Abstract
Hypoxemia is a defining feature of acute respiratory distress syndrome (ARDS), an often-fatal complication of pulmonary or systemic inflammation, yet the resulting tissue hypoxia, and its impact on immune responses, is often neglected. In the present study, we have shown that ARDS patients were hypoxemic and monocytopenic within the first 48 h of ventilation. Monocytopenia was also observed in mouse models of hypoxic acute lung injury, in which hypoxemia drove the suppression of type I interferon signaling in the bone marrow. This impaired monopoiesis resulted in reduced accumulation of monocyte-derived macrophages and enhanced neutrophil-mediated inflammation in the lung. Administration of colony-stimulating factor 1 in mice with hypoxic lung injury rescued the monocytopenia, altered the phenotype of circulating monocytes, increased monocyte-derived macrophages in the lung and limited injury. Thus, tissue hypoxia altered the dynamics of the immune response to the detriment of the host and interventions to address the aberrant response offer new therapeutic strategies for ARDS.
Subject terms: Monocytes and macrophages, Acute inflammation, Mucosal immunology, Haematopoiesis
Mirchandani and colleagues show that tissue hypoxia alters the number and phenotype of circulating monocytes and their differentiation into lung macrophages, with important implications for the resolution of inflammation in the lung.
Main
ARDS, a clinical syndrome defined by bilateral opacites on chest imaging denoting the presence of lung inflammation, blood hypoxemia with tissue hypoxia and the requirement for positive-pressure ventilation, has a mortality rate of up to 40%1,2. Despite decades of research, effective therapies for ARDS remain elusive, with much of the treatment research focusing on modulating the inflammatory injury, because its persistence is a poor prognostic indicator3,4. However, the effects of blood hypoxemia and the ensuing tissue hypoxia on the persistence of inflammation in ARDS have not been fully investigated.
Macrophages have a key role in driving tissue inflammation resolution. In mice, lung macrophages can be subdivided into SiglecF+ alveolar macrophages (AMs) and SiglecF– interstitial macrophages (IMs), which inhabit distinct anatomical niches. Under homeostatic conditions, AMs largely self-renew5,6 whereas IMs require recruitment of circulating monocytes7,8. Evidence is emerging for key roles for IMs in inflammation regulation9,10 and repair11.
In the present study, we investigated whether hypoxemia associated with ARDS, including severe COVID-19 disease, affected the accumulation of lung monocyte-derived macrophages (MDMs) and whether this, in turn, impacted the resolution of inflammation. We found that patients with ARDS had clinical evidence of persistent hypoxemia despite ventilatory support and were profoundly monocytopenic during the first 48 h of ventilation, an observation replicated in mouse models of hypoxic acute lung injury (ALI). We further showed that systemic responses to tissue hypoxia suppressed type I interferon (IFN) signaling and fundamentally altered bone marrow hematopoiesis, with ensuing consequences for the phenotype and number of monocytes in the blood and accumulation of MDMs in the lung during ALI. This, in turn, led to the persistence of inflammation. Critically, targeting this pathway with the monocyte and macrophage growth factor colony-stimulating factor 1 (CSF-1) corrected these hypoxia-mediated changes and drove inflammation resolution.
Results
ARDS is characterized by monocytopenia and an altered immune phenotype
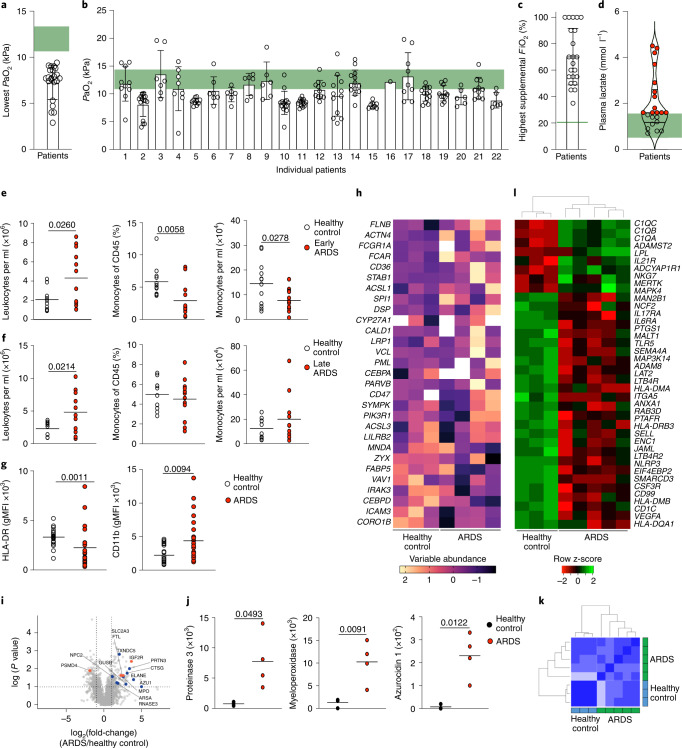
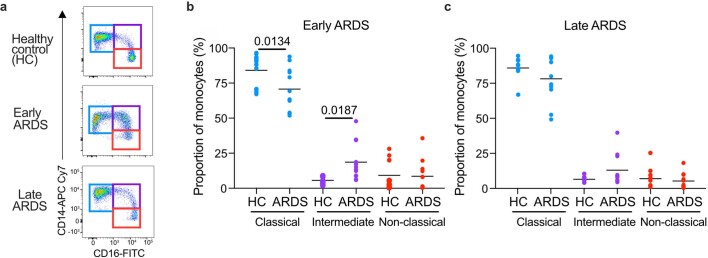
Despite the heterogeneity of the etiologies leading to ARDS, one defining feature is blood hypoxemia and tissue hypoxia. We first characterized the arterial oxygen partial pressure (PaO2) in patients with moderate-to-severe ARDS. Patients were sampled within 1 week of a known insult or new or worsening respiratory symptoms in accordance with the Berlin criteria, namely, if they had bilateral opacities on a chest radiograph and evidence of respiratory failure with a PaO2/FiO2 (fraction inspired O2) < 200 mmHg and positive end-expiratory pressure >5 cmH2O. Patients with ARDS had a range of etiologies and associated pathogens (Table 1) and showed clinically important (Fig. 1a) and sustained (Fig. 1b) blood hypoxemia over a 24-h period despite supplementary oxygen therapy (Fig. 1c) and ventilatory support. In 13 of the 22 patients, this was associated with an elevated circulating lactate (Fig. 1d), indicating ongoing tissue hypoxia. To characterize the kinetics of circulating leukocyte populations, we sampled blood from ventilated patients <48 h from diagnosis of ARDS and commencement of positive-pressure ventilation (hereafter early ARDS) or 48 h to 7 d from diagnosis (late ARDS). Healthy donors were used as controls because tissue hypoxia is a common feature of critically unwell patients. Early ARDS patients had elevated circulating leukocyte counts (Fig. 1e), but significantly lower proportions and numbers of circulating monocytes (Fig. 1e) compared with controls. In late ARDS patients, circulating leukocyte numbers remained elevated compared with controls (Fig. 1f), but monocyte frequency and counts were equivalent (Fig. 1f). In early ARDS, we detected an increase in the proportion of CD14+CD16+ intermediate monocytes at the expense of classical CD14++CD16− monocytes (Extended Data Fig. 1a–c)12. ARDS monocytes, irrespective of timepoint, had a lower expression of the major histocompatibility class II (MHC-II) marker HLA-DR (human leukocyte antigen-DR) and higher expression of CD11b compared with healthy controls (Fig. 1g). Proteomic analysis of sorted blood CD14++CD16− ARDS monocytes indicated changes in the abundance of proteins with transcripts that had been reported to be sensitive to hypoxic culture in human monocytes13 when compared with healthy controls (Fig. 1h). We also observed a significant increase in secretory-granule content in ARDS CD14++CD16− monocytes (Fig. 1i,j) which was associated with altered expression of hypoxia-regulated proteins, including SLC2A314, IGFR215, PSMD416 and FTL17. NanoString analysis identified a specific transcriptional signature in ARDS monocytes (Fig. 1k), with 41 genes differentially expressed compared with healthy controls (Fig. 1l). Notably, four MHC complex genes (HLA-DMA, HLA-DMB, HLA-DQA1 and HLA-DRB3), important for antigen-presenting function, as well as genes associated with monocyte adhesion and extravasation (SELL18), transendothelial migration (CD9919) and LPS signaling (MAP2K420) and MAP3K1421) were significantly downregulated in ARDS monocytes (Fig. 1l). Thus, ARDS affected both the transcriptomic and the protein signature of blood monocyte
Table 1.
ARDS patient cohort clinical characteristics and demographics
| Early ARDS (n = 11) | Late ARDS (n = 11) | |
|---|---|---|
| Age (years) | 58.8 (±11.5) | 56.9 (±13.6) |
| Proportion females—number (%) | 6 (55) | 5 (45) |
| Body mass index (kg m−2) | 35.8 (±11.6) | 31.1 (±4.2) |
| APACHE2a score | 20.4 (±7.2) | 18.8 (±8.6) |
| Pulmonary ARDS—number (%) | 9 (82) | 7 (64) |
| Pulmonary aetiologiesb—number (%) | ||
| Positive bacterial culture | 4 (36) | 4 (36) |
| Positive viral PCRc | 2 (18) | 1 (9) |
| Positive mycology | 2 (18) | 2 (18) |
| No positive microbiology samples | 2 (18) | 1 (9) |
| Extrapulmonary aetiologies—number (%) | ||
| Fecal peritonitis | 1 (9) | |
| Mediastinal soft-tissue infection | 1 (9) | |
| Bacteremia | 1 (9) | |
| Biliary sepsis | 1 (9) | |
| Retropharyngeal abscess | 1 (9) | |
| Noninfective | 1 (9) | |
| Index of tissue hypoxia | ||
| Reference PaO2: 11.1–14.4 kPad | ||
| Lowest PaO2 in hospitalization preceding sampling (kPa) | 4.38 (±1.72) | 6.42 (±2.23) |
| Lowest PaO2 24 h before sampling (kPa) | 6.69 (±2.11) | 8.15 (±1.90) |
| Reference FiO2: 21% | ||
| Highest FiO2 in 24 h before sampling (%) | 68.1 (±21.2) | 72.3 (±22.3) |
| Reference arterial lactate: 0.5–1.6 mmol l−1d | ||
| Highest lactate within 24 h of sampling (mmol l−1) | 2.45 (±1.28) | 1.56 (±0.96) |
| Receipt of organ-supportive therapies—number (%) | ||
| Invasive mechanical ventilation | 7 (64) | 7 (64) |
| Vasopressors | 7 (64) | 6 (55) |
| Renal replacement therapy | 1 (9) | 1 (9) |
| Receipt of additional medications—number (%) | ||
| Dexamethasonee | 0 (0) | 0 (0) |
| Lopinavir or ritonavir | 1 (9) | 0 (0) |
| Tocilizumab | 0 (0) | 0 (0) |
| Hydroxychloroquine | 1 (9) | 1 (9) |
Patient cohort and clinical characteristics demonstrate heterogeneity of etiology and evidence of clinically significant ongoing hypoxemia.
± data values refer to mean ± s.d.
aAcute Physiology and Chronic Health Evaluation Score 2 (APACHE2).
bOne patient in each group returned mixed fungal and bacterial cultures, which could not be causatively differentiated.
cTwo patients in the ‘Early’ group and one patient in the ‘Late’ group were SARS-CoV-2 positive.
dReference ranges as indicated by the local health board (NHS Lothian).
eNo other corticosteroids were administered.
Fig. 1. Patients with ARDS are monocytopenic early in the disease with phenotypically distinct circulating monocytes.
a,b, Lowest (a) and all (b) partial pressures of oxygen (PaO2) from clinical arterial blood samples from ARDS patients, 24 h preceding research blood sampling (green: normal range). c,d, Highest recorded FiO2 (c) and highest recorded arterial plasma lactate level within 24 h of research sampling (d) in ARDS patients (red samples: lactate ≥upper limit of normal; green: normal local reference (0.5–1.6 mmol l−1)). e, Blood leukocyte counts, monocyte proportions and monocyte counts from ARDS patients, collected within 48 h of diagnosis (early ARDS) and a healthy volunteer cohort (HC). f, Blood leukocyte count, monocyte proportions and monocyte counts from ARDS patients collected between 48 h and 7 d (late ARDS) and HC. g, Monocyte HLA-DR and CD11b expression in HC and ARDS patients. h, CD14++CD16− classical monocyte proteomic data from ARDS patients, relative to HC, for proteins associated with a human monocyte, in vitro hypoxic gene signature13. i, Classical (CD14++CD16−) monocyte proteome volcano plot from HC and ARDS patients. Significantly upregulated granule-associated proteins in ARDS patients versus HC (blue), a sample of known hypoxia-regulated proteins (orange). j, Classical monocytes proteinase 3, myeloperoxidase and azurocidin 1 copy numbers in HC and ARDS patients. k,l, Pearson’s correlation (k) and heatmap (l) of differentially expressed genes from HC and ARDS patient blood monocytes. Data in a–c are mean ± s.d. expressed as median (e,f) or shown as mean (g,j). In a,c–g and j each datapoint represents one patient/HC; in b, each datapoint represents one independent clinical sample. Statistical testing used was: unpaired, two-tailed Student’s t-test (e–f and j) and Mann–Whitney U-test (g).
Extended Data Fig. 1. Monocyte sub-populations are altered early in ARDS.
(a) Representative plots and proportions of monocyte sub-populations based on CD14 and CD16 expression early and (b) late (c). Each data point = one individual patient/ healthy donor control (HC), b, c Data+mean, one-way ANOVA with Holm-Sidak post-test.
