Abstract
Background
COVID-19 can cause multiple organ damages as well as metabolic abnormalities such as hyperglycemia, insulin resistance, and new onset of diabetes. The insulin/IGF signaling pathway plays an important role in regulating energy metabolism and cell survival, but little is known about the impact of SARS-CoV-2 infection. The aim of this work was to investigate whether SARS-CoV-2 infection impairs the insulin/IGF signaling pathway in the host cell/tissue, and if so, the potential mechanism and association with COVID-19 pathology.
Methods
To determine the impact of SARS-CoV-2 on insulin/IGF signaling pathway, we utilized transcriptome datasets of SARS-CoV-2 infected cells and tissues from public repositories for a wide range of high-throughput gene expression data: autopsy lungs from COVID-19 patients compared to the control from non-COVID-19 patients; lungs from a human ACE2 transgenic mouse infected with SARS-CoV-2 compared to the control infected with mock; human pluripotent stem cell (hPSC)-derived liver organoids infected with SARS-CoV-2; adipose tissues from a mouse model of COVID-19 overexpressing human ACE2 via adeno-associated virus serotype 9 (AAV9) compared to the control GFP after SARS-CoV-2 infection; iPS-derived human pancreatic cells infected with SARS-CoV-2 compared to the mock control. Gain and loss of IRF1 function models were established in HEK293T and/or Calu3 cells to evaluate the impact on insulin signaling. To understand the mechanistic regulation and relevance with COVID-19 risk factors, such as older age, male sex, obesity, and diabetes, several transcriptomes of human respiratory, metabolic, and endocrine cells and tissue were analyzed. To estimate the association with COVID-19 severity, whole blood transcriptomes of critical patients with COVID-19 compared to those of hospitalized noncritical patients with COVID-19.
Results
We found that SARS-CoV-2 infection impaired insulin/IGF signaling pathway genes, such as IRS, PI3K, AKT, mTOR, and MAPK, in the host lung, liver, adipose tissue, and pancreatic cells. The impairments were attributed to interferon regulatory factor 1 (IRF1), and its gene expression was highly relevant to risk factors for severe COVID-19; increased with aging in the lung, specifically in men; augmented by obese and diabetic conditions in liver, adipose tissue, and pancreatic islets. IRF1 activation was significantly associated with the impaired insulin signaling in human cells. IRF1 intron variant rs17622656-A, which was previously reported to be associated with COVID-19 prevalence, increased the IRF1 gene expression in human tissue and was frequently found in American and European population. Critical patients with COVID-19 exhibited higher IRF1 and lower insulin/IGF signaling pathway genes in the whole blood compared to hospitalized noncritical patients. Hormonal interventions, such as dihydrotestosterone and dexamethasone, ameliorated the pathological traits in SARS-CoV-2 infectable cells and tissues.
Conclusions
The present study provides the first scientific evidence that SARS-CoV-2 infection impairs the insulin/IGF signaling pathway in respiratory, metabolic, and endocrine cells and tissues. This feature likely contributes to COVID-19 severity with cell/tissue damage and metabolic abnormalities, which may be exacerbated in older, male, obese, or diabetic patients.
Keywords: SARS-CoV-2 infection, COVID-19, Insulin/IGF signaling pathway, IRF1, Interferone, Cytokine, Lung, Liver, Adipose tissue, Pancreatic cell, Older age, Male sex, Obesity, Diabetes, Tissue damage, Multiorgan dysfunction, Insulin signaling, Insulin resistance, Metabolic abnormality, Dihydrotestosterone, Dexamethasone
Graphical abstract
1. Introduction
The pandemic of a novel β-coronavirus SARS-CoV-2 infection, termed coronavirus disease 2019 (COVID-19), has resulted in over 518,000,000 infection cases and 6,250,000 deaths, leading to a global health crisis. The influences of COVID-19 vary among individuals, and severe outcomes are associated with host health states, such as age, sex, obesity, and diabetes [1]. Most patients present mild to moderate symptoms, which accounts for 85–90 % of the patients, whereas approximately 10–15 % develop severe pneumonia. Approximately 5 % of infected people progress to serious stages with respiratory failure, septic shock, and acute respiratory distress syndrome (ARDS) [2]. The infection causes host cell and tissue damage and, in some cases, it triggers metabolic abnormalities, such as insulin resistance, hyperglycemia, abnormal lipid control, abnormal amino acid control, and new onset of diabetes [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13]. However, the molecular mechanisms have not yet been fully explained.
Insulin/IGF signaling plays a critical role in the regulation of energy metabolism [14], [15], [16], [17]. Ligand/receptor interactions transduce cellular signaling through IRS/PI3K, and the downstream AKT/mTOR/MAPK pathway and related molecules mediate subsequent biological processes for energy uptake and utilization [14], [15], [16], [17]. Impairment of the insulin/IGF signaling pathway in metabolic organs and tissue triggers abnormal insulin/IGF responses, so-called insulin resistance, and causes pathological development of metabolic syndromes, such as insulin resistance, hyperglycemia, hyperlipidemia, obesity, and diabetes [14], [15], [16], [17], [18], [19]. Systemic knockout of insulin receptor in mice triggers early onset of diabetes and ultimately death caused by ketoacidosis [20], [21]. Blockade of the insulin/IGF signaling pathway in adipose tissue causes loss of adipose tissue and severe metabolic syndrome [22]. Insulin/IGF signaling is also critical in the functions of pancreatic beta cells; specific knockout of the insulin receptor in beta cells leads to a remarkable reduction in insulin secretion and the development of diabetes mellitus [23].
Insulin/IGF signaling is also important for cellular survival and the control of cell damage and cell death in various organs and tissues, including the lung [24], [25], [26], [27], [28]. The downstream pathway regulates antiapoptotic programs, thus managing cellular damage and enhancing survival [24], [25], [26], [27], [28]. Abnormal insulin sensitivity, called insulin resistance, is associated with lung dysfunction in humans [29], [30], [31], [32]. Endothelial restoration of insulin receptor (IR) rescues vascular impairment of IR heterogenic knockout [33]. Defects in the IGF-1 receptor lead to lethal neonatal respiratory distress with severe lung abnormalities and increased cellular apoptosis [34]. IGF-1 therapy is effective in attenuating lipopolysaccharide (LPS)-induced lung injury [35]. IGFs and IGF receptors are elevated during lung injury and tissue repair, and their blockade inhibits alveolar epithelial proliferation [35], [36]. The significance of insulin/IGF signaling in the control of lung function as well as cell and tissue damage is well understood, but the impact of SARS-CoV-2 infection and the association with COVID-19 pathologies has not been reported.
Advances in bioanalysis technology have accumulated an enormous amount of biological information from a wide range of research fields, which is open for other research uses (e.g., GEO datasets and GTEx project) [37], [38]. Since the outbreak of COVID-19, there have been intensive studies on the pathology of SARS-CoV-2, which has also generated a substantial amount of unanalyzed biological data. Here, we gathered, reanalyzed, and compiled this dispersed biological information and found a novel pathological feature of COVID-19.
2. Material and methods
2.1. Material information
All material information utilized in this study is described in Table 1 .
Table 1.
Material information.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Human COVID-19 lung tissue | Gene Expression Omnibus (GEO) | GSE150316 |
| Human COVID-19 liver organoids | Gene Expression Omnibus (GEO) | GSE151803 |
| IFNb-treated mouse primary hepatocytes | Gene Expression Omnibus (GEO) | GSE149497 |
| Golden hamster liver tissue fed HFHS | Gene Expression Omnibus (GEO) | GSE175943 |
| TNF/IL1B-treated mouse primary hepatocytes | Gene Expression Omnibus (GEO) | GSE19272 |
| Mouse COVID-19 lung tissue | Gene Expression Omnibus (GEO) | GSE154104 |
| Mouse COVID-19 adipose tissue | Gene Expression Omnibus (GEO) | GSE162113 |
| Human COVID-19 pancreatic cells | Gene Expression Omnibus (GEO) | GSE165890 |
| IFN-treated human primary bronchial epithelial cells | Gene Expression Omnibus (GEO) | GDS1256 |
| IFN-treated mouse primary adipocytes | Gene Expression Omnibus (GEO) | GSE110236 |
| IFN-treated human pancreatic islets | Gene Expression Omnibus (GEO) | GSE124810 |
| IRF1-overexpressed human hepatoma Huh-7 cells | Gene Expression Omnibus (GEO) | GSE26817 |
| IRF1 knockout human bronchial epithelial BEAS-2B cells | Gene Expression Omnibus (GEO) | GSE114284 |
| IRF1 knockout human PH5CH8 hepatocytes | Gene Expression Omnibus (GEO) | GSE115198 |
| IRF1 knockout human monocytic THP-1 cells | Gene Expression Omnibus (GEO) | GSE147313 |
| M. avium-infected mouse lung tissue | Gene Expression Omnibus (GEO) | GSE11809 |
| Human xenograft tumor luCaP35 | Gene Expression Omnibus (GEO) | GDS4120 |
| Mouse prostate with testosterone replacement | Gene Expression Omnibus (GEO) | GDS2562 |
| Human epithelial LNCaP cells | Gene Expression Omnibus (GEO) | GSE152254 |
| Human obese adipocytes | Gene Expression Omnibus (GEO) | GDS3602 |
| IL6-treated human SGBS adipocytes | Gene Expression Omnibus (GEO) | GSE179347 |
| IRF1 ChIP-Seq | Gene Expression Omnibus (GEO) | GSM1239465 |
| Mouse pancreatic β cells | Gene Expression Omnibus (GEO) | GSE33647 |
| IL1B-treated rat pancreatic INS-1rαβ cells | Gene Expression Omnibus (GEO) | GDS4332 |
| Human whole blood from critical COVID-19 patients | Gene Expression Omnibus (GEO) | GSE172114 |
| DHT-treated human epithelial LNCaP cells | Gene Expression Omnibus (GEO) | GSE114052 |
| DEX-treated human subcutaneous adipose tissue | Gene Expression Omnibus (GEO) | GSE167250 |
| Humanized mouse lung tissue | Gene Expression Omnibus (GEO) | GSE200561 |
| TNF-and/or IFNb-treated human DC | Gene Expression Omnibus (GEO) | GSE134209 |
