Significance
Although G-protein–coupled receptors (GPCRs) control vast physiological pathways, their activation remains chemically and physically enigmatic. Our osmotic stress studies of the visual receptor rhodopsin have redefined the standard model of GPCR signaling by revealing the essential role of bulk water. We show results consistent with a large number of water molecules flooding the rhodopsin interior during activation to stabilize the effector binding conformation. These results suggest a model of GPCR activation in which the receptor becomes solvent-swollen upon formation of the active state. We thus demonstrate the mechanism whereby water acts as a powerful allosteric modulator of a pharmacologically important membrane protein family.
Keywords: GPCR, osmotic stress, rhodopsin, sponge model, structural water
Abstract
The Rhodopsin family of G-protein–coupled receptors (GPCRs) comprises the targets of nearly a third of all pharmaceuticals. Despite structural water present in GPCR X-ray structures, the physiological relevance of these solvent molecules to rhodopsin signaling remains unknown. Here, we show experimental results consistent with the idea that rhodopsin activation in lipid membranes is coupled to bulk water movements into the protein. To quantify hydration changes, we measured reversible shifting of the metarhodopsin equilibrium due to osmotic stress using an extensive series of polyethylene glycol (PEG) osmolytes. We discovered clear evidence that light activation entails a large influx of bulk water (∼80–100 molecules) into the protein, giving insight into GPCR activation mechanisms. Various size polymer osmolytes directly control rhodopsin activation, in which large solutes are excluded from rhodopsin and dehydrate the protein, favoring the inactive state. In contrast, small osmolytes initially forward shift the activation equilibrium until a quantifiable saturation point is reached, similar to gain-of-function protein mutations. For the limit of increasing osmolyte size, a universal response of rhodopsin to osmotic stress is observed, suggesting it adopts a dynamic, hydrated sponge-like state upon photoactivation. Our results demand a rethinking of the role of water dynamics in modulating various intermediates in the GPCR energy landscape. We propose that besides bound water, an influx of bulk water plays a necessary role in establishing the active GPCR conformation that mediates signaling.
G-protein–coupled receptors (GPCRs) are essential cellular signal transducers involved in the progression of countless human diseases, making them attractive drug targets (1–5). In recent years, considerable attention has been devoted to structural studies of active and inactive conformations of GPCRs, occasionally with binding partners, by X-ray crystallography (6, 7) and cryogenic electron microscopy (cryo-EM) (8, 9). Such studies provide valuable information on key residues and structural motifs involved in several stages of GPCR signaling. At the same time, these structural studies are incomplete, reducing the dynamic conformational ensembles occupied by the various receptor states to a single representative structure selected by the crystallographic or cryogenic conditions (10, 11). However, GPCR activation is thought to be more complex than a binary or bimodal switch model (12), wherein the receptor adopts only on or off conformations modulated by the binding of a ligand (13). Alternative activation mechanisms of GPCRs involve a ternary complex model (14, 15) and an ensemble of states (16, 17) as described by an energy landscape model (ELM) (18–22). In addition, more recent studies provide evidence that the soft matter surrounding these receptors, including water and membrane lipids, plays crucial roles in shaping the energy landscape of GPCR activation (23–28).
Because crystallography and cryo-EM are conducted under dehydrating conditions, and because bulk-phase water molecules are inherently disordered, we cannot rely on these techniques alone to accurately depict hydration states of a protein molecule. As an example, we consider the case of bovine rhodopsin, the archetypal GPCR responsible for dim-light vision which adopts a dynamic equilibrium between two metastable intermediates—the inactive metarhodopsin-I (MI) and active metarhodopsin-II (MII)—in response to light (29). Although crystal structures are available for dark (30) and photoactivated MII (31) states of rhodopsin, both of these show very few internal water molecules. Such structures are insufficient to indicate the true rhodopsin-associated water volume and dynamics. We hypothesized that the metarhodopsin equilibrium following isomerization of the retinal ligand is hydration modulated, e.g., due to large-scale helical movements (29, 32) which are coupled to a flood of bulk water into the protein core (33, 34). Supporting this hypothesis is recent evidence from small-angle and quasi-elastic neutron scattering studies of rhodopsin (35–37), which describe the MII state as solvent-swollen (35). Additional computational (5, 38, 39) and hydroxyl radical methods (40) have suggested internal water pathways extending through the rhodopsin interior. Following Frauenfelder et al. (41), we connected water to a hierarchical energy landscape in which the protein dynamics involve transitions among different tiers or substates that are coupled (slaved) to the hydration shell or bulk water. To this end, we aimed to monitor changes to the metarhodopsin equilibrium upon selective perturbation of the water corresponding to the substates of the energy landscape. This strategy can be realized using force-based methods—hydrostatic pressure to probe core shell hydration (42, 43), and osmotic pressure to probe the bulk solvent (44, 45). Yet despite significant interest into the role of bound water in GPCR function (5, 46), there has been little such direct experimental evidence detailing the functional role of bulk water in GPCR activation.
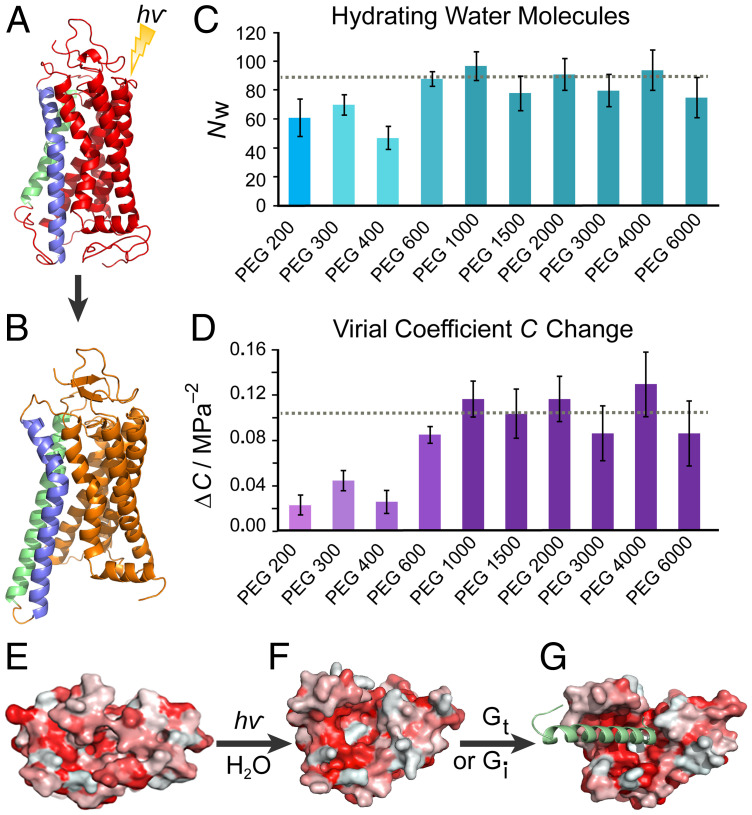
Here, we describe an experimental approach that provides complementary and essential new insights into the surprising role of bulk water in GPCR activation. To observe large-scale hydration-coupling interactions that are typically invisible to structural techniques such as X-ray crystallography (Fig. 1 A and B) or cryo-EM, we used an osmotic stress approach that directly shows a bulk influx of water stabilizing the active rhodopsin conformation (34, 47). A universal osmotic response is demonstrated for a range of excluded osmolytes, revealing that approximately 80–100 water molecules hydrate rhodopsin during MII formation, forcing the intracellular side of the receptor to open for transducin (Gt) binding (Fig. 1 C and D). We detail how our results may be reconciled with the alternate perspective of crowding and the parallel approach of using hydrostatic pressure. A picture is proposed where rhodopsin becomes solvent-swollen during activation (Fig. 1 E–G) to recruit G-protein binding. Finally, we point out that the activated state of the GPCR is not unlike a dynamic, hydrated sponge with substantial penetration of waters into the protein core.
