Abstract
The COVID-19 pandemic has strained healthcare systems. Sensitive, specific, and timely COVID-19 diagnosis is crucial for effective medical intervention and transmission control. RT-PCR is the most sensitive/specific, but requires costly equipment and trained personnel in centralized laboratories, which are inaccessible to resource-limited areas. Antigen rapid tests enable point-of-care (POC) detection but are significantly less sensitive/specific. CRISPR-Cas systems are compatible with isothermal amplification and dipstick readout, enabling sensitive/specific on-site testing. However, improvements in sensitivity and workflow complexity are needed to spur clinical adoption. We outline the mechanisms/strategies of major CRISPR-Cas systems, evaluate their on-site diagnostic capabilities, and discuss future research directions.
Keywords: CRISPR-Cas, diagnostics, infectious diseases, RNA/DNA detection, Cas12/13, point-of-care
Current COVID-19 diagnostic platforms
The current pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has inflicted ~280 million infections and 5.4 million deaths (December 2021). In view of the high human-to-human transmissibility of SARS-CoV-2, and the fact that ~20% of infected people are asymptomatic [1,2], rapid and accurate diagnostic tests deployable on-site at the point-of-need will be crucial for timely transmission control and treatment. Such rapid on-site tests can be used for community-level screening at medical facilities, work sites, air/sea customs, or pop-up/mobile testing sites before subsequent confirmatory tests at central clinical laboratories. This will increase diagnostic reach by enabling more tests to be conducted at a faster pace. Reduced testing at clinical laboratories will reduce the strain on healthcare resources, which can be better diverted to medical intervention and patient care. Furthermore, when used at immigration checkpoints, schools, trade and convention centers, such accurate on-site tests will enable safe resumption of travel, education, work, social, and economic activities.
Three major biomarkers are employed for the diagnosis of COVID-19 [3]: the genomic RNA of the virus, four major structural and functional proteins/antigens – namely the spike (S), nucleocapsid (N), envelope (E), and membrane (M) proteins – and the neutralizing antibodies (IgM/IgG) in the infected host produced as part of the humoral immune response against SARS-CoV-2.
There are several major testing platforms for diagnosing COVID-19, namely reverse transcription PCR (RT-PCR), lateral flow rapid test kits (RTKs), and sequencing. RT-PCR, which detects the SARS-CoV-2 RNA sequences, is the current gold standard for diagnosing COVID-19 owing to its high sensitivity/specificity. However, it requires instrumentation for nucleic acid extraction and thermal cyclers with fluorescence readout capabilities, skilled personnel to conduct and interpret the test, and takes ~1.5–2 h in a centralized laboratory setting. Lateral flow test kits are fast and user-friendly, can be completed within 15–20 min, and do not require instrumentation for readout because the results are presented as visible colorimetric bands. Such test kits detect the SARS-CoV-2 nucleocapsid and/or spike antigens. However, lateral flow test kits are much less sensitive than RT-PCR, and may lead to significant false-negative readings. Although sequencing is a powerful tool in understanding the dynamics of outbreaks and in discovering variants, it is even more costly and time-consuming for routine testing than RT-PCR. Details on the kinetics of COVID-19 infection and performance of current diagnostic techniques are found in Box 1 .
Box 1. Current detection platforms for COVID-19 diagnostics – RT-PCR and RTK.
Figure I shows an estimated kinetic profile for the SARS-CoV-2. The median incubation period of the SARS-CoV-2 is ~5 days (range 4–14 days) [42,43], and viral load peaks 0–3 days post symptom onset (DPO). Lower respiratory tract samples may remain RT-PCR-positive until 39 DPO [44], although this could be due to viral shedding instead of true infectivity [45]. The immune response of the patient in the form of neutralizing IgM/IgG antibodies appears at ~7 DPO and is detectable in 90–100% of patients by 12–14 DPO [46], when the infectivity of the virus begins to be inhibited by the antibodies. However, antibodies have limited application in detecting infectious patients because they become detectable only in the later phase of the disease when the window to stem viral transmission has closed.
Current commercial SARS-CoV-2 RT-PCR tests have very high sensitivity (detection limit 101–103 copies/ml) [47., 48., 49.] and hence a wide detection window (Figure I and Table I). Antigen testing using RTKs have a relatively short testing window mainly during the first week of symptoms, which marks the infectious period in most patients [50,51]. In general, RTKs provide a sensitivity of 65–89% compared to RT-PCR; this value increases to >90% only when testing samples with Ct values less than 25 [50,52., 53., 54., 55.]. Given this level of sensitivity, RTKs are useful to detect patients who shed large amounts of viral RNA; however, their use will be limited in detecting asymptomatic or presymptomatic patients, where RTK lags RT-PCR by 1–2 days before testing positive. Their lower sensitivity than RT-PCR also makes them unable to confirm negative results in the very early and later phases of COVID-19 infection. As such, RTK can only serve to supplement RT-PCR testing as initial screening tools, unless more work is carried out to increase its sensitivity, such as employing catalytic/fluorescent nanoparticles, crosslinking polymers, electrochemistry, or Raman spectroscopy [56., 57., 58., 59., 60.]. Hence, cost-effective, rapid, and sensitive detection of asymptomatic or presymptomatic cases, which contribute to a significant proportion of virus transmission, remains a challenge.
Figure I.
Kinetic profile for SARS-CoV-2 infection and estimated expression levels of various biomarkers.
Table I.
