Abstract
By the year 2050, the world's elderly population may increase exponentially, raising the rate of disease characteristic of this group, such as prostate cancer (PCa) and benign prostatic hyperplasia (BPH). Prostate disorders have a multifactorial etiology, especially age and genetic factors. Currently, PCa is the second most frequent neoplasm in the male population worldwide. The fibromodulin gene encodes a small leucine-rich proteoglycan (SLRP) which acts in the collagen fibrillogenesis pathway, cell adhesion, and modulation of TGF-β signaling pathways, which has been recently associated with PCa. The present study sequenced the coding region of the FMOD in a sample of 44 PCa, 90 BPH, and 82 controls from a Brazilian population, and the results identified 6 variants: 2 missenses (p.(Tyr42Ser) and p.(Pro24Ala)); 3 synonymous (p.(His253=), p.(Asn353=), and p.(Glu79=)); and 1 intronic (c.980-114A>G). Of these, p.(Tyr42Ser), p.(Pro24Ala), and p.(Asn353=) are rare variants, and p.(Tyr42Ser) was predicted as potential pathogenic by the algorithms used here, in addition to not being observed in controls, suggesting that may be a potential biomarker for development of PCa and BPH. In conclusion, we identified for the first time, in Brazilian individuals with PCa and BPH, a potentially pathogenic variant in the analysis of FMOD gene. Further studies are needed to investigate the deleterious effect of this variant on the structure and/or function of the FMOD protein.
1. Introduction
Prostate cancer (PCa) is the most common noncutaneous cancer in men worldwide, with an estimated 1,600,000 cases and 366,000 deaths per year [1]. PCa is the second most frequent tumor in American men, estimating 248,530 new cases and 34,130 deaths in 2021 [2]. According to Brazilian National Cancer Institute (INCA), PCa is the second most common cancer among Brazilian men and, for 2021, more than 65,840 new cases were expected [3].
Screening for detection and monitoring of PCa is one of the most controversial question in the field of urology. This is because the primary tests commonly used in the early detection of neoplasia are the digital rectal examination (DRE) and the PSA test (prostate specific antigen). Additionally, other tests are necessary to confirm the diagnosis, as the transrectal ultrasound (TRUS) and biopsy [4, 5].
Although the association between the density of prostate-specific antigen (PSA), benign prostatic hyperplasia (BPH), and PCa is consolidated in the literature [6, 7], several studies involving potentially more sensitive and specific biomarkers have been developed with the aim of improving the detection and monitoring of these pathologies and eliminating the limitations inherent to PSA [8, 9].
The PSA is not specific for prostate cancer; many benign conditions can elevate PSA levels, such as simple urinary tract inflammation or even BPH, leading to false-positive results [10, 11]. Some studies have also revealed that some individuals with PCa may have PSA levels below the reference threshold (4.0 ng/mL), thus leading to false-negative results, demonstrating that there are still no reliable values in PCa investigation [12]. While there is no evidence that the inflammatory process or BPH leads to PCa, it is possible for some individuals to develop such conditions and also prostate cancer [13].
Cancer is characterized by abnormal cell proliferation due to loss of control of cell division and the apoptosis process, leading to tumor formation [14, 15]. Any cell type needs to inhabit the extracellular matrix (ECM) and interact with its components that can play important roles in tumorigenesis [16]. Several small leucine-rich proteoglycans (SLRP) that are part of ECM, as class I biglycan, decorin, fibromodulin (FMOD), and class II lumican, favor the collagen fibrillogenesis regulation [17]. Among these, the proteoglycan FMOD has emerged with interesting particularities that involve several crucial biological processes, as angiogenesis, migration, and apoptosis [16].
Some studies have been indicated that fibromodulin may modulate certain signaling pathways, such as vascular endothelial growth factor (VEGF), fibroblast growth factor-2 (FGF-2), and especially, transforming growth beta (TGF-β) [18]. Moreover, fibromodulin also acts in the initiation and progression of pathologies like B-cell chronic lymphocytic leukemia [19].
