Abstract
The prolonged transmission of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus in the human population has led to demographic divergence and the emergence of several location-specific clusters of viral strains. Although the effect of mutation(s) on severity and survival of the virus is still unclear, it is evident that certain sites in the viral proteome are more/less prone to mutations. In fact, millions of SARS-CoV-2 sequences collected all over the world have provided us a unique opportunity to understand viral protein mutations and develop novel computational approaches to predict mutational patterns. In this study, we have classified the mutation sites into low and high mutability classes based on viral isolates count containing mutations. The physicochemical features and structural analysis of the SARS-CoV-2 proteins showed that features including residue type, surface accessibility, residue bulkiness, stability and sequence conservation at the mutation site were able to classify the low and high mutability sites. We further developed machine learning models using above-mentioned features, to predict low and high mutability sites at different selection thresholds (ranging 5–30% of topmost and bottommost mutated sites) and observed the improvement in performance as the selection threshold is reduced (prediction accuracy ranging from 65 to 77%). The analysis will be useful for early detection of variants of concern for the SARS-CoV-2, which can also be applied to other existing and emerging viruses for another pandemic prevention.
Keywords: COVID-19, SARS-CoV-2, Mutation, Protein mutability, Machine learning
Abbreviations: SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; ACE2, Angiotensin-converting enzyme 2; HIV, Human immunodeficiency virus; ML, Machine learning; VOI, Variants of interest; VOC, Variants of concern; SNP, Single nucleotide polymorphism; PSSM, Position specific scoring matrix; ROC, Receiver operating characteristic; AUC, Area under the curve; SVM, Support vector machine; LOOCV, Leave-one-out cross-validation; WHO, World Health Organization; PWM, Position weight matrix; IC, Information content; SMO, Sequential minimal optimization; MOI, Mutations of interest; MOC, Mutations of concern
Graphical abstract
1. Introduction
The Coronavirus pandemic (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has emerged as a global pandemic affecting more than 494 million people worldwide and resulting in around 6.1 million deaths (https://covid19.who.int/; accessed on April 11, 2022). The SARS-COV-2 virus is around 30,000 base pairs long single-stranded RNA virus that targets the human ACE2 receptor for fusion with the human cell membrane [1,2]. The viral genome encodes four structural proteins namely, spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins that are considered of high therapeutic value [3]. In a short span of COVID-19 emergence, the scientific community has developed several potential anti-SARS-CoV-2 therapeutics by targeting the viral protein(s) [[4], [5], [6], [7], [8], [9], [10], [11]].
Almost two years into the pandemic, several variants of the SARS-CoV-2 virus have emerged all around the world. Viruses naturally have high mutation rates, which provide critical diversity for natural selection to screen variants with better transmission and survival according to the environment [12,13]. One such example, “mutation D614G in spike protein,” is studied extensively. It enhances viral replication in human airway passage tissues and lung epithelial cells by increasing the infectivity and stability of virions [14]. Some of the new strains of SARS-CoV-2, for example, Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2) or Omicron (B.1.1.529) are more transmissible than the original one [15,16]. Currently, Delta and Omicron variants are dominant strains in circulation. The Delta variants had relatively fewer mutation sites in the Spike protein (T19, E156, L452, T478, D614, P681 and D950) compared to the Omicron variants, which had more than 30 mutation sites in the spike protein. Previously reported Alpha, Beta and Gamma variants had few overlapping mutation sites (N501, K417 and D614) in the spike protein and these variants are no longer in circulation.
As of April 2022, a significant part of the population is vaccinated in most of the countries. Although, there is a possibility of existing or emerging strains to be vaccine-resistant. It is an utmost priority for the scientific community to observe and understand these new strains to terminate the pandemic as early as possible. A deeper understanding of the mutation can lead to early prediction of the viral variants and subsequently in silico protocols can be utilized to identify/design drugs and vaccines [[17], [18], [19]].
In the recent advancement in the mutational study of the SARS-CoV-2, Garvin et al. [20] used an artificial intelligence approach to find the mutational hotspots in SARS-CoV-2 genome to provide insights into drug development and surveillance strategies to combat the current and future pandemics. Other studies have compared the binding interface [21,22] and mutational pattern in the SARS-CoV-2 with other similar coronaviruses [23]. Several studies have looked into the evolving geographic diversity of the SARS-CoV-2 [[24], [25], [26], [27], [28]]. Sen et al. [29] have analyzed the structural malleability of viral proteins that may lead to comorbidities. Researchers have also studied the effect of mutation on binding affinity of the known SARS-CoV-2 specific antibodies [30]. The analysis of protein site conservation/mutability was mainly limited to HIV viruses before pandemic to identify immunogens for vaccine [31,32] due to the unavailability of relatively large-scale datasets. Similarly, there have been few studies exploring the conservation of SARS-CoV-2 proteome using multiple sequence alignment to identify the vaccine targets [33,34]. However, these studies depend on prior knowledge of viral variants and may not be effective for the prediction in new viruses. A recent study used unsupervised probabilistic models using direct coupling analysis (DCA) to predict SARS-CoV-2 mutable and constrained positions, which incorporate pairwise epistatic terms and all known coronavirus genomes to allow selective pressure for coronaviruses [35].
The ongoing pandemic is changing the mutation dynamics of the SARS-CoV-2 proteome on a daily basis. However, not all protein sites observe an equal mutation rate in the population. The observed mutability of protein sites can be potentially attributed to the combined effect of intrinsic physicochemical parameters [36] and effect on the transmission/survival of the virus [13]. The intrinsic physicochemical parameters (such as residue composition, surface accessibility, local stability, residue contacts, hydrophobicity, etc.) predicted from the viral sequence-structure information, are well characterized in several studies such as aggregation [37,38], stability [39,40], function [[41], [42], [43]], binding [44], etc. These are the inherent properties of the sequence and therefore expected to be applicable to protein site mutability of all multicellular organisms including virus species, over a large time frame [45]. On the other hand, protein-protein interaction and potential residue modifications affecting biological processes are important for preservation of mutation. However, these phenomena cannot be generalized and are specific to organisms [46,47].
