Background:
The ability to diagnose preeclampsia clinically is suboptimal. Our objective was to validate a novel multianalyte assay and characterize its performance, when intended for use as an aid to rule-out preeclampsia.
Methods:
Prospective, multicenter cohort study of pregnant individuals presenting between 280/7 and 366/7 weeks’ with preeclampsia-associated signs and symptoms. Individuals not diagnosed with preeclampsia after baseline evaluation were enrolled in the study cohort, with those who later developed preeclampsia, classified as cases and compared with a negative control group who did not develop preeclampsia. Individuals with assay values at time of enrollment ≥0.0325, determined using a previously developed algorithm, considered at risk. The primary analysis was the time to develop preeclampsia assessed using a multivariate Cox regression model.
Results:
One thousand thirty-six pregnant individuals were enrolled in the study cohort with an incidence of preeclampsia of 30.3% (27.6%–33.2%). The time to develop preeclampsia was shorter for those with an at-risk compared with negative assay result (log-rank P<0.0001; adjusted hazard ratio of 4.81 [3.69–6.27, P<0.0001]). The performance metrics for the assay to rule-out preeclampsia within 7 days of enrollment showed a sensitivity 76.4% (67.5%–83.5%), negative predictive value 95.0% (92.8%–96.6%), and negative likelihood ratio 0.46 (0.32–0.65). Assay performance improved if delivery occurred <37 weeks and for individuals enrolled between 28 and 35 weeks.
Conclusions:
We confirmed that a novel multianalyte assay was associated with the time to develop preeclampsia and has a moderate sensitivity and negative likelihood ratio but high negative predictive value when assessed as an aid to rule out preeclampsia within 7 days of enrollment.
Registration:
The study was registered on Clinicaltrials.gov (Identifier NCT02780414).
Keywords: biomarkers, hypertension, preeclampsia
Novelty and Relevance.
What Is New?
The ability to diagnose preeclampsia clinically is suboptimal. We sought to validate a novel multianalyte assay and characterize its performance, when intended for use as an aid to rule-out preeclampsia in pregnant individuals presenting between 280/7 to 366/7 weeks’ with preeclampsia-associated signs and symptoms.
What Is Relevant?
We validated a novel multi-analyte assay, which together with the gestational age at sample collection, was associated with the time to develop preeclampsia, had moderate sensitivity and likelihood ratios, but a high negative predictive value for development of preeclampsia within 7 days of enrollment.
Clinical/Pathophysiological Implications?
An assay of 8 biomarkers associated with angiogenic imbalance, placental and trophoblast dysfunction, hypoxia, and inflammation and immune regulation was validated as an aid for physicians evaluating pregnant individuals presenting with signs and symptoms of preeclampsia.
Preeclampsia is a pregnancy-specific multisystem disorder that affects 3% to 8% of pregnancies and remains a leading cause of maternal and neonatal morbidity and mortality.1,2 In the United States, it is the third leading cause of maternal death, and worldwide >70 thousand pregnant individuals die from preeclampsia-related causes each year.3 Recent data suggest that preeclampsia contributes to racial and ethnic disparities, with Black pregnant individuals 2 to 3× more likely to die from preeclampsia compared with White individuals.4–6
Preeclampsia is traditionally diagnosed using the criteria of hypertension along with proteinuria, or in the absence of the latter, clinical symptoms, laboratory abnormalities, or evidence of end-organ injury.1 Although hypertension is a hallmark of preeclampsia, other hypertensive diseases also can occur during pregnancy and must be differentiated in order for appropriate management to occur.1 Relying on clinical criteria is suboptimal mainly because obtaining blood pressure and proteinuria is subject to errors during collection (cuff size, position, activity) and can fluctuate during the observation period,7,8 and relying on maternal symptoms may be problematic in clinical practice because of their subjective nature.1,9,10 Because the specificity of these criteria is poor, pregnant individuals in whom there is a suspicion for preeclampsia—but who may not actually have it—may be admitted for monitoring and delivered prematurely.
While the pathogenesis of preeclampsia is not completely understood, it is thought to be related to impaired early placental development,11–13 with associated abnormalities in angiogenesis, endothelial and hypoxic injury, oxidative stress, and inflammation.11,14–17 These derangements ultimately lead to the clinical manifestations of the disease. Based on these pathophysiologic pathways, a molecular assay was developed by Progenity, Inc, intended to be used as an aid to rule-out preeclampsia.18 It includes 8 markers associated with angiogenic imbalance (free and total placental growth factor, soluble FMS-like tyrosine kinase-1, soluble endoglin), placental and trophoblast dysfunction (fibroblast growth factor-21 and decorin), hypoxia (kidney injury molecule-1), and inflammation and immune regulation (cluster of differentiation-274, or PD-L1 [programmed death-ligand 1]).18,19
The objective of this study was to validate the assay and characterize its performance, when intended for use as an aid to rule-out preeclampsia in pregnant individuals presenting with preeclampsia-associated signs and symptoms between 280/7 and 366/7 weeks.
Methods
De-identified data that support the findings of this study are available from the sponsor upon reasonable request.
Study Population
Pregnant individuals 18 to 45 years old, with singleton, nonanomalous pregnancies between 280/7 weeks and 366/7 weeks gestation, who presented to triage, labor and delivery, or an outpatient setting to rule-out preeclampsia were eligible for enrollment in this multicenter prospective cohort study. We excluded from enrollment those who had known fetal genetic or major malformations; received dialysis; or multifetal gestations. Anomalous pregnancies were excluded as they may affect pregnancy management and neonatal outcomes and may be associated with genetic conditions that could impact biomarkers levels. From those enrolled, we also excluded from subsequent analysis those without a baseline sample, with a sample that could not be analyzed, and with active cancer or history of cancer with current status unknown, due to potential effects of cancer on angiogenic biomarkers.