Fig. 1.
Rhodopsin activation in lipid membranes entails a large influx of bulk water into the protein core. (A) Dark-state rhodopsin has a closed, dehydrated conformation of the 7-transmembrane helical bundle (30). (B) Open metarhodopsin-II conformation is adopted after retinal photoisomerization with outward-tilted TM6 (blue) and extended TM5 (green) helices following pH-dependent breakage of the Glu134–Arg135 salt bridge (31). (C) Large numbers of water molecules (~80–100) enter rhodopsin as determined by osmotic stress data for various-sized polyethylene glycols (PEGs). Smaller PEGs show smaller apparent water influx than more excluded large PEGs. The number of water molecules determined by the universal large osmolyte response is indicated by the dotted line. (D) Increase in virial coefficient ΔC for volume versus osmotic pressure accompanies rhodopsin photoactivation. Larger ΔC is determined in the exclusion limit of larger PEGs with the dotted line as the universal response. (E) Viewed from the intracellular side, dark-state rhodopsin is closed with residues colored by hydrophobicity (red, hydrophobic; white, hydrophilic). (F) The MII state shows an opened hydrophilic cavity (white) to accommodate the influx of bulk water which (G) widens as the C-terminal peptide of the G-protein (here Gi) is docked (8).
Results
Biophysical approach to measuring hydration changes in GPCRs.
We selected polyethylene glycol (PEG) as the primary osmolyte due to its relative inertness and ability to induce high osmotic pressures. Rhodopsin, the class-A namesake GPCR, provides a convenient case study due to its photoactive nature (SI Appendix, Fig. S1). Its photoactivation is typically portrayed by the following general reaction scheme (29, 48): that is, photon absorption triggers isomerization of the 11-cis retinal inverse agonist in dark-state rhodopsin (Rho) to all-trans, culminating in formation of the photoactivated intermediates MI and MII (29). Protonation of the conserved E(D)RY motif stabilizes the active MII conformation, to which the G-protein transducin can bind and which is differentiated from inactive MI by the deprotonated retinylidene Schiff base (31).
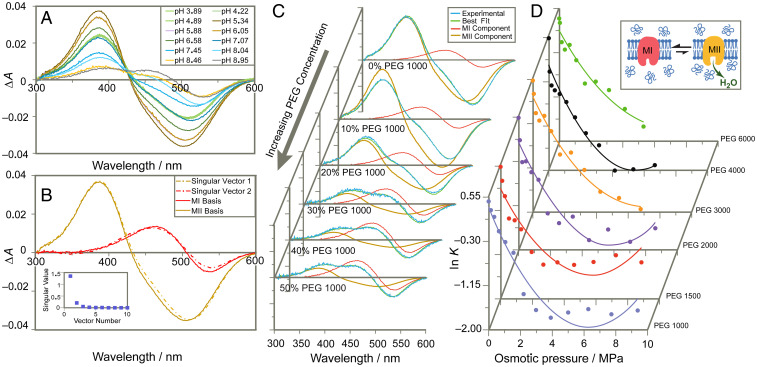
To directly monitor the fraction of deprotonated Schiff base corresponding to the MII state of rhodopsin in various osmotic conditions (49), we used ultraviolet (UV)–visible spectroscopy of rhodopsin in native retinal disk membranes (RDM) (48). Unlike osmotic pressure which governs movement of water into or out of a cell, we consider here the osmotic stress across a virtual membrane that defines the protein-water interface from which the osmotically active solute is excluded. Our experiment directly assays the difference in associated water volume of the rhodopsin molecule in various conformations versus the bulk solution (44). Quantification of relative MI and MII fractions relied on the technique of basis spectral fitting (48). Using this method, UV-visible difference spectra (light-activated minus dark state) for rhodopsin under MI and MII-favoring conditions were collected as basis spectra (Fig. 2 A and B). Linear combinations were fit to experimental difference spectra to determine the apparent equilibrium constant K = [MII]/[MI] and fraction of rhodopsin in the MII state θ in various equilibrated PEG environments (Fig. 2 C and D and SI Appendix, Fig. S2).
Fig. 2.
UV-visible difference spectroscopy reveals shift in equilibrium to inactive MI state upon osmotic dehydration. (A) Experimental difference UV-visible spectra of rhodopsin (light-activated minus dark state) show mixtures of inactive MI and active MII at varying pH conditions. (B) Singular-value decomposition yields two major singular vectors corresponding to experimental MI and MII basis spectra isolated by temperature and pH (pH 9.2 and 5 °C for MI, pH 5.0 and 15 °C for MII). (C) Increasing concentrations of osmolyte PEG 1000 shift spectrum toward MI component. (D) For large-PEG osmolytes the logarithm of the metarhodopsin equilibrium constant (K = [MII]/[MI]) varies approximately quadratically with applied osmotic pressure yielding the change in hydration and virial coefficient C for rhodopsin activation under standard-state conditions.
Thermodynamic formulation of osmotic pressure in terms of protein hydration.
Osmotic pressure Π is expected to behave similarly to hydrostatic pressure P by altering the molar Gibbs free energy associated with a protein conformation where . Because the molar hydration volume is itself dependent on osmotic pressure, we may express it as the virial expansion for constant temperature T, where B, C, etc. are virial coefficients characteristic of a specific hydrated protein state. Upon integrating with respect to Π and considering the MI to MII transition, we can derive the standard change in Gibbs free energy perturbed by osmotic pressure, , where ΔG° is the standard free energy change under zero osmotic pressure (see SI Appendix). Notably, the first-order term gives the change in protein hydration volume ΔV° between MI and MII under standard-state (zero osmotic pressure) conditions. We may then relate the change in free energy to the apparent equilibrium constant K = [MII]/[MI] by ΔG° = −RT ln K, so that the logarithm of the equilibrium constant becomes a second-degree polynomial of Π:
| [1] |
where Ko is the unperturbed equilibrium constant. Experimentally, we observe that large molar mass polymer osmolytes universally back shift the metarhodopsin equilibrium to the MI state, where the linear dependency is proportional to the MI–MII volume change in accordance with Eq. 1. Curvature in the empirical relationship between osmotic pressure and ln K may indicate a change in compressibility of the hydrated protein volume, or increasing quinary interactions of the polymers with the hydrophilic domains of the receptor during the MI to MII transition (Fig. 2D) (50). Fitting the data with a quadratic function according to Eq. 1 yields the number of hydrating water molecules per mole rhodopsin NW under standard-state conditions, based on the partial molar volume of water , and using the relation for the MI–MII transition.
