Significance
Daily rhythms in the molecular clock, in calcium, and in electrical activity all interact to support the functions of circadian pacemaker neurons. However, the regulatory mechanisms that unify these properties are not defined. Here, we utilize the cellular resolution of the Drosophila circadian neural circuit with technological improvements in light-sheet imaging. We report that individual Drosophila pacemakers display two cophasic rhythms of daily calcium fluctuations. We previously described the first: slow changes in intracellular calcium. The second involves high-frequency calcium fluctuations that depend on the function of the T-type calcium channel. We propose that the fast rhythms, emerging sequentially across the 24-h day, correspond to spontaneous electrical activity patterns displayed by different pacemaker groups.
Keywords: Drosophila, calcium, circadian rhythms, ITPR, T-type calcium channel
Abstract
Circadian pacemaker neurons in the Drosophila brain display daily rhythms in the levels of intracellular calcium. These calcium rhythms are driven by molecular clocks and are required for normal circadian behavior. To study their biological basis, we employed genetic manipulations in conjunction with improved methods of in vivo light-sheet microscopy to measure calcium dynamics in individual pacemaker neurons over complete 24-h durations at sampling frequencies as high as 5 Hz. This technological advance unexpectedly revealed cophasic daily rhythms in basal calcium levels and in high-frequency calcium fluctuations. Further, we found that the rhythms of basal calcium levels and of fast calcium fluctuations reflect the activities of two proteins that mediate distinct forms of calcium fluxes. One is the inositol trisphosphate receptor (ITPR), a channel that mediates calcium fluxes from internal endoplasmic reticulum calcium stores, and the other is a T-type voltage-gated calcium channel, which mediates extracellular calcium influx. These results suggest that Drosophila molecular clocks regulate ITPR and T-type channels to generate two distinct but coupled rhythms in basal calcium and in fast calcium fluctuations. We propose that both internal and external calcium fluxes are essential for circadian pacemaker neurons to provide rhythmic outputs and thereby, regulate the activities of downstream brain centers.
Circadian rhythms in multiple aspects of cellular physiology help organisms across taxa, from unicellular cyanobacteria to multicellular animals, adapt to environmental day–night changes (1, 2). In mammals, neurons in the hypothalamic suprachiasmatic nucleus (SCN) show circadian rhythms in gene expression, intracellular calcium, neural activity, and other cellular properties (3). Circadian rhythms in SCN neuronal outputs coordinate circadian rhythms in other cells throughout the body and generate behavioral rhythms (4). The rhythms of SCN neuronal outputs can be generated cell intrinsically by the negative transcription/translation feedback loop of core clock genes as a molecular clock, which then generates 24-h oscillations in a series of genes (1, 5–7). These gene oscillations then regulate different aspects of membrane physiology, such as the expression levels of channels for potassium, sodium, and calcium (8–12). The mechanisms by which the molecular clockworks coordinate complex membrane physiology to generate neural activity rhythms within individual circadian pacemakers remain to be defined.
Calcium signaling regulates many cellular processes, such as neural excitability, neurotransmitter release, and gene expression (13). Cytoplasmic calcium can be regulated from extracellular calcium influx as well as from intracellular calcium stored in the endoplasmic reticulum (ER) and mitochondria (14). Studies on SCN neurons in vitro (15, 16) and recently, in vivo (17) measured circadian calcium rhythms (CCRs) in SCN neurons. Some studies suggested that calcium rhythms were driven by neuronal firing and voltage-gated calcium channels (15, 18), while others suggested they were driven by intracellular stores via the ER channel ryanodine receptor (RyR) (16). These alternative hypotheses may derive from the technical differences in the various studies, including the details of in vitro preparations, but also, due to a lack of single-cell resolution in the calcium measurements.
In Drosophila, circadian pacemaker neurons also show clock-driven CCRs (19). The dynamics can be resolved across all five major pacemaker groups (the small ventral lateral neurons [s-LNv], the large ventral lateral neurons [l-LNv], the dorsal lateral neurons [LNd], the group #1 dorsal neurons [DN1], and the group #3 Dorsal Neurons [DN3]), and each group exhibits distinct and sequential daily peak phases. Within such groups, the rhythms can be measured in single identified cells (19). The multihour phase diversity exhibited by this network requires a series of delays effected by environmental light and by noncell-autonomous modulation mediated by different neuropeptides (20). Precisely how neuropeptide signaling regulates calcium activity in pacemaker neurons over long (many-hour) durations is unknown. To begin to understand these critical mechanisms of pacemaker modulation, we begin by addressing the cellular and molecular basis of pacemaker calcium rhythms with physiological, genetic, and behavioral measures.
In this study, we again used in vivo calcium imaging at single-cell resolution, here using a high-speed light-sheet microscope termed OCPI-2 (21); the acronym OCPI stands for objective-coupled planar illumination. OCPI-2 represents a fundamental technical advance because it permits sampling frequencies to capture stacks of large tissue volumes, without compromising photon efficiency or spatial resolution. Whereas OCPI-1 methods permitted us to sample a whole-brain volume once every 10 min across the 24-h day, OCPI-2 methods permit us to sample volumetrically at rates as high as 5 Hz. Thus, we simultaneously measured both basal calcium levels and fast calcium fluctuations at single-cell resolution over entire 24-h durations. We found circadian rhythmicity in both measures. We consider the fast fluctuations to represent events closely coupled to neuronal firing, as have previous studies conducted in much more restricted temporal durations (i.e., not circadian) (e.g., refs. 22–26). In all the Drosophila pacemaker neurons we studied, these two layers of calcium rhythms shared the same daily temporal pattern (i.e., they were cophasic). To gain insights into the mechanism of these patterns, we exploited the fact that in Drosophila, many calcium channels are encoded by single genes (14), and we used genetics to study the roles of individual channels in generating daily pacemaker calcium rhythms. Here, we present results of experiments in which we knocked down RNAs encoding different calcium channels selectively in all or a subset of pacemakers. We evaluated the impact of individual channels in setting both slow daily changes in basal calcium levels and in fast fluctuations. Finally, we measured PERIOD (PER) protein staining levels and behavior to determine which channels provide feedback to the molecular clock and which are required for normal circadian output from the pacemaker network.
Results
The Rhythms of Slow and Fast Calcium Activity Changes Show Similar Daily Patterns.
Previously, we reported that five major groups of circadian pacemaker neurons each exhibit daily calcium rhythms with distinct phases (19). These results stand in apparent contrast to descriptions of synchronous daily electrical activity rhythms among three of these groups: s-LNv, l-LNv, and DN1 (12, 27, 28). The electrical activity rhythms were recorded ex vivo from different brains isolated at four to six different time points of the day. In contrast, we measured calcium rhythms in vivo by scanning individual flies every 10 min for 24 h. Because of the close peak phases of calcium rhythms in s-LNv, l-LNv, and DN1 (between late night to midmorning) (19) and because of the coarse sampling of electrophysiological studies, it is not certain whether calcium and electrical activity patterns are in fact distinct. To help clarify apparent differences in results derived from the two sets of studies, we began by performing short-term continuous in vivo calcium imaging (1-Hz volumetric rate) on fly brains that were exposed acutely before each imaging experiment at five different times of day. We focused on the LNd because this group has a phase of calcium rhythms most distinct from those of s-LNv, l-LNv, and DN1. In addition, the daily electrophysiological activity pattern of LNd has not previously been reported. We found that ∼0.1-Hz calcium fluctuations peaked at around the same ZT8 to ZT12 (ZT, Zeitgeber Time), at which time this pacemaker group shows peak intensity in its daily calcium rhythm (Fig. 1A and SI Appendix, Fig. S1). The time course of “fast” (by circadian standards) calcium fluctuations in the late day suggests they might be caused by the calcium influx that occurs during single action potentials or bursts of them (Fig. 1A) (23, 24, 26). This result suggested that one or more LNd pacemakers exhibit a daily rhythm in electrical neural activity that is roughly cophasic with this pacemaker group’s slow daily calcium rhythm.