| SARS-CoV-2 RT PCR test | Reaction volume (μl) | LOD copies/reaction | LOD copies/ml | Antigen RTK | 50% Detection limit |
95% Detection limit |
||
|---|---|---|---|---|---|---|---|---|
| RNA copies/ml | Ct value | RNA copies/ml | Ct value | |||||
| Beijing Wantai SARS-CoV-2 RT-PCR Kit | 40 | 2.0 | 50 | Healgen | 3.8 × 103 | 33.2 | 2.3 × 106 | 24.4 |
| Roche Cobas® SARS-CoV-2 | 100 | 5.2 | 52 | Innova | 2.3 × 104 | 30.7 | Not determined | |
| Abbott Alinity m SARS-CoV-2 assay (Alinity m system) | Unknown (closed system) | 100 | R-Biopharm | 3.1 × 104 | 30.3 | 2.1 × 106 | 24.6 | |
| NeuMoDx SARS-CoV-2 Assay (NeuMoDx 96 and 288 Molecular Systems) | 30 | 4.5 | 150 | Abbott | 5.1 × 104 | 29.6 | 3.5 × 106 | 23.8 |
| Cellbae TEPAT 1.0 SARS-CoV-2 Multiplex RT-PCR Kit |
25 | 6.0 | 240 | Roche-SD Biosensor | 5.5 × 104 | 29.5 | 6.0 × 106 | 23.1 |
| Cepheid Xpert Xpress SARS-CoV-2 Test (GeneXpert Dx or GeneXpert Infinity Systems) | Unknown (closed system) | 250 | Spring Healthcare | 3.8 × 105 | 26.8 | Not determined | ||
| HDPCR SARS-CoV-2 Assay (Applied Biosystems QuantStudio 12K System) | 20 | 5.0 | 250 | E25Bio | 4.3 × 105 | 26.7 | ||
| SD Biosensor STANDARD M nCoV Real-Time Detection Kit | 30 | 7.5 | 250 | Encode | 6.3 × 105 | 26.2 | ||
| CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel | 20 | 6.3 | 316 | SureScreen-F | 8.9 × 105 | 25.7 | ||
| Luminex ARIES SARS-CoV-2 Assay | 50 | 16.7 | 333 | Nal von minden | 1.9 × 106 | 24.7 | 9.3 × 106 | 22.5 |
| BioMérieux SARS-COV-2 R-GENE® | 25 | 9.5 | 380 | SureScreen-V | 2.1 × 106 | 24.5 | Not determined | |
| DiaSorin Molecular Simplexa COVID-19 Direct (Liaison MDX System) | 100 | 50.0 | 500 | Coris BioConcept | 5.0 × 106 | 23.4 | 2.9 × 107 | 21.0 |
| QIAstat-Dx Respiratory SARS-CoV-2 Panel | 60 | 30.0 | 500 | RapiGEN | 4.0 × 108 | 17.3 | 1.6 × 1010 | 12.3 |
| BioFire Respiratory Panel 2.1 (RP2.1) (BioFire FilmArray System) | Unknown (closed system) | 500 | ||||||
| Altona RealStar® SARS-CoV-2 RT-PCR | 30 | 18.8 | 625 | |||||
| EUROIMMUN AG EURORealTime SARS-CoV-2 | 20 | 20.0 | 1000 | |||||
| ThermoFisher TaqPath™ COVID-19 CE- IVD RT-PCR Kit |
20 | 25.0 | 1250 | |||||
| Seegene Inc. Allplex™ 2019-nCoV Assay | 25 | 104.2 | 4167 | |||||
Alt-text: Box 1
Competitiveness of CRISPR-Cas versus RT-PCR and RTKs
Given the inadequate sensitivity of RTKs to detect asymptomatic/presymptomatic patients, who may still be infectious, and that RT-PCR usually requires a central laboratory setting with dedicated facilities, equipment, and trained personnel, there is a need for a rapid on-site test that combines the speed and simplicity of a RTK with the sensitivity and specificity of RT-PCR. Isothermal nucleic acid amplification techniques, such as reverse transcription-recombinase polymerase amplification (RT-RPA) [4] and reverse transcription loop-mediated isothermal amplification (RT-LAMP) [5], reduce the requirement for equipment as compared to RT-PCR, and are able to provide on-site RNA detection capabilities. However, such techniques suffer from non-specific amplification that can lead to reduced clinical specificity [6., 7., 8.]. Their sensitivity can be increased by utilizing sequence/target-specific modules, such as fluorescent probes, oligo strand-displacement probes, or guide nucleotides [9., 10., 11., 12., 13., 14.]. CRISPR-Cas-based diagnostics have emerged as a robust next-generation diagnostic platform (Box 2 ). By combining CRISPR with isothermal techniques and a lateral flow readout, CRISPR has the potential to marry the advantages of RT-PCR with RTK, while overcoming the shortfalls of both platforms.
Box 2. Overview of the CRISPR system.
CRISPR is a microbial adaptive immune system that can be found in ~50% of prokaryotes and ~90% of archaeal organisms [61]. This immune system displays an endonuclease activity that is specific against invading nucleic acids from bacteriophages, transposons, or plasmids [24]. The endonuclease system consists of two main components: (i) a guide RNA (gRNA) that directs the endonuclease activity to a specific sequence, and (ii) an effector CRISPR-associated (Cas) protein that cleaves the target DNA. CRISPR-Cas systems are divided into two classes on the basis of the design principles of the effector proteins [62]. Class 1 systems utilize multiple effector proteins to bind to different parts of the gRNA and the DNA, and to cut the target DNA. In contrast, class 2 systems have a single multi-domain protein that performs all activities. CRISPR-based diagnostics typically adopt class 2 systems because they are simpler to produce. Cas enzymes in the class 2 systems include Cas9, Cas12a/b, Cas13, and Cas14. In Cas12a and Cas13, the gRNA only consists of a short RNA, known as CRISPR RNA (crRNA), which has a segment complementary to the target DNA. In addition to the crRNA, Cas9, Cas12b, and Cas14 also need a second short RNA, known as tracrRNA, to recognize the crRNA. In most biotechnology applications, however, the crRNA and the tracrRNA are joined together into a single guide RNA (sgRNA). A final important feature of class 2 systems is that the target, double-stranded (ds)DNA or RNA needs to have a PAM, which is a short sequence next to the targeted sequence. CRISPR-Cas immune systems in bacteria and archaea record prior infections as spacers within the CRISPR arrays of each system. A CRISPR array consists of DNA repeats and spacers, and the spacers all share a common PAM sequence. In wild-type systems, the lack of PAM in the arrays circumvents the problem of autoimmunity because target sequences in the host genome are not adjacent to a PAM. However, in biotechnology applications, the necessity of the PAM sequence constrains the selection of target sequences. It is worth noting that, although Cas14 belongs to class 2 systems, the need for a PAM has not yet been demonstrated [63]. Currently, CRISPR-Cas systems have been utilized in various applications such as gene editing, and imaging and detection of nucleic acids [64., 65., 66.].
Cas9 enzyme was used in one of the first CRISPR-based diagnostic systems [39., 40., 41.,67,68], and is characterized by its high specificity and ability to differentiate single-base mismatches. Pardee and coworkers utilized a detection system incorporating a toehold switch sensor for colorimetric detection of the Zika virus (ZIKV), and a Cas9 enzyme for discriminating between American and African ZIKV strains. The specific Cas9 target cleavage was incorporated after the last step of a nucleic acid sequence-based amplification (NASBA). The endonuclease incorporation gave rise to an assay named NASBA-CRISPR cleavage (NASBACC) [40]. In short, an isothermal technique is used to amplify target RNA sequences into dsDNA using a primer designed to attach a toehold trigger sequence to the dsDNA. In the presence of an appropriate PAM in the target sequence, the dsDNA is cleaved by Cas9, removing the trigger. Alternatively, in the absence of the PAM, the toehold trigger remains intact, thus activating sensor H and downstream translation of the enzyme LacZ. This generates a colorimetric signal because LacZ catalyzes the conversion of a yellow substrate (chlorophenol red-β-D-galactopyranoside) substrate into a purple product (chlorophenol red). Although this method is sensitive (low femtomolar sensitivity), specific (able to differentiate between different ZIKV strains), and suitable for on-site detection (colorimetric signal via naked eye reading as well as lyophilization-compatible reagents), the RNA amplification (2 h) and sensor H (1 h) reactions require a 3 h runtime, which is even longer than a typical RT-PCR assay. Hence, a faster amplification and signal generation technique is required, especially for virus detection during a pandemic.