FMOD function goes far beyond the regulation of collagen fibrillogenesis and cell adhesion. Fibromodulin is able to inhibit VEGF expression, suggesting that proteoglycan regulates other signaling pathways besides TGF-β [16, 20]. FMOD is also considered an efficient and fundamental angiogenic factor in lung cancer, in wound healing process and optical as well as skin diseases [18, 21, 22]. Therefore, FMOD functions are mainly related to angiogenesis process, suggesting as a potential therapeutic target to cancer and also other conditions associated with abnormal angiogenesis [21, 23].
Due to its product be detected in body fluids as urine, blood, or prostatic secretions, the fibromodulin coding gene has great potential to be used as a new biomarker to diagnosis of patients with benign or malignant prostate tumors [19, 24]. Therefore, to understand FMOD influence in several physiological processes to development of PCa is very necessary. Up to our knowledge, screening of FMOD gene is the first study in a sample with PCa and BPH in Brazil. Thus, we hope that the results of this study will contribute to the improvement of the diagnosis of these prostatic disorders.
2. Material and Methods
2.1. Study Population
This cross-sectional observational study included 216 unrelated male patients, aged plus 55 years from Rio de Janeiro State, Brazil. All were examined by urologists and submitted to DRE, PSA, and biopsy tests. Individuals considered healthy for this study (mean age of 60.82 ± 9.41) had a PSA level < 4 ng/mL, an unaltered rectal examination, and no other history of neoplasia. The diagnosis of PCa was made based on the altered DRE, the PSA quantification (>4 ng/mL), and the positive biopsy for malignancy. The diagnosis of BPH was made based on the altered clinical characteristics, the PSA quantification (>4 ng/mL), and the altered prostate volume.
Our cohort was divided into 44 with PCa, 90 with BPH, and 82 healthy controls. The Research Ethics Committee of the National Cancer Institute José Alencar Gomes da Silva (CAE: 88510618.8.30025274) and the University of Grande Rio (CAE: 8810618.8.0005253) approved the research protocol and the free and informed consent term, obtained from all participants.
2.2. Molecular Analyses
Genomic DNA was isolated from peripheral blood leukocytes using the QIAamp DNA Blood (Qiagen, Hilden, Germany). To assess the presence of variants in the FMOD gene, we amplified two exons and intron-exon boundaries by the polymerase chain reaction (PCR) technique using 6 pairs of oligonucleotides (IDT-Integrated DNA Technologies) (Supplementary Material Table S1), followed by Sanger sequencing. The conditions used in the PCR are shown in Supplementary Material Table S2.
The PCR products were purificated using the enzyme ExoSAP-IT (Applied Biosystems, Vilnius, Lithuania). Then, these products were sequenced using the Big Dye Terminator v3.1 kit (Applied Biosystems, Austin, TX, USA) and processed on an ABI 3130 Genetic Analyzer (Applied Biosystems Inc. US). The sequences were aligned with the reference sequence (ENST00000354955.4) and were analyzed by BioEdit Sequence Alignment Editor software v7.2.6.1 (Isis Pharmaceuticals).
2.3. Bioinformatic Tools
The identified FMOD variants were investigated in order to ascertain their previous occurrence in public databases, such as the Online Archive of Brazilian Mutations (http://abraom.ib.usp.br/index.php), project database 1000 Genomes (http://www.internationalgenome.org), Clinvar (http://www.ncbi.nlm.nih.gov/clinvar/), dbSNP (https://www.ncbi.nlm.nih.gov/), ExAC Browser (http://exac.broadinstitute.org), Human Genome Mutation Database (HGMD) (https://pubmed.ncbi.nlm.nih.gov/), and PubMed (https://pubmed.ncbi.nlm.nih.gov/).
To assess the potential functional impact of the identified missense variants, we used in silico prediction tools including the SIFT4G [25], PolyPhen-2 [26], PROVEAN [27], WS-SNPs & GO [28], MutPred [29], SNAP [30], Fathmm [31], M-PCA [32], mutation assessor [33], PANTHER-PSEP [34], mutation taster [35], and Revel [36].
2.4. Statistics Analysis
We used IBM® SPSS software (V.22.0) for descriptive and inferential analyses. In the case of normally distributed variables, comparisons were made between groups using the Student's t and ANOVA tests (for variables: age, body mass index, and prostate weight); subsequently, Student-Newman-Keuls (SNK) post hoc analyses were performed to complement ANOVA. In relation to PSAT scores, we had observe its nonnormal distribution, being influenced by extremely high values observed in three PCa patients, so the comparisons among groups were made using Kruskal-Wallis nonparametric test. All these inferences were made at the 5% level of significance.