In this work, we have analyzed the mutation information from “2019 Novel Coronavirus Resource (2019nCoVR, https://bigd.big.ac.cn/ncov)” to understand the intrinsic sequence-structure factors that affect the mutability of proteins with respect to reference SARS-CoV-2 proteome from Wuhan. The initial analysis to understand the physicochemical parameters affecting mutability of protein sites in the whole proteome and each protein is done at 30% selection threshold (top 30% high and low mutation sites based on mutant isolate count). We observed that physicochemical features such as residue type, surface accessibility, residue bulkiness, stability and conservation can distinguish the sites with high and low mutation frequency. Further, we developed machine learning (ML) models using these features to classify the high and low mutation sites at different selection thresholds ranging from 5 to 30% and obtained model accuracy in the range of 65–76.7%. It was observed that increasing the confidence level of low and high mutability sites (i.e. lowering the selection threshold) improves the prediction performance of the ML models. We further observed that the physicochemical features of the mutation sites in variant of concern (VOC) and interest (VOI) are potential causes of their higher mutation rate (VOC and VOI information collected in June 2021). The study provides significant insights into the viral mutability, which can be helpful in early detection of potentially harmful viral variants for SARS-CoV-2 and other infectious viruses.
2. Methods
2.1. Dataset preparation
We have downloaded the variance annotation dataset from the 2019 Novel Coronavirus Resource (https://bigd.big.ac.cn/ncov/variation/annotation) [48] in June 2021. The non-synonymous single nucleotide polymorphism (SNP) entries were considered in the analysis. The structures of the SARS-CoV-2 proteins were obtained from (https://zhanglab.ccmb.med.umich.edu/COVID-19/) [49]. The physicochemical features of the mutation sites were analyzed by classifying the mutation sites at 30% selection threshold, where top 30% mutation sites with high isolate count were considered “high mutability sites” and bottom 30% of the mutation sites with low isolate count were considered “low mutability sites”. The remaining 40% in the middle were considered ambiguous to be classified in any category. The number of mutation sites and cutoff of isolate count for 30% selection threshold are given in Table S1. A separate dataset at 10% selection threshold was also prepared to verify the observations of 30% selection threshold.
2.2. Collection of sequence and structural-based features
Collection of biologically-relevant features is an important step in machine learning model development and statistical analysis of complex biological problems [[50], [51], [52], [53]]. We collected several sequence and structure features for the viral proteins from various sources and custom scripts. Briefly, features include relative accessible surface area (rASA) [54], all atom residue depth [55], surrounding hydrophobicity (within heavy atoms contact distance of 5 Å) (https://www.iitm.ac.in/bioinfo/pdbparam/index.html), sequence based physicochemical and energetic features [56], residue type (polar, non-polar and charged) and contacting residues information (within the heavy atom contact distance of 5 Å). The sequence-based features were average values of tripeptides occurring at the mutation site along with one residue on each side. The position specific scoring matrix (PSSM) profiles were generated for each viral protein position using “blastpgp” on the “UniRef90” database [57]. Further, above-mentioned features were filtered based on inter-property correlation (r ≤ 0.8) and statistical difference in mean value of low and high mutability sites (p-value≤ 10–11, at 30% selection threshold). The final dataset contained 21 sequence and structure-based features (Table S2).
2.3. Feature selection and development of machine learning (ML) models
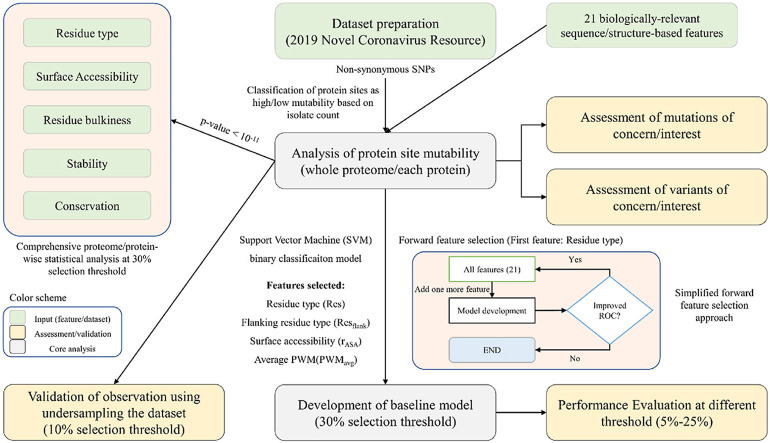
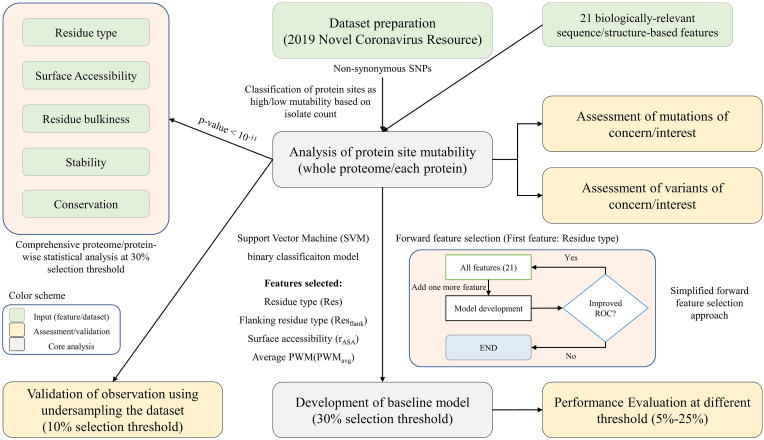
We used a forward feature selection approach to select the optimal number of features in the baseline model. Firstly, we selected the best performing feature in the ML model based on Area under the ROC curve (AUC). Further, features were added one by one until the best performance (AUC) of the model was reached (Fig. 1 ). We restricted to a maximum of six features to avoid overfitting and the final model contains four features with a balance between the number of features and performance. The baseline ML model was developed at 30% selection threshold using “support vector machine (SVM)” and linear kernel in Weka 3.8.6 [58]. The SVM based parameter “BuildLogisticModels” was kept “True” and “optimal complexity parameter (c)" was optimized to 2.0 to obtain the best performance. The rest of the parameters were kept default. The final selected model was also trained on selection thresholds ranging from 5% to 25% to observe change in performance upon undersampling.
Fig. 1.
Workflow illustrating the steps followed in the current study.
2.4. Performance evaluation
The performance of the model at 30% selection threshold was evaluated primarily using area under the ROC (receiver operating characteristic) curve. We have also included following performance measures for the final ML model:
| (1) |
| (2) |
| (3) |
where TP, TN, FP and FN are the number of true positives, true negatives, false positives and false negatives, respectively. In our study, low mutability sites are considered positive and high mutability sites are considered negative class. The robustness of the model was evaluated using 10-fold and leave-one-out cross-validation (LOOCV). The 10-fold cross-validation was performed 100 times while randomizing the dataset each time. In leave-one-out cross-validation, the regression model was trained on n-1 data points and tested on the remaining one data point, recursively.
2.5. Analysis of variants of concern (VOCs) and variant of interest (VOIs)
The list containing mutations in VOCs and VOIs of SARS-CoV-2 virus (designated by WHO as of June 2021) were obtained from https://outbreak.info/. In addition, we have also collected the list of mutations of interest and mutation of concern. The radar plot for these mutation(s) was plotted using Matplotlib library [59] in python.
