Abstract
The prevalence of the hemolytic enterotoxin complex HBL was determined in all species of the Bacillus cereus group with the exception of Bacillus anthracis. hblA, encoding the binding subunit B, was detected by PCR and Southern analysis and was confirmed by partial sequencing of 18 strains. The sequences formed two clusters, one including B. cereus and Bacillus thuringiensis strains and the other one consisting of Bacillus mycoides, Bacillus pseudomycoides, and Bacillus weihenstephanensis strains. From eight B. thuringiensis strains, the enterotoxin gene hblA could be amplified. Seven of them also expressed the complete HBL complex as determined with specific antibodies against the L1, L2, and B components. Eleven of 16 B. mycoides strains, all 3 B. pseudomyoides strains, 9 of 15 B. weihenstephanensis strains, and 10 of 23 B. cereus strains carried hblA. While HBL was not expressed in the B. pseudomycoides strains, the molecular assays were in accordance with the immunological assays for the majority of the remaining strains. In summary, the hemolytic enterotoxin HBL seems to be broadly distributed among strains of the B. cereus group and relates neither to a certain species nor to a specific environment. The consequences of this finding for food safety considerations need to be evaluated.
The Bacillus cereus group comprises four valid species, namely B. cereus, Bacillus thuringiensis, Bacillus mycoides, and Bacillus anthracis (25, 40, 46). Recently, a Bacillus pseudomycoides (38) and a psychrotolerant Bacillus weihenstephanensis (32) were described. These species share a significant degree of similarity as determined by DNA-DNA hybridization studies and sequence analysis of the 16S rDNA (4) and 23S rDNA (3). In addition, the analysis of a variety of biochemical characters indicates the closeness of these strains. However, there are some physiological parameters which are considered unique for the different species of the B. cereus group. B. anthracis expresses the anthrax toxin, which is encoded by the plasmids pXO1 and pXO2 (36), and can be identified by a randomly amplified polymorphic DNA marker (15). B. thuringiensis produces an insecticidal toxin and is widely used to control lepidopterans in agriculture and forestry programs (19, 23, 31). These plasmids can easily be transformed into B. cereus, making B. cereus look like B. anthracis or B. thuringiensis (5, 24). Little is known about B. mycoides (20) and B. pseudomycoides (38). Their most remarkable phenotype is the rhizobial growth pattern on agar plates, and they can be distinguished from one another by their fatty acid composition (38). The newly described B. weihenstephanensis is defined by its ability to grow at and below temperatures of 7°C (32). The psychrotolerant B. weihenstephanensis can be discriminated from the mesophilic B. cereus by sequence differences in the 16S rDNA (32, 48) and the major cold shock protein gene cspA (21).
B. cereus is known to produce an emetic toxin, which causes vomiting, and at least three different enterotoxins, which cause diarrhea (for a review, see reference 26). The emetic toxin is named cereulide and consists of a ring structure of three repeats of four amino and/or oxy acids (d-O-Leu-d-Ala-l-O-Val-l-Val)3. Chemically, it is closely related to the ionophore valinomycin (1).
The best investigated enterotoxin is HBL, a three-component hemolysin (7, 8). It consists of two lytic components (L1 and L2) and a binding component (B). This toxin has hemolytic, dermonecrotic, and vascular permeability activities and is considered the primary virulence factor in B. cereus diarrhea (9). The hbl operon includes the three known proteins, the lytic components L2 and L1, encoded by hblC and hblD, and the binding component B, encoded by hblA (27, 42). Another enterotoxin gene, bceT, has been cloned and sequenced (2). It encodes only one component. This toxin exhibits Vero cell cytotoxicity and was positive in vascular permeability assays, but is not hemolytic. Besides these two enterotoxins, B. cereus produces another three-component enterotoxin, called NHE (34, 35). The three components are different from the HBL. The toxin is nonhemolytic, but it has also been involved in food poisoning (35).
There have been some indications that toxins are produced by B. thuringiensis and B. mycoides. Budarina and coworkers (11) found a hemolysin II that was more characteristic for B. thuringiensis than for B. cereus. Damgaard (16) observed diarrheal enterotoxin production by strains of B. thuringiensis isolated from commercial B. thuringiensis-based insecticides. Beattie and Williams (6) detected toxigenic strains of B. thuringiensis and B. mycoides in a cytotoxicity assay. The prevalence of B. cereus-type enterotoxins in natural isolates of B. thuringiensis was also observed by Perani et al. (39). In addition, B. thuringiensis strains were isolated during a gastroenteritis outbreak in Canada (29). However, the exact nature of the toxic compounds is not very well understood in some of these studies. Also, there is, to our knowledge, no information available about the production of toxins in B. pseudomycoides and B. weihenstephanensis. Toxin production of the widely used B. thuringiensis and the prevalent soil bacterium B. mycoides are of considerable importance for food microbiology. Therefore, this study focuses on the production of the hemolytic enterotoxin complex HBL in B. cereus, B. thuringiensis, B. mycoides, B. pseudomycoides, and B. weihenstephanensis at both the genetic and phenotypic levels.
MATERIALS AND METHODS
Bacterial strains.
All bacteria used in this study are listed in Table 1. Strains were obtained from different culture collections, such as the Deutsche Sammlung für Mikroorganismen, Braunschweig, Germany; the American Type Culture Collection, Rockville, Md.; the Centre de reference pour virus bacteriens, Felid d’Herbelle Universite Laval, Quebec, Canada; the Northern Regional Research Center, now the National Center for Agricultural Utilization Research (Agricultural Research Service Culture Collection, U. S. Department of Agriculture, Peoria, Ill.); the Weihenstephan Collection, Freising, Germany; and the Weihenstephan Bacillus Collection, Freising, Germany. In addition, strains were isolated from a variety of natural environments, including soil, milk, and other foods. These strains were isolated on B. cereus-selective polymyxin-pyruvate-egg yolk-mannitol-bromthymol blue (PEMBA) agar, containing 1 g of peptone, 10 g of mannitol, 2 g of sodium chloride, 0.1 g of magnesium sulfate, 2.5 g of disodium hydrogen phosphate, 0.25 g of potassium dihydrogen phosphate, 0.12 g of bromthymol blue, 10 g of sodium pyruvate, and 105 IU of polymyxin per liter (DIN 10198). The strains were identified as members of the B. cereus group based upon their morphological and biochemical characteristics (API 50CH and API 50CHB, bioMerieux). For many strains, the 16S rDNA was sequenced (32, 41).
