Significance
Missense mutations in the TP53 gene, encoding the p53 tumor suppressor, are very frequent in human cancer. Some of those mutations, particularly the more common (“hotspot”) ones, not only abrogate p53’s tumor suppressor activities but also endow the mutant protein with oncogenic gain of function (GOF). We report that p53R273H, the most common p53 mutant in pancreatic cancer, interacts with the SQSTM1/p62 protein to accelerate the degradation of cell adhesion proteins. This enables pancreatic cancer cells to detach from the epithelial sheet and engage in individualized cell migration, probably augmenting metastatic spread. By providing insights into mechanisms that underpin mutant p53 GOF, this study may suggest ways to interfere with the progression of cancers bearing particular p53 mutants.
Keywords: mutant p53, p62, protein–protein interaction, migration, cell adhesion
Abstract
Missense mutations in the p53 tumor suppressor abound in human cancer. Common (“hotspot”) mutations endow mutant p53 (mutp53) proteins with oncogenic gain of function (GOF), including enhanced cell migration and invasiveness, favoring cancer progression. GOF is usually attributed to transcriptional effects of mutp53. To elucidate transcription-independent effects of mutp53, we characterized the protein interactome of the p53R273H mutant in cells derived from pancreatic ductal adenocarcinoma (PDAC), where p53R273H is the most frequent p53 mutant. We now report that p53R273H, but not the p53R175H hotspot mutant, interacts with SQSTM1/p62 and promotes cancer cell migration and invasion in a p62-dependent manner. Mechanistically, the p53R273H-p62 axis drives the proteasomal degradation of several cell junction–associated proteins, including the gap junction protein Connexin 43, facilitating scattered cell migration. Concordantly, down-regulation of Connexin 43 augments PDAC cell migration, while its forced overexpression blunts the promigratory effect of the p53R273H-p62 axis. These findings define a mechanism of mutp53 GOF.
Mutations in the TP53 gene, which encodes the p53 tumor suppressor protein, are very common in human cancer (1, 2). In the majority of cases, these are missense mutations within the DNA binding domain (DBD) of p53, which abrogate the ability of the wild-type (wt) p53 protein to act as a sequence-specific transcriptional activator. Loss of wtp53’s function as a sequence-specific transcription factor and, consequently, as a tumor suppressor is key to the selective advantage conferred by TP53 mutations during cancer initiation and progression (3–5). In addition, at least some of the many cancer-associated p53 missense variants, particularly the more common “hotspot” mutants, can actively contribute to cancer by gain-of-function (GOF) mechanisms (6–11).
Structurally, most of the cancer-associated p53 missense mutants fall into two main classes: DNA contact mutants and structural/conformational mutants (12). DNA contact mutants carry substitutions in one of the several amino acid residues involved directly in sequence-specific DNA binding, without incurring substantial changes in the overall structure of the DBD. Structural mutants, on the other hand, result in substantial distortion of the conformation of the p53 DBD, exposing residues that are normally buried within the DBD and leading to extensive changes in the composition of proteins that interact with the misfolded p53 protein (8). The extent of conformational distortion varies among different missense mutants; moreover, some DNA contact mutants can also cause partial local distortion of the DBD structure (13).
Increased migration and invasiveness are common properties of cancer cells and are often associated with more aggressive tumor features (14, 15). Notably, mutant p53 (mutp53) proteins can promote the migration and invasiveness of many different cancer cell types, and this is believed to be a major contributor to mutp53 GOF activity (16). Numerous studies have addressed the mechanistic basis for the promigratory and proinvasive effects of mutp53, revealing that this can be achieved via altered expression of relevant genes as well as via direct protein–protein interactions (17–22). On the other hand, SQSTM1/p62 can promote cell migration by up-regulating the transcription function of Twist1 (23), and it also stabilizes vimentin by physical binding (24). Both events lead to enhanced cell proliferation, migration, and metastasis.
In the present study, we set out to explore mechanisms of mutp53 GOF by identifying interaction partners of mutp53. Specifically, we focused on the DNA contact mutant p53R273H, one of the most common p53 hotspot mutants and the most frequent TP53 mutant in pancreatic cancer. Here, we report that p53R273H binds selectively to the polyubiquitin-binding protein SQSTM1/p62. The cross-talk between p53R273H and p62, which does not rely on transcriptional regulation by mutp53, leads to proteasomal degradation of several cell adhesion–associated proteins, resulting in loss of cell–cell junctions and enhancement of cancer cell scattered migration and invasion.
Results
p53R273H Interacts with p62 via Its CTD.
To gain insight into mechanisms of action of GOF p53 missense mutants, we chose the PANC-1 cell line as our experimental system. PANC-1 cells are derived from a human pancreatic ductal adenocarcinoma (PDAC) tumor and harbor the p53 hotspot mutant p53R273H, which is the most frequent p53 mutant in PDAC. Mutp53 GOF is mediated mainly via interactions of mutp53 with other cellular proteins, most of which are involved in regulation of gene expression (12, 17, 25–33). We therefore set out to characterize the p53R273H interactome in PANC-1 cells. To that end, we immunoprecipitated (IP) the endogenous mutp53 from PANC-1 cells, as well as from PANC-1-derived p53 knockout (p53KO) cells, serving as negative control (CTRL) (SI Appendix, Fig. S1A). IPs were subjected to analysis by mass spectrometry (MS); putative mutp53 interactors were defined as significantly enriched (absolute fold change ≥1.4, P ≤ 0.05, unique peptides ≥2) in IPs from PANC-1 cells, compared to the isogenic p53KO cells (SI Appendix, Table S1 and Dataset S1). This analysis identified the polyubiquitin-binding protein SQSTM1/p62 as a putative p53R273H interactor (Fig. 1A).
Fig. 1.
p53R273H interacts with p62 via its CTD. (A) Volcano plot of the top hits (red) in immunoprecipitation and mass spectrometry analysis of p53R273H IPs from CTRL PANC-1 cells, compared to p53KO PANC-1 cells. Top hits were defined as having absolute fold change ≥1.4 (log2 fold-change [FC] ≥ 0.49), P ≤ 0.05, and unique peptides ≥2. (B) PANC-1 cell lysates were IP with p53-specific antibodies (Santa Cruz FL-393) or IgG CTRL, followed by WB analysis with antibodies against p62 (Abcam 56416); inputs are shown on the left. (C) PANC-1 cell lysates underwent IP with p62-specific antibodies (Santa Cruz D-3) or IgG CTRL, followed by WB analysis with antibodies against p53 (p53-HRP). (D) PLA between p62 and p53, performed on CTRL and shp53 cells using the Duolink PLA kit (Sigma) with tetramethylrhodamine-5-isothiocyanate (TRITC) as a probe (red). Counterstaining was with DAPI (blue) and phalloidin-FITC (green) to visualize nuclear DNA and F-actin, respectively. Quantification of PLA foci is shown below, ****P < 0.0001 (Student’s t test), based on counting of 40 cells. (Scale bar, 10 μm.) (E) PANC-1 cells stably expressing shRNA directed against the 3′UTR of the endogenous p53 mRNA (3′UTR shp53) were transiently transfected to express wtp53, p53R175H, p53R273H or p53R248W. Cell lysates were subjected to IP with antibodies against p62 (Santa Cruz D-3), followed by WB analysis with antibodies against p53 (p53-HRP) or p62 (Santa Cruz D-3). (F) PANC-1 cells stably expressing shRNA directed against the 3′UTR of the endogenous p53 mRNA (3′UTR shp53) were stably transduced to overexpress V5-tagged intact p53R273H (FL) or a p53R273H mutant in which residues 331–335 have been replaced by Ala (5A). Cell lysates underwent IP with antibodies against p62 (Santa Cruz D-3), followed by WB analysis with antibodies against p62 (Santa Cruz D-3) or p53 (anti rabbit V5 CST #13202). RASGAP was used as a loading CTRL. (G) PANC-1 cells as in F were subjected to PLA with antibodies against the V5 tag (reactive with the V5-tagged overexpressed mutp53) and p62. Nuclei were visualized by DAPI staining. (Scale bar, 10 μm.)
To validate the interaction, we immunoprecipitated endogenous p53R273H from PANC-1 whole-cell extracts, followed by Western blot (WB) analysis with p62 antibodies. As seen in Fig. 1B, p62 was, indeed, specifically coprecipitated with mutp53. The presence of a p62-mutp53 complex was also validated by a reciprocal experiment, employing p62 IP followed by WB analysis of p53 (Fig. 1C). Furthermore, proximity ligation assay (PLA) also confirmed the interaction and revealed that it occurs predominantly in the cytoplasm (Fig. 1D). Likewise, p62 coprecipitated with p53R273H and, to a lesser extent, with another hotspot DNA contact mutant, p53R248W, when these p53 mutants were transiently expressed in p53-silenced PANC-1 cells (Fig. 1E). Notably, neither the structural mutant p53R175H nor wtp53 bound robustly to p62 under the same conditions (Fig. 1E). Hence, binding to p62 is p53 mutant specific.