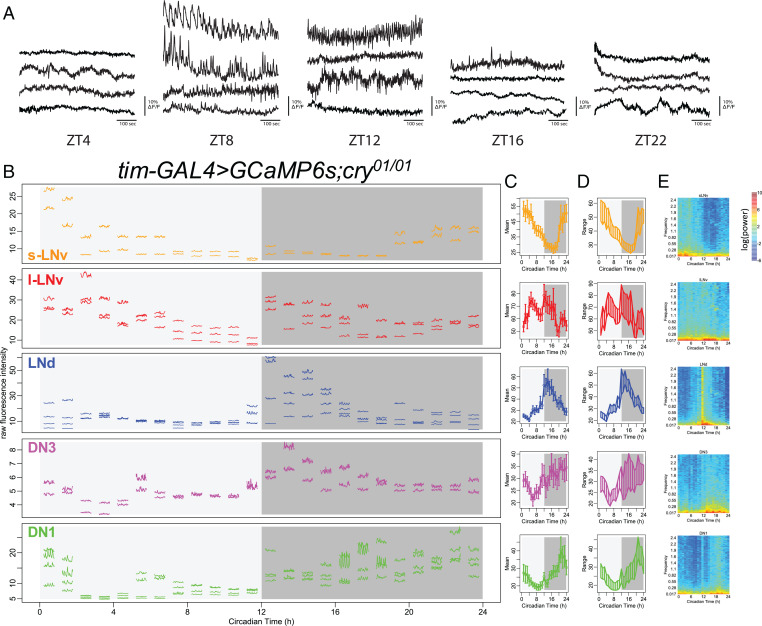
Fig. 1.
Daily pattern of fast calcium activity in circadian pacemaker neurons. (A) Representative calcium activity traces of LNd recorded at 1 Hz from five different times of day: ZT4, ZT8, ZT12, ZT16, and ZT22. For each time point, the fly’s brains are acutely exposed for short-term imaging. (B) Raw calcium fluorescence intensity traces from one representative fly. Each segmented trace is 1-min activity of a single neuron recorded at 5 Hz; 24 segments are recorded over 24 h with 1-h intervals. Activity traces of five circadian pacemaker groups are plotted in separate panels and color coded. For each group, only cells that can be tracked throughout all 24 recording sessions are showed. In this specific fly, three s-LNv, four l-LNv, three LNd, four DN3, and four DN1 can be reliably tracked across 24 recording sessions. Circadian pacemaker neurons exhibit daily modulation in basal calcium level and in the frequency of fast calcium spikes. (C) Daily pattern of mean calcium intensity over the 1-min recording session at each of the 24 time points averaged across all six flies studied. Error bars denote SEM. (D) Daily patterns of the range of calcium transients over the 1-min recording session at each of the 24 time points averaged across all six flies studied. (E) Daily pattern of the power spectrum over the 1-min recording session at each of 24 time points averaged across all six flies studied.
Because the slow and fast calcium LNd rhythms are synchronous as measured, it is formally possible that one rhythm is downstream of the other; for example, the slow calcium rhythm could be the consequence of a rhythm in the fast. Alternatively, these two processes could be completely distinct. To better understand the relationships between the two and better describe their phases across the entire network, we performed a series of short-term (1-min) high-frequency (5-Hz) in vivo calcium imaging episodes at 1-h intervals using the light-sheet microscope dual-channel Objective Coupled Planar Illumination (OCPI-2) (21). In so doing, we tracked both slow basal calcium level and fast calcium fluctuations in the same individual neurons from all five major circadian pacemaker groups; we collected these data consecutively from single brains for entire 24-h durations (Fig. 1B). To ensure minimal disruption to the circadian clocks due to repeated optical scanning, we used cry01 flies for these experiments, which are null for the internal photosensitive protein CRYPTOCHROME (29). On average, all circadian neuron groups displayed slow calcium rhythms comparable with those we previously reported (19), except for l-LNv, which showed additional daily calcium activation peaks right after the time of lights off and again just before lights on. Nevertheless, all pacemaker groups displayed daily changes in the minimal calcium level, demonstrating that their basal calcium levels cycle with a daily rhythm (Fig. 1 C and D). We found that within all five pacemaker groups, changes in basal calcium levels and in fast calcium fluctuations shared similar daily patterns; when basal calcium levels were high within a single pacemaker group, that group also exhibited larger-amplitude fast calcium fluctuations (SI Appendix, Fig. S3). Power spectrum analysis clearly revealed that, for individual neurons within each pacemaker group, calcium activity at all frequency domains increased when the basal calcium level was high (Fig. 1E). We asked whether the change in the incidence of high-frequency fluctuations, measured using the genetically-encoded calcium reporter GCaMP6s could have a technical basis: specifically, whether it derives from a higher level of photon shot noise due to the higher baseline intensity. In order to normalize the effect of shot noise, we also calculated the intensity of the calcium signal as the square root of photon number collected from an individual region of interest (ROI). In this analysis, we still found daily rhythms in fast calcium fluctuations (SI Appendix, Fig. S2). These results support the hypothesis that, in each circadian pacemaker group, fast calcium fluctuations exhibit a daily rhythmic pattern that is cophasic with a slow daily rhythm in basal calcium levels.
An RNAi Screen to Identify Potential Contributions of Different Calcium Channels.
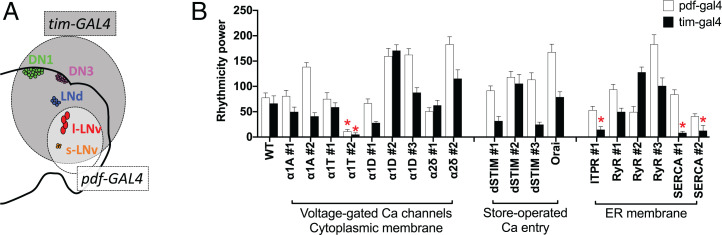
The observations described above support the conclusion that for individual pacemakers, slow and fast calcium activities covary across the day. Yet, these observations do not reveal whether the two rhythms are mechanistically linked or represent independent functions. To identify the sources for different calcium rhythms and ask about their relatedness, we used RNA interference (RNAi) methods to knock down different calcium channels. We performed a limited screen for calcium channels, including three subtypes of α1-subunits and one type of α2δ-subunits of voltage-gated calcium channels. In addition, we tested two types of store-operated calcium entry, dSTIM and dOrai, and two types of calcium channels on the ER, RyR and inositol trisphosphate receptor (Itpr); finally, we included the sarco/endoplasmic reticulum calcium pump (SERCA). By knocking down these genes selectively in circadian pacemaker neurons using tim-GAL4 or in a subset of eight pacemaker neurons that express the pigment dispersing factor neuropeptide (PDF) using pdf-GAL4, we first tested whether any of these genes are required for normal circadian behavioral rhythms. We found evidence for the involvement of three (Fig. 2 and Table 1) as indicated by increases in the percentage of arrhythmic (AR) flies tested under constant darkness (DD). Reduced expression of the α1T channel, which encodes the α1-subunit for the T-type voltage-gated calcium channel, caused the strongest behavioral arrhythmicity when driven by either pdf-GAL4 or tim-GAL4 (65 and 83%, respectively) with one of the two RNAi lines tested (KK100082). Likewise, knockdown of expression of the SERCA calcium pump caused strong arrhythmicity in two different RNAi lines. Yet, knocking down SERCA with the stronger RNAi line in all circadian pacemakers by tim-GAL4 also shortened the flies’ life spans; 69% of flies died during behavioral experiments. Knockdown of another calcium channel on the ER membrane, Itpr, also affected the circadian rhythm in behavior when driven by tim-GAL4. These behavioral deficits suggested that α1T, SERCA, and ITPR might be involved in the regulation of calcium rhythms in circadian pacemaker neurons.
Fig. 2.
RNAi screening for calcium channels required for circadian rhythms. (A) Schematic of the five pacemaker groups in the brain superimposed with a Venn diagram of expression driven by tim-GAL4 (all groups) and pdf-GAL4 (s- and l-LNV groups). (B) Summary of behavioral screening for calcium channel knockdowns that reduced average rhythm strength in locomotor activity under DD (Table 1). *P < 0.05 (two-way ANOVA followed by Dunnett's multiple comparisons test).
Table 1.