3. Result and discussion
3.1. Analysis of the dataset
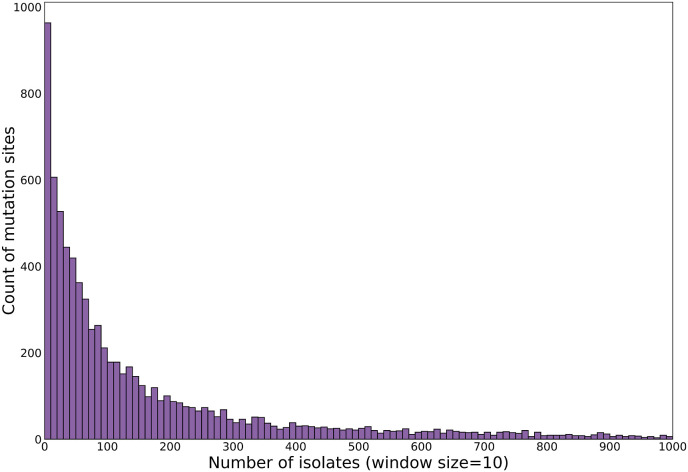
We analyzed 8673 protein sites in SARS-CoV-2 proteome containing at least one mutation among 1079273 isolates (Fig. 1). Firstly, we plotted a histogram for the number of mutation sites in the whole SARS-CoV-2 proteome with respect to their isolate counts (Fig. 2 ). The histogram represented approximately 90% of the protein sites with less than 1000 isolate count and showed an exponential decay curve, where more than 950 protein sites had less than 10 mutant isolates and only 14 protein sites had mutant isolate count of more than 100,000. The higher isolate count generally denotes mutation in the protein site at an early stage of the pandemic and incorporation of the mutation in all major viral variants.
Fig. 2.
A histogram plotted for the number of isolates observed with respect to number of mutation sites. Approximately 90% of the mutation sites have less than 1000 isolates containing mutation, although the highest isolate count is 1,079,273.
3.2. Role of sequence and structure-based features on site mutability
We have analyzed several sequence and structure-based features to classify the low and high mutability of the protein sites. We first filtered the features based on the “statistical significance” and “low inter-property correlation” selection criteria (as described in Collection of sequence and structural-based features in Methods section). The features, capable of distinguishing the low and high mutability of protein sites, are further classified into five general categories and discussed in detail. It is also important to note that size of viral proteins (corresponding to number of mutation sites) vary greatly (ranging from 30 to 1757 residues), which can lead to less to no statistical significance in some protein-wise results (Table S1).
3.3. Residue type
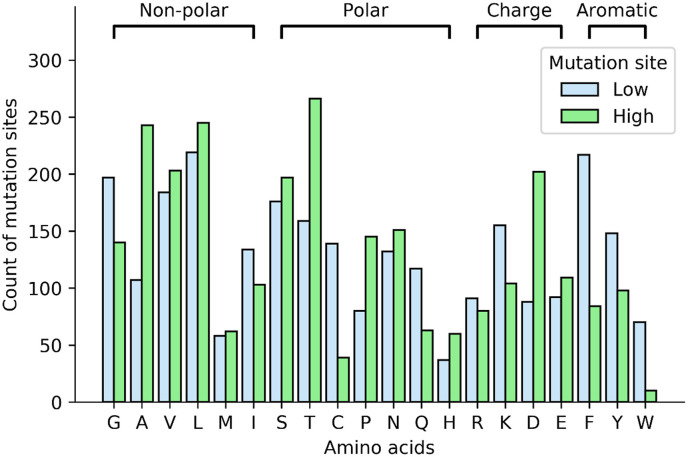
We observed that residue-type is the most capable feature to classify the high and low mutability of protein sites in the SARS-CoV-2 proteome (Fig. 3 ). The proportion of bulky aromatic residues (F, Y, W) is significantly higher in the low mutability sites. The positively charged residues (R, K) occur frequently at low mutability sites whereas negatively charged residues (D, E) are present more in high mutability sites. Gly (G), a smaller amino acid with similar physicochemical features as Ala (A) has surprisingly higher frequency in the low mutability. Overall, Ala (A), Cys (C), Asp (D), Phe (F), Trp(W) amino acids have more than 2-fold difference in the frequency in the low and high mutability sites. The probable reason for low mutation frequency observed for bulky and small amino acids is likely to be due to the loss of interactions and steric hindrance, respectively. The mutability of the charge residues is mainly dependent on the environment. However, intravirion environment (RNA) and overall viral surface is negatively charged leading to higher mutation rate in negatively charged residues for better stability [60]. Mutations in R, G, C and W residues have been linked to higher probability of disease-causing mutations in humans [61]. A similar observation in SARS-CoV-2 virus indicates that mutations in these residues may also lead to decrease in fitness of the virus.
Fig. 3.
Amino acid frequency in low and high mutation sites class.
