Abstract
The USH2A variant c.2276 G > T (p.(Cys759Phe)) has been described by many authors as a frequent cause of autosomal recessive retinitis pigmentosa (arRP). However, this is in contrast with the description of two asymptomatic individuals homozygous for this variant. We therefore assessed pathogenicity of the USH2A c.2276 G > T variant using extensive genetic and functional analyses. Whole genome sequencing and optical genome mapping were performed for three arRP cases homozygous for USH2A c.2276 G > T to exclude alternative genetic causes. A minigene splice assay was designed to investigate the effect of c.2276 G > T on pre-mRNA splicing, in presence or absence of the nearby c.2256 T > C variant. Moreover, an ush2ap.(Cys771Phe) zebrafish knock-in model mimicking human p.(Cys759Phe) was generated and characterized using functional and immunohistochemical analyses. Besides the homozygous c.2276 G > T USH2A variant, no alternative genetic causes were identified. Evaluation of the ush2ap.(Cys771Phe) zebrafish model revealed strongly reduced levels of usherin expression at the photoreceptor periciliary membrane, increased levels of rhodopsin localization in the photoreceptor cell body and decreased electroretinogram (ERG) b-wave amplitudes compared to wildtype controls. In conclusion, we confirmed pathogenicity of USH2A c.2276 G > T (p.(Cys759Phe)). Consequently, cases homozygous for c.2276 G > T can now receive a definite genetic diagnosis and can be considered eligible for receiving future QR-421a-mediated exon 13 skipping therapy.
Subject terms: Mutation, Next-generation sequencing, Genetic variation, Animal breeding, Disease genetics
Introduction
Pathogenic variants in USH2A explain in general 8-19% of cases with autosomal recessive retinitis pigmentosa (arRP) (OMIM: #613809)1,2, 57–90% of Usher syndrome type 2 (USH2) (OMIM: #276901) cases and ~50% of Usher syndrome (USH) cases3,4. To date, more than 2,000 unique variants have been reported in USH2A, most of which are rare. One of the exceptions is the c.2276 G > T variant (hg19/GRCh37: NM_206933.2; g.216420460 C > A; p.(Cys759Phe)) which was first described by Rivolta et al.5. It appeared to be one of the most frequently detected putatively pathogenic variant in USH2A and is therefore regarded to be of high clinical significance. The variant was identified both in cases with arRP and in cases with USH2, although it has a higher reported prevalence in arRP (25.4% of USH2A variants observed in a USH2A cohort) than in USH2 (2.8% of variants)6. Further studies confirmed that USH2A c.2276 G > T is prevalent, representing ~4.5% of all disease-causing alleles in cohorts of arRP cases7,8. In the general population, the overall allele frequency (AF) of c.2276 G > T is 0.097% (273/282,114 alleles, gnomAD v3.1) and 0.14% in the non-Finnish European population (182/128,602 alleles)9. No individuals homozygous for the c.2276 G > T variant have been reported in gnomAD (https://gnomad.broadinstitute.org/variant/1-216420460-C-A?dataset=gnomad_r2_1). Also, the variant is classified as pathogenic by the ClinVar expert panel10 and was recently reported to be significantly enriched in homozygous state, and in compound heterozygosity with a protein-truncating USH2A allele, in a cohort of 982 non-Asian arRP probands after analysis of next-generation sequencing data11. These data suggest putative pathogenicity of c.2276 G > T.
The findings described above are in contrast with the description of two unaffected family members of arRP cases (family S23) that were reported to be homozygous for this variant12. Extensive genetic testing of family S23 revealed a homozygous variant in PDE6B (NM_000283.3: c.1678C > T; p.(Arg560Cys); gnomAD AF: 0.0015%) that fully segregated with arRP in this family. This further strengthened the claim that USH2A c.2276 G > T may not be pathogenic and resulted in the recommendation to re-evaluate all families and cases with this variant13. Until today, arRP cases that are homozygous for USH2A c.2276 G > T therefore do not receive a conclusive genetic diagnosis in a number of diagnostic centers in Europe and the USA. Importantly, a therapeutic approach with antisense oligonucleotides that induce USH2A exon 13 skipping (QR-421a) is currently being evaluated in a Phase I/II clinical trial (ClinicalTrials.gov Identifier: NCT03780257). arRP cases carrying the c.2276 G > T variant, which also resides in USH2A exon 13, could be potentially eligible for receiving this treatment when reaching the market, in case this variant is proven to be pathogenic14.
With the aim to affirm pathogenicity of the c.2276 G > T (p.(Cys759Phe)) variant in USH2A, we implemented a comprehensive array of genetic and functional tests including whole genome sequencing (WGS) for three arRP cases homozygous for the USH2A c.2276 G > T variant and optical genome mapping (OGM) in two of these cases to exclude hidden structural variants. A minigene splicing assay was performed to determine a potential effect of this variant on USH2A pre-mRNA splicing, as well as molecular modeling of the effect of the p.(Cys759Phe) variant on the structure of the associated protein domain. Furthermore, we confirmed a pathogenic effect through a thorough phenotypic assessment of a generated zebrafish knock-in model, ush2ap.(Cys771Phe), that mimics the human USH2A c.2276 G > T (p.(Cys759Phe)) variant.
Results
Whole genome sequencing of arRP cases homozygous for USH2A c.2276 G > T does not reveal additional pathogenic variants
WGS was performed for three individuals with arRP (cases I, II and III), that previously underwent diagnostic exome sequencing and were shown to be homozygous for the USH2A c.2276 G > T variant. To exclude other causes of arRP in these cases, such as intronic variants, structural variants and variants in regulatory elements, a total of 14,343 exonic and intronic single nucleotide variants (SNVs) were evaluated in all 63 previously published arRP-associated genes15. Of these SNVs, 1187 have an AF ≤ 1% in gnomAD and our in-house exome sequencing database containing variants identified in 24,488 individuals. No (likely) pathogenic homozygous or compound heterozygous SNVs were observed, with the exception of USH2A c.2276 G > T (Supplementary Table 1).
All SNVs in USH2A and 200 kilobases upstream and downstream of USH2A (n = 1615) were extracted from WGS data and the number of alleles carrying each SNV was extracted. From chr1:216632415 (35 kb upstream of USH2A) to chr1:216247667 (USH2A intron 27), the majority of SNVs are shared amongst all alleles in the three cases, indicating a shared haplotype (Supplementary Fig 1). SNVs between chr1:216241286 (USH2A intron 30) and chr1:216211989 (USH2A intron 32) are shared amongst four alleles. In total, 1117 SNVs within the genomic region of USH2A were evaluated. Seven SNVs have an AF ≤ 1% and are shared amongst all three cases and were further investigated as potential variants of interest (Supplementary Table 2). Of these seven variants, c.2276 G > T, c.784 + 9428 A > G and c.4628-23020_4628-23007del were identified in a homozygous state in all cases, whereas the synonymous variant c.2256 T > C (p.(His752 = ), gnomAD AF: 0.071%) was homozygous in cases I and III and heterozygous in case II. None of the intronic variants, either shared amongst all cases or unique, identified in USH2A were predicted to have an effect on splicing (>0.1) upon assessment using SpliceAI prediction software16 or were predicted to have a strength of 75% and to result in a minimal increase of 2% in strength for two of the following algorithms: SpliceSiteFinder-like17, MaxEntScan18, NNSPLICE19 and GeneSplicer20, as previously established21. All variants were therefore considered irrelevant.