Human p53 (393 amino acids) consists of two N-terminal transactivation domains (residues 1–61; SI Appendix, Fig. S1B), a proline-rich domain (residues 64–91), a DBD/core domain (residues 94–292), a tetramerization domain (TD; residues 325–355), and a C-terminal regulatory domain (CTD; residues 356–393) (34). Of these, only the DBD and TD are structured, as determined for wtp53 (Protein Data Bank [PDB]: 4HJE and PDB: IC26, respectively), while the other domains form largely intrinsically disordered or unstructured regions (34). The crystal structure of the core domain of p53R273H (PDB: 4IJT) indicates that it largely resembles that of wtp53. However, the structures of full-length (FL) wtp53 and p53R273H are yet to be determined experimentally. Therefore, we modeled the FL structures of the wt and p53R273H in silico, using the structures of their core domains as templates. The modeled structures of wtp53 and p53R273H are largely similar (root-mean-square deviation: 3.03Å) (SI Appendix, Fig. S1C); a relatively greater structural stability of p53R273H over wtp53 may be attributed to the presence of helical regions in p53R273H corresponding to regions Ser15-Lys24 and Pro359-His368 (SI Appendix, Fig. S1C). Due to the close structural similarities, we used the modeled structure of FL wtp53 for predicting interaction sites with p62. p62 comprises three major biologically important domains: the structured N-terminal PB1 (residues 1–20) and ZZ (double zinc finger domain, residues 21–120) domains and the disordered Ubiquitin-associated (UBA) domain (residues 343–486). The PB1 domain is crucial for the oligomerization of p62 (PDB: 6TGY, 2KKC) (35), whereas the ZZ domain is essential for the interaction of p62 with other proteins. We performed molecular docking of the modeled structure of wtp53 and the crystal structure of p62 (PDB: 2K6Q) (details in Materials and Methods). This in silico analysis suggested that p53 may employ its C-terminal region, including the TD and CTD, to bind the ZZ domain of p62 via salt bridge interactions. The most probable interacting pairs of residues are displayed in SI Appendix, Fig. S1D. Based on these predictions, we replaced residues 331–335 of p53R273H with alanines to generate p53R273H-5A (SI Appendix, Fig. S1E). V5-tagged p53R273H and p53R273H-5A were then stably overexpressed in PANC-1 cells in which the endogenous mutp53 had been knocked down with short hairpin RNA (shRNA) directed against the 3′ untranslated region (UTR) of p53 messenger RNA (mRNA). Remarkably, although both proteins were expressed at similar levels (SI Appendix, Fig. S1F), the binding of p53R273H-5A to p62 was greatly reduced relative to intact (FL) p53R273H, as revealed by both coIP (Fig. 1F) and PLA (Fig. 1G), supporting the in silico predictions.
p62 Promotes Migration and Invasion of Cancer Cells Harboring p53R273H.
Enhanced cell migration and invasion are common manifestations of oncogenic processes, including GOF activities of mutp53 (21). Therefore, to explore functional consequences of the p62-p53R273H interaction, we evaluated the effect of silencing p62 or p53R273H on PANC-1 cell migration and invasion. To that end, PANC-1 cells were depleted of p62, mutp53, or both by lentiviral transduction of the corresponding shRNAs (SI Appendix, Fig. S2A), and migration was then measured by gap closure (“scratch”) and transwell migration assays. As shown in Fig. 2A and SI Appendix, Fig. S2B, down-regulation of either p62 or mutp53 attenuated cell migration in both assays. Moreover, the effect of depleting both proteins together was not greater than that of depleting either one alone (SI Appendix, Fig. S2B), suggesting a common mechanism of action. Likewise, silencing of either p53R273H or p62 decreased significantly transwell invasion through a Matrigel-coated membrane (Fig. 2B). Importantly, while ectopic expression of FL p53R273H rescued the migration defect of PANC-1 cells in which the endogenous mutp53 had been silenced, the p53R273H-5A mutant, deficient in p62 binding, failed to do so (Fig. 2C). This is consistent with the conjecture that interaction with p62 is required for p53R273H in order to promote migration of these cells, although it remains possible that the 5A mutation also abolishes other mutp53 interactions, particularly those that depend on p53 tetramerization, and such interactions may also contribute to cell migration. In a complementary approach, we overexpressed a p62-green fluorescent protein (GFP) fusion protein in CTRL and mutp53 knockdown PANC-1 cells (SI Appendix, Fig. S2C), followed by a gap-closure assay. As seen in Fig. 2D and SI Appendix, Fig. S2D, while p62 overexpression increased the migration of CTRL cells, it failed to do so after p53R273H knockdown, implying that the migration-promoting effect of p62 requires the presence of mutp53. Together, these observations strongly suggest that the cross-talk between p53R273H and p62 contributes to the migratory phenotype of these pancreatic cancer cells. In agreement with this notion, PLA analysis of PANC-1 cells undergoing gap closure revealed that p53R273H-p62 interactions were more prominent in the migrating front than in the center of the culture (Fig. 2E).
Fig. 2.
p62 promotes migration and invasion of cancer cells harboring p53R273H. (A and B) (Top) Representative images of transwell migration (A) and invasion (B) assays (t = 18 h) of PANC-1 cells stably transfected with CTRL shRNA, p53 shRNA (shp53), or p62 shRNA (shp62). (Bottom) Quantification of area covered by migrating/invading cells, from three independent biological repeats, normalized to CTRL. Error bars indicate SEM. Statistical significance was determined using one-way ANOVA and Tukey’s post hoc test of the indicated comparisons, *P < 0.05, **P < 0.01, ***P < 0.001. (C) Quantification of transwell migration (t = 18 h) of 3′UTR shp53 PANC-1 cells stably transfected with p53R273H (FL) or p53R273H-5A, area covered by migrating cells, from three independent biological repeats, normalized to nontransfected cells (−). Statistical analysis as in A and B. *P < 0.05, NS, not significant. (D) Quantification of gap closure assays of CTRL and shp53 PANC-1 cells without or with stable overexpression of GFP-tagged p62, from three independent biological repeats. Statistical analysis as in A and B. *P < 0.05, **P < 0.01. (E) PANC-1 cells were subjected to a gap closure assay for 8 h. Cells were then fixed and subjected to PLA analysis of p62-p53R273H interaction. Shown are representative sections from the migrating front (yellow box) and the center of the monolayer (red box). Yellow arrow indicates direction of migration. Right: Quantification of PLA foci, based on counting the average numbers of foci/cell in four different fields in each indicated region. ***P < 0.001 (Student’s t test). (Scale bar, 10 μm.) (F) CTRL HCT116 cells (expressing endogenous wtp53; wt), p53KO HCT116 cells, or KO cells ectopically expressing p53R273H or p53R175H were stably transduced with either CTRL shRNA or shRNA directed against p62. Cells were seeded in 12-well plates containing ibidi culture inserts. Gaps were created by removing the insert at t = 0, and cells were allowed to migrate for 24 h. Shown is quantification of percent of gap covered, from two independent biological repeats, for three fields in each condition. Error bars represent SEM. Statistical analysis as in A and B. *P < 0.05. (G) GFP tagged PANC-1 cells expressing CTRL shRNA, p53 shRNA, or p62 shRNA were injected into the tail vein of NSG mice (0.1 million cells/mouse). Lung metastases were evaluated 2 wk postinjection. (Top) Representative images of lung metastases. (Bottom) Total area of metastases at the lung surface (calibrated units), as quantified with ImageJ (n = 5 mice per group). Error bars represent SEM. Statistical analysis as in A and B. **P < 0.01, ***P < 0.001. (Scale bar, 0.5 cm.)
To extend our findings beyond PANC-1 cells, we next examined the consequences of p62 silencing in a panel of isogenic HCT116 cells. HCT116 cells are derived from a human colorectal cancer and harbor wtp53. We employed HCT116 cells in which the endogenous p53 had been deleted by CRISPR/Cas9 (36) (kind gift of Karen Vousden, Francis Crick Institute, London, UK), followed by reconstitution with either p53R273H or p53R175H (37). As seen in Fig. 2F, both mutants enhanced the migration of the p53 KO cells. Silencing of p62 (SI Appendix, Fig. S2E) abrogated the promigratory effect of p53R273H (Fig.2F and SI Appendix, Fig. S2F), confirming its dependence on p62 also in HCT116 cells. In contrast, p62 depletion did not reduce the migration of parental HCT116 cells or p53KO cells. Importantly, p62 depletion did not compromise the promigratory effect of p53R175H (Fig. 2F and SI Appendix, Fig. S2F), implying that p53R175H promotes migration by a different, p62-independent mechanism. Together with the fact that p53R175H does not significantly interact with p62 (Fig. 1E), this further supports the notion that the physical interaction with p62 is required for p53R273H in order to enhance cancer cell migration.
Higher expression of p53R273H has been associated with poor prognosis and, likewise, for p62 (38–40). We therefore injected CTRL, p62-depleted, and p53-depleted PANC-1 cells orthotopically into the pancreas of nonobese diabetic/severe combined immunodeficiency-gamma (NSG) mice and monitored tumor growth. As seen in SI Appendix, Fig. S2G, depletion of either p53 or p62 delayed tumor growth significantly, relative to mice injected with CTRL cells. Moreover, to evaluate the impact of mutp53 and p62 on metastatic colonization, GFP-tagged PANC-1 cells stably expressing shRNA against p53, p62, or CTRL shRNA were injected into the tail vein of NSG mice, and lung metastases were monitored 2 wk after injection. As seen in Fig. 2G, depletion of either mutp53 or p62 resulted in a significantly reduced area of lung metastases, relative to the CTRL (Fig. 2G). Hence, both mutp53 and p62 augment the tumorigenicity and metastatic capacity of PANC-1 cells.
