Abstract
Plasmid pGT232 (5.1 kb), an indigenous plasmid of Lactobacillus reuteri 100-23, was determined, on the basis of nucleotide and deduced protein sequence data, to belong to the pC194-pUB110 family of plasmids that replicate via the rolling-circle mechanism. The minimal replicon of pGT232 was located on a 1.7-kb sequence consisting of a double-strand origin of replication and a gene encoding the replication initiation protein, repA. An erythromycin-selectable recombinant plasmid containing this minimal replicon was stably maintained (>97% erythromycin-resistant cells) without antibiotic selection in an L. reuteri population under laboratory growth conditions but was poorly maintained (<33% resistant cells) in the L. reuteri population inhabiting the murine gastrointestinal tract. Stable maintenance (>90% resistant cells) of pGT232-derived plasmids in the lactobacillus population in vivo required an additional 1.0-kb sequence which contained a putative single-strand replication origin (SSO). The SSO of pGT232 is believed to be novel and functions in an orientation-specific manner.
Lactobacilli are common inhabitants of the gastrointestinal tract of mice, rats, pigs, fowl, and humans (30). They are utilized in the preparation of probiotics that are administered as dietary supplements to farm animals (feed additives) and humans (Acidophilus-type milk products). Consumption of the probiotic product is believed to influence the normal microflora of the digestive tract, conferring microecological, physiological, and immunological benefits on the consumer (10). However, improved probiotics of reliable efficacy and scientific validity are required (29). Genetic modification of intestinal strains of lactobacilli so that they produce novel products (e.g., immunogenic peptides) when inhabiting the gastrointestinal tract is one such approach (29). An early goal in these studies is to derive plasmid-based expression vectors for use with gastrointestinal species of lactobacilli. The construction of recombinant plasmids based on such vectors could be used in preliminary studies to investigate the efficacy of genetically modified lactobacilli in influencing the mammalian host. Prerequisite for success in these studies is the derivation of small multipurpose plasmid vectors that will be maintained, both structurally and segregationally, by the lactobacillus population when inhabiting the relatively harsh and highly competitive environment of the gastrointestinal tract. It is essential, therefore, to study the biology of lactobacillus plasmids with respect to their replication and maintenance both in vitro and in vivo.
We have chosen the mouse as the experimental animal model for preliminary studies and specifically a unique colony of mice, termed reconstituted lactobacillus-free (RLF) mice (32). These animals, maintained in isolators by gnotobiotic technology, harbor a gastrointestinal microflora equivalent to that of conventional mice (32). Unlike conventional mice, they do not harbor lactobacilli as gastrointestinal inhabitants. Single strains of Lactobacillus spp. can therefore be studied in relation to the microecology of the gastrointestinal tract by using these mice (30).
We describe here details of an indigenous plasmid, pGT232, detected in Lactobacillus reuteri 100-23 (31). This strain originated in the digestive tract of a rat (31), colonizes the murine digestive tract at a high population level, and is amenable to genetic modification by electrotransformation (18). We provide evidence that the double-strand replication origin (DSO) and the gene (repA) encoding the replication protein are sufficient for stable maintenance of a pGT232-based recombinant plasmid in laboratory culture (in vitro) but that the presence of the putative single-strand replication origin (SSO) of pGT232 is essential for stable maintenance of the recombinant plasmid in lactobacilli growing in the gastrointestinal tract of the murine host (in vivo).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Details of the bacterial strains and plasmids used in this study are given in Table 1. Escherichia coli DH5αF′ was used in all cloning experiments and was cultured at 37°C in Luria broth (Difco Laboratories, Detroit, Mich.). Lactobacillus strains were cultured anaerobically at 37°C in Lactobacilli MRS medium (Difco). Lactococcus lactis was cultivated in GM17 broth (M17 broth base [Difco] supplemented with 0.5% [wt/vol] glucose) at 30°C aerobically. Solidified media contained 1.5% (wt/vol) agar. When required, ampicillin was added to the culture media at a concentration of 50 μg/ml, erythromycin was added at 500 μg/ml (E. coli) or 50 μg/ml (lactobacilli), and chloramphenicol was added at 5 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or referencea |
|---|---|---|
| Strains | ||
| Escherichia coli DH5αF′ | Cloning host; F′ endA1 hsdR17 (rK− mK+) supE44 thi-1 λ− recA1 gyrA96 relA1 deoR Δ(lacZYA-argF)U169 φ80dlacZΔM15 | 12 |
| L. reuteri DSM 20016 | Type strain of L. reuteri, plasmid free; electrotransformation host | DSM |
| L. reuteri 100-23 | Rodent gastrointestinal isolate | 37 |
| L. reuteri 100-23C | Plasmid-free derivative of 100-23; electrotransformation host | 18 |
| L. reuteri NH103 | 100-23C harboring pNCKH103 | This study |
| L. reuteri NH104 | 100-23C harboring pNCKH104 | This study |
| L. reuteri NH105 | 100-23C harboring pNCKH105 | This study |
| L. reuteri NH107 | 100-23C harboring pNCKH107 | This study |
| L. reuteri NH108 | 100-23C harboring pNCKH108 | This study |
| L. reuteri WX1 | 100-23C harboring pWX1 | This study |
| L. acidophilus ATCC 4356 | Electrotransformation host | ATCC |
| L. fermentum ATCC 14931 | Electrotransformation host | ATCC |
| L. gasseri ATCC 33323 | Electrotransformation host | ATCC |
| L. helveticus ATCC 15009 | Electrotransformation host | ATCC |
| L. salivarius subsp. salicinius ATCC 11742 | Electrotransformation host | ATCC |
| L. lactis MG1363 | Electrotransformation host | 11 |
| L. lactis MG1363(pCP12) | MG1363 harboring pCP12 | 20 |
| Plasmids | ||
