Abstract
Melanoma is a kind of skin cancer caused by the irregular development of pigment-producing cells. Since melanoma detection efficiency is limited to different factors such as poor contrast among lesions and nearby skin regions, and visual resemblance among melanoma and non-melanoma lesions, intelligent computer-aided diagnosis (CAD) models are essential. Recently, computational intelligence (CI) and deep learning (DL) techniques are utilized for effective decision-making in the biomedical field. In addition, the fast-growing advancements in computer-aided surgeries and recent progress in molecular, cellular, and tissue engineering research have made CI an inevitable part of biomedical applications. In this view, the research work here develops a novel computational intelligence-based melanoma detection and classification technique using dermoscopic images (CIMDC-DIs). The proposed CIMDC-DI model encompasses different subprocesses. Primarily, bilateral filtering with fuzzy k-means (FKM) clustering-based image segmentation is applied as a preprocessing step. Besides, NasNet-based feature extractor with stochastic gradient descent is applied for feature extraction. Finally, the manta ray foraging optimization (MRFO) algorithm with a cascaded neural network (CNN) is exploited for the classification process. To ensure the potential efficiency of the CIMDC-DI technique, we conducted a wide-ranging simulation analysis, and the results reported its effectiveness over the existing recent algorithms with the maximum accuracy of 97.50%.
1. Introduction
Computational intelligence (CI) and its wide-ranging applications in biomedical engineering offer oncology, genetic data, genomics, bio-mimetic systems, ontologies construction, protein structure prediction, biomedical data analysis, and biomedical electronics [1]. CI is the study of the design of an intelligent agent, that is a system that can act wisely: they do what they think is applicable for the goal and circumstance; they are flexible to goals and changing environments; they learn from experience, and they make proper selections given finite computation and perceptual limitations. In addition, the rapidly increasing advancement in computer-aided research and surgeries in cellular, tissue, molecular, and engineering makes CI an inevitable part of biomedical applications [2]. The CI paradigm renders more advantages to for enhancing and maintaining the area of biomedical engineering. Skin malignant growth contrasted with other kinds of tumors is a major factor that makes a serious medical illness [3–5]. In earlier years, melanoma was an uncommon malignant growth, but nowadays the total cases of melanoma are increasing dramatically.
To identify skin cancer quickly at the beginning and resolve the abovementioned problems, there is a comprehensive study solution by proposing a computer image analysis algorithm [6]. Most of the algorithmic solution was parametric, which means they needed information to be distributed normally [7]. Since the nature of information could not be controlled, this method will be inadequate to precisely identify the disease. But the nonparametric solution does not depend on the constraints that the information is in standard distribution format. With current advancements in software and hardware techniques, DL is emerging as an effective mechanism for learning features [8]. Feature engineering is a procedure of extracting and determining features by human expertise which is a time-consuming and cumbersome task. The DL method removes the necessity for feature engineering since it is capable of learning and extracting meaningful features automatically from the raw information [9].
DL has transformed various areas, particularly computer vision. In biomedical engineering, DL demonstrates a considerable achievement in the present study. Automatic classification of skin lesions utilizing images is a difficult process because of the fine-grained variation in the appearance of skin lesions. A deep convolution neural network (DCNN) shows potential for highly variable and general tasks through several fine-grained object classes [10]. Outfitted with a deep neural network, the mobile device possibly extends the reach of dermatologists outside of the hospital. The CNN accomplishes performance on par with each testing expert through both tasks, which demonstrates an AI proficiency to classify skin lesions with a level of competencies compared with dermatologists [11].
This study introduces an effective CI-based melanoma detection and classification using the dermoscopic images (CIMDC-DIs) technique. The proposed CIMDC-DI model encompasses bilateral filtering based noise reduction with fuzzy k-means (FKM) clustering based image segmentation as a preprocessing step. Besides, NasNet-based feature extractor with stochastic gradient descent is applied for feature extraction. Finally, realizing the significance of the parameter optimization in enhancing the model performance [12, 13], the manta ray foraging optimization (MRFO) algorithm with cascaded neural network (CNN) is exploited for the classification process. To ensure the better outcomes of the CIMDC-DI technique, a wide-ranging simulation analysis was carried out and the results are assessed under distinct aspects.
2. Related Works
Lai et al. [7] presented the technique which combines genomics data, a disease network, and the DL technique for classifying melanoma patients to prognosis, evaluating the influence of genomic features on the classifier, and offering interpretation to impactful features. It combined genomics data with a melanoma network and executed the AE method for identifying subgroups from TCGA melanoma patients. This technique employs community identified from the network to efficiently decrease the dimensionality of genomics data as to patient score profiles. Lafraxo et al. [14] propose a CCN-based deep learning model to automate the classification of benign or malignant skin lesions in dermoscopy images. Furthermore, the performance of the model is improved by utilizing three techniques such as data augmentation, regularization, and dropout to avoid overfitting.