TABLE 1.
Strains and phenotypes used in this study
| Straina | Sourcea | Reference | cryIAb | hblAc | L1d | L2d | Bd | Cytotoxicitye | Hemolysisf |
|---|---|---|---|---|---|---|---|---|---|
| B. thuringiensis | |||||||||
| WS2734T | DSM2046T | 32 | + | + | + | + | + | 279 | + |
| WS2621 | HER1357 | H.-W. Ackermann, Universite Laval, Quebec, Canada | + | + | + | + | + | 758 | + |
| WS2623 | HER1078 | H.-W. Ackermann, Universite Laval, Quebec, Canada | + | + | ± | ± | ± | <10 | − |
| WS2627 | HER1410 | H.-W. Ackermann, Universite Laval, Quebec, Canada | − | + | + | + | + | 83 | + |
| WS2629 | HER1418 | H.-W. Ackermann, Universite Laval, Quebec, Canada | + | + | ± | + | + | 56 | + |
| WSBC28001 | Soil from Poland (PO1) | 33 | + | + | ± | + | + | 175 | + |
| WSBC28002 | Soil from Poland (PO2) | 33 | + | + | + | + | + | 321 | + |
| WSBC28021 | Soil from Poland (PO3) | 33 | + | + | + | + | + | 204 | + |
| B. mycoides | |||||||||
| WS2641T | DSM2048T | 32 | − | + | + | + | + | 339 | + |
| WSBC10276 | Pasteurized milk from Germany | 32 | − | + | + | ± | + | 118 | + |
| WSBC10277 | Pasteurized milk from Germany | 32 | − | + | + | ± | + | 205 | + |
| WSBC10278 | Pasteurized milk from Germany | 32 | − | − | + | + | + | 182 | + |
| WSBC10279 | Pasteurized milk from Germany | 32 | − | + | + | + | + | 100 | + |
| WSBC10359 | Soil from Germany | 48 | − | + | + | + | + | 214 | + |
| WSBC10360 | Soil from Germany | 48 | − | + | − | ± | − | 62 | ± |
| WSBC10361 | Soil from Germany | 48 | − | + | − | − | − | 61 | + |
| WSBC10294 | Soil from Germany | 48 | − | + | + | + | + | 374 | + |
| WSBC10256 | Soil from Denmark (S38, 1) | 17 | − | + | + | + | + | 120 | + |
| WSBC10257 | Soil from Denmark (S38, 2) | 17 | − | + | + | + | + | 306 | ± |
| WSBC10258 | Soil from Denmark (S38, 3) | 17 | − | + | + | + | + | 212 | + |
| WSBC10291 | Kurkuma root from Thailand | 48 | − | + | + | ± | − | <10 | ± |
| WSBC10292 | Kurkuma root from Thailand | 48 | − | − | + | − | − | 193 | ± |
| WSBC10293 | Kurkuma root from Thailand | 48 | − | − | ± | − | − | <10 | ± |
| WSBC10366 | Kurkuma root from Thailand | 48 | − | − | − | − | − | <10 | ± |
| B. pseudomycoides | |||||||||
| WS3118T | Soil from United States (NRRL B-617T) | 38 | − | + | + | − | − | <10 | + |
| WS3119 | Soil from United States (NRRL NRS-322) | 38 | − | + | − | − | − | <10 | + |
| WS3120 | Soil from United States (NRS BD-10) | 38 | − | + | − | − | − | <10 | + |
| B. weihenstephanensis | |||||||||
| WSBC10201 | Pasteurized milk from Germany | 32 | − | − | ± | − | − | <10 | + |
| WSBC10202 | Pasteurized milk from Germany | 32 | − | + | ± | − | − | 22 | + |
| WSBC10204T | Pasteurized milk from Germany | 32 | − | ± | ± | ± | ± | 21 | + |
| WSBC10205 | Pasteurized milk from Germany | 32 | − | + | ± | + | + | <10 | + |
| WSBC10206 | Pasteurized milk from Germany | 32 | − | + | ± | − | − | 42 | + |
| WSBC10207 | Pasteurized milk from Germany | 32 | − | + | + | − | ± | 385 | + |
| WSBC10208 | Pasteurized milk from Germany | 32 | − | + | − | − | − | <10 | + |
| WSBC10209 | Pasteurized milk from Germany | 32 | − | + | + | − | − | 96 | + |
| WSBC10211 | Pasteurized milk from Germany | 32 | − | + | + | + | ± | <10 | + |
| WSBC10311 | Soil from Germany | 48 | − | − | + | + | − | 44 | + |
| WSBC10363 | Soil from Germany | 48 | − | − | + | − | − | 60 | + |
| WSBC10364 | Soil from Germany | 48 | − | − | ± | − | − | 17 | ± |
| WSBC10297 | Soil from Germany | 48 | − | + | ± | + | ± | 15 | + |
| WSBC10365 | Soil from Germany | 48 | − | + | ± | − | ± | 35 | + |
| WSBC10316 | Milk powder from Germany (48) | 49 | − | − | ± | − | − | <10 | + |
| B. cereus | |||||||||
| WSBC10027 | Pasteurized milk from Germany | 32 | − | + | ± | + | ± | <10 | + |
| WSBC10028 | Pasteurized milk from Germany | 32 | − | + | + | + | + | 149 | + |
| WSBC10030 | Pasteurized milk from Germany | 32 | − | − | ± | − | − | <10 | + |
| WSBC10032 | Pasteurized milk from Germany | 32 | − | − | + | − | − | 200 | + |
| WSBC10034 | Pasteurized milk from Germany | 32 | − | + | ± | + | ± | <10 | + |
| WSBC10035 | Pasteurized milk from Germany | 32 | − | − | + | − | − | <10 | + |
| WSBC10039 | Pasteurized milk from Germany | 32 | − | + | + | ± | − | 82 | + |
| WSBC10040 | Pasteurized milk from Germany | 32 | − | + | + | − | − | 102 | + |
| WSBC10041 | Pasteurized milk from Germany | 32 | − | − | + | − | − | <10 | + |