| AR ChIP-Seq | Gene Expression Omnibus (GEO) | GSM3577148 |
| GR ChIP-Seq | Gene Expression Omnibus (GEO) | GSM3579033 |
| Human lung, liver, and pancreas tissues | Genotype Tissue Expression (GTEx) | N.A |
| 1000 genome project | https://asia.ensembl.org/ | N.A |
| Experimental materials | ||
| Anti-IRF1 Antibody (D5E4) | Cell signaling | #8478 |
| Anti-Phospho-AKT (Ser473) (D9E) | Cell signaling | #4060 |
| Anti-total AKT Antibody (C67E7) | Cell signaling | #4691 |
| Anti-b-Actin Antibody (AC-15) | Sigma | A5441 |
| Anti-ISG15 Antibody (F-9) | Santa Cruz | sc-166755 |
| px459v2 plasmid | Addgene | #62988 |
| px459v2-Negative Control gRNA | This paper | N.A |
| px459v2-IRF1 gRNA | This paper | N.A |
| pEF1a-Empty | This paper | N.A |
| pEF1a-IRF1-FLAG | This paper | N.A |
| Recombinant Murine TNF-α | PeproTech | 315-01A |
| Dihydrotestosterone (DHT) | Selleck | S4757 |
| Insulin | Sigma | I5500 |
| Dexamethasone (DEX) | Sigma | D1756 |
| Lipofectimine 3000 | Thermo Fisher Scientific | N.A |
| Software and algorithms | ||
| GEO2R | https://www.ncbi.nlm.nih.gov/geo/geo2r/ | N.A |
| Microsoft Excel | https://www.microsoft.com/ | N.A |
| Integrative Genomics Viewer | https://software.broadinstitute.org/software/igv/ | N.A |
| ChIP-Atlas | https://chip-atlas.org/ | N.A |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | https://www.genome.jp/kegg/ | N.A |
| REACTOME | https://reactome.org/ | N.A |
| Toppgene functional analysis software (ToppFun) | https://toppgene.cchmc.org/enrichment.jsp | N.A |
| JMP Pro 15.2.1 software | https://www.jmp.com/ | N.A |
| CHOPCHOP CRISPR gRNA design tool | http://chopchop.cbu.uib.no/ | N.A |
| Benchiling CRISPR gRNA design tool | https://www.benchling.com/crispr | N.A |
| eQTL Dashboard | https://www.gtexportal.org/ | N.A |
2.2. Transcriptome dataset analysis
The gene expression data of human tissues were obtained from Genotype Tissue Expression (GTEx) (Lonsdale et al., 2013). The gene expression datasets related to COVID-19, IRF1, AR, and GR were obtained from the Gene Expression Omnibus (GEO) (Edgar et al., 2002) and analyzed by the GEO2R analysis tool or Microsoft Excel. For human COVID-19 lung tissue (GSE150316), lung tissue samples from NegControl1, 2, 3, 4, and 5 were utilized for the analysis of the control group. Average gene expression of multiple lung tissue samples from high viral cases 1, 5, 8, 9, and 11 in addition to gene expression of lung tissue samples from NYC cases A, C, and D were utilized for the analysis of the COVID-19 group. For the mouse COVID-19 lung tissue (GSE154104), all gene expression levels were normalized to GAPDH, and average gene expression levels of each day's post-infection (DPI) group were utilized to display heatmap images. For the analysis of mouse COVID-19 adipose tissue (GSE162113), samples collected on Day 3 after administration of SARS-CoV-2 were utilized. Additionally, interferon-treated human primary bronchial epithelial cells (GDS1256), interferon-treated mouse primary adipocytes datasets (GSE110236), interferon-treated human pancreatic islets (GSE124810), IRF1-overexpressing human hepatoma Huh-7 cells (GSE26817), IRF1 knockout-related human bronchial epithelial BEAS-2B cells (GSE114284), IRF1 knockout-related human monocytic THP-1 cells (GSE147313), IRF1 knockout-related human hepatocyte PH5CH8 cells (GSE115198), and M. avium-infected mouse lung tissue (GSE11809) were included. The average gene expression of each group in M. avium-infected mouse lung tissue (GSE11809) was utilized to display heatmap images. The following data were also included: Human COVID-19 Liver organoids (GSE151803); castration and testosterone replacement-related mouse prostate (GDS2562); mouse castration and human xenograft LuCaP35 tumor (GDS4120); human epithelial LNCaP cells treated with the AR antagonist, enzalutamide (GSE152254); golden hamster liver tissue fed HFHS (GSE175943); IFNb-treated gouse primary hepatocytes (GSE149497); TNF/IL1B-treated mouse primary hepatocytes (GSE19272); human adipocytes from lean and obese subjects (GDS3602); human SGBS adipocytes treated with IL6 (GSE179347); mouse pancreatic β cells from control and ob/ob mice (GSE33647); rat pancreatic INS-1rαβ cells treated with IL1B (GDS4332); whole blood samples from hospitalized noncritical and critical COVID-19 patients (GSE172114); human prostate epithelial LnCaP cells treated with DHT (GSE114052); human subcutaneous adipose tissues treated with DEX (GSE88966); human islets treated with DEX (GSE167250; study id: Isl10 samples); and humanized mouse lung tissue infected with SARS-CoV-2 with/without DEX (GSE200561; irf1: DPI14 samples; insulin/IGF signaling molecules: DPI28 samples). The average gene expression of each group in human subcutaneous adipose tissues (GSE88966) and humanized mouse lung tissue (GSE200561) were utilized to display heatmap images.
2.3. ChIP-Seq source and analysis
ChIP-Seq datasets of human IRF1 (GSM1239465), AR (GSM3577148), and GR (GSM3579033) were analyzed from ChIP-Atlas [39] and visualized by Integrative Genomics Viewer (IGV) [40].
2.4. Heatmap generation
Heatmap images were generated by the web-based heatmapping tools, Heatmapper [41] and Morpheus.
2.5. Pathway analysis
Pathway analysis for Kyoto Encyclopedia of Genes and Genomes (KEGG) [42] and REACTOME [43] was performed by utilizing ToppGene functional analysis software (ToppFun).
2.6. Cellular experiments
HEK293T or Calu3 cells were maintained in high glucose DMEM supplemented with 10 % FBS and 1 % Penicillin streptomycin. HEK293T or Calu3 cells was plated in 12-well or 24-well dish plate and utilized for experiments when 80–95 % confluent. For Insulin signaling assay, cells were stimulated/infected 24 h before treatment of 100 nM insulin at specific time points.
2.7. Plasmid transfection
80–95 % confluent HEK293T or Calu3 cells were transfected with the expression or CRISPR/Cas9 plasmid by utilizing lipofectamine3000 transfection reagent following manufacturer instructions.
2.8. IRF1 overexpression
IRF1 gene (C-terminal 3xFLAG tag inserted) was cloned in an expression plasmid (pEF1a). The plasmid was transiently transfected in HEK293T cells by lipofectamine3000 transfection reagent. 24–48 h after transfection, cells were utilized for insulin signaling experiments.
2.9. IRF1 knockout cell establishment
Specific guide RNA (gRNA) for IRF1 gene was designed by web-based gRNA designing tools, such as CHOPCHOP, Benchiling, and CRISPOR. The gRNA for non-targeting or IRF1 gene was cloned into CRISPR/Cas9 plasmid (px459v2; puromycin selection plasmid) [44]. These plasmids were transiently transfected in HEK293T cells, 24 h after transfection, the cells were selected by puromycin for 5 days. The selected HEK293T cells were utilized for insulin signaling experiments.
2.10. Statistics
All data are expressed as the mean ± SEM. Differences between two groups were evaluated for statistical significance by Student's t-test. The effects of castration and testosterone replacement on IRF1 gene expression in the mouse prostate were tested by one-way analysis of variance (ANOVA) followed by the Tukey–Kramer test. Aging effects on IRF1 gene expression in human lung tissues were assessed by the Wilcoxon test. A P value of <0.05 indicated a statistically significant difference. JMP Pro 15.2.1 software was used in all statistical analyses.
3. Results
3.1. COVID-19 impairs the insulin/IGF signaling pathway of the host lung, liver, adipose tissue, and pancreatic cells
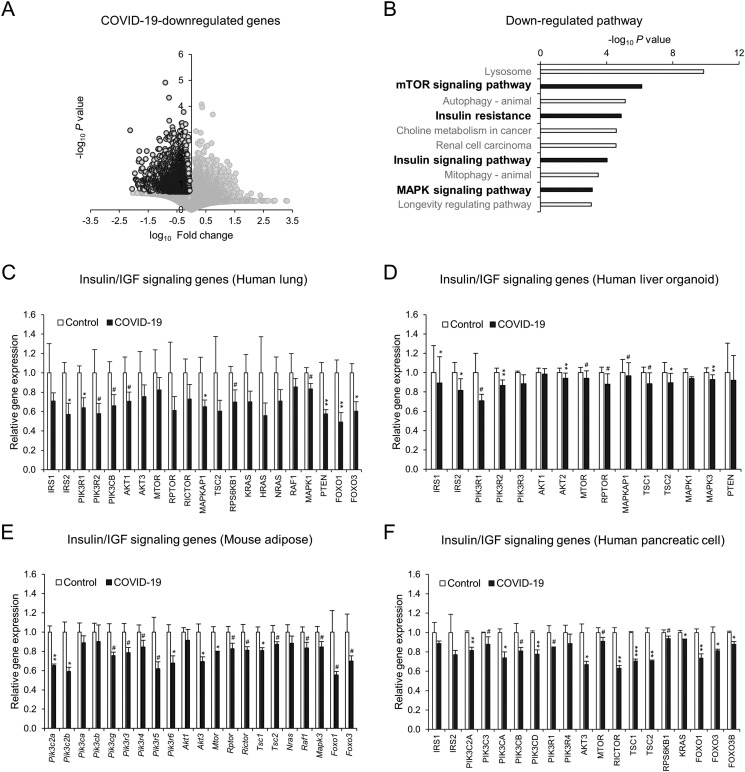
To understand the impact of SARS-CoV-2 infection on the host, we first analyzed the transcriptome datasets of autopsy lungs from COVID-19 patients compared to the control from non-COVID-19 patients with pulmonary pathologies, such as pneumonia [45]. Infection with SARS-CoV-2 led to remarkable alterations in the gene expression of the autopsy lungs of COVID-19 patients (Fig. 1A). The downregulated genes were significantly associated with insulin signaling/resistance, mTOR signaling and MAPK signaling in pathway analysis (Fig. 1B). Detailed gene expression analysis showed a wide range of impairments in insulin/IGF signaling molecules, including IRS, PI3K, AKT, mTOR, MAPK, PTEN, and FOXO (Fig. 1C), without changes in immune cell markers (sFig. 1), ruling out the immune cell compositional impact. Similar impairments in insulin/IGF signaling pathway genes were observed in the lung tissues of a COVID-19 mouse model (sFig. 2A), a human ACE2 (hACE2) transgenic mouse infected with SARS-CoV-2 [46]. SARS-CoV-2 not only infects the respiratory track but also other organs and cells, including liver [47], [48], adipose tissue [49], [50], [51], and pancreas [52], [53], [54]. We analyzed liver organoids derived from a human pluripotent stem cell (hPSC) infected with SARS-CoV-2 [48], adipose tissues from a mouse model of COVID-19 overexpressing human ACE2 via adeno-associated virus serotype 9 (AAV9) and infected with SARS-CoV-2 [55] as well as iPS-derived human pancreatic cells infected with SARS-CoV-2 [56]. SARS-CoV-2 infection markedly downregulated insulin/IGF signaling pathway genes in hPSC-derived human liver organoid (Fig. 1D), AAV9-hACE2-transfected mouse adipose tissue (Fig. 1E), and iPS-derived human pancreatic cells (Fig. 1F).
Fig. 1.
SARS-CoV-2 infection (COVID-19) impairs the insulin/IGF signaling pathway in the host lung, liver, adipose tissue, and pancreatic cells. A–C: Transcriptome analysis of human autopsy lung tissues from control non-COVID-19 patients with other pulmonary pathologies and COVID-19 patients (GSE150316: Control n = 5, COVID-19 n = 8). Volcano plot (A), pathway analysis of COVID-19-downregulated genes (B: fold change<1, p value<0.2), and gene expression of insulin/IGF signaling-related molecules (C). D–F: Gene expression of insulin/IGF signaling-related molecules in human liver organoids (D: GSE151803, SARS-CoV-2 infection, Control n = 6, COVID-19 n = 6), adipose tissue (E: GSE162113, 3 days post-SARS-CoV-2 infection, Control AAV9-GFP n = 3, COVID-19 AAV9-hACE2 n = 3) and iPS-derived pancreatic cells (F: GSE165890, Control Mock infection n = 3, COVID-19 SARS-CoV-2 infection n = 3). Data represent the mean ± SEM. #p < 0.25, *p < 0.05, **p < 0.01, and ***p < 0.001.