Shorfuzzaman [15] presented an explainable CNN-based stacked ensemble infrastructure for detecting melanoma skin cancer at previous phases. During the stacking ensemble infrastructure, the transfer learning (TL) method was utilized in which several CNN submodels which carry out similar classifier tasks are collected. A novel technique named meta-learner utilizes every submodel forecast and creates the last forecast outcomes. Kim et al. [16] presented a novel unsupervised technique for hair extraction and estimated it on a real-world melanoma data set. During the generative adversarial learning infrastructure, hair feature is considered with coarse-grained label easily utilizing a binary classification. Besides, an essential feature of lesions was well-maintained by minimized L1-norm reconstruction loss dependent upon Laplace noise assumptions.
In [17], a melanoma segmentation method dependent upon DL was presented. In conjunction with post-processed methods, the presented modified U-net network that is extremely effectual from lesions segmentation is established. Hagerty et al. [18] introduced the technique which integrates convention image processing with DL by fusing the feature in the individual approaches. It is offered that two approaches, with distinct error profiles, are synergistic. The convention image processing arm utilizes 3 handcrafted biologically simulated image processing elements and one medicinal data element. In [19], a novel proposal of the DCNN method to classifier skin lesions as malignant and benign on dermoscopic images was presented by creating several connecting blocks for allowing huge feature data to flow directly with the network. All the blocks of the network utilize distinct parameters like the number of kernels, filter size, and stride for extracting low- and high-level feature data in lesions.
3. The Proposed Model
This study developed a novel CIMDC-DI approach for melanoma identification and classification using dermoscopic images. The presented CIMDC-DI model involves BF-enabled noise reduction, FKM segmentation, NasNet feature extraction, CNN classifier, and MRFO parameter optimization. The utilization of the MRFO algorithm assists in the effectual choice of parameter values involved in the CNN model. Figure 1 illustrates the overall block diagram of CIMDC-DI technique.
Figure 1.
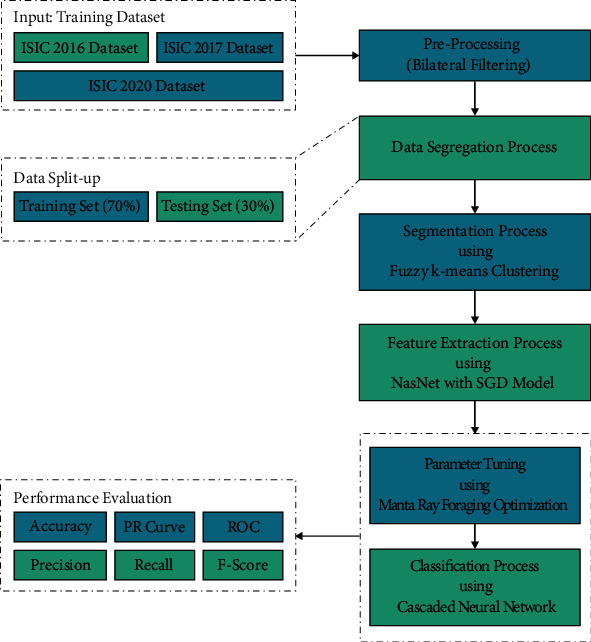
Overall block diagram of CIMDC-DI technique.
3.1. Bilateral Filtering
At the primary level, the BF technique is used to eradicate the occurrence of noise in dermoscopic images. Dermoscopic images comprise noises like Gaussian, salt pepper noise, and so on [20]. Extracting the noise preserves the data similar to the input data. The BF approach was utilized to denoise this input image. Without utilizing the smoothing edge, the spatial weight averaging was executed by BF. This filtering combines two Gaussian filters to accomplish filtering both in spatial and intensity domain; another one is functioning. In order to weight, both the intensity as well as spatial distances was utilized. The BF output at pixel place p is explained as follows:
| (1) |
where S(p) implies the pixel spatial neighbourhood F(p), N denotes the normalized constant, and εe and εr define the parameters governing weighted from the domains of intensity and spatial begin fall off.
| (2) |
The BFs are utilized in texture removal, tone mapping, volumetric denoising, and another application as denoising the images. It creates easy conditions for down-sampling the vital methods and attaining acceleration by expressing under this augmented space by the 2 modest nonlinearity, and the BF was executed as easy linear convolution.