3. Results
3.1. Basic Clinical Characteristic
This study comprised 216 individuals, stratified into 44 PCa patients, 90 with BPH, and 82 healthy controls, whose clinical characteristics are showing in Table 1. As expected, we observed that PSAt, PSAf, and prostate weight were statistically different among groups.
Table 1.
Comparative analysis of quantitative variables between sample groups.
| Variables | Control | BPH | PCa | p | p1 | p2 | p3 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Values | N | Values | N | Values | |||||
| Age (years) | 82 | 60.82 ± 9.41 | 90 | 69.80 ± 8.12 | 44 | 71.27 ± 7.64 | <0.01 | <0.05 | <0.05 | >0.05 |
| BMI (kg) | 75 | 26.12 ± 3.35 | 84 | 26.26 ± 3.93 | 24 | 27.35 ± 3.83 | 0.509 | >0.05 | >0.05 | >0.05 |
| PSAT (ng/mL) | 82 | 0.00 ± 0.87 | 90 | 2.54 ± 3.76 | 44 | 11.52 ± 16.95 | <0.01 | — | — | — |
| PSAF (ng/mL) | 82 | 0.00 ± 0.23 | 90 | 0.43 ± 0.69 | 44 | 0.00 ± 1.29 | <0.01 | — | — | — |
| Prostate weight (g) | 21 | 30.15 ± 9.87 | 52 | 49.28 ± 21.54 | 20 | 46.30 ± 26.30 | 0.947 | <0.05 | <0.05 | >0.05 |
Note: values indicate median ± interquartile range; p is the probabilistic value used to measure the significance of differences between the means of variables. p1 is the probabilistic value used to measure the significance of differences between the control and BPH groups. p2 is the probabilistic value used to measure the significance of differences between the control and PCa groups. p3 is the probabilistic value used to measure the significance of differences between the BPH and PCa groups. Some variables were not passed on in part of the studied group. PSAT: total prostate specific antigen; PSAF: free prostate specific antigen; SBP: systolic blood pressure; DBP: diastolic blood pressure; BMI: body mass index; source: elaborated by the author.
Table 1 presents the results of the comparisons made between the three sample groups.
3.2. FMOD Molecular Screening
In this study, we sequenced the coding region of the FMOD gene in 134 probands and 82 controls from a sample from Rio de Janeiro. Our results showed 6 variants, in which 2 were missenses [(p. (Tyr42Ser) and p. (Pro24Ala)], 3 synonymous (p. His253=, p. Asn353=, and p. Glu79=), and 1 intronic (c.980-114A>G) (Table 2). The electropherograms are presented in Figure 1.
Table 2.
Variants found in the molecular analysis of the FMOD gene.
| Gene location | SNP ID | c.DNA | Protein | Type of change | MAF | No. of individuals with alteration | ||
|---|---|---|---|---|---|---|---|---|
| Control | BPH | PCa | ||||||
| Exon 2 | rs115908597 | c.125A>C | p.(Tyr42Ser) | Missense | <0.01 | 0 | 2 | 1 |
| Exon 2 | rs139299015 | c.70C>G | p.(Pro24Ala) | Missense | <0.01 | 1 | 0 | 0 |
| Intron 2-3 | rs1891180 | c.980-114A>G | — | Intronic | 0.60 | 82 | 74 | 44 |
| Exon 2 | rs77856193 | c.759C>T | p.(His253=) | Synonym | <0.01 | 1 | 0 | 2 |
| Exon 2 | rs7543148 | c.237G>A | p.(Glu79=) | Synonym | 0.40 | 66 | 70 | 44 |
| Exon 3 | rs145901742 | c.1059C>T | p.(Asn353=) | Synonym | 0.04 | 0 | 0 | 1 |
Representation of the variants found through the automatic sequencing of the FMOD gene, with its nitrogenous base and/or amino-acid base exchange characteristics, in addition to the number of individuals found with the alteration in each of the sample groups. Source: prepared by the author.
Figure 1.