| WSBC10042 | Pasteurized milk from Germany | 32 | − | − | + | − | − | 352 | + |
| WSBC10362 | Soil from Germany | 48 | − | + | + | + | ± | 29 | + |
| WSBC10310 | Soil from Germany | 48 | − | + | + | ± | ± | <10 | ± |
| WSBC10246 | Soil from Denmark (S38, 21) | 17 | − | − | + | − | − | <10 | + |
| WSBC10247 | Soil from Denmark (S38, 22) | 17 | − | − | + | − | − | 333 | + |
| WSBC10248 | Soil from Denmark (S38, 23) | 17 | − | − | + | − | − | 714 | + |
| WSBC 10249 | Soil from Denmark (S38, 24) | 17 | − | + | + | + | + | <10 | + |
| WSBC10252 | Soil from Denmark (S38, 27) | 17 | − | − | + | − | − | <10 | + |
| WSBC10312 | Kurkuma root from Thailand | 48 | − | + | + | + | + | 625 | + |
| WSBC10367 | Red rice from Thailand | 48 | − | − | ± | − | − | <10 | + |
| WSBC10313 | Milk powder from Germany (3) | 49 | − | − | ± | − | − | 84 | + |
| WSBC10314 | Milk powder from Germany (4) | 49 | − | − | + | + | + | 250 | + |
| WSBC10315 | Milk powder from Germany (6) | 49 | − | + | + | + | + | 88 | + |
| WSBC10368 | Milk powder from Germany (10) | 49 | − | − | ± | + | + | 116 | + |
DSM, Deutsche Sammlung für Mikroorganismen und Zellkulturen; HER, Centre de reference pour virus bacteriens; Felix d’Herbelle Universite Laval; PO, Institute of Microbiology, Wroclaw University; NRRL, Northern Regional Research Laboratory (ARS, Agricultural Research Service Culture Collection); WS, Weihenstephan Collection; WSBC: Weihenstephan Bacillus cereus Collection. B. thuringiensis strains were designated according to the provider’s information. The presence of the crystal toxin CryA was confirmed by using the multiplex PCR assay of Bourque et al. (10). B. mycoides strains are characterized by their mycoid growth on agar plates, regardless of the minimum growth temperature. B. pseudomycoides strains were designated according to the provider’s information (38). B. weihenstephanensis strains grwo at 7°C (32) and exhibit a psychrotolerant signature in their 16S rDNA (48). The remaining strains of the group are designated B. cereus.
The assay was done twice. +, positive in both assays; −, negative in both assays.
The assay was done up to 10 times. +, positive in at least 80% of the assays; −, negative in at least 80% of the assays.
The assays were done once or twice. +, good reaction; ±, weak reaction, −, no reaction.
The assay was done once or twice. Cytotoxicity is given in arbitrary units (18). The bacterial dilution was determined that gave a 50% reduction in the survival rate of the Vero cells.
The assay was done three times. +, strong beta hemolysis in three assays; ±, weak beta hemolysis in three assays; −, no beta hemolysis in three assays.
PCR and sequence analysis.
A multiplex PCR assay was performed identifying the three different cryIA genes (10). The sequence of the B. cereus hblA (27) was obtained from the EMBL and GenBank databases (L20441). The PCR was performed according to the method of Mäntynen and Lindström (37). The PCR fragments were purified following the Qiagen PCR purification protocol (Qiagen, Hilden, Germany). DNA sequence determination (Sequiserve, Vaterstetten, Germany) was performed using the Dyedeoxy Terminator Cycle Sequencing kit (Applied Biosystems, Weiterstadt, Germany), and the reaction products were analyzed by using an Applied Biosystems model 373A automatic DNA sequencer according to the manufacturer’s instructions. The cryIA gene was sequenced by using the primers 5′-TCG GAA AAT GTG CCC AT-3′ [cryIA(b))] and 5′-GGG ACT GCA GGA GTG AT-3′ [cryIA(c))]. The hblA gene was sequenced by using the primer 5′-GCT AAT GTA GTT TCA CCT GTA GCA AC-3′. The obtained hblA sequences were used to construct a similarity network with the neighbor-joining method (47).
Detection of the hblA gene by Southern hybridization.
Chromosomal DNA was digested with EcoRI. The fragments were separated on a 1.2% agarose gel and blotted onto nitrocellulose. A Southern analysis (44) was performed using a digoxygenin-labelled oligonucleotide probe directed against B. cereus hblA. This probe was derived from a 883-bp fragment that was obtained from strain WS2629 by PCR with the primers 5′-GCT AAT GTA GTT TCA CCT GTA GCA AC-3′ and 5′-AAT CAT GCC ACT GCG TGG ACA TAT AA-3′. The hybridization temperature was 65°C.
Immunological detection of the L1, L2, and B components of the HBL complex.