Experimental ARDS reproduces monocytopenia and phenotypic alterations
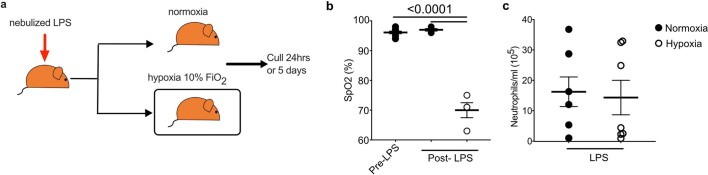
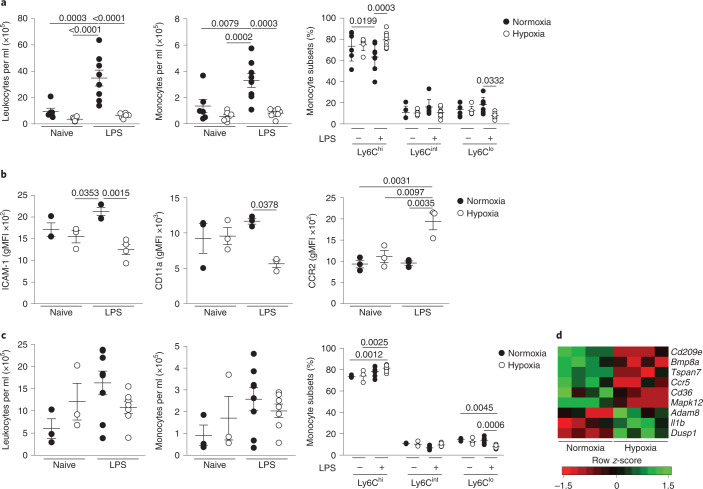
We next investigated whether the circulating monocyte profile alterations observed in ARDS patients were replicated by the induction of hypoxemia in a mouse model of ALI (Extended Data Fig. 2a). Mice exposed to 10% FiO2 demonstrated equivalent levels of hypoxemia to patients with ARDS (Extended Data Fig. 2b). Administration of nebulized lipopolysaccharide (LPS) induced an increase in circulating leukocytes and CD115+CD11b+ monocytes (Fig. 2a) in mice housed in normoxia for 24 h, relative to naive mice, yet this increase was absent in LPS-challenged mice housed in hypoxia (10% FiO2) immediately post-LPS for 24 h. A selective loss of nonclassical CD115+CD11b+Ly6Clo monocytes in LPS-challenged hypoxic mice (Fig. 2a) relative to normoxic LPS-challenged mice was observed. Blood CD115+CD11b+Ly6Chi classical monocytes from hypoxic LPS-challenged mice had decreased expression of the adhesion molecules intercellular adhesion molecule (ICAM)-1 and CD11a and increased expression of the CCR2 chemokine receptor, compared with normoxic counterparts (Fig. 2b). The absolute number of circulating leukocytes, CD115+CD11b+ monocytes (Fig. 2c) and Ly6G+CD11b+ neutrophils (Extended Data Fig. 2c) in the blood of normoxic or hypoxic LPS-treated mice normalized at day 5, although the proportion of Ly6Clo monocytes remained contracted in hypoxic LPS-challenged mice compared with normoxic LPS-challenged controls (Fig. 2c). We also observed persistent alterations in the transcriptome of Ly6Chi monocytes in hypoxic LPS-challenged mice (Fig. 2d), including decreased expression of the chemokine receptor Ccr5 and the scavenger receptor Cd36, which are markers of monocyte maturity22 and increased expression of Il1b, an inflammatory cytokine associated with poor outcomes in ARDS23. Thus, in mice with ALI, hypoxia drives alterations in monocyte numbers and phenotype that parallel those described in ARDS patients.
Extended Data Fig. 2. A hypoxic environment induces hypoxaemia with equivalent circulating neutrophil.
(a) Schematic of normoxic and hypoxic LPS-induced ALI. (b) Oxygen saturations in mice were measured in mice at baseline pre-LPS nebulisation (pre-LPS) and 6 hours post-LPS (N LPS- mice housed in normoxia post-LPS, H LPS- mice housed in hypoxia post-LPS). (Pre-LPS n = 6, N LPS n = 3, H LPS n = 3). (c) Blood was collected from mice treated with LPS and placed in normoxia (N LPS) or hypoxia 10% (H LPS) for 5 days and circulating neutrophils quantified by flow cytometry (Live Singles CD45+Ly6G+CD11b+). b, c Each data point represents an individual mouse. Data shown as mean±SEM. c two pooled independent experiments. Statistical testing performed using one-way ANOVA with Tukey’s multiple comparisons test.
Fig. 2. Hypoxic acute lung injury replicates early monocytopenia in mice and alters the circulating monocyte phenotype.
a, Blood leukocyte counts, monocyte counts and proportion of blood monocyte subgroups in naive or LPS-treated mice housed in normoxia or hypoxia for 24 h. b, Classical monocyte (CD115+CD11b+Ly6Chi) surface expression of ICAM, CD11a and CCR2 at 24 h post-LPS. c, Blood leukocyte counts, monocyte counts and proportions of monocyte sub-populations in naive or LPS-treated mice housed in normoxia or hypoxia for 5 d post-LPS. d, Differentially expressed genes in circulating classical monocytes from LPS-treated mice housed in normoxia or hypoxia for 5 d. Data represent the mean ± s.e.m. Data for a and c are pooled from two independent experiments. b is representative of 2 experiments (n=3-4/ group). Each datapoint represents an individual mouse. Statistical testing: one-way ANOVA with Tukey’s multiple comparison test (a and b).
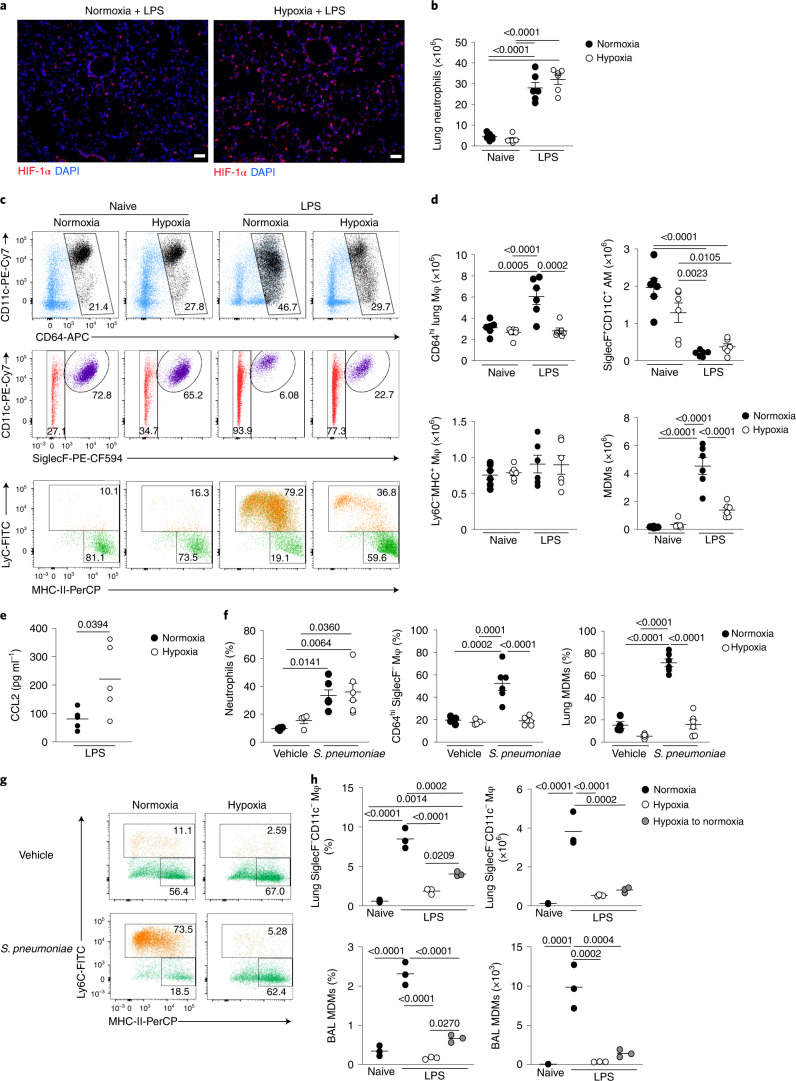
Tissue hypoxia prevents accumulation of lung MDMs
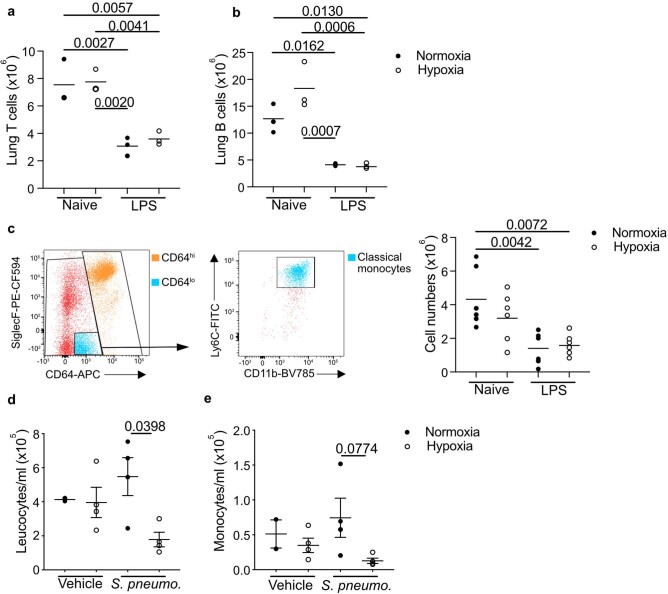
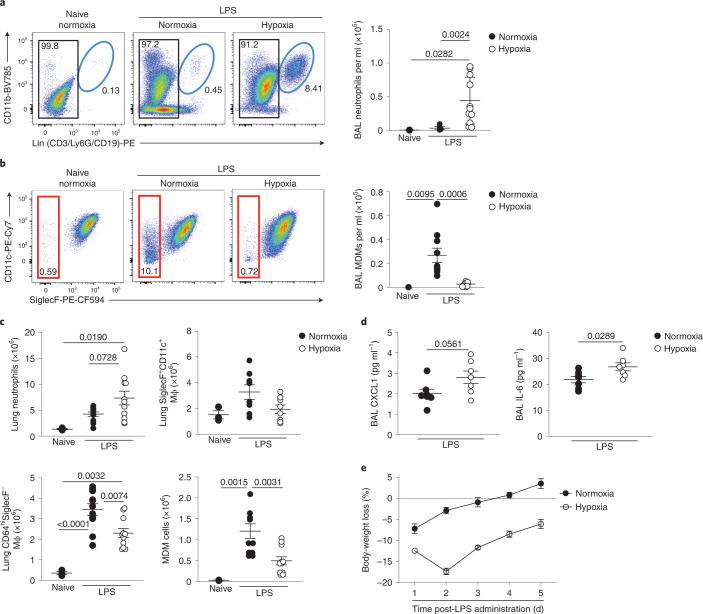
Next we explored the effect of hypoxemia on LPS-induced inflammation in the lung. Consistent with tissue hypoxia, hypoxia-inducible factor 1α (HIF-1α) protein was most highly expressed in the lungs of hypoxic LPS-treated mice compared with normoxic LPS controls (Fig. 3a). LPS challenge significantly increased lung Ly6G+CD11b+ neutrophil numbers in hypoxia- or normoxia-housed mice compared with naive controls (Fig. 3b), with equivalent numbers of CD3+CD19−MHC-II− T cells and CD3−CD19+MHC-II+ B cells in the lungs of these mice (Extended Data Fig. 3a,b). LPS challenge led to a significant expansion of the Lin–CD45+CD64hi macrophage compartment in normoxia- but not hypoxia-housed mice (Fig. 3c,d). AMs are CD64hiSiglecF+CD11c+, whereas CD64hiSiglecF– macrophages almost exclusively comprise IMs in health, and include inflammation-elicited, parenchymal or alveoli-localized, CD64hiSiglecF–Ly6C+ MDMs after injury. The number of CD64hiSiglecF+CD11c+ macrophages was equivalent in LPS-challenged mice housed in hypoxia or normoxia (Fig. 3c,d). Hypoxia significantly blunted the LPS-mediated expansion of the CD64hiSiglecF– macrophage compartment observed in normoxic mice (Fig. 3c,d), an effect that appeared to be entirely attributable to the absence of CD64hiSiglecF–Ly6C+ MDMs in LPS-treated hypoxic mice (Fig. 3c,d). The number of CD64hiSiglecF–Ly6C−MHC-II+ macrophages was similar in all treatments (Fig. 3c,d). The reduction in the number of CD64hiSiglecF–Ly6C+ MDMs occurred despite elevated amounts of the monocyte chemoattractant CCL2 in the alveoli of LPS-treated hypoxic mice (Fig. 3e), and similar numbers of CD64loCD11b+Ly6C+ monocytes in the lungs of hypoxic and normoxic LPS-treated mice (Extended Data Fig. 3c). These observations suggested that monocytes recruited to the lung after LPS treatment did not convert to CD64hiSiglecF–Ly6C+ MDMs in hypoxia. The contribution from intravascular cells to lung cell counts could not be completely excluded from these data.
Fig. 3. Systemic hypoxia hampers expansion of the CD64hiSiglecF− macrophage niche in ALI and S. pneumoniae infection.
a, Representative lung immunofluorescence HIF-1α and DAPI expression from LPS-challenged mice, housed in normoxia or hypoxia for 24 h. Scale bar, 50 μm. b, Absolute numbers of live lung neutrophils in naive or LPS-challenged mice housed in normoxia (N) or hypoxia (H) for 24 h (n = 6 per group). c, Representative dot plots of the CD64hi macrophage compartment (top), CD64hiSiglecF+CD11c+ AMs (middle) and Ly6C and MHC-II expression by CD64hiSiglecF− macrophages (bottom) in mice as in b. d, Absolute number of CD64hi macrophages (Mφ), CD64hiSiglecF+CD11c+ AMs, Ly6C−MHC-II+ lung macrophages and CD64hiSiglecF−Ly6C+ MDMs as in b. e, BAL CCL2 levels from LPS-challenged mice housed in normoxia or hypoxia for 24 h. f, Frequencies of neutrophils among total lung leukocytes, of CD64hiSiglecF− macrophages among lung CD64hi macrophages and MDMs among CD64hiSiglecF− macrophages in mice inoculated with S. pneumoniae (n = 6 per group) or vehicle control (Veh, n = 4 per group) and housed in normoxia or hypoxia until 24 h post-inoculation. g, Representative plots of CD64hiSiglecF− macrophages in mice as in f. h, Frequency of lung CD64hiSiglecF− macrophages among total leukocytes, absolute numbers of lung CD64hiSiglecF− macrophages, proportion of BAL MDMs and absolute numbers of BAL MDMs in naive or LPS-challenged mice housed either in normoxia or hypoxia for 48 h or for 24 h in hypoxia, followed by 24 h of normoxia (hypoxia to normoxia) (n = 3 per group). Data represent the mean ± s.e.m. Data in a represent n = 3 per group; data in b–e are pooled from two independent experiments; data in g represent two independent experiments. Each datapoint represents an individual mouse. Statistical testing for b,d,f and h is by one-way ANOVA with Tukey’s multiple comparison test and for e by unpaired, two-tailed Student’s t-test.
