Abstract
We are investigating glycosyl hydrolases from new psychrophilic isolates to examine the adaptations of enzymes to low temperatures. A β-galactosidase from isolate BA, which we have classified as a strain of the lactic acid bacterium Carnobacterium piscicola, was capable of hydrolyzing the chromogen 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) at 4°C and possessed higher activity in crude cell lysates at 25 than at 37°C. Sequence analysis of a cloned DNA fragment encoding this activity revealed a gene cluster containing three glycosyl hydrolases with homology to an α-galactosidase and two β-galactosidases. The larger of the two β-galactosidase genes, bgaB, encoded the 76.8-kDa cold-active enzyme. This gene was homologous to family 42 glycosyl hydrolases, a group which contains several thermophilic enzymes but none from lactic acid bacteria. The bgaB gene from isolate BA was subcloned in Escherichia coli, and its enzyme, BgaB, was purified. The purified enzyme was highly unstable and required 10% glycerol to maintain activity. Its optimal temperature for activity was 30°C, and it was inactivated at 40°C in 10 min. The Km of freshly purified enzyme at 30°C was 1.7 mM, and the Vmax was 450 μmol · min−1 · mg−1 with o-nitrophenyl β-d-galactopyranoside. This cold-active enzyme is interesting because it is homologous to a thermophilic enzyme from Bacillus stearothermophilus, and comparisons could provide information about structural features important for activity at low temperatures.
The ability of microorganisms to adapt to extreme environments raises important questions about the changes in enzyme structure necessary for activity under extreme conditions. For example, psychrophiles living in cold environments need enzymes which remain catalytically active at low temperatures. Current information suggests that enzymes have evolved with limited temperature ranges that provide activity at either high or low temperature but not at both (4). Thus, it is of fundamental interest to explore the parameters that establish the temperature range in which an enzyme is active. One approach would be to obtain data on a large collection of related enzymes, which would allow the sequences and enzymology of enzymes naturally adapted to different thermal optima to be compared. One difficulty with this approach is that there is no comprehensive database containing information on both gene sequences and enzyme attributes. Some papers present enzyme properties but do not contain sequence data (2, 7, 29). Others report sequences but often contain rudimentary enzymatic characterization (20, 32). The biochemical characterization and evolutionary relatedness of these enzymes is rarely discussed in the same forum.
One goal of our work is to provide information on both the biochemistry and the phylogeny of cold-active enzymes, which can be used for comparison with their higher-temperature counterparts. β-Galactosidases are good candidates for building a database for comparisons because they have been studied extensively and have been characterized in a number of organisms. β-Galactosidases are members of a broad group of enzymes, the glycosyl hydrolases, which are responsible for the breakdown of a variety of saccharides. Glycosyl hydrolases have been classified by hydrophobic cluster analysis into 64 families according to sequence similarity (16–18). Four different families of glycosyl hydrolases contain enzymes with β-galactosidase activity. About 25% of the β-galactosidase sequences available in GenBank are from lactic acid bacteria such as Streptococcus, and all of these fall into family 1 or 2 of Henrissat’s classification. Family 1 includes the LacG β-galactosidases and phospho-β-galactosidases, as well as some β-glucosidases. These enzymes are found in Lactococcus lactis (10), Streptococcus mutants (31), and Lactobacillus casei (13). The second group, family 2, contains the well-studied Escherichia coli LacZ enzyme (23) plus β-galactosidases from Streptococcus thermophilus (35) and an isozyme from L. lactis (GenBank accession no. X80037). None of the lactic acid bacteria enzymes show sequence similarity to family 35 or 42, the other β-galactosidase groups.
Lactic acid bacteria have been the focus of extensive research because of their value in the food-processing industry (15, 26). One of the most recent taxonomic additions to the lactic acid bacteria group is the genus Carnobacterium (8, 9), which was first isolated from refrigerated meat products. This genus is physiologically similar to Lactobacillus but differs in certain characteristics, such as the inability to grow on acetate agar and a higher tolerance to oxygen and high pH (33). A 16S rRNA sequence analysis demonstrates that Carnobacterium forms a distinct phylogenetic clade within the lactic acid bacteria (9). Most research on this genus has focused on production and regulation of bacteriocins (19, 27); however, no work has been reported on hydrolases or other metabolic enzymes. Because lactic acid bacteria have been such an integral part of food chemistry, Carnobacterium species are a logical source for the discovery of new catalysts.
We are interested in comparing enzymes within the different families of glycosyl hydrolases in our investigation of cold-active glycosidases from psychrophilic organisms. As part of this investigation, we have isolated a large collection of psychrophilic bacteria and are studying genes from these organisms which encode cold-active enzymes. One isolate, designated Carnobacterium piscicola BA, contained a fragment with two genes, bgaB and bgaC, encoding β-galactosidases. Characterization of the purified BgaB protein showed that the enzyme was thermolabile and had an optimal temperature of activity 20°C below that of E. coli LacZ (25). Furthermore, analysis of the deduced primary amino acid sequence of BgaB indicates that it belongs to the family 42 glycosidases and is phylogenetically related to an enzyme from a thermophile, Bacillus stearothermophilus (20). Its cold activity and similarity to a thermophilic enzyme made the BgaB enzyme of special interest for characterization. Examination of several related enzymes with incremental differences in temperature optima may lead to an understanding of how an enzyme’s “thermostat” for activity is established.
MATERIALS AND METHODS
Isolation and characterization of C. piscicola BA.
C. piscicola BA was obtained from a farm field treated with whey. Samples were taken in late winter and transported and stored at 4°C to increase the probability of finding psychrophilic microorganisms. C. piscicola BA was chosen for study because it hydrolyzed the chromogen 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal; United States Biological, Swampscott, Mass.) as an indicator of glycosidase activity and grew at 4°C on Trypticase soy agar (Becton-Dickinson, Cockeysville, Md.).
