Significance
The genetic basis of isolated microtia, a congenital abnormality of the external ear, is poorly understood. Indigenous American (Amerindigenous) populations have the highest reported incidence of microtia. Here, we use whole genome sequencing to study microtia in Latin American families and identify a common microtia risk allele that is enriched among individuals with Amerindigenous ancestry. This allele is located in a regulatory region governing the expression of Roundabout 1 (ROBO1) and Roundabout 2 (ROBO2) in induced pluripotent stem cell–derived neural crest cells and is associated with a complex repeat sequence. These results identify a shared genetic basis for isolated microtia and other craniofacial abnormalities and account for, at least in part, the increased incidence of microtia in Amerindigenous populations.
Keywords: microtia, craniofacial microsomia, ancestry
Abstract
Microtia is a congenital malformation that encompasses mild hypoplasia to complete loss of the external ear, or pinna. Although the contribution of genetic variation and environmental factors to microtia remains elusive, Amerindigenous populations have the highest reported incidence. Here, using both transmission disequilibrium tests and association studies in microtia trios (parents and affected child) and microtia cohorts enrolled in Latin America, we map an ∼10-kb microtia locus (odds ratio = 4.7; P = 6.78e-18) to the intergenic region between Roundabout 1 (ROBO1) and Roundabout 2 (ROBO2) (chr3: 78546526 to 78555137). While alleles at the microtia locus significantly increase the risk of microtia, their penetrance is low (<1%). We demonstrate that the microtia locus contains a polymorphic complex repeat element that is expanded in affected individuals. The locus is located near a chromatin loop region that regulates ROBO1 and ROBO2 expression in induced pluripotent stem cell–derived neural crest cells. Furthermore, we use single nuclear RNA sequencing to demonstrate ROBO1 and ROBO2 expression in both fibroblasts and chondrocytes of the mature human pinna. Because the microtia allele is enriched in Amerindigenous populations and is shared by some East Asian subjects with craniofacial malformations, we propose that both populations share a mutation that arose in a common ancestor prior to the ancient migration of Eurasian populations into the Americas and that the high incidence of microtia among Amerindigenous populations reflects the population bottleneck that occurred during the migration out of Eurasia.
Microtia and anotia are mild hypoplasia and complete absence of the pinna (external ear or auricle) that diminish localization, amplification, and transmission of sound resulting in hearing loss (Fig. 1A). Microtia can occur in isolation, as a component of neural crest syndromes (1, 2), or in the context of craniofacial microsomia (CFM) in which hypoplasia of facial bone, musculature, and soft tissues arise from maldevelopment of first and second pharyngeal arch-derived structures (3). Genetic causes for microtia in the context of syndromic phenotypes have been identified (4–8); however, a genetic cause has remained elusive in the majority of isolated cases (9). While environmental exposures such as high altitude are proposed as predisposing factors (10, 11), the co-occurrence of microtia with other developmental abnormalities, studies of microtia in monozygotic and dizygotic twins (12), and evidence of increased risk in certain ancestry groups (11, 13, 14) indicate genetic contributions. Indigenous populations in the Americas have among the highest reported prevalence of isolated microtia, with observed rates ranging from 8 to 20/10,000 individuals compared to an estimated global prevalence of ∼2/10,000 (11, 13–16). In addition, admixed Latin American populations have been historically underrepresented in genomic studies (17). Capitalizing on these observations, we studied whole genome sequences of Latin American microtia probands and parents (trios) with Indigenous American (Amerindigenous) ancestry to investigate the genetic basis of ancestry-associated microtia risk.
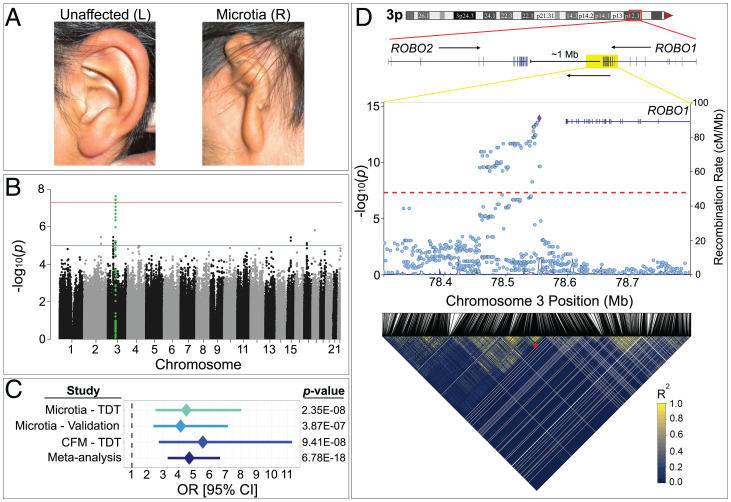
Fig. 1.
Association of a locus on chromosome 3p12.3 with craniofacial abnormalities. (A) Unilateral grade 3 microtia in a subject from Ecuador demonstrating malformation of the pinna (microtia) and absence of the ear canal (aural atresia) and the unaffected contralateral ear. (B) Manhattan plot of the genome-wide TDT in 73 microtia proband–parent trios demonstrates a single locus at chromosome 3p12.3 (green) exceeding the threshold for genome-wide significance (P < 5E-08, red line) and five other suggestive associations (P < 1E-05, blue line). (C) Forest plot of the association between 3p12.3 and microtia by TDT and validation case control analysis, association in GMKF-CFM by TDT, and metaanalysis using a fixed-effects model. (D) Genomic context of the associated region and LocusZoom plot of combined microtia/GMKF-CFM TDT results at chr3p12. The lead SNP (rs9682630, purple) is ∼50 kb 3′ of ROBO1. LD structure in microtia and GMKF-CFM trios demonstrates a 10 kb minimal microtia locus (asterisk) with strongest association.
Results
A Locus Associated with Microtia in Latin Americans of Amerindigenous Ancestry Maps between ROBO1 and ROBO2.