The mutp53-p62 Axis Down-Regulates Cell Adhesion Proteins.
To identify molecular mechanisms whereby the cross-talk between p53R273H and p62 might enhance migration, we performed global proteome analysis on PANC-1 cells depleted of either p62 or mutp53. Analysis of differentially regulated proteins relative to CTRL cells (absolute fold change ≥1.4, P ≤ 0.05) upon stable knockdown of either p62 (shp62) or mutp53 (shp53) identified 64 proteins whose steady-state levels increased upon mutp53 silencing (down-regulated by mutp53) and 23 that decreased upon mutp53 silencing (up-regulated by mutp53), and, likewise, 194 proteins that increased upon p62 silencing (down-regulated by p62) and 105 that decreased upon p62 silencing (up-regulated by p62) (SI Appendix, Table S2 and Dataset S2). Analysis of the data by Database for Annotation, Visualization, and Integrated Discovery revealed that p62 knockdown resulted in up-regulation of many mitochondrial proteins (SI Appendix, Table S3), consistent with the involvement of p62 in mitophagy (41) and mitochondrial clustering (42). Likewise, many lysosomal proteins were up-regulated. Terms related to exosomes and vesicular transport were also enriched. These cellular processes are in agreement with the canonical biological functions of p62 (43–45), implying that these functions are also maintained by p62 in PANC-1 cells.
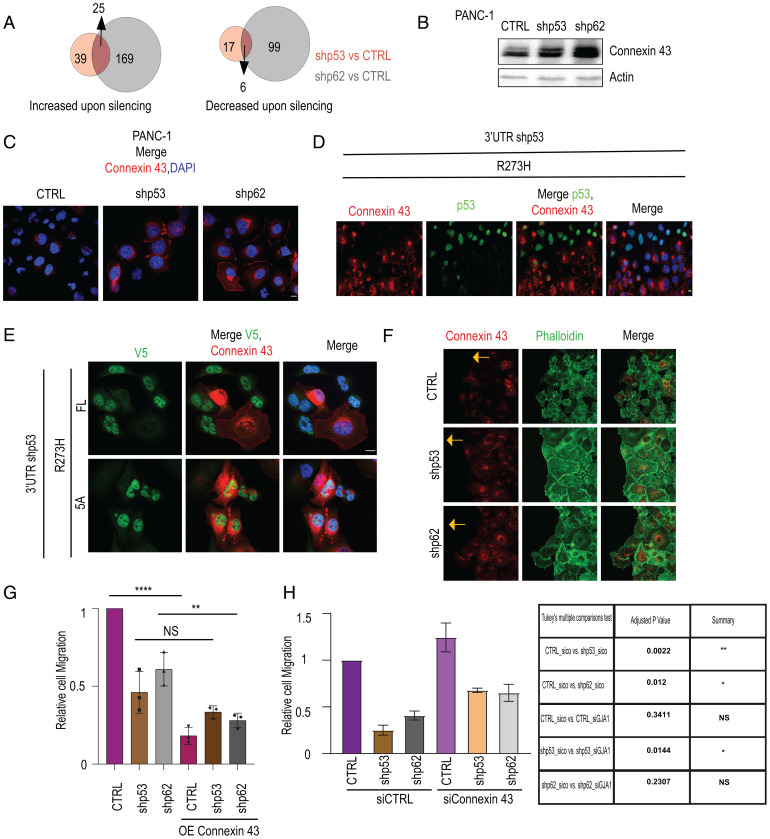
We next focused on proteins whose levels are affected similarly by both mutp53 and p62, marking them as potential downstream effectors of the mutp53-p62 axis. As shown in Fig. 3A, out of 64 and 194 proteins down-regulated by mutp53 and p62, respectively (increased upon silencing of either mutp53 or p62), 25 proteins were shared. Conversely, 6 proteins were up-regulated by both. Notably, several proteins involved in cell adhesion or its regulation—including Connexin 43 (GJA1), Filamin A (FLNA), Vinculin (VCL), and Zyxin (ZYX)—were up-regulated upon knockdown of either mutp53 or p62 (SI Appendix, Table S2).
Fig. 3.
The p53R273H-p62 axis down-regulates proteins associated with cell adhesion. (A) Total proteome analysis by LC-MS/MS was performed on PANC-1 cells stably expressing p53 shRNA, p62 shRNA, or CTRL shRNA. The Venn diagrams show the numbers of proteins increased upon knockdown of either p53 (beige) or p62 (gray) compared to CTRL (absolute fold change ≥ 1.4, P ≤ 0.05), and likewise for proteins decreased upon knockdown of p53 or p62, from two biological repeats. Numbers of overlapping proteins are indicated by arrows. (B) WB analysis of Connexin 43 in CTRL, shp62, and shp53 PANC-1 cells. Actin was used as loading CTRL. (C) IF analysis of Connexin 43 (red) in CTRL, shp62, and shp53 PANC-1 cells. Cells were counterstained with DAPI (blue) to visualize nuclei. (Scale bar, 10 μm.) (D) 3′UTR shp53 PANC-1 cells (see SI Appendix, Fig. S1F) were transiently transfected with DNA encoding p53R273H. Then, 72 h later, cells were fixed and stained for Connexin 43 (red) and p53 (green) and counterstained with DAPI. Shown is a field in which some cells overexpress ectopic p53R273H while the rest have only residual endogenous p53R273H. (Scale bar, 10 μm.) (E) IF imaging of Connexin 43 (red) and V5 (green) in 3′UTR shp53 PANC-1 cells stably expressing V5-tagged p53R273H (FL) or p53R273H-5A (5A). (Scale bar, 10 μm.) (F) IF imaging of the migrating front (indicated by yellow arrow) of CTRL, shp62, and shp53 PANC-1 cells, stained for Connexin 43 (red) and phalloidin-FITC to visualize F-actin (green). (Scale bar, 10 μm.) (G) CTRL, shp62, and shp53 PANC-1 cells, either nontransfected or 48 h after transfection with a Connexin 43 expression plasmid, were subjected to a transwell migration assay for 18 h. Area covered by migrating cells was calculated from three independent biological repeats, normalized to CTRL. Error bars indicate SEM. One-way ANOVA and Tukey’s post hoc test of the indicated comparisons are shown. **P < 0.01, ****P < 0.0001. (H) CTRL, shp62, and shp53 PANC-1 cell cultures were transfected with the indicated siRNAs. Then, 36 h later, cells were subjected to a transwell migration assay for 18 h. Area covered by migrating cells was calculated from two independent biological repeats and normalized to CTRL. Error bars indicate SEM. One-way ANOVA and Tukey’s post hoc test of the indicated comparisons are shown in the table (right). *P < 0.05; **P < 0.01.
In agreement with the total proteome analysis, WB analysis (Fig. 3B) and confocal microscopy (Fig. 3C) confirmed elevated steady-state levels of the gap junction protein Connexin 43 upon p62 knockdown and, to a somewhat lesser extent, mutp53 knockdown. Importantly, whereas Connexin 43 was hardly visible in the junctional regions of CTRL PANC-1 cells, the presence of Connexin 43 at cell–cell junctions was greatly enhanced by depletion of either p62 or mutp53 (Fig. 3C). Likewise, up-regulation of Filamin A upon p62 knockdown was validated with two different p62 shRNAs (SI Appendix, Fig. S3A). ZYX was also up-regulated, albeit modestly, upon mutp53 or p62 knockdown (SI Appendix, Fig. S3B). Up-regulation of Connexin 43 and Filamin A by p53 knockdown was validated with two different p53 shRNAs (SI Appendix, Fig. S3C). Conversely, transient transfection of p53-silenced cells with p53R273H revealed preferential attenuation of Connexin 43 staining in cells with abundant p53R273H overexpression (Fig. 3D). In contrast to their altered steady-state protein levels, the mRNA levels of GJA1 (encoding Connexin 43) and FLNA (encoding Filamin A) were not significantly affected by depletion of either p53R273H or p62 (SI Appendix, Fig. S3 D and E), arguing that both p53R273H and p62 down-regulate these proteins translationally or posttranslationally.
Remarkably, whereas stable overexpression of intact p53R273H (FL; SI Appendix, Fig. S3F) resulted in reduced Connexin 43 levels, the 5A mutant did not exert such an effect. Concordantly, in the p53R273H-transduced culture, robust Connexin 43 staining was evident only in cells that failed to overexpress mutp53 (FL, Fig. 3F), whereas cells overexpressing p53R273H-5A maintained conspicuous Connexin 43 staining. Hence, mutation of p53 residues 331–335, which compromises the interaction of p53R273H with p62, abrogates both the ability of p53R273H to promote cell migration and its ability to down-regulate Connexin 43, suggesting that these three features are interconnected. Thus, down-regulation of Connexin 43 by p53R273H might augment cancer cell migration.