The rhythm strength and period of locomotor activity under DD of flies expressing calcium channel RNAi transgenes in PDF neurons or all clock neurons
| Gene | CG no. | Genotype* | N † | AR, % | Period | pwr | Wid | SNR** | ACT day | ACT night | ACT cycle |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | — | pdf > dcr2 | 24 | 4 | 24.8 | 81.3 | 6.0 | 1.6 | 23.7 | 27.9 | 25.8 |
| tim > dcr2 | 14 | 21 | 24.2 | 66.3 | 5.1 | 1.6 | 20.4 | 10.2 | 15.3 | ||
| α1A cac | 43,368 | tim > JF02572 | 16 | 0 | 24.1 | 49.8 | 4.5 | 0.5 | 19.9 | 10.7 | 15.3 |
| pdf > JF02572 | 16 | 6 | 24.5 | 81.0 | 4.1 | 0.7 | 16.4 | 16.1 | 16.2 | ||
| pdf > kk104178 | 16 | 0 | 25.4 | 138.5 | 8.6 | 3.1 | 13.9 | 28.7 | 21.3 | ||
| tim > kk104178 | 14 | 7 | 24.4 | 41.1 | 4.9 | 1.0 | 14.8 | 21.0 | 17.9 | ||
| α1T | 15,899 | pdf > JF02150 | 14 | 7 | 24.5 | 75.1 | 4.9 | 1.5 | 22.1 | 15.6 | 18.8 |
| tim > JF02150 | 16 | 0 | 23.6 | 59.0 | 4.4 | 1.1 | 14.3 | 2.4 | 8.4 | ||
| pdf > KK100082 | 23 | 65 | 24.4 | 21.4 | 3.0 | 0.7 | 14.4 | 11.9 | 13.2 | ||
| tim > KK100082 | 23 | 83 | 24.2 | 13.2 | 2.0 | 0.4 | 13.5 | 12.5 | 13.0 | ||
| α1D | 4,894 | pdf > JF01848 | 14 | 0 | 24.3 | 66.7 | 4.4 | 0.9 | 18.7 | 13.4 | 16.1 |
| tim > JF01848 | 16 | 0 | 24.3 | 28.1 | 4.1 | 0.5 | 14.3 | 5.7 | 10.0 | ||
| pdf > GD1737 | 16 | 0 | 24.7 | 159.8 | 6.4 | 2.4 | 19.8 | 10.2 | 15.0 | ||
| tim > GD1737 | 23 | 0 | 24.4 | 170.8 | 6.3 | 2.3 | 18.4 | 14.8 | 16.6 | ||
| pdf > HMS00294 | 16 | 0 | 24.5 | 162.5 | 5.4 | 1.9 | 25.7 | 30.2 | 27.9 | ||
| tim > HMS00294 | 16 | 0 | 24.0 | 87.9 | 4.1 | 0.8 | 19.7 | 7.9 | 13.8 | ||
| α2δ | 12,295 | pdf > JF01825 | 16 | 6 | 24.3 | 50.8 | 4.6 | 0.7 | 27.1 | 18.0 | 22.6 |
| tim > JF01825 | 16 | 13 | 23.9 | 62.7 | 5.9 | 1.0 | 25.0 | 13.7 | 19.4 | ||
| pdf > KK101267 | 16 | 0 | 24.7 | 183.1 | 6.2 | 2.6 | 17.0 | 21.3 | 19.1 | ||
| tim > KK101267 | 15 | 0 | 24.0 | 115.5 | 4.9 | 1.4 | 20.7 | 11.8 | 16.2 | ||
| Orai | 11,430 | pdf > HMC03562 | 15 | 0 | 24.6 | 167.6 | 6.3 | 2.5 | 10.4 | 14.2 | 12.3 |
| tim > HMC03562 | 20 | 10 | 23.6 | 78.9 | 4.1 | 0.8 | 10.6 | 4.2 | 7.4 | ||
| pdf > uas-dOrai | 13 | 0 | 24.9 | 108.9 | 5.6 | 1.0 | 24.8 | 26.2 | 25.5 | ||
| tim > uas-dOrai | 15 | 0 | 24.3 | 115.4 | 5.7 | 1.2 | 16.1 | 8.8 | 12.4 | ||
| dSTIM | 9,126 | pdf > GLC01785 | 16 | 6 | 24.2 | 91.8 | 7.4 | 2.0 | 21.4 | 18.6 | 20.0 |
| tim > GLC01785 | 15 | 27 | 23.7 | 31.9 | 3.6 | 0.7 | 13.3 | 6.0 | 9.7 | ||
| pdf > KK102366 | 23 | 0 | 25.4 | 118.3 | 6.2 | 1.2 | 11.4 | 12.6 | 12.0 | ||
| tim > KK102366 | 22 | 5 | 23.9 | 105.5 | 4.8 | 1.6 | 11.9 | 12.6 | 12.2 | ||
| pdf > JF02567 | 16 | 0 | 24.5 | 113.5 | 5.3 | 1.3 | 23.5 | 24.5 | 24.0 | ||
| tim > JF02567 | 12 | 25 | 23.8 | 24.7 | 3.3 | 0.6 | 7.3 | 3.4 | 5.3 | ||
| pdf > uas-dSTIM | 15 | 0 | 24.3 | 129.4 | 5.8 | 1.2 | 23.6 | 11.5 | 17.5 | ||
| tim > uas-dSTIM | 14 | 0 | 23.7 | 137.1 | 5.3 | 1.4 | 21.1 | 6.2 | 13.7 | ||
| SERCA | 3,725 | pdf > JF01948 | 14 | 0 | 25.2 | 84.1 | 6.4 | 1.3 | 33.6 | 35.2 | 34.4 |
| tim > JF01948 | 12 | 50 | 25.3 | 12.7 | 2.8 | 1.4 | 13.1 | 10.1 | 11.6 | ||
| pdf > KK107371 | 16 | 13 | 24.5 | 40.9 | 4.4 | 0.7 | 28.3 | 29.1 | 28.7 | ||
| tim > KK107371 ‡ | 15 | 73 | 23.7 | 65.4 | 3.4 | 0.7 | 14.9 | 8.7 | 11.8 | ||
| RyR | 10,844 | pdf > HM05130 | 15 | 0 | 24.7 | 94.0 | 6.2 | 1.7 | 19.0 | 19.1 | 19.0 |
| tim > HM05130 | 16 | 6 | 24.0 | 49.8 | 4.6 | 0.8 | 17.4 | 6.7 | 12.0 | ||
| pdf > JF03381 | 39 | 28 | 24.4 | 55.2 | 3.4 | 0.7 | 16.5 | 11.7 | 14.1 | ||
| tim > JF03381 | 28 | 7 | 23.9 | 128.1 | 5.3 | 1.4 | 19.1 | 10.1 | 14.6 | ||
| pdf > KK101716 | 13 | 8 | 24.7 | 165.1 | 6.6 | 2.5 | 15.1 | 17.1 | 16.1 | ||
| tim > KK101716 | 16 | 0 | 24.8 | 101.1 | 4.9 | 1.1 | 12.1 | 17.6 | 14.8 | ||
| Itpr | 1,063 | pdf > itprRNAi#1 | 36 | 11 | 24.2 | 51.7 | 4.0 | 0.8 | 18.9 | 18.1 | 19.4 |
| tim > itprRNAi#1 | 20 | 50 | 24.4 | 14.7 | 2.8 | 0.9 | 9.0 | 5.6 | 9.7 |
*All genotypes include UAS-dcr2 if it is not mentioned (UAS = Upstream Activating Sequence). CG no.: coding gene number in fly genome; AR: arrhythmic; pwr: power of rhythmicity by chi-square periodogram; wid: width of rhythmicity peak by chi-square periodogram; ACT: averaged locomotor activity level.
†N = flies alive at the end of the testing period.
‡In this genotype, 69% of flies died during the behavioral experiment.
** SNR = Signal to Noise Ratio
Slow Calcium Rhythms Require inositol trisphosphate receptor (ITPR).