Physiological testing of C. piscicola BA was performed with API test strips (Bio-Mérieux Vitek, Inc., Hazelwood, Mo.), and substrate testing of carbohydrate fermentation was done with phenol red broth (10 g of proteose peptone, 5 g of NaCl, 0.018 g of phenol red/liter) at an incubation temperature of 25°C. Cell walls were prepared according to the short method (34), and amino acid analyses were completed at the Protein and Carbohydrate Structure Facility at the University of Michigan.
Phylogenetic analyses used sequence data from the 16S rRNA gene, amplified from C. piscicola BA chromosomal DNA by PCR (Techne Progene thermocycler, Cambridge, England) with primers FD1 and rP2 (39) designed to regions of ghe 16S gene conserved among eubacteria. Ready To Go PCR beads (Amersham Pharmacia Biotech, Piscataway, N.J.) and hot start conditions were used. Amplification was for 30 cycles, with a melting temperature of 94°C for 1.5 min, an annealing temperature of 55°C for 1.5 min, and an elongation temperature of 72°C for 1.5 min. The amplified fragment was sequenced at the ABI automated fluorescence sequencing facility at the Pennsylvania State University. Identification of similar sequences was determined by BLAST sequence analysis at the Ribosomal Database Project web page (Center for Microbial Ecology, Michigan State University) and the GenBank database at the National Center for Biotechnology Information. RNA sequences were compiled in a preliminary alignment with the program MEGALIGN (DNAStar, Inc.) via Clustal V, and alignments were then optimized by eye with the Eyeball Sequence Editor (ESEE) (5). Phylogenetic trees were constructed from these alignments with the program Phylip (12).
Cloning β-galactosidase genes from C. piscicola BA.
Chromosomal DNA was extracted from C. piscicola BA by the Puregene kit (Gentra, Minneapolis, Minn.) protocol for DNA isolation from gram-positive organisms. Chromosomal DNA was subjected to a partial Sau3AI digest, and 2- to 6-kb fragments were purified from a 0.7% agarose gel, using the Bioclean gel extraction kit (United States Biological). DNA fragments were ligated (Epicentre Fast Link ligase; Epicentre Technologies, Madison, Wis.) into a phosphatase-treated plasmid vector (shrimp alkaline phosphatase; Amersham Life Sciences, Arlington Heights, Ill.). The vector, pΔα, was constructed from pUC18 by deletion of the α fragment of the lacZ β-galactosidase normally found on that plasmid (37). Recombinant plasmids were transformed into E. coli DH5α cells and incubated at 37°C on Luria-Bertani agar (10 g of tryptone, 5 g of yeast extract, 10 g of NaCl, 15 g of Bacto Agar per liter) with 100 μg of ampicillin (Fisher Biotech, Fairlawn, N.J.)/ml, 100 μg of X-Gal (United States Biological)/ml, and 0.1 mM isopropyl β-d-thiogalactopyranoside (IPTG; Fisher Biotech). After 16 h, the plates were transferred to an 18°C incubator.
Restriction mapping and sequencing of cloned genes.
Plasmid DNA was isolated from transformants which demonstrated hydrolytic activity on X-Gal. These plasmids were digested with restriction enzymes (Promega Life Sciences, Madison, Wis.) to construct plasmid maps. Both ends of each DNA insert were sequenced with primers designed for conserved regions on the vector DNA. Complete double-stranded sequences of insert regions were obtained by primer walking (The Nucleic Acid Facility, The Pennsylvania State University) and alignment of overlapping DNA sequence with the program ESEE (5).
Subcloning and expression of the bgaB gene product.
The bgaB gene was amplified by PCR, using primers which created unique restriction sites at either end of the gene. The forward primer contained an engineered NdeI site and had the 5′ to 3′ sequence TTTCATATGTTACAGC. The reverse primer had an engineered EcoRI restriction site and had the 5′ to 3′ sequence GACACTAGGAATTCTCCCC. PCR conditions were identical to those used for amplification of the 16S gene, with the exception that the extension time was 2.5 min at 72°C. The PCR product was ligated into the expression vector pET22b (Stratagene Cloning Systems, La Jolla, Calif.) and transformed into MC1061 DE3 cells (λDE3 lysogenization kit; Novagen, Madison, Wis.). Expression of bgaB was induced by adding IPTG to a final concentration of 1 mM. Cell pellets from IPTG-induced cultures were resuspended in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) plus 10% (vol/vol) glycerol before being lysed with a French pressure cell at 1,000 lb/in2. Resolubulization experiments to recover active enzyme from cellular inclusion bodies (80% of overproduced protein) were unsuccessful. Soluble enzyme was collected from the supernatant of the crude cell lysate and used for all subsequent experiments.
Enzyme purification of overexpressed bgaB gene product.
