Abstract
Phospholipid fatty acid (PLFA) analysis of a soil microbial community was coupled with 13C isotope tracer analysis to measure the community’s response to addition of 35 μg of [13C]toluene ml of soil solution−1. After 119 h of incubation with toluene, 96% of the incorporated 13C was detected in only 16 of the total 59 PLFAs (27%) extracted from the soil. Of the total 13C-enriched PLFAs, 85% were identical to the PLFAs contained in a toluene-metabolizing bacterium isolated from the same soil. In contrast, the majority of the soil PLFAs (91%) became labeled when the same soil was incubated with [13C]glucose. Our study showed that coupling 13C tracer analysis with PLFA analysis is an effective technique for distinguishing a specific microbial population involved in metabolism of a labeled substrate in complex environments such as soil.
Toluene is a widespread pollutant that biodegrades rapidly relative to many other organic pollutants in both oxic and anoxic environments. Toluene degradation pathways have been studied almost exclusively in Pseudomonas species (23, 30, 38, 39, 43); however, other gram-negative and gram-positive toluene-degrading bacteria (1, 12, 19, 21, 27, 33, 35) and fungi (36, 40) have been described. Despite extensive research on toluene biodegradation in soil and groundwater (5, 6, 11, 32, 34) and in pure cultures isolated from such environments, previous studies have provided little evidence directly linking the disappearance of the pollutant to specific populations of microorganisms in complex environments. The ability to identify populations actively involved in biodegradation in natural environments would greatly improve our ability to model pollutant degradation. Most kinetic models require the physiological characteristics and population densities of the organisms carrying out biodegradation (10, 31), which clearly requires knowledge of the population(s) involved in the process.
The polar lipid fraction of soils and other environmental samples is composed primarily of the phospholipid fatty acids (PLFA) of the viable microorganisms present (37). For complex matrices such as soil, PLFA analysis has been shown to be a valuable tool for detecting changes in microbial communities in response to heavy-metal pollution (13, 25, 42), hydrocarbon pollution (26, 29, 31), and different agricultural management regimes (3, 41). Recently, PLFA analysis has been combined with 13C-labeled substrates and compound-specific isotope analysis in order to link indigenous populations with metabolism of specific substrates via PLFA biomarkers (2).
Our laboratory has previously investigated the biodegradation of toluene, as well as toluene-induced trichloroethylene cometabolism, by indigenous microbial populations in a surface soil (Yolo silt loam) (11, 22). Development of a model describing the coupled diffusive transport and biodegradation of toluene and trichloroethylene in columns of Yolo silt loam indicated the need for a better understanding of the microbial populations responsible for toluene degradation (8, 10). The primary objective of our study was to identify the indigenous population(s) responsible for toluene disappearance in Yolo silt loam. To this end we employed traditional culture-based approaches as well as PLFA and 13C-PLFA methods not affected by the biases associated with laboratory cultivation.
MATERIALS AND METHODS
Chemicals.
Reagent grade toluene and chemicals used in the phospholipid extraction and purification steps (all Fisher Optima grade) were obtained from Fisher Scientific (Fair Lawn, N.J.). Ring-labeled [13C]toluene (99 atom%) and uniformly labeled [13C]glucose (99 atom%) were obtained from Isotech Inc. (Miamisburg, Ohio). All solutions were prepared with NANOpure water (Series 550; Barnstead/Thermolyne Corp., Dubuque, Iowa).
Soil.
The top 15 cm of Yolo silt loam (31% sand, 50% silt, 19% clay) was collected from a fallow agricultural plot at the Student Farm of the University of California, Davis. Samples were passed through a 2-mm-pore-size sieve, and the soil moisture was brought to 14% (determined gravimetrically). The soil was stored in double polyethylene bags at 4°C to preserve biological activity. The organic-matter content was determined to be 1.81% by the Walkley-Black wet-combustion method. Total mineral N was 59 μg g of soil−1, and mineral P was 19 μg g of soil−1. All analyses were performed by the Division of Agriculture and Natural Resources Analytical Laboratory at the University of California, Davis.
Biodegradation assays.