Bacteria were grown at 32°C in CGY (7) medium containing 2% caseine, 0.6% yeast, 0.2% (NH4)2SO4, 1.4% K2HPO4, 0.6% KH2PO4, 0.1% sodium citrate, and 0.2% MgSO4, supplemented with 1% glucose for 6 h. EDTA (1 mM) was added at the time of harvesting. Cell-free supernatants, obtained by centrifugation (10,000 × g at 4°C for 20 min), followed by filtration through 0.2-μm-pore-size Millipore filters, were used in the enzyme immunoassay. For the determination of individual components of the HBL in cell-free supernatants, the microtiter plates were coated with serial dilutions of the supernatants. The enzyme immunoassay, based on monoclonal antibodies, was performed according to the method of Dietrich et al. (18). Antibody 2A3 was specific for the B component, the antibodies 1A12 and 8B12 were specific for the L2 component, and antibody 1C2 was specific for the L1 protein. Free protein binding sites of the plates were blocked with phosphate-buffered saline containing sodium caseinate (30 g/liter) for 30 min. Subsequently, 100 μl of the respective purified monoclonal antibody (2 μg/ml) were added, and the plates were developed as described (18).
Determination of the cytotoxicity.
Cytotoxicity of the cell-free supernatants was determined by measuring cell proliferation and cell viability using Vero cells (18). Growth medium and diluent consisted of Eagle minimum essential medium (Biochrom KG, Berlin, Germany) with Earle salts supplemented with 1% calf serum and 2 mM glutamine. The activity was tested as serial dilutions in microtiter plates. Cell-free supernatant (0.1 ml) was added to 0.1 ml of the Vero cells, and the plates were incubated for 24 h at 37°C in a 5% CO2 atmosphere. Cell Proliferation Reagent WST-1 (10 μl) (Roche Diagnostics, Mannheim, Germany) was added to 0.1 ml of the above suspension, and the plates were incubated for another hour under the same conditions. The absorbance was determined at 450 nm, and the 50% inhibitory concentration was calculated as described (18).
Determination of the hemolytic activity.
The hemolytic activity was determined at 30°C on blood agar plates (Merck, Darmstadt, Germany) containing 23 g of peptone, 5 g of NaCl, 14 g of agar, and 65 ml of sheep blood per liter. Escherichia coli JM109 and Bacillus subtilis JH642 were used as negative controls.
Nucleotide sequence accession numbers.
The EMBL accession no. for the sequences reported in this paper are AJ243147 through AJ243164.
RESULTS
Detection of cryIA by PCR.
A PCR was performed to test the B. cereus group strains for the presence of the B. thuringiensis crystal toxin gene cryIA (for a revision on the crystal toxin nomenclature, see reference 14). The assay simultaneously detects cryIA(a), cryIA(b), and cryIA(c) and was developed to detect B. thuringiensis (10). The PCR was performed twice with each strain, and four of the obtained PCR fragments were sequenced. Seven of the eight B. thuringiensis strains were confirmed as such. The strains WS2734, WS2621, and WS2623 contained CryIA(b). Strains WS2629, WSBC28001, WSBC28002, and WSBC28021 contained CryIA(c). Only WS2627 did not yield a PCR product (Table 1).
The remaining B. mycoides, B. pseudomycoides, B. weihenstephanensis, and B. cereus strains did not contain CryIA. The B. mycoides strains were classified according to their mycoid growth on agar plates. The B. pseudomycoides strains were designated according to the provider’s information (38). These strains were all mesophilic. The psychrotolerant B. weihenstephanensis strains were identified by their ability to grow at 4°C (32) and by a PCR assay directed against the major cold shock gene cspA (21) or the 16S rDNA (48). The remaining mesophilic strains which did not exhibit mycoid growth and did not possess cryIA were designated B. cereus (Table 1).
Detection of the hblA by PCR.
A PCR was performed according to the method of Mäntynen and Lindström (37) to test the B. cereus group strains for the presence of the hemolytic enterotoxin binding component gene hblA. The PCR was performed up to eight times with each strain (Table 1). All of the eight B. thuringiensis strains, most of which also possessed the crystal toxin gene cryIA, produced the 883-bp fragment in the PCR. Similarly, the majority of the B. mycoides (69%), all of the B. pseudomycoides (100%), and most of the B. weihenstephanensis (60%) strains turned out positive in the PCR. In addition, some of the B. cereus strains (43%) produced the 883-bp fragment, indicating the presence of hblA in these strains. With regard to the environment, 68% of the strains isolated from milk and 68% of the strains isolated from soil were positive for hblA.
Detection of the hblA by Southern hybridization.
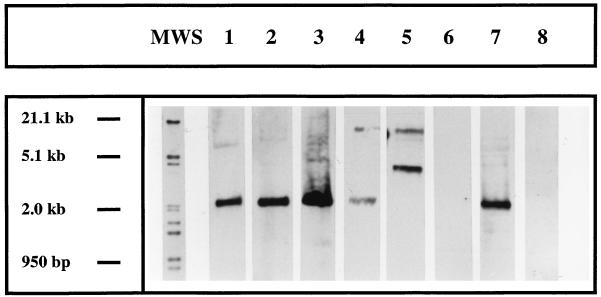
The presence or absence of hblA was confirmed by Southern hybridization. The genomic DNAs of five B. thuringiensis strains, two B. mycoides strains, two B. pseudomycoides strains, six B. weihenstephanensis strains, and six B. cereus strains were digested with EcoRI and were analyzed by Southern hybridization by using a probe directed against the hblA from strain WS2629. Figure 1 shows the hybridization patterns for a selection of these strains. Strains WS2621, WS2623, WS2641, WSBC10278, and WSBC10027 showed one strong signal at about 2 kb, and strain WSBC10204 showed a strong signal at about 5 kb. Occasionally, several weak bands were detected as well. WSBC10201 and WSBC10042 did not yield a signal. The remaining strains yielded signals in the Southern analysis which were in agreement with the data obtained by PCR (data not shown).
FIG. 1.
Southern blot analysis of selected strains. The genomic DNA was digested with EcoRI. Southern analysis was performed using a DIG-labelled oligonucleotide probe recognizing the B. cereus hblA. Lane 1, WS2621; lane 2, WS2623; lane 3, WS2641; lane 4, WSBC10278; lane 5, WSBC10204; lane 6, WSBC10201; lane 7, WSBC10027; lane 8, WSBC10042. MWS, molecular weight standard. Molecular size is given in kilobases.