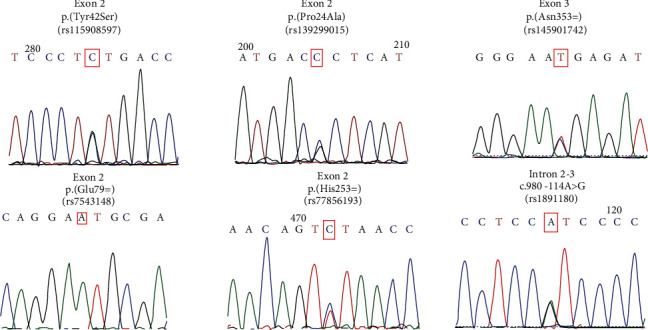
Electropherograms referring to alterations found in the FMOD gene tracking.
The missense variant p.(Tyr42Ser) was present in 2 hyperplastic patients and 1 neoplastic patient. This variant was absent in the control group. The p.(Pro24Ala) variant was observed in heterozygosity in a control patient. Regarding the synonymous variants, p.Asn353= was found in heterozygosity in 1 neoplastic proband, and p.His253= was found in two neoplastic patients and one control in heterozygosity. The p.Glu79= was common in our sample, which was found in 44 neoplastic probands, 70 hyperplastic, and 66 controls. This variant was observed in heterozygosity and homozygosity state. Additionally, the intronic variation c.980-114A>G was observed in 44 neoplastic, 74 hyperplastic, and 82 healthy individuals. Therefore, both p.Glu79= and c.980-114A>G variants were characterized as polymorphism.
The p.(Tyr42Ser) is presented in exon 2 and is characterized by the exchange of the amino acid tyrosine for serine at position 42 of the protein. In our sample, we identified this change in heterozygosity in 3 individuals, of which two were BPH group aged 72 and 74 years and in one individual was the PCa group aged 75 years. The tyrosine residue at position 42 of fibromodulin is evolutionarily conserved across species (Figure 2). The SIFT4G Prediction, PolyPhen-2_HVAR, MutPred2, SNAP, PANTHER-PSEP, and mutation taster tools (Table 3) predicted this variant to be probably harmful. Tyrosine and serine are polar amino acids that have no charge, but tyrosine contains a phenol group attached to its side chain while serine has a hydroxylated methyl group. On the other hand, the p.(Pro24Ala) located in exon 2 of the FMOD gene results in the exchange of the amino acid proline to alanine at position 24 of the protein. This variant, found in a 57-year-old control individual, was predicted as benign by almost all silico tools used (Table 3).
Figure 2.
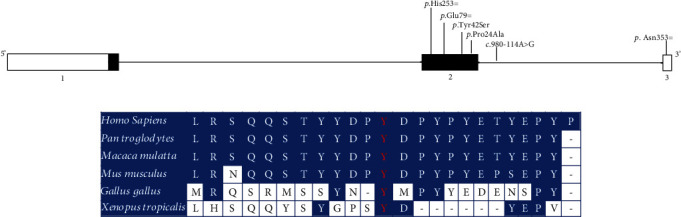
Schematic representation of the FMOD gene and protein domains. (a) Alterations found are pointed out in the gene scheme. (b) Mutation taster alignment of the FMOD between species.
Table 3.
In silico prediction of missense changes identified in the FMOD gene.