Triplicate 50-g (dry weight) soil microcosms were spiked with 10, 20, 50, or 100 μg of toluene ml of soil solution−1 (accounting for partitioning into the vapor phase and sorption to the soil particles; Henry’s law constant, 0.27; sorption partition coefficient [Koc], 200 [11]) and incubated at 25°C in the dark. Biodegradation was measured by gas chromatographic (GC) analysis of headspace concentrations as described earlier (22). When toluene was no longer detectable in the microcosms, samples were frozen for subsequent PFLA analysis. For [13C]toluene incubations, the initial toluene concentration in the microcosms was 35 μg of [13C]toluene ml−1. The microcosms were destructively sampled at 0, 95.5, and 119 h after toluene addition. All treatments were performed in triplicate, and [13C]toluene-amended samples were compared with sterile (gamma-irradiated) and toluene-free controls. For [13C]glucose incubations, 4,000 μg of [13C]glucose g−1 was added to 25 g (dry weight) of soil in enough water to bring gravimetric moisture levels to 25%. Samples were incubated at 25°C for 40 h prior to destructive sampling.
PLFA analyses.
PLFA analyses were performed as previously described (3). Briefly, soil lipids and microbial cultures were extracted with a one-phase chloroform-methanol-water extractant, fractionated according to polarity by column chromatography on silica gel columns, and derivatized by mild alkaline methanolysis to form polar lipid-derived fatty acid methyl esters (FAME). The FAME were separated on a Hewlett-Packard (HP) 6890 gas chromatograph equipped with a 25-meter HP Ultra 2 capillary column (internal diameter, 0.20 mm; film thickness, 0.33 μm) with H2 carrier gas and a programmed temperature increase from 170 to 260°C at 2°C/min. GC-mass spectrometry (MS) analyses were performed with a Varian Star 3400CX gas chromatograph (30-meter J&W DB-5 capillary column; internal diameter, 0.25 mm; film thickness, 0.25 μm) coupled to a Varian Saturn 4D ion trap MS. Chromatographic peaks were quantified based on a 19:0 internal standard by GC with flame ionization detection. Double-bond locations in monounsaturated PLFA were confirmed by GC-MS of their dimethyldisulfide adducts (7). Identification of PLFA is based on comparison of retention times with authentic standards and on mass spectral analysis. Polar lipid-derived FAME were also analyzed by capillary GC-combustion-isotope ratio MS (GC-C-IRMS). Carbon isotopic compositions of individual polar lipid FAME in each soil extract were determined with an HP 6890 gas chromatograph (HP-5 capillary column; 30 m; internal diameter, 0.32 mm; film thickness, 0.25 μm) connected via a Europa ORCHID on-line combustion interface to a Europa Geo 20/20 MS operating in continuous-flow mode. The carrier gas was He, and the oven temperature program was 20°C/min from 45 to 140°C, 1.5°C/min to 210°C, and 20°C/min to 250°C. The atom percent 13C (moles of 13C/moles of 12C × 100) values for individual FAME peaks were calculated with respect to a standard CO2 reference gas injected at the beginnings and ends of analytical runs. The values reported are averages of three replicate microcosm PLFA extracts. The natural abundance of 13C is 1.11% (15).
The following fatty acid nomenclature is used: a total number of carbon atoms:number of double bonds, followed by the position of the double bond from the methyl (ω) end of the molecule (e.g., 17:1ω8). cis and trans geometry is indicated by the suffixes c and t. The prefixes a and i refer to anteiso- and isobranching, 10me indicates a methyl group on the 10th carbon atom from the carboxyl end of the molecule, the positions of hydroxy (OH) groups are noted, and cy indicates cyclopropane fatty acids.
Bacterial isolation and identification.
Toluene-degrading bacterial cultures were isolated from Yolo silt loam by enrichment on toluene. One gram (dry weight) of soil was added to 99 ml of mineral salts medium (MSM) (22) supplemented with 200 μg of toluene ml−1 as the sole carbon source. The culture was incubated at 25°C in the dark with shaking. After the culture became turbid (approximately 5 days), 1 ml was transferred to a second 99 ml of MSM containing 200 μg of toluene ml−1. This was repeated for a total of four transfers. The final liquid culture was streaked onto 0.1× tryptic soy agar (TSA) and incubated at 25°C. Individual colonies were tested in liquid culture for the ability to utilize toluene as a sole carbon source. Subsequent culturing of toluene-degrading strains was accomplished in toluene-amended MSM under the conditions described above.