| pGT232 | Indigenous cryptic plasmid from L. reuteri 100-23, 5.1 kb | 31 |
| pUC19 | Cloning vector, Apr, 2.7 kb | 39 |
| pUC29 | Cloning vector, Apr, 2.7 kb | 2 |
| pBluescript II SK | Cloning vector, Apr, 3.0 kb | Stratagene |
| pTRK95 | Source of erythromycin resistance (ermGT) gene; Apr Cmr Emr, 7.5 kb | 33 |
| pNCKH100 | Derivative of pUC29, bla (Apr) gene replaced with ermGT, Emr, 4.0 kb | This study |
| pNCKH101 | pNCKH100 containing the 3.2-kb BclI fragment from pGT232 cloned into the BamHI site, Emr, 7.2 kb | This study |
| pNCKH102 | pNCKH100 containing the 3.1-kb XbaI-NcoI fragment from pGT232, Emr, 7.1 kb | This study |
| pNCKH103 | pNCKH100 containing the 2.7-kb SalI-NcoI fragment from pGT232, Emr, 6.7 kb | This study |
| pNCKH103ΔBam | Derivative of pNCKH103; internal BamHI site within the repA gene eliminated by digestion and subsequent end filling with Klenow fragment, Emr, 6.7 kb | This study |
| pNCKH104 | pNCKH100 containing the 1.7-kb PvuII-NcoI fragment from pGT232 cloned into the SmaI-NcoI sites, Emr, 5.7 kb | This study |
| pNCKH105 | pNCKH103 derivative containing the 1.9-kb AccI-NcoI fragment from pGT232, Emr, 5.9 kb | This study |
| pNCKH106 | pNCKH100 containing the 1.7-kb BglI-NcoI fragment from pGT232 cloned into the SmaI-NcoI sites, Emr, 5.7 kb | This study |
| pNCKH107 | pNCKH105 containing the 0.7-kb SalI-EcoRI fragment from pGT232 (pBS-SalEco) cloned into the SalI-PstI sites, Emr, 6.6 kb | This study |
| pNCKH108 | pNCKH106 containing the 0.7-kb SalI-EcoRI fragment from pGT232 (pBS-SalEco) cloned into the SalI-PstI sites, Emr, 6.4 kb | This study |
| pWX1 | pNCKH104 containing the 1.0-kb SalI-PvuII putative SSO fragment from pNCKH103 cloned into the MluI-SalI sites, Emr, 6.7 kb | This study |
| pFX3 | Broad-host-range cloning vector, Cmr, 4.3 kb | 38 |
| pNH-Sal | pUC29 containing pGT232 cloned into the SalI site, Apr, 7.8 kb | This study |
| pBS-SalEco | pBluescript containing the 0.7-kb SalI-EcoRI fragment from pGT232 (pNH-Sal), Apr, 3.7 kb | This study |
| pWX2 | pNH-Sal derivative containing the 1.4-kb ClaI-Sau3AI cat-194 (Cmr) fragment from pFX3, Apr Cmr, 9.2 kb | This study |
| pWC1 | Indigenous pC194-pUB110-type rolling-circle plasmid from L. lactis subsp. cremoris 2204, 2.8 kb | 20 |
| pCP12 | Derivative of pWC1 containing the cat-194 gene, Cmr, 3.9 kb | 20 |
DSM, Deustche Sammlung von Mikroorganismen und Zellkulturen; ATCC, American Type Culture Collection.
Genetic techniques.
DNA manipulation methods, agarose gel electrophoresis, and plasmid detection and purification techniques for E. coli were used as described by Sambrook et al. (24). Restriction endonucleases, Klenow DNA polymerase, and T4 DNA ligase (Boehringer Mannheim, Mannheim, Germany) were used according to the supplier’s instructions. Small-scale plasmid extracts were prepared from lactobacilli as described by Tannock et al. (34), and large-scale purifications were done by cesium chloride-ethidium bromide density gradient centrifugation (18). Recombinant DNA molecules were introduced into E. coli, lactobacilli, and L. lactis by electrotransformation (6, 18, 36).
Nucleotide sequencing.
A restriction map of plasmid pGT232 was prepared, and a library of fragments, produced by digestion with various combinations of enzymes, was generated in pUC19 or pUC29. Inserts were sequenced by the dideoxy chain-termination method (25) with the appropriate sequencing kits recommended for use in combination with either an Applied Biosystems 373A (Applied Biosystems, Foster City, Calif.) or a Li-Cor DNA4200 (Li-Cor Inc., Lincoln, Nebr.) automated DNA sequencing system. Computer-assisted assembly and analysis of the nucleotide sequence were accomplished with GeneJockey (BioSoft, Cambridge, United Kingdom) and the GCG version 8.0 software package (Genetics Computer Group, Madison, Wis.). Computer-assisted alignment of the Rep protein sequences was carried out with the MegAlign module of the Lasergene software package (DNAStar Inc., Madison, Wisc.).
Construction of plasmids pNCKH100 and pWX2.
The replicon probe vector pNCKH100 was constructed by replacing the β-lactamase (Apr) gene of pUC29 (removed by digestion with DraI and SspI) with a 2.2-kb end-filled BamHI fragment from pTRK95 (33) containing ermGT (erythromycin resistance determinant) of pGT633. Plasmid pWX2, which was used in the single-stranded DNA (ssDNA) experiments (described below), was derived by insertion of a 1.4-kb ClaI-Sau3AI fragment from pFX3 (38) containing the chloramphenicol resistance (cat-194) gene into the EcoRV site of pNH-Sal, one of the clones used in the determination of the pGT232 nucleotide sequence.
In vitro and in vivo stability of the pGT232 replicon in L. reuteri populations.
To determine the in vitro stability of the pGT232 replicon, L. reuteri 100-23C transformants harboring pNCKH103 or pNCKH104 were subcultured daily (1/300 volume for each subculture) for 7 days in antibiotic-free medium, estimated to be approximately 60 generations of growth. Serial 10-fold dilutions were prepared from the cultures at intervals and diluted, and 0.1-ml aliquots were spread on Lactobacilli MRS agar plates. After 48 h of incubation, colonies (100 to 300 per plate) were replica plated onto MRS agar and MRS agar containing 50 μg of erythromycin per ml. This permitted the proportion of erythromycin-resistant lactobacilli relative to the total population to be determined.
L. reuteri 100-23C transformants containing either pNCKH103 or pNCKH104 were used to inoculate mice belonging to a unique colony of lactobacillus-free (RLF) mice (32). Groups of RLF mice were inoculated with a culture of the appropriate recombinant lactobacillus as described previously (18). The animals were killed 14 days after inoculation, and lactobacilli were cultured quantitatively, by previously described methods (18), from the forestomach and cecum of each mouse. Comparison of lactobacillus populations enumerated on selective medium with or without erythromycin showed the proportion of CFU in the population that harbored the pGT232-based recombinant plasmid.
