatitis C virus. J Virol. 2001;75(12):5550–8.. doi: 10.1128/JVI.75.12.5550-5558.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ye B, Liu X, Li X, et al. . T-cell exhaustion in chronic hepatitis B infection: current knowledge and clinical significance. Cell Death Dis. 2015;6(3):e1694. doi: 10.1038/cddis.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng H-Yi, Zhang Mi, Yang C-X, et al. . Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17(5):541–3.. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fried JA, Ramasubbu K, Bhatt R, et al. . The variety of cardiovascular presentations of COVID-19. Circulation. 2020;141(23):1930–6.. doi: 10.1161/CIRCULATIONAHA.120.047164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liao M, Liu Y, Yuan J, et al. . Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26(6):842–4.. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 14. Wilk AJ, Rustagi A, Zhao NQ, et al. . A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26(7):1070–6.. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buck MD, O'sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med. 2015;212(9):1345–60.. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Macosko EZ, Basu A, Satija R, et al. . Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–14.. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang X, Lan Y, Xu J, et al. . CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47:D721–8.. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou Y, Zhou B, Pache L, et al. . Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopez R, Regier J, Cole MB, et al. . Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018;15(12):1053–8.. doi: 10.1038/s41592-018-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eraslan G, Simon LM, Mircea M, et al. . Single-cell RNA-seq denoising using a deep count autoencoder. Nat Commun. 2019;10(1):390. doi: 10.1038/s41467-018-07931-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tian T, Wan Ji, Song Qi, et al. . Clustering single-cell RNA-seq data with a model-based deep learning approach. Nature Machine Intelligence. 2019;1(4):191–8.. doi: 10.1038/s42256-019-0037-0. [DOI] [Google Scholar]
- 22. Liu Q, Chen S, Jiang R, et al. . Simultaneous deep generative modelling and clustering of single-cell genomic data. Nature Machine Intelligence. 2021;3(6):536–44.. doi: 10.1038/s42256-021-00333-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen S, Yan G, Zhang W, et al. . RA3 is a reference-guided approach for epigenetic characterization of single cells. Nat Commun. 2021;12(1):2177. doi: 10.1038/s41467-021-22495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen X, Chen S, Song S, et al. . Cell type annotation of single-cell chromatin accessibility data via supervised Bayesian embedding. Nature Machine Intelligence. 2022;4(2):116–26.. doi: 10.1038/s42256-021-00432-w. [DOI] [Google Scholar]
- 25. Efremova M, Vento-Tormo M, Teichmann SA, et al. . CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484–506.. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 26. Ren X, Wen W, Fan X, et al. . COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184(23):1895–1913 e1819.. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robilotti EV, Babady NE, Mead PA, et al. . Determinants of COVID-19 disease severity in patients with cancer. Nat Med. 2020;26(8):1218–23.. doi: 10.1038/s41591-020-0979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20(5):269–70.. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang C, Wang Y, Li X, et al. . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet North Am Ed. 2020;395(10223):497–506.. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seo W, Jerin C, Nishikawa H. Transcriptional regulatory network for the establishment of CD8(+) T cell exhaustion. Exp Mol Med. 2021;53(2):202–9.. doi: 10.1038/s12276-021-00568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med. 2020;383(23):2255–73.. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aibar S, González-Blas CB, Moerman T, et al. . SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(1):1083–6.. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mclane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol. 2019;37(1):457–95.. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 34. Bories G, Colin S, Vanhoutte J, et al. . Liver X receptor activation stimulates iron export in human alternative macrophages. Circ Res. 2013;113(11):1196–205.. doi: 10.1161/CIRCRESAHA.113.301656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turnquist C, Ryan BM, Horikawa I, et al. . Cytokine storms in cancer and COVID-19. Cancer Cell. 2020;38(5):598–601.. doi: 10.1016/j.ccell.2020.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang Y, Li Y, Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 2015;6(6):e1792. doi: 10.1038/cddis.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang H, Zhang Q, Xu M, et al. . CCL2-CCR2 axis recruits tumor associated macrophages to induce immune evasion through PD-1 signaling in esophageal carcinogenesis. Mol Cancer. 2020;19(1):41. doi: 10.1186/s12943-020-01165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hao Q, Vadgama JV, Wang P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun Signal. 2020;18(1):82. doi: 10.1186/s12964-020-00589-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blackburn SD, Shin H, Haining WN, et al. . Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37.. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu Yi-Ru, Imrichova H, Wang H, et al. . Disturbed mitochondrial dynamics in CD8(+) TILs reinforce T cell exhaustion. Nat Immunol. 2020;21(12):1540–51.. doi: 10.1038/s41590-020-0793-3. [DOI] [PubMed] [Google Scholar]
- 41. Luther SA, Cyster JG. Chemokines as regulators of T cell differentiation. Nat Immunol. 2001;2(2):102–7.. doi: 10.1038/84205. [DOI] [PubMed] [Google Scholar]
- 42. Tang Y, Liu J, Zhang D, et al. . Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Group RC, Horby P, Lim WS, et al. . Dexamethasone in hospitalized patients with COVID-19. N Engl J Med. 2021;384:693–704.. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lu J, Zhong H, Chu T, et al. . Role of anlotinib-induced CCL2 decrease in anti-angiogenesis and response prediction for nonsmall cell lung cancer therapy. Eur Respir J. 2019;53:1801562. doi: 10.1183/13993003.01562-2018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.