Toluene-degrading bacterial strains were initially identified by whole-cell FAME analysis with the MIDI system (24). One strain, designated YT-2, was further characterized by 16S ribosomal DNA (rDNA) sequencing performed at the University of California, Davis, Automated Sequencing Facility with the eubacterial primers 357F and 1406R (18). Both strands of the 16S gene were sequenced. Analysis of the sequences obtained was performed with the ALIGN-SEQUENCE algorithm from the National Science Foundation Ribosomal Database Project (20).
Nucleotide sequence accession number.
The 16S rDNA sequence has been deposited in GenBank (accession no. AF148495).
RESULTS AND DISCUSSION
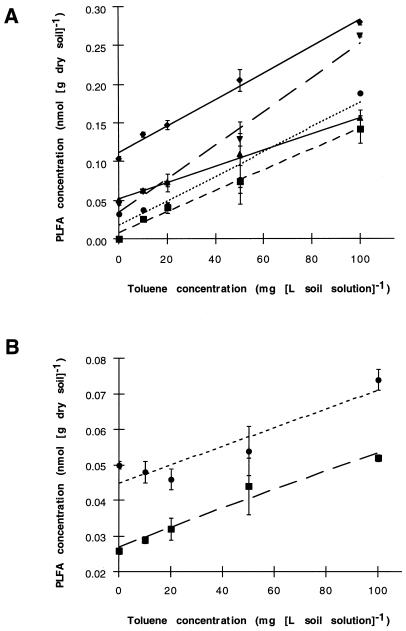
In initial experiments, we used PLFA analysis to detect changes in soil microbial populations in response to toluene additions. As toluene disappeared from Yolo silt loam (initial concentrations ranged from 10 to 100 μg of toluene ml of soil solution−1 or 35 to 350 μg g of dry soil−1), a subset of 10 PLFA were enriched. These fatty acids were 14:0, 15:1* (* denotes an unknown double-bond location), 15:0, 16:1ω6, 17:1ω8, 17:0, 10me17:0, i18:0, 10me18:0, and 10me19:0. Fatty acids 15:1*, i18:0, and 10me19:0 were detected only in microcosms that received the highest concentration (100 μg ml−1) of toluene. The concentrations of the remaining seven PLFA were strongly correlated with the initial toluene concentration, as shown in Fig. 1. The sum of mass increases in these 10 PLFA after incubation with 100 μg of toluene ml−1 was equivalent to 14.4% of the total soil PLFA pool, suggesting that toluene-degrading populations represented a large component of the microbial community after incubation with toluene. A small subset of soil PLFA decreased significantly (P < 0.05) after exposure to 100 μg of toluene ml−1 (Table 1). The 18:2ω6,9 fatty acid is commonly considered a biomarker for eukaryotic organisms (14), while the 16:1ω11c, 16:1ω5c, iso 17:1*, and cyclopropyl 19:0 fatty acids are widely distributed among soil bacterial groups (28).
FIG. 1.
PLFA enriched during soil incubations with successively higher initial toluene concentrations (±1 standard deviation; n = 3 microcosms except 100 μg ml−1, where n = 2). (A) Fatty acids 15:0 (----●----), 16:1ω6 (— ■– -), 17:1ω8 (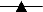 ), 10me17:0 (—▾- -), and 10me18:0 (—⧫—). (B) Fatty acids 14:0 (--●--) and 17:0 (—■ -). The fatty acid nomenclature follows that in reference 3; ∗, unidentified double-bond location. The r2 values for linear correlations shown are greater than 0.95 with the exception of 14:0 (r2 = 0.89).
), 10me17:0 (—▾- -), and 10me18:0 (—⧫—). (B) Fatty acids 14:0 (--●--) and 17:0 (—■ -). The fatty acid nomenclature follows that in reference 3; ∗, unidentified double-bond location. The r2 values for linear correlations shown are greater than 0.95 with the exception of 14:0 (r2 = 0.89).
TABLE 1.