3.2. IRF1 mediates COVID-19-associated impairments in the insulin/IGF signaling pathway
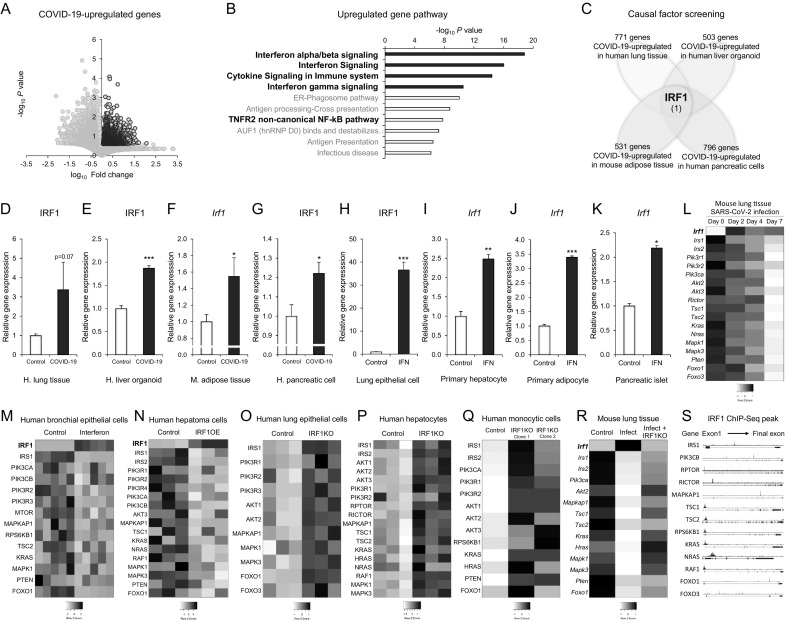
We next investigated the mechanism of COVID-19-associated impairments on the insulin/IGF signaling pathway in the autopsy lung of COVID-19 patients and SARS-CoV-2-infected hPSC-derived human liver organoid, AAV9-hACE2-transfected mouse adipose tissue, and iPS-derived human pancreatic cells. An uncontrolled innate immune response and excessive production of proinflammatory cytokines are hallmarks of SARS-CoV-2 infection [57]. COVID-19-upregulated genes were associated with interferon, cytokine and TNF signaling pathways in human autopsy lungs (Fig. 2A and B). To identify the key causal factor, we listed and compiled upregulated genes in the human autopsy lungs, hPSC-derived human liver organoid, AAV9-hACE2-transfected mouse adipose tissue, and iPS-derived human pancreatic cells of COVID-19 transcriptomes (Fig. 2C) as follows: COVID-19-upregulated genes in human autopsy lung tissue (728 genes), liver organoid (503 genes), mouse adipose tissue (531 genes), and human pancreatic cells (796 genes). The comparative analysis led to only one common gene (Fig. 2C), interferon regulatory factor 1 (IRF1), which is a crucial mediator of interferon and cytokine responses [58].
Fig. 2.
IRF-1 mediates COVID-19-associated impairments in the insulin/IGF signaling pathway. A and B: Transcriptome analysis of human autopsy lung tissues from control patients (non-COVID-19 with other pulmonary pathologies) and COVID-19 patients (GSE150316: Control n = 5, COVID-19 n = 8). Volcano plot (A) and pathway analysis of COVID-19-upregulated genes (B; fold change>1, p value<0.25). C: Schematic diagram of causal factor screening (human lung GSE150316: fold change>3, p value<0.25; human liver organoids GSE151803: fold change>1.15, p value<0.05; mouse adipose tissue GSE162113: fold change>1.2, p value <0.1; and human pancreatic cells GSE165890: fold change>1.2, t-test<0.1). D-G: Gene expression of the IRF1 gene in human autopsy lung tissues (D: GSE150316, Control n = 5, COVID-19 n = 8), human liver organoids (E: GSE151803, SARS-CoV-2 infection, Control n = 6, COVID-19 n = 6), mouse adipose tissue (F: GSE162113, 3 days post-SARS-CoV-2 infection, Control AAV9-GFP n = 3, COVID-19 AAV9-hACE2 n = 3), and iPS-derived pancreatic cells (G: GSE165890, Control Mock infection n = 3, COVID-19 SARS-CoV-2 infection n = 3). H-K: Relative gene expression of IRF1 in primary human bronchial epithelial cells (H: GDS1256, control n = 5, interferon gamma 24-hour treatment n = 5), mouse primary hepatocytes (I: GSE14949, control n = 3, interferon beta treatment n = 3), mouse primary adipocytes (J: GSE110236, control n = 2, interferon beta treatment n = 2), and human pancreatic islets (K: GSE124810, control n = 3, interferon alpha treatment n = 3). L: Heatmap image of average IRF1 and insulin/IGF signaling pathway genes in mouse lung tissues at Days 0, 2, 4, and 7 after SARS-CoV-2 infection (GSE154104). M: Heatmap image of IRF1 and insulin/IGF signaling pathway genes in primary human bronchial epithelial cells treated with interferon gamma for 24 h (GDS1256). N: Heatmap image of IRF1 and insulin/IGF signaling pathway genes in human hepatoma Huh-7 cells with or without IRF1 overexpression (GSE26817). O-Q: Heatmap images of insulin/IGF signaling pathway genes in human bronchial epithelial BEAS-2B cells (O: GSE114284), human PH5CH8 hepatocytes (P: GSE115198), human monocytic THP-1 cells (Q: GSE147313), and wild-type control and IRF1 knockout cells. R: Heatmap image of average IRF1 and insulin/IGF signaling pathway genes in M. avium-infected mouse lung tissue with or without IRF1 knockout (GSE11809). S: Human IRF1 ChIP-Seq peaks in the gene regions of insulin/IGF signaling molecules (GSM1239465). Data represent the mean ± SEM. #p < 0.25, *p < 0.05, **p < 0.01, and ***p < 0.001.
Detailed gene expression analysis showed that SARS-CoV-2 infection increased the gene expression of IRF1 in the human autopsy lungs, hPSC-derived human liver organoid, AAV9-hACE2-transfected mouse adipose tissue, and iPS-derived human pancreatic cells (Fig. 2D–G and sFig. 2B). The induction of IRF1 was recapitulated by interferon treatments in human primary bronchial epithelial cells (Fig. 2H), mouse primary hepatocytes (Fig. 2I), mouse primary adipocytes (Fig. 2J), and pancreatic islets (Fig. 2K). SARS-CoV-2 infection caused early phase induction of I rf 1 at Day 2 and late phase significant impairments of the insulin/IGF signaling pathway at Day 7 after infection in the lung of hACE2-transgenic mouse (Fig. 2L), suggesting the time course effect of COVID-19. Interferon gamma treatment in human primary bronchial epithelial cells resulted in significant induction of IRF1, which was associated with the downregulation of insulin/IGF signaling pathway genes (Fig. 2M).
IRF1 functions as both a transcriptional activator and repressor to control interferon and cytokine responses [58]. Overexpression of IRF1 in human hepatoma Huh-7 cells [59] decreased the gene expression of the insulin/IGF signaling pathway (Fig. 2N). Knockout of the IRF1 gene in various SARS-CoV-2 infectable human cell types, including human bronchial epithelial BEAS-2B cells [60], human PH5CH8 hepatocytes [61], and human monocytic THP-1 cells [62], enhanced gene expression of the insulin/IGF signaling pathway (Fig. 2O–Q). A similar result was observed in mouse lungs infected with Mycobacterium avium (M. avium), a mouse model of respiratory diseases [63]; infection increased the gene expression of I rf and impaired that of the insulin/IGF signaling pathway in the lung, which was attenuated by I rf 1 knockout (Fig. 2R). IRF1 ChIP-Seq analysis revealed direct binding in the gene regions of insulin/IGF signaling molecules (Fig. 2S). These data suggested that IRF1 might contribute to the impairment of insulin/IGF signaling.
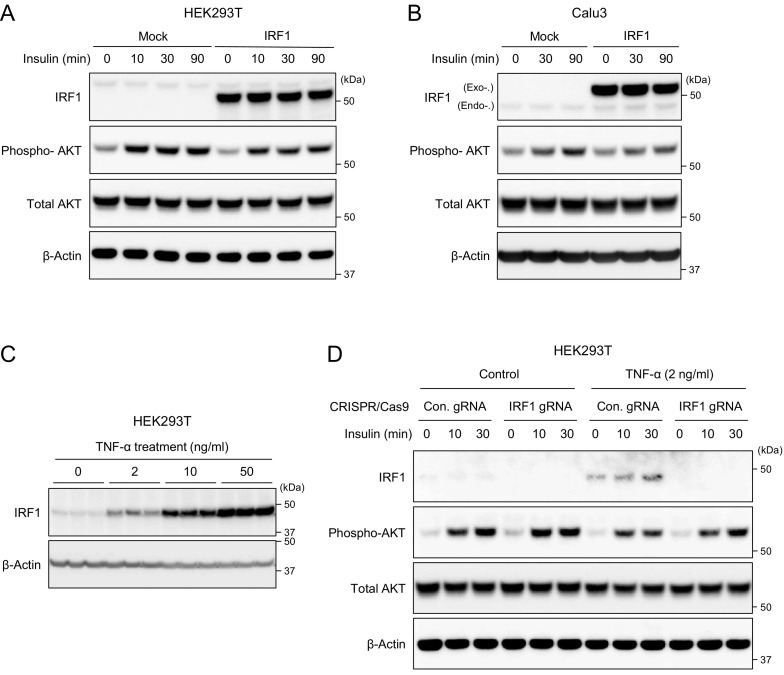
To evaluate whether IRF1 activation indeed impairs insulin signaling in human cells, we established IRF1 gain-of-function models, in which FLAG-tagged IRF1 was cloned in an expression plasmid, transiently transfected in HEK293T and human epithelial Calu3 cells, and then estimated insulin-mediated AKT phosphorylation by western blotting. Overexpression of IRF1 gene in HEK293T cells, which properly increased IRF1 and the target ISG15 protein expressions (sFig. 2), impaired insulin-induced AKT phosphorylation in human HEK293T cells (Fig. 3A). Similar results were observed in human epithelial cell line Calu3; the overexpression of IRF1 blocked insulin-mediated AKT phosphorylation in Calu3 cells (Fig. 3B). These results clearly showed that IRF1 gene activation indeed impairs the insulin signaling in human cells.
Fig. 3.
IRF1 gain-and loss-of function models in human cells. A and B: Western blot image of IRF1, phospho-Akt, total Akt, and β-Actin in IRF1-FLAG overexpressed HEK293T (A) or Calu3 (B) cells treated with insulin (100 nM) at the indicated time points. C: Western blot image of IRF1 and β-Actin in HEK293T treated with TNF-α with the indicated concentrations for 24 h. D: Western blot image of IRF1, phospho-Akt, total Akt, and β-Actin in HEK293T transfected with negative control non-targeting gRNA (Con. gRNA) and IRF1 gene targeting gRNA (IRF1 gRNA) cloned CRISPR-Cas9 plasmid (px459v2) and treated with TNF-α for 24 h followed by insulin (100 nM) treatment at the indicated time points.
We also validated the impact of IRF1 in a loss-of-function model, in which IRF1 knockout HEK293T cells were generated by CRISPR-Cas9 system and evaluated the insulin-mediated AKT phosphorylation under inflammatory condition. As we described in Fig. 2, interferon, cytokine, and TNF responses were associated with the induction of IRF1 gene expression. The treatment of TNF-α in HEK293T cells significantly increased IRF1 protein expression in a dose dependent manner (Fig. 3C). The induction of IRF1 was significantly associated with the impaired insulin-mediated phosphorylation, which were in part rescued by the IRF1 knockout (Fig. 3D). The data suggested that IRF1 repression could ameliorate inflammatory response-related insulin resistance in human cells, potentially in COVID-19 pathology.