Potential USH2A regulatory elements were determined based on a database containing predicted cis-regulatory elements identified in post-mortem human retina tissue by Cherry et al.22, epigenetic data derived from mouse photoreceptor cells23, data on open chromatin structures derived from the mouse inner ear24 and DNA-methylation data derived from mice cochlear sensory epithelium25. The Cherry and Genehancer databases were employed to assess putative enhancers that associate with the putative USH2A promoter region22,26. The ten most prominent regions that were identified in at least two out of these five databases, including the predicted promoter region, were selected (Supplementary Table 3). Seven, mainly homozygous and shared, SNVs are present in these ten selected potential regulatory regions of USH2A (Supplementary Table 4). None of these variants has an AF ≤ 1% and these variants are therefore considered benign.
Eight heterozygous variants with an AF < 1% were detected within PDZD7, a previously described modifier for USH2A-associated retinal disease27 (Supplementary Table 5). None of the variants were shared amongst cases. Seven variants were intronic or synonymous, were not predicted to have an effect on splicing, and therefore were considered benign. One variant is located in the 3’UTR and has an AF of 1.04% in the non-Finnish European population. This variant is located in a G-stretch and therefore unlikely to have any effect.
Variants c.2276 G > T and c.2256 T > C do not affect USH2A pre-mRNA splicing
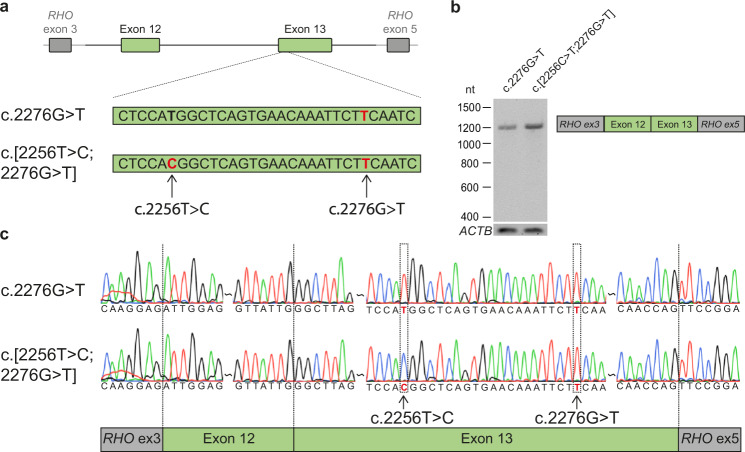
Although c.2276 T > C was not predicted to have an effect on splicing, we wanted to address such an effect. Therefore, we performed minigene splice assays in HEK293T cells to assess a potential effect of the variant on USH2A pre-mRNA splicing either alone or in combination with the nearby synonymous variant c.2256 T > C (p.(His752 = )) present in cis (Fig. 1). The c.2256 T > C variant was already shown to reside in cis with the c.2276 G > T variant when it was first reported5. No indications for alternative splicing were observed and Sanger sequencing confirmed that both exons 12 and 13 were correctly incorporated in the mRNA. Therefore, a pathogenic effect on USH2A pre-mRNA splicing resulting from c.2276 G > T and from the complex allele c.[2256 T > C;2276 G > T] was excluded in HEK293T cells.
Fig. 1. Minigene splice assays for variants c.2256 C > T and c.2276 G > T.
a A minigene splice assay was performed with a construct spanning from USH2A intron 11 to intron 13 (6,814 nt), containing either c.2276 G > T or both variants (c.[2256 T > C;2276 G > T]). b A single RT-PCR product of 1112 nt was observed after expression of both splice vectors, indicative of the incorporation of USH2A exons 12 and 13 between RHO exons 3 and 5 in both transcripts. c Sanger sequencing confirmed that USH2A exon 12 and exon 13 were correctly incorporated in the mRNA.
No pathogenic structural variants were identified in arRP-associated genes
The three studied cases were assessed for the presence of putatively pathogenic structural variants (SVs) or copy number variants (CNVs) in 63 arRP-associated genes, including USH2A. In case I, a large inversion was detected encompassing the entire USH2A gene. However, the breakpoints are located more than 18 Mb up- and downstream of the USH2A gene and its predicted regulatory regions. Moreover, this SV was found in 43% of the Wellderly population database (https://ega-archive.org/studies/EGAS00001002306/) containing 1022 alleles. Therefore, a putative pathogenic or modifying effect on USH2A expression was considered highly unlikely. Five heterozygous deletions encompassing known arRP-associated genes were identified. The regions of these deletions were manually inspected using the Integrative Genomics Viewer software (v2.4.11). In all five regions that were predicted to be heterozygously deleted, several heterozygous SNVs were identified, indicating the presence of two alleles. Therefore, these deletions are likely false positive calls present in the WGS data.
To enable a more accurate detection of chromosomal aberrations that potentially could have been missed by our short-read WGS approach, we employed OGM for case II and III. A total of 6761 and 6739 possible SVs were detected in cases II and III, respectively. Evaluation of these SVs did not reveal any rare (<1% AF in control samples) or unique SVs within or in the vicinity of genes associated with arRP.
Modeling the structural effect of the p.(Cys759Phe) variant on laminin–epidermal growth factor domain 5
The p.(Cys759Phe) variant affects the third cysteine residue within the fifth of ten consecutive laminin-epidermal-growth-factor (EGF Lam) domains that are predicted to be present in usherin28. EGF Lam domains typically contain eight cysteine residues that interact in a pairwise fashion (Cys1 + Cys3; Cys2 + Cys4; Cys5 + Cys6; Cys7 + Cys8) to form four covalent disulfide bonds, that are essential for protein folding and stability. Twenty-nine of the 80 cysteine residues in the usherin EGF Lam domains have been reported to be mutated (USH2A LOVD variation database, https://www.lovd.nl/USH2A; consulted July 13, 2021), indicating that these residues are crucial for usherin function. We assessed the effect of the p.(Cys759Phe) substitution on the structure of EGF Lam domain 5 by molecular modeling (Supplementary Fig 2). The loss of a covalent cysteine bond is expected to cause local destabilization of the domain structure. Furthermore, Cys747 is present as an unpaired cysteine residue containing a reactive-free thiol group that can induce unwanted multimerization or crosslinking with other proteins29. Based on this model, we consider the p.(Cys759Phe) variant detrimental for usherin folding and function.
