Significance
Middle East respiratory syndrome coronavirus (MERS-CoV) causes highly lethal respiratory disease. MERS-CoV encodes innate immune antagonists, including accessory proteins NS4a and NS4b, unique to the merbeco lineage of betacoronaviruses, and the nsp15 protein endoribonuclease (EndoU), conserved among all coronaviruses. While mutation of each antagonist protein alone has little effect on innate immunity, infections with recombinant MERS-CoVs with mutations of EndoU in combination with either NS4a or NS4b activate innate signaling pathways and are attenuated for replication. Our data indicate that EndoU and accessory proteins NS4a and NS4b together suppress innate immunity during MERS-CoV infection, to optimize viral replication. This is in contrast to SARS-CoV-2 which activates these pathways and is thus more vulnerable to host innate immune responses.
Keywords: MERS-CoV, endonuclease U, innate immune antagonism, RNase L, PKR
Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) emerged into humans in 2012, causing highly lethal respiratory disease. The severity of disease may be, in part, because MERS-CoV is adept at antagonizing early innate immune pathways—interferon (IFN) production and signaling, protein kinase R (PKR), and oligoadenylate synthetase/ribonuclease L (OAS/RNase L)—activated in response to viral double-stranded RNA (dsRNA) generated during genome replication. This is in contrast to severe acute respiratory syndrome CoV-2 (SARS-CoV-2), which we recently reported to activate PKR and RNase L and, to some extent, IFN signaling. We previously found that MERS-CoV accessory proteins NS4a (dsRNA binding protein) and NS4b (phosphodiesterase) could weakly suppress these pathways, but ablation of each had minimal effect on virus replication. Here we investigated the antagonist effects of the conserved coronavirus endoribonuclease (EndoU), in combination with NS4a or NS4b. Inactivation of EndoU catalytic activity alone in a recombinant MERS-CoV caused little if any effect on activation of the innate immune pathways during infection. However, infection with recombinant viruses containing combined mutations with inactivation of EndoU and deletion of NS4a or inactivation of the NS4b phosphodiesterase promoted robust activation of dsRNA-induced innate immune pathways. This resulted in at least tenfold attenuation of replication in human lung–derived A549 and primary nasal cells. Furthermore, replication of these recombinant viruses could be rescued to the level of wild-type MERS-CoV by knockout of host immune mediators MAVS, PKR, or RNase L. Thus, EndoU and accessory proteins NS4a and NS4b together suppress dsRNA-induced innate immunity during MERS-CoV infection in order to optimize viral replication.
Middle East respiratory syndrome coronavirus (MERS-CoV) first emerged in 2012, in Saudi Arabia (1), and was the second of three zoonotic coronaviruses to emerge into humans in the 21st century, following severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002 and preceding SARS-CoV-2 in 2019. MERS-CoV is a highly pathogenic betacoronavirus, of the merbeco lineage, that has caused 890 deaths in 2,585 laboratory-confirmed cases (as of February 2022) according to the World Health Organization, amounting to a case fatality rate of over 34%. MERS-CoV circulates in its natural reservoir, dromedary camels, and closely related viruses have been found in bats, suggesting that MERS-CoV descended from a bat virus (2–7).
Coronavirus genomes are large positive-sense single-stranded RNA (+ssRNA) with a 5′ cap and 3′ polyA tail. Specifically, the MERS-CoV genome is 30,119 nucleotides in length. The 5′ two-thirds of the coronavirus genome encodes the conserved replicase proteins in ORF1a and ORF1b, involved in several processes such as RNA replication and transcription, RNA capping and processing, proteolytic processing of replicase proteins, and host innate immune antagonism. The 3′ one-third of the genome encodes the structural genes and the unique, lineage-specific accessory genes, which are not necessary for viral replication but play important roles in immune evasion and viral pathogenesis.
Early in infection, coronaviruses establish transcription/replication complexes within the endoplasmic reticulum-associated double-membrane vesicles. Replication of the coronavirus genome RNA and transcription of messenger RNAs (mRNAs) takes place in these complexes and generates double-stranded RNA (dsRNA) intermediates. This dsRNA provides a pathogen associate molecular pattern (PAMP) that is detected by several different host pattern recognition receptors (PRRs), leading to activation of innate immune antiviral pathways. Melanoma differentiation-associated protein 5 (MDA5) is a PRR that detects coronavirus dsRNA (8) and induces signaling through mitochondrial antiviral-signaling protein (MAVS), which leads to activation of interferon (IFN) regulatory factors, and transcription of type I and III IFNs (9). IFNs are produced and secreted by the infected cell and can signal in autocrine or paracrine fashion. Downstream Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling and phosphorylation leads to expression of several hundred IFN-stimulated genes (ISGs) which produce an antiviral state. Protein kinase R (PKR) is another cytoplasmic PRR activated by dsRNA. Upon sensing dsRNA, PKR autophosphorylates and then phosphorylates translation initiation factor eIF2α, leading to translational arrest (10). Oligoadenylate synthetases (OAS1-3), upon detecting dsRNA, produce 2’,5′-oligoadenylates (2-5A) that activate the host enzyme ribonuclease L (RNase L) which cleaves viral and host ssRNA (11, 12). Activation of each of these pathways leads to restriction of virus replication and spread. RNase L and PKR activation can promote IFN production, cellular stress, inflammation, and/or apoptotic death (13–16). Many viruses, including coronaviruses, have evolved various mechanisms for counteracting these pathways. Importantly, each of the three pathways can be activated independently of the others (17–19); therefore, if IFN signaling is inhibited during infection as is the case for some viruses, the OAS/RNase L or PKR pathways can still be activated. In addition, since OASs and PKR are ISGs, the PKR and OAS/RNase L pathways can be further up-regulated by IFN.
Coronavirus accessory proteins contribute to differences between the three lineages of betacoronaviruses. The merbeco virus MERS-CoV is particularly efficient at shutting down host innate immune pathways, including IFN signaling, PKR, and OAS/RNase L, while the sarbecovirus, SARS-CoV-2, activates PKR and RNase L and weakly activates IFNs (18). Two accessory proteins encoded by MERS-CoV have been shown to play important roles in this innate immune antagonism (20–22). NS4a and NS4b are both encoded on mRNA4 by ORF4a and ORF4b, respectively. NS4a is a dsRNA-binding protein and, as such, has been reported to block IFN induction and PKR activation (21, 23–27). NS4b has at least two functional domains, a phosphodiesterase (PDE) and an amino terminal nuclear localization signal (NLS). The PDE degrades 2-5A, preventing activation of RNase L, and weakly inhibits IFN production (21, 22). The amino terminal NLS confers expression mostly in the nucleus and has been reported to block NFκB translocation to the nucleus (22, 28–31). Previously, we found that a recombinant mutant MERS-CoV ablated for NS4a expression induced low levels of phosphorylated PKR and no detectable phosphorylation of eIF2α, as well as a low-level of type I and III IFN expression (21), and a recombinant MERS-CoV with an inactivated NS4b PDE led to mild activation of RNase L and IFN induction (21, 22). Notably, neither the embecoviruses (for example, human coronavirus [HCoV]-OC43 or murine coronavirus [MHV]) nor sarbeco lineages (for example, SARS-CoV-2) encode a dsRNA-binding protein. While the sarbecoviruses do not express a PDE, the embecoviruses do encode a PDE in the NS2 protein. However, in contrast to NS4b, this PDE lacks an NLS and thus is expressed completely in the cytoplasm (32). These differences in accessory proteins between CoVs likely contribute to differences in pathogenesis and immune evasion.
Nonstructural protein 15 (nsp15) is a conserved coronavirus replicase protein encoded in ORF1b that contains an endoribonuclease (EndoU) domain with two catalytic histidine residues. Murine coronavirus (MHV) EndoU has been shown to limit dsRNA accumulation, and consequently reduces IFN production and signaling as well as activation of OAS/RNase L and PKR pathways during infection of murine bone marrow–derived macrophages (33, 34). While the exact mechanism of action of EndoU is not completely understood, two different mechanisms have been reported. One report concludes that EndoU cleaves genomic RNA to limit the production of dsRNA (35), and the other concludes that EndoU degrades polyU from the 5′ end of negative strand RNA, eliminating a PAMP for sensors (36). The role of MERS-CoV EndoU activity in antagonizing innate immunity has yet to be examined during infection of human cells.
Using a group of recombinant mutant viruses, we investigated the role of EndoU, alone and in combination with NS4a or NS4b, in innate immune antagonism during MERS-CoV infection. In addition to investigating immune evasion by MERS-CoV in the lung-derived cell line A549DPP4, which expresses MERS-CoV receptor dipeptidyl peptidase 4, and Calu-3, we extended the study of these host–pathogen immune interactions to primary patient-derived nasal epithelial cells in air–liquid interface cultures. Our findings indicate that MERS-CoV EndoU together with its accessory proteins effectively shut down innate immune pathways to optimize replication.