3.4. Surface accessibility
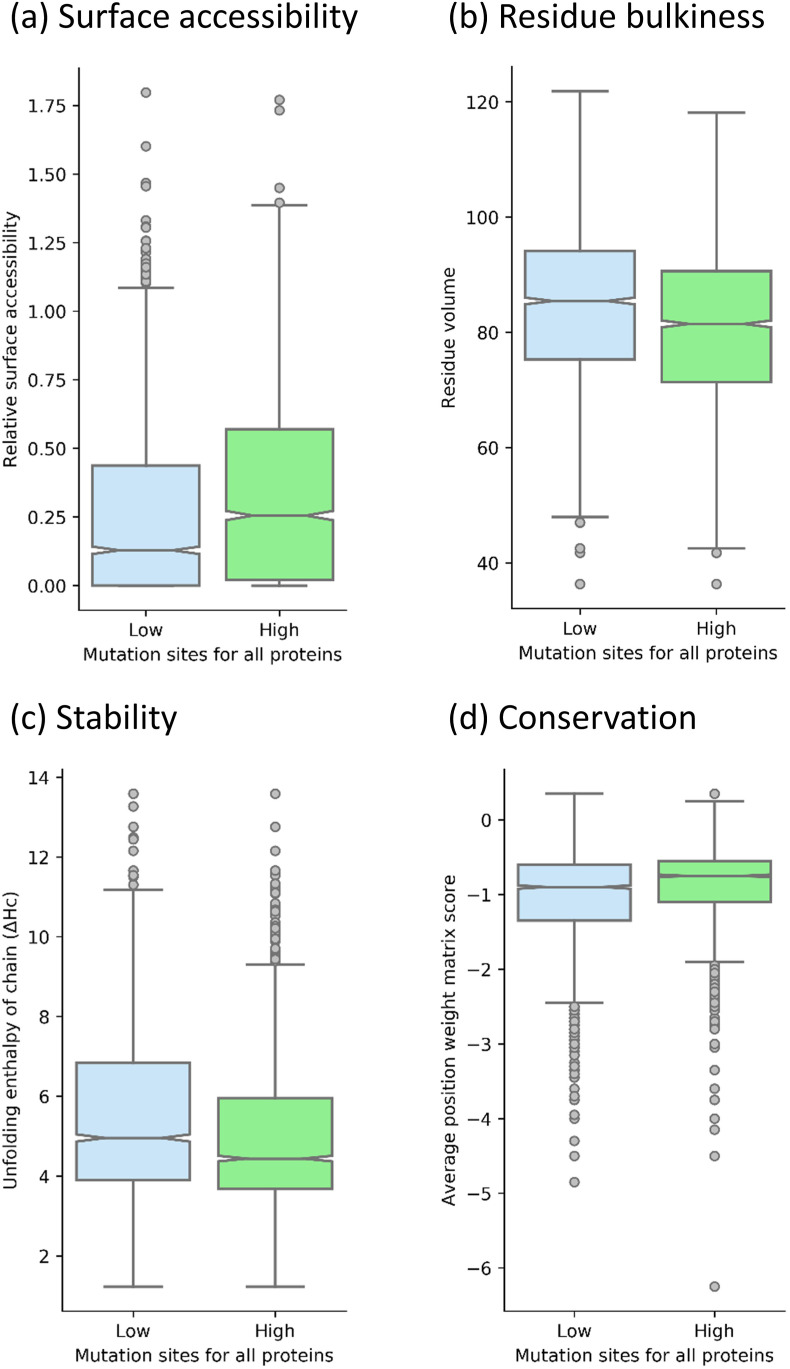
The Relative accessible surface area (rASA) feature calculated from Dictionary of Secondary Structure of Proteins (DSSP) [54] is an important feature to identify mutation sites with high and low mutation rates (Figs. 4a and S1a). In the SARS-CoV-2 proteins, it was observed that high mutability sites also have higher relative accessible surface area and vice versa, for most of the large proteins. The observation is reasonable as most residues in small proteins are surface accessible due to small size. The proteins including E, M, nsp4, nsp6, nsp7, nsp8 and ORF6 showed an opposite or no trend for surface accessibility. The features related to buriedness (such as number of contacts at 5 Å distance and residue depth) also supports the observation of relative accessible surface area (data not shown). Surface accessibility is also important for the interaction with the environment including self/host proteins [62]. Therefore, mutability of these sites can significantly affect the survival or transmission of the virus. It is also important to note that surface accessibility alone is not sufficient to predict the mutability of protein sites as only a small percentage of the surface accessible sites interact with other molecules.
3.5. Residue bulkiness
The residue type analysis showed that bulky residues such as aromatic residues are highly preferred in the low mutability sites in the SARS-CoV-2 proteome. The extended analysis using residue volume feature (AAindex id: BIGC670101) supported the observation for all amino acids (Fig. 4 b). Similar trend was also observed in each protein (Fig. S1b). However, the p-values from the t-test showed relatively less statistically significant outcomes among other major features discussed. Other related features such as molecular weight also showed that low mutability sites have higher molecular weight and vice versa (data not shown). The bulky amino acids most likely show low mutational frequency due to higher contact order and biosynthesis cost [[63], [64], [65]]. This also explains the observation that bulky aromatic groups such as Tyr are preferred at protein interaction sites and are less likely to mutate [21,66,67].
Fig. 4.
Major features under the category of surface accessibility, residue bulkiness, stability of mutation site and conservation of the mutation site (p-value<10−11).
3.6. Stability of the mutation site
Understandably, we observed that locally stable protein sites are less likely to mutate to avoid destabilization of the protein structure [68]. We calculated the local average stability of the mutation site and one flanking residue on each side using unfolding enthalpy of the chain (ΔHc). The feature showed that low mutability sites are more stable compared to high mutability sites (Fig. 4c). The protein-wise analysis also showed similar results except for E, nsp10 and ORF6 proteins (Fig. S1c).
3.7. Conservation of the mutation site
Residue conservation is directly linked to the mutability of the amino acids. A residue is likely to be conserved in observed protein if it is conserved in the closest homologous proteins. Therefore, we used several sequence conservation related features derived from the position-specific scoring matrix (PSSM). The average value of the 20 amino acids in the position weight matrix (PWM) was able to classify the high and low mutation sites (Fig. 4d) and it is more negative for specific dominant mutations. The higher chances of random mutations shift the average value of the PWM matrix towards the positive scale. We observed that high mutability sites also have higher values of average PWM, which is consistent in all SARS-CoV-2 proteins except N, nsp1 and nsp7 (Fig. S1d). Therefore, these high mutability sites are more prone to be replaced by any other amino acids. On the other hand, low mutability sites prefer only self (or specific) mutations and are considered relatively more conserved. We have also analyzed the information content (IC) parameter in the PSSM file, which measures the probability of a given PWM to be different from the uniform distribution. Expectedly, we observed a weak negative correlation (−0.14) between isolate count and information content (Fig. S2a). The high mutability sites are expected to have more uniform distribution of possible mutations leading to decrease in information content [69]. However, it is not sufficient to differentiate high and low mutability sites alone (Fig. S2b).
3.8. Analysis for high and low mutability sites using 10% selection threshold
The above-discussed major features are also calculated for an undersampled dataset to observe consistency of the results. The isolate count cutoffs and number of mutation data are reevaluated at 10% selection threshold (881 protein sites in low mutability class and 867 protein sites in high mutability class), which in turn also reduced the dataset size for each protein and restricted statistically significant observations. However, the observations with the undersampled dataset at 10% selection threshold were the same as the observation at 30% selection threshold. In summary, the residue type at the mutation site showed higher presence of Gly, positively charged and aromatic residues in the low mutability sites (Fig. S3). High mutability sites also observed lower values for residue bulkiness and stability features and higher values for surface accessibility and conservation features (Fig. S4).