Soil PLFA that decreased significantly (P < 0.05) after exposure to toluene
| Fatty acida | Amt [nmol (SD)]b
|
|
|---|---|---|
| 0 μg of toluene ml−1 | 100 μg of toluene ml−1 | |
| 16:1ω11c | 0.102 (0.003) | 0.090 |
| 16:1ω5c | 0.218 (0.003) | 0.191 |
| Iso 17:1* | 0.249 (0.004) | 0.221 |
| 18:2ω6,9 | 0.185 (0.003) | 0.108 |
| Cyclopropyl 19:0 | 0.304 (0.001) | 0.260 |
Nomenclature follows that in reference 3. *, unknown double-bond location.
Values are averages (n = 3 for 0 μg of toluene ml−1; n = 2 for 100 μg of tuluene ml−1).
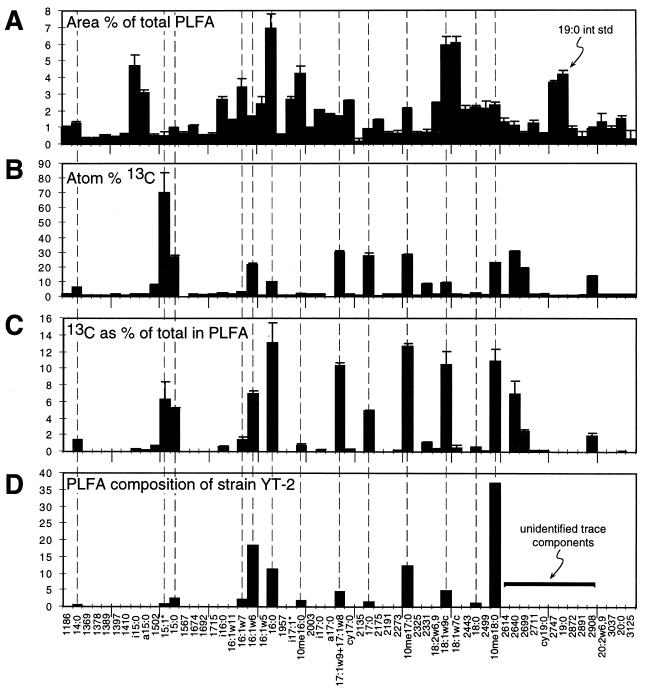
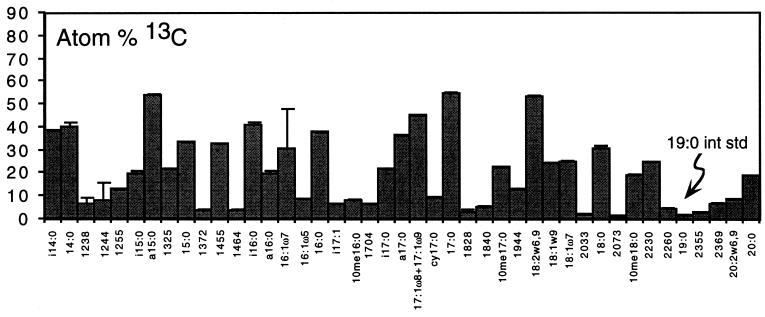
To directly measure the flow of carbon from toluene into microbial populations responsible for biodegradation, we incubated Yolo silt loam with 35 μg of ring-labeled [13C]toluene ml−1 and analyzed the community PLFA patterns. Toluene amendment resulted in a striking 13C enrichment of a small subset of the soil PLFA (Fig. 2A and B). The [13C]toluene results contrasted with a similar incubation of Yolo silt loam with [13C]glucose, an easily metabolized carbon substrate, in which most of the soil PLFA detected were highly enriched in 13C (Fig. 3). In the [13C]glucose incubation, 39 of a total of 43 PLFA were highly labeled (with >3 atom% 13C). In contrast, in the [13C]toluene incubation, 96% of 13C detected in all PLFA (59 fatty acids) was contained in 16 highly labeled fatty acids (Fig. 2B and C).
FIG. 2.
Results of [13C]toluene-labeling study in soil microcosms (±1 standard deviation; n = 3 microcosms). (A) PLFA area percentages. The initial toluene concentration (99 atom% 13C in the toluene ring) was 30 mg liter of soil solution−1. The microcosms were incubated for 119 h. int std, internal standard. (B) Atom percent 13C values for individual PLFA shown in panel A. (C) Carbon-13 tracer content of each soil PLFA as a percentage of total 13C enrichment in all PLFA. (D) PLFA composition of strain YT-2 grown on mineral medium plus toluene. The PLFA present in YT-2 grown on 0.1× tryptic soy broth were identical. Unidentified PLFA are denoted by their retention times in seconds.