Functional assessment of the p.(Cys759Phe) variant in an ush2a zebrafish knock-in model
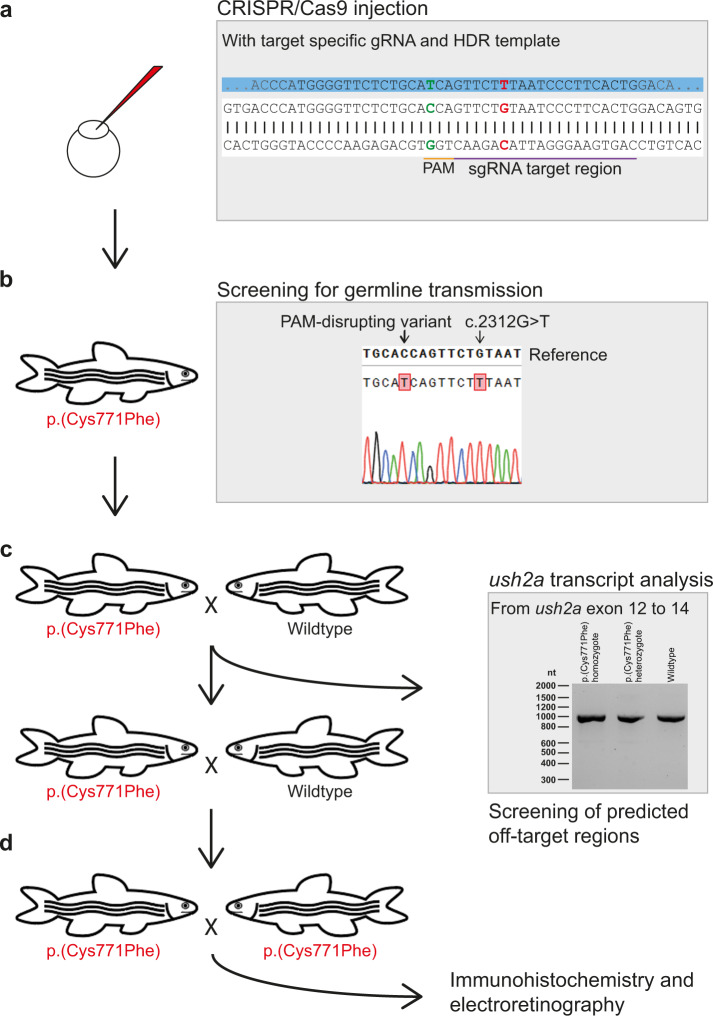
In order to assess the effect of the usherin p.(Cys759Phe) variant on visual function, we used CRISPR/Cas9 technology to generate a zebrafish p.(Cys771Phe) knock-in model, which is the equivalent of the human usherin p.(Cys759Phe) variant. Zebrafish has previously been shown to be a relevant model for USH2A-associated arRP30,31. Fertilized eggs (n = 197) at a single-cell stage were injected with a CRISPR/Cas9 mixture consisting of Cas9 endonuclease and a target specific sgRNA (Fig. 2a). In addition, a 126 nt oligonucleotide homology directed DNA-repair (HDR) template was included in the mixture to enable incorporation of the c.2312 G > T variant (p.(Cys771Phe); ENSDART00000086201.4). The HDR template furthermore contains a silent variant (c.2304 C > T; p.(His768 = )) that disrupts the protospacer-adjacent motif (PAM) and prevents repeated Cas9 cleavage after introduction of c.2312 G > T.
Fig. 2. Generation of the usherinp.(Cys771Phe) zebrafish model.
a Eggs were injected at a one-cell stage with CRISPR/Cas9 mix containing both the target specific single guide RNA (sgRNA) and the 126 nt homology directed repair (HDR) template (partially depicted, in blue) for incorporation of the c.2312 G > T (p.(Cys771Phe)) variant (in red) and protospacer-adjacent motif (PAM, in orange) disrupting variant (c.2304 C > T, in green). The sgRNA target region is depicted in purple. b Once zebrafish were at reproductive age, eggs of the initially injected zebrafish were screened for transmission of the c.2312 G > T and PAM disturbing variant. Three out of ten zebrafish that were screened transmitted the c.2312 G > T variant to their offspring. c Crossbreeding of a c.2312 G > T (p.(Cys771Phe)) zebrafish with a wildtype zebrafish was performed for two generations to reduce unforeseen off-target effects. After the first crossbreeding, genomic DNA was screened for predicted off-target effects of our CRISPR-Cas9 strategy and RNA of homozygous larvae was screened from exon 12 to 14 for deviations on transcript level. d Two p.(Cys771Phe) zebrafish were crossbred with each other to produce homozygous zebrafish. The phenotype of five-day-old larvae was then investigated with immunohistochemistry and electroretinography.
The offspring (F1) of ten individual adult F0 zebrafish was screened for germline transmission of variant c.2312 G > T after cross-breeding with strain-matched wildtypes32. Three out of ten screened F0 zebrafish successfully transmitted the variant to their progeny. We did not detect any alterations in ush2a exon 13 other than c.2312 G > T and c.2304 C > T, compared to DNA from wildtype zebrafish (Supplementary Fig 3). In order to minimize the presence of potential CRISPR/Cas9-induced off-target modifications, two generations of outbreeding with strain-matched wildtypes were performed (F2), followed by an inbreeding of two heterozygous zebrafish. The resulting model is homozygous for both the variant of interest (c.2312 G > T; p.(Cys771Phe)) and the silent PAM-disrupting variant (c.2304 C > T; p.(His768 = )) (Fig. 2b). Transcript analysis on homozygous c.2312 G > T larvae was performed to identify potential effects on ush2a pre-mRNA splicing resulting from the introduction of the ush2a c.2312 G > T and c.2304 C > T. RT-PCR from exons 12 to 14 on larval mRNA of amplicons did not reveal any alternative splicing events or other sequence alterations in homozygous knock-in zebrafish as compared to their wildtype siblings, indicating that both introduced in ush2a exon 13 do not have an effect on splicing (Fig. 2c), which was validated by Sanger sequencing.