| Prediction tool | p.(Tyr42Ser) Y42S | p.(Pro24Ala) P24A | Results | Website | Ref. | ||
|---|---|---|---|---|---|---|---|
| Score | Prediction | Score | Prediction | ||||
| SIFT4G predictions | 0.004 | Deleterious | 0.339 | Tolerated | Score range: 0 to 1 (≤0.05 damaging/>0.05 tolerated). | https://sift.bii.a-star.edu.sg/www/SIFTdbSNP.html | [25] |
| PolyPhen-2_HVAR | 0.831 | Probably damaging | <0.01 | Benign | Score range: 0 (benign) to 1 (damaging). Probably damaging, possibly damaging or benign. | http://genetics.bwh.harvard.edu/pph2/index.shtml | [26] |
| PROVEAN | -0.86 | Neutral | -0.56 | Neutral | Default score threshold: -2.5 (≤-2.5 deleterious/>-2.5 neutral). | http://provean.jcvi.org/ | [27] |
| WS-SNPs & GO | 0.280 | Neutral | 0.106 | Neutral | Score range: 0 to 1 (probability score: >0.5 disease-associated). | http://snps.biofold.org/snps-and-go/ | [28] |
| MutPred2 | 0.625 | Possibly pathogenic | 0.209 | Neutral | Score range: 0 to 1 (general pathogenicity score: ≥0.50). | http://mutpred.mutdb.org/#qform | [29] |
| SNAP | 2 | Effect | -54 | Neutral | Score: -100 to 100 (≥1 effect). | http://www.rostlab.org/services/SNAP | [30] |
| FATHMM | 0.52 | Tolerated | 0.67 | Tolerated | Pathogenicity threshold: <0 (dano). >0 (Tolerado). | http://fathmm.biocompute.org.uk/inherited.html | [31] |
| M-CAP | ∗ | ∗ | 0.003 | Benign | Pathogenicity threshold: >0.025. | http://bejerano.stanford.edu/mcap/ | [32] |
| Mutation assessor | 1.04 | Low impact | 0.345 | Neutral | Score cutoff: 0.8 neutral and low impact/1.9 low impact and medium impact/3.5 medium impact and high impact. | http://mutationassessor.org/r3/ | [33] |
| PANTHER-PSEP | 455 | Possibly pathogenic | 91 | Possibly benign | Length of time: >450 my probably damaging/450 my > time > 200 my possibly damaging/<200 my probably benign. | http://www.pantherdb.org/tools/csnpScore.do | [34] |
| Mutation taster | 0.999999999606647-A | Disease causing | 0.999999999606647-P | Polymorphism | Prediction: A. Disease causing: probably deleterious/D. disease causing automatic: deleterious/N. polymorphism: probably harmless/P. polymorphism automatic: harmless. | http://www.mutationtaster.org/ | [35] |
| Revel | 0.168 | Benign | 0.081 | Benign | Score range: 0 to 1 (>0.50 likely disease causing/<0.50 likely benign). | https://sites.google.com/site/revelgenomics/downloads | [36] |
Notes: ∗MCAP-MCAP scoring is not available for some alleles; location 1 : 203317274. Abbreviation: my: millions of years.
4. Discussion
In recent years, changes in mortality and age pyramid structure of the global population in recent years has resulted in a substantial increase in complex diseases [37]. This scenario brings a progression of numerous diseases, including cancer, which is one of the main causes of death in several countries [38, 39]. With respect to PCa, the incidence rates by age increase sharply from the age of 45 years [40]. Despite the numerous efforts of the world medical community to raise awareness of prostate cancer, there is still great resistance from the male population about the care of their health.
Mutations or polymorphisms in many genes are described in the literature as influencing the development of PCa and possibly BPH [41–48]. The FMOD gene, which encodes fibromodulin, plays an important role in the regulation of collagen fibrillogenesis, angiogenesis, reprogramming of human fibroblasts into pluripotent cells, and modulation of TGF-β activity and is associated with metastatic phenotypes. Interestingly, until recently, this gene had not been associated with prostatic pathologies [19, 24].
Bettin and Reyes (2016) showed the strong interaction between prostate cancer and FMOD, revealing that the gene is highly expressed in human PCa cells, whereas in hyperplastic cells, there is no increase expression. In our research, FMOD gene was sequenced in 216 individuals. A total of six alterations were observed p.(Tyr42Ser), p.(Pro24Ala), p.His253=, p.Asn353=, and c.980-114A>G, of which p.(Tyr42Ser) was rare and predicted to be pathogenic by our in silico analyses. It is important to emphasize that none of these alterations were associated with PCa or BPH and neither with any other pathology, prior to our study.
The missense p.(Tyr42Ser) alteration was identified in individuals with prostatic alterations, presenting a frequency in our population of 1.6%. In contrast, the p.(Pro24Ala) was found in 1 individual of the control group. One of the synonymous variants found was p.His253=, located in exon 2 of the FMOD gene and characterized by the exchange of a cytosine for a thymine (c.759C>T). This change was observed in the study sample in three individuals, with two of them allocated in the PCa group (57 and 74 years) and one in the control group (65 years) representing 1,4% of its frequency in our study.