Huang and coworkers developed a novel Cas9 detection system based on isothermal exponential amplification reaction (CAS-EXPAR) and nicking endonuclease (NEase)-mediated nucleic acid amplification for fast (<1 h), specific (can detect a single-base mismatch), and highly sensitive (detection limit of 0.82 amol) detection of reverse transcribed total L. monocytogenes RNA as well as DNA methylation [41]. Their method merges the benefits of the site-specificity of Cas9 and the rapid amplification kinetics of EXPAR. In their assay, the gRNA recognizes and binds to the PAM sequence in the target sequence. The gRNA–Cas9 complex then cleaves the target and gives rise to product X. In the subsequent EXPAR isothermal amplification, product X goes through a cyclic amplification to generate a large amount of dsDNA, and consequently fluorescence signal using SYBR Green I dye. The authors determined the detection limit to be 0.82 amol, which is equivalent to ~500 copies/ml (Ct ~36.0), similar to that of most commercial RT-PCR tests (see Table I in Box 1). However, the fluorescence signal generated at the detection limit is very close to that of the blank and may lead to false negative results because it would be difficult to differentiate from a negative control. This would be problematic if the patient is in his/her incubation period and has a low viral load – the patient would not be isolated in a timely manner, leading to potential transmission of the virus to others. Furthermore, the use of fluorescence signal requires dedicated equipment for signal detection, and thus has the same weakness as RT-PCR.
Most CRISPR-Cas assays for COVID-19 utilize class 2 systems, such as Cas12 [8,18,21,33,69., 70., 71., 72.] and Cas13 [7,22,73,74], and employ simple instrumentation that aids cost-efficient POC testing (Figure IA). In addition to cleavage upstream of the PAM site, Cas12 and Cas13 enzymes display non-specific trans cleavage of single-stranded nucleic acids upon target recognition (Figure 1, middle panel). Cas12 cleaves single-stranded DNA (ssDNA) whereas Cas13 cleaves ssRNA. In the presence of a target sequence, the Cas enzyme also carries out collateral cleavage of bystander reporter nucleic acid molecules, and a fluorescein dye-quencher probe is employed for fluorescence readout, and the reporter nucleotide sequence is labeled with both fluorescein and biotin for visual dipstick readout (Figure IB). In the absence of target sequence and hence absence of reporter molecule cleavage, the entire biotin–fluorescein reporter is captured on the control line (immobilized with streptavidin). When gold nanoparticles (Au NPs) conjugated with anti-fluorescein antibody flow past the control line, the Au NPs accumulate there because of the binding of anti-fluorescein antibody to fluorescein. Hence, a visible signal is only generated at the control line. In the presence of the target sequence in a positive sample, cleavage of the biotin-fluorescein reporter causes the fluorescein end to flow towards and be captured at the test line (immobilized with anti-fluorescein antibody). When anti-fluorescein-Au NPs flow past the test line, the Au NPs accumulate there because of binding of the anti-fluorescein antibody to fluorescein, generating visible signal at both the control and test lines. This strategy is similar for all dipstick readouts for CRISPR-Cas detection systems.
Figure I.
CRISPR-Cas assay for COVID-19 detection.
(A) Simple instrumentation needed for POC testing. (B) Mechanism for visual signal readout using a lateral flow dipstick.
Alt-text: Box 2
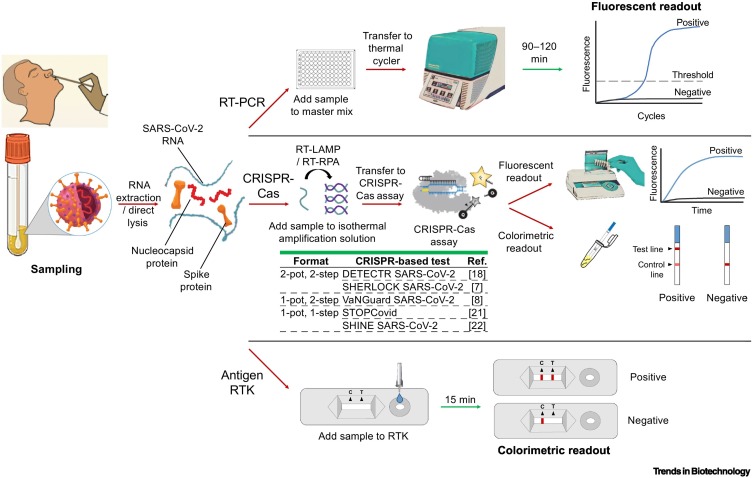
Several assay features affect whether the assay will be adopted by clinicians in a centralized laboratory setting or by healthcare workers on-site at a POC. These features include sample/specimen type (e.g., direct sample or DNA/RNA extracts), turnaround time, throughput, infrastructure/personnel requirements, and the ability for operator to walk away once the assay has started (Figure 1 ). The assay features in turn affect the complexity of the assay and its wider use. Low-complexity tests such as RTKs can be used at the POC, whereas moderate- to high-complexity tests such as RT-PCR assays must be performed in a laboratory for reliable and robust confirmatory testing. RT-PCR is the gold standard for molecular testing because it is the most sensitive/specific and has high throughput capability – able to test 96 (or 384) samples in a single run. Although its turnaround time is relatively long (1.5–2 h), operators can walk away to perform other tasks once the assay has started, and only return at the end of the assay to view/analyze the results. However, the need for trained personnel and specialized equipment requires RT-PCR to be conducted in a laboratory setting. On the other hand, RTKs have much lower sensitivity/specificity and can only run one sample per device, although it is possible to run several devices in parallel to increase throughput. After loading the sample onto an RTK, operators can also leave it unattended during the short run time (15–20 min) and read the results visually afterwards without any instrumentation. Such user-friendly features make RTK a popular choice for POC testing.
Figure 1.
Comparison of the workflows and assay conditions for COVID-19 detection platforms.
Schematic outlining the workflows of RT-PCR, CRISPR-Cas assay, and rapid test kits (RTKs); red and green arrows represent hands-on and hands-free processes, respectively. Formats for CRISPR-Cas assays include (i) a two-pot, two-step format in which isothermal amplification is first performed in one tube and the amplified products are then transferred to a second tube for the CRISPR-Cas assay; (ii) a one-pot, two-step format where the first isothermal amplification is performed in a tube and CRISPR-Cas reagents are then transferred to the same tube; (iii) a one-pot, one-step format in which isothermal amplification and the CRISPR-Cas reaction are performed simultaneously in a single tube.
Although CRISPR-Cas assays are more suitable than RT-PCR for POC testing, and have higher sensitivity than RTKs, their main disadvantage is the added hands-on time and human intervention in their workflow (Table 1 ). In this review we outline the mechanisms and strategies of various major CRISPR-Cas detection systems based on Cas9/12/13. Beyond the background information and detection principles of CRISPR assays (that have been covered elsewhere [15., 16., 17.]), we assess the feasibility of these systems as rapid, on-site diagnostic tests, based on the features mentioned above, with a focus on workflow complexity, signal readout, and detection limit, and compare them to current diagnostic platforms. We have also standardized the limit of detection (LOD) of different commercial RT-PCR methods and RTKs, as well as of different CRISPR-based assays from various laboratories in terms of RNA copies/ml and cycle threshold (Ct) values, allowing direct comparison of their sensitivity from a clinical point of view. This analysis could be particularly valuable to researchers and clinicians, and is currently lacking in the literature. Furthermore, we give an overview of the current shortfalls of CRISPR-based assays, which constitute more hands-on steps and user interventions than RT-PCR and RTKs. We also propose solutions and outline future development directions, such as developing CRISPR-based assays into one-step, one-pot systems to spur their translation to the bedside and adoption by clinicians.
Table 1.