Examination of plasmid extracts for the presence of ssDNA plasmid intermediates.
Plasmid DNA was extracted from lactobacilli and lactococci by the method of Tannock et al. (34) with the following modifications: (i) cells were washed and resuspended in 10 mM Tris-HCl (pH 6.9), (ii) the lysis solution consisted of a mixture of lysozyme (6 mg/ml) and mutanolysin (40 μg/ml), and (iii) sodium hydroxide was omitted from the procedure. Plasmid DNA prepared after lysis of bacterial cells under neutral pH conditions has been reported to be more suitable for detection of ssDNA intermediates (35). The positive control used in these experiments was plasmid pCP12, a derivative of pWC1, a pC194-pUB110-type plasmid isolated from Lactococcus lactis subsp. cremoris (20). In some experiments, plasmid DNA extracts were adjusted to pH 4.5 and incubated with 10 U of S1 nuclease (Boehringer Mannheim) for 15 min at 37°C. DNA samples were separated by agarose gel electrophoresis and subsequently transferred onto nylon membranes (Hybond-N+; Amersham) by vacuum blotting. Hybridization experiments were carried out as described by Sambrook et al. (24) with radioactively labelled fragments of pGT232 or pWC1 plasmid DNA as probes.
Nucleotide sequence accession number.
The complete nucleotide sequence of pGT232 has been deposited in the GenBank database under accession no. U21859.
RESULTS
Nucleotide sequence analysis.
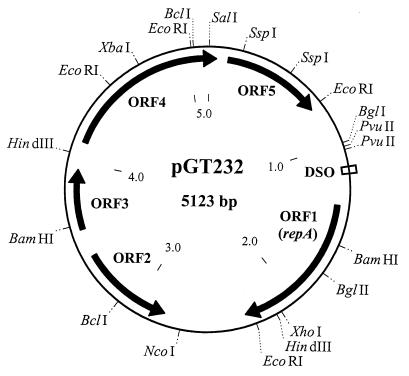
A restriction map of plasmid pGT232 is shown in Fig. 1. The complete nucleotide sequence of pGT232 totalled 5,123 bp and possessed a mol% G+C content of 37%, which was slightly lower than that reported for the L. reuteri chromosome (40 to 42% [16]). Numbering of the sequence starts from the first nucleotide of the unique SalI site. A total of five open reading frames (ORFs; designated ORF1 to ORF5) that were capable of encoding polypeptides greater than 100 amino acids (aa) in length were located (Fig. 1). The location and sizes of the encoded polypeptides are as follows: ORF1 (nucleotides [nt] 1370 to 2308, 312 aa), ORF2 (nt 3405 to 2818 [complementary strand], 195 aa), ORF3 (nt 3588 to 3962, 124 aa), ORF4 (nt 4132 to 44 [spans the SalI site], 343 aa), and ORF5 (nt 121 to 745, 208 aa).
FIG. 1.
Restriction map of pGT232. The restriction sites used for subcloning of fragments for nucleotide sequencing purposes are displayed. The five ORFs described in the text are also shown. The nucleotide sequence coordinates and the predicted size of the encoded polypeptide for each ORF are provided in the text. repA, gene encoding the replication initiation protein.
These ORFs and the complete pGT232 DNA sequence were compared at the level of the nucleotide and deduced amino acid sequences to the GenBank and Swiss-Prot/PIR databases by using the BLAST algorithm (1). The highest alignment scores were obtained with plasmids detected in Lactobacillus species: pLEM5 from L. fermentum (8), pC30il and pLP1 from L. plantarum (3, 28), pLAB1000 from L. hilgardii (15), p353-2 from L. pentosus (17), and pLC88 from L. casei (GenBank accession no. U31333). These plasmids all belong to the recently defined Lactobacillus subgroup of the pC194-pUB110 family of plasmids that replicate via a rolling-circle mechanism (27).
Two distinct regions of homology, both within a 1.4-kb segment of pGT232 DNA, were observed (Fig. 2). The first region (nt 1086 to 1130) had 77 to 80% identity with the DSO and contained a highly conserved 12-bp sequence, 5′-CTTATCTTGATA-3′ (nt 1092 to 1193), which is present in virtually all members of the pC194-pUB110 family (27). The replication protein (Rep) is believed to initiate replication of the plasmid DNA by cleaving the bond between the guanine and adenine residues (Fig. 2) (27).
FIG. 2.
Nucleotide sequence of the functional regions of pGT232. The numbering of the nucleotide sequence (on the left) starts from the unique SalI site. The deduced amino acid sequence (numbered on the right) of the putative replication initiation (Rep) protein encoded by ORF1 (repA) is shown beneath the nucleotide sequence in single-letter code. The DSO (nt 1083 to 1093) is underlined. The putative ribosome-binding site (AGAGG) and the start codon of repA are denoted by the double underlining and superscription, respectively. Opposing arrows delineate IR sequences (IR1 to IR6) capable of forming imperfect stem-loop structures (see text). Filled circles indicate the start and end points of each IR, and the dots indicate mismatches. The BamHI site that was modified to give rise to a mutated repA is in bold italics. The relevant restriction endonuclease sites used in the construction of plasmids pNCKH103 through pNCKH108 are also indicated. The conserved amino acid motifs in the Rep protein corresponding to the proposed divalent ion-binding site (HWHVHILLMV, residues 162 to 171) and the active site (EVSKYQVKDYD, residues 223 to 233) are indicated by the dotted underlining. The putative rho-independent transcription terminator of repA is indicated by the GC-rich IR sequence (T) followed by the run of four thymine residues.