Results
Construction of Recombinant MERS-CoVs and Replication Kinetics in VeroCCL81, A549DPP4, and Calu-3 Cells.
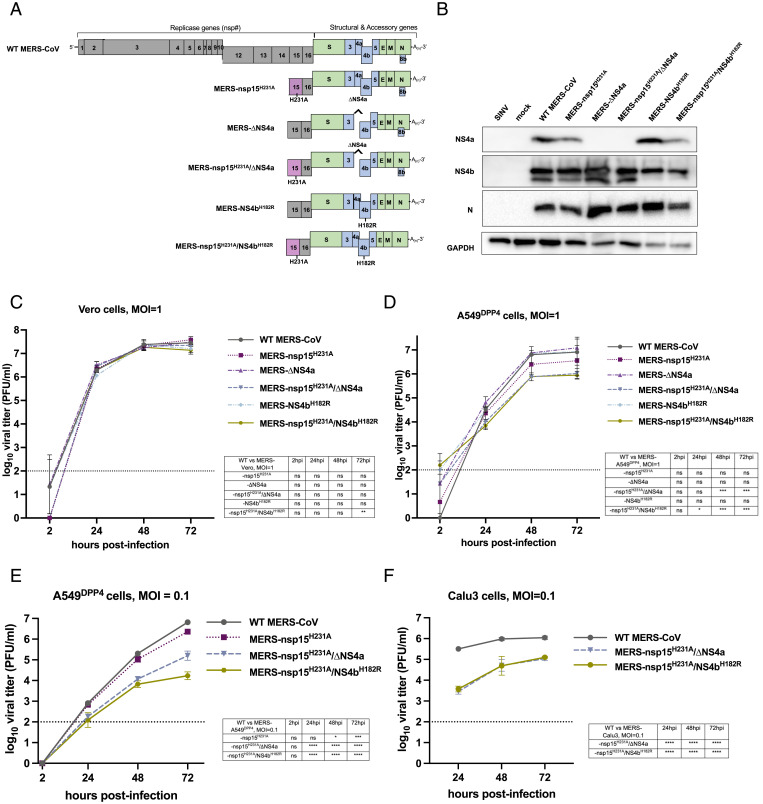
In order to study the effects of EndoU activity on the dsRNA-induced antiviral innate immune pathways during MERS-CoV infection, several recombinant mutants were constructed using the Lambda Red recombination system and a bacterial artificial chromosome (BAC) that encodes the full-length MERS-CoV genome (37). Recombinant viruses (summarized in Fig. 1A) were MERS-CoV-nsp15H231A with an amino acid substitution inactivating EndoU, MERS-CoV-ΔNS4a with interruption of NS4a expression by insertion of two termination codons at positions 11 and 12, MERS-CoV-NS4bH182R with an amino acid substitution inactivating the PDE, and double mutants MERS-CoV-nsp15H231A/ΔNS4a and MERS-CoV-nsp15H231A/NS4bH182R. We confirmed by Western blot that MERS-ΔNS4a and MERS-nsp15H231A/ΔNS4a infection produced undetectable expression of NS4a protein (Fig. 1B). As we observed previously, an NS4bH182R mutant expressed less NS4b than wild-type (WT) MERS-CoV (Fig. 1B) (21).
Fig. 1.
Recombinant MERS-CoV design and replication kinetics. (A) Diagram of the MERS-CoV genome including the replicase locus (encoding 16 nsps), structural genes, and accessory genes is shown along with a diagram of each recombinant mutant constructed using the BAC reverse genetics system. (B) A549DPP4 cells were infected at MOI = 5 with the indicated viruses, and protein lysates were harvested at 24 hpi. Expression of NS4a and NS4b was determined by SDS/PAGE and Western blot. (C) Vero cells were infected in triplicate at MOI = 1 with the indicated viruses. Supernatants were collected at the indicated hpi, and infectious virus was quantified by plaque assay. (D) A549DPP4 cells were infected in triplicate with the indicated viruses at MOI = 1, and replication was quantified as in C. Data in C and D are from one representative of three independent experiments. (E) A549DPP4 and (F) Calu-3 cells were infected in triplicate with the indicated viruses at MOI = 0.1, and replication was quantified as in C. All data in C–F are displayed as means ± SD. The dotted line in C–F indicates the limit of detection of the plaque assay. Statistical significance of each recombinant virus compared to WT MERS-CoV was calculated by repeated measures two-way ANOVA: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. Data that were not statistically significant are labeled ns.
We compared replication of the mutant viruses with WT MERS-CoV initially in African Green Monkey kidney Vero CCL81 (Vero) cells, defective in IFN production. Cells were infected at a multiplicity of infection (MOI) of 1, and supernatant samples were harvested at 2, 24, 48, and 72 h postinfection (hpi). Quantification of infectious virus was completed by standard viral plaque assay on Vero cells as previously described (18). Mutant viruses replicated to similar levels as WT MERS-CoV in three independent experiments (Fig. 1C), indicating the absence of inherent replication defects. To investigate the effects of dsRNA-induced innate immune activity on replication of these viruses, A549DPP4 cells (with intact IFN, PKR, and RNase L pathways) were infected at an MOI of 1, and infectious virus was quantified as above. At 48 and 72 hpi, we observed an approximate 1 log10 plaque-forming units per mL decrease in infectious virus produced in the MERS- nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R mutants compared to WT MERS-CoV that reached statistical significance (Fig. 1D). Interestingly, mutation of the EndoU catalytic site alone, MERS-nsp15H231A, resulted in a small but not statistically significant defect in infectious virus production. No differences were noted in MERS-ΔNS4a or MERS-NS4bH182R replication. This is consistent with our previous finding of only mild effects on viral replication in a mutant lacking expression of either of these two accessory proteins. To further investigate this replication defect, A549DPP4 cells were also infected at an MOI of 0.1 with MERS-nsp15H231A, MERS-nsp15H231A/ΔNS4a, and MERS-nsp15H231A/NS4bH182R (Fig. 1E). The attenuation of the viruses compared to WT MERS-CoV at an MOI of 0.1 was similar to what was observed at an MOI of one. Furthermore, the replication defects of MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R compared to WT MERS-CoV at an MOI of 0.1 were observed in Calu-3 cells, a different lung carcinoma cell line. Thus, the loss of EndoU activity combined with loss of NS4a expression or NS4b PDE activity led to attenuation of virus replication in two lung-derived cells lines that have intact innate immune responses.
dsRNA Accumulation Is Increased When EndoU Is Inactive during MERS-CoV Infection.
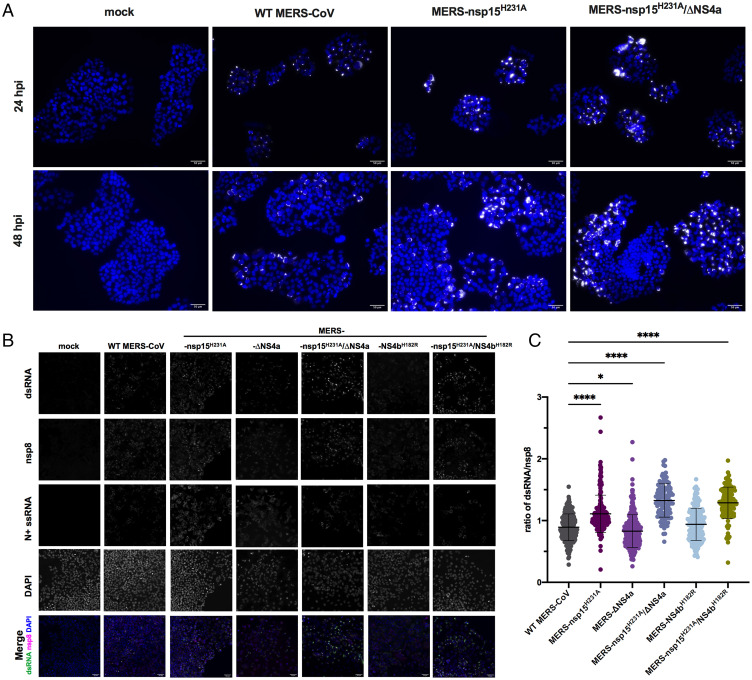
Next, we sought to examine production of dsRNA during infection with MERS-CoV recombinants containing catalytic inactivation mutations in EndoU (nsp15H231A) compared with WT MERS-CoV. Detection of dsRNA was assessed by immunofluorescence (IF) assay using the monoclonal antibody J2 directed against dsRNA and imaged using widefield microscopy. In six independent experiments, we observed increased expression of dsRNA when EndoU was inactivated, including during infection with MERS-nsp15H231A and MERS-nsp15H231A/ΔNS4a compared to WT MERS-CoV (Fig. 2A), consistent with previous findings for inactivation of MHV EndoU (33, 36). To quantify this observation and to extend it to the other MERS-CoV mutants, we infected A549DPP4 at an MOI of five on glass coverslips and fixed with 4% paraformaldehyde. We performed combined fluorescent in situ hybridization (FISH)/IF using J2 antibody to detect dsRNA, antiserum against the viral primase, nsp8, a component of the viral polymerase complex and therefore a marker for virus replication/transcription complexes (38), and oligonucleotide probes to detect nucleocapsid (N)+ssRNA to define the cytoplasm. We observed brighter staining of dsRNA in MERS-nsp15H231A, MERS-nsp15H231A/ΔNS4a, and MERS-nsp15H231A/NS4bH182R compared to WT MERS-CoV at 48 hpi (Fig. 2B). Fiji software was used for quantification, as described in detail in Materials and Methods. Briefly, due to extensive cell-to-cell fusion (syncytia), we were unable to outline individual infected cells for quantification. To quantify dsRNA, we outlined infected areas of images (syncytia and single cells) using nucleocapsid RNA staining to define the cytoplasm and called them regions of interest (ROIs). We measured the mean gray value (MGV) of fluorescence signal of dsRNA or nsp8 within each ROI and recorded the ratio of dsRNA MGV over nsp8 MGV for each ROI. These ratios were compared across infection with mutant and WT viruses at 48 hpi in A549DPP4 cells (Fig. 2C). We observed statistically significant increases in dsRNA/nsp8 ratios in MERS-nsp15H231A, MERS-nsp15H231A/ΔNS4a, and MERS-nsp15H231A/NS4bH182R compared to WT MERS-CoV, but not in MERS-ΔNS4a or MERS-NS4bH182R, at 48 hpi.