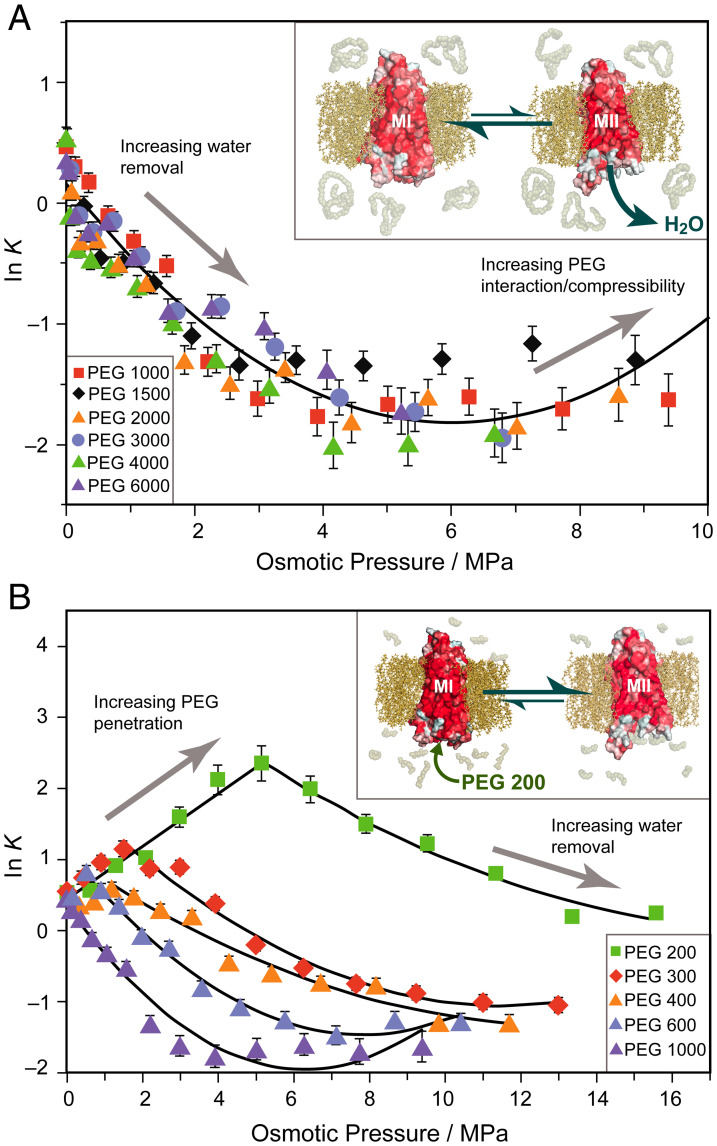
For each large PEG osmolyte with relative molar mass (Mr) between 1,000 and 6,000 Da, an increase of ∼80–100 hydrating water molecules was calculated for the MI–MII transition from the initial decrease of ln K with osmotic pressure (Figs. 1C and 2D). This number is much greater than the few structural waters typically seen in protein crystallography (51). Similarly, based on the curvature of the osmotic stress dependencies for each size osmolyte (Figs. 1D and 2D), we calculate virial coefficient changes ΔC on the order of 0.1 MPa−2, estimated to be equivalent to changes in osmotic compressibility of ∼0.01 MPa−1. The universal response of the metarhodopsin equilibrium to applied osmotic pressure obtained for various large PEGs (Fig. 3A) strongly implies the effects are colligative and not due to specific interactions with the protein. Although PEG osmolytes also increase sample viscosity, this property of solutions is a descriptor of kinetics, whereas here we consider the thermodynamic equilibrium which appears stable over the course of the experiment. Furthermore, if viscosity were the primary factor at work, we should not expect such drastically different results between small PEGs and large PEGs, which are similarly effective viscogens. From the osmolyte response, we determine a total number of ∼80–100 water molecules entering the rhodopsin interior under standard-state conditions. These results are consistent with Fourier-transform infrared data suggesting increased water accessibility in the MII state (52, 53). Similarly, they correspond to the ∼70 water molecules modeled by μs-scale molecular dynamics simulations that flood rhodopsin following induced retinal isomerization from the dark state (38), exhibiting microscopic reversibility (54) with no approximations of additional disorder (55).
Fig. 3.
Large and small molar mass osmolytes affect rhodopsin differently and show emergence of a universal trend for excluded polymers. (A) Natural logarithm of the MI–MII equilibrium constant (K = [MII]/[MI]) has approximately second-order relationship to osmotic pressure of large PEGs. A universal trend arises for PEGs of Mr between 1,000 and 6,000 Da with the linear term proportional to change in hydration. Inset: Metarhodopsin equilibrium is shifted to the MI (closed) state by large polymer osmolytes which are entropically excluded and dehydrate the protein. (B) Initially, small osmolytes (PEG 200–PEG 600) forward shift ln K to the MII state. A saturation effect is observed beyond which the equilibrium is back shifted to MI resembling the large osmolyte behavior. As PEG size increases, the trend behaves more like the universal behavior. Inset: Small osmolytes such as PEG 200 penetrate the transducin binding cavity and stabilize the open active MII state until it is saturated with small PEGs.
Osmolyte effects on rhodopsin depend on exclusion from the hydrated protein core.
Polymers of differing molar mass face varying degrees of entropic exclusion from a protein cavity. Larger polymers have greater conformational entropy, making their confinement within a tightly closed cavity highly unfavorable in terms of the entropy reduction. The interface between the receptor and surroundings acts as a virtual Gibbs dividing surface analogous to a semipermeable membrane, allowing the movement of water but preventing the penetration of large polymer osmolytes. However, for smaller osmolytes, a different effect is expected. Due to their lower conformational entropy in solution, these osmolytes face a decreased entropic penalty for penetrating the hydrated protein interior. Hence, the Gibbs dividing surface is permeable to small osmolytes, allowing an equal chemical potential of water on both sides of the interface. By consequence, nonosmotic effects should prevail and modulate the receptor equilibrium for low concentrations of small PEGs. These trends have been observed previously in analogous experiments with small osmolytes, which give only a partial view of colligative effects on rhodopsin (56). Recent static structures of rhodopsin in complex with transducin or Gi solved by cryo-EM (8, 57) indicate a Gα-C-terminal volume of ∼1,500 Å3, equivalent to ∼5 molecules of PEG 200, while not even a single molecule of PEG 1500 (volume > 2,000 Å3) can be accommodated neglecting entropic confinement. Likewise, the 1,500-Å3 transducin terminus is equivalent in volume to ∼50 water molecules, contrasting the minimal core shell hydration water observed in the same structures (8, 57). Much of this water could be displaced by transducin as the Gα-C terminus forms specific interactions with residues in the rhodopsin interior (58).
Experimentally, the metarhodopsin equilibrium is found to behave differently in the presence of small PEGs (200–600 Da) as compared to large PEGs. Initial addition of small PEGs shifts ln K linearly to the MII state up to a critical saturation point (Fig. 3B and SI Appendix, Figs. S3–S5). Midsized PEGs achieve this critical value at lower concentrations and a lower maximum MII fraction compared to the smallest-Mr PEGs. In the higher-concentration regime above this critical value, the metarhodopsin equilibrium more closely resembles the large-Mr PEGs. Fitting this latter tendency of the osmotic pressure curve as the second component of a piecewise function, we calculated associated thermodynamic values for the MI–MII transition (i.e., the magnitude of the hydration event and change in the system virial coefficient). Initially, small osmolytes penetrate the transducin binding cavity of rhodopsin and stabilize the MII state through nonosmotic, chemical (quinary) interactions. As more osmolytes crowd the transducin binding cleft, they preclude the entry of additional polymer osmolytes as seen for large PEGs. Hence, osmotic effects become the dominant equilibrium-modulating forces beyond receptor saturation. The critical saturation point varies with polymer size, because smaller osmolytes are capable of greater packing inside rhodopsin and reach saturation at higher concentrations.