The second region of homology (nt 1408 to 2210) corresponded to the region encompassing ORF1 and displayed significant similarity (42 to 63%) to the sequences of pC194-pUB110-type Rep proteins characterized from the above-mentioned Lactobacillus species. Furthermore, the presence of two motifs (EVSKYDVKDYD and HQHVHILLM [Fig. 2]) which conformed to the consensus sequence of the highly conserved active site region of pC194-type Rep proteins (19, 27) and a proposed divalent metal ion binding region (14), respectively, was noted. A putative ribosome-binding site, AGAGG (nt 1357 to 1361), was located 8 bp upstream of the potential start codon (ATG) of ORF1 (Fig. 2). It was expected that a promoter-like sequence would exist upstream of the ribosome-binding site of ORF1. However, examination of the sequence data did not reveal sequences which conformed to consensus Lactobacillus promoters (22). An inverted repeat (IR) sequence (nt 2579 to 2608) which resembles a rho-independent transcription terminator was present downstream of ORF1 (Fig. 2). ORF1 has been subsequently designated repA. The organization of these two replication-associated elements (DSO and repA) was similar to that of other rolling-circle plasmids (13, 27).
The repA gene is essential for plasmid replication, and Rep protein activity can be supplied in trans.
To confirm experimentally that repA was solely responsible for the replication of pGT232, a frameshift mutation was introduced into the BamHI site located within the repA gene of pNCKH103 by digestion and subsequent end filling. Plasmid pNCKH103 consisted of pNCKH100 (see Materials and Methods) containing the 2.7-kb SalI-NcoI fragment of pGT232. The loss of the BamHI site was confirmed by restriction endonuclease analysis. The resulting plasmid, pNCKH103ΔBam, predicted to lead to production of a truncated RepA polypeptide, was used to electrotransform L. reuteri DSM 20016 and 100-23C. Erythromycin-resistant transformants were not obtained, confirming the essential role of repA in plasmid replication.
Furthermore, when wild-type L. reuteri 100-23 was transformed with pNCKH103ΔBam, Emr colonies were obtained. Both the native and recombinant forms of pGT232 were present in plasmid extracts of these transformants, indicating that RepA function was provided in trans by pGT232 for replication of pNCKH103ΔBam. In contrast, when the replication-proficient plasmid pNCKH103 was introduced into L. reuteri 100-23, only the recombinant plasmid was observed in plasmid extracts. This can be explained by the incompatibility between the identical replication functions of native pGT232 and pNCKH103 and the selective advantage of the latter plasmid in the presence of antibiotic selection. This “curing” phenomenon was consistent with those described by Bringel et al. (4) and by Posno et al. (21) with derivatives of pLP1 and p353-2, respectively.
Location of the pGT232 minimal replicon.
A replicon probe vector, pNCKH100 (see Materials and Methods), was used to determine the minimal replicon of pGT232, usually defined as the smallest fragment of DNA permitting autonomous replication of the plasmid. In this study, we have extended this definition to include stable maintenance of the plasmid replicon under in vitro and in vivo growth conditions. The naturally plasmid-free strain L. reuteri DSM 20016 was used in preliminary electrotransformation experiments with plasmids pNCKH101, pNCKH102, pNCKH103, and pNCKH104. Erythromycin-resistant transformants were obtained with all four plasmids. The minimal replicon of pGT232 appeared to be contained within the 1.7-kb PvuII-NcoI fragment in pNCKH104.
Plasmids pNCKH103 (containing the 2.7-kb SalI-NcoI fragment) and pNCKH104 were subsequently used to electrotransform L. reuteri 100-23C (Table 2). Selected L. reuteri 100-23C transformants harboring pNCKH103 and pNCKH104, designated L. reuteri NH103 and NH104, respectively, were used in subsequent experiments to determine the stability of the recombinant plasmids in the L. reuteri host growing under laboratory conditions or when inhabiting the gastrointestinal tract of RLF mice.
TABLE 2.
In vitro and in vivo maintenance of pGT232-based recombinant plasmids in L. reuteri 100-23C
| Strain and expt (nf) | Plasmid maintenance
|
||||||
|---|---|---|---|---|---|---|---|
| In vitro for no. of daysbc
|
In vivoa
|
||||||
| 1 | 4 | 7 | Forestomach
|
Cecum
|
|||
| Lactobacillus populationd | % Emre | Lactobacillus populationd | % Emre | ||||
| NH103 | |||||||
| I (5) | 100 | 99.3 | 98.4 | 8.7 (0.1) | 90.8 (5.9) | 7.8 (0.1) | 98.4 (1.4) |
| II (8) | 100 | 99.2 | 99.3 | 8.8 (0.1) | 95.3 (2.5) | 8.1 (0.1) | 90.3 (2.8) |
| NH104 | |||||||
| I (6) | 100 | 100 | 100 | 8.9 (0.1) | 36.2 (9.5) | 8.4 (0.03) | 30.7 (7.0) |
| II (7) | 97.7 | 99.2 | 98.1 | 8.7 (0.1) | 30.0 (9.3) | 7.8 (0.3) | 32.3 (9.9) |
| NH105 | |||||||
| I (5) | 93.2 | 100 | 100 | 8.9 (0.1) | 79.8 (2.4) | 8.3 (0.1) | 93.2 (4.2) |
| NH107 | |||||||
| I (5) | 100 | 100 | 100 | 8.7 (0.1) | 77.7 (6.1) | 8.1 (0.2) | 68.0 (10.5) |
| NH108 | |||||||
| I (5) | 94.8 | 98.8 | 100 | 8.9 (0.1) | 65.1 (10.6) | 8.4 (0.4) | 44.7 (13.5) |
| II (5) | NDg | ND | ND | 7.8 (0.4) | 62.6 (11.1) | 7.8 (0.2) | 61.1 (5.3) |
| WX1 | |||||||
| I (8) | 100 | 96.7 | 91.7 | 8.9 (0.1) | 30.1 (7.9) | 8.3 (0.2) | 25.7 (6.3) |
Strains were associated with mice for a period of 14 days.
Number of days cultured in the absence of antibiotic selection (daily transfers [1/300 volume] to fresh medium were made).
Expressed as a percentage of Emr colonies over the total number of colonies.
Expressed as mean log10 CFU per gram of organ (standard error of the mean).
Mean percentage of Emr colonies recovered over the total number of colonies (standard error of the mean).
Number of mice in the experiment; applicable only to in vivo plasmid maintenance experiments.
ND, not determined.
In vitro and in vivo maintenance of the pGT232 replicon in lactobacillus populations.