Current advantages and disadvantages of CRISPR-Cas assays compared to RT-PCR and RTKs, and solutions that have been developed to overcome such limitations
| Advantages – comparing CRISPR-Cas to RT-PCR and RTKs | |
|---|---|
| Isothermal: no need for thermal cycling as compared to RT-PCR | |
| Sensitivity is significantly higher than for RTKs | |
| Amenable to both fluorescent (for higher sensitivity) and colorimetric (for on-site detection) readouts | |
| Can be completed within 30–60 min as compared to RT-PCR (90–120 min) | |
| Disadvantages | Solution |
| Sensitivity is lower than RT-PCR for most CRISPR-Cas assays | Magnetic bead-based sample preparation to achieve comparable Ct values to RT-PCR (STOPCovid [21]) |
| Early CRISPR-Cas assays require two operating temperatures because of temperature mismatch between initial isothermal amplification and the subsequent CRISPR-Cas reaction | Use of an engineered Cas enzyme (enAsCas12a), which works over a wide temperature range (37–65°C) (VaNGuard SARS-CoV-2 [8]) |
| Requires additional human interventions in the workflow, such as transferring solution between the isothermal amplification and the CRISPR-Cas assay, and opening of tubes to insert dipsticks for colorimetric readout | Use of thermal stable AapCas12b, which is active at up to 65°C, enabling mixing with RT-LAMP reagents (STOPCovid [21]) Comprehensive and thorough optimization of assay reagents and conditions to enable a one-pot, one-step assay (SHINE SARS-CoV-2 [22]) |
| Using a dipstick colorimetric readout requires longer incubation times for lower viral titers, is less amenable to high-throughput testing, and introduces a risk of sample cross-contamination and exposure to technicians when opening sample tubes | Use of a portable in-tube fluorescent reader, which can image multiple samples in parallel, increase sensitivity, and reduce cross-contamination and personnel exposure (SHINE SARS-CoV-2 [22]) |
CRISPR-Cas systems for COVID-19 detection
Chen and coworkers were among the first to report a Cas12-based system [14]. They used Cas12a from Lachnospiraceae bacteria (LbCas12a) for guide RNA (gRNA)-directed site-specific double-stranded (ds)DNA cleavage and subsequent non-specific single-stranded (ss)DNA degradation. To improve sensitivity, the authors integrated isothermal amplification via RPA to create a DNA-detection platform, known as DNA endonuclease-targeted CRISPR trans reporter (DETECTR), for rapid and accurate detection of human papillomaviruses (HPVs) 16 and 18. Crude DNA extracts from complex samples, such as human anal swabs, were subjected to RPA for 10 min at 37°C before being transferred to the LbCas12a assay, which was also incubated at 37°C. On its own, the LbCas12a assay could detect 109 copies/ml within 30–60 min. When combined with RPA, the sensitivity improved to 102 copies/ml.
Chen and coworkers later adapted the DETECTR assay for the detection of SARS-CoV-2 [18]. This version of the DETECTR assay accepted RNA extracts from nasopharyngeal or oropharyngeal swab samples in universal transport medium (UTM). Next, RT-LAMP amplified target envelope (E) and nucleoprotein (N) genes of SARS-CoV-2 at 62°C for 20–30 min using a standard heat block. Subsequently, the LbCas12a assay consisting of gRNAs targeting the E, N, and human RNase P (internal control) genes was performed at 37°C for 10 min on another heat block (to reduce the time taken for temperature ramping down). Finally, the results were visualized on a dipstick for low-cost, on-site detection. The assay could detect 104 RNA copies/ml with signal saturation in 10–15 min. As compared to the Centers for Disease Control (CDC) SARS-CoV-2 N gene qRT-PCR assay, the DETECTR achieved a 95% positive predictive value (PPV) and a 100% negative predictive value (NPV). Key advantages of DETECTR include simple equipment for isothermal amplification, fast turnaround time (30–40 min), and inexpensive dipstick for signal reporting, as compared to RT-PCR which requires ~120 min, an expensive thermal cycler with fluorescence optics, and trained personnel. Nonetheless, the temperature mismatch between RT-LAMP and Cas12 assay (62°C vs. 37°C) requires the use of two heat blocks, and operators need to transfer 2 μl of amplicon from the RT-LAMP step to Cas reagents. Hence, the workflow requires more human intervention than the automated thermal cycling employed for RT-PCR.
Ooi and coworkers utilized an engineered E174R/S542R/K548R variant of AsCas12a (by Kleinstiver and coworkers), named enAsCas12a [19], which is active over a wide temperature range (37–65°C), enabling their assay to be performed together with RT-LAMP on a single heat block in a more streamlined workflow [8]. The authors also discovered that using hybrid DNA–RNA guides enhances on-target signal, compared to conventional gRNAs, while minimizing off-target activity that increases background signal. Their VaNGuard (variant nucleotide guard) test is also amenable to direct application of nasopharyngeal samples, without RNA purification/extraction, and signal readout on a dipstick. The VaNGuard test is a quasi-one-pot reaction in which, instead of the usual procedure of adding LAMP reaction solution to the CRISPR reaction, CRISPR reaction is transferred into the LAMP tube. This helps to utilize all the LAMP products, thus increasing sensitivity, and there is no need for temperature change. Nevertheless, additional human intervention is still required, making the test less seamless than would be desired. The optimized assay could be completed within 30 min (22 min for RT-LAMP, 5 min for the enAsCas12a reaction, and 2 min for dipstick testing). The VaNGuard test is specific, and does not crossreact with other coronaviruses and respiratory viruses.
Subsequently, the authors validated the VaNGuard test with RNA extracts of clinical nasopharyngeal swabs from 45 COVID-19 patients and 30 healthy individuals. Using a dipstick readout, the test showed unambiguous positive results for samples with Ct values <33.32 (103 copies/ml). Lastly, the authors explored the possibility of testing non-purified clinical samples to reduce user complexity and testing duration by ≥15 min. Patient swabs in UTM were treated with proteinase K and heated for RNase inactivation before the VaNGuard test was performed. Not surprisingly, the cut-off Ct value between a positive and negative sample was less clearcut with this approach. The test would return a positive or negative result for Ct values between 29 and 32. The mean Ct value detectable decreased to 30.0 (104 copies/ml) compared to tests performed on RNA extracts.
Patchsung and coworkers reported clinical validation of their SARS-CoV-2 SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) assay using Cas13a enzyme (Leptotrichia wadei) [7]. The SHERLOCK assay consisted of two steps: (i) RT-RPA amplification reaction at 42°C for 25 min, followed by (ii) Cas13 detection at 37°C for 30 min. Similar to DETECTR, there was a temperature mismatch (unlike the original SHERLOCK [20], which utilized 37°C for the RT-RPA step), and a need to transfer RT-RPA amplicons between the isothermal amplification and Cas detection steps. The results of the Cas13 reaction could also be visualized either on dipsticks or by fluorescence readout.
The authors performed clinical validation on 154 nasopharyngeal and throat swab samples (81 COVID-19 positive samples with N gene of Ct 11–37 and 73 negative samples). Both dipstick and fluorescence readout achieved 100% specificity, while obtaining 87.7% and 96.3% sensitivity, respectively. Samples with Ct <32 were 100% detected via fluorescence and lateral flow readout. For samples of Ct >33, detection with dipsticks was much less reliable, with a false negative rate of 73% (vs. 27% for fluorescence). This was attributed to the need for a critical concentration of cleaved RNA to accumulate sufficient gold nanoparticles (Au NPs) to generate a visible colorimetric signal. SHERLOCK could detect 103 copies/ml and achieved 100% PPV, and 96% and 88% NPV for the fluorescence and dipstick readouts, respectively. Clearly, more work needs to be done to enhance the sensitivity of dipstick readout, as well as to simplify CRISPR-Cas systems to a one-step, one-pot platform for a more user-friendly and seamless workflow.