Similar to the gap junction protein Connexin 43, the tight junction protein ZO-1 also localized more robustly to the plasma membrane upon knockdown of either p53R273H or p62 (SI Appendix, Fig. S3G). The ability of mutp53 to decrease the presence of ZO-1 at cell–cell junctions has already been observed by Muller et al. (21), who reported that mutp53 drives random cell migration (21, 22). Indeed, PANC-1 cells undergoing gap closure displayed a scattered migrating front when stained for F-actin (CTRL; Fig. 3F), presumably owing to the loosening of cell–cell junctions, whereas the migrating front of cells depleted of either mutp53 or p62 displayed a contiguous pattern indicative of robust cell–cell junctions (Fig. 3F).
To test more directly whether levels of junctional proteins dictate the migratory phenotype of PANC-1 cells, we transiently (48 h) overexpressed Connexin 43 in PANC-1 cells as well as in their p62-silenced and mutp53-silenced derivatives (SI Appendix, Fig. S3H), followed by a transwell migration assay. As seen in Fig. 3G, overexpression of Connexin 43 strongly abrogated the migration of PANC-1 cells, regardless of mutp53 or p62 status. Interestingly, Connexin 43 overexpression increased the presence of ZO-1 at the plasma membrane (SI Appendix, Fig. S3I), suggesting that modulation of Connexin 43 levels may also impact the recruitment of other proteins to cell–cell junctions. In a complementary approach, we transiently knocked down Connexin 43 in PANC-1 cells, as well as in their p62 and mutp53-silenced derivatives (SI Appendix, Fig. S3J), followed by a transwell migration assay. As seen in Fig. 3H, silencing of Connexin 43 restored, to some extent, the migratory capacity of cells depleted of either p62 or mutp53. However, restoration was only partial, implying that the promigratory effect of the mutp53-p62 axis involves additional processes besides enhanced Connexin 43 degradation. In fact, additional activities of mutp53, including enhanced integrin recycling (22) and interaction with other proteins (17), are known to contribute to the migratory capacity of cancer cells.
In sum, our findings suggest that the p53R273H-p62 axis may promote scattered cell migration by down-regulating the steady-state levels of several proteins involved in cell adhesion.
p62 and p53R273H Promote the Proteasomal Degradation of Cell Adhesion Proteins.
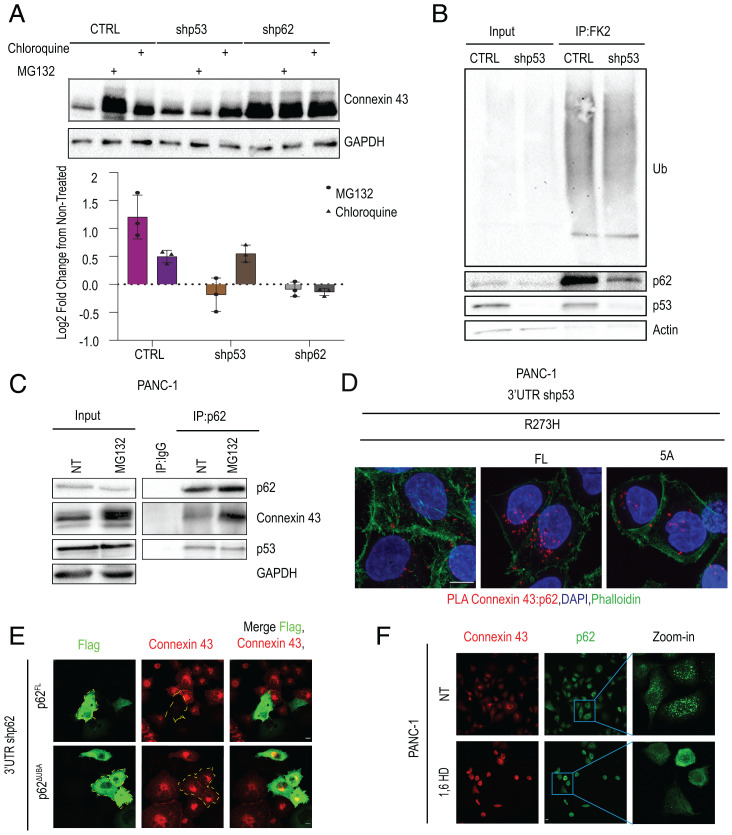
Connexin 43, Filamin A, VCL, and ZYX are all heavily ubiquitinated and are subject to degradation by either the lysosome or the proteasome (46–48). We therefore wished to determine whether mutp53 and p62 reduce the steady-state levels of Connexin 43 by promoting its degradation, and—if so—by which mechanism. To that end, PANC-1 cells and their p53- and p62-depleted derivatives were treated for 4 h with either chloroquine or MG132 to inhibit lysosomal or proteasomal degradation, respectively. As seen in Fig. 4A, both inhibitors increased markedly the steady-state levels of Connexin 43 in CTRL PANC-1 cells. Hence, Connexin 43 is rapidly degraded in these cells by both proteasomal and lysosomal processes. However, MG132 had a more pronounced effect than chloroquine, implying that most of the degradation takes place in the proteasome. Remarkably, the stabilizing effect of MG132 was not observed in cells depleted of either mutp53 or p62, whereas chloroquine still stabilized Connexin 43 in p53-depleted cells but not in p62-depleted cells, where Connexin 43 was very stable even without treatment. Overall, these observations argue that mutp53 selectively promotes the proteasomal degradation of Connexin 43 in PANC-1 cells, while p62 is required for both proteasomal and lysosomal degradation.
Fig. 4.
p53R273H and p62 promote proteasomal degradation of Connexin 43. (A) CTRL, shp62, and shp53 PANC-1 cells were treated for 4 h with either MG132 (10 µM) or chloroquine (25 µM) or were left untreated. (Top) Cell lysates were subjected to WB analysis for Connexin 43 and GAPDH as loading CTRL. (Bottom) Quantification of change in Connexin 43 abundance, relative to untreated cells, from three biological repeats. (B) Extracts of CTRL and shp53 PANC-1 cells were IP with FK2 antibody, which binds mono- and polyubiquitinated proteins, followed by SDS-PAGE (7% polyacrylamide) and WB analysis with the indicated antibodies. Separate aliquots of the same IPs were resolved on another 7% polyacrylamide gel, followed by WB analysis with FK2, anti-p53 (p53-HRP), anti-p62 (Santa Cruz D-3), and anti-beta actin as loading CTRL. (C) CTRL PANC-1 cells were treated for 4 h with MG132 (10 µM) or were left untreated. Cell lysates underwent IP with p62-specific antibodies (Santa Cruz D-3) or IgG CTRL, followed by WB analysis with antibodies against Connexin 43 and p53 (p53-HRP). (D) PLA between p62 and Connexin 43 in 3′UTR shp53 PANC-1 cells stably expressing p53R273H (FL) or p53R273H-5A (5A). Counterstaining with DAPI (blue) and phalloidin-FITC (green) to visualize F-actin. (Scale bar, 10 μm.) (E) PANC-1 cells with stable p62 knockdown (using 3′UTR targeted shRNA) were transiently transfected with FLAG-tagged FL p62 or p62ΔUBA. Then, 48 h later, cells were fixed and stained with anti-FLAG (green) and anti-Connexin 43 (red) antibodies. Yellow borders indicate p62-overexpressing cells. (Scale bar, 10 μm.) (F) PANC-1 cells were treated with 1,6-HD or 1,6-HD (3.5% solution) for 5 min or left untreated (NT), fixed, and stained for Connexin 43 (red) and p62 (green). (Scale bar, 10 μm.)
In the unmodified state of p62, dimerization of two UBA domains restricts their ability to bind ubiquitinated proteins (49). Efficient recognition of ubiquitinated cargo by p62, therefore, requires that this autoinhibitory interaction be relieved, opening the UBA for binding of ubiquitin chains on target proteins. We thus hypothesized that mutp53 binding might somehow increase the accessibility of p62’s UBA domain, enabling it to bind more avidly to ubiquitinated proteins. In agreement with this assumption, immunoprecipitation of total cellular monoubiquitinated and polyubiquitinated proteins by the FK2 antibody revealed that while depletion of mutp53 did not alter significantly the overall extent of protein ubiquitination (Fig. 4 B, Top), it greatly reduced the amount of p62 that coprecipitated with the ubiquitinated proteins (Fig. 4B, p62 panel). Thus, p53R273H promotes the binding of p62 to ubiquitinated proteins.
p62 has already been reported to bind ubiquitinated Connexin 43, albeit in the context of lysosomal, rather than proteasomal, degradation (50). To also validate binding in PANC-1 cells, we immunoprecipitated p62, followed by WB analysis with Connexin 43 and p53 antibodies. As seen in Fig. 4C, Connexin 43 was, indeed, specifically coprecipitated with p62. We also performed PLA on these two proteins. As seen in Fig. 4D, the close proximity of p62 and Connexin 43 could, indeed, be confirmed. Remarkably, this colocalization was significantly enhanced by overexpression of intact p53R273H but not p53R273H-5A, consistent with the conjecture that binding of mutp53 to p62 promotes efficient association of p62 with Connexin 43 (Fig. 4D and SI Appendix, Fig. S4A).
p62 binds ubiquitinated Connexin 43 via its UBA domain (51). To assess more directly the involvement of the UBA domain in the accelerated degradation of Connexin 43, PANC-1 cells in which the endogenous p62 had been silenced by 3′UTR-targeted shRNA were transiently transfected with DNA encoding FLAG-tagged FL p62 or a deletion mutant lacking the UBA domain (SI Appendix, Fig. S4B). As expected, cells reconstituted with FL p62 displayed markedly reduced Connexin 43 levels (Fig. 4E and SI Appendix, Fig. S1C). Importantly, the delta-UBA mutant failed to down-regulate Connexin 43, despite being abundantly expressed within the transfected cells (Fig. 4E). Thus, the p62 UBA domain is essential for its ability to drive proteasomal degradation of Connexin 43.