FIG. 3.
Results of [13C]glucose-labeling study in soil microcosms (±1 standard deviation; n = 3 replicate extractions). The atom percent 13C values for individual PLFA are shown. The retention times cannot be directly compared to those in Fig. 2 due to slight modifications of the GC temperature program. int std, internal standard.
When subsamples of Yolo silt loam that had been enriched with toluene were plated onto 0.1× TSA, only gram-positive cells with a single colony morphology, a buff-colored irregular form, were found to metabolize toluene. This result was observed for three independent enrichments. This same colony morphology was the sole morphotype on TSA plates spread with solutions obtained from the most dilute most probable number (MPN) bottles (22) of toluene degraders in Yolo silt loam (data not shown). Whole-cell fatty acid analysis of selected colonies from both soil enrichments and MPN bottles yielded FAME profiles most similar to high-G+C-content, gram-positive nocardioform actinomycetes. One isolate, designated strain YT-2, from the toluene enrichment of Yolo silt loam was chosen for further phylogenetic analysis.
Whole-cell fatty acid analysis of strain YT-2 yielded a FAME profile that was 35% similar to that of a Gordona sp. (formerly designated Rhodococcus). The small-subunit rDNA sequence of strain YT-2 was 88% similar to that of a Rhodococcus sp. Interestingly, those soil PLFA which increased in mass during the toluene incubations (Fig. 1) closely matched the PLFA found in strain YT-2 when it was grown on toluene. Thus, 97.9% of the mass increase in PLFA enriched in soil during incubation with toluene (100 μg ml−1) was associated with the same PLFAs that are contained in strain YT-2.
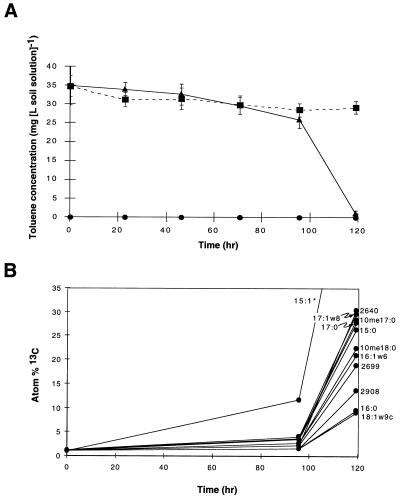
As in our initial soil incubations with unlabeled toluene, enriched fatty acids in [13C] toluene incubations matched those found in strain YT-2. The major PLFA components of strain YT-2 (Fig. 2D) contained 85% of the total 13C enrichment in soil PLFA after incubation with [13C]toluene (Fig. 2C). The three additional highly 13C-labeled PLFA, all with retention times longer than that of 10me18:0, may correspond to trace PLFA components in strain YT-2 but have not yet been identified. The period in which 13C enrichment in soil PLFA occurred was the same period during which [13C]toluene disappeared from the soil microcosms (Fig. 4).
FIG. 4.
Timing of [13C]toluene disappearance and 13C labeling of soil PLFA. (A) Toluene disappearance measured by GC of microcosm headspace. ■, sterile control; ▴, [13C]toluene added; ●, no toluene added. The error bars indicate ±1 standard deviation (n = 3 microcosms). (B) Atom percent 13C in soil PLFA measured by GC-C-IRMS. The PLFA analyses and fatty acid nomenclature are as described in the legends to Fig. 1 and 2. Unidentified PLFA are denoted by their retention times in seconds.