3.2. FKM-Based Image Segmentation
In order to identify the lesion regions in the dermoscopic images, the FKM technique has been exploited. The segmentation is employed by an FKM on the extracted set of the melanoma cancer for separating the healthy pixel in the melanoma pixel [21]. The major reason for selecting the FKM over K‐means clustering is that K‐means clustering is the hard kind of clustering in which one instance belongs to a single cluster; however, in FKM, one instance belongs to one or more clusters; hence, it performs well for overlapped information. The FKM approach separates the image into regions in which R1(I=1,2.3) is associated with the cluster centered Cr. FKM has the fuzzy or soft relation among ROI and image and reduces distortion:
| (3) |
Here, k refers the amount of clusters whereas f indicates fuzzifier parameter that manipulates the data point and resulting cluster, and bij ∈ [0,1] characterizes the relations among the data points and clusters, whereas gij characterizes the Euclidean distance amongst data points and clusters.
3.3. Feature Extraction
At the time of generating feature vectors, the segmented images are fed into the NasNet model. CNN comprises input and output layers with many hidden convolutional layers. The NASNet model is inspired by Neural Architecture Search (NAS) model [22], which exhibits high flexibility and scalability in terms of computation resources and parameters. It has been trained on the chosen ImageNet database which and is optimized. It comprises a collection of filters, which are then employed to RGB pixel values of the image via sliding window manner. The dot product of filters and input pixels is determined. The feature map is reached in a 2-dimension activation map of the filter. Figure 2 illustrates the structure of cascaded NN.
Figure 2.
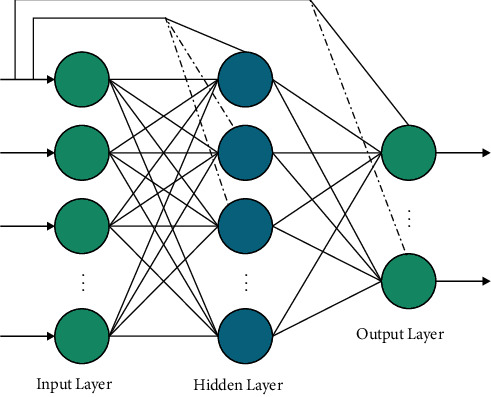
Structure of cascaded neural network.
It learned the features need to be activated if identified features in the input are attained. Then, the convolutional function is carried out on all feature maps. It enables CNN in learning various feature map weights and biases. Then, max-pooling operation can be utilized for reducing the feature map size. Next to every convolutional layer, subsampling layer is attained which enables to reduction of the size of the convolutional map.
3.4. Image Classification
Finally, the MRFO-CNN model receives the features and assigns appropriate class labels to the dermoscopic images. The perceptron connection which is created amongst input as well as output is process of direct relation but in FFNN connection considered among input as well as output is indirect connection [23]. The connection is nonlinear from the shape with activation function under the hidden layer. Once the connection procedure on perceptron and multilayer network was joined, afterward the network with direct connections amongst input as well as output layers was produced, moreover the connection indirectly. The network made in this connection pattern is termed CNN. The formulation developed in CNN approach is demonstrated as follows:
| (4) |
where fi refers the activation function in the input to output layers and ωii implies the weight in the input to output layers. Once the bias is extra for input layer and the activation function of each neuron under the hidden layer is fh, then
| (5) |
The optimal parameter adjustment of the CNN model is performed by the use of the MRFO algorithm [24]. The MRFO algorithm is stimulated by the fascinating behaviour of the manta rays (MRs). It considers three distinct MR processes like a chain, cyclone, and somersault foraging in providing an efficient optimization approach for identifying solutions to distinct optimization issues. In the MRFO algorithm, the location of the agents can be upgraded at each round via the optimal location of the prey with many planktons, which can be represented as follows:
| (6) |
where a signifies weight coefficient, r displays an arbitrary number in [0, 1], χbestd(t) denotes specific density of plankton, χid(t) means location of ith and χi−1d(t) the (i − 1)th representatives at iteration t in dth dimension, and α can be attained using the following equation:
| (7) |
The animals create a long bait chain and swim nearer to the bait in case of fixing planktons. The process of storm can be defined in the following:
| (8) |
where T indicates maximum iteration, β displays weight coefficient, and r1 describes arbitrary numbers among 0 and 1. Bait can be considered as a reference at the time of accomplishing optimal solutions randomly. The storm process can be attained by improving the exploration process in obtaining proper arbitrary location solutions. It can be defined as follows:
| (9) |
where χrandd signifies arbitrary location solution and Ld indicates lower and Ud implies higher constraint of the dth dimension. The feeding position can be considered as a pivot via somersault foraging. The agents look for somersault and hinge to other locations. Therefore, the positions are considered for attaining optimal positions. Then, it can be represented using the following equation:
| (10) |
where S describes somersault bait and amounts to 2 and r2 and r3 display arbitrary numbers. The chaos gets reduced by reducing the distance of the individual planktons. Hence, the somersault forage range reduced with an increase in iterations. In order to choose optimal parameters of the CNN model, the MRFO algorithm has accomplished an objective function, representing a positive integration for implying maximum performance. In this case, the error rate is treated as the fitness function and the solution with minimal error is considered an optimal one. It can be defined as follows:
| (11) |
4. Performance Validation
In this section, the experimental validation of the proposed model is performed using three challenging benchmark datasets [25], as shown in Table 1. The results are inspected with 70% of training data and 30% of testing data. A few sample images are demonstrated in Figure 3.