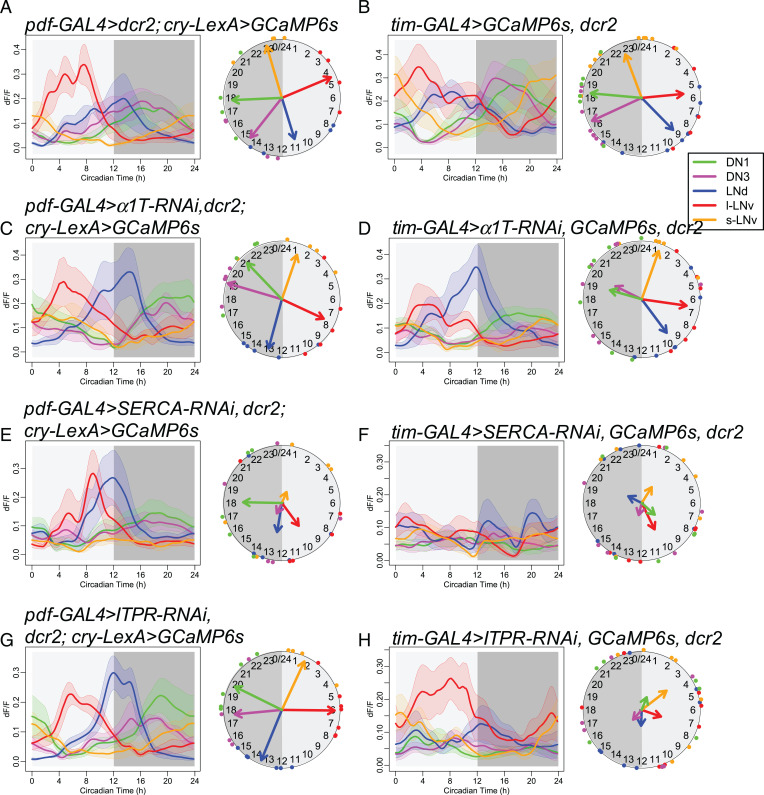
We then asked whether the α1T, SERCA, and ITPR channels that regulate circadian behavior also influence calcium rhythms. We measured GCaMP6 fluorescence during in vivo 24-h recordings in Drosophila knockdowns in all circadian neurons or in just the subset of PDF-positive ones (Fig. 3). Although knocking down α1T caused the strongest behavioral deficits, the slow calcium rhythms of all pacemaker neuron groups in these flies were similar to those in the control genotypes. The amplitude of calcium rhythms in flies with α1T knocked down in all pacemaker neurons showed a nonsignificant trend of decrease to 59.3% on average (SI Appendix, Fig. S5A), while their activity phases were still normal (Fig. 3D and SI Appendix, Fig. S4). In contrast, when SERCA was knocked down in PDF neurons (Fig. 3E) or in all circadian neurons (Fig. 3F) (using the stronger RNAi line KK107371), the slow calcium activities of these neurons were largely AR. The amplitudes of calcium fluctuations were decreased to 37.8% on average (SI Appendix, Fig. S3), and the coherence was lost within groups (Rayleigh test, P > 0.2). Likewise, the calcium rhythms were still normal when Itpr was knocked down in PDF neurons (Fig. 3G) but became largely AR when Itpr was knocked down in all circadian neurons (Fig. 3H). In the latter case, the coherence of peak phase was lost within groups (Rayleigh test, P > 0.1), consistent with the behavioral phenotypes of the two manipulations for Itpr RNAi. Together, these results implicate SERCA and IP3R channel activities as essential for slow calcium rhythms and suggest that the ER may be a key calcium source for the daily fluctuations of the basal calcium levels in circadian pacemaker neurons.
Fig. 3.
RNAi screening for calcium channels required for CCRs. (A and B) Daily calcium activity rhythms of five major circadian pacemaker groups in control flies for (A) pdf-GAL4–driven knockdown (n = 4 flies) and (B) tim-GAL4–driven knockdown (n = 12 flies) in the first day under DD. (C and D) Daily calcium activity rhythms are normal in (C) pdf-GAL4–driven α1T knockdown (KK100082) flies (n = 5 flies) and (D) tim-GAL4–driven α1T knockdown flies (n = 7 flies). (E and F) Daily calcium activity rhythms are partially impaired in (E) pdf-GAL4–driven SERCA knockdown (KK107371) flies (n = 5 flies) and completely impaired in (F) tim-GAL4–driven SERCA knockdown flies (n = 6 flies). (G and H) Daily calcium activity rhythms are normal in (G) pdf-GAL4–driven Itpr knockdown flies (n = 7 flies) and impaired in (H) tim-GAL4–driven Itpr knockdown flies (n = 7 flies).
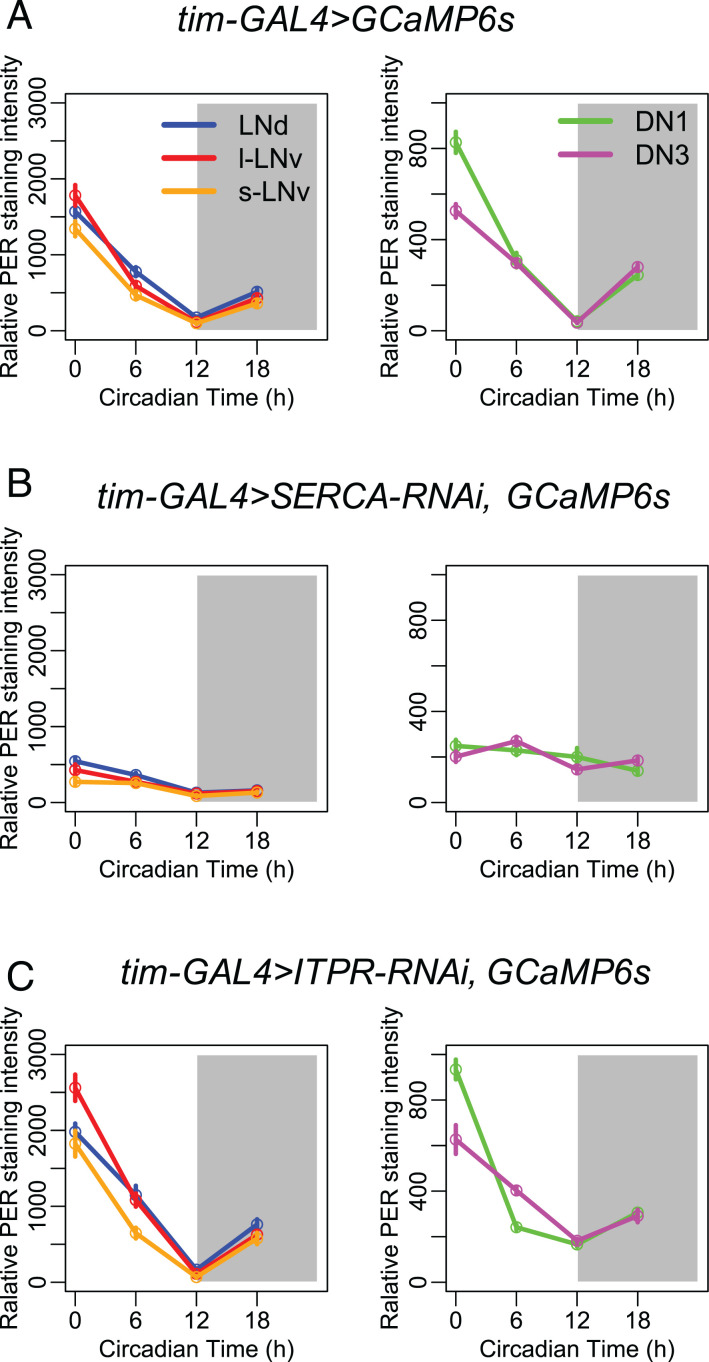
Because the slow calcium rhythms are driven by molecular clock gene oscillations (19), we asked whether the molecular clock controls slow calcium rhythms by regulating SERCA and/or ITPR levels. If SERCA and Itpr are downstream of the molecular clock, knocking down these genes would affect calcium rhythms and behavior but not affect the molecular clock itself. We examined PER protein levels in all five circadian pacemaker groups at four 12-h light:12-h dark (LD) time points, and we found that PER cycling appeared robust in Itpr knockdown flies but was clearly diminished in SERCA knockdown flies (Fig. 4). Therefore, in this system, only Itpr appears to operate principally downstream of the molecular clock and is necessary to generate daily rhythms in basal calcium levels.
Fig. 4.
PER protein rhythms of control flies and flies with SERCA or Itpr knocked down in all pacemaker neurons. (A) Averaged PER protein staining intensity at four different time points (ZT0, ZT6, ZT12, and ZT18) in five groups of circadian pacemaker neurons from control flies. (B) PER protein rhythms are diminished when knocking down SERCA in all pacemaker neurons by tim-GAL4 (KK107371). (C) PER protein rhythms are robustly cycling when knocking down Itpr in all pacemaker neurons by tim-GAL4 (n > 3 flies at each time point for each genotype).