Extended Data Fig. 3. T cell and B cells are equivalent post-LPS and Streptococcus pneumoniae infection in hypoxia leads to leukopenia and monocytopenia.
T cells (a) (Live, singles, CD45+, Lin+ (CD3/CD19/Ly6G) MHCII− CD11b−) and B cells (b) (Live, singles, CD45+, Lin+ (CD3/CD19/Ly6G) MHCII+ CD11b−) were quantified in lung digests from mice housed in normoxia (N) or 10% FiO2 hypoxia (H) for 24 hours, left naïve or nebulised with LPS. Mice were inoculated with Streptococcus pneumoniae (Strep) or vehicle (Veh) intratracheally (i.t.) and housed in normoxia (N) or hypoxia (H) until 24 hours post-i.t. (c) Representative dot plots of gating strategy for classical monocytes in the lung gated on Singles Live CD45 + Lin- lung cells and associated counts in the lung 24 hours post-LPS challenge and housed in normoxia (N) and hypoxia (H). Blood cell counts and (c) monocyte counts mice challenged with vehicle or strep pneumoniae and housed in normoxia (N) or hypoxia (H) for 24 hours. Each point represents and individual mouse. Data shown as mean±SEM. Statistical testing performed using one-way ANOVA with Tukey’s multiple comparisons test.
To examine the effects of hypoxia on monocyte–macrophage dynamics in a model of severe streptococcal pneumonia, C57/BL6J mice were inoculated with D39 Streptococcus pneumoniae or vehicle and housed in either hypoxia or normoxia after a 4-h recovery period. Reduced blood leukocyte and monocyte counts were detected in infected mice housed in hypoxia compared with normoxia (Extended Data Fig. 3d,e). Although the number of lung neutrophils was equivalent in hypoxic and normoxic S. pneumoniae-infected mice (Fig. 3f), the accumulation of CD64hiSiglecF– macrophages was reduced (Fig. 3f), with a particular absence of CD64hiSiglecF–Ly6C+ MDMs (Fig. 3f,g).
To determine whether hypoxia altered the monocyte–macrophage lung compartment directly, we returned LPS-challenged hypoxic mice to normoxic conditions, for a further 24 h, after 24 h of hypoxia. Mice returned to normoxia showed a significant increase in the proportion and number of CD64hiSiglecF– macrophages and bronchoalveolar lavage (BAL) MDMs, compared with mice that remained in hypoxia (Fig. 3h). Together, these data indicated that hypoxia directly induced sustained changes in the lung macrophage compartment during various inflammatory challenges.
Hypoxia directly alters hematopoiesis
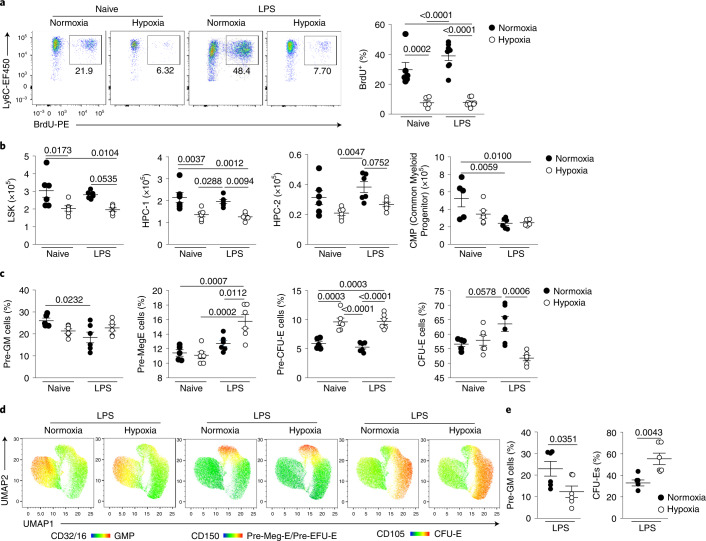
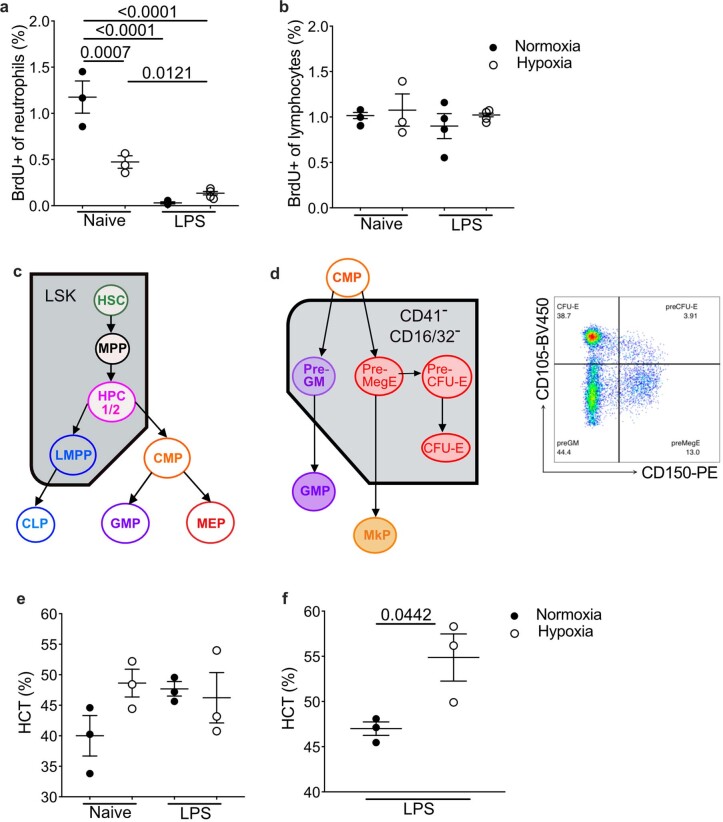
To determine the mechanism by which hypoxia regulated the number of circulating monocytes, we measured bone marrow (BM) output by pulsing naive or LPS-challenged mice, housed in hypoxia or normoxia, with bromodeoxyuridine (BrdU) 12 h post-LPS12,24. Hypoxemic LPS-challenged mice had a 80% reduction in the proportion of BrdU+CD115+CD11b+Ly6Chi monocytes compared with normoxic LPS-challenged counterparts (Fig. 4a). LPS equally reduced the proportion of blood BrdU+ neutrophils in hypoxic and normoxic mice compared with naive controls (Extended Data Fig. 4a), with a similar frequency of BrdU+ lymphocytes in all samples (Extended Data Fig. 4b). Examination of the BM stem cell compartment (Extended Data Fig. 4c) 24 h post-hypoxic exposure indicated a reduction in absolute numbers of Lin–Sca-1+Kit+ (LSK) cells in hypoxic mice compared with normoxic mice independent of LPS treatment (Fig. 4b), with a specific reduction in the CD48+CD150− HPC-1 and CD48+CD150+ HPC-2 hematopoietic progenitor cells (HPCs; Fig. 4b), which have restricted multipotency25. Irrespective of oxygenation, LPS treatment reduced the absolute number of Lin−cKit+Sca1−CD127−CD16/32−CD34+ common myeloid progenitor cells (CMPs) compared with naive control mice (Fig. 4b).
Fig. 4. Systemic hypoxia alters BM hematopoiesis toward increased erythropoiesis.
a, Representative dot plots gated on live CD45+Lin−(CD3/CD19/Ly6G)CD115+Ly6Chi cells and proportion of BrdU+ monocytes in naive or LPS-challenged mice housed in normoxia or hypoxia for 24 h and pulsed with BrdU for the last 12 h (naive, n = 5–6; LPS treated, n = 8 in). b, Absolute numbers of BM LSK cells, CD48+CD150− HPC-1, CD48+CD150+ HPC-2 (n = 6 per group) and Lin−cKit+Sca1−CD127−CD16/32−CD34+ CMPs (n = 5–6 group) in mice as in a. c, Proportion of BM pre-GMs, pre-Meg-E, pre-CFU-E and CFU-E CD41−CD32/16− cells in mice as in a (n = 6 per group). d,e, Representative UMAP analysis of BM cells gated on live CD45+Lineage−Sca1−C-Kit+CD41−CD32/16− cells (d) and summary data of proportions of pre-GM and CFU-E cells (e) measured in the BM of mice treated with LPS and housed in normoxia (N) or hypoxia (H) for 5 d (n = 6 per group). Data are shown as the mean ± s.e.m. Each datapoint represents an individual mouse. Data in a–e are pooled from two independent experiments. Statistical testing: for a–c is by one-way ANOVA with Tukey’s multiple comparison test and for e by unpaired, two-tailed Student’s t-test.
Extended Data Fig. 4. Impact of hypoxia on bone marrow cell egress and composition.
(a) BrdU+ blood lymphocytes (CD3 and CD19+) and (b) BrdU+ blood neutrophil proportion in mice treated with LPS and housed in normoxia (N) and hypoxia (H) for 24 hours (Naïve N and H n = 3, LPS N and H n = 4). Data representative of 2 experiments (c) Schematic showing hematopoietic hierarchy with Lin−Sca−C-Kit+ (LSK) compartment and progenitors (HSC-hematopoietic stem cell, MPP- multipotent progenitor, HPC- hematopoietic progenitors, CMP- common myeloid progenitor, LMPP- lymphoid-primed multipotent progenitor, CLP- common lymphoid progenitor, GMP- granulocyte/monocyte progenitor, MEP-Megakaryocyte/erythrocyte progenitor). (d) Schematic showing erythrocytosis and monopoiesis (Pre-GM-pre-granulocyte/monocyte precursor, Pre-MegE - megakaryocyte-erythrocyte precursor, Pre-CFU-E - pre-colony forming unit erythroid, CFU-E - Colony forming unit erythroid, GMP – granulocyte-monocyte precursor, MkP- megakaryocyte precursor) gating strategy for bone marrow common myeloid progenitor progeny on CD41−CD16/32− cells. (e) Blood hematocrit at 24 hours (n = 3/ group, data representative of 2 experiments) or (f) 5 days in mice treated with LPS and housed in normoxia (N) and hypoxia (H) (n = 3/group, data representative of 2 experiments) was measured. a, b Mean±SD. Statistical testing performed using one-way ANOVA with Tukey’s multiple comparisons test, e, f Mean±SD. f, statistical testing performed using unpaired two-sided t-test.
As erythrocytes and monocytes originate from CMPs26 (Extended Data Fig. 4d), we investigated whether hypoxia shifted hematopoiesis in favor of red blood cell (RBC) production by measuring the effect of hypoxia on CMP progeny (Lin−cKit+CD41−CD32/16−) 24 h after LPS challenge26. Hypoxia did not affect the number of CD150−CD105− pre-granulocyte/monocyte precursors (pre-GMs) (Fig. 4c), but significantly increased the proportion of CD150+CD105− megakaryocyte-erythroid precursor (pre-Meg-E) cells (Fig. 4c), and led to an increase in downstream CD150+CD105+ pre-colony-forming unit erythroid precursor (pre-CFU-E) cells (Fig. 4c). Although hematocrit values were equivalent at this 24-h timepoint (Extended Data Fig. 4e), BM CFU-E cell proportions were reduced in LPS-treated hypoxic mice compared with normoxic counterparts (Fig. 4c), suggesting increased red cell egress. The hematocrit of LPS-treated hypoxic mice was increased compared with normoxic controls at day 5 (Extended Data Fig. 4f). UMAP (Uniform Manifold Approximation and Projection) analysis of Lin−cKit+CD41−CD32/16− cells within the BM compartment post-LPS indicated that skewing of the CMPs toward erythropoiesis was achieved by day 5 (Fig. 4d,e). Taken together, these data demonstrated that hypoxia altered hematopoiesis, reducing monocyte BM output.
Hypoxia suppresses type I IFN signaling
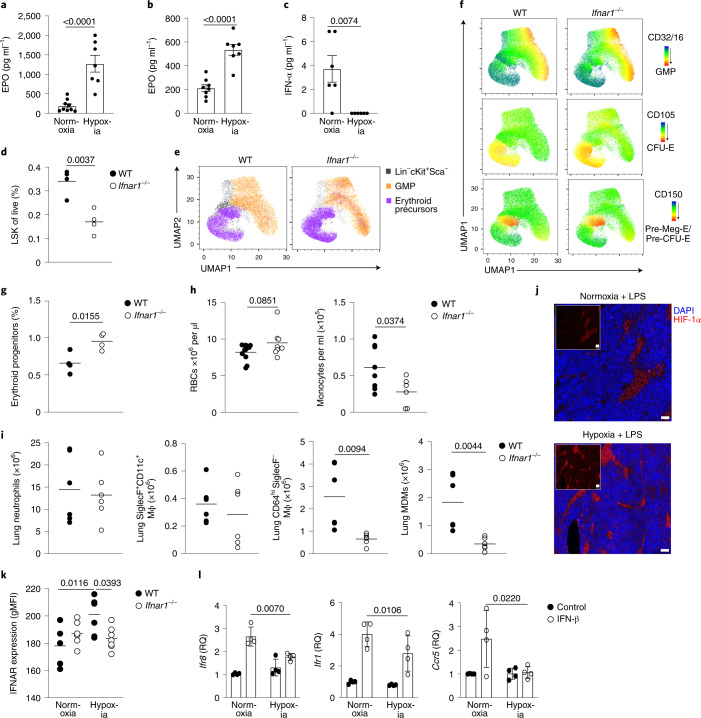
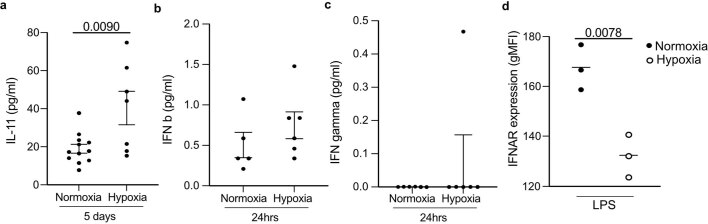
Next, we investigated the mechanism by which hypoxia suppressed monopoiesis in mice with ALI. Erythropoietin (EPO), an RBC growth factor, was significantly increased at 24 h (Fig. 5a) and day 5 post-LPS in hypoxia-housed compared with normoxia-housed mice (Fig. 5b). Interleukin (IL)-11, a hypoxia-responsive megakaryocyte and hematopoietic growth factor27,28, was also increased at day 5 post-LPS in hypoxic compared with normoxic mice (Extended Data Fig. 5a). Type I IFN (IFN-α and IFN-β) and type II IFN (IFN-γ) are known drivers of emergency monopoeisis29,30. Although IFN-β and IFN-γ were equivalent in hypoxic and normoxic LPS-treated mice, IFN-α was markedly reduced in hypoxic mice 24 h post-LPS challenge compared with normoxic counterparts (Fig. 5c and Extended Data Fig. 5b,c). Ifnar1–/– mice, which lack the type I IFN receptor, had a contraction of the LSK compartment (Fig. 5d) and increased proportion of the megakaryocyte/erythrocyte progenitor (MEP)/pre-CFU-E/CFU-E erythroid progenitors 24 h after LPS treatment during normoxia (Fig. 5e–g). Then, 5 d post-LPS challenge, normoxic Ifnar1–/– mice had enhanced numbers of circulating RBCs and were significantly monocytopenic compared with wild-type (WT) controls (Fig. 5h). In addition, normoxic Ifnar1–/– mice had similar numbers of neutrophils and CD64hiSiglecF+ macrophages (Fig. 5i), but reduced numbers of CD64hiSiglecF– macrophages and MDMs (Fig. 5i) in the lung 24 h post-LPS, compared with WT controls.