Based on the pattern of PLFA enriched in toluene degradation experiments and PLFA present in strain YT-2, we hypothesize that the microorganisms actively degrading toluene in Yolo silt loam were either strain YT-2 or related high-G+C-content, gram-positive strains with very similar PLFA compositions. PLFA with midchain branching (10me-) are narrowly distributed among bacteria and are characteristic of a subset of high-G+C-content, gram-positive bacteria that includes Rhodococcus and other nocardioform and filamentous actinomycetes (4, 17). PLFA with midchain branching are also found in certain gram-negative, anaerobic, sulfate-reducing proteobacteria typified by the genus Desulfobacter (16). However, sulfate-reducing bacteria are unlikely to have been active under the oxic conditions of our incubations. PLFA with midchain branching have not been identified in other bacterial groups or in archaeal or eucaryal domains. Well-studied bacterial toluene degraders, such as Pseudomonas and related genera, have PLFA compositions characterized by cyclopropyl (cy17:0 and cy19:0) and straight-chain fatty acids of even chain length (16:1 and 18:1), and do not contain methyl-branched PLFA (28). Cyclopropyl and straight-chain fatty acids of even chain length are common in soils but were not enriched in our experiments, with the exception of specific fatty acids also identified in strain YT-2 (16:1ω6 and 18:1ω9c). Terminally branched, odd-chain PLFA (a/i15:0, a/i17:0) are also common in soils and in diverse bacterial groups, including many gram-negative and gram-positive lineages (28). Terminally branched, odd-chain PLFA were not enriched in our experiments (Fig. 2). In addition, polyunsaturated PLFA, biomarkers for eukaryotes such as fungi (14), either decreased (18:2ω6,9), showed no changes (20:2ω6,9), or were not detected in our analyses.
Our study showed that coupling 13C tracer analysis with PLFA analysis was a very effective technique for distinguishing the particular microbial population involved in metabolism of the labeled substrate. Given that toluene degradation is widespread among multiple prokaryotic lineages as well as some fungal strains, it was surprising that only a small subset of PLFA associated with a single bacterial population became labeled with 13C following incubation with [13C]toluene. It is possible that under other environmental conditions, e.g., higher or lower initial toluene concentrations or different moisture or temperature regimes, other populations in Yolo silt loam might become involved in toluene degradation. Experiments performed under a variety of incubation conditions will explore whether this soil has a greater diversity of toluene-degrading populations than those we measured under our experimental conditions.
In summary, PLFA analysis showed that the addition of small (micromolar) amounts of a specific carbon and energy source, such as toluene, can cause a rapid and dramatic shift in the population structure of a highly complex soil microbial community. The application of 13C-PLFA tracer analysis supported the idea that this community shift is due in part to the growth of a population of toluene-degrading bacteria. These PLFA data are consistent with MPN determinations (22) and kinetic model calculations (9), indicating that toluene-degrading microbial populations in Yolo silt loam increase substantially with exposure to 20 μg of toluene ml−1. Future studies will evaluate the feasibility of using biomarker lipids to estimate the densities of biodegrading populations to calibrate or validate biodegradation kinetic models.
ACKNOWLEDGMENTS
We thank Egbert Schwartz and Mary Ann Bruns for useful discussion and gratefully acknowledge the assistance of John Newman and Angela Dickens.
This study was supported by the NIEHS Superfund Basic Research Program (2P42 ES04699), the EPA Center for Ecological Health Research, the UC Toxic Substances Training Program in Ecotoxicology, the DOE Joint Program in Bioavailability, and a fellowship from the Kearney Foundation for Soil Science.
REFERENCES
- 1.Biegert T, Fuchs G, Heider J. Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur J Biochem. 1996;238:661–668. doi: 10.1111/j.1432-1033.1996.0661w.x. [DOI] [PubMed] [Google Scholar]
- 2.Boschker H T S, Nold S C, Wellsbury P, Bos D, de Graaf W, Pel R, Parkes R J, Cappenberg T E. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature. 1998;392:801–805. [Google Scholar]
- 3.Bossio D A, Scow K M, Gunapala N, Graham K J. Determinants of soil microbial communities: effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb Ecol. 1998;36:1–12. doi: 10.1007/s002489900087. [DOI] [PubMed] [Google Scholar]
- 4.Bousfield I J, Smith G L, Dando T R, Hobbs G. Numerical analysis of total fatty acid profiles in the identification of coryneform, nocardioform and some other bacteria. J Gen Microbiol. 1983;129:375–394. [Google Scholar]
- 5.Chapelle F H, Bradley P M, Lovely D R, Vroblesky D A. Measuring rates of biodegradation in a contaminated aquifer using field and laboratory methods. Ground Water. 1996;34:691–698. [Google Scholar]
- 6.Corseuil H X, Hunt C S, Santos R C F D, Alvarez R J J. The influence of the gasoline oxygenate ethanol on aerobic and anaerobic BTX biodegradation. Water Res. 1998;32:2065–2072. [Google Scholar]
- 7.Dunkelblum E, Tan S H, Silk P J. Double-bond location in monounsaturated fatty acids by dimethyl disulfide derivatization and mass spectrometry. J Chem Ecol. 1985;11:265–277. doi: 10.1007/BF01411414. [DOI] [PubMed] [Google Scholar]
- 8.El-Farhan Y H. Ph.D. dissertation. University of California, Davis; 1997. [Google Scholar]
- 9.El-Farhan, Y. H., D. E. Rolston, and K. M. Scow. Unpublished data.