The results of IRF1 gain-and loss-of-function experiments are well align with previous research that IRF1 expression is involved not only in tissue damage and cell death in association with inflammatory responses but also in metabolic disorders in metabolic and endocrine tissues and cells such as adipose tissue and pancreas [64], [65], [66], [67], [68], [69], suggesting the possible involvement in COVID-19-related tissue/cell damage and metabolic abnormalities.
3.3. IRF1 gene expression is highly relevant to risk factors for severe COVID-19
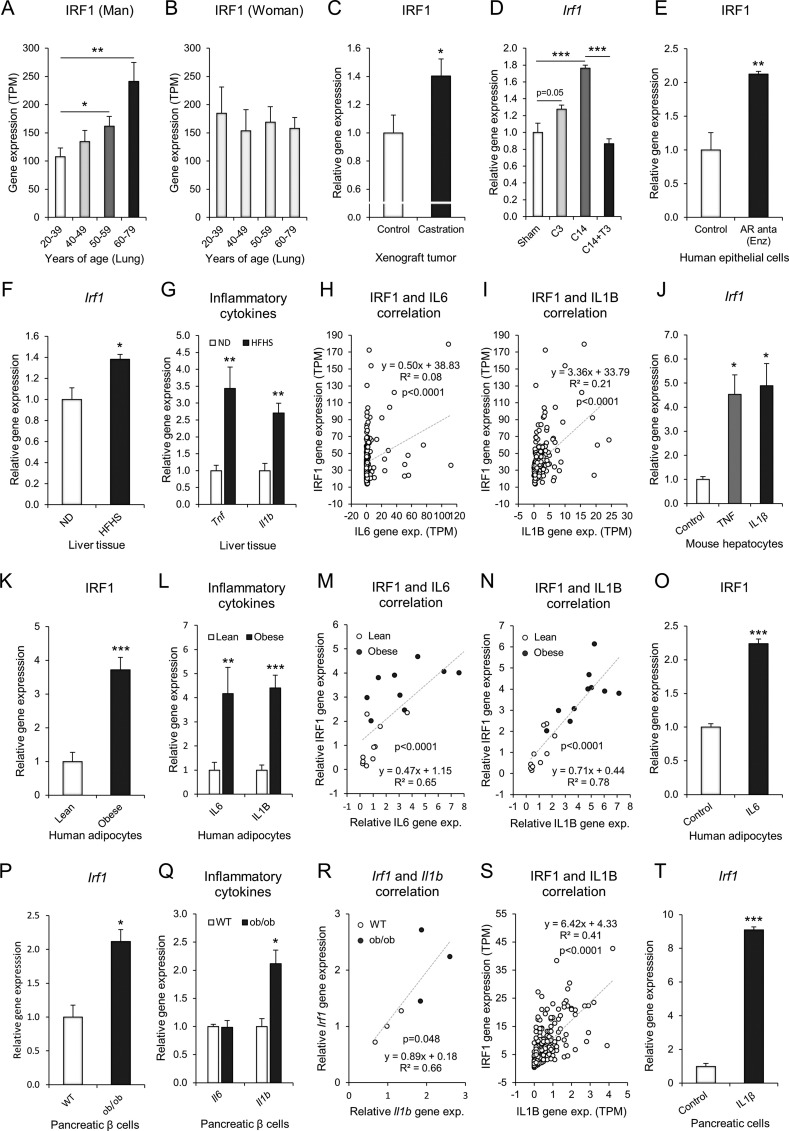
We next analyzed the expression features of IRF1 associated with risk factors for severe COVID-19, such as aging, male sex, obesity, and diabetes. The IRF1 gene was significantly increased in human lung tissues with aging, specifically in men (Fig. 4A), and the expression was slightly decreased in women but was not significant (Fig. 4B). The sexual dimorphic differences suggested that aging-related androgen deprivation in men might be associated with increased IRF1 gene expression. In fact, androgen deprivation by castration in mouse increased IRF1 gene expression in a xenograft tumor model (Fig. 4C). In a mouse model of castration and testosterone replacement [70], there was a significant increase in I rf 1 gene expression 14 days after castration in the prostate tissue, which was rescued by 3 days of testosterone replacement (Fig. 4D). Pharmacological antagonism of androgen receptor (AR) by enzalutamide (Enz) markedly increased the gene expression of IRF1 in cultured human epithelial cells (Fig. 4E). In an obese diabetic model of golden hamster model [44], high-fat and high-sugar diet (HFHS) for 16 weeks significantly upregulated the expression of I rf 1 gene in the liver (Fig. 4F), which was associated with the induction of inflammatory cytokines genes, such as Tnf and Il1b (Fig. 4G); Il6 gene was not found in the dataset. In human liver tissue [71], the gene expression of IRF1 was significantly correlated with that of IL6 and IL1B gene (Fig. 4H and I). The treatment of mouse primary hepatocytes with either TNF or IL1β, an inflammatory response model of hepatocytes [72], significantly up-regulated the gene expression of Irf1, suggesting the direct involvement in I rf1 regulation in mouse primary hepatocytes (Fig. 4J). Collectively, these data suggested that SARS-CoV-2 infection could affects hepatic IRF1 gene expression, which was significantly associated with interferon/cytokine responses and obese/obesogenic diet conditions. In obese human [73], The IRF1 gene expression was significantly upregulated in the human adipocytes (Fig. 4K). The induction of the IRF1 gene was associated with that of the IL6 and IL1B inflammatory cytokine genes (Fig. 4L). The gene expression of IRF1 showed significant correlations with that of IL6 or IL1B (Fig. 4M and N). IL6 treatment in cultured human adipocytes, a mimetic model of inflammatory human adipocytes in vitro [74], significantly increased the gene expression of IRF1 (Fig. 4O), suggesting direct involvement in the regulation of the IRF1 gene. In an obese diabetic mouse model [75], the gene expression of Irf 1 was markedly increased in the pancreatic beta cells of ob/ob mice (Fig. 4P). The induction of the I rf 1 gene was associated and correlated with that of the I l 1 b gene (Fig. 4Q and R). A similar correlation of IRF1 and IL1B gene expression was observed in human pancreas samples (Fig. 4S). Treatment of rat pancreatic INS-1rαβ cells with IL1B significantly increased the gene expression of I rf 1 (Fig. 4T). These results suggested that hormonal and inflammatory conditions in older age, male sex, obesity, and diabetes are relevant to the upregulation of IRF1 expression in the lung, liver, adipose tissue, and pancreatic cells.
Fig. 4.
IRF1 gene expression is highly relevant to risk factors for severe COVID-19. A-B: Transcriptome analysis of the IRF1 gene in humans (GTEx). Age-dependent IRF1 gene expression in lung tissues of male (A: 20–39 years of age, n = 37; 40–49 years of age n = 47; 50–59 years of age, n = 109; 60–79 years of age, n = 93) and female (B: 20–39 years of age, n = 20; 40–49 years of age n = 29; 50–59 years of age, n = 36; 60–79 years of age, n = 56) subjects. C: Relative gene expression of IRF1 in human xenograft tumor luCaP35 with or without castration (GDS4120, control n = 5, castration n = 5). D: Relative gene expression of IRF1 in mouse prostate (GDS2562) of sham, 3 days after castration (C3, n = 5), 14 days after castration (C14, n = 5), and C14 + 3 days of testosterone replacement (C14 + T3, n = 5). E: Relative gene expression of IRF1 in human epithelial LNCaP cells (GSE152254, control n = 4, AR antagonist enzalutamide 1 μM 24 h; Enz n = 4). F and G: Relative gene expression of Irf1 (F), Tnf, and Il1b (G) in the liver tissues of golden hamster (GSE175943, ND: Normal diet n = 4, HFHS: High-fat and high-sugar diet n = 3). H and I: Correlation analysis of the IRF1 gene with IL6 gene (H) or IL1B gene (I) in the human liver tissues (GTEx, n = 226). J: Relative gene expression of Irf1 in mouse primary hepatocytes (GSE19272, control n = 3, TNF treatment n = 3, IL1B treatment n = 3). K and L: Relative gene expression of IRF1 (K), IL6, and IL1B (L) in human adipocytes (GDS3602, lean n = 10, obese n = 10). M and N: Correlation analysis of the IRF1 gene with the IL6 (M) or IL1B (N) gene in human adipocytes (GDS3602, lean n = 10, obese n = 10). O: Relative gene expression of IRF1 in human SGBS adipocytes (GSE179347, control n = 3, IL6 treatment n = 3). P and Q: Relative gene expression of Irf1 (P), Il6, and Il1b (Q) in mouse pancreatic β cells (GSE33647, control n = 3, ob/ob n = 3). R: Correlation analysis of the Irf1 gene with the Il1b gene in mouse pancreatic β cells (GSE33647, control n = 3, ob/ob n = 3). S: Correlation analysis of the IRF1 gene with the IL1B gene in the human pancreas (GTEx, n = 247). T: Relative gene expression of Irf1 in rat pancreatic INS-1rαβ cells (GDS4332, control n = 3, IL1B treatment n = 3). Data represent the mean ± SEM. #p < 0.25, *p < 0.05, **p < 0.01, and ***p < 0.001.
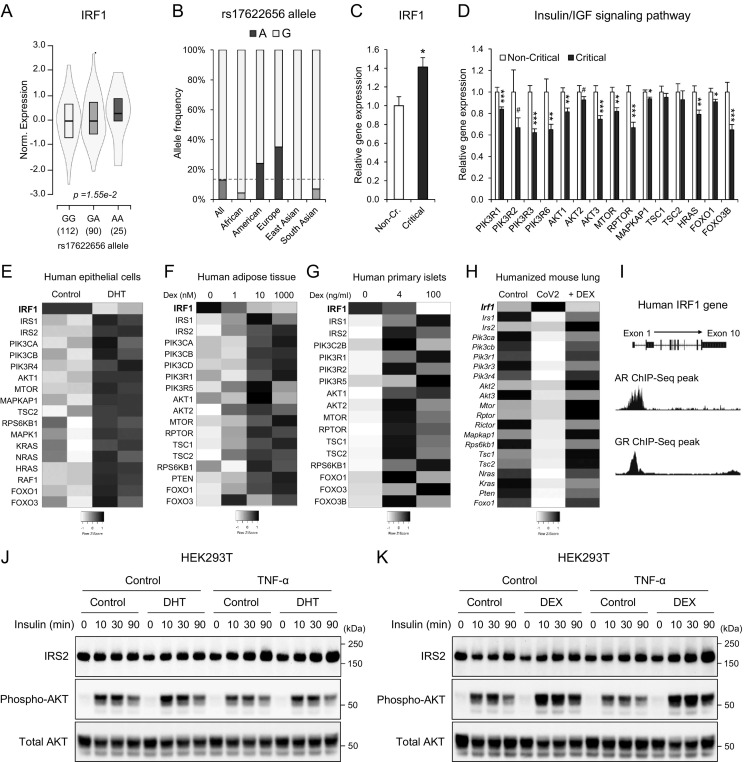
3.4. Higher IRF1 gene expression by the intron variant rs17622656-A might be associated with the prevalence of COVID-19 in American and European populations
In a recent study, a single nucleotide polymorphism (SNP) rs17622656 in an intron region of IRF1 gene was found to be associated with the prevalence of COVID-19 [76], but the expressional impact on IRF1 was not evaluated. Here, we analyzed the impact of rs17622656 on IRF1 gene expression by expression quantitative trait loci (eQTL) analysis in GTEx datasets. The expression of IRF1 was significantly upregulated or tended to be increased by rs17622656-A SNP in human skin, spleen, and adipose tissue (Fig. 5A and sFig. 4). We also investigated the frequency of rs17622656-A allele in 1000 genome project and found that the rs17622656-A SNP was frequently found in American and European population, 24 % and 35 % respectively (Fig. 5B). These results suggested that higher basal expression of IRF1 by genetic SNP rs17622656-A might contribute to COVID-19 prevalence and be possibly associated with the devastating impact in these countries.