Fig. 5. Hypoxia regulates type I IFN responses, hindering lung CD64hiSiglecF− macrophage expansion in response to LPS.
a,b, Serum EPO levels in mice challenged with LPS and housed in normoxia or hypoxia for 24 h (n = 9 normoxia, n = 7 hypoxia) (a) or 5 d (n = 7 per group) (b). c, Serum IFN-α in in mice challenged with LPS and housed in normoxia or hypoxia for 24 h (n = 6 per group). d, Proportion of LSK cells in the BM of WT or Ifnar1−/− mice 24 h post-LPS challenge (n = 4 per group). e, Manual gating of erythroid precursors and granulocyte/macrophage progenitors (GMPs) in LSK cells displayed on UMAP projection in mice as in d. f, Representative expression of CD32/16 (GMP marker) and CD150/CD105 (erythroid progenitor-associated markers) in LSK cells from mice as in d using the Pronk gating strategy26 displayed on UMAP projection. g, Proportion of erythroid progenitor cells (combined MEPs, pre-CFU-E and CFU-E cells) in WT and Ifnar1−/− BM 24 h post-LPS (n = 4 per group). h, Peripheral RBCs (n = 10 WT, n = 8 knockout) and monocyte counts at day 5 post-LPS in WT and Ifnar1−/− mice. i, Neutrophils, CD64hiSiglecF+CD11c+ macrophages, CD64hiSiglecF− macrophages and MDM numbers in the lungs of WT and Ifnar1−/− mice 24 h post-LPS. j, Representative HIF-1α and DAPI expression in the femoral BM in mice challenged with LPS and housed in normoxia (N LPS) or hypoxia (H LPS) for 24 h. Scale bar, 20 μm. k, IFNAR expression in the BM LSK in naive (n = 5–6 per group) or LPS-treated mice (n = 6 per group) housed in normoxia or hypoxia for 24 h. l, Fold change in Quantitative PCR of Irf8, Irf1 and Ccr5 expression (normalized to actin-β, relative quantification) in BM cells from naive mice cultured in normoxia or hypoxia for 4 h ± IFN-β (n = 3 per group) relative to untreated normoxia control. Data represent the mean ± s.e.m. All datapoints represent individual mice. Statistical testing for a–d and g–i was by unpaired, two-sided Student’s t-test, for k by one-way ANOVA with Tukey’s multiple comparison test and for l by two-way ANOVA with Šídák's multiple comparison post-test. The data in a–c and d–i represent two independent experiments, and represent n = 3 per group in j and two independent pooled experiments in l.
Extended Data Fig. 5. Hypoxia elevates circulating IL-11 levels without altering IFN beta and IFN gamma levels and reducing IFNAR expression in circulating classical monocytes.
(a) Serum IL-11, (b) IFN beta and (c) IFN gamma level mice treated with nebulised LPS and housed in normoxia (N) or hypoxia (H), for 24 hours or 5 days (as indicated on figure), were measured by ELISA as per manufacturers’ instructions. Data mean±SEM. Each data point represents an individual mouse. (d) IFNAR expression was measured by flow cytometry in classical blood monocytes in mice treated with nebulized LPS and housed in either normoxia (N LPS) or hypoxia (H LPS) for 24 hours. a, d, Statistical testing unpaired two-sided t-test.
Increased HIF-1α stabilization was observed in the BM of hypoxic, LPS-treated, mice compared with normoxic counterparts (Fig. 5j), in keeping with tissue hypoxia. As hypoxia can alter type I IFN signaling in cancer31, we next investigated interferon-α/β receptor (IFNAR) expression in the BM. LSK cells from hypoxic mice 24 h post-LPS treatment did not upregulate IFNAR expression, compared with normoxic mice (Fig. 5k). IFNAR expression was also reduced in blood CD115+CD11b+Ly6Chi monocytes from hypoxic LPS-challenged mice compared with normoxic counterparts (Extended Data Fig. 5d). BM cells from naive mice cultured in vitro, in hypoxia (1% O2), showed significant blunting of type I IFN-mediated Irf8, Irf1 and Ccr5 expression (Fig. 5l). Taken together, these findings suggested that hypoxia directly altered the immune response through local and systemic mechanisms.
Loss of monocyte recruitment associated with persistence of inflammation
We subsequently investigated the longer-term effects of systemic hypoxia on the myeloid compartment and inflammation resolution. Infiltrating neutrophils promote vascular injury, protein leak and alveolar epithelial damage in ARDS, and drive deleterious inflammatory responses in murine models of hypoxic ALI32,33. 5 d after LPS challenge, normoxic mice had very few neutrophils within the bronchoalveolar space whereas hypoxic mice showed evidence of ongoing inflammation with significant bronchoalveolar neutrophilia (Fig. 6a). Hypoxic LPS-treated mice had a reduction in the number of bronchoalveolar CD64hiSiglecF– MDMs compared with normoxic counterparts (Fig. 6b). The total number of lung Ly6G+CD11b+ neutrophils was greater in hypoxic compared with normoxic mice at day 5 post-LPS (Fig. 6c) and, although the number of CD64hiSiglecF+CD11c+ macrophages had returned to baseline (Fig. 6c), the non-AM CD64hiSiglecF– macrophages remained contracted (Fig. 6c), largely as a consequence of fewer CD64hiSiglecF–Ly6C+ MDMs (Fig. 6c). In addition, the BAL from hypoxic mice at this timepoint had higher CXCL1 and IL-6 (Fig. 6d), parameters reported to be elevated in ARDS nonsurvivors23. Moreover, hypoxic mice showed more sustained weight loss at day 5 post-LPS, compared with the normoxia-housed controls (Fig. 6e). Collectively, these data indicated that hypoxia-induced monocytopenia was associated with persistent lung inflammation.
Fig. 6. Ongoing CD64hiSiglecF− macrophage expansion failure is associated with inflammation persistence in hypoxic ALI.
a,b, Representative dot plots and absolute numbers of BAL neutrophils (a) and CD45+Ly6G−CD64hiSiglecF− MDMs gated on CD45hi cells (b) in mice treated with LPS and housed in normoxia or hypoxia for 5 d. c, Lung neutrophils, CD64hiSiglecF+CD11c+ macrophages, CD64hiSiglecF− macrophages and CD64hiSiglecF−Ly6C+ MDM numbers in LPS-challenged mice housed in normoxia or hypoxia for 5 d. d, BAL CXCL1 and IL-6 levels in mice treated with LPS and housed in normoxia (N) or hypoxia (H) for 5 d (n = 6 N LPS, n = 7 H LPS). e, Daily weight changes from baseline in LPS-challenged mice housed in normoxia or hypoxia for 5 d (n = 4 per group). Data are shown as the mean ± s.e.m. Each datapoint represents an individual mouse. Statistical testing for a–c was by one-way ANOVA with Tukey’s multiple comparison test and for d by unpaired, two-tailed Student’s t-test. Data in a–c were pooled from three independent experiments and in d from two independent experiments.
CSF-1 accelerates the resolution of lung inflammation in hypoxia
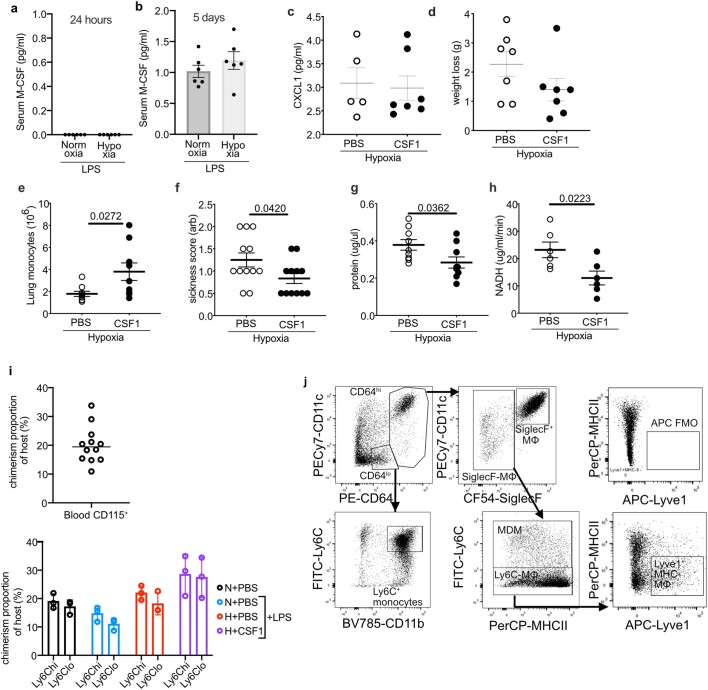
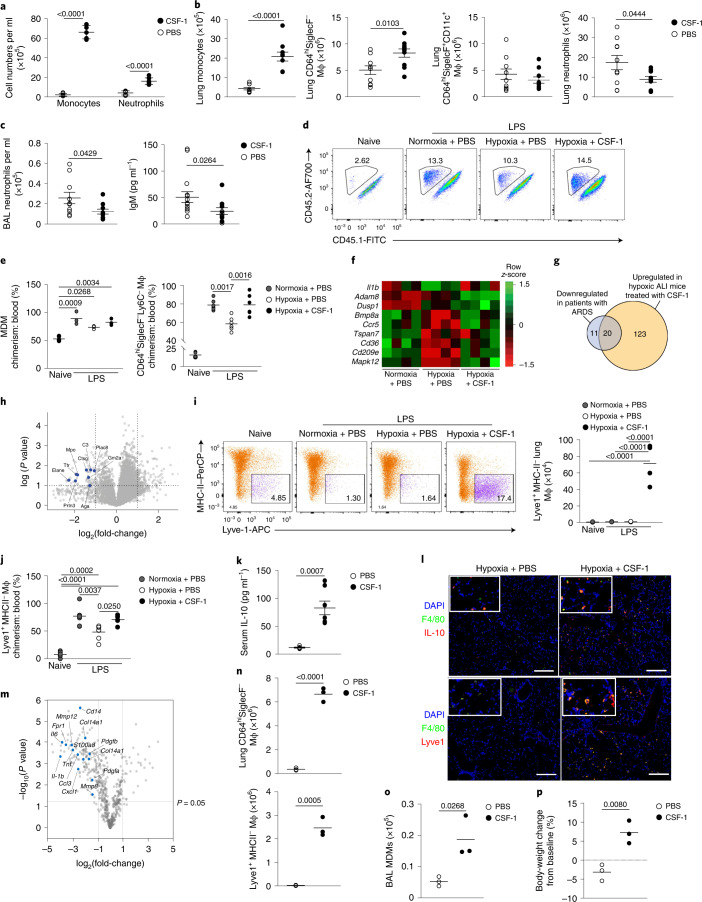
Finally, we tested whether increasing the number of monocytes during hypoxia facilitated inflammation resolution. LPS-challenged hypoxic mice with absent (24 h) or low (5 d) baseline levels of macrophage-colony-stimulating factor (M-CSF) (Extended Data Fig. 6a,b) were treated with four daily injections of CSF-1–Fc fusion protein34 or phosphate-buffered saline (PBS). CSF-1–Fc markedly increased the number of CD115+CD11b+ monocytes and moderately increased the number of Ly6G+CD11b+ neutrophils in the blood at day 5 compared with PBS (Fig. 7a). CSF-1–Fc-treated, hypoxic, LPS-challenged mice also had increased numbers of CD64loCD11b+Ly6C+ monocytes and CD64hiSiglecF– macrophages in the lung compared with mice receiving PBS (Fig. 7b). The number of CD64hiSiglecF+CD11c+ macrophages was not affected (Fig. 7b). Importantly, the absolute number of Ly6G+CD11b+ neutrophils in the lung tissue (Fig. 7b) and BAL (Fig. 7c) was reduced in CSF-1–Fc-treated, hypoxic, LPS-challenged mice at day 5, despite equivalent levels of CXCL1 in the BAL between CSF-1–Fc-treated and PBS-treated mice (Extended Data Fig. 6c). Hypoxic LPS-challenged mice treated with CSF-1–Fc had reduced weight loss (Extended Data Fig. 6d) and reduced immunoglobulin (Ig)M levels in the BAL fluid at day 5 (Fig. 7c) compared with PBS counterparts, suggesting reduced alveolar inflammation and vascular leak. To test these observations in a model of virally-induced epithelial injury, C57/BL6J mice were inoculated with influenza A virus (PR8) and placed in hypoxia immediately after. CSF-1–Fc or PBS was administered 12 h and 36 h post-PR8 challenge. Hypoxic PR8-infected mice treated with CSF-1–Fc had increased numbers of CD64loCD11b+Ly6Chi monocytes in the lung (Extended Data Fig. 6e) and improved physiological outcomes (Extended Data Fig. 6f) compared with mice receiving PBS. This was associated with a significant reduction in BAL protein levels, a marker of lung injury (Extended Data Fig. 6g) and lactate dehydrogenase activity, as an indicator of cellular damage (Extended Data Fig. 6h).
