Significance
This study reveals that antiandrogen therapy induces viral mimicry responses that are crucial for antitumor activity. H3K9 trimethylation to silence endogenous repeat elements is essential for regaining heterochromatin stability and progression to antiandrogen resistance in prostate cancer. We found that the H3K9 trimethylation machinery is linked to poor outcomes in men with prostate cancer. Blockade of this epigenetic axis can resensitize drug-resistant tumors and elicit cytotoxic interferon responses. Antiandrogen timing and regulation of the H3K9 methylation–endogenous repeat elements– interferon axis should be considered in the development of novel epigenetic therapies and immunotherapeutic strategies for prostate cancer.
Keywords: prostate cancer, hormonal therapy, androgens, enzalutamide, epigenetics
Abstract
Antiandrogen strategies remain the prostate cancer treatment backbone, but drug resistance develops. We show that androgen blockade in prostate cancer leads to derepression of retroelements (REs) followed by a double-stranded RNA (dsRNA)-stimulated interferon response that blocks tumor growth. A forward genetic approach identified H3K9 trimethylation (H3K9me3) as an essential epigenetic adaptation to antiandrogens, which enabled transcriptional silencing of REs that otherwise stimulate interferon signaling and glucocorticoid receptor expression. Elevated expression of terminal H3K9me3 writers was associated with poor patient hormonal therapy outcomes. Forced expression of H3K9me3 writers conferred resistance, whereas inhibiting H3K9-trimethylation writers and readers restored RE expression, blocking antiandrogen resistance. Our work reveals a drug resistance axis that integrates multiple cellular signaling elements and identifies potential pharmacologic vulnerabilities.
Targeting androgen receptor (AR) signaling constitutes the backbone of treatment strategies for advanced prostate cancer (1). Despite the initial response to androgen-deprivation therapy (ADT), achieved by medical or surgical castration, patients eventually progress to castration-resistant prostate cancer (CRPC), frequently through mechanisms reinstating AR activity (2, 3). Next-generation antiandrogens such as the androgen synthesis inhibitor, abiraterone, or the potent AR blocker, enzalutamide (Enz), have significantly improved the overall survival of CRPC patients; however, resistance to these agents eventually occurs and leads to lethality (4–6). Among various mechanisms, elevated levels of glucocorticoid receptor (GR) compensates for the reduced AR-signaling and confers resistance to antiandrogen therapies. In order to bypass androgen blockade, GR is thought to partially take over the transcriptional landscape of AR and regulate gene expression to promote tumor progression (7).
Unbiased genetic and chemical screening approaches have the potential to identify unexpected underlying mechanisms of drug resistance and can enable the discovery of clinically targetable drug-resistance pathways (8, 9). Validation-based insertional mutagenesis (VBIM) is one such genetic screening tool, which, by use of lentiviral vectors (LVs), inserts a cytomegalovirus (CMV) promoter in random genomic locations in a population of cells, generating libraries of millions of cells, each with a unique integration site (10). By isolating cells harboring a mutant phenotype and identifying the VBIM insertion sites, novel genes can be linked with the phenotype of interest. Here, we applied this rigorous screening strategy to identify mechanisms of Enz resistance and found that epigenetic modification of repeat elements (REs) is important for the progression to antiandrogen resistance in CRPC.
Transcriptional silencing of REs in normal somatic cells is achieved through DNA methylation (5-methylcytosine) and repressive histone marks (methylation of H3K9 and H4K20 residues) that are enriched in constitutive heterochromatin (11). Among these, H3K9me1 and H3K9me2 are dynamically regulated by H3K9 dimethyltransferases, euchromatic histone-lysine N-methyltransferase 1 and 2 (EHMT1 and EHMT2). Mono- and dimethylation of the H3K9 also helps in the deposition of the DNA methylation marks at the CpG islands, further aiding the gene silencing (12). H3K9me3 modifications are catalyzed by histone methyltrasferases SUV39H1, SUV39H2, and SETDB1. Aberrant RE regulation leads to their transcriptional activation and formation of dsRNAs that elicit IFN-mediated viral mimicry responses. Tumor cells employ endogenous or acquired strategies to evade treatment-induced viral mimicry states that cause immune responses or undermine their genomic stability and fitness (13). A variety of these repeat elements are differentially expressed in prostate cancer (14, 15); however, there are no previous studies to support the notion of RE perturbation in response to anti-AR treatment in prostate cancer cells. Our studies suggest that increased H3K9me3 synthesis inhibits the detrimental activation of REs that is central for the growth inhibitory effects of Enz in CRPC. In summary, combining epigenetic and antiandrogen therapies, we present a promising strategy for overcoming drug resistance and immune evasion in prostate cancer.
Results
Forward Genetics Approach Identifies Increased H3K9 Methylation as a Driver of Enz Resistance.
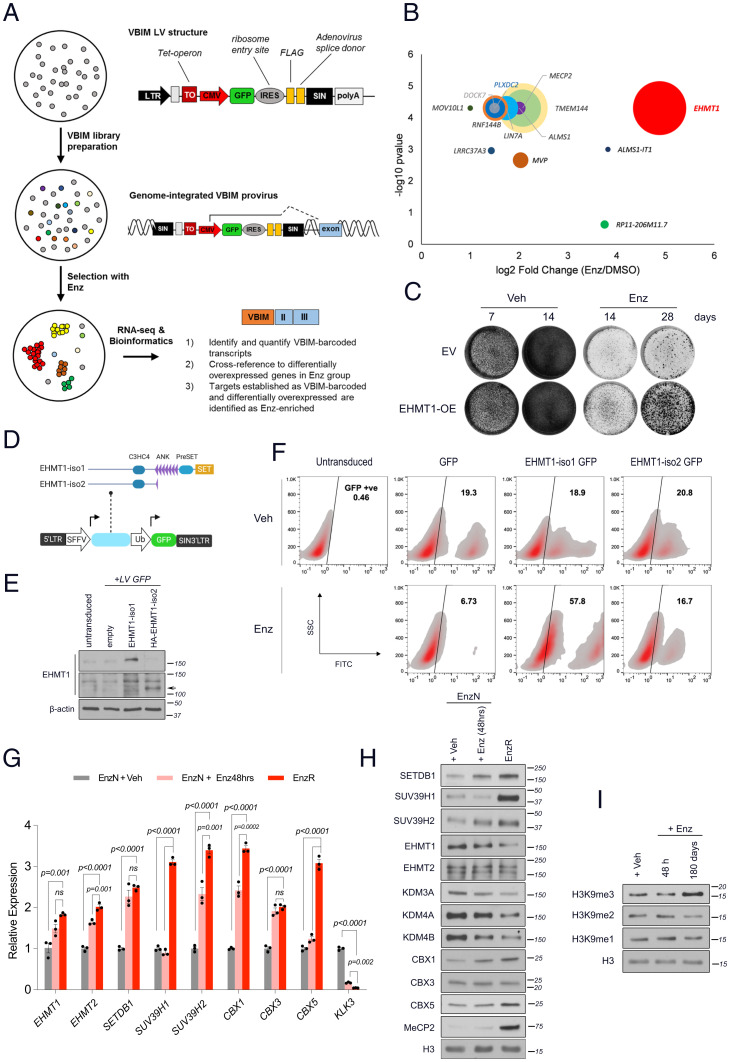
To search for novel drivers of resistance to antiandrogens, we performed a positive selection screen using VBIM LVs (Fig. 1A). The VBIM strategy readily generates libraries of millions of cells, with each infected cell having one or more unique integration sites, potentially leading to overexpression of full or truncated gene transcripts or of antisense RNAs (10). We generated a library of 6 × 106 human prostate LNCaP-VBIM cells, which were then expanded and treated continuously for over 60 d with vehicle or Enz (10 µM), in the presence of dehydroepiandrosterone (DHEA [10 nM]) to mimic human adrenal physiology. This drug selection identified the histone methyltransferase EHMT1 as the top Enz-enriched gene, with a 26-fold increase over vehicle-treated cells (Fig. 1B, Dataset S1). The in-frame insertion site of the VBIM promoter mapped to exon 2 of the EHMT1 gene, excluding only the first six N-terminal amino acids (SI Appendix, Fig. S1 A–C). To validate this finding, we generated stable cell lines that were infected with empty- or EHMT1-overexpressing (OE) lentiviral vectors and treated them with Enz or vehicle for 28 d. Forced EHMT1 expression in the absence of Enz caused no noticeable increase in cell growth as compared with control cells, but it conferred substantial resistance to Enz treatment (Fig. 1C), therefore demonstrating a specific advantage only under conditions of drug treatment.