The trial was conducted at 20 academic and community-based medical centers (Supplemental Material) across the United States between 2016 and 2020. The Institutional Review Boards at all the participating sites approved the study protocol. All individuals provided written informed consent. The study was registered on Clinicaltrials.gov before start of the study.
Study Design and Intervention
Individuals presenting to rule-out preeclampsia (based on new-onset elevated blood pressure if previously normotensive, or worsening preexisting hypertension, new-onset proteinuria or worsening of preexisting proteinuria, and other signs or symptoms that necessitated evaluation to rule out preeclampsia) were evaluated according to the standard of care at individual centers. Those not diagnosed with preeclampsia at the baseline evaluation were considered the main study cohort. In this cohort, those who later developed preeclampsia before or at delivery were classified as cases. The time (in days) from the baseline sampling to diagnosis of preeclampsia was recorded. Conversely, individuals who did not develop preeclampsia by delivery represented the negative control group, and the time from baseline sampling (in days) to either delivery or loss to follow-up was recorded. On the contrary, eligible individuals who were found to have preeclampsia at the time of baseline evaluation (or within 24 hours of presentation if a 24-hour urine was collected) were enrolled in a positive preeclampsia control group.
Pregnancy management (eg, antenatal testing, management of preeclampsia, inpatient versus outpatient management, timing, and mode of delivery) was left to the discretion of the treating clinician and performed per standard of care at the respective participating institutions. All data were collected or abstracted by research coordinators at the clinical centers. No pregnant individual, care provider, or investigator had access to the results of the biomarkers assay, which was run at the completion of the study.
Study Outcome
The primary outcome for this study was the time to develop preeclampsia, irrespective of disease severity, however, individuals who delivered without developing preeclampsia were censored at the time of their delivery. Preeclampsia was defined according to the 2013 criteria set by the American College of Obstetricians and Gynecologists,20 (the active guidelines at the time the study was designed conducted; Supplemental Material). Adjudication rules and process were set a priori (Supplemental Material).
Sample Collection and Assay Development
Blood samples were collected at baseline and shipped to a central laboratory facility (Progenity Inc; Ann Arbor, MI) where they were analyzed in batch at the completion of the study. The assay was developed by Progenity, Inc. (San Diego, CA) using a separate cohort of individuals who were evaluated for preeclampsia.18,19 Details in the Supplemental Material.
For this study, the assay results are reported based on a locked algorithm that includes the 8 biomarkers and gestational age at the time of blood draw. This algorithm was previously developed and optimized (data not included, but available upon reasonable request from the sponsor), with a cutoff determined to be 0.0325 during a robust cut point determination process aiming for a 90% sensitivity using a subset of the training data. Individuals with values below 0.0325 are considered at reduced risk for preeclampsia and considered to have a negative result. Conversely, those with values at or greater than the cutoff are considered at increased risk for preeclampsia events and reported to be at risk. There was no cutoff for individual biomarkers for classification decision-making, and the dichotomization was determined solely based on the cutoff on the model score generated by the algorithm. All laboratory staff were masked to clinical outcomes.
Statistical Analysis
The study was originally designed to enroll up to 1541 individuals into the study cohort and 250 in the positive preeclampsia control group. This sample size was calculated using Cox Proportional Hazards Regression, conservative estimates of both performance (minimum hazard ratio [HR], 1.4) and prevalence for preeclampsia (20%), an R2 for other covariates of 0.10, 80% power, and alpha of 0.05. However, due to the higher than anticipated incidence of preeclampsia, an unplanned one-time subsequent sample size assessment indicated that a sample size of at least 1036 in the study cohort would provide >90% power at a HR of at least 1.6.
Out of 1730 individuals enrolled, samples from 70 of the positive preeclampsia control group were randomly selected to assist in the development and optimization of the algorithm and are not part of this current analysis (Figure S1). The algorithm was then locked. The sponsor divided the remaining 1660 individuals into a prevalidation set (n=356) and a validation set (n=1,304; Figure S1). The 356 individuals in the prevalidation set were selected randomly with the same gestational age distribution as the overall study and were used in an additional unplanned assay robustness analysis, which did not lead to changes in the algorithm.
Analyses reported in this article were performed by a statistician who had full access to the data and was independent of the sponsor, and the results were reviewed for accuracy and conformance to the prespecified statistical analysis plan by one of the authors (M. Sarno) who had full access to all the data in the study and takes responsibility for the integrity of the conducted data analyses. Statistical analyses were performed using JMPro 16.0 (SAS, Cary, NC).
Data were reported using descriptive statistics with mean and SD or median and interquartile range for continuous measurements and number (%) for categorical variables. Continuous variables were compared using Wilcoxon rank-sum and categorical variables using the χ2 or Fisher exact tests, as appropriate. The primary analysis was based on Cox proportional hazard regression and reported a HR with its associated 2-sided Wald 95% CI. The model employed a 2-group time-to-event analysis that compared the days to a preeclampsia diagnosis between individuals with at risk and negative test results. We calculated the HR for the test to indicate the hazard of preeclampsia in individuals with an at-risk test result compared with the hazard of preeclampsia in those with a negative test result using the time to develop preeclampsia or delivery, whichever occurs first. Individuals who did not develop preeclampsia were censored at delivery or if lost to follow-up. The prespecified multivariate Cox regression model for the primary analysis adjusted for age (years, continuous), race and ethnicity (self-reported), gestational age at enrollment (weeks, continuous), body mass index (kg/m2, continuous), prior history of preeclampsia, and history of diabetes.