Our results further show that the apparent thermodynamic parameters of the MI–MII transition have an expected property of osmolyte saturation. With smaller PEGs, a slightly lower hydration volume is measured, combined with a substantially smaller virial coefficient change ΔC. Because small PEG replaces protein-associated water molecules in the low-concentration range, fewer waters are available for withdrawal from the protein in the osmotic regime, requiring higher osmotic pressures to generate similar volume reductions. Consequently, we find a less negative first-order term in the osmotic regime, and thus lower apparent hydration volume for the MI–MII transition for the small PEGs versus the better excluded large PEGs (Fig. 1C). In addition, the stabilizing effect of PEG penetration is observed in the smaller virial coefficient change ΔC for small PEGs: these small osmolytes penetrate and stabilize the protein in the lower-concentration range, decreasing the variability in properties such as osmotic compressibility for the high-concentration regime (Fig. 1D). Strongly interacting PEGs should experience fewer variations in number within rhodopsin compared to water, thereby reducing the stochastic volume fluctuations that may give a higher compressibility, or virial coefficient C, in the MII state.
Rhodopsin activation due to conserved ERY motif protonation is shifted depending on osmolyte size.
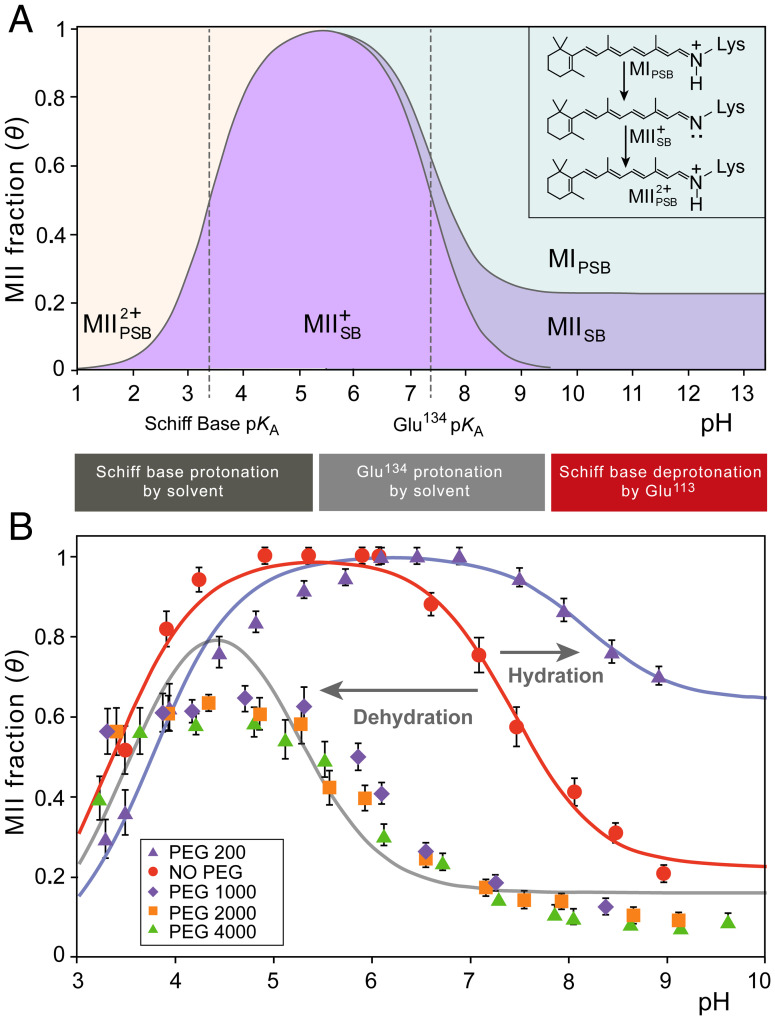
Another striking influence of PEG is its effect on the apparent pKA of Glu134 following deprotonation of the Schiff base and breakage of its ionic lock with Glu113 (31, 48, 59). Protonation of Glu134 within the conserved ERY motif couples to disrupting another ionic lock involving Arg135 and Glu247, which stabilizes the outward-tilted TM6 conformation of MII, or MIIH+ in the protonated case (48). Accordingly, the pH-dependent equilibrium between MI with protonated Schiff base (PSB) and the MII state with a deprotonated Schiff base (SB) is described by an extended titration curve. The apparent pKA of this curve is given by Glu134 (Fig. 4A) and is thought to be modulated by ionic strength and membrane surface potential (60) plus specific ion binding effects (61). Selectively stabilizing either the closed MI or open MII states impacts this pKA as well as the protein environment around the Schiff base (e.g., hydration within the protein). Apart from the pKA describing the rhodopsin activation, the pH titration of rhodopsin also demonstrates a nonzero alkaline endpoint at higher temperatures due to thermally activated MII substates (48). In an extended mechanism, the MII state corresponds to MIIa and MIIb substates, where both have a deprotonated Schiff base and absorb maximally at 380 nm even at high pH (48, 62). Another factor is the additional pKA under acidic conditions that reflects the protonation of the Schiff base in MIIH+ (63). Below this pKA, the apparent MII fraction determined by UV-visible spectroscopy decreases (Fig. 4A). The phenomenological pH-titration curve thus consists of two pKA values and an alkaline endpoint, which we model by an extended Henderson-Hasselbalch equation for the MII fraction θ (see SI Appendix for derivation):
| [2] |
Fig. 4.
Osmotic pressure induces large shifts of pH-dependent activation of rhodopsin. (A) Influences of pH on rhodopsin are described by phenomenological Henderson-Hasselbalch equation involving two pKA values and an alkaline endpoint. The states are distinguished by having a protonated or deprotonated Schiff base (PSB or SB, indicated by a subscript), while a superscript indicates the charge relative to MI. The lower pKA (designated pKA,SB) reflects pH-dependent protonation of the retinal Schiff base (SB), which lowers the apparent MII fraction detected by UV-visible spectroscopy. The higher pKA value (designated pKA,Glu) reflects protonation of Glu134 in the E(D)RY motif to stabilize the fully active MII conformation. The alkaline endpoint at higher pH corresponds to MII substates that persist at higher temperatures even when Glu134 is fully deprotonated. (B) Applied osmotic stress stemming from large osmolytes (50% wt/wt at T = 15 °C) back shifts the rhodopsin activation titration curve from pKA = 7.4 to 5.2. At 30% wt/wt PEG 200 (T = 15 °C) the titration curve is maximally forward shifted to a pKA of 8.2 favoring the active MII state. Also observed is an osmolyte effect on the alkaline endpoint: small osmolytes stabilize the open MII conformation even when Glu134 is fully deprotonated, increasing the alkaline endpoint, whereas dehydrating large osmolytes decrease the deprotonated MII population and thus the alkaline endpoint.
Here, θalk designates the alkaline endpoint (apparent MII fraction at high pH), while pKA,SB reflects the MIIH+ Schiff base protonation equilibrium and pKA,Glu reflects the Glu134 protonation step (Fig. 4A). Titration data for the apparent MII fraction of rhodopsin in solutions of four different large-Mr PEGs were used to determine the model parameters (Fig. 4B and SI Appendix, Fig. S6). These results revealed a striking pKA,Glu shift from 7.4 to 5.2 in the presence of 50% (wt/wt) large polymer osmolytes.