Another synonymous variant p.Asn353= (c.1059C>T) found in exon 3 was observed in heterozygosis in an individual with PCa aged 64 years, being absent in the healthy individuals analyzed. This synonymous alteration was identified in individuals with prostatic alterations, presenting a frequency in our population of 0.49%. However, even though the p.His253= and p.Asn353= variants do not present amino acid alterations according to the Human Splicing Finder tool, the base exchange may be located in the splicing site, so we cannot rule out its potential modifying effect on the protein. In this context, further functional studies are necessary to elucidate it.
The other two alterations, p.Glu79= and c.980-114A>G, showed a polymorphic behavior, being very frequent in our sample. The frequency of the p.Glu79= polymorphism in our study was 70.67%, whereas the frequency of the c.980-114A>G was 61.54%. However, after performing genotypic and allelic association analyzes, no significant associations were found for any of the groups analyzed (p > 0.05) in relation to these variants. It is to our knowledge that the p.Glu79= variant was previously studied in relation to other pathologies, as myopia and the risk of tearing the anterior cruciate ligament; however, it has not been associated with such diseases [49, 50].
In conclusion, we identified a potentially pathogenic variant in FMOD gene in a cohort with Brazilian individuals with PCa and BPH. As far as we know, this is the first study to screen part of the FMOD gene, analyzing its association with prostate cancer and benign prostatic hyperplasia. We believe that the functional studies are needed to better understand the role of the alterations found here, especially the missense ones, in the mentioned pathologies and their contribution to the clinical profile of patients.
Acknowledgments
The authors would like to thank the patients and their families, the collaborating centers Duque de Caxias Polyclinic-Health/Unigranrio and the Brazilian National Tumor Bank (BNT/INCA) and the Oswaldo Cruz Institute's DNA Sequencing Platform (PDTIS/FIOCRUZ) for DNA sequencing. This study was partially supported by the Higher Education Personnel Improvement Coordination–CAPES (finance code 001, 2022), Foundation for Research Support of the State of Rio de Janeiro in Brazil–FAPERJ (no. E-26/010.001634/2019), Oswaldo Cruz Institute (IOC/Fiocruz) (scholarships), and National Institute of Metrology, Quality and Technology (Inmetro) (student grant).
Data Availability
The present study deals with the tracking of the FMOD gene, in Brazilian individuals with PCa and BPH that showed new allelic variants, among which one potentially pathogenic. We believe that this information can significantly contribute to expanding our biological knowledge, prevention, early diagnosis, and treatment of prostate cancer and benign prostatic hyperplasia.
Conflicts of Interest
The authors declare no potential conflicts of interest regarding the publication of this paper.
Supplementary Materials
All information presented in this study are products of analysis of polymorphisms identified by PCR and Sanger sequencing, which are available in the article and in the supplementary material (S1, S2, and S3).
References
- 1.Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA: a Cancer Journal for Clinicians . 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.ACS-American Cancer Society. Cancer Statistics Center. http://cancerstatisticscenter.cancer.org .
- 3.Silva J. A. G. Instituto Nacional de Câncer José Alencar Gomes da Silva . 2011;118 [Google Scholar]
- 4.Ilic D., Neuberger M. M., Djulbegovic M. D. P., Dahm P., Cochrane Urology Group Screening for prostate cancer. Cochrane Database of Systematic Reviews . 2013;2013, article CD004720(1) doi: 10.1002/14651858.CD004720.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigau M., Olivan M., Garcia M., et al. The present and future of prostate cancer urine biomarkers. International Journal of Molecular Sciences . 2013;14(6):12620–12649. doi: 10.3390/ijms140612620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irani J., Levillain P., Goujon J. M., Bon D., Doré B., Aubert J. Inflammation in benign prostatic hyperplasia: correlation with prostate specific antigen value. The Journal of Urology . 1997;157(4):1301–1303. doi: 10.1016/S0022-5347(01)64957-7. [DOI] [PubMed] [Google Scholar]
- 7.Vasavada S. R., Dobbs R. W., Kajdacsy-Balla A. A., Abern M. R., Moreira D. M. Inflammation on prostate needle biopsy is associated with lower prostate cancer risk: a meta-analysis. The Journal of Urology . 2018;199(5):1174–1181. doi: 10.1016/j.juro.2017.11.120. [DOI] [PubMed] [Google Scholar]
- 8.Cerrato A., Bedia C., Capriotti A. L., et al. Untargeted metabolomics of prostate cancer zwitterionic and positively charged compounds in urine. Analytica Chimica Acta . 2021;1158, article 338381 doi: 10.1016/j.aca.2021.338381. [DOI] [PubMed] [Google Scholar]