Sequence analysis.
The 883-bp PCR fragment was confirmed as hblA by sequence analysis. The sequence was obtained for three B. thuringiensis strains, four B. mycoides strains, three B. pseudomycoides strains, four B. weihenstephanensis strains, and four B. cereus strains. All sequences shared a significant degree of homology with the B. cereus L20441 sequence (27). Single-base-pair substitutions occurred in 55 positions of the 640 bp examined, with no more than a 30-bp difference between any two strains.
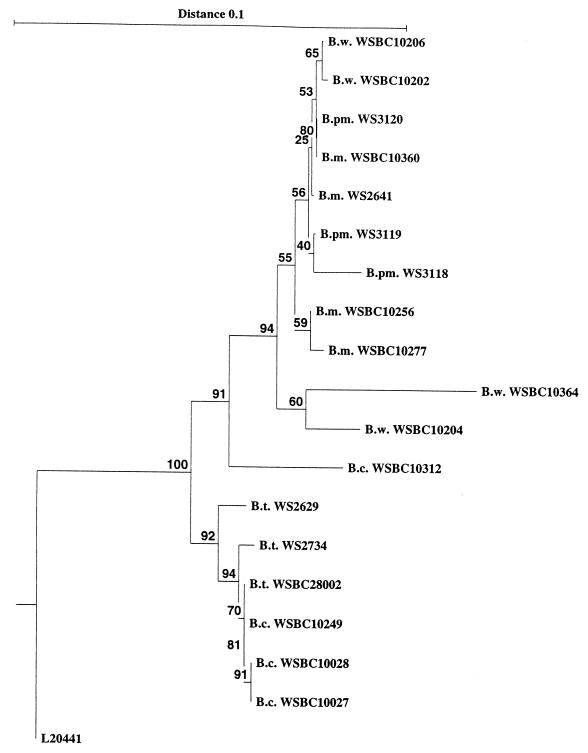
The hblA sequences were used to construct a similarity network (Fig. 2). The three B. thuringiensis strains and three of the four B. cereus strains formed one similarity cluster. The remaining B. mycoides, B. pseudomycoides, and B. weihenstephanensis strains formed a second cluster which also contained the B. cereus strain WSBC10312. We were not able to resolve individual groups within these two clusters of strains.
FIG. 2.
Similarity network based on the nucleotide sequences of the B component of the hemolytic enterotoxin of the B. thuringiensis (B. t.) strains WS2734, WS2629, and WSBC28002; the B. mycoides (B. m.) strains WS2641, WSBC10277, WSBC10360, and WSBC10256; the B. pseudomycoides (B. pm.) strains WS3118, WS3119, and WS3120; the B. weihenstephanensis (B. w.) strains WSBC10202, WSBC10204, WSBC10206, and WSBC10364; and the B. cereus (B. c.) strains WSBC10027, WSBC10028, WSBC10249, and WSBC10312 as compared to the B. cereus L20441 (26). The neighbor-joining method (38) was used, and bootstrap analysis was performed with 100 repeats. For strain designations see Table 1.
Most of the observed base pair substitutions were in the third position of their respective codon and did not affect the amino acid sequence. Therefore, the deduced amino acid sequences of the strains differed only in 18 positions. A conserved region was located between amino acids 51 and 106. A variable region was found between amino acids 167 and 208. Within this region, there were four positions where the B. cereus and B. thuringiensis strains differed from the B. mycoides, B. pseudomycoides, and B. weihenstephanensis strains: positions 167 (Y to V), 184 (H to Q), 194 (E to D), and 208 (D to E).
Immunological assays.
The individual components of the HBL complex were determined with specific monoclonal antibodies (18). Seven of the eight B. thuringiensis strains expressed all three subunits (Table 1). The exception was strain WS2623, which also exhibited weak cytotoxicity. The majority of the B. mycoides strains also expressed the L1, L2, and B component of the HBL complex. Strain WSBC10278, which was negative in the PCR but positive in the Southern analysis, also expressed the three subunits and exhibited cytotoxicity. The three B. pseudomycoides strains turned out negative for the L1, L2, and B proteins of the HBL complex and exhibited no detectable cytotoxicity. The majority of the B. weihenstephanensis strains only exhibited weak cytotoxicity and weak expression levels of the HBL complex. The exceptions were strains WSBC10207 and WSBC10209, which exhibited high levels of the L1 component and high cytotoxicity. The detection of the HBL components for the B. cereus strains was in agreement with the molecular data for most strains. The majority of the strains that were positive for hblA in the PCR also possessed detectable amounts of the three HBL components. The majority of the strains that were negative in the PCR did not possess the HBL proteins.
Hemolytic activity.
The hemolytic activity of the strains was tested on blood agar plates (Table 1). The control strains E. coli and B. subtilis did not show hemolysis on blood agar plates. The majority of the B. cereus group strains exhibited β hemolysis, confirming the presence of a hemolytic enterotoxin. Two strains (WSBC10364 and WSBC10365) showed a discontinuous pattern of hemolysis on the plates. Hemolysis started at a few millimeters distance from the colony, forming a ring around the bacteria. The B. thuringiensis strain WS2623 was the only strain that did not show hemolysis. All the other strains which were positive for hblA in the PCR exhibited hemolysis on the blood agar plates. The strains that were negative for hblA in the PCR were also able to hemolyse erythrocytes. Interestingly, the hemolytic activity of many B. mycoides strains was weaker than that of the other strains. This may be due to the rhizobial growth of these strains yielding lower cell densities on the surface of the agar plate. Particularly strains WSBC10291, WSBC10292, WSBC10293, and WSBC10366 exhibited extremely weak hemolysis.