Extended Data Fig. 6. Circulating CSF1 is unchanged in hypoxic ALI and exogenous CSF1 improves injury outcomes altering the CD64hiSiglecF− Mϕ phenotype.
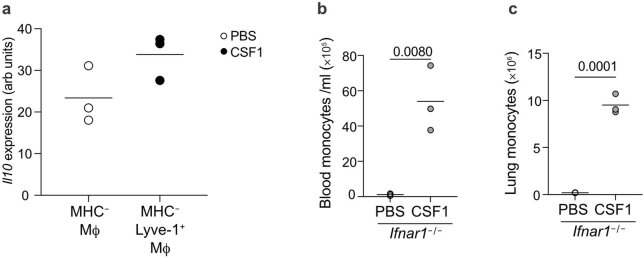
Serum MCSF (CSF1) from LPS-challenged mice housed in normoxia (N) or hypoxia (H) for 24 hours (a) or 5 days (b) was measured. (c) BAL CXCL1 measured in LPS-challenged mice housed in hypoxia for 5 days and treated with PBS or CSF1-Fc (H CSF1). (d) weight loss from baseline in hypoxic LPS-induced ALI treated with PBS (H PBS) or CSF1-Fc (H CSF1). (e) Lung monocyte numbers, (f) arbitrary sickness scores, (g) BAL protein and (h) LDH activity (as measured by NADH) measured at 48 hours in mice with virally-induced ALI housed in hypoxia and treated with PBS or CSF1-Fc (H CSF1). (i) Baseline blood chimerism (proportion of donor cells relative to host) of circulating monocytes in lung-protected chimeras prior to ALI induction and chimerism of monocytes based on Ly6C expression post-LPS. (j) Lung cDC1 (gated on Alive CD45+Lin-CD64−CD11c+Cd103+) chimerism and counts and (k) cDC2 (gated on Alive CD45+Lin-CD64−CD11c+Cd103− CD11b+) chimerism and counts. Chimerism relative to blood monocyte chimerism. (l) Il10 expression was measured by NanoString platform analysis in MHC-lung macrophages from LPS-challenged mice housed in hypoxia for 5 days and treated with PBS and compared to Lyve1+MHCII- of LPS-challenged mice, housed in hypoxia and treated with CSF1-Fc. (m) Representative dot plots of lung digests showing gating strategy for identification of the different monocyte and macrophage populations in the lung gated on Live Singles CD45+Ly6G− cells, and including Lyve1+MHC-CD64hiSiglecF−Mϕ, and associated APC FMO control including Lyve1+MHC-CD64hiSiglecF−Mϕ, and associated APC FMO control. Ifnar−/− (KO) mice were nebulised with LPS and treated with PBS (KO PBS) or CSF1-Fc (KO CSF1). Mice were sacrificed on day 5 and (n) blood monocyte and (o) lung Ly6C+ monocytes were quantified by flow cytometry. c, d Mean±SD, e-h mean±SEM. c-h, j, k 2 pooled experiments, l representative of 3 experiments, n, o representative of 2 experiments. Statistical testing e-h, n, o unpaired two-sided t-test, k, one-way ANOVA with Tukey’s multiple comparisons test.
Fig. 7. CSF-1 rescues the hypoxic monocytopenia driving inflammation resolution.
a–c, Monocyte and neutrophil counts in the blood (a), monocyte, CD64hiSiglecF− macrophage, CD64hiSiglecF+ CD11c+ macrophage and neutrophil counts in the lung (b), and absolute numbers of neutrophils and IgM titers in the BAL (c) in hypoxic LPS-challenged mice treated with four daily injections of PBS or CSF-1–Fc. d,e, Representative lung CD64hiSiglecF−Ly6C+ MDM CD45.2 and CD45.1 expression (d) and CD64hiSiglecF−Ly6C+ MDM:blood monocyte chimerism and CD64hiSiglecF−Ly6C− macrophage:blood monocyte chimerism (e) in lung-protected, naive or LPS-challenged mice that are normoxia or hypoxia housed and treated with PBS or CSF-1–Fc. f, Differentially expressed genes in PBS- or CSF-1–Fc-treated, LPS-challenged mice in Ly6Chi blood monocytes at day 5 post-LPS challenge. g, Overlap between differentially downregulated genes in ARDS blood monocytes and genes upregulated in CSF-1–Fc-treated mice relative to PBS-treated mice. h, Comparison of Ly6Chi blood monocyte proteomes from hypoxic, LPS-challenged, CSF-1–Fc-treated mice relative to the PBS-treated counterparts (granule-associated proteins identified). i, Representative Lyve1 expression and number of lung CD64hiSiglecF−Ly6C−Lyve1+MHC-II− macrophages in naive or LPS-challenged mice housed in normoxia or hypoxia and treated with PBS or CSF-1–Fc for 5 d. j, Chimerism of CD64hiSiglecF−Ly6C−Lyve1+MHC-II− macrophages to blood monocytes in lung-protected, LPS-challenged chimeras housed in normoxia or hypoxia, treated with PBS or CSF-1–Fc. k,l, Serum IL-10 (k) and representative tiled immunofluorescence of lung sections stained for F4/80, IL-10 and DAPI (l), and F4/80, Lyve1 and DAPI in LPS-challenged mice housed in hypoxia for 5 d and treated with CSF-1–Fc or PBS. Scale bar, 200 μm. m, Differentially regulated genes in CD64hiSiglecF−Lyve1+MHC-II− macrophages from LPS-challenged, CSF-1–Fc-treated mice relative to CD64hiSiglecF−MHC-II− macrophages from PBS-treated counterparts, housed in hypoxia for 5 d. n–p, Total lung CD64hiSiglecF− macrophages and CD64hiSiglecF−Ly6C−Lyve1+MHC-II− macrophages (n), BAL MDMs (o) and body-weight change (relative to baseline) (p) in LPS-challenged Ifnar1−/− mice treated with PBS (Ifnar1−/− PBS) or CSF-1–Fc (Ifnar1−/− CSF-1) for 5 d. Data represent the mean ± s.e.m. Each datapoint represents an individual mouse. Statistics for b,c,k and n–p were by unpaired, two-sided Student’s t-test, for i and j by a one-way ANOVA with Tukey’s post-test and for e by a two-tailed Mann–Whitney U-test following D’Agostino and Pearson’s normality test. For b and c data are pooled from three independent experiments, and for f,k,j and n–p data are pooled from two independent experiments. In g all genes have a fold-change >1, except H2-DMa, H2-DMb2, IL-17Ra and Nlrp3 where the fold-change is >0.5 and P < 0.05.
To dissect the mechanism by which CSF-1–Fc accelerated inflammation resolution, we tracked the ontogeny of CD64hiSiglecF– macrophages in CD45.1+CD45.2+ C57BL6 mice reconstituted with CD45.2+ BM cells after lung-protected, single-dose irradiation. Then 8 weeks after BM reconstitution (Extended Data Fig. 6i), the mice were challenged with LPS, housed in normoxia or hypoxia for 5 d and treated with PBS or CSF-1–Fc daily. In keeping with increased recruitment from the blood, the proportion of CD64hiSiglecF–Ly6C+ MDMs derived from donor CD45.2+ BM cells in LPS-treated mice paralleled that seen in the blood (Fig. 7d,e). In the absence of LPS challenge, the proportion of CD45.2+CD64hiSiglecF−Ly6C− macrophages relative to the CD45.2+CD115+CD11b+ blood monocyte pool was ~20%, indicating that maintenance of this subset was dependent on blood monocytes (Fig. 7e)35. In LPS-treated mice housed in normoxia, the proportion of CD45.2+CD64hiSiglecF−Ly6C− macrophages relative to CD45.2+CD115+CD11b+ blood monocytes was ~80% (Fig. 7e), suggesting that the expansion of this population during inflammation was predominantly through recruitment of blood monocytes. The chimerism of this population was ~60% in hypoxic, LPS-challenged, PBS-treated mice and ~80% in hypoxic, LPS-challenged mice treated with CSF-1–Fc (Fig. 7e), indicating that CSF-1–Fc replenished the number of CD64hiSiglecF– lung macrophages in LPS-treated hypoxic mice predominantly through increased recruitment of circulating CD115+CD11b+ monocytes.
In LPS-treated hypoxic mice, CSF-1–Fc treatment also normalized the hypoxic suppression of the type I IFN-associated gene Ccr5, without enhancing expression of Il1b (Fig. 7f). CSF-1–Fc significantly induced the expression of a number of genes that were suppressed in the ARDS patient samples, such as Itga5, Cd99, Sell and Anxa36 (Fig. 7g and Supplementary Table 1). A proteomic survey showed reduced abundance of secretory-granule proteins in circulating Ly6Chi monocytes of LPS-challenged, hypoxic, CSF-1-treated compared with PBS-treated counterparts (Fig. 7h), suggesting that CSF-1 altered their phenotype toward a less inflammatory profile. Increased numbers of lung MHC-II–Lyve-1+ macrophages, a subset of repair-associated macrophages reported in bleomycin-induced lung injury11, were observed in CSF-1–Fc-treated, hypoxic, LPS-challenged mice compared with their PBS counterparts (Fig. 7i and Extended Data Fig. 6j), with increased chimerism noted in the CSF-1–Fc-treated mice in the lung-protected chimera model (Fig. 7j)
To explore how CSF-1–Fc-mediated expansion of CD64hiSiglecF− macrophages facilitated neutrophil clearance, we investigated the expression of known mediators of efferocytosis in the blood and lung37. IL-10 was elevated in the serum of CSF-1–Fc-treated, hypoxic, LPS-challenged mice compared with the PBS-treated counterparts (Fig. 7k). Immunofluorescence staining showed increased numbers of IL-10-expressing interstitial F4/80+ cells (Fig. 7l and Extended Data Fig. 7a), and F4/80+Lyve1+ macrophages (Fig. 7l and Extended Data Fig. 7b) in the lung of hypoxic, LPS-challenged, CSF-1–Fc-treated mice compared with their PBS-treated counterparts. Sorted CD64hiSiglecF−MHC-II−Lyve1+ macrophages from these CSF-1–Fc-treated mice expressed the Il-10 transcript (Extended Data Fig. 8a). Transcriptional profiling indicated that CD64hiSiglecF−MHC-II−Lyve1+ macrophages from CSF-1–Fc-treated, LPS-challenged, hypoxic mice had lower expression of archetypal inflammatory genes (Il1b, Il6, Tnf and Il18 and S100a11) and genes associated with lung fibrosis (Mmp8, Mmp12, Col14a1, Fpr1, Pdgfa and Pdgfb) relative to CD64hiSiglecF−MHC-II− from PBS-treated counterparts. Furthermore, genes reported to be enriched in severe SARS-CoV2 infection in BAL monocytes or macrophages, such as CD14, CCL3 and S100A8 (ref. 38), were suppressed in the CD64hiSiglecF−MHC-II−Lyve1+ macrophages from the CSF-1–Fc-treated hypoxic mice compared to CD64hiSiglecF−MHC-II− macrophages from the PBS-treated counterparts (Fig. 7m), indicating that CSF-1 expanded the number of IL-10-producing macrophages and enabled resolution of hypoxia-driven inflammation.
Extended Data Fig. 7. Lung immunohistochemistry tiled separate-channel images with negative controls.
(a, b) Representative images of separate channels for stained tissue and negative controls (no primary antibody) for the indicated antibodies in lung sections from mice housed in hypoxia post-LPS and treated with either PBS or CSF1 as indicated (n = 3/ group). Scale bar represents 200μm.
Extended Data Fig. 8. Treatment with CSF-1-Fc elevates II10 expression in Lyve1+MHCII− macrophages and increases blood and lung monocyte numbers in Ifnar1–/– mice.
(a) II10 expression was measured by NanoString platform analysis in MHC− lung macrophages from LPS-challenged mice housed in hypoxia for 5 days and treated with PBS and compared to Lyve1+MHCII− of LPS-challenged micehoused in hypoxia and treated with CSF-1-Fc. (b, c) Ifnar1−/− mice were nebulized with LPS and treated with PBS or CSF-1-Fc. Mice were sacrificed on day (b) blood monocyte and (c) lung Ly6C+ monocytes were quantified by flow cytometry. (a–c) representative of 2 experiments. Statistical testing (a–c) unpaired, two-sided Student’s t-test.
Finally, we asked whether CSF-1 was sufficient to overcome the loss of CD64hiSiglecF–Ly6C+ MDMs and CD64hiSiglecF− macrophages observed in the absence of type I IFN signaling. Normoxic LPS-challenged Ifnar1 −/− mice treated with CSF-1–Fc (four daily injections) had increased numbers of CD115+CD11b+ monocytes in the blood, Ly6Chi monocytes (Extended Data Fig. 8b,c) and CD64hiSiglecF− macrophages in the lung (Fig. 7n) compared with those receiving PBS. In addition, these mice had increased lung CD64hiSiglecF−Lyve1+ macrophages (Fig. 7n) and BAL-recovered CD64hiSiglecF– MDM numbers (Fig. 7o), with enhanced return to baseline body weight at day 5 post-LPS challenge (Fig. 7p) compared with LPS-challenged, PBS-treated Ifnar1−/− mice.
Discussion
In the present study, we showed that monocyte recruitment and conversion into lung macrophages are required to drive inflammation resolution in hypoxic ALI. Hypoxemic mice with ALI demonstrated an increase in erythropoiesis, with an associated reduction in monopoiesis, monocytopenia and failure to expand the MDM and non-AM CD64hiSiglecF−macrophage compartment in the lung. In the context of prioritizing the preservation of tissue oxygen delivery, increased erythropoiesis makes physiological sense, such as in adaptation to altitude, where monocytopenia had been reported as early as 1969 (ref. 39). However, when engagement of an effective innate immune response is also required, our data demonstrated that hypoxia-induced immune changes observed in early disease have long-term consequences for inflammation resolution, such as persistence of neutrophilic inflammation, a well-known poor prognostic feature of ARDS3.
Monocytes are professional phagocytes and key mediators of the restoration of homeostasis. There is increasing appreciation that the phenotype of circulating monocytes can be predetermined by systemic cues affecting their BM progenitors40. The presence of a specific phenotypic profile in ARDS patients’ circulating monocytes, irrespective of the sampling timepoint, would be in keeping with alterations within their progenitors. It will be important, in future work, to explore whether the absence of systemic IFN-α, and suppressed IFNAR expression on BM LSK and circulating monocytes, as observed in our mouse model of hypoxic ALI and in hypoxemic patients with severe SARS-Cov2 infection41,42, may be sufficient to drive the phenotypic and functional changes observed in the circulating monocytes of a hypoxic ARDS cohort.