To achieve this, Joung and coworkers developed SHERLOCK testing in one pot (STOPCovid) [21], which combined RNA extraction, RT-LAMP, and CRISPR-Cas detection at a single temperature in <60 min. Nasal swab samples were dipped into the extraction buffer for lysis and concentration of SARS-CoV-2 RNA with magnetic beads (15 min). Utilizing magnetic beads enabled viral RNA from the entire swab sample (~600× that afforded by the CDC test) to be available during detection. After removing extraction buffer supernatant, STOPCovid reaction mixture was added and incubated at 60°C for 45 min (fluorescence readout) or 80 min (dipstick readout). STOPCovid utilized Cas12b from Alicyclobacillus acidiphilus (AapCas12b), which was active up to 65°C and amenable to RT-LAMP operating temperatures of 55–70°C, enabling a one-pot isothermal amplification Cas reaction. Only standard laboratory equipment is necessary to run STOPCovid, making it suitable for use in low-resource settings. The authors conducted blind testing of 202 COVID-19-positive and 200 SARS-CoV-2-negative nasopharyngeal swab samples, and obtained 93.1% sensitivity and 98.5% specificity. Using magnetic beads for RNA concentration constitutes a more complex sample preparation than thermal/chemical lysis of direct RT-PCR assays, and may be too tedious for on-site testing. Nonetheless, it enabled a good detection limit of 33 RNA copies/ml (Ct 40.3) and 83 RNA copies/ml (Ct 38.4) for fluorescence and dipstick readouts, respectively, which is comparable to RT-PCR, making STOPCovid the most sensitive CRISPR-Cas assay reported to date.
Arizti-Sanz and coworkers developed the SHINE (streamlined highlighting of infections to navigate epidemics) assay [22], which was also based on SHERLOCK, but adapted to detecting viral RNA directly from patient samples without extraction using the HUDSON (heating unextracted diagnostic samples to obliterate nucleases) method [23] to inactivate nucleases and lyse viral particles. Similarly, the authors integrated RT-RPA with Cas13 into one step, and signal could be detected by dipsticks or an in-tube fluorescent readout. To develop the one-step SHINE assay, the authors added reagents, such as RNase H (increases RT efficiency), poly(U) reporter, and SuperScript IV RT, as well as using optimized conditions including pH, monovalent salt, Mg2+, and primer concentrations. The detection limit of the optimized assay was 104 copies/ml spiked into HUDSON-treated UTM and 103 copies/ml spiked into HUDSON-treated saliva using the in-tube fluorescent readout, and the entire workflow took <1 h. Clinical validation with 30 positive patients and 20 negative samples revealed 90.0% sensitivity and 100% specificity. It was found that SHINE could only detect samples with >106 RNA copies/ml; three RT-PCR-positive samples that had ~106 copies/ml were negative on SHINE. Further analytical studies showed that SHINE was unable to detect samples with viral titers of <105 copies/ml, whereas samples with titers ranging 105–106 copies/ml could be detected in one or more technical replicates, and samples with titers >106 copies/ml were detected in all replicates. This corroborates the clinical study results. Although in the right direction, it might be premature for extraction-free, direct detection using CRISPR-Cas systems at present because the detection limit achieved is similar to that of RTKs, and RTKs are less expensive, easier, and faster to perform.
Concluding remarks and future perspectives
As of December 2021, oral antiviral treatments for COVID-19, such as those from Pfizer and Merck, are still in Phase 2/3 clinical trials and have yet to receive Emergency Use Authorization (EUA) from the US Food and Drug Administration (FDA). In addition, vaccination rate is still picking up globally, but with shortages in more resource-limited countries, and early and widespread diagnosis remains the best route to control/contain the transmission of SARS-CoV-2. Even for diagnostic tests, many low- and middle-income countries, especially rural areas, are struggling with stable access to reliable diagnostic tests. Within a few years, CRISPR-Cas detection systems should become clinically viable tests for fast, affordable, sensitive, and specific diagnostics. Given the simple instrumentation required to run the assay and read the signal, CRISPR-Cas systems have good prospects of large-scale implementation, especially at the POC. This is evident because the SHERLOCK SARS-CoV-2 assay received EUA from the US FDA early in the pandemic (May 2020). Since then, the Mammoth DETECTR Boost high-throughput diagnostic kit (in collaboration with Agilent) also obtained EUA in January 2022. Table 2 gives a summary of the major CRISPR-Cas systems. Although we report the estimated Ct values obtained from CRISPR-Cas systems that utilize RNA extraction, the amount of RNA extracted can vary considerably depending on the operator. This variation, for published data in particular, might be skewed towards higher RNA concentrations and would not reflect a higher assay sensitivity. A fairer benchmark would be to first spike a defined amount of RNA into the clinical sample, then measure the amount of RNA obtained from the extraction and report the Ct values obtained, or to transfer a portion of the clinical sample directly into the molecular assay, in other words, ‘strict’ direct application of the clinical sample.
Table 2.
Characteristics of reported CRISPR-Cas detection systems
| Cas enzyme | Name | Isothermal technique | Signal readout | Detection limit (molarity) | Detection limit (copies/ml) | Estimated Ct value | Input sample type | Refs |
|---|---|---|---|---|---|---|---|---|
| Cas9 | CRISDAa | SIBAb | Fluorescence | 6.7 × 10−18 | 4.0 × 103 | 33.1 | DNA extract | [39] |
| CRISPR/Cas9 ZIKV |
NASBA | Colorimetric | 1.0 × 10−15 | 6.0 × 105 | 26.2 | RNA extract | [40] | |
| CAS-EXPAR | EXPAR | Fluorescence | 8.2 × 10−19 | 4.9 × 102 | 36.0 | Synthetic target without complex sample background | [41] | |
| Cas12 | DETECTR HPV |
RPA | Fluorescence | 1.0 × 10−18 | 6.0 × 102 | 35.7 | Synthetic target without complex sample background | [14] |
| DETECTR SARS-CoV-2 |
RT-LAMP | Colorimetric Fluorescence |
1.7 × 10−17 | 1.0 × 104 | 31.9 | RNA extract | [18] | |
| 8.5 × 10−16 | 5.0 × 105 | 26.5 | Direct clinical sample | |||||
| VaNGuard SARS-CoV-2 |
RT-LAMP | Colorimetric Fluorescence |
3.3 × 10−18 | 2.0 × 103 | 34.1 | RNA extract | [8] | |
| 6.6 × 10−17 | 4.0 × 104 | 30.0 | Direct clinical sample | |||||
| STOPCovid | RT-LAMP | Colorimetric Fluorescence |
5.5 × 10−20 | 3.3 × 10 | 40.3 | Direct, but sample was concentrated by >100-foldc | [21] | |
| Cas13 | SHERLOCK SARS-CoV-2 |
RT-RPA | Colorimetric Fluorescence |
4.2 × 10−18 | 2.5 × 103 | 33.5 | RNA extract | [7] |
| SHINE SARS-CoV-2 |
RT-RPA | Colorimetric Fluorescence |
1.7 × 10−16 | 1.0 × 105 | 28.7 | Direct clinical sample | [22] |
Not reviewed in extensor in this article.