Fig. 5.
The association of higher IRF1 gene expression with the prevalence and severity of COVID-19 and the potential therapeutic strategies. A: rs17622656 eQTL analysis on IRF1 gene expression in human spleen of GTEx datasets. B: rs17622656 allele frequency of global populations in 1000 genome project. C and D: Relative gene expression of IRF1 (C) and insulin/IGF signaling pathway molecules (D) in whole blood samples from hospitalized noncritical and critical COVID-19 patients (GSE172114, noncritical n = 23, critical n = 46). E: Heatmap image of human epithelial LNCaP cells treated with or without DHT (GSE114052). F: Heatmap image of cultured human subcutaneous adipose tissues treated with the indicated dexamethasone (DEX) concentrations (GSE88966). G: Heatmap image of cultured human primary islets treated with the indicated dexamethasone (DEX) concentrations (GSE167250). H: Heatmap image of lung tissue in humanized mouse infected with SARS-CoV-2 with/without dexamethasone (DEX) intervention (GSE200561; Control: non-infected; SARS-CoV-2 infection: CoV2: SARS-CoV-2 infection + Dexamethasone: + DEX,). I: Human AR and GR ChIP-Seq peaks in the promoter regions of the IRF1 gene (AR: GSM3577148, GR: GSM3579033). J and K: Western blot image of IRS2, phospho-Akt, and total Akt in HEK293T cells treated with TNF-α (2 ng/ml) with/without DHT (100 nM; J) or DEX (100 nM; K) for 24 h followed by insulin response (100 nM) at the indicated time points. Data represent the mean ± SEM. #p < 0.25, **p < 0.01, and ***p < 0.001.
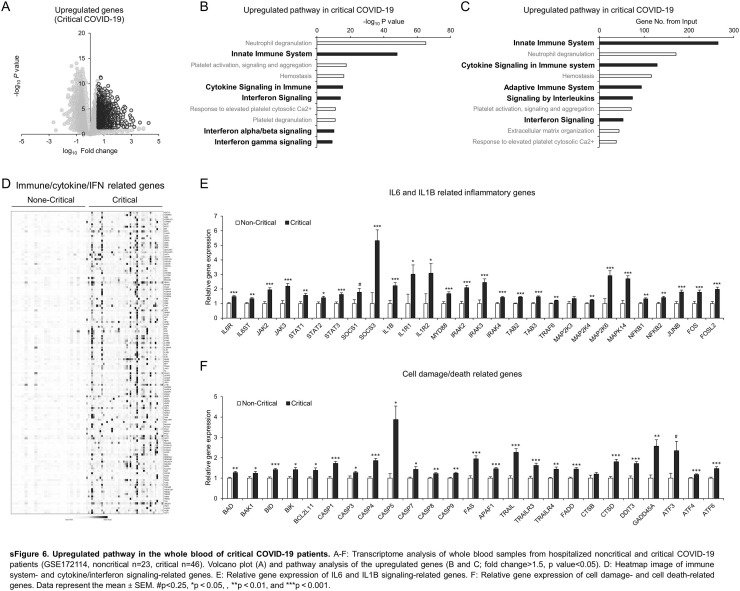
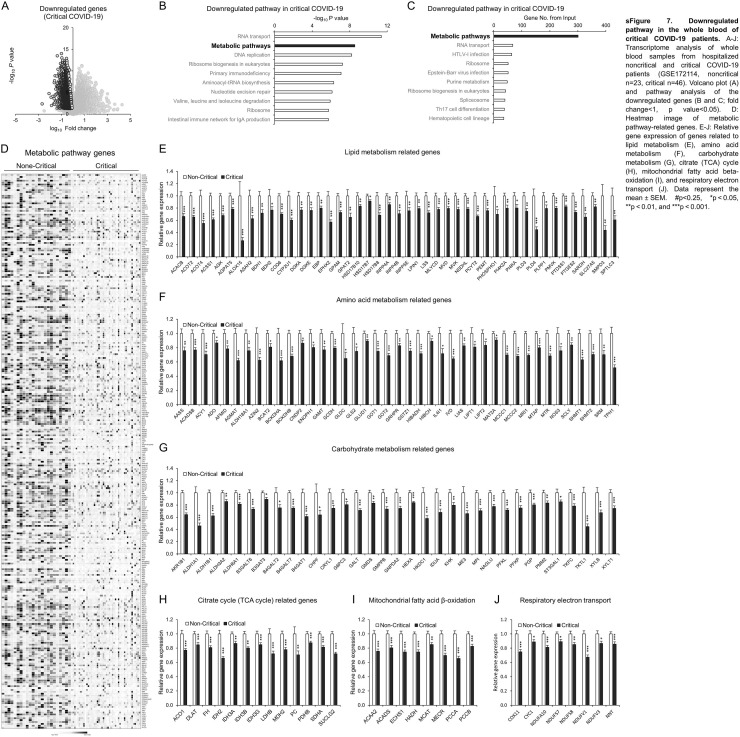
3.5. Critical COVID-19 is characterized by exacerbated cell damage, cell death and impaired metabolic pathways
The whole blood transcriptome is a useful tool to monitor systemic disease, health status, and local pathophysiological activity in various cells and tissues [77], [78]. To understand the signature of critical COVID-19, we reanalyzed the whole blood transcriptome of a well-controlled, young, comorbidity-free, hospitalized COVID-19 patient cohort divided into two groups [79] as follows: 46 critical patients who were hospitalized in the intensive care unit (ICU) with symptoms of ARDS and respiratory failure requiring mechanical ventilation; and 23 noncritical patients who stayed at a noncritical care ward [79]. As expected, the upregulated genes in critical COVID-19 patients compared to noncritical patients were significantly associated with innate/adaptive immune system, cytokine signaling pathways, and interferon signaling pathways (sFig. 6A–D). Detailed gene expression analysis showed exacerbated IL6 and IL1B signaling as well as cell damage- and cell death-related genes (sFig. 6E and F). Of note, the downregulated genes in critical COVID-19 patients were significantly associated with the metabolic pathway (Supplemental Fig. 7A–D). The detailed analysis indicated that a wide range of metabolic genes, which were categorized into lipid, amino acid, carbohydrate metabolism, citrate (TCA) cycle, mitochondrial fatty acid beta-oxidation, and respiratory electron transport, was remarkably defective in the whole blood of critical COVID-19 patients (sFig. 7E–J).
sFig. 6.
Upregulated pathway in the whole blood of critical COVID-19 patients. A–F: Transcriptome analysis of whole blood samples from hospitalized noncritical and critical COVID-19 patients (GSE172114, noncritical n = 23, critical n = 46). Volcano plot (A) and pathway analysis of the upregulated genes (B and C; fold change>1.5, p value<0.05). D: Heatmap image of immune system- and cytokine/interferon signaling-related genes. E: Relative gene expression of IL6 and IL1B signaling-related genes. F: Relative gene expression of cell damage- and cell death-related genes. Data represent the mean ± SEM. #p < 0.25, *p < 0.05, **p < 0.01, and ***p < 0.001.
sFig. 7.
Downregulated pathway in the whole blood of critical COVID-19 patients. A–J: Transcriptome analysis of whole blood samples from hospitalized noncritical and critical COVID-19 patients (GSE172114, noncritical n = 23, critical n = 46). Volcano plot (A) and pathway analysis of the downregulated genes (B and C; fold change<1, p value<0.05). D: Heatmap image of metabolic pathway-related genes. E–J: Relative gene expression of genes related to lipid metabolism (E), amino acid metabolism (F), carbohydrate metabolism (G), citrate (TCA) cycle (H), mitochondrial fatty acid beta-oxidation (I), and respiratory electron transport (J). Data represent the mean ± SEM. #p < 0.25, *p < 0.05, **p < 0.01, and ***p < 0.001.
3.6. IRF1-mediated impairment of the insulin/IGF signaling pathway is associated with critical COVID-19 and potential therapeutic approaches
We next estimated whether the induction of IRF1 and the impairment of the insulin/IGF signaling pathway are associated with critical COVID-19. The gene expression of IRF1 was significantly increased and that of insulin/IGF signaling molecules was downregulated in the whole blood of critical COVID-19 patients (Fig. 5C and D). These results suggested that IRF1-associated impairment in the insulin/IGF signaling pathway may critically contribute to the outcome of COVID-19 patients with exacerbated cell damage, cell death, and metabolic abnormalities.
We finally investigated potential therapeutic strategies that may inhibit the expression of the IRF1 gene and enhance that of insulin/IGF signaling pathway genes in SARS-CoV-2 infectable cells and tissues. Consistent with the results of aging-associated IRF1 regulation (Fig. 4A–E), treatment of cultured human epithelial cells with dihydrotestosterone (DHT), an active metabolite of testosterone, significantly reduced IRF1 gene expression, which was associated with increased gene expression of insulin/IGF signaling molecules (Fig. 5E). Glucocorticoids are well established for their anti-inflammatory properties and are widely used in the treatment of various pulmonary diseases, including COVID-19. Dexamethasone treatment, a synthetic analog of cortisol, significantly reduced IRF1 gene expression and enhanced insulin/IGF signaling pathway genes in cultured human adipose tissue (Fig. 5F) and human primary pancreatic islets (Fig. 5G). Of note, in the lung of a humanized mouse COVID-19 model [80], SARS-CoV-2 infection significantly upregulated IRF1 gene expression and impaired insulin/IGF signaling pathway genes, which were effectively rescued by DEX treatment (Fig. 5H). ChIP-Seq analyses of androgen receptor (AR) and glucocorticoid receptor (GR; NR3C1) indicated the significant bindings in the gene regions of IRF1 (Fig. 5I), suggesting direct transcriptional regulation.
To evaluate whether DHT and DEX could ameliorate insulin signaling under inflammatory conditions in human cells, we established a general inflammatory condition in HEK293T cells by the treatment of TNF-α and estimated DHT and DEX efficacy on insulin signaling. HEK293T cells were treated with TNF-α in the presence or absent of 100 nM DHT or DEX for 24 h, followed by insulin response at the indicated time points, 0-, 10-, 30-, and 90-min. Both DHT and DEX treatment significantly enhanced insulin signaling in all insulin time points and clearly reversed the TNF-α-mediated AKT phosphorylation impairment, which was in part associated with IRS2 and AKT protein levels (Fig. 5J and K). Collectively, these data suggested that both DEX and DHT could enhance insulin signaling in human cells under inflammatory conditions.