Fig. 1.
Forward genetics selection with VBIM identifies increased H3K9 methylation as a possible mechanism of Enz resistance. (A) Schematic representation of the validation-based insertional mutagenesis (VBIM)-seq workflow. (B) Bubble plot representing the top VBIM-enriched genes in response to Enz (10 µM) treatment of VBIM-infected LNCaP cells for 72 d. Circle size proportionately correlates with the FPKM value of the associated gene in the vehicle-treated samples. (C) Proliferation assay of EV or full-length EHMT1-overexpressing LNCaP cells treated with vehicle (DMSO) or Enz (10 µM) for the indicated times. Results shown are representative of three biological repeats. (D) Schematic representation of EHMT1 isoform 1 (iso1: full length) and isoform 2 (iso2: lacking the catalytic SET domain); the corresponding genes were cloned into a double cassette lentiviral vector coexpressing GFP under a separate ubiquitin promoter (Ub). (E) Immunoblots showing the expression of the indicated proteins in lentivirally transduced LNCaP cell populations that harbor ∼20% cells expressing GFP, EHMT1 iso1, or HA-EHMT1 iso2. Arrow indicates the position of HA-EHMT1-iso2. (F) Representative contour plots of a flow cytometry–based competition assay showing the distribution of EHMT1 iso1-GFP or iso2-GFP expressing cells versus the GFP-expressing control cells. Cells were treated with DMSO or Enz (5 µM) for 80 d before the flow analysis. The percentage of GFP-positive (GFP +ve) cells is shown for each group. (G) RT-qPCR and (H) immunoblot analyses comparing the relative expression of H3K9 methyltransferases and readers in Enz-naïve and -resistant LNCaP cells that were treated with either Enz (10 µM) or DMSO (Veh) for the indicated times. Transcript levels in G were normalized first to RPLP0 and then to vehicle-treated cells for all comparisons. Significance was calculated using two-way ANOVA; error bars represent SD. Results are representative of three biological repeats, performed in triplicate. ns, not significant. (I) Immunoblots comparing the indicated histones in histone extracts purified from LNCaP cells treated with Enz (5 µM) or DMSO (Veh), as indicated, for 180 d. SIN, self-inactivating; SFFV, spleen focus-forming virus; SET, Su(var)3-9, enhancer-of-zeste and trithorax; ANK, ankyrin repeat; C3HC4, C3HC4 type zinc finger.
We next asked whether the catalytic activity of EHMT1 was required for its induction of resistance. To this end, we performed a flow cytometry-based competition assay using a double-cassette LV vector, coexpressing GFP with isoform 1 (full length) or isoform 2 (lacking the catalytic domain) of EHMT1 (Fig. 1 D and E and SI Appendix, Fig. S1D). Although iso2 was depleted of activity, it surprisingly retained residual activity, possibly because of inducing multimeric H3K9MT complex (16). Cell populations containing ∼20% GFP-positive cells were generated and treated with Enz or vehicle. Consistent with the VBIM analysis, at day 80, treatment with Enz led to a threefold increase in cells overexpressing full-length EHMT1 (57.8%) as compared with vehicle-treated cells (18.9%). Neither the percentage of GFP-only control cells nor that of the overexpressing noncatalytic EHMT1 cells increased with Enz treatment over baseline levels (Fig. 1F). Notably, our VBIM search also pulled out the methyl CpG-binding protein 2 (MeCP2) as a highly Enz-enriched hit (Fig. 1B), which was further validated by immunoblotting (Fig. 1H). MeCP2 has been shown to link H3K9 methylation to DNA methylation and, together with H3K9me3, is concentrated in heterochromatin regions (17, 18). Together, these data suggested that the enzymatic H3K9 methyltransferase activity of EHMT1 is critical for rendering cancer cells resistant to Enz.
EnzR Cells Up-Regulate H3K9 Trimethyl Transferases and Readers.
Speculating a role for H3K9 methylation in mediating drug resistance, we next evaluated the changes in expression levels of the specific H3K9 methyltransferases, demethylases, and readers in Enz-naïve (EnzN) versus short-term (48 h) Enz-treated and Enz-resistant (EnzR) cells. EnzR cells were established by continuously treating cells with 10 µM Enz for at least 6 mo. First, RT-qPCR analysis showed a significant increase in mRNA levels of all H3K9MTs and methyl readers in chronic Enz-treated relative to EnzN cells (Fig. 1G). We then evaluated the protein levels of H3K9me regulators in chromatin-extracted fractions by immunoblotting. Compared with control cells, EnzR cells had substantial increases in the levels of H3K9 di- and trimethyltransferases (SETDB1, SUV39H1, and SUV39H2) and H3K9-methyl readers (CBX1 and CBX5), accompanied by down-regulation of H3K9 demethylases (Fig. 1H). Despite a modest induction of mRNA levels for EHMT1 and EHMT2 upon Enz treatment, we did not detect any increase in protein levels within chromatin extracts. We speculate that although EHMT1 protein may not be up-regulated in the natural course of Enz treatment, its enforced expression in a VBIM assay catalyzes the pioneering methylation reactions that provide substrate for trimethyl writers or facilitates the assembly and activation of “H3K9MT megacomplexes” (16). A subset of the EHMT1, EHMT2, SETDB1, and SUV39H1 methyltransferases form H3K9MT multimeric complexes that demonstrate functional interdependency between subunits and together are recruited to heterochromatic regions (16). Indeed, as detected by immunoblotting, we observed a global increase in H3K9me3, the hallmark of heterochromatin, following chronic but not short-term exposure to Enz (Fig. 1I). This observation was verified by mass spectrometry for histone posttranslational modifications in LNCaP cells, which also revealed Enz-induced accumulation of H4K20me3, another histone mark enriched in heterochromatic regions (SI Appendix, Fig. S2 A and B). Elevation in H3K9me3 levels following continued Enz treatment was similarly observed by immunoblotting in two other human prostate cancer cell lines, LAPC4 and VCaP (SI Appendix, Fig. S2 C and D). Chromatin immunoprecipitation (ChIP)-PCR further confirmed the accumulation of H3K9me3 on the genomic loci of retroelements in the heterochromatin region of EnzR prostate cancer cell lines (SI Appendix, Fig. S2 E and F). Consistent with histone tail modifications, cells chronically treated with Enz demonstrate a significant enrichment for heterochromatin-associated gene signatures (SI Appendix, Fig. S2G). Together, these results reveal that persistent AR blockade dictates concerted changes in the H3K9 methylation machinery, leading to increased H3K9me3 synthesis and heterochromatin assembly.
The H3K9 Methylation Pathway Mediates Progression to Enz Resistance.