Fast Calcium Fluctuations Require α1T Calcium Channels and ITPR.
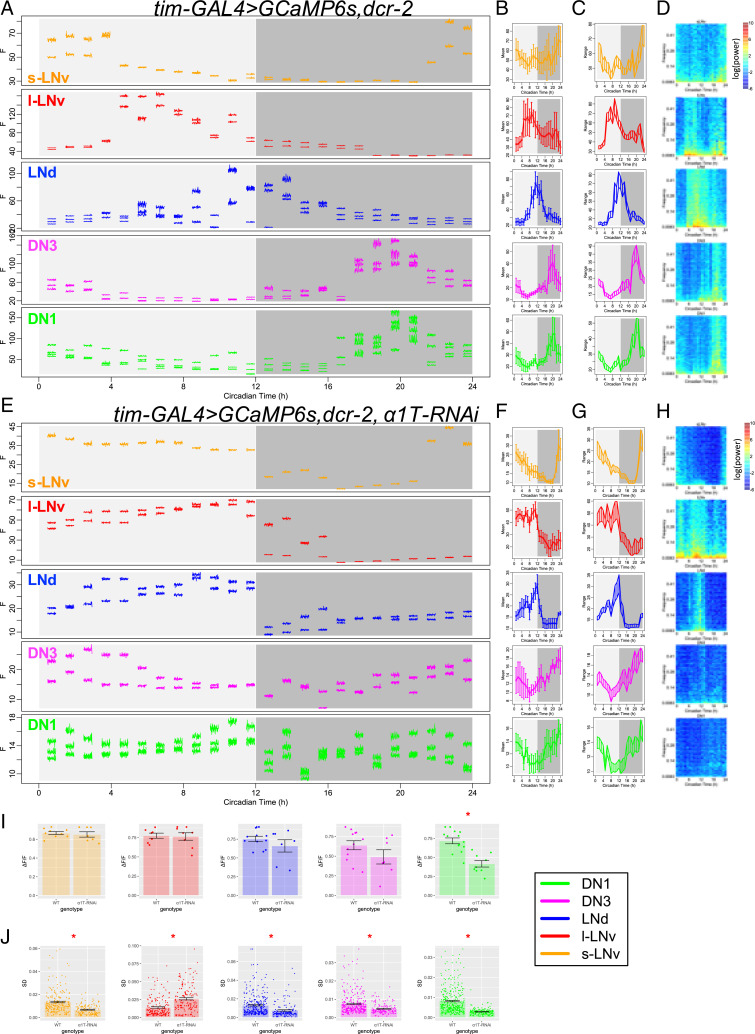
Knocking down the RNA for α1T voltage-gated calcium channels in pacemaker neurons impaired circadian rhythms in behavior but did not affect circadian rhythmicity in basal calcium levels within those neurons. Therefore, we next asked whether α1T may underlie the circadian rhythm of fast calcium fluctuations in pacemakers. To test this, we again performed imaging across a series of short-term (2-min) high-frequency (1-Hz) calcium measurements on the same flies for a 24-h day with 1-h intervals (similar to Fig. 1 yet with a slightly lower sampling rate and using flies that were wild type for cry). By comparing control Drosophila with those with α1T knocked down in all circadian pacemaker neurons by tim-GAL4, we found that knocking down α1T did not affect the daily rhythms in the basal (slow) calcium level in any pacemaker group except for the DN1, which showed a reduction in the day–night difference of the basal calcium level (Fig. 5 A–I and SI Appendix, Fig. S6). These high-frequency measures of slow basal calcium levels largely conform with those obtained with the slow-frequency (every 10 min) recording sessions (cf Fig. 3D). However, the high-frequency recording revealed that fast calcium fluctuations were significantly reduced in all circadian pacemaker neurons of the α1T knockdown flies, except for l-LNv (Fig. 5J and SI Appendix, Fig. S7). That specific pacemaker group instead displayed higher levels of fast calcium fluctuations. These results indicated that, at least in the majority of circadian pacemaker neurons, α1T is required for strong daily rhythms in fast calcium fluctuations and that the rhythm of fast calcium fluctuations can be selectively impaired.
Fig. 5.
α1T knockdown reduces fast calcium fluctuations. (A–D) As in Fig. 1 A–D, (A) raw calcium fluorescence intensity traces from one representative control fly are shown. Each segmented trace is 2-min activity recorded at 1 Hz. Averaged daily patterns of (B) mean calcium intensity, (C) the range of calcium transient, and (D) the power spectrum (n = 4 flies). (E–H) As in A–D, raw calcium activity traces from one representative fly with α1T knockdown (KK100082) in all pacemaker neurons and averaged daily patterns of mean, range, and power spectrum in this genotype (n = 4 flies) are shown. (I) Bar plots of the daily range of calcium signal for individual neurons of five pacemaker groups between control flies and α1T knockdown flies. The daily variation of DN1 calcium reduces in α1T knockdown flies (t test). *P < 0.05. (J) Bar plots of SDs of calcium signal in each recording session for individual neurons of five pacemaker groups between control flies and α1T knockdown flies. The SDs of fast calcium fluctuations in s-LNv, LNd, DN3, and DN1 are smaller, while that in l-LNv is larger in α1T knockdown flies than that in control (t test). *P < 0.05.
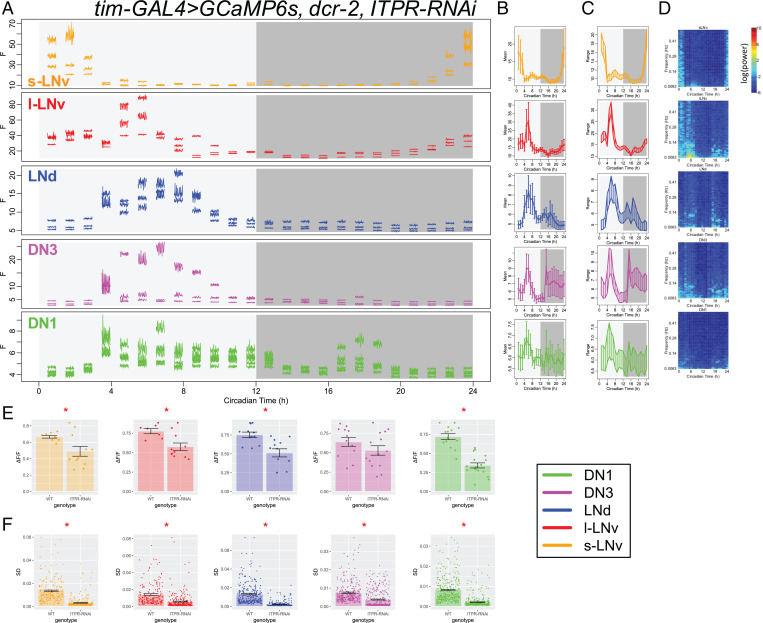
We then asked whether the fast calcium fluctuations are also affected in flies with Itpr knocked down in all pacemaker neurons. We performed the same high-frequency calcium imaging and analysis in Itpr knockdown flies (Fig. 6 and SI Appendix, Fig. S8) as in α1T knockdown flies. The daily variation of pacemaker calcium was greatly reduced in Itpr knockdown flies (Fig. 6E), which largely recapitulated previous observations with less frequent sampling (cf Fig. 3H). In addition, fast calcium fluctuations were significantly reduced in all circadian pacemaker neurons of the Itpr knockdown flies (Fig. 6F and SI Appendix, Fig. S7). These results suggested that rhythms in basal calcium levels, regulated by ITPR, may be necessary for rhythms in fast calcium fluctuations.
Fig. 6.
Itpr knockdown reduces fast calcium fluctuations. (A–D) As Fig. 1 A–D, (A) raw calcium fluorescence intensity traces from one representative fly with Itpr knockdown in all pacemaker neurons are shown. Each segmented trace is 2-min activity recorded at 1 Hz. Averaged daily patterns of (B) mean calcium intensity, (C) the range of calcium transient, and (D) the power spectrum (n = 5 flies). (E) Bar plots of the daily range of calcium signal for individual neurons of five pacemaker groups between control flies and Itpr knockdown flies. The daily variation of l-LNv, LNd, and DN1 calcium reduces in Itpr knockdown flies (t test). *P < 0.05. (F) Bar plots of SDs of calcium signal in each recording session for individual neurons of five pacemaker groups between control flies and Itpr knockdown flies. The SDs of fast calcium fluctuations in all circadian pacemaker neuron groups are smaller in Itpr knockdown flies than that in control (t test). *P < 0.05.