4. Discussion
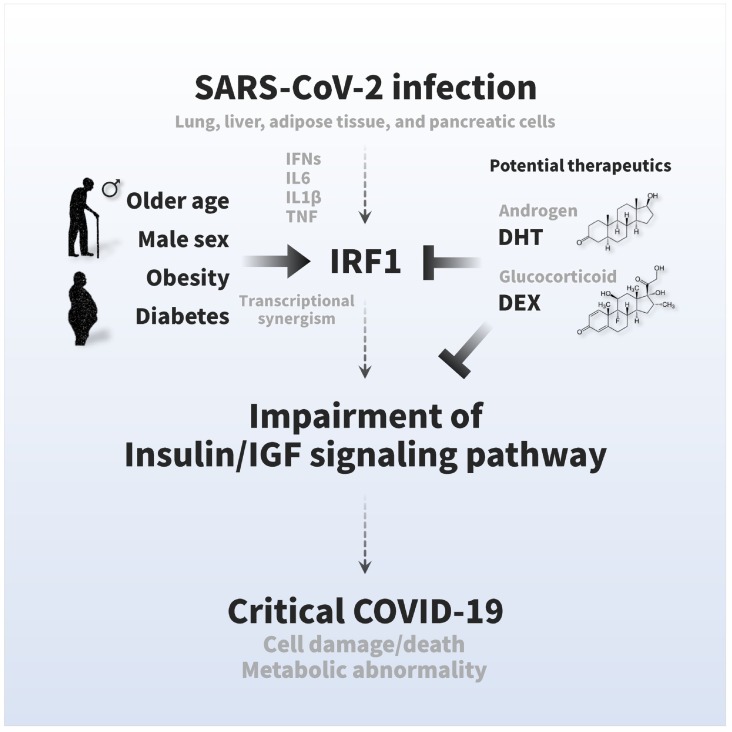
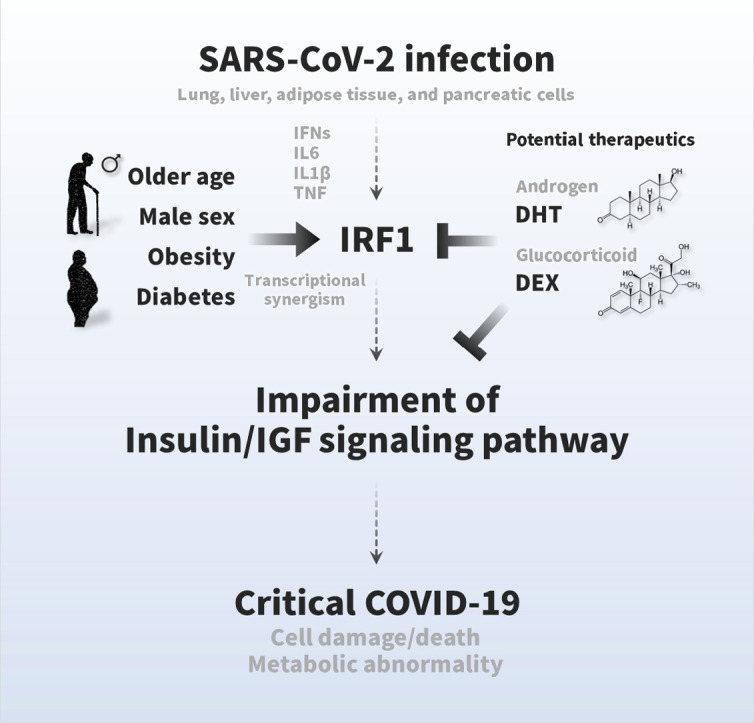
SARS-CoV-2 infection causes multiple tissue damage and, in some cases, metabolic abnormalities, but the molecular mechanism has not yet been elucidated. Here, we found a novel pathological feature of COVID-19, namely, an impairment of the insulin/IGF signaling pathway, and an association with the critical outcome. SARS-CoV-2 infection impaired a wide range of insulin/IGF signaling pathways in the host lung, liver, adipose tissue, and pancreatic cells (Fig. 1). The impairments were associated with excessive interferon responses, in which IRF1 likely attributed to the impairment of the insulin/IGF signaling pathway (Fig. 2, Fig. 3). The expression of IRF1 was highly relevant to older age, male sex, obesity, and diabetes with induction of inflammatory cytokines, such as IL6, IL1B, and TNF (Fig. 4). Also, IRF1 intron variant rs17622656-A, which was frequently found in American and European populations, was associated with the prevalence of COVID-19, suggesting the genetic involvement on these countries (Fig. 5A and B; sFig. 4). The higher basal IRF1 by older age, obesity, diabetes and/or genetic variant might intensify the upregulation of IRF1 in response to SARS-CoV-2 infection (sFig. 5), which potentially contribute to the pathology of COVID-19. In fact, the higher expression of IRF1 and the lower expression of insulin/IGF signaling pathway molecules were significantly associated with critical outcomes in COVID-19 patients (Fig. 5C and D), which were associated with exacerbated IL6 signaling, exacerbated IL1B signaling, cell damage, cell death, and metabolic abnormalities (sFig. 6, sFig. 7). Hormonal interventions, such as dihydrotestosterone and dexamethasone, were effectively lowered the gene expression of IRF1 and enhanced that of the insulin/IGF signaling pathway in SARS-CoV-2 infectable cells and tissues (Fig. 5E–G) and improved the cellular signaling, suggesting therapeutic potential for DHT and DEX. The present study provides the first scientific evidence that SARS-CoV-2 impairs the insulin/IGF signaling pathway not only in the respiratory tract but also in metabolic and endocrine cells and tissues. Higher basal IRF1 expression by pathological (older age, male sex, obesity, and diabetes) and/or genetic (IRF1 intron variant rs17622656-A) reasons in respiratory, metabolic, and/or endocrine organs might contribute to synergistic upregulation of IRF1 in response to SARS-CoV-2 infection, which may make the people more vulnerable to COVID-19 in association with tissue damage and metabolic abnormalities. We hope that the pathological features and findings would be a molecular basis for a better understanding of COVID-19 pathologies and finding future therapeutic approaches (Fig. 6).
Fig. 6.
Graphical summary of the pathogenesis of COVID-19 symptoms and therapeutic strategies. The insulin/IGF signaling pathway plays an important role in many biological processes, such as energy metabolism and cell survival. SARS-CoV-2 infection impairs transcriptional expression of the insulin/IGF signaling pathway in the host lung, liver adipose tissue, and pancreatic cells, which is likely attributed to interferon regulatory factor 1 (IRF1). The pathological trait is aggravated in whole blood, a systemic indicator, of critical patients with COVID-19 with exacerbated cell damage, cell death, and metabolic abnormalities, which could be ameliorated by androgen (DHT) and/or glucocorticoid (DEX) interventions. Higher basal IRF1 expression by pathological (older age, male sex, obesity, and diabetes) and/or genetic (IRF1 intron variant rs17622656-A) reasons in respiratory, metabolic, and/or endocrine organs might contribute to synergistic upregulation of IRF1 in response to SARS-CoV-2 infection, which may make the people more vulnerable to COVID-19.
Multiple organ failure critically contributes to the severity and mortality of COVID-19, but the molecular mechanisms have not been fully explained. In the present study, we showed that SARS-CoV-2 infection impaired the gene expression of the insulin/IGF signaling pathway in the lung tissue sample of COVID-19 patients and various COVID-19 experimental models including liver, adipose tissue, and pancreatic cells (Fig. 1). The pathological features were associated with critical COVID-19 outcome (Fig. 5) as well as exacerbated cell damage and cell death pathways (sFig. 6). SARS-CoV-2 also infects many other cells, tissues, and organs, including the kidney, heart, liver, brain, and blood cells [81], [82]. Collectively, impairment of the insulin/IGF signaling pathway in infected cells and tissues might contribute to the development of multiorgan dysfunction, and further clinical research must be performed to address these points.
Insulin/IGF signaling plays a central role in the regulation of systemic energy homeostasis [14], [15]. Emerging studies have shown that COVID-19, in some cases, predisposes patients to develop metabolic complications during or after the recovery process, such as insulin resistance, hyperglycemia, hyperlipidemia, and new onset of diabetes [13]. In the present study, we showed that SARS-CoV-2 infection caused impairment of the insulin/IGF signaling pathway in important metabolic tissues and endocrine cells (Fig. 1), which was associated with downregulation of various metabolic pathways, such as lipid metabolism, amino acid metabolism, carbohydrate metabolism, citrate (TCA) cycle, beta oxidation, and respiratory electron transport (Supplemental Fig. 7). This finding might be helpful to understand metabolic abnormalities in COVID-19.
Older age, male sex, obesity, and diabetes are well-established risk factors for the severity and mortality of COVID-19 [1], but the molecular mechanisms have not been fully elucidated. In the present study, we showed that the expression of IRF1 was relevant to these risk factors, aggravated with aging in male lung tissue, and augmented by obesity and diabetic conditions in liver, adipose tissue, and pancreatic islets (Fig. 4). The high expression of IRF1 in COVID-19 patients with older age, male sex, obesity, and diabetes may exacerbate the impairment of the insulin/IGF signaling pathway, which may potentially contribute to severe tissue damage and metabolic abnormalities, thereby leading to critical outcomes in COVID-19 (Fig. 6).
In the present study, we showed that hormonal interventions, such as dihydrotestosterone and dexamethasone, inhibited the gene expression of IRF1 and enhanced that of the insulin/IGF signaling pathway presumably via direct transcriptional regulation and effectively ameliorate TNF-mediated insulin resistance in HEK293T cells (Fig. 5). These results were well aligned with COVID-19 clinical research. In a recent clinical study [83], they showed that the serum testosterone levels were significantly lower in severe COVID-19 patients compared to mild-moderate COVID-19 patients (median 85.1 vs 315 ng/dl; p < 0.001, respectively), in COVID-19 patients in need of intensive care compared to COVID-19 patients who did not need intensive care (median, 64.0 vs 286 ng/dl; p < 0.001, respectively), and in COVID-19 patients who deceased compared to survivors (median, 82.9 vs 166 ng/dl; p < 0.001, respectively), suggesting the importance in COVID-19 pathology.
Dexamethasone (DEX) is well known for its antagonistic effect of immune and inflammatory responses and clinically used to alleviate the symptoms of COVID-19; Dexamethasone treatment also effectively lowered the mortality in critical COVID-19 patients [84], [85]. It directly suppresses the downstream of immune responses in the cells and effectively ameliorate the acute exacerbation of COVID-19-meidated inflammation and the consequential cell death and tissue damage. Inflammation is also well known to trigger insulin resistance in various cells and tissues and contribute to the development of metabolic abnormalities. Thus, the inhibition of immune and inflammatory response by dexamethasone could be a therapeutic approach to ameliorate insulin resistance and metabolic abnormalities such as in case of COVID-19. In the present study, we showed the significant improvement of insulin signaling by DEX treatment. Previous studies, which shows glucocorticoid-mediated insulin resistance in vitro, utilized 10-fold higher dosage of dexamethasone (1 μM) than our biological experiments (100 nM), performed in different cell types, and explained by different mechanisms such as glucose transport and mitochondria defects. Therefore, further physiological, pathological, and pharmacological experiments and clinical estimations are required to fully understand their true natures in future study. Both testosterone and glucocorticoid treatment may be a therapeutic approach to prevent the severity and mortality of COVID-19 patients, but great care should be taken to decide appropriate dosage, time, and duration of DEX and DHT interventions depending on each patient's condition, diseases background, severity, and progress of COVID-19. Also, further clinical studies are needed to validate the efficacy.
For the past decades, technological advances in bioinformatics analysis, such as microarrays and RNA-Seq, have accumulated a considerable amount of biological information from various research fields. These data have been uploaded to public databases and made available for scientific use. In the present study, we utilized these datasets by collecting, reanalyzing, and compiling them in view of COVID-19 pathology. The easy access and reproducible results (data sources already deposited) were the strengths of the present study, but many limitations still need to be addressed in future work. The present study was based on analyses of transcriptome datasets; thus, detailed tissue- and cell-specific studies are needed to elucidate this matter in other clinical and basic research fields. The roles of androgens in the pathogenesis of COVID-19 are still controversial. Other studies have suggested that androgens may increase susceptibility to SARS-CoV-2 infection [86], [87]. Therefore, further clinical investigations are needed to evaluate this hormone. We hope that this study will be helpful for better comprehension of COVID-19 pathology and for identifying therapeutic approaches.