Strand-invasion based amplification (SIBA) – a type of isothermal amplification.
A nasal swab was dipped in 400 μl of extraction buffer that contained magnetic beads. After applying a magnet to retain the nucleic acids associated to the magnetic beads and aspiration of extraction buffer, a 50 μl STOPCovid reaction aliquot was added to the beads. By this procedure, the input RNA might be increased by >100-fold because all the RNA in the extraction buffer can be utilized. Thus, strictly speaking, this is not direct application of a clinical sample for the STOPCovid assay because a substantial concentration procedure was performed and added to the entire assay duration.
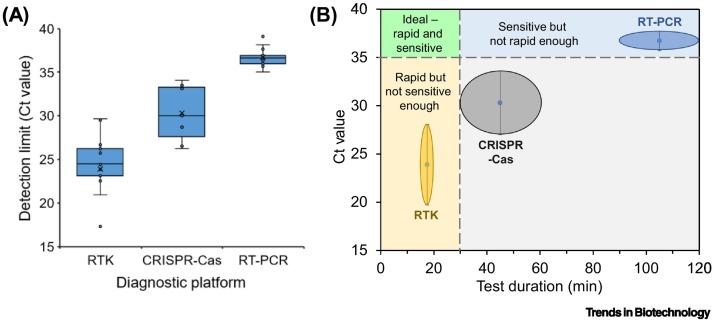
With a detection limit approaching that of the RT-PCR gold standard (Figure 2 ), as well as lateral flow dipstick and in-tube fluorescence reader signal readouts, CRISPR-Cas systems are well poised to become rapid, on-site confirmatory diagnostic tests. The detection limit of antigen by RTKs is on the average 3.4 × 106 copies/ml (~Ct 23.9), which is orders of magnitude away from that of RT-PCR (2.9 × 102 copies/ml, ~Ct 36.7). On the other hand, CRISPR-Cas systems have a detection limit of 3.1 × 104 copies/ml (~Ct 30.3), which is about two orders of magnitude higher than that of RT-PCR. Combined with the fact that CRISPR-Cas systems can be completed in as fast as 30–60 min and detected via a simple colorimetric readout, this new diagnostic platform may gain increasing acceptance as an alternative to RT-PCR and antigen RTKs.
Figure 2.
Comparison of rapid test kit (RTK), CRISPR-Cas, and RT-PCR platforms.
(A) Detection limits of different diagnostic platforms: PCR, CRISPR-Cas, and antigen RTK. To give a better sense of the detection limit of CRISPR-Cas assays in real-world applications, we have only included cycle threshold (Ct) values (Table 2 and Table I in Box 1) obtained from input samples that were either a direct sample or an extracted RNA/DNA sample. We excluded Ct values derived from synthetic targets because this would not reflect the actual scenario in field applications. We have excluded concentrated direct samples because their Ct values are a combination of the intrinsic Ct value of the molecular assay and the Ct units gained from the sample concentration step. (B) Schematic illustration of how the various diagnostic platforms compare to each other in terms of detection limit and test duration. Ct 35 is taken as the sensitivity cut-off because viral load is very low for samples with Ct >35, and most RT-PCR tests have a sensitivity of Ct ≥35. In addition, 30 min is the upper limit for any practical on-site test because most RTKs have a test duration of 15–20 min.
Nonetheless, CRISPR-based detection systems face several challenges and drawbacks (see Outstanding questions). By itself, CRISPR detection is probably only as sensitive as a RTK (105–106 copies/ml) [24], and needs to be paired with an amplification process such as RT-RPA or RT-LAMP to boost its sensitivity to RT-PCR levels. This increases the complexity of the assay because most CRISPR-Cas systems for COVID-19 detection require a two-step process, namely isothermal amplification before conducting the CRISPR assay.
Outstanding questions.
How can the design of CRISPR-Cas assays be made more efficient and streamlined?
Can the signal intensity of dipstick readouts be made more sensitive for more accurate testing and reduced reliance on fluorescence readouts?
Is it possible to make CRISPR-Cas systems more streamlined and reduce the amount of human intervention, perhaps via integration of functional modules?
What optimizations and/or engineering of CRISPR-Cas reagents/enzymes will be necessary to enable and improve RNA extraction-free detection?
Alt-text: Outstanding questions
Thorough optimization of reagents, enzymes, and assay conditions will be necessary for the development of one-step, one-pot, CRISPR-Cas assays. A relative drawback is the need for a protospacer adjacent motif (PAM) sequence next to the target sequence, limiting the number of targets [25], and makes the assay design not as straightforward as RT-PCR. Perhaps the most troublesome part of the design of a CRISPR-Cas assay is that, even with plenty of PAMs within the desired target region, the problem of selecting a highly specific and active gRNA/target remains. In fact, prediction of good targets in terms of specificity is an active line of research in which high-throughput experiments and machine learning have converged to score how specific a target sequence is [26,27]. Currently, most CRISPR-Cas assay developments involve a brute-force approach in which several target regions with many target sequences are screened and evaluated. For example, STOPCovid authors evaluated four target regions in the SARS-CoV-2 genome, each with an average of 15 possible targets. Thus, even if CRISPR-Cas assays might be more user-friendly, they are more difficult to test and develop than RT-PCR.
Furthermore, the detection of more than one target is necessary to provide wider coverage of the SARS-CoV-2 genome, especially because more variants are emerging. Multiplex assays are also useful to detect co-occurring pathogens or to differentiate between pathogens that have similar symptoms (e.g., SARS-CoV-2 and influenza virus). The development of three- or four-plex CRISPR-Cas detection system will become more and more crucial. To achieve this, Gootenberg and coworkers developed SHERLOCKv2, which was capable of four-channel single-reaction multiplexing using four orthogonal CRISPR enzymes (PsmCas13b, LwaCas13a, CcaCas13b, and AsCas12a) whose cleavage activities could be independently measured with four fluorescent dyes (FAM, TEX, Cy5, and HEX) [28]. Briefly, the authors selected the CRISPR enzymes with specific cleavage sequence preferences by evaluating collateral cleavage activity across RNA homopolymer and then dinucleotide motifs, namely AU (LwaCas13a), UC (CcaCas13b), AC (LbaCas13a), and GA (PsmCas13b). By incorporating these specific cleavage motifs into the reporter sequences with different fluorophore–quencher combinations, the authors could identify the presence of specific targets in a single reaction based on the fluorophore signal, for example Zika virus (ZIKV) in the HEX channel and dengue virus (DENV) in the FAM channel. To expand the in-sample multiplexing capability, AsCas12a was utilized for the detection of dsDNA. Jiao and coworkers [29] also developed LEOPARD, a Cas9 multiplexing assay using an exciting gRNA discovery method: in addition to crRNAs, mRNAs can also hybridize to trans-activating CRISPR RNAs (tracrRNAs) and create functional gRNAs. They called this type of crRNA ‘noncanonical crRNA’ (ncrRNA). Their multiplexing idea was to create tracrRNAs against mRNAs of different pathogens. Together in solution, these molecules would self-complement to create pathogen-specific gRNAs that would de facto contain a pathogen sequence for downstream detection. Indeed, the authors created an assay containing different types of pathogenic dsDNAs as signal molecules. When a sample was added with any combination of the mRNAs of the different pathogens, the corresponding hybrid gRNAs were formed and specifically cleaved their cognate dsDNA signal molecules. Using LEOPARD, the authors demonstrated the parallel detection of nine different RNA fragments associated with respiratory viruses. These advances would certainly be crucial in helping CRISPR-Cas assays to overcome their design limitations and expand beyond the detection of SAR-CoV-2.