Performance metrics including sensitivity, specificity, positive predictive value, negative predictive value (NPV), positive likelihood ratio, negative likelihood ratio, and accuracy were calculated along with their associated 2-sided 95% CIs. NPV and positive predictive value are reported using the study population prevalence as well as theorized population prevalences of 2.7%, 10%, and 20% to compare performance across studies using Bayes Rule. In addition, we estimated the false negative rate of the assay in a cross-sectional manner using the positive preeclampsia control group. Performance analyses in the study cohort were also performed to assess the ability of the assay to rule out preeclampsia for 7 and 14 days. For this analysis, individuals were divided into those who developed the outcome (preeclampsia case within the rule-out window; positive status) and everyone else (those who did not develop preeclampsia or developed preeclampsia outside of the rule-out window; negative status). Additional prespecified analyses were performed for those who developed the outcome overall and in the 7- and 14-day rule-out window but who delivered <37 weeks, using the clinical center’s diagnosis of preeclampsia (n=1042), and for individuals who were enrolled between 28 and 35 weeks’ gestation. The analysis using the clinical center’s diagnosis of preeclampsia represents the real-word performance of the assay. Post hoc analyses for added benefit, including a computation of the Net Reclassification Improvement,21 were also performed following unblinding and completion of all prespecified statistical analyses (Supplemental Materials).
All statistical tests were 2-tailed and performed at the 5% significance level.
Results
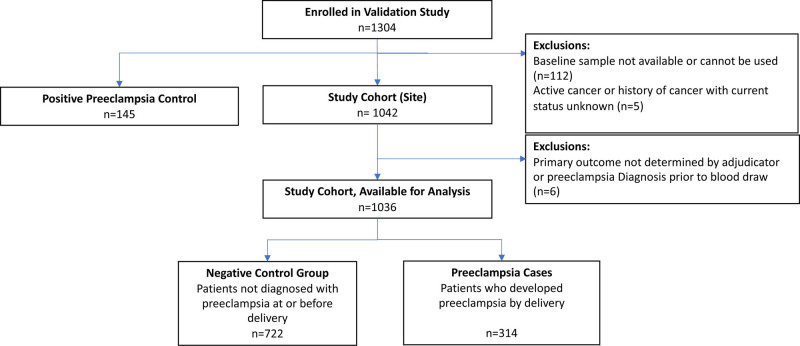
Of 1304 individuals, 117 were excluded (112 for no usable samples and 5 for active cancer or history of cancer with current status unknown), leaving 1042 individuals in the study cohort and 145 in the positive preeclampsia control group. In addition, 6 individuals from the cohort were excluded due to inability to determine the primary outcome by the adjudication committee (Figure 1). The demographic and clinical characteristics of the positive preeclampsia control group (n=145) are summarized in Table S1.
Figure 1.
Study flow chart.
Baseline characteristics of the study cohort (n=1036) are summarized in Table S2. Individuals were enrolled at 33.6±2.4 weeks gestation, most commonly for new-onset hypertension (50% in the cohort). The cohort was diverse with 30.9% self-identified as non-Hispanic Black. Moreover, the cohort represented a high-risk population with 35.5% being nulliparous, 30.5% having a history of preeclampsia, 36.3% chronic hypertension, and 11.4% pregestational diabetes. Maternal and neonatal outcomes of study cohort participants overall and then of preeclampsia cases and negative control group are summarized in Table 1 and demonstrate that cases were more likely to have worse maternal and neonatal outcomes compared with the negative control group.
Table 1.
Maternal and Neonatal Outcomes of Study Cohort (n=1036) Overall, and Then of Preeclampsia Cases (n=314) and Negative Control Group (n=722)
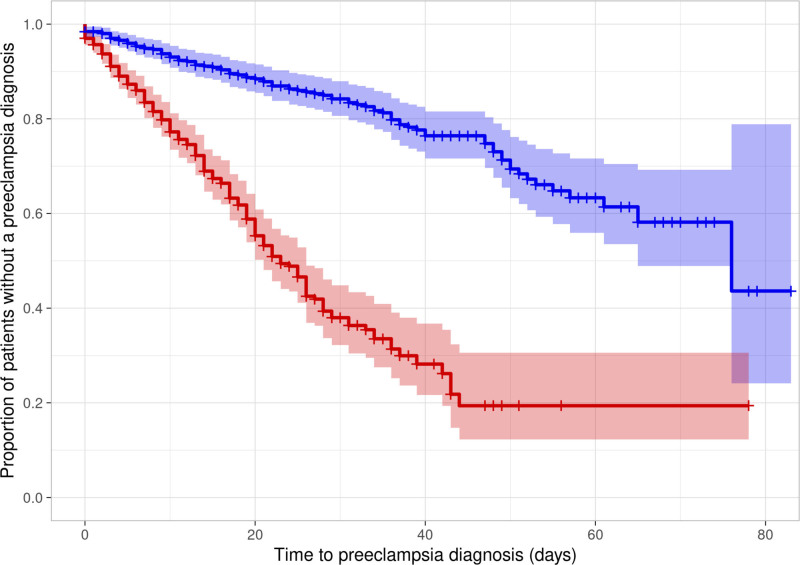
Using the final adjudicated diagnoses, the overall rate of preeclampsia in the study cohort was 30.3% (95% CI, 27.6%–33.2%), and the median (5th–95th percentiles) time to develop preeclampsia was 51 (47–65) days. The concordance in preeclampsia diagnosis between the clinical sites and the adjudication committee was 94.6%. Figure 2 represents the survival curve plotting the proportion of individuals without preeclampsia diagnosis by assay status (at-risk or negative). Overall, the median (5th–95th percentiles) time to develop preeclampsia was 23 (20–26) days for those with at-risk assay result compared with 76 (65–incalculable) days for those with negative assay test result (P<0.0001). The association between the test status and time to develop preeclampsia was assessed with the univariable Cox modeling with a univariable HR of 4.65 (95% CI, 3.60–6.00; P<0.0001). The primary model which a priori adjusted for age, gestational age at enrollment, non-Hispanic Black race/ethnicity, body mass index, diabetes, and history of preeclampsia, demonstrated the adjusted HR was 4.81 (95% CI, 3.69–6.27; P<0.0001), indicating that the assay is associated with the time to develop preeclampsia. In other models adjusting for gestational age at enrollment and body mass index with or without race, the association remained similar (Table S3). No significant difference in the model’s observed associations was demonstrated using the outcome of preeclampsia as determined by clinical centers (adjusted HR, 5.18 [95% CI, 3.97–6.75]) instead of by the adjudication committee. Among the 145 individuals in the positive preeclampsia cohort, the false negative rate of the assay was 25.5% (19.1%–33.2%).
