Abstract
Nontuberculous mycobacterium (NTM) infections are increasing in the USA and have a high cost burden associated with treatment. Thus, it is necessary to understand what changes could be contributing to this increase in NTM disease rate. Water samples from 40 sites were collected from around the USA. They represented three water types: groundwater disinfected with chlorine and surface water disinfected with chlorine or monochloramine. Two methods, culture and qPCR, were used to measure M. avium and M. intracellulare. Heterotrophic bacteria and NTM counts were also measured. M. avium and M. intracellulare were molecularly detected in 25% (73/292) and 35% (102/292) of samples. The mean concentrations of M. avium and M. intracellulare were 2.8 × 103 and 4.0 × 103 genomic units (GU) L−1. The Northeast sites had the highest sample positively rate for both M. avium and M. intracellulare. The highest NTM counts and M. avium concentrations were observed in the surface water treated with chloramine. Geographic location and source water/disinfectant type were observed to significantly influence M. avium and M. intracellulare occurrence rates. These studies can help improve public health risk management by balancing disinfectant treatments and diverse microbial loads in drinking water.
Keywords: Culture, Drinking water, Mycobacterium avium complex, M. avium, M. intracellulare, qPCR, Water quality
Introduction
Nontuberculous mycobacteria (NTM), i.e., species of mycobacteria not belonging to the Mycobacterium tuberculosis complex, are bacteria that occupy natural and human-engineered environments (Falkinham 2009). There are over 150 recognized species of NTM, with many species considered clinically significant such as the following: M. avium complex (MAC), M. kansasii, M. abscessus, M. chelonae, M. fortuitum, M. genavense, M. gordonae, M. haemophilum, M. immunogenum, M. malmoense, M. marinum, M. mucogenicum, M. nonchromogenicum, M. scrofulaceum, M. simiae, M. smegmatis, M. szulgai, M. terrae complex, M. ulcerans, and M. xenopi (Griffith et al. 2007). Of the twenty clinically significant species, M. avium and M. intracellulare (Parte et al. 2020) which belong within the Mycobacterium avium complex (MAC) are particularly pathogenic and have higher percentage of the health burden than other NTM species (Butler and Crawford 1999; Donohue 2018; Good 1980). Mycobacterium avium and M. intracellulare cause respiratory, soft tissue, skin, lymph, and systemic infections (WHO 2004).
NTM infections are not reportable to the Centers for Disease Control and Prevention’s (CDC) National Notifiable Diseases Surveillance System (NNDSS). Therefore, the national NTM infection trends are unknown. However, a few localized epidemiological studies have observed NTM disease prevalence rate increasing (Adjemian et al. 2012; Donohue 2021; Donohue and Wymer 2016; Kendall and Winthrop 2013; Marras and Daley 2002; Prevots et al. 2010). In the 1980s, NTM disease prevalence was estimated to be 1.3 per 100,000 persons compared to 2015 estimates of 6.78 per 100,000 persons (O’Brien et al. 1987; Winthrop et al. 2020). There are approximately 50,000 to 80,000 individuals affected by NTM lung infections (Donohue 2018; Strollo et al. 2015), and the United States (US) spends approximately 1.5 billion per year on treatment and hospitalization of NTM infections (Collier et al. 2021). Among the NTM infections (CDC 2017), the MAC species are the etiological agent that causes the most infections in the US (Donohue 2018; Henkle et al. 2015; Prevots et al. 2010).
The observed increase in NTM infection rate is most likely multifactorial, including factors such as improved reporting procedures (electronic state disease surveillance systems), newer data sources (Healthcare and hospitalization records, and International Classification of Disease (ICD) codes, etc.), and an increase in disease awareness in the physician community.
The first comprehensive survey of NTM in drinking water at locations throughout the US was conducted (Covert et al. 1999). That survey provided baseline occurrence information that can be compared to present-day and future occurrence data. Drinking water has been investigated as a source of human exposure to NTMs but using culture methods (Aronson et al. 1999; Donohue et al. 2015; Falkinham et al. 2001; von Reyn et al. 1993).
The limitations of culture methods for isolation of mycobacteria from environmental samples are well-known: decontamination of water samples reduces recovery, a minimum 8-week incubation is required to form M. avium and M. intracellulare colonies, overgrowth of non-target organisms on the plate can result in lost NTM counts, colonies require downstream identification, and results are only semi-quantitative. The method of quantification using quantitative polymerase chain reaction (qPCR) assays can overcome some of these limitations. Quantitative polymerase chain reaction assays are designed for many levels of taxonomic specificity (e.g., genus, species, sub-species). Quantitative polymerase chain reaction assays are quantitative and rapid, can detect multiple physiological states (e.g., alive, dead, viable but not culturable), and rarely result in data gaps due to sample loss (e.g., overgrowth or inhibition from the presence of other organisms).
Nontuberculous mycobacterium are environmental bacteria recovered routinely from both water and soil samples. Thus, both ecological sources have been implicated in disseminating disease (De Groote et al. 2006; Griffith et al. 2007; Prevots and Marras 2015; Tzou et al. 2020). Due to M. avium and M. intracellulare’s higher health burden in the US and the need to accurately detect their presences, the goals of the present study were twofold: to evaluate M. avium and M. intracellulare-specific qPCR assays for rapid, quantitative detection of these organisms in drinking water and, in combination with a culture method, to characterize M. avium and M. intracellulare occurrence and their density (concentration) by geography and water type.
Material and methods
Bacterial strains and growth conditions
M. avium American Type Culture Collection (ATCC) ATCC® 700,897 and M. intracellulare ATCC® 13,950 were grown in 10 mL Middlebrook 7H9 broth containing albumin dextrose catalase (ADC) enrichment (BD, Franklin Lakes, NJ) and 2 mg L−1 mycobactin J (Allied Monitor, Fayette, MO) for preparation of genomic DNA for generating standard curves. Cultures were incubated in the dark at 37 °C with 10% CO2 for at least 2 weeks or until visible growth was observed. Cells were pelleted, and genomic DNA was purified using the Wizard® Genomic DNA Purification Kit (Promega Corporation, Madison, WI).
Mycobacterium avium ATCC® 700,897 and M. intracellulare ATCC® 13,950 were propagated in 10 mL Middlebrook 7H9 broth (BD) containing the above amendments and incubated similarly for preparation of cell stocks which were used to determine the analytical sensitivity of the method. After four weeks incubation, cells were pelleted by centrifugation and washed three times with sterile phosphate-buffered saline (PBS), pH 7.0, and resuspended in 5 mL PBS. Cells stocks were prepared by dividing the cell suspension into 200 μL volumes and freezing aliquots at − 20 °C. Cells stocks were enumerated by spread plating serial dilutions on Middlebrook 7H10 agar (BD) containing OADC enrichment and mycobactin J in triplicate.
M. avium and M. intracellulare taxonomy note
Within the Mycobacterium avium complex (MAC), Mycobacterium avium is divided into four subspecies: M. avium subsp. avium, subsp. silvaticum, subsp. hominissuis, and subsp. paratuberculosis, of which only subsp. hominissuis (Rindi et al. 2018) and paratuberculosis (Scanu et al. 2007) are associated with human illness. Several new species have been proposed: M. chimaera (Tortoli et al. 2004), M. timonense, M. marseilense, and M. bouchedurhonense (Ben Salah et al. 2009) species. These species are highly similar to M. intracellulare. In 2013, Wallace et al. suggested that M. intracellulare isolates recovered from water are M. chimaera species (Wallace et al. 2013). In 2017, the whole genome of M. chimaera Fl-0169 (type strain) was sequenced (Pfaller et al. 2017). This genome sequence was > 98% homologous to that of M. intracellulare ATCC-13950. Therefore, M. chimaera (genetically) is not an independent species within the MAC complex but a subspecies of M. intracellulare (Tateishi et al. 2021).
qPCR assays
qPCR specificity and analytical sensitivity
Mycobacterium avium and M. intracellulare qPCR assays were designed to detect all subspecies of M. avium and M. intracellulare (Pfaller et al. 2017; Wallace et al. 2013). The separate assays use the same forward primer and probe. However, the M. avium and M. intracellulare reverse primers differ by 6 base pairs (bp). Primer and probe sequences for the M. avium and M. intracellulare qPCR assays have been characterized previously for detection of these organisms in biofilm (Chern et al. 2015). Mycobacterium avium and M. intracellulare primers and probes sequences are in Table 1. The DNA primers and probe target the 16S rRNA gene.
Table 1.
Primer and probe sequences for M. avium and M. intracellulare qPCR assays
| Target | Sequence (5′ to 3′) | Reference |
| M. avium (16S rRNA) | MAF: GGGTGAGTAACACGTGTGCAA MAR: CCAGAAGACATGCGTCGTGA MAP: FAM-TGCACTTCGGGATAAGCCTGGGAAA-TAMRA |
Chern et al. (2014) |
| M. intracellulare (16S rRNA) | MAF: GGGTGAGTAACACGTGTGCAA MIR: CCACCTAAAGACATGCGACTAAA MAP: FAM-TGCACTTCGGGATAAGCCTGGGAAA-TAMRA |
Chern et al. (2014) |
Chern et al. 2014. J Water Health. 10.2166/wh.2014.060
Specificity of the qPCR assays was evaluated in silico and in vitro and reported in Chern et al. (2015). The M. avium assay detects all four subspecies of M. avium. The M. intracellulare assay detects the M. intracellulare subspecies chimaera. Both analyses suggest the assays are 100% specific for their respective targets.