The pH titration curves for rhodopsin activation are analogous to pharmacological dose–response curves (39, 48) due to changing ligand binding affinity by 2–3 orders of magnitude. This observed pKA,Glu shift implies a model in which large osmolytes dehydrate the receptor to shift its equilibrium to the closed MI state, stabilizing the intact ionic lock and favoring a lower fraction of MII. An apparent MII fraction of unity is never achieved for these conditions, due in part to the MIIH+ Schiff base protonation region of the titration curve, which begins to overlap with the principal MI–MIIH+ region. Strikingly, large-Mr PEGs of different molar mass have identical behavior with a nearly equal pKA,Glu shift, thus reinforcing the universal osmotic effect by large polymers. Near the alkaline endpoint of the titration, pH values >9.5 induced base-catalyzed retinal hydrolysis following photoactivation, yielding the apoprotein opsin. A quantifiable effect of PEG on the alkaline endpoint was extrapolated where large polymer osmolytes lowered the alkaline endpoint. Our results indicate that large PEGs destabilize the more-open MII state under high-pH conditions where Glu134 is fully deprotonated, consistent with the pKA shift that reflects favorability of the closed MI state under osmotic pressure. For small PEGs, the opposite effect is observed, with forward shifting of the metarhodopsin equilibrium by intermediate concentrations of small PEGs (Fig. 4B). With a concentration of 30% (wt/wt) PEG 200, for example, the titration curve incurs a substantial pKA,Glu shift from 7.4 to 8.2 opposite to that observed for large PEGs. In total, there is a pKA,Glu difference of 3 units between the large and small PEG titration curves, showing the dramatic influence that osmolyte size poses for receptor hydration and proton transfer (64), with consequent activation. This magnitude of pKA shift is analogous to that observed for constitutive mutations of rhodopsin or retinoid antagonists (39, 65).
Discussion
Our findings recast water as an important player in the activation of GPCRs such as rhodopsin (34, 38). Using osmotic stress from large polymer osmolytes (e.g., PEG 1000–6000), we obtain results consistent with a large influx of ∼80–100 water molecules into rhodopsin during formation of the active MII state. These direct experimental results are supported by microsecond-scale molecular dynamics (MD) simulations (38, 39), as well as indirect evidence of small-angle neutron scattering (35) and radiolytic protein footprinting showing reorganization of internal waters during rhodopsin activation (40, 46). The osmotic stress methodology primarily investigates the larger volume bulk water associated with the protein, in contrast to experimental techniques which assay structural or bound water molecules (66). While previous hydroxyl radical footprinting experiments did not observe labeling from bulk water, this is possibly because of limited exchange between bound and bulk waters over the experimental timescale (40). In such experiments, tightly bound structural waters are better poised to label internal protein residues upon hydroxyl radical formation. Hence, we view the results as consistent with and complementary to our proposal that various classes of water affect GPCR activation. According to our interpretation, osmotic stress is a weak force that mainly acts on bulk water, whereas hydration shell or structural water is affected by hydrostatic pressure perturbation or hydroxyl radical labeling.
Although the protein hydration shell typically couples to more localized motions, here we are interested in the global dynamics accompanying GPCR activation (29, 32). Within the unified model of protein dynamics proposed by Frauenfelder et al. (41), these motions are coupled to bulk solvent movements. The molecular picture afforded by MD simulations provides a possible explanation for this functional hydration mechanism, which we refer to as a sponge model. Water penetration into the GPCR has been described as the formation of a continuous internal water channel upon receptor activation (38, 67), whereby bound water movements may be implicated in the GPCR allosteric link between the cytoplasmic face and ligand binding pocket (46, 68–70). While previous osmotic stress studies have suggested that MI is the more hydrated conformation based on only small (penetrating) osmolytes (56), here, we show these experimental results may be reconciled with the differing behaviors of large (excluded) cosolutes to arrive at the self-consistent interpretation of rhodopsin hydration upon MII formation. Correspondingly, we find an entropic stabilization by bulk water which creates a dramatic expansion of microstates in the hydrated active receptor, rather than a purely enthalpic stabilization of the GPCR active state by the hydration shell.
In view of our results, we propose a model of a hydration-mediated GPCR activation mechanism, which stands in stark contrast to the current understanding of the role of water in GPCR function. Our model directly challenges interpretations of internal water molecules gathered by X-ray crystallography for dark-state versus MII-state rhodopsin (30, 31), as well as osmotic stress data for small, penetrable osmolytes (56). We observe that rhodopsin activation is tightly coupled (slaved) to a bulk water influx, so that osmotic dehydration by large, excluded polymers inhibits GPCR activation. Small osmolytes with lower intrinsic conformational entropy generate an alternative, nonosmotic trend in modulating rhodopsin activation. They penetrate inside the receptor and stabilize the active (expanded) receptor state through interactions with the transducin binding cleft until reaching a quantifiable saturation point. In the limit of increasing osmolyte size, GPCR activation follows a universal dependence on osmotic stress, similar to that observed for ion channel opening (44).
Integration of osmotic pressure and hydrostatic pressure effects on metarhodopsin equilibrium.
We next show how further insights into GPCR activation are obtained by integrating osmotic pressure data with hydrostatic pressure effects. Both hydrostatic pressure and osmotic pressure backshift the metarhodopsin equilibrium to the preactive MI state, but for different reasons (71, 72). Because hydrostatic pressure entails a thermodynamically closed system, the perturbation detects the change in molar protein volume (i.e., density) of the system, but not changes in water molecule number. For rhodopsin, these changes in density can involve the collapsing or penetration of water into small protein cavities or voids, or alternatively a higher-density solvation shell versus the bulk (73). By contrast, osmotic pressure involves an open system, which is separated from the surroundings by a semipermeable Gibbs dividing surface (44). Hence, osmotic pressure involves changes to protein hydration, so that both methods are complementary to one another (74). By analogy to pressure effects on protein folding reactions (42, 75), MI can be seen as more tightly packed, which minimizes void volume or forces small amounts of solvent into internal cavities (42, 75–78). Conversely, in MII there may be an increase in number of water molecules, giving a solvent-swollen state (35, 37). While the active MII state is less dense (71, 72), it is also more hydrated than its precursor MI state, with greater penetration of water molecules into the protein (38, 54). The MII state is less tightly packed in its formation of a large internal cavity for G-protein binding, which in tandem with the high-density hydration shell accommodates the larger hydration volume. Water molecules previously forced into restricted MI cavities may find greater entropic stabilization in the larger, nascent hydration cavity of MII. The two force-based methods can thus be unified to give a more comprehensive picture of the rhodopsin changes in a hydrated lipid membrane upon light activation (39).
Osmotic pressure informs volumetric fluctuations and cosolute quinary interactions in the active GPCR state.
Additional information regarding GPCR activation is obtained from the dependence of the active MII fraction on osmotic pressure. Modeled as changes in the virial coefficient ΔC, the curvature term may result from an apparent change in compressibility of the hydrated protein (see SI Appendix), related to increasing volume fluctuations in the dynamic, hydrated conformation of the active receptor (37). Analogous compressibility-governed behavior of enzyme-catalyzed equilibria has been found in the case of large hydrostatic pressures (43). For rhodopsin under osmotic stress, we could gather from the sign of the second-order term an increase in osmotic compressibility, , which is related to fluctuations of the hydration volume V of the protein system (79). Hence, a greater osmotic compressibility in the solvent-swollen active MII state indicates greater volumetric fluctuations, consistent with a hydrated sponge-like state having variations in how water packs within the protein. Greater fluctuations in the active-state GPCR would indicate a greater number of available microstates for the entropy-stabilized active receptor, reflecting similar observations by quasi-elastic neutron scattering (37).
Alternatively, the change in virial coefficient may be explained by competing quinary interactions of the large polymers with the active MII state (80). Similar to the interactions with low concentrations of small PEGs that drive the equilibrium forward to MII (39, 47), large PEGs may also experience chemical, or quinary interactions with the protein, albeit to a lesser extent (80). As the concentration of polymer osmolytes in the surrounding bulk solvent nears its saturating limit, the entropic penalty for osmolyte penetration and interaction with the receptor is reduced, yielding polymer stabilization of the active GPCR that cancels the osmotic effect. A combination of both specific quinary interactions and compressibility changes between the two states may likely be involved in the ΔC apparent curvature term. As a second-order approximation, our model of the deviation from linearity may exclude higher-order terms describing either the changes in compressibility or quinary interactions with the polymer osmolyte (see SI Appendix). Although not trivial to mathematically separate these terms, both types of contributions qualitatively confirm the essential behavior expected of a dynamic and solvent-swollen active receptor conformation.