Figure 2.
Survival curves (with 95% confidence margin) plotting the proportion of individuals without preeclampsia diagnosis vs the time to develop preeclampsia (in days), by assay status (at risk red or negative blue). Those who did not develop preeclampsia were censored at delivery or if lost to follow-up.
Table 2 summarizes the performance metrics of the assay. For the outcome of preeclampsia within 7 days of enrollment, the incidence of preeclampsia was 10.2% (95% CI, 8.5%–12.2%), and the cumulative percentage of participants who did not develop preeclampsia was significantly higher in the negative assay test compared with the at-risk test group (adjusted HR, 3.84 [95% CI, 2.42–6.09]; P<0.0001), with a sensitivity 76.4% (95% CI, 67.5%–83.5%), specificity 51.5% (48.3%–54.7%), negative likelihood ratio 0.46 (0.32–0.65), and a NPV 95.0% (95% CI, 92.8%–96.6%). For an outcome of preeclampsia at any time in pregnancy, the assay had a sensitivity of 69.4% (95% CI, 64.1%–74.3%) and a NPV 81.0% (95% CI, 77.3%–84.1%) with an accuracy of 60.4% (95% CI, 57.4%–63.4%). Sensitivity, NPV, and negative likelihood ratio slightly improved for those who ultimately delivered <37 weeks. Assay performance was overall similar when using preeclampsia as determined by the clinical centers (Table 3). Performance slightly improved for individuals enrolled between 28 and 35 weeks’ gestation (Table S4). Test accuracy for the entire cohort is reported in Table S4. Test heterogeneity for individual sites (Supplemental Materials) was demonstrated graphically using a forest plot (Figure S2) with the results indicating that test accuracy was similar across the study sites.
Table 2.
Performance (95% CI) Test Characteristics for Various Outcomes Based on Adjudication Committee Assessments and the Prespecified Cutoff
Table 3.
Performance (95% CI) Test Characteristics for Various Outcomes Based on Clinical Centers Assessments (n=1042)
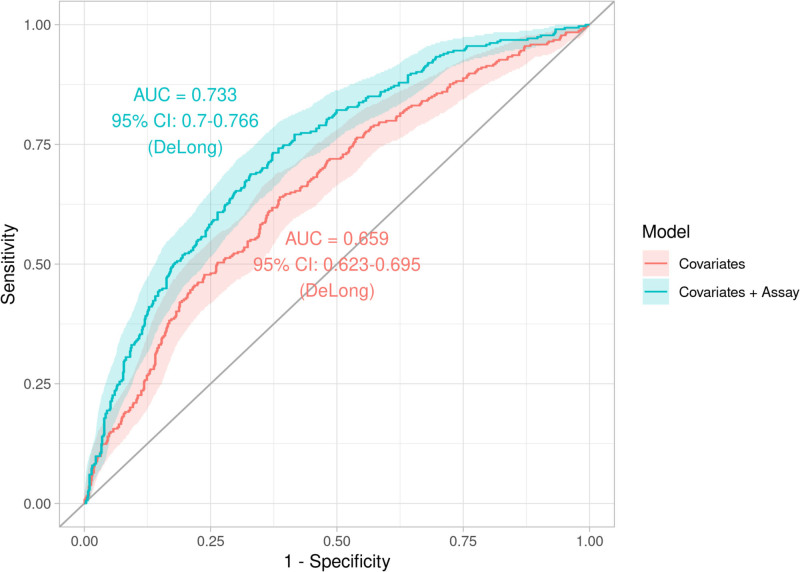
Lastly, post hoc added benefit analyses (methods in Supplemental Materials)21,22 indicated significant improvement of predictive power by including the assay in addition to clinical covariates. The log-likelihood analysis indicated accuracy improvement (P<0.0001), whereas the comparison of receiver operating curve curves indicated an approximate 11% increase in area under the curve in the primary model (area under the curve, 0.733 [95% CI, 0.700–0.766]) as compared with the model using clinical covariates (area under the curve, 0.659 [95% CI, 0.623–0.695]; P<0.0001). (Figure 3) Last, the net reclassification index calculation resulted in an 10.98% (95% CI, 3.77%–8.19%) net improvement in predictive categorization (P=0.003).