Analytical sensitivity of the complete method was evaluated using M. avium ATCC® 700,897 and M. intracellulare ATCC® 13,950 cell stocks prepared as described above and thawed at room temperature. Ten-fold serial dilutions of cells were prepared from stocks ranging from 104 to a theoretical 101 cells mL−1 and spiked into five 1-L tap water samples. Samples were processed identically to drinking water samples described below. Analytical sensitivity is defined as the lowest approximate number of M. avium and M. intracellulare cells that could be detected in a 1-L water sample by qPCR 100% of the time.
qPCR assay characteristics, specificity, and method sensitivity
The master standard curves generated for M. avium and M. intracellulare qPCR assays demonstrated that the quantification results were linear over at least 5 log10 of cells and, most importantly, accurate at low target concentrations, which is what is typically encountered in a treated drinking water sample. The amplification efficiency (E) was estimated from the slope of the standard curve using the formula [E =10(−1/slope) − 1] and was 95 for M. avium and 96 for M. intracellulare qPCR assays. Limits of detection (LOD) and quantification (LOQ) were determined by Chern et al. (2014). The LOD of both assays was 10 genomic units per qPCR reaction and LOQ of both assays was 10 genomic units per qPCR reaction.
Analytical sensitivity of the complete method was determined by spiking known quantities of M. avium or M. intracellulare target cells into five different 1-L tap samples each. The method provided consistent detection (5/5) of either M. avium or M. intracellulare in a spiked sample of 102 cells L−1. In a spiked sample of 101 cells L−1, M. avium and M. intracellulare were detected 60% of the time.
Drinking water sample collection
In 2009–2010, drinking water was collected three times during the first year of the study and once the following year from 40 locations (10 private residences and 30 commercial buildings) located in 25 states, one district, and one US territory. Either one or two cold water taps were sampled at each location for a total of 73 taps and a total of 292 drinking water samples, analyzed by culture and qPCR, for a total of 876 analyses. The tap types in this study were kitchen sink, bathroom sink, drinking water fountains, and refrigerator door. The taps included in the study were supplied with drinking water from untreated private wells or from treatment plants that ranged in size from small to large (population served 3,301 to ≥ 100,000 persons), using a variety of source waters (surface water, groundwater, or mixed surface and groundwater) and disinfection regimes. Table 2 describes the type of source water and disinfectant used for water supplied to the taps surveyed in this study.
Table 2.
Sites in this study characterized by source water and disinfectant type
| Source water type | Disinfectant | Number of sites |
|---|---|---|
| Groundwater | None | 2 |
| Groundwater | Chlorine | 8 |
| Groundwater | Chloramine | 0 |
| Surface water | None | 0 |
| Surface water | Chlorine | 16 |
| Surface water | Chloramine | 13 |
| Surface water | Chlorine dioxide | 1 |
Four liters of cold tap water was collected from each tap after a 15 s flush into 1-L sterile polypropylene bottles according to Sects. 9060A and B of 23rd Edition of Standard Methods for the Examination of Water and Wastewater, except no preservative was included. Samples were transported back to the laboratory on ice and stored at 4 °C, and processed within 48 h. Of the 4 L of water collected per tap, 2 L was analyzed by culture, and 2 L was used for qPCR as described below (Donohue et al. 2015).
Sample processing, DNA extraction, and qPCR analyses
Two liters of drinking water sample was analyzed with M. avium and M. intracellulare qPCR assays using a bead beating method described previously (Beumer et al. 2010). Briefly, the water sample was vacuum filtered (MilliporeSigma, Burlington, MA) through a 47.0 mm, 0.45 μm polycarbonate membrane (Whatman Inc., Piscataway, NJ), and the membrane was transferred aseptically to a sterile 2 mL microcentrifuge tube with an O-ring cap containing 0.3 g of 0.1 mm glass beads (BioSpec Products Inc., Bartlesville, OK). Cells trapped on the membrane were lysed by adding 500 μL Tissue and Cell Lysis Solution (Lucigen Corporation, Middleton, WI) and beaten in a Mini-Bead beater (BioSpec Products) for 3 min on “homogenize” setting.
After a 5-min cooldown in ice, the lysate was pipetted into a new 1.5 mL microcentrifuge tube, and 2 μL of proteinase K (50 μg/μL) (Lucigen Corporation) was added followed by incubation at 65 °C in a water bath for 15 min. Next, 2 μL of RNase A (5 μg/μL) (Lucigen Corporation) was added to the mixture and incubated at 37 °C for 30 min. Subsequently, 350 μL of MPC Protein Precipitation Reagent (Lucigen Corporation) was added to precipitate the cellular proteins. The resulting supernatant was transferred to a microcentrifuge tube with an equal volume of ice-cold isopropanol (~ − 4 °C) and centrifuged at 10,000 × g for 10 min. The isopropanol was poured off, and the resulting DNA pellet washed with 500 μL of ice-cold (~ − 4 °C) 70% ethanol. Samples were centrifuged, and the ethanol was removed. The DNA pellets were re-suspended in 150 μL of molecular biology grade water (Corning Inc., Corning, NY) and stored at − 80 °C until analyzed.
Mycobacterium avium and M. intracellulare qPCR assays were performed on 10 μL of DNA extract/template. Reactions were performed in triplicate. All reactions were carried out in an ABI Prism® 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). A method control was performed at the time of filtration. A 100 mL sterile molecular biology grade water (Corning Inc) was prepared and processed along with incoming samples. An internal positive control (Life Technologies Corp.® TaqMan® Exogenous Internal Positive Control; Grand Island, NY) was added to every reaction to detect PCR inhibition. Positive qPCR assay controls included target DNA standards of known concentration. Negative qPCR assay controls included sterile H2O added to the qPCR reaction rather than the DNA template. A sample was considered positive if the quantification cycle (Cq) of two of three replicate reactions was < 40, and if both negative and method controls were negative (undetermined). A sample was considered inhibited if the internal positive control Cq deviated more ± 2.5 units.
A master standard curve was generated from six independent sets of tenfold serially diluted genomic DNA purified from the M. avium and M. intracellulare reference strains described above. Each dilution series contained eight standards run in triplicate for a total of 18 Cq measurements per standard. Linear regression was performed on Cq versus the log10 genomic units, and a line equation was generated. Estimated M. avium and M. intracellulare quantities were extrapolated from Cq measurements input into the line equations obtained from the master standard curves.
Sample processing and Mycobacterium culture analysis
Two liters of drinking water was analyzed by culture. The sample was shaken vigorously by hand, each liter was divided into two 500 mL aliquots, placed in sterile plastic bottles, and treated with 0.04% cetylpyridinium chloride (w/v, final concentration) (MilliporeSigma, Burlington, MA) (Glover et al. 1994). Samples were shaken and incubated at room temperature for 30 min. Each 500 mL was vacuum filtered through a 47 mm, 0.45 μm membrane black-gridded HABG047S6 filter (MilliporeSigma) and placed on Middlebrook 7H10 agar (BD) containing 500 μg L−1 cycloheximide (MilliporeSigma) and 2 mg L−1 mycobactin J (Allied Monitor Inc., Fayette, MO). Plates were incubated in sealed plastic bags at 37 °C in a 10% CO2 atmosphere for a minimum of 8 weeks and inspected weekly (Donohue et al. 2015). Colonies that fit the morphologies described for mycobacteria in Glover et al. were selected for identification (Glover et al. 1994). One to five representative isolates (dependent on the sample’s colony-forming unit (CFU) densities and morphology) were selected for identification by Sanger sequencing.
DNA extraction for Sanger sequencing
DNA was extracted from each isolate using Wizard DNA Cleanup System for extractions, following the manufacturer’s instructions (Promega, Madison, WI). DNA extracts were first quantified using a Nanodrop™ spectrophotometer (Thermo Scientific, Waltham, MA) and were subsequently stored at − 80 °C.
Sanger sequencing PCR
The 16S rRNA gene was amplified by PCR and sequenced for subsequent isolate identification. Amplification of the 16S rRNA gene was performed with primers 8F and 1492R (Lane 1991). The amplification hsp65 gene was performed using primers HSPF3 and HSPF4 (Kim et al. 2005). Amplification was done with a 25 μL reaction volume, containing 2.5 μL 10 × PCR buffer, 1.25 μL of each 10 μM primer, 1.5–2.0 μL of 25 mM MgCl2 solution, 2.0–2.5 μL of 10 mM dNTPs, 1 U/μL Taq polymerase, 13.1–13.3 μL dH2O, and 2 μL DNA. The thermocycle for each PCR reaction was an initial 5 min denaturation step at 95 °C, followed by an additional 30 s at 95 °C. DNA was annealed at 50 °C for 60 s with an extension for 30 s at 72 °C plus a terminal 1 cycle at 72 °C for 5 min.