Discussion
In this study, we used in vivo 24-h high-frequency calcium imaging and genetic screening to study the cellular biology of daily calcium rhythms in circadian pacemaker neurons of Drosophila. We found that the calcium rhythm is in fact a composite; it reflects daily fluctuations in both a slow component (basal levels) and a fast one (high-frequency fluctuations). We interpret fast calcium fluctuations as representations of calcium dynamics that occur as neurons fire single action potentials or bursts of them. While it is not sufficient to resolve single action potentials, GCaMP6-induced fluorescence is a good index of neuronal electrical activity (e.g., refs. 24 and 26). For individual identified pacemakers, these two calcium rhythms share the same daily pattern, yet distinct calcium sources appear to contribute differentially to these two rhythms. An extracellular calcium influx, through plasma membrane calcium channels that include the α1T subunit, is critical for the fast calcium fluctuations. In contrast, calcium fluxes from the ER via the channel ITPR are required for both the slow rhythms in the basal calcium levels and the fast ones. Importantly, both channels are essential for normal circadian behavior. Thus, the molecular clocks may drive circadian rhythms in pacemaker neuron output by regulating different calcium sources to generate coordinate but distinct rhythms in its calcium activities.
CCRs are widespread across taxa (15, 30). Calcium rhythms are required for circadian pacemaker functions in both rodents and Drosophila (31, 32). Studies on mammalian circadian pacemakers in the SCN are controversial regarding the temporal relationship between the CCR and rhythms in electrical activity, such as in spontaneous firing rate (SFR) and in resting membrane potential (RMP). Recordings from SCN slice cultures showed that the phases of CCR in individual pacemakers are diverse and could be different from the populational phase of SFR rhythms (16, 33). However, the populational SFR phase is composed of many diverse phases of SFR rhythms on the individual cell level (34); it is unclear whether SFR phases align with the phases of CCR. Imaging with both voltage sensor and calcium sensors in SCN slices, Brancaccio et al. (35) concluded that RMP rhythms and CCR were in phase, yet Enoki et al. (18) concluded that RMP rhythms and CCR were in phase in the ventral SCN but that in dorsal SCN, the CCR phase led the RMP rhythms by ∼2 h. Because the voltage sensor signal measured from dorsal SCN may derive from the neural processes of ventral SCN neurons, the cellular interpretation of these results is unclear. Another source for the inconsistency might be the culture conditions; in fact, when SCN neurons were recorded in vivo by photometry, the rhythms in fast calcium activity were in phase with slow calcium rhythms (17). In general, comparisons of population rhythms and rhythms in single cells are not easily reconciled. Our recordings that tracked pacemaker neurons from different identified groups in vivo for 24 h showed that at the single cell level, slow calcium rhythms (CCRs) were unambiguously in phase with rhythms in fast calcium fluctuations; the latter likely reflects rhythms in SFR (Fig. 1).
To measure the fast calcium fluctuations, we employed a high-frequency light-sheet scanning microscope termed OCPI-2 (21). OCPI-2 resolved limitations present in previous microscope designs that created a bottleneck in terms of the rate at which tissue volumes can be repeatedly scanned. Importantly, it does so without compromising photon efficiency or spatial resolution. Its use here was instrumental in letting us sample the whole-brain volume at frequencies as high as 5 Hz periodically throughout the day. As before (19), we were mindful that the wavelength and amount of total incident light may activate pacemaker neurons and alter molecular clocks (36). We used cry01 flies to avoid the direct light responses of pacemaker neurons and found that all pacemaker groups displayed slow calcium rhythms, with patterns that were comparable with those we previously reported (19), except for that of l-LNv (Fig. 1). That group displayed an additional calcium peak in the early evening and another just before lights on. Because l-LNv innervates the optic lobes and receives large-scale visual inputs (37), we speculate that the repeated optical scanning might anomalously activate l-LNv in the evening via visual systems. In the later experiments, when we reduced the illumination duration per hour from 31.5 s (Fig. 1) (7-ms exposure time, 15 frames per stack, 5 Hz for 1 min) to 2.4 s (Figs. 5 and 6) (1-ms exposure time, 20 frames per stack, 1 Hz for 2 min), l-LNv did not show the additional evening peaks; this was true even when measured in flies wild type for cry. Shorter durations of light exposure permit all pacemaker groups to display slow calcium rhythms (Fig. 5A) with phases similar to those obtained with the slow-frequency (every 10-min) recording sessions (cf Fig. 3B and ref. 19). Therefore, the concern to avoid technical artifacts when imaging from light-sensitive pacemaker appears especially acute in the case of l-LNv. Nevertheless, by carefully tuning the illumination intensity for calcium imaging, we propose that it is possible to monitor normal slow calcium rhythms and fast calcium fluctuations from the same individual pacemaker neurons.
The causal relationships between clock gene rhythms, calcium rhythms, and electrical activity rhythms in the SCN remain generally unresolved. Treating SCN slices with Tetrodotoxin (TTX, to block Na-dependent action potentials) diminished SFR rhythms (16), partially affected RMP rhythms and CCRs (18, 33, 38), and slowly affected clock gene rhythms over several days (39). Dispersed SCN cells in vitro showed a TTX-resistant CCR, suggesting that CCR is driven by clock gene rhythms (40). Thus, the variation in CCR sensitivity to TTX treatment might be caused by the degree to which clock gene rhythms in vitro become progressively dysfunctional. In Drosophila, our findings suggested that clock gene rhythms drive two components of the CCR—both basal calcium levels and fast calcium fluctuations—via circadian regulation of the ER channel ITPR and membrane voltage-gated calcium channel α1T. Both channels might then contribute to SFR and RMP rhythms. Similarly, in SCN pacemakers, pharmacologically blocking another ER channel RyR affected both CCR and SFR rhythms (16), suggesting that rhythms in basal calcium levels are regulated by calcium from ER and are required for fast electric activity rhythms. In addition, SCN pacemakers also showed a circadian rhythm in fast calcium activity mediated by L-type voltage-gated calcium channels (8). Pharmacologically blocking these membrane channels affected SFR rhythms and in some case, affected CCR (16, 18). In our studies, manipulating a membrane voltage-gated calcium channel in all or a subset of pacemakers selectively affected rhythms in fast calcium fluctuations, which likely reflected SFR rhythms and thus, impaired circadian outputs; however, it did not significantly affect the slow rhythms in basal calcium levels (Fig. 5). Manipulating the ER calcium channel IP3R in all pacemakers affected rhythms in both slow and fast calcium rhythms. Therefore, in parallel to mammalian SCN neurons, Drosophila circadian pacemakers generate calcium rhythms by regulating both ER and extracellular calcium sources. Since our results suggest little or no role for the RyR channel (Fig. 2B), the daily rhythmic regulation in fly pacemakers likely acts on a different set of ER and cytoplasmic membrane channels from those in mammalian pacemakers.
We also presented RNAi evidence implicating the Itpr and Ca-alpha1T genes in pacemaker cell calcium fluxes. However, for each, only a single RNAi proved effective in reducing rhythmic power and increasing percentage of arrhythmicity. Corroboration for our findings is provided by a prior report (41), which found that the same RNAi line we used decreases Itpr RNA levels. Likewise, we find corroboration in a prior report by Jeong et al. (42), who described a specific Ca-alpha1T insertion line as a protein null mutation that exhibited reduced percentage of rhythmicity and power in DD but without a change in circadian period or in the daily per rhythm. They concluded that Ca-alpha1T expression does not affect the central clock mechanism and surmised instead that it likely affects the output of the circadian system, specifically via a requirement for the channel to permit normal physiological activation of pacemaker neurons. That work provides independent genetic confirmation of our RNAi results, and its conclusions align precisely with our own.