The following are the supplementary data related to this article.
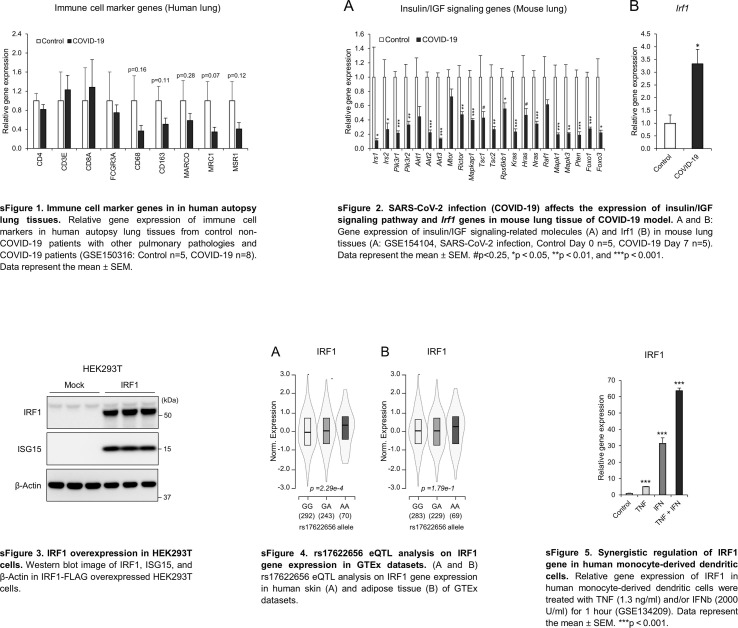
sFig. 1. Immune cell marker genes in in human autopsy lung tissues. Relative gene expression of immune cell markers in human autopsy lung tissues from control non-COVID-19 patients with other pulmonary pathologies and COVID-19 patients (GSE150316: Control n = 5, COVID-19 n = 8). Data represent the mean ± SEM.
sFig. 2. SARS-CoV-2 infection (COVID-19) affects the expression of insulin/IGF signaling pathway and Irf1 genes in mouse lung tissue of COVID-19 model. A and B: Gene expression of insulin/IGF signaling-related molecules (A) and Irf1 (B) in mouse lung tissues (A: GSE154104, SARS-CoV-2 infection, Control Day 0 n = 5, COVID-19 Day 7 n = 5). Data represent the mean ± SEM. #p < 0.25, *p < 0.05, **p < 0.01, and ***p < 0.001.
sFig. 3. IRF1 overexpression in HEK293T cells. Western blot image of IRF1, ISG15, and β-Actin in IRF1-FLAG overexpressed HEK293T cells.
sFig. 4. rs17622656 eQTL analysis on IRF1 gene expression in GTEx datasets. (A and B) rs17622656 eQTL analysis on IRF1 gene expression in human skin (A) and adipose tissue (B) of GTEx datasets.
sFig. 5. Synergistic regulation of IRF1 gene in human monocyte-derived dendritic cells. Relative gene expression of IRF1 in human monocyte-derived dendritic cells were treated with TNF (1.3 ng/ml) and/or IFNb (2000 U/ml) for 1 h (GSE134209). Data represent the mean ± SEM. ***p < 0.001.
Abbreviations
- COVID-19
Coronavirus disease-2019
- SARS-CoV-2
Severe acute respiratory syndrome coronavairus-2
- IRF1
Interferon regulatory factor 1
Data availability
Materials for transcriptome analysis in this paper are publicly available at Gene Expression Omnibus (GEO: https://www.ncbi.nlm.nih.gov/geo/) with accession numbers described in Table 1. Any additional information required to reanalyze the data reported in this paper is available from the corresponding author upon reasonable request.
Authors' relationships and activities
All authors listed in this paper declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Funding
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
CRediT authorship contribution statement
Jihoon Shin: Conceptualization, Data curation, Formal analysis, Investigation,
Methodology, Writing-Original draft preparation.
Shinichiro Toyoda: Formal analysis, Discussion.
Shigeki Nishitani: Formal analysis, Discussion.
Toshiharu Onodera: Formal analysis, Discussion.
Shiro Fukuda: Formal analysis, Discussion.
Shunbun Kita: Formal analysis, Discussion.
Atsunori Fukuhara: Methodology, Formal analysis, Writing-review.
Iichiro Shimomura: Methodology, Writing-review and editing, Supervision.
Acknowledgments
We would like to thank all the members of the Department of Metabolic Medicine, Graduate School of Medicine, Osaka University.
References
- 1.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Team C.C.-R. Severe outcomes among patients with coronavirus disease 2019 (COVID-19) - United States, February 12-March 16, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montefusco L., Ben Nasr M., D’Addio F., Loretelli C., Rossi A., Pastore I., et al. Acute and long-term disruption of glycometabolic control after SARS-CoV-2 infection. Nat Metab. 2021;3:774–785. doi: 10.1038/s42255-021-00407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayres J.S. A metabolic handbook for the COVID-19 pandemic. Nat Metab. 2020;2:572–585. doi: 10.1038/s42255-020-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J., Wang X., Chen J., Zuo X., Zhang H., Deng A. COVID-19 infection may cause ketosis and ketoacidosis. Diabetes Obes Metab. 2020;22:1935–1941. doi: 10.1111/dom.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldman N., Fink D., Cai J., Lee Y.-N., Davies Z. High prevalence of COVID-19-associated diabetic ketoacidosis in UK secondary care. Diabetes Res Clin Pract. 2020;166 doi: 10.1016/j.diabres.2020.108291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee Y.J., SJH Ng, Yeoh E. Diabetic ketoacidosis precipitated by Covid-19 in a patient with newly diagnosed diabetes mellitus. Diabetes Res Clin Pract. 2020;164 doi: 10.1016/j.diabres.2020.108166. 108166- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchand L., Pecquet M., Luyton C. Type 1 diabetes onset triggered by COVID-19. Acta Diabetol. 2020;57:1265–1266. doi: 10.1007/s00592-020-01570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebekozien O.A., Noor N., Gallagher M.P., Alonso G.T. Type 1 diabetes and COVID-19: preliminary findings from a multicenter surveillance study in the U.S. Diabetes Care. 2020;43 doi: 10.2337/dc20-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollstein T., Schulte D.M., Schulz J., Glück A., Ziegler A.G., Bonifacio E., et al. Autoantibody-negative insulin-dependent diabetes mellitus after SARS-CoV-2 infection: a case report. Nat Metab. 2020;2:1021–1024. doi: 10.1038/s42255-020-00281-8. [DOI] [PubMed] [Google Scholar]
- 11.Accili D. Can COVID-19 cause diabetes? Nat Metab. 2021;3:123–125. doi: 10.1038/s42255-020-00339-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T., Du Z., Zhu F., Cao Z., An Y., Gao Y., et al. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metwally A.A., Mehta P., Johnson B.S., Nagarjuna A., Snyder M.P. COVID-19-induced new-onset diabetes: trends and technologies. Diabetes. 2021;70(12):2733–2744. doi: 10.2337/dbi21-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boucher J., Kleinridders A., Kahn C.R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronald Kahn C. Insulin resistance, insulin insensitivity, and insulin unresponsiveness: a necessary distinction. Metabolism. 1978;27:1893–1902. doi: 10.1016/s0026-0495(78)80007-9. [DOI] [PubMed] [Google Scholar]
- 16.Cheatham B., Kahn C.R. Insulin action and the insulin signaling network. Endocr Rev. 1995;16:117–142. doi: 10.1210/edrv-16-2-117. [DOI] [PubMed] [Google Scholar]
- 17.Boucher J., Kleinridders A., Kahn C.R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamemoto H., Kadowaki T., Tobe K., Yagi T., Sakura H., Hayakawa T., et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 19.Brüning J.C., Michael M.D., Winnay J.N., Hayashi T., Hörsch D., Accili D., et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 20.Accili D., Drago J., Lee E.J., Johnson M.D., Cool M.H., Salvatore P., et al. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat Genet. 1996;12:106–109. doi: 10.1038/ng0196-106. [DOI] [PubMed] [Google Scholar]
- 21.Kitamura T., Kahn C.R., Accili D. Insulin receptor knockout mice. Annu Rev Physiol. 2003;65:313–332. doi: 10.1146/annurev.physiol.65.092101.142540. [DOI] [PubMed] [Google Scholar]
- 22.Softic S., Boucher J., Solheim M.H., Fujisaka S., Haering M.-F., Homan E.P., et al. Lipodystrophy due to adipose tissue–specific insulin receptor knockout results in progressive NAFLD. Diabetes. 2016;65:2187–2200. doi: 10.2337/db16-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulkarni R.N., Brüning J.C., Winnay J.N., Postic C., Magnuson M.A., Kahn C.R. Tissue-specific knockout of the insulin receptor in pancreatic β cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96:329–339. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 24.Peruzzi F., Prisco M., Dews M., Salomoni P., Grassilli E., Romano G., et al. Multiple signaling pathways of the insulin-like growth factor 1 receptor in protection from apoptosis. Mol Cell Biol. 1999;19:7203–7215. doi: 10.1128/mcb.19.10.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conti E., Carrozza C., Capoluongo E., Volpe M., Crea F., Zuppi C., et al. Insulin-like growth Factor-1 as a vascular protective factor. Circulation. 2004;110:2260–2265. doi: 10.1161/01.CIR.0000144309.87183.FB. [DOI] [PubMed] [Google Scholar]
- 26.Yang S., Lin L., Chen J.-X., Lee C.R., Seubert J.M., Wang Y., et al. Cytochrome P-450 epoxygenases protect endothelial cells from apoptosis induced by tumor necrosis factor-α via MAPK and PI3K/Akt signaling pathways. Am J Physiol Heart Circ Physiol. 2007;293:H142–H151. doi: 10.1152/ajpheart.00783.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi C., Booth L.N., Murphy C.T. Insulin-like peptides and the mTOR-TFEB pathway protect Caenorhabditis elegans hermaphrodites from mating-induced death. eLife. 2019;8 doi: 10.7554/eLife.46413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Sahra I., Dirat B., Laurent K., Puissant A., Auberger P., Budanov A., et al. Sestrin2 integrates akt and mTOR signaling to protect cells against energetic stress-induced death. Cell Death Differ. 2013;20:611–619. doi: 10.1038/cdd.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiengo A., Fadini G.P., Avogaro A. The metabolic syndrome, diabetes and lung dysfunction. Diabetes Metab. 2008;34:447–454. doi: 10.1016/j.diabet.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Klein O.L., Krishnan J.A., Glick S., Smith L.J. Systematic review of the association between lung function and type 2 diabetes mellitus. Diabet Med. 2010;27:977–987. doi: 10.1111/j.1464-5491.2010.03073.x. [DOI] [PubMed] [Google Scholar]
- 31.Goldman M.D. Lung dysfunction in diabetes. Diabetes Care. 2003;26:1915–1918. doi: 10.2337/diacare.26.6.1915. [DOI] [PubMed] [Google Scholar]
- 32.Forno E., Han Y.-Y., Muzumdar R.H., Celedón J.C. Insulin resistance, metabolic syndrome, and lung function in US adolescents with and without asthma. Journal of Allergy and Clinical Immunology. 2015;136 doi: 10.1016/j.jaci.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sengupta A., Patel P.A., Yuldasheva N.Y., Mughal R.S., Galloway S., Viswambharan H., et al. Endothelial insulin receptor restoration rescues vascular function in male insulin receptor haploinsufficient mice. Endocrinology. 2018;159:2917–2925. doi: 10.1210/en.2018-00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epaud R., Aubey F., Xu J., Chaker Z., Clemessy M., Dautin A., et al. Knockout of insulin-like growth Factor-1 receptor impairs distal lung morphogenesis. PLOS ONE. 2012;7 doi: 10.1371/journal.pone.0048071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz K., Wasnik S., Abdipour A., Bi H., Wilson S.M., Tang X., et al. The effects of insulin-like growth factor I and BTP-2 on acute lung injury. Int J Mol Sci. 2021;22:5244. doi: 10.3390/ijms22105244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narasaraju T.A., Chen H., Weng T., Bhaskaran M., Jin N., Chen J., et al. Expression profile of IGF system during lung injury and recovery in rats exposed to hyperoxia: a possible role of IGF-1 in alveolar epithelial cell proliferation and differentiation. J Cell Biochem. 2006;97:984–998. doi: 10.1002/jcb.20653. [DOI] [PubMed] [Google Scholar]