L. reuteri NH104 was cultured in the absence of erythromycin in Lactobacilli MRS broth for 7 days (daily subculture to fresh medium) without significant loss of the plasmid (Table 2). In contrast, the plasmid was poorly maintained in lactobacilli when these were inhabiting the gastrointestinal tract of mice (Table 2), as shown by the low percentage of cells in the population harboring pNCKH104 (average, <33%). Marked variation in the proportion of the lactobacillus population harboring pNCKH104 was also observed between mice. The 1.7-kb PvuII-NcoI fragment therefore did not, by our definition, constitute the minimal replicon of pGT232.
Conversely, pNCKH103, which contained an extra 1.0 kb of pGT232 DNA compared with pNCKH104, was maintained to the same extent (>90% of cells in the population harbored the plasmid) both in vitro and in vivo in the lactobacillus populations (Table 2). The proportion of the lactobacillus population harboring pNCKH103 was similar in each mouse. Isolates of lactobacilli characterized after the in vitro and in vivo experiments had plasmid profiles identical to that of the stock culture of L. reuteri NH103. It thus appeared that the 2.7-kb SalI-NcoI fragment represented the true minimal replicon of pGT232.
Examination of the nucleotide sequence of the 1.0-kb SalI-PvuII fragment revealed a series of six imperfect IR sequences (designated IR1 to IR6) capable of forming stem-loop structures (Fig. 2). The calculated free energy (ΔG°) values (in kilocalories per mole) for IR1 to IR6 are −52.4, −5.8, −24.4, −2.0, −14.6, and −11.3, respectively. Stem-loop structures are characteristic of a third replication-associated element present on rolling-circle plasmids, the SSO. SSO elements serve to facilitate the conversion of single-stranded plasmid DNA intermediates (formed during the rolling-circle replicative cycle) to their dsDNA forms and may play an important role in segregational stability of the plasmids (22).
Further delineation of the pGT232 SSO.
In order to determine which of the potential IRs (or combination of IRs) was responsible for SSO function, and hence in vivo segregational stability, an approach similar to that taken by Seegers et al. (26), in which restriction endonucleases were utilized to selectively remove IRs in the rolling-circle plasmid pWVO1, was undertaken. A set of plasmids lacking either IR1 (pNCKH105 and pNCKH107) or IR1 to IR3 (pNCKH106 and pNCKH108) was constructed (Table 1). Plasmids pNCKH107 and pNCKH108 were derived from pNCKH105 and pNCKH106, respectively, to take into account the possibility that the region of DNA upstream of IR1 (the 0.7-kb SalI-EcoRI fragment [Fig. 2]), which did not appear to contain any potential stem-loop structures, might contribute to SSO function in pGT232. These plasmids were subsequently used to electrotransform L. reuteri 100-23C. For unknown reasons, Emr transformants were not obtained with pNCKH106. Selected Emr transformants harboring pNCKH105, pNCKH107, and pNCKH108 were designated L. reuteri NH105, NH107, and NH108, respectively, and were used for in vitro and in vivo experiments. More than 94% of the Lactobacillus population retained the respective recombinant plasmids under in vitro culture conditions (Table 2), which was consistent with experiments involving pNCKH103 and pNCKH104. At least 70% of lactobacillus cells recovered from mice still harbored the recombinant plasmids lacking IR1 (pNCKH105 and pNCKH107 [Table 2]). The difference in the proportion of lactobacilli harboring the two plasmid types was not significant. On the other hand, a smaller proportion (61 to 65%) of cells harbored pNCKH108, which lacked IR1 to IR3 (Table 2).
Taken together, these results suggest that (i) SSO function was localized within the 0.3-kb EcoRI-PvuII portion containing IR1 to IR4 (Fig. 2); (ii) full segregational stability was not solely dependent on a single IR (for example, IR1) but rather on the presence of at least IR1 to IR4, as observed with L. reuteri carrying pNCKH103; and (iii) the regions of DNA upstream of IR1 did not appear to contribute to segregational stability, as evidenced by the experiments involving L. reuteri NH105 and NH107.
The pGT232 SSO is orientation specific.
It has been firmly established that SSO sequences of rolling-circle plasmids must be located on the same DNA strand as the DSO and Rep elements and that the SSOs function only when they are in a particular orientation, i.e., SSOs are strand and orientation specific (13, 22). One way of determining whether the 1.0-kb SalI-PvuII fragment contained a putative SSO was to construct a derivative of pNCKH103 in which the orientation of the SalI-PvuII fragment was reversed. This was accomplished by cloning the SalI-PvuII fragment from pNCKH103 into the MluI (end filled with Klenow fragment) and SalI sites of pNCKH104. This resulted in plasmid pWX1, which was essentially pNCKH103 with the exception that the 1.0-kb SalI-PvuII fragment was now in the reverse orientation. This “flipped SSO” in pWX1 was confirmed by nucleotide sequencing. Plasmid pWX1 was introduced into L. reuteri 100-23C by electrotransformation, and one of the transformants, designated L. reuteri WX1, was subjected to the same in vitro and in vivo experiments as carried out with strains NH103 and NH104. As shown in Table 2, at least 90% of the population were still Emr after daily subculture for 7 days in antibiotic-free medium. However, only 25 to 30% of the Lactobacillus population was Emr after L. reuteri WX1 had been associated with RLF mice for 14 days (Table 2), an observation consistent with those obtained with L. reuteri NH104.
Together, these results demonstrate that the true minimal replicon of pGT232 was the 2.7-kb SalI-NcoI fragment and that the putative SSO of pGT232 was localized to a 0.3-kb EcoRI-PvuII region which contained a series of IRs capable of forming stem-loop structures.
Detection of ssDNA intermediates.
One of the characteristic features of rolling-circle replication is the generation of ssDNA plasmid intermediates. These are usually observed in agarose gels as fast-migrating bands compared to the corresponding double-stranded forms of the plasmid. Upon treatment with S1 nuclease, which degrades single-stranded nucleic acid forms, these ssDNA bands are no longer visible. Detection of these ssDNA intermediates provides definitive proof of the replicative mode of any rolling-circle plasmid (13). Despite several attempts, we were unable to detect the presence of ssDNA intermediates in plasmid DNA extracted from L. reuteri 100-23 or L. reuteri DSM 20016 hosts harboring pNCKH103 or pNCKH104 (data not shown). This suggested that, in the homologous (L. reuteri) hosts, the conversion of ssDNA to dsDNA under conditions of laboratory culture was very efficient.