Fig. 2.
The dsRNA expression is increased when EndoU is inactive during MERS-CoV infection. (A) Representative dsRNA staining in MERS-CoV infection: A549DPP4 cells were infected at MOI = 5 and fixed at 24 and 48 hpi with 4% paraformaldehyde and subjected to IF to detect dsRNA by 20× widefield microscopy. Nuclei were stained by Hoechst, shown in blue, and dsRNA was stained by J2, shown in white. (Scale bar, 50 μm.) (B) Representative images for quantification of dsRNA in A549DPP4 cells, mock infected (24 hpi), or MERS-CoV infected at MOI = 5 and fixed 48 hpi are shown with all single channels (dsRNA, nsp8, N+ssRNA, DAPI). Merged image shows DAPI (blue), nsp8 (magenta), and dsRNA (green). (Scale bar, 50 μm.) The dsRNA and nsp8 were stained by IF using J2 and anti-nsp8 serum, N+ssRNA was stained by FISH, and nuclei were stained by Hoechst (DAPI). (C) The dsRNA was quantified by graphing the ratio of dsRNA MGV over nsp8 MGV in each infected ROI. An ROI was defined by thresholding using the Otsu method for N+ssRNA staining in Fiji. Each dot represents the ratio of dsRNA/nsp8 for that ROI. Five to seven representative fields of widefield microscopy at 20× with 1.5× zoom were analyzed per condition. Data shown are from one representative of two independent experiments, and mean plus SD is shown. Statistical significance compared to WT MERS-CoV was determined by one-way ANOVA. *P ≤ 0.05; ****P ≤ 0.0001. Data that were not statistically significant are not labeled.
MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R Induce IFN and ISG Expression.
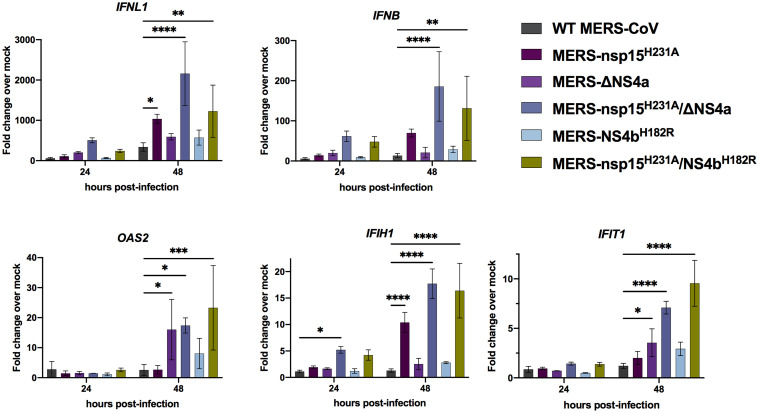
To investigate whether activation of the type I/III IFN pathways was increased during infection in the absence of EndoU activity, we infected A549DPP4 cells at an MOI of 5 with the same panel of MERS-CoV recombinant mutant viruses and collected total intracellular RNA at 24 and 48 hpi (Fig. 3). We used qRT-PCR to quantify mRNA expression of select IFN genes (IFNL1 and IFNB) and ISGs as compared to mock infected cells. We found minimal expression of IFNs or ISGs over mock infected cells with infection of any of the viruses at 24 hpi. However, at 48 hpi, expression of IFNL1 and IFNB (over mock infected) remains minimal in WT infected cells and is induced to modest and variable extents in cells infected by the single mutants compared to WT infected cells; in contrast, IFN mRNAs as well as ISG mRNAs are significantly induced in double-mutant MERS-nsp15H231A/ΔNS4a– and MERS-nsp15H231A/NS4bH182R–infected cells compared to WT MERS-CoV infection. Similarly, expression (over mock infected) of select ISG mRNAs, IFIH1, OAS2, and IFIT1, was also significantly increased in RNA from cells infected with MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R compared to RNA from WT MERS-CoV–infected cells. Additionally, ISG expression was also significantly increased during infection with single mutant MERS-nsp15H231A or MERS-ΔNS4a infection alone, compared to WT-MERS-CoV at 48 hpi, although less than in double-mutant infected cells.
Fig. 3.
MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R induce IFN/ISG expression in A549DPP4 cells. A549DPP4 cells were mock infected or infected in triplicate at MOI = 5. Total RNA was harvested at 24 and 48 hpi, and expression of IFNL1, IFNB, IFIH1, OAS2, and IFIT1 mRNAs was quantified by qRT-PCR and expressed as fold change over mock infected using the 2−Δ(ΔCT) formula. Data are displayed as means ± SD. Data are from one representative of three independent experiments. Statistical significance compared to WT MERS-CoV was calculated by two-way ANOVA: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. Data that were not statistically significant are not labeled.
RNase L and PKR pathways are activated by MERS-CoV double mutants.
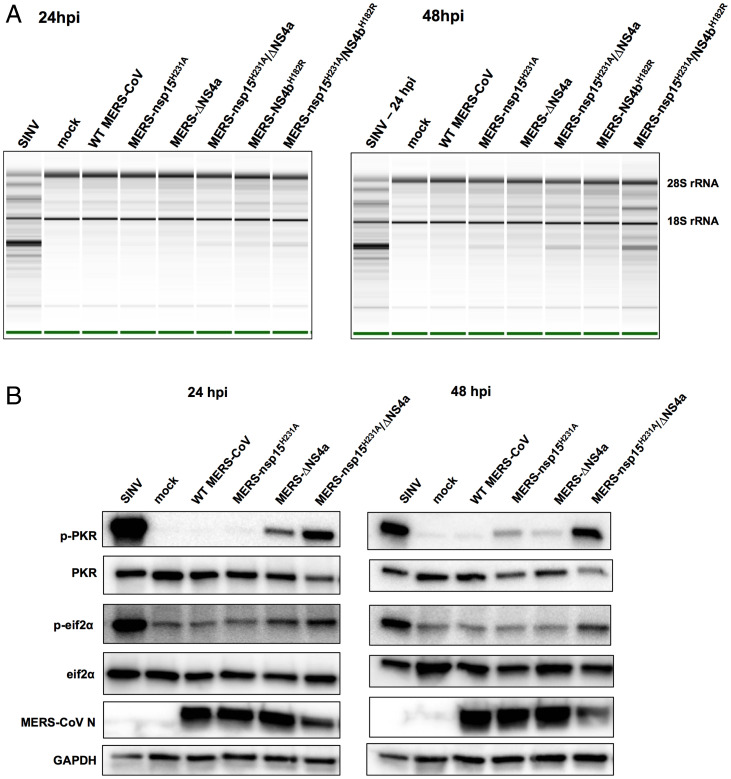
We next investigated whether RNase L was activated in the absence of EndoU catalytic activity alone or in combination with loss of NS4a expression or NS4b PDE activity. Thus, we infected A549DPP4 cells with single- and double-mutant MERS-CoV as well as WT MERS-CoV at an MOI of 5, harvested total cellular RNA at 24 and 48 hpi, and determined RNase L activation by cleavage of ribosomal RNA (rRNA) as indicated by the Agilent RNA Nano 6000 assay (Fig. 4A). Sindbis virus (SINV), an unrelated alphavirus that robustly activates RNase L, served as a positive control. As we reported previously, RNase L was weakly activated during infection with MERS-NS4bH182R in the absence of PDE activity but not by MERS-ΔNS4a (21, 22). We found that inactivation of EndoU alone (MERS-nsp15H231A) also did not lead to strong RNase L activation in A549DPP4 cells and that MERS- nsp15H231A/ΔNS4a infection weakly activated RNase L as evidenced by slight 28S and 18S rRNA degradation. In contrast, MERS-nsp15H231A/NS4bH182R infection promoted rRNA degradation by 48 hpi, indicating more robust RNase L activation in the absence of both EndoU and NS4b PDE enzymatic activities. This contrasts with previous findings in MHV infection of bone marrow–derived macrophages where inactivation of either the PDE or EndoU results in robust activation of RNase L and extreme attenuation of replication, to be discussed below.