Figure 3.
Receiver-operating characteristic curves (with 95% confidence margin) of the predictive probabilities of the models with only clinical covariates and the model including clinical covariates and the assay. AUC indicates area under the curve.
Discussion
Using a large cohort of high-risk pregnant individuals presenting with preeclampsia associated signs and symptoms between 280/7 and 366/7 weeks, we validated a novel multi-analyte assay, which together with the gestational age at sample collection, was demonstrated to be associated with the time to develop preeclampsia and had a net reclassification improvement of ≈11%. The assay demonstrated a moderate sensitivity and likelihood ratios, but a high NPV for development of preeclampsia within 7 days after enrollment. Test performance was improved for individuals enrolled between 28 and 35 weeks and for those who developed preeclampsia within 7 days of enrollment and delivered <37 weeks.
The Cox regression model remained highly significant after adjusting for multiple covariates suggesting the strength and independence of the assay in its association with preeclampsia. The inclusion of race as a socially constructed variable was a priori planned in the multivariable analysis. Also, the algorithm to assess the risk status of preeclampsia, including the determination of the cutoff, was done without the inclusion of race, to avoid perpetuating health disparities, as we are cognizant of the racial disparities in preeclampsia rates and morbidities, due to social drivers, in the United States. The outcomes of 6 individuals could not be adjudicated by the adjudication committee due to missing data and inability to obtain the data from the sites. However, these 6 individuals were included in the analysis using preeclampsia diagnosis as determined by the clinical center, and there was excellent concordance (94.6%) between preeclampsia diagnosed by providers at the sites and preeclampsia confirmed by the adjudication committee. Lastly, while other studies evaluating preeclampsia biomarkers assays focused on PlGF or sFlt-1/PlGF, the current assay included eight biomarkers associated with pathophysiologic pathways of preeclampsia including angiogenic imbalance (free and total placental growth factor, soluble FMS-like tyrosine kinase-1, soluble endoglin), placental and trophoblast dysfunction (fibroblast growth factor-21 and decorin), hypoxia (kidney injury molecule 1), and inflammation and immune regulation (cluster of differentiation-274 or programmed death-ligand-1).18,19
This is one of the larger studies evaluating an assay as an aid to rule out preeclampsia in at-risk pregnant individuals presenting with signs and symptoms. In a prospective multicenter study from the United Kingdom, Chappell et al23 demonstrated that, among individuals with suspected preeclampsia enrolled between 20 and 35 weeks, a low plasma PlGF concentration (<5th percentile for gestation; <100 pg/mL) had a high sensitivity (96% [95% CI, 89%–99%]) and a high NPV (98% [95% CI, 93%–99.5%]) for the development of preeclampsia requiring delivery within 14 days. In the Preeclampsia Triage by Rapid Assay Trial, which included individuals with signs or symptoms of preeclampsia between 20 and 35 weeks’ gestation, a low PlGF (≤100 pg/mL) had have a HR of 7.17 (95% CI, 5.08–10.13) for time to delivery, 92.5% specificity and 90.3% NPV for preeclampsia and delivery within 14 days, however that study had a prevalence of preeclampsia of 71.4%.24 Our assay had excellent NPV (95.2% [95% CI, 93.0%–96.8%]), but lower sensitivity, in ruling out preeclampsia within 14 days when the individual delivered before 37 weeks. Zeisler et al25 showed that in individuals between 240/7 and 366/7 weeks’, an sFlt-1:PlGF ratio cutoff of 38, has a NPV of 99.3% (95% CI, 97.9%–99.9%) for preeclampsia within 1 week of presentation. However, the prevalence of preeclampsia in that cohort was extremely low at 2.7%, indicating a low-risk population. When setting the prevalence at 2.7% (similar to the Zeisler cohort), our assay achieved a NPV of 98.5% (95% CI, 97.0%–99.3%) for preeclampsia at any time in pregnancy (Table S5). Moreover, our assay had a NPV of 95.0% (95% CI, 92.8%–96.6%) to rule out preeclampsia within 1 week, with a prevalence of preeclampsia of 10.2%. When evaluating the entire cohort, the prevalence of preeclampsia in Zeisler’s cohort was 17.8%, lower than the rate of 30.3% in our cohort, which is more representative of a high-risk US population. However, among the 145 individuals in the positive preeclampsia cohort in our study, the false negative rate of the assay was 25.5% (19.1%–33.2%). This is higher than the other assays and should be considered in any future study evaluating the clinical utility of the assay and its incorporation in clinical practice.
With the limitations of clinical parameters in diagnosing or ruling out preeclampsia,1 a need exists to develop laboratory assays that can assist providers and complement clinical assessments. The utility of such assays has been shown in a cluster stepped-wedge trial from the United Kingdom which demonstrated that the availability of a rapid PlGF assay results reduced the time to preeclampsia confirmation in individuals presenting with suspected preeclampsia and was associated with a lower frequency of maternal adverse outcomes.26
This study was designed to collect specimens for the validation of a rule-out assay and was neither developed to be a diagnostic test or function as a clinical utility study. Thus, further studies should be conducted to determine the clinical utility of this assay vis-a-vis the current model of care in the United States especially with a net reclassification improvement of ≈11%, its ability to reduce time to diagnosis of preeclampsia, or improve clinical outcomes. In view of the results, future studies should also focus on the utility of the 7-day rule out window for the assay, and the interpretation of a negative or at risk test in high-risk individuals in the context of clinical standards of care, especially with a high false negative rate of 25.5%. In addition, we did not perform a cost analysis or evaluate whether incorporation of the assay would lead to reduced utilization of health care resources or improved outcomes.