Sequence analysis and isolate identification
Genotyping-by-sequencing analysis of both forward and reverse PCR products was accomplished on an Applied Biosystems™ 3730xl DNA Analyzer using ABI’s BigDye® Terminator v3.1 Cycle Sequencing Kit (Life Technologies, Grand Island, NY). Consensus sequences were generated on BioEdit Sequence Alignment Editor Version 7.1.3.0 for each gene using the forward and reverse sequence (Hall 2011). Two approaches were used to identify a colony. Initially, consensus sequences of both PCR products were submitted into the National Center for Biotechnology Information’s (NCBI) nucleotide non-redundant (nr) BLAST search to determine percent homology to known species. Next, a tree comparison using Phylogenetic Analysis Using Parsimony (PAUP)* 4.0 software was performed (Swofford 2002). The software compared NTM type strains 16S rRNA gene and hsp65 sequences to the unknown NTM isolates gene sequences. The 16S rRNA gene sequences were given GenBank accession numbers KU172693-KU173100. The hsp65 sequences have the following GenBank accession numbers MW451671-MW451717, MW523685-MW523777, MW566384-MW566432, MW874724-MW874829, and MW874663-MW4874723. By nucleotide BLAST searches and PAUP sequence alignment, the M. avium isolates were ≥ 98% similar to M. avium subsp. hominissuis. The M. intracellulare isolates were ≥ 98% similar to M. intracellulare subsp. chimaera.
In addition to qPCR, there were four key differences between this study and Covert et al. (1999). In the present study, water samples were collected after a 15 s flush, not 1–2 min.
Additionally, no sodium thiosulfate was included in the sample bottles. It was reported that better recoveries of mycobacteria were achieved if the chlorine residual remains in the sample (Thomson et al. 2008). Also, membranes were rinsed with sterile molecular biology-grade water rather than Standard Methods Buffer. Isolates obtained by culture were identified by sequencing of 16S rRNA and hsp65 genes.
Heterotrophic plate counts
Upon the sample’s arrival into the laboratory, two 100 μL aliquots were removed for heterotrophic plate count (HPC) analysis (Standard Method 9215B). For each water sample, a 100 μL aliquot of water was aseptically spread-plated onto a R2A agar plate (BD; Franklin Lake, NJ) and incubated at 25 °C for 7 days in the dark. The limit of detection for the HPC method is 1 CFU/100 μL. CFUs were counted and recorded on day 7.
Data and statistical analysis
The qPCR’s Cq values were transformed to genomic units using the master standard curve. The genomic unit number per replicate was average for each positive sample. A culture sample was considered positive if a colony’s identity was determined to be either M. avium or M. intracellulare. The culture results were not quantitative.
MAC detection frequency was determined if one or both species were identified on the agar plated or if the sample was positive for one or both qPCR assays. Mycobacterium avium complex concentrations were calculated by summing the M. avium and M. intracellulare concentrations. Statistical significance between detection frequencies was determined by chi-square analysis in SigmaPlot 14.0 (SYSTAT, San Jose, CA) with a significance level of alpha = 0.05. Statistical significance among the concentrations were evaluated using the Mann–Whitney U rank sum test and one-way ANOVA with a significance level of alpha = 0.05.
Results
Occurrence of M. avium and M. intracellulare in tap water in the USA
M. avium and M. intracellulare were detected in 95% (38/40) of sites in this study by one or both detection methods. Five percent (2/40) of sites either had no detection by either method or M. avium/M. intracellulare concentrations were below the limit of detection (Fig. 1). Most sites/taps were either M. avium or M. intracellulare positive at least once among the four sampling events. The number of samples per regional area was Northeast n = 28, South n = 132, Midwest n = 76, and West n = 56. There were slight geographic differences observed in the number of samples positive per regional area. The Northeast locations had the highest sampling positivity rate by either one or both methods for both M. avium and M. intracellulare, 36% (10/28) and 54% (15/28), respectively. The Southern region was a close second with an M. avium and M. intracellulare sampling positivity rate of 33% (44/132) and 39% (52/132), respectively. Mycobacterium avium’s lowest sampling positivity rate was in the Western region, 21% (12/56). Mycobacterium intracellulare’s lowest sampling positivity rate was observed in the Midwest 22% (17/76). Mycobacterium intracellulare’s rate difference between the Northeast (highest) and the Midwest (lowest) was significant, χ2: P = 0.005.
Fig. 1.
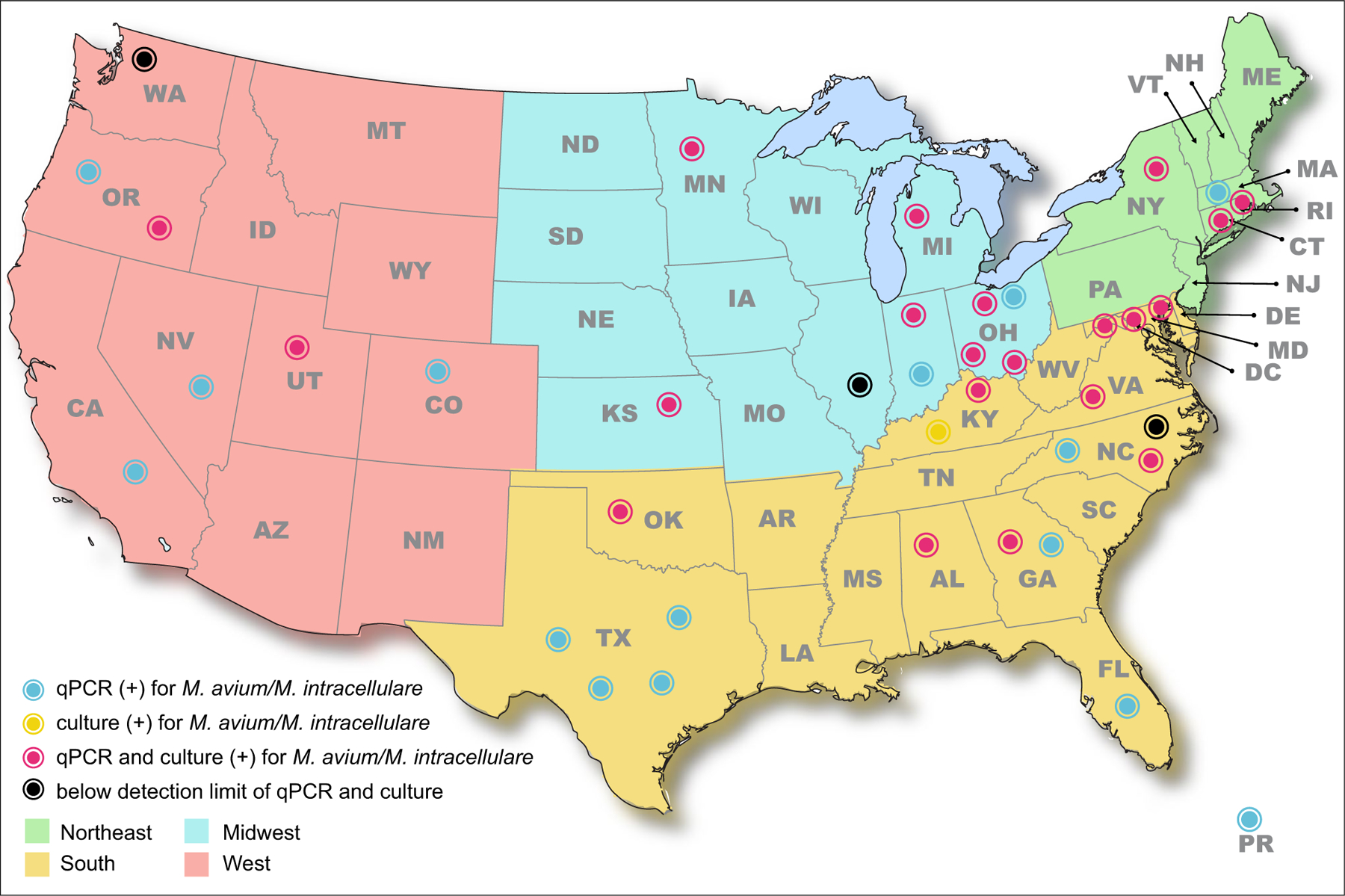
Geographical distribution of sites positive for M. avium and/or M. intracellulare by qPCR, culture, or both
Mycobacterium avium complex was culture recovered from 18% (48/272) of all samples. Of the samples that were NTM positive, MAC was detected in 23% (48/211). Mycobacterium avium and M. intracellulare isolates were culture recovered from 14% (37/272) and 6% (16/272) of samples. Of the samples that yielded M. avium and M. intracellulare isolates, only 8% (4/48) had both M. avium and M. intracellulare species.