An alternative approach was to introduce a pGT232-based recombinant plasmid into a heterologous host. It has been reported that SSO sequences are usually host specific and that their function, if any, in a foreign bacterial host is relatively inefficient, resulting in accumulation of ssDNA intermediates (13). A number of Lactobacillus ATCC strains and L. lactis MG1363 (Table 1) were selected for electrotransformation experiments. These strains have been shown to be amenable to electrotransformation with plasmid pFX3 (18a). Initially, pNCKH103 was used as the test plasmid, but during subsequent experimentation, it was found that the ermGT gene was not expressed in these strains (data not shown).
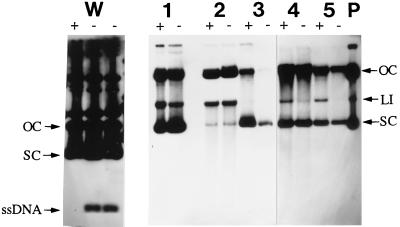
Plasmid pWX2 (see Materials and Methods) was then used to electrotransform the aforementioned strains. In addition to the three L. reuteri strains, Cmr transformants were obtained with Lactobacillus salivarius subsp. salicinius ATCC 11742 and L. lactis MG1363 with electrotransformation frequencies ranging from 1.2 × 103 (L. reuteri 100-23C) to 2.0 × 105 (L. lactis) Cmr transformants per μg of plasmid DNA. The presence of pWX2 in representative Cmr transformants was confirmed by plasmid profiling. ssDNA intermediates could not be detected in plasmid DNA extracts prepared from the L. reuteri strains or L. salivarius or L. lactis transformants, as no discernible differences were observed between plasmid DNA samples with and without S1 nuclease treatment (Fig. 3). In contrast, ssDNA plasmid intermediates could be detected in plasmid preparations of the positive control plasmid pCP12 from the L. lactis MG1363 host (Fig. 3). Plasmid pCP12 is a derivative of pWC1, a pC194-pUB110-type plasmid isolated from L. lactis subsp. cremoris (20).
FIG. 3.
Detection of ssDNA plasmid intermediates. Plasmid DNA was extracted from L. lactis and Lactobacillus strains carrying either pCP12 or pWX2. The extracts were then either treated (+) or not treated (−) with S1 nuclease. Plasmid extracts containing pWC1 and pGT232-based recombinant plasmids were blotted onto nylon membranes and probed with radioactively labelled pWC1 and pWX2 plasmid DNA, respectively. Lanes: W, L. lactis MG1363(pCP12); 1, L. salivarius subsp. salicinius ATCC 11742(pWX2); 2, L. reuteri DSM 20016(pWX2); 3, L. lactis MG1363(pWX2); 4, L. reuteri 100-23(pWX2); 5, L. reuteri 100-23C(pWX2); P, plasmid pWX2 purified from E. coli (positive control). The different topological forms of double-stranded plasmid DNA (supercoiled [SC], linear [LI], and open circular [OC]) are indicated. The position of the ssDNA intermediate of pCP12 is also shown.
DISCUSSION
The overall aim of the work described in this study was to utilize the genetic information obtained from an indigenous plasmid of an intestinal strain of L. reuteri to develop plasmid vectors that were structurally and segregationally stable within the Lactobacillus population under laboratory growth conditions and, more importantly, in the environment of the murine gastrointestinal tract.
The 5.1-kb indigenous plasmid pGT232 of L. reuteri 100-23 was sequenced completely and, on the basis of sequence homology of the DSO and the replication protein, shown to be a member of the pC194-pUB110 family of rolling-circle plasmids. Many small (<10-kb) plasmids isolated from a variety of bacterial species have been shown to replicate by this mechanism (13, 22, 23). The pC194-pUB110 plasmid family represents the largest and most diverse collection of rolling-circle plasmids, consisting of at least 25 members isolated from gram-positive and gram-negative species (27). Based on homology of the RepA proteins, the plasmids isolated from the other lactobacilli mentioned in this study fall into a Lactobacillus subgroup of the family (27). On the same basis, pGT232 can now be included in this subgroup.
During the course of determining the minimal replicon of pGT232, it was found that although autonomous replication and stable maintenance of a pGT232-based recombinant plasmid in L. reuteri cultured in vitro required only 1.7 kb of pGT232 DNA (pNCKH104), a 2.7-kb fragment (pNCKH103) was required for stable maintenance of the recombinant plasmid in the Lactobacillus population when inhabiting the gastrointestinal tract of mice. A striking difference between pNCKH103 and pNCKH104 was the presence of a series of IR sequences capable of forming stem-loop structures contained within the extra 1.0 kb of DNA. It seemed likely that this region of IRs constituted the SSO region of pGT232, which contributed to the segregational stability of pNCKH103. This initial hypothesis was supported by the experiments involving plasmid pWX1, which yielded results similar to those obtained with pNCKH104. As SSOs are orientation dependent (5, 13), this result would be anticipated since pWX1 would essentially contain a nonfunctional SSO.
In a further attempt to delineate which of the IR sequences were responsible for SSO function in pGT232, an experimental approach similar to that conducted by Seegers et al. (26) with the SSO of the lactococcal plasmid pWVO1 was undertaken. It was initially believed that IR1, which was capable of forming the largest and most energetically favorable (ΔG° = −52.4 kcal/mol) stem-loop structure, would constitute the SSO of pGT232. However, from the in vivo experiments involving plasmids pNCKH105 and pNCKH107, in which both lacked IR1, considerable segregational stability (i.e., maintenance of the plasmid by >70% of the Lactobacillus population [Table 2] was observed. It was expected that if IR1 was solely responsible for segregational stability of pGT232, experiments conducted with pNCKH105 and pNCKH107 would generate values similar to that obtained with plasmid pNCKH104. Furthermore, the in vivo experiments involving L. reuteri harboring pNCKH108 demonstrated the contributions of IR2 and IR3 to segregational stability, in which pNCKH108 was maintained in a lower proportion, as opposed to a similar level, than that of pNCKH105 or pNCKH107. These results strongly suggested that a single IR did not confer full SSO function under in vivo conditions but that rather a combination of IR1 to IR4 was required.