Expanded numbers of airway monocytes and macrophages in the BAL of patients with severe SARS-Cov2 infection have been reported38, although it is unclear whether this expansion is promoting or limiting disease pathogenesis. An important limitation of our present study is the inability to sample the lung macrophage compartment in patients with ARDS. Transcriptomic survey of the airway monocytes and macrophages in patients with severe SARS-Cov2 infection identified an enrichment of markers of immaturity, inflammatory proteins and cytokines38. We show that treatment with CSF-1 increased the number of CD64hiSiglecF− macrophages and monocyte differentiation toward CD64hiSiglecF−MHC−Lyve1+ macrophages, a cell type that has tissue-repair roles in various disease contexts43, including in the lung11. In addition, CSF-1–Fc suppressed the expression of several genes reported to be enriched in the BAL of patients with severe SARS-Cov2 infection38. These findings, compounded by the effect of CSF-1–Fc on accelerating inflammation resolution and reducing lung injury, underscored the therapeutic potential of CSF-1 in ARDS. The use of other growth factors has been trialed in ARDS without success, with systemic-44 or lung-delivered45 granulocyte–macrophage colony-stimulating factor (GM-CSF) failing to improve ARDS mortality. Although GM-CSF plays a key role in AM homeostasis, its pleotropic nature means that it can also act as a neutrophil chemoattractant and growth factor46, with neutrophils pathogenic in the context of ARDS and hypoxia promoting neutrophil survival and proinflammatory function32,33. On the other hand, monocytes and MDMs can directly inhibit neutrophil-mediated damage to the host47. In our system, CSF-1–Fc treatment expanded the CD64hiSiglecF− macrophage compartment and led to an increase in lung IL-10+ macrophages, with a concomitant increase in systemic IL-10, and reduced numbers of neutrophils in the airspace and the lung. Macrophages are known to release IL-10 on efferocytosis, which drives the resolution of inflammation37. Furthermore, IL-10-producing lung IMs are key regulators of both allergic8,9 and endotoxin-mediated lung injury10. These findings strengthen the case of the therapeutic potential of CSF-1 in human ARDS.
Methods
Resources availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, A.M. (Ananda.Mirchandani@ed.ac.uk).
Human healthy control blood donors
Patients with ARDS were recruited and informed consent obtained directly or by proxy under the ‘META-CYTE’ study (17/SS/0136/AM01) and ‘ARDS-NEUT’ study (20/SS/0002), as approved by the Scotland A Research Ethics Committee. Samples were also obtained under the ‘Effects of Critical Illness on the Innate Immune System’ study as approved by Health Research Authority (REC no. 18/NE/0036).
All healthy participants gave written informed consent in accordance with the Declaration of Helsinki principles, with ACCORD Medical Ethics Research Committee approval for the study of healthy human volunteers through the University of Edinburgh Centre for Inflammation Research blood resource (15-HV-013).
Up to 20–40 ml of whole blood was collected into citrate tubes and up to 10 million cells were stained for flow cytometry assessment and sorting. Briefly, the whole blood was treated with red cell lysis buffer (Invitrogen) and cells counted before staining for flow cytometry. Cells were incubated with anti-CD16/32 Fc block (2:50) for 30 min, followed by staining for 30 min with antibodies (Table 2) followed by a wash with FACS buffer (PBS + 2% fetal calf serum (FCS)). DAPI (1:1,000) was added before flow cytometry to determine live cells. Monocytes were identified as Singles Dapi−CD45+ nongranulocyte Lin(CD3/CD56/CD19±CD66b)− HLADR+ CD14+ and/or CD16+ cells.
Table 2.
List of antibodies
| Antibody | Clone no. | Catalog no. | Lot no. | Fluorophore | Source | Dilution |
|---|---|---|---|---|---|---|
| CD16 | eBioCD16 | 1-9161-71 | 4304474 | FITC | eBioscience | 1:20 |
| CD3 | OKT3 | 317308 | B256076 | PE | BioLegend | 1:80 |
| CD56 | HCD56 | 318306 | B252053 | PE | BioLegend | 1:80 |
| CD19 | HIB19 | 302254 | B227178 | PE | BioLegend | 1:200 |
| CCR2 | K036C2 | 357212 | B260108 | PE/Cy7 | BioLegend | 1:80 |
| ICAM | HCD54 | 322718 | B193832 | AF647 | BioLegend | 1:80 |
| CD45 | 2D1 | 368514 | B248834 | AF700 | BioLegend | 1:20 |
| CD14 | M5E2 | 301820 | B274258 | APC/Cy7 | BioLegend | 1:20 |
| HLA-DR | L243 | 307624 | B278326 | Pacific Blue | BioLegend | 1:20 |
| CD66b | G10F5 | 305106 | B278603 | PE | BioLegend | 1:20 |
| SiglecF | E50-2440 | 552126 | 7058859 | PE | BD Biosciences | 1:200 |
| CD11b | M1/70 | 101256 | B238075 | PE Dazzle | BD Biosciences | 1:400 |
| CD11b | M1/70 | 101243 | B253527 | BV785 | Biolegend | 1:200 |
| MHC-II | M5.114.15.2 | 107624 | B267551 | PerCP | eBioscience | 1:200 |
| Epcam | G8.8 | 118230 | B251914 | APCCy7 Fire | BioLegend | 1:200 |
| CD3 | 17A2 | 100244 | B198733 | BIOTIN | BioLegend | 1:200 |
| CD3 | 17A2 | 100213 | B261416 | Pacific Blue | BioLegend | 1:200 |
| CD3 | 17A2 | 100229 | B282101 | BV650 | BioLegend | 1:200 |
| CD3 | 17A2 | 100206 | B210714 | PE | BioLegend | 1:200 |
| CD19 | 6D5 | 115541 | B242632 | BV650 | BioLegend | 1:200 |
| CD19 | 6D5 | 115504 | B244881 | Biotin | BioLegend | 1:200 |
| CD19 | 6D5 | 115526 | B265435 | Pacific Blue | BioLegend | 1:200 |
| CD19 | 6D5 | 115508 | B223615 | PE | BioLegend | 1:200 |
| CD103 | 2E7 | 121433 | BV605 | BioLegend | 1:400 | |
| Ly6G | 1A8 | 127604 | B218526 | Biotin | BioLegend | 1:200 |
| Ly6G | 1A8 | 127608 | B221647 | PE | BioLegend | 1:200 |
| Ly6G | 1A8 | 127628 | B280589 | BV421 | BioLegend | 1:200 |
| Ly6G | 1A8 | 135512 | B213676 | AF488 | BioLegend | 1:200 |
| Lyve-1 | ALY7 | 50-0443-82 | 2205461 | eFluor 660 | eBioscience | 1:200 |
| CD115 | AFS98 | 135510 | B211309 | APC | BioLegend | 1:200 |
| CD115 | AFS98 | 128006 | B217035 | FITC | BioLegend | 1:200 |
| Ly6C | HK1.4 | 128032 | B232012 | BV421 | BioLegend | 1:200 |
| Pan-CD45 | 30-F11 | 103128 | B274307 | AF700 | BioLegend | 1:200 |
| CD11c | N418 | 117318 | B222652 | PE/Cy7 | BioLegend | 1:200 |
| CD11c | N418 | 117352 | B218048 | APC/Fire750 | Biolegend | 1:200 |
| CD64 | X54-5/7.1 | 139304 | B191540 | PE | BioLegend | 1:200 |
| CD64 | X54-5/7.1 | 139306 | B207411 | APC | BioLegend | 1:200 |
| CD4 | H129.19 | 553649 | Biotin | BD Biosciences | 1:1,600 | |
| CD5 | 53-7.3 | 553019 | Biotin | BD Biosciences | 1:800 | |
| CD5 | 53-7.3 | 100603 | B254317 | Biotin | BioLegend | 1:200 |
| CD8a | 53 -6.7 | 553029 | Biotin | BD Biosciences | 1:800 | |
| CD11b | M1/70 | 101256 | B238075 | Biotin | BD Biosciences | 1:200 |
| CD45R/B220 | RA3-6B2 | 553086 | Biotin | BD Biosciences | 1:200 | |
| Ter119 | TER-119 | 116204 | B295203 | Biotin | BioLegend | 1:200 |
| Ter119 | TER-119 | 553672 | Biotin | BD Biosciences | 1:50 | |
| Gr-1/Ly-6G/C | RB6-8C5 | 553125 | Biotin | BD Biosciences | 1:100 | |
| CD117/cKit | 2B8 | 105811 | B249345 | APC | BioLegend | 1:200 |
| Sca-1/Ly- | E13-161.7 | 122506 | FITC | BioLegend | 1:200 | |
| CD48 | HM48-1 | 103406 | PE | BioLegend | 1:500 | |
| CD150 | 12F12.2 | 115914 | PECy7 | BioLegend | 1:200 | |
| CD71 | RI7217 | 113807 | PE | BioLegend | 1:500 | |
| Fc Block CD16/32 | 93 | 101320 | B295040 | BioLegend | 1:100 | |
| Streptavidin | – | 405232 | B251688 | BV650 | BioLegend | 1:1,000 |
| Streptavidin | – | Pacific Blue | BD biosciences | |||
| LIVE/DEAD Fixable Aqua | – | L34957 | 2068285 | UV650 | Life Technologies or BioLegend | 1:50–1:100 |
| CD45.2 | 104 | 109822 | B252126 | AF700 | BioLegend | 1:200 |
| CD45.1 | A20 | 110741 | B253101 | BV510 | BioLlegend | 1:200 |
| Sca.1 | D7 | 108129 | B262926 | BV510 | BioLegend | 1:200 |
| CD150 | TC15-12F12.2 | 115903 | PE | BioLegend | 1:200 | |
| CD105 | MJ7/18 | 120412 | B245562 | PacBlue | BioLegend | 1:200 |
| CD41 | MWReg30 | 133927 | B268849 | APCCy7 | BioLegend | 1:200 |
| IFNAR | MAR1-5A3 | 127325 | B286788 | PECy7 | BioLegend | 1:200 |
| CD5 | 53-7.3 | 100603 | B254317 | Biotin | BioLegend | 1:200 |
| Ly6G | 1A8 | 127604 | B218529 | Biotin | Biolegend | 1:200 |
| B220 | RA3-6B2 | 103204 | B288658 | Biotin | BioLegend | 1:200 |
| CD11b | M1/70 | 562287 | CF594 | BioLegend | 1:200 | |
| F480 | CI:A3-1 | ab6640 | Purified | Abcam | 1:100 | |
| IL-10 | JES5-2A5 | ab189392 | Purified | Abcam | 1:100 | |
| LYVE-1 | Polyclonal | 103-PA50AG | Purified | ReliaTech GmbH | 1:200 | |
| HIF-1α | Polyclonal | NB100-479 | Purified | Novus Biotech | 1:00 | |
| CD11b | M1/70 | 101243 | B287244 | BV785 | BioLegend |
Samples obtained from April 2020 were fixed before acquisition given the potential for SARS-Cov2 dissemination. Briefly, 1 μl of Zombie Aqua fixable viability dye (stock 1:20 dilution) was added to 100 μl of whole blood for 15 min at room temperature in the dark. Then, 2 μl of Fc Block was added for a further 30 min, on ice. Samples were then stained as above and fixed/lysed using BD FACS Lyse for 10 min at room temperature. The sample was then resuspended in 300 μl of FACS buffer and 50 μl of Countbright beads added (Thermo Fisher Scientific) before acquisition.
Mice
Male C57/BL6J mice aged 8–15 weeks were purchased from Envigo or Charles River. Ifnar1−/− (ifnar tm/agt) mice were obtained from J.S. who purchased them originally from the Jackson Laboratory. Animal experiments were conducted in accordance with the UK Home Office Animals (Scientific Procedures) Act of 1986 with local ethical approval.
Mouse LPS ALI model
Mice were treated with nebulized LPS (3 mg) and then housed in normoxia or hypoxia (10% O2) immediately thereafter for up to 5 d. Mice were treated daily (days 1–4 post-LPS), by subcutaneous injection, with PBS or 0.75 mg kg−1 of porcine CSF-1 fused to the Fc region of porcine IgG1a (generated by David Hume), prior to cull on day 5.
D39 S. pneumoniae infection
Mice were anesthetized and 107 colony-forming units (c.f.u.) (or vehicle) was delivered in 50 μl of PBS via intratracheal intubation. After reversal of the anesthetic and a period of recovery, the mice remained in normoxia or were placed in hypoxia.
Influenza A (PR8) virally induced ALI model
Mice were lightly anesthetized using isoflurane and 20 plaque-forming units (p.f.u.) of PR8 influenza A virus in Dulbecco’s modified Eagle’s medium (DMEM) was inoculated intranasally. After 1 h of recovery time, mice were placed in hypoxia for 48 h. Subcutaneous PBS or CSF-1–Fc injections (as above) at 12 h and 36 h were administered. Sickness scores were determined using methods described previously32.
Lung and alveolar cell sampling
Mice were culled with an overdose of intraperitoneal anesthetic (Euthetal) followed by blood collection from the inferior vena cava. Alveolar leukocytes were collected by BAL, then mice were perfused lightly with PBS through the heart, before harvesting the lung tissue. On occasion, the lower limbs were harvested for BM leukocyte assessment (see below).
Tissue leukocytes were extracted from surgically dissociated lung tissue by enzymatic digestion with 2 ml of enzyme mix (RPMI with 0.625 mg ml−1 of Collagenase D (Roche), 0.85 mg ml−1 of Collagenase V (Sigma-Aldrich), 1 mg ml−1 of dispase (Gibco, Invitrogen) and 30 U ml−1 of DNase (Roche Diagnostics GmbH)) for 45 min at 37 °C in a shaking incubator. The digest material was passed through a 100-μm cell strainer with the addition of FACS buffer (PBS with 0.5% BSA/2% FCS and 0.02 mM EDTA). Cell pellets were treated with red cell lysis buffer (Sigma-Aldrich) and washed in FACS buffer. The resulting cell suspension was subsequently passed through a 40-μm strainer before cell counting using a Casey TT counter (Roche). Single-cell suspensions (5 million cells per sample) were then stained for flow cytometry. BAL samples were counted before staining for flow cytometry.