DNA was removed from crude extracts by incubating lysate on ice in the presence of 0.225% (vol/vol) polyethyleneimine. After centrifugation to remove the precipitated DNA, the sample was dialyzed at 4°C against 0.025 M diethanolamine buffer (pH 9.5) plus 10% (vol/vol) glycerol in preparation for chromatofocusing. The enzyme was able to withstand a pH range from 9.5 to 6.0 provided that glycerol was present in the buffers during dialysis and subsequent purification. The dialyzed sample was then injected onto a Mono P chromatofocusing column by using the AKTA protein purification system (Pharmacia Biotech, Uppsala, Sweden). A chromatofocusing gradient was created by running ice-cold Polybuffer 96, 10% (vol/vol) glycerol, pH 6.0 (Pharmacia), over the column after the protein was loaded. In the resulting gradient, BgaB protein eluted between pH 6.9 and 6.5 (the predicted pI of the protein is 6.69). A gel filtration column, Sephadex G-75 (Sigma Chemical Co., St. Louis, Mo.), was required to remove Polybuffer from the purified protein, since this buffer interfered with the protein determination. Column fractions collected in Z buffer with 10% (vol/vol) glycerol were dialyzed against polyethylene glycol 8000 to concentrate the protein sample before subsequent assays. All subsequent assays for enzyme characterization were performed with the purified protein. Verification of the deduced amino acid sequence was performed by N-terminal sequencing of the purified protein (Macromolecular Core Facility, Hershey Medical Center, Hershey, Pa.).
Characterization of the BgaB enzyme.
Triplicate assays were performed with the chromogen ONPG (o-nitrophenyl β-d-galactopyranoside) (28). The substrate specificity was tested with p-nitrophenyl substrates (Sigma). The concentrations of product formed were measured at 420 nm with a Hewlett-Packard diode array spectrophotometer. One unit of activity is defined as 1 μmol of ortho-nitrophenyl product released per min, and specific activity is expressed as micromoles of ortho-nitrophenyl produced per minute per milligram of protein. Protein concentration was determined with the Bio-Rad (Hercules, Calif.) protein assay dye reagent concentrate with bovine serum albumin as a standard. The thermostability and thermal dependency of activity assays were carried out in an Isotemp refrigerated circulator waterbath (Fisher Scientific) with thermal accuracy of ±0.05°C. Kinetic analysis was performed with the Enzyme Kinetics package from Trinity Software (Campton, N.H.).
Nucleotide sequence accession number.
The GenBank accession numbers of the C. piscicola BA bgaB and 16S rRNA gene sequences are AF184246 and AF184247, respectively.
RESULTS
Characterization of C. piscicola BA.
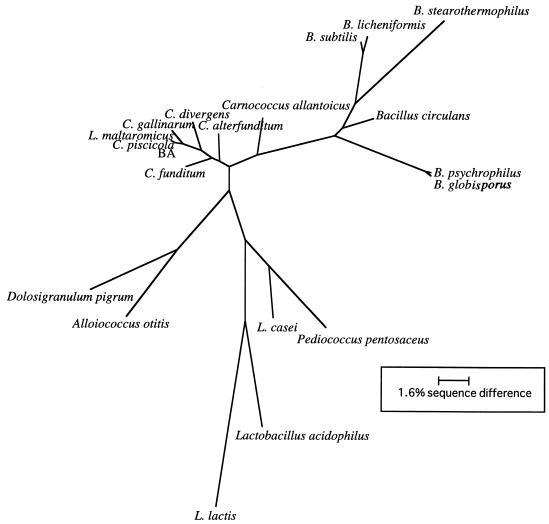
C. piscicola BA was selected from several other isolates because it grew at 4°C and hydrolyzed X-Gal. In addition, initial enzyme assays with crude extracts from this organism indicated the presence of a cold-active β-galactosidase. Because of our interest in this organism, we wanted to examine its phylogenetic relationships and physiological characteristics. We amplified the 16S rRNA gene by PCR, determined its sequence, and examined its phylogenetic relationships by the maximum-likelihood method (Fig. 1). Trees constructed by maximum parsimony were congruent with this analysis (data not shown). C. piscicola BA clustered with the lactic acid bacteria and was most closely related to C. piscicola and Lactobacillus maltaromicus within the Carnobacterium clade. The 16S rRNA sequences of these organisms differed by 3 or fewer nucleotides.
FIG. 1.
Phylogenetic analysis of the sequence from the 16S rRNA gene from isolate BA. 16S rRNA sequences from all other organisms are from the Ribosomal Database Project. Sequences from GenBank were also analyzed for similarity to isolate BA 16S rRNA, but their inclusion did not change the position of isolate BA within the Carnobacterium clade. Phylogenetic relationships are based on MegAlign alignments, optimized in ESEE and analyzed with the program Phylip. Sequence differences between C. piscicola BA and its closest relatives are too small (2 to 3 bp) to show on this scale. B., Bacillus; C., Carnobacterium; L., Lactobacillus.
Gram-stained preparations of C. piscicola BA showed that it was a short, gram-positive rod which did not form spores. The strain was characterized physiologically by examining carbohydrate fermentation and cell wall structure. We also examined growth on acetate agar, since the inability to utilize acetate is one of the key features distinguishing the genus Carnobacterium from Lactobacillus. The results (Table 1) showed the presence of meso-diaminopimelic acid (mDAP) linkage in the cell wall and were consistent with the classification of C. piscicola BA as part of the Carnobacterium genus. Other physiological traits of C. piscicola BA were similar, but not identical, to members of the lactic acid bacteria (Table 1). C. piscicola BA differed from the type strain of C. piscicola in its ability to ferment sorbitol, but it also differed from L. maltaromicus because it could ferment raffinose. Another possible distinguishing feature was its habitat. Neither C. piscicola nor L. maltaromicus has been isolated from soil; however, L. maltaromicus has been found in milk. It is possible that C. piscicola BA was introduced into our soil samples as part of the whey treatment.
TABLE 1.