From the results of in vitro plasmid stability experiments, the putative SSO region was not required for stable maintenance of the plasmid in the absence of antibiotic selection under laboratory conditions. These results might be explained by differences in the extent of nonspecific conversion of single-stranded replication intermediates to their double-stranded plasmid forms under the different culture conditions (13). An example where nonspecific conversion is purported to play a role in the maintenance of rolling-circle plasmids is the replication of plasmids pC194 and pLS1 in Bacillus subtilis (5). The palA SSO present in these plasmids is not functional in B. subtilis, but it is thought that alternative sites of conversion of ssDNA to dsDNA exist, thus allowing replication in that host (5). In the case of L. reuteri harboring pNCKH104, such a nonspecific conversion mechanism may exist, but the extent of nonspecific conversion may be influenced by the conditions of growth. Complex (rich) culture media such as MRS provide the bacterial cells with an abundant supply of nutrients. Under these conditions, it is possible that there are sufficient components in the bacterial host’s replication machinery, in combination with the nonspecific conversion mechanisms, to compensate for the lack of a specific SSO. However, in the gastrointestinal tract the bacterial host confronts environmental conditions that are suboptimal for rapid growth (nutrient limitation and direct competition with other bacterial species [9]). The limited supply of replication-associated components under these conditions could lower the rate of nonspecific ssDNA conversion, eventually leading to segregational instability of pNCKH104. The fully functional SSO region present in pNCKH103 would ensure that specific and efficient conversion of ssDNA to dsDNA could occur despite diminished nutrient supply, hence, the much better segregational stability of this plasmid in vivo. Additionally, or alternatively, the replication of pNCKH104 doubtless places an additional metabolic weight on the lactobacilli. This would not create problems for the bacteria when cultured in a rich medium, but in the gastrointestinal tract, jettisoning of the recombinant plasmid could provide a growth advantage to the cells. Thus, the nutrient-limited conditions in the gastrointestinal tract would select for a reduced copy number of the plasmid and, in the absence of efficient conversion of ssDNA to dsDNA, would result in an increased rate of plasmid loss. While these views remain speculative, our results clearly demonstrated that plasmid pNCKH103 contained the genetic elements sufficient for structural and segregational stability in vivo and thus was a suitable pGT232-based vector for use in future research.
ssDNA plasmid intermediates were not detected in any of the bacterial hosts electrotransformed with pWX2. Since pWX2 presumably contained an intact SSO region, and assuming the conversion signal was highly efficient, such a result was not unexpected at least with the L. reuteri hosts. It is possible that even in the heterologous hosts (L. salivarius and L. lactis), the SSO region was recognized and the conversion of ssDNA to dsDNA was highly efficient. A similar result was obtained by Pillidge et al. (20) in that ssDNA intermediates were not observed when pCP12 was introduced into Enterococcus faecalis as opposed to the native L. lactis host. To fully explain the results we have obtained with the ssDNA experiments, detailed information pertaining to (i) the exact host factors and mechanisms involved in SSO recognition and initiation of the process of conversion of ssDNA to dsDNA and (ii) the similarity of these host factors in the heterologous hosts compared to the L. reuteri hosts is required. Host enzymes such as DNA polymerases I and III have been implicated in these processes (7), which could serve as suitable starting points for such studies.
Our study shows that in vitro observations of recombinant plasmid maintenance in a Lactobacillus population do not necessarily predict the fate of the molecule in vivo. It emphasizes the need for the results of molecular genetic studies to be investigated in relation to natural systems which are their microecological or biotechnological targets. Elegant molecular constructions that function well under laboratory conditions may be ill suited for use in an applied situation. Clearly, with respect to the utilization of the gastrointestinal microflora as a source of microbes for the development of in vivo delivery systems, there is a need for linkage between molecular biology and microecology (30).
ACKNOWLEDGMENTS
We thank V. Benes and C. Pillidge for providing plasmids pUC29 and pWC1-pCP12, respectively.
The support of the Anderson and Telford Trusts (Dunedin) and the New Zealand Dairy Research Institute (Palmerston North) is gratefully acknowledged. N.C.K.H. was the recipient of a Blair postgraduate scholarship.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Benes V, Hostomsky Z, Arnold L, Paces V. M13 and pUC vectors with new unique restriction sites for cloning. Gene. 1993;130:151–152. doi: 10.1016/0378-1119(93)90360-f. [DOI] [PubMed] [Google Scholar]
- 3.Bouia A, Bringel F, Frey L, Kammerer B, Belarbi A, Guyonvarch A, Hubert J-C. Structural organization of pLP1, a cryptic plasmid from Lactobacillus plantarum CCM 1904. Plasmid. 1989;22:185–192. doi: 10.1016/0147-619x(89)90001-2. [DOI] [PubMed] [Google Scholar]
- 4.Bringel F, Frey L, Hubert J-C. Characterization, cloning, curing and distribution in lactic acid bacteria of pLP1, a plasmid from Lactobacillus plantarum CCM 1904, and its use in shuttle vector construction. Plasmid. 1989;22:193–202. doi: 10.1016/0147-619x(89)90002-4. [DOI] [PubMed] [Google Scholar]
- 5.Dempsey L A, Zhao A C, Khan S A. Localization of the start sites of lagging-strand replication of rolling-circle plasmids from gram-positive bacteria. Mol Microbiol. 1995;15:679–687. doi: 10.1111/j.1365-2958.1995.tb02377.x. [DOI] [PubMed] [Google Scholar]
- 6.Dower W J, Miller J F, Ragsdale C W. High-efficiency transformation of E. coli by high-voltage electroporation. Nucleic Acids Res. 1988;16:6127–6147. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espinosa M, del Solar G, Rojo F, Alonso J C. Plasmid rolling circle replication and its control. FEMS Microbiol Lett. 1995;130:111–120. doi: 10.1111/j.1574-6968.1995.tb07707.x. [DOI] [PubMed] [Google Scholar]
- 8.Fons M, Hégé T, Ladiré M, Raibaud P, Ducluzeau R, Maguin E. Isolation and characterization of a plasmid from Lactobacillus fermentum conferring erythromycin resistance. Plasmid. 1997;37:199–203. doi: 10.1006/plas.1997.1290. [DOI] [PubMed] [Google Scholar]
- 9.Freter R. Mechanisms of bacterial colonization of the mucosal surfaces of the gut. In: Roth J A, editor. Virulence mechanisms of bacterial pathogens. Washington, D.C.: American Society for Microbiology; 1988. pp. 45–60. [Google Scholar]
- 10.Fuller R. History and development of probiotics. In: Fuller R, editor. Probiotics. The scientific basis. London, United Kingdom: Chapman and Hall; 1992. pp. 1–8. [Google Scholar]
- 11.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant S G N, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruss A, Ehrlich S D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989;53:231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilyina T V, Koonin E V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Josson K, Soetart P, Michiels F, Joos H, Mahillon J. Lactobacillus hilgardii plasmid pLAB1000 consists of two functional cassettes commonly found in other gram-positive organisms. J Bacteriol. 1990;172:3089–3099. doi: 10.1128/jb.172.6.3089-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kandler O, Weiss N. Genus Lactobacillus Beijerinck 1901. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1209–1234. [Google Scholar]
- 17.Leer R, van Luijk N, Posno M, Pouwels P H. Structural and functional analysis of two cryptic plasmids from Lactobacillus pentosus MD353 and Lactobacillus plantarum ATCC 8014. Mol Gen Genet. 1992;234:265–274. doi: 10.1007/BF00283847. [DOI] [PubMed] [Google Scholar]
- 18.McConnell M A, Mercer A A, Tannock G W. Transfer of plasmid pAMβ1 between members of the normal microflora inhabiting the murine digestive tract and modification of the plasmid in a Lactobacillus reuteri host. Microb Ecol Health Dis. 1991;4:343–355. [Google Scholar]
- 18a.McMillan, B., and G. W. Tannock. Unpublished observations.