Blood and BM sampling
Mouse blood and BM were treated with RBC lysis buffer (BioLegend) before counting and staining for flow cytometry (Table 2).
Hematopoietic cell assessment was performed using both hind legs, which were crushed using a pestle and mortar until a homogeneous cell suspension was achieved, or flushed though using a 32G needle. Cells were collected in cold FACS buffer and filtered through a 70-μm nylon strainer (BD Falcon, catalog no. 352340). Cells were treated with RBC lysis buffer (BioLegend) before staining.
Tissue-protected chimeras
C57BL/6J CD45.1+CD45.2+ mice aged 6–8 weeks were anesthetized and irradiated with a single dose of 9.5-Gy γ-irradiation, with all but the hind legs and lower abdomen protected by a 5cm lead shield. The next day, the mice received 2 × 106–5 × 106 BM cells from CD45.2+ C57BL/6J by intravenous injection. The chimerism of blood monocytes (proportion of donor cells) was determined by flow cytometry in each individual mouse at day 5 and the chimerism in the lung macrophage populations (as described in the figures) was divided by this reference value, thereby determining the proportion of the cells that were of blood ontogeny.
Flow cytometry
Mouse cells were treated with α-CD16/32 Fc block (eBioscience) (1:100) before staining with antibodies (Table 2). Relevant fluorescence − 1 samples were used as controls. Zombie Aqua fixable viability dye (BioLegend) was used before Fc Block to exclude dead cells from digest samples or DAPI for single-cell suspensions.
Cells were acquired on the LSRFortessa (Becton Dickinson) or sorted on an Aria II or Fusion machine (Becton Dickinson). Compensation was performed using BD FACSDiva software and data analyzed in FlowJo v.10 or FCS Express 7 for t-distributed stochastic neighbor embedding analysis.
Gating strategies
Human monocytes: Singles Dapi−CD45+non-granulocyte Lin(CD3/CD56/CD19/CD66b)− HLADR+ CD14+ and/or CD16+ cells.
Mouse blood monocytes: Singles Dapi−CD45+ Lin(CD3/CD19/ Ly6G)−CD115+CD11b+Ly6Chi, Ly6Cint or Ly6C−.
Mouse blood neutrophils: Singles, Dapi−CD45+Ly6G+CD11b+Ly6Cint.
Mouse lung/BAL alveolar macrophages: Singles, Zombie Aqua−CD45+Lin (CD3/CD19/Ly6G)−CD64hiSiglecF+CD11c+.
Mouse lung interstitial /BAL inflammatory macrophages: Singles, Zombie Aqua−CD45+Lin (CD3/CD19/Ly6G)−CD64hiSiglecF−CD11c+/− then Ly6C+/−MHC-II+/−.
Lung classical monocytes: Singles, Zombie Aqua−SinglesCD45+Lin (CD3/CD19/Ly6G)−CD64lo CD11b+Ly6C+.
Lung/BAL neutrophils: Singles, Aqua or Dapi−, CD45+CD11b+Ly6G+.
Lung cDC1 subset: Zombie Aqua-Singles CD45+CD11chi,CD103+, CD64−MHC-II+.
BM HSPC SLAM analysis Alive: Singles LK (Lin-cKit+) and LSK (Lin-cKit+Sca-1+) cells. LSK cells were further sub-gated on hematopoietic stem cells (HSCs: LSK CD48−CD150+), multipotent progenitors (MPPs: LSK CD48−CD150−), HPC-1 (LSK CD48+CD150−) and HPC-2 (LSK CD48+CD150+).
BM erythroid progenitors based on Pronk analysis26: Singles, Dapi or Aqua−, Lin−, CD11b−, cKit+, Sca1−, CD32/16−, CD41−, CD105+ or CD150+ (pre-Meg-E CD150+CD105−, pre-CFU-E CD150+CD105+, CFU-E CD150−CD105+).
Further gating strategy information can be made available on request.
BAL/serum cytokine/chemokine quantification
BAL and serum supernatants were collected and stored at −80 °C until use. Cytokine and chemokine levels were measured using an MSD V-plex plate per the manufacturer’s instructions.
Lung injury measurements
IgM BAL levels were measured using the Ab133047 Abcam kit as per the manufacturer’s instructions.
BAL lactate dehydrogenase activity (measured as colorimetric reduction of NAD to NADH) was performed using Ab102526 (Abcam) as per the manufacturer’s instructions.
BAL total protein was measured using Pierce BCA Assay (Thermo Fisher Scientific) as per the manufacturer’s instructions.
In vitro BM culture
Naive WT C57BL/6 BM was obtained by flushing the femoral and tibial bones and RBCs were lysed. Cells were cultured in hypoxia (FiO2 1%) or normoxia (FiO2 21%) with conditioned DMEM for 1 h before the addition of IFN-β 10 ng ml−1 (RnD 8234-MB-010) for a further 3 h. Cell pellets were collected and QIAGEN RLT buffer added (containing 10 μl ml−1 of 2-mercaptoethanol). Pellets were snap-frozen and stored at −80 °C for RNA extraction.
RNA isolation and relative quantification
RNA was isolated from BM cells using the genomic DNA eliminator solution for purification of total RNA (RNeasy Plus Mini Kit, QIAGEN). Complementary DNA was synthesized using AMV reverse transcriptase with random primers (Promega). TaqMan gene expression assays (Applied Biosystems, Thermo Fisher Scientific) and PrimeTime qPCR Probe Assays (IDT) were used for relative quantification of cDNA using SDS 2.4 (Thermo Fisher Scientific) and normalized to ACTB expression.
Immunohistochemistry
Murine paraffin-embedded blocks were prepared from lungs fixed via the trachea with 10% buffered formalin. The lung sections were stained with anti-IL-10 (catalog no. ab189392, Abcam), anti-F4/80 (catalog no. ab6640, Abcam) or isotype control after deparaffinization and antigen retrieval. Antigen retrieval was performed by microwave heating in citric acid-based, antigen-unmasking solution (Vector, catalog no. H-3300-250). The following were used: tyramide signal amplification (TSA) plus system amplification (catalog no. NEL744B001KT, Perkin Elmer) and autofluorescence quenching with TrueView (Vector, catalog no. SP-8400). The nuclei were stained with DAPI (catalog no. 422801, Sigma-Aldrich). Images were obtained using EVOS FL Auto 2 (Invitrogen). All image acquisition and processing steps were performed using the same settings for both sample groups.
The lung sections were stained with anti-mouse LYVE-1 (catalog no. 103-PA50AG, ReliaTech GmbH) and anti-mouse F4/80 (catalog no. ab6640, Abcam) overnight at 4 °C after deparaffinization and antigen retrieval. Antigen retrieval was performed by microwave heating in citric acid-based, antigen-unmasking solution (Vector, catalog no. H-3300-250). The following were used: TSA plus system amplification (catalog no. NEL744B001KT, Perkin Elmer) and autofluorescence quenching with TrueView (Vector, catalog no. SP-8400) according to the manufacturer’s instructions. The nuclei were stained with DAPI (catalog no. 422801, Sigma-Aldrich). Images were acquired using a EVOS FL Auto 2 (Invitrogen).
All image acquisition and processing steps were performed using the same settings for both sample groups.
The nCounter NanoString platform analysis
For human monocytes, 5,000 HLADR++ cells were sorted using the aforementioned human monocyte gating strategy directly into 2 μl of RLT buffer using a BD Fusion Sorter (patients 4–8 were sampled). 5,000 mouse classical monocytes were sorted from mice treated with LPS and housed in normoxia, hypoxia and hypoxia + CSF-1 gating on single DAPI−CD45+Lin−CD115+Ly6Chi cells into 2 μl of RLT. Cell pellets were vortexed and centrifuged before immediate freezing until ready for processing. NanoString gene expression plates of human and mouse myeloid inflammation were run as per the manufacturer’s instructions at the University of Edinburgh HTPU Centre within the MRC Institute of Genetics and Molecular Medicine/Cancer Research UK Edinburgh Centre.
Proteomic analysis
Sorted classical monocytes were processed for proteomics using the ‘in-cell digest’, as described by Kelly et al.48, resuspended in digestion buffer (0.1 M triethylammonium bicarbonate + 1 mM MgCl2) and digested with benzonase (>99%, Millipore) for 30 min at 37 °C, followed by trypsin (Thermo Fisher Scientific, 1:50 w:w protein) overnight at 37 °C. A second aliquot of trypsin (1:50) was subsequently added and incubated at 37 °C for 4 h. A minimum of 25 ng of trypsin was added. Digests were acidified and desalted using StageTips49 and subjected to either tip-based fractionation or direct analysis by liquid chromatography–tandem mass spectroscopy (LC–MS/MS).
After digestion, and to generate the reference spectral library, peptides were subjected to reverse-phase, high pH, tip fractionation following the general guidelines described by Rappsilber et al.49. In brief, tips for fractionation were made using three SDB-XC disks (Merck) per tip. The tip was cleaned and conditioned using, sequentially, methanol, 80% acetonitrile (MeCN) (Thermo Fisher Scientific) in 0.1% NH4OH (v:v), and 0.1% NH4OH (52 mM) (v:v). Peptides, also resuspended in 0.1% NH4OH, pH 10, were spun through the SDB-XC disks and the flow-through was collected, acidified and concentrated on C-18 StageTips before being subjected to MS analysis. Fractionation was then achieved by sequential elution with 7%, 14%, 21%, 28%, 35%, 55% and 80% MeCN in 0.1% NH4OH. Fractions were then dried at ambient temperature (Concentrator 5301, Eppendorf) and prepared for MS analysis by resuspension in 6 μl of 0.1% trifluoroacetic acid (TFA).
Data-dependent acquisition LC–MS analyses were performed on an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific) coupled, on-line, to an Ultimate 3000 HPLC (Dionex, Thermo Fisher Scientific). Peptides were separated on a 50-cm (2-µm particle size) EASY-Spray column (Thermo Fisher Scientific), which was assembled on an EASY-Spray source (Thermo Fisher Scientific) and operated constantly at 50 °C. Mobile phase A consisted of 0.1% formic acid in LC–MS-grade water and mobile phase B consisted of 80% acetonitrile and 0.1% formic acid. Peptides were loaded on to the column at a flow rate of 0.3 μl min−1 and eluted at a flow rate of 0.25 μl min−1 according to the following gradient: 2–40% mobile phase B in 120 min and then to 95% in 11 min. Mobile phase B was retained at 95% for 5 min and returned back to 2% a minute after until the end of the run (160 min in total).
The spray voltage was set at 2.2 kV and the ion capillary temperature at 280 °C. Survey scans were recorded at 60,000 resolution (scan range 400–1,600 m/z) with an ion target of 1.0 × 106 and injection time of 50 ms. MS2 was performed in the orbitrap (resolution at 15,000), with an ion target of 5.0 × 104 and higher-energy C-trap dissociation (HCD) fragmentation50 with a normalized collision energy of 27. The isolation window in the quadrupole was 1.4 Thomson. Only ions with a charge between 2 and 6 were selected for MS2. Dynamic exclusion was set at 60 s. The cycle time was set at 3 s.
Samples subjected to data-independent acquisition (DIA) were prepared for MS analysis by resuspension in 0.1% TFA. MS analyses were performed on an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific). LC conditions (instrumentation, column and gradient) were the same as described above.
Survey scans were performed at 15,000 resolution, with a scan range of 350–1,500 m/z, maximum injection time 50 ms and AGC target 4.5 × 105. MS/MS DIA was performed in the orbitrap at 30,000 resolution with a scan range of 200–2,000 m/z. The mass range was set to ‘normal’, the maximum injection time to 54 ms and the AGC target to 2.0 × 105. The inclusion mass list with the corresponding isolation windows is shown in Table 3. Data for both survey and MS/MS scans were acquired in profile mode. A blank sample (0.1% TFA, 80% MeCN, 1:1 v:v) was run between each sample to avoid carryover.
Table 3.
List of masses, default charge states and isolation windows used for data independent acquisition
| m/z | z | Time start (min) | Time stop (min) | Isolation window (m/z) |
|---|---|---|---|---|
| 410 | 3 | 0 | 155 | 20 |
| 430 | 3 | 0 | 155 | 20 |
| 450 | 3 | 0 | 155 | 20 |
| 470 | 3 | 0 | 155 | 20 |
| 490 | 3 | 0 | 155 | 20 |
| 510 | 3 | 0 | 155 | 20 |
| 530 | 3 | 0 | 155 | 20 |
| 550 | 3 | 0 | 155 | 20 |
| 570 | 3 | 0 | 155 | 20 |
| 590 | 3 | 0 | 155 | 20 |
| 610 | 3 | 0 | 155 | 20 |
| 630 | 3 | 0 | 155 | 20 |
| 650 | 3 | 0 | 155 | 20 |
| 670 | 3 | 0 | 155 | 20 |
| 690 | 3 | 0 | 155 | 20 |
| 710 | 3 | 0 | 155 | 20 |
| 730 | 3 | 0 | 155 | 20 |
| 750 | 3 | 0 | 155 | 20 |
| 770 | 3 | 0 | 155 | 20 |
| 790 | 3 | 0 | 155 | 20 |
| 820 | 3 | 0 | 155 | 40 |
| 860 | 3 | 0 | 155 | 40 |
| 910 | 3 | 0 | 155 | 60 |
| 970 | 3 | 0 | 155 | 60 |
MS raw data files were processed using Spectronaught v.14.7.201007.47784 with either a human or a mouse reference FASTA sequence from UniProt, using default search parameters. The resulting protein-level data were analyzed using R v.3.5.0. Protein parts per million (p.p.m.) intensities were calculated by dividing the mean p.p.m. intensities between conditions (for example, for the human monocyte samples, ARDS patients and healthy controls), the P values were calculated using a Student’s t-test on log(transformed p.p.m. intensities). Proteins were designated as significantly changing if they showed P values <0.05 and fold-changes exceeding 1.96 s.d. away from the mean (that is, z-score >1.96). Only proteins that were quantified in all samples are shown in the volcano plot.