Strengths of this study include enrollment at 20 clinical study sites of a large geographically and racially diverse cohort in the United States. Testing of the samples was performed in a blinded manner at a central laboratory ensuring rigorous quality control. Analyses were performed using the final adjudicated preeclampsia diagnosis and the clinical center’s diagnosis of preeclampsia.
Despite the large cohort, we were limited in our ability to assess meaningfully whether the assay performs differently in subgroups of the population (eg, those with medical co-morbidities). The primary outcome of the study was the time to develop preeclampsia irrespective of its severity, therefore the performance of the test as an aid in the diagnosis of preeclampsia with severe features alone is not available. However, 73% of preeclampsia cases in the cohort had severe features. In addition, the study was not powered to detect differences in rates of individual clinical outcomes such as placental abruption or postpartum hemorrhage. The rates of these and other outcomes are similar to what is reported in prior similar trials and what is expected in a high-risk cohort.23,25–27 Although study enrollment was conducted between 2016 and 2020, the specimens were banked, and the analysis was not complete until July 2021 while the laboratory was developing and optimizing the assay and laboratory processes. The study enrolled a larger number than were in the final analysis, since samples from 70 individuals from the positive preeclampsia control group were used to optimize the assay, and an additional 365 from the study cohort were later used by the sponsor in a prevalidation cohort to ensure the robustness of the algorithm in divergent populations; both originally unplanned analyses. Moreover, the incorporation of this assay in clinical flow should be developed further, and a point-of-care rapid test may enhance its clinical value. The individual analytes were not analyzed separately, and their individual performance was not available. Therefore, we are not able to compare their individual performance s vis-à-vis the composite multianalyte assay or known published literature on performance of sFlt1 and PlGF measured on other platforms.23,25,26 We cannot comment on the added performance benefit of the multianalyte assay versus its individual analytes’ performance, especially PlGF. Last, we did not evaluate the value of repeated testing and did not evaluate the performance of the assay in ruling out adverse pregnancy outcomes, as this study was underpowered for such an analysis.
Conclusions
Using a large cohort of high-risk pregnant individuals, we have confirmed that a novel multianalyte assay is associated with the time to develop preeclampsia and has a moderate sensitivity and negative likelihood ratio but high NPV when assessed as an aid to rule out preeclampsia within 7 days of enrollment.
Perspectives
Preeclampsia is a pregnancy-specific multisystem disorder that affects 3% to 8% of pregnancies and remains a leading cause of maternal and neonatal morbidity and mortality, with significant racial and ethnic disparities. Relying on clinical criteria to diagnose preeclampsia is suboptimal because they are subjective, may be subject to errors during collection, and can fluctuate during the observation period. Because the specificity of these criteria is poor, pregnant individuals in whom there is a suspicion for preeclampsia—but who may not actually have it—may be admitted for monitoring and delivered prematurely. A significant need exists to develop laboratory assays that can assist providers and complement clinical assessments.
This study validated the performance of a multi-analyte test among pregnant people presenting with signs and symptoms of preeclampsia between 280/7 and 366/7 weeks’ gestation and demonstrated that the assay was associated with the time to develop the disease and has moderate sensitivity and likelihood ratios, but a high NPV for development of preeclampsia within 7 days after enrollment.
Article Information
Acknowledgments
We thank all the individuals for participating in the study and all the research staff for working tirelessly to recruit individuals and collect outcomes (Supplemental Material). M.M. Costantine drafted the article, assisted by M. Sarno, who provided substantial scientific input to the drafting and revision of this article with regard to its scientific content and form, reviewed the statistical analysis, and approved the final article as submitted. A.T. Bombard and M. Sarno were involved in study concept origination, study design, and protocol development. P. Oberoi, M. Cooper, and A. Mazloom led the assay development and analysis teams. All the authors contributed to critical aspects of the conduct of this research including assessment of individuals recruitment; monitoring center performance; oversight of data quality; and acquisition, analysis, and interpretation of data. All the authors provided significant intellectual contribution to the drafting and revision of this article with regard to scientific content and form, and approved the final article as submitted.
Sources of Funding
The project was supported by funds from Progenity Inc, San Diego, CA, which were paid to the institutions where the study was conducted. Progenity was involved in study design and has final access to the data and takes public responsibility for its accuracy. A statistician independent from the trial sponsor also had full access to the primary data and provided data analyses. Results of the analyses were reviewed for accuracy and conformance to the prespecified statistical analysis plan by one of the authors (M. Sarno) who also had full access to all the data in the study and takes responsibility for the integrity of the conducted data analyses. Deidentified data that support the findings of this study are available from the sponsor upon reasonable request.
Disclosures
M.M. Costantine, B. Sibai, A.T. Bombard, M.R. Chavez, and M. Sarno have been paid as consultants for Progenity for work not related to the publication of this article. C. Grotegut is affiliated with Nixxi, a biotech startup that is producing risk stratification tools for pregnancy. P. Oberoi, M. Cooper, S. Lockton, and A. Mazloom are employees of Progenity. The preeclampsia test described in the present paper is the subject of published and unpublished patent applications in the U.S. and other jurisdictions, see for example, Patent Cooperation Treaty (PCT) patent publication numbers WO2019055661 and WO2021113710, which are both owned by Progenity, Inc. The other authors report no conflicts.