- 19.Noirot-Gros M F, Bidnenko V, Ehrlich S D. Active site of the replication protein of the rolling circle plasmid pC194. EMBO J. 1994;13:4412–4420. doi: 10.1002/j.1460-2075.1994.tb06761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillidge C J, Cambourn W M, Pearce L E. Nucleotide sequence and analysis of pWC1, a pC194-type rolling circle replicon in Lactococcus lactis. Plasmid. 1996;35:131–140. doi: 10.1006/plas.1996.0015. [DOI] [PubMed] [Google Scholar]
- 21.Posno M, Leer R J, van Luijk N, van Giezen M J F, Heuvelmans P T H M, Lokman B C, Pouwels P H. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl Environ Microbiol. 1991;57:1822–1828. doi: 10.1128/aem.57.6.1822-1828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pouwels P H, Leer R J. Genetics of lactobacilli: plasmids and gene expression. Antonie Leeuwenhoek. 1993;64:85–107. doi: 10.1007/BF00873020. [DOI] [PubMed] [Google Scholar]
- 23.Pridmore D, Stefanova T, Mollet B. Cryptic plasmids from Lactobacillus helveticus and their evolutionary relationship. FEMS Microbiol Lett. 1994;124:301–306. doi: 10.1111/j.1574-6968.1994.tb07300.x. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seegers J F M L, Zhao A C, Meijer W J J, Khan S A, Venema G, Bron S. Structural and functional analysis of the single-strand origin of replication from the lactococcal plasmid pWVO1. Mol Gen Genet. 1995;249:43–50. doi: 10.1007/BF00290234. [DOI] [PubMed] [Google Scholar]
- 27.Seery L T, Nolan N C, Sharp P M, Devine K M. Comparative analysis of the pC194 group of rolling circle plasmids. Plasmid. 1993;30:185–196. doi: 10.1006/plas.1993.1051. [DOI] [PubMed] [Google Scholar]
- 28.Skaugen M. The complete nucleotide sequence of a small cryptic plasmid from Lactobacillus plantarum. Plasmid. 1989;22:175–179. doi: 10.1016/0147-619x(89)90028-0. [DOI] [PubMed] [Google Scholar]
- 29.Tannock G W. Genetic manipulation of gut microorganisms. In: Fuller R, editor. Probiotics. The scientific basis. London, United Kingdom: Chapman and Hall; 1992. pp. 181–207. [Google Scholar]
- 30.Tannock G W. Microecology of the gastrointestinal tract in relation to lactic acid bacteria. Int Dairy J. 1995;5:1059–1070. [Google Scholar]
- 31.Tannock G W, Savage D. Detection of plasmids in gastrointestinal strains of lactobacilli. Proc Univ Otago Med Sch. 1985;63:29–30. [Google Scholar]
- 32.Tannock G W, Crichton C, Welling G W, Koopman J P, Midtvedt T. Reconstitution of the gastrointestinal microflora of lactobacillus-free mice. Appl Environ Microbiol. 1988;54:2971–2975. doi: 10.1128/aem.54.12.2971-2975.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tannock G W, Luchansky J B, Miller L, Connell H, Thode-Andersen S, Mercer A A, Klaenhammer T R. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid. 1994;31:60–71. doi: 10.1006/plas.1994.1007. [DOI] [PubMed] [Google Scholar]
- 34.Tannock G W, Fuller R, Pedersen K. Lactobacillus succession in the piglet digestive tract demonstrated by plasmid profiling. Appl Environ Microbiol. 1990;56:1310–1316. doi: 10.1128/aem.56.5.1310-1316.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vujcic M, Topisirovic L. Molecular analysis of the rolling-circle replicating plasmid pA1 of Lactobacillus plantarum A112. Appl Environ Microbiol. 1993;59:274–280. doi: 10.1128/aem.59.1.274-280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wells J M, Wilson P W, Le Page R W F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 37.Wesney E, Tannock G W. Association of rat, pig and fowl biotypes of lactobacilli with the stomach of gnotobiotic mice. Microb Ecol. 1979;5:35–42. doi: 10.1007/BF02010576. [DOI] [PubMed] [Google Scholar]
- 38.Xu F, Pearce L, Yu P-L. Construction of a family of lactococcal vectors for gene cloning and translational fusion. FEMS Microbiol Lett. 1991;77:55–60. doi: 10.1111/j.1574-6968.1991.tb04321.x. [DOI] [PubMed] [Google Scholar]
- 39.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]