Gene expression analysis
Normalization of data was carried out using the geNorm selection of housekeeping genes function on NanoString nCounter analysis software. The resulting log2(normalized values) were used in subsequent analyses. Differential genes (‘DE genes’) were defined as genes with log2(fold-change) > 1, P < 0.05 across sample groups. Hierarchical clustering of sets of DE genes was carried out using Euclidian and Ward methods based on Pearson’s correlation values across transcriptional scores. The z-score scalar normalization of data was applied to the data before plotting as heatmaps. Analyses, including the drawing of heatmaps and volcano plots, were carried out in R using the package ggplot2 (https://cran.r-project.org/web/packages/ggplot2/index.html). Analysis of datasets was carried out by Thomson Bioinformatics, Edinburgh, UK.
Quantification, statistical analysis and reproducibility
Statistical tests were performed using Prism 8.00 and 9.0.2 software (GraphPad Software Inc.) (specific tests detailed in figure legends). Significance was defined as a P < 0.05 (after correction for multiple comparisons where applicable). Sample sizes (with each n number representing a different blood donor for human cells or an individual mouse for animal experiments) are shown in each figure.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41590-022-01216-z.
Supplementary information
Supplementary Table 1.
Acknowledgements
We thank D. Hume for providing the CSF-1–Fc used in these experiments. We thank Thomson Bioinformatics, Edinburgh, UK, for analyzing the NanoString data. Flow cytometry data were generated with support from the QMRI Flow Cytometry and Cell Sorting Facility, University of Edinburgh. We thank the Royal Infirmary of Edinburgh Critical Care Research Team for their assistance in recruiting, consenting and obtaining samples from patients with ARDS. This work was funded by a Wellcome Trust Senior Clinical fellowship awarded to S.R.W. (grant nos. 098516 and 209220), Wellcome Trust Post-doctoral Training Clinical Fellowship (grant no. 110086) and a Wellcome Trust iTPA grant (no. PIII052) awarded to A.S.M., and was partly funded by UK Research and Innovation/National Institute for Health and Care Research (NIHR) through the UK Coronavirus Immunology Consortium. C.C.B holds a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (grant no. 206234/Z/17/Z). K.K.’s laboratory is supported by grants from Cancer Research UK (nos. C29967A/14633 and C29967/A26787), MRC, the Bart’s Charity, the Kay Kendall Leukaemia Fund and Blood Cancer UK. A.J.S. is an NIHR Senior Investigator. The views expressed in this article are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted manuscript version from this submission.
Extended data
Author contributions
A.S.M., S.J.J., C.C.B., H.L., K.K., S.J.J., M.K.W. and S.R.W. conceived and designed the experiments. A.S.M, C.C.B., H.L., P.C., F.M, M.A.S.-G., L.R., T.M., S.A., R.D., E.R.W., G.C., L.D., D.G., I.H., J.S., A.Z., L.C., D.L., V.K., C.S., N.J.P. and P.S.-L. performed the experiments. A.S.M., C.C.B., M.A.S.-G., H.L., T.L. and G.C. analyzed the data. A.S.M., C.C.B., S.J.J., H.L., K.K., S.J.F., C.P., P.J.R., M.K.W. and S.R.W. interpreted the data. S.C, K.M.M., J.S., A.J.R., A.J.S. and D.H.D. facilitated obtaining patient samples. A.S.M. S.J.J., N.H., M.K.W. and S.R.W. helped obtain funding. J.K.B. and C.L. provided reagents. J.S. provided Ifnar1 knockout mice. A.S.M., S.J.J., C.C.B., H.L., K.K., C.W.P., S.J.F., M.K.W. and S.R.W. wrote the manuscript.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Ioana Visan, in collaboration with the Nature Immunology editorial team. Peer reviewer reports are available.
Data availability
The present study did not generate new unique reagents. All NanoString data shown in this manuscript have been deposited in the Gene Expression Omnibus (GEO) at accession nos.: GSE200429, GSE200549, GSE200558. All proteomic data generated in this project have been deposited in Pride at accession nos.: PXD033151. Analyses, including the drawing of heatmaps and volcano plots, were carried out in R using the package ggplot2 (https://cran.r-project.org/web/packages/ggplot2/index.html). Analysis of datasets was carried out by Thomson Bioinformatics, Edinburgh, UK.
Competing interests
A.S.M., S.R.W., M.K.W., S.J.F. and S.J.J. have filed a patent for the use of CSF-1 as a therapy in ARDS with the University of Edinburgh (PCT/GB/2020/051184).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Stephen J. Jenkins, Calum C. Bain.
Change history
7/19/2022
A Correction to this paper has been published: 10.1038/s41590-022-01286-z
Extended data
Extended data are available for this paper at 10.1038/s41590-022-01216-z.
Supplementary information
The online version contains supplementary material available at 10.1038/s41590-022-01216-z.
References
- 1.Bellani G, et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. J. Am. Med. Assoc. 2016;315:788–800. doi: 10.1001/jama.2016.0291. [DOI] [PubMed] [Google Scholar]
- 2.Fan E, et al. An official American Thoracic Society/European Society of intensive care medicine/society of critical care medicine clinical practice guideline: mechanical ventilation in adult patients with acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2017;195:1253–1263. doi: 10.1164/rccm.201703-0548ST. [DOI] [PubMed] [Google Scholar]
- 3.Steinberg KP, et al. Evolution of bronchoalveolar cell populations in the adult respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 1994;150:113–122. doi: 10.1164/ajrccm.150.1.8025736. [DOI] [PubMed] [Google Scholar]
- 4.Park WY, et al. Cytokine balance in the lungs of patients with acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2001;164:1896–1903. doi: 10.1164/ajrccm.164.10.2104013. [DOI] [PubMed] [Google Scholar]
- 5.Tarling JD, Lin HS, Hsu S. Self-renewal of pulmonary alveolar macrophages: evidence from radiation chimera studies. J. Leukoc. Biol. 1987;42:443–446. doi: 10.1002/jlb.42.5.443. [DOI] [PubMed] [Google Scholar]
- 6.Guilliams M, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedoret D, et al. Lung interstitial macrophages alter dendritic cell functions to prevent airway allergy in mice. J. Clin. Invest. 2009;119:3723–3738. doi: 10.1172/JCI39717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabatel C, et al. Exposure to bacterial CpG DNA protects from airway allergic inflammation by expanding regulatory lung interstitial macrophages. Immunity. 2017;46:457–473. doi: 10.1016/j.immuni.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Zhou B, et al. The angiocrine Rspondin3 instructs interstitial macrophage transition via metabolic-epigenetic reprogramming and resolves inflammatory injury. Nat. Immunol. 2020;21:1430–1443. doi: 10.1038/s41590-020-0764-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakarov, S. et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science363, eaau0964 (2019). [DOI] [PubMed]
- 12.Patel AA, et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J. Exp. Med. 2017;214:1913–1923. doi: 10.1084/jem.20170355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosco MC, et al. Hypoxia modifies the transcriptome of primary human monocytes: modulation of novel immune-related genes and identification of CC-chemokine ligand 20 as a new hypoxia-inducible gene. J. Immunol. 2006;177:1941–1955. doi: 10.4049/jimmunol.177.3.1941. [DOI] [PubMed] [Google Scholar]
- 14.Mimura I, et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol. Cell Biol. 2012;32:3018–3032. doi: 10.1128/MCB.06643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen WK, et al. CREB negatively regulates IGF2R gene expression and downstream pathways to inhibit hypoxia-induced H9c2 cardiomyoblast cell death. Int. J. Mol. Sci. 2015;16:27921–27930. doi: 10.3390/ijms161126067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abu-El-Rub, E. et al. Hypoxia-induced shift in the phenotype of proteasome from 26S toward immunoproteasome triggers loss of immunoprivilege of mesenchymal stem cells. Cell Death Dis. 11, 419 (2020). [DOI] [PMC free article] [PubMed]
- 17.Sammarco MC, Ditch S, Banerjee A, Grabczyk E. Ferritin L and H subunits are differentially regulated on a post-transcriptional level. J. Biol. Chem. 2008;283:4578–4587. doi: 10.1074/jbc.M703456200. [DOI] [PubMed] [Google Scholar]
- 18.Xu H, Manivannan A, Crane I, Dawson R, Liversidge J. Critical but divergent roles for CD62L and CD44 in directing blood monocyte trafficking in vivo during inflammation. Blood. 2008;112:1166–1174. doi: 10.1182/blood-2007-06-098327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schenkel AR, Mamdouh Z, Chen X, Liebman RM, Muller WA. CD99 plays a major role in the migration of monocytes through endothelial junctions. Nat. Immunol. 2002;3:143–150. doi: 10.1038/ni749. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Q, et al. The role of mitogen-activated protein kinase phosphatase-1 in the response of alveolar macrophages to lipopolysaccharide: attenuation of proinflammatory cytokine biosynthesis via feedback control of p38. J. Biol. Chem. 2005;280:8101–8108. doi: 10.1074/jbc.M411760200. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharyya S, Borthakur A, Dudeja PK, Tobacman JK. Lipopolysaccharide-induced activation of NF-κB non-canonical pathway requires BCL10 serine 138 and NIK phosphorylations. Exp. Cell Res. 2010;316:3317–3327. doi: 10.1016/j.yexcr.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aggarwal, N. R., King, L. S. & D’Alessio, F. R. Diverse macrophage populations mediate acute lung inflammation and resolution. Am. J. Physiol. Lung Cell. Mol. Physiol.306, L709–L725 (2014). [DOI] [PMC free article] [PubMed]
- 23.Meduri GU, et al. Persistent elevation of inflammatory cytokines predicts a poor outcome in ARDS: plasma IL-1β and IL-6 levels are consistent and efficient predictors of outcome over time. Chest. 1995;107:1062–1073. doi: 10.1378/chest.107.4.1062. [DOI] [PubMed] [Google Scholar]
- 24.Bain CC, et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6C hi monocyte precursors. Mucosal Immunol. 2013;6:498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pronk CJH, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Paul SR, et al. Molecular cloning of a cDNA encoding interleukin 11, a stromal cell-derived lymphopoietic and hematopoietic cytokine. Proc. Natl Acad. Sci. USA. 1990;87:7512–7516. doi: 10.1073/pnas.87.19.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onnis B, Fer N, Rapisarda A, Perez VS, Melillo G. Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells. J. Clin. Invest. 2013;123:1615–1629. doi: 10.1172/JCI59623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lasseaux C, Fourmaux MP, Chamaillard M, Poulin LF. Type I interferons drive inflammasome-independent emergency monocytopoiesis during endotoxemia. Sci. Rep. 2017;7:16935. doi: 10.1038/s41598-017-16869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Bruin AM, et al. IFNγ induces monopoiesis and inhibits neutrophil development during inflammation. Blood. 2012;119:1543–1554. doi: 10.1182/blood-2011-07-367706. [DOI] [PubMed] [Google Scholar]
- 31.Katlinski KV, et al. Inactivation of interferon receptor promotes the establishment of immune privileged tumor microenvironment. Cancer Cell. 2017;31:194–207. doi: 10.1016/j.ccell.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, A. A. R. et al. Hypoxia determines survival outcomes of bacterial infection through HIF-1α-dependent reprogramming of leukocyte metabolism. Sci. Immunol. 2, eaal2861 (2017). [DOI] [PMC free article] [PubMed]
- 33.Watts ER, et al. Hypoxia drives murine neutrophil protein scavenging to maintain central carbon metabolism. J. Clin. Invest. 2021;131:e134073. doi: 10.1172/JCI134073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gow DJ, et al. Characterisation of a novel Fc conjugate of macrophage colony-stimulating factor. Mol. Ther. 2014;22:1580–1592. doi: 10.1038/mt.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bain CC, et al. Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat. Commun. 2016;7:ncomms11852. doi: 10.1038/ncomms11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheikh MH, Solito E. Annexin A1: uncovering the many talents of an old protein. Int. J. Mol. Sci. 2018;19:1045. doi: 10.3390/ijms19041045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung EY, et al. Interleukin-10 expression in macrophages during phagocytosis of apoptotic cells Is mediated by homeodomain proteins Pbx1 and Prep-1. Immunity. 2007;27:952–964. doi: 10.1016/j.immuni.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 39.Hannon JP, Shields JL, Harris CW. Effects of altitude acclimatization on blood composition of women. J. Appl. Physiol. 1969;26:540–547. doi: 10.1152/jappl.1969.26.5.540. [DOI] [PubMed] [Google Scholar]
- 40.Yáñez A, et al. Granulocyte-monocyte progenitors and monocyte-dendritic cell progenitors independently produce functionally distinct monocytes. Immunity. 2017;47:890–902.e4. doi: 10.1016/j.immuni.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas C, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hadjadj J, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alivernini S, et al. Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis. Nat. Med. 2020;26:1295–1306. doi: 10.1038/s41591-020-0939-8. [DOI] [PubMed] [Google Scholar]
- 44.Paine R, et al. A randomized trial of recombinant human granulocyte-macrophage colony stimulating factor for patients with acute lung injury. Crit. Care Med. 2012;40:90–97. doi: 10.1097/CCM.0b013e31822d7bf0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herold S, et al. Inhaled granulocyte/macrophage colony-stimulating factor as treatment of pneumonia-associated acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2014;189:609–611. doi: 10.1164/rccm.201311-2041LE. [DOI] [PubMed] [Google Scholar]
- 46.Laan M, et al. A role of GM-CSF in the accumulation of neutrophils in the airways caused by IL-17 and TNF-α. Eur. Respir. J. 2003;21:387–393. doi: 10.1183/09031936.03.00303503. [DOI] [PubMed] [Google Scholar]
- 47.Grainger JR, et al. Inflammatory monocytes regulate pathologic responses to commensals during acute gastrointestinal infection. Nat. Med. 2013;19:713–721. doi: 10.1038/nm.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelly, V., Al-Rawi, A., Lewis, D., Kustatscher, G. & Ly, T. Low cell number proteomic analysis using in-cell protease digests reveals a robust signature for cell cycle state classification. Mol. Cell Proteomics 21, 100169 (2022). [DOI] [PMC free article] [PubMed]
- 49.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 50.Olsen JV, et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Data Availability Statement
The present study did not generate new unique reagents. All NanoString data shown in this manuscript have been deposited in the Gene Expression Omnibus (GEO) at accession nos.: GSE200429, GSE200549, GSE200558. All proteomic data generated in this project have been deposited in Pride at accession nos.: PXD033151. Analyses, including the drawing of heatmaps and volcano plots, were carried out in R using the package ggplot2 (https://cran.r-project.org/web/packages/ggplot2/index.html). Analysis of datasets was carried out by Thomson Bioinformatics, Edinburgh, UK.