Osmotic stress effects are a direct assay of hydration changes and are compatible with cosolute crowding models.
Thus far, we have considered the introduction of polymer cosolutes purely from the viewpoint of osmotic stress, which is a consequence of the Gibbs-Duhem equation for systems in equilibrium and directly assays hydration states (45). Naturally arising osmotic effects of cosolutes on macromolecules can also be recovered from popular alternative perspectives such as crowding. In this framework, large cosolutes exert steric forces on macromolecules to restrict their volume occupancy (81, 82). Due to exclusion from the macromolecular environment, these particles create depletion forces that attract multiple proteins or structural motifs together (83, 84), which are themselves a consequence of osmotic stress (84). Steric interactions that constrict volume in crowded environments have a first-order effect on the molar free energy of the macromolecule according to , where M denotes the molar concentration of perfectly excluded polymer solutes and is the molar hydration volume as before (82). Using the van’t Hoff law of ideal solutions , a first-order approximation for osmotic pressures, we can recast this expression to the equivalent osmotic stress relation, . Thus, crowding and osmotic stress effects are mathematically equivalent to first order. It is likely that higher-order terms in either picture can be reconciled, although computationally challenging to demonstrate due to significant overlap of the two approaches (45). While either framework may be applicable to a particular situation, we show here the utility of the osmotic stress approach toward directly measuring hydration changes of rhodopsin as an archetypal GPCR.
Evidence for a hydration-modulated energy landscape model of GPCR activation.
Our discoveries enlarge the scope of the role that soft matter plays in key biological phenomena. Already having detailed the influences of membrane curvature stress in GPCR function (25, 28), we now describe an equally powerful biological influence in the form of the bulk solvent (85). It is important to appreciate the fundamental distinction between these two: the osmotic stress effect must be a result of protein dehydration apart from osmotic dehydration of the lipid bilayer. Membrane dehydration alone would produce an effect opposite to that observed in this study, by decreasing the area per lipid and increasing the membrane thickness, which are known to favor active MII (23, 28, 63). Chemical interactions of PEG with lipids are thought to be relatively weak or nonspecific (86) and they promote negative spontaneous curvature of lipid monolayers (87), thereby also tending to favor MII unlike the current results. Consequently, both effects must be weaker than the osmotic stress effect on the protein itself that we observe experimentally. Our findings offer insights into how GPCRs, like rhodopsin, produce their high-fidelity rapid signal amplification. To achieve this biological outcome, there must be ways to allosterically modulate the binding affinity of the G-protein at different stages of its cycle (88–90). For rhodopsin-based signaling, the occurrence of a hydrated, sponge-like active state is consistent with an energy landscape mechanism (ELM) for receptor activation. The thermodynamic parameters are in accord with a partial unfolding of the receptor protein in analogy to hydrostatic-pressure effects on globular proteins. None of these conclusions can be deduced from X-ray crystal structures or cryo-EM structures alone, thus requiring physicochemical studies such as force-based methods as described herein. The hydration-activation mechanism has natural implications in GPCR pharmacology and drug design.
To summarize, we have experimentally shown the existence and functional role of a large hydration event coupled to activation of the GPCR rhodopsin. Accordingly, we propose a hydration-mediated sponge-like mechanism with potential implications for biological signal amplification. The GPCR activation is but one event in a sequence of signal transduction, the next being the binding of the G-protein and receptor-catalyzed exchange of GTP for GDP. Interestingly, hydration plays a key role in lipid bilayer dynamics that could possibly also have crucial implications for GPCRs like rhodopsin. Cooperativity of membrane lipids and hydration can potentially affect the energy landscape of GPCR activation. Further studies of hydration effects on GTP-GDP exchange may present important clues as to the complete mechanism of receptor-catalyzed G-protein binding and release. Whether the water interacts synergistically with lipids to affect receptor function, and whether it regulates the binding and unbinding of the G-protein transducin, remain as intriguing questions.
Materials and Methods
Retinal disk membrane purification.
Retinal disk membranes (RDM) were isolated from bovine retinas and characterized by UV-visible spectroscopy as previously described (SI Appendix, Fig. S1) (91). The RDM were suspended with concentrations of 10 μM rhodopsin in 67 mM bis-Tris propane (BTP) buffer with 130 mM NaCl and 2 mM MgCl2 at pH 7.5. The osmolal contributions of the buffer and salts were assumed to be negligible in comparison to the PEG osmotic pressure. PEG was added in 0–60% wt/wt concentrations and lightly sonicated with the RDM for equilibration (SI Appendix, Fig. S2).
UV-visible spectroscopy and data analysis.
Absorption spectra for suspensions of RDM containing 10 μM of rhodopsin plus the osmolyte were recorded with a Varian Cary-50 UV-visible spectrophotometer (Palo Alto, CA). Throughout the experiment, the temperature was kept constant at 15 °C. A 1-mm pathlength cuvette placed close to the detector was used to minimize light scattering. The UV-visible absorption spectra were collected from 270 to 650 nm for both the dark-state protein and rhodopsin exposed to a green LED (528 nm) actinic light source for 7 s. A scattering correction was performed using the equation to account for scattering changes after bleaching. Here, ΔAcorr is the corrected amplitude of the difference spectrum, ΔA is the experimental difference amplitude, a is the fitting constant, λ is the wavelength, and b is an additive constant to correct for baseline shifts (92). The fraction of photoactivated protein in the MII state (θ) was calculated by fitting the experimental difference spectrum by a linear combination of MI and MII basis spectra using least-squares regression. Here, the coefficients c1 and c2 of each basis vector component yield the relative concentrations of either species. The MI basis spectrum was recorded for RDM suspended in pH 9.2 buffer at 5 °C, while the MII basis spectrum was recorded for RDM suspended in pH 5.0 buffer at 15 °C. Spectral lineshapes of the basis spectra were confirmed mathematically through singular value decomposition of pH titration spectra in multiple osmotic environments.
Calculation of number of water molecules.
Osmotic pressure values were calculated from universal fitting curves to model the experimental osmotic pressure data (93) as reported in Cohen et al. (49). The number of waters involved in the rhodopsin activation process was calculated from the dependency of ln K on osmotic pressure (44). A second-order polynomial (Eq. 1) was fit to the osmotic regimes of the data by a nonlinear least-squares algorithm using MATLAB R2018a (for small PEGs, including those curves in the decreasing regions above the identified saturation point). A similar approach was used in fitting the extended Henderson-Hasselbalch curve (Eq. 2) describing the pH dependence of the rhodopsin activation.
Supplementary Material
Acknowledgments
We thank J. Somers, G. Fitzwater, A. Amihan, and C. Webber for assistance with sample preparation and data collection, K. Bao for constructing the actinic LED light source, and D. Harries, A. Parsegian, and H. Petrache for discussions. A.V.S. acknowledges partial support from St. Petersburg State University (51142660). This research was supported by grants to M.F.B. from the NIH (R01EY026041) and NSF (MCB 1817862 and CHE 1904125).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117349119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. Experimental data can be accessed at the University of Arizona Research Data Repository (DOI: 10.25422/azu.data.19686555).