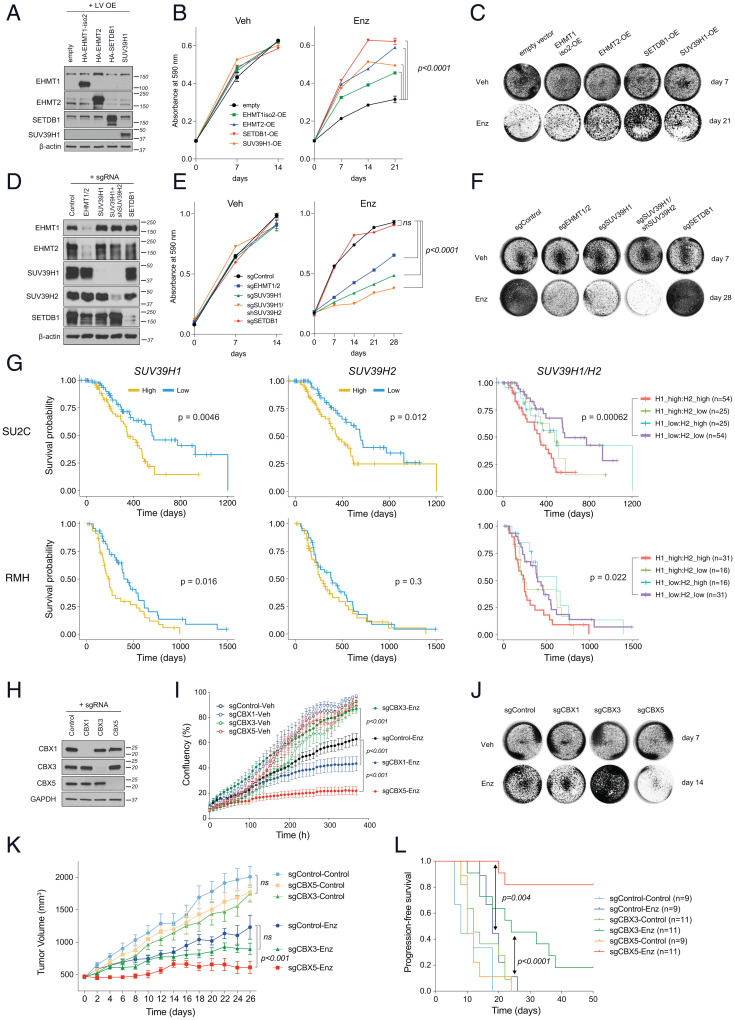
To directly interrogate the requirement for H3K9MTs and methyl readers in the development of the Enz-resistant phenotype, we generated LNCaP cell lines stably overexpressing or lacking these components and examined their function following extended Enz treatment. Compared with control cells (empty vector [EV]), forced expression of each of the EHMT1, EHMT2, SUV39H1, and SETDB1 enzymes individually (Figs. 1C and 2A and SI Appendix, Fig. S3A) was sufficient to confer Enz resistance, as determined by colony formation assays; no noticeable differences in growth were detected when cells were treated with vehicle (Fig. 2 B and C). In contrast, CRISPR- or shRNA-mediated depletion of EHMT 1 and 2 together (sgEHMT1/2), SUV39H1 alone, or in combination with SUV39H2 (sgSUV39H1/shSUV39H2) substantially sensitized cells to Enz (Fig. 2 D–F and SI Appendix, Fig. S3B). The added Enz sensitivity for double SUV39H1/2 knockdown as compared with SUV39H1-only knockdown cells, suggests that some functional redundancy between these enzymes may exist in their functions as terminal enzymes required for H3K9 trimethylation (Fig. 2F and SI Appendix, Fig. S3B). Consistent with gene-silencing outcomes, treatment with very low concentrations of the SUV39H1 inhibitor, chaetocin (2.5 to 10 nM) ablated the growth of Enz-resistant cells, when combined with Enz and in a dose-dependent manner (SI Appendix, Fig. S4 A and B). Although overexpression of SETDB1 robustly established the EnzR phenotype (Fig. 2C), its inactivation did not sensitize LNCaP cells to Enz (Fig. 2F). This is consistent with the absence of H3K9me2/3 changes in the SETDB1 knockout (KO) cells (SI Appendix, Fig. S3).
Fig. 2.
The H3K9 methylation pathway is essential for progression to antiandrogen resistance. (A–C) Immunoblots showing the expression of ectopically expressed H3K9 methyltransferases (A), proliferation analysis (B), and crystal violet-stained representative images (C) of LNCaP cells that were lentivirally transduced to overexpress the H3K9 methyltransferases after treatment with Enz (10 µM) or DMSO (Veh). Cells in the Enz group were pretreated with Enz for 21 d before seeding. (D–F) Immunoblots (D), cell growth analysis (E), and representative crystal violet stained images (F) of LNCaP cells that were lentivirally transduced to express guide RNAs targeting the indicated H3K9 methyltransferase or a nontargeting control gRNA. (E and F) Enz group cells were pretreated with Enz (10 µM) for 28 d before seeding at 105 and 2 × 105/well for Veh and Enz treatments, respectively. (G) Kaplan–Meier plots of estimated survival time based on the transcript levels of SUV39H1 and SUV39H2 in mCRPC tumor biopsies from SU2C (n = 158) and RMH (n = 94) cohorts. P values were calculated from a Cox proportional hazards model to determine differences in outcome between patients with high (above median = orange) and low (below median = blue) expression. On the Right, patients were placed in one of four groups based on high or low expression of SUV39H1 and SUV39H2 combined. (H–J) Immunoblots showing CBX knockout efficacy by CRISPR (H), cell growth as monitored by IncuCyte live cell imaging (I), and representative crystal violet stained images (J) of LNCaP cells that were lentivirally transduced to express sgRNAs, targeting the indicated CBX proteins or a nontargeting control sgRNA, as indicated in H. Results represent mean ± SEM, n = 3; significance was determined using an unpaired t test. Enz cells in H were pretreated with Enz (10 µM) for 90 d before initiation of the experiment. Cells were seeded at 105 and 2 × 105/well for Veh and Enz treatments, respectively. (K) Tumor growth and (L) progression-free survival analyses of subcutaneous xenografts established from LNCaP cells that were lentivirally transduced to express sgRNAs targeting CBX3, CBX5, or a nontargeting (control) sgRNA, as indicated, and engrafted into surgically orchiectomized NSG mice supplemented with DHEA pellets to mimic human physiology and treated with Enz or control chow.
To clinically interrogate whether activity of the terminal enzymes SUV39H1/SUV39H2 is associated with Enz response, we investigated CRPC clinical outcomes in two independent clinical cohorts from Stand Up To Cancer (SU2C) and the Royal Marsden Hospital (RMH). High SUV39H1 expression was associated with significantly worse survival in both cohorts (Fig. 2G), whereas the expression of SUV39H2 was significantly associated with poor outcomes in one of the cohorts. The combined expression analysis of these H3K9me3 enzymes supports the functional requirement for at least one, and perhaps both, for resistance. Further evaluation of these cohorts provides no evidence supporting a role for EHMT1/2 specifically in patient outcomes (SI Appendix, Fig. S5). The identification of EHMT1 in our forward genetics experiment therefore is probably indicative of the role of downstream H3K9MT enzymes in mediating resistance.
H3K9MTs also catalyze lysine methylation on many nonhistone substrates (19). To corroborate that the emergence of Enz resistance relied upon their H3K9-methylation activities, we generated cells stably expressing H3 harboring a lysine 9-to-methionine mutation (H3K9M). This dominant-negative mutant sequesters H3K9MTs at heterochromatin nucleation sites, impedes their H3K9 methylation activity on wild-type H3 (H3WT), and thus blocks heterochromatin expansion and assembly (20, 21). Immunoblot analysis of whole cell extracts from LNCaP and VCaP cells with the H3K9M transgene showed that H3K9M expression led to markedly lower levels of H3K9me2 and H3K9me3, without detectable changes in H3K9me1 or H3K9ac (SI Appendix, Fig. S4 C and F). In both LNCaP (SI Appendix, Fig. S4 D and E) and VCaP cells (SI Appendix, Fig. S4G), introducing the H3K9M transgene enhanced Enz-mediated growth suppression, as compared with H3WT.
Trimethylated H3K9 marks exert their effects by recruiting their specific chromobox (CBX) readers (22, 23). We therefore extended our functional studies to analysis of H3K9me readers, CBX1, CBX3, and CBX5, which are found in heterochromatic structures. To this end, we generated LNCaP cells that were transduced with lentiviruses to inactivate these readers individually (or transduced with a nontargeting control) using CRISPR-Cas9 (Fig. 2H). We performed cell growth analyses on both Enz-naïve and Enz-pretreated (for 90 d) cells to determine their short or adaptive response to Enz. In both conditions, CBX5 depletion strongly blocked the proliferation of tumor cells treated with Enz (Fig. 2I and SI Appendix, Fig. S4H), but not vehicle, whereas the loss of CBX1 had milder effects that only manifested after longer exposure to Enz (>90 d) (Fig. 2 I and J). Although CBX3 deletion did not confer any additional vulnerability to Enz compared with control cells for up to 8 wk, to our surprise, longer treatment gave rise to an enhanced resistant phenotype of hyperproliferative tumor cells in vitro (Fig. 2 I and J). Recent reports have documented that each H3K9me3 reader has a distinct role in heterochromatin organization and that depletion of each can result in redistribution of the other effector proteins (24). In our models, deletion of CBX3 may have reorganized heterochromatin structure in favor of CBX5-regulated pericentric heterochromatin assembly, accelerating drug resistance in response to chronic exposure to Enz. Furthermore, whereas CBX5 primarily locates to pericentric heterochromatin, CBX3 is also found in euchromatic regions and participates in gene silencing (25). Thus, like loss of SETDB1, which also targets gene coding regions, CBX3 silencing may additionally activate expression of a gene or a subset of genes favoring a drug-resistant phenotype.