- 9.Salciccia S., Capriotti A. L., Laganà A., et al. Biomarkers in prostate cancer diagnosis: from current knowledge to the role of metabolomics and exosomes. International Journal of Molecular Sciences . 2021;22(9):p. 4367. doi: 10.3390/ijms22094367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimakakos A., Armakolas A., Koutsilieris M. Novel tools for prostate cancer prognosis, diagnosis, and follow-up. BioMed Research International . 2014;2014:9. doi: 10.1155/2014/890697.890697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abrate A., Lughezzani G., Gadda G. M., et al. Clinical use of [-2]proPSA (p2PSA) and its derivatives (%p2PSA and prostate health index) for the detection of prostate cancer: a review of the literature. Korean Journal of Urology . 2014;55(7):436–445. doi: 10.4111/kju.2014.55.7.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts M. J., Schirra H. J., Lavin M. F., Gardiner R. A. Metabolomics: a novel approach to early and noninvasive prostate cancer detection. Korean Journal of Urology . 2011;52(2):79–89. doi: 10.4111/kju.2011.52.2.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruno S. M., Falagario U. G., d’Altilia N., et al. PSA density help to identify patients with elevated PSA due to prostate cancer rather than intraprostatic inflammation: a prospective single center study. Frontiers in Oncology . 2021;11:1–8. doi: 10.3389/fonc.2021.693684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.American Cancer Society – ACS (2016) Cancer facts & figures 2016. Atlanta: American Cancer Society . https://www.cancer.org/
- 15.Hollstein M., Alexandrov L. B., Wild C. P., Ardin M., Zavadil J. Base changes in tumour DNA have the power to reveal the causes and evolution of cancer. Oncogene . 2017;36(2):158–167. doi: 10.1038/onc.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pourhanifeh M. H., Mohammadi R., Noruzi S., et al. The role of fibromodulin in cancer pathogenesis: implications for diagnosis and therapy. Cancer Cell International . 2019;19(1):157–159. doi: 10.1186/s12935-019-0870-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang C. T., Halper J. Proteoglycans and diseases of soft tissues. Advances in Experimental Medicine and Biology . 2021;1348:127–138. doi: 10.1007/978-3-030-80614-9_5. [DOI] [PubMed] [Google Scholar]
- 18.Ao Z., Yu S., Qian P., et al. Tumor angiogenesis of SCLC inhibited by decreased expression of FMOD via downregulating angiogenic factors of endothelial cells. Biomedicine & Pharmacotherapy . 2017;87:539–547. doi: 10.1016/j.biopha.2016.12.110. [DOI] [PubMed] [Google Scholar]
- 19.Reyes N., Benedetti I., Bettin A., Rebollo J., Geliebter J. The small leucine rich proteoglycan fibromodulin is overexpressed in human prostate epithelial cancer cell lines in culture and human prostate cancer tissue. Cancer Biomarkers . 2016;16(1):191–202. doi: 10.3233/CBM-150555. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Z., Jian J., Velasco O., et al. Fibromodulin enhances angiogenesis during cutaneous wound healing. Plastic and Reconstructive Surgery Global Open . 2014;2(12):1–10. doi: 10.1097/GOX.0000000000000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adini I., Ghosh K., Adini A., et al. Melanocyte-secreted fibromodulin promotes an angiogenic microenvironment. The Journal of Clinical Investigation . 2014;124(1):425–436. doi: 10.1172/JCI69404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kontos C. D. More than skin deep: connecting melanocyte pigmentation and angiogenic diseases. The Journal of Clinical Investigation . 2014;124(1):76–79. doi: 10.1172/JCI73559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jian J., Zheng Z., Zhang K., et al. Fibromodulin promoted in vitro and in vivo angiogenesis. Biochemical and Biophysical Research Communications . 2013;436(3):530–535. doi: 10.1016/j.bbrc.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettin A., Reyes I., Reyes N. Gene expression profiling of prostate cancer-associated genes identifies fibromodulin as potential novel biomarker for prostate cancer. The International Journal of Biological Markers . 2016;31(2):e153–e162. doi: 10.5301/jbm.5000184. [DOI] [PubMed] [Google Scholar]