Immunofluorescence (IF) imaging of MG132-treated cells revealed stabilization of Connexin 43 within 2 h of exposure to the proteasome inhibitor (SI Appendix, Fig. S4D), with further increase by 4 h. By 6 h, Connexin 43 was already readily visible at cell–cell junctions. Interestingly, the MG132-stabilized Connexin 43 colocalized initially with the Golgi apparatus marker p115 (SI Appendix, Fig. S4D), suggesting that it accumulates at, or very close to, the Golgi. A similar pattern of stabilization and gradual localization to the plasma membrane after MG132 treatment was observed for ZYX, a focal adhesion protein (SI Appendix, Fig. S4E). Together, our observations imply a role for p53R273H, through its interaction with p62, in down-regulating cell junction proteins via selective proteasomal degradation.
Recently, p62 was reported to be an essential constituent of nuclear liquid-liquid phase separation condensates, which contain proteasomes and many other components of the ubiquitin-proteasome system (UPS) and carry out proteasomal degradation of short-lived nuclear proteins, such as c-Myc, as well as of unassembled proteasome subunits (52). Cytosolic p62 condensates have also been reported to sequester ubiquitinated proteins and promote their degradation (53). Indeed, when we applied limiting amounts of anti-p62 antibody to IF of PANC-1 cells, we could readily observe cytoplasmic p62 condensates (SI Appendix, Fig. S4F). These condensates also contained both K48- and K63-based ubiquitin chains, implying the presence of ubiquitinated proteins (SI Appendix, Fig. S4F). Importantly, when we treated PANC-1 cells with 1,6-hexanediol (1,6-HD), an aliphatic alcohol that disrupts liquid droplets (54), Connexin 43 became markedly up-regulated concurrently with a transition of p62 from condensates to a diffuse cytoplasmic distribution (Fig. 4F). Thus, degradation of Connexin 43 by the UPS may rely on p62 condensates.
Discussion
Mutp53 GOF is exerted largely through protein–protein interactions (8, 10). Identification of mutp53 interactors and elucidation of the functional implications of such interactions, therefore, holds potential for approaches to interfere with mutp53 GOF in cancer. In the present study, we identify p62/SQSTM1 as a mutp53 interactor, which associates selectively with the DNA contact mutant p53R273H but not with the structural mutant p53R175H. wtp53 has already been reported to bind p62 (55–57); considering that the structure of p53R273H is very similar to that of wtp53, whereas p53R175H is structurally destabilized (13), it is perhaps not surprising that p62 interacts with p53R273H but not with p53R175H. We further show that the interaction with p62 is required for the ability of p53R273H to promote cancer cell migration and invasion in PANC-1 cells and propose that this is due, at least in part, to the role of the mutp53-p62 axis in driving the ubiquitin-dependent proteasomal degradation of Connexin 43 and possibly additional cell junction proteins.
Previous studies have already described several molecular mechanisms that underpin mutp53-driven cell migration. In most cases, this relies on the ability of mutp53 to interact with particular nuclear transcription factors and modulate the expression of genes whose products affect migration. In contrast, the mechanism described herein is posttranslational and relies on the binding of mutp53 to a cytoplasmic protein that is a key regulator of protein degradation. Conceivably, the mechanisms whereby mutp53 augments cell migration and invasion are largely cell context dependent. Furthermore, whereas some of those mechanisms may be shared by a broad panel of p53 mutants (22, 58–60) others may be more restricted to particular types of mutants (61–63), as illustrated also in this study. Remarkably, in the same cells where p62 was necessary for the promigratory effect of p53R273H, it was entirely dispensable for the effect of p53R175H (Fig. 2F and SI Appendix, Fig. S2F). Of note, p53R175H has recently been shown to selectively augment oncogenic RAS signaling by modulating alternative splicing (64); it will be of interest to determine whether this accounts for the p62-independent promigratory effect of p53R175H in HCT116 cells, which carry a KRAS mutation.
Our observations are consistent with a model wherein p53R273H, upon binding to the ZZ domain of p62, increases the accessibility of p62’s UBA domain; this could take place through an allosteric mechanism or perhaps by eliciting a change in the posttranslational modifications (PTMs) of the UBA domain. This may facilitate the binding of ubiquitinated Connexin 43 and, presumably, additional proteins to p62, eventually targeting those proteins to proteasomal degradation. In line with that model, the UBA domain of p62 is required for enhanced degradation of Connexin 43 in PANC-1 cells. It is presently unknown whether mutp53 must remain bound to p62 throughout this process or, alternatively, whether it acts in a “hit and run” manner (e.g., by instigating particular PTMs).
The cardinal roles of p62 in protein degradation through autophagy and lysosomal targeting have been extensively documented (65, 66). However, p62 can also target ubiquitinated proteins to the proteasome. Thus, p62 was reported to promote c-Jun degradation via the UPS (67). Likewise, p62 shuttles polyubiquitinated tau proteins for proteasomal degradation (68) and promotes the proteasomal degradation of FLNA and RAD51 in the nucleus in response to DNA damage (69). P62 also promotes the interaction of TrkA with the proteasome, although this actually results in deubiquitination and lysosomal degradation (70). Our observations (Fig. 4F and SI Appendix, Fig. S4F) now suggest that cytoplasmic p62-containing condensates may play a role in UPS-mediated Connexin 43 degradation.
Unexpectedly, we observed that proteasome inhibition resulted in rapid accumulation of Connexin 43 in close proximity to the Golgi apparatus. A substantial portion of newly synthesized Connexin 43 was reported to be eliminated by endoplasmic reticulum (ER)-associated protein degradation (ERAD) without reaching the plasma membrane (71). Our observation raises the intriguing possibility that in other contexts, the proteasomal demise of Connexin 43 might take place in conjunction with the Golgi, recently reported to be associated with active proteasomes (72). Alternatively, Connexin 43 degradation might take place in the ER or elsewhere in the cytoplasm, but MG132-stabilized Connexin 43 translocates rapidly to the vicinity of the Golgi. Of note, this is not the first link between mutp53 and the Golgi; Capaci et al. (58) have recently reported that mutp53, through transcriptional regulation of miR-30d, can alter the structure and function of the Golgi, leading to elevated secretion of a plethora of promigratory, prometastatic factors (58).
The relationship between Connexin 43 levels and cancer is complex and multifaceted. Depending on biological context, Connexin 43 can exert either tumor-suppressive or oncogenic effects (73, 74). This also applies to the impact of such changes on cancer cell migration and invasion. Generally speaking, cell migration can occur in two major modes: single (amoeboid or mesenchymal) and collective (75). Single-cell migration is associated with attenuation of cell–cell junctions, enabling cells to detach and move away from the epithelial monolayer. In contrast, collective cell migration relies on robust cell–cell junctions, which enable effective cell–cell communication and coordinated movement of a group of cells (75). Thus, down-regulation of Connexin 43 and additional cell junction proteins, as observed in the current study, is expected to promote tumor aggressiveness specifically in situations where tumor spread is achieved through single-cell migration. Importantly, while the vast majority of PDAC cell migration studies have characterized this process only in vitro, intravital microscopy employing a mouse PDAC model has revealed that the cancer cells migrate in vivo as individual single cells or, less often, as a stream of noncohesive single cells (76). Plausibly, the impact of mutants such as p53R273H, through the molecular mechanism uncovered in the present study, will greatly depend on the particular mode of migration exercised by the cancer cells in which the p53 mutation emerges and more broadly on the role of cell junctions in the biology of the particular cancer cells.
Altogether, the findings reported in this study present a mode of mutp53 GOF, which does not involve modulation of gene expression and, thus, is unlikely to be revealed by approaches that rely on gene expression profiling. Moreover, these findings further illustrate the multiplicity and diversity of the mechanisms through which mutp53 may impact cancer and the importance of the cell context in conjunction with the identity of the particular p53 mutant that is expressed in this context.
Materials and Methods
Cell Lines and Culture Conditions.
All cell lines were maintained at 37 °C with 5% CO2. PANC-1 cells were cultured in Dulbecco’s modified Eagle’s medium (Biological Industries [BI]), and HCT116 cells were grown in McCoy’s 5A (Sigma). All culture media were supplemented with 10% fetal bovine serum (FBS; BI) and 1% penicillin-streptomycin (BI). All cell lines were tested negative for Mycoplasma before proceeding with the experiments. HCT116 TP53 KO cells were a kind gift from Karen Vousden. Stable expression of pEF1alpha-p53R273H IRES-EGFP (R273H) and pEF1alpha-p53R175H IRES-EGFP (R175H) in HCT116 TP53 KO cells was done as described (36, 37).
In Vitro KO of mutp53 in PANC-1 Cells.
Mutp53 KO clones of PANC-1 cells were generated using the CRISPR/Cas9 system. PANC-1 cells were transfected with plasmids encoding TP53-specific guide RNA (77) as well as Cas9 and GFP (transient for sorting). After 48 h, transfected cells were sorted by fluorescence-activated cell sorting (FACS) in order to obtain single cells and grow them into colonies. KO of mutp53 was verified by WB analysis and genomic DNA sequencing.