Comparison of the physiological features of isolate BA with those of other organisms of the lactic acid bacteria group
| Test | Resultsa
|
|||
|---|---|---|---|---|
| Isolate BA | C. piscicolab | L. maltaromicusc | L. acidophilusc | |
| Cell wall linkage | mDAP-direct | mDAP-direct | mDAP-direct | Lys-D-Asp |
| Catalase | − | − | − | − |
| Esculin | + | + | NDd | + |
| Urease | − | − | ND | ND |
| Glucose | + | + | + | + |
| Ribose | + | + | + | − |
| Xylose | − | − | − | − |
| Mannose | + | + | + | + |
| Maltose | + | + | + | + |
| Lactose | + | + | + | + |
| Galactose | + | + | + | + |
| Trehalose | + | + | + | De |
| Sorbitol | + | − | + | + |
| Mellibiose | + | + | + | D |
| Raffinose | + | + | − | D |
| Inulin | + | + | ND | ND |
| Mannitol | + | + | + | − |
| Acetate | − | − | ND | + |
Cloning and sequencing β-galactosidase genes from C. piscicola BA.
Two transformants from a chromosomal library of C. piscicola BA were able to hydrolyze X-Gal at 37°C, and two more were discovered after the plates were shifted to 18°C. Analysis of plasmid DNA from these transformants indicated that each carried a uniquely sized fragment of chromosomal DNA. Sequencing showed that each contained part of the same gene sequence (bgaC), which had homology to a β-galactosidase gene from Xanthomonas manihotis. Upstream of this gene was another open reading frame (ORF) with homology to the bgaB gene from B. stearothermophilus. A third ORF, which coded for a putative α-galactosidase, was detected upstream of bgaB in three of the transformants. These genes are clustered, and the absence of a sequence resembling a known promoter prior to bgaB or bgaC suggests that the three genes form an operon. The two β-galactosidase genes were both relatively small compared to those of the lacZ family, which typically encode subunits of over 110,000 Da. The bgaB gene was 2 kb long and encoded a 668-amino-acid protein. A putative ribosomal binding site (RBS) was apparent 8 bases before the initiating methionine codon and was selected on the basis of sequence similarity to other RBSs from lactic acid bacteria and the absence of any other possible RBSs upstream. The gene had a high mol% A+T, characterized by several series of As or Ts throughout the sequence and by a predominant A-T codon usage bias, reflecting the low G+C mol% (33.0 to 37.2) of the Carnobacterium genus.
The finding of contiguous genes that could all encode glycosidases was interesting. In order to analyze each in depth, we separated them to examine the genes independently. Two of these genes have been subcloned, and their hydrolase activities have been demonstrated with chromogenic substrates. Because initial work suggested that the bgaB gene product had a lower temperature optimum than the bgaC product, it was selected for further characterization.
Phylogenetic analysis of bgaB.
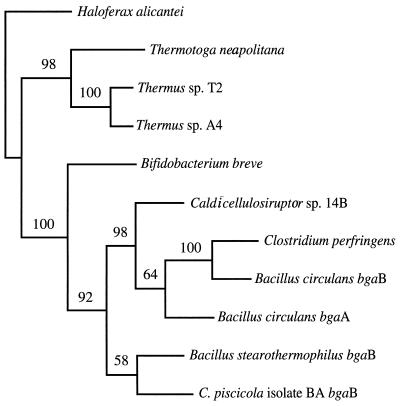
The complete sequence of the bgaB gene was examined for similarity to β-galactosidase genes cloned from other organisms. Two of the sequences that were found have been grouped by Henrissat and Bairoch into family 42 based on sequence similarity (17), and there is a high probability that bgaB from isolate BA also belongs in this group. Both of these enzymes are from thermophilic organisms, bgaB from B. stearothermophilus and a β-galactosidase from thermophilic isolate NA10. Interestingly, there are several thermophilic enzymes with sequences showing homology to sequences of this family. They include genes cloned from Caldicellulosiruptor sp., Thermus sp., and Thermotoga maritima.
Sequence alignments of 11 homologous genes, including bgaB from C. piscicola BA, were optimized by eye in ESEE. These alignments were imported into the PAUP (36) and Phylip (12) programs to examine phylogenetic relationships (Fig. 2). From the diagram, it appears that sequences from the deep-branching eubacteria cluster together and may form a separate family of β-galactosidases. Of all of these sequences, bgaB appears most closely related to the B. stearothermophilus bgaB gene, with 49% sequence identity.
FIG. 2.
Phylogenetic analysis of the bgaB gene, based on its deduced amino acid sequence. Sequences for β-galactosidases from GenBank were aligned with MegAlign and ESEE. Phylogenetic trees were constructed using the program PAUP. Haloferax alicantei was used as the outgroup for this analysis. Bootstrap values were calculated based on 100 replicates. Sequence sources were as follows; Haloferax alicantei (22), Thermus sp. A4 (GenBank accession no. BAA28362), Thermus sp. T2 (38), Thermotoga neapolitana (GenBank accession no. AAC24217), Bifidobacterium breve (GenBank accession no. E05040), Caldicellulosiruptor sp. 14B (GenBank accession no. CAA10365), Clostridium perfringens pbg (GenBank accession no. BAA08485), Bacillus circulans bgaB (GenBank accession no. AAA22260), Bacillus circulans bgaA (GenBank accession no. AAA22258), B. stearothermophilus bgaB (20).
Although genes with similarity to family 42 enzymes have been found in other organisms, it is not clear how prevalent they are because the database is limited to those that have been cloned and sequenced. The availability of genomic sequences provides an independent method of asking about the prevalence or scarcity of these genes. Therefore, to determine whether genes similar to bgaB exist in other organisms that had not been examined specifically for these genes, we searched the available genomic databases for sequences with homology to bgaB. From an analysis of 17 completed genome sequences, genes with similarity to bgaB were detected in the Bacillus subtilis and T. maritima genomes. In the database containing unfinished genomes, raw sequence with similarity to bgaB was found in the genome of Yersinia pestis. This analysis shows that although sequences with similarity to those of family 42 can be found in other organisms, they are not ubiquitous among bacteria or archaea.