References
- 1.Stevens R. C., et al. , The GPCR Network: A large-scale collaboration to determine human GPCR structure and function. Nat. Rev. Drug Discov. 12, 25–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latorraca N. R., Venkatakrishnan A. J., Dror R. O., GPCR dynamics: Structures in motion. Chem. Rev. 117, 139–155 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Hilger D., Masureel M., Kobilka B. K., Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 25, 4–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weis W. I., Kobilka B. K., The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 87, 897–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkatakrishnan A. J., et al. , Diverse GPCRs exhibit conserved water networks for stabilization and activation. Proc. Natl. Acad. Sci. U.S.A. 116, 3288–3293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen S. G. F., et al. , Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang Y., et al. , Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang Y., et al. , Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 558, 553–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du J., et al. , Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature 594, 589–593 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Srivastava A., Nagai T., Srivastava A., Miyashita O., Tama F., Role of computational methods in going beyond X-ray crystallography to explore protein structure and dynamics. Int. J. Mol. Sci. 19, 3401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renaud J.-P., et al. , Cryo-EM in drug discovery: Achievements, limitations and prospects. Nat. Rev. Drug Discov. 17, 471–492 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Park P. S. H., Lodowski D. T., Palczewski K., Activation of G protein-coupled receptors: Beyond two-state models and tertiary conformational changes. Annu. Rev. Pharmacol. Toxicol. 48, 107–141 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leff P., The two-state model of receptor activation. Trends Pharmacol. Sci. 16, 89–97 (1995). [DOI] [PubMed] [Google Scholar]
- 14.De Lean A., Stadel J. M., Lefkowitz R. J., A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled beta-adrenergic receptor. J. Biol. Chem. 255, 7108–7117 (1980). [PubMed] [Google Scholar]
- 15.Rovati G. E., et al. , The DRY motif and the four corners of the cubic ternary complex model. Cell. Signal. 35, 16–23 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Hofmann K. P., et al. , A G protein-coupled receptor at work: The rhodopsin model. Trends Biochem. Sci. 34, 540–552 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Kenakin T., Theoretical aspects of GPCR-ligand complex pharmacology. Chem. Rev. 117, 4–20 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Bhattacharya S., Vaidehi N., Computational mapping of the conformational transitions in agonist selective pathways of a G-protein coupled receptor. J. Am. Chem. Soc. 132, 5205–5214 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Deupi X., Kobilka B. K., Energy landscapes as a tool to integrate GPCR structure, dynamics, and function. Physiology (Bethesda) 25, 293–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Struts A. V., Salgado G. F. J., Brown M. F., Solid-state 2H NMR relaxation illuminates functional dynamics of retinal cofactor in membrane activation of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 108, 8263–8268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alhadeff R., Vorobyov I., Yoon H. W., Warshel A., Exploring the free-energy landscape of GPCR activation. Proc. Natl. Acad. Sci. U.S.A. 115, 10327–10332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harpole T. J., Delemotte L., Conformational landscapes of membrane proteins delineated by enhanced sampling molecular dynamics simulations. Biochim. Biophys. Acta 1860, 909–926 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Soubias O., Gawrisch K., The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim. Biophys. Acta 1818, 234–240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chawla U., et al. , A usual G-protein-coupled receptor in unusual membranes. Angew. Chem. Int. Ed. Engl. 55, 588–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown M. F., Soft matter in lipid-protein interactions. Annu. Rev. Biophys. 46, 379–410 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Lorent J. H., et al. , Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 8, 1219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turoverov K. K., et al. , Stochasticity of biological soft matter: Emerging concepts in intrinsically disordered proteins and biological phase separation. Trends Biochem. Sci. B 44, 716–728 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Fried S. D. E., et al. , Membrane curvature revisited–the archetype of rhodopsin studied by time-resolved electronic spectroscopy. Biophys. J. 120, 440–452 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada T., Ernst O. P., Palczewski K., Hofmann K. P., Activation of rhodopsin: New insights from structural and biochemical studies. Trends Biochem. Sci. 26, 318–324 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Okada T., et al. , The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 342, 571–583 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Choe H. W., et al. , Crystal structure of metarhodopsin II. Nature 471, 651–655 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Altenbach C., Kusnetzow A. K., Ernst O. P., Hofmann K. P., Hubbell W. L., High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl. Acad. Sci. U.S.A. 105, 7439–7444 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown M. F., Chawla U., Perera S. M., Struts A. V., Role of membrane lipids in activating G-protein-coupled receptors. Biophys. J. 106, 434a (2014). [Google Scholar]
- 34.Chawla U., Perera S. M. D. C., Struts A. V., Pitman M. C., Brown M. F., Hydration mediated G-protein-coupled receptor activation. Biophys. J. 110, 83a (2016). [Google Scholar]
- 35.Perera S. M. D. C., et al. , Small-angle neutron scattering reveals energy landscape for rhodopsin photoactivation. J. Phys. Chem. Lett. 9, 7064–7071 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Perera S. M. D. C., Chawla U., Brown M. F., Powdered G-protein-coupled receptors. J. Phys. Chem. Lett. 7, 4230–4235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shrestha U. R., et al. , Quasi-elastic neutron scattering reveals ligand-induced protein dynamics of a G-protein-coupled receptor. J. Phys. Chem. Lett. 7, 4130–4136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leioatts N., et al. , Retinal ligand mobility explains internal hydration and reconciles active rhodopsin structures. Biochemistry 53, 376–385 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chawla U., et al. , Activation of the G-protein-coupled receptor rhodopsin by water. Angew. Chem. Int. Ed. Engl. 60, 2288–2295 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Angel T. E., Gupta S., Jastrzebska B., Palczewski K., Chance M. R., Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 106, 14367–14372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frauenfelder H., et al. , A unified model of protein dynamics. Proc. Natl. Acad. Sci. U.S.A. 106, 5129–5134 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Royer C. A., Revisiting volume changes in pressure-induced protein unfolding. Biochim. Biophys. Acta 1595, 201–209 (2002). [DOI] [PubMed] [Google Scholar]
- 43.McCoy J., Hubbell W. L., High-pressure EPR reveals conformational equilibria and volumetric properties of spin-labeled proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 1331–1336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parsegian V. A., Rand R. P., Rau D. C., Macromolecules and water: Probing with osmotic stress. Methods Enzymol. 259, 43–94 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Parsegian V. A., Rand R. P., Rau D. C., Osmotic stress, crowding, preferential hydration, and binding: A comparison of perspectives. Proc. Natl. Acad. Sci. U.S.A. 97, 3987–3992 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Angel T. E., Chance M. R., Palczewski K., Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 106, 8555–8560 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fried S. D. E., et al. , G-protein-coupled receptor activation mediated by internal hydration. Biophys. J. 116, 207a (2019). [Google Scholar]