To identify genetic loci associated with microtia we determined the whole genome sequence of 73 microtia proband–parent trios with isolated microtia (∼85% unilateral and ∼15% bilateral) (Table 1 and SI Appendix, Figs. S1 and S2) and performed genome-wide transmission disequilibrium tests (TDTs). None of the microtia probands had either de novo or transmitted loss of function (LoF) variants in genes previously associated with microtia in humans (Materials and Methods and SI Appendix, Supplementary text). TDT is a family-based association test that evaluates transmission of an allele to affected probands at greater than expected frequency and is robust to confounding by population stratification. A single locus was significantly associated with microtia (P < 5E-08) on chromosome 3p12.3 (lead single nucleotide polymorphisms [SNP] rs1444472, P = 2.35E-08, odds ratio [OR] = 4.5), and five other loci had suggestive associations (P < 1E-05; Fig. 1B; SI Appendix, Fig. S3; and Dataset S1). We next performed a validation study in a case-control cohort, using mixed-model association testing in Latin American probands from three independent microtia cohorts from the United States and Latin America to validate the significant and suggestive associations identified by TDT (Materials and Methods and SI Appendix, Fig. S4). Only the SNPs on chromosome 3p12.3 exceeded the Bonferroni adjusted P value threshold of 5.6E-04 (SNP rs34486998, P = 3.87E-07, approximate beta = 1.47) (Fig. 1C; SI Appendix, Fig. S4; and Dataset S2).
Table 1.
Demographics of the microtia cohort
| Country of recruitment | |||
|---|---|---|---|
| Ecuador | Colombia | United States | |
| Subjects | 171 | 193 | 25 |
| No. affected (%) | 108 (63) | 69 (36) | 22 (88) |
| No. of complete trios | 14 | 58 | 1 |
| % male | 56 | 55 | 68 |
| % affected males | 64 | 64 | 73 |
| Microtia laterality | |||
| % right | 52 | 53 | 44 |
| % left | 29 | 32 | 31 |
| % bilateral | 19 | 15 | 25 |
Given the overlap between isolated microtia and CFM phenotypes (18), we asked if there is a shared genetic basis for these malformations. TDT analyses in 68 CFM proband–parent trios sequenced as part of the Gabriella Miller Kids First Program (referred to as GMKF-CFM) that included a large cohort from Latin America (19) (SI Appendix, Fig. S2) replicated the association on 3p12.3 (rs13071898, chr3:78554399, P = 9.41E-08, OR = 5.5; Fig. 1C; SI Appendix, Fig. S5; and Dataset S3). Metaanalysis of the independent TDT and validation studies using a fixed-effects model demonstrated increased statistical significance of the association at 3p12.3 with microtia and CFM (P = 6.78E-18, OR = 4.7; Fig. 1C). A combined TDT of all microtia and GMKF-CFM trios (n = 141) identified the most significant association at rs9682630 (chr3:78555137, overtransmission of the reference/ancestral G allele) (P = 1.19E-14, OR = 4.9; Fig. 1D and Dataset S4). Among three microtia subjects and one GMKF-CFM proband with Amerindigenous ancestry, we observed three separate recombination events that define a shared 10-kb region (chr3: 78546526 to 78555137, referred to as the minimal microtia locus [MIC locus]) among microtia/GMKF-CFM subjects (Fig. 1D and SI Appendix, Fig. S6 A and B). The MIC locus resides between the ROBO1 and ROBO2 genes, ∼50 kb from the 3′ end of ROBO1 and 900 kb from the 3′ end of ROBO2 (Fig. 1D).
The Amerindigenous MIC Allele Is Associated with CFM in a Chinese Cohort.
A prior publication reported an ∼100-kb locus on chromosome 3p12.3 that is associated with CFM in a Chinese cohort (20). The coincident chromosomal region for a CFM and MIC locus, together with epidemiologic evidence of heightened incidence of microtia specifically in Amerindigenous, Latin American, and Asian populations (11, 21), implied that a shared risk haplotype may reside within these ancestry groups. Moreover, as genetic studies indicate that Amerindigenous populations are descendants of a “first wave” of migration from Eurasia (22, 23), we hypothesized that a mutation arose in a common ancestor and is shared within microtia/CFM loci in probands from both Asia and Latin America. To test this model, we first confirmed that the same 10-kb haplotype identified by TDT in Latin American microtia/GMKF-CFM trios (Fig. 1D) was associated with the larger CFM locus identified in China (Dataset S5). We next compared the frequency of common haplotypes at the MIC locus across populations from the 1000 Genomes Project Phase 3 (1000G) and Human Genome Diversity Project (HGDP) datasets. The 10-kb haplotype associated by TDT was found in all global populations and had the highest frequency in Latin Americans with Amerindigenous ancestry (PEL 26%) and in microtia and GMKF-CFM probands (50%) (Fig. 2A). We denote this 10-kb haplotype as the MIC haplotype.
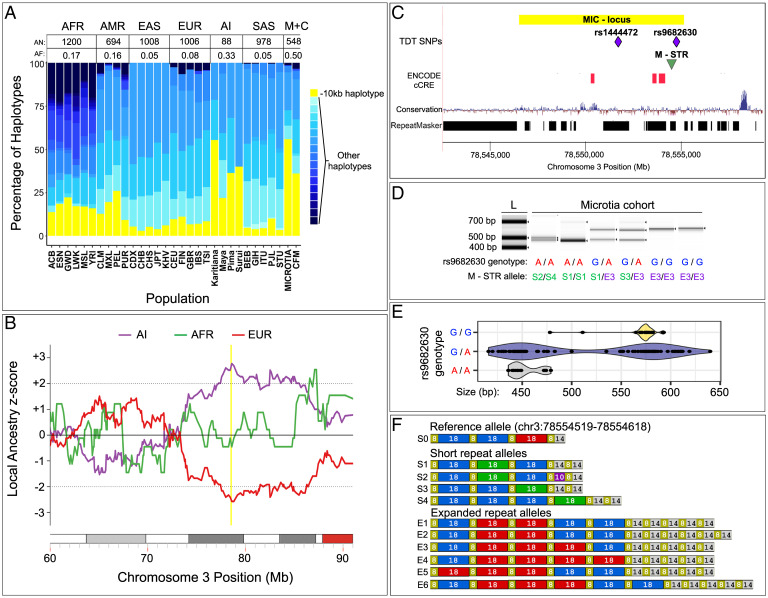
Fig. 2.
Population frequency and ancestry of the MIC haplotype and associated polymorphic repeat elements. (A) Frequency of the 10 kb MIC haplotype in probands and global reference populations from the HDGP and 1000G datasets demonstrates enrichment in Amerindigenous ancestry. Haplotype number (AN) and frequencies (AF) across various superpopulations are shown. (B) Local ancestry analysis of a 30-Mb region on chromosome 3p12.3 in affected individuals demonstrates enrichment (z = +2.52) of Amerindigenous ancestry in a 135-kb region overlying the associated MIC locus (yellow). (C) Schematic showing the relative position of microtia associated SNPs, vertebrate sequence conservation, and the location of repetitive sequence elements within the MIC locus. (D and E) PCR amplification of a repeat element demonstrates larger fragments in carriers of microtia-associated SNPs. (F) Diagram of repeat alleles detected in the microtia probands and family members by sequencing. See SI Appendix, Fig. S10 for repeat sequence. AFR, African; AMR, Latin American; EAS, East Asian; EUR, European; AI, Amerindigenous; SAS, Southeast Asian; M+C, microtia and GMKF-CFM probands. See SI Appendix, Supplementary text, for additional abbreviations.