DISCUSSION
The taxonomy of the B. cereus group is still under investigation, and contradictory results have been obtained. Based upon their high similarity, it has been repeatedly proposed that all species of the B. cereus group be merged into a single species (3, 4, 12, 30, 40). The major reason for maintaining a complex nomenclature (as seen in the 1980 approved list of bacterial names, reference 43) is the economic and medical importance of individual species. In this study, the taxonomy as described by Lechner et al. (32) and Nakamura (38) was applied, and the five groups of B. cereus strains were considered separate species.
The existence of the enterotoxin HBL, which is considered unique for B. cereus, was investigated among the different species of this group. The hblA gene was identified by different molecular biological assays. For all but two of the strains tested, PCR and Southern analyses led to the same result. It turned out that the majority of the strains investigated which were isolated from a variety of natural habitats possessed hblA. hblA thus appears to be broadly distributed among all the species of the B. cereus group, regardless of the environment or geographic origin.
Southern analysis confirmed the data obtained by PCR, with two exceptions. Strains WSBC10278 and WSBC10204, which were PCR negative for hblA, were positive in the Southern analysis. This may be due to a mutation in the binding site of one of the primers leading to a false-negative result in the PCR. The long probe used for the hybridization is more likely to bind despite occasional point mutations. This finding indicates that the detection of pathogenic B. cereus group strains by PCR alone (37) may lead to false-negative results and that a thorough investigation of the strains is necessary to make a clear statement.
Sequence analysis of 18 strains of the B. cereus group revealed two clusters of strains. One contained B. thuringiensis and a majority of the B. cereus strains, and the other cluster contained the B. mycoides, B. pseudomycoides, and B. weihenstephanensis strains as well as one B. cereus strain. These differences were located to the region between amino acids 167 and 208 in the deduced amino acid sequence. The two groups were also evident when 16S rDNA was analyzed (32).
The immunological assays confirmed the molecular data with few exceptions. B. thuringiensis WS2623 was positive for hblA in the PCR but was negative for the L1, L2, and B components and was not cytotoxic or hemolytic. This indicates that the hblA gene is present in this strain, but it may be incomplete or silent. B. mycoides WSBC10278 was negative for hblA in the PCR but was positive in the Southern analysis. This strain was positive for L1, L2, and B, and it was cytotoxic and hemolytic. Most likely, one of the primer binding sites was modified. Strains WSBC10360 and WSBC10361, which were positive in the PCR, were negative in the immunological assay. The three B. pseudomycoides strains were all positive for hblA in the PCR and positive in the Southern analysis. None of them exhibited any detectable amount of the HBL proteins. They were also not cytotoxic. However, hemolysis was good. Apparently, the toxin genes are not expressed in these strains. The majority of the B. weihenstephanensis strains expressed little or no HBL complex and were only slightly cytotoxic even though they possessed the hblA gene. The immunoassays for the B. cereus strains were in agreement with the molecular data with only few exceptions. The majority of the strains that were PCR positive for hblA were also positive for L1, L2, and B in the immunological assay, and the majority of the strains that were negative for hblA were also negative for L1, L2, and B.
For many strains (i.e., WSBC10292, WSBC10207, WSBC10209, WSBC10032, WSBC10039, WSBC10040, WSBC10042, WSBC10247, and WSBC10248), the L1 component was detected even though L2 and B were not present. Some of these strains were highly cytotoxic. An explanation for these results could be the cross-reactivity of the L1 antibody with the 39-kDa component of the nonhemolytic but cytotoxic NHE (18), which shows some degree of amino acid homology to the L1 compound of HBL (35).
The determination of the hemolytic activity on blood agar plates led to interesting results. All but one of the strains that were positive for hblA exhibited hemolytic activity. This is in agreement with evidence that several hemolytic factors are expressed in B. cereus group bacteria: cereolysin (13), sphingomyelinase (45), cereolysin AB (22), or cereolysin-like hemolysin (28).
In summary, we detected the B. cereus enterotoxin HBL in all species of the B. cereus group investigated, even in B. thuringiensis that is used as a biological insecticide. A certain discrepancy between the different methods was noticed, indicating that a thorough investigation is required to detect pathogenic B. cereus strains, particularly for B. thuringiensis strains. The consequences of this finding for food safety regulations need to be evaluated.
ACKNOWLEDGMENTS
We thank H.-W. Ackermann (Felix d’Herbelle Universite Laval, Quebec, Canada), C. Wiebe (Bundesanstalt für Milchforschung, Kiel, Germany), P. Damgaard (Denmark), L. Nakamura (U. S. Department of Agriculture, Peoria, Ill.), R. Mayr (this laboratory), and F. von Stetten (this laboratory) for providing strains; W. Metzger (Sequiserve) for performing the sequence analysis; and H. Hermann (this laboratory) for excellent technical assistance.
This work was supported in part by the Deutsche Forschungsgemeinschaft (grants Sche 316/3-1 and Ma 1702/3-1).