The RNAi knockdown experiments indicate a role for the ER calcium channel SERCA in supporting slow calcium rhythms in Drosophila pacemakers and behavioral rhythms. However, SERCA knockdown also produced high rates of lethality and among survivors, strong effects on the PER molecular oscillation. We conclude that SERCA, which maintains the ER cytoplasmic calcium gradient, is essential for the normal physiology of the cells but that these additional phenotypes precluded an assessment of its precise role in circadian rhythmicity. In contrast to the situation with SERCA, our results support a hypothesis that ITPR is a crucial actuator of the molecular clock. Previous transcriptomic analysis also supports that possibility; in circadian neurons, Itpr displays rhythmic expression, while SERCA does not (SI Appendix, Fig. S9) (43). Knocking down Itpr in all circadian neurons (with tim-GAL4) generally caused stronger deficits in both behavioral rhythms and calcium rhythms than in just the PDF-positive neurons (with pdf-GAL4). One possibility might be that the RNAi expression level is lower when driven by pdf-GAL4 than when driven by tim-GAL4. However, in SI Appendix, Fig. S5A, knocking down Itpr in all circadian neurons caused stronger deficits in the amplitude of calcium rhythms of non–PDF-positive neurons (LNd, DN1, and DN3) than in those of PDF-positive neurons (s-LNv and l-LNv). Hence, a difference in the vulnerability to IP3R disruption between PDF-negative and PDF-positive neurons might provide an additional or alternative explanation for why the pdf-GAL4–driven knockdown of Itpr affected neither calcium rhythms nor behavior. We also note that the literature provides examples, wherein a single RNAi line produces stronger behavioral effects when coupled with pdf-Gal4 than with tim-GAL4 (e.g., ref. 44). Such findings suggest that the final result in such experiments will represent a mixture of GAL4 driver line strength and the differential contributions of the UAS:Responder gene product across different positions within an affected neural circuit.
Finally, the RNAi knockdown of the plasma membrane calcium channel α1T indicated a role for voltage-gated T-type calcium channels in the final rhythmic output of the pacemakers. Consistent with a role in the presumed output pathway, impairing the rhythm of fast calcium activity strongly affected circadian behavior but did not affect the molecular clock (42) or the slow calcium rhythms. T-type channels play a crucial role in other pacemakers, such as the Sino-Atrial node of the mammalian heart (45, 46). Their conduction in the hyperpolarized state and closure at more depolarized potentials are central to their role in generating bursting dynamics with periods much longer than the membrane time constant (47). Given the power spectrum of the fluctuations we observed, it seems possible that α1T channels play a similar role in the fast (∼0.1-Hz) fluctuations of Drosophila circadian neurons. Remarkably, the expression of α1T also displays a circadian rhythm, with distinct phases in different groups of pacemaker neurons (SI Appendix, Fig. S9) (43). Collectively, these results suggest that Itpr and α1T channel activity are together critical to produce clock regulation of rhythms in both slow and fast calcium activities of key pacemaker neurons.
Materials and Methods
Fly Stocks.
Flies were reared on standard yeast-supplemented cornmeal/agar food at room temperature. After eclosion, male flies were entrained under LD cycles at 25 °C for at least 3 d. The tim > GCaMPS6s; cry01/01 flies were entrained under LD for more than 6 d.
The following fly lines were previously described: tim(UAS)-GAL4 (48), pdf-GAL4 (49), cry-LexA (20), UAS-GCaMP6s, and LexAop-GCaMP6s (24). UAS-dSTIM and UAS-dOrai (50) were gifts from G. Hasan, National Center For Biological Sciences, Bengaluru, India. UAS-Itpr RNAi (41) was a gift from Mark Wu, Johns Hopkins University, Baltimore, MD. Stable lines UAS-dcr2; tim(UAS)-GAL4; UAS-GCaMP6s and pdf-GAL4;UAS-dcr2;cry-LexA,LexAop-GCaMP6s were created for RNAi screening of calcium channels. The cry-LexA line was a gift from F. Rouyer, National Center for Scientific Research (France), Gyf, Paris, France.
RNAi lines were obtained from the Bloomington Drosophila Stock Center (BDSC) , the Vienna Drosophila Resource Center (VDRC), and the Tokyo Stock Center: two lines for cac (CG43368): UAS-KK101478-RNAi (VDRC 104168) and UAS-JF02572-RNAi (BDSC 27244); two lines for α1T (CG15899): UAS-KK100082-RNAi (VDRC 108827) and UAS-JF02150-RNAi (BDSC 26251); three lines for α1D (CG4894): UAS-GD1737-RNAi (VDRC 51491), UAS-JF01848-RNAi (BDSC 25830), and UAS-HMS00294-RNAi (BDSC 33413); two lines for α2δ (CG12295): UAS-KK101267-RNAi (VDRC 18569) and UAS-JF01825-RNAi (BDSC 25807); one line for Orai (CG11430) UAS-HMC03562-RNAi (BDSC 53333); three lines for dSTIM (CG9126): UAS-KK102366-RNAi (VDRC 106256), UAS-GLC01785-RNAi (BDSC 51685), and UAS-JF02567-RNAi (BDSC 27263); two lines for SERCA (CG3725): UAS-KK107371-RNAi (VDRC 107446) and UAS-JF01948-RNAi (BDSC 25928); and three lines for RyR (CG19844): UAS-KK101716-RNAi (VDRC 109631), UAS-HM05130-RNAi (BDSC 28919), and UAS-JF03381-RNAi (BDSC 29445).
In Vivo Fly Preparations and Calcium Imaging.
The fly surgery followed procedures previously described (19, 20). Flies were first anesthetized by CO2 and immobilized by inserting the neck into a narrow cut in an aluminum foil base. A portion of the dorsoanterior cuticle on one side of the head, an antenna, and a small part of one compound eye were then removed. For slow calcium rhythm measurements, imaging was conducted with a custom horizontal-scanning Objective Coupled Planar Illumination microscope (51). The scanning was done by moving the stage horizontally every 10 min for 24 h. Each scan contained 20 to 40 separate frames with a step size of 5 to 10 μm. For fast calcium rhythm measurements, imaging was conducted with a custom high-speed OCPI-2 microscope (21). Each scanning session involved moving the objective using a piezo motor at 1 to 5 Hz for 1 to 2 min. The same scans were then repeated on the same specimens every hour for 24 h. During both slow and fast imaging modes, fresh haemolymph-like solution (HL3 saline) was continuously perfused (0.1 to 0.2 mL/min).
Locomotor Activity Rhythm.
The Trikinetics Drosophila Activity Monitor system was used to monitor the locomotor activity rhythms of individual flies; 4- to 6-d-old male flies were monitored for 6 d under LD cycles and then, for 9 d under DD conditions. The circadian rhythmicity and periodicity were measured by χ2 periodograms with a 95% confidence cutoff and SNR analysis (52). Arrhythmicity was defined by a power value (χ2 power at the best period) less than 10, a width lower than one, and a period less than 18 h or more than 30 h.
Immunocytochemistry.
The flies were entrained for 6 d under LD and dissected at ZT0, ZT6, ZT12, and ZT18. After dissection in ice-cold, calcium-free saline, fly brains were fixed for 15 min in 4% paraformaldehyde containing 7% (vol/vol) picric acid in phosphate-buffered saline. Primary antibodies were rabbit anti-PER [1:5,000; provided by M. Rosbash, Brandeis University, Waltham, MA (53)]. Secondary antisera were Cy3-conjugated (1:1,000; Jackson Immunoresearch). Images were taken on the Olympus FV1200 confocal microscope. PER protein immunostaining intensity was measured in ImageJ-based Fiji (54).
Imaging Data Analysis.
Calcium imaging data were acquired by custom software, Imagine (51), and preprocessed using custom scripts in Julia 0.6 to produce nonrigid registration, alignment, and maximal projection along the z axis. The images were then visualized and analyzed in ImageJ-based Fiji by manually selecting ROIs over individual cells or groups of cells and measuring the intensity of ROIs over time. Slow calcium activity was analyzed as described previously (19, 20). Fast calcium activity in each scanning session was analyzed similarly. Between sequential scanning sessions, the ROIs for individual neurons were manually corrected for position drifts. For the calcium signal of each ROI in each session, the mean of calcium intensity (e.g., Fig. 1C) was measured. The range of calcium transients (e.g., Fig. 1D) was measured by the average maximal calcium intensity at each time point and by the average minimal calcium intensity at each time point. The power spectrum (e.g., Fig. 1E) was generated by fast Fourier transform. Then, the calcium signal was filtered by a high-pass filter (1/15 Hz), and the SD of calcium changes was measured. Calcium activity trace analysis and statistics were performed using R 3.3.3 and Prism 8 (GraphPad).