As Latin Americans have varying degrees of genetic admixture from ancestral European, African, and Amerindigenous populations (24), we next performed local ancestry analysis at the MIC locus. Local ancestry inference identified relative enrichment of Amerindigenous ancestry at the MIC locus in microtia and GMKF-CFM probands (z score = +2.52; Fig. 2B). We then genotyped nearby short tandem repeats (STRs) to identify ancestry-informative alleles. Four alleles containing a CATTT motif at an STR located ∼25 kb away from and in linkage disequilibrium (LD) with SNPs in the MIC haplotype (chr3: 78525532; SI Appendix, Fig. S7) demonstrated enrichment in Amerindigenous and Latin American haplotype carriers and differentiated haplotypes in African from non-African populations (SI Appendix, Fig. S7C). Based on this observation we next asked if the ancestry-related STR alleles might be absent in African individuals and enriched in Amerindigenous/East Asians due to ancient hominin introgression and did not identify the MIC locus as a segment of potential introgression (25) (SI Appendix, Fig. S8).
The High Frequency of the MIC Allele in Amerindigenous Populations.
Because subjects with severe craniofacial malformations might be expected to have reduced reproductive fitness, we explored genetic mechanisms that might explain the higher-than-expected population frequency of the MIC haplotype. We considered two hypotheses to explain the high haplotype frequency in Latin Americans with Amerindigenous ancestry: 1) balancing selection (i.e., forces, other than the microtia/CFM phenotype, are selecting for the MIC allele) or 2) that the population bottleneck that occurred during migration out of Eurasia altered the MIC allele frequency in the Amerindigenous populations (26). We observed that haplotypes carrying the sentinel microtia SNP identified by combined TDT (rs9682630, G, ancestral allele) are usually associated with an extended haplotype in Latin American subjects from 1000G, compared with haplotypes with the derived A allele (SI Appendix, Fig. S9 A and B), but do not demonstrate evidence of positive selective pressure, or balancing selection, by several methodologies (SI Appendix, Supplementary Methods and Fig. S9 C–E).
The Low-Penetrant MIC Allele Contains a Polymorphic Repeat Element.
The MIC allele has low penetrance (i.e., a small fraction of MIC allele carriers develop microtia) as evident from the 63 TDT transmission events from phenotype negative parents to microtia probands (Dataset S1). Furthermore, the predominance of unilateral pinna malformations in 85% of MIC haplotype carriers suggests incomplete penetrance with respect to the contralateral unaffected ear in a majority of cases (Table 1). Despite the fact that the MIC allele increases the risk of microtia 4.9-fold (Fig. 1C), we estimate the penetrance of the MIC allele to be <1% based on estimates of microtia prevalence and MIC allele frequency in the Latin American population.
We next sought to identify candidate causal variants within the MIC locus. Comparison of whole genome sequence, assessed by next-generation paired end short read sequence, revealed no variation in SNPs, insertions/deletions, or structural variants across the MIC locus among probands and MIC haplotype carriers from reference populations (SI Appendix, Fig. S6). To identify additional genetic variants associated with microtia, we performed PCR amplification across the 10-kb microtia locus and noted a polymorphic repeat element located near the lead TDT SNPs (M-STR; Fig. 2C). PCR amplification across the M-STR region demonstrated longer PCR product in carriers of the MIC haplotype than observed in noncarriers (Fig. 2 D and E). Sequencing of M-STR PCR products demonstrated polymorphic repeat alleles consisting of repeat elements ranging from 22 to 26 base pairs in size and expanded M-STR alleles ∼120 bp greater in length in phase with the microtia haplotype (Fig. 2 E and F and SI Appendix, Fig. S10).
ROBO1 and ROBO2 Are Expressed in the Pinna.
The MIC locus is located between the ROBO1 and ROBO2 genes, which mediate signaling to negatively regulate cell adhesion and promote cellular migration in development and disease (27). ROBO signaling is critical to craniofacial neural crest cell (NCC) properties (28), and ROBO1 and ROBO2 are expressed in embryonic murine Wnt1-positive cranial NCCs that migrate to the pharyngeal arches and contribute to craniofacial structures (29). To better define the roles of ROBO1 and ROBO2 in the human pinna, we first performed single nuclear RNA sequencing (snRNA-seq) of three human pinnae, two with microtia and one with normal morphology. Clustering of nuclei based on differential gene expression demonstrated presence of multiple cell types that contribute to cartilage and soft tissue (Fig. 3A and SI Appendix, Fig. S11). ROBO1/ROBO2 transcripts are broadly expressed in multiple cell types of the pinna, with most prominent expression in chondrocytes and fibroblasts (Fig. 3B). We also assessed ROBO1/ROBO2 expression in reported human embryonic stem cells differentiated into NCC and into NCC-derived chondrocytes (30). ROBO1 is expressed at higher levels than ROBO2 in NCC, and these differences are accentuated in NCC-derived chondrocytes (SI Appendix, Fig. S11C).
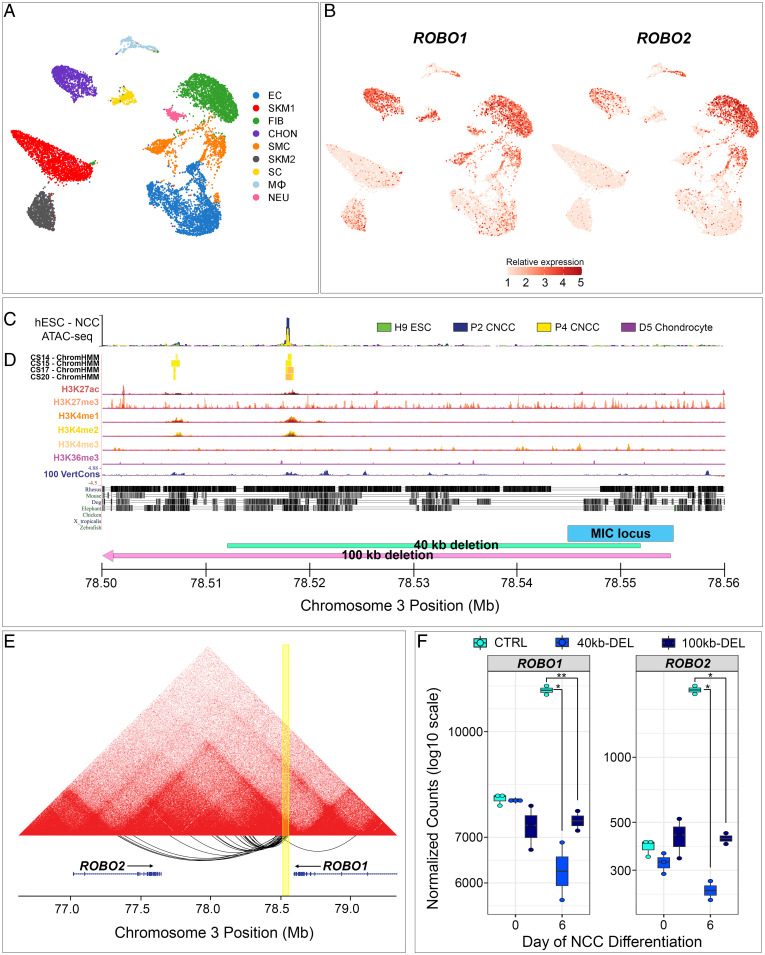
Fig. 3.