Forty-five percent (130/292) of samples were positive for MAC (either M. avium and/or M. intracellulare) by qPCR. Mycobacterium avium was detected by qPCR in 25% (73/292) of tap samples. The mean concentration of M. avium in positive samples was 2.8 × 103 genomic units L−1 water and the median concentration was 1.8 × 102 genomic units L−1 (Table 3). Forty samples contained approximately < 10–100 genomic units L−1, twenty-two samples contained approximately 101–1,000 genomic units L−1, and eleven samples contained > 1,001 genomic units L−1. Mycobacterium avium was detected by culture in 12% (36/292) of all tap samples.
Table 3.
Mean and median concentrations of M. avium and M. intracellulare in tap samples positive by qPCR
| Target | Mean of qPCR positives (genomic units L−1) | Median of qPCR positives (genomic units L−1) |
|---|---|---|
| M. avium | 2.8 × 103 | 1.8 × 102 |
| M. intracellulare | 4.0 × 103 | 2.1×102 |
Mycobacterium intracellulare was detected by qPCR in 35% (102/292) of tap samples. The mean concentration of M. intracellulare positive samples were 4.0 × 103 genomic units L−1 and the median concentration was 2.0 × 102 genomic units L−1 (Table 3). Fifty-six samples contained approximately < 10–100 genomic units L−1, twenty-nine samples contained approximately 101–1,000 genomic units L−1, and seventeen samples contained > 1,001 genomic units L−1. Mycobacterium intracellulare was detected by culture in 5% (16/292) of all tap samples.
There was little agreement between the qPCR and culture positive results for M. avium and M. intracellulare detection (Fig. 2). Mycobacterium avium was detected in 92 samples by either culture or qPCR. There was only an 18% (17/92) agreement between the two methods for M. avium detection. Mycobacterium intracellulare was detected in 108 samples by either culture or qPCR. Culture and qPCR only agreed in 10% (10/108) of the samples. Approximately, 33 to 36% of the M. avium and M. intracellulare qPCR-positive samples could not be verified by culture. These sample plates were overgrown with bacteria or mold, or the PCR product sequence was corrupt (> 30% of base pairs are undetermined by the sequencer and assigned as N).
Fig. 2.
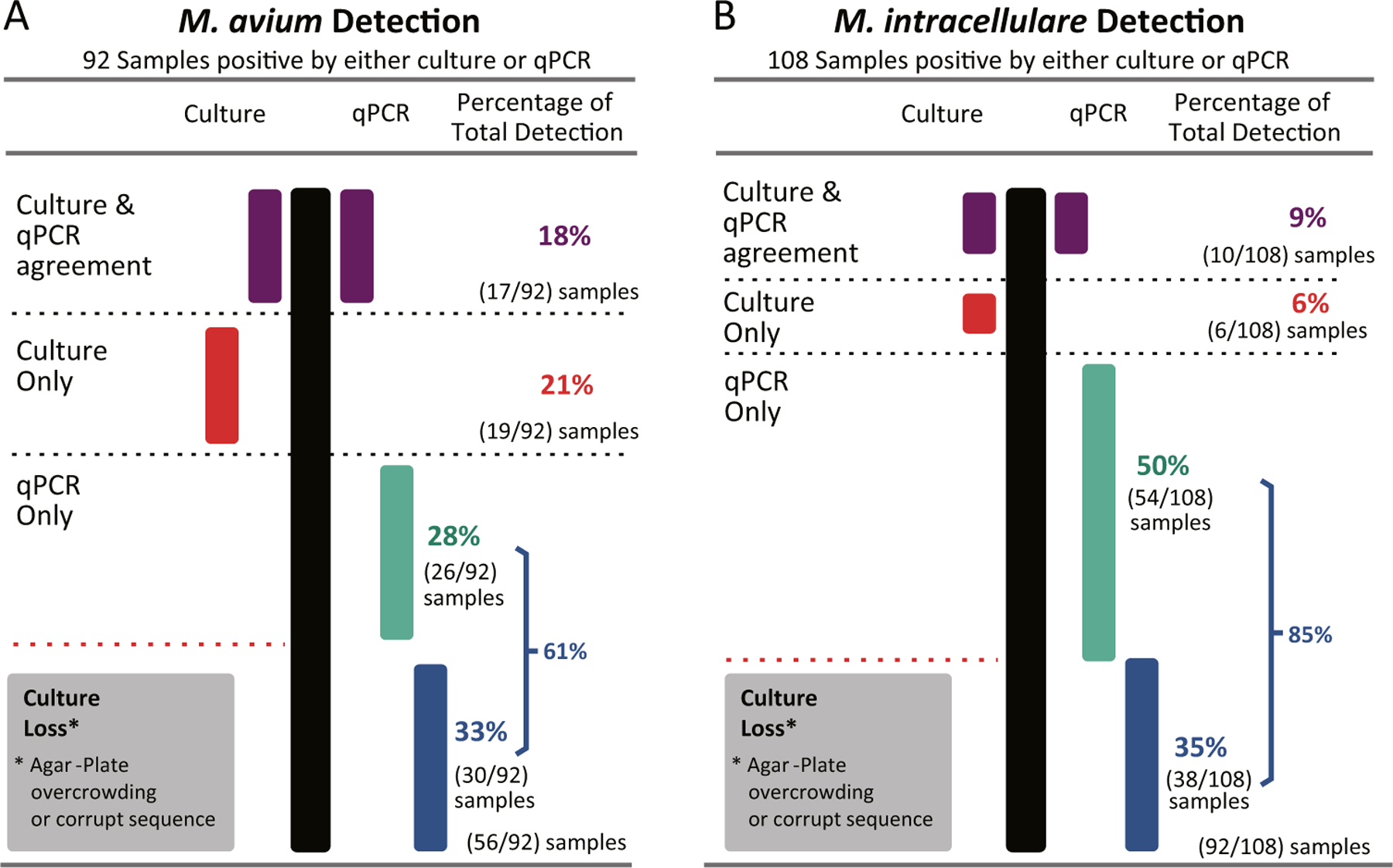
Agreement and disagreement between culture and qPCR for detection of M. avium and M. intracellulare
Persistence at taps
The persistence of M. avium and M. intracellulare was evaluated by sampling each tap on four separate occasions (three times in 2009 and once in 2010) to determine if detections were sporadic or constant over time, as shown in Fig. 3. Figure 3 is a waffle chart representing M. avium and M. intracellulare persistence. Based on the qPCR results, M. avium was less persistent in the tap water than M. intracellulare. The culture method results indicated that few taps were positive for MAC, but M. avium was more persistent than M. intracellulare.
Fig. 3.
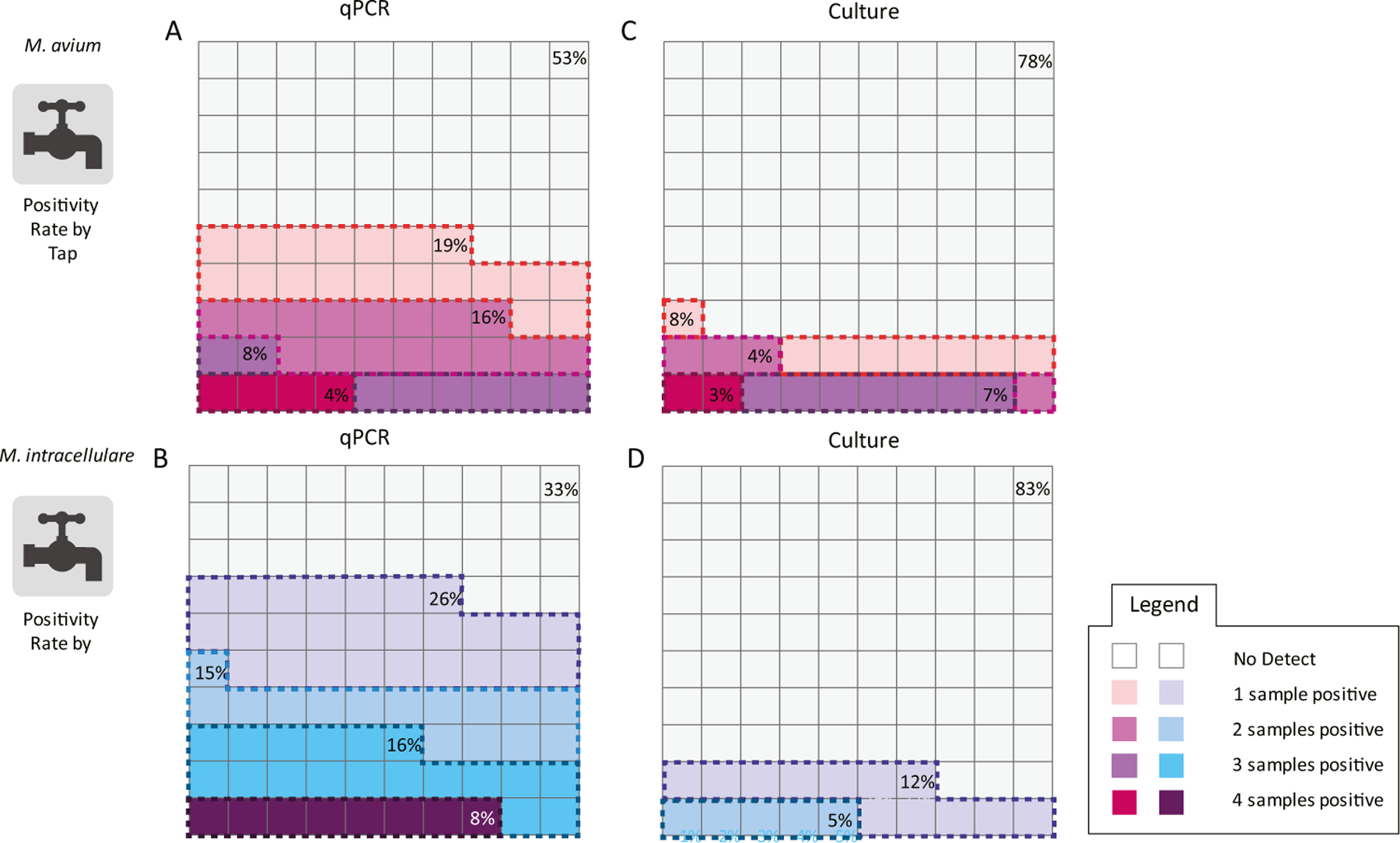
Waffle charts depicting tap positivity rate of M. avium (MA) A by qPCR and B by culture and M. intracellulare (MI) C by qPCR and D by culture
Occurrence of MAC in taps with different source waters and disinfectants
Taps surveyed in this study were supplied with water of four main types: untreated groundwater (16 samples), chlorinated-groundwater (64 samples), chlorinated-surface water (116 samples), and chloraminated surface water (88 samples). The eight chlorine dioxide samples were not included in this analysis for there was no detection of M. avium or M. intracellulare. Mycobacterium avium complex qPCR detection frequencies were the highest in surface water treated with chloramine 59% (52/88), followed by untreated groundwater 55% (9/16), next groundwater treated with chlorine 45% (29/64), and lastly in surface water treated with chlorine 34% (40/116) (Fig. 4). The MAC detection frequency in chloraminated surface water was significantly higher than chlorinated-surface water, χ2: P = 0.006, but there was no significance between the other source water types and disinfectants.