Fig. 4.
RNase L and PKR are activated in the absence of EndoU activity combined with loss of accessory proteins during MERS-CoV infection. (A) A549DPP4 cells were mock infected or infected at MOI = 5, and total cellular RNA was harvested at 24 and 48 hpi. The rRNA degradation was assessed on an Agilent Bioanalyzer. 28S and 18S rRNA positions are indicated. Data are from one representative of three independent experiments. (B) A549DPP4 cells were mock infected or infected at MOI = 5 and cell lysates were harvested at 24 and 48 hpi. Proteins were separated by SDS/PAGE and immunoblotted with antibodies against phosphorylated PKR (p-PKR), PKR, phosphorylated eIF2α (p-eIF2α), eIF2α, MERS-CoV nucleocapsid (N), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are from one representative of four (24 hpi) or two (48 hpi) independent experiments.
We also examined induction of the PKR pathway when EndoU is inactivated alone or in combination with loss of NS4a expression. Thus, A549DPP4 cells were infected with the single and double MERS-CoV mutants at an MOI of 5, and protein lysates were harvested at 24 and 48 hpi and processed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) followed by Western blotting for detection of PKR activation (Fig. 4B). As observed previously, PKR activation was not detected during infection with WT MERS-CoV; however, MERS-ΔNS4a infection induced weak phosphorylation of PKR but not its downstream mediator eIF2α (21, 23, 25). In addition, inactivation of EndoU alone, in infection with MERS-nsp15H231A, did not induce strong activation of PKR. Only mild phosphorylation of PKR was observed at 48 hpi, late in infection, but not at the earlier 24-hpi time point, and no phosphorylation of eIF2α was detected. However, during infection with double-mutant MERS-nsp15H231A/ΔNS4a, strong activation of PKR was observed at both 24 and 48 hpi, indicated by phosphorylation of PKR and eIF2α.
Knockout of Key Host Immune Mediators Rescues Attenuation Observed in MERS-CoV Recombinants.
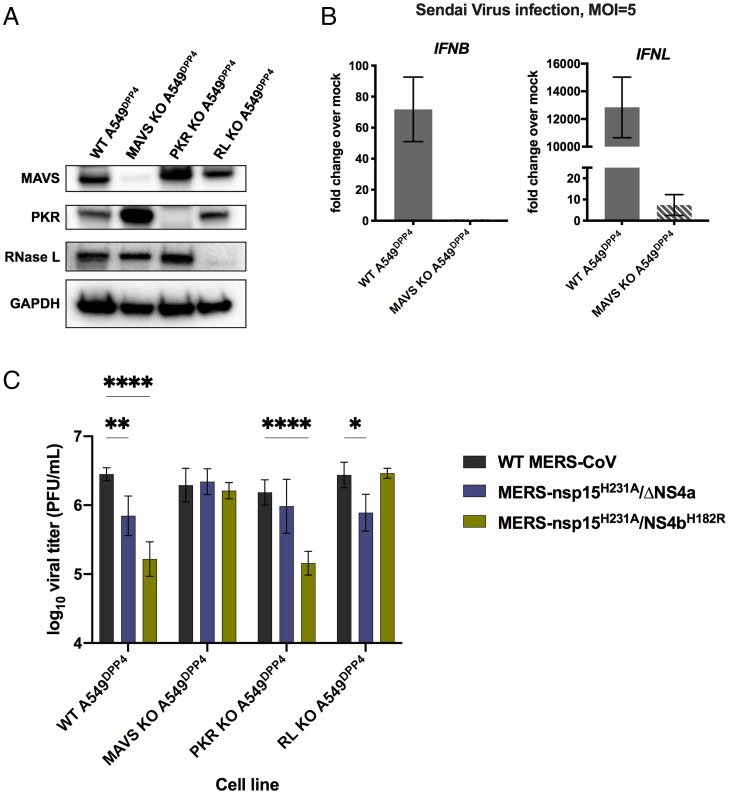
Due to replication defects observed in A549DPP4 cells and the strong innate immune-stimulatory phenotype observed during MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R infection, we sought to determine whether knockout (KO) of a key mediator in each of the three dsRNA-induced antiviral pathways would rescue replication of these recombinant viruses. We had previously constructed A549DPP4 RNase L KO cells (21), and here we additionally generated A549DPP4 MAVS KO cells and A549DPP4 PKR KO cells to abrogate the host RNase L, IFN, and PKR pathways, respectively. KO of these host immune mediators was confirmed via Western blotting (Fig. 5A). We further confirmed KO of the IFN pathway in A549DPP4 MAVS KO cells via qRT-PCR in the context of Sendai virus (Cantell strain) infection (39), a paramyxovirus that induces very high levels of IFNs, showing that both type I (IFNB) and type III (IFNL1) expressions were almost completely eliminated in infected A549DPP4 MAVS KO cells compared to WT A549DPP4 cells (Fig. 5B). We compared replication of WT MERS-CoV with double mutants MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R in the WT, PKR KO, and MAVS KO A549DPP4 cells, along with previously described A549DPP4 RNase L KO cells (Fig. 5C) (21). Cells were infected at an MOI of 1, and supernatant samples were harvested at 48 hpi. Quantification of infectious virus was completed by plaque assay. In WT A549DPP4 cells, we observed a significant attenuation in viral replication in both viral recombinants compared to WT MERS-CoV, as expected (Fig. 1C). KO of PKR in A549DPP4 cells rescued MERS-nsp15H231A/ΔNS4a replication to WT MERS-CoV levels, while MERS-nsp15H231A/NS4bH182R remained attenuated, consistent with strong activation of the PKR pathway by MERS-nsp15H231A/ΔNS4a (Fig. 4B). KO of RNase L in A549DPP4 cells rescued replication of MERS-nsp15H231A/NS4bH182R lacking the PDE activity, while MERS-nsp15H231A/ΔNS4a remained attenuated, consistent with our observation of stronger activation of RNase L by MERS-nsp15H231A/NS4bH182R (Fig. 4A). In MAVS KO cells, replication of both double-mutant recombinant viruses (MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R) was rescued to WT MERS-CoV levels. This suggests the particular importance of the IFN pathway in limiting MERS-CoV replication in A549DPP4 cells, as elimination of this pathway via MAVS KO was sufficient to restore replication of both recombinant viruses to WT levels.
Fig. 5.
KO of innate immune pathways rescues attenuation in MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R. (A) Cell lysates of WT A549DPP4 as well as MAVS KO, PKR KO, and RNase L KO A549DPP4 were collected, and proteins were separated by SDS/PAGE and immunoblotted with antibodies against MAVS, PKR, RNase L, and GADPH. (B) WT A549DPP4 and MAVS KO A549DPP4 cells were infected with Sendai virus at MOI = 5, and total cellular RNA was harvested at 18 hpi. Expression of IFNL1 and IFNB was quantified by qRT-PCR, and CT values were normalized to beta-actin and shown as fold change over mock using the formula 2-Δ(ΔCT). Data are displayed as means ± SD. (C) A549DPP4 cell lines: WT, MAVS KO, PKR KO, and RNase L KO were infected in triplicate at MOI = 1, and supernatant samples were collected at 48 hpi. Viral replication was quantified by plaque assay. Data are displayed as means ± SD. Statistical significance of differences in viral replication for each recombinant virus compared to WT MERS-CoV was calculated by repeated measures two-way ANOVA: *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001. Data that were not statistically significant are not labeled. Data are from one representative of three independent experiments.
Infection of Primary Nasal Epithelial Cell Cultures with Mutant MERS-CoV Recombinants Reproduces Attenuation and Immune-Stimulatory Phenotype Observed in A549DPP4 Cells.