The MIC locus is in a ROBO1/ROBO2 regulatory element. (A) Uniform manifold approximation and projection plot of cell types identified by snRNA-seq of the human pinna. EC, endothelial cell; SKM, skeletal muscle; FIB, fibroblast; CHON, chondrocytes; SMC, smooth muscle; SC, satellite cell; Mϕ, macrophage; NEU, neuronal. (B) ROBO1 and ROBO2 are expressed in multiple cell types of the human pinna, with highest expression in chondrocytes and fibroblasts. (C) ATAC-seq in hESC and hESC-derived NCC and chondrocytes and (D) epigenetic profiling in developing human craniofacial tissue demonstrate an active enhancer element ∼30 kb from the MIC locus. (E) Profiling of chromatin conformation in undifferentiated hiPSCs by HiC demonstrates that the MIC locus (yellow) lies within a TAD domain encompassing ROBO1 and ROBO2 and has chromatin–chromatin interactions with ROBO1 and the second intron of ROBO2. (F) Expression of ROBO1 and ROBO2 transcripts from undifferentiated (day 0) and NCC differentiated (day 6) control hiPSCs and hiPSCs harboring a 40- or 100-kb heterozygous deletion overlapping the MIC locus. *P < 0.05, **P < 0.01.
Defining Regulatory Sequences and Topologically Associating Domains near the MIC Locus.
We considered if a regulatory element within the MIC locus influenced either or both ROBO1 and ROBO2 expression. Previous studies, using transposase-accessible chromatin sequencing (ATAC-seq), identified a region of open chromatin in human embryonic stem cell (hESC)–derived NCC that is absent from undifferentiated hESC and chondrocytes (30) (Fig. 3C). This open chromatin region overlaps with a craniofacial enhancer element identified in an atlas of human embryonic craniofacial epigenetic regulatory histone modifications (31) (Fig. 3D) . Together these studies implicate a region ∼30 kb from the MIC locus as an important epigenetic regulatory region in human craniofacial development. We assessed whether this putative human regulatory region influenced ROBO1 and/or ROBO2 expression using Hi-C profiling of chromatin conformation (32) in undifferentiated human-induced pluripotent stem cells (hiPSCs). The MIC locus lies at the boundaries of a topologically associating domain (TAD) that exhibits chromatin–chromatin interactions with intronic regions of both ROBO1 and ROBO2 genes (Fig. 3E). Using CRISPR/Cas9 genome editing, we then generated hiPSC-derived NCCs harboring either a heterozygous 40- or 100-kb deletion (Fig. 3D and SI Appendix, Fig. S12) that overlaps the MIC locus and the nearby putative epigenetic regulatory element. hiPSCs with heterozygous deletion of this region differentiated into NCCs and expressed many established marker transcripts in wild-type hiPSC-derived NCCs (SI Appendix, Fig. S13 A and B) but failed to induce either ROBO1 or ROBO2 transcripts in NCCs (Fig. 3F). Further, the absence of appropriate ROBO1/ROBO2 expression attenuated developmental down-regulation of CDH1 (E-cadherin; SI Appendix, Fig. S13C), which is required for an epithelial-to-mesenchymal transition phenotype and NCC migration (33).
Discussion
Using both TDTs and genome wide association studies (GWAS), we demonstrate that one allele (designated the MIC allele) of a 10-kb region located between ROBO1 and ROBO2 significantly increases risk for microtia in Latin Americans of Amerindigenous ancestry. We suggest that the MIC allele carries a polymorphic repeat element that perturbs ROBO1 and ROBO2 gene expression in the developing and formed pinna.
The high frequency of craniofacial malformations and microtia among some East Asian and Latin American populations has been noted for some time (11, 34, 35). We demonstrate here that ∼50% of Latin Americans of Amerindigenous ancestry with microtia share the same MIC allele (P = 6.78E-18, OR = 4.7; Fig. 1C). Further, this MIC allele is shared with some East Asian subjects with craniofacial malformations. Because population genetic studies and other demographic analyses have demonstrated that Amerindigenous populations descended from Eurasians who migrated across the Bering Strait into North America, we speculate that a founder mutation arose in Eurasia and was carried to North America by the ancestors of Amerindigenous populations. Multiple studies estimate that between 150 and 2,000 Eurasians migrated to North America, creating a severe population bottleneck (36). The higher frequency of the MIC allele in the Amerindigenous populations than in East Asian populations may reflect this population bottleneck (i.e., a higher fraction of early immigrants carried the MIC allele to North America than would be expected based on the population prevalence in Asia). Additional studies of whole genome sequence from microtia subjects of East Asian and Amerindigenous ancestry will be important to explore this hypothesis further.
A definitive causal variant(s) within the MIC locus was not uncovered by analyses of whole genome short read paired end sequencing. However, subsequent analyses of PCR-amplified fragments from the microtia locus did identify a complex, polymorphic repeat element associated with an expanded repeat in microtia probands (Fig. 2). We hypothesize that this expanded repeat element may perturb gene expression regulatory functions of this region and that either stochastic effects or instability of the repeat element resulting in somatic mosaicism may contribute to the asymmetric phenotypes observed in a majority of microtia subjects.