Supplementary Material
Acknowledgments
We thank Cody Greer for building the OCPI-2 microscope, Weihua Li for experimental support, the laboratories of T.E.H. and P.H.T. for advice, and the Washington University Center for Cellular Imaging for technical support. Gaiti Hasan, Mark Wu, Francois Rouyer, Michael Rosbash, the Bloomington Stock Center, the Vienna Drosophila Resource Center, and the Tokyo Stock Center provided fly stocks and reagents. Portions of this work were developed from the doctoral thesis of X.L. The work was supported by the Washington University McDonnell Center for Cellular and Molecular Neurobiology and by NIH Grants R01 NS068409 (to T.E.H.), R01 DP1 DA035081 (to T.E.H.), R24 NS086741 (to T.E.H. and P.H.T.), R01 NS099332 (to P.H.T.), and R01 GM127508 (to P.H.T.).
Footnotes
Competing interest statement: T.E.H. has a patent on OCPI microscopy. The authors have no other financial interests or positions to declare.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109969119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Dunlap J. C., Molecular bases for circadian clocks. Cell 96, 271–290 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Herzog E. D., Neurons and networks in daily rhythms. Nat. Rev. Neurosci. 8, 790–802 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Welsh D. K., Takahashi J. S., Kay S. A., Suprachiasmatic nucleus: Cell autonomy and network properties. Annu. Rev. Physiol. 72, 551–577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohawk J. A., Green C. B., Takahashi J. S., Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35, 445–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welsh D. K., Logothetis D. E., Meister M., Reppert S. M., Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Panda S., et al. , Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Colwell C. S., Linking neural activity and molecular oscillations in the SCN. Nat. Rev. Neurosci. 12, 553–569 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennartz C. M., de Jeu M. T. G., Bos N. P. A., Schaap J., Geurtsen A. M. S., Diurnal modulation of pacemaker potentials and calcium current in the mammalian circadian clock. Nature 416, 286–290 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Itri J. N., Michel S., Vansteensel M. J., Meijer J. H., Colwell C. S., Fast delayed rectifier potassium current is required for circadian neural activity. Nat. Neurosci. 8, 650–656 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitts G. R., Ohta H., McMahon D. G., Daily rhythmicity of large-conductance Ca2+-activated K+ currents in suprachiasmatic nucleus neurons. Brain Res. 1071, 54–62 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Meredith A. L., et al. , BK calcium-activated potassium channels regulate circadian behavioral rhythms and pacemaker output. Nat. Neurosci. 9, 1041–1049 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flourakis M., et al. , A conserved bicycle model for circadian clock control of membrane excitability. Cell 162, 836–848 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berridge M. J., Neuronal calcium signaling. Neuron 21, 13–26 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Chorna T., Hasan G., The genetics of calcium signaling in Drosophila melanogaster. Biochim. Biophys. Acta 1820, 1269–1282 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Colwell C. S., Circadian modulation of calcium levels in cells in the suprachiasmatic nucleus. Eur. J. Neurosci. 12, 571–576 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda M., et al. , Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron 38, 253–263 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Jones J. R., Simon T., Lones L., Herzog E. D., SCN VIP neurons are essential for normal light-mediated resetting of the circadian system. J. Neurosci. 38, 7986–7995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enoki R., et al. , Synchronous circadian voltage rhythms with asynchronous calcium rhythms in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U.S.A. 114, E2476–E2485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang X., Holy T. E., Taghert P. H., Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science 351, 976–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang X., Holy T. E., Taghert P. H., A series of suppressive signals within the Drosophila circadian neural circuit generates sequential daily outputs. Neuron 94, 1173–1189.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greer C. J., Holy T. E., Fast objective coupled planar illumination microscopy. Nat. Commun. 10, 4483 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pologruto T. A., Yasuda R., Svoboda K., Monitoring neural activity and [Ca2+] with genetically encoded Ca2+ indicators. J. Neurosci. 24, 9572–9579 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaksi E., Friedrich R. W., Reconstruction of firing rate changes across neuronal populations by temporally deconvolved Ca2+ imaging. Nat. Methods 3, 377–383 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Chen T.-W., et al. , Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Streit A. K., Fan Y. N., Masullo L., Baines R. A., Calcium imaging of neuronal activity in Drosophila can identify anticonvulsive compounds. PLoS One 11, e0148461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenberg D. S., et al. , Accurate action potential inference from a calcium sensor protein through biophysical modeling. bioRxiv [Preprint] (2018). 10.1101/479055 (Accessed 29 November 2018). [DOI]
- 27.Cao G., Nitabach M. N., Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J. Neurosci. 28, 6493–6501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheeba V., et al. , Pigment dispersing factor-dependent and -independent circadian locomotor behavioral rhythms. J. Neurosci. 28, 217–227 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolezelova E., Dolezel D., Hall J. C., Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics 177, 329–345 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knight M. R., Campbell A. K., Smith S. M., Trewavas A. J., Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352, 524–526 (1991). [DOI] [PubMed] [Google Scholar]
- 31.Lundkvist G. B., Kwak Y., Davis E. K., Tei H., Block G. D., A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J. Neurosci. 25, 7682–7686 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrisingh M. C., Wu Y., Lnenicka G. A., Nitabach M. N., Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J. Neurosci. 27, 12489–12499 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enoki R., et al. , Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U.S.A. 109, 21498–21503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.VanderLeest H. T., et al. , Seasonal encoding by the circadian pacemaker of the SCN. Curr. Biol. 17, 468–473 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Brancaccio M., Maywood E. S., Chesham J. E., Loudon A. S., Hastings M. H., A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 78, 714–728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fogle K. J., Parson K. G., Dahm N. A., Holmes T. C., CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science 331, 1409–1413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashmore L. J., Sehgal A., A fly’s eye view of circadian entrainment. J. Biol. Rhythms 18, 206–216 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Hong J. H., Jeong B., Min C. H., Lee K. J., Circadian waves of cytosolic calcium concentration and long-range network connections in rat suprachiasmatic nucleus. Eur. J. Neurosci. 35, 1417–1425 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi S., et al. , Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Noguchi T., et al. , Calcium circadian rhythmicity in the suprachiasmatic nucleus: Cell autonomy and network modulation. eNeuro 4, ENEURO.0160-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S., Liu Q., Tabuchi M., Wu M. N., Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165, 1347–1360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeong K., et al. , Ca-α1T, a fly T-type Ca2+ channel, negatively modulates sleep. Sci. Rep. 5, 17893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abruzzi K. C., et al. , RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Genet. 13, e1006613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kula-Eversole E., et al. , Phosphatase of regenerating liver-1 selectively times circadian behavior in darkness via function in PDF neurons and dephosphorylation of TIMELESS. Curr. Biol. 31, 138–149.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vassort G., Talavera K., Alvarez J. L., Role of T-type Ca2+ channels in the heart. Cell Calcium 40, 205–220 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Ono K., Iijima T., Cardiac T-type Ca(2+) channels in the heart. J. Mol. Cell. Cardiol. 48, 65–70 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Perez-Reyes E., Molecular physiology of low-voltage-activated t-type calcium channels. Physiol. Rev. 83, 117–161 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Blau J., Young M. W., Cycling vrille expression is required for a functional Drosophila clock. Cell 99, 661–671 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Renn S. C., Park J. H., Rosbash M., Hall J. C., Taghert P. H., A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Agrawal N., et al. , Inositol 1,4,5-trisphosphate receptor and dSTIM function in Drosophila insulin-producing neurons regulates systemic intracellular calcium homeostasis and flight. J. Neurosci. 30, 1301–1313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holekamp T. F., Turaga D., Holy T. E., Fast three-dimensional fluorescence imaging of activity in neural populations by objective-coupled planar illumination microscopy. Neuron 57, 661–672 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Levine J. D., Funes P., Dowse H. B., Hall J. C., Signal analysis of behavioral and molecular cycles. BMC Neurosci. 3, 1–25 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanewsky R., et al. , Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: Mapping elements of the PER protein involved in circadian cycling. J. Neurosci. 17, 676–696 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.