Table 1.
Dataset details.
| ISIC 2016 dataset | |||
| Class | Training samples | Testing samples | Total samples |
| Melanoma | 108 | 42 | 150 |
| Benign | 102 | 48 | 150 |
| Total | 210 | 90 | 300 |
|
| |||
| ISIC 2017 dataset | |||
| Class | Training samples | Testing samples | Total samples |
| Melanoma | 140 | 60 | 200 |
| Benign | 140 | 60 | 200 |
| Total | 280 | 120 | 400 |
|
| |||
| ISIC 2020 dataset | |||
| Class | Training samples | Testing samples | Total samples |
| Melanoma | 179 | 71 | 250 |
| Benign | 171 | 79 | 250 |
| Total | 350 | 150 | 500 |
Figure 3.
Sample images.
Figure 4 portrays the set of three confusion matrices attained by the CIMDC-DI model on three datasets. On the ISIC2016 dataset, the CIMDC-DI model has recognized 41 images of melanoma and 47 images of benign. Moreover, on the ISIC2017 dataset, the CIMDC-DI algorithm has recognized 55 images of melanoma and 62 images of benign. Furthermore, on the ISIC2017 dataset, the CIMDC-DI approach has recognized 67 images of melanoma and 77 images of benign.
Figure 4.
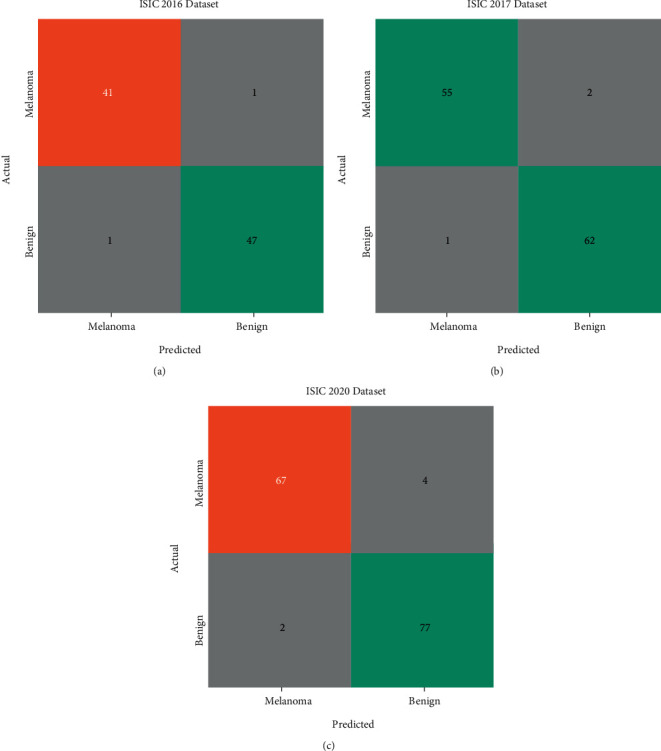
Confusion matrix of the CIMDC-DI model on three datasets.
Table 2 provides detailed melanoma classification outcomes of the CIMDC-DI model on the ISIC2016 dataset. The results indicated that the CIMDC-DI model has reported effectual outcomes on both training and testing datasets. For instance, with 70% of the training dataset, the CIMDC-DI model has resulted in average accuy, precn, recal, and Fscore of 97.78%, 97.77%, 97.77%, and 97.77%, respectively. Besides, with 30% of the testing dataset, the CIMDC-DI (Table 3) model has accomplished average accuy, precn, recal, and Fscore of 94.29%, 94.36%, 94.36%, and 94.29%, respectively.
Table 2.
Result analysis of CIMDC-DI technique on ISIC 2016 dataset
| Class labels | Accuracy | Precision | Recall | F-score |
|---|---|---|---|---|
| Training (70%) | ||||
| Melanoma | 97.78 | 97.62 | 97.62 | 97.62 |
| Benign | 97.78 | 97.92 | 97.92 | 97.92 |
| Average | 97.78 | 97.77 | 97.77 | 97.77 |
|
| ||||
| Testing (30%) | ||||
| Melanoma | 94.29 | 97.06 | 91.67 | 94.29 |
| Benign | 94.29 | 91.67 | 97.06 | 94.29 |
| Average | 94.29 | 94.36 | 94.36 | 94.29 |
Table 3.