We propose that the MIC allele mediates pinna/craniofacial malformations by perturbing NCC expression of both ROBO1 and ROBO2. ROBO signaling promotes NCC induction, delamination, epithelial–mesenchymal transition, and migration that enable the development of craniofacial structures (37). We demonstrate here that human chondrocytes and fibroblasts of pinna obtained from subjects with and without microtia express both ROBO1 and ROBO2; however, their role in these cell types is unclear. Previous studies both in humans and in murine models have implicated either ROBO1 and/or ROBO2 in microtia/craniofacial development. That is, genetic ablation of both Robo1/Robo2 in mice causes cranial-facial abnormalities (28). Two studies of humans with ear malformations have implicated these genes in pinna development. Damaging variants in HOXA2, a homeobox transcription factor that broadly regulates Robo/Slit signaling, were shown to cause microtia in a large family (38). In addition, Shaffer et al. (39) performed a large GWAS metaanalysis and identified a ROBO1 SNP, not in linkage disequilibrium with MIC allele, as associated with normal morphologic variation in earlobe attachment. Damaging variants in ROBO1 and ROBO2 are associated with pituitary stalk interruption syndrome and congenital abnormalities of the kidney and urinary tract, respectively (40, 41), and have not been associated with craniofacial abnormalities. Interrogation of Robo1 and Robo2 function in cNCCs and differentiated chondrocytes using cell-specific conditional knockouts in model systems will provide important information on the relative contribution of these gene paralogs to craniofacial phenotypes and the developmental processes sensitive to their perturbation. Furthermore, studies of common and rare genetic variants affecting ROBO signaling are expected to be informative to the complex and fundamental roles of NCC in human embryogenesis and to defining mechanisms that regulate ROBO expression in these cells.
The intergenic region between ROBO1 and ROBO2, where the MIC allele is located, is not known to carry sequences that regulate expression of these genes. However, we demonstrate that NCCs derived from hiPSC cells lacking 40 kb flanking the MIC allele express less ROBO1 and ROBO2 than isogenic NCC cells with two normal alleles (Fig. 3). Although the mechanism by which the MIC allele perturbs ROBO1 and ROBO2 expression is not certain, we note that the MIC locus is located at the boundary of chromatin loops that contact both ROBO1 and ROBO2. Changes in repeat length might alter the chromatin loops and thereby perturb gene expression. Further characterization of these complex repeat elements in affected and unaffected individuals may elucidate a causal role.
In summary, we conclude that the founder mutation that created the MIC allele arose more than 20,000 y ago in a Eurasian ancestor and is likely responsible for the high incidence of craniofacial malformation and microtia observed both in East Asian and Amerindigenous populations today. The MIC allele was likely carried to North America by ancestors of Indigenous Americans who migrated from Eurasia. The high incidence of microtia in Latin America reflects the high frequency of the MIC allele among Latin Americans of Amerindigenous ancestry, which arose during the migration-associated population bottleneck. We suggest that the MIC allele, which contains a highly polymorphic repeat element, predisposes to microtia by altering chromatin structure flanking the ROBO1 and ROBO2 genes and thereby perturbs ROBO1 and ROBO2 gene expression. The low penetrance of the MIC allele and the common occurrence of unilateral rather than bilateral microtia likely reflect the relatively subtle changes it imparts on ROBO1 and ROBO2 expression.
Materials and Methods
Human Subjects.
Patients and unaffected relatives were identified at collaborating sites in South America (Fundacion Tierra Nueva, Hospital Un Canto a la Vida in Quito, Ecuador, and the Clinica del niño Empresa Social del Estado Luis Carlos Galan Sarmiento in Bogota, Colombia) and the United States (Vanderbilt University Medical Center [Nashville, TN] and the Massachusetts Eye and Ear Infirmary [Boston, MA] in partnership with Brigham and Women’s Hospital [Boston, MA]). Study subjects were assessed by history, physical examination, and audiogram by a multidisciplinary team including an otolaryngologist and audiologist. A diagnosis of microtia was made based on clinical evidence and graded on scale of 1 to 4 in accordance with the Marx microtia classification scale. Subjects were selected for isolated microtia in the absence of syndromic features, and patients with syndromic forms of microtia, such as Treacher-Collins syndrome, CHARGE (coloboma, heart defects, choanal atresia, growth retardation, genital abnormalities, and ear abnormalities) syndrome, branchio-oto-renal syndrome, DiGeorge, or Kabuki syndrome, were excluded from the study. Informed written consent was obtained from all participants or their legal guardians in accordance with the protocols approved by The Partners Human Research Committee, Institutional Review Board of Partners Healthcare, Brigham and Women’s Hospital, Boston, MA. Sex and phenotype data are available with genomic datasets in dbGaP and at https://kidsfirstdrc.org. Peripheral blood samples were collected for DNA extraction.
DNA Sequencing, Data Processing, and Genotyping.
Preparation of PCR-free whole genome sequencing (WGS) libraries and WGS (Illumina, 150-bp paired-end) with 30× average coverage was performed at the Broad Institute (Cambridge, MA). Reads were aligned to the hg38 reference genome using the Burrows–Wheeler Aligner (BWA-MEM) and processed in accordance with Genome Analysis Toolkit (GATK; Broad Institute) workflow best practices (42). Briefly, sequence alignments underwent BaseQuality Score Recalibration (GATK) and marking of duplicates (MarkDuplicates; Picard), followed by individual-level variant calling with HaplotypeCaller (GATK). Individual samples were then joint-genotyped, and variants underwent variant quality score recalibration log odds ratio scoring (VQSR-LOD). Final SNP and indel (insertions or deletions < 50 base pairs) calls were filtered with the PASS filter from VQSR-LOD scoring, read depth ≥10, and genotype quality ≥20 and subsequently phased with Eagle (v2.4.1) using the 1000G reference panel. Genomic structural variants (SVs) greater than 50 base pairs were genotyped using a GATK-SV (43) workflow in Terra. Briefly, initial structural variant evidence was collected using the tools Manta, WHAM, GATK-gCNV, cnMOPs, and MELT. SV evidence was then merged across all samples, and joint genotyping of SVs across the entire cohort was performed by GATK-SV as previously described (43).
Study Design.
Based on the size of the microtia cohort and the availability of trios, we elected to perform a two-tiered analysis. The microtia cohort was split into a discovery cohort consisting of 73 complete proband–parent trios for linkage/association analysis and a separate validation cohort of nontrio probands, unrelated unaffected individuals, and additional control subjects. We elected to use the TDT to identify linkage and association which mitigates, at least in part, the effects of population stratification in the analysis (44). Genome-wide significant and suggestive associations identified by TDT underwent validation by association testing in the case-control dataset.
Identification of Rare, LoF Variants.