Conserved sequences.
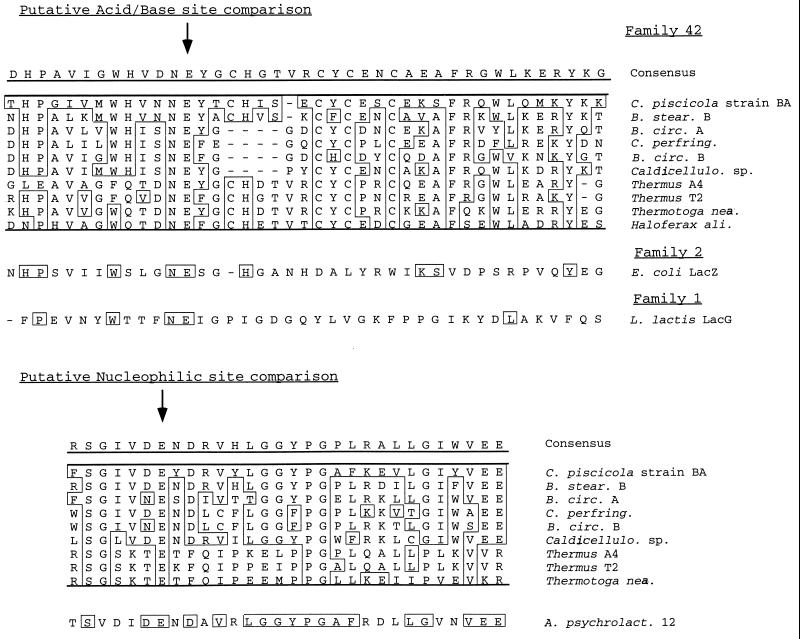
When comparing the deduced amino acid sequence of bgaB with others similar to those of family 42, we made the observation that there were three highly conserved regions shared among all of the sequences in our analysis. These sequences may indicate putative active sites for the enzyme. For the E. coli LacZ enzyme, the mechanism of hydrolysis involves a proton donor (a glutamic acid residue, preceded by an asparagine) and a nucleophile (also a glutamic acid). The L. lactis enzyme and LacS from Sulfolobus sulfotaricus (both family 1 enzymes) have been shown through inhibitor studies to require these specific residues for catalysis as well. Seven other glycosyl hydrolases are also believed to require the same catalytic residues, and these enzymes have been grouped collectively into the GH-A clan (11). Primary sequence comparison reveals some slight similarity among conserved regions of different families, as shown for the acid-base site diagrammed in (Fig. 3). A second conserved region is shown in Fig. 3, which may be the nucleophilic site. This region does not show homology to any of the other families of β-galactosidases, however. The change of the nucleophilic site of family 42 enzymes may mean that this family has a different substrate for activity than others of the GH-A clan. It may also indicate that bgaB from isolate BA shares only partial common ancestry with GH-A but may have arisen through the recombination of a GH-A gene with one from another family.
FIG. 3.
Conserved sequences found for glycosyl hydrolases. The alignment of the proposed acid-base site is compared to the similar sequences of LacG from L. lactis (10) and with the E. coli LacZ enzyme (23). The sequence of a conserved region which may contain the putative nucleophilic site does not demonstrate homology to either LacZ or the family 1 β-galactosidases and is compared to the B7-12 enzyme from Arthrobacter psychrolactophilus (14).
Purification of the BgaB enzyme.
Purification of the BgaB enzyme was difficult due to the sensitivity of the protein to standard purification conditions. Many methods resulted in a substantial loss of activity, including the use of hydrophobic interaction resins and isoelectric focusing. The enzyme also demonstrated significant sensitivity to salts, which prohibited using salt gradients to elute from columns. For example, the enzyme lost 67% of its activity after 5 min in 0.5 M NaCl. Similar results were seen with KCl and to a lesser extent with (NH4)2SO4 (data not shown). The enzyme was rapidly inactivated during storage in crude lysate at 4°C. After considerable experimentation, we found that the presence of 10% glycerol stabilized BgaB, and we were able to purify the enzyme by chromatofocusing, followed by gel filtration to remove the Polybuffer. The enzyme was at least 95% pure as demonstrated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). N-terminal sequencing of the purified protein was performed, and the resulting amino acid sequence showed translation initiation at an unprocessed methionine. The sequence MLQQKKLFYG was identical to the sequence deduced from DNA.
Characterization of the BgaB enzyme.
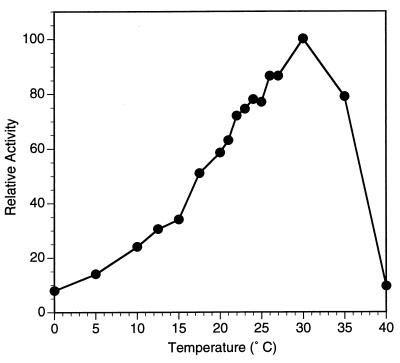
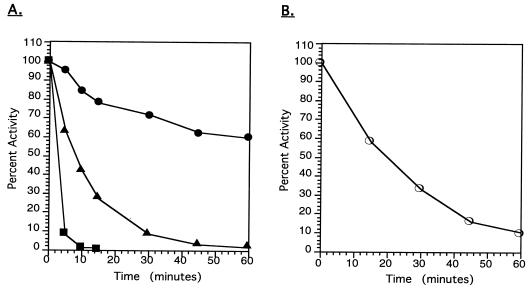
Purified BgaB had an optimal temperature much lower than those of related glycosyl hydrolases. Based on thermal dependence of activity experiments with ONPG as a substrate, the optimal temperature of the purified enzyme was 30°C (Fig. 4). This optimum is at least 25°C lower than that reported for the B. stearothermophilus enzyme (to which it is most similar). When the purified enzyme was tested for stability in the presence of 10% (vol/vol) glycerol, it proved very thermolabile, losing almost half of its activity at 30°C within 1 h and becoming completely inactivated at 40°C within 10 min (Fig. 5A). In the absence of glycerol, the enzyme is extremely unstable even with low-temperature incubation, with an 85% loss of activity at 20°C after 1 h (Fig. 5B).