- 37.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., et al. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., et al. ChIP-atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018;19 doi: 10.15252/embr.201846255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desai N., Neyaz A., Szabolcs A., Shih A.R., Chen J.H., Thapar V., et al. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat Commun. 2020;11:6319. doi: 10.1038/s41467-020-20139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wanner N., Andrieux G., Badia I.M.P., Edler C., Pfefferle S., Lindenmeyer M.T., et al. Molecular consequences of SARS-CoV-2 liver tropism. Nat Metab. 2022;4:310–319. doi: 10.1038/s42255-022-00552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27(125–36) doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poma A.M., Bonuccelli D., Giannini R., Macerola E., Vignali P., Ugolini C., et al. COVID-19 autopsy cases: detection of virus in endocrine tissues. J Endocrinol Invest. 2021;45:209–214. doi: 10.1007/s40618-021-01628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reiterer M., Rajan M., Gómez-Banoy N., Lau J.D., Gomez-Escobar L.G., Ma L., et al. Hyperglycemia in acute COVID-19 is characterized by insulin resistance and adipose tissue infectivity by SARS-CoV-2. Cell Metab. 2021;33 doi: 10.1016/j.cmet.2021.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin J., Toyoda S., Nishitani S., Fukuhara A., Kita S., Otsuki M., et al. Possible involvement of adipose tissue in patients with older age, obesity, and diabetes with coronavirus SARS-CoV-2 infection (COVID-19) via GRP78 (BIP/HSPA5): significance of hyperinsulinemia management in COVID-19. Diabetes. 2021;70(12):2745–2755. doi: 10.2337/db20-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Müller J.A., Groß R., Conzelmann C., Krüger J., Merle U., Steinhart J., et al. SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas. Nat Metab. 2021;3:149–165. doi: 10.1038/s42255-021-00347-1. [DOI] [PubMed] [Google Scholar]
- 53.Qadir M.M.F., Bhondeley M., Beatty W., Gaupp D.D., Doyle-Meyers L.A., Fischer T., et al. SARS-CoV-2 infection of the pancreas promotes thrombofibrosis and is associated with new-onset diabetes. JCI Insight. 2021;6 doi: 10.1172/jci.insight.151551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu C.-T., Lidsky P.V., Xiao Y., Lee I.T., Cheng R., Nakayama T., et al. SARS-CoV-2 infects human pancreatic β cells and elicits β cell impairment. Cell Metab. 2021;33(8):1565–1576. doi: 10.1016/j.cmet.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S., Ma F., Yokota T., Garcia G., Jr., Palermo A., Wang Y., et al. Metabolic reprogramming and epigenetic changes of vital organs in SARS-CoV-2-induced systemic toxicity. JCIInsight. 2021;6 doi: 10.1172/jci.insight.145027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaharuddin S.H., Wang V., Santos R.S., Gross A., Wang Y., Jawanda H., et al. Deleterious effects of SARS-CoV-2 infection on human pancreatic cells. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.678482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaninov N. In the eye of the COVID-19 cytokine storm. Nat Rev Immunol. 2020;20 doi: 10.1038/s41577-020-0305-6. 277- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 59.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Panda D., Gjinaj E., Bachu M., Squire E., Novatt H., Ozato K., et al. IRF1 maintains optimal constitutive expression of antiviral genes and regulates the early antiviral response. Front Immunol. 2019;10:1019. doi: 10.3389/fimmu.2019.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forero A., Ozarkar S., Li H., Lee C.H., Hemann E.A., Nadjsombati M.S., et al. Differential activation of the transcription factor IRF1 underlies the distinct immune responses elicited by type I and type III interferons. Immunity. 2019;51 doi: 10.1016/j.immuni.2019.07.007. 451-64.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song R., Gao Y., Dozmorov I., Malladi V., Saha I., McDaniel M.M., et al. IRF1 governs the differential interferon-stimulated gene responses in human monocytes and macrophages by regulating chromatin accessibility. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aly S., Mages J., Reiling N., Kalinke U., Decker T., Lang R., et al. Mycobacteria-induced granuloma necrosis depends on IRF-1. J Cell Mol Med. 2009;13:2069–2082. doi: 10.1111/j.1582-4934.2008.00470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsung A., Stang M.T., Ikeda A., Critchlow N.D., Izuishi K., Nakao A., et al. The transcription factor interferon regulatory factor-1 mediates liver damage during ischemia-reperfusion injury. Am J Physiol Gastrointest Liver Physiol. 2006;290 doi: 10.1152/ajpgi.00460.2005. [DOI] [PubMed] [Google Scholar]
- 65.Moore F., Naamane N., Colli M.L., Bouckenooghe T., Ortis F., Gurzov E.N., et al. STAT1 is a master regulator of pancreatic {beta}-cell apoptosis and islet inflammation. J Biol Chem. 2011;286:929–941. doi: 10.1074/jbc.M110.162131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kano A., Haruyama T., Akaike T., Watanabe Y. IRF-1 is an essential mediator in IFN-gamma-induced cell cycle arrest and apoptosis of primary cultured hepatocytes. Biochem Biophys Res Commun. 1999;257:672–677. doi: 10.1006/bbrc.1999.0276. [DOI] [PubMed] [Google Scholar]
- 67.Friesen M., Camahort R., Lee Y.K., Xia F., Gerszten R.E., Rhee E.P., et al. Activation of IRF1 in human adipocytes leads to phenotypes associated with metabolic disease. Stem Cell Rep. 2017;8:1164–1173. doi: 10.1016/j.stemcr.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakazawa T., Satoh J., Takahashi K., Sakata Y., Ikehata F., Takizawa Y., et al. Complete suppression of insulitis and diabetes in NOD mice lacking interferon regulatory factor-1. J Autoimmunol. 2001;17:119–125. doi: 10.1006/jaut.2001.0531. [DOI] [PubMed] [Google Scholar]
- 69.Pavlovic D., Chen M.C., Gysemans C.A., Mathieu C., Eizirik D.L. The role of interferon regulatory factor-1 in cytokine-induced mRNA expression and cell death in murine pancreatic beta-cells. Eur Cytokine Netw. 1999;10:403–412. [PubMed] [Google Scholar]
- 70.Wang X.D., Wang B.E., Soriano R., Zha J., Zhang Z., Modrusan Z., et al. Expression profiling of the mouse prostate after castration and hormone replacement: implication of H-cadherin in prostate tumorigenesis. Differentiation. 2007;75:219–234. doi: 10.1111/j.1432-0436.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- 71.Consortium GT The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sparna T., Retey J., Schmich K., Albrecht U., Naumann K., Gretz N., et al. Genome-wide comparison between IL-17 and combined TNF-alpha/IL-17 induced genes in primary murine hepatocytes. BMC Genomics. 2010;11:226. doi: 10.1186/1471-2164-11-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee Y.H., Nair S., Rousseau E., Allison D.B., Page G.P., Tataranni P.A., et al. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima indians: increased expression of inflammation-related genes. Diabetologia. 2005;48:1776–1783. doi: 10.1007/s00125-005-1867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Balliu B., Carcamo-Orive I., Gloudemans M.J., Nachun D.C., Durrant M.G., Gazal S., et al. An integrated approach to identify environmental modulators of genetic risk factors for complex traits. Am J Hum Genet. 2021;108:1866–1879. doi: 10.1016/j.ajhg.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vivas Y., Martinez-Garcia C., Izquierdo A., Garcia-Garcia F., Callejas S., Velasco I., et al. Early peroxisome proliferator-activated receptor gamma regulated genes involved in expansion of pancreatic beta cell mass. BMC Med Genomics. 2011;4:86. doi: 10.1186/1755-8794-4-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rusmini M., Uva P., Amoroso A., Tolomeo M., Cavalli A. How genetics might explain the unusual link between malaria and COVID-19. Front Med (Lausanne). 2021;8 doi: 10.3389/fmed.2021.650231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Basu M., Wang K., Ruppin E., Hannenhalli S. Predicting tissue-specific gene expression from whole blood transcriptome. ScienceAdvances. 2021;7 doi: 10.1126/sciadv.abd6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mohr S., Liew C.-C. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med. 2007;13:422–432. doi: 10.1016/j.molmed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 79.R Carapito R Li J Helms C Carapito S Gujja V Rolli et al n.d.Identification of driver genes for critical forms of COVID-19 in a deeply phenotyped young patient cohort. Sci Transl Med.0:eabj7521. [DOI] [PubMed]
- 80.Zhang D., Hu Q., Liu X., Ji Y., Chao H.P., Liu Y., et al. Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat Commun. 2020;11:2089. doi: 10.1038/s41467-020-15815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Müller J.A., Groß R., Conzelmann C., Krüger J., Merle U., Steinhart J., et al. SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas. NatMetab. 2021;3:149–165. doi: 10.1038/s42255-021-00347-1. [DOI] [PubMed] [Google Scholar]
- 83.Cinislioglu A.E., Cinislioglu N., Demirdogen S.O., Sam E., Akkas F., Altay M.S., et al. The relationship of serum testosterone levels with the clinical course and prognosis of COVID-19 disease in male patients: a prospective study. Andrology. 2022;10:24–33. doi: 10.1111/andr.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Group RC. Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ye Z., Wang Y., Colunga-Lozano L.E., Prasad M., Tangamornsuksan W., Rochwerg B., et al. Efficacy and safety of corticosteroids in COVID-19 based on evidence for COVID-19, other coronavirus infections, influenza, community-acquired pneumonia and acute respiratory distress syndrome: a systematic review and meta-analysis. Can Med Assoc J. 2020;192:E756–E767. doi: 10.1503/cmaj.200645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salciccia S., Del Giudice F., Eisenberg M.L., Mastroianni C.M., De Berardinis E., Ricciuti G.P., et al. Testosterone target therapy: focus on immune response, controversies and clinical implications in patients with COVID-19 infection. Ther Adv Endocrinol Metab. 2021;12 doi: 10.1177/20420188211010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Samuel R.M., Majd H., Richter M.N., Ghazizadeh Z., Zekavat S.M., Navickas A., et al. Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men. Cell Stem Cell. 2020;27 doi: 10.1016/j.stem.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Materials for transcriptome analysis in this paper are publicly available at Gene Expression Omnibus (GEO: https://www.ncbi.nlm.nih.gov/geo/) with accession numbers described in Table 1. Any additional information required to reanalyze the data reported in this paper is available from the corresponding author upon reasonable request.