Fig. 4.
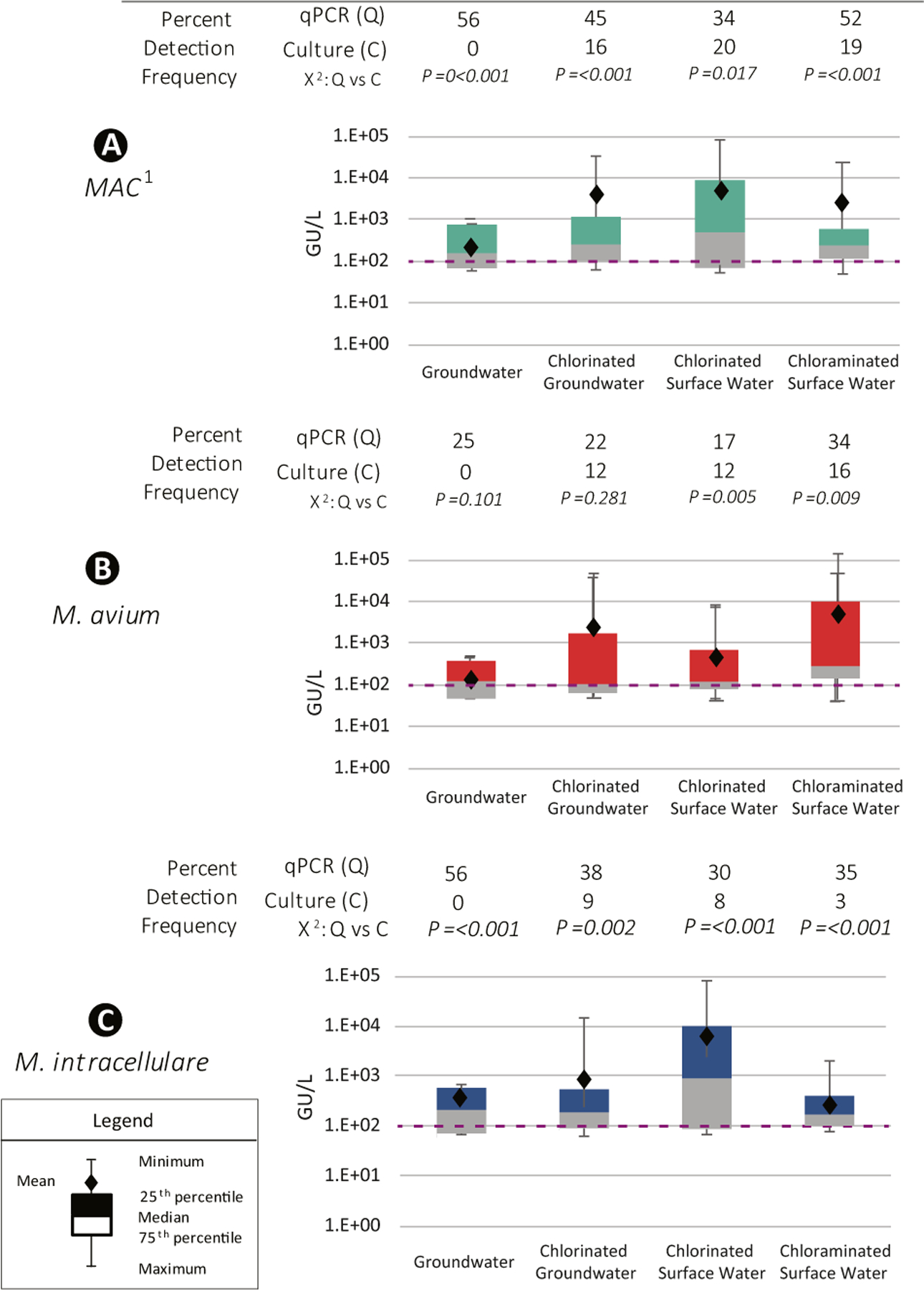
Percent detection by qPCR and culture, and box and whisker plots of concentrations (genomic units (GU)/L) for A MAC, B M. avium, and C M. intracellulare by source water and disinfectant. 1MAC, M. avium and/or M. intracellulare. Chi-square test (χ2) results compared qPCR and culture detection frequencies. The dotted orange line demarks the 102 genomic units (GU)/L
Mycobacterium avium’s highest detection rate was observed in chloraminated surface water samples 34% (30/88), followed by untreated groundwater 25% (4/16), then in chlorinated groundwater 22% (20/64), and lastly in chlorinated-surface water 17% (20/116). Mycobacterium avium detection frequency in chloraminated surface water was significantly higher than chlorinated-surface water, χ2: P = 0.006, but there was no significance between the other source water types and disinfectants.
Mycobacterium intracellulare’s highest detection rate was observed in untreated groundwater 56% (9/16), followed by chlorinated-groundwater 38% (24/64), then chloraminated surface water 35% (31/88), and lastly by chlorinated-surface water 30% (35/116). There were no statistical differences detected between M. intracellulare detection frequencies among four source water and disinfectant groups. Additionally, M. avium and M. intracellulare concentrations did not differ significantly between source water and disinfectant type (Fig. 4).
Interesting culture results showed a different detection frequency profile. Mycobacterium avium complex culture detection frequency was near equivalent between the four source water + disinfectant groups. The highest culture detection frequency was in chlorinated-surface water samples 19% (22/116), followed by chloraminated surface water 19% (17/88), next chlorinated-groundwater 16% (10/64), and lastly groundwater 0% (0/16). Mycobacterium avium culture detection ranking are as follows: chloraminated surface water 16% (14/88), chlorinated groundwater 13% (8/64), chlorinated surface water 12% (14/116), and groundwater 0% (0/16). Mycobacterium intracellulare culture detection frequency rankings were the following: chlorinated groundwater 9% (6/64), chlorinated surface water 8% (9/116), chloraminated surface water 3% (3/88), and groundwater 0% (0/16).
MAC‑positive samples, heterotrophic bacteria, and NTM counts by source waters and disinfectants
Positive M. avium and M. intracellulare samples’ heterotrophic bacteria and NTM counts were analyzed by source water and disinfection type (Fig. 5). The green circles are the positive samples’ heterotrophic bacteria concentration (CFU/500 mL), and the grey squares are the positive samples’ NTM concentration (CFU/500 mL). The green (HPC) and grey (NTM) bands represent mean (upper edge) and median (lower edge) concentrations. The rose (M. avium) or blue (M. intracellulare) bands represents the positive sample’s mean and median concentration. The purple dotted line represents the National Primary Drinking Water Regulation (NPDWR) HPC Maximum Contaminant Level (MCL) at distribution’s entry point 500 CFU/mL (2.5 × 105 CFU/500 mL). Depending on the source water and disinfectant, HPC, NTM, and M. avium/M. intracellulare concentrations shift. For the M. avium–positive samples, the highest mean-median concentration for heterotrophic bacteria is observed in the chlorinated surface water (Fig. 5B). However, the highest NTM counts and M. avium concentrations were observed in the chloraminated surface water (Fig. 5C). Mycobacterium avium concentrations can be a log greater than the NTM counts in chloraminated surface water (Fig. 5C).