- 25.Vaser R., Adusumalli S., Leng S. N., Sikic M., Ng P. C. SIFT missense predictions for genomes. Nature Protocols . 2016;11(1):1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 26.Adzhubei I. A., Schmidt S., Peshkin L., et al. A method and server for predicting damaging missense mutations. Nature Methods . 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi Y., Chan A. P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics . 2015;31(16):2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capriotti E., Calabrese R., Fariselli P., Martelli P. L., Altman R. B., Casadio R. WS-SNPs&GO: a web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics . 2013;14(Supplement_3) doi: 10.1186/1471-2164-14-s3-s6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pejaver V., Urresti J., Lugo-Martinez J., et al. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nature Communications . 2020;11(1):p. 5918. doi: 10.1038/s41467-020-19669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bromberg Y., Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Research . 2007;35(11):3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shihab H. A., Gough J., Cooper D. N., et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Human Mutation . 2013;34(1):57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jagadeesh K. A., Wenger A. M., Berger M. J., et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nature Genetics . 2016;48(12):1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 33.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Research . 2011;39(17):e118–e143. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang H., Thomas P. D. PANTHER-PSEP: predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics . 2016;32(14):2230–2232. doi: 10.1093/bioinformatics/btw222. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz J. M., Cooper D. N., Schuelke M., Seelow D. Mutationtaster2: mutation prediction for the deep-sequencing age. Nature Methods . 2014;11(4):361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 36.Ioannidis N. M., Rothstein J. H., Pejaver V., et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. American Journal of Human Genetics . 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.World Health Organization. World health statistics 2016: monitoring health for the SDGs sustainable development goals. World Health Organization . 2016;3(2):356–369. [Google Scholar]
- 38.DESA. U. N. United Nations Department of Economic and Social Affairs. Population Division, World Population Prospects . 2019.
- 39.Pilleron S., Soto-Perez-de-Celis E., Vignat J., et al. Estimated global cancer incidence in the oldest adults in 2018 and projections to 2050. International Journal of Cancer . 2021;148(3):601–608. doi: 10.1002/ijc.33232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merriel S. W. D., Funston G., Hamilton W. Prostate cancer in primary care. Advances in Therapy . 2018;35(9):1285–1294. doi: 10.1007/s12325-018-0766-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amundadottir L. T., Sulem P., Gudmundsson J., et al. A common variant associated with prostate cancer in European and African populations. Nature Genetics . 2006;38(6):652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 42.Al Olama A. A., Kote-Jarai Z., Giles G. G., et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nature Genetics . 2009;41(10):1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 43.Eeles R. A., Kote-Jarai Z., Al Olama A. A., et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nature Genetics . 2009;41(10):1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gudmundsson J., Sulem P., Gudbjartsson D. F., et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nature Genetics . 2009;41(10):1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takata R., Akamatsu S., Kubo M., et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nature Genetics . 2010;42(9):751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 46.Haiman C. A., Chen G. K., Blot W. J., et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nature Genetics . 2011;43(6):570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kote-Jarai Z., Al O. A. A., Giles G. G., et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nature Genetics . 2011;43(8):785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eeles R. A., Al O. A. A., Benlloch S., et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nature Genetics . 2013;45(4):385–391. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mannion S., Mtintsilana A., Posthumus M., et al. Genes encoding proteoglycans are associated with the risk of anterior cruciate ligament ruptures. British Journal of Sports Medicine . 2014;48(22):1640–1646. doi: 10.1136/bjsports-2013-093201. [DOI] [PubMed] [Google Scholar]
- 50.Lin H. J., Wan L., Tsai Y., et al. Sclera-related gene polymorphisms in high myopia. Molecular Vision . 2009;15:p. 1655. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All information presented in this study are products of analysis of polymorphisms identified by PCR and Sanger sequencing, which are available in the article and in the supplementary material (S1, S2, and S3).
Data Availability Statement
The present study deals with the tracking of the FMOD gene, in Brazilian individuals with PCa and BPH that showed new allelic variants, among which one potentially pathogenic. We believe that this information can significantly contribute to expanding our biological knowledge, prevention, early diagnosis, and treatment of prostate cancer and benign prostatic hyperplasia.