Four potential off-target regions for the used sgRNA were identified within the zebrafish genome using the Cas-OFFinder webtool33. F2 larvae derived from either of the three germline-positive F0 founder zebrafish (fish 1–3) were screened for the presence of potential off-target edits by PCR and Sanger sequencing (Supplementary Table 6). One SNV was identified (heterozygous in fish 1 and homozygous in fish 2 and 3) in the vicinity of the predicted off-target region on chromosome 20 (chr20:41755382 G > A (GRCz10)). However, the variant was located 86 nt upstream of the predicted off-target site and labeled as a known SNV in the ENSEMBL zebrafish genome browser. The SNV was therefore considered irrelevant in our screening. A second SNV (chr5:40167334 G > A (GRCz10)) was identified exactly at the predicted off-target site in heterozygous state in fish 1. We therefore decided to exclude this zebrafish from further studies. For a predicted off-target site on chromosome 17 (chr17:25767450 (GRCz10)), no sequence aberrations were observed. We failed to amplify the region of interest for a fourth potential off-target site located deep within intron3 of the myo18b gene (KX421389) at chromosome 10 (chr10:44374669 (GRCz10)).
Introduction of p.(Cys771Phe) results in reduced usherin expression levels, pronounced rhodopsin mislocalization and reduced ERG traces
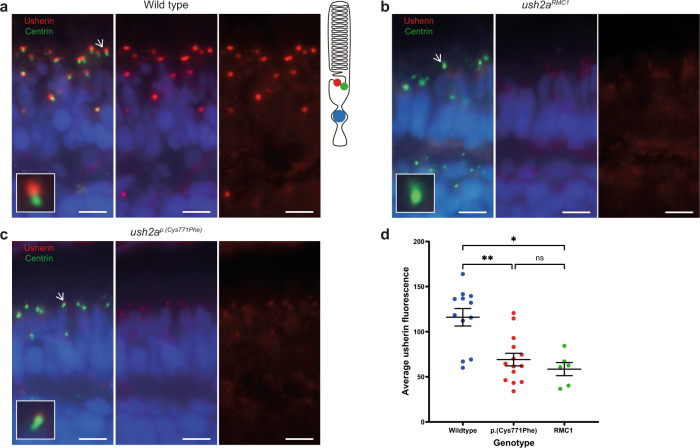
To detect functional effects of the p.(Cys771Phe) substitution, we evaluated the expression and subcellular localization of usherin using cryosections of ush2ap.(Cys771Phe) larval eyes (5 days post-fertilization (dpf); n = 14 eyes of 7 larvae), as well as of strain- and age-matched wildtypes (n = 12 eyes of 6 larvae) and usherin knock-outs (ush2armc1) (n = 6 eyes of 3 larvae). As was previously described30, usherin localizes to the photoreceptor periciliary membrane adjacent to the basal body and connecting cilium marker centrin in wildtype larvae (Fig. 3a), and is absent from retinal cryosections of ush2armc1 knock-out larvae (Fig. 3b). In our ush2ap.(Cys771Phe) mutants, the usherin signal intensity at the periciliary membrane appeared significantly decreased and no clear mislocalization of mutant usherin protein at other regions within the larval zebrafish retina was observed (Fig. 3c). These results suggest that the p.(Cys771Phe) variant significantly reduces usherin expression in the zebrafish photoreceptors (Fig. 3d).
Fig. 3. Reduced expression level of usherinp.(Cys771Phe) at the photoreceptor periciliary membrane.
a In wildtype zebrafish larval eyes (n = 12 eyes), usherin (red signal) localizes at the photoreceptor periciliary membrane adjacent to the basal body and connecting cilium marker centrin (green signal) as shown by the schematic representation of a photoreceptor on the right. A magnification of one photoreceptor (indicated by an arrow) is depicted in the inlay. b In ush2armc1 knock-out larvae usherin is not detectable (n = 6 eyes). c Localization of usherin at the photoreceptor periciliary membrane was strongly reduced in eyes of ush2ap.(Cys771Phe) larvae (n = 14 eyes) as compared to wildtype. d A Kruskal–Wallis test was performed based on the average of the mean grey value for usherin adjacent to each centrin spot and confirmed a significant decrease of usherin localization adjacent to centrin for the usherinp.(Cys771Phe) and the usherinRMC1 models. The average grey value per retinal section was plotted in a scatter plot (mean ± SEM). Nuclei are stained with DAPI (blue signal). Scale bar: 5 µM. **p: 0.0094, *p: 0.0107, ns not significant.
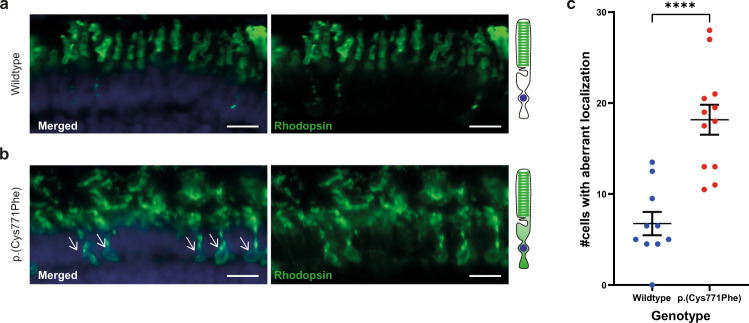
As rhodopsin was previously found to be partially mislocalized towards the photoreceptor cell body of ush2a zebrafish knock-out models31, we also investigated the localization of rhodopsin in our ush2ap.(Cys771Phe) knock-in model. Cryosections were made from wildtype (n = 10 eyes of 5 larvae) and ush2ap.(Cys771Phe) larval eyes (n = 12 eyes of 6 larvae). We observed a significant increase in the number of photoreceptor cells with aberrant rhodopsin localization in ush2ap.(Cys771Phe) as compared to matched wildtype controls (unpaired Student’s t test (two-tailed); p < 0.0001; t: 5.318, df: 20) (Fig. 4).
Fig. 4. Rhodopsin localization to the photoreceptor cell body in the ush2ap.(Cys771Phe) zebrafish.
a Rhodopsin (green signal) localizes to the rod photoreceptor outer segments in wildtype larval eyes. A visual representation of a photoreceptor is shown on the right. b Aberrant localization of rhodopsin to the photoreceptor cell body was observed in ush2ap.(Cys771Phe) larvae (indicated by white arrows), which was observed in a significantly higher number of photoreceptors of ush2ap.(Cys771Phe) larvae as compared to wildtype larvae (c; unpaired t test). Nuclei are stained with DAPI (blue signal). Scale bar: 10 µM. ****p < 0.0001.