In addition, visualization on lateral flow dipsticks is generally less sensitive than a fluorescence readout. Thus, enhancing the sensitivity of lateral flow assays [30] needs to be conducted. Alternatively, devices that allow integration of sample preparation, isothermal/CRISPR-Cas assays, and portable, inexpensive, and robust signal readouts are needed. Several CRISPR-Cas systems utilizing microfluidic and portable fluorescence reader, such as a volumetric bar-chart chip [31], all-in-one dual CRISPR-Cas12a (AIOD-CRISPR) assay [32], CRISPR fluorescence detection system (FDS) assay [33], Cas13-based, rugged, equitable, scalable testing (CREST) assay [34], and minimally instrumented SHERLOCK (miSHERLOCK) assay [35], have been developed to fulfill this need.
Finally, as the need for more streamlined and faster testing increases, RT-PCR assay developers have been moving towards direct, extraction-free RNA detection. Although efforts have been made in this direction with CRISPR-Cas systems [8,22], the detection limit achieved is much worse than that of CRISPR-Cas systems utilizing RNA extracted from patient samples. Therefore, more development is needed in this area if CRISPR-Cas systems are to be competitive. A relatively recent study by Lee and coworkers introduced a simplified sample preparation method, S-PREP, as an alternative to full RNA extraction, in their SHERLOCK-based assay to detect Plasmodium species in malaria [36]. The authors optimized various detergents, thermal lysis, and chemical deactivation protocols for their S-PREP, as well as RPA primer and gRNA selection. Patient samples (whole blood or dried blood spots) underwent S-PREP for 10 min at 95°C. Prepared samples were then added to lyophilized SHERLOCK reagents, and the signal could be read using fluorescence or dipstick. The authors achieved LODs of 360 and 60 000 parasites/ml for 60 and 30 min reactions, respectively. Although the 60 min reaction reaches the LOD goal of the WHO for asymptomatic carriage, this CRISPR-Cas assay is still 1.6–3.8 orders of magnitude away from the RT-PCR LOD of 9.3–22 parasites/ml [37,38].
Although we have indicated the estimated Ct values for different diagnostic platforms, these serve only as a limited proxy to compare the detection limits of diagnostic kits. Determination based on standard molecular copies per reaction, or standardized comparisons from the same amount of patient samples, are better forms of comparison that should be undertaken. The field would benefit from sharing the clinical performance of approved CRISPR-Cas-based diagnostics under the FDA EUA framework (or its equivalents in other territories). Paired studies against RT-PCR of such kits by independent clinical laboratories would be important in assessing the suitability of use of new CRISPR-Cas systems. This is particularly important because of (i) the lower sensitivity of CRISPR-Cas systems compared to confirmatory RT-PCR, and (ii) the longer turnaround time (hence, reduced on-site applicability) of CRISPR-Cas systems compared to cost-effective RTKs (Figure 2B). Hence, although major advances have been made with CRISPR-Cas systems, it will be crucial to further improve CRISPR-Cas sensitivity and/or turnaround time, as well as to develop a true one-pot system that does not require additional human intervention during the assay runtime. Otherwise, the use of CRISPR-Cas systems may fall into an unenviable grey area where they are adopted neither as confirmatory clinical tests, because of insufficient accuracy, nor as on-site screening tests, because of longer turnaround time and higher assay complexity.
Tackling the above challenges would enable more widespread deployment of CRISPR-Cas detection systems at POC, customs, and ports of entry, as well as other venues of socioeconomic interest to help different countries to return to a new normal way of life with what seems more and more likely to be endemic COVID-19.
Acknowledgments
Acknowledgments
This work was supported by the Deputyship for Research and Innovation, Saudi Arabia Ministry of Education through International Collaboration Grand Challenge Grant (Project #1095); Cellbae Pte Ltd; COVID-19 Research Fund GAP Funding (NHIC-COV19-2005010) (National Health Innovation Centre, National Medical Research Council, Singapore Ministry of Health); Central Research Fund (CRF, ATR) Award (Biomedical Research Council, A*STAR); NanoBio Lab (A*STAR Infectious Diseases Labs and IMRE, A*STAR); and King Abdulaziz University.
Declaration of interests
J.H.S., E.B., and M.N.A.R. are management members, whereas J.K.C.C. and H-M.C. are members, of Cellbae Pte Ltd which manufactures RT-PCR assays for COVID-19 diagnostics. The other authors declare no competing interests.
Glossary
- CRISPR-Cas
a unique genomic element originating in bacteria and archaea that functions as an adaptive immune defense system against invading bacteriophages or foreign nucleic acids. The system consists of a short, repeated DNA array (clustered regularly interspaced short palindromic repeats, CRISPR) and CRISPR-associated proteins (Cas).
- Cycle threshold (Ct) value
the number of cycles required for the fluorescent signal to exceed background levels in a PCR reaction. Ct values are inversely proportional to the amount of target nucleic acid in the sample (i.e., the lower the Ct level the greater the amount of target nucleic acid present in the sample). Ct values ≤36 are clear positives. Results of suspect, inconclusive/weak positive have Ct values ranging from 37 to 40. This indicates minimal amounts of target nucleic acid which could represent early or late infection, or environmental contamination.
- Limit of detection (LOD)
the lowest concentration of an analyte in a sample that can be consistently detected with a stated probability, typically 95% certainty.
- Protospacer-adjacent motif (PAM) sequence
a short DNA sequence directly adjacent to the DNA sequence that is targeted by the CRISPR effectors.