Result analysis of CIMDC-DI technique on ISIC 2017 dataset.
| Class labels | Accuracy | Precision | Recall | F-score |
|---|---|---|---|---|
| Training (70%) | ||||
| Melanoma | 96.79 | 99.26 | 94.41 | 96.77 |
| Benign | 96.79 | 94.44 | 99.27 | 96.80 |
| Average | 96.79 | 96.85 | 96.84 | 96.79 |
|
| ||||
| Testing (30%) | ||||
| Melanoma | 97.50 | 98.21 | 96.49 | 97.35 |
| Benign | 97.50 | 96.88 | 98.41 | 97.64 |
| Average | 97.50 | 97.54 | 97.45 | 97.49 |
Table 3 depicts a brief melanoma classification outcome of the CIMDC-DI technique on ISIC 2017 dataset. The results exposed that the CIMDC-DI algorithm has reported effectual outcomes on both training and testing datasets.
For instance, with 70% of the training dataset, the CIMDC-DI methodology has resulted in average accuy, precn, recal, and Fscore of 96.79%, 96.85%, 96.84%, and 96.79% correspondingly. Finally, with 30% of the testing dataset, the CIMDC-DI technique has accomplished an average accuy, precn, recal, and Fscore of 97.50%, 97.54%, 97.45%, and 97.49%, respectively.
Table 4 offers detailed melanoma classification outcomes of the CIMDC-DI technique on ISIC 2020 dataset. The results demonstrated that the CIMDC-DI method has reported effectual outcomes on both training and testing datasets. For instance, with 70% of training dataset, the CIMDC-DI model has resulted in average accuy, precn, recal, and Fscore of 93.14%, 93.16%, 93.18%, and 93.14% correspondingly. Also, with 30% of testing dataset, the CIMDC-DI method has been able average accuy, precn, recal, and Fscore of 96%, 96.08%, 95.92%, and 95.98% correspondingly.
Table 4.
Result analysis of CIMDC-DI technique on ISIC 2020 dataset.
| Class labels | Accuracy | Precision | Recall | F-score |
|---|---|---|---|---|
| Training (70%) | ||||
| Melanoma | 93.14 | 94.80 | 91.62 | 93.18 |
| Benign | 93.14 | 91.53 | 94.74 | 93.10 |
| Average | 93.14 | 93.16 | 93.18 | 93.14 |
|
| ||||
| Testing (30%) | ||||
| Melanoma | 96.00 | 97.10 | 94.37 | 95.71 |
| Benign | 96.00 | 95.06 | 97.47 | 96.25 |
| Average | 96.00 | 96.08 | 95.92 | 95.98 |
A brief precision-recall examination of the CIMDC-DI model on three datasets is portrayed in Figure 5. By observing the figure, it is noticed that the CIMDC-DI model has accomplished maximum precision-recall performance under three datasets.
Figure 5.
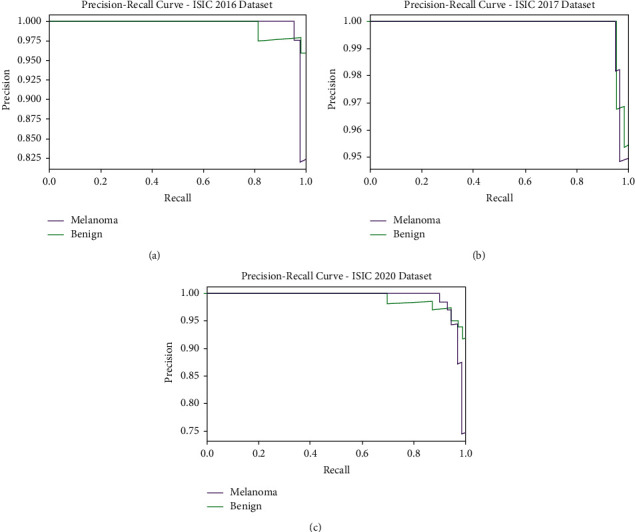
Precision-recall analysis of CIMDC-DI technique under three datasets.
Figure 6 portrays a clear ROC investigation of the CIMDC-DI model on three datasets. The figure portrays that the CIMDC-DI model has resulted in proficient results with maximum ROC values under distinct class labels.
Figure 6.