Fig. 5.
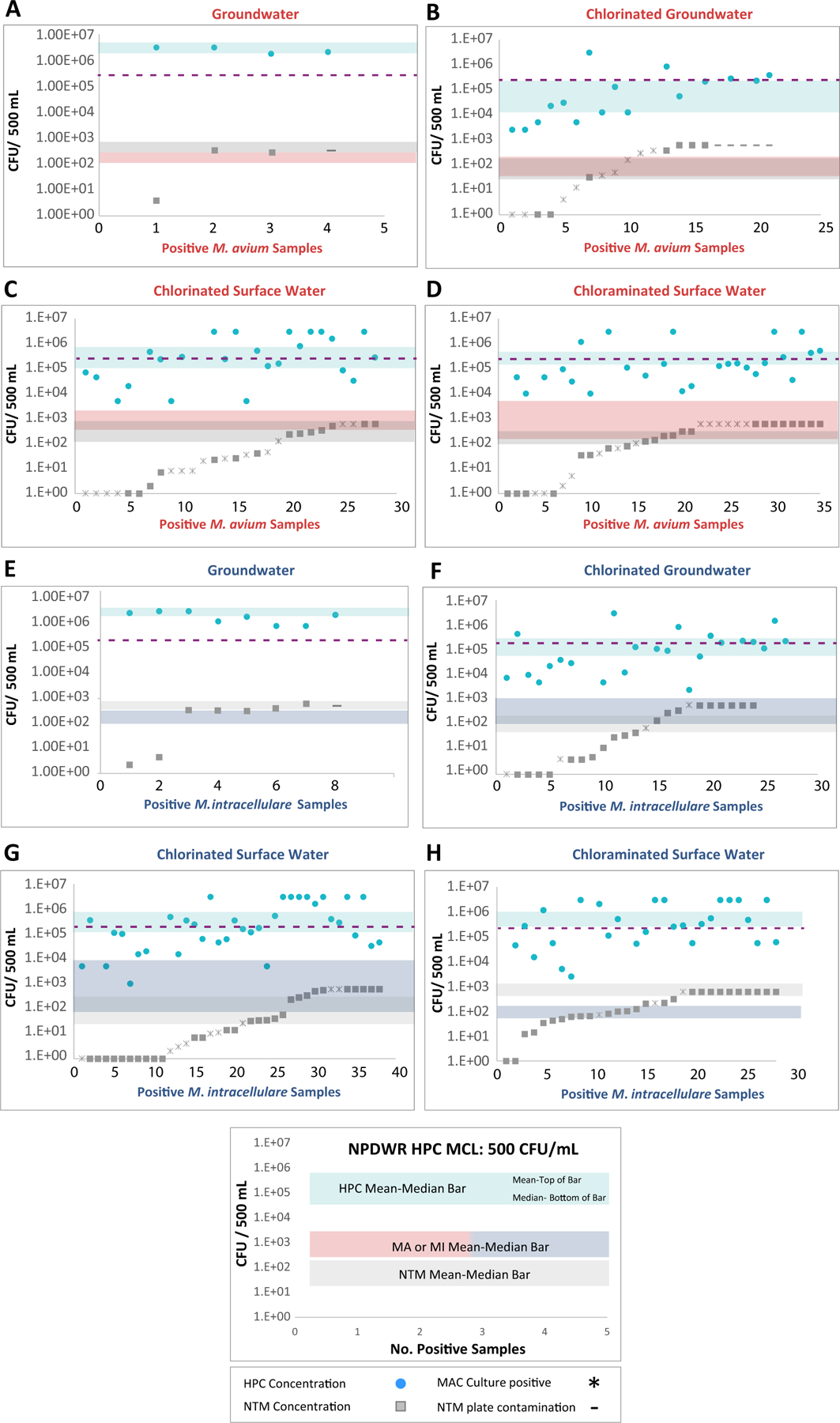
Positive M. avium and M. intracellulare samples’ heterotrophic bacteria and NTM counts by source water and disinfection type. The purple dotted line represents the National Primary Drinking Water Regulation (NPDWR) HPC Maximum Contaminant Level (MCL) at distribution entry point 500 CFU/mL (2.5 × 105 CFU/500 mL)
The broadest mean-median bar for M. intracellulare was observed in chlorinated surface water (Fig. 5E). The M. intracellulare concentrations are about 1.5 logs higher than the average NTM concentration. In chlorinated surface water, the M. intracellulare concentration appears to be suppressed by the other NTM species and the heterotrophic bacteria. Mycobacterium intracellulare concentrations are approximately half a log lower than the NTM average. Overall, Fig. 5 shows that there are significant interplays among bacterial species that are driven by source water and disinfectant.
Discussion
The present study analyzed 292 water samples from across the US, using the culture method (Donohue et al. 2015) and qPCR. In this study, at the sample analysis level, regional differences were observed in the samples’ positivity rate. The Northeast’s higher sample positivity rate is supported by Gerbert et al.’s biofilm research and Spaulding et al.’s analysis of the geographic distribution of clinical isolates (Gebert et al. 2018; Spaulding et al. 2017). Mycobacterium intracellulare was detected more frequently than M. avium (35% and 31%, respectively) by qPCR, while M. avium was detected more frequently than M. intracellulare by culture (13% and 6%, respectively).
In the Covert et al., (1999) NTM survey, NTMs were recovered in drinking water samples from across the US, including 105 taps samples, of which < 1% (1/105) were positive for M. avium and 5% (5/105) were positive for M. intracellulare using a culture method. The authors concluded that MAC organisms might not be ubiquitous in tap water in the US In the current survey, the M. avium culture detection frequency was significantly greater (χ2: P = < 0.001) than the frequency reported by Covert et al. (1999). In comparison, M. intracellulare culture detection frequency had not changed significantly from the 1999 survey results (Covert et al. 1999).
In general, there were more samples positive by qPCR than by culture. This result is due to the specificity of each approach. Quantitative PCR is highly specific for the precise detection of M. avium and M. intracellulare. The culture method is a general approach that will recover most NTM species. Rasanen et al. (2013) compared a modification of the culture method used in this study to their Mycobacterium genus–specific qPCR assay to detect mycobacteria in 48 drinking water samples from three different sources in Finland. The authors found a good correlation in detection frequency between the two methods but significant differences in the quantities detected, with qPCR detecting higher concentrations in some samples. In the Rasanen et al. (2013) study, the culture and qPCR detection frequency were positively correlation because both methods detected multiple species of mycobacteria. Since both methods detected multiple species of mycobacteria (Rasanen et al. 2013).
In this study, the qPCR and culture method did not agree with each other. There are several potential reasons for the disagreement between methods, including different targets (DNA vs. cells), cells were non-culturable or dead, and split samples (the unequal partitioning of microorganisms). The qPCR and culture methods analyze different amounts of water (culture: 500 mL per plate and qPCR: 200 mL per reaction), and methods have different sensitivities (culture method sensitivity was not characterized). Additionally, only a small number of colonies per culture plate were sequenced and identified. In general, however, regardless of the targeted microorganism, culture and molecular techniques rarely corroborate each other’s results.
Previous studies have shown a considerable range of occurrence of MAC in drinking water, using culture and qPCR methods. A survey conducted by Hilborn et al. (2006) involving 56 tap samples originating from two different drinking water treatment plants found that 54% of samples were positive for M. avium using the culture method (Hilborn et al. 2006). This study also found three different M. avium genotypes persisted at three different taps for 18–26 months. The Hilborn et al. (2006) study also demonstrated that M. avium and M. intracellulare persisted in specific taps over time (Hilborn et al. 2006). Feazel et al. (2009) examined 14 potable water samples collected from showers in homes in the US using an M. avium–specific qPCR assay and 93% (13/14) were positive, though concentrations were not provided. These samples were obtained from two bathroom showers in private residences located in Denver and New York City, and the paper does not specify if the water came from either the hot or cold plumbing (Feazel et al. 2009). Mycobacterium avium might have colonized the plumbing, resulting in the high frequency of positive samples, or the author’s qPCR method may be more sensitive than others.
There are very few studies that investigated M. avium and M. intracellulare occurrence by source water type: surface water and groundwater. In the King et al. (2016) study that compared 25 utility’s source and treated water, M. avium and M. intracellulare were not detected in treated groundwater samples (0/3) (King et al. 2016). In comparison, M. avium and M. intracellulare were detected in 32% (7/22) of the treated surface water samples. Mycobacterium avium and M. intracellulare average concentrations at the distribution entry point were 2.1 × 103 and 8.0 × 102 genomic units/L (King et al. 2016). In the King et al. (2016) paper, M. avium, and M. intracellulare detection frequencies were higher in chloramine-treated surface water samples (samples taken at the entry point to distribution) than in chlorine-treated surface water samples; chloraminated surface water: MA 33% (3/9) and MI: 44% (4/9); verse chlorinated surface water: MA 23% (3/13) and MI: 23% (3/13).