FIG. 4.
Thermal dependency of activity of purified enzyme BgaB. Enzyme solutions were stored in Z buffer plus 10% glycerol. Activity was assayed for 2 to 10 min in a reaction mixture containing Z buffer without glycerol and with ONPG. The 100% specific activity value is 393 U/mg.
FIG. 5.
Stability of purified BgaB enzyme activity at different temperatures. (A) Enzyme stored in Z buffer containing 10% glycerol was incubated at 30 (●), 35 (▴), or 40°C (■), and aliquots were removed at various times and assayed at 20°C. The 100% specific activity was 179.5 U/mg. (B) Enzyme was incubated at 20°C in Z buffer in the absence of glycerol. Aliquots of enzyme were removed at different times and assayed for activity at 20°C.
Because Carnobacterium species can grow anaerobically, we considered the possibility that the instability of the BgaB enzyme was due to a sensitivity to oxygen. To test this, we assayed the enzyme under anaerobic conditions. The enzyme lost all activity under these conditions even though a control with a β-galactosidase from Kluyveromyces lactis was active under anaerobic conditions (data not shown). Activity of the anaerobic BgaB enzyme was restored when it was subsequently assayed under aerobic conditions.
A study of the substrate specificity of purified BgaB was performed by comparing enzymatic activity on a variety of chromogenic p-nitrophenyl (pNP) analogs. All reactions with pNP β-galactosidase, pNP α-galactoside, pNP β-mannoside, pNP β-fucoside, pNP β-arabanoside, pNP β-xyloside, pNP β-galacturonide, pNP β-glucuronide, pNP β-lactoside, and pNP β-cellobioside were performed at 20°C. The reactions were done with purified protein which had been stored at −20°C and had a specific activity of 19.8 U/mg with ONPG (which contains a β-1-4 sugar linkage). Of the 10 pNP substrates tested, only 3 showed any detectable hydrolysis: pNP β-galactoside (12.2 U/mg), pNP β-fucoside (1.23 U/mg), and pNP β-galacturonide (0.04 U/mg).
Freshly purified enzyme was used to determine the Km, Vmax, and catalytic constant (kcat) values at 20, 25, and 30°C. The Vmax was highest at 30°C, with a value of 450 μmol · min−1 · mg−1 the Km was 1.7 mM, and the kcat was 588 (s−1). The Km was lowest at 25°C, with a value of 1.04 mM, though the value for all three temperatures tested was very close.
DISCUSSION
We are exploring the phenomenon of cold activity in glycosyl hydrolases by characterizing enzymes from organisms adapted to low temperature. The organism described here was isolated from whey-treated Pennsylvania farmlands in late winter. On the basis of 16S rRNA and physiological results, we have determined that the isolate BA is a strain of C. piscicola rather than a separate species, but it retains differences that distinguish it from the type strain. The finding that isolate BA is a member of the lactic acid bacteria is of special interest because β-galactosidases from these organisms make up about 25% of the β-galactosidase gene sequences available in GenBank. The finding of a previously undiscovered β-galactosidase gene from a lactic acid bacterium is especially valuable because others have studied these enzymes extensively for their potential use in the dairy industry.
The fragment encoding β-galactosidase activity cloned from isolate BA had several unique features. First, it contains a putative α-galactosidase gene followed by two contiguous genes encoding different β-galactosidases. No other fragment with a similar arrangement of genes has been reported for any organism, including other lactic acid bacteria. In addition, neither of the β-galactosidase genes is homologous to the LacZ family; in E. coli it is the lacZ gene which encodes the enzyme responsible for lactose utilization. Thus, these enzymes may have functions other than growth with lactose as a carbon source.
We have tested the substrate specificity of the BgaB glycosidase to explore its function. The highest activity was found with the lactose analogs ONPG and pNP β-galactoside, with a low activity with pNP β-fucoside and pNP β-galacturonide. It is possible that BgaB hydrolyzes long-chain sugars, such as exopolysaccharide capsules of other organisms, or it may function in synthesis rather than degradation. Other β-galactosidases have transglycosylation activities that produce oligosaccharides of four or more sugar moieties (30) or are used in the glycosylation of nucleotides (6). Although the function of BgaB is not known, its presence on a fragment containing other glycosidase genes may provide clues. For example, future work will examine the possibility that the three genes are part of a previously undiscovered operon for the degradation of oligosaccharides containing both α and β linkages.
A second surprising feature is that both β-galactosidase genes, bgaB and bgaC, demonstrate homology to gene families not found among other lactic acid bacteria (families 42 and 35, respectively). Family 42, which presently contains up to 13 sequences, does not have any complete gene sequences originating from the lactic acid bacteria group. In Leuconostoc lactis, an ORF has been identified with homology to those of family 42, a pseudogene fragment located upstream of the LacS transporter coding region. This codes for a truncated 95-amino-acid protein which is not known to have a function in the cell. It will be interesting to determine whether other genes similar to those cloned from isolate BA remain undiscovered in other lactic acid bacteria or if they are rare.