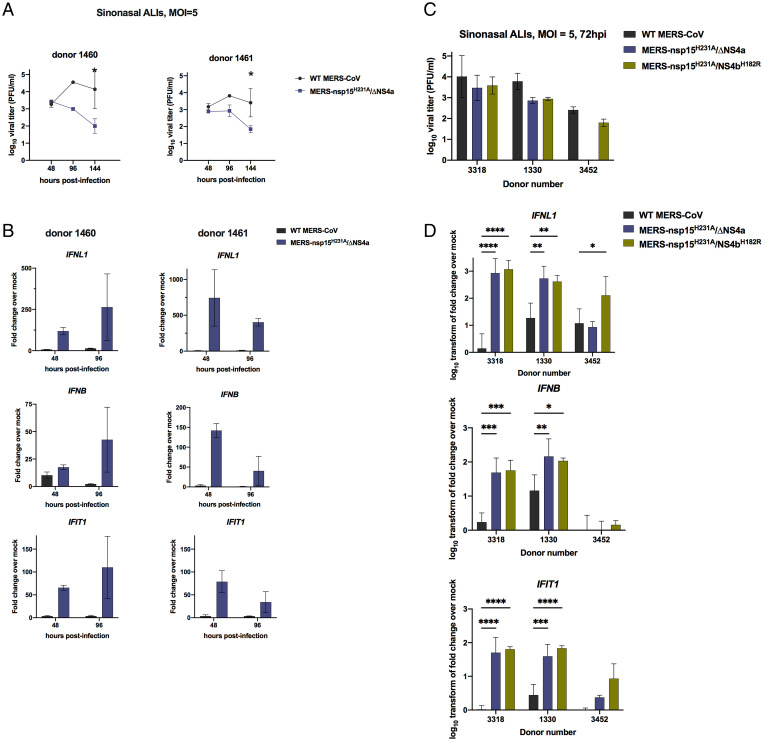
Given the robust activation of dsRNA-induced antiviral innate immune pathways that we observe in A549DPP4 cells upon infection with MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R, we sought to compare these viral mutants with WT MERS-CoV in a primary nasal epithelial cell culture system. We grew and differentiated patient-derived nasal samples at an air–liquid interface (ALI) (40) in order to recapitulate the in vivo airway epithelium in terms of cell types, including epithelial cells with beating cilia and mucus-producing goblet cells, the latter targeted by MERS-CoV (41, 42). We previously showed that these nasal ALI cultures can be productively infected by MERS-CoV (18). To investigate the kinetics of viral replication as well as innate immune induction in these cultures, we infected nasal ALI cultures derived from two independent donors with WT MERS-CoV and MERS-nsp15H231A/ΔNS4a at an MOI of 5. We collected airway surface liquid (ASL) at 48, 96, and 144 hpi, and collected total cellular RNA at 48 and 96 hpi. Shed virus in ASL samples was quantified by plaque assay, and total RNA was used for qRT-PCR for analysis of IFN and ISG expression at each time point. We observed significant reductions in viral replication in MERS-nsp15H231A/ΔNS4a compared to WT MERS-CoV in both donors at late time points (Fig. 6A), consistent with observations in A549DPP4 cells. MERS-nsp15H231A/ΔNS4a infection induced significantly more IFN (both IFNL1 and IFNB) and a representative ISG (IFIT1) expression compared to WT MERS-CoV in nasal ALI cultures (Fig. 6B). We next infected nasal ALI cultures derived from three additional donors with WT MERS-CoV, MERS-nsp15H231A/ΔNS4a, and MERS-nsp15H231A/NS4bH182R to query whether the immune-stimulatory phenotype of both of the double mutants observed in A549DPP4 cells could be replicated in this primary epithelial culture system. We observed decreases in viral shedding with both recombinant viruses compared to WT MERS-CoV in two of three donors at 72 hpi (Fig. 6C) and hypothesize that the defect in viral replication would be amplified in these cultures at later time points (as observed in Fig. 6A). We also observed significantly increased induction of IFNB and IFNL1 mRNA expression in nasal ALIs infected with both recombinant viruses compared to WT MERS-CoV, as well as significantly increased induction of the ISG, IFIT1 (Fig. 6D). We observed variability in IFN responses across donors, likely due to inherent differences between donors in permissiveness to infection and host immune response. Overall, these data demonstrate that the increased innate immune responses observed during infection of A549DPP4 cells with both MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R compared to WT MERS-CoV, as well as the attenuation in these viral recombinants, can be replicated in primary nasal epithelial cultures. This highlights the physiologic relevance of innate immune evasion by MERS-CoV in the upper airway.
Fig. 6.
MERS-nsp15H231A/ΔNS4a and MERS-nsp15H231A/NS4bH182R are attenuated and induce IFN/ISGs in primary nasal epithelial cells. (A) Nasal air–liquid interface (ALI) cultures derived from two independent donors were infected in duplicate on the apical cell surface at MOI = 5. Apically released virus was collected at 48, 96, and 144 hpi and quantified by plaque assay. (B) After apically released virus was collected from each transwell at the indicated time, total cellular RNA was collected, and IFN and ISG mRNA expression was quantified via qRT-PCR. CT values were normalized to 18S rRNA and shown as fold change over mock using the formula 2-Δ(ΔCT). (C) Nasal ALI cultures from three independent donors were infected apically in triplicate at MOI = 5, and apically released virus was collected at 72 hpi for quantification by viral plaque assay. (D) After collecting apically shed virus, total cellular RNA was collected, and IFN and IFIT1 mRNA expression was quantified via qRT-PCR as detailed above using the 2-Δ(ΔCT) method. Data were then log10 transformed and displayed as mean ± SD. Statistical significance of differences were calculated by repeated measures two-way ANOVA: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. Data that were not statistically significant are not labeled.
Discussion
The timing of IFN response has been shown to be a crucial factor in determining the outcome of MERS-CoV infection in a mouse model (43). While administration of type I IFN within 1 d post infection protected mice from lethal infection, delayed treatment with IFN failed to inhibit viral replication and resulted in increased ISG and inflammatory cytokine gene expression, with mice eventually succumbing to lethal pneumonia (43). More recently, during the COVID-19 pandemic, various genome-wide association studies have identified variants in genes with innate immune and antiviral functions that are associated with COVID-19 susceptibility as well as more-severe clinical phenotypes such as respiratory failure. These include genes encoding type I IFN receptor (IFNAR2), cytokines and antiviral signaling pathways (TYK2), and 2’-5′-oligoadenylate synthetases (OAS), which are key for induction of RNase L (reviewed in ref. 44). This makes it crucial to understand how coronavirus antagonist proteins interact with host antiviral pathways to delay or prevent the induction of IFN and related antiviral responses.
Coronaviruses encode multiple antagonists of host innate immune responses. These include replicase encoded proteins such as nsp15 that are conserved among all coronaviruses, and accessory proteins that are unique for each lineage of coronaviruses, including NS4a and NS4b for MERS-CoV and related merbecoviruses. We have found that MERS-CoV is highly effective at shutting down dsRNA-induced antiviral pathways—IFN signaling, OAS/RNase L, and PKR—compared to coronaviruses of other lineages including the sarbecovirus, SARS-CoV-2. Indeed, WT MERS-CoV induces minimal IFN and ISGs, and fails to activate either the OAS/RNase L or PKR pathways, both of which are activated during infection with SARS-CoV-2 (18). We demonstrate here that this is due to expression of NS4a and NS4b, lineage-specific proteins, in combination with the conserved nsp15.
We sought to build on our understanding of the interactions of MERS-CoV accessory proteins NS4a and NS4b with host dsRNA-induced innate immune pathways, by assessing their activities in combination with the nsp15 endoribonuclease. EndoU activity has been shown to limit accumulation of dsRNA and thereby block induction of IFN, OAS/RNase L, and PKR in the context of other coronavirus infections including murine coronavirus MHV, HCoV 229E, and porcine epidemic diarrhea virus (33, 34, 45), but the role of EndoU in innate immune evasion during MERS-CoV infection has not been investigated before. Thus, to further understand MERS-CoV antagonism of innate immune pathways and how the effects of EndoU may be combined with those of viral accessory proteins, we constructed and used a series of recombinant mutant viruses, either with mutations in single antagonist genes (NS4a, NS4b, or nsp15) or with combinations of mutations in nsp15 and either NS4a or NS4b.
We found that only combined inactivation of EndoU with ablation of NS4a expression or inactivation of the PDE domain of NS4b caused a significant replication defect in MERS-CoV (Fig. 1). We propose that these MERS-CoV–encoded innate immune antagonist proteins play overlapping roles by reducing accumulation of dsRNA (EndoU), reducing sensing of dsRNA by PRRs (NS4a), and directly antagonizing RNase L activity (NS4b PDE), leading to attenuation of replication when mutated in combination. In addition to their direct antiviral activities, the dsRNA-induced innate immune pathways are proapoptotic (13–16). Thus, we hypothesize that apoptosis is induced earlier, or to a greater extent, during infection with the mutant viruses due to the increased induction of these pathways and may also contribute to further attenuation of MERS-CoV mutant replication.
To add to the physiologic relevance of this study, we infected primary cultures of patient-derived nasal epithelial cells with WT MERS-CoV and double mutants. The nasal cavity is the primary site encountered by respiratory viruses like MERS-CoV, and these cultures reproduce various features and cell types of the in vivo airway (including mucus-producing goblet cells targeted by MERS-CoV, as well as ciliated epithelial cells). Thus, it presents an optimal system in which to study viral immune evasion. We found that both double mutants are attenuated for replication and induce the IFN pathway significantly as compared to WT MERS-CoV in this culture system (Fig. 6). Thus, MERS-CoV nsp15 EndoU, NS4a, and NS4b play pivotal roles in evading host innate immunity in the upper airways as well as in lung-derived cell lines.