SiRNA, Transfections, and Treatments.
siRNA-mediated knockdown of Connexin 43 was done by using Dharmacon ON-TARGETplus SMARTpool with Dharmafect#1 transfection reagent (Dharmacon) at a final concentration of 30 nmol/L. The medium containing oligonucleotides and reagents was replaced after 6 h.
All plasmid transfections were done using the jetPEI DNA transfection reagent (Polyplus Transfection). The final DNA amount was 2 μg per well in a 6-well dish, and the transfection medium was replaced after 5 h. Cells were collected either 48 or 72 h after transfection for analysis by WB or IF. pCMV-wt, R175H, R273H, and R248W were used for transient overexpression (Fig. 1E). pCB6-R273H (used in Fig. 3D) was a generous gift from Karen Vousden. FLAG-tagged FL(wt) p62 and p62ΔUBA plasmids were as described (78). Plenti6.3/hCx43-stop (#27383) was from Addgene.
For lentiviral infections, PANC-1 cells were infected with recombinant lentiviruses based on pLKO.1-puro (CTRL), TRCN0000010814 (Sigma), and TRCN0000430110 (Sigma) to produce shRNA against the 3′UTR of endogenous TP53 and SQSTM1 mRNA, respectively. Virus-containing supernatants were collected 36 h after transfection and were filtered and supplemented with 4 µg/mL polybrene (Sigma). Then, 48 h after infection, p53 knockdown cells were selected with puromycin, and protein expression was confirmed by WB analysis. PANC-1 CTRL and shp53 cells were also infected with pFUGWm-GFP and pFUGWm-p62-GFP as described above, except that no selection was employed, and protein expression was confirmed by WB analysis. PANC-1 cells expressing pFUGWm-GFP were further FACS sorted for obtaining pure GFP population for in vivo studies. PANC-1 shp53 (3′UTR) cells were infected with recombinant lentiviruses based on pLEX306-p53R273H-cV5-Neo (FL) and pLEX306-p53R273H-PentaAla-cV5-Neo (5A). Then, 48 h after infection, p53-overexpressing cells were selected with G418 (InvivoGen), and protein expression was confirmed by WB analysis.
To generate stable cell lines with double knockdown of mutp53 and p62, PANC-1 cells were first subjected to retroviral infection. Retroviral packaging was performed by jetPEI-mediated transfection of HEK293 Phoenix cells with pBABE-Blast-pshEmpty (CTRL) or pBABE-Blast-pshp53 DNA together with a plasmid encoding the vesicular stomatitis virus G (VSV G) envelope protein. Virus-containing supernatants were collected 24 and 36 h after transfection and were filtered and supplemented with 4 µg/mL polybrene (Sigma). Blasticidin (InvivoGen) selection was initiated 48 h after infection and continued for a week. The cells were then subjected to lentiviral infection with recombinant lentiviruses based on pLKO1-puro (CTRL) or TRCN0000007237 (Sigma, targeting p62 mRNA), puromycin selection was done as above, and knockdown efficiency was assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and WB analysis. When indicated, cells were treated with MG132 (Sigma, 10 µM) or chloroquine (Sigma, 25 µM). Treatment with 3.5% 1,6-HD (Sigma-Aldrich- 240117) was for 5 min.
Lentiviral-Based p53 Overexpression Constructs.
Expression plasmids for V5-tagged p53R273H (FL) or p53R273H-5A (5A) were generated using the Gateway cloning system. In brief, each open reading frame was first inserted as an entry clone into the pDONR223 vector (79) and then was cloned into the pLEX306-cV5-Neo expression vector (modified from pLEX_306, Addgene plasmid #41391) by substituting the puromycin N-acetyl-transferase with amino 3′-glycosyl phosphotransferase (Neo) using the attL and attR (LR) recombination reaction, following manufacturer's instruction (Invitrogen). To generate p53R273H-5A, residues 331–335 of p53R273H were mutated to alanine using PCR-based site-directed mutagenesis in the entry clone.
Cell Extracts, IP, and WB Analysis.
Cells were lysed in Nonidet P-40 lysis buffer (150 mM NaCl, 50 mM Tris pH8.0, 1% Nonidet P-40 supplemented with phosphatase inhibitor mixtures 2 and 3 [Sigma] and protease inhibitor mix [Sigma]). Protein concentration was estimated by the Pierce BCA Protein Assay kit (Thermo Scientific). IP was performed as described by Di Agostino et al. (31). For IP, we used anti-p53 rabbit polyclonal antibody (Santa Cruz FL-393), anti-p62 mouse monoclonal antibody (Santa Cruz D-3), anti-ubiquitinylated proteins antibody clone FK2 (Merck, 04-263), anti-immunoglobulin G (IgG) rabbit polyclonal (R&D Systems, AB-105-C; Sigma-Aldrich,12-370), and anti-IgG mouse monoclonal (Abcam, ab124055; Sigma-Aldrich,12-371). The following antibodies were used for WB: anti-p53 hybridoma (mix of PAb421 and PAb1801), p53 horseradish peroxidase (HRP)-conjugated antibody (R&D Systems, HAF1355), anti-p62 (Santa Cruz D-3 and Abcam 56416), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Millipore, MAB374 and Cell Signaling 14C10 #2118), anti-RAS GTPase activating protein (RASGAP) (Santa Cruz SC-63, B4F8), anti-Connexin 43 (Abcam, ab11370), anti-V5 (ThermoFisher, R960-25, CST #13202), anti-ubiquitinated proteins (Merck, clone FK2, 04-263), anti-beta actin (Merck, clone 4, MAB1505), anti-Filamin A (Abcam, ab76289), anti-ZYX (kindly provided by Prof. Benjamin Geiger, made by Antibody Production Laboratory of the Department of Biological Services, Weizmann Institute of Science), and anti-FLAG (Merck-Sigma F7425). Conjugated anti-mouse or anti-rabbit secondary antibodies were from Jackson ImmunoResearch. Imaging and quantification were performed using ChemiDoc MP Imager with Image Lab 4.1 software (Bio-Rad).
MS Analysis and Data Processing.
IPs were subjected to on-bead tryptic digestion, followed by a desalting step. The resulting peptides were analyzed using nanoflow liquid chromatography (nanoAcquity) coupled to high-resolution, high-mass-accuracy MS (Orbitrap Fusion Lumos). Each sample was analyzed on the instrument separately in a random order in discovery mode. Raw data were processed with MaxQuant v1.6.0.16. The data were searched with the Andromeda search engine against the human proteome database appended with common laboratory protein contaminants and the following modifications: fixed modification, cysteine carbamidomethylation; variable modifications, methionine oxidation, asparagine, and glutamine deamidation, protein N-terminal acetylation. Quantitative comparisons were calculated using Perseus v1.6.0.7. Decoy hits were filtered out. All the identified peptides were filtered with 1% false discovery rate (FDR), at least two identified peptides, and mass accuracy.
For total proteome, proteins were subjected to proteolysis and digested by trypsin. The tryptic peptides were desalted using C18 tips (Top tip, Glygen) dried and resuspended in 0.1% formic acid. The resultant peptides were analyzed by liquid chromatography–mass spectrometry/mass spectrometry (LC-MS/MS) on Q-Exactive HFX (Thermo) and identified by MaxQuant software 1.5.2.8 for peak picking and identification using the Andromeda search engine, searching against the human proteome from the Uniprot database with mass tolerance of 6 ppm for the precursor masses and the fragment ions. Oxidation on methionine and protein N terminus acetylation were accepted as variable modifications, and carbamidomethyl on cysteine was accepted as a static modification. The minimal peptide length was set to six amino acids, and a maximum of two mis-cleavages was allowed. The data were quantified by label-free analysis using the same software. Peptide- and protein-level FDRs were filtered to 1% using the target-decoy strategy. Protein tables were filtered to eliminate the identifications from the reverse database, common contaminants, and single peptide identifications.
RNA Isolation and Quantitative Real-Time PCR Analysis.
NucleoSpin kit (Macherey Nagel) was used for RNA isolation. In total, 1 μg of each RNA sample was reverse transcribed using Luna Universal qPCR Master Mix (New England Biolabs). Real-time qPCR was performed using SYBR Green PCR Supermix (Invitrogen) with a StepOne real-time PCR instrument (Applied Biosystems). For each gene, values for the standard curve were measured, and the relative quantity was normalized to GAPDH mRNA. Primers were as follows:
SQSTM1: fw: CCAAGCTCTCTGGACCCCT; rv: CTGGGAGAGGGACTCAATCA
TP53: fw: CCCAAGCAATGGATGATTTGA; rv: GGCATTCTGGGAGCTTCATCT
GAPDH: fw: GGAAGGTGAAGGTCGGAGTC;
rv: TGAGGTCAATGAAGGGGTCA
GJA1: fw: TCAAGCCTACTCAACTGCTGG; rv: TGTTACAACGAAAGGCAGACTG
FLNA: fw: ACACAGCCATGGTGTCCTG; rv: TACTCCGGGGCCGTATACTT
IF and Confocal Microscopy.