The predicted effects of SNPs and indels on protein-coding genes were annotated with SnpEff, and variants annotated as frame_shift, stop_gained, start_lost, splice_acceptor_variant, and splice_donor_variant were designated as LoF variants. Rare variants were identified by a maximum population allele frequency (Popmax AF) less than 1E-04 in gnomAD. We then restricted analysis to missense and LoF variants in 33 genes associated with syndromic and isolated microtia in previously published studies (SI Appendix, Supplementary text) and evaluated 73 microtia proband–parent trios for transmitted and de novo LoF variants. Predicted functional effects of genomic SVs were annotated using svtk. LoF SVs were defined as deletions overlapping at least a single exon, insertions with exons, inversions with a single or both breakpoints within a gene, and duplications with both breakpoints within exons. To maximize sensitivity, the unfiltered SV call set was evaluated for SVs predicted to result in LoF of the 33 genes in the 73 trios. No transmitted or de novo LoF variants (SNVs/indels/SVs) in the 33 candidate genes were identified.
TDT.
A dataset of common biallelic SNPs was generated using PLINK (v1.9) with variant filtering criteria of an in-cohort minor allele frequency (MAF) ≥ 0.05, genotype missingness less than 5%, and Hardy–Weinberg P value cutoff of 1E-06. After filtering, 5,146,268 biallelic SNPs remained for testing. This dataset was then used in the implementation of TDT in PLINK v1.9. SNPs exceeding a statistical threshold of P < 5E-08 were considered statistically significant, and SNPs with P < 1E-05 were considered suggestive associations.
Validation Study in a Case-Control Dataset.
We generated a case-control cohort containing nontrio microtia probands and unaffected individuals of Latin American ancestry in the GMKF Microtia cohort, unaffected parents from the Gabriella Miller Kids First Orofacial Cleft (OFC) cohort from Colombia (45), and Latin American subjects from Peru (PEL) and Colombia (CLM) and of Mexican ancestry (MXL) from 1000G dataset. We merged genotype data from microtia probands and controls and filtered for variants with an MAF ≥ 0.05, genotype missingness < 5%, and Hardy–Weinberg P value cutoff of 1E-06 using PLINK v1.9. The GENESIS software package was then used to perform principal component analysis with PC-Air to account for cryptic familial relatedness (SI Appendix, Fig. S4) and kinship analysis using PC-Relate. We removed individuals with a kinship threshold greater than 0.02. The resulting dataset contained 85 probands with microtia (7 from Colombia, 72 from Ecuador, and 6 from the United States) and 780 unaffected control subjects (6 GMKF-Microtia from Colombia, 31 GMKF-Microtia from Ecuador, 504 GMKF-OFC from Colombia, and 239 1000G samples [MXL, PEL, and CLM]). Ninety SNPs with P < 1E-05 identified by TDT (Dataset S1) were used for association testing in the validation cohort. We then used mixed model association testing implemented in the GENESIS package to perform score tests, with sex and the first six principal components from PC-Air as fixed effect covariates (SI Appendix, Fig. S4B), and a genetic relationship matrix using kinship coefficient estimates from PC-Relate as random effects in the null model. A Bonferroni-corrected P value of 5.6E-04 was considered statistically significant based on the number of SNPs tested.
TDT for CFM.
We performed genotyping and filtering as described above on chromosome 3 from 68 parent–proband trios from the GMKF CFM cohort (19). A 500-kb region (chr3: 78300000 to 78800000) was then used for TDT as previously described. A total of 963 common variants were tested, and a Bonferroni corrected P value of 5.19E-05 was considered statistically significant.
Meta-analysis of TDT and Validation Study.
A meta-analysis of independent TDT and validation studies was performed using a fixed effects model described by Kazeem and Farrall (46) with a modified version of the catmap package in R (47).
Global Ancestry Analysis.
Common biallelic SNPs (MAF > 0.05) from 1000G, HGDP Amerindigenous individuals, and the microtia cohort WGS were merged in PLINK v1.9 and linkage disequilibrium (LD) pruned using the parameters –indep-pairwise 50 10 0.1, yielding a dataset of 226,185 SNPs. This dataset was then used to run ADMIXTURE (v1.3) with a K = 4 based on population history and previously reported admixture analyses (48).
Local Ancestry Analysis.
Local ancestry was inferred from phased genotypes using RFMIX (49) with three EM iterations, a node size of 5, and the reanalyze reference option. A balanced reference panel of Amerindigenous, European, and African ancestry consisting of samples from the 1000G Peruvian (PEL) and HGDP Amerindigenous populations, European (CEU), and African (YRI) populations. A case-only local ancestry analysis was then performed as previously described (50). A z-score comparing the mean local ancestry and chromosome-wide (chr3) mean ancestry in cases was computed for each ancestry block across a 30-Mb region of chr3.
Estimation of Penetrance.
Penetrance of the MIC allele in Latin American populations was estimated as previously described (51). Briefly, MIC allele frequency in Latin American microtia probands was obtained from our dataset and from Latin American subjects from 1000G AMR population as a comparison group. A range of population-level disease risk for microtia in Latin America was obtained from Luquetti et al. (11) and Castilla and Orioli (15).
STR Genotyping from WGS.
STRs within a 100-kb region overlapping the MIC locus were joint genotyped across individuals from 1000G, HGDP, and the microtia/CFM cohorts using HipSTR (52). Phased single nucleotide variants were used to inform phasing of STR alleles using the –snp-vcf option. Alleles with a posterior probability of correct phasing >0.9 were then used to determine if STR alleles were in phase with the MIC haplotype. We identified Human_STR_954934 (rs1170220055, chr3: 78525532 to 78525595) containing ATTTT repeats as ancestry informative by comparing allele frequencies in reference populations. Accuracy of genotyping of the CATTT insertion alleles was validated by visual inspection of WGS alignments, absence of Mendelian errors in trios, and Sanger sequencing in one proband.
RNA-Sequencing.
RNA was isolated from tissues and cells using the TRIzol (Thermo Fisher) method, and libraries were prepared as previously described (53). Transcripts were quantified using Salmon (1.2.1) with an hg38 transcript index from GENCODE (v35). Gene counts were then generated from transcript abundance using tximport, and differential gene expression analysis was performed with DESeq2. A t test was used to compare normalized counts across cell types and conditions, and nominal P values are reported. Two to three independent differentiations (replicates) were used per hiPSC-NCC genotype and condition analyzed.
Generation of iPS Cell Lines.