Sensing of viral dsRNA is the first step in inducing activation of all three pathways. We observed increased accumulation of dsRNA (Fig. 2) during MERS-CoV infection when the EndoU catalytic domain was inactivated, similar to findings in the MHV system (33, 34). However, while EndoU limits accumulation of dsRNA, the accessory proteins are required to more completely suppress the host pathways responding to dsRNA. A mutant with catalytically inactive EndoU induced a modest increase in IFN/ISG mRNA responses. However, a double mutant with inactivated EndoU in combination with either ablation of NS4a expression or inactivation of NS4b PDE catalytic activity significantly increased induction of IFN and ISG mRNA expression late in infection of both lung-derived A549 cells and primary nasal cells (Figs. 3 and 6).
As with IFN/ISG induction, expression of both enzymatically active EndoU and an accessory protein are required to fully block activation of the PKR and OAS/RNase L pathways. While, in the absence of NS4a expression, infection promoted mild activation of PKR (21), infection with the mutant lacking both NS4a and EndoU activity strongly activated the PKR pathway (Fig. 4B). Interestingly, inactivation of EndoU alone did not promote significant phosphorylation of PKR, indicating that NS4a plays a more significant role in blocking induction of this pathway. We observed the most robust activation of RNase L during infection with MERS-CoV expressing inactive EndoU as well as inactive PDE. However, the binding of NS4a to dsRNA may also contribute to blocking activation of the OAS/RNase L pathway as suggested by the somewhat increased activation of RNase L during infection with nsp15/NS4a double mutant compared to the single nsp15 mutant infection (Fig. 4A). This role for NS4a in antagonizing OAS/RNase L induction during infection had not been previously appreciated.
In contrast to the current findings, we previously reported that inactivation of either the NS2 PDE or EndoU promoted robust RNase L activation in the MHV system (46). The differences observed between MHV and MERS-CoV infection suggest that the PDE and/or EndoU may function differently in the two viral systems. MERS-CoV NS4b is localized mostly in the nucleus due to its NLS, and, as such, may be less effective as a PDE than the MHV NS2 which lacks an NLS and is completely cytoplasmic. In the MHV system, when expressed in the nucleus, a PDE (AKAP7) was unable to effectively prevent activation of RNase L and rescue virus replication of mutant MHV-NS2H126R with an inactive PDE, but, when the same PDE was expressed in the cytoplasm, it prevented activation of RNase L and restored replication of MHV-NS2H126R to WT MHV levels (47). Additionally, our MHV studies were carried out in bone marrow–derived macrophages which express very high levels of OAS genes (17, 48) which may lead to increased activation of RNase L, and both antagonists may be necessary to prevent RNase L activation in a cell-type-dependent manner. These findings highlight the importance of studying the function of these proteins in the context of different coronavirus infections and different cell types.
Utilizing A549DPP4 KO cells, we found that abrogation of specific immune pathways can rescue viral attenuation of each of the double mutants to WT MERS-CoV levels. While PKR KO rescued only MERS-nsp15H231A/ΔNS4a replication, RNase L KO rescued only MERS-nsp15H231A/NS4bH182R, which was consistent with these recombinant viruses robustly activating each of these pathways. Interestingly, replication by both of these double-mutant viruses was rescued to WT MERS-CoV levels in MAVS KO cells, suggesting that the IFN signaling pathway is particularly important in limiting MERS-CoV replication. Indeed, in a MERS-CoV mouse model, early IFN induction after infection is protective against lethal pneumonia (43). Finally, there was no detectable enhancement of replication of WT MERS-CoV in the absence of MAVS (Fig. 5C), consistent with its almost complete shutdown of IFN production (Figs. 2 and 6). These observations underscore the importance of antagonism of these pathways by MERS-CoV, and provide further evidence that these host immune pathways play pivotal roles in limiting CoV replication in the airway.
In addition to NS4a and NS4b, the MERS-CoV genome encodes accessory proteins in ORF3 and ORF5. A mutant recombinant MERS-CoV with deletions in ORF3 through ORF5 was attenuated for replication in airway cells and induced increased IFN response and robust inflammation (49) as well as RNase L activation (22) consistent with our data herein. Moreover, the ORF5 encoded protein has been identified as a potential NFκB antagonist (30, 49). We hypothesize that deletion of ORF3 through ORF5 combined with inactivation of EndoU would severely attenuate virus replication and strongly induce innate immune responses.
There are many unanswered questions about interactions of virus with host innate immunity during infection by highly pathogenic coronaviruses. Different lineages of betacoronaviruses encode distinct groups of accessory proteins that confer differences in host–virus interactions. One example of homologous proteins in two different betacoronavirus lineages is the PDEs of merbecoviruses (NS2) and embecoviruses (NS4b); however, these proteins differ in that only NS4b contains an NLS and has a second function in host antagonism (21, 22, 28–31, 50). The SARS-CoV-2 genome encodes sarbecovirus lineage-specific accessory proteins encoded in ORF3a, ORF3b, ORF6, ORF7a, and ORF9b, which have all been reported to be IFN antagonists (51, 52). Most notably, the ORF6 encoded protein of SARS-CoV and SARS-CoV-2 blocks STAT1/STAT2 translocation into the nucleus, thus impairing induction of ISGs (53, 54). However, it is worth noting that many of these immune evasion activities of SARS-CoV-2 accessory proteins have largely been characterized in overexpression systems, and thus need to be confirmed in the setting of infection using viral mutants. Indeed, a mutant SARS-CoV-2 with a deletion of ORF6 is attenuated in a mouse model, supporting its role in host antagonism (55).
Additionally, unlike WT MERS-CoV, WT SARS-CoV-2 activates the dsRNA-induced innate immune pathways (IFN, PKR, and OAS/RNase L) and is restricted in replication by RNase L (18). We hypothesize that loss of EndoU activity during SARS-CoV-2 infection may lead to more robust activation of these pathways and detrimental effects on replication and pathogenesis than mutation of EndoU alone during MERS-CoV infection did. This hypothesis is currently under investigation. Given its high conservation across CoV lineages, EndoU activity of nsp15 is an attractive antiviral drug target for current and future highly pathogenic coronaviruses.
Materials and Methods
Recombinant Viruses.
Recombinant MERS-CoV viruses were made using lambda red recombination with the MERS-CoV BAC as previously described (37, 56, 57). The following primers were made to generate the mutations (bolded) to create the recombinant viruses:
MERS-nsp15H231A:
F 5′-GTGATGTTTTCATTAAGAAGTATGGCTTGGAAAACTATGCTTTTGAGGCCGTAGTCTATGGAGACTT-3′
R 5′-CGCCTAACGTAGTATGAGAGAAGTCTCCATAGACTACGGCCTCAAAAGCATAGTTTTCCA-3′
MERS-ΔNS4a: (amino acids 11 and 12 replaced by stop codons)
F 5′-GAACTCTATGGATTACGTGTCTCTGCTTAATCAAATTTGATAGAAGTACCTTAACTCACC-3′
R 5′-TGTACAAACAAGTAGTATACGGTGAGTTAAGGTACTTCTATCAAATTTGATTAAGCAGAG-3′
MERS-NS4bH182R:
F 5′-GTTCAGGGATTTTCCCTTTACCATAGTGGCCTCCCTTTACGTATGTCAATCTCTAAATTG-3′
R 5′-GTAACATCATCCAGTGCATGCAATTTAGAGATTGACATACGTAAAGGGAGGCCACTATGG-3′
MERS-nsp15H231A/ΔNS4a: the same mutations as above combined on one BAC
MERS-nsp15H231A/NS4bH182R: the same mutations as above combined on one BAC
All amplified KanI using the following additions onto the above primers:
F 5′-AGGATGACGACGATAAGTAGGG-3′ R 5′-GCCAGTGTTACAACCAATTAACC-3′.
Passage 0 (P0) viruses were recovered by Invitrogen Lipofectamine 2000 transfection reagent on African Green Monkey VeroCCL81 cells using 1.25 μg of BAC DNA. Cells were monitored for cytopathic effect, and cells and supernatant were harvested 5 d to 7 d after transfection. P0 virus was freeze thawed, and cells were removed by centrifugation and passaged onto Vero CCL81 cells to generate P1 stock. A P2 stock was generated by using a low MOI (0.05) of P1 stock and infecting Vero CCL81 cells, freeze thawed to harvest, cells were removed, and supernatant was used as the stock virus for experiments. Quantification of infectious virus was done by standard viral plaque assay on Vero CCL81 cells (18). WT and mutant MERS-CoV genomes were fully sequenced by Illumina sequencing of total RNA from highly infected cell samples. Consensus sequences for each virus were generated and aligned with the MERS-CoV HCoV-EMC/2012 reference genome (National Center for Biotechnology Information Reference Sequence: NC_019843.3) All mutant viruses were found to have the correct engineered mutations.
Sindbis virus Girdwood (G100) (SINV) was obtained from Mark Heise, University of North Carolina, Chapel Hill, NC, and prepared as previously described (58). Sendai virus was obtained from Carolina Lopez, Washington University in St. Louis, St. Louis, MO.