3.9. Machine-learning model development
We further developed a machine-learning model to assess the ability of the intrinsic physicochemical features to predict low and high mutability sites in SARS-CoV-2 proteome. The baseline model is developed at 30% selection threshold (top and bottom 30% of the mutation sites selected based on mutant isolate count) as discussed below:
3.10. Development of baseline model
We used a forward feature selection approach to select the optimal number of features in the baseline model (Fig. S5). We observed the best model performance (area under the ROC curve: 0.71) with four features and SMO (Sequential Minimal Optimization) algorithm, a SVM (Support Vector Machine) based method for the classification (see Methods section for more detail). SVM based models have been extensively used in the biological problem due to better interpretability, learnability and generalization [[70], [71], [72]]. The selected feature in the SVM model includes residue at the mutation site (residue type; Res), residues flanking the mutation site (Resflank), relative accessible surface area (rASA) and average value of position weight matrix (PWMavg). Further optimization of the model parameters revealed the accuracy of 65% with sensitivity of 62.4% and specificity of 67.7% with ROC value of 0.711 for the training dataset (Table 1 ). The performance of the model was further rigorously tested using different performance measures including 10-fold cross-validation with randomization (average ROC of 0.646 ± 0.002 after 100 iteration) and n-fold cross-validation (ROC = 0.648). The analysis showed that the developed model is robust (Table 1).
Table 1.
Performance of the baseline model at 30% selection threshold.
| Performance Measure | Accuracy | Sensitivity | Specificity | ROC |
|---|---|---|---|---|
| Training dataset | 65 | 62.4 | 67.7 | 0.711 |
| Leave-one-out cross-validation | 60.03 | 57.9 | 62.2 | .648 |
| 10-fold cross-validationa | 60.2 ± 0.34 | 57.6 ± 0.51 | 62.7 ± 0.46 | 0.646 ± 0.002 |
The average values are listed along with standard deviation from 100 iterations after randomizing data each time.
We further analyzed the importance of each feature in the ML model. The features “Res” and “Resflank” significantly reduce the performance of the model upon elimination (ROC 0.652 and 0.649, respectively). On the other hand, “Res” feature showed the best performance (ROC = 0.621) when only one feature was used in the model. Therefore, we concluded that “Residue type (Res)” feature is the most important feature for the classification of the low and high mutability of protein sites (Table S3).
3.11. Performance of the machine learning model at different selection threshold
The baseline model developed at 30% selection threshold was further tested on other thresholds ranging from 5 to 25%. We observed that the performance of the model increases as the selection threshold decreases (Table 2 ). The correlation between area under the ROC curve and selection threshold was also high (r2 = 0.95; Fig. S6). This is mainly due to the fact that decreasing the selection threshold proportionally setup more stringent conditions for mutations to be assigned to either low or high mutation sites, thus improving the confidence level.
Table 2.
Performance of the baseline model at different selection threshold range.
| Selection threshold | Dataset |
Performance measures |
|||||
|---|---|---|---|---|---|---|---|
| Total mutation sites | Low mutability sites | High mutability sites | Accuracy | Sensitivity | Specificity | ROC | |
| 5 | 864 | 430 | 434 | 76.7 | 76.5 | 77 | 0.84 |
| 10 | 1748 | 881 | 867 | 72.8 | 73 | 72.5 | 0.795 |
| 15 | 2589 | 1288 | 1301 | 69.9 | 68.3 | 71.5 | 0.761 |
| 20 | 3453 | 1718 | 1735 | 68.4 | 66.5 | 70.3 | 0.747 |
| 25 | 4357 | 2187 | 2170 | 66.8 | 66.1 | 67.5 | 0.73 |
| 30 | 5204 | 2600 | 2604 | 65 | 62.4 | 67.7 | 0.711 |
3.12. Case study: analysis of variants/mutation of concern/interest
The physicochemical features including surface accessibility, residue bulkiness, stability and conservation were analyzed for the mutation/variants of concern and interest with respect to average value of all protein sites (Table 3 ). There was a total of nine protein sites in spike protein containing at least one mutation of interest (L18, K417, N439, L452, S477, S494, N501, P681) or concern (E484). These mutation sites were considered as high mutability sites, where they are expected to have higher than average values for surface accessibility and conservation features and lower than the average values for residue bulkiness and stability features. Among the nine mutations of interest and concern in the spike protein, six mutations (E484, K417, N439, S477, N501, P681) satisfied the criteria for all four features. The remaining three mutation sites L18, S494 and L452 satisfy the criteria for 3, 2 and 1 features, respectively.
Table 3.
The features related to mutation probability analyzed for the mutation of concern and mutation of interest.
| Mutation sites of concern/interest | Surface accessibility | Residue bulkiness | Stability of the mutation site | Conservation of the mutation site |
|---|---|---|---|---|
| S:E484 | 1.05 | 68.7 | 3.46 | −0.4 |
| S:L18 | 0.07 | 82.97 | 3.92 | −0.65 |
| S:K417 | 0.64 | 81.13 | 2.94 | −0.25 |
| S:N439 | 0.39 | 68.77 | 4.36 | −0.45 |
| S:L452 | 0.29 | 111.47 | 10.84 | −0.25 |
| S:S477 | 0.96 | 54.13 | 3.82 | −0.35 |
| S:S494 | 0.63 | 86.93 | 8.23 | −0.55 |
| S:N501 | 0.41 | 61.07 | 3.1 | −0.3 |
| S:P681 | 0.61 | 79.2 | 4.6 | −0.85 |
| Average | 0.3 | 82.98 | 5.16 | −0.94 |
Note: The average values are calculated from the mutation sites considered in the current study of SARS-CoV-2 proteome. These mutations of concern/interest are expected to be present at the high mutability sites. The features that do not follow the observed trend in the study are highlighted.
The list of mutations obtained from https://outbreak.info/.
Mutation of concern (MOC): S:E484K.
Mutation of interest (MOI): S:L18F; S:K417N; S:K417T; S:N439K; S:L452R; S:S477N; S:S494P; S:N501Y; S:P681H; S:P681R.
A further extended analysis was carried out for all mutation sites in the proteome of SARS-CoV-2 variants of concern (VOC) and interest (VOI), and the results are presented in Figs. S7 and S8, respectively. The analysis of the four physicochemical features showed higher mutability in most protein sites of the four VOCs: Delta (B.1.617.2), Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) and two VOIs: Lambda (C.37), Mu (B.1.621). The VOCs (average: ∼76.4%) had a relatively higher number of mutation sites satisfying 3 or more features compared to VOIs (average: ∼64%) (Table 4 ). Therefore, higher chances of mutation in these protein sites lead to emergence of new variants that improved the fitness of the virus in terms of better survivability and more transmissibility.
Table 4.