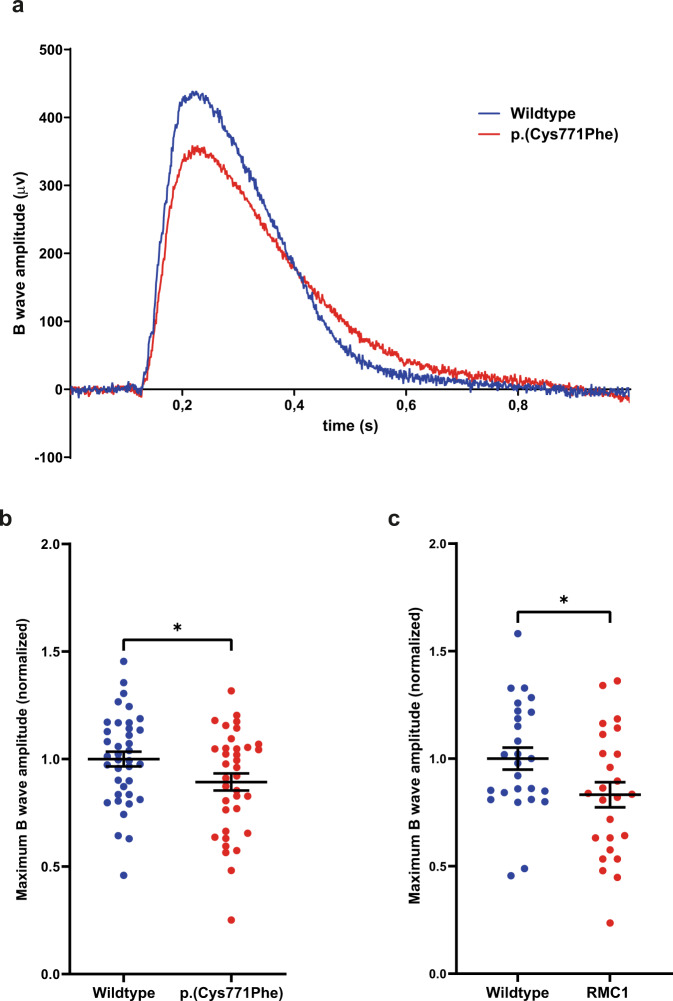
Finally, ERG responses were recorded for both wildtype (n = 38 from three biological replicates) and p.(Cys771Phe) (n = 36 from three biological replicates) larvae at 5 dpf. A significant decrease in maximum B wave amplitudes (~11%) was observed in the p.(Cys771Phe) larvae as compared to wildtype larvae (unpaired Students t test (two-tailed); p: 0.0445; t: 2.045; df: 72), which is indicative of an impaired visual function (Fig. 5b). ERG responses were also recorded for ush2armc1 knock-out larvae (5 dpf; n = 25 from three biological replicates) and age- and strain-matched wildtype siblings (n = 26 from three biological replicates). In the knock-out model a similar significant decrease in maximum B wave amplitude of ~15% compared to age- and strain-matched wildtype larvae was observed (unpaired Student’s t test (two-tailed); p: 0.0345; t: 2.175, df: 49) (Fig. 5c).
Fig. 5. Electroretinogram recordings show that ush2ap.(Cys771Phe) mutants are vision impaired.
a Representative electroretinograms of a p.(Cys771Phe) zebrafish and a wildtype sibling at 5 days post-fertilization. b The maximum B wave amplitude was significantly lower in p.(Cys771Phe) zebrafish as compared to wildtype siblings (unpaired t test). The average wildtype amplitude was normalized to 1. Each datapoint corresponds to recordings from an individual larvae (mean ± SEM). *p: 0.0445. c A comparative analysis of ush2armc1 knock-out larvae and age- and strain-matched wildtype larvae was shown to result in a similar and significant decrease in maximum B wave amplitude. Again, the average wildtype amplitude was normalized to 1. Each datapoint corresponds to recordings from an individual larvae (unpaired t test, mean ± SEM). *p: 0.0345.
Discussion
Here, we confirmed the pathogenicity of the USH2A c.2276 G > T (p.(Cys759Phe)) variant, based on comprehensive genetic analyses, protein modeling and functional assessment of a zebrafish knock-in model. We did not identify any other potentially pathogenic variants within the haplotype containing c.2276 G > T, neither did we identify bi-allelic variants in other arRP-associated genes. With protein modeling we showed that the substitution of phenylalanine for cysteine affects one of four disulfide bridges essential for a proper folding of EGF Lam domain 5, which is indicative of the pathogenic nature of p.(Cys759Phe). Finally, we evaluated the effect of the p.(Cys759Phe) variant on visual function after scrutinizing the ush2ap.(Cys771Phe) zebrafish knock-in model. We observed significantly reduced levels of usherin p.(Cys771Phe) at the zebrafish photoreceptor periciliary membrane and increased levels of aberrantly localized rhodopsin, which was previously shown to be a hallmark of ush2a-associated retinopathy in a zebrafish knock-out model31. Moreover, ERG recordings demonstrated that visual function was impaired in ush2ap.(Cys771Phe) zebrafish larvae. These results are similar to the reduced B wave amplitudes recorded in the previously published ush2armc1 knock-out model (Fig. 5c) and similar to the previously published ERG recordings on ush2a knock-out zebrafish models shown by Han et al. and Toms et al., confirming pathogenicity of the p.(Cys771Phe) variant31,34.
Our conclusion that c.2276 G > T is pathogenic is in agreement with the analyses provided by many other groups, but does not correspond with the conclusion of Bernal et al12. In their paper, they reported two healthy individuals of family S23 that are homozygous for c.2276 G > T. As a result, Bernal et al. recommended a genetic re-evaluation of families with this variant. A panel of 26 IRD-associated genes was subsequently analyzed and the absence of the deep-intronic c.7595–2144 A > G variant in USH2A was verified by Sanger sequencing in affected and unaffected individuals of family S2313. With this approach, a homozygous variant in PDE6B (c.1678C > T (p.(Arg560Cys)) was identified in affected family members. The variant is equivalent to mouse Pde6b c.1678C > T (p.(Arg560Cys), which has been identified as the causal variant in a previously reported mouse model for retinal degeneration35. In contrast, the healthy homozygous carriers of USH2A c.2276 G > T were shown to be heterozygous for the variant in PDE6B. Currently, we can only speculate about a possible explanation for the discrepancy between our findings and the findings of Bernal and colleagues. Reanalysis of family S23 with current genetic screening methods, such as WGS and OGM, would be recommendable. A first hypothesis could be an undetected and novel (deep) intronic variant resulting in the in-frame excision of USH2A exon 13 in these individuals. Skipping of specifically USH2A exon 13 from the transcript was recently shown to result in the production of a functional usherin protein14. A second hypothesis is the presence of modifier variants. PDZD7 is a known disease-aggravating modifier of USH2A-associated retinal disease27. However, to date no protective modifiers of USH2A-associated disease have been reported, in contrast to what has been reported for other disorders. For example, a homozygous deletion of the SMN1 gene results in a spinal muscular atrophy (SMA) phenotype, however asymptomatic individuals have also been reported having the same homozygous deletion as their affected siblings36. Unaffected individuals exhibit increased levels of PLS3 which rescues the detrimental effects on axon growth that underly SMA and as a consequence PLS3 was suggested to act as a protective modifier for SMA. Also in sensory disorders, a similar protective effect has been observed in a mouse model for GRXCR2-associated hearing loss37. Grxcr2-deficient mice showed pronounced hearing loss and disorganized stereocilia. However, reduced levels of taperin, which has also been linked to deafness, in Grxcr2-deficient mice restored their hearing and stereocilia morphology.