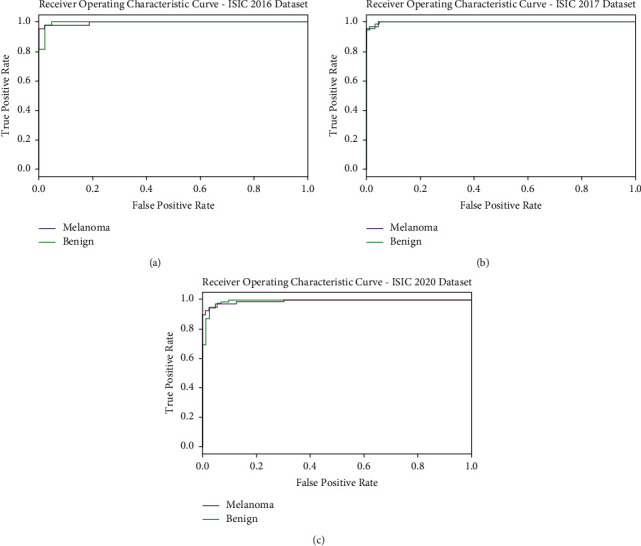
ROC analysis of CIMDC-DI technique under three datasets.
Table 5 and Figure 7 report the comparative classification outcomes of the CIMDC-DI model with recent models on training phase [26]. The experimental outcomes indicated that the CIMDC-DI technique has outperformed other models in terms of different metrics. On examining the outcome with respect to accuy, the CIMDC-DI algorithm has gained higher accuy of 96.79% whereas the VGG16, Inception v3, Xception, Inception ResNetV2, and DenseNet121 models have reached lower accuy of 90.90%, 88.03%, 91.01%, 93.14%, and 93.30% correspondingly. Besides, on investigating (Table 5) the outcome in terms of precn, the CIMDC-DI technique has gained higher precn of 96.85% whereas the VGG16, Inception v3, Xception, Inception ResNetV2, and DenseNet121 models have reached lower precn of 88.94%, 90.28%, 90.88%, 89.27%, and 92.94%, respectively. Followed by, on investigating the outcome with respect to recal, the CIMDC-DI model has gained higher recal of 96.84% whereas the VGG16, Inception v3, Xception, Inception ResNetV2, and DenseNet121 models have reached lower recal of 89.79%, 89.29%, 92.70%, 89.96%, and 91.46% correspondingly.
Table 5.
Comparative analysis of CIMDC-DI technique with recent algorithms on training phase.
| Training phase | |||
|---|---|---|---|
| Methods | Accuracy | Precision | Recall |
| VGG16 model | 90.90 | 88.94 | 89.79 |
| InceptionV3 model | 88.03 | 90.28 | 89.29 |
| Xception model | 91.01 | 90.88 | 92.70 |
| Inception ResnetV2 model | 93.14 | 89.27 | 89.96 |
| DenseNet121 model | 93.30 | 92.94 | 91.46 |
| CIMDC-DI | 96.79 | 96.85 | 96.84 |
Figure 7.

Comparative analysis of CIMDC-DI technique on training phase.
Table 6 and Figure 8 demonstrate the comparative classification outcomes of the CIMDC-DI technique with recent techniques in testing phase. The experimental outcomes indicated that the CIMDC-DI system has outperformed other models in terms of different metrics. On investigating the outcome with respect to accuy, the CIMDC-DI methodology has gained higher accuy of 97.50% whereas the VGG16, Inception v3, Xception, Inception ResNetV2 (Table 6), and DenseNet121 methodologies have gained minimal accuy of 92.75%, 89.61%, 90.49%, 92.80%, and 91.40% correspondingly.
Table 6.
Comparative analysis of CIMDC-DI technique with recent algorithms on testing phase.
| Testing phase | |||
|---|---|---|---|
| Methods | Accuracy | Precision | Recall |
| VGG16 model | 92.75 | 91.25 | 91.58 |
| InceptionV3 model | 89.61 | 88.50 | 89.24 |
| Xception model | 90.49 | 91.16 | 91.36 |
| Inception ResnetV2 model | 92.80 | 90.53 | 92.99 |
| DenseNet121 model | 91.40 | 91.00 | 93.21 |
| CIMDC-DI | 97.50 | 97.54 | 97.45 |
Figure 8.
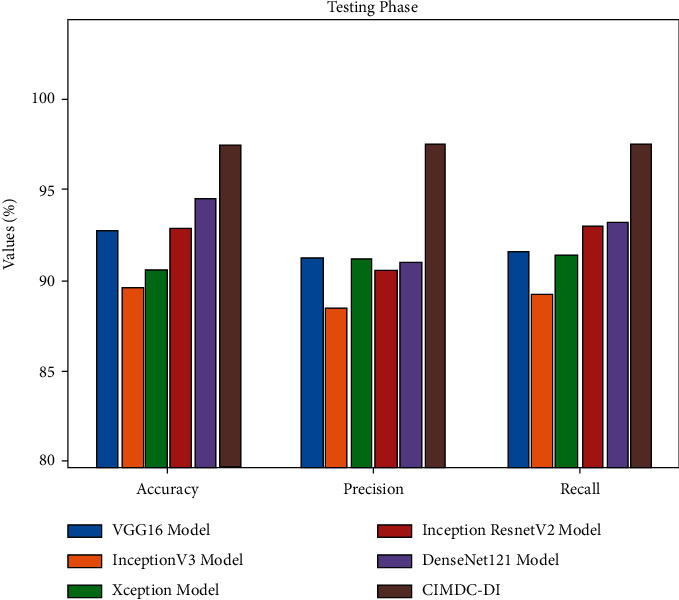
Comparative analysis of the CIMDC-DI technique on testing phase.