- 10.El-Farhan Y H, Scow K M, de Jonge L W, Rolston D E, Moldrup P. Coupling transport and biodegradation of toluene and trichloroethylene in unsaturated soils. Water Res. 1998;34:437–445. [Google Scholar]
- 11.Fan S, Scow K. Biodegradation of trichloroethylene and toluene by indigenous microbial populations in soil. Appl Environ Microbiol. 1993;59:1911–1918. doi: 10.1128/aem.59.6.1911-1918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fries M R, Zhou J, Chee-Sanford J, Tiedje J M. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frostegard A, Tunlid A, Baath E. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl Environ Microbiol. 1993;59:3605–3617. doi: 10.1128/aem.59.11.3605-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood J L, Russell N J. Lipids in plants and microbes. London, England: George, Allen & Unwin; 1984. [Google Scholar]
- 15.Hoefs J. Stable isotope geochemistry. 2nd ed. Berlin, Germany: Springer-Verlag; 1980. [Google Scholar]
- 16.Kohring L L, Ringelberg D B, Devereux R, Stahl D A, Mittelman M W, White D C. Comparison of phylogenetic relationships based on phospholipid fatty acid profiles and ribosomal RNA sequence similarities among dissimilatory sulfate-reducing bacteria. FEMS Microbiol Lett. 1994;119:303–308. doi: 10.1111/j.1574-6968.1994.tb06905.x. [DOI] [PubMed] [Google Scholar]
- 17.Kroppenstedt R M. Fatty acid and menaquinone analysis of actinomycetes and related organisms. In: Goodfellow M, Minnikin D E, editors. Chemical methods in bacterial systematics. Orlando, Fla: Academic Press; 1985. pp. 173–199. [Google Scholar]
- 18.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 19.Lovely D R, Lonergan D J. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism GS-15. Appl Environ Microbiol. 1990;56:1858–1864. doi: 10.1128/aem.56.6.1858-1864.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maidak B L, Olsen G L, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malachowsky K J, Phelps T J, Teboli A B, Minnikin D E, White D C. Aerobic mineralization of trichloroethylene, vinyl chloride, and aromatic compounds by Rhodococcus species. Appl Environ Microbiol. 1994;60:542–548. doi: 10.1128/aem.60.2.542-548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mu D Y, Scow K M. Effect of trichloroethylene (TCE) and toluene concentrations on TCE and toluene biodegradation and the population density of TCE and toluene degraders in soil. Appl Environ Microbiol. 1994;60:2661–2665. doi: 10.1128/aem.60.7.2661-2665.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen R H, Kukor J J, Kaphammer B. A novel toluene-3-monooxygenase pathway cloned from Pseudomonas pickettii PKO1. J Bacteriol. 1994;176:3749–3756. doi: 10.1128/jb.176.12.3749-3756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paisley R, editor. MIS whole cell fatty acid analysis by gas chromatography. Newark, Del: MIDI, Inc.; 1995. [Google Scholar]
- 25.Pennanen R, Frostegard A, Fritze H, Baath E. Phospholipid fatty acid composition and heavy metal tolerance of soil microbial communities along two heavy metal-polluted gradients in coniferous forests. Appl Environ Microbiol. 1996;62:420–428. doi: 10.1128/aem.62.2.420-428.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phelps T J, Ringleberg D, Hedrick D, Davis J, Fliermans C B, White D C. Microbial biomass and activities associated with subsurface environments contaminated with chlorinated hydrocarbons. Geomicrobiol J. 1988;6:157–170. [Google Scholar]
- 27.Rabus R, Nordhaus R, Ludwig W, Widdel F. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol. 1993;59:1444–1451. doi: 10.1128/aem.59.5.1444-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratledge C, Wilkinson S G, editors. Microbial lipids. Vol. 1. London, England: Academic Press; 1988. [Google Scholar]