Although not identified so far, protective modifiers of USH2A-associated disease might exist and similar mechanisms as described for SMA and deafness could protect against USH2A c.2276 G > T-associated arRP. Either a negative modifier or an unknown pathogenic variant on the c.2276 G > T haplotype could result in disease in the majority of cases with c.2276 G > T-associated arRP, but these could be absent in family S23. DuPont and colleagues identified a shared haplotype of 199 kilobases, spanning from USH2A exon 14 to exon 25, that could harbor a pathogenic variant other than c.2276 G > T11. Based on our WGS data, we also determined a haplotype by identifying SNVs that are shared amongst all three cases in our study (Supplementary Fig 1). A ‘homozygosity block’ was observed with SNVs that are shared amongst all alleles and that spans the region from 35 kb upstream of USH2A to intron 27. This supports the conclusion from DuPont and co-workers of a shared haplotype containing c.2276 G > T. However, they suggest a possible recombination causing c.2276 G > T to be no longer linked with the true disease-causing variant in the family of Bernal et al. 2003. To the best of our ability, we excluded the presence of pathogenic variants other than c.2276 G > T in the cases in our study with both WGS and OGM, and demonstrated that a possible recombination is highly unlikely.
Our study provides indisputable evidence that the USH2A c.2276 G > T; p.(Cys759Phe) variant is pathogenic, which can be further demonstrated in the future by treatment of arRP cases homozygous for USH2A c.2276 G > T with QR-421a14. Future studies will be needed to unravel the exact mechanism of disease underlying arRP caused by the p.(Cys759Phe) variant in usherin.
Methods
Whole genome sequencing
DNA of three unrelated probands (I, II and III), clinically diagnosed with arRP and homozygous for c.2276 G > T, was obtained by the Radboud University Medical Center (Nijmegen, the Netherlands). Written informed consent was received from all individuals, adherent to the tenets of the Declaration of Helsinki and as approved by the local ethics committee of the Radboud University Medical Center Nijmegen, as an amendment to the approval by the local ethics committee of the Rotterdam Eye Hospital (MEC-2010-359; OZR protocol no. 2009-32). The clinical status of all cases is listed in Supplementary Table 7. WGS data processing and variant calling was performed using genome assembly 37 (hg19), as published previously38.
WGS data of the three cases were evaluated for variants in 63 genes described to be associated with arRP (Supplementary Table 8) (Retnet; visited May 21, 2021)15, PDZD727 and potential USH2A regulatory regions22–26. SNVs were considered potentially pathogenic if they had an overall gnomAD AF ≤ 1%9 and were either (1) a stop gain variant, frameshift variant or canonical splice site variant, (2) a missense variant with either; Grantham score ≥ 80 (range: 5–215), CADD_PHRED ≥ 15 (range: 0–99) or PhyloP ≥ 2.7 or 3) a putative splice-modulating variant with a spliceAI score ≥ 0.1 (default settings, range 0–1)16 or a score ≥ 75% with a minimal increase of 2% in strength21 for two of the following algorithms: SpliceSiteFinder-like17, MaxEntScan18, NNSPLICE19 and GeneSplicer20.
CNVs and SVs that passed our quality filter and had a high quality score (≥500/1000) were prioritized if these affect the aforementioned 63 genes associated with arRP. OGM was performed for cases II and III, as described previously39,40. Variants with an AF ≤ 1% in 107 control samples and that overlap genes that are known to be involved in arRP (Supplementary Table 8) were evaluated.
Minigene splice assay
USH2A exons 12 and 13 and ~1000 nt of intronic sequence upstream of exon 12 and downstream of exon 13 were amplified from genomic DNA of case II (heterozygous for c.2256 T > C (NM_206933.2; p.(His752 = )), homozygous for c.2276 G > T) (primers 5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCcctccaaatgtgaagagctgg-3’(forward); 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTGtgcagtggtccttctccttag-3’(reverse)). Both c.2276 G > T and c.[2256 T > C;2276 G > T] fragments were individually cloned between RHO exons 3 and 5 of an adapted pCI-NEO vector, with Gateway® cloning technology (Thermo Fisher Scientific, Carlsbad (CA), USA), as described previoiusly41. USH2A cDNA was amplified (primers 5’-CGGAGGTCAACAACGAGTCT-3’(forward); 5’-AGGTGTAGGGGATGGGAGAC-3’(reverse)) as well as ACTB cDNA as loading control (primers 5’-ACTGGGACGACATGGAGAAG-3’(forward); 5’-TCTCAGCTGTGGTGGTGAAG-3’(reverse)). Both samples and loading control were processed in parallel.
Modeling the fifth laminin–epidermal growth factor like domain
A homology model of the fifth EGF Lam domain was created using the YASARA & WHAT IF Twinset homology modeling module42,43. A hybrid model of residues 747–792 (PDB files 5LF2) was created representing Laminin beta 2 from the rat, with a sequence identity of 46%44.
Zebrafish maintenance and husbandry
The animal experiments were approved by the Radboud University Institutional Review Board of the Centrale Commissie Dierproeven (AVD103002017945). All experiments were carried out in accordance with European guidelines on animal experiments (2010/63/EU). Wildtype Tüpfel Long fin (TLF) zebrafish were used. All animals were maintained and raised by standard methods45.
CRISPR/Cas9 approach for the generation of an ush2ap.(Cys771Phe) knock-in zebrafish model
A constant single guide RNA (sgRNA-1, AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC), a target specific single guide RNA (sgRNA-2, CCGCTAGCTAATACGACTCACTATAGAGTGAAGGGATTACAGAACGTTTTAGAGCTAGAAATAGCAAG) and a 126 nt oligonucleotide template (TTGTAACTATGGCTTCAAATTCCTCAATCACACCAATCCCGATGGTTGCATTTCCTGTGGCTGTGACCCATGGGGTTCTCTGCATCAGTTCTTTAATCCCTTCACTGGACAGTGTGAGTGCAAAGC) for HDR were designed, generated and micro-injected in zebrafish zygotes as previously described32. The resulting ush2ap.(Cys771Phe) allele is deposited as ush2armc016 in the ZFIN database (www.zfin.org).
Genotyping
Larvae were collected from separate breeding pairs 1 dpf. Zebrafish samples, being either individual embryos (1 dpf), larval tail fins (5 dpf) or adult tail fins, were lysed in 25 µl (embryos, larval tails) or 75 µl (adult tail fins) lysis buffer (0.2 mM EDTA, 40 mM NaOH) at 95 °C for 20 min and then neutralized with 2.5 µl or 7.5 µl 1 M Tris-HCl (pH 8). Obtain DNA was used as input for a PCR using standard cycling conditions to amplify the genomic region of ush2a exon 13 (primers 5’-TTCCTCAATCACACCAATCCCG-3’(forward); 5’-TGCCTCTGAGATCACACTGA-3’(reverse)). Fragments were analyzed using Sanger sequencing.