To further investigate the in vitro results that showed opposing roles for CBX5 and CBX3 in progression to Enz resistance, we assessed their behavior in xenograft tumors. In vivo, CBX5 depletion robustly sensitized LNCaP xenografts to Enz with near-complete blockade of tumor growth (Fig. 2K). We did not detect a significant difference in tumor size between control and CBX3 KO cells up to 28 d of Enz treatment; however, extended monitoring revealed that progression-free survival significantly increased in CBX3-deficient xenografts (Fig. 2L). It seems plausible that the drug-resistant state seen with CBX3 KO cells in vitro is a late-stage phenomenon requiring longer treatment periods. Nevertheless, in vivo, the absence of CBX3 appears to enhance sensitivity to Enz (Fig. 2L). Collectively, these results suggest that AR blockade induces an accumulation of H3K9me3 in regions that are likely to be recognized by multiple CBX family readers, and distinct H3K9me readers play specific context-dependent roles under Enz treatment.
H3K9me3 Modifications in EnzR Cells Accumulate over Repeat Elements.
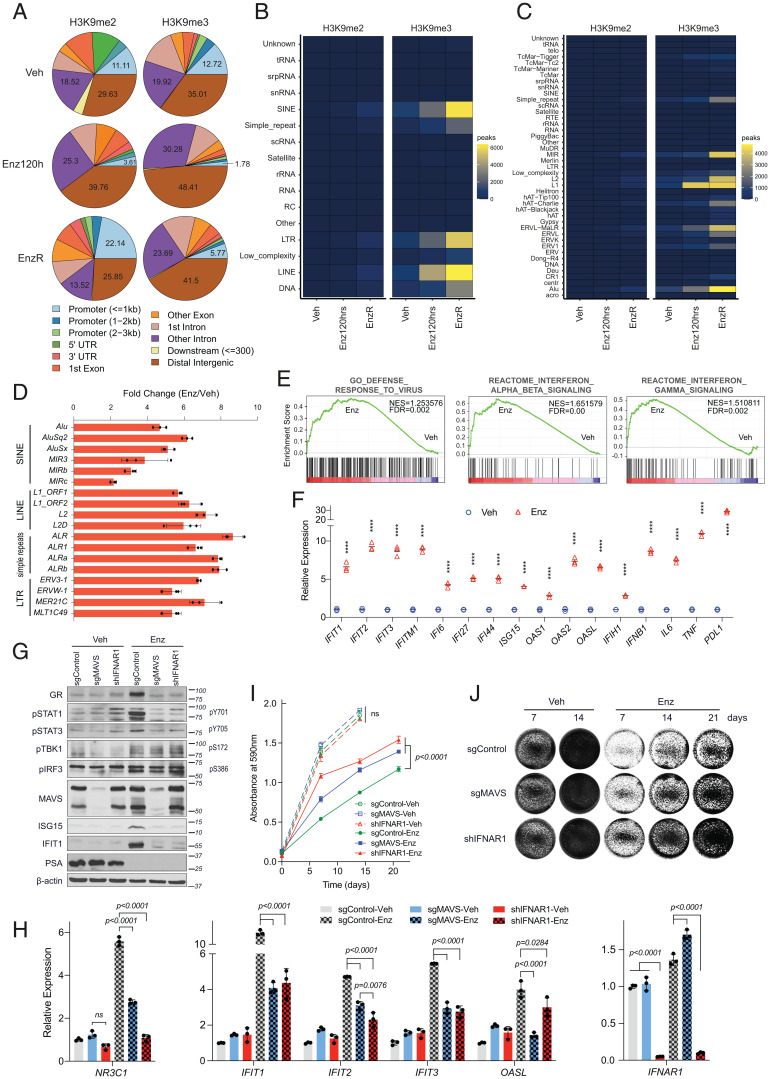
Following the phenotypic interrogations, we sought to determine the chromatin localization of H3K9me2 and H3K9me3 in parental and EnzR cells by chromatin immunoprecipitation followed by sequencing (ChIP-seq). Cells treated with Enz for 120 h were also included to control for any epigenetic alterations that might be induced by short-term treatment. The total number of peaks identified in EnzR chromatin increased considerably relative to parental cells for both H3K9me2 (21,479 versus 3,023) and H3K9me3 (45,996 versus 32,184). The identified peaks were filtered to determine stringent signatures of peaks associated with different treatment conditions by applying a minimum threshold of a 1.5-fold change in the intensity count. Condition-specific peaks were annotated against genomic features. A 2-fold elevation in the number of H3K9me2 peaks that mapped to promoter regions specifically occurred in EnzR compared with parental cells (Fig. 3A). In the case of H3K9me3, however, both short- and long-term Enz-treated cells showed an increase in peak counts primarily mapping to intronic and intergenic regions (Fig. 3A). Given the functional requirement for H3K9me3-mediated heterochromatin formation in EnzR survival and a wealth of reports demonstrating that H3K9me3 is central to silencing of REs (26), we sought to annotate each set of peaks against repeat features of the human genome. This uncovered a robust sequential increase in the number of 120-h Enz-treated and EnzR-specific H3K9me3 peaks that mapped to short and long interspersed nuclear elements (SINEs and LINEs, respectively), long terminal repeats (LTRs), and simple repeat classes of REs (Fig. 3 B and C). More specifically, Alu and MIR subfamilies from the SINE and L2 sequences from the LINE group were the top REs identified within H3K9me3-occupied regions of EnzR cells (Fig. 3C). This result led us to hypothesize that increased H3K9 methylation in EnzR cells may be a compensatory mechanism to overcome any long-term deleterious effects associated with hyperactivation of REs immediately following AR blockade. Supporting this notion, analysis of the RE transcript levels showed a clear induction of various RE subtypes upon Enz treatment (Fig. 3D), which was accompanied by innate immune responses including expression of interferon (IFN) and IFN-stimulated genes (ISGs) (Fig. 3 E and F and SI Appendix, Figs. S6 A and B, S7 A and B, and S8 A and B). Pathway analyses on a cohort of matched pre- and post-ADT biopsies (27) further corroborated that activation of the JAK/STAT and IL6 inflammatory responses ensues following AR-targeted therapies (SI Appendix, Fig. S8C).
Fig. 3.
Enz treatment leads to viral mimicry responses induced by expression of repetitive elements. (A–C) Analysis of H3K9me2 and H3K9me3 ChIP-seq experiments performed on Enz-naïve LNCaP cells treated with DMSO (Veh) or Enz for 120 h and on EnzR cells. Peaks specific to each treatment condition were separated using a 1.5-fold change threshold as compared to both the other conditions. Pie charts showing the genomic annotations of differentially regulated peak sets (A). Annotations were generated using the ChIPSeeker R package. Heatmaps showing the number of peaks mapping to different repeat classes (B) and families (C) in each condition. (D) RT-qPCR analyses of the indicated retroelement transcripts in LNCaP cells treated with DMSO or Enz (10 µM) for 14 d. All transcript levels were normalized to RPLP0 levels. Results are representative of three biological repeats, performed in triplicates. (E) Gene set enrichment analyses showing the significant induction of antiviral and IFN responses in Enz (5 µM) versus DMSO treated cells for 85 d. (F) RT-qPCR results showing the relative expression of the indicated interferon-stimulated genes in cells treated with Enz or vehicle as in D. ****P < 0.0001, calculated by one-way ANOVA. (G and H) Immunoblots (G) and RT-qPCR analysis (H) of LNCaP cells that were lentivirally transduced with sgRNA targeting MAVS, shRNA targeting IFNAR1, or a nontargeting control sgRNA. Cells were cultured with DMSO (Veh) or Enz (5 µM) for 40 d in G or 21 d in H. Results represent mean ± SD, n = 3; significance was calculated using one-way ANOVA. ns, not significant. (I) Cell growth analysis of sgMAVS and shIFNAR1-transduced LNCaP cells treated with Veh or Enz (5 µM), determined by a colorimetric crystal violet assay. Treatment endpoints were compared using an unpaired t test. (J) Representative images of each group at the indicated time points. Cells in the Enz group were pretreated with Enz for 28 d before seeding.
Enz-Activated Endogenous dsRNAs Trigger IFN Stimulation and Cell Death.