Many M. avium and M. intracellulare occurrence surveys have noticed that the disinfectant impacts detection frequencies and concentration. In a survey of tap water from two chloramine systems, M. avium was detected in only 9% of samples from one system and 11% of samples from the other system by qPCR, though the characteristics of each system were different, including differences in the size of each system and its geographic location. The average sample’s M. avium concentration in the two chloramine systems were 1.9 × 103 and 3.8 × 104 gene copies L−1(Wang et al. 2012). Waak et al. (2019) tested two chloramine systems in the Northern area of the USA and did not detect MAC by qPCR. This could have been due to the small amount of DNA extract (1 μL) utilized in their qPCR reactions (Waak et al. 2019). In the present study, the qPCR reaction contained 10 μL of DNA extract (representing 200 mL of water collected) per reaction. In the King et al. (2016) paper, MAC, M. avium, and M. intracellulare detection frequencies were higher in chloramine-treated surface water samples (samples taken at the entry point to distribution) than in chlorine-treated surface water samples (44% and 33% (chloraminated surface water); MAC and MI: 4/9, MA: 3/9; versus 23% (chlorinated surface water); MAC, MA, and MI 3/13).
In a chlorine- versus chloramine-treated drinking water study, M. avium’s detection frequency was significantly higher in chloramine-treated water than chlorine (chloramine: 22% (29/74) versus chlorine: 12% (26/105); P = 0.02) and M. intracellulare’s concentration was significantly different (chloramine: 9.3 × 102 cell equivalence (CE) L−1 versus chlorine: 3.5 × 102 CE L−1; P = 0.02) depending on disinfectant utilized (Donohue et al. 2019). In the present study, MAC was detected more frequently in chloraminated surface water samples (59%) than chlorinated surface water or groundwater samples (34% and 45%, respectively).
NTM infections are increasing in the USA and have a high-cost burden associated with treatment. Thus, it is necessary to understand what environmental changes may have occurred that could be contributing to this increase in disease rate. This drinking water study provides new insights into M. avium and M. intracellulare occurrence. The M. avium and M. intracellulare occurrence stories are different depending on which method is used. Culture indicates that the M. avium detection rate has increased and that M. intracellulare detection rate has remained the same (comparison to Cover et al. 1999). By culture, these species are not affected by source water/disinfectant types. Whereas qPCR shows that M. intracellulare occurs more often than what culture suggests. Quantitative PCR indicates that source water/disinfectant type influences both M. avium and M. intracellulare detection rate and cell density (concentration). The molecular approach also reveals the source water/disinfectant type’s impact by providing a finer occurrence resolution. Since these bacteria may impact human health, it is important to get the clearest image of their occurrence in the built environment. By knowing where these species occur in potable water, water treatment and water management strategies can be optimized to potentially reduce disease transmission.
Key points.
M. avium (MA) culture rate increased significantly: 1% (1999) to 13%.
Culture versus qPCR method: 13% vs 31% for MA and 6% vs 35% for MI.
The results of each method type tell two different stories of MA and MI occurrence.
Footnotes
Declarations
Ethics approval and consent to participate Not applicable.
Consent for publication Not applicable.
Competing interests The authors declare no competing interests.
Publisher's Disclaimer: Disclaimer The United States Environmental Protection Agency through its Office of Research and Development funded and managed the research described here. It has been subjected to Agency’s administrative review and approved for publication. The views expressed in this paper are those of the author and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency.
Data availability
Raw data will be publicly accessible at data.gov. search: author’s name.
References
- Adjemian J, Olivier KN, Seitz AE, Holland SM, Prevots DR (2012) Prevalence of nontuberculous mycobacterial lung disease in US Medicare beneficiaries. Am J Respiratory Critical Care Med 185(8):881–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson T, Holtzman A, Glover N, Boian M, Froman S, Berlin OG, Hill H, Stelma G Jr (1999) Comparison of large restriction fragments of Mycobacterium avium isolates recovered from AIDS and non-AIDS patients with those of isolates from potable water. J Clin Microbiol 37(4):1008–1012. 10.1128/JCM.37.4.1008-1012.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Salah I, Cayrou C, Raoult D, Drancourt M (2009) Mycobacterium marseillense sp. nov., Mycobacterium timonense sp. nov. and Mycobacterium bouchedurhonense sp. nov., members of the Mycobacterium avium complex. Int J Syst Evol Microbiol 59(11):2803–2808. 10.1099/ijs.0.010637-0 [DOI] [PubMed] [Google Scholar]
- Beumer A, King D, Donohue M, Mistry J, Covert T, Pfaller S (2010) Detection of Mycobacterium avium subsp. paratuberculosis in drinking water and biofilms by quantitative PCR. App Environ Microbiol 76(21):7367–70. 10.1128/AEM.00730-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler WR, Crawford JT (1999) Nontuberculous mycobacteria reported to the public health laboratory information system by state public health laboratories United States, 1993–1996. Centers for Disease Control and Prevention, National Center for Infectious Diseases, Georgia, p 1–51 [Google Scholar]
- CDC (2017) Counil of State and Terriorial Epidemiology for reporting of NTM infections and the standardized case definitions. Standardized Case Definition for Extrapulmonary Nontuberculous Mycobacteria Infections. Centers for Disease Control and Prevention, Atlanta, GA, p 1–12 [Google Scholar]
- Chern EC, King D, Haugland R, Pfaller S (2015) Evaluation of quantitative polymerase chain reaction assays targeting Mycobacterium avium M intracellulare and M avium subspecies paratuberculosis in drinking water biofilms. J water health 13(1):131–9. 10.2166/wh.2014.060 [DOI] [PubMed] [Google Scholar]
- Collier SA, Deng L, Adam EA, Benedict KM, Beshearse EM, Black-stock AJ, Bruce BB, Derado G, Edens C, Fullerton KE, Gargano JW, Geissler AL, Hall AJ, Havelaar AH, Hill VR, Hoekstra RM, Reddy SC, Scallan E, Stokes EK, Yoder JS, Beach MJ (2021) Estimate of burden and direct healthcare cost of infectious waterborne disease in the United States. Emerg Infect Dis 27(1):140–149. 10.3201/eid2701.190676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert TC, Rodgers MR, Reyes AL, Stelma GN Jr (1999) Occurrence of nontuberculous mycobacteria in environmental samples. Appl Environ Microbiol 65(6):2492–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groote MA, Pace NR, Fulton K, Falkinham JO 3rd (2006) Relationships between Mycobacterium isolates from patients with pulmonary mycobacterial infection and potting soils. Appl Environ Microbiol 72(12):7602–7606. 10.1128/AEM.00930-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue MJ (2018) Increasing nontuberculous mycobacteria reporting rates and species diversity identified in clinical laboratory reports. BMC infectious diseases [DOI] [PMC free article] [PubMed]
- Donohue MJ (2021) Epidemiological risk factors and the geographical distribution of eight Mycobacterium species. BMC Infect Dis 21(1):258. 10.1186/s12879-021-05925-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue MJ, Mistry JH, Donohue JM, O’Connell K, King D, Byran J, Covert T, Pfaller S (2015) Increased frequency of nontuberculous mycobacteria detection at potable water taps within the United States. Environ Sci Technol 49(10):6127–6133. 10.1021/acs.est.5b00496 [DOI] [PubMed] [Google Scholar]
- Donohue MJ, Vesper S, Mistry J, Donohue JM (2019) Impact of chlorine and chloramine on the detection and quantification of Legionella pneumophila and Mycobacterium species. App environ microbiol 85(24) 10.1128/AEM.01942-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue MJ, Wymer L (2016) Increasing prevalence rate of nontuberculous mycobacteria infections in five states, 2008–2013. Ann Am Thorac Soc 13(12):2143–2150. 10.1513/AnnalsATS.201605-353OC [DOI] [PubMed] [Google Scholar]
- Falkinham JO 3rd (2009) Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J Appl Microbiol 107(2):356–367. 10.1111/j.1365-2672.2009.04161.x [DOI] [PubMed] [Google Scholar]