An ∼100-kb-long microtia-associated locus (chr3: 78459959 to 78555332) and a smaller 40-kb-long region (chr3: 78512902 to 78551748) encompassing overlapping LD blocks (Fig. 1D) were targeted for deletion using CRISPR-Cas9. Guide RNAs (gRNAs) targeting each end of the 100- and 40-kb block were designed using Benchling. CRISPR RNA (crRNA), trans-activating CRISPR RNA (tracrRNA), and Cas9 enzyme were purchased from Integrated DNA Technologies (IDT). crRNAs were individually hybridized with tracrRNA and Cas9 enzyme to form ribonucleoprotein complexes (RNP) according to the manufacturer’s instruction. Equal volume of RNPs targeting each end of the 100- or 40-kb block were mixed before transfection. Mixed RNPs were delivered to hiPSCs using the Human Stem Cell Nucleofector Kit (Lonza). Single cell colonies were grown, expanded, and genotyped. Cells were genotyped by 1) PCR amplifying across the targeted region and 2) PCR amplifying heterozygous SNPs within the targeted deletion sites, and were verified using Sanger sequencing (SI Appendix, Fig. S12C). Cell lines carrying a large deletion were selected, further expanded, and used for the subsequent experiments. gRNA sequences and genotyping primers are listed in the SI Appendix, Key Resources Table.
Generation of iPSC-Derived NCC.
Cells were then differentiated down an NCC lineage using a STEMDiff Neural Crest Differentiation Kit from StemCell Technologies. Cells were analyzed at day 6 of differentiation, and NCC phenotype was confirmed by up-regulation of established NCC markers (SI Appendix, Fig. S13).
Chromatin Conformation Profiling by Hi-C.
Undifferentiated hiPSCs (two biological replicates per time point) were harvested using TrypLE. Cells were crosslinked with 2% Formaldehyde for 10 min and then quenched with 0.125 M Glycine for 15 min. Cells were then snap-frozen in liquid nitrogen. In situ high-throughput chromatin conformation capture (Hi-C) was done as previously described (54). Sequencing reads were aligned with BWA-MEM -SP5M. Aligned reads were then parsed and sorted using pairtools (https://github.com/open2c/pairtools). Uniquely mapped reads were selected, and PCR duplicates were removed. Biological replicates were merged using HOMER. Significant interactions and Hi-C files were created using HOMER. Hi-C data were visualized using University of California Santa Cruz (UCSC) browser.
Targeted Sequencing of STRs.
We designed PCR primers to amplify a polymorphic region located at chr3: 78554380 to 78554771 and performed PCR amplification in 25 µL reactions using 10 to 50 ng of input genomic DNA using a protocol optimized for amplifying repeat sequences (55). PCR products were fragmented using an New England Biolabs (NEB) Fragmentation Kit, and Illumina sequencing libraries were prepared. Libraries were sequenced in a MiSeq using 250-bp paired-end reads. Adapter-trimmed single-end sequencing reads were analyzed by direct visualization using highlight (v1.103) to aid in analysis of repeat motifs and by aligning single-end reads to a custom STR allele reference sequence. STR allele genotypes were confirmed by identifying supporting evidence from 30× WGS data, evaluating transmission in trios and duos, and comparison to expected PCR product sizes.
Supplementary Material
Acknowledgments
We extend our gratitude to the patients and families of the microtia and CFM communities for enabling this research. Funding was provided through the John S. LaDue Memorial Fellowship (D.Q.), NIH Gabriella Miller Kids First Research Program Grant X01 HL145690-01 (J.G.S.), Fondation Leducq Grant 16-CVD03 (J.G.S. and C.E.S.), and a Howard Hughes Medical Institute grant (C.E.S.).
Footnotes
Reviewers: J.H., Case Western Reserve University School of Medicine; B.M., Albert Einstein College of Medicine.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2203928119/-/DCSupplemental.
Data Availability
All WGS datasets were sequenced as part of the NIH Gabriella Miller Kids First Program and are available via controlled access from dbGaP (accession nos. phs002172 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002172.v1.p1), phs002130 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002130.v1.p1), and phs001420 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001420.v1.p1) and at https://kidsfirstdrc.org. Human pinna and hiPSC RNA-seq gene expression data are available at the National Center for Biotechnology Information Sequence Read Archive and Gene Expression Omnibus (GSE202442). All software programs used are publicly available and are is listed in SI Appendix, Key Resources Table.
References
- 1.Gendron C., Schwentker A., van Aalst J. A., Genetic advances in the understanding of microtia. J. Pediatr. Genet. 5, 189–197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander N. L., Kini S. D., Liu Y. C., Cardiac anomalies in microtia patients at a tertiary pediatric care center. Int. J. Pediatr. Otorhinolaryngol. 136, 110211 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Brandstetter K. A., Patel K. G., Craniofacial microsomia. Facial Plast. Surg. Clin. North Am. 24, 495–515 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Abdelhak S., et al. , A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat. Genet. 15, 157–164 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Anonymous; The Treacher Collins Syndrome Collaborative Group, Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat. Genet. 12, 130–136 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Hoskins B. E., et al. , Transcription factor SIX5 is mutated in patients with branchio-oto-renal syndrome. Am. J. Hum. Genet. 80, 800–804 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tekin M., et al. , Homozygous mutations in fibroblast growth factor 3 are associated with a new form of syndromic deafness characterized by inner ear agenesis, microtia, and microdontia. Am. J. Hum. Genet. 80, 338–344 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alasti F., et al. , A mutation in HOXA2 is responsible for autosomal-recessive microtia in an Iranian family. Am. J. Hum. Genet. 82, 982–991 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown K. K., et al. , HOXA2 haploinsufficiency in dominant bilateral microtia and hearing loss. Hum. Mutat. 34, 1347–1351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castilla E. E., Lopez-Camelo J. S., Campaña H., Altitude as a risk factor for congenital anomalies. Am. J. Med. Genet. 86, 9–14 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Luquetti D. V., Leoncini E., Mastroiacovo P., Microtia-anotia: A global review of prevalence rates. Birth Defects Res. A Clin. Mol. Teratol. 91, 813–822 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artunduaga M. A., et al. , A classic twin study of external ear malformations, including microtia. N. Engl. J. Med. 361, 1216–1218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zernotti M. E., Curet C. A., Cortasa S., Chiaraviglio M., Di Gregorio M. F., Congenital Aural Atresia prevalence in the Argentinian population. Acta Otorrinolaringol. Esp. 70, 32–35 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Nelson S. M., Berry R. I., Ear disease and hearing loss among Navajo children–A mass survey. Laryngoscope 94, 316–323 (1984). [DOI] [PubMed] [Google Scholar]
- 15.Castilla E. E., Orioli I. M., Prevalence rates of microtia in South America. Int. J. Epidemiol. 15, 364–368 (1986). [DOI] [PubMed] [Google Scholar]
- 16.Jaffe B. F., The incidence of ear diseases in the Navajo Indians. Laryngoscope 79, 2126–2134 (1969). [DOI] [PubMed] [Google Scholar]