Cell Lines.
Vero CCL81 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) + 10% fetal bovine serum (FBS), sodium pyruvate, and Hepes. Human A549DPP4 cells were cultured in RPMI 1640 (Gibco catalog # 11875) supplemented with 10% FBS and penicillin-streptomycin. Human Calu-3 cells (clone HTB-55) were cultured in mininum essential media (MEM) supplemented with 20% FBS without antibiotics. A549DPP4 and A549DPP4 RNASEL KO cells were previously described (21). A549DPP4 PKR KO cells were constructed using the same Lenti-CRISPR system and guide RNA sequences as previously described (11, 59). A549DPP4 MAVS KO cells were constructed using the same Lenti-CRISPR system as previously described (59) but with the following guide RNAs: sgMAVS-2 forward: 5′-CACCGGGGTCTCCTGGACAGGCATG-3′ and sgMAVS-2 reverse: 5′-AAACCATGCCTGTCCAGGAGACCCC-3′
Nasal ALI Cultures.
Primary epithelial cultures were derived from human nasal mucosal specimens acquired from residual clinical materials obtained during surgery performed through the Department of Otorhinolaryngology (Head and Neck Surgery) at the University of Pennsylvania after informed consent was obtained. Patients with history of systemic disease or on immunosuppressive medications were excluded as previously described (32). Acquisition and use of these nasal specimens have been approved by the University of Pennsylvania Institutional Review Board (protocol #800614). Cultures were grown and differentiated on transwell inserts as previously described (32). Once fully differentiated (∼6 wk postseeding), basal media was replaced and the cultures were washed apically with PBS three times prior to infection apically with MERS-CoV.
MERS-CoV Infections and Titration.
Viruses were diluted in serum-free DMEM and added to cells for adsorption for 1 h at 37 °C. Cells were washed three times with phosphate-buffered saline (PBS) (for growth curves and RNA, no washes for protein or IF samples) and fed with RPMI + 2% FBS for A549DPP4 or DMEM + 2% FBS for Vero CCL81; 150 μL of supernatant was collected at the times indicated and stored at −80 °C for titration by plaque assay on Vero CCL81 cells as previously described (18). All infections and virus manipulations were conducted in a biosafety level 3 laboratory using appropriate personal protective equipment and protocols.
IF and FISH Staining.
At indicated times post infection, cells were fixed with 4% paraformaldehyde for 30 min at room temperature. Cells were then washed three times with PBS and then dehydrated with increasing amounts of ethanol: 50%, 70%, and 100% for 5 min each. Samples were stored at 100% ethanol at −20 °C overnight. Cells were rehydrated with 70% and then 50% ethanol for 2 min each, washed with PBS once, and then permeabilized for 10 min with PBS + 0.1% Triton-X100. Cells were then blocked in PBS + 0.1% Triton-X100 and 2% bovine serum albumin (BSA) containing RNaseOUT (Invitrogen catalog #10777019) for 30 min at room temperature. Primary antibodies were diluted in block buffer and incubated at room temperature for 1 h. Cells were washed three times with PBS and then incubated at room temperature for 30 min with secondary antibodies diluted in block buffer. All steps following secondary antibody addition were carried out in the dark. Cells were washed three times with PBS. For IF only, nuclei were stained with DAPI diluted in PBS for 5 min at room temperature, and coverslips were mounted onto slides using Invitrogen ProLong Diamond Antifade Mounting reagent for analysis by widefield microscopy. For combined IF/FISH staining, cells were washed three times with PBS (as stated above) and then fixed again with 4% paraformaldehyde for 10 min at room temperature. Cells were washed with 2× standard saline citrate (SSC) for 5 min and then 5 min with FISH wash buffer (10% formaldehyde in 2× SSC). FISH probes complementary to the N gene of MERS-CoV (see list below) conjugated to CAL Fluor Red 610 (Stellaris custom order) were diluted 1:50 in nuclease-free water.
Probes used for imaging MERS-CoV N+ssRNA (5′ to 3):
TTTTGGATTACGTCCTCTAC; AGTGTTATTTGGTGCAGCTC; AAGCCCAGTGTACCAAGAG;
CCAGGTGGAAAGGTAAGAGG; TTGGCATTAAGAGGTACACC; AATTTTTCTGTCCTGTCTC;
CAGTTGCTTAATTCCATTCC;TCCAGTGTAGTAGAAGTACC;TTAACAGCCCGGAATGGGAG; ATGGACCCAAACGATGCCAT; TCCCAAAAGTTGAAGGAGCA; AATCATTGTTAGGGTTCCG;
GGGCGCGAATTGTGTAACAA; CCCTCAATGTGGAAGTTTTT;TGATTGACTATTGCCTCCAG;
TTAAGCTAGAGGCTCTTGAA; CTAGATCTGGAAGAGTTTCT; AGATGGACCTGGAGAAGTG;
AAGTAGATCACCTCCTACTG;GCTTGTAGTCTGTTCAGAAG;CTTTACTTTGCCAGACTCAA; TGATTACTTTTGGCTGCGAT; AACTTTTGGTGGAAGTGCGC; AGACCAAAAGCTTGCACCA;
GAAGATCACCAAAGTTTCCC; CAGTGCCGAGTTTATTCAA; TCAGCAATTTGGGGCCAACG;
AAAAGCACTGGCTGTAGGAG; GTTTAAATTGCGACATACCC; CATCATTGTTCTGATGGGT;
ACCGAAGGAAGTACACAGGG; TCAAGTTTAATGGCTCCACT; GGTTCAGACATTTGGTCTG;
TGAGTGATGCTACCTTGCAC;AACACTTGGACGGGTGCGAG; ACATCAATCATTGGACCAGG.
Probes were diluted 1:50 in hybridization buffer (10% wt/vol dextran sulfate in 10% formaldehyde in 2× SSC) and incubated with samples overnight at 37 °C in a humidity chamber. Samples were washed with FISH wash buffer for 30 min at 37 °C, nuclei were stained with DAPI diluted in FISH wash buffer for 5 min at room temperature, and then cells were washed for 5 min with 2× SSC. Coverslips were mounted onto slides as stated above. The dsRNA was detected using commercial monoclonal antibody J2 (Scions) at 1:500, and nsp8 was detected using anti-nsp8 rabbit serum at 1:500 (obtained from Mark Denison, Vanderbilt University, Nashville, TN). Secondary antibodies were diluted 1:1,000, all highly cross-adsorbed IgG (H+L) from Invitrogen: goat anti-mouse AF488 (catalog #A11029) and goat anti-rabbit AF647 (catalog #A32733).
Widefield microscopy was done using either Nikon Eclipse Ti2 using a Nikon 20× objective and NikonDS-Qi1Mc-U3 12-bit camera or Nikon Eclipse Ti2 using a Nikon 20× objective and Hamamatsu digital camera C13440. Fiji was used for quantification of dsRNA: N+ssRNA staining was used to generate a mask by setting a threshold using the Otsu method. These thresholded images were used as a mask to mark infected ROIs. These ROIs, due to syncytia formation, marked infected areas of cells rather than singular cells. The MGV of the fluorescence signal of dsRNA or nsp8 was measured within each ROI across several images per infection condition. Five to seven fields of view at 20× with 1.5× zoom were analyzed for each condition. The ratio of MGV of dsRNA over MGV of nsp8 was recorded for each individual ROI and plotted using GraphPad Prism software.
Western Blotting.
Cells were washed once with ice-cold PBS, and lysates were harvested at indicated times post infection with lysis buffer (1% Nonidet P-40, 2 mM EDTA, 10% glycerol, 150 mM NaCl, 50 mM Tris HCl) supplemented with protease inhibitors (Roche: cOmplete mini EDTA-free protease inhibitor) and phosphatase inhibitors (Roche: PhosStop easy pack). After 5 min, lysates were harvested, incubated on ice for 20 min, and centrifuged for 20 min at 4 °C, and supernatants were mixed 3:1 with 4× Laemmli sample buffer. Samples were heated at 95 °C for 5 min, then separated on 4 to 15% SDS/PAGE, and transferred to polyvinylidene difluoride membranes. Blots were blocked with 5% nonfat milk or 5% BSA in TBST and probed with the antibodies as listed in SI Appendix, Table S1, diluted in the same block buffer. Blots were visualized using Thermo Scientific SuperSignal west chemiluminescent substrates (catalog #34095 or 34080). Blots were probed sequentially with antibodies and, in between antibody treatments, were stripped using Thermo Scientific Restore Western blot stripping buffer for 1 h at room temperature (catalog #21059).
qRT-PCR.