The mutation sites of variants of concern and interest analyzed for four physicochemical features.
| Variant of concern | Mutation sites | Number of features satisfying criteria |
||||
|---|---|---|---|---|---|---|
| 4 | 3 | 2 | 1 | 0 | ||
| Delta (B.1.617.2) | 24 | 11 (45.8%) | 7 (29.2%) | 3 (12.5%) | 2 (8.3%) | 1 (4.2%) |
| Alpha (B.1.1.7) | 19 | 10 (52.6%) | 4 (21.1%) | 5 (26.3%) | 0 (0%) | 0 (0%) |
| Beta (B.1.351) | 16 | 8 (50%) | 4 (25%) | 2 (12.5%) | 2 (12.5%) | 0 (0%) |
| Gamma (P.1) | 22 | 13 (59.1%) | 5 (22.7%) | 3 (13.6%) | 1 (4.5%) | 0 (0%) |
| Variant of interest | ||||||
| Lambda (C.37) | 19 | 7 (36.8%) | 5 (26.3%) | 4 (21.1%) | 3 (15.8%) | 0 (0%) |
| Mu (B.1.621) | 20 | 11 (55%) | 2 (10%) | 6 (30%) | 1 (5%) | 0 (0%) |
As per the study, the satisfactory criteria for the feature is: 1. High mutability sites are likely to have higher than average value for surface accessibility and conservation, and vice versa.
2. High mutability sites are likely to have lower than average value for residue bulkiness, and stability, and vice versa.
3.13. Potential applications
The study will improve our understanding of intrinsic physicochemical parameters affecting mutability of the viral proteome, which in combination with the virus-specific biological features (such as important binding/cleavage sites) can be used to predict the potential future mutations leading to improvement in survivability, infectivity or lethality of the virus. The intrinsic parameters discussed here can be used as a starting point for in silico prediction of future variants of any pathogen. Moreover, exposed protein sites with less probability of mutation can be used as immunogens for vaccine development or potential epitopes for antibody-based therapeutics.
4. Conclusion
In this study, we have provided insights into the mutability of SARS-CoV-2 proteome from the perspective of intrinsic sequence-structure-based features. The study highlights the role of surface accessibility, residue bulkiness, stability and evolutionary conservation in determining the mutational probability of a protein site. The major advantage of the study is that it does not require any priori information other than the sequence and structure information of the virus of concern. The study leverages the large-scale mutational data (1079273 viral isolates of SARS-CoV-2) to predict the protein sites that are less or more prone to mutations. The study also focuses on the robustness of the inference by utilizing different selection thresholds, as the reference dataset is changing daily. Although, it is also important to note that the study has some limitations such as mutations are considered mutually independent, deletions/insertions are excluded and biological/functional aspects are not considered. Moreover, mutations are considered only with respect to reference Wuhan strain due to lack of real-time mutation data, which may lead to biases towards the early mutations in the SARS-CoV-2 genome. A more sophisticated time series analysis based on real-time viral mutation, effect of concurrent mutations and role of the biologically relevant protein sites can be explored further for greater understanding of viral protein mutability. The dataset/features used in the study can be obtained from the GitHub repository (https://github.com/puneetrawat/COVID_Mutation_Site).
Author contribution
Puneet Rawat: Conceptualization; Formal Analysis; Data Curation; Investigation; Methodology; Writing – Original Draft Preparation. Divya Sharma: Data Curation; Methodology. Medha Pandey: Methodology. R. Prabakaran: Investigation. M. Michael Gromiha: Conceptualization; Funding Acquisition; Supervision; Writing – Review & Editing.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgement
We thank the Department of Biotechnology and Indian Institute of Technology Madras for computational facilities and the Ministry of human resource and development (MHRD) for HTRA scholarship to DS and MP. This work is partially supported by the Robert Bosch Center for Data Science and Artificial Intelligence (RBCDSAI), Indian Institute of Technology Madras, India to MMG (Project no: CR1718CSE001RBEIBRAV).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105708.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W., Sun Z., Liu F., Wu K., Zhong B., Mei Y., Zhang W., Chen Y., Li Y., Shi M., Lan K., Liu Y. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microb. Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yesudhas D., Srivastava A., Gromiha M.M. COVID-19 outbreak: history, mechanism, transmission, structural studies and therapeutics. Infection. 2021;49:199–213. doi: 10.1007/s15010-020-01516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabie A.M. Two antioxidant 2,5-disubstituted-1,3,4-oxadiazoles (CoViTris2020 and ChloViD2020): successful repurposing against COVID-19 as the first potent multitarget anti-SARS-CoV-2 drugs. New J. Chem. 2021;45:761–771. doi: 10.1039/d0nj03708g. [DOI] [Google Scholar]
- 5.Zhang S., Amahong K., Sun X., Lian X., Liu J., Sun H., Lou Y., Zhu F., Qiu Y. The miRNA: a small but powerful RNA for COVID-19, Brief. Bioinformation. 2021;22:1137–1149. doi: 10.1093/bib/bbab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabie A.M. Discovery of Taroxaz-104: the first potent antidote of SARS-CoV-2 VOC-202012/01 strain, J. Mol. Struct. 2021;1246 doi: 10.1016/j.molstruc.2021.131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabie A.M. Cyanorona-20: the first potent anti-SARS-CoV-2 agent, Int. Immunopharmacology. 2021;98 doi: 10.1016/j.intimp.2021.107831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Rabie A.M. Teriflunomide: a possible effective drug for the comprehensive treatment of COVID-19, Curr Res Pharmacol Drug Discov. 2021;2 doi: 10.1016/j.crphar.2021.100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Y.-W., Xie Y., Tang L.-S., Pu D., Zhu Y.-J., Liu J.-Y., Ma X.-L. Therapeutic targets and interventional strategies in COVID-19: mechanisms and clinical studies. Signal Transduct. Targeted Ther. 2021;6:317. doi: 10.1038/s41392-021-00733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niknam Z., Jafari A., Golchin A., Danesh Pouya F., Nemati M., Rezaei-Tavirani M., Rasmi Y. Potential therapeutic options for COVID-19: an update on current evidence. Eur. J. Med. Res. 2022;27:6. doi: 10.1186/s40001-021-00626-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabie A.M. Potent inhibitory activities of the adenosine analogue cordycepin on SARS-CoV-2 replication. ACS Omega. 2022;7:2960–2969. doi: 10.1021/acsomega.1c05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 14.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.-Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Oosterhout C., Hall N., Ly H., Tyler K.M. COVID-19 evolution during the pandemic – implications of new SARS-CoV-2 variants on disease control and public health policies. Virulence. 2021;12:507–508. doi: 10.1080/21505594.2021.1877066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahase E. Delta variant: what is happening with transmission, hospital admissions, and restrictions? BMJ. 2021;373 doi: 10.1136/bmj.n1513. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z., Bogdan P., Nazarian S. An in silico deep learning approach to multi-epitope vaccine design: a SARS-CoV-2 case study. Sci. Rep. 2021;11:3238. doi: 10.1038/s41598-021-81749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawat P., Sharma D., Srivastava A., Janakiraman V., Gromiha M.M. Exploring antibody repurposing for COVID-19: beyond presumed roles of therapeutic antibodies. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-89621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galanis K.A., Nastou K.C., Papandreou N.C., Petichakis G.N., Pigis D.G., Iconomidou V.A. Linear B-cell epitope prediction for in silico vaccine design: a performance review of methods available via command-line interface, int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22063210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garvin M.R., T Prates E., Pavicic M., Jones P., Amos B.K., Geiger A., Shah M.B., Streich J., Felipe Machado Gazolla J.G., Kainer D., Cliff A., Romero J., Keith N., Brown J.B., Jacobson D. Potentially adaptive SARS-CoV-2 mutations discovered with novel spatiotemporal and explainable AI models. Genome Biol. 2020;21:304. doi: 10.1186/s13059-020-02191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawat P., Jemimah S., Ponnuswamy P.K., Gromiha M.M. Why are ACE2 binding coronavirus strains SARS-CoV/SARS-CoV-2 wild and NL63 mild? Proteins. 2021;89:389–398. doi: 10.1002/prot.26024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity, Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. .e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matyášek R., Kovařík A. Mutation patterns of human SARS-CoV-2 and bat RaTG13 coronavirus genomes are strongly biased towards C>U transitions, indicating rapid evolution in their hosts, genes. 2020. 11. [DOI] [PMC free article] [PubMed]
- 24.Chitranshi N., Gupta V.K., Rajput R., Godinez A., Pushpitha K., Shen T., Mirzaei M., You Y., Basavarajappa D., Gupta V., Graham S.L. Evolving geographic diversity in SARS-CoV2 and in silico analysis of replicating enzyme 3CLpro targeting repurposed drug candidates. J. Transl. Med. 2020;18:278. doi: 10.1186/s12967-020-02448-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercatelli D., Giorgi F.M., Geographic, Distribution Genomic. Of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta A., Banerjee S., Das S. Significance of geographical factors to the COVID-19 outbreak in India. Model Earth Syst Environ. 2020:1–9. doi: 10.1007/s40808-020-00838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prabakaran R., Jemimah S., Rawat P., Sharma D., Gromiha M.M. A novel hybrid SEIQR model incorporating the effect of quarantine and lockdown regulations for COVID-19, Sci. Rep. 2021;11 doi: 10.1038/s41598-021-03436-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha I., Ghosh N., Sharma N., Nandi S. Hotspot mutations in SARS-CoV-2. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.753440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen S., Dey A., Bandhyopadhyay S., Uversky V.N., Maulik U. Understanding structural malleability of the SARS-CoV-2 proteins and relation to the comorbidities, Brief. Bioinformation. 2021 doi: 10.1093/bib/bbab232. [DOI] [PubMed] [Google Scholar]
- 30.Sharma D., Rawat P., Janakiraman V., Gromiha M.M. Elucidating important structural features for the binding affinity of spike - SARS-CoV-2 neutralizing antibody complexes. Proteins. 2021 doi: 10.1002/prot.26277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferguson A.L., Mann J.K., Omarjee S., Ndung’u T., Walker B.D., Chakraborty A.K. Translating HIV sequences into quantitative fitness landscapes predicts viral vulnerabilities for rational immunogen design. Immunity. 2013;38:606–617. doi: 10.1016/j.immuni.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahirel V., Shekhar K., Pereyra F., Miura T., Artyomov M., Talsania S., Allen T.M., Altfeld M., Carrington M., Irvine D.J., Walker B.D., Chakraborty A.K. Coordinate linkage of HIV evolution reveals regions of immunological vulnerability. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11530–11535. doi: 10.1073/pnas.1105315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed S.F., Quadeer A.A., McKay M.R. COVIDep: a web-based platform for real-time reporting of vaccine target recommendations for SARS-CoV-2. Nat. Protoc. 2020;15:2141–2142. doi: 10.1038/s41596-020-0358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarmarkovich M., Warrington J.M., Farrel A., Maris J.M. Identification of SARS-CoV-2 vaccine epitopes predicted to induce long-term population-scale immunity. Cell Rep Med. 2020;1 doi: 10.1016/j.xcrm.2020.100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.J. Rodriguez-Rivas, G. Croce, M. Muscat, M. Weigt, Epistatic models predict mutable sites in SARS-CoV-2 proteins and epitopes, (n.d.). 10.1101/2021.12.11.472202. [DOI] [PMC free article] [PubMed]
- 36.Hecht M., Bromberg Y., Rost B. News from the protein mutability landscape. J. Mol. Biol. 2013;425:3937–3948. doi: 10.1016/j.jmb.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 37.Rawat P., Prabakaran R., Kumar S., Michael Gromiha M. AggreRATE-Pred: a mathematical model for the prediction of change in aggregation rate upon point mutation. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz764. [DOI] [PubMed] [Google Scholar]
- 38.Prabakaran R., Rawat P., Thangakani A.M., Kumar S., Gromiha M.M. Protein aggregation: in silico algorithms and applications. Biophys. Rev. 2021;13:71–89. doi: 10.1007/s12551-021-00778-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marabotti A., Scafuri B., Facchiano A. Predicting the stability of mutant proteins by computational approaches: an overview. Briefings Bioinf. 2021;22 doi: 10.1093/bib/bbaa074. [DOI] [PubMed] [Google Scholar]
- 40.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan K.P., Kanitkar T.R., Kwoh C.K., Madhusudhan M.S. Packpred: predicting the functional effect of missense mutations. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.646288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong J., Luo Y., Mou M., Fu J., Zhang Y., Xue W., Xie T., Tao L., Lou Y., Zhu F. Convolutional neural network-based annotation of bacterial type IV secretion system effectors with enhanced accuracy and reduced false discovery. Briefings Bioinf. 2020;21:1825–1836. doi: 10.1093/bib/bbz120. [DOI] [PubMed] [Google Scholar]