REFERENCES
- 1.Agata N, Mori M, Ohta M, Suwan S, Ohtani I, Isobe M. A novel dodecadepsipeptide, cereulide, isolated from Bacillus cereus causes vacuole formation in HEp-2 cells. FEMS Microbiol Lett. 1994;121:31–34. doi: 10.1111/j.1574-6968.1994.tb07071.x. [DOI] [PubMed] [Google Scholar]
- 2.Agata N, Ohta M, Arakawa Y, Mori M. The bceT gene of Bacillus cereus encodes an enterotoxic protein. Microbiology. 1995;141:983–988. doi: 10.1099/13500872-141-4-983. [DOI] [PubMed] [Google Scholar]
- 3.Ash C, Collins M D. Comparative analysis of 23S ribosomal RNA gene sequences of Bacillus anthracis and emetic Bacillus cereus determined by PCR-direct sequencing. FEMS Microbiol Lett. 1992;94:75–80. doi: 10.1016/0378-1097(92)90586-d. [DOI] [PubMed] [Google Scholar]
- 4.Ash C A, Farrow J A E, Dorsch M, Stackebrandt E, Collins M D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rDNA. Int J Syst Bacteriol. 1991;41:343–346. doi: 10.1099/00207713-41-3-343. [DOI] [PubMed] [Google Scholar]
- 5.Battisti L, Green B D, Thorne C B. Mating system for transfer of plasmids among Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. J Bacteriol. 1985;162:543–550. doi: 10.1128/jb.162.2.543-550.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beattie S H, Williams A G. Detection of toxigenic strains of Bacillus cereus and other Bacillus spp. with an improved cytotoxicity assay. Lett Appl Microbiol. 1999;28:221–225. doi: 10.1046/j.1365-2672.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- 7.Beecher D J, Wong A C L. Improved purification and characterization of hemolysin BL, a hemolytic dermonecrotic vascular permeability factor from Bacillus cereus. Infect Immun. 1994;62:980–986. doi: 10.1128/iai.62.3.980-986.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beecher D J, Wong A C L. Tripartite hemolysin BL from Bacillus cereus. Hemolytic analysis of component interaction and model for its characteristic paradoxical zone phenomenon. J Biol Chem. 1997;272:233–239. doi: 10.1074/jbc.272.1.233. [DOI] [PubMed] [Google Scholar]
- 9.Beecher D J, Schoeni J L, Wong A C L. Enterotoxin activity of hemolysin BL from Bacillus cereus. Infect Immun. 1995;63:4423–4428. doi: 10.1128/iai.63.11.4423-4428.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourque S N, Valero J R, Mercier J, Lavoie M C, Levesque R C. Multiplex polymerase chain reaction for detection and differentiation of the microbial insecticide Bacillus thuringiensis. Appl Environ Microbiol. 1993;59:523–527. doi: 10.1128/aem.59.2.523-527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budarina Z I, Sinev M A, Mayorov S G, Tomashevski A Y, Shmelev I V, Kuzmin N P. Hemolysin II is more characteristic of Bacillus thuringiensis than Bacillus cereus. Arch Microbiol. 1994;161:252–257. doi: 10.1007/BF00248701. [DOI] [PubMed] [Google Scholar]
- 12.Carlson C R, Kolsto A-B. A complete physical map of a Bacillus thuringiensis chromosome. J Bacteriol. 1993;175:1053–1060. doi: 10.1128/jb.175.4.1053-1060.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowell J L, Grushoff-Kosyk P S, Bernheimer A W. Purification of cereolysin and the electrophoretic separation of the active (reduced) and inactive (oxidized) forms of the purified toxin. Infect Immun. 1976;14:144–154. doi: 10.1128/iai.14.1.144-154.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crickmore N, Zeigler D R, Feitelson J, Schnepf E, Van Rie J, Lereclus D, Baum J, Dean D H. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal protein. Microbiol Mol Biol Rev. 1998;62:807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daffonchio D, Borin S, Frova G, Gallo R, Mori E, Fani R, Sorlini C. A randomly amplified polymorphic DNA marker specific for the Bacillus cereus group is diagnostic for Bacillus anthracis. Appl Environ Microbiol. 1999;65:1298–1303. doi: 10.1128/aem.65.3.1298-1303.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damgaard P. Diarrhoeal enterotoxin production by strains of Bacillus thuringiensis isolated from commercial Bacillus thuringiensis-based insecticides. FEMS Immunol Med Microbiol. 1995;12:245–250. doi: 10.1111/j.1574-695X.1995.tb00199.x. [DOI] [PubMed] [Google Scholar]
- 17.Damgaard P H, Jacobsen C S, Soerensen J. Development and application of a primer set for specific detection of Bacillus thuringiensis and Bacillus cereus in soil using magnetic capture hybridization and PCR amplification. System Appl Microbiol. 1996;19:436–441. [Google Scholar]
- 18.Dietrich R, Fella C, Strich S, Märtlbauer E. Production and characterization of monoclonal antibodies against the hemolysin BL enterotoxin complex produced by Bacillus cereus. Appl Environ Microbiol. 1999;65:4470–4474. doi: 10.1128/aem.65.10.4470-4474.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falcon L A. Use of bacteria for microbial control. In: Burges H D, Hussey N W, editors. Microbial control of insects and mites. New York, N.Y: Academic Press, Inc.; 1971. pp. 67–95. [Google Scholar]
- 20.Flugge C. Die Mikroorganismen. Leipzig, Germany: Vogel; 1886. [Google Scholar]
- 21.Francis K P, Mayr R, von Stetten F, Stewart G S A B, Scherer S. Discrimination of psychrotolerant and mesophilic strains of the Bacillus cereus group by PCR targeting of major cold shock protein genes. Appl Environ Microbiol. 1998;64:3525–3529. doi: 10.1128/aem.64.9.3525-3529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilmore M S, Cruz-Rodz A L, Leimeister-Wachter M, Kreft J, Goebel W. A Bacillus cereus cytolytic determinant, cereolysin AB, which comprises the phospholipase C and sphingomyelinase genes: nucleotide sequence and genetic linkage. J Bacteriol. 1989;171:744–753. doi: 10.1128/jb.171.2.744-753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez J M, Jr, Dulmage H T, Carlton B C. Correlation between specific plasmids and delta-endotoxin production in Bacillus thuringiensis. Plasmid. 1981;5:351–365. doi: 10.1016/0147-619x(81)90010-x. [DOI] [PubMed] [Google Scholar]