- 29.Ringelberg D B, Davis J D, Smith G A, Pfiffner S M, Nichols P D, Nickels J S, Henson J M, Wilson J T, Yates M, Kampbell D H, Read H W, Stocksdale T T, White D C. Validation of signature polarlipid fatty acid biomarkers for alkane-utilizing bacteria in soils and subsurface aquifer materials. FEMS Microbiol Ecol. 1989;62:39–50. [Google Scholar]
- 30.Shields M S, Montgomery S O, Chapman P J, Cuskey S M, Pritchard P H. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium G4. Appl Environ Microbiol. 1989;55:1624–1629. doi: 10.1128/aem.55.6.1624-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith G A, Nickels J S, Kerger B D, Davis J D, Collins S P, Wilson J T, McNabb J F, White D C. Quantitative characterization of microbial biomass and community structure in subsurface material: a prokaryotic consortium responsible to organic contamination. Can J Microbiol. 1986;32:104–111. [Google Scholar]
- 32.Stapleton R D, Savage D C, Sayler G S, Stagey G. Biodegradation of aromatic hydrocarbons in an extremely acidic environment. Appl Environ Microbiol. 1998;64:4180–4184. doi: 10.1128/aem.64.11.4180-4184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tay S T L, Hemond H F, Polz M F, Cavanaugh C M, Dejesus I, Krumholz L R. Two new mycobacterium strains and their role in toluene degradation in a contaminated stream. Appl Environ Microbiol. 1998;64:1715–1720. doi: 10.1128/aem.64.5.1715-1720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsao C-W, Song H-G, Bartha R. Metabolism of benzene, toluene, and xylene hydrocarbons in soil. Appl Environ Microbiol. 1998;64:4924–4929. doi: 10.1128/aem.64.12.4924-4929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warhurst A M, Clarke K F, Hill R A, Holt R A, Fewson C A. Metabolism of styrene by Rhodococcus rhodochrous NCIMB 13259. Appl Environ Microbiol. 1994;60:1137–1145. doi: 10.1128/aem.60.4.1137-1145.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber F J, Hage K C, de Bont J A M. Growth of the fungus Cladosporium sphaerospermum with toluene as the sole carbon and energy source. Appl Environ Microbiol. 1995;61:3562–3566. doi: 10.1128/aem.61.10.3562-3566.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White D C, Findlay R H. Biochemical markers for measurement of predation effects on the biomass, community structure, nutritional status, and metabolic activity of microbial biofilms. Hydrobiologia. 1988;159:119–132. [Google Scholar]
- 38.Whited G M, Gibson D T. Toluene-4-monooxygenase, a three-component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J Bacteriol. 1991;173:3010–3016. doi: 10.1128/jb.173.9.3010-3016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worsey M J, Williams P A. Metabolism of toluene and the xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadav J S, Reddy C A. Degradation of benzene, toluene, ethylbenzene, and xylenes (BTEX) by the lignin-degrading basidiomycete Phanaerochaete chrysosporium. Appl Environ Microbiol. 1993;59:756–762. doi: 10.1128/aem.59.3.756-762.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zelles L, Bai Q Y, Beck T, Beese F. Signature fatty acids in phospholipids and lipopolysaccharides as indicators of microbial biomass and community structure in agricultural soils. Soil Biol Biochem. 1992;24:317–323. [Google Scholar]
- 42.Zelles L, Bai Q Y, Ma R X, Rackwitz R, Witner K, Beese F. Microbial biomass, metabolic activity and nutritional status determined from fatty acid patterns and poly-hydroxybutyrate in agriculturally-managed soils. Soil Biol Biochem. 1994;26:439–446. [Google Scholar]
- 43.Zylstra G J, McCombie W R, Gibson D T, Finette B A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988;54:1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]