- 43.Hong J., Luo Y., Zhang Y., Ying J., Xue W., Xie T., Tao L., Zhu F. Protein functional annotation of simultaneously improved stability, accuracy and false discovery rate achieved by a sequence-based deep learning. Briefings Bioinf. 2020;21:1437–1447. doi: 10.1093/bib/bbz081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delgado J., Radusky L.G., Cianferoni D., Serrano L. FoldX 5.0: working with RNA, small molecules and a new graphical interface. Bioinformatics. 2019;35:4168–4169. doi: 10.1093/bioinformatics/btz184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan S., Wu G. Application of neural network to predict mutations in proteins from influenza A viruses - a review of our approaches with implication for predicting mutations in coronaviruses. J. Phys. Conf. Ser. 2020;1682 [Google Scholar]
- 46.Wargo A.R., Kurath G. Viral fitness: definitions, measurement, and current insights. Curr. Opin. Virol. 2012;2:538–545. doi: 10.1016/j.coviro.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Domingo E., de Ávila A.I., Gallego I., Sheldon J., Perales C. Viral fitness: history and relevance for viral pathogenesis and antiviral interventions. Pathog. Dis. 2019;77 doi: 10.1093/femspd/ftz021. [DOI] [PubMed] [Google Scholar]
- 48.Zhao W.-M., Song S.-H., Chen M.-L., Zou D., Ma L.-N., Ma Y.-K., Li R.-J., Hao L.-L., Li C.-P., Tian D.-M., Tang B.-X., Wang Y.-Q., Zhu J.-W., Chen H.-X., Zhang Z., Xue Y.-B., Bao Y.-M. The 2019 novel coronavirus resource. Yi Chuan. 2020;42:212–221. doi: 10.16288/j.yczz.20-030. [DOI] [PubMed] [Google Scholar]
- 49.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction, Nat. Methods. 2014;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F., Zhou Y., Zhang Y., Yin J., Qiu Y., Gao J., Zhu F. POSREG: proteomic signature discovered by simultaneously optimizing its reproducibility and generalizability. Briefings Bioinf. 2022;23 doi: 10.1093/bib/bbac040. [DOI] [PubMed] [Google Scholar]
- 51.Yang Q., Li B., Tang J., Cui X., Wang Y., Li X., Hu J., Chen Y., Xue W., Lou Y., Qiu Y., Zhu F. Consistent gene signature of schizophrenia identified by a novel feature selection strategy from comprehensive sets of transcriptomic data. Briefings Bioinf. 2020;21:1058–1068. doi: 10.1093/bib/bbz049. [DOI] [PubMed] [Google Scholar]
- 52.Tang J., Wang Y., Fu J., Zhou Y., Luo Y., Zhang Y., Li B., Yang Q., Xue W., Lou Y., Qiu Y., Zhu F. A critical assessment of the feature selection methods used for biomarker discovery in current metaproteomics studies, Brief. Bioinformation. 2020;21:1378–1390. doi: 10.1093/bib/bbz061. [DOI] [PubMed] [Google Scholar]
- 53.Tang J., Mou M., Wang Y., Luo Y., Zhu F. MetaFS: performance assessment of biomarker discovery in metaproteomics. Briefings Bioinf. 2021;22 doi: 10.1093/bib/bbaa105. [DOI] [PubMed] [Google Scholar]
- 54.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 55.Tan K.P., Nguyen T.B., Patel S., Varadarajan R., Madhusudhan M.S. Depth: a web server to compute depth, cavity sizes, detect potential small-molecule ligand-binding cavities and predict the pKa of ionizable residues in proteins. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt503. W314–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawashima S., Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 2000;28:374. doi: 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Witten I.H., Frank E., Hall M.A., Pal C. Data mining: practical machine learning tools and techniques. Morgan Kaufmann. 2016 [Google Scholar]
- 59.Hunter, Matplotlib: A 2D Graphics Environment. vol. 9. 2007. pp. 90–95. [Google Scholar]
- 60.Michen B., Graule T. Isoelectric points of viruses. J. Appl. Microbiol. 2010;109:388–397. doi: 10.1111/j.1365-2672.2010.04663.x. [DOI] [PubMed] [Google Scholar]
- 61.Vitkup D., Sander C., Church G.M. The amino-acid mutational spectrum of human genetic disease, Genome Biol. 2003;4:R72. doi: 10.1186/gb-2003-4-11-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lins L., Thomas A., Brasseur R. Analysis of accessible surface of residues in proteins. Protein Sci. 2003;12:1406–1417. doi: 10.1110/ps.0304803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bohórquez H.J., Suárez C.F., Patarroyo M.E. Publisher Correction: mass & secondary structure propensity of amino acids explain their mutability and evolutionary replacements. Sci. Rep. 2018;8:4273. doi: 10.1038/s41598-018-21981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lehmann J., Libchaber A., Greenbaum B.D. Fundamental amino acid mass distributions and entropy costs in proteomes. J. Theor. Biol. 2016;410:119–124. doi: 10.1016/j.jtbi.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 65.Seligmann H. Cost-minimization of amino acid usage. J. Mol. Evol. 2003;56:151–161. doi: 10.1007/s00239-002-2388-z. [DOI] [PubMed] [Google Scholar]
- 66.Akbar R., Robert P.A., Pavlović M., Jeliazkov J.R., Snapkov I., Slabodkin A., Weber C.R., Scheffer L., Miho E., Haff I.H., Haug D.T.T., Lund-Johansen F., Safonova Y., Sandve G.K., Greiff V. A compact vocabulary of paratope-epitope interactions enables predictability of antibody-antigen binding. Cell Rep. 2021;vol. 34 doi: 10.1016/j.celrep.2021.108856. [DOI] [PubMed] [Google Scholar]
- 67.Mason D.M., Friedensohn S., Weber C.R., Jordi C., Wagner B., Meng S.M., Ehling R.A., Bonati L., Dahinden J., Gainza P., Correia B.E., Reddy S.T. Optimization of therapeutic antibodies by predicting antigen specificity from antibody sequence via deep learning. Nat Biomed Eng. 2021 doi: 10.1038/s41551-021-00699-9. [DOI] [PubMed] [Google Scholar]
- 68.Faure G., Koonin E.V. Universal distribution of mutational effects on protein stability, uncoupling of protein robustness from sequence evolution and distinct evolutionary modes of prokaryotic and eukaryotic proteins. Phys. Biol. 2015;12 doi: 10.1088/1478-3975/12/3/035001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ishida T., Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm363. W460–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Q., Li B., Chen S., Tang J., Li Y., Li Y., Zhang S., Shi C., Zhang Y., Mou M., Xue W., Zhu F. MMEASE: online meta-analysis of metabolomic data by enhanced metabolite annotation, marker selection and enrichment analysis. J. Proteonomics. 2021;232 doi: 10.1016/j.jprot.2020.104023. [DOI] [PubMed] [Google Scholar]
- 71.Rawat P., Kumar S., Michael Gromiha M. An in-silico method for identifying aggregation rate enhancer and mitigator mutations in proteins. Int. J. Biol. Macromol. 2018;118:1157–1167. doi: 10.1016/j.ijbiomac.2018.06.102. [DOI] [PubMed] [Google Scholar]
- 72.Li B., Tang J., Yang Q., Li S., Cui X., Li Y., Chen Y., Xue W., Li X., Zhu F. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017;45:W162–W170. doi: 10.1093/nar/gkx449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.