Supplementary Material
Nonstandard Abbreviations and Acronyms
- HR
- hazard ratio
- NPV
- negative predictive value
- PD-L1
- programmed death-ligand 1
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/HYPERTENSIONAHA.122.19038.
For Sources of Funding and Disclosures, see page 1523.
Contributor Information
Baha Sibai, Email: Baha.M.Sibai@uth.tmc.edu.
Allan T. Bombard, Email: allan@nvbpartners.com.
Mark Sarno, Email: mjsarno@visionclinicalresearch.com.
Holly West, Email: hawest@UTMB.EDU.
David M. Haas, Email: dahaas@iupui.edu.
Alan T. Tita, Email: atita@uabmc.edu.
Michael J. Paidas, Email: mjpaglia@geisinger.edu.
Erin A.S. Clark, Email: Erin.Clark@hsc.utah.edu.
Kim Boggess, Email: kboggess@med.unc.edu.
Chad Grotegut, Email: cgrotegu@wakehealth.edu.
William Grobman, Email: William.Grobman@osumc.edu.
Emily J Su, Email: emily.su@cuanschutz.edu.
Irina Burd, Email: iburd@jhmi.edu.
George Saade, Email: george.macones@austin.utexas.edu.
Martin R. Chavez, Email: Martin.Chavez@nyulangone.org.
Michael J. Paglia, Email: mjpaglia@geisinger.edu.
Audrey Merriam, Email: audrey.merriam@yale.edu.
Carlos Torres, Email: torres@rocob.com.
Mounira Habli, Email: Mounira_Habli@trihealth.com.
Georges Macones, Email: george.macones@austin.utexas.edu.
Tony Wen, Email: t.wen@ufl.edu.
James Bofill, Email: fulham1955@yahoo.com.
Anna Palatnik, Email: apalatnik@mcw.edu.
Rodney K. Edwards, Email: Rodney-Edwards@ouhsc.edu.
Sina Haeri, Email: SinaHaeri@gmail.com.
Pankaj Oberoi, Email: Poberoi306@gmail.com.
Amin Mazloom, Email: aminreza.mazloom@gmail.com.
Matthew Cooper, Email: Matthew.Cooper@progenity.com.
Steven Lockton, Email: slockton@gmail.com.
Gary D. Hankins, Email: ghankins@utmb.edu.
References
- 1. Gestational hypertension and preeclampsia: ACOG practice bulletin, number 222. Obstet Gynecol. 2020;135:e237–e260. doi: 10.1097/AOG.0000000000003891 [DOI] [PubMed] [Google Scholar]
- 2.Firoz T, Sanghvi H, Merialdi M, von Dadelszen P. Pre-eclampsia in low and middle income countries. Best Pract Res Clin Obstet Gynaecol. 2011;25:537–548. doi: 10.1016/j.bpobgyn.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 3.Say L, Chou D, Gemmill A, Tunçalp Ö, Moller AB, Daniels J, Gülmezoglu AM, Temmerman M, Alkema L. Global causes of maternal death: a WHO systematic analysis. Lancet Glob Health. 2014;2:e323–e333. doi: 10.1016/S2214-109X(14)70227-X [DOI] [PubMed] [Google Scholar]
- 4.MacKay AP, Berg CJ, Atrash HK. Pregnancy-related mortality from preeclampsia and eclampsia. Obstet Gynecol. 2001;97:533–538. doi: 10.1016/s0029-7844(00)01223-0 [DOI] [PubMed] [Google Scholar]
- 5.Hoyert DL, Miniño AM. Maternal mortality in the united states: changes in coding, publication, and data release, 2018. Natl Vital Stat Rep. 2020;69:1–18. [PubMed] [Google Scholar]
- 6.Rossen LM, Womack LS, Hoyert DL, Anderson RN, Uddin SFG. The impact of the pregnancy checkbox and misclassification on maternal mortality trends in the united states, 1999-2017. Vital Health Stat 3. 2020:1–61. [PubMed] [Google Scholar]
- 7.Hurrell A, Webster L, Chappell LC, Shennan AH. The assessment of blood pressure in pregnant women: pitfalls and novel approaches. Am J Obstet Gynecol. 2022;226(2S):S804–S818. doi: 10.1016/j.ajog.2020.10.026 [DOI] [PubMed] [Google Scholar]
- 8.Fishel Bartal M, Lindheimer MD, Sibai BM. Proteinuria during pregnancy: definition, pathophysiology, methodology, and clinical significance. Am J Obstet Gynecol. 2022;226(2S):S819–S834. doi: 10.1016/j.ajog.2020.08.108 [DOI] [PubMed] [Google Scholar]
- 9.Sperling JD, Dahlke JD, Huber WJ, Sibai BM. The role of headache in the classification and management of hypertensive disorders in pregnancy. Obstet Gynecol. 2015;126:297–302. doi: 10.1097/AOG.0000000000000966 [DOI] [PubMed] [Google Scholar]
- 10.Ives CW, Sinkey R, Rajapreyar I, Tita ATN, Oparil S. Preeclampsia-pathophysiology and clinical presentations: JACC state-of-the-art review. J Am Coll Cardiol. 2020;76:1690–1702. doi: 10.1016/j.jacc.2020.08.014 [DOI] [PubMed] [Google Scholar]
- 11.Smith DD, Costantine MM. The role of statins in the prevention of preeclampsia. Am J Obstet Gynecol. 2022;226(2S):S1171–S1181. doi: 10.1016/j.ajog.2020.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redman CW, Sargent IL. Latest advances in understanding preeclampsia. Science. 2005;308:1592–1594. doi: 10.1126/science.1111726 [DOI] [PubMed] [Google Scholar]