Transcripts originated from REs are processed to self dsRNA, a common pathogen-associated molecular pattern, which upon binding to MDA5 or RIG-I activates MAVS, leading to the phosphorylation of STATs and the activation of type-I IFN signaling (28–30). Indeed, long-term Enz treatment induced dsRNA formation (SI Appendix, Fig. S9 A and B) followed by phosphorylation of STAT1 and STAT3, paired with expression of ISGs. These IFN responses were completely blocked by knockdown of MAVS or IFNAR1 (Fig. 3 G and H). Importantly, disruption of dsRNA sensing also blunted the sensitivity of cells to Enz in MAVS knockdown and, more markedly, in IFNAR1 knockdown cells, indicating that viral mimicry responses play a major role in Enz-mediated cytotoxicity (Fig. 3 I and J). Inhibition of ADAR1, which catalyzes adenosine-to-inosine editing of dsRNA resulting in destabilization (31), augments Enz-induced dsRNA accumulation and blocks resistance (SI Appendix, Fig. S10 A–D).
Enz-Induced GR Up-Regulation Is Triggered by dsRNA-Mediated IFN Stimulation.
Following AR blockade in prostate cancer, the expression of GR is up-regulated alongside a metabolic mechanism that permits GR stimulation (7, 32, 33). The GR activation axis has been implicated as a resistance mechanism in patients with CRPC (7, 34). Remarkably, we found that blocking the IFN cascade also potently abrogated the up-regulation of both GR protein (Fig. 3G) and transcript, NR3C1 (Fig. 3H). To further examine whether dsRNA-mediated IFN activation could be a source of GR up-regulation, we transfected cells with increasing concentrations of synthetic dsRNA (poly I:C) or dsDNA (poly dA:dT), commonly used as viral mimetics, and measured their ISG and GR levels. In LNCaP cells, poly I:C, but not poly dA:dT, robustly and in a dose-dependent manner stimulated the transcription of canonical ISGs, which mirrored GR expression, both at the protein (SI Appendix, Fig. S11A) and mRNA levels (SI Appendix, Fig. S11B). Our findings are further supported by analysis of transcriptomic data from the SU2C cohort of patients with metastatic CRPC, in which GR levels positively correlate with a panel of ISGs and inversely correlate with H3K9MTs (SI Appendix, Fig. S12 A and B). The latter observation is consistent with our hypothesis that GR expression is associated with heterochromatin status of CRPC tumors. Taken together, these data lead us to propose that GR up-regulation is a downstream event of Enz-mediated IFN signaling and may serve as a marker of treatment-induced RE activation and stress.
H3K9me3 Regulates Enz-Induced REs in Resistance.
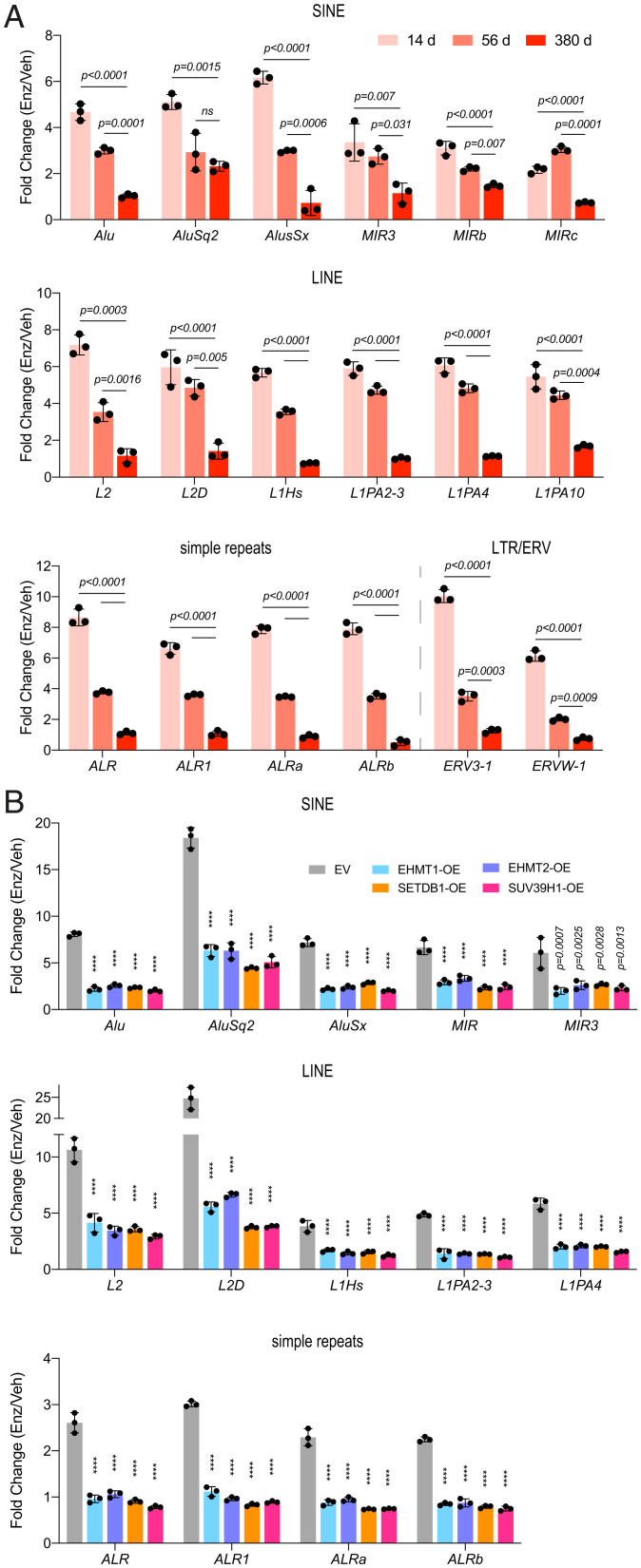
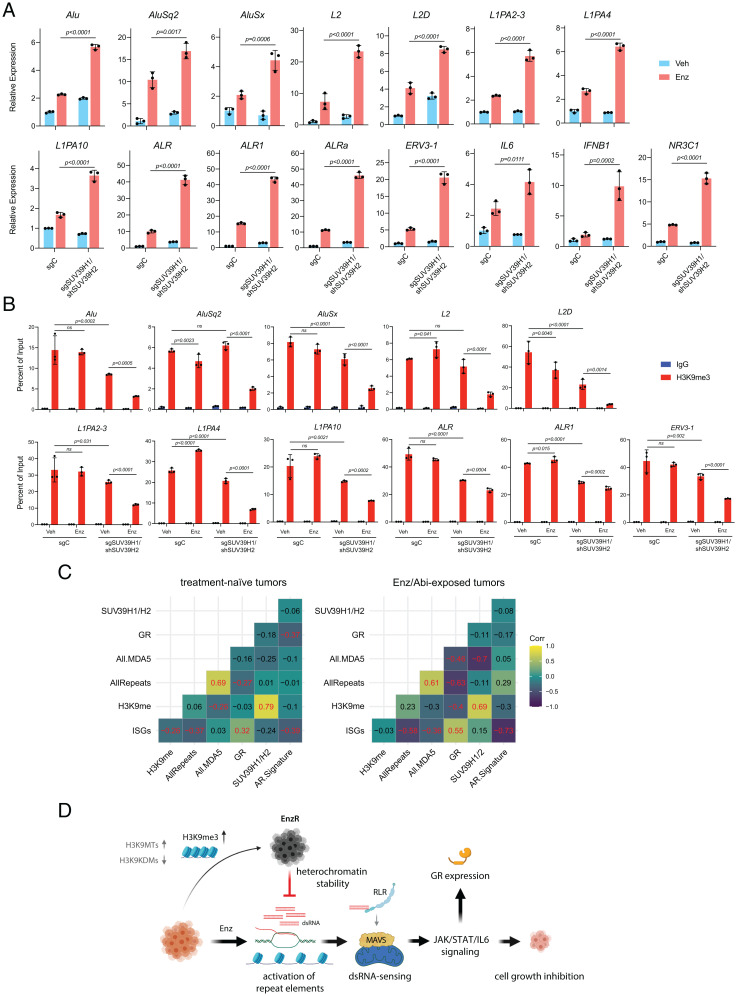
Our data suggest a model in which epigenetic mechanisms, including H3K9 hypermethylation, that facilitate the silencing of REs are required for progression to antiandrogen resistance. We therefore first asked whether the observed increase in H3K9me3 peaks across RE subtypes in EnzR cells corresponded to a decrease in their expression. A time-course qPCR analysis of RE transcripts showed a robust induction at day 14 post-Enz treatment that progressively diminished to the point that EnzR cells (treated for 380 d) displayed levels of RE expression similar to control cells (Fig. 4A). Next, we interrogated the role of H3K9MTs in regulating treatment-induced repeat RNAs. Consistently, individual overexpression of EHMT1, EHMT2, SUV39H1, or SETDB1 was sufficient to phenocopy EnzR cells and significantly repressed Enz-activated RE subtypes that belonged to SINE, LINE, or simple repeat families (Fig. 4B). In contrast, combined depletion of SUV39H1 and SUV39H2, which produced the greatest susceptibility to Enz (Fig. 2 E and F), resulted in substantial treatment-induced RE expression relative to control cells (Fig. 5A). ChIP experiments further confirmed that SUV39H1/H2 KD cells could not compensate for Enz-induced depletion of H3K9me3 modifications across REs (Fig. 5B), which manifested in their excessive derepression upon long-term treatment. Conversely, we did not detect a considerable loss of H3K9me3 enriched across REs in intact cells treated with Enz (Fig. 5B). Additionally, genetic or pharmacological inhibition of H3K9MTs also led to derepression of ISGs and REs in multiple EnzR cell lines (SI Appendix, Figs. S13 A and B and S14 A and B). Supporting our in vitro findings, analysis of REs in metastatic CRPC tumors revealed a highly significant negative correlation (r = −0.697, P = 1.3 × 10−5) between levels of SUV39H1/H2 and MDA5-binding repeat RNAs in patients exposed to the antiandrogens Enz or abiraterone (Fig. 5C). The subset of RE transcripts that bind to and stimulate MDA5 constitute the immunogenic REs (35).