Of special importance for future comparisons of cold-active enzymes to their higher-temperature counterparts is the finding that the BgaB protein is closely related to thermophilic enzymes and shows 49% amino acid identity with the BgaB enzyme of B. stearothermophilus. The BgaB enzyme from isolate BA has an optimum around 30°C (Fig. 5), which is at least 25°C below that of the B. stearothermophilus enzyme. In addition, the B. stearothermophilus enzyme maintains 80% of its activity after incubation at 70°C for 30 min (21). We have examined trends in amino acid composition to determine whether changes proposed by others to be characteristics of either cold activity or thermal stability were noted when the isolate BA and B. stearothermophilus genes were compared. The BgaB protein from isolate BA does show an increase in lysine compared to arginine, a decrease in proline, and an increase in serine compared to the protein encoded by the B. stearothermophilus gene. Although such changes have been postulated as mechanisms for maintaining activity at low temperature, whether they are indeed responsible is not clear, since some studies with thermophilic enzymes found that trends in amino acid composition do not always reflect thermostability (1, 3).
One difficulty in determining the features important for setting the temperature range for an enzyme is that many of the comparisons have to be made with distantly related genes because data on closely related genes are unavailable. Unfortunately, it is difficult to separate changes due to other evolutionary factors from those responsible for temperature differences when comparing phylogenetically distant genes. In addition, the database for protein comparisons is limited, especially for cold-active enzymes. Comparisons of these few proteins may not be sufficient to highlight the differences associated with temperature from average variations in protein structure. We have analyzed the properties of several proteins with α/β barrel structures and found that the variation in amino acid composition within a group of thermophilic enzymes most often fits within the average variation found for enzymes from mesophiles (28a).
A considerably larger database of structures and biochemical properties for phylogenetically related thermophilic and cold-active enzymes is needed to help identify the features that set an enzyme’s thermostat. The BgaB enzyme from isolate BA is an ideal candidate for further structural studies. It is a cold-active glycosidase which is likely to have an α/β barrel structure that can be compared to other α/β barrel glycosyl hydrolases in the database. The gene has a 49% identity with its counterpart from B. stearothermophilus, so comparisons of more phylogenetically related proteins can be made. Because structural comparisons may be particularly useful for the BgaB enzyme, future investigations to crystallize it and determine its X-ray structure are planned.
ACKNOWLEDGMENTS
We thank G. Ferry and A. Phillips for helpful discussions and suggestions. We also thank J. Loveland-Curtze for help with cell wall analysis and other members of our laboratory, especially K. Gutshall and P. Sheridan, for their expertise in technical advice and data analysis.
This work was supported by Department of Energy grant DE-FG02-93ER20117 from the Division of Energy Biosciences.
REFERENCES
- 1.Adams M W W, Perler F B, Kelly R M. Extremozymes: expanding the limits of biocatalysis. Biotechnology. 1995;13:662–668. doi: 10.1038/nbt0795-662. [DOI] [PubMed] [Google Scholar]
- 2.Berger J-L, Lee B H, Lacroix C. Purification, properties and characterization of a high-molecular-mass β-galactosidase isoenzyme from Thermus aquaticus YT-1. Biotechnol Appl Biochem. 1997;25:29–41. [Google Scholar]
- 3.Bohm G, Jaenicke R. Relevance of sequence statistics for the properties of extremophilic proteins. Int J Peptide Protein Res. 1994;43:97–106. doi: 10.1111/j.1399-3011.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 4.Brenchley J E. Psychrophilic microorganisms and their cold-active enzymes. J Ind Microbiol. 1996;17:432–437. [Google Scholar]
- 5.Cabot E. The Eyeball Sequence Editor (ESEE), version 1.09d. Rochester, N.Y: University of Rochester; 1990. [Google Scholar]
- 6.Campbell J A, Davies G J, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–942. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi Y J, Kim I H, Lee B H, Lee J S. Purification and characterization of β-galactosidase from alkalophilic and thermophilic Bacillus sp. TA-11. Biotechnol Appl Biochem. 1995;22:191–201. [Google Scholar]
- 8.Collins M D, Farrow J A E, Phillips B A, Ferusu S, Jones D. Classification of Lactobacillus divergens, Lactobacillus piscicola, and some catalase negative, asporogenous, rod-shaped bacteria from poultry in a new genus, Carnobacterium. Int J Syst Bacteriol. 1987;37:310–316. [Google Scholar]
- 9.Collins M D, Rodrigues U, Ash C, Aguirre M, Farrow J A E, Martinez-Murcia A, Phillips B A, Williams A M, Wallbanks S. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol Lett. 1991;77:5–12. [Google Scholar]
- 10.De Vos W M, Gasson M J. Structure and expression of the Lactococcus lactis gene for phospho-β-galactosidase (lacG) in Escherichia coli and L. lactis. J Gen Microbiol. 1989;135:1833–1846. doi: 10.1099/00221287-135-7-1833. [DOI] [PubMed] [Google Scholar]