References
- 1.Nogrady B. What the data say about asymptomatic COVID infections. Nature. 2020;587:534–535. doi: 10.1038/d41586-020-03141-3. [DOI] [PubMed] [Google Scholar]
- 2.Pollock A.M., Lancaster J. Asymptomatic transmission of COVID-19. BMJ. 2020;371:m4851–m4852. [Google Scholar]
- 3.Ghaffari A., et al. COVID-19 serological tests: how well do they actually perform. Diagnostics. 2020;10:453. doi: 10.3390/diagnostics10070453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piepenburg O., et al. DNA detection using recombination proteins. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Notomi T., et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D.G., et al. Two methods for increased specificity and sensitivity in loop-mediated isothermal amplification. Molecules. 2015;20:6048–6059. doi: 10.3390/molecules20046048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patchsung M., et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 8.Ooi K.H., et al. An engineered CRISPR-Cas12a variant and DNA–RNA hybrid guides enable robust and rapid COVID-19 testing. Nat. Commun. 2021;12:1739. doi: 10.1038/s41467-021-21996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori Y., et al. Sequence specific visual detection of LAMP reactions by addition of cationic polymers. BMC Biotechnol. 2006;6:3. doi: 10.1186/1472-6750-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips E.A., et al. Strand displacement probes combined with isothermal nucleic acid amplification for instrument-free detection from complex samples. Anal. Chem. 2018;90:6580–6586. doi: 10.1021/acs.analchem.8b00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Baar M.P., et al. One-tube real-time isothermal amplification assay to identify and distinguish human immunodeficiency virus type 1 subtypes A, B, and C and circulating recombinant forms AE and AG. J. Clin. Microbiol. 2001;39:1895–1902. doi: 10.1128/JCM.39.5.1895-1902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abudayyeh O.O., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353 doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.East-Seletsky A., et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J.S., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., et al. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 16.van Dongen J.E., et al. Point-of-care CRISPR/Cas nucleic acid detection: recent advances, challenges and opportunities. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nouri R., et al. CRISPR-based detection of SARS-CoV-2: a review from sample to result. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broughton J.P., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleinstiver B.P., et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gootenberg J.S., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joung J., et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 2020;383:1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arizti-Sanz J., et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020;11:5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myhrvold C., et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaminski M.M., et al. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021;5:643–656. doi: 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- 25.Rahimi H., et al. CRISPR systems for COVID-19 diagnosis. ACS Sens. 2021;6:1430–1445. doi: 10.1021/acssensors.0c02312. [DOI] [PubMed] [Google Scholar]
- 26.Doench J.G., et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chuai G., et al. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018;19:80. doi: 10.1186/s13059-018-1459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gootenberg J.S., et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiao C., et al. Noncanonical crRNAs derived from host transcripts enable multiplexable RNA detection by Cas9. Science. 2021;372:941–948. doi: 10.1126/science.abe7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soh J.H., et al. Strategies for developing sensitive and specific nanoparticle-based lateral flow assays as point-of-care diagnostic device. Nano Today. 2020;30 [Google Scholar]
- 31.Shao N., et al. CRISPR-Cas12a coupled with platinum nanoreporter for visual quantification of SNVs on a volumetric bar-chart chip. Anal. Chem. 2019;91:12384–12391. doi: 10.1021/acs.analchem.9b02925. [DOI] [PubMed] [Google Scholar]
- 32.Ding X., et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ning B., et al. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauch J.N., et al. A scalable, easy-to-deploy protocol for Cas13-based detection of SARS-CoV-2 genetic material. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.02402-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Puig H., et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee R.A., et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl. Acad. Sci. U. S. A. 2020;117:25722–25731. doi: 10.1073/pnas.2010196117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imwong M., et al. Numerical distributions of parasite densities during asymptomatic malaria. J. Infect. Dis. 2016;213:1322–1329. doi: 10.1093/infdis/jiv596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabias B., et al. Sensitive real-time PCR detection of Plasmodium falciparum parasites in whole blood by erythrocyte membrane protein 1 gene amplification. Malar. J. 2019;18:116. doi: 10.1186/s12936-019-2743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W., et al. A CRISPR-Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018;9:5012. doi: 10.1038/s41467-018-07324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pardee K., et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 41.Huang M., et al. Clustered regularly interspaced short palindromic repeats/Cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal. Chem. 2018;90:2193–2200. doi: 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- 42.Guan W.J., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C., et al. SARS-CoV-2-positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Ann. Intern. Med. 2020;172:832–834. doi: 10.7326/M20-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGrath B.A., et al. Tracheostomy in the COVID-19 era: global and multidisciplinary guidance. Lancet Respir. Med. 2020;8:717–725. doi: 10.1016/S2213-2600(20)30230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J., et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visseaux B., et al. Evaluation of the RealStar(R) SARS-CoV-2 RT-PCR kit RUO performances and limit of detection. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaffaf T., Ghafar-Zadeh E. COVID-19 diagnostic strategies. Part I: nucleic acid-based technologies. Bioengineering. 2021;8:49–77. doi: 10.3390/bioengineering8040049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sohni Y. Variation in LOD across SARS-CoV-2 assay systems: need for standardization. Lab. Med. 2020;52:107–115. [Google Scholar]
- 50.Corman V.M., et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study. Lancet Microbe. 2021;2:e311–e319. doi: 10.1016/S2666-5247(21)00056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 52.Pickering S., et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests and association with detection of infectious virus in clinical specimens: a single-centre laboratory evaluation study. Lancet Microbe. 2021;2:e461–e471. doi: 10.1016/S2666-5247(21)00143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaaskelainen A.E., et al. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. J. Clin. Virol. 2021;137 doi: 10.1016/j.jcv.2021.104785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kohmer N., et al. The comparative clinical performance of four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. J. Clin. Med. 2021;10:328–338. doi: 10.3390/jcm10020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young S., et al. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS antigen point-of-care test. J. Clin. Microbiol. 2020;59 doi: 10.1128/JCM.02338-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao Z., et al. Platinum-decorated gold nanoparticles with dual functionalities for ultrasensitive colorimetric in vitro diagnostics. Nano Lett. 2017;17:5572–5579. doi: 10.1021/acs.nanolett.7b02385. [DOI] [PubMed] [Google Scholar]
- 57.Loynachan C.N., et al. Platinum nanocatalyst amplification: redefining the gold standard for lateral flow immunoassays with ultrabroad dynamic range. ACS Nano. 2018;12:279–288. doi: 10.1021/acsnano.7b06229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soh J.H., et al. Distinct bimodal roles of aromatic molecules in controlling gold nanorod growth for biosensing. Adv. Funct. Mater. 2017;27 [Google Scholar]
- 59.Tran V., et al. Rapid, quantitative, and ultrasensitive point-of-care testing: a portable SERS reader for lateral flow assays in clinical chemistry. Angew. Chem. Int. Ed. Engl. 2019;58:442–446. doi: 10.1002/anie.201810917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Y., et al. Paper-based microfluidic electrochemical immunodevice integrated with nanobioprobes onto graphene film for ultrasensitive multiplexed detection of cancer biomarkers. Anal. Chem. 2013;85:8661–8668. doi: 10.1021/ac401445a. [DOI] [PubMed] [Google Scholar]
- 61.Westra E.R., et al. The ecology and evolution of microbial CRISPR-Cas adaptive immune systems. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2019;374 doi: 10.1098/rstb.2019.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koonin E.V., Makarova K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2019;374 doi: 10.1098/rstb.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harrington L.B., et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cong L., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen B., et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheth R.U., et al. Multiplex recording of cellular events over time on CRISPR biological tape. Science. 2017;358:1457–1461. doi: 10.1126/science.aao0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang T., et al. An RNA-guided Cas9 nickase-based method for universal isothermal DNA amplification. Angew. Chem. Int. Ed. Engl. 2019;58:5382–5386. doi: 10.1002/anie.201901292. [DOI] [PubMed] [Google Scholar]
- 68.Hajian R., et al. Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019;3:427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X., et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci. Bull. (Beijing) 2020;65:1436–1439. doi: 10.1016/j.scib.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang Z., et al. Ultra-sensitive and high-throughput CRISPR-powered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ali Z., et al. iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo L., et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6:34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ponce-Rojas J.C., et al. A fast and accessible method for the isolation of RNA, DNA, and protein to facilitate the detection of SARS-CoV-2. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.02403-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fozouni P., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]