Fig. 4.
H3K9MTs repress Enz-induced activation of retroelements. (A) RT-qPCR analyses showing the induction of the indicated transcripts from SINE, LINE, simple repeat, and ERV families in LNCaP cells treated with Enz (10 µM) for 14, 56, or 380 d, as compared with the respective Veh-treated controls. Endpoints were compared using a one-way ANOVA. (B) Enz/Veh fold-change induction levels of the indicated repeat element transcripts in LNCaP cells stably overexpressing the EV or H3K9-methyltransferases (OE), following treatment with Enz (5 µM) or vehicle for 40 d. P values were calculated using two-way ANOVA; error bars represent SD. Results are a representative of two biological repeats performed in triplicate. ****P < 0.001.
Fig. 5.
SUV39H1 and SUV39H2 mediate the repression of antiandrogen-induced retroelements. (A) RT-qPCR analysis showing the transcript levels of the indicated RE subtypes, IL6, IFNB1, and NR3C1 and (B) ChIP-qPCR analysis of H3K9me3 levels at the indicated REs in control (sgC) versus double SUV39H1/SUV39H2 knockdown (KD) (sgSUV39H1/shSUV39H2) LNCaP cells treated with Enz (10 µM) or DMSO (Veh) for 31 d. Error bars represent SD and significance was calculated using two-way ANOVA followed by Sidak’s multiple comparisons test. Results are representative of three biological repeats performed in triplicate. (C) Correlograms show Pearson's correlations, pairwise, between ssGSEA scores for various gene signatures for Enz- or abiaterone-exposed (n = 31) and naïve (n = 61) mCRPC transcriptional profiles derived from polyA RNA-seq data. Correlations significant at P < 0.05 are highlighted in red font in the correlograms. “H3K9me” signature was derived from the expression levels of H3K9me writers and readers. (D) Proposed model. Enz treatment induces the derepression of the repeat elements. This is followed by processing of their transcripts into dsRNAs that can bind to and activate RIG-I–like receptors (RLR), leading to MAVS activation. MAVS activation triggers IFN-mediated viral mimicry responses, resulting in growth inhibition and cytotoxicity in CRPC cells. GR up-regulation is a marker of the Enz-induced inflammatory signaling pathway. EnzR cells epigenetically adapt to evade viral mimicry through increased deposition of repressive marks such as H3K9-trimethylation on specific REs and inhibiting their activation.
Like RE levels, GR expression was consistently and strongly inversely correlated with the status of H3K9MT (SI Appendix, Fig. S15 A–D). Overexpression of all H3K9MTs effectively blocked GR induction (SI Appendix, Fig. S15 A and B), whereas their knockdown, most robustly with loss of SUV39H1, augmented Enz-induced GR activity (SI Appendix, Fig. S15 C and D). Together, these data suggest that H3K9me3-mediated heterochromatin formation over REs is a major driver of Enz resistance.
Discussion
In summary, the work presented here suggests that antiandrogen-resistant prostate cancer employs a developmental mechanism that promotes repression of REs to preserve heterochromatin homeostasis. REs are transcriptionally silenced through collective function of DNA and histone methyl marks such as H3K9me3, H4K20me3, and H3K27me3, which generate a compact chromatin over repeat regions (36). The compensatory potential of these processes is observed in normal development as well as in malignancy. For instance, H3K9me3-mediated heterochromatinization of repeat sequences maintains genomic integrity during developmental stages where those regions become temporarily hypomethylated (37, 38). Similarly, across multiple cancer types, there is increasing evidence for H3K9me3- or H3K27me3-mediated repression of treatment-induced REs as a major chemoresistance mechanism (39, 40). Such repressive histone modifications are likely to preserve heterochromatin stability in response to treatment-induced DNA and/or histone hypomethylation (41).
In the context of AR-antagonist therapy, we found that H3K9 methyltransferases and readers are crucial for drug resistance. A limitation of our work is that the mechanistic studies were mainly performed in a single cell line model. Nevertheless, H3K9me3 accumulation occurs with Enz resistance across three cell line models and our data align with a study using patient-derived xenografts that suggests CBX5-dependent heterochromatin formation mediates transdifferentiation of prostate adenocarcinomas to treatment-resistant neuroendocrine tumors (42). This correlation raises the possibility that RE suppression by H3K9 methylation remains essential even when prostate cancer loses AR expression.
Much work has focused on the role of the H3K27 trimethyltransferase EZH2 in driving treatment resistance and neuroendocrine differentiation (43). This raises the question of whether those effects are partially achieved through H3K27me3-mediated gain of heterochromatin over repeat regions. Support for this proposition comes from a recent study in which EZH2 inhibition sensitizes tumor cells to checkpoint inhibitor therapies through activation of dsRNA-induced ISGs in mouse and human-derived prostate cancer organoids (44). Interestingly, EZH2 inhibition within the context of Enz treatment is also shown to substantially increase GR expression (45). These studies, together, align with our model in which GR up-regulation is triggered by activation of REs that stems from the loss of repressive heterochromatin marks.
We also show that GR up-regulation is downstream of the IFN signaling that is induced by AR blockade. GR has been demonstrated to inhibit type I IFN responses (46). Therefore, it is plausible that GR up-regulation is required to restrain the cytotoxicity of IFN or to transform acute unfavorable IFN signaling to chronic prosurvival signaling or the IFN-related DNA damage resistance signature, which mediates resistance to treatment across different cancer types (47, 48).
An emerging body of evidence has revealed the epigenetic repression of REs as an important mechanism of treatment resistance across various cancer types and therapeutic modalities (28, 29, 40, 49). RE expression in cancer cells is highly immunogenic, and cancers that exhibit hyperactivation of these elements often need to coevolve immune-suppressive mechanisms for survival. These mechanisms, in turn, render the cancers vulnerable to therapeutic immune checkpoint blockade. In contrast to REs, GR activation is routinely associated with immunosuppression. Interestingly, for reasons that have been thus far unclear, CRPCs are not highly responsive to these immunotherapies, indicating an innately nonimmunogenic state. Perhaps, the prominent repression of immunogenic REs and the simultaneous activation of immunosuppressive GR in cells that have been exposed to extended Enz treatment, underlies these immunotherapy-resistant properties in CRPC. Thus, combination therapies employing immune-checkpoint inhibitors either with GR antagonists or inhibitors of H3K9MTs could be of clinical relevance.
Methods
Cell Lines.