- 13.Staff AC, Fjeldstad HE, Fosheim IK, Moe K, Turowski G, Johnsen GM, Alnaes-Katjavivi P, Sugulle M. Failure of physiological transformation and spiral artery atherosis: their roles in preeclampsia. Am J Obstet Gynecol. 2022;226(2S):S895–S906. doi: 10.1016/j.ajog.2020.09.026 [DOI] [PubMed] [Google Scholar]
- 14.Rana S, Burke SD, Karumanchi SA. Imbalances in circulating angiogenic factors in the pathophysiology of preeclampsia and related disorders. Am J Obstet Gynecol. 2022;226(2S):S1019–S1034. doi: 10.1016/j.ajog.2020.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branch DW, Mitchell MD, Miller E, Palinski W, Witztum JL. Pre-eclampsia and serum antibodies to oxidised low-density lipoprotein. Lancet. 1994;343:645–646. doi: 10.1016/s0140-6736(94)92639-5 [DOI] [PubMed] [Google Scholar]
- 16.Rana S, Lemoine E, Granger JP, Karumanchi SA. Preeclampsia: pathophysiology, challenges, and perspectives. Circ Res. 2019;124:1094–1112. doi: 10.1161/CIRCRESAHA.118.313276 [DOI] [PubMed] [Google Scholar]
- 17.Sargent IL, Germain SJ, Sacks GP, Kumar S, Redman CW. Trophoblast deportation and the maternal inflammatory response in pre-eclampsia. J Reprod Immunol. 2003;59:153–160. doi: 10.1016/s0165-0378(03)00044-5 [DOI] [PubMed] [Google Scholar]
- 18.Cooper M, Mazloom A, Obrochta C, Wapner R, Rosen T, Bombard A. Performance of a novel multi-biomarker rule out preeclampsia test: a prospective verification study. Am Coll Obstet Gynecol Annual Virtual Meeting, April 30-May 2, 2021; Poster number 999251. [Google Scholar]
- 19.Hendershot JM, Abbasi M, Fortier L, Benn M, Giacobone C, Del Mastro R, Bahrami-Samani E, Mazloom AR, Oberoi P. Analytical validation of a novel panel of biomarkers for a test for preeclampsia. J Pharm Biomed Anal. 2022;214:114729. doi: 10.1016/j.jpba.2022.114729 [DOI] [PubMed] [Google Scholar]
- 20.Hypertension in pregnancy. Report of the american college of obstetricians and gynecologists’ task force on hypertension in pregnancy. Obstet Gynecol. 2013;122:1122–1131. [DOI] [PubMed] [Google Scholar]
- 21.Moons KG, de Groot JA, Linnet K, Reitsma JB, Bossuyt PM. Quantifying the added value of a diagnostic test or marker. Clin Chem. 2012;58:1408–1417. doi: 10.1373/clinchem.2012.182550 [DOI] [PubMed] [Google Scholar]
- 22.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 23.Chappell LC, Duckworth S, Seed PT, Griffin M, Myers J, Mackillop L, Simpson N, Waugh J, Anumba D, Kenny LC, et al. Diagnostic accuracy of placental growth factor in women with suspected preeclampsia: a prospective multicenter study. Circulation. 2013;128:2121–2131. doi: 10.1161/CIRCULATIONAHA.113.003215 [DOI] [PubMed] [Google Scholar]
- 24.Barton JR, Woelkers DA, Newman RB, Combs CA, How HY, Boggess KA, Martin JN, Jr, Kupfer K, Sibai BM; PETRA (Preeclampsia Triage by Rapid Assay) Trial. Placental growth factor predicts time to delivery in women with signs or symptoms of early preterm preeclampsia: a prospective multicenter study. Am J Obstet Gynecol. 2020;222:259.e1–259.e11. doi: 10.1016/j.ajog.2019.09.003 [DOI] [PubMed] [Google Scholar]
- 25.Zeisler H, Llurba E, Chantraine F, Vatish M, Staff AC, Sennström M, Olovsson M, Brennecke SP, Stepan H, Allegranza D, Dilba P, Schoedl M, Hund M, Verlohren S. Predictive value of the sFlt-1:PlGF ratio in women with suspected preeclampsia. N Engl J Med. 2016;374:13–22. doi: 10.1056/NEJMoa1414838 [DOI] [PubMed] [Google Scholar]
- 26.Duhig KE, Myers J, Seed PT, Sparkes J, Lowe J, Hunter RM, Shennan AH, Chappell LC; PARROT trial group. Placental growth factor testing to assess women with suspected pre-eclampsia: a multicentre, pragmatic, stepped-wedge cluster-randomised controlled trial. Lancet. 2019;393:1807–1818. doi: 10.1016/S0140-6736(18)33212-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koopmans CM, Bijlenga D, Groen H, Vijgen SM, Aarnoudse JG, Bekedam DJ, van den Berg PP, de Boer K, Burggraaff JM, Bloemenkamp KW, et al. ; HYPITAT study group. Induction of labour versus expectant monitoring for gestational hypertension or mild pre-eclampsia after 36 weeks’ gestation (HYPITAT): a multicentre, open-label randomised controlled trial. Lancet. 2009;374:979–988. doi: 10.1016/S0140-6736(09)60736-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.