Screening for CRISPR/Cas9-induced off-target editing
Cas-OFFinder was employed to identify potential off-target regions for the used sgRNA, using cut-off values of <3 mismatches and <2 bulges33. A genomic PCR was performed to amplify all identified regions (Supplementary Table 6), and amplicons were screened with Sanger sequencing.
Transcript analysis
Total RNA was isolated from larval heads of an F1-incross using the RNeasy Micro kit according to manufacturer’s instructions (Qiagen, Hilden, Germany). 200 µg of total RNA was used for cDNA synthesis with the SuperScript IV Reverse Transcriptase kit (Thermo Fisher Scientific, Waltham (MA), USA). PCR was performed on cDNA according to standard protocol, from ush2a exon 12 to exon 14 (primers are listed in Supplementary Table 6), followed by Sanger sequencing.
Immunohistochemistry
Cryosections (7 µM) of unfixed larvae (5 dpf) were treated, stained and imaged as described previously14,46 with primary antibodies directed against zebrafish usherin (rabbit anti-usherin-C (1:500; #27640002; Novus Biological, Centennial (CO), USA) and centrin (mouse anti-centrin (1:500; #04-1624; Millipore, Burlington (MA), USA)). The following secondary antibodies were used: goat anti-rabbit Alexa Fluor® 568 (1:800, A11011, Molecular Probes, Eugene (OR), USA) and goat anti-mouse Alexa Fluor® 488 (1:800, A11029, Molecular Probes, Eugene (OR), USA) and DAPI (1:800, D1306; Molecular Probes, Eugene (OR), USA).
For rhodopsin, cryosections (7 µM) of PFA 4% fixed larvae (5 dpf) treated, stained and imaged as published previously31,47. Primary antibody: mouse anti-rhodopsin (1:2000, Clone 4D2; NBP2-59690, Novus Biological, Centennial (CO), USA), goat anti-mouse Alexa Fluor® 488 (1:800, AA11029, Molecular Probes, Eugene (OR), USA) and DAPI (1:800, D1306; Molecular Probes, Eugene (OR), USA).
Quantification of usherin and rhodopsin localization
Usherin localization at the photoreceptor periciliary membrane was quantified using an automated Fiji (v.1.51n) script as previously published30. Rhodopsin mislocalization was quantified by manually scoring the number of photoreceptor cells per retinal cryosection with clear rhodopsin localization in the photoreceptor cell body. All images were blinded, randomized and scored independently by two researchers.
Electroretinograms
ERG recordings were performed on isolated larval eyes (5 dpf) as previously described30. Larvae were dark-adapted for at least 30 min prior to testing. Isolated eyes were stimulated with a light pulse with an intensity of 6000 lux. Electrical signals were captured and subsequently amplified using an electrode that was positioned at the center of the cornea.
Statistical analysis
All statistical analyses were performed using PRISM software (v9.0.0). Average scores were calculated and normality was checked, followed by an unpaired two-tailed Student’s t test (rhodopsin localization assay and ERG recordings) or a Kriskal–Wallis test followed by a Dunn’s multiple comparison test (usherin quantification).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors gratefully acknowledge the individuals who participated in this study, the Radboud University Zebrafish Facility (RU ZF Facility), and the Radboud Technology Center Genomics for their assistance with whole genome sequencing and data analysis as well as the optical genome mapping experiments. This work was funded by VELUX Stiftung (Project number: 1129 to F.P.M.C., H.K., S.R. & E.v.W.) and Stichting Zeldzame Ziekten Fonds (to E.d.V & E.v.W.).
Author contributions
J.R., S.R., E.v.W., H.K., F.P.M.C. and E.d.V. designed the study; J.R., E.v.B., S.B., M.A., T.P., J.O., K.N., H.V., M.G.R., S.R. and E.v.W. performed experiments and were involved in data analysis; C.G., G.D.N.A. and J.C.G. contributed with bioinformatic support; C.H.Z.L., J.J.C.v.L.V., C.W.O., L.H.W. and C.B.H. contributed by collecting cases and with clinical evaluation of the cases; S.R., E.v.W., H.K., F.P.M.C. and E.d.V. were involved in funding acquisition; J.R., S.R. and E.v.W.wrote the original draft; E.d.V., S.R., E.v.W., H.K. and F.P.M.C. contributed by reviewing and editing the manuscript. All authors read and approved the manuscript.
Data availability
Data are available upon request. All whole genome sequencing variants that were considered to be potentially pathogenic are available in the supplementary data. Pathogenic variant data are uploaded to the Leiden Open Variation Database (https://databases.lovd.nl/shared/genes/USH2A). All other whole genome sequencing data are subject to controlled access because these may compromise research participant privacy. These data may become available upon a data transfer agreement approved by the local ethics committee and can be obtained from corresponding author S.R. upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Susanne Roosing, Email: Susanne.Roosing@radboudumc.nl.
Erwin van Wijk, Email: Erwin.vanWyk@radboudumc.nl.
Supplementary information
The online version contains supplementary material available at 10.1038/s41525-022-00306-z.