- Falkinham JO 3rd, Norton CD, LeChevallier MW (2001) Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other Mycobacteria in drinking water distribution systems. Appl Environ Microbiol 67(3):1225–1231. 10.1128/AEM.67.3.1225-1231.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR (2009) Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci U S A 106(38):16393–16399. 10.1073/pnas.0908446106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert MJ, Delgado-Baquerizo M, Oliverio AM, Webster TM, Nichols LM, Honda JR, Chan ED, Adjemian J, Dunn RR, Fierer N (2018) Ecological analyses of mycobacteria in showerhead biofilms and their relevance to human health. mBio 9(5) 10.1128/mBio.01614-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover N, Holtzman A, Aronson T, Froman S, Berlin OGW, Dominguez P, Kunkel KA, Overturf G, Stelma GN Jr, Smith C, Yakrus MA (1994) The isolation and identification of Mycobacterium avium complex (MAC) recovered from Los Angeles potable water, a possible source of infection in AIDS patients. I J Environ Health Res 4:63–72 [Google Scholar]
- Good RC (1980) From the Center for Disease Control. Isolation of nontuberculous mycobacteria in the United States 1979. J Infect Dis 142(5):779–83 [DOI] [PubMed] [Google Scholar]
- Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K, Subcommittee ATSMD, American Thoracic S, Infectious Disease Society of A (2007) An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175(4):367–416. 10.1164/rccm.200604-571ST [DOI] [PubMed] [Google Scholar]
- Hall T (2011) BioEdit: An important software for molecular biology. GERF Bull Biosci 2(1):60–61 [Google Scholar]
- Henkle E, Hedberg K, Schafer S, Novosad S, Winthrop KL (2015) Population-based incidence of pulmonary nontuberculous mycobacterial disease in Oregon 2007 to 2012. Ann Am Thorac Soc 12(5):642–647. 10.1513/AnnalsATS.201412-559OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilborn ED, Covert TC, Yakrus MA, Harris SI, Donnelly SF, Rice EW, Toney S, Bailey SA, Stelma GN Jr (2006) Persistence of nontuberculous mycobacteria in a drinking water system after addition of filtration treatment. Appl Environ Microbiol 72(9):5864–5869. 10.1128/AEM.00759-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall BA, Winthrop KL (2013) Update on the epidemiology of pulmonary nontuberculous mycobacterial infections. Semin Respir Crit Care Med 34(1):87–94. 10.1055/s-0033-1333567 [DOI] [PubMed] [Google Scholar]
- Kim H, Kim SH, Shim TS, Kim MN, Bai GH, Park YG, Lee SH, Chae GT, Cha CY, Kook YH, Kim BJ (2005) Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int J Syst Evol Microbiol 55(Pt 4):1649–1656. 10.1099/ijs.0.63553-0 [DOI] [PubMed] [Google Scholar]
- King DN, Donohue MJ, Vesper SJ, Villegas EN, Ware MW, Vogel ME, Furlong EF, Kolpin DW, Glassmeyer ST, Pfaller S (2016) Microbial pathogens in source and treated waters from drinking water treatment plants in the United States and implications for human health. Sci Total Environ 562:987–995. 10.1016/j.scitotenv.2016.03.214 [DOI] [PubMed] [Google Scholar]
- Lane DJ (1991) Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY [Google Scholar]
- Marras TK, Daley CL (2002) Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin Chest Med 23(3):553–567 [DOI] [PubMed] [Google Scholar]
- ÓBrien RJ, Geiter LJ, Snider DE Jr (1987) The epidemiology of nontuberculous mycobacterial diseases in the United States Results from a national survey. Am Rev Respiratory Dis 135(5):1007–14 [DOI] [PubMed] [Google Scholar]
- Parte AC, Carbasse JS, Meier-Kolthoff JP, Reimer LC, Goker M (2020) List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol 70(11):5607–5612. 10.1099/ijsem.0.004332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller S, Tokarev V, Kessler C, McLimans C, Gomez-Alvarez V, Wright J, King D, Lamendella R (2017) Draft genome sequence of Mycobacterium chimaera type strain Fl-0169. Genome Announc 5(8) 10.1128/genomeA.01620-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevots DR, Marras TK (2015) Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin Chest Med 36(1):13–34. 10.1016/j.ccm.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevots DR, Shaw PA, Strickland D, Jackson LA, Raebel MA, Blosky MA, Montes de Oca R, Shea YR, Seitz AE, Holland SM, Olivier KN (2010) Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med 182(7):970–976. 10.1164/rccm.201002-0310OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasanen NH, Rintala H, Miettinen IT, Torvinen E (2013) Comparison of culture and qPCR methods in detection of mycobacteria from drinking waters. Can J Microbiol 59(4):280–286. 10.1139/cjm-2012-0695 [DOI] [PubMed] [Google Scholar]
- Rindi L, Lari N, Garzelli C (2018) Virulence of Mycobacterium avium subsp hominissuis human isolates in an in vitro macrophage infection model. Int J Mycobacteriol 7(1):48–52. 10.4103/ijmy.ijmy_11_18 [DOI] [PubMed] [Google Scholar]
- Scanu AM, Bull TJ, Cannas S, Sanderson JD, Sechi LA, Dettori G, Zanetti S, Hermon-Taylor J (2007) Mycobacterium avium sub-species paratuberculosis infection in cases of irritable bowel syndrome and comparison with Crohn’s disease and Johne’s disease: common neural and immune pathogenicities. J Clin Microbiol 45(12):3883–3890. 10.1128/JCM.01371-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaulding AB, Lai YL, Zelazny AM, Olivier KN, Kadri SS, Prevots DR, Adjemian J (2017) Geographic distribution of nontuberculous mycobacterial species identified among clinical isolates in the United States, 2009–2013. Ann Am Thorac Soc 14(11):1655–1661. 10.1513/AnnalsATS.201611-860OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strollo SE, Adjemian J, Adjemian MK, Prevots DR (2015) The burden of pulmonary nontuberculous mycobacterial disease in the United States. Ann Am Thorac Soc 12(10):1458–1464. 10.1513/AnnalsATS.201503-173OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10 edn. Sinauer Associates, Cary, NC [Google Scholar]
- Tateishi Y, Ozeki Y, Nishiyama A, Miki M, Maekura R, Fukushima Y, Nakajima C, Suzuki Y, Matsumoto S (2021) Comparative genomic analysis of Mycobacterium intracellulare: implications for clinical taxonomic classification in pulmonary Mycobacterium avium-intracellulare complex disease. BMC Microbiol 21(1):103. 10.1186/s12866-021-02163-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson R, Carter Robyn, Gilpin Chris, Coulter Chris, Hargreaves Megan (2008) Mycobacterium avium Mycobacterium intracellulare. Appl Environ Microbiol 74(10):3094–3098. 10.1128/AEM.02009-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoli E, Rindi L, Garcia MJ, Chiaradonna P, Dei R, Garzelli C, Kroppenstedt RM, Lari N, Mattei R, Mariottini A, Mazzarelli G, Murcia MI, Nanetti A, Piccoli P, Scarparo C (2004) Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int J Syst Evol Microbiol 54(4):1277–1285. 10.1099/ijs.0.02777-0 [DOI] [PubMed] [Google Scholar]
- Tzou CL, Dirac MA, Becker AL, Beck NK, Weigel KM, Meschke JS, Cangelosi GA (2020) Association between Mycobacterium avium complex pulmonary disease and mycobacteria in home water and soil. Ann Am Thorac Soc 17(1):57–62. 10.1513/AnnalsATS.201812-915OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Reyn CF, Waddell RD, Eaton T, Arbeit RD, Maslow JN, Barber TW, Brindle RJ, Gilks CF, Lumio J, Lahdevirta J (1993) Isolation of Mycobacterium avium complex from water in the United States, Finland, Zaire, and Kenya. J Clin Microbiol 31(12):3227–3230. 10.1128/jcm.31.12.3227-3230.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waak MB, LaPara TM, Halle C, Hozalski RM (2019) Nontuberculous mycobacteria in two drinking water distribution systems and the role of residual disinfection. Environ Sci Technol 53(15):8563–8573. 10.1021/acs.est.9b01945 [DOI] [PubMed] [Google Scholar]
- Wallace RJ Jr, Iakhiaeva E, Williams MD, Brown-Elliott BA, Vasireddy S, Vasireddy R, Lande L, Peterson DD, Sawicki J, Kwait R, Tichenor WS, Turenne C, Falkinham JO 3rd (2013) Absence of Mycobacterium intracellulare and presence of Mycobacterium chimaera in household water and biofilm samples of patients in the United States with Mycobacterium avium complex respiratory disease. J Clin Microbiol 51(6):1747–1752. 10.1128/JCM.00186-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Edwards M, Falkinham JO 3rd, Pruden A (2012) Molecular survey of the occurrence of Legionella spp Mycobacterium spp Pseudomonas aeruginosa and amoeba hosts in two chloraminated drinking water distribution systems. App Environ Microbiol 78(17):6285–94. 10.1128/AEM.01492-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2004) Pathogenic mycobacteria in water: a guide to public health consequences, monitoring and management. International Water Association Publishing, London, UK [Google Scholar]
- Winthrop KL, Marras TK, Adjemian J, Zhang H, Wang P, Zhang Q (2020) Incidence and prevalence of nontuberculous mycobacterial lung disease in a large US managed care health plan 2008–2015. Ann Am Thor Soc 17(2):178–185. 10.1513/AnnalsATS.201804-236OC [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data will be publicly accessible at data.gov. search: author’s name.