At indicated times post infection, cells were lysed with buffer RLT Plus (Qiagen RNeasy Plus #74136), and RNA was extracted following the prescribed protocol. RNA was reverse transcribed into complementary DNA (cDNA) with a High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems). The cDNA was amplified using specific qRT-PCR primers, iQ SYBR Green Supermix (Bio-Rad), and the QuantStudio 3 PCR system (Thermo Fisher). Fold changes in mRNA level compared to mock infected samples were calculated using the formula 2-Δ(ΔCt)(ΔCt = Ctgene of interest − Ct18S) and expressed as fold infected/mock-infected. Primer sequences are as listed in SI Appendix, Table S2.
Analyses of RNase L-Mediated rRNA Degradation.
RNA was harvested with buffer RLT plus (Qiagen RNeasy Plus #74136) and analyzed on an RNA chip with an Agilent Bioanalyzer using the Agilent RNA 6000 Nano Kit and its prescribed protocol (catalog #5067-1511).
Statistical Analysis.
Plotting of data and statistical analysis were performed using GraphPad Prism software (GraphPad Software, Inc.). Statistical significance was determined by comparing mutant viruses to WT MERS-CoV using repeated measures two-way ANOVA for viral replication curves and qRT-PCR and by one-way ANOVA for dsRNA quantification. Displayed significance is determined by P value, where * = P < 0.05; ** = P < 0.01; *** = P < 0.001; and **** = P < 0.0001; ns = not significant, and, in some figures, ns is not displayed on the graph.
Supplementary Material
Acknowledgments
We thank the members of the S.R.W. lab for feedback and discussion of this project, Joshua Hatterschide (University of Pennsylvania) for assistance with microscopy image analysis, Rudragouda Channappanavar (Oklahoma State University) for construction of MERS-nsp15H231A, Nicholas Parenti for performing the Bioanalyzer assays, Mark Heise (University of North Carolina, Chapel Hill) for SINV, and Carolina Lopez (Washington University in St. Louis) for Sendai virus. This work was supported by NIH Grants R01AI140442 (S.R.W.), P20 GM113117 (A.R.F.), K22 AI134993 (A.R.F.), P01AI060699 (S.P.), and RO1AI129269 (S.P.); Department of Veterans Affairs Merit Review 1-I01-BX005432-01 (N.A.C. and S.R.W.); the Penn Center for Research on Coronaviruses and Other Emerging Pathogens (S.R.W.); and start-up funds from the University of Kansas (A.R.F.). D.M.R. was supported, in part, by NIH grant T32 AI055400, and C.E.C. was supported, in part, by NIH grant T32 NS007180.
Footnotes
Competing interest statement: S.R.W. is on the Scientific Advisory Boards of Immunome, Inc and Ocugen, Inc. N.A.C. consults for GSK, AstraZeneca, Novartis, Sanofi/Regeneron, and Oyster Point Pharmaceuticals and has US patent “Therapy and Diagnostics for Respiratory Infection” (10,881,698 B2, WO20913112865) and a licensing agreement with GeneOne Life Sciences. A.R.F. consults with Deciphera Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123208119/-/DCSupplemental.
Data Availability
All data are included in the manuscript and/or SIAppendix .
References
- 1.Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D., Fouchier R. A., Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Khalafalla A. I., et al. , MERS-CoV in upper respiratory tract and lungs of dromedary camels, Saudi Arabia, 2013-2014. Emerg. Infect. Dis. 21, 1153–1158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reusken C. B. E. M., et al. , Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerg. Infect. Dis. 20, 1370–1374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wernery U., et al. , A phylogenetically distinct Middle East respiratory syndrome coronavirus detected in a dromedary calf from a closed dairy herd in Dubai with rising seroprevalence with age. Emerg. Microbes Infect. 4, e74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman V. M., et al. , Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 88, 11297–11303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geldenhuys M., et al. , A metagenomic viral discovery approach identifies potential zoonotic and novel mammalian viruses in Neoromicia bats within South Africa. PLoS One 13, e0194527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anthony S. J., et al. , Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. MBio 8, e00373-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth-Cross J. K., Bender S. J., Weiss S. R., Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 82, 9829–9838 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehwinkel J., Gack M. U., RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 20, 537–551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hur S., Double-stranded RNA sensors and modulators in innate immunity. Annu. Rev. Immunol. 37, 349–375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., et al. , Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. eLife 6, e25687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverman R. H., Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81, 12720–12729 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakrabarti A., et al. , RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe 17, 466–477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang R., Tang D., PKR-dependent inflammatory signals. Sci. Signal. 5, pe47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malathi K., Dong B., Gale M. Jr., Silverman R. H., Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448, 816–819 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou A., et al. , Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16, 6355–6363 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birdwell L. D., et al. , Activation of RNase L by murine coronavirus in myeloid cells is dependent on basal Oas gene expression and independent of virus-induced interferon. J. Virol. 90, 3160–3172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., et al. , SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A. 118, e2022643118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Dong B., Wei Z., Silverman R. H., Weiss S. R., Activation of RNase L in Egyptian Rousette bat-derived RoNi/7 cells is dependent primarily on OAS3 and independent of MAVS signaling. MBio 10, e02414-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kindler E., Thiel V., Weber F., Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv. Virus Res. 96, 219–243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comar C. E., et al. , Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. MBio 10, e00319-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thornbrough J. M., et al. , Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. MBio 7, e00258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa K., Narayanan K., Wada M., Makino S., Inhibition of stress granule formation by Middle East respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J. Virol., 92, 00902–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niemeyer D., et al. , Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 87, 12489–12495 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabouw H. H., et al. , Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog. 12, e1005982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siu K.-L., et al. , Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J. Virol. 88, 4866–4876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Yang Y., et al. , Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 5, 17554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu D. K. W., et al. , MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc. Natl. Acad. Sci. U.S.A. 115, 3144–3149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canton J., et al. , MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection. PLoS Pathog. 14, e1006838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y., et al. , The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 4, 951–961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews K. L., Coleman C. M., van der Meer Y., Snijder E. J., Frieman M. B., The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J. Gen. Virol. 95, 874–882 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman R. H., Weiss S. R., Viral phosphodiesterases that antagonize double-stranded RNA signaling to RNase L by degrading 2-5A. J. Interferon Cytokine Res. 34, 455–463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kindler E., et al. , Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 13, e1006195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng X., et al. , Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U.S.A. 114, E4251–E4260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ancar R., et al. , Physiologic RNA targets and refined sequence specificity of coronavirus EndoU. RNA 26, 1976–1999 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hackbart M., Deng X., Baker S. C., Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. U.S.A. 117, 8094–8103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fehr A. R., Bacterial artificial chromosome-based lambda red recombination with the I-SceI homing endonuclease for genetic alteration of MERS-CoV. Methods Mol. Biol. 2099, 53–68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.te Velthuis A. J., van den Worm S. H., Snijder E. J., The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 40, 1737–1747 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Genoyer E., López C. B., Defective viral genomes alter how Sendai virus interacts with cellular trafficking machinery, leading to heterogeneity in the production of viral particles among infected cells. J. Virol. 93, e01579-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohanski M. A., et al. , Solitary chemosensory cells are a primary epithelial source of IL-25 in patients with chronic rhinosinusitis with nasal polyps. J. Allergy Clin. Immunol. 142, 460–469.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee R. J., et al. , Bitter and sweet taste receptors regulate human upper respiratory innate immunity. J. Clin. Invest. 124, 1393–1405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee R. J., et al. , T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J. Clin. Invest. 122, 4145–4159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Channappanavar R., et al. , IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 129, 3625–3639 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velavan T. P., et al. , Host genetic factors determining COVID-19 susceptibility and severity. EBioMedicine 72, 103629 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng X., et al. , Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J. Virol. 93, e02000-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao L., et al. , Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11, 607–616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gusho E., et al. , Murine AKAP7 has a 2′,5′-phosphodiesterase domain that can complement an inactive murine coronavirus ns2 gene. MBio 5, e01312-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao L., et al. , Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J. Virol. 87, 8408–8418 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menachery V. D., et al. , MERS-CoV accessory ORFs play key role for infection and pathogenesis. MBio 8, e00665-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldstein S. A., et al. , Lineage A betacoronavirus NS2 proteins and the homologous torovirus Berne pp1a carboxy-terminal domain are phosphodiesterases that antagonize activation of RNase L. J. Virol. 91, e02201-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chazal N., Coronavirus, the king who wanted more than a crown: From common to the highly pathogenic SARS-CoV-2, is the key in the accessory genes? Front. Microbiol. 12, 682603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bai C., Zhong Q., Gao G. F., Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 65, 280–294 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frieman M., et al. , Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81, 9812–9824 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miorin L., et al. , SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U.S.A. 117, 28344–28354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silvas J. A., et al. , Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 human ACE2 transgenic mice. J. Virol. 95, e0040221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Almazán F., et al. , Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. MBio 4, e00650-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fehr A. R., et al. , The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 89, 1523–1536 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suthar M. S., Shabman R., Madric K., Lambeth C., Heise M. T., Identification of adult mouse neurovirulence determinants of the Sindbis virus strain AR86. J. Virol. 79, 4219–4228 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y., et al. , Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. U.S.A. 113, 2241–2246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and/or SIAppendix .