References
- 1.Perea-Romero I, et al. Genetic landscape of 6089 inherited retinal dystrophies affected cases in Spain and their therapeutic and extended epidemiological implications. Sci. Rep. 2021;11:1526. doi: 10.1038/s41598-021-81093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stone EM, et al. Clinically focused molecular investigation of 1000 consecutive families with inherited retinal disease. Ophthalmology. 2017;124:1314–1331. doi: 10.1016/j.ophtha.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jouret G, et al. Genetics of Usher syndrome: new insights from a meta-analysis. Otol. Neurotol. 2019;40:121–129. doi: 10.1097/MAO.0000000000002054. [DOI] [PubMed] [Google Scholar]
- 4.Toualbi L, Toms M, Moosajee M. USH2A-retinopathy: from genetics to therapeutics. Exp. Eye Res. 2020;201:108330. doi: 10.1016/j.exer.2020.108330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivolta C, Sweklo EA, Berson EL, Dryja TP. Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am. J. Hum. Genet. 2000;66:1975–1978. doi: 10.1086/302926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierrache LH, et al. Visual prognosis in USH2A-associated retinitis pigmentosa is worse for patients with Usher Syndrome Type IIa Than for those with nonsyndromic retinitis pigmentosa. Ophthalmology. 2016;123:1151–1160. doi: 10.1016/j.ophtha.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 7.Aller E, et al. Genetic analysis of 2299delG and C759F mutations (USH2A) in patients with visual and/or auditory impairments. Eur. J. Hum. Genet. 2004;12:407–410. doi: 10.1038/sj.ejhg.5201138. [DOI] [PubMed] [Google Scholar]
- 8.Seyedahmadi BJ, Rivolta C, Keene JA, Berson EL, Dryja TP. Comprehensive screening of the USH2A gene in Usher syndrome type II and non-syndromic recessive retinitis pigmentosa. Exp. Eye Res. 2004;79:167–173. doi: 10.1016/j.exer.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landrum MJ, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DuPont M, Jones EM, Xu M, Chen R. Investigating the disease association of USH2A p.C759F variant by leveraging large retinitis pigmentosa cohort data. Ophthalmic Genet. 2018;39:291–292. doi: 10.1080/13816810.2017.1418388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernal S, et al. Mutations in USH2A in Spanish patients with autosomal recessive retinitis pigmentosa: high prevalence and phenotypic variation. J. Med. Genet. 2003;40:e8. doi: 10.1136/jmg.40.1.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pozo MG, et al. Re-evaluation casts doubt on the pathogenicity of homozygous USH2A p.C759F. Am. J. Med. Genet. A. 2015;167:1597–1600. doi: 10.1002/ajmg.a.37003. [DOI] [PubMed] [Google Scholar]
- 14.Dulla K, et al. Antisense oligonucleotide-based treatment of retinitis pigmentosa caused by USH2A exon 13 mutations. Mol. Ther. 2021;29:2441–2455. doi: 10.1016/j.ymthe.2021.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daiger, S. R., BJF.; Greenberg, J.; Christoffels, A.; Hide, W. RetNet, the Retinal Information Network, https://sph.uth.edu/RetNet/ (1998).
- 16.Jaganathan K, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e524. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 17.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 19.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 20.Pertea M, Lin X, Salzberg SL. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29:1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sangermano R, et al. Deep-intronic ABCA4 variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Genet. Med. 2019;21:1751–1760. doi: 10.1038/s41436-018-0414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherry TJ, et al. Mapping the cis-regulatory architecture of the human retina reveals noncoding genetic variation in disease. Proc. Natl Acad. Sci. USA. 2020;117:9001–9012. doi: 10.1073/pnas.1922501117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mo A, et al. Epigenomic landscapes of retinal rods and cones. Elife. 2016;5:e11613. doi: 10.7554/eLife.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muthu, V. et al. Genomic architecture of Shh-dependent cochlear morphogenesis. Development146, 10.1242/dev.181339 (2019). [DOI] [PMC free article] [PubMed]
- 25.Yizhar-Barnea O, et al. DNA methylation dynamics during embryonic development and postnatal maturation of the mouse auditory sensory epithelium. Sci. Rep. 2018;8:17348. doi: 10.1038/s41598-018-35587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fishilevich, S. et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford)2017, 10.1093/database/bax028 (2017). [DOI] [PMC free article] [PubMed]
- 27.Ebermann I, et al. PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J. Clin. Invest. 2010;120:1812–1823. doi: 10.1172/JCI39715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutten JW, et al. Therapeutic NOTCH3 cysteine correction in CADASIL using exon skipping: in vitro proof of concept. Brain. 2016;139:1123–1135. doi: 10.1093/brain/aww011. [DOI] [PubMed] [Google Scholar]
- 30.Dona M, et al. Usherin defects lead to early-onset retinal dysfunction in zebrafish. Exp. Eye Res. 2018;173:148–159. doi: 10.1016/j.exer.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toms M, et al. Clinical and preclinical therapeutic outcome metrics for USH2A-related disease. Hum. Mol. Genet. 2020;29:1882–1899. doi: 10.1093/hmg/ddaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Vrieze, E. et al. Efficient Generation of Knock-In Zebrafish Models for Inherited Disorders Using CRISPR-Cas9 Ribonucleoprotein Complexes. Int. J. Mol. Sci.22, 10.3390/ijms22179429 (2021). [DOI] [PMC free article] [PubMed]
- 33.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han S, et al. Knockout of ush2a gene in zebrafish causes hearing impairment and late onset rod-cone dystrophy. Hum. Genet. 2018;137:779–794. doi: 10.1007/s00439-018-1936-6. [DOI] [PubMed] [Google Scholar]
- 35.Chang B, et al. Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cGMP phosphodiesterase gene. Vis. Res. 2007;47:624–633. doi: 10.1016/j.visres.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oprea GE, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C, Luo N, Tung CY, Perrin BJ, Zhao B. GRXCR2 regulates taperin localization critical for stereocilia morphology and hearing. Cell Rep. 2018;25:1268–1280.e1264. doi: 10.1016/j.celrep.2018.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fadaie, Z. et al. BBS1 branchpoint variant is associated with non-syndromic retinitis pigmentosa. J. Med. Genet., 10.1136/jmedgenet-2020-107626 (2021). [DOI] [PubMed]
- 39.Neveling K, et al. Next-generation cytogenetics: Comprehensive assessment of 52 hematological malignancy genomes by optical genome mapping. Am. J. Hum. Genet. 2021;108:1423–1435. doi: 10.1016/j.ajhg.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mantere T, et al. Optical genome mapping enables constitutional chromosomal aberration detection. Am. J. Hum. Genet. 2021;108:1409–1422. doi: 10.1016/j.ajhg.2021.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sangermano R, et al. Photoreceptor progenitor mRNA analysis reveals exon skipping resulting from the ABCA4 c.5461-10T→C mutation in stargardt disease. Ophthalmology. 2016;123:1375–1385. doi: 10.1016/j.ophtha.2016.01.053. [DOI] [PubMed] [Google Scholar]
- 42.Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA-a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 43.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-V. [DOI] [PubMed] [Google Scholar]
- 44.Pulido D, Briggs DC, Hua J, Hohenester E. Crystallographic analysis of the laminin β2 short arm reveals how the LF domain is inserted into a regular array of LE domains. Matrix Biol. 2017;57-58:204–212. doi: 10.1016/j.matbio.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 46.Slijkerman, R. et al. Poor Splice-Site Recognition in a Humanized Zebrafish Knockin Model for the Recurrent Deep-Intronic c.7595-2144A>G Mutation in USH2A. Zebrafish 15, 597–609 (2018). [DOI] [PubMed]
- 47.Corral-Serrano JC, et al. C2orf71a/pcare1 is important for photoreceptor outer segment morphogenesis and visual function in zebrafish. Sci. Rep. 2018;8:9675. doi: 10.1038/s41598-018-27928-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request. All whole genome sequencing variants that were considered to be potentially pathogenic are available in the supplementary data. Pathogenic variant data are uploaded to the Leiden Open Variation Database (https://databases.lovd.nl/shared/genes/USH2A). All other whole genome sequencing data are subject to controlled access because these may compromise research participant privacy. These data may become available upon a data transfer agreement approved by the local ethics committee and can be obtained from corresponding author S.R. upon reasonable request.