- 48.Mahalingam M., Martínez-Mayorga K., Brown M. F., Vogel R., Two protonation switches control rhodopsin activation in membranes. Proc. Natl. Acad. Sci. U.S.A. 105, 17795–17800 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen J. A., Podgornik R., Hansen P. L., Parsegian V. A., A phenomenological one-parameter equation of state for osmotic pressures of PEG and other neutral flexible polymers in good solvents. J. Phys. Chem. B 113, 3709–3714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knowles D. B., et al. , Chemical interactions of polyethylene glycols (PEGs) and glycerol with protein functional groups: Applications to effects of PEG and glycerol on protein processes. Biochemistry 54, 3528–3542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huber T., Botelho A. V., Beyer K., Brown M. F., Membrane model for the G-protein-coupled receptor rhodopsin: Hydrophobic interface and dynamical structure. Biophys. J. 86, 2078–2100 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishimura S., Sasaki J., Kandori H., Lugtenburg J., Maeda A., Structural changes in the lumirhodopsin-to-metarhodopsin I conversion of air-dried bovine rhodopsin. Biochemistry 34, 16758–16763 (1995). [DOI] [PubMed] [Google Scholar]
- 53.Rath P., DeGrip W. J., Rothschild K. J., Photoactivation of rhodopsin causes an increased hydrogen-deuterium exchange of buried peptide groups. Biophys. J. 74, 192–198 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng J., Brown M. F., Mertz B., Retinal flip in rhodopsin activation? Biophys. J. 108, 2767–2770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martínez-Mayorga K., Pitman M. C., Grossfield A., Feller S. E., Brown M. F., Retinal counterion switch mechanism in vision evaluated by molecular simulations. J. Am. Chem. Soc. 128, 16502–16503 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Mitchell D. C., Litman B. J., Effect of protein hydration on receptor conformation: Decreased levels of bound water promote metarhodopsin II formation. Biochemistry 38, 7617–7623 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Gao Y., et al. , Structures of the rhodopsin-transducin complex: Insights into G-protein activation. Mol. Cell 75, 781–790 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheerer P., et al. , Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Arnis S., Hofmann K. P., Two different forms of metarhodopsin II: Schiff base deprotonation precedes proton uptake and signaling state. Proc. Natl. Acad. Sci. U.S.A. 90, 7849–7853 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y., Botelho A. V., Martinez G. V., Brown M. F., Electrostatic properties of membrane lipids coupled to metarhodopsin II formation in visual transduction. J. Am. Chem. Soc. 124, 7690–7701 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Vogel R., Siebert F., Conformation and stability of α-helical membrane proteins. 1. Influence of salts on conformational equilibria between active and Inactive states of rhodopsin. Biochemistry 41, 3529–3535 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Zaitseva E., Brown M. F., Vogel R., Sequential rearrangement of interhelical networks upon rhodopsin activation in membranes: The Meta IIa conformational substate. J. Am. Chem. Soc. 132, 4815–4821 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Botelho A. V., Huber T., Sakmar T. P., Brown M. F., Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys. J. 91, 4464–4477 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knight C., Voth G. A., The curious case of the hydrated proton. Acc. Chem. Res. 45, 101–109 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Vogel R., et al. , Agonists and partial agonists of rhodopsin: Retinal polyene methylation affects receptor activation. Biochemistry 45, 1640–1652 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Pal S., Chattopadhyay A., Hydration dynamics in biological membranes: Emerging applications of terahertz spectroscopy. J. Phys. Chem. Lett. 12, 9697–9709 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Yuan S., Filipek S., Palczewski K., Vogel H., Activation of G-protein-coupled receptors correlates with the formation of a continuous internal water pathway. Nat. Commun. 5, 4733 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Ahuja S., Smith S. O., Multiple switches in G protein-coupled receptor activation. Trends Pharmacol. Sci. 30, 494–502 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Kimata N., et al. , Retinal orientation and interactions in rhodopsin reveal a two-stage trigger mechanism for activation. Nat. Commun. 7, 12683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vickery O. N., et al. , Intracellular transfer of Na+ in an active-state G-protein-coupled receptor. Structure 26, 171–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lamola A. A., Yamane T., Zipp A., Effects of detergents and high pressures upon the metarhodopsin I–metarhodopsin II equilibrium. Biochemistry 13, 738–745 (1974). [DOI] [PubMed] [Google Scholar]
- 72.Attwood P. V., Gutfreund H., The application of pressure relaxation to the study of the equilibrium between metarhodopsin I and II from bovine retinas. FEBS Lett. 119, 323–326 (1980). [DOI] [PubMed] [Google Scholar]
- 73.Martin D. R., Matyushov D. V., Dipolar nanodomains in protein hydration shells. J. Phys. Chem. Lett. 6, 407–412 (2015). [DOI] [PubMed] [Google Scholar]
- 74.Mallikarjunaiah K. J., Kinnun J. J., Petrache H. I., Brown M. F., Flexible lipid nanomaterials studied by NMR spectroscopy. Phys. Chem. Chem. Phys. 21, 18422–18457 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Hummer G., Garde S., García A. E., Paulaitis M. E., Pratt L. R., The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 1552–1555 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mittal J., Hummer G., Static and dynamic correlations in water at hydrophobic interfaces. Proc. Natl. Acad. Sci. U.S.A. 105, 20130–20135 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Richards F. M., The interpretation of protein structures: Total volume, group volume distributions and packing density. J. Mol. Biol. 82, 1–14 (1974). [DOI] [PubMed] [Google Scholar]
- 78.Akasaka K., Probing conformational fluctuation of proteins by pressure perturbation. Chem. Rev. 106, 1814–1835 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Peitzsch R. M., Reed W. F., High osmotic stress behavior of hyaluronate and heparin. Biopolymers 32, 219–238 (1992). [DOI] [PubMed] [Google Scholar]
- 80.Shkel I. A., Knowles D. B., Record M. T. Jr., Separating chemical and excluded volume interactions of polyethylene glycols with native proteins: Comparison with PEG effects on DNA helix formation. Biopolymers 103, 517–527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Minton A. P., The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 276, 10577–10580 (2001). [DOI] [PubMed] [Google Scholar]
- 82.Minton A. P., Models for excluded volume interaction between an unfolded protein and rigid macromolecular cosolutes: Macromolecular crowding and protein stability revisited. Biophys. J. 88, 971–985 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zosel F., Soranno A., Buholzer K. J., Nettels D., Schuler B., Depletion interactions modulate the binding between disordered proteins in crowded environments. Proc. Natl. Acad. Sci. U.S.A. 117, 13480–13489 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stadmiller S. S., Pielak G. J., Protein-complex stability in cells and in vitro under crowded conditions. Curr. Opin. Struct. Biol. 66, 183–192 (2021). [DOI] [PubMed] [Google Scholar]
- 85.Dill K. A., Bromberg S., Stigter D., Molecular Driving Forces: Statistical Thermodynamics in Biology, Chemistry, Physics, and Nanoscience (Garland Science, 2010). [Google Scholar]
- 86.Arnold K., Zschoernig O., Barthel D., Herold W., Exclusion of poly(ethylene glycol) from liposome surfaces. Biochim. Biophys. Acta 1022, 303–310 (1990). [DOI] [PubMed] [Google Scholar]
- 87.Liu Y., Agudo-Canalejo J., Grafmüller A., Dimova R., Lipowsky R., Patterns of flexible nanotubes formed by liquid-ordered and liquid-disordered membranes. ACS Nano 10, 463–474 (2016). [DOI] [PubMed] [Google Scholar]
- 88.Dror R. O., et al. , Structural basis for nucleotide exchange in heterotrimeric G proteins. Science 348, 1361–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Flock T., et al. , Universal allosteric mechanism for Gα activation by GPCRs. Nature 524, 173–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oldham W. M., Van Eps N., Preininger A. M., Hubbell W. L., Hamm H. E., Mapping allosteric connections from the receptor to the nucleotide-binding pocket of heterotrimeric G proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 7927–7932 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Struts A. V., Chawla U., Perera S. M. D. C., Brown M. F., Investigation of rhodopsin dynamics in its signaling state by solid-state deuterium NMR spectroscopy. Methods Mol. Biol. 1271, 133–158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van de Hulst H. C., Light Scattering by Small Particles (Dover Publications, New York, 1981). [Google Scholar]
- 93.Harries D., Rand R. P., Osmotic pressure data. https://scholars.huji.ac.il/danielharries/book/osmotic-stress-data. Accessed 26 April 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. Experimental data can be accessed at the University of Arizona Research Data Repository (DOI: 10.25422/azu.data.19686555).