- 24.Gonzales J M, Jr, Brown B J, Carlton B C. Transfer of Bacillus thuringiensis plasmids coding for δ-endotoxin among strains of Bacillus thuringiensis and Bacillus cereus. Proc Natl Acad Sci USA. 1982;79:6951–6955. doi: 10.1073/pnas.79.22.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon R E, Haynes W C, Pang C H N. The genus Bacillus. U.S. Washington, D.C.: Department of Agriculture; 1973. [Google Scholar]
- 26.Granum P E, Lund T. Bacillus cereus and its food poisoning toxins. FEMS Microbiol Lett. 1997;157:223–228. doi: 10.1111/j.1574-6968.1997.tb12776.x. [DOI] [PubMed] [Google Scholar]
- 27.Heinrichs J H, Beecher D J, MacMillan J D, Zilinskas B A. Molecular cloning and characterization of the hblA gene encoding the B component of hemolysin BL from Bacillus cereus. J Bacteriol. 1993;175:6760–6766. doi: 10.1128/jb.175.21.6760-6766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda T, Shiba A, Seo S, Yamamoto J, Matsuyama J, Miwatani T. Identity of hemolysins produced by Bacillus thuringiensis and Bacillus cereus. FEMS Microbiol Lett. 1991;79:205–210. doi: 10.1016/0378-1097(91)90087-q. [DOI] [PubMed] [Google Scholar]
- 29.Jackson S G, Goodbrand R B, Ahmed R, Kasatiya S. Bacillus cereus and Bacillus thuringiensis isolated in a gastroenteritis outbreak investigation. Lett Appl Microbiol. 1995;21:103–105. doi: 10.1111/j.1472-765x.1995.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 30.Kaneko T, Nozaki R, Aizawa K. Deoxyribonucleic acid relatedness between Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Microbiol Immunol. 1978;22:639–641. doi: 10.1111/j.1348-0421.1978.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 31.Kronstadt J W, Schnepf H E, Whiteley H R. Diversity of locations for Bacillus thuringiensis crystal protein genes. J Bacteriol. 1983;154:419–428. doi: 10.1128/jb.154.1.419-428.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lechner S, Mayr R, Francis K P, Prüß B M, Kaplan T, Wießner-Gunkel E, Stewart G S A B, Scherer S. Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int J Syst Bacteriol. 1998;48:1373–1382. doi: 10.1099/00207713-48-4-1373. [DOI] [PubMed] [Google Scholar]
- 33.Lonc E, Lecadet M-M, Lachowicz T M, Panek E. Description of Bacillus thuringiensis wratislaviensis (H-47), a new serotype originating from Wroclaw (Poland), and other Bt soil isolates from the same area. Lett Appl Microbiol. 1997;24:467–473. [Google Scholar]
- 34.Lund T, Granum P E. Characterization of a non-hemolytic enterotoxin complex from Bacillus cereus isolated after a foodborne outbreak. FEMS Microbiol Lett. 1996;141:151–156. doi: 10.1111/j.1574-6968.1996.tb08377.x. [DOI] [PubMed] [Google Scholar]
- 35.Lund T, Granum P E. Comparison of biological effect of the two different enterotoxin complexes isolated from three different strains of Bacillus cereus. Microbiology. 1997;143:3329–3336. doi: 10.1099/00221287-143-10-3329. [DOI] [PubMed] [Google Scholar]
- 36.Makino S I, Iinuma-Okada Y, Maruyama T, Ezaki T, Sasakawa C, Yoshikawa M. Direct detection of Bacillus anthracis DNA in animals by polymerase chain reaction. J Clin Microbiol. 1993;312:547–551. doi: 10.1128/jcm.31.3.547-551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mäntynen V, Lindström K. A rapid PCR-based DNA test for enterotoxic Bacillus cereus. Appl Environ Microbiol. 1998;64:1634–1639. doi: 10.1128/aem.64.5.1634-1639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura L K. Bacillus pseudomycoides sp. nov. Int J Syst Bacteriol. 1998;48:1031–1035. doi: 10.1099/00207713-48-3-1031. [DOI] [PubMed] [Google Scholar]
- 39.Perani M, Bishop A H, Vaid A. Prevalence of β-exotoxin, diarrhoeal toxin and specific δ-endotoxin in natural isolates of Bacillus thuringiensis. FEMS Microbiol Lett. 1998;160:55–60. doi: 10.1111/j.1574-6968.1998.tb12890.x. [DOI] [PubMed] [Google Scholar]
- 40.Priest F G, Goodfellow M, Todd C. A numerical classification of the genus Bacillus. J Gen Microbiol. 1988;134:1847–1882. doi: 10.1099/00221287-134-7-1847. [DOI] [PubMed] [Google Scholar]
- 41.Prüß B M, Francis K P, von Stetten F, Scherer S. Correlation of 16S ribosomal DNA signature sequences with temperature dependent growth rates of mesophilic and psychrotolerant strains of the Bacillus cereus group. J Bacteriol. 1999;181:2624–2630. doi: 10.1128/jb.181.8.2624-2630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan P A, MacMillan J D, Zilinskas B A. Molecular cloning and characterization of the genes encoding the L1 and L2 components of hemolysin BL from Bacillus cereus. J Bacteriol. 1997;179:2551–2556. doi: 10.1128/jb.179.8.2551-2556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skerman V B D, McGowan V, Sneath P H A. Approved list of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. [Google Scholar]
- 44.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:742–744. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 45.Tomita M, Taguchi I R, Ikezawa H. Molecular properties and kinetic studies on sphingomyelinase of Bacillus cereus. Biochim Biophys Acta. 1982;704:90–99. doi: 10.1016/0167-4838(82)90135-2. [DOI] [PubMed] [Google Scholar]
- 46.Turnball P C B, Kramer J M. Bacillus. In: Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: American Society for Microbiology; 1991. pp. 296–303. [Google Scholar]
- 47.Van de Peer Y, de Wachter R. Treecon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 48.von Stetten F, Francis K P, Lechner S, Neuhaus K, Scherer S. Rapid discrimination of psychrotolerant and mesophilic strains of the Bacillus cereus group by PCR targeting of 16S rDNA. J Microbiol Methods. 1998;34:99–106. [Google Scholar]
- 49.Wiebe C, Hammer P. Zum Vorkommen von B. cereus in einem Milchtrocknungsbetrieb—Phänotypische Charakterisierung der isolierten Stämme. Berlin, Germany: Milchkonferenz, Deutsche Gesellschaft für Milchwissenschaft; 1997. [Google Scholar]