In addition, on examining the outcome in terms of precn, the CIMDC-DI approach has gained higher precn of 97.54% whereas the VGG16, Inception v3, Xception, Inception ResNetV2, and DenseNet121 models have achieved decreased precn of 91.25%, 88.50%, 91.16%, 90.53%, and 91% correspondingly. At the same time, on inspecting the outcome with respect to recal, the CIMDC-DI model has gained higher recal of 97.45% whereas the VGG16, Inception v3, Xception, Inception ResNetV2, and DenseNet121 models have reached lower recal of 91.58%, 89.24%, 91.36%, 92.99%, and 93.21% correspondingly.
After observing the detailed results and discussion, it is ensured that the CIMDC-DI model has accomplished maximum outcome on melanoma identification and classification using dermoscopic images.
5. Conclusion
In this study, a novel CIMDC-DI algorithm was developed for melanoma identification and classification using dermoscopic images. The presented CIMDC-DI model involves BF-enabled noise reduction, FKM segmentation, NasNet feature extraction, CNN classifier, and MRFO parameter optimization. The utilization of the MRFO algorithm assists in the effectual choice of parameter values involved in the CNN model. To ensure the better outcomes of the CIMDC-DI technique, a wide-ranging simulation analysis was implemented and the results are assessed under distinct aspects. A wide-ranging simulation analysis was executed, and the results reported the betterment over the recent methods with the maximum accuracy of 97.50%. Thus, the CIMDC-DI model can be exploited as a proficient tool for real-time melanoma classification. In the future, the CIMDC-DI model can be extended to the incorporation of DL-assisted segmentation approaches. Besides, a fusion-based ensemble classifier model can be developed for melanoma classification.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (IF-PSAU-2021/01/18550).
Data Availability
Data sharing is not applicable to this article as no datasets were generated during the current study.
Ethical Approval
This article does not contain any studies with human participants performed by any of the authors.
Consent
Not applicable.
Conflicts of Interest
The authors declare that they have no conflicts of interest to report regarding the present study.
References
- 1.Stiff K. M., Franklin M. J., Zhou Y., Madabhushi A. T. J. Artificial Intelligence and Melanoma: A Comprehensive Review of Clinical, Dermoscopic, and Histologic Applications. Pigment Cell & Melanoma Research . 2022;35(2):203–211. doi: 10.1111/pcmr.13027. [DOI] [PubMed] [Google Scholar]
- 2.Elansary I., Ismail A., Awad W. Medical Informatics and Bioimaging Using Artificial Intelligence . Cham, Switzerland: Springer; 2022. Efficient classification model for melanoma based on convolutional neural networks; pp. 15–27. [DOI] [Google Scholar]
- 3.Hosseinzadeh Kassani S., Hosseinzadeh Kassani P. A comparative study of deep learning architectures on melanoma detection. Tissue and Cell . 2019;58:76–83. doi: 10.1016/j.tice.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Kaur R., GholamHosseini H., Sinha R., Lindén M. Melanoma classification using a novel deep convolutional neural network with dermoscopic images. Sensors . 2022;22(3):p. 1134. doi: 10.3390/s22031134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaiyapuri T., Dutta A. K., Punithavathi H., et al. Intelligent deep-learning-enabled decision-making medical system for pancreatic tumor classification on CT images. Healthcare . 2022;10(4):p. 677. doi: 10.3390/healthcare10040677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dildar M., Akram S., Irfan M., et al. Skin cancer detection: a review using deep learning techniques. International Journal of Environmental Research and Public Health . 2021;18(10):p. 5479. doi: 10.3390/ijerph18105479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai X., Zhou J., Wessely A., et al. A disease network‐based deep learning approach for characterizing melanoma. International Journal of Cancer . 2022;150(6):1029–1044. doi: 10.1002/ijc.33860. [DOI] [PubMed] [Google Scholar]
- 8.Brinker T. J., Hekler A., Enk A. H., et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. European Journal of Cancer . 2019;113:47–54. doi: 10.1016/j.ejca.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Adegun A., Viriri S. Deep learning techniques for skin lesion analysis and melanoma cancer detection: a survey of state-of-the-art. Artificial Intelligence Review . 2021;54(2):811–841. doi: 10.1007/s10462-020-09865-y. [DOI] [Google Scholar]