- 17.Sirugo G., Williams S. M., Tishkoff S. A., The missing diversity in human genetic studies. Cell 177, 1080 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Keogh I. J., Troulis M. J., Monroy A. A., Eavey R. D., Kaban L. B., Isolated microtia as a marker for unsuspected hemifacial microsomia. Arch. Otolaryngol. Head Neck Surg. 133, 997–1001 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Luquetti D. V., et al. , MYT1 role in the microtia-craniofacial microsomia spectrum. Mol. Genet. Genomic Med. 8, e1401 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y. B., et al. , Genome-wide association study identifies multiple susceptibility loci for craniofacial microsomia. Nat. Commun. 7, 10605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarskaia L. A., et al. , [The structure and diversiity of hereditary pathology in Sakha Republic (Yakutia)]. Genetika 40, 1530–1539 (2004). [PubMed] [Google Scholar]
- 22.Reich D., et al. , Reconstructing Native American population history. Nature 488, 370–374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raghavan M., et al. , Genomic evidence for the Pleistocene and recent population history of Native Americans. Science 349, aab3884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryc K., et al. , Colloquium paper: Genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc. Natl. Acad. Sci. U.S.A. 107 (suppl. 2), 8954–8961 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Browning S. R., Browning B. L., Zhou Y., Tucci S., Akey J. M., Analysis of human sequence data reveals two pulses of archaic Denisovan admixture. Cell 173, 53–61.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wall J. D., et al. , Genetic variation in Native Americans, inferred from Latino SNP and resequencing data. Mol. Biol. Evol. 28, 2231–2237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blockus H., Chédotal A., Slit-Robo signaling. Development 143, 3037–3044 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Li Y., et al. , Robo signaling regulates the production of cranial neural crest cells. Exp. Cell Res. 361, 73–84 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Minoux M., et al. , Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science 355, eaal2913 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Long H. K., et al. , Loss of extreme long-range enhancers in human neural crest drives a craniofacial disorder. Cell Stem Cell 27, 765–783.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilderman A., VanOudenhove J., Kron J., Noonan J. P., Cotney J., High-resolution epigenomic atlas of human embryonic craniofacial development. Cell Rep. 23, 1581–1597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin F., et al. , A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scarpa E., et al. , Cadherin switch during EMT in neural crest cells leads to contact inhibition of locomotion via repolarization of forces. Dev. Cell 34, 421–434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris J., Källén B., Robert E., The epidemiology of anotia and microtia. J. Med. Genet. 33, 809–813 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forrester M. B., Merz R. D., Descriptive epidemiology of anotia and microtia, Hawaii, 1986-2002. Congenit. Anom. (Kyoto) 45, 119–124 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Fagundes N. J. R., et al. , How strong was the bottleneck associated to the peopling of the Americas? New insights from multilocus sequence data. Genet. Mol. Biol. 41(1, suppl. 1)206–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordero D. R., et al. , Cranial neural crest cells on the move: Their roles in craniofacial development. Am. J. Med. Genet. A. 155A, 270–279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geisen M. J., et al. , Hox paralog group 2 genes control the migration of mouse pontine neurons through slit-robo signaling. PLoS Biol. 6, e142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaffer J. R., et al. ; 23andMe Research Team, Multiethnic GWAS reveals polygenic architecture of earlobe attachment. Am. J. Hum. Genet. 101, 913–924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bashamboo A., Bignon-Topalovic J., Moussi N., McElreavey K., Brauner R., Mutations in the human ROBO1 gene in pituitary stalk interruption syndrome. J. Clin. Endocrinol. Metab. 102, 2401–2406 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Lu W., et al. , Disruption of ROBO2 is associated with urinary tract anomalies and confers risk of vesicoureteral reflux. Am. J. Hum. Genet. 80, 616–632 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poplin R., et al. , Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv [Preprint] (2017). https://www.biorxiv.org/content/10.1101/201178v3. Accessed 1 May 2020.
- 43.Collins R. L., et al. ; Genome Aggregation Database Production Team; Genome Aggregation Database Consortium, A structural variation reference for medical and population genetics. Nature 581, 444–451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spielman R. S., McGinnis R. E., Ewens W. J., Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am. J. Hum. Genet. 52, 506–516 (1993). [PMC free article] [PubMed] [Google Scholar]
- 45.Mukhopadhyay N., et al. , Whole genome sequencing of orofacial cleft trios from the Gabriella Miller Kids First Pediatric Research Consortium identifies a new locus on chromosome 21. Hum. Genet. 139, 215–226 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazeem G. R., Farrall M., Integrating case-control and TDT studies. Ann. Hum. Genet. 69, 329–335 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Nicodemus K. K., Catmap: Case-control and TDT meta-analysis package. BMC Bioinformatics 9, 130 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Homburger J. R., et al. , Genomic insights into the ancestry and demographic history of South America. PLoS Genet. 11, e1005602 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maples B. K., Gravel S., Kenny E. E., Bustamante C. D., RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 93, 278–288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montana G., Pritchard J. K., Statistical tests for admixture mapping with case-control and cases-only data. Am. J. Hum. Genet. 75, 771–789 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minikel E. V., et al. ; Exome Aggregation Consortium (ExAC), Quantifying prion disease penetrance using large population control cohorts. Sci. Transl. Med. 8, 322ra9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willems T., et al. , Genome-wide profiling of heritable and de novo STR variations. Nat. Methods 14, 590–592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma A., et al. ; Pediatric Cardiac Genomics Consortium, GATA6 mutations in hiPSCs inform mechanisms for maldevelopment of the heart, pancreas, and diaphragm. eLife 9, e53278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao S. S., et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Long A., et al. , Somatic instability of the expanded GAA repeats in Friedreich’s ataxia. PLoS One 12, e0189990 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All WGS datasets were sequenced as part of the NIH Gabriella Miller Kids First Program and are available via controlled access from dbGaP (accession nos. phs002172 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002172.v1.p1), phs002130 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002130.v1.p1), and phs001420 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001420.v1.p1) and at https://kidsfirstdrc.org. Human pinna and hiPSC RNA-seq gene expression data are available at the National Center for Biotechnology Information Sequence Read Archive and Gene Expression Omnibus (GSE202442). All software programs used are publicly available and are is listed in SI Appendix, Key Resources Table.