LNCaP, VCaP, and 293T cells were purchased from the American Type Culture Collection and cultured in RMPI-1640 (LNCaP) or Dulbecco's Modified Eagle Medium (DMEM) (VCaP and 293T), with 10% fetal bovine serum (FBS), 2 mM L-glutamine and 1% penicillin–streptomycin (PenStrep). LAPC4 cells were generously provided by Charles Sawyers, Howard Hughes Medical Institute and Memorial Sloan Kettering Cancer Center and were maintained in Iscove’s modified Dulbecco’s medium (IMDM) with 10% FBS and the above-mentioned antibiotics. V16D and 49FENZR cells were a kind gift from Amina Zoubeidi, Senior Research Scientist at Vancouver Prostate Centre, University of British Columbia and were cultured in RMPI-1640 with 10% FBS and 1% PenStrep. For culturing 49FENZR cells, 10 µM enzalutamide was supplemented to the media. VCaP EnzR cells were commercially purchased (Sigma-Aldrich, Cat. No. SCC421) and cultured in 20 µM enzalutamide. All cell lines were authenticated by Labcorp every 6 mo and frequently tested for mycoplasma contamination using primers 5′-ACACCATGGGAGCTGGTAAT-3′ and 5′-GTTCATCGACTTTCAGACCCAAGGCAT-3′. For functional experiments, cells were treated with dimethyl sulfoxide (DMSO) or enzalutamide (Medivation) in media supplemented with 10 nM DHEA. The EHMT1/2 inhibitor, UNC0638 (Cayman, Cat. No. 10734) and SUV39H1 inhibitor, F5446 (Aobious, Cat. No. AOB17639) were used for treating the EnzR cells.
Xenograft Studies.
All mouse studies were performed in compliance with a protocol approved by the Institutional Animal Care and Use Committee of the Cleveland Clinic Lerner Research Institute. Male NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) (6 to 8 wk old) mice were obtained from the Cleveland Clinic Biological Resources Unit. Sample size was determined based on our prior CRPC xenograft studies to allow for statistical comparisons (30, 31). LNCaP cells expressing guide RNAs targeting CBX3 or CBX5 or a nontargeting control sgRNA were injected subcutaneously with Matrigel (BD Biosciences) into the flank of mice (107 cells/mouse). Once tumors reached 100 mm3 (length × width × height × 0.52), mice were surgically castrated and implanted with a 5 mg 90-d sustained-release DHEA pellet (Innovative Research of America) to mimic the human adrenal synthesis of DHEA in CRPC patients. When tumor volumes reached 450 to 500 mm3, mice injected with each cell type, were arbitrarily divided into two groups, one to be fed chow containing Enz (62.5 mg/kg) and the other chow without Enz (control diet). Tumor measurements were performed every other day and progression-free survival was defined as a twofold increase in tumor size (1,000 mm3) from treatment start. Numbers of mice in each treatment group that reached tumor size required for treatment initiation are shown in Fig. 2L. The statistical differences in survival were calculated with Kaplan–Meier analysis using a log-rank test in Prism 9.1.
Flow Cytometry–Based Competition Assay.
The 5 × 105 LNCaP cells were seeded in six-well plates and 24 h later, transduced with incremental volumes (1, 2, 4, 8, 16, 32, and 64 µL) of 100× concentrated lentiviral particles encoding EHMT1-iso1, or EHMT1-iso2, or empty vector together with GFP. At 72 h posttransduction, cells were trypsinized and replated for expansion. A small portion of harvested cells was washed and resuspended in 1× phosphate buffered saline (PBS) and analyzed by flow cytometry for the percentage of fluorescent protein–expressing cells. Cells transduced with dilutions that resulted in an ∼20% GFP-positive population were treated with DMSO or 5 µM Enz for over 60 d and subsequently analyzed for changes in the GFP-positive population. Data were collected by MACSQant Analyzer 10 (Miltenyi Biotech) and analyzed using FlowJo 10.6.2. Forward and side scatter gating criteria to exclude debris and select for GFP-positive cells were uniformly applied to all samples.
VBIM Library.
VBIM-LVs (Fig. 1A) were originally constructed as three separate plasmids, each harboring a different FLAG-splice donor (SD 1, 2, and 3) site to allow for targeting of the three different reading frames upon insertion to the genome (10). Lentiviral particles produced from each VBIM construct were titrated on LNCaP cells by flow cytometry analysis for GFP expression. Subsequently, 2 × 106 LNCaP cells were separately transduced with each of the VBIM-SD 1, 2, or 3 lentiviruses at a multiplicity of infection of 3 to ensure at least one VBIM integration per cell. Forty-eight hours later, cells transduced with each of the three VBIM-LVs were pooled together and expanded. The final LNCaP-VBIM library was then divided into two populations: treated with DMSO or Enz (10 µM), both in the presence of 10 nM DHEA for 73 d. Finally, total RNA was extracted from cells and subjected to RNA-sequencing (RNA-seq) analysis.
VBIM-Seq Data Analysis.
Sequencing reads generated from the samples were trimmed and quality control checked with a modified version of Cutadapt (50). This modified version identifies reads with the VBIM tag (5′-CCACCATGGATTACAAGGATGACGACGATAAGAATTCTT-3′) and separates the reads containing the insertion sequence tag from reads without. Reads identified with the VBIM sequence were aligned to the human genome (GRCh38) using Bowtie 2 using the “very-sensitive local” mode. The alignment files were filtered for mapping quality >5, and intervals less than 50 base pairs apart were merged with bedtools. Resulting intervals with read counts >5 were annotated using HOMER and kept as putative VBIM insertion sites. Reads without the insertion tag were separately aligned using Bowtie 2 and differential expression analysis was done between the experimental groups using Cufflinks. Significantly differentially expressed genes with q value <0.05 were then cross-referenced to the putative VBIM insertion sites and confirmed visually using the Integrated Genome Viewer.
Analysis of Repeat Element Expression.
Kallisto was used to quantify transcripts against a joint reference of GRCh38 Ensembl transcripts and Repbase consensus sequences for repeats. Quantification estimates were then normalized to library sequence depth (counts per million) and used for downstream analyses. Single-sample GSEA (ssGSEA) was used to quantify the activity of gene sets compared to genes outside the gene set within a sample using the GSVA R package.
Custom gene sets were defined using 1) all repeat elements quantified using the Kallisto pipeline above and 2) repeats known to bind MDA5 in protection assays from colorectal cancer cell lines treated with a DNA demethylating agent, coupled with ADAR knockdown, previously published in ref. 33. ssGSEA scores were estimated for both.
A 22-gene ISG signature was produced using an amended version of a previously published signature (28). Glucocorticoid receptor transcription was used to measure GR induction. An H3K9 methylation signature was defined using methyltransferases (SUV39H1, SUV39H2, SETDB1, EHMT1, and EHMT2) and readers (CBX1, CBX3, and CBX5), and a separate signature was defined for SUV39H1/SUV39H2 alone since these genes showed prognostic association in antiandrogen-treated mCRPC patients.
Pearson’s correlations between ssGSEA scores were computed separately for enzalutamide/abiraterone naïve and exposed patients and were visualized using the ggcorrplot package. Significant correlations were defined at P < 0.05.
ChIP-qPCR.
ChIP was performed using the SimpleChIP Plus Sonication Chromatin IP Kit (Cell Signaling Technology, Cat. No. 56383) according to the manufacturer’s guidelines. Briefly, the cells were cross-linked with 1% formaldehyde, followed by quenching with 1× glycine. The cells were scraped in 1× PBS with protease inhibitor mixture (PIC) and pelleted at 1,000 × g at 4 °C for 5 min and resuspended in 1× ChIP Sonication Cell Lysis Buffer with PIC. This was followed by incubation on ice for 10 min and centrifugation at 5,000 × g at 4 °C for 5 min. The pellet was then resuspended in 1× ChIP Sonication Cell Lysis Buffer with PIC and incubated on ice for 5 min. Next, the cells were pelleted and resuspended in 1× ChIP Sonication Nuclear Lysis Buffer with PIC, incubated on ice for 10 min, followed by sonication using the Bioruptor (Diagenode) to obtain ∼500 bp DNA fragments and clarified by centrifugation at 21,000 × g for 10 min at 4 °C. The sonicated chromatin (∼10 to 15 µg) was incubated with 4 µg of either primary H3K9me3 antibody (Active Motif, No. 39062) or isotype control antibodies overnight at 4 °C. The next day, the Protein G magnetic beads were added to the lysates and incubated for 2 to 4 h. The beads were then washed three times with low salt wash buffer with PIC and once with high salt wash buffer with PIC. The immunocomplex was eluted in 1× ChIP elution buffer with gentle vortexing (1,200 rpm) at 65 °C for 30 min. The eluted chromatin and 1% input were reverse cross-linked by treating the samples with NaCl and RNase A for 30 min at 37 °C followed by adding Proteinase K and incubating for 2 h at 65 °C. The DNA purification was performed using the spin columns, and ChIP-qPCR was performed using primers mentioned in SI Appendix, Table S2.