- 11.Durand P, Lehn P, Callebaut I, Fabrega S, Henrissat B, Mornon J-P. Active-site motifs of lysosomal acid hydrolases: invariant features of clan GH-A glycosyl hydrolases deduced from hydrophobic cluster analysis. Glycobiology. 1997;7:277–284. doi: 10.1093/glycob/7.2.277. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J. PHYLIP (Phylogeny Inference Package). Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 13.Gosalbes M J, Monedero V, Alpert C-A, Perez-Martinez G. Establishing a model to study the regulation of the lactose operon in Lactobacillus casei. FEMS Microbiol Lett. 1997;148:83–89. doi: 10.1111/j.1574-6968.1997.tb10271.x. [DOI] [PubMed] [Google Scholar]
- 14.Gutshall K R, Trimbur D E, Kasmir J J, Brenchley J E. Analysis of a novel gene and β-galactosidase isozyme from a psychrotrophic Arthrobacter isolate. J Bacteriol. 1995;177:1981–1988. doi: 10.1128/jb.177.8.1981-1988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammes W P, Tichaczek P S. The potential of lactic acid bacteria for the production of safe and wholesome food. Z Lebensm Unters Forsch. 1994;198:193–201. doi: 10.1007/BF01192595. [DOI] [PubMed] [Google Scholar]
- 16.Henrissat B. Enzymology of cell-wall degredation. Biochem Soc Trans. 1998;26:153–156. doi: 10.1042/bst0260153. [DOI] [PubMed] [Google Scholar]
- 17.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 19.Herbin S, Mathieu F, Brule F, Branlant C, Lefebvre G, Lebrihi A. Characteristics and genetic determinants of bacteriocin activities produced by Carnobacterium piscicola CP5 isolated from cheese. Curr Microbiol. 1997;35:319–326. doi: 10.1007/s002849900262. [DOI] [PubMed] [Google Scholar]
- 20.Hirata H, Fukazawa T, Negoro S, Okada H. Structure of a β-galactosidase gene of Bacillus stearothermophilus. J Bacteriol. 1986;166:722–727. doi: 10.1128/jb.166.3.722-727.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirata H, Negoro S, Okada H. High production of thermostable beta-galactosidase of Bacillus stearothermophilus in Bacillus subtilis. Appl Environ Microbiol. 1985;49:1547–1549. doi: 10.1128/aem.49.6.1547-1549.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes M L, Scopes R K, Moritz R L, Simpson R J, Englert C, Pfeifer P, Dyall-Smith M L. Purification and analysis of an extremely halophilic β-galactosidase from Haloferax alicantei. Biochim Biophys Acta. 1997;1337:276–286. doi: 10.1016/s0167-4838(96)00174-4. [DOI] [PubMed] [Google Scholar]
- 23.Kalnins A, Otto K, Ruther U, Muller-Hall B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2:593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kandler O, Weiss N. Regular, nonsporing gram-positive rods. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: Williams and Wilkins; 1986. pp. 1208–1260. [Google Scholar]
- 25.Loveland J, Gutshall K, Kasmir J, Prema P, Brenchley J E. Characterization of psychrotrophic microorganisms producing β-galactosidase activities. Appl Environ Microbiol. 1994;60:12–18. doi: 10.1128/aem.60.1.12-18.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKay L L, Baldwin K A. Applications for biotechnology: present and future improvements in lactic acid bacteria. FEMS Microbiol Rev. 1990;87:3–14. doi: 10.1111/j.1574-6968.1990.tb04876.x. [DOI] [PubMed] [Google Scholar]
- 27.Meltivier A, Pilet M F, Dousset X, Sorokine O, Anglade P, Zagorec M, Piard J C, Marion D, Cenatiempo Y, Fremaux C. Divercin V41, a new bacteriocin with two disulphide bonds produced by Carnobacterium divergens V41: primary structure and genome organization. Microbiology. 1998;144:2837–2844. doi: 10.1099/00221287-144-10-2837. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28a.Panasik, N., J. E. Brenchley, and G. K. Farber. Unpublished results.
- 29.Rahim K A A, Lee B H. Production and characterization of β-galactosidase from psychrotrophic Bacillus subtilis KL88. Biotechnol Appl Biochem. 1991;13:246–256. [Google Scholar]
- 30.Rahim K A A, Lee B H. Specificity, inhibitory studies, and oligosaccharide formation by β-galactosidase from psychrotrophic Bacillus subtilis KL88. J Dairy Sci. 1991;74:1773–1778. doi: 10.3168/jds.S0022-0302(91)78341-0. [DOI] [PubMed] [Google Scholar]
- 31.Rosey E L, Stewart G C. Nucleotide and deduced amino acid sequence of the lacR, lacABCD, and lacEF genes encoding the repressor, tagatose 6-phosphate gene cluster, and sugar-specific phosphotransferase system components of the lactose operon in Streptococcus mutans. J Bacteriol. 1992;174:6159–6170. doi: 10.1128/jb.174.19.6159-6170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito T, Kato K, Suzuki T, Iigima S, Kobayashi T. Nucleotide sequence of the lacN gene encoding thermostable β-galactosidase of a thermophilic anaerobe, strain NA10. J Ferment Bioeng. 1992;73:51–53. [Google Scholar]
- 33.Schillinger U, Wood J B, Holzapfel W H, editors. The genera of the lactic acid bacteria. Vol. 2. Glasgow, United Kingdom: Chapman & Hall; 1995. p. 398. [Google Scholar]
- 34.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroeder C J, Robert C, Lenzen G, McKay L L, Mercenier A. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricus β-galactosidase sequences. J Gen Microbiol. 1991;137:369–380. doi: 10.1099/00221287-137-2-369. [DOI] [PubMed] [Google Scholar]
- 36.Swofford D. PAUP (Phylogenetic Analysis Using Parsimony), version 3.1.1. Washington, D.C.: Laboratory of Molecular Systematics, Smithsonian Institution; 1993. [Google Scholar]
- 37.Trimbur D E, Gutshall K R, Prema P, Brenchley J E. Characterization of a psychrotrophic Arthrobacter gene and its cold-active β-galactosidase. Appl Environ Microbiol. 1994;60:4544–4552. doi: 10.1128/aem.60.12.4544-4552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vian A, Carrascosa A V, Garcia J L, Cortes E. Structure of the β-galactosidase gene from Thermus sp. strain T2: expression in Escherichia coli and purification in a single step of an active fusion protein. Appl Environ Microbiol. 1998;64:2187–2191. doi: 10.1128/aem.64.6.2187-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]