For staining, cells were washed in saline containing phosphate-buffered saline (PBS; BI), fixed in formaldehyde (4%), and permeabilized in PBS with 0.1% Triton X-100. Next, blocking in blocking solution (Sigma) was performed for 30 min, followed by incubation with a primary antibody in antibody diluent containing FBS (Sigma), and then with Alexa fluorophore-conjugated secondary antibodies (Alexa Fluor 488, Alexa Fluor 594) and DAPI. Z- stack images were taken using an LSM 800 (Zeiss) confocal microscope with 40/60× objective oil immersion.
The following antibodies were used for IF: p62 (Santa Cruz D-3), p53 (DO-1, Santa Cruz SC-126 and CM1, BioLegend-SIG-3520), Connexin 43 (Abcam, ab11370), V5 (ThermoFisher, R960-25), and FLAG (Merck-Sigma, F1804). F-actin was visualized with phalloidin-fluorescein isothiocyanate (FITC) (Sigma P5282), ZYX (kindly provided by Prof. Benjamin Geiger, made by Antibody Production Laboratory of the Department of Biological Services, Weizmann Institute of Science), anti-K48 (Merck, 05-1307), and anti-K63 (Merck, 05-1308). Monoclonal anti-p115 was kindly provided by Prof. Sima Lev (Weizmann Institute of Science, Israel).
In Situ PLA and Quantification of PLA Foci.
PLA was performed as per Xiao et al. (33). Briefly, cells were fixed with 4% PFA for 15 min and permeabilized with 0.1% Triton for 5 min. PLA was performed using the DuoLink In Situ PLA Detection Kit (DUO92101, Sigma). Imaging was done as described above for IF. To quantify PLA foci, confocal images were processed using ImageJ as in Law et al. (80). Z-stack projections for multicolor confocal images were generated using ImageJ. Channels were split, and individual grayscale binary images were generated for nuclei (DAPI), cytoplasm (phalloidin minus DAPI), and foci. Binary images for DAPI and phalloidin were used to mark cell nucleus and boundaries to generate regions of interest (ROIs). For each ROI within the binary image, foci were quantified using the “Find Maxima” tool in ImageJ (noise tolerance = constant for the same set of images, output type = maxima within tolerance).
Gap Closure Assays.
For gap closure (“scratch”) assays, 50,000 PANC-1 cells were plated in ibidi culture inserts (2 well, ibidi GmbH, Martinsried, Germany). Cells were allowed to adhere overnight, and then inserts were removed and cultures were washed with PBS to remove floating cells. Fresh medium was provided before imaging at T = 0 h. Gap closure was imaged at 0 and 18 h (PANC-1) or 24 h (HCT116) with a Nikon Eclipse Ti-E microscope at ×4 magnification, capturing at least three fields for each condition. The percent gap closure was calculated as per Shreberk-Shaked et al. (81).
Where indicated, PANC-1 cells were plated in ibidi culture inserts on top of microscope coverslips to visualize the migrating front, and a gap was generated as above. Cells were allowed to migrate for 8 h and fixed and subjected to IF or PLA.
Transwell Migration and Invasion Assays.
Migration assays were performed using a transwell system (8-μm pore size; Costar). In brief, 60,000 PANC-1 cells in 0.02% FBS-containing medium were seeded in the upper chamber, while the lower chamber was filled with 650 microliters of culture medium supplemented with 20% FBS. Cells were allowed to migrate for 18 h. Then, cells on the lower surface of the chamber were fixed with 4% paraformaldehyde and stained with crystal violet for 10 min. Stained cells were imaged with an Olympus SZX16 microscope at ×1 magnification, capturing the entire field for each condition. Crystal violet-stained areas were quantified with ImageJ.
For invasion assays, 60,000 cells were seeded in transwell chambers precoated with Matrigel (Costar). All other details were as above.
In Silico Analysis (Structural Modeling and Molecular Docking).
The structures of FL wtp53 and p53R273H were modeled using the I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/), employing the available structures of the DBDs (core domain) of wtp53 (PDB:4HJE) and p53R273H (PDB:4JIT) as templates. The best-fit model for both proteins was selected based on C-score and on further examination of the stereochemical parameters by SAVES server (SAVES v5.0) (https://www.acronymfinder.com/Structure-Analysis-and-Verification-Server-(University-of-California%2c-Los-Angeles)-(SAVES).html). The FATCAT (https://fatcat.godziklab.org/) server was used for pairwise structural alignment of the modeled structures with the available PDB structures. Molecular docking between wtp53 and p62 was performed using the PatchDock server (https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php). Interactions across the top-scoring models (ranked according to a geometric shape complementarity score and corresponding ΔG values) were shortlisted for experimental validation and functional characterization. Figures and illustrations were made using PyMol.
Mice.
Animal husbandry, ethical handling of mice, and all animal work was carried out according to guidelines approved by the Canadian Council on Animal Care and under protocols approved by the Centre for Phenogenomics Animal Care Committee (18-0272H) or Weizmann Institutional Animal Care and Use Committee. Suspensions of PANC-1 CTRL, shp53, and shp62 cells were prepared at a concentration of 10^6 cells in 50 μL PBS and kept on ice with occasional agitation. The 8-wk-old NOD-SCID mice were anesthetized using Isoflurane. A 1-cm incision was made at the left abdominal flank, slightly medial to the splenic silhouette. The entire pancreatic body and the spleen were gently pulled out and exposed to the outside of the peritoneal cavity. A needle was inserted into the tail of the pancreas and passed into the pancreas head. Then, 50-μL cell suspension was slowly injected while withdrawing the needle. The pancreas and spleen were carefully returned back to the peritoneal cavity, and the abdominal muscle layer and skin layer were sequentially closed with sutures and staples. After cell implantation, all mice were monitored at least twice weekly for tumor growth and general signs of morbidity. Tumors started ∼40 d post-surgery.
For tail vein injection, 10^5 cells were resuspended in 100 microliter PBS before being injected into the tail vein of NSG mice. Lungs were harvested 2 wk post-injection and visualized in a fluorescent microscope.
Statistical Analysis.
Independent biological replicates were performed, and group comparisons were done as detailed in the figure legends. P values below 0.05 were considered significant. Statistical analysis was performed using the Graph-Pad Prism 9.1.0 software. Statistical significance between two experimental groups is indicated by asterisks; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, NS, not significant.
Supplementary Material
Acknowledgments
We are grateful to Karen Vousden for insightful comments and suggestions and to Varda Rotter, Yifat Merbl, and Avital Eisenberg for inspiring scientific discussions. We thank Noa Wigoda and Gilgi Friedlander for statistical help and Yishai Levin and Alon Savidor of the Crown Genomics Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science, for MS.
This work was supported in part by a grant from the Canadian Institutes of Health Research, the International Development Research Centre, the Israel Science Foundation and the Azrieli Foundation, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Thompson Family Foundation, Anat and Amnon Shashua, and the Moross Integrated Cancer Center. M.O. is incumbent of the Andre Lwoff chair in molecular biology.
Footnotes
Reviewers: C.P., Columbia University; and A.L., Institute for Advanced Study.
Competing interest statement: A.L. is the founder, director, and consultant of PMV Pharma, whose goal is to produce small molecules that reactivate mutant p53 proteins to wild-type functions. While PMV works with R273H mutations, it does nothing at this time with gain-of-function phenotypes, the topic of this manuscript.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119644119/-/DCSupplemental.
Data Availability
Proteomics data have been deposited in ProteomeXchange Consortium (PXD029518) (82). All other study data are included in the article and/or supporting information.