Survival Analysis of mCRPC Patient Datasets.
Two mCRPC cohorts (SU2C-Prostate Cancer Foundation [PCF] and RMH) were analyzed for differences in survival based on H3K9MT expression levels using Kaplan–Meier analysis. A total of 158 mCRPC transcriptomes (polyA), generated by the international SU2C/PCF Prostate Cancer Dream Team, were reanalyzed. Transcriptomes were aligned to the human reference genome (GRCh38/hg38) using TopHat2 (version 2.0.7). Gene expression as fragments per kilobase of transcript per million mapped reads (FPKM) was calculated using Cufflinks. Overall survival was considered as the time period from biopsy to death for all samples (SU2C, n = 158 and RMH n = 94). Hazard ratios and P values were calculated from a Cox proportional hazards model.
Statistical Methods.
Statistical analysis and data visualization were performed in GraphPad Prism 9.1 or R. For comparisons between two groups, Student’s t test was used. For comparisons between more than two groups with one variable, one-way ANOVA was used and for those with more than one variable, two-way ANOVA was used. All tests were two tailed. Details of the statistical tests are indicated in the figure legends, and all bioinformatics analyses are described in their respective Methods section.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Cancer Institute (R01CA172382 to N.S., R01CA236780 to N.S., R01CA261995 to N.S., R01CA249279 to N.S., 2P01CA062220 to G.R.S., R50CA251961 to M. Berk and T32 CA094186 to S.K.), the Prostate Cancer Foundation (to N.S.), the National Institute for Allergy and Infectious Disease (R01AI135922 to R.H.S.), and the Department of Defense (W81XWH-20-1-0137 to N.S.). The de Bono laboratory are funded by Cancer Research UK, Movember, Prostate Cancer UK, the Prostate Cancer Foundation, US Department of Defense, and the UK Medical Research Council. This work used a Leica SP8 confocal microscope that was purchased with funding from NIH Scientific Interest Group Grant 1S10OD019972-01. We acknowledge Ricky Chan, Belinda Willard, Shuvojit Banerjee, and Matthew Booker for help with RNA-seq analysis and histone protein analysis.
Footnotes
Competing interest statement: N.S. has been a consultant for Janssen Pharmaceuticals and has laboratory research supported by Bristol Myers Squibb (BMS).
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114324119/-/DCSupplemental.
Data Availability
The sequencing data for this study has been submitted to the NCBI Gene Expression Omnibus (GEO) under the accession number GSE202335. All study data are included in the article and/or supporting information.
References
- 1.Desai K., McManus J. M., Sharifi N., Hormonal therapy for prostate cancer. Endocr. Rev. 42, 354–373 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai C., Heemers H., Sharifi N., Androgen signaling in prostate cancer. Cold Spring Harb. Perspect. Med. 7, a030452 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Einstein D. J., Arai S., Balk S. P., Targeting the androgen receptor and overcoming resistance in prostate cancer. Curr. Opin. Oncol. 31, 175–182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran C., et al. , Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beer T. M., et al. ; PREVAIL Investigators, Enzalutamide in metastatic prostate cancer before chemotherapy. N. Engl. J. Med. 371, 424–433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher H. I., et al. ; AFFIRM Investigators, Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 367, 1187–1197 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Arora V. K., et al. , Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 155, 1309–1322 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerry C. J., Schreiber S. L., Chemical probes and drug leads from advances in synthetic planning and methodology. Nat. Rev. Drug Discov. 17, 333–352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shalem O., Sanjana N. E., Zhang F., High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16, 299–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu T., et al. , Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc. Natl. Acad. Sci. U.S.A. 106, 16339–16344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allis C. D., Jenuwein T., The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Epsztejn-Litman S., et al. , De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat. Struct. Mol. Biol. 15, 1176–1183 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishak C. A., De Carvalho D. D., Reactivation of endogenous retroelements in cancer development and therapy. Annu. Rev. Cancer Biol. 4, 159–176 (2020). [Google Scholar]
- 14.Steiner M. C., et al. , Locus-specific characterization of human endogenous retrovirus expression in prostate, breast, and colon cancers. Cancer Res. 81, 3449–3460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attermann A. S., Bjerregaard A.-M., Saini S. K., Grønbaek K., Hadrup S. R., Human endogenous retroviruses and their implication for immunotherapeutics of cancer. Ann. Oncol. 29, 2183–2191 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Fritsch L., et al. , A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol. Cell 37, 46–56 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Fuks F., et al. , The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278, 4035–4040 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Li C. H., et al. , MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature 586, 440–444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanouette S., Mongeon V., Figeys D., Couture J. F., The functional diversity of protein lysine methylation. Mol. Syst. Biol. 10, 724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayaram H., et al. , S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3. Proc. Natl. Acad. Sci. U.S.A. 113, 6182–6187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan C. M., et al. , A histone H3K9M mutation traps histone methyltransferase Clr4 to prevent heterochromatin spreading. eLife 5, 1–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannister A. J., et al. , Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T., Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Bosch-Presegué L., et al. , Mammalian HP1 isoforms have specific roles in heterochromatin structure and organization. Cell Rep. 21, 2048–2057 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Maison C., Almouzni G., HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5, 296–304 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Allshire R. C., Madhani H. D., Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19, 229–244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long X., et al. , Immune signature driven by ADT-induced immune microenvironment remodeling in prostate cancer is correlated with recurrence-free survival and immune infiltration. Cell Death Dis. 11, 779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiappinelli K. B., et al. , Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015). Correction in: Cell 164, 1073 (2016); Cell 169, 361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roulois D., et al. , DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hur S., Double-stranded RNA sensors and modulators in innate immunity. Annu. Rev. Immunol. 37, 349–375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liddicoat B. J., et al. , RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J., et al. , Aberrant corticosteroid metabolism in tumor cells enables GR takeover in enzalutamide resistant prostate cancer. eLife 6, e20183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., et al. , Hexose-6-phosphate dehydrogenase blockade reverses prostate cancer drug resistance in xenograft models by glucocorticoid inactivation. Sci. Transl. Med. 13, eabe8226 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isikbay M., et al. , Glucocorticoid receptor activity contributes to resistance to androgen-targeted therapy in prostate cancer. Horm. Cancer 5, 72–89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehdipour P., et al. , Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 588, 169–173 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Rowe H. M., Trono D., Dynamic control of endogenous retroviruses during development. Virology 411, 273–287 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Liu S., et al. , Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 28, 2041–2055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsui T., et al. , Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Deblois G., et al. , Epigenetic switch-induced viral mimicry evasion in chemotherapy-resistant breast cancer. Cancer Discov. 10, 1312–1329 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Guler G. D., et al. , Repression of stress-induced LINE-1 expression protects cancer cell subpopulations from lethal drug exposure. Cancer Cell 32, 221–237.e13 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Hon G. C., et al. , Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 22, 246–258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ci X., et al. , Heterochromatin protein 1α mediates development and aggressiveness of neuroendocrine prostate cancer. Cancer Res. 78, 2691–2704 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Davies A. H., Beltran H., Zoubeidi A., Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat. Rev. Urol. 15, 271–286 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Morel K. L., et al. , EZH2 inhibition activates a dsRNA–STING–interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nat. Cancer 2, 444–456 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah N., et al. , Regulation of the glucocorticoid receptor via a BET-dependent enhancer drives antiandrogen resistance in prostate cancer. eLife 6, 1–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flammer J. R., et al. , The type I interferon signaling pathway is a target for glucocorticoid inhibition. Mol. Cell. Biol. 30, 4564–4574 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weichselbaum R. R., et al. , An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 18490–18495 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheon H., et al. , IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 32, 2751–2763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen J. Z., et al. , FBXO44 promotes DNA replication-coupled repetitive element silencing in cancer cells. Cell 184, 352–369.e23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data for this study has been submitted to the NCBI Gene Expression Omnibus (GEO) under the accession number GSE202335. All study data are included in the article and/or supporting information.