- 10.Sankarapandian S., Kohn S., Spurrier V., et al. A pathology deep learning system capable of triage of melanoma specimens utilizing dermatopathologist consensus as ground truth. Proceedings of the IEEE/CVF International Conference on Computer Vision; 2021; Montreal, Canada. pp. 629–638. [DOI] [Google Scholar]
- 11.Raza R., Zulfiqar F., Tariq S., Anwar G. B., Sargano A. B., Habib Z. Melanoma classification from dermoscopy images using ensemble of convolutional neural networks. Mathematics . 2022;10(1):p. 26. [Google Scholar]
- 12.Vaiyapuri T., Dutta A. K., Sikkandar M. Y., et al. Design of metaheuristic optimization-based vascular segmentation techniques for photoacoustic images. Contrast Media and Molecular Imaging . 2022;2022:12. doi: 10.1155/2022/4736113.4736113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaiyapuri T., Alaskar H. Whale optimization for wavelet-based unsupervised medical image segmentation: application to CT and MR images. International Journal of Computational Intelligence Systems . 2020;13(1):941–953. doi: 10.2991/ijcis.d.200625.001. [DOI] [Google Scholar]
- 14.Lafraxo S., Ansari M. E., Charfi S. MelaNet: An Effective Deep Learning Framework for Melanoma Detection Using Dermoscopic Images. Multimedia Tools and Applications . 2022;81(11):1–25. [Google Scholar]
- 15.Shorfuzzaman M. An Explainable Stacked Ensemble of Deep Learning Models for Improved Melanoma Skin Cancer Detection. Multimedia Systems . 2021:1–15. doi: 10.1145/3469841. [DOI] [Google Scholar]
- 16.Kim D., Hong B. W. Unsupervised feature elimination via generative adversarial networks: application to hair removal in melanoma classification. IEEE Access . 2021;9 doi: 10.1109/access.2021.3065701.42610 [DOI] [Google Scholar]
- 17.Araújo R. L., Ricardo de Andrade L. R., Rodrigues J. J., e Silva R. R. Automatic segmentation of melanoma skin cancer using deep learning. Proceedings of the 2020 IEEE International Conference on E-Health Networking, Application & Services (HEALTHCOM); March 2021; Shenzhen, China. IEEE; pp. 1–6. [DOI] [Google Scholar]
- 18.Hagerty J. R., Stanley R. J., Almubarak H. A., et al. Deep learning and handcrafted method fusion: higher diagnostic accuracy for melanoma dermoscopy images. IEEE journal of biomedical and health informatics . 2019;23(4):1385–1391. doi: 10.1109/JBHI.2019.2891049. [DOI] [PubMed] [Google Scholar]
- 19.Adegun A. A., Viriri S. FCN-based DenseNet framework for automated detection and classification of skin lesions in dermoscopy images. IEEE Access . 2020;8 doi: 10.1109/access.2020.3016651.150377 [DOI] [Google Scholar]
- 20.Vaiyapuri T., Alaskar H., Sbai Z., Devi S. GA-based multi-objective optimization technique for medical image denoising in wavelet domain. Intelligent & Fuzzy Systems . 2021:1–14. [Google Scholar]
- 21.Nawaz M., Mehmood Z., Nazir T., et al. Skin cancer detection from dermoscopic images using deep learning and fuzzy k‐means clustering. Microscopy Research and Technique . 2022;85(1):339–351. doi: 10.1002/jemt.23908. [DOI] [PubMed] [Google Scholar]
- 22.Adedoja A., Owolawi P. A., Mapayi T. Deep learning based on nasnet for plant disease recognition using leave images. Proceedings of the 2019 International Conference on Advances in Big Data, Computing and Data Communication Systems (icABCD); August 2019; Winterton, South Africa. IEEE; pp. 1–5. [DOI] [Google Scholar]
- 23.Li C., Guo J., Porikli F., Fu H., Pang Y. A cascaded convolutional neural network for single image dehazing. IEEE Access . 2018;6 doi: 10.1109/access.2018.2818882.24877 [DOI] [Google Scholar]
- 24.Feng J., Luo X., Gao M., Abbas A., Xu Y. P., Pouramini S. Minimization of energy consumption by building shape optimization using an improved Manta-Ray Foraging Optimization algorithm. Energy Reports . 2021;7:1068–1078. doi: 10.1016/j.egyr.2021.02.028. [DOI] [Google Scholar]
- 25.ISIC. The 2020 Live Challenge is open. 2020. https://challenge.isic-archive.com/data/
- 26.Naeem A., Farooq M. S., Khelifi A., Abid A. Malignant melanoma classification using deep learning: datasets, performance measurements, challenges and opportunities. IEEE Access . 2020;8 doi: 10.1109/access.2020.3001507.110575 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated during the current study.