References
- 1.Donehower L. A., et al. , Integrated analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep. 28, 1370–1384.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hainaut P., Pfeifer G. P., Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb. Perspect. Med. 6, 1–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boutelle A. M., Attardi L. D., p53 and tumor suppression: It takes a network. Trends Cell Biol. 31, 298–310 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laptenko O., Prives C., p53: Master of life, death, and the epigenome. Genes Dev. 31, 955–956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnoud T., Indeglia A., Murphy M. E., Shifting the paradigms for tumor suppression: Lessons from the p53 field. Oncogene 40, 4281–4290 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oren M., Rotter V., Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2, a001107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller P. A. J., Vousden K. H., Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 25, 304–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim M. P., Lozano G., Mutant p53 partners in crime. Cell Death Differ. 25, 161–168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mantovani F., Collavin L., Del Sal G., Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 26, 199–212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bargonetti J., Prives C., Gain-of-function mutant p53: History and speculation. J. Mol. Cell Biol. 11, 605–609 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baugh E. H., Ke H., Levine A. J., Bonneau R. A., Chan C. S., Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 25, 154–160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed-Pastor W. A., Prives C., Mutant p53: One name, many proteins. Genes Dev. 26, 1268–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joerger A. C., Fersht A. R., Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 77, 557–582 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Novikov N. M., Zolotaryova S. Y., Gautreau A. M., Denisov E. V., Mutational drivers of cancer cell migration and invasion. Br. J. Cancer 124, 102–114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller P. A. J., Vousden K. H., Norman J. C., p53 and its mutants in tumor cell migration and invasion. J. Cell Biol. 192, 209–218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffill C. R., et al. , Mutant p53 interactome identifies nardilysin as a p53R273H-specific binding partner that promotes invasion. EMBO Rep. 13, 638–644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arjonen A., et al. , Mutant p53-associated myosin-X upregulation promotes breast cancer invasion and metastasis. J. Clin. Invest. 124, 1069–1082 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeudall W. A., et al. , Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinogenesis 33, 442–451 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Ahn J. H., Kim T. J., Lee J. H., Choi J. H., Mutant p53 stimulates cell invasion through an interaction with Rad21 in human ovarian cancer cells. Sci. Rep. 7, 9076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller P. A. J., et al. , Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene 32, 1252–1265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller P. A. J., et al. , Mutant p53 drives invasion by promoting integrin recycling. Cell 139, 1327–1341 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Li S. S., et al. , p62/SQSTM1 interacts with vimentin to enhance breast cancer metastasis. Carcinogenesis 38, 1092–1103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiang L., et al. , Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc. Natl. Acad. Sci. U.S.A. 111, 9241–9246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Hwang C. I., et al. , Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc. Natl. Acad. Sci. U.S.A. 108, 14240–14245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Do P. M., et al. , Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 26, 830–845 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Agostino S., et al. , Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 10, 191–202 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Stambolsky P., et al. , Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell 17, 273–285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amelio I., et al. , p53 mutants cooperate with HIF-1 in transcriptional regulation of extracellular matrix components to promote tumor progression. Proc. Natl. Acad. Sci. U.S.A. 115, E10869–E10878 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim M. P., et al. , Oncogenic KRAS recruits an expansive transcriptional network through mutant p53 to drive pancreatic cancer metastasis. Cancer Discov. 11, 2094–2111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Agostino S., et al. , YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins. EMBO Rep. 17, 188–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S., et al. , Mutant p53 drives clonal hematopoiesis through modulating epigenetic pathway. Nat. Commun. 10, 5649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao G., et al. , Gain-of-function mutant p53 R273H interacts with replicating DNA and PARP1 in breast cancer. Cancer Res. 80, 394–405 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joerger A. C., Fersht A. R., The tumor suppressor p53: From structures to drug discovery. Cold Spring Harb. Perspect. Biol. 2, a000919 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobi A. J., et al. , Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake. Nat. Commun. 11, 440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blagih J., et al. , A role for p53 in the adaptation to glutamine starvation through the expression of SLC1A3. Cell Metab. 28, 721–736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotler E., et al. , A systematic p53 mutation library links differential functional impact to cancer mutation pattern and evolutionary conservation. Mol. Cell 71, 178–190.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Zhu L., et al. , Cytoplasmic SQSTM1/ P62 accumulation predicates a poor prognosis in patients with malignant tumor. J. Cancer 9, 4072–4086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu L., Zeng J., Evaluation of K-ras and p53 expression in pancreatic adenocarcinoma using the cancer genome atlas. PLoS One 12, e0181532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruan H., et al. , The prognostic value of p62 in solid tumor patients: A meta-analysis. Oncotarget 9, 4258–4266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geisler S., et al. , PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Narendra D., Kane L. A., Hauser D. N., Fearnley I. M., Youle R. J., p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6, 1090–1106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones S. M., Crosby J. R., Salamero J., Howell K. E., A cytosolic complex of p62 and rab6 associates with TGN38/41 and is involved in budding of exocytic vesicles from the trans-Golgi network. J. Cell Biol. 122, 775–788 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aparicio R., Rana A., Walker D. W., Upregulation of the autophagy adaptor p62/SQSTM1 prolongs health and lifespan in middle-aged drosophila. Cell Rep. 28, 1029–1040.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao B., et al. , p62-Mediated mitochondrial clustering attenuates apoptosis induced by mitochondrial depolarization. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1308–1317 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Razinia Z., et al. , The E3 ubiquitin ligase specificity subunit ASB2α targets filamins for proteasomal degradation by interacting with the filamin actin-binding domain. J. Cell Sci. 124, 2631–2641 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teckchandani A., Cooper J. A., The ubiquitin-proteasome system regulates focal adhesions at the leading edge of migrating cells. eLife 5, 1–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ribeiro-Rodrigues T. M., Catarino S., Pinho M. J., Pereira P., Girao H., Connexin 43 ubiquitination determines the fate of gap junctions: Restrict to survive. Biochem. Soc. Trans. 43, 471–475 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Isogai S., et al. , Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J. Biol. Chem. 286, 31864–31874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falk M. M., Kells R. M., Berthoud V. M., Degradation of connexins and gap junctions. FEBS Lett. 588, 1221–1229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fong J. T., et al. , Internalized gap junctions are degraded by autophagy. Autophagy 8, 794–811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu A., Cohen-Kaplan V., Avni N., Livneh I., Ciechanover A., p62-containing, proteolytically active nuclear condensates, increase the efficiency of the ubiquitin-proteasome system. Proc. Natl. Acad. Sci. U.S.A. 118, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen-Kaplan V., Livneh I., Avni N., Cohen-Rosenzweig C., Ciechanover A., The ubiquitin-proteasome system and autophagy: Coordinated and independent activities. Int. J. Biochem. Cell Biol. 79, 403–418 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Yasuda S., et al. , Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Yang Y., et al. , Cytoplasmic DAXX drives SQSTM1/p62 phase condensation to activate Nrf2-mediated stress response. Nat. Commun. 10, 3759 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang J. H., et al. , Renal cell carcinoma escapes death by p53 depletion through transglutaminase 2-chaperoned autophagy. Cell Death Dis. 7, e2163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korolchuk V. I., Mansilla A., Menzies F. M., Rubinsztein D. C., Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell 33, 517–527 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Capaci V., et al. , Mutant p53 induces Golgi tubulo-vesiculation driving a prometastatic secretome. Nat. Commun. 11, 3945 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lakoduk A. M., et al. , Mutant p53 amplifies a dynamin-1/APPL1 endosome feedback loop that regulates recycling and migration. J. Cell Biol. 218, 1928–1942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian M., et al. , A mutant p53/let-7i-axis-regulated gene network drives cell migration, invasion and metastasis. Oncogene 34, 1094–1104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun S., et al. , Hotspot mutant p53-R273H inhibits KLF6 expression to promote cell migration and tumor metastasis. Cell Death Dis. 11, 595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klemke L., et al. , The gain-of-function p53 R248W mutant promotes migration by STAT3 deregulation in human pancreatic cancer cells. Front. Oncol. 11, 642603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lv T., et al. , p53-R273H promotes cancer cell migration via upregulation of neuraminidase-1. J. Cancer 11, 6874–6882 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Escobar-Hoyos L. F., et al. , Altered RNA splicing by mutant p53 activates oncogenic RAS signaling in pancreatic cancer. Cancer Cell 38, 198–211.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lamark T., Svenning S., Johansen T., Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays Biochem. 61, 609–624 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Dikic I., Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Yu W., Wang B., Zhou L., Xu G., Endoplasmic reticulum stress-mediated p62 downregulation inhibits apoptosis via c-Jun upregulation. Biomol. Ther. (Seoul) 29, 195–204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Babu J. R., Geetha T., Wooten M. W., Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J. Neurochem. 94, 192–203 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Hewitt G., et al. , SQSTM1/p62 mediates crosstalk between autophagy and the UPS in DNA repair. Autophagy 12, 1917–1930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geetha T., Seibenhener M. L., Chen L., Madura K., Wooten M. W., p62 serves as a shuttling factor for TrkA interaction with the proteasome. Biochem. Biophys. Res. Commun. 374, 33–37 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.VanSlyke J. K., Musil L. S., Dislocation and degradation from the ER are regulated by cytosolic stress. J. Cell Biol. 157, 381–394 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eisenberg-Lerner A., et al. , Golgi organization is regulated by proteasomal degradation. Nat. Commun. 11, 409 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mulkearns-Hubert E. E., Reizes O., Lathia J. D., Connexins in cancer: Jekyll or Hyde? Biomolecules 10, 1–30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bonacquisti E. E., Nguyen J., Connexin 43 (Cx43) in cancer: Implications for therapeutic approaches via gap junctions. Cancer Lett. 442, 439–444 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Wu J. S., et al. , Plasticity of cancer cell invasion: Patterns and mechanisms. Transl. Oncol. 14, 100899 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beerling E., Oosterom I., Voest E., Lolkema M., van Rheenen J., Intravital characterization of tumor cell migration in pancreatic cancer. Intravital 5, e1261773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang D., et al. , Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 538, 118–122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cohen-Kaplan V., et al. , p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 113, E7490–E7499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rual J. F., et al. , Human ORFeome version 1.1: A platform for reverse proteomics. Genome Res. 14 (10B), 2128–2135 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Law A. M. K., et al. , Andy’s Algorithms: New automated digital image analysis pipelines for FIJI. Sci. Rep. 7, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shreberk-Shaked M., et al. , A division of labor between YAP and TAZ in non-small cell lung cancer. Cancer Res. 80, 4145–4157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.T. Ziv, M. Oren, Crosstalk between Mutant p53 and p62/SQSTM1 Augments Cancer Cell Migration by Promoting the Degradation of Cell Adhesion Proteins. ProteomeXchange Consortium via the PRIDE database. https://www.ebi.ac.uk/pride/archive/projects/PXD029518/. Deposited 11 February 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data have been deposited in ProteomeXchange Consortium (PXD029518) (82). All